Abstract
Background
Cancer is a genetic and epigenetic disease that involves inactivation of tumor suppressor genes and activation of proto-oncogenes. All-trans retinoic acid (ATRA) is an isomer of retinoic acid involved in the onset of differentiation and apoptosis of a number of normal and cancer cells, functioning as an anti-cancer agent in several neoplasms. Ectopic changes in the expression of certain microRNAs (miRNAs) occur in response to ATRA, leading to phenotypic alterations in neoplastic cell lines. Moreover, the modulation of miRNA patterns upon ATRA-treatment may represent an effective chemopreventive and anti-cancer therapy strategy. The present systematic review was performed to provide an overview of the modulation of ATRA-induced miRNA expression in different types of neoplastic cells and identify the efficacy of intervention factors (i.e., concentration and duration of treatment) and how they influence expression profiles of oncogenesis-targeting miRNAs.
Methods
A systematic search was conducted according to the PRISMA statement via the US National Library of Medicine MEDLINE/PubMed bibliographic search engine.
Results
The search identified 31 experimental studies involving human cell lines from nine different cancer types (neuroblastoma, acute myeloid leukemia, breast cancer, lung cancer, pancreatic cancer, glioma, glioblastoma, embryonal carcinoma, and colorectal cancer) treated with ATRA at concentrations ranging from 10− 3 μmol/L to 102 μmol mol/L for 24 h to 21 days.
Conclusion
The concentrations used and the duration of treatment of cancer cells with ATRA varied widely. The presence of ATRA in the culture medium of cancer cells was able to modulate the expression of more than 300 miRNAs, and inhibit invasive behavior and deregulated growth of cancer cells, resulting in total tumor remission in some cases. ATRA may thus be broadly effective for neoplasm treatment and prevention, although these studies may not accurately represent in vivo conditions. Additional studies are required to elucidate ATRA-induced miRNA modulation during neoplasm treatment.
Keywords: Cancer, All-trans retinoic acid, miRNA, Expression modulation
Background
Cancer is the third leading cause of death worldwide, representing a considerable public health burden [1]. It is anticipated that the impact of this disease will correspond to approximately 26 million new cancer cases per year by 2030 [1]. The initiation and progression of cancer is mainly driven by genetic and epigenetic alterations in DNA and histones that result in the inactivation of tumor suppressor genes or activation of proto-oncogenes [2].
Retinoic acid (RA) is the major bioactive metabolite of retinol or vitamin A and serves as a potent regulator of cell growth, differentiation, and matrix formation of various cell types during embryogenesis [3, 4]. All-trans retinoic acid (ATRA) is an isomer of RA that exhibits dose-dependent effects on differentiation and apoptosis of a number of normal and cancer cells [5]. ATRA has also been shown to function as an anti-cancer agent in several neoplasms, such as gastric cancer [6], breast cancer [7, 8] leukemia [9–12], nephroblastoma [13], melanoma [14], lung cancer [15], and neuroblastoma [16, 17].
miRNAs are endogenous, small, non-coding RNAs that regulate gene expression by binding to their target mRNAs, leading to degradation and/or translational repression [5]. These molecules have been extensively associated with cancer development, as they play important roles in regulating biological processes such as differentiation, cell proliferation, apoptosis, epithelial-mesenchymal transition, cancer metastasis, and angiogenesis [18]. miRNAs can function as oncogenes or tumor suppressors, and their abnormal expression has already been identified in both solid and hematopoietic tumors. Thus, miRNA profiling is a promising strategy for cancer diagnosis and prognosis [19].
Several studies have shown that ectopic expression of certain miRNAs, which are upregulated in response to ATRA treatment, is sufficient to produce phenotypic changes that are typically induced by ATRA [20, 21]. Furthermore, the potential role of miRNAs induced by ATRA in modulating cancer cells has been demonstrated in different cellular contexts [22–29]. Thus, the aim of our systematic review is to provide an overview of the modulation of ATRA-induced miRNA expression in different types of human cancer.
