Significance
The endoplasmic reticulum (ER) deteriorates with age and fails to mount an effective stress response against misfolded proteins (UPRER), leading to protein folding disorders. However, preconditioning the ER using a mild ER stress (ER hormesis) can protect against future insults. We show that dietary restriction, an intervention that protects against protein misfolding disorders and increases life span across species, uses ER hormesis as a mechanism of action. Simply mimicking the ER hormesis in Caenorhabditis elegans by transient treatment with pharmacological reagents leads to delayed age-onset failure of UPRER, better capacity to process misfolded proteins, and increased life span. We also show that this process may be conserved in a mammalian cellular model of neurodegenerative disease.
Keywords: endoplasmic reticulum, dietary restriction, hormesis, life span, aging
Abstract
Unfolded protein response (UPR) of the endoplasmic reticulum (UPRER) helps maintain proteostasis in the cell. The ability to mount an effective UPRER to external stress (iUPRER) decreases with age and is linked to the pathophysiology of multiple age-related disorders. Here, we show that a transient pharmacological ER stress, imposed early in development on Caenorhabditis elegans, enhances proteostasis, prevents iUPRER decline with age, and increases adult life span. Importantly, dietary restriction (DR), that has a conserved positive effect on life span, employs this mechanism of ER hormesis for longevity assurance. We found that only the IRE-1–XBP-1 branch of UPRER is required for the longevity effects, resulting in increased ER-associated degradation (ERAD) gene expression and degradation of ER resident proteins during DR. Further, both ER hormesis and DR protect against polyglutamine aggregation in an IRE-1–dependent manner. We show that the DR-specific FOXA transcription factor PHA-4 transcriptionally regulates the genes required for ER homeostasis and is required for ER preconditioning-induced life span extension. Finally, we show that ER hormesis improves proteostasis and viability in a mammalian cellular model of neurodegenerative disease. Together, our study identifies a mechanism by which DR offers its benefits and opens the possibility of using ER-targeted pharmacological interventions to mimic the prolongevity effects of DR.
A vast majority of the secreted as well as membrane proteins fold and mature in the endoplasmic reticulum (ER) before they are exported to their destinations (1). The protein folding capacity of the ER is carefully monitored and calibrated by 3 conserved signal transducers, collectively called the unfolded protein response (UPRER) regulators, that ensure protein homeostasis (proteostasis) and proper cellular function. This is achieved by activating the proteostasis network (PN) consisting of molecular chaperons, protein degradation machinery, and stress response pathways that act to resolve consequences of protein misfolding in the ER (2). In metazoan ER, the 3 arms of UPRER, namely IRE1, PERK, and ATF6, function in parallel to trigger the PN and counteract ER stress by 1) increasing folding capacity through the expression of various chaperons, 2) attenuating translation to reduce protein load in ER, 3) regulating IRE1-dependent decay of mRNA (RIDD), or by 4) activating ER-associated proteasomal degradation (ERAD) to remove terminally misfolded proteins (3–5).
Aging is characterized by a catastrophic collapse of the proteostasis network due to the loss of protein quality-control machineries across the cellular compartments (2, 6–8). With age, the ER structure begins to deteriorate and the ER health fails as it is unable to mount an optimal UPRER in response to external stress (induced UPRER [iUPRER]) (6, 8–10). Expression levels of key ER mRNA and proteins as well as activities of ER resident proteins, including BiP, PDI, calnexin, and GRP94, are known to decline with age (11–13). As a possible consequence, many diseases of protein misfolding, including Alzheimer’s, Parkinson’s, and Huntington’s diseases, are age-onset disorders.
The flagging cellular proteostasis can be improved simply by exposing cells or organisms to moderate stress before they encounter any acute stress. This process is called hormesis and has been shown to be beneficial to life span and health (14–16). Hormesis can be affected in a compartment-specific manner in a cell. For example, in Caenorhabditis elegans, Drosophila, and human fibroblasts, hormetic heat shock up-regulates cytosolic molecular chaperones that further protect against acute heat stress (17–19). In C. elegans, glucose restriction increases mitochondrial respiration and reactive oxygen species (ROS) production, which results in protection against oxidative stress and enhanced longevity by a process termed mitohormesis (20). Mitohormesis has also been implicated in other long-lived mutants in worms (21, 22) and fly (23). Similarly, ER hormesis has been observed in Drosophila carrying mutations associated with ER protein folding/degradation machinery that induce cytoprotective response toward subsequent ER insults (24, 25). In mammalian cells, ER stress preconditioning protects against brain ischemia and attenuates heart ischemia/reperfusion injury (26, 27). ER preconditioning is also neuroprotective in Drosophila and mice models of Parkinson’s disease (28). Recently, mutation in the C. elegans heterochromatin protein like-2 (HPL-2), was shown to produce a hormetic induction of stress resistance dependent on the IRE-1 branch of UPRER (29). Further, overexpression of xbp-1s (the transcription factor otherwise induced in response to ER stress) in neurons has been shown to increase longevity in a cell nonautonomous manner (9), suggesting a possible role of ER hormesis in life span regulation.
Dietary restriction (DR) is a conserved intervention that can delay proteostasis, activate cytoprotective pathways, and increase life span across species (30). It has been suggested that DR may act as a mild stress that enhances life span through hormesis (31). The fact that glucose restriction, that is akin to DR, increases life span through mitohormesis attests to this (20). However, the involvement of ER hormesis in DR is not known. Here, using 2 genetic models of DR, we show that dietary restriction transiently up-regulates UPRER during early larval development, possibly as a result of reduced protein glycosylation. Mimicking this response with a pharmacological glycosylation inhibitor, tunicamycin (Tm) or a nonhydrolyzable glucose analog (2-deoxyglucose), increases life span. Both transient UPRER and increased life span depend on the ER stress sensor, IRE-1. We show that the transient ER stress leads to better iUPRER at adulthood and better proteostasis. Consequently, ERAD genes are up-regulated by DR, leading to efficient degradation of ER resident proteins and thus contributes toward enhanced proteostasis in adults and increased longevity. The DR-specific FOXA transcription factor PHA-4 transcriptionally regulates UPRER during early larval development as well as the increase in ER protein processing genes during adulthood in DR worms, justifying its central role in DR-associated longevity assurance. Finally, we found that transient ER stress improves proteostasis in C. elegans as well as in a mammalian cell culture model of polyglutamine aggregation-induced proteotoxicity. Thus, our study elucidates a mechanism by which DR positively affects life span and shows that transient nutrient restriction or ER-directed pharmacological intervention during development may be an effective intervention to treat protein misfolding disorders as well as increase life span.
Results
DR Triggers a Transient Up-Regulation of UPRER during Early Larval Development.
