Abstract
This perspective addresses recent advances in lipid transport across the Gram-negative inner and outer membranes. While we include a summary of previously existing literature regarding this topic, we focus on the maintenance of lipid asymmetry (Mla) pathway. Discovered in 2009 by the Silhavy group [J. C. Malinverni, T. J. Silhavy, Proc. Natl. Acad. Sci. U.S.A. 106, 8009–8014 (2009)], Mla has become increasingly appreciated for its role in bacterial cell envelope physiology. Through the work of many, we have gained an increasingly mechanistic understanding of the function of Mla via genetic, biochemical, and structural methods. Despite this, there is a degree of controversy surrounding the directionality in which Mla transports lipids. While the initial discovery and subsequent studies have posited that it mediated retrograde lipid transport (removing glycerophospholipids from the outer membrane and returning them to the inner membrane), others have asserted the opposite. This Perspective aims to lay out the evidence in an unbiased, yet critical, manner for Mla-mediated transport in addition to postulation of mechanisms for anterograde lipid transport from the inner to outer membranes.
Keywords: glycerophospholipid, Mla, outer membrane, LPS, lipoprotein
Lipid bilayers are the ubiquitous and fundamental basis of cellular envelopes across all domains of life. While the constituent lipids vary across genera, the principle remains the same: Hydrophobic membranes serve to compartmentalize cellular contents and separate them from surrounding milieu (1). In eukaryotes, lipid transport from sites of biosynthesis to end destinations are varied and complex. The operation of transporting lipids across an aqueous environment requires shielding of hydrophobic acyl chains. This is possible largely through vesicular trafficking or via a protein escort (2). Importantly, these processes all occur within the cytoplasm, providing critical accessibility to intracellular ATP and activated precursor pools.
Prokaryotic cells typically couple terminal steps of lipid biosynthesis to the inner leaflet of the inner membrane (3, 4). This eliminates any requirement for shielding, as the lipid is already incorporated into the bilayer, with enzymes having access to the necessary energy pools. Glycerophospholipids (GPLs) must flip from the inner leaflet to the outer leaflet of the membrane readily and rapidly to sustain growth (5). GPL flipping in lipid bilayers is an intrinsically slow process in the absence of accessory proteins (6). Remarkably, GPLs are flipped at rates upwards of 30,000 times faster in vivo than occurs in vitro (7, 8). Vesicles reconstituted with protein extracts enhanced flipping of lipids independent of energy, lending to the generalized hypothesis that inner membrane transmembrane helices are capable of facilitating nonspecific flipping (9, 10). In contrast, eukaryotic cells possess 3 classes of enzymes that flip lipids across membranes: Scramblases, flippases, and floppases (5). While scramblases nonspecifically mix lipids without energy, flippases and floppases are ATP-dependent enzymes with specific substrates to direct asymmetry within the membrane. Dedicated flippases have been identified for unique bacterial lipids, lipid A and bactoprenol-linked precursors, but no generalized GPL flippases or scramblases have been discovered in bacteria (11–15).
Transbilayer movement is only one of the challenges that Gram-negative bacteria face in maintaining their cell envelope, as they have a second, outer membrane organelle that enshrouds the cytoplasm, peptidoglycan, and periplasm, which is devoid of energy sources (16, 17). Unlike the cytoplasmic membrane, this outer membrane exhibits stringent lipid asymmetry (18). While the inner leaflet is composed of GPLs, the outer leaflet consists of lipopolysaccharide (LPS) (18) (Fig. 1). This lipid asymmetry is fundamental for the outer membranes’ function as a barrier to the environment, preventing diffusion of not only large polar molecules, but also from lipophilic compounds (18).
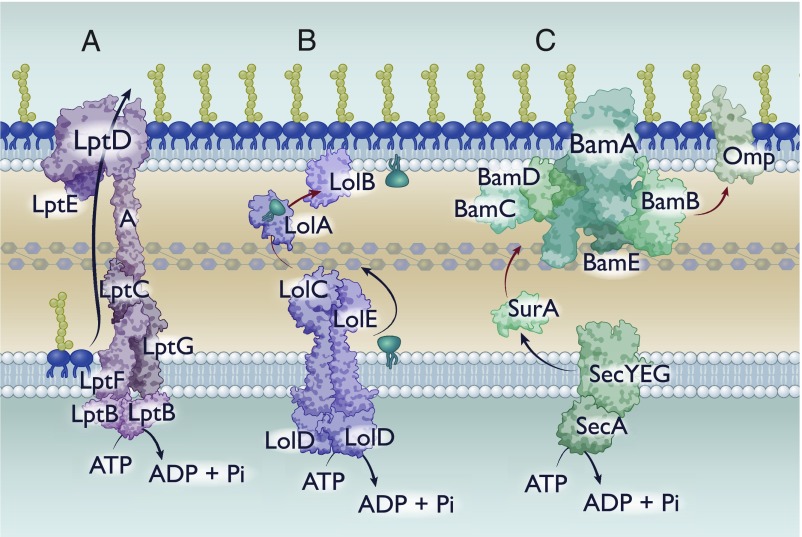
Fig. 1.
Known mechanisms of hydrophobic substrate transfer across the periplasm. Cartoon depictions of hydrophobic substrate transfer across the periplasm. Black arrows indicate energy-dependent processes. Red arrows represent transfer that must occur without energy. (A) Lpt-mediated transfer of LPS. (B) Lol-mediated transfer of lipoproteins. (C) Chaperone-mediated transport of hydrophobic proteins.
While this outer membrane is highly beneficial and essential for Gram-negative bacteria, it has raised a critically important question in cell envelope biology. How do cells transport biomolecules from the point of synthesis (inner membrane) across an aqueous periplasm to the destination (outer membrane) and maintain the necessary lipid asymmetry in the absence of accessible ATP?
This Perspective aims to address recent work that has expanded our understanding of how bacteria achieve intermembrane transport. While we address recent work on major systems of lipid and hydrophobic protein transport across the periplasm, the primary focus of this perspective is the maintenance of lipid asymmetry (Mla) pathway that has been implicated in both retrograde and anterograde GPL transport. There has been an element of controversy surrounding the directionality and function of this pathway. By laying out the data for both sides of the argument, we hope to allow readers to make educated and informed decisions on the functionality of Mla.
Mechanisms to Accommodate Different Cargo Across the Periplasm
Transport across the periplasm requires mechanisms that can accommodate intrinsic properties, such as hydrophobicity. In that context, proteins and lipids have different degrees to which transport must be facilitated from the inner to outer membranes (19). Over the course of several decades, research has identified and characterized some of these transport pathways, although significant knowledge gaps remain. Analysis of these systems has yielded commonalities and parameters that guide our pursuit of additional periplasmic transport systems in Gram-negative bacteria.
LPS Transport via the LPS Transport Pathway System.
One of the major lipid species that requires facilitated transport to the outer membrane is LPS. LPS has 3 moieties: The lipid A anchor, core sugars, and the O-antigen, a variable, polymeric repeat of sugars (18). The conserved biosynthetic machinery that synthesizes the lipid A anchor has been extensively characterized (18). In Escherichia coli we have significant knowledge of the enzymes involved in biosynthesis and assembly of the core and O-antigen carbohydrate domains; however, sugar composition is highly variable even within a given species (20). Lipid A biosynthesis begins in the cytoplasm and later steps are performed by inner membrane-anchored enzymes, including the addition of all core sugars (18). The intact lipid A with attached core oligosaccharide is flipped via a dedicated flippase, MsbA, to the periplasmic side of the inner membrane (18). In organisms with O-antigen, the polymer is added in the periplasm from its isoprenoid carrier (18), at which point the entire LPS molecule requires transport to the outer membrane.
