Significance
One origin of cancer arises from defects in the homologous recombination (HR) DNA repair pathway, which results in oncogenic genomic instability. Normal cells prefer HR for repair of stressed replication forks because it is highly conservative. The back-up pathway for repair of stressed replication forks, alternative non-homologous end-joining (aNHEJ), can result in chromosomal instability. Normal cells use microRNA (miR) 223-3p to down-regulate aNHEJ and thus promote HR, which protects genomic stability. However, those cancers that arise from inherited or acquired defects in HR components, such as the BRCA1 breast and ovarian cancers, silence miR223-3p to enhance aNHEJ repair of stressed replication forks. Reconstituting expression of miR223-3p in these cancers causes synthetic lethality, and is a potential therapeutic modality.
Keywords: oncogenesis, DNA repair, replication fork, microRNA
Abstract
Defects in DNA repair give rise to genomic instability, leading to neoplasia. Cancer cells defective in one DNA repair pathway can become reliant on remaining repair pathways for survival and proliferation. This attribute of cancer cells can be exploited therapeutically, by inhibiting the remaining repair pathway, a process termed synthetic lethality. This process underlies the mechanism of the Poly-ADP ribose polymerase-1 (PARP1) inhibitors in clinical use, which target BRCA1 deficient cancers, which is indispensable for homologous recombination (HR) DNA repair. HR is the major repair pathway for stressed replication forks, but when BRCA1 is deficient, stressed forks are repaired by back-up pathways such as alternative nonhomologous end-joining (aNHEJ). Unlike HR, aNHEJ is nonconservative, and can mediate chromosomal translocations. In this study we have found that miR223-3p decreases expression of PARP1, CtIP, and Pso4, each of which are aNHEJ components. In most cells, high levels of microRNA (miR) 223–3p repress aNHEJ, decreasing the risk of chromosomal translocations. Deletion of the miR223 locus in mice increases PARP1 levels in hematopoietic cells and enhances their risk of unprovoked chromosomal translocations. We also discovered that cancer cells deficient in BRCA1 or its obligate partner BRCA1-Associated Protein-1 (BAP1) routinely repress miR223-3p to permit repair of stressed replication forks via aNHEJ. Reconstituting the expression of miR223-3p in BRCA1- and BAP1-deficient cancer cells results in reduced repair of stressed replication forks and synthetic lethality. Thus, miR223-3p is a negative regulator of the aNHEJ DNA repair and represents a therapeutic pathway for BRCA1- or BAP1-deficient cancers.
DNA replication is not a smooth and continuous process, but rather prone to interruptions (1, 2). DNA damage or nucleotide depletion can induce stalling of the replication machinery (3, 4), which can result in replication stress and subsequent fork collapse, with disassociation of the replication apparatus (5, 6). As such, replication stress is a common etiology of genomic instability, resulting in either cell death or neoplastic transformation (4, 6–8). HR is the most common pathway for stressed replication fork repair and restart (5, 7, 8). The rate-limiting step in HR is the nucleolytic resection of the 5′ strand at a DNA free end, initiated when BRCA1/CtIP replaces 53BP1/RIF1/Shieldin at the site of DNA damage (9–12). After 5′ strand resection (13–16), BRCA2 then loads the RAD51 recombinase onto the 3′ single-stranded DNA for invasion of the sister chromatid (17–19), which results in Holliday junction formation that is later resolved by MUS81 and SLX4 (20, 21). Germline or acquired mutations in BRCA1 or its interacting partner BAP1 result in defects in the HR pathway, and subsequent replication fork instability and oncogenesis (22–25). HR-deficient cancers become addicted to other repair pathways to resolve replication fork stress such as aNHEJ (26–30). This addiction is a singular weakness of these cancer cells, and can be targeted with therapeutic agents (8, 25, 29, 30), generating synthetic lethality. This synthetic lethality underlies the efficacy of PARP1 inhibitors in breast, ovarian, and prostate cancers that harbor inherited or acquired mutations in BRCA1 (31, 32).
PARP1 can initiate aNHEJ by displacing the Ku complex from the DNA double-strand break (33, 34). CtIP, Pso4, and Mre11 then mediate limited 5′ end resection, followed by DNA polymerase theta promoting microhomology annealing (27–29). DNA ligase 1 or 3 with XRCC1 then promote ligation of the free DNA ends (27, 33). Pharmacologic inhibitors of PARP1 lock it onto DNA, generating obstacles that stall replication forks, which in BRCA1- or BAP1-mutant cells cannot be repaired by HR (31, 32, 35). PARP1 inhibition also prevents the use of the aNHEJ back-up repair pathway to resolve the replication forks stalled by PARP1 locked on DNA, resulting in fork collapse and cell death (8, 25, 27). While aNHEJ promotes repair of stressed replication forks in BRCA1/BAP1-defective cells, it comes at a cost; aNHEJ can mediate chromosomal translocations (33, 34). This occurs when aNHEJ erroneously anneals and then ligates free DNA ends on distinct chromosomes (26, 33, 34). The three FDA-approved PARP1 inhibitors, olaparib, rucaparib, and niraparib, have each shown impressive response rates in HR-deficient cancers (36–41). However, these agents can occasionally lead to myelodysplastic syndrome, and resistance can develop (30, 41, 42). In this study, we found that the microRNA species miR223-3p is a negative regulator of aNHEJ, likely to restrain aNHEJ from generating aberrant chromosomal translocations that would lead to genomic instability. Importantly, we discovered that cancer cells deficient in HR repress miR223-3p strand selection and instead express miR223-5p. Reconstituting expression of miR223-3p is synthetically lethal to BRCA1- and BAP1-defective cancer cells, demonstrating its potential as an effective cancer therapeutic agent.