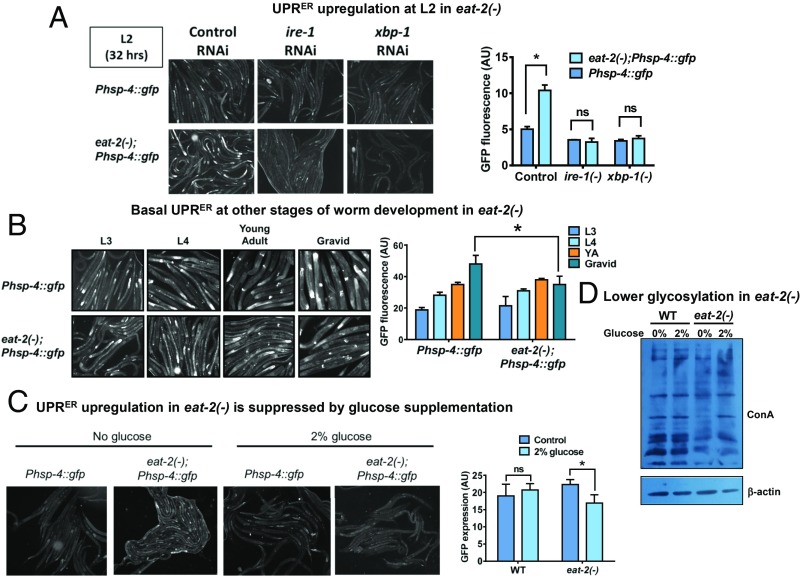
In order to elucidate the kinetics of UPRER during nutrient stress, we evaluated the levels of UPRER in the 2 genetic paradigms of DR, namely eat-2(ad1116) (32) and drl-1 RNAi (33). For this, we used the Phsp-4:gfp(zcIs4) transgenic strain, where the promoter of hsp-4 is transcriptionally fused to gfp and GFP fluorescence gives a quantitative readout of UPRER (34, 35). We compared the GFP fluorescence in Phsp-4:gfp(zcIs4) and eat-2(ad1116);Phsp-4:gfp(zcIs4) at 32 to 35 h after eggs were placed on the plates (egg synchronization) and found that DR triggered a transient up-regulation of UPRER specifically during the L2 larval stage of eat-2(ad1116) (Fig. 1A). The transient up-regulation was observed irrespective of the way the eggs were obtained, either by bleaching or by synchronized egg lay of gravid adult worms (SI Appendix, Fig. S1A). The basal UPRER was found to be the same in WT or DR worms in other larval stages, suggesting that only an early-life transient up-regulation of UPRER is specific to conditions that mimic DR (Fig. 1B). Likewise, we grew Phsp-4:gfp(zcIs4) on control or drl-1 RNAi and found that DR implemented by drl-1 knockdown (KD) triggered a similar transient up-regulation of UPRER specifically during the L2 larval stage (SI Appendix, Fig. S1 B and C). Importantly, this response was found to be ER specific as the cytosolic [Phsp-16.2:gfp(dvIs20)] or mitochondrial [Phsp-6:gfp(zcIs9)] stress response reporters were not affected (SI Appendix, Fig. S1D). This early and transient up-regulation of the ER stress response was found to be dependent on the IRE-1–XBP branch of UPRER as eat-2(ad1116);Phsp-4:gfp(zcIs4) grown on ire-1 or xbp-1 RNAi were unable to show similar response (Fig. 1A). Also, ire-1(zc14);Phsp-4:gfp(zcIs4) worms grown on drl-1 RNAi failed to mount the transient UPRER at L2 (SI Appendix, Fig. S1E). Further, 3 other ER resident proteins, ER chaperons calnexin ortholog cnx-1 and enpl-1 as well as ER oxido-reductin ortholog ero-1 are transcriptionally up-regulated in eat-2(ad1116) (SI Appendix, Fig. S2A) with distinct temporal kinetics. Together, an early and transiently high basal UPRER in eat-2(ad1116) mutants and on drl-1 knockdown suggests higher ER stress levels at initial larval life in worms undergoing a DR-like condition since hatching.
Fig. 1.
DR triggers a transient up-regulation of UPRER early in life. (A) Representative images of transient UPRER of Phsp-4:gfp(zcIs4) and eat-2(ad1116);Phsp-4:gfp(zcIs4) worms at L2 (32 h after egg synchronization) grown on control, ire-1, or xbp-1 RNAi. Densitometric quantification shows the up-regulation of UPRER on control RNAi but not on ire-1 and xbp-1 RNAi. Average of 2 biological replicates is shown. Error bars show SD. *P < 0.05 by 2-way ANOVA. (B) Representative images of Phsp-4:gfp(zcIs4) and eat-2(ad1116);Phsp-4:gfp(zcIs4) at indicated larval stages and in gravid adults. Densitometric quantification shows no up-regulation in basal UPRER in these stages in eat-2(ad1116) compared to WT. Basal UPRER is down-regulated in eat-2(ad1116) in gravid adults. Average of 2 biological replicates is shown. Error bars show SD. *P < 0.05 by Student’s t test. YA, young adult. (C) Representative images of transient UPRER of Phsp-4:gfp(zcIs4) and eat-2(ad1116);Phsp-4:gfp(zcIs4) L2 stage worms grown in presence or absence of 2% glucose. The transient UPRER is suppressed in presence of glucose in eat-2(ad1116);Phsp-4:gfp(zcIs4). Average of 3 biological replicates is shown. Error bars show SD. *P < 0.05 by 2-way ANOVA. ns, nonsignificant. (D) Concavalin A Western analysis of WT or eat-2(ad1116) with or without 2% glucose supplementation. WT has more glycosylated proteins compared to eat-2(ad1116). Glucose supplementation increases the levels of glycosylated proteins. Quantified Western blot with different amounts of loaded proteins is presented in SI Appendix, Fig. S2B.
DR may lead to depletion in the availability of glucose for metabolism that may cause the observed up-regulation of basal UPRER. If that is the case, supplementing glucose to worms undergoing DR should suppress the transient UPRER up-regulation. We used media supplemented with 2% glucose and grew the Phsp-4:gfp(zcIs4) or eat-2(ad1116);Phsp-4:gfp(zcIs4) worms on them. We observed that the transient UPRER up-regulation is mitigated on glucose-containing media in eat-2(ad1116) (Fig. 1C), suggesting that glucose deprivation may cause the early ER stress during DR. Glucose is essential for protein glycosylation and hence ER proteostasis; depletion could decrease protein glycosylation and increase ER stress. Consistent with this hypothesis, we found that eat-2(ad1116) worms have lower levels of protein glycosylation compared to WT, using a concavalin A (ConA) Western assay (Fig. 1D and SI Appendix, Fig. S2B for quantification). The level of ConA signal increases when the worms are supplemented with glucose. Finally, we show that supplementing Phsp-4:gfp(zcIs4) with 2-deoxyglucose (2DG), a nonhydrolyzable glucose analog that can bring about transient glucose restriction and is often used to initiate DR, leads to increased expression of GFP that is suppressed by addition of glucose (SI Appendix, Fig. S2C). Together, DR triggers a transient up-regulation of UPRER, possibly as a result of glucose restriction.
Life Span Extension by DR Is Dependent on IRE-1.
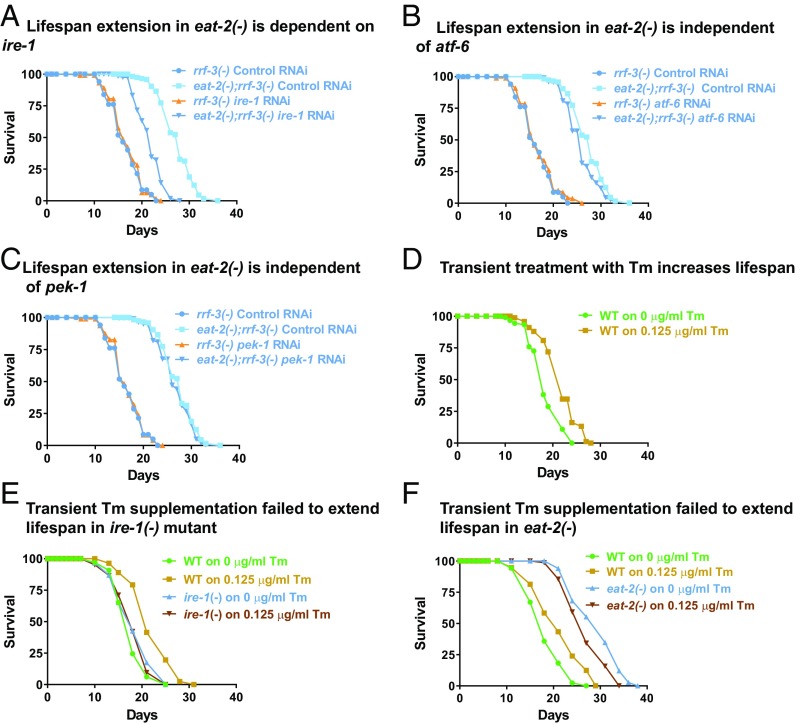
Knocking down the eat-2 gene or implementing bacterial dilution-mediated dietary restriction (BDR) has been shown to decrease proteotoxicity and enhance longevity in adult worms (36). Since in higher eukaryotes, ER is responsible for folding a considerable part of the proteome, we investigated the importance of the UPRER machinery in proteoprotective, longevity benefits conferred by DR. Toward this end, life span analysis was performed in 2 genetic mimics of DR, in the presence or absence of ire-1, atf-6, or pek-1, the ER membrane proteins that function to sense misfolded protein stress. Interestingly, knocking down ire-1 led to a significant suppression in life span of eat-2(ad1116) (Fig. 2A and SI Appendix, Table S1) while the life span extension by drl-1 KD was completely abolished in an ire-1 mutant ire-1(v33) (SI Appendix, Fig. S3A and Table S1). However, the other signal sensors, i.e., pek-1 and atf-6 were found to be dispensable for both the genetic paradigms of DR-mediated longevity (Fig. 2 B and C and SI Appendix, Fig. S3 B and C and Table S1). It may be noted that the RNAi interventions efficiently decreased transcript levels of ire-1, atf-6, as well as pek-1 in WT and eat-2(ad1116) (SI Appendix, Fig. S3D). In order to enhance the RNAi efficiency in the eat-2 mutant, the rrf-3 mutant background was used (37). These data suggest that specific UPRER signaling through IRE-1 is required for eat-2 and drl-1 KD-mediated life span extension.
