Significance
Transposable elements (TE) are abundant in plant genomes, comprising almost 80% of the sunflower genome. In a process known as RNA-dependent DNA methylation, plants use 24-nt siRNAs to keep TEs silenced and prevent transposition. Here, we report that a TE-derived IR element, located within the HaWRKY6 locus in the sunflower genome, can regulate the expression of the neighboring gene by changing the structure of the chromatin. Our findings represent a remarkable, dynamic, and self-limited mechanism of gene regulation wherein several regulatory pathways converge. Considering the poor conservation and mobile nature of TEs, the identified mechanism has substantial evolutionary implications.
Keywords: small RNAs, chromatin loops, DNA methylation, sunflower
Abstract
Transposable elements (TEs) are extremely abundant in complex plant genomes. siRNAs of 24 nucleotides in length control transposon activity in a process that involves de novo methylation of targeted loci. Usually, these epigenetic modifications trigger nucleosome condensation and a permanent silencing of the affected loci. Here, we show that a TE-derived inverted repeat (IR) element, inserted near the sunflower HaWRKY6 locus, dynamically regulates the expression of the gene by altering chromatin topology. The transcripts of this IR element are processed into 24-nt siRNAs, triggering DNA methylation on its locus. These epigenetic marks stabilize the formation of tissue-specific loops in the chromatin. In leaves, an intragenic loop is formed, blocking HaWRKY6 transcription. While in cotyledons (Cots), formation of an alternative loop, encompassing the whole HaWRKY6 gene, enhances transcription of the gene. The formation of this loop changes the promoter directionality, reducing IR transcription, and ultimately releasing the loop. Our results provide evidence that TEs can act as active and dynamic regulatory elements within coding loci in a mechanism that combines RNA silencing, epigenetic modification, and chromatin remodeling machineries.
Given their sessile nature, plants need to adjust their growth patterns depending on external stimuli. Such adaptive responses are orchestrated by the expression and repression of specific genes. In plants, small RNAs (sRNAs) can control gene expression both at transcriptional and at posttranscriptional levels (1). Transcriptional gene silencing is triggered and controlled by 24-nt heterochromatic siRNAs (het-siRNAs) (1). In plants, sRNAs are produced by the catalytic action of Dicer-like (DCL) type III ribonucleases after the recognition of dsRNA precursors. DCL3 produces het-siRNAs using RNA polymerase IV (RNAPIV) transcripts, converted into dsRNAs by the RNA-dependent RNA polymerase 2 (RDR2), as a template. Het-siRNAs are preferentially loaded into ARGONAUTE 4 (AGO4) to trigger, either in cis and/or in trans, DNA methylation in a process known as RNA-directed DNA methylation (RdDM) (2). This process, commonly observed in TEs, normally induces changes in the chromatin state, leading to nucleosome condensation and stable silencing of the targeted loci (1, 2). Besides this control over the density of nucleosomes, a dynamic fluctuation in the 3D chromatin conformation, known as genome topology, also modulates gene expression in transcriptional hubs. Chromatin folding can lead to both local and long-distance chromatin loop formation (3, 4). Local loops joining 5′ and 3′ ends of a gene have been proposed to allow efficient recycling of the RNA polymerase II (RNAPII) from the termination site back to the promoter in a process known as gene looping (5, 6). Repressive loops are also frequent as is the case of the intragenic loop formed between the promoter region and the first intron of FLOWERING LOCUS C (FLC), stably repressing its expression (7). In animals, it has been found that gene loops can also affect the directionality of transcription by forcing RNAPII to move in one direction (8).
Plants contain large amounts of repetitive sequences and TEs in their genomes, ranging from ∼20% of the Arabidopsis thaliana genome (9) to extremes, such as sunflower with a ∼78% of its genome represented by these elements (10). TEs are DNA segments that can insert into new chromosomal locations, often duplicating themselves in the process. Miniature IR TEs (MITEs), first described in plants, are short (50–500 bp) nonautonomous TEs with terminal IRs, predominantly inserted in gene-rich regions and affecting the expression of neighboring genes (11).
Here, we show that a transcribed sunflower MITE, located 600 bp upstream from the transcription start site of the HaWRKY6 gene, influences the chromatin 3D conformation of this locus. The expression of this IR leads to the production of het-siRNAs that trigger RdDM of the MITE-containing region of the gene, which in turn serves as an anchor point to stabilize the formation of chromatin loops in the locus. We identified 2 short-range chromatin interactions, modulated by expression of the MITE. The first one, comprising the whole HaWRKY6 gene, mediates gene looping and enhances its transcription. A second loop, comprising the HaWRKY6 regulatory region up to its fourth intron, represses expression of the gene, probably by blocking the movement of RNAPII through the junction. The formation of each loop appears to be tissue specific and dependent on additional DNA methylation signatures in the locus. Additionally, the formation of these loops changes RNAPII directionality, potentially reducing the transcription of the IR region, het-siRNA production, and, ultimately, releasing the loops. Our findings represent a remarkably dynamic mechanism of gene regulation where several regulatory pathways, including sRNA silencing, epigenetic regulation, and chromatin remodeling, converge.
Results
An IR-Derived Noncoding RNA (ncRNA) Is Transcribed from the HaWRKY6 Proximal Regulatory Region.