Transporting this molecule requires shielding of the hydrophobic acyl chains on the lipid anchor while allowing enough space and exposure to accommodate variable hydrophilic sugars in the core. Over the past decade and a half, several groups have identified and characterized the major protein components of the LPS transport (Lpt) pathway (21). The Lpt pathway consists of 6 proteins, each of which is essential for lipid transport although certain mutants are viable in organisms other than E. coli (22, 23). At the inner membrane is the LptB2CFG complex, which is an ATP-binding cassette (ABC) transporter. This complex then interacts with a periplasmic LptA protein, with a hydrophobic groove that can fit the lipid A molecule (21, 24). Finally, at the outer membrane is a β-barrel (LptD) and a lipoprotein (LptE) (25) (Fig. 1A). In solving the crystal structure of the LptDE complex and generating protein variants at critical residues, it was demonstrated that LPS molecules are deposited directly into the outer leaflet of the outer membrane: The initial establishment of lipid asymmetry in the outer membrane (26). How the cell couples a cytoplasmic ATPase with periplasmic and outer membrane components remained an integral question. Recently, it has been posited that monomers of LptA form a bridge across the periplasm, connecting the inner membrane LptB2CFG to the outer membrane LptDE (21) (Fig. 1A). This model was experimentally validated by the Kahne group using an elegant in vitro liposome transfer assay, demonstrating that LPS transfer between vesicles that contained either the outer membrane or inner membrane components required soluble LptA bridging (27). With this model, a connected Lpt system mediates continuous transport of LPS molecules driven by an inner membrane ABC transporter.
Lipoprotein Transport via the Localization of Lipoprotein System.
Lipoproteins are N-terminally acylated proteins that serve a wide array of functions in bacterial physiology. These lipoproteins can be anchored in the inner membrane facing the periplasm or in the outer membrane with either inner or outer leaflet topology (28). These lipoproteins are integral components of multiple molecular machines important for folding outer membrane proteins, transport of lipophilic substrates across the periplasm, assembly and remodeling of peptidoglycan, and cell division (29–33). After synthesis in the cytoplasm and transport to the periplasmic face of the inner membrane by the Sec pathway, these proteins undergo significant posttranslational processing (28). A characteristic consensus motif at the N terminus, known as a lipobox, directs the enzyme Lgt to transfer diacylglycerol from phosphatidylglycerol to the sulfhydryl group of a conserved cysteine residue at the C-terminal end of the lipobox (28). This facilitates cleavage of lipobox residues by a dedicated peptidase, resulting in the apolipoprotein with diacylated N-terminal cysteine (28). The last step in lipoprotein maturation involves the transfer of an acyl chain from GPLs to the amino group of the cysteine, resulting in the mature triacylated lipoprotein (28).
The acylation of the lipoprotein results in a hydrophobic anchor that tethers the protein to the inner membrane. However, most lipoproteins reside in the outer membrane: Transport of these lipoproteins is mediated through the localization of lipoprotein (Lol) pathway (28). The Lol pathway consists of a dedicated ABC transporter in the inner membrane (LolCDE), a periplasmic chaperone (LolA), and an outer membrane lipoprotein (LolB) (34). Mature lipoproteins destined for the outer membrane are extracted from the inner membrane via LolCDE and transferred to LolA (35) (Fig. 1B). LolA diffuses across the periplasm and interacts with LolB to transfer the lipoprotein to the inner leaflet of the outer membrane via an undetermined mechanism that must rely on differential affinity or alterations in protein topology (36) (Fig. 1B). Certain lipoproteins, once reaching the outer membrane, flip to the extracellular facing side (28). While key mechanisms have been identified that translocate lipoproteins to face the extracellular environment, a generalized mechanism remains to be elucidated (37–40).
Outer Membrane Protein Chaperones.
Another constituent of the outer membrane is outer membrane proteins (Omps). These β-barrel proteins vary in size and function, yet they all exhibit similar antiparallel β-sheet topologies. Bacterial Omps fold such that the hydrophobic residues face the exterior, allowing for intercalation into the membrane (41). By folding in this manner, it generally creates a hydrophilic lumen that can function as nonspecific porins or substrate-specific transporters (42). Omps that have alternative functions are typically significantly smaller and lack an accessible lumen (43).
Because of the hydrophobicity of Omps, unassisted diffusion across the aqueous periplasm results in rapid and irreversible aggregation of the unfolded peptide (44). To prevent this, periplasmic chaperones sequester or shield hydrophobic regions of Omps, preventing premature (mis)folding (45, 46) (Fig. 1C). Protected by these periplasmic chaperones, Omps are transported to the outer membrane prior to being released to and folded by the Bam complex.
The Bam complex is an outer membrane complex that mediates the folding and insertion of Omps into the membrane and has been the subject of several reviews (29, 47). The 2 essential proteins of this complex, BamA and BamD, are a β-barrel and lipoprotein, respectively (48, 49). Additional accessory proteins are thought to mediate folding or substrate specificity, but are neither fully conserved nor essential for function across Gram-negative bacteria (47). Bam is thought to facilitate folding by altering membrane dynamics in concurrence with directed insertion of the oriented Omp peptide (29, 47). Although this model requires experimental validation, a functional Bam system is necessary for proper folding and localization of Omps in vivo.
Discovery of and Phenotypes Associated with the Mla System
In the process of maintaining 2 membranes, the outer membrane is at the forefront of extracellular assaults. Perturbation of the outer membrane can significantly disrupt the lipid asymmetry promoting flipping of GPLs to the bacterial surface. Symmetric GPL rafts compromise the membrane to bulkier and hydrophobic molecules (50). As such, the cell inevitably requires removal of GPLs from the outer leaflet of the outer membrane. Multiple mechanisms have been identified to date that are implicated in this process. These include the outer membrane phospholipase PldA, which deacylates mislocalized GPLs at both the sn-1 and sn-2 positions (Fig. 2A) (51). Another is the palmitoyl transferase enzyme PagP, which acylates lipid A using a palmitate from a GPL donor as a substrate (52). Thorough structural analysis reveals that the active sites of these enzymes are only accessible to GPLs in the outer leaflet (43).
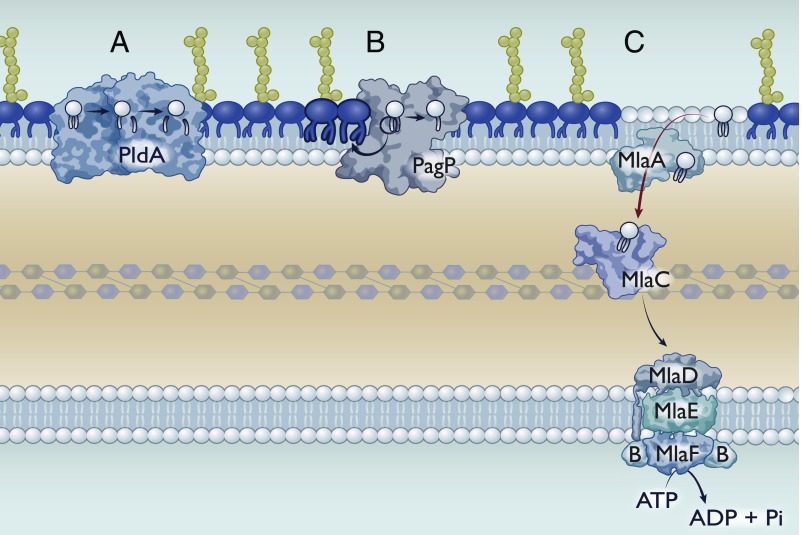
Fig. 2.