Fig. 2.
Ire-1 is required for longevity assurance by DR and transient Tm supplementation. (A) Life span extension in eat-2(ad1116) mutant is suppressed on knocking down ire-1. (B) Atf-6 knockdown has little effect on eat-2(ad1116) life span. (C) Pek-1 knockdown has no effect on life span extension in eat-2(ad1116) mutant. (D) Transient supplementation of Tm during initial larval stage is sufficient to extend adult life span. WT worms were bleached and eggs were added to M9 buffer containing bacterial feed and supplemented with 0.125 μg/mL Tm for the first 24 h and later scored for adult life span. As a control, worms were treated with 0.05% DMSO (Tm = 0 μg/mL). (E) Tm-mediated increase in life span is dependent on ire-1. WT and ire-1(v33) mutants were supplemented with Tm (0.125 μg/mL) for the initial 24 h and later scored for adult life span. (F) Tm supplementation cannot further increase life span of eat-2(ad1116). Complete life span data are presented in SI Appendix, Table S1.
Early and Transient Up-Regulation of UPRER Is Sufficient for Life Span Extension.
Since we observed the transient up-regulation of UPRER, we asked whether it plays a causal role in life span extension, as observed during DR. For this, we mimicked the transient UPRER up-regulation using an external supplementation of a small dose of Tm for 24 h during hatching of the eggs in liquid culture. After the drug was washed off, the worms were grown on solid nematode growth media (NGM) and postadult life span was recorded. Interestingly, we found that exposing WT larvae to 0.125 μg/mL of Tm during the first 24 h of its postembryonic life led to significant extension of life span (Fig. 2D). Similar to DR, this longevity effect was completely dependent on ire-1 (Fig. 2E). This observation supports the role of hormesis, triggered by an early ER stress as a mechanism of DR-mediated life span enhancement, in the models that we tested. Importantly, pharmacological ER stress could not further extend the life span of eat-2(ad1116) worms (Fig. 2F), suggesting that these 2 interventions use similar adaptive response to retard aging. Additionally, transiently supplementing 2DG was also capable of significantly increasing life span, in an ire-1–dependent manner (SI Appendix, Fig. S3 E and F). These results suggest that a hormetic dose of ER stress may be responsible for increased life span observed during DR.
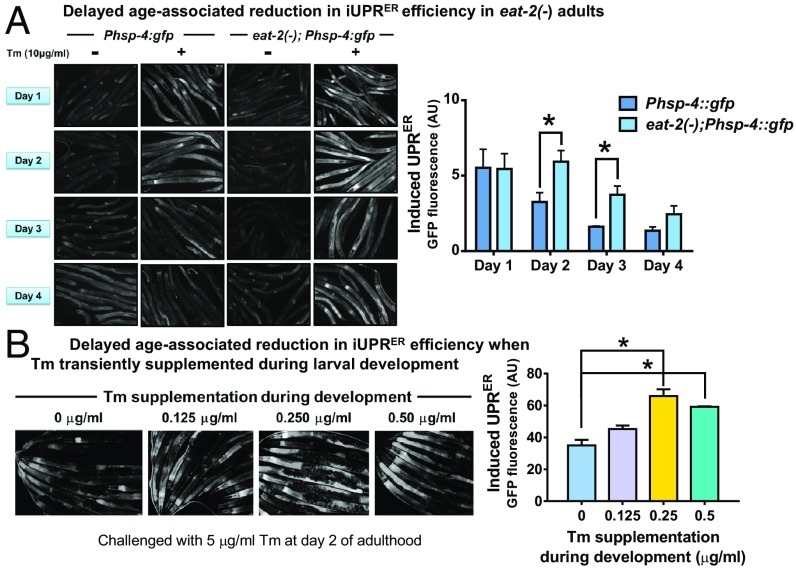
DR and a Hormetic Dose of ER Stress Delays the Age-Related Decline in iUPRER Efficiency.
The efficiency of the ER stress response decreases with age, resulting in a progressively dysfunctional ER. Not only the levels of various UPRER target genes decrease in old rats and mice, the efficiency of mounting UPRER in response to various insults (iUPRER) also decreases with age (9, 38, 39). We asked whether the iUPRER efficiency is maintained in the genetic models of DR on different days of adulthood. As previously reported, we also observed a decline in iUPRER efficiency on the second and third days of adulthood in WT worms, respectively (Fig. 3A [dark blue bars], SI Appendix, Fig. 3 S4A [dark green bars]). Importantly, this decline was significantly delayed in DR worms as both eat-2(ad1116);Phsp-4:gfp as well as Phsp-4:gfp worms grown on drl-1 RNAi displayed higher inductions of GFP fluorescence following acute Tm treatment (Fig. 3A [light blue bars] and SI Appendix, Fig. 3 S4A [light green bars]). The DR worms also maintained low basal levels of UPRER similar to that observed in daf-2(e1370) (40) (SI Appendix, Fig. S4 B and C).
Fig. 3.
DR delays age-associated reduction in iUPRER efficiency. (A) eat-2(ad1116) mutants have delayed age-associated decline in iUPRER efficiency. Representative images of Phsp-4:gfp and eat-2(ad1116);Phsp-4:gfp worms with/without acute Tm treatment (10 μg/mL for 6 h) on different days of adulthood. Graph represents normalized fold induction in GFP fluorescence after 6 h of Tm treatment in WT and eat-2(ad1116) on different days of adulthood, averaged over 3 biological repeats. (Normalization was performed with the basal GFP fluorescence [basal UPRER] for individual experiments). (B) Transient Tm supplementation increases iUPRER efficiency at day 2 of adulthood. Representative images of day 2 adult Phsp-4:gfp(zcIs4) worms grown on control RNAi, treated with different concentrations of Tm for 24 h after egg synchronization and further challenged with Tm (5 μg/mL for 6 h) on day 2 of adulthood. Graph represents normalized fold induction in GFP fluorescence averaged over 2 biological repeats. Error bars are SEM. * represents P < 0.05, Student’s t test.
Since we observed that transient ER stress using Tm at early postembryonic stage increased life span, we asked whether this treatment could also lead to a better iUPRER efficiency with age. Toward this end, we checked the iUPRER efficiency of transiently Tm-treated worms on day 2 of adulthood and found that these worms displayed higher iUPRER efficiency (Fig. 3B). These observations suggest that ER in DR worms are maintained in a healthier state and are more potent in combating external stressors even at a later age, when the WT ER efficiency typically declines. Importantly, hormetic up-regulation of the ER stress response during early developmental life leads to this increased ER efficiency contributing toward enhanced longevity during DR.
DR Increases the Expression of Genes Responsible for Protein Homeostasis within ER.
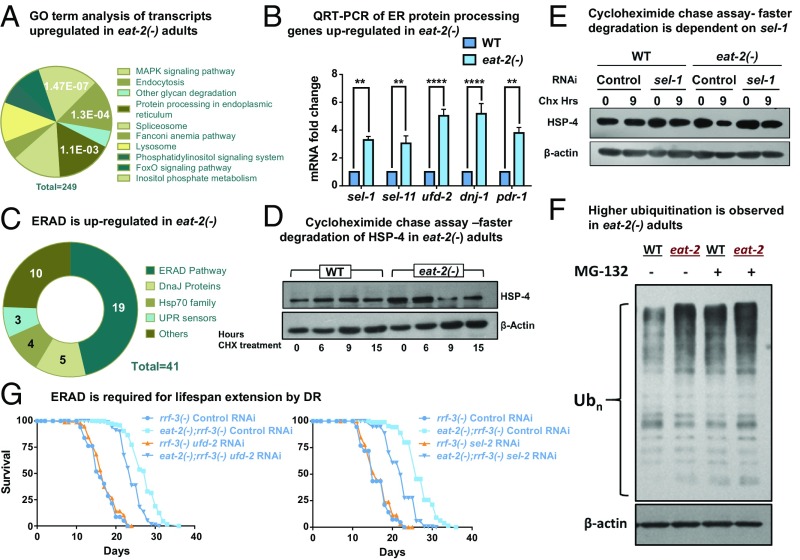
Our data suggest that DR may help to partly combat the proteostasis collapse that occurs within the first 2 d of adult life (41), through a hormetic up-regulation of UPRER during development. In an effort to better understand this process during DR, we assessed the changes in the global transcriptome profile of wild type and eat-2(ad1116) at day 1 of adulthood (42, 43). We determined the Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for the genes that were up-regulated >2-fold (P value ≤0.05) in eat-2(ad1116) mutant as compared to WT and found that genes associated with protein folding functions within the ER were significantly enriched in this set (Fig. 4A). A total of 41 transcripts comprising the proximal UPR sensors (ire-1, atf-6, and pek-1), hsp-70 family members, J-domain proteins, and ERAD components were up-regulated (SI Appendix, Table S2). Many of these genes are well-known UPRER targets. These results suggested that DR leads to a transcriptional reprogramming that supports a more efficient ER.