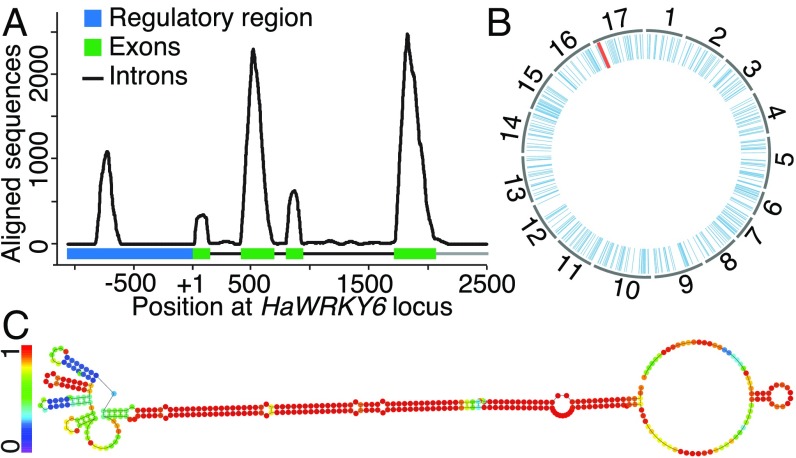
We identified the sunflower gene HaWRKY6 as a recently evolved target of the conserved miRNA396 regulatory network controlling the plant response to temperature damage (12). Aiming to identify the HaWRKY6 transcriptional start site (TSS) and its promoter region, we aligned sunflower expressed sequence tags (ESTs) to the complete gene locus. Introns and exons were quickly recognized as well as expressed regions (Fig. 1A). Interestingly, we detected a discrete expressed region within the regulatory region of the gene located between 600 and 800 bp upstream from the TSS (Fig. 1A). Sunflower RNA sequencing analysis confirmed the expression of this region and revealed the existence of an alternative splicing event involving exon 2 (SI Appendix, Fig. S1). Sequence analysis of the expressed region on the HaWRKY6 promoter revealed the absence of any protein ORF either in the sense or in the antisense orientation, thus, defining it as a noncoding (nc) transcript which we designated ncRNA-W6 (ncW6) (SI Appendix, Fig. S1). A sequence alignment against the sunflower genome (13) revealed multiple copies of ncW6 across the genome, while no alignments were detected against the A. thaliana genome (Fig. 1B). These features suggested that this sequence might be a sunflower-specific transposon. Similar to MITEs, the ncW6 is a short 260-bp sequence, does not encode a transposase (nonautonomous element), and possesses terminal IRs (SI Appendix, Fig. S1), suggesting that it is a member of the MITE family of transposons. An in silico analysis revealed that the ncW6 folds into a highly stable structure with a long dsRNA stem typical of regulatory ncRNAs, MITEs, and miRNAs precursors (Fig. 1C).
Fig. 1.
An IR-derived ncRNA is transcribed from the HaWRKY6 proximal promoter. (A) Alignment of the HaWRKY6 locus against a sunflower ESTs database. (B) Alignment of a ncW6 sequence against the 17 sunflower chromosomes. The red line shows the position of the ncW6, and the blue lines are similar copies. (C) Secondary structure of the ncW6. Base pair probabilities are noted from 0 (purple) to 1 (red).
Abundant sRNAs Are Produced from the ncW6 Transcript.
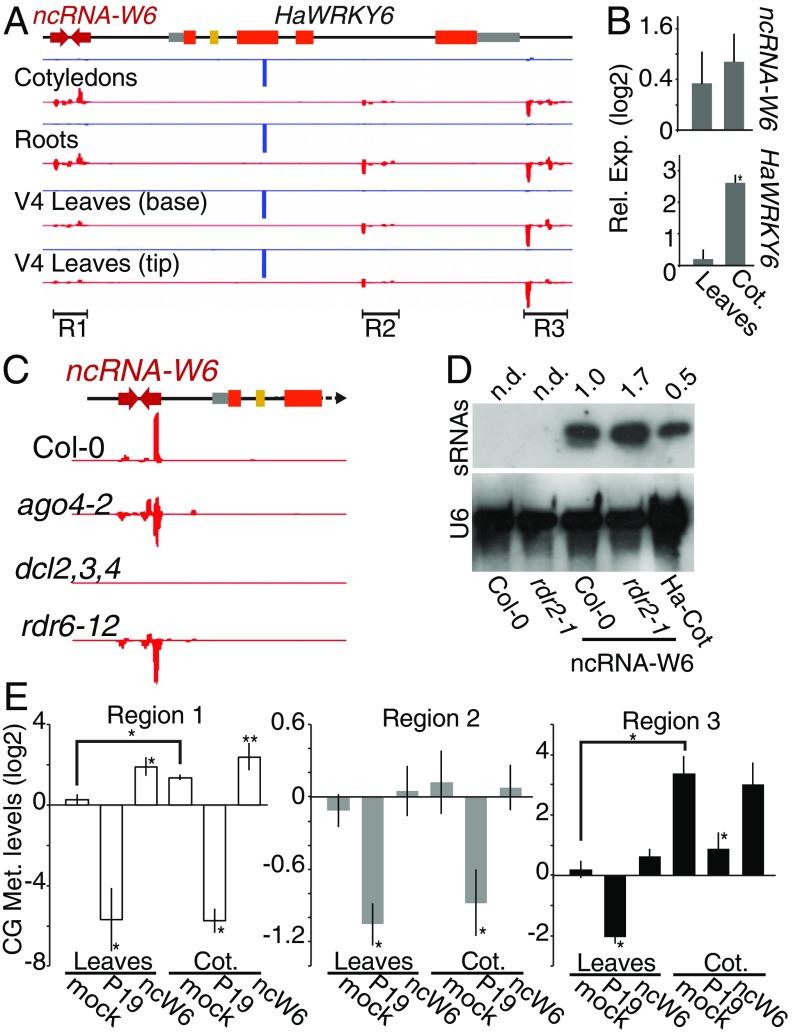
The transposonic nature of the ncW6 and the dsRNA structure of its transcript suggested that it could act as a precursor for sRNAs that could potentially modulate the epigenetic landscape of the region. sRNAs sequencing of different sunflower tissues showed a clear peak of 21-nt sRNAs mapping to the recently evolved miR396 target site in the third exon (second exon of the alternatively spliced HaWRKY6 transcript) (12) (Fig. 2A). We also found abundant 24-nt sRNAs mapping to the ncW6 region, hereafter referred to as region 1. Interestingly, the normalized abundance of these 24-nt sRNAs was higher in Cots and roots than in leaves (Fig. 2A). Despite multiple similar copies of this MITE found in the sunflower genome (Fig. 1B), 24-nt sRNAs mapping to the ncW6 sequence were mostly unique suggesting that they are produced from this nc transcript (SI Appendix, Fig. S2A). The similar levels of the ncW6 transcript in Cots compared to leaves (Fig. 2B), even when considerably more sRNAs are produced in this tissue, suggested that either the transcript is rapidly processed into 24-nt siRNAs or that the generated siRNAs are able to repress its locus in a negative feedback loop. We also detected even amount of 24-nt sRNAs mapping