Asymmetry maintenance mechanisms in Gram-negative bacteria. Gram-negative bacteria have multiple mechanisms to maintain asymmetry, although their presence or absence is species-dependent. (A) PldA is an outer membrane phospholipase that sequentially degrades GPLs. Its active site is exposed to the outer leaflet of the outer membrane, such that it can only degrade mislocalized GPLs. PldA can remove both the sn-1 and sn-2 fatty acids. (B) PagP is an outer membrane palmitoyltransferase that catalyzes the transfer of the sn-1 palmitate from a GPL to lipid A, resulting in a hepta-acylated lipid A species. The resulting lyso-GPL could be either removed via an unknown mechanism or degraded by PldA. (C) The Mla system for GPL transport. Importantly, we have depicted Mla as mediating retrograde transport; however, this perspective addresses evidence for both retrograde and anterograde transport.
A third proposed system, the Mla pathway, is markedly different in that it does not rely on enzymatic Omps at all. Instead, the Mla system commonly consists of 6 proteins: 1/2) A dedicated ABC transporter complex (MlaEF), 3) a cytoplasmic accessory protein (MlaB), 4) a membrane-anchored periplasmic protein (MlaD), 5) a periplasmic chaperone (MlaC), and 6) an outer membrane lipoprotein (MlaA) (53) (Fig. 2C). This system was identified in E. coli as mutants defective in the Mla pathway were hypersensitive to detergent (53).
Sensitivity to detergents and hydrophobic compounds is a hallmark of outer membrane perturbation. Spontaneous suppressors for detergent sensitivity of mla mutants mapped to the promoter region of pldA (53). Each suppressor elevated pldA transcript levels, suggesting that increased PldA overcomes Mla defects (53). Additionally, a double mutant defective for both Mla and PldA is severely defective for outer membrane integrity (53). Taken together, these data contribute to the hypothesis that Mla performs a functionally redundant role to PldA, providing strong evidence for Mla’s involvement in maintaining lipid asymmetry in the outer membrane (Fig. 2). Additional evidence for Mla’s function in lipid transport was demonstrated when expression of the MlaBCDEF operon in trans suppressed major lipid asymmetry defects associated with tol-pal mutants in E. coli (54). Work in Salmonella enterica also identified increased outer membrane GPL levels in tol-pal mutants, hinting to the potential that tol-pal plays an undefined role in outer membrane lipid homeostasis (55).
In 2016, a suppressor for antibiotic sensitivity in an E. coli mutant with reduced LPS levels mapped to mla, which resulted in an MlaA protein with an internal deletion of 2 residues (56). In an otherwise WT background, this gain-of-function allele (mlaA*) renders cells sensitive to detergents, hydrophobic antibiotics, and causes rapid cell death in stationary phase (56). Further characterization of the stationary cell-death pathway showed excessive outer membrane vesiculation and shrinking of the inner membrane. Based on these phenotypes, Sutterlin et al. proposed that MlaA* facilitates energy-independent rapid diffusion of GPLs from the inner to the outer leaflet of the outer membrane. Curiously, suppressors of the cell-death phenotype of mlaA* were null-alleles of pldA. While the ΔpldA mlaA* strains were still sensitive to hydrophobic antibiotics, they lacked the cell-death phenotype. These data suggest that in an mlaA* genotype, PldA hyperactivity generates a significant number of free fatty acids and lyso-species from outer membrane, outer leaflet GPLs. While residual lyso-species could exhibit detergent-like properties, it has subsequently been shown that PldA-derived fatty acids also stimulate LPS production. In WT bacteria, increased LPS synthesis is beneficial as increased PldA activity would serve as a putative signal that outer membrane asymmetry is compromised. However, in the mlaA* background, additional LPS was shown to further destabilize the outer membrane (57). Unsurprisingly, inactivation of pldA alleviated cell death but did not restore asymmetry in the presence of MlaA* (56).
This hypothesis was supported when the structure of MlaA was solved by van den Berg and colleagues (58). The intact MlaA protein is an α-helical lipoprotein with a thickness of a single leaflet, ∼20 Å, and contains an amphipathic pore through which GPL transit likely occurs. In addition, by occupying the entirety of the inner leaflet of the outer membrane, MlaA restricts lateral diffusion of GPLs from the inner leaflet which have no access to the amphipathic transit pore. In contrast, the MlaA* variant has an internal deletion of N26 and F27 in the first α-helix, which is predicted to disrupt the helix. Disruption of this first helix would clear the road for lateral access to the pore directly from the inner leaflet, no longer restricting lateral diffusion of inner leaflet lipids into the amphipathic pore of MlaA (58).
In addition to the crystal structure, Chng and colleagues (33, 59) used in vivo cross-linking and in vivo photocross-linking methodologies to identify interprotein interactions of Mla components. Focusing on MlaA, they found that it interacts with OmpC trimers. This interaction, while functionally still elusive, is important for activity. Altering residues for OmpC trimerization or OmpC–MlaA interactions appeared to abolish Mla activity, suggesting that MlaA binding to OmpC or other trimeric Omps is important for activity in stationary phase (33). The MlaA structure was solved in complex with trimeric OmpF; however, overexpression of the protein was carried in a strain not producing OmpC (59). Furthermore, the interaction between MlaA and OmpC appears to be a specific one, as deletion of OmpF under low osmolarity conditions [where it would be more abundant than OmpC (60)] had no effect on lipid asymmetry (33). Additionally, immunoprecipitation evidence strongly suggests that inner membrane MlaBDEF proteins form a biologically relevant complex in vivo (61). This was later supported by cryoelectron microscopy structures of the Mla inner membrane complex from both E. coli and Acinetobacter baumannii (62, 63).
A critical experiment for Mla’s implication in lipid transport was the ability for Mla proteins to bind GPLs. Three proteins are predicted to bind lipids for the system to function: MlaA, MlaC, and MlaD. MlaA facilitates the diffusion of GPLs from the outer leaflet through a central cavity. Using coarse-grained and atomistic simulations, it was computationally demonstrated that GPLs could partially traverse the amphipathic pore of MlaA (58). It remains unclear what pushes lipids through the entire cavity. MlaC, the soluble periplasmic chaperone, readily binds GPLs when purified from whole cells (62, 64). Crystal structures of MlaC reveal a hydrophobic pocket capable of binding 2 acyl-chains, which could accommodate 2 of the 3 major GPL species in most bacteria (62, 65). Curiously, one deposited MlaC structure from Pseudomonas putida (PDB ID code 4FCZ) has electron density that could be modeled to have cardiolipin bound to the hydrophobic pocket, suggesting a degree of flexibility in the hydrophobic pocket (62). In each case, head groups were solvent-exposed, removing head group as a specificity factor in regards to MlaC binding and transport (62).