Fig. 4.
ER protein processing machinery is transcriptionally up-regulated in DR adults. (A) Pie chart showing KEGG pathway/GO analysis of the transcripts up-regulated in eat-2(ad1116) worms as compared to WT at day 1 of adulthood. Forty-one transcripts pertaining to protein processing functions in ER were up-regulated (out of a total of 249 up-regulated genes), according to the analysis performed using the DAVID functional annotation tool. The listed pathways have P < 0.05 and FDR < 10 (Bonferroni corrected P = 0.0011). (B) QRT-PCR analysis to detect the expression of ER protein processing genes reports significant up-regulation in eat-2(ad1116) adults as compared to WT at day 1. Error bars indicate SEM over 3 independent biological replicates. **P < 0.01, ****P < 0.0001, Student’s t test. (C) ERAD genes are up-regulated in eat-2(ad1116) worms. Pie chart representing classification of the 41 up-regulated ER genes. Approximately 50% (19 out of 41) of up-regulated genes function in the ERAD. (D) ER resident protein HSP-4 is degraded faster in eat-2(ad1116). Day 1 adult WT and eat-2(ad1116) worms were treated with cycloheximide (2 mg/mL) for the indicated time and HSP-4 levels were detected by Western blot. (E) HSP-4 degradation is partly abrogated when sel-1 is knocked down. Day 1 adult WT and eat-2(ad1116) worms grown on control or sel-1 RNAi were treated with cycloheximide (2 mg/mL) for the indicated time and HSP-4 levels were detected by Western blot. (F) The eat-2 mutant has higher levels of ubiquitinated proteins. Ubiquitinated proteins were detected using a polyubiquitin antibody in WT and eat-2(ad1116) adults with or without proteasomal inhibitor, MG132 treatment. β-Actin was used as a measure of equal loading of protein. (G) DR-mediated longevity is dependent on ERAD genes ufd-2 and sel-1. Life span extension in eat-2(ad1116) worms was partially suppressed on knocking down ufd-2 or sel-1 using RNAi. Complete life span data are present in SI Appendix, Table S1.
We performed quantitative real-time PCR (QRT-PCR) analysis to validate our sequencing results. The mRNA levels of 5 of the UPR target genes, sel-1, sel-11, ufd-2, pdr-1, and dnj-1, show up-regulation in eat-2 mutant worms as compared to WT (Fig. 4B). This increase in expression starts early in development in eat-2 mutant worms undergoing DR and is sustained until adulthood (SI Appendix, Fig. S5A). Similarly, implementing DR by knocking down drl-1 also led to a significant increase in the mRNA level of these UPR target genes during adulthood, suggesting conserved up-regulation of this pathway in different DR paradigms (SI Appendix, Fig. S5B). We further identified that the up-regulation of these UPR genes is dependent on the UPR sensor ire-1, in both the eat-2 and drl-1 paradigms (SI Appendix, Fig. S5 C and D).
ERAD in DR Worms Efficiently Degrades ER Proteins.
ERAD is an adaptive mechanism of the ER that regulates protein homeostasis within the organelle (44). It involves degradation of the misfolded proteins so as to prevent their accumulation inside the ER lumen. In the DR transcriptome mentioned above, genes pertaining to the ERAD pathway formed almost half of the enriched ER gene set (19 out of 41 up-regulated transcripts) (Fig. 4C and SI Appendix, Table S2). This indicated that DR in the 2 paradigms that we tested, may transcriptionally up-regulate ERAD, possibly to maintain proteostasis within the organelle at adulthood. To validate this, we performed a cycloheximide chase assay in WT and eat-2(ad1116) worms on day 1 of adulthood to probe the turnover kinetics of an ER resident protein (Fig. 4D). Protein abundance at a particular time is a cumulative result of active protein synthesis and degradation. Cycloheximide is a protein synthesis inhibitor and supplementation of this drug followed by Western blotting allows for specific reporting of the rate of protein degradation. We compared the levels of HSP-4, an ER resident protein, after cycloheximide treatment in WT and eat-2 mutant and found a faster degradation of HSP-4 in DR worms (Fig. 4D) that was abrogated on sel-1 knockdown using RNAi (Fig. 4E). SEL-1 (ortholog of mammalian HRD3) is a vital component of HRD-3/HRD-1 E3 ubiquitin ligase complex (44–47). This indicates a more competent ERAD machinery operating within DR worms as compared to their wild-type counterpart. Proteins destined for degradation are affixed with a polyubiquitin chain that serves as a degradation signal. Therefore, we assessed the changes in ubiquitination profile of WT and DR worms, by performing a Western blot using a polyubiquitin antibody. Consistently with a robust ERAD, we observed higher ubiquitination of proteins in eat-2(ad1116), suggesting that more proteins are destined for degradation under DR (Fig. 4F). Better ERAD is also indicative of a healthy ER that may contribute to the increase in longevity of DR worms.
Life Span Extension by DR Is Dependent on ERAD Genes.
As we found evidence of an up-regulated ERAD during DR, we next asked whether it is required for promoting increased longevity. We performed life span analysis of WT and eat-2(ad1116) worms on bacteria expressing RNAi against 2 of the up-regulated ERAD genes, ufd-2 and sel-1. UFD-2 functions in complex with CDC48 to regulate the length of polyubiquitin chains attached to proteins (44–47). We found that life span extension of eat-2(ad1116) was partially suppressed on knocking down ufd-2 or sel-1 (Fig. 4G and SI Appendix, Table S1), supporting their important role in DR-mediated longevity. The inability to observe a complete repression of DR life span is possibly due to the existence of different ERAD branches and redundancy in the functions performed by them. Altogether, DR promotes a healthy ER that can mount an effective ER stress response with age while efficiently degrading ER resident proteins to maintain ER homeostasis and increase life span.
FOXA Transcription Factor PHA-4 Controls Important Aspects of ER Homeostasis.
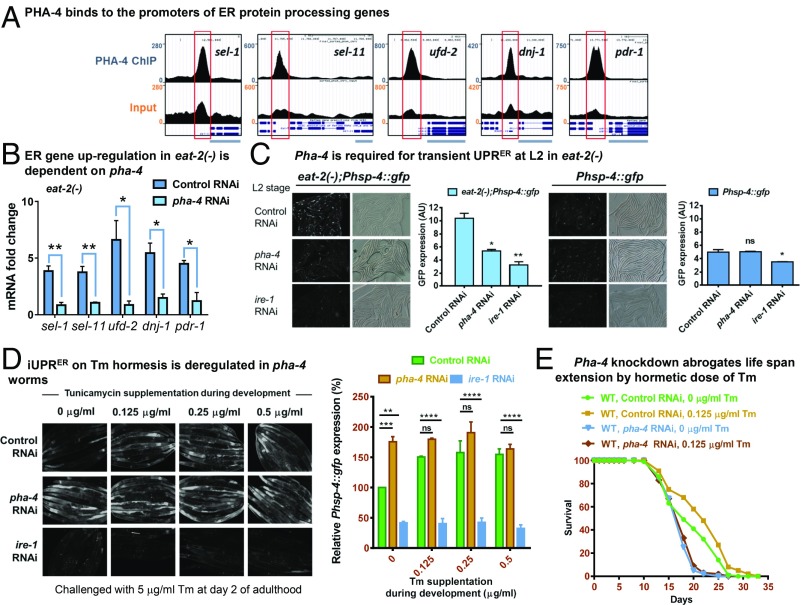
The C. elegans FOXA transcription factor PHA-4 is required for increased life span that is observed in multiple genetic as well as nongenetic paradigms of DR (33, 43, 48, 49). Hence, we asked whether up-regulation of the ER protein processing genes in an eat-2 mutant worm is dependent on PHA-4. Using metadata analysis of published PHA-4 ChIP-seq data (43, 50), we found that PHA-4 binds to the promoter proximal sites of all of the 5 ER protein processing genes that are up-regulated in eat-2(ad1116) (Fig. 5A). QRT-PCR analysis showed that knocking down pha-4 significantly suppressed the induction of these genes in eat-2(ad1116) at young adult stage (Fig. 5B). This suggests that DR transcriptionally up-regulates genes that maintain protein folding homeostasis in the ER during early adulthood, in a manner dependent on ER sensor ire-1 and DR-specific FOXA transcription factor PHA-4.