to other 2 regions in the HaWRKY6 locus: one within the fourth HaWRKY6 intron and the other downstream from its 3′ UTR, hereinafter named regions 2 and 3, respectively (Fig. 2A). Aiming to determine whether the biogenesis mechanism of the ncW6 sRNAs follows the canonical het-siRNA pathway, we transformed A. thaliana dcl2/3/4 and ago4-2 mutants with a copy of ncW6 and quantified sRNAs by sRNA-seq. The lack of sunflower mutants in these genes forced us to use the model plant species. The analysis revealed almost undetectable levels of ncW6-derived sRNAs in dcl2/3/4, confirming their canonical origin (Fig. 2C). As expected, the mutation of AGO4, the effector in the RdDM pathway guiding the DNA methylation, did not affect the production of sRNA in the region (Fig. 2C). The generation of these sRNAs was still observed in rdr6-12 mutants, eliminating the possibility that they are produced by the transgene-silencing pathway (Fig. 2C). Moreover, the ncW6 was able to produce 24-nt sRNAs in rdr2-1 mutants (Fig. 2D). This indicates that the extensive sequence complementarity and stable folding of the ncW6 could be processed into sRNA production in an RDR2 and probably a RNAPIV/V independent pathway. The RDR2-independent origin of these 24-nt siRNAs made us wonder whether they can still trigger DNA methylation on the parental locus. To this end, we first performed bisulfite sequencing to identify and map DNA methylation of the endogenous HaWRKY6 locus in Cots and leaves, organs showing differential sRNA accumulation. The assay showed that sunflower Cots accumulate more 5-methyl cytosine in the asymmetrical context CHH, in the ncW6, and in region 3 than leaves, while such a difference is less pronounced in region 2 (SI Appendix, Fig. S2 B and C). To confirm this result and quantify such differences, we checked the DNA methylation status in the same samples by Chop-qPCR. We confirmed that the HaWRKY6 locus is methylated in the 3 analyzed regions (Fig. 2E). Once again, we found differential methylation in the ncW6 region and in region 3 but not in region 2 between tested tissues (Fig. 2E). The large difference in region 3 methylation detected by HpaII Chop-qPCR even when the CG context was nearly fully methylated in both tissues (HpaII restriction site, CCGG) possibly reflected a stronger enzyme inhibition caused by the additional methylation of the first cytosine of the site, fully methylated in Cots but not in leaves. Aiming to confirm that the siRNAs derived from the ncW6 trigger the DNA methylation on its locus, we transiently transformed sunflower Cots and leaves with constructs constitutively expressing an additional copy of the ncW6 or the viral suppressor protein P19. Even though P19 has a special affinity for DCL4-derived 21-nt siRNAs, it has been shown that it can also bind endogenous 24-nt siRNAs (14, 15) and that this sRNA population is strongly reduced in plants expressing this protein (16). As expected, the 35S::ncW6 construct increased the levels of 24-nt siRNAs while 35S::P19 reduced them (SI Appendix, Fig. S2D). Accordingly, the methylation of the endogenous ncW6 regions is increased or reduced, respectively (Fig. 2E). The 35S::P19 construct also produced a demethylation of the whole HaWRKY6 locus as expected from a general suppressor of the siRNA pathway (Fig. 2E). These results confirmed that the methylation of the locus depends on the RdDM pathway and that the ncW6-derived siRNAs locally trigger it.