Because MlaC is the soluble chaperone, it presumably interacts with both MlaA and the inner membrane MlaBDEF complex to complete transfer of a GPL. Using biolayer interferometry, MlaC interacts with both MlaA and MlaBDEF (62). These interactions were subsequently confirmed by in vivo photocross-linking (64). These interactions must be transient as MlaC is not able to be stably purified in complex with either set of proteins.
Finally, MlaD is predicted to bind lipids with its mammalian cell entry (MCE) domain (discussed below). MlaD forms a hexameric structure associated with the MlaBEF ABC-transporter, with the MCE domain exposed to the periplasm (61, 62). Like MlaC, MlaD binds GPLs in vivo, making it the likely inner membrane candidate for handling lipid transfer in the Mla operon (62).
The Directionality of Mla: Reconciling Genetics and Biochemistry
The genetics and structural work on the Mla system has supported a role as a lipid transporter involved in preserving outer membrane lipid asymmetry by removing GPLs from the outer leaflet. This hypothesis has yet to be completely validated biochemically in vitro. Several groups have valiantly worked to directly monitor GPL transport; however, attempts to do so have been met with varying degrees of success as reconstitution of this system into in vitro liposomes is not trivial. The Chng group monitored lipid transfer using holo-MlaC (bound with GPL) and apo-MlaD (not GPL bound) or the inverse scenario of apo-MlaC (not GPL bound) and holo-MlaD (GPL bound) (64). Strikingly, holo-MlaC was unable to transfer lipid to apo-MlaD, yet transfer was seen from holo-MlaD to apo-MlaC (64). At a first approximation, this would run in stark contrast to the retrograde transport model. However, such an interpretation fails to consider the biological relevance of the ABC-transporter components MlaEF, which were excluded from this initial in vitro study. Assuming the retrograde model, MlaC requires an incredibly strong affinity for its substrate in order to extract GPLs from MlaA in the absence of energy. If this affinity is greater than that of MlaD, transfer will never be seen in vitro without the ABC-transporter complex and ATP to provide energy.
In a similar vein, Knowles and colleagues (66) used purified apo- or holo-MlaC and apo or holo-MlaD to monitor transfer of lipid. Corroborating the former result, they found that MlaD rapidly transfers lipid to MlaC, but the inverse was never seen in vitro. In this case, they were able to reconstitute the ABC-transport complex into liposomes. Under these conditions, the authors saw transfer from MlaD to MlaC independent of ATP hydrolysis (66). Knowles and colleagues conclude that ATPase activity must not be directly related to Mla’s role in GPL trafficking, but rather an alternative function in the pathway. While possible, this hypothesis underappreciates the well-established function of ABC-transporters, which would suggest that observed in vitro transfer from MlaD to MlaC is an artifact. The ABC-transporter complex is highly conserved across all Mla systems, which provides strong evolutionary evidence that its function is biologically important (67), and deletion of the ATPase (MlaF) or expression of catalytically null mutants exhibit identical phenotypes to other Mla-null mutants (53, 63). Critically, no work has focused on lipid transfer between MlaA and MlaC in either direction. With these caveats in mind, there is not conclusive in vitro biochemical evidence that confirms directionality of transfer.
Earlier this year, a report from Miller and colleagues (63) suggested that the Mla system in A. baumannii functions in an anterograde direction in the A. baumannii strain ATCC 17978. Their conclusion relies on an in vivo pulse-chase assay using 2-13C acetate to label GPLs, coupled with separation of membranes and LC-MS/MS analysis of lipids. From these data, the authors determined relative ratios of labeled/unlabeled lipids in the inner and outer membranes. In theory, this methodology allows for monitoring of de novo synthesis and localization of newly synthesized GPLs. Using this technique, Miller and colleagues (63) report an accumulation of labeled lipids in the inner membrane with a corresponding decrease of labeled lipids in the outer membrane in multiple mla mutants in A. baumannii. Unfortunately, there are 2 major caveats to this interpretation. The first is that it is unclear how these cells are viable with the significant reduction of GPLs observed in the outer membrane. The second was the omission of analysis of the lipid content of outer membrane vesicles (OMVs).
Multiple groups have shown that defects in Mla result in increased shedding of OMVs across multiple organisms, including Neisseria meningitidis (68), Haemophilus influenzae (67), Vibrio cholerae (67), and A. baumannii in our hands. Vesicle formation is varied and complex, yet one route of formation is through the accumulation of GPL rafts (a symmetric bilayer patch) in the outer membrane (69). By producing more OMVs, GPL abundance in the outer membrane during the pulse-chase experiment would be severely underestimated as cells would be shedding GPLs from the outer leaflet at rates disproportionate to that of the inner membrane. It is entirely possible that LC-MS/MS analysis of OMVs from the mla mutants could explain this discrepancy in the pulse-chase assay, which could then yield more conclusive support for their model or alternatively support the retrograde model. Unfortunately, without these data a confident interpretation is hindered.
Additional in vivo evidence published by our group last year supports the retrograde model of Mla in A. baumannii (70). At the time, we were studying A. baumannii’s ability to grow in the total absence of lipid A, a molecule that is typically essential for growth of Gram-negative organisms. A. baumannii and certain species produce lipooligosaccharide (LOS), which attaches a few sugars to the core in lieu of O-antigen. By growing in the absence of lipid A, these LOS-deficient bacteria produce an outer membrane which is predominantly GPLs (70–73). Unsurprisingly, LOS-deficient A. baumannii exhibit a severe growth defect (70). Using experimental evolution, we evolved 10 different populations of A. baumannii across multiple strains until they grew to near WT levels. Seven of 10 populations accumulated disruptive mutations in mla genes and the second most-abundant mutation was in pldA (70). We validated these results by showing that deletion of these 2 pathways improved fitness in LOS-deficient A. baumannii independent of any evolution experiment (70). In the context of a LOS-deficient outer membrane, we interpreted these results to favor the model of retrograde transport. Without LOS, the major lipid species in the outer membrane must be GPLs. We hypothesized that upon LOS-deficiency, Mla and PldA function as they have evolved to do, which is to degrade or remove GPLs from the outer leaflet of the outer membrane. Typically, this is beneficial as the gaps could be replaced by LOS, preserving asymmetry. In cells without LOS, Mla and PldA would constantly remove GPLs from the outer leaflet only to be replaced by more GPLs, creating an endless loop of constitutive degradation. Removal of these 2 systems would allow the outer membrane to achieve homeostasis between the leaflets of the lipid bilayer. One of the major limitations of this study was the inability to confidently separate inner and outer membranes in A. baumannii. While membrane separations work robustly for E. coli and S. enterica, its applicability to other organisms including N. meningitidis, Helicobacter pylori, and V. cholerae have been met with limited to zero success (25, 74–76). Separation depends on the protein–lipid ratio and the lipid composition, although the process itself is far from formulaic. Until Miller and colleagues (63) published membrane separations of A. baumannii, this had not been successfully achieved for this organism.
Despite significant advances in our understanding of the Mla system of lipid transport, it is clear the field is far from a conclusive decision on its directionality. While a majority of data support the retrograde model, there is limited data to suggest the opposite, a question that will only be solved with further analysis.