Fig. 5.
PHA-4 regulates diverse aspect of ER homeostasis during DR. (A) University of California, Santa Cruz browser view of PHA-4/FOXA peaks on promoter proximal regions of ER protein processing genes, as determined by ChIP-seq analysis of the unc-119(ed3)III;wgIs37(OP37) strain; data mined from MODENCODE and reanalyzed using our bioinformatic pipeline. Red boxes indicate the promoter regions where peaks are observed. (Upper) PHA-4 ChIP using anti-GFP antibody. (Lower) Input DNA. Light blue bars represent a portion of the gene of interest. (B) QRT-PCR analysis was used to compare ER protein processing genes between eat-2(ad1116) and WT adults grown on control or pha-4 RNAi. Graph shows significant suppression of the up-regulated genes on pha-4 knockdown. Average of 3 biological replicates is shown. (C) Knocking down pha-4 by RNAi prevents transient UPRER up-regulation at L2 (32 h postegg synchronization) in eat-2(ad1116). Ire-1 RNAi is used as control. Quantification shown on Right of each figure. Average of 2 replicates is shown. (D) iUPRER is deregulated on pha-4 knockdown. Phsp-4:gfp(zcIs4) worms, grown on control, pha-4, or ire-1 RNAi, with or without treatment with hormetic dose of Tm were challenged with 5 μg/mL of Tm at day 2 of adulthood. Quantification is provided on Right side. Average of 2 replicates is shown. Error bars are SEM in all cases, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Student’s t test. ns, nonsignificant. (E) The life span of WT worms given a hormetic dose of Tm is dependent on PHA-4. WT worms grown on control or pha-4 RNAi were supplemented with Tm (0.125 μg/mL) for the initial 24 h and later scored for adult life span. Complete life span data are presented in SI Appendix, Table S1.
Next, we asked whether PHA-4/FOXA is involved in any other aspect of ER homeostasis. For that, we knocked down pha-4 using RNAi is hsp-4::gfp or eat-2(ad1116)::hsp-4::gfp and measured the transient up-regulation of ER stress early during development as seen in the genetic paradigms of DR. We found that the eat-2(ad1116) worms lose the capability to mount the transient response at L2 stage when pha-4 is knocked down (Fig. 5C), while there was no effect of pha-4 RNAi on hsp-4::gfp in WT background. Next, we asked whether PHA-4 regulates the iUPRER at adulthood when the worms are preconditioned at L2 with a hormetic dose of ER stress using Tm. Intriguingly, in absence of preconditioning, the adult pha-4 knockdown worms mounted an iUPRER (on acute 5 mg/mL tunicamycin treatment) (Fig. 5D; compare panels under 0 μg/mL column) that was much higher than the control RNAi worms. On the other hand, while the control RNAi worms mounted an even higher iUPRER when given Tm hormesis (compare panels in control RNAi row), pha-4 RNAi worms were not able to increase the iUPRER response, suggesting that the process is deregulated in absence of PHA-4. Consequently, knocking down PHA-4 suppressed the Tm hormesis-mediated life span extension (Fig. 5E; compare panels in control RNAi). Coupled to many other DR-specific functions of PHA-4, these observations justify the important role of this transcription factor in DR-mediated life span extension.
DR Alleviates Polyglutamine Aggregation in an ire-1- and ERAD-Dependent Manner.
In the context of protein homeostasis, a better health span will translate into fewer deleterious proteins accumulating or aggregating within the cell. Proteins that carry a stretch of polyglutamine residue over a threshold length are known to aggregate with age. These form the basis for various polyglutamine expansion disorders like Huntington’s and spinocerebellar ataxias (51). The exposed hydrophobic patches formed by multiple glutamine residues, typically more than 35, increases the chances of protein aggregation. Inefficient degradation of these proteins with increasing age exacerbates aggregation and associated diseases (52, 53). Supporting this is the observation that impaired ERAD and ER stress are primary events that augment polyglutamine toxicity (54). C. elegans is used as a model organism to study polyglutamine aggregation owing to the availability of fluorescent reporters that help in visualizing aggregate formation in different tissues, in real time.
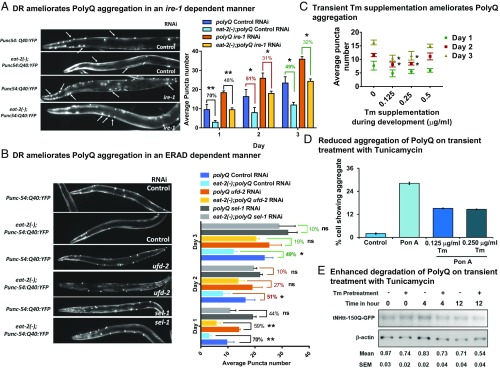
In order to evaluate the role of DR in augmenting the ERAD and its physiological consequences, we used a reporter that expresses a stretch of 40 glutamines (Q40) tagged with a yellow fluorescent protein in C. elegans muscle (55). We crossed this reporter strain with eat-2(ad1116) to generate mutant worms carrying both the reporter transgene and the eat-2 mutation [hereafter, referred to as eat-2(−);polyQ]. We observed that incorporating eat-2 mutation led to 50 to 70% suppression in the number of aggregates at all age points, supporting the earlier observation of alleviation in the age-associated increase in polyQ aggregation during DR (Fig. 6A) (36). Additionally, the SDS-soluble form of polyQ protein was more in eat-2 mutant worms compared to WT, suggesting that partitioning of polyQ into SDS-insoluble aggregates decreased in eat-2 worms (SI Appendix, Fig. S6A). This may exclude the possibility that lower level of polyQ expression in eat-2 mutants led to decrease in puncta numbers. Knocking down drl-1 also decreased the number of aggregates observed in the reporter worm with age, suggesting a conserved phenomenon of DR-mediated reduction in aggregation (SI Appendix, Fig. S6B). Since ire-1 was a requirement for DR-mediated longevity, we therefore asked whether this beneficial effect conferred by DR is also dependent on this gene. We observed that the decrease in polyQ aggregation observed in eat-2 mutant worms was abrogated on ire-1 knockdown (Fig. 6A). Supporting this was the observation that SDS-soluble polyQ protein decreased in the eat-2(ad1116) with ire-1 RNAi knockdown (SI Appendix, Fig. S6A). Similarly, ire-1 mutants failed to suppress the number of polyQ aggregates on drl-1 knockdown at different days of adulthood, suggesting a dependence on ire-1 branch of UPRER for maintenance of proteostasis (SI Appendix, Fig. S6B). It may be noted that the puncta numbers increased in wild type when ire-1 is knocked down or mutated, showing its role in proteostasis.
Fig. 6.