Fig. 2.
ncW6 generates epigenetically active sRNAs. (A) Alignment of sRNA sequencing reads to the HaWRKY6 locus in samples of different sunflower tissues; 24 nt (red) mapped to regions 1–3 and 21 nt (blue) corresponding to ha-miR396 reads mapped to the miRNA-target site in the gene. (B) ncW6 and HaWRKY6 transcript levels measured by RT-qPCR in sunflower Cots relative to leaves. (C) Alignment of sRNAs to a 35S::ncW6 construct transformed into Col-0, ago 4–2, dcl2,3,4, and rdr6-12 A. thaliana mutant plants. (D) RNA blots detecting ncW6-derived sRNAs in A. thaliana wild-type (Col-0) and rdr2-1 mutant plants control or transformed with 35S::ncW6 or sunflower Cots (Ha-Cot) as the positive control. U6 was used as a loading control and signal intensity calculated using ImageJ. Not detected signal (n.d.). (E) HpaII HaWRKY6 Chop-qPCR analysis of mock sunflower samples, plants overexpressing the silencing suppressor P19 (P19) as a sRNA decoy, or the ncW6. Digestion efficiency was quantified by qPCR with primers spanning restriction sites in the sRNAs mapping regions and normalized to an undigested region. Error bars show 2 × SEM, P values of less than 0.05 (**) or 0,01 (*) in a 2-tailed unpaired t test were considered significant.
The ncW6 Modulates Alternative Loop Formation at the HaWRKY6 Locus.
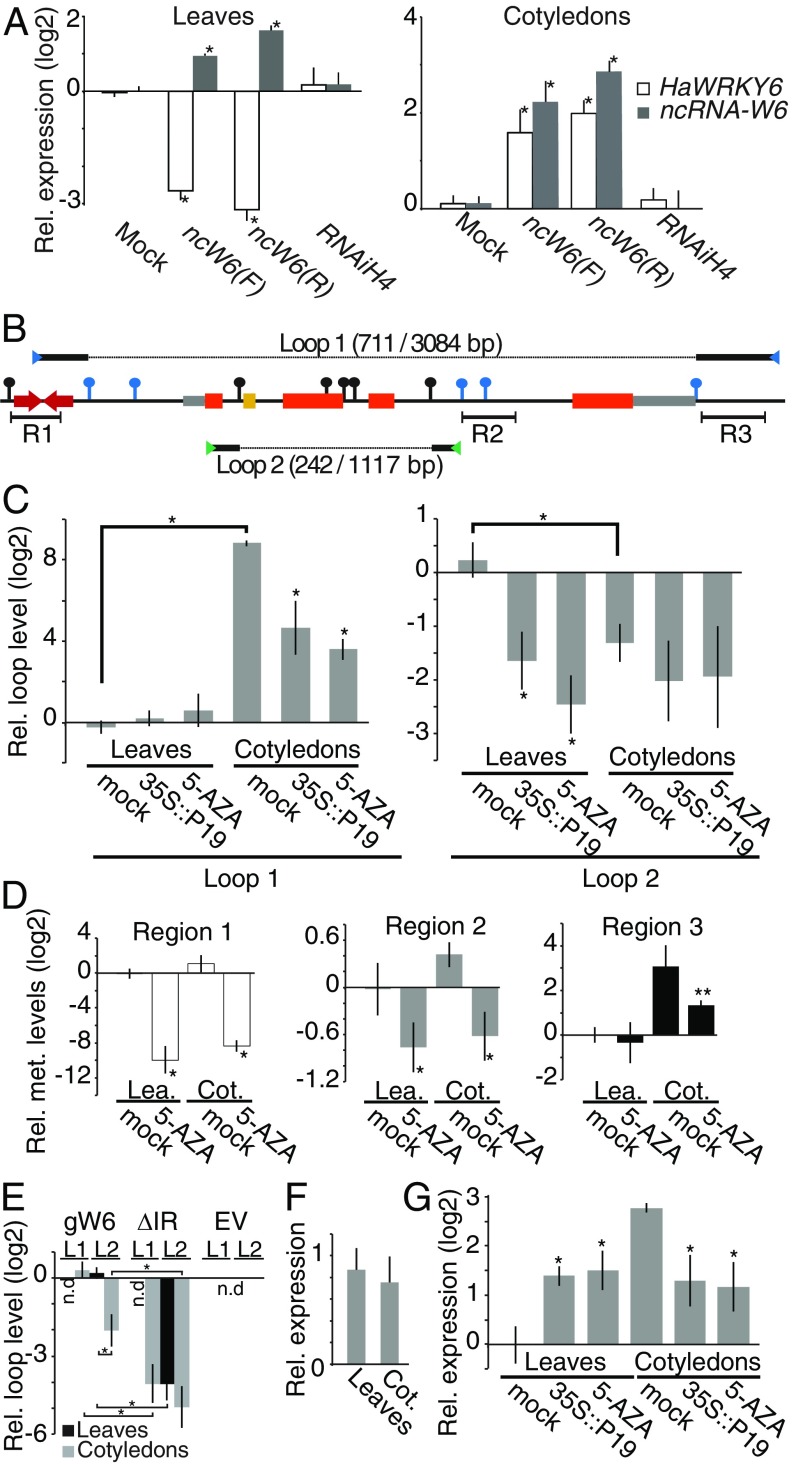
MITEs are often found close to genes where they can affect their expression (17, 18). To test whether ncW6 affects the expression of this locus in sunflower, we transiently overexpressed the ncRNA in sunflower leaves and measured the expression of both ncW6 and HaWRKY6. The results showed that the ectopic accumulation of the ncW6 was sufficient to reduce the abundance of the endogenous HaWRKY6 (Fig. 3A). Interestingly, when the transient expression was performed in Cots, we observed the opposite behavior, i.e., an increment of HaWRKY6 along with the high ncW6 levels (Fig. 3A). Such opposite regulatory behaviors would not be expected if methylation directly regulates transcription of the locus. It is worth noting that stronger DNA methylation correlates with high HaWRKY6 expression, which is noncanonical for the RdDM pathway. However, besides directly influencing transcription, DNA methylation could also influence chromatin conformation to affect gene expression in a different way (4). Given the methylation patterns along the HaWRKY6 locus, we wondered whether the methylation of the ncW6 region could serve as an anchor point to stabilize interactions between this region and the methylated sites in regions 2 and 3. To test this hypothesis, we performed chromosome conformation capture (3C) assays in samples extracted from sunflower leaves and Cots with primers designed to explore all possible combinations of interactions between the methylated regions. Using this approach, we detected a chromatin loop linking the ncW6 region and the methylated region 3 in Cots (Fig. 3B and SI Appendix, Fig. S3). The abundance of this loop, hereinafter called “Loop 1” (L1) was significantly higher in Cots than in leaves, which showed nearly undetectable levels (Fig. 3C). This observation is in agreement with the higher degree of DNA methylation and ncW6-derived sRNAs mapping to regions 1 and 3 in Cots compared to leaves (Fig. 2 A and E and SI Appendix, Fig. S2 A–C). We also found a second chromatin loop between the ncW6 region and the methylated region 2 (hereinafter