Results
MiR223-3p Down-Regulates aNHEJ Components.
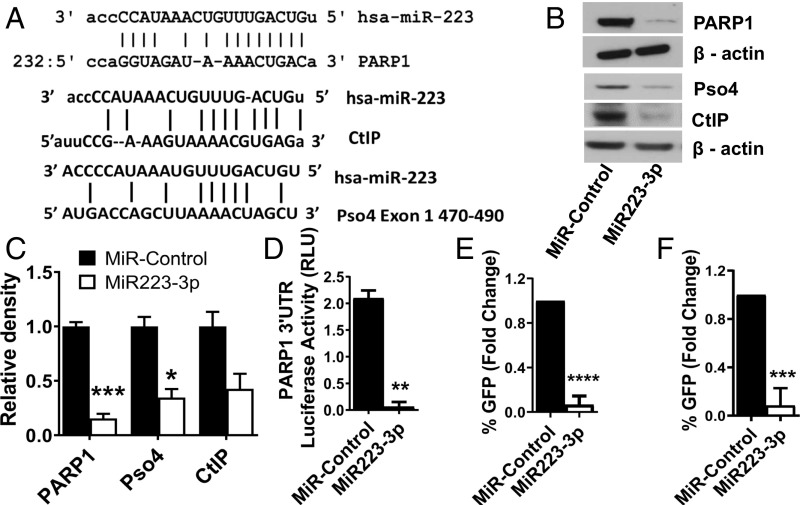
MiR223-3p has a strong seed sequence match for the 3′ UTR of PARP1, the 3′ UTR of CtIP, and exon 1 of Pso4, each of which is important for aNHEJ (33, 34) (Fig. 1A). Consistent with this, transfection of miR223-3p in multiple cell types markedly decreased PARP1, CtIP, and Pso4 protein levels by an average of fivefold, 2.5-fold, and twofold, respectively (P value < 0.001, <0.05, >0.05, respectively), but did not affect the levels of other aNHEJ components such as Ligase III, Pol Q, and XRCC1 (Figs. 1 B and C and SI Appendix, Fig. S1 C and D). However, we observed that the expression levels of PARP1 mRNA were not affected by miR223-3p (SI Appendix, Fig. S1A), suggesting that miR223-3p is a translational repressor. To confirm that miR223-3p mediates translational repression, we performed luciferase assays where the reporter was fused to PARP1 3′UTR. When the reporter construct is cotransfected with the miR223-3p, we found a 20-fold decrease in the luciferase activity in cells transfected with the miR223-3p compared with control cells transfected with a scrambled microRNA (Fig. 1D, P value < 0.01). We did not observe cleaved PARP1 indicative of apoptosis activation upon transfection of miR223-3p in any cell line (SI Appendix, Fig. S1B). We measured aNHEJ after transfection with miR223-3p using two reporter systems, EJ2-GFP (43) and MMEJ-GFP (8). We found that when both of these reporter systems were transfected with miR223-3p, aNHEJ repair efficiency was reduced 10-fold (Fig. 1 E and F, P value < 0.001).
Fig. 1.
MiR223-3p down-regulates aNHEJ components PARP1, CtIP, and Pso4 in mammalian cells. (A) Pairing of miR223-3p to the 3′UTR region of PARP1 mRNA and CtIP mRNA, and to exon 1 of PSO4. (B) Western blot showing the levels of PARP1, CtIP, and Pso4 upon miR223-3p transfection in MDA-MB-436 cells. (C) Densitometry of Western blots showing levels of aNHEJ components (n = 3). (D) Luciferase reporter of PARP1 3′ UTR upon miR223-3p mimic transfection. (E and F) Measurement of aNHEJ in the EJ2-GFP (E) and MMEJ-GFP (F) cell reporter systems after transfection of miR223-3p. GFP-expressing cells indicate productive aNHEJ repair. (*-P < 0.05, **-P < 0.01, ***-P < 0.001, ****-P < 0.0001 for all figures).
MiR223-3p Represses Chromosomal Translocation in Hematopoietic Cells.
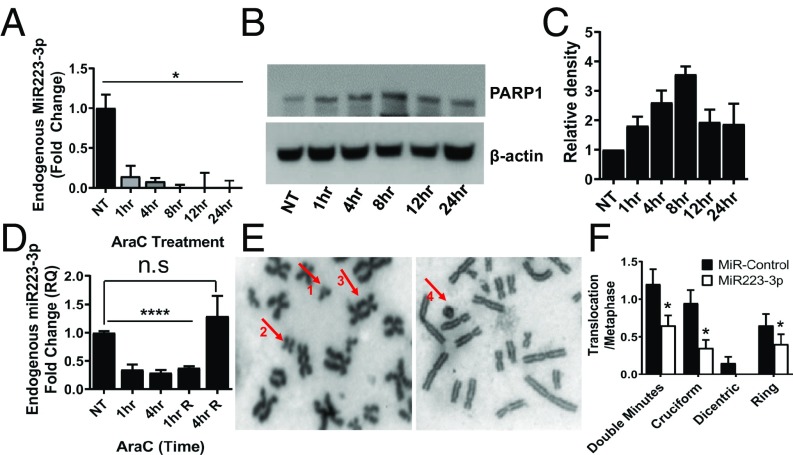
MiR223-3p is highly expressed in hematopoietic cells and is involved in the regulation of granulopoiesis (44). However, the functional relationship between miR223-3p and the maintenance of genomic stability in the hematopoietic system has not been defined. We first studied the change in miR223-3p expression levels in hematopoietic cell lines in response to replication stress. HL-60 cells (HR proficient) treated with 50 µM Ara-C rapidly decreased the expression of endogenous miR223-3p by 10-fold at 1 h after treatment as measured by qRT-PCR (Fig. 2A). This decrease in miR223-3p upon replication stress is accompanied by an increase in PARP1 protein levels by twofold (Fig. 2 B and C). MiR223-3p levels returned to baseline when the cells were released from Ara-C (Fig. 2D). It is certainly possible that there were transcriptional regulatory changes in PARP1 from Ara-C, and PARP1 changes were from that and not miR223-3p reduction.