MCE Domain Proteins: Established and Predicted Roles
One additional source of insight to the function of Mla resides in the established roles of other MCE domain-containing proteins in both prokaryotes and eukaryotes. The first MCE domain protein was initially characterized by a landmark study in 1993 that identified a region of the Mycobacterium tuberculosis genome that conferred the ability, when heterologously expressed in E. coli, to invade and persist within HeLa cells (77). A subsequent study in closely related Rhodococcus sp. identified up-regulated genes during growth with cholesterol, one of which was the mce4ABCDEF gene cluster (78). Deletion of the mce4 operon abolished growth on cholesterol. In corroboration with this result, an mce4 deletion was shown to be defective for cholesterol uptake (79). This lends insight as to why MCE mutants in certain Actinobacteria may have abrogated macrophage survival, as cholesterol is thought to be important for cell entry for mycobacterial species (78). Regardless of the functional role of cholesterol for invasion (80), the abundance and number of MCE domain proteins in M. tuberculosis is thought to facilitate the uptake of various types of lipids, including palmitic acid, which serves as an important energy source for M. tuberculosis throughout infection (81). It is unlikely that these MCE domain proteins are exclusively involved in mediating membrane biogenesis as a sextuple mutant of all 6 operons in the closely related Mycobacterium smegmatis exhibited no major alterations to cellular lipids (82).
MCE domain proteins have been identified not just in Actinobacteria, but also broadly throughout diderm Gram-negative bacterial species, albeit at a lower copy number. This is curious as intracellular invasion is not a conserved lifestyle for many Gram-negative species. This lends itself to the hypothesis that MCE domain proteins may play a more global role in cell envelope biology in nonmycobacterial species.
Indeed, this appears to be the case. E. coli has 3 detectable MCE domain proteins—MlaD, PqiB, and YebT—each encoded in separate operons (83). Each of these proteins copurify with the major GPLs from E. coli, suggesting that all could have a role in lipid transport in Gram-negatives. From genetic analyses, MlaD appears to be dominant as an mla mutant appears to have identical phenotypes to the triple mla, pqi, yeb mutant with respect to sensitivity to detergents (62). Viability of the triple deletion removes the possibility that these systems function collectively as redundant anterograde transporters, a key point when considering directionality of transport. Structural work showed that each of the 3 MCE operons transcribe proteins that have varying numbers of MCE domains and radically different quaternary structures (62). Each of these proteins form symmetric hexamers with the MCE domains stacked vertically (62). MlaD, which has one MCE domain, remains in close association with the ABC-transporter MlaEF (62). YebT has 7 MCE domains that form 7 stacked rings, resulting in a ∼230-Å-long tube capable of spanning the periplasm (62). Finally, PqiB, which has 3 MCE domains, forms 3 stacked rings with the C termini forming a needle-like structure that again could span ∼230 Å (62). The quaternary structures suggest independent mechanisms for transporting substrates; however, the mechanism by which transport occurs through the Pqi and Yeb complexes is elusive. Neither is associated with an ABC transporter, making it more likely that different MCE domains within the given polypeptide have different affinities for substrate or conformational changes in vivo to push substrate via peristaltic action (83). Although shown to bind GPLs, there is no direct biochemical or genetic evidence as to the function of Pqi and Yeb in Gram-negative bacteria. Interestingly, despite A. baumannii having a homologous Pqi system, it was never mutated in the evolution of LOS-deficient A. baumannii, suggesting it is not redundant with the Mla system (70).
Outside of bacteria, MCE domain-containing proteins have been found in the chloroplasts of plants (84, 85). Chloroplast organelles are self-replicative diderms, having an inner and outer membrane (86). Plant chloroplasts are capable of de novo phosphatidic acid (PA) synthesis in the plastid or can import PA from the endoplasmic reticulum (ER), with the origin of the lipid distinguishable by the chain length at the sn-2 position (84). PA serves as a precursor for numerous galactolipids important for chloroplast membranes. In Arabidopsis thaliana, mutations in trigalactosyldiacylglycerol (tgd) 1 to 3 were shown to significantly alter plastid lipids (84, 85, 87). TGD1 and -3 are an inner membrane permease and an ATPase, respectively, resembling the ABC transporter in bacteria (87). TGD4 is a PA-binding protein that localizes to the outer membrane of the chloroplast (88). Finally, tgd2 encodes for an MCE domain-containing protein. Like the bacterial Mla system, TGD1 to -3 form a complex in the inner membrane (Fig. 3).
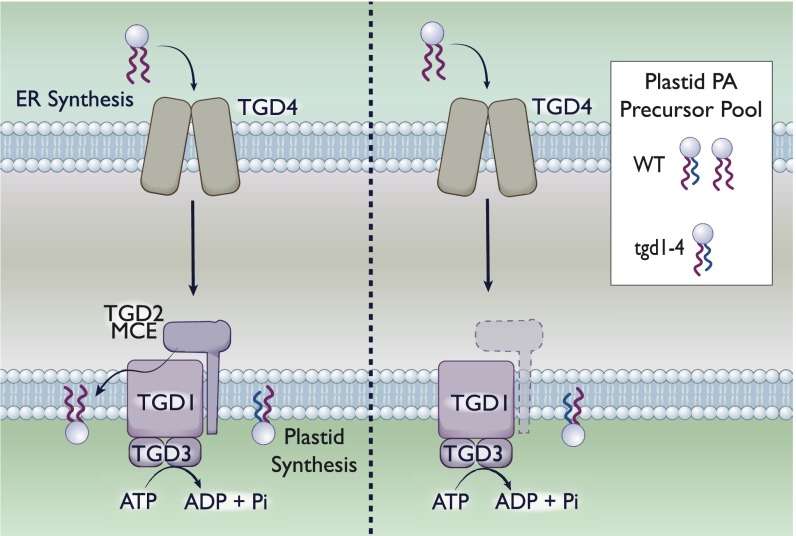
Fig. 3.
MCE domain protein TGD2 mediates PA transport in chloroplasts. The tgd1-4 genes in plant chloroplasts encode for multiple components of a PA import complex. The sn-2 position of PA differs depending on its origin: ER-derived PA has 18 carbon fatty acids, whereas plastid-derived PA has 16 carbon fatty acids, although the degree of unsaturation can vary. (Left) With the TGD1–4 system intact, the plastid is capable of both de novo synthesis and import of PA. As such, the PA precursor pool contains a roughly equivalent mixture (for A. thaliana) of both species. (Right). A deletion of any tgd1-4 genes eliminates the ability of the plastid to import PA from the ER. As such, the resultant pool of PA in the plastid is de novo-synthesized. We are depicting the deletion of tgd; however, a deletion of any individual component has been demonstrated to eliminate PA import.
In the absence of any of the 4 TGD proteins, the PA pool was derived primarily from the plastid, lacking ER-derived PA (84, 85, 87) (Fig. 3). This accumulation strongly suggests that in the absence of this system, chloroplasts are incapable of importing exogenous lipids from the ER. Taking these data into consideration, we find that it is highly likely that the TGD system is involved in import of PA from the outer membrane to the inner membrane within chloroplasts, supporting a retrograde model of transport similar to that proposed for Mla.