called “Loop 2” [L2]) (Fig. 3B and SI Appendix, Fig. S3). Unlike L1, this second loop appeared to be more abundant in leaves than in Cots (Fig. 3C). Such alternative loop formation in leaves goes along with a drastic reduction of CHH methylation in region 3, which could cause the use of region 2, that is more uniformly methylated between tissues as an alternative anchor point (Fig. 2E and SI Appendix, Fig. S2B). To confirm that the methylation of these regions allows the formation of tissue-specific loops in the chromatin, we treated sunflower plants with 5-aza-2′-deoxycytidine (5-AZA), which inhibits DNA methyltransferase activity, resulting in DNA demethylation. Chop-qPCR analysis of leaves and Cots of treated plants showed a drastic reduction in DNA methylation in all 3 regions of the HAWRKY6 locus when compared to control plants (Fig. 3D). We also made use of the 35S::P19 transgenic tissues that showed reduced DNA methylation in the HaWRKY6 locus (Fig. 2E). Both treatments induced the opening of L1 and L2 in the tested tissues (Fig. 3C). These results confirm that the stability of both loops depends on the methylation of these regions of the locus. In addition we cloned 2 HaWRKY6 genomic constructs, one comprising the whole locus (from regions 1–3) and a second one that excludes the ncW6 encoding region. We used these constructs to transform sunflower leaves and Cots transiently, and we tested their capacity to form chromatin loops using primers designed to bind vector-specific sequences, thus, avoiding measuring the endogenous HaWRKY6 locus. Supporting the importance of the IR in the loop formation, we mainly detected L1 and L2 formation when the plants were transformed with the artificial construct comprising the whole locus (Fig. 3E).
Fig. 3.
The ncW6 modulates chromatin topology through alternative loop formation at the HaWRKY6 locus. (A) HaWRKY6 and ncW6 transcript levels in transiently transformed sunflowers expressing the ncW6 cloned in each orientation. A nonrelated hairpin structure (RNAiH4) was used as a negative control. (B) Schematic of the HaWRKY6 genomic region. HindIII (black pins) and MspI (blue pins) restriction sites. R1–R3 note sRNA mapping regions. Blue and green arrowheads indicated primers used to detect L1 and L2, respectively. Solid lines on the top and bottom of the scheme show the obtained sequence after 3C ligation while the dashed line indicates the missing sequence. The obtained and undigested sequence length is given in brackets. (C) Quantification of loop formation by 3C-qPCR in leaves and Cots of mock controls and plants treated with 5-AZA or expressing P19. (D) Chop-qPCR of genomic DNA from mock or treated leaves and Cots to quantified DNA methylation in region 1 (white), regions 2 (gray), and region 3 (black). (E) Loop formation (L1 and L2) in leaves and Cots transformed with a full-length copy of the HaWRKY6 genomic locus (gW6), a version excluding the ncW6 (ΔIR) or an empty vector (EV). (F) HaWRKY6 promoter activity as measured by RT-qPCR quantification of transiently transformed sunflower expressing the reporter gene GUS under the HaWRKY6 promoter. (G) HaWRKY6 transcript levels as measured by RT-qPCR in samples extracted from 5-AZA-treated or transiently transformed sunflower tissues. In all panels, error bars represent 2 × SEM, P values of less than 0.05 (**) or 0,01 (*) in a 2-tailed unpaired t test were considered significant.