Fig. 2.
MiR22-3p represses chromosomal translocation in hematopoietic cells. (A) QRT-PCR showing the endogenous levels of miR223-3p at different time points after Ara-C treatment in HL-60 cells. (B) Western analysis showing the PARP1 protein levels at different time points after Ara-C treatment in HL60 cells. (C) Quantitation of the Western blots showing relative levels of PARP1 in HL-60 cells after Ara-C treatment. (D) QRT-PCR showing levels of endogenous miR223-3p in HL-60 cells at different time points after release from Ara-C treatment. (E) Representative confocal metaphase images showing chromosomal translocation phenotypes in Jurkat cells after VP16 exposure (1- Double minutes, 2- Cruciform structure, 3- Dicentric chromosomes, and 4- Ring chromosome). (F) Percentage of cells showing different chromosomal translocation phenotypes in Jurkat cells treated with VP16 with or without prior transfection of miR223-3p.
We next investigated the occurrence of chromosomal translocations following miR223-3p-mediated down-regulation of aNHEJ. We treated Jurkat cells (HR proficient) with VP16, known to induce chromosomal translocations (34), and then scored metaphases for the translocation phenotypes, dicentric, ring, cruciform, and double minute chromosomes. We found that transfecting miR223-3p decreased PARP1 protein levels and decreased chromosomal translocation events compared with the control group (Fig. 2 E and F), consistent with the known role of PARP1 in aNHEJ-mediated chromosomal translocations (3, 34).
MiR223-Deleted Mice Exhibit Increased Chromosomal Aberrations.
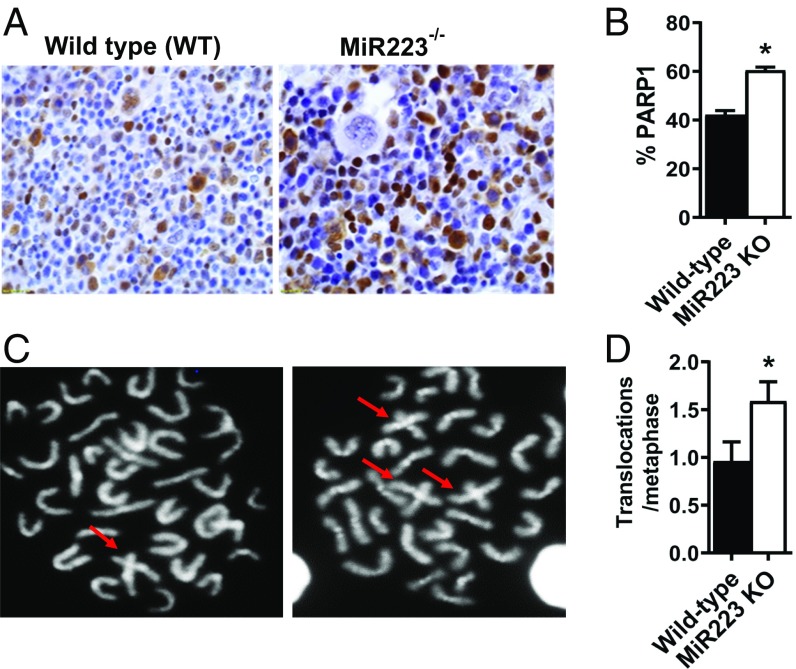
Loss of miR223 has been reported in AML patients (44), which could be one origin of leukemogenic translocations. We hypothesized that the genetic loss of miR223 would result in increased chromosomal translocations due to increased aNHEJ in hematopoietic cells. Immunohistology of bone marrow from mice with homozygous deletion of the miR223 locus demonstrated that there was an increased fraction of hematopoietic cells expressing PARP1 protein compared with wild-type (WT) controls (Fig. 3 A and B. P value < 0.05) (43). Further, miR223−/− hematopoietic cells had a twofold increase in unprovoked hematopoietic cell chromosomal translocations compared with the WT (Fig. 3 C and D. P value < 0.05), consistent with a role for miR223-3p in maintaining genomic stability.
Fig. 3.
MiR223 KO mice exhibit increased unprovoked chromosomal aberrations. (A) Representative images for PARP1 protein assessed by immunohistology in the bone marrow of miR223 wild-type (WT) and genetically deleted mice. (B) Percentage of PARP1-expressing cells in the bone marrow of miR223−/− and WT mice. (C) Representative confocal images showing metaphase chromosomes in the MiR223−/− and WT mouse bone marrow. Cruciform structures indicative of chromosomal fusions are shown with arrows. (D) Percentage of chromosomal aberrations per metaphase in MiR223−/− and WT mice.
MiR223-3p Induces Synthetic Lethality in HR-Deficient Cancers.