Dynamic Membranes: The Ebb and Flow of Lipid Transport
Despite significant advancements in lipid biosynthesis and regulation, we still lack critical, in-depth knowledge of how various lipid species are transported between membranes of the bacterial cell. To date, our understanding of LPS and lipoprotein transport far exceed our understanding of GPL transport. Recent structural work by multiple groups provide additional insight into how LPS molecules are queued in place and ultimately loaded onto the periplasmic transport bridge in an ATP-dependent fashion (89, 90). In a similar note, we have a clear biological understanding of how the LolA lipoprotein carrier is loaded with lipoproteins in an energy-dependent fashion (35). Curiously, in the case of lipoprotein transport, there is compelling biological evidence that alternative mechanisms exist in E. coli, and presumably other organisms, to mediate lipoprotein transfer in the absence of the periplasmic or outer membrane Lol components (91).
A hallmark paper by Nicholas Jones and Mary Jane Osborn (92) experimentally demonstrated that exogenously incorporated GPLs were capable of reversible transport from the outer to inner membranes, but this trait was not evident for LPS molecules, which were retained in the outer membrane. With the benefit of hindsight, it is likely that this finding was due at least in part to the Mla system. Around a similar time, it was shown by Manfred Bayer via electron microscopy that plasmolyzed E. coli cells exhibited zones of adhesion between the inner membrane and outer membrane (93). These proposed zones of adhesion, or “Bayer’s junctions,” were hypothesized to be responsible for the diffusion of lipidic material between the 2 membranes. While it is clear now that these junctions do not mediate LPS transport, their function in GPL transport remains inconclusive.
One of the most consequential questions in the field is: What mechanisms carry out anterograde transport of GPLs? Any system playing this role must fulfill certain criteria, including—but not limited to—substrate specificity and an optimized rate of transport. Bacterial size is remarkably homogeneous, which suggests that biosynthesis and transport are intimately linked. Because cell size is determined by the membrane, it makes sense that these processes are either coupled or have integrated feedback. When rates of LPS transport were severely decreased, suppressors of lethality mapped to FabH, which subsequently decreased GPL biosynthesis to match the growth rate of the LPS-defective mutant (94). Conversely, if GPL biosynthesis is increased via overexpression of FadR in trans, E. coli cell size practically doubled (95). This finding suggests that mechanisms of GPL anterograde transport accommodate abnormal rates of biosynthesis to approximately twice that of WT. There may be an absolute limit to the rate of GPL transport capping at the level to support cells twice the size of WT, because with FadR overexpression the authors found invaginations of the inner membrane via transmission electron microscopy (indicative of excess GPLs in the inner membrane). However, these findings do not impede interpretation of cell size (95).
A useful thought experiment, then, is to hypothesize mechanisms capable of anterograde transport while accommodating a range in rate of transport and targeted substrates. With this mentality, the most flexible system would be Bayer’s junctions (Fig. 4). These fusions between the outer leaflet of the inner membrane and the inner leaflet of the outer membrane would be broadly accommodative to lipid headgroups. However, these junctions rely on passive diffusion rates between the membranes with no obvious control on directionality. Additionally, mechanisms would be required to exclude certain lipids like LPS and lipoproteins from diffusing via this method. It is also possible that these junctions are formed with the assistance of yet to be identified accessory proteins that function as gate keepers.
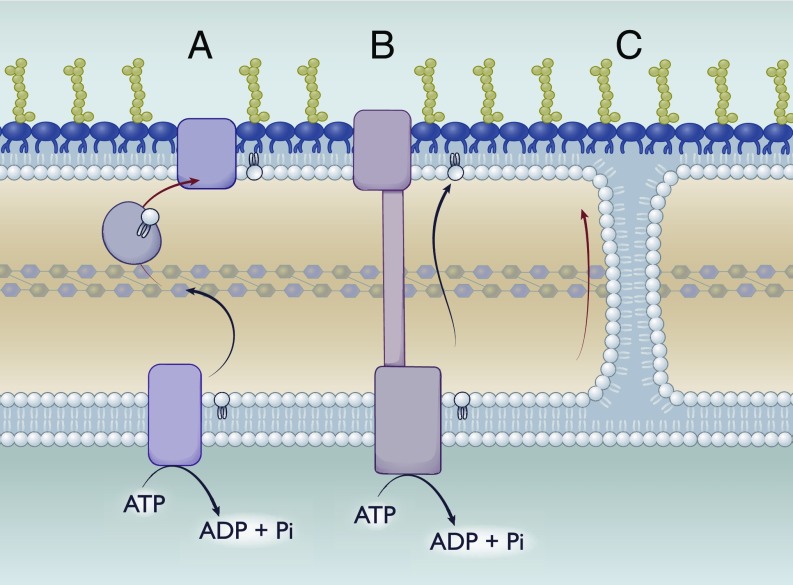
Fig. 4.
The quest for anterograde transport: Potential mechanisms. Possible mechanisms to mediate transport of GPLs from the inner membrane to the outer membrane. (A) Transport using a periplasmic chaperone. Similar to Lol, transport could be mediated via a periplasmic chaperone that would bind to GPLs either nonspecifically or in a head group-dependent manner. (B) Transport via a periplasmic-spanning bridge. Similar to the Lpt system, this model would rely on a continuous protein bridge consisting of an inner membrane complex, periplasmic components, and an outer membrane partner. (C) Passive diffusion via Bayer’s junctions. Bayer’s junctions have been proposed to form between the outer leaflet of the inner membrane and the inner leaflet of the outer membrane. These junctions could allow for the passive diffusion of GPLs between both inner and outer membranes continuously.
A second approach would be a system analogous to the Lpt system: That is, a protein bridge across the periplasm linked to an ABC-transporter (Fig. 4). Theoretically, this system could utilize ATP to continuously drive GPLs to the inner leaflet of the outer membrane while matching the rate of the Lpt system. Similar to the Bayer’s junctions, mechanisms would be required to exclude LPS from being transported erroneously.
Finally, transport could be mediated via soluble periplasmic chaperones. In effect, this would look like the Lol system (Fig. 4). An ABC-transporter in the inner membrane could drive transfer to periplasmic chaperones, which would then deposit lipid to the outer membrane, presumably in conjunction with an accessory protein. While this system could be highly adaptable to work exclusively for certain lipids by having multiple periplasmic chaperone proteins, it would also be significantly rate-limited, having a 1:1 lipid:protein stoichiometry.
Of course, while this list of putative transport mechanisms is far from exhaustive, it presents what we would argue are 3 likely candidates for anterograde lipid transport in Gram-negative bacteria based on the existing systems that have been identified to date.
Concluding Remarks
Bacterial cell envelope biologists have pursued mechanisms of transport for lipophilic substrates for decades. While significant progress has been made, particularly in the characterization of the Lpt system, much remains to be determined. One system, the Mla lipid transport system, has been the subject of controversy regarding its directionality of transport. While its initial identification and subsequent genetic characterization in numerous organisms, including A. baumannii by our laboratory, have led to the conclusion that it functions in a retrograde manner, there is a smaller subset of evidence suggesting the opposite. When contemplating the directionality of the Mla system, it is important to consider the following:
-
1)
The outer membrane phospholipase PldA has been identified as a suppressor for 2 independent mla phenotypes. Notably, the PldA active site lies at the bacterial surface and only cleaves GPLs in the outer leaflet of the outer membrane.
-
2)
Overexpression of pldA suppresses permeability defects of mla mutants.
-
3)
Deletion of pldA suppresses the cell death phenotype conferred by the mlaA* allele.