It has been reported that chromatin loops that contain both a gene regulatory region and the whole transcriptional unit as does L1 in Cots promote a more efficient usage of RNAPII enhancing the locus transcription in a process known as gene looping (5, 6). Interestingly, when we expressed a GUS reporter gene under the HaWRKY6 promoter in sunflower Cots and leaves, we detected similar expression levels in both tissues (Fig. 3F) even when the HaWRKY6 transcripts were considerably more abundant in Cots than in leaves (Fig. 2B). This result suggested that L1 could produce a gene-looping phenomenon in Cots, hinting at a L1-mediated positive regulatory role over HaWRKY6 expression. The formation of ‘‘intragenic loops,” such as L2 in leaves, could affect RNAPII processivity resulting in a decrease in the transcription rate (7). The fact that L2 is more abundant in leaves, tissue where the promoter construct is active but the HaWRKY6 transcript is hardly detectable, suggests that L2 functions to limit expression by such a mechanism. Supporting this interpretation, we found that disrupting the loop formation with P19 or 5-AZA treatments enhanced HaWRKY6 transcription in leaves where L2 is more abundant and repressed it in Cots where L1 predominates (Fig. 3G).
The HaWRKY6 Promoter Controls the Expression of Both HaWRKY6 and ncW6.
The position of the ncW6 upstream of the HaWRKY6 locus suggests that the same regulatory region could act as a bidirectional promoter of the divergent genes as has been reported (19). A bioinformatic prediction revealed TATA boxes at both ends of the cloned region, compatible with bidirectional transcription (SI Appendix, Fig. S4A). To test such a possibility, we cloned a long and a short version of the HaWRKY6 promoter (890 bp and 623 bp upstream from the TSS, respectively) including or not the ncW6, respectively (SI Appendix, Fig. S4A). Both promoter versions were cloned in the sense and antisense directions upstream from the GUS coding sequence and transformed into A. thaliana (Col-0) plants. GUS staining was then used to score the activity of the promoter. We observed that both versions of the HaWRKY6 promoter, independent of the orientation, direct the expression of the reporter gene in hypocotyls, Cots, leave veins, petioles, cauline leaves, as well as in flowers (SI Appendix, Fig. S4B). It is worth mentioning that both versions and orientations of the promoter share the same expression pattern and similar levels, supporting the hypothesis of a similar bidirectional activity (SI Appendix, Fig. S4B). A transient transformation of sunflower Cots with the same promoter constructs supported the observations in A. thaliana, showing similar expression levels of all tested constructs (SI Appendix, Fig. S4C).
Chromatin L1 Changes the Transcription Directionality of the HaWRKY6 Locus in Cots.
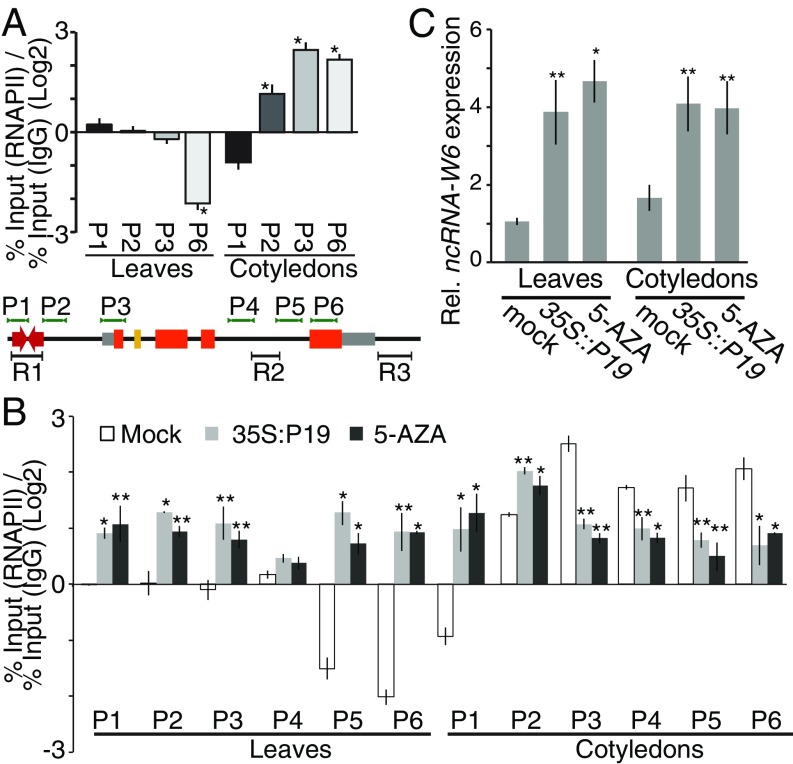
The phenomenon of divergent transcription as the case of the HaWRKY6 locus and its neighbor ncW6 is common to most active promoters in diverse organisms (20, 21). Recently, it was reported that gene looping plays an important role in regulating the bidirectional activity of promoter regions, reducing the production of divergently transcribed ncRNAs (8). The fact that transcriptional activity of the HaWRKY6 promoter appeared similar in both orientations (SI Appendix, Fig. S2 B and C), but only the HaWRKY6 is actively transcribed in Cots (Fig. 2B), suggests that formation of the alternative chromatin loop could modulate promoter directionality. In this context, the formation of L1 in Cots could restrict transcription of ncW6 and push RNAPII within the gene loop encompassing the HaWRKY6 locus. To test this theory, we assessed RNAPII deposition across the HaWRKY6 locus by chromatin immunoprecipitation (ChIP)-qPCR to determine transcription directionality in Cots and leaves. Results showed that, in Cots, RNAPII deposition in the promoter region (P2), an increased density over the HaWRKY6 transcription start site (P3), and even levels across the gene body (P4-P6) as expected for active transcription (Fig. 4A). However, we observed that the RNAPII is depleted in the ncW6 region (P1) indicating reduced transcription in the opposite direction and suggesting impairment of the bidirectional promoter by L1 (Fig. 4 A and B). In leaves, we found low but homogeneous levels of RNAPII in regions P1–P4 and a marked decrease in RNAPII density in regions P5 and P6, suggesting blockage of transcription at intron 4 by the intragenic L2 (Fig. 4A). To confirm the influence of both chromatin loops in promoter directionality on transcription, we repeated the RNAPII occupancy assay in plants treated with 5-AZA or