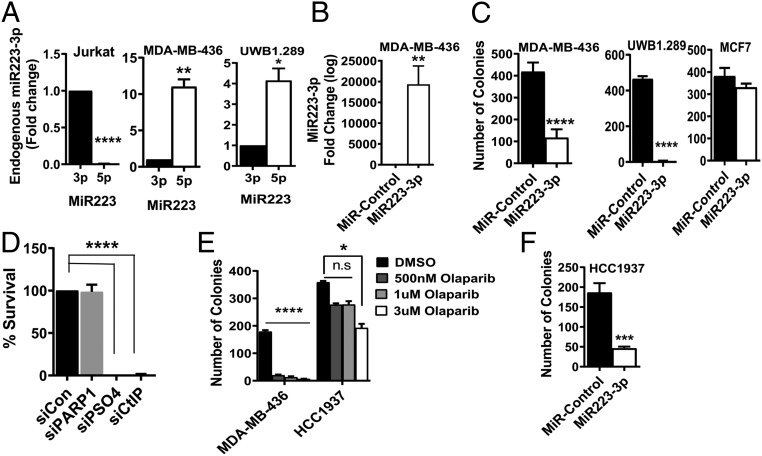
Given that miR223-3p decreased aNHEJ efficiency, we hypothesized that HR-deficient cancers would not tolerate its expression. BRCA1 is in a complex with BAP1, and both are required for HR (24, 45, 46). We found that all BRCA1- and BAP1-deficient cancer cells tested in this study suppressed expression of miR223-3p and instead processed the 5p form of miR223 to maturity (Fig. 4A and SI Appendix, Fig. S2A), opposite of all HR-proficient cell lines examined (Fig. 4A and SI Appendix, Fig. S2B). Continued expression of miR223-3p in HR-deficient cancers would repress aNHEJ, which would decrease those cells’ ability to repair replication stress (25–29). Significantly, when we repressed BRCA1 in the HR-proficient breast cancer cell line MCF7, miR223-3p levels declined as well (SI Appendix, Fig. S2D). We next studied whether reconstitution of miR223-3p in BRCA1- or BAP1-deficient cancer cells could induce synthetic lethality. We found that a single transfection of miR223-3p was highly cytotoxic to BRCA1-deficient breast and ovarian cancer cells, and to BAP1-mutant malignant pleural mesothelioma (MPM) cells (Fig. 4 B and C and SI Appendix, Fig. S2C). Specifically, a single transfection of 25 nM miR223-3p resulted in fourfold decrease in clonogenicity in BRCA1-mutant MDA-MB-436 breast cancer cells (Fig. 4C. P value < 0.0001), 40-fold decrease in BRCA1-mutant UWB1.289 ovarian cancer cells (Fig, 4C. P value < 0.0001) and 20-fold decrease in H2452 BAP1-mutant mesothelioma cells (SI Appendix, Fig. S2C, P value < 0.001). Injecting BRCA1-mutant MDA-MB-436 luciferase cells transfected with miR223-3p into NSG mice showed significantly decreased tumor growth compared with the mice with miR-control transfected cells (SI Appendix, Fig. S3C). Reconstituting miR223-3p in MDA-MB-436 increased the fraction of cells in S-phase (SI Appendix, Fig. S4B). However, transfecting miR223-3p into the HR-proficient MCF7 breast cancer cells did not appreciably alter their clonal cell survival (Fig. 4C). Depleting BRCA1 in MCF7 cells followed by miR223-3p transfection resulted in significant decrease in the clonal survival (SI Appendix, Fig. S3B). BRCA1-mutant HCC1937 cells are resistant to relatively high concentrations of olaparib (42) (Fig. 4E). However, a single transfection of 25 nM miR223-3p resulted in a fourfold decrease in clonogenicity in the HCC1937 cells (Fig. 4F, P value < 0.001).
Fig. 4.
Reconstitution of miR223-3p induces synthetic lethality in HR-deficient cancers. (A) QRT-PCR comparing the levels of miR223-3p and miR223-5p in Jurkat cells (HR-replete), MDA-MB-436 (BRCA1-mutant breast cancer cell line), and UWB1.289 (BRCA1-mutant ovarian cancer cell line). (B) QRT-PCR showing the expression of miR223-3p following transfection with miR223-3p in MDA-MB-436 breast cancer cells. (C) Clonogenic survival assay showing the number of colonies in MDA-MB-435, UWB1.289, and MCF7 (HR-replete breast cancer) cells upon transfection of miR223-3p. (D) Clonogenic survival assay in MDA-MB-436 after depletion of PARP1, CtIP, and PSO4. (E) Comparison of number of colonies in MDA-MB-436, and HCC-1937 (BRCA1-mutant breast cancer) cells in the presence of olaparib. (F) Clonogenic survival assay in HCC1937 cells after miR223-3p transfection.
Reconstituting miR223-3p Blocks Stressed Replication Fork Repair in BRCA1- or BAP1-Deficient Cancer Cells.