-
4)
MlaA has an amphipathic pore only accessible from the outer leaflet of the outer membrane, suggesting that GPL delivery to the inner leaflet of the outer membrane by Mla is not feasible.
-
5)
Evidence for anterograde lipid transfer (MlaD to MlaC) in vitro occurred regardless of ATP hydrolysis by MlaF, yet MlaF is required for function in vivo.
-
6)
LOS-deficient A. baumannii with a symmetric GPL bilayer outer membrane rapidly select for nonfunctional alleles of mla and pldA.
-
7)
MCE domain proteins mediate retrograde lipid transfer in other bacterial species and plant chloroplasts.
Even with the potential of new evidence that more strongly indicates anterograde transport, Mla is not essential, indicating other anterograde GPL transporters exist. While we remain confident in our conclusions, the field necessitates further empirical studies of the Mla lipid transport system to conclusively determine the directionality of the Mla system.
Acknowledgments
We thank the PNAS editorial board for the invitation to write this Perspective and Dr. Brent Simpson for insightful comments. This work was funded by NIH Grants AI138576, AI129940, and AI076322 (to M.S.T.) and by NSF Graduate Research Fellowship 049347-06 (to M.J.P.). We apologize for any work that was unable to be cited due to space restrictions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Lombard J., Once upon a time the cell membranes: 175 years of cell boundary research. Biol. Direct 9, 32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voelker D. R., Organelle biogenesis and intracellular lipid transport in eukaryotes. Microbiol. Rev. 55, 543–560 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohlenkamp C., Geiger O., Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Bell R. M., Mavis R. D., Osborn M. J., Vagelos P. R., Enzymes of phospholipid metabolism: Localization in the cytoplasmic and outer membrane of the cell envelope of Escherichia coli and Salmonella typhimurium. Biochim. Biophys. Acta 249, 628–635 (1971). [DOI] [PubMed] [Google Scholar]
- 5.Pomorski T. G., Menon A. K., Lipid somersaults: Uncovering the mechanisms of protein-mediated lipid flipping. Prog. Lipid Res. 64, 69–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai J., Pagano R. E., Measurement of spontaneous transfer and transbilayer movement of BODIPY-labeled lipids in lipid vesicles. Biochemistry 36, 8840–8848 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Rothman J. E., Kennedy E. P., Rapid transmembrane movement of newly synthesized phospholipids during membrane assembly. Proc. Natl. Acad. Sci. U.S.A. 74, 1821–1825 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huijbregts R. P. H., de Kroon A. I. P. M., de Kruijff B., Rapid transmembrane movement of newly synthesized phosphatidylethanolamine across the inner membrane of Escherichia coli. J. Biol. Chem. 273, 18936–18942 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Kol M. A., de Kroon A. I. P. M., Killian J. A., de Kruijff B., Transbilayer movement of phospholipids in biogenic membranes. Biochemistry 43, 2673–2681 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Hrafnsdóttir S., Menon A. K., Reconstitution and partial characterization of phospholipid flippase activity from detergent extracts of the Bacillus subtilis cell membrane. J. Bacteriol. 182, 4198–4206 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz N., Lipid flippases for bacterial peptidoglycan biosynthesis. Lipid Insights 8 (suppl. 1), 21–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doerrler W. T., Gibbons H. S., Raetz C. R. H., MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 279, 45102–45109 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Yan A., Guan Z., Raetz C. R. H., An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J. Biol. Chem. 282, 36077–36089 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rick P. D., et al. , Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid intermediate in the assembly of enterobacterial common antigen. J. Biol. Chem. 278, 16534–16542 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Liu D., Cole R. A., Reeves P. R., An O-antigen processing function for Wzx (RfbX): A promising candidate for O-unit flippase. J. Bacteriol. 178, 2102–2107 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Petris S., Ultrastructure of the cell wall of Escherichia coli and chemical nature of its constituent layers. J. Ultrastruct. Res. 19, 45–83 (1967). [DOI] [PubMed] [Google Scholar]
- 17.Stock J. B., Rauch B., Roseman S., Periplasmic space in Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 252, 7850–7861 (1977). [PubMed] [Google Scholar]
- 18.Simpson B. W., Trent M. S., Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 17, 403–416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doerrler W. T., Lipid trafficking to the outer membrane of gram-negative bacteria. Mol. Microbiol. 60, 542–552 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Silipo A., Molinaro A., The diversity of the core oligosaccharide in lipopolysaccharides. Subcell. Biochem. 53, 69–99 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Okuda S., Sherman D. J., Silhavy T. J., Ruiz N., Kahne D., Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat. Rev. Microbiol. 14, 337–345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bojkovic J., et al. , Characterization of an Acinetobacter baumannii lptD deletion strain: Permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J. Bacteriol. 198, 731–741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bos M. P., Tommassen J., The LptD chaperone LptE is not directly involved in lipopolysaccharide transport in Neisseria meningitidis. J. Biol. Chem. 286, 28688–28696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran A. X., Trent M. S., Whitfield C., The LptA protein of Escherichia coli is a periplasmic lipid A-binding protein involved in the lipopolysaccharide export pathway. J. Biol. Chem. 283, 20342–20349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bos M. P., Tefsen B., Geurtsen J., Tommassen J., Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl. Acad. Sci. U.S.A. 101, 9417–9422 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Y., et al. , Lipopolysaccharide is inserted into the outer membrane through an intramembrane hole, a lumen gate, and the lateral opening of LptD. Structure 23, 496–504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman D. J., et al. , Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359, 798–801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konovalova A., Silhavy T. J., Outer membrane lipoprotein biogenesis: Lol is not the end. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malinverni J. C., Silhavy T. J., Assembly of outer membrane β-Barrel proteins: The Bam complex. EcoSal Plus, 10.1128/ecosalplus.4.3.8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Botos I., et al. , Structural and functional characterization of the LPS transporter LptDE from gram-negative pathogens. Structure 24, 965–976 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang M.-J., Yakhnina A. A., Bernhardt T. G., NlpD links cell wall remodeling and outer membrane invagination during cytokinesis in Escherichia coli. PLoS Genet. 13, e1006888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama Si., Yokota N., Tokuda H., A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 16, 6947–6955 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeow J., et al. , The architecture of the OmpC-MlaA complex sheds light on the maintenance of outer membrane lipid asymmetry in Escherichia coli. J. Biol. Chem. 293, 11325–11340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grabowicz M., Lipoproteins and their trafficking to the outer membrane. EcoSal Plus, 10.1128/ecosalplus.ESP-0038-2018 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Taniguchi N., Tokuda H., Molecular events involved in a single cycle of ligand transfer from an ATP binding cassette transporter, LolCDE, to a molecular chaperone, LolA. J. Biol. Chem. 283, 8538–8544 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukahara J., Mukaiyama K., Okuda S., Narita S., Tokuda H., Dissection of LolB function–Lipoprotein binding, membrane targeting and incorporation of lipoproteins into lipid bilayers. FEBS J. 276, 4496–4504 (2009). [DOI] [PubMed] [Google Scholar]
- 37.d’Enfert C., Ryter A., Pugsley A. P., Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 6, 3531–3538 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hooda Y., et al. , Slam is an outer membrane protein that is required for the surface display of lipidated virulence factors in Neisseria. Nat. Microbiol. 1, 16009 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Konovalova A., Perlman D. H., Cowles C. E., Silhavy T. J., Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc. Natl. Acad. Sci. U.S.A. 111, E4350–E4358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho S.-H., et al. , Detecting envelope stress by monitoring β-barrel assembly. Cell 159, 1652–1664 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Kleinschmidt J. H., den Blaauwen T., Driessen A. J., Tamm L. K., Outer membrane protein A of Escherichia coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry 38, 5006–5016 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Nikaido H., Porins and specific diffusion channels in bacterial outer membranes. J. Biol. Chem. 269, 3905–3908 (1994). [PubMed] [Google Scholar]