expressing P19. In leaves, the opening of the loops by these treatments produced an increment in RNAPII occupancy evenly across the locus with a reversion in the polymerase density toward the end of the locus after methylation region 2 (Fig. 4B). In Cots, the same treatments produced an even reduction of polymerase across the HaWRKY6 coding region and increased polymerase occupancy in the ncW6 region, suggesting reactivation of the promoter bidirectionality in this tissue (Fig. 4B). Further support for this interpretation is provided by the increase in ncW6 transcript levels following 5-AZA treatment and P19 plants (Fig. 4C). These results imply that gene looping suppresses promoter bidirectionality in Cots, supporting the idea that this type of chromatin interaction enhances the directionality of transcription as shown for Saccharomyces cerevisiae (8).
Fig. 4.
Chromatin looping in the HaWRKY6 locus changes the transcription directionality of the HaWRKY6 promoter. (A and B) ChIP of RNAPII follow by qPCR quantification. RNAPII profile across the HaWRKY6 locus in sunflower Cots and leaves (A) or in plants treated with 5-AZA or expressing P19 (B). A diagram of the locus and primers used (P1–P6) is shown at the bottom of A. (C) ncW6 transcript levels as measured by RT-qPCR in plants treated with 5-AZA expressing P19. Error bars represent 2 × SEM, P values of less than 0.05 (**) or 0,01 (*) in a 2-tailed unpaired t test were considered significant.
Discussion
Exploring the HaWRKY6 regulatory region, we identified a transcribed MITE-like TE (ncW6) capable of forming a hairpin structure. Sunflower sRNA-seq analysis revealed 24-nt sRNAs that map to the ncW6 sequence and are substantially more abundant in Cots than in leaves. Usually, sRNA-dependent DNA methylation of TEs triggers histone modifications to stably repress their expression (22, 23). However, the HaWRKY6 locus does not appear to be regulated by this canonical mechanism since the largest sRNA accumulation and DNA methylation are observed in those tissues with the highest locus expression. There are numerous examples in which heterochromatic silencing of TEs influence expression of nearby genes without a permanent silencing, including agouti and Axin loci in mouse (24), FLC (25), and FWA (26) in A. thaliana, but the underlying mechanisms are not well understood. A recent report describes a methyl-DNA–binding complex that promotes expression of proximal genes upon recruitment to methylated regions (27). DNA methylation can also influence chromatin architecture by promoting interaction between methylated sequences (28, 29). Our analyses revealed that enrichment of DNA methylation in flanking regions of HaWRKY6 is associated with gene loop formation and elevated gene transcription. In contrast, low methylation at these regions, especially in region 3, together with a stable methylation signature in intron 4, correlates with the formation of an intragenic loop in leaves and repression of the gene. RNAPII occupancy assays revealed that the stabilization of the chromatin loops not only affects HaWRKY6 transcription, but also modifies the directionality of its promoter, ultimately reducing ncW6 transcription, sRNA production, and locus methylation, in turn, resulting in loop release (Fig. 5). This implies that the methylation of regions 2 and 3 could dictate which loop is formed, but the dynamic methylation of region 1 modulates them. It could also be expected that additional factors, such as changes in transcription rates, also influence which loop forms. On top of this, ha-miR396 also controls, in a temperature- and salicylic acid-dependent manner, the abundance of HaWRKY6 (12), giving the system an additional layer of complexity.
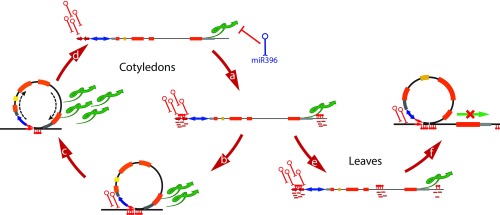
Fig. 5.
A proposed model for the IR-mediated dynamic regulation of the HaWRKY6 locus. In Cots, transcription of the bidirectional HaWRKY6 promoter yields a hairpin ncW6 transcript, which is processed into siRNAs that trigger RdDM on its own locus (a). An additional DNA methylated region downstream from the HaWRKY6 3′UTR stabilizes the formation of a loop encompassing the whole locus (b), potentially promoting an efficient RNAPII usage and transcription of the HaWRKY6 gene (c). Methylation of the ncW6 sequence and loop formation are associated with a change in the direction of transcription, which, in turn, reduces sRNA abundance and methylation, resulting in loop dissolution (d). In leaves, the methylation of a region within the fourth intron, together with reduced methylation of region 3, (e) triggers the formation of an intragenic chromatin loop that blocks RNAPII transcription (f). The red pins show methylated regions.