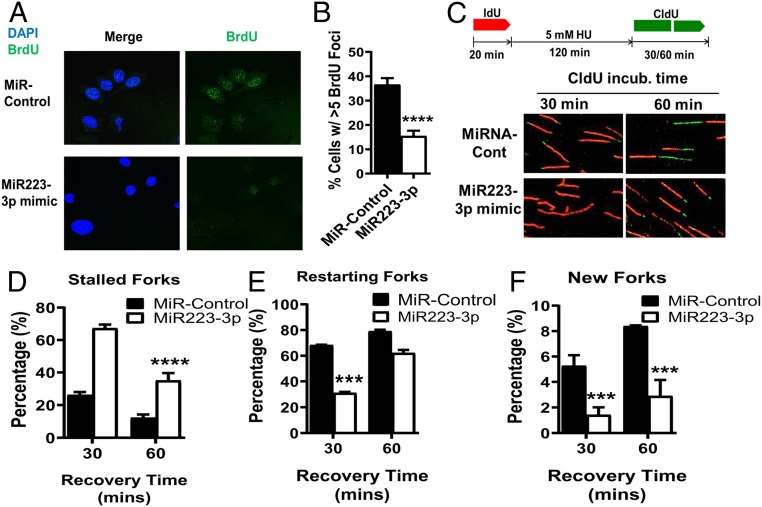
We next measured the fraction of active replication forks in BRCA1- or BAP1-deficient cancer cells after transfection with miR223-3p. First, we assessed BrdU incorporation in BRCA1-deficient breast cancer MDA-MB-436 cells (Fig. 5 A and B) and in BAP1-mutant MPM cells (H2452) (SI Appendix, Fig. S4A). We found that the MDA-MB-436 cells transfected once with miR223-3p had a threefold decrease in BrdU foci compared with controls (Fig. 5 A and B, P value < 0.0001), while the H2452 cells exhibited a fourfold decrease (SI Appendix, Fig. S4A, P value < 0.001). The decrease in BrdU foci reflects perturbation of DNA replication, suggesting that miR223-3p interferes with replication forks by decreasing the capability of aNHEJ to manage replication stress in BRCA1- and BAP1-deficient cells (42, 43, 47). We used DNA fiber analysis to assess repair and restart of stressed replication forks in the above BRCA1- and BAP1-deficient cancer cells after transfection of miR223-3p (Fig. 5C) (9, 14). Replication was stressed by deoxynucleotide depletion with hydroxyurea (HU) (1–4). The percentage of stalled forks were significantly higher in the BRCA1-deficient MDA-MB-436 cells and BAP1-deficient H2452 cells transfected with miR223-3p (Fig. 5D and SI Appendix, Fig. S4C). After release from replication stress, reconstitution of miR223-3p resulted in a twofold decrease in the initial repair and restart of stalled replication forks in MDA-MB-436 cells (Fig. 5E, P value < 0.001) and a twofold decrease in the H2452 cells (SI Appendix, Fig. S4C, P value < 0.001). MiR223-3p reconstitution also decreased the fraction of new forks starting after release from stress that stem from firing from normally dormant origins by fivefold in MDA-MB-436 cells (Fig. 5F, P value < 0.001) and by fourfold in H2452 cells (SI Appendix, Fig. S4C, P value < 0.001). However, the MDA-MB-436 cells exposed to miR223-3p ultimately recovered their ability to restart stalled replication forks to the same extent as controls. There were fewer new forks in the miR223-3p-reconstituted MDA-MB-436 cells than controls at all time points, implying a role for aNHEJ components in the firing of new replication origins (Fig. 5F). Nearly identical findings were seen in the BAP1-mutant H2452 mesothelioma cells after reconstitution of miR223-3p (SI Appendix, Fig. S4C), indicating that BAP1-deficient cancers also require aNHEJ for stressed replication fork repair, and thus are sensitive to miR223-3p restoration.
Fig. 5.
Reconstitution of miR223-3p in HR-deficient MDA-MB-436 breast cancer cells results in delayed repair and restart of stressed replication forks. (A) Representative confocal immunofluorescence microscopic images of BrdU from scrambled control and miR223-3p-reconstituted MDA-MB-436 cells exposed to BrdU for 30 min. (B) Fraction of BrdU-positive cells in control and miR223-3p reconstituted MDA-MB-436 cells. (C) Experimental protocol and representative images of DNA fiber assays from control and miR223-3p-transfected MDA-MB-436 cells pulse-labeled with IdU for 20 min (red), treated with HU for 120 min, and then pulse-labeled with CldU (green) for 30 and 60 min. Analysis of stalled replication forks (D), restarted forks (E), and initiation of new replication forks (F) by DNA fiber analysis.
MiR223-3p Reconstitution Induces Genomic Instability in BRCA1- and BAP1-Deficient Cells.
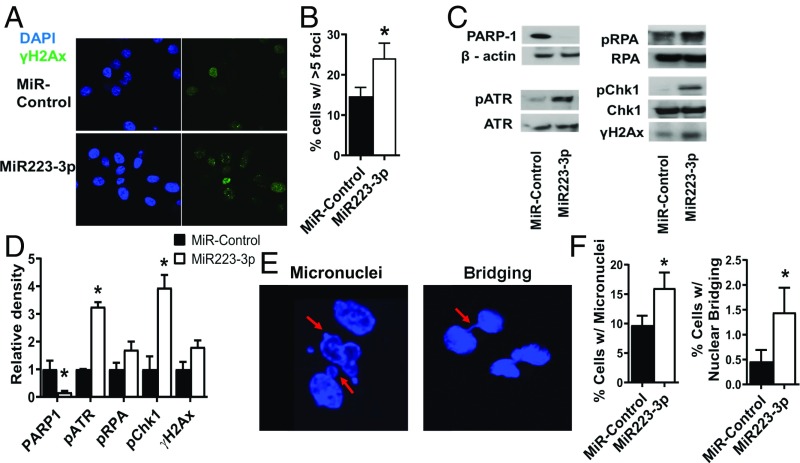
We tested whether miR223-3p-induced BRCA1- or BAP1-deficient cell delay of stressed replication fork repair and restart led to genomic instability. Unrepaired stressed replication forks can result in unresolved fork cleavage and ultimately nucleolytic destruction of fork structures (47, 48). Cleaved and/or degraded replication forks can be identified by the presence of γH2Ax (9, 49). Therefore, we measured the number of γH2Ax foci using immunofluorescence microscopy in the BRCA1-mutant MDA-MB-436 and BAP1-mutant H2452 cells. We found that the miR223-3p-reconstitued in these cancer cells had a twofold increase in the accumulation of γH2Ax foci (Fig. 6 A and B, P value < 0.05, SI Appendix, Fig. S5A). We also found increased phosphorylation of ATR, RPA and Chk1 by Western blot, again consistent with the accumulation of stressed replication forks (Fig. 6 C and D). These data indicate that restoration of miR223-3p in BRCA1- and BAP1-deficient cancer cells results in replication fork structural damage. Genomic instability arising from unrepaired stressed replication forks can also be measured by abnormal nuclear structures such as micronuclei and bridging. These nuclear aberrancies are the result of mitotic catastrophe, which occurs when aberrantly resolved damaged fork structures result in fused chromosomes via classical NHEJ (9, 50). Acentric chromosomes are retained in the parent cell after mitosis, and are seen as micronuclei. Dicentric chromosomes form a nuclear bridge between daughter cells (50, 51). We measured these nuclear abnormalities in BRCA1-deficient MDA-MB-436 breast cancer cells and in BAP1-deficient H2452 mesothelioma cells. In the MDA-MB-436 cells, there was a twofold increase in nuclear bridging (Fig. 6 E and F, P value < 0.05) and a fourfold increase in micronuclei upon miR223-3p transfection in MDA-MB-436 cells (Fig. 6 E and F, P value < 0.05). In the H2452 cells, there was an eightfold increase in nuclear bridging (P value < 0.01) and a twofold increase in micronuclei (P value < 0.001) upon miR223-3p transfection (SI Appendix, Fig. S5B).