- 43.Bishop R. E., Structural biology of membrane-intrinsic beta-barrel enzymes: Sentinels of the bacterial outer membrane. Biochim. Biophys. Acta 1778, 1881–1896 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebie Tan A., Burgess N. K., DeAndrade D. S., Marold J. D., Fleming K. G., Self-association of unfolded outer membrane proteins. Macromol. Biosci. 10, 763–767 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiffrin B., et al. , Skp is a multivalent chaperone of outer-membrane proteins. Nat. Struct. Mol. Biol. 23, 786–793 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sklar J. G., Wu T., Kahne D., Silhavy T. J., Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21, 2473–2484 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagan C. L., Silhavy T. J., Kahne D., β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Kim S., et al. , Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Malinverni J. C., et al. , YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61, 151–164 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Nikaido H., Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rangl M., et al. , Real-time visualization of phospholipid degradation by outer membrane phospholipase a using high-speed atomic force microscopy. J. Mol. Biol. 429, 977–986 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Bishop R. E., et al. , Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 19, 5071–5080 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malinverni J. C., Silhavy T. J., An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 8009–8014 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shrivastava R., Jiang X., Chng S.-S., Outer membrane lipid homeostasis via retrograde phospholipid transport in Escherichia coli. Mol. Microbiol. 106, 395–408 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Masilamani R., Cian M. B., Dalebroux Z. D., Salmonella tol-pal reduces outer membrane glycerophospholipid levels for envelope homeostasis and survival during bacteremia. Infect. Immun. 86, e00173-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sutterlin H. A., et al. , Disruption of lipid homeostasis in the gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. U.S.A. 113, E1565–E1574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.May K. L., Silhavy T. J., The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. MBio 9, e00379-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abellón-Ruiz J., et al. , Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat. Microbiol. 2, 1616–1623 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Chong Z.-S., Woo W.-F., Chng S.-S., Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol. Microbiol. 98, 1133–1146 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Slauch J. M., Silhavy T. J., Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J. Mol. Biol. 210, 281–292 (1989). [DOI] [PubMed] [Google Scholar]
- 61.Thong S., et al. , Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. eLife 5, e19042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ekiert D. C., et al. , Architectures of lipid transport systems for the bacterial outer membrane. Cell 169, 273–285.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamischke C., et al. , The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. eLife 8, e40171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ercan B., Low W.-Y., Liu X., Chng S.-S., Characterization of interactions and phospholipid transfer between substrate binding proteins of the OmpC-mla system. Biochemistry 58, 114–119 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Huang Y. M., et al. , Molecular dynamic study of MlaC protein in gram-negative bacteria: Conformational flexibility, solvent effect and protein-phospholipid binding. Protein Sci. 25, 1430–1437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hughes G. W., et al. , Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol. 10.1038/s41564-019-0481-y (2019). [DOI] [PubMed] [Google Scholar]
- 67.Roier S., et al. , A novel mechanism for the biogenesis of outer membrane vesicles in gram-negative bacteria. Nat. Commun. 7, 10515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baarda B. I., Zielke R. A., Le Van A., Jerse A. E., Sikora A. E., Neisseria gonorrhoeae MlaA influences gonococcal virulence and membrane vesicle production. PLoS Pathog. 15, e1007385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toyofuku M., Nomura N., Eberl L., Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17, 13–24 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Powers M. J., Trent M. S., Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc. Natl. Acad. Sci. U.S.A. 115, E8518–E8527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moffatt J. H., et al. , Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Powers M. J., Trent M. S., Expanding the paradigm for the outer membrane: Acinetobacter baumannii in the absence of endotoxin. Mol. Microbiol. 107, 47–56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boll J. M., et al. , A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A. 113, E6228–E6237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Osborn M. J., Munson R., Separation of the inner (cytoplasmic) and outer membranes of gram-negative bacteria. Methods Enzymol. 31, 642–653 (1974). [DOI] [PubMed] [Google Scholar]
- 75.Sabarth N., et al. , Identification of surface proteins of Helicobacter pylori by selective biotinylation, affinity purification, and two-dimensional gel electrophoresis. J. Biol. Chem. 277, 27896–27902 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Chakrabarti S. R., Chaudhuri K., Sen K., Das J., Porins of Vibrio cholerae: Purification and characterization of OmpU. J. Bacteriol. 178, 524–530 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arruda S., Bomfim G., Knights R., Huima-Byron T., Riley L. W., Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 261, 1454–1457 (1993). [DOI] [PubMed] [Google Scholar]
- 78.Van der Geize R., et al. , A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pandey A. K., Sassetti C. M., Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gilk S. D., et al. , Bacterial colonization of host cells in the absence of cholesterol. PLoS Pathog. 9, e1003107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nazarova E. V., et al. , Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. eLife 6, e26969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klepp L. I., et al. , Impact of the deletion of the six mce operons in Mycobacterium smegmatis. Microbes Infect. 14, 590–599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Isom G. L., et al. , MCE domain proteins: Conserved inner membrane lipid-binding proteins required for outer membrane homeostasis. Sci. Rep. 7, 8608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Awai K., Xu C., Tamot B., Benning C., A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc. Natl. Acad. Sci. U.S.A. 103, 10817–10822 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu C., Fan J., Froehlich J. E., Awai K., Benning C., Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17, 3094–3110 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kirk J. T., Chloroplast structure and biogenesis. Annu. Rev. Biochem. 40, 161–196 (1971). [DOI] [PubMed] [Google Scholar]
- 87.Roston R. L., Gao J., Murcha M. W., Whelan J., Benning C., TGD1, -2, and -3 proteins involved in lipid trafficking form ATP-binding cassette (ABC) transporter with multiple substrate-binding proteins. J. Biol. Chem. 287, 21406–21415 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Z., Anderson N. S., Benning C., The phosphatidic acid binding site of the Arabidopsis trigalactosyldiacylglycerol 4 (TGD4) protein required for lipid import into chloroplasts. J. Biol. Chem. 288, 4763–4771 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Owens T. W., et al. , Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567, 550–553 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y., Orlando B. J., Liao M., Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 567, 486–490 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grabowicz M., Silhavy T. J., Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc. Natl. Acad. Sci. U.S.A. 114, 4769–4774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jones N. C., Osborn M. J., Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J. Biol. Chem. 252, 7405–7412 (1977). [PubMed] [Google Scholar]
- 93.Bayer M. E., Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J. Struct. Biol. 107, 268–280 (1991). [DOI] [PubMed] [Google Scholar]
- 94.Yao Z., Davis R. M., Kishony R., Kahne D., Ruiz N., Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 109, E2561–E2568 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vadia S., et al. , Fatty acid availability sets cell envelope capacity and dictates microbial cell size. Curr. Biol. 27, 1757–1767.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]