Usually RdDM is initiated from RNAPIV/V TE transcripts in a RDR-dependent process (30). However, it is possible that RNAPII could transcribe TEs inserted near genes in divergent orientation (19, 31). IRs transcribed by RNAPII can produce 24-nt sRNAs and trigger spreading of DNA methylation over the target locus and silence it (32). Accordingly, we showed that the transcription of ncW6 is controlled by the HaWRKY6 bidirectional promoter in a RNAPII-dependent manner to produce sRNA in DCL2,3,4-dependent and RDR-independent pathways. Taken together, these features define ncW6 as an autonomous regulatory element. A prediction of IRs within different distance windows from every annotated sunflower TSS revealed that these elements are rather common in the vicinity of genes (SI Appendix, Fig. S5). Furthermore, we found that siRNAs mapped to over half of these predicted IRs and that sRNA abundance changes upon drought stress (SI Appendix, Fig. S5), suggesting a regulatory role for these IRs similar to what we have described for the ncW6 locus.
Our results define a dynamic and complex mechanism of transcriptional regulation for the HaWRKY6 locus. In view of the abundance of intergenic TEs in large genomes, we envision that these elements have a more pervasive regulatory role than previously thought. Although most of the TEs in plants are inactive due to both epigenetic regulation and their propensity to decay into defective forms, current knowledge positions these elements as regulatory elements fine-tuning gene expression and having substantial effects on the surrounding genomic neighborhood.
Materials and Methods
The materials and methods used in this study are described in detail in SI Appendix, SI Materials and Methods, including plant materials, RNA and sRNA analyses, DNA methylation profiling, chromatin loop detection and quantification, immunoblot analysis, and computational analysis.
Supplementary Material
Acknowledgments
We would like to thank Dr. Detlef Weigel for the valuable support for this work. This work was supported by grants from ANPCyT, HFSP, the Max Planck Society, ICGEB. A.L.A., F.D.A. and P.A.M. are members of CONICET; D.G., D.A.C., and A.H.T. are fellows of the same institution.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequencing data are available at the European Nucleotide Archive (ENA) PRJEB28614.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903131116/-/DCSupplemental.
References
- 1.Borges F., Martienssen R. A., The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matzke M. A., Kanno T., Matzke A. J., RNA-directed DNA methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66, 243–267 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Kadauke S., Blobel G. A., Chromatin loops in gene regulation. Biochim. Biophys. Acta 1789, 17–25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez-Granados N. Y., et al. , Put your 3D glasses on: Plant chromatin is on show. J. Exp. Bot. 67, 3205–3221 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Crevillén P., Sonmez C., Wu Z., Dean C., A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 32, 140–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan-Wong S. M., Wijayatilake H. D., Proudfoot N. J., Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 23, 2610–2624 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D. H., Sung S., Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell. 40, 302–312.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan-Wong S. M., et al. , Gene loops enhance transcriptional directionality. Science 338, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed I., Sarazin A., Bowler C., Colot V., Quesneville H., Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res. 39, 6919–6931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staton S. E., et al. , The sunflower (Helianthus annuus L.) genome reflects a recent history of biased accumulation of transposable elements. Plant J. 72, 142–153 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Wessler S. R., Bureau T. E., White S. E., LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5, 814–821 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Giacomelli J. I., Weigel D., Chan R. L., Manavella P. A., Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 195, 766–773 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Badouin H., et al. , The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546, 148–152 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Kontra L., et al. , Distinct effects of p19 RNA silencing suppressor on small RNA mediated pathways in plants. PLoS Pathog. 12, e1005935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papp I., et al. , Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132, 1382–1390 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton A., Voinnet O., Chappell L., Baulcombe D., Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu C., et al. , Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol. Biol. Evol. 29, 1005–1017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen J., et al. , Translational repression by a miniature inverted-repeat transposable element in the 3′ untranslated region. Nat. Commun. 8, 14651 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q., et al. , Searching for bidirectional promoters in Arabidopsis thaliana. BMC Bioinformatics 10 (suppl. 1), S29 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X., Sharp P. A., Divergent transcription: A driving force for new gene origination? Cell 155, 990–996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacadie S. A., Ibrahim M. M., Gokhale S. A., Ohler U., Divergent transcription and epigenetic directionality of human promoters. FEBS J. 283, 4214–4222 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Law J. A., Jacobsen S. E., Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volpe T., Martienssen R. A., RNA interference and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 3, a003731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michaud E. J., et al. , Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 8, 1463–1472 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Liu P. P., et al. , Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52, 133–146 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Soppe W. J., et al. , The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell 6, 791–802 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Harris C. J., et al. , A DNA methylation reader complex that enhances gene transcription. Science 362, 1182–1186 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ariel F., et al. , Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 55, 383–396 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Feng S., et al. , Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell 55, 694–707 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigman M. J., Slotkin R. K., The first rule of plant transposable element silencing: Location, location, location. Plant Cell 28, 304–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trinklein N. D., et al. , An abundance of bidirectional promoters in the human genome. Genome Res. 14, 62–66 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panda K., et al. , Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 17, 170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.