Fig. 6.
MiR223-3p induces mitotic catastrophe in HR-deficient MDA-MB-436 breast cancer cells. (A) Representative confocal immunofluorescence microscopic images of γH2Ax foci after reconstitution of miR223-3p. (B) Analysis of percentage of cells >5 γ-H2Ax foci in control and miR223-3p-reconstituted cells. (C) Western analysis and (D) Relative densitometric measurements (n = 3) showing the levels of various DNA damage response proteins such as phosphorylated ATR, phosphorylated CHK1, and phosphorylated RPA in control and miR223-3p reconstituted MDA-MB-436 cells. (E) Representative DAPI-stained confocal microscopic images showing abnormal nuclear structures such as micronuclei (Left) and bridging (Right) in MDA-MB-436 cells with reconstitution of miR223-3p. (F) Analysis of percentage of cells with micronuclei and bridging.
Discussion
Faithful repair of damaged replication forks is critical for maintaining genomic stability and suppressing oncogenesis (4, 6). The proper choice of which DNA repair pathway to use to repair and restart damaged replication forks is crucial to conserve sequence integrity (27, 52). HR repair and restart of damaged replication forks is conservative, while aNHEJ repair has risk of aberrant ligation of free DNA ends at such forks, resulting in chromosomal translocations (33, 34). Such chromosomal abnormalities lead to mitotic catastrophe and cell death, or neoplastic transformation and cancer (51, 53). Thus, it is not surprising that cells negatively regulate aNHEJ to promote the choice of HR to repair stressed replication forks. In this study, we identified miR223-3p as a repressor of aNHEJ pathway repair efficiency by down-regulating expression of aNHEJ components. MiR223-3p is highly expressed in most normal tissues, especially in hematopoietic cells which have a high proliferative rate, and thus face a higher risk of replication stress (44). This heightened risk of replication stress can lead to genomic instability if such stress is not properly managed by efficient and conservative fork repair pathways such as HR.
We have previously demonstrated reduced aNHEJ and decreased chromosomal translocation when PARP1 is inhibited (34). We demonstrate here that miR223-3p functions to maintain genomic stability by decreasing use of aNHEJ. However, when there is extensive replication stress requiring full deployment of all relevant repair pathways, levels of miR223-3p decrease. This increases PARP1 levels and promotes aNHEJ activity to address the repair of damaged replication forks. In miR223−/− mice, hematopoietic cells accumulate unprovoked chromosomal translocations, indicating that miR223-3p maintains steady state genomic stability in vivo. Conversely, provoked chromosomal translocations are decreased after over-expression of miR223-3p in cell lines. Interestingly, miR223-3p−/− mice develop dysplastic hematopoiesis (44), similar to myelodysplasia in humans, which could be the result of genomic instability from unregulated use of aNHEJ in stressed replication fork repair (33, 34).
Interestingly, BRCA1- and BAP1-deficient cancer cells have greatly decreased expression of the miR223-3p strand, and instead process premiR223 to miR223-5p (54, 55). Since miR223-3p inhibits aNHEJ, and aNHEJ replaces HR in processing stressed replication forks in BRCA1-deficient cancers, it is unlikely that such cancers could originate in the first place without repressing miR223-3p. A single transfection of miR223-3p induced synthetic lethality in BRCA1- and BAP1-deficient malignancies, likely by triggering the destruction of required aNHEJ components CtIP and Pso4 (Fig. 4). SiRNA reduction of PARP1 levels did not result in BRCA1-deficient cell cytotoxicity (Fig. 4D), consistent with the finding that PARP1 inhibitors trap PARP1 on DNA, and one mechanism of cancer resistance to such inhibitors is reducing PARP1 levels (35). Reconstituting miR223-3p in BRCA1- and BAP1-resistant cells led to slowed repair of stressed replication forks, but ultimately the stressed forks recovered and restarted. However, we and others have shown that even a short delay in stressed replication fork repair results in cell death (3, 9). In cells lacking both HR and aNHEJ, cleaved replication forks can be fused via 53BP1-dependent classical NHEJ to give rise to the micronuclei and chromosomal bridges as seen here in the BRCA1- and BAP1-deficient miR223-3p-reconstituted cells (50, 56).