Suppression of polyQ aggregation with age in DR is dependent on ire-1 and ERAD. (A) Representative images showing polyQ aggregates in Punc-54:polyQ40:yfp and eat-2(ad1116);Punc-54:polyQ40:yfp worms on control or ire-1 RNAi at day 2 of adulthood. Suppression of polyQ aggregation in eat-2 mutants is partly abrogated on knocking down ire-1. Average puncta number per worm from 3 biological replicates is plotted. Error bars show SD. (B) Representative images showing polyQ aggregates for Punc-54:polyQ40:yfp and eat-2(ad1116);Punc-54:polyQ40:yfp worms on control RNAi or RNAi against ERAD genes (ufd-2 or sel-1) at day 2 of adulthood. Suppression of polyQ aggregation in eat-2 mutants is partially abrogated on knocking down ufd-2 and sel-1. Average of 3 biological replicates is plotted. Error bars show SD. (C) PolyQ aggregates in Punc-54:polyQ40:yfp worms on control RNAi that have been exposed to different concentrations of Tm during early larval development. Graph represents number of aggregates in Punc-54:polyQ40:yfp worms on different days of adulthood. Worms exposed to 0.125 µg/mL and 0.25 µg/mL Tm during larval development show significant reduction in the number of aggregates on day 2 and day 3. Average of 3 biological replicates is plotted. Error bars show SEM. Representative images shown in SI Appendix, Fig. S6C. *P < 0.05, **P < 0.01 as determined by Student’s t test; ns, nonsignificant. (D) HD150Q cells were pretreated with tunicamycin for 5 h, washed, and then incubated with Pon A for 24 h (which was then washed off) to induce the expression of mutant huntingtin-GFP. At 36 h postinduction, cells were observed under fluorescence microscope to assess the formation of mutant huntingtin aggregates. Representative images are presented in SI Appendix, Fig. S7A. (E) Cells were treated as above and after washing off Pon A, chased following cycloheximide treatment for different time periods. The lysates were subjected to immunoblot analysis of mutant huntingtin-GFP using GFP antibody. Data were normalized with β-actin. Values are mean ± SEM of 3 independent experiments.
Finally, to further evaluate the role of ERAD machinery in DR-induced suppression of polyglutamine aggregation, eat-2(-);polyQ worms were grown on control, ufd-2, or sel-1 RNAi. We found that eat-2(ad1116) worms grown on ERAD gene RNAi had less suppression of polyQ puncta (Fig. 6B) and a lesser level of SDS-soluble polyQ protein (SI Appendix, Fig. S6A) compared to control RNAi-grown worms. This suggests that DR leads to a healthier ER that reduces polyglutamine aggregation in adult worms in a manner dependent on ire-1 and ERAD. It may be noted that the suppression of aggregation is not complete either due to the efficiency of the RNAi or due to redundant pathways that may be involved.
A Hormetic Dose of ER Stress, during Early Life, Suppresses polyQ Aggregation.
We reported that an atypical up-regulation of UPRER in larvae contributes toward increased life span during DR. We showed that pharmaceutically mimicking the transient UPRER up-regulation during early larval life augmented the iUPRER efficiency with age and increased life span. To determine whether such hormetic intervention can protect against age-related proteostasis collapse, we exposed WT worms with a range of concentrations to Tm only during the start of larval development, as mentioned before. We compared the untreated and Tm-treated polyQ worms for the number of aggregates on different days of adulthood and witnessed a suppression in the treated worms (Fig. 6C and SI Appendix, Fig. S6C). Moreover, this phenomenon was also found to be dependent on the proximal UPR sensor ire-1, as in ire-1(v33);Q40:yfp worms treated with a hormetic dose of Tm, no reduction in puncta was observed (SI Appendix, Fig. S6D). Together these observations suggest that DR triggers ER hormesis to improve proteostasis and ameliorate polyglutamine aggregation during adult life and extend longevity.
A Hormetic Dose of ER Stress Is Able to Improve Proteostasis in Mammalian Cells.
Since a hormetic dose of ER stress has beneficial effects in worms in terms of better proteostasis, we next asked whether such an intervention will be beneficial in a mammalian model of protein aggregation. For this, we used an inducible cell line system that overexpresses polyglutamine using ponasterone A (Pon A) (56). The cells were pretreated with 0.125 or 0.25 µg/mL of Tm for 5 h, washed and induced with Pon A for 36 h. We find that pretreatment with Tm leads to significant abrogation of polyQ150 aggregation (Fig. 6D and SI Appendix, Fig. S7A). Western blot analysis revealed that Tm pretreatment leads to decreased levels of polyQ150 but increased levels of ER-localized HSP70 (SI Appendix, Fig. S7B). Levels of IRE-1α and several ER chaperones as well as apoptotic marker CHOP were up-regulated by Tm pretreatment, suggesting that the UPRER is up-regulated (SI Appendix, Fig. S7C). Cycloheximide chase experiment showed that the polyQ is degraded faster on Tm pretreatment (Fig. 6E). Finally, the Tm preconditioning led to higher viability of the polyQ150-expressing cells on day 5 post-Pon A induction, as determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (SI Appendix, Fig. S7D). Together, these experiments show that Tm hormesis may also provide beneficial effects in a mammalian model of protein aggregation, similar to worms, although the molecular mechanisms need to be elucidated.
Discussion
Proteostasis collapse has long been associated with incidences of various diseases of protein aggregation. The catastrophic collapse of cellular proteostasis marks the commencement of the aging process (8). Thus, interventions that can delay the onset of the collapse has positive effects on health and longevity. Here, we show that DR effectively delays proteostasis collapse by maintaining robust iUPRER and ERAD during adulthood, leading to increased life span. This is partially mediated by a sublethal dose of ER stress early during development that primes the ER for better function later in life. We also show that the mechanism maybe conserved in a mammalian cell culture model of protein aggregation. Since a sublethal ER stress generated by DR is able to confer health and life span benefits at adulthood, this mechanism may be categorized as hormesis.
DR has long been argued as a case of hormesis (57–59). While DR confers health and life span benefits, extended periods of DR or starvation may be detrimental (58). Even in C. elegans, BDR produces a typical bell-shaped curve with ad libitum and lowest dilution-fed worms showing no life span benefits (48, 60). Incidentally, the mechanisms of hormesis in case of DR mostly point toward mitochondrial metabolism. Glucose restriction, another mode of DR that also increases life span, works through a process of mitohormesis involving ROS-mediated up-regulation of cellular detoxification machinery (20). Additionally, lowering insulin-IGF1 signaling that is akin to reduced glucose metabolism requires mitohormesis to increase life span (21). Although, IRE-1 was found to be involved in life span regulation during DR, the mechanism was not linked to ER hormesis (61). Our study now elucidates how ER hormesis functions during DR, adding to the list of known mechanisms by which the conserved life span-extending intervention of diet restriction works.
DR can be implemented in a variety of ways. Classically, in the widely used C. elegans genetic model (eat-2) and the relatively newer paradigm (drl-1), DR is initiated at hatching; these paradigms may be termed developmental DR. On the contrary, nongenetic methods of DR are usually implemented during adulthood. As the C. elegans in culture gets ad libitum access to food, unlike in the wild, genetic DR paradigms may actually represent optimum nutrition to the organism from early development. Rapid cell division and active metabolism during the early growth period may be sensitive to the developmental DR regime due to glucose restriction. This may result in the transient ER stress that we observed in the genetic models of DR. This assumption is supported by the fact that glycosylated proteins are fewer during DR and supplementation of glucose abrogates the transient ER stress (this study) as well as reduces life span (62, 63). We also show that a nonhydrolyzable form of glucose induces ER stress early in life that is suppressed by glucose supplementation. This transient intervention also increases life span. Since glucose restriction is known to alter mitochondrial dynamics, it will be interesting to study whether mitochondria may have a role to play in ER hormesis during DR.
We show that exposing worms to an early transient ER stress is able to increase life span and improve proteostasis in adulthood by the process of ER hormesis. It appears that a cellular memory is created by the sublethal ER stress during development that helps maintain a prolongevity transcriptional status. In support of this, we observe that the expression of the ERAD genes are increased early in the eat-2 mutant worms and maintained into adulthood, even when the basal ER stress is low during DR. In future, the nature of the memory needs to be deciphered. It is possible that the memory is at the level of chromatin and transcription, involving DNA or histone modifications. Indeed, in the case of mitohormesis involving C. elegans mitochondrial dysfunction (64, 65), histone lysine methylases like jmjd-1.2 and jmjd-3.2 are essential (66). This assumption is also supported by observations where DR has been shown to affect chromatin modifications (67) and that ER stress can modify the epigenome (68).
We have shown here that DR as well ER hormesis prevents decline of iUPRER efficiency that occurs with age. We observe that the basal ER stress levels are lower and that the organism can mount a robust iUPRER when challenged. The basal ER stress levels were also found to be lower in an insulin-IGF1 signaling mutant (40). We have shown earlier that ROS generated due to protein misfolding stress is responsible for attenuation of induced UPRER in yeast and C. elegans (34). It is possible that lower levels of ROS that is generated in eat-2 and daf-2 mutants due to increased levels of ROS detoxification system genes may help the system to maintain a poised ER (33, 34, 40, 48, 69, 70).
The FOXA transcription factor PHA-4 plays a central role in DR-mediated longevity in C. elegans. It appears that the PHA-4 controls many prolongevity aspects of DR. PHA-4 has been shown to control the expression of the majority of the mRNAs as well as miRNAs up-regulated during DR (42, 43). It transcriptionally regulates the expression of genes coding for chromatin modifiers required for modulation of gene expression (49), xenobiotic detoxification pathway components (33), the superoxide dismutase system (48), as well as those involved in the splicing and nonsense-mediated decay pathway (43). In this study, we show that PHA-4 regulates the expression of the ERAD component genes, the transient UPRER at L2, as well as modulates iUPRER at adulthood. These finding show that the transcription factor controls diverse aspects of the regulatory network that provides prolongevity benefits of DR, qualifying as the central regulator of this process.
In conclusion, our study provides insights into the various cellular mechanisms used by DR to increase health and life span, showing the role of ER hormesis in maintaining proteostasis in adulthood.
Materials and Methods
Detailed experimental procedures are provided in SI Appendix. Unless otherwise mentioned, all of the C. elegans strains were maintained and propagated at 20 °C on Escherichia coli OP50 using standard procedures (71). For life span assays, 50 to 60 young adult worms grown on RNAi bacteria since hatching were transferred to similar RNAi plates in triplicates and overlaid with fluorodeoxyuridine (FudR) to a final concentration of 0.1 mg/mL of agar (72). After day 7 of adulthood, when all sick, sluggish, and slow dwelling worms were removed, numbers of dead worms were scored every alternate day and plotted as % survival against the number of days. Statistical analysis for survival was conducted using Mantel–Cox log rank test using Oasis software available at http://sbi.postech.ac.kr/oasis. For measurement of basal UPRER during larval development, Phsp-4:gfp(zcIs4) and eat-2(ad1116);Phsp-4:gfp(zcIs4) eggs were hatched on control RNAi. Fifty L3, L4, young adult or day 1 gravid adult worms were immobilized on glass slides coated with 2% agarose using 20 mM sodium azide and visualized under an Axio-imager M2 epifluorescent microscope (Carl Zeiss, Germany) equipped with a monochromatic camera lens (MRm) and GFP filter set. Fluorescence of 20 worms at different time points was quantified using NIH ImageJ software and represented as arbitrary units (AU). For measurement of induced UPRER efficiency, worms were allowed to grow as above until adulthood and then transferred onto plates overlaid with FuDR to a final concentration of 0.1 mg/mL. At each successive day (day 1 until day 3 of adulthood), ∼100 worms were transferred to plates supplemented with 5 or 10 µg/mL Tm and incubated for 6 h at 20 °C. After 6 h, worms were visualize as above. Average fluorescence of treated worms was normalized to untreated worms and plotted as normalized GFP-fold induction at different days of adulthood. For Tm hormesis treatment, eggs were resuspended in 100 to 200 μL of 1× M9 buffer and treated with 0.125 µg/mL (see SI Appendix for details of media preparation). Eggs were left on rotation in the Tm-bacteria mixture at a slow speed for 24 h. Following this, the hatched L1 larvae were washed 3 to 4 times with 1× M9 buffer to remove traces of Tm and added onto seeded control RNAi plates containing no drug. They were allowed to grow until adulthood when 100 worms belonging to each treatment regime were transferred onto FuDR containing RNAi plates, and life span was scored as mentioned above.
For RNA-seq analysis, synchronized day 1 gravid worms [WT and eat-2(ad1116)] grown on E. coli OP50 were collected using M9 buffer after washing thrice to remove bacteria. Total RNA was isolated from these worm pellets using the TRIzol method. A RNA-seq library was subsequently constructed by TruSeq RNA Library Prep Kit v2 (Illumina) and sequencing was performed using the Illumina Genome Analyzer. The RNA-seq data are available at the National Center for Biotechnology Information (NCBI) with the BioProject ID PRJNA342407 (42, 43). For the cycloheximide chase experiments, day 1 adult WT and eat-2(ad1116) worms grown on control RNAi were washed from plates to remove adhered bacteria and added to 1× S-basal buffer containing 2 mg/mL cycloheximide (Sigma-Aldrich). The experiment was performed in 24-well plates with each well containing 500 µL of drug-buffer solution with gentle agitation. At each time point, including at 0 h, worms were washed thrice with 1 ×M9 buffer to remove traces of cycloheximide and resuspended in 100 µL of 5% Triton X-100 solution containing 1×-EDTA free protease inhibitor mixture (Sigma-Aldrich). Cell lysates, prepared from these samples, were run on a denaturing SDS/PAGE, and HSP-4 levels were detected at each time point by Western blot analysis. Ubiquitination assay was performed on day 1 adult WT (Bristol N2) or eat-2(ad1116) worms grown on control RNAi. The worms were washed to remove adherent bacteria and added to 1× S-basal buffer with or without 100 µM of MG-132, a proteasomal inhibitor. The worms were incubated in these solutions in 24-well plates for 6 h at 20 °C with gentle shaking. They were later washed thrice to remove traces of MG-132. The worm pellets were then resuspended in 100 µL of 5% Triton X-100 solution containing 1× EDTA-free protease inhibitor mixture (Sigma-Aldrich). Cell lysates, prepared from these samples were run on denaturing SDS/PAGE and blotted using a polyubiquitin antibody (catalog no. BML-PW8810; Enzo Lifesciences). For aggregation assay, the polyQ reporter strain was crossed with ire-1(v33) and eat-2(ad1116) mutants to assess the polyQ aggregation profile in these mutants as compared to WT. All of the strains were grown on control or test-RNAi until adulthood and then transferred onto RNAi plates overlaid with FuDR to a final concentration of 0.1 mg/mL. At each successive day from day 1 until day 3 of adulthood, 25 worms of each strain (grown on a particular RNAi) were mounted onto 2% agarose slides and visualized using an Axio-Imager M2 epifluorescent microscope using the YFP filter set. Total number of bright spots (puncta or aggregates) on the worm body wall were counted and average number of aggregates for >20 worms was plotted. For cell culture experiments, HD150Q cells were cultured in DMEM with 10% FBS and antibiotics containing 0.4 mg/mL zeocin and 0.4 mg/mL G418 (geneticin). During experiments, cells were plated onto 6-well tissue culture plates at subconfluent density and on the following day, they were treated with different concentrations of tunicamycin (0.125 and 0.250 μg/mL) for 5 h, washed, and then treated with ponasterone A (1 µM) to induce the expression of mutant huntingtin. After 36 h of ponasterone A treatment, cells were observed under fluorescence microscope followed by collection and processing of the cells for immunoblotting experiments. Aggregate formation was manually counted under a fluorescence microscope (∼500 cells in each group) and the cells containing more than one aggregate were considered to have a single aggregate. MTT assay was used to evaluate the viability of the cells on Tm hormesis.
Supplementary Material
Acknowledgments
We thank all members of the Molecular Aging Laboratory and the K.C. laboratory for their support. We are grateful to the Department of Biotechnology (DBT), Government of India, for a generous infrastructure grant for establishment of the National Institute of Immunology Next Generation Sequencing core facility. This project was partly funded by the Ramalingaswami Fellowship (BT/HRD/35/02/12/2008) and National Bioscience Award for Career Development (BT/HRD/NBA/38/04/2016) to A.M., Department of Science and Technology (DST)-Science and Engineering Research Board (SERB) (EMR/2014/000377) and core funding from National Institute of Immunology. K.C. is a recipient of Swarnajayanti Fellowship from DST-SERB (DST/SJF/LSA-01/2015-16). L.M. was supported by Council of Scientific and Industrial Research (CSIR)-Junior Research Fellowship (JRF), M.C. and U.R. by DBT-JRF. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900055116/-/DCSupplemental.