Because BRCA1- and BAP1-deficient malignancies are addicted to aNHEJ to cope with replication stress, miR223-3p holds promise as a therapeutic agent for such cancers. Indeed, reconstituting miR223-3p led to synthetic lethality even in BRCA1-mutant cells that were resistant to the PARP1 inhibitor olaparib (Fig. 4). Thus, MiR223-3p could be an important therapy in BRCA1- or BAP1-deficient cancers, especially those that develop resistance to PARP1 inhibitors (57, 58). In addition, since miR223-3p is normally expressed in most tissues, it would be well tolerated by normal cells compared with the clinical PARP1 inhibitors, which have some gastrointestinal and hematologic toxicities (35–40). Notably, BAP1 mutations occur in difficult to treat malignancies, such as MPM, renal cell carcinoma, uveal melanoma, and cholangiocarcinoma (24, 45, 46). In summary, miR223-3p negatively regulates aNHEJ, and is repressed in BRCA1- and BAP1-deficient cancers. Its reconstitution is synthetically lethal to these cancers, and it would therefore be an anti-neoplastic therapeutic agent.
Methods and Materials
Cell Culture.
Cells and the EJ2-GFP and MMEJ-GFP aNHEJ reporter cells were cultured as described (8, 9, 41). For inducing DNA damage, cells were treated with VP16 from Sigma Aldrich (Cat # E1383-25MG) and Cytosine B-d-arabinofuranoside (Ara-C) from Sigma Aldrich (Cat # C6645).
Animal Care.
MiR223 knock-out (KO) mice (Cat # 013198), wild-type (WT) mice (Cat # 002014), and NSG mice (Stock # 005557) were purchased from the Jackson Laboratory. Housing of the animals and all experimental procedures were approved by the Institutional Animal Care and Research Advisory Committee of the University of Florida, Gainesville and University of Texas Health Science Center at San Antonio.
Antibodies.
These are defined in the SI Appendix in the supplemental materials section.
MiRNA and SiRNA.
MirVana miR223-3p microRNA mimic was purchased from Ambion Life Technologies (Cat # 4464067). Small interfering RNA (siRNA) against BRCA1 (Cat # l-003461-00-0020), PARP1 (Cat # l-006656-03-0005), PSO4 (Cat # l-004668-00-0005), and CtIP (Cat # M-011376-00-0005) were purchased from Dharmacon. All were transfected using Lipofectamine RNAiMAX from Life Technologies (Cat # 13778–150) at 25 nM.
Luciferase Assay for Assessing PARP1 3′ UTR mRNA Stability.
PARP1 3′ UTR plasmid was transfected using Lipofectamine, and cells were collected and assessed for luciferase activity using Luc-Pair Dual Luciferase assay kit from GeneCopoeia (Cat # LPFR-P010). Firefly luciferase activity is normalized against Renilla luciferase.
Survival Assays.
Clonogenic survival assays were performed as described (9, 49).
Cytogenetics.
Structural aberrations in Giemsa-stained metaphase chromosomes were scored as described (25, 34).
Immunohistology of Mouse Marrow.
Femurs from WT and KO mice were dissected and fixed in 10% neutral buffered formalin for at least 4 d. The samples were then processed for H&E and immunohistochemical staining with PARP1 antibody (Santa Cruz Biotechnology, cat# sc-8007).
Nuclear Structure Assays and DNA Damage Foci.
Confocal analysis of DAPI-stained nuclear structural abnormalities such as micronuclei and nuclear bridging were assessed as described (9). Confocal immunofluorescence γH2Ax foci assays were performed as described (9).
Replication Fork Analysis.
Active replication forks were measured using immunofluorescent detection of BrdU foci after DNA denaturation as described (9). DNA fiber analysis was performed to measure replication fork arrest and restart in cells transfected with either scramble control or 25 nM miR223-3p mimic as described (9, 49).
Quantitative Real-Time PCR.
CDNA conversion was performed using the first-strand cDNA synthesis kit for MiRNA from Origene (Cat # HP100042). QRT-PCR for miR223 was performed on a 7900HT Fast Real37-Time PCR system (ABI) according to Origene protocol, and ΔCT values were calculated. The primers used had the following sequences: MiR223-3p - forward (5′TGTCAGTTTGTCAAATACC) and reverse (5′GAACATGTCTGCGTATCTC), MiR223-5p - forward (5′CGTGTATTTGACAAGCTG) and reverse (5′GAACATGTCTGCGTATCTC). U6 RNA was used as an endogenous control.
Murine s.c. Xenograft Detection.
One million BRCA1-deficient MDA-MB-436-luciferase breast cancer cells were transfected with either miR-Control or miR223-3p mimic at a concentration of 100 nM and then injected s.c. into the flank of mice (n = 6/group). Bioluminescent tumor signals were obtained by a Xenogen IVIS Spectrum imaging system (Perkin-Elmer) once every week after isoflurane anesthesia as per IACUC protocol.
Supplementary Material
Acknowledgments
This work was supported by NIH (RO1 CA205224 and R01 GM109645 to R.H., RO1 ES007061 and RO1 CA220123 to P.S., and RO1 CA197796 to S.B.); the Korean Institute for Basic Science (IBS-R022-A1-2017 to O.S.); and the Cancer Prevention and Research Institute of Texas Research Training Award (RP17035 to G.S.).
Footnotes
Conflict of interest statement: R.H. has equity in Dialectic Therapeutics, which holds a licensing agreement for use of miR223-3p as a cancer therapeutic agent.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903150116/-/DCSupplemental.