References
- 1.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Labbadia J., Morimoto R. I., Proteostasis and longevity: When does aging really begin? F1000Prime Rep. 6, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernales S., Papa F. R., Walter P., Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22, 487–508 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Schröder M., Kaufman R. J., The mammalian unfolded protein response. Annu. Rev. Biochem. 74, 739–789 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Maurel M., Chevet E., Tavernier J., Gerlo S., Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 39, 245–254 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Taylor R. C., Dillin A., Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 3, a004440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Zvi A., Miller E. A., Morimoto R. I., Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. U.S.A. 106, 14914–14919 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor R. C., Dillin A., XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 153, 1435–1447 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinds J. W., McNelly N. A., Dispersion of cisternae of rough endoplasmic reticulum in aging CNS neurons: A strictly linear trend. Am. J. Anat. 152, 433–439 (1978). [DOI] [PubMed] [Google Scholar]
- 11.Naidoo N., Ferber M., Master M., Zhu Y., Pack A. I., Aging impairs the unfolded protein response to sleep deprivation and leads to proapoptotic signaling. J. Neurosci. 28, 6539–6548 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain S. G., Ramaiah K. V., Reduced eIF2alpha phosphorylation and increased proapoptotic proteins in aging. Biochem. Biophys. Res. Commun. 355, 365–370 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Brown M. K., Naidoo N., The endoplasmic reticulum stress response in aging and age-related diseases. Front. Physiol. 3, 263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gems D., Partridge L., Stress-response hormesis and aging: “That which does not kill us makes us stronger”. Cell Metab. 7, 200–203 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Mattson M. P., Hormesis defined. Ageing Res. Rev. 7, 1–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumsta C., Chang J. T., Schmalz J., Hansen M., Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat. Commun. 8, 14337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rattan S. I., Hormetic modulation of aging and longevity by mild heat stress. Dose Response 3, 533–546 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristensen T. N., Sørensen J. G., Loeschcke V., Mild heat stress at a young age in Drosophila melanogaster leads to increased Hsp70 synthesis after stress exposure later in life. J. Genet. 82, 89–94 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Fonager J., Beedholm R., Clark B. F., Rattan S. I., Mild stress-induced stimulation of heat-shock protein synthesis and improved functional ability of human fibroblasts undergoing aging in vitro. Exp. Gerontol. 37, 1223–1228 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Schulz T. J., et al. , Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 6, 280–293 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Zarse K., et al. , Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 15, 451–465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S. J., Hwang A. B., Kenyon C., Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 20, 2131–2136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owusu-Ansah E., Song W., Perrimon N., Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell 155, 699–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griciuc A., et al. , Inactivation of VCP/ter94 suppresses retinal pathology caused by misfolded rhodopsin in Drosophila. PLoS Genet. 6, e1001075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendes C. S., et al. , ER stress protects from retinal degeneration. EMBO J. 28, 1296–1307 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mollereau B., et al. , Adaptive preconditioning in neurological diseases - therapeutic insights from proteostatic perturbations. Brain Res. 1648, 603–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang P. L., et al. , Preinduced molecular chaperones in the endoplasmic reticulum protect cardiomyocytes from lethal injury. Ann. Clin. Lab. Sci. 34, 449–457 (2004). [PubMed] [Google Scholar]
- 28.Fouillet A., et al. , ER stress inhibits neuronal death by promoting autophagy. Autophagy 8, 915–926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozlowski L., Garvis S., Bedet C., Palladino F., The Caenorhabditis elegans HP1 family protein HPL-2 maintains ER homeostasis through the UPR and hormesis. Proc. Natl. Acad. Sci. U.S.A. 111, 5956–5961 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speakman J. R., Mitchell S. E., Caloric restriction. Mol. Aspects Med. 32, 159–221 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Masoro E. J., The role of hormesis in life extension by dietary restriction. Interdiscip. Top. Gerontol. 35, 1–17 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Lakowski B., Hekimi S., The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 95, 13091–13096 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chamoli M., Singh A., Malik Y., Mukhopadhyay A., A novel kinase regulates dietary restriction-mediated longevity in Caenorhabditis elegans. Aging Cell 13, 641–655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maity S., et al. , Oxidative homeostasis regulates the response to reductive endoplasmic reticulum stress through translation control. Cell Rep. 16, 851–865 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Calfon M., et al. , IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Steinkraus K. A., et al. , Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell 7, 394–404 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmer F., et al. , Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12, 1317–1319 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Naidoo N., The endoplasmic reticulum stress response and aging. Rev. Neurosci. 20, 23–37 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Naidoo N., ER and aging-Protein folding and the ER stress response. Ageing Res. Rev. 8, 150–159 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Henis-Korenblit S., et al. , Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc. Natl. Acad. Sci. U.S.A. 107, 9730–9735 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labbadia J., Morimoto R. I., Repression of the heat shock response is a programmed event at the onset of reproduction. Mol. Cell 59, 639–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandit A., Jain V., Kumar N., Mukhopadhyay A., PHA-4/FOXA-regulated microRNA feed forward loops during Caenorhabditis elegans dietary restriction. Aging (Albany N.Y.) 6, 835–855 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabrez S. S., Sharma R. D., Jain V., Siddiqui A. A., Mukhopadhyay A., Differential alternative splicing coupled to nonsense-mediated decay of mRNA ensures dietary restriction-induced longevity. Nat. Commun. 8, 306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruggiano A., Foresti O., Carvalho P., Quality control: ER-associated degradation: Protein quality control and beyond. J. Cell Biol. 204, 869–879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janiesch P. C., et al. , The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat. Cell Biol. 9, 379–390 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Sasagawa Y., Yamanaka K., Ogura T., ER E3 ubiquitin ligase HRD-1 and its specific partner chaperone BiP play important roles in ERAD and developmental growth in Caenorhabditis elegans. Genes Cells 12, 1063–1073 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R., OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10, 272–282 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panowski S. H., Wolff S., Aguilaniu H., Durieux J., Dillin A., PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447, 550–555 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Singh A., et al. , A chromatin modifier integrates insulin/IGF-1 signalling and dietary restriction to regulate longevity. Aging Cell 15, 694–705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong M., et al. , Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 6, e1000848 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao J., Diamond M. I., Polyglutamine diseases: Emerging concepts in pathogenesis and therapy. Hum. Mol. Gen. 16 R115–R123 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Saez I., Vilchez D., The mechanistic links between proteasome activity, aging and age-related diseases. Curr. Genomics 15, 38–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vilchez D., Saez I., Dillin A., The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 5, 5659 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Duennwald M. L., Lindquist S., Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 22, 3308–3319 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brignull H. R., Morley J. F., Garcia S. M., Morimoto R. I., Modeling polyglutamine pathogenesis in C. elegans. Methods Enzymol. 412, 256–282 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Jana N. R., Zemskov E. A., Wang Gh Nukina N., Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum. Mol. Genet. 10, 1049–1059 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Masoro E. J., Hormesis and the antiaging action of dietary restriction. Exp. Gerontol. 33, 61–66 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Le Bourg E., Hormesis, aging and longevity. Biochim. Biophys. Acta 1790, 1030–1039 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Ristow M., Schmeisser K., Mitohormesis: Promoting health and lifespan by increased levels of reactive oxygen species (ROS). Dose Response 12, 288–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mair W., Panowski S. H., Shaw R. J., Dillin A., Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS One 4, e4535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen D., Thomas E. L., Kapahi P., HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 5, e1000486 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee S. J., Murphy C. T., Kenyon C., Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 10, 379–391 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlotterer A., et al. , C. elegans as model for the study of high glucose- mediated life span reduction. Diabetes 58, 2450–2456 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang W., Hekimi S., A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 8, e1000556 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hekimi S., Lapointe J., Wen Y., Taking a “good” look at free radicals in the aging process. Trends Cell Biol. 21, 569–576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Merkwirth C., et al. , Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell 165, 1209–1223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y., Daniel M., Tollefsbol T. O., Epigenetic regulation of caloric restriction in aging. BMC Med. 9, 98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dicks N., Gutierrez K., Michalak M., Bordignon V., Agellon L. B., Endoplasmic reticulum stress, genome damage, and cancer. Front. Oncol. 5, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Honda Y., Honda S., The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 13, 1385–1393 (1999). [PubMed] [Google Scholar]
- 70.Vanfleteren J. R., De Vreese A., The gerontogenes age-1 and daf-2 determine metabolic rate potential in aging Caenorhabditis elegans. FASEB J. 9, 1355–1361 (1995). [DOI] [PubMed] [Google Scholar]
- 71.Stiernagle T., Maintenance of C. elegans. WormBook 11, 1–11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hosono R., Mitsui Y., Sato Y., Aizawa S., Miwa J., Life span of the wild and mutant nematode Caenorhabditis elegans. Effects of sex, sterilization, and temperature. Exp. Gerontol. 17, 163–172 (1982). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.