References
- 1.Sirbu B. M., Cortez D., DNA damage response: Three levels of DNA repair regulation. Cold Spring Harb. Perspect. Biol. 5, a012724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortez D., Preventing replication fork collapse to maintain genome integrity. DNA Repair (Amst.) 32, 149–157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carr A. M., Lambert S., Replication stress-induced genome instability: The dark side of replication maintenance by homologous recombination. J. Mol. Biol. 425, 4733–4744 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Allen C., Ashley A. K., Hromas R., Nickoloff J. A., More forks on the road to replication stress recovery. J. Mol. Cell Biol. 3, 4–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petermann E., Helleday T., Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 11, 683–687 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Berti M., Vindigni A., Replication stress: Getting back on track. Nat. Struct. Mol. Biol. 23, 103–109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeman M. K., Cimprich K. A., Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaheen M., Allen C., Nickoloff J. A., Hromas R., Synthetic lethality: Exploiting the addiction of cancer to DNA repair. Blood 117, 6074–6082 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Wu Y., et al. , EEPD1 rescues stressed replication forks and maintains genome stability by promoting end resection and homologous recombination repair. PLoS Genet. 11, e1005675 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anand R., Ranjha L., Cannavo E., Cejka P., Phosphorylated CtIP functions as a Co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell 64, 940–950 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Noordermeer S. M., et al. , The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560, 117–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dev H., et al. , Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 20, 954–965 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Symington L. S., Gautier J., Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Kim H. S., Williamson E. A., Nickoloff J. A., Hromas R. A., Lee S. H., Metnase mediates loading of exonuclease 1 onto single strand overhang DNA for end resection at stalled replication forks. J. Biol. Chem. 292, 1414–1425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H. S., et al. , Endonuclease EEPD1 is a gatekeeper for repair of stressed replication forks. J. Biol. Chem. 292, 2795–2804 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomimatsu N., et al. , Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst.) 11, 441–448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sung P., Robberson D. L., DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82, 453–461 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Sung P., Klein H., Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7, 739–750 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Sung P., Krejci L., Van Komen S., Sehorn M. G., Rad51 recombinase and recombination mediators. J. Biol. Chem. 278, 42729–42732 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Chen X. B., et al. , Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8, 1117–1127 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Muñoz I. M., et al. , Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell 35, 116–127 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Bryant H. E., et al. , Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Farmer H., et al. , Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Testa J. R., et al. , Germline BAP1 mutations predispose to malignant mesothelioma. Nat. Genet. 43, 1022–1025 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srinivasan G., et al. , Synthetic lethality in malignant pleural mesothelioma with PARP1 inhibition. Cancer Chemother. Pharmacol. 80, 861–867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arana M. E., Seki M., Wood R. D., Rogozin I. B., Kunkel T. A., Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 36, 3847–3856 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceccaldi R., Rondinelli B., D’Andrea A. D., Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood R. D., Doublié S., DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair (Amst.) 44, 22–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mladenov E., Iliakis G., Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat. Res. 711, 61–72 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Lupo B., Trusolino L., Inhibition of poly(ADP-ribosyl)ation in cancer: Old and new paradigms revisited. Biochim. Biophys. Acta 1846, 201–215 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Ledermann J. A., PARP inhibitors in ovarian cancer. Ann. Oncol. 27 (suppl. 1), i40–i44 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Chan D. A., Giaccia A. J., Harnessing synthetic lethal interactions in anticancer drug discovery. Nat. Rev. Drug Discov. 10, 351–364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iliakis G., Murmann T., Soni A., Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 793, 166–175 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Wray J., et al. , PARP1 is required for chromosomal translocations. Blood 121, 4359–4365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murai J., et al. ; Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors , Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufman B., et al. , Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 33, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coleman R. L., et al. ; ARIEL3 investigators , Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 390, 1949–1961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott L. J., Niraparib: First global approval. Drugs 77, 1029–1034 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Tutt A., et al. , Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet 376, 235–244 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Taneja S. S., Re: DNA-repair defects and olaparib in metastatic prostate cancer. J. Urol. 195, 925–928 (2016). [DOI] [PubMed] [Google Scholar]
- 41.De Felice F., Tombolini V., Marampon F., Musella A., Marchetti C., Defective DNA repair mechanisms in prostate cancer: Impact of olaparib. Drug Des. Devel. Ther. 11, 547–552 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parameswaran B., et al. , Damage-induced BRCA1 phosphorylation by Chk2 contributes to the timing of end resection. Cell Cycle 14, 437–448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennardo N., Cheng A., Huang N., Stark J. M., Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trissal M. C., DeMoya R. A., Schmidt A. P., Link D. C., MicroRNA-223 regulates granulopoiesis but is not required for HSC maintenance in mice. PLoS One 10, e0119304 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arzt L., Quehenberger F., Halbwedl I., Mairinger T., Popper H. H., BAP1 protein is a progression factor in malignant pleural mesothelioma. Pathol. Oncol. Res. 20, 145–151 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Carbone M., et al. , BAP1 cancer syndrome: Malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J. Transl. Med. 10, 179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexander J. L., Orr-Weaver T. L., Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev. 30, 2241–2252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeeles J. T., Poli J., Marians K. J., Pasero P., Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 5, a012815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hromas R., et al. , The endonuclease EEPD1 mediates synthetic lethality in RAD52-depleted BRCA1 mutant breast cancer cells. Breast Cancer Res. 19, 122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fenech M., et al. , Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 26, 125–132 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Vitale I., Galluzzi L., Castedo M., Kroemer G., Mitotic catastrophe: A mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 12, 385–392 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Hills S. A., Diffley J. F., DNA replication and oncogene-induced replicative stress. Curr. Biol. 24, R435–R444 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Castedo M., et al. , Cell death by mitotic catastrophe: A molecular definition. Oncogene 23, 2825–2837 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Meijer H. A., Smith E. M., Bushell M., Regulation of miRNA strand selection: Follow the leader? Biochem. Soc. Trans. 42, 1135–1140 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Choo K. B., Soon Y. L., Nguyen P. N., Hiew M. S., Huang C. J., MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J. Biomed. Sci. 21, 95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bunting S. F., et al. , 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edwards S. L., et al. , Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451, 1111–1115 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Montoni A., Robu M., Pouliot E., Shah G. M., Resistance to PARP-inhibitors in cancer therapy. Front. Pharmacol. 4, 18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.