Methods
This review was written in accordance with the PRISMA statement [30].
Search strategy
A systematic search of the published literature form January 1, 2007 to November 12, 2018 was undertaken using the US National Library of Medicine MEDLINE/PubMed (www.ncbi.nlm.nih.gov/pubmed) bibliographic search engine. Multiple PubMed searches were conducted using the following keyword combinations “(miRNA OR microRNA) AND (“retinoic acid” OR “vitamin A” OR retinol) AND (cancer OR neoplasm OR tumor).” The studies were then compiled into a single database and duplicates removed. An initial screening was performed by assessing the title and abstract. After reading the selected studies, the references section of each text was analyzed for additional relevant studies.
Study selection
For this review, we selected studies written in the English language that evaluated changes in the expression profile of miRNAs in neoplastic human cell lines after treatment with ATRA. The exclusion criteria were other systematic reviews on the subject, bioinformatics analyses, short communications, and supporting information.
Data extraction
Data extraction was conducted by one reviewer (LL) and verified by the other authors. Extracted data included cancer type, cell type analyzed, conditions of ATRA treatment (dose and duration), miRNAs evaluated, and significant findings. For studies that determined ATRA-induced miRNA expression at multiple time points, the values obtained on the last day were considered. All studies are grouped according to the type of cancer.
Results
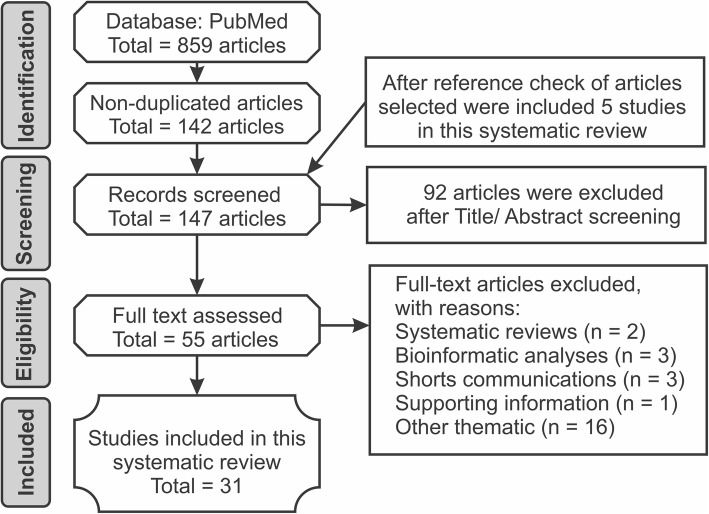
After conducting searches using 18 combinations of key terms, we identified 859 studies. A total of 147 studies were identified through database searching. After screening the abstracts, 92 studies were excluded, and 55 full-text studies were accessed and assessed for eligibility. Reasons for study exclusion included studies describing ATRA-mediated effects not related to miRNA expression or mentioning genes that exhibited modifications induced by ATRA without describing the miRNA involved, systematic reviews, bioinformatic analysis, short communications, and supporting information (Fig. 1).
Fig. 1.
Flow of information through the different phases of the systematic review regarding the modulation of ATRA-induced expression of miRNAs in different types of neoplastic cells
The characteristics and findings of the 31 studies included in this systematic review are summarized in Tables 1, 2, 3, 4 and 5 according to cancer type. In particular, nine different types of cancer were addressed, neuroblastoma (n = 12), acute myeloid leukemia (n = 9), breast carcinoma (n = 3), lung cancer (n = 2), pancreatic cancer (n = 1), glioma (n = 1), glioblastoma (n = 1), embryonal carcinoma (n = 1), and colorectal cancer (n = 1). All studies were based on human neoplastic cell lines treated with ATRA, followed by evaluation of changes in miRNA expression patterns. One study utilized human and animal cell lines [32]; however, only results related to human cells were included in this review. The results have been grouped according to cancer type and are presented below.
Table 1.
miRNAs regulated by ATRA in NB cell lines
| Study (reference and country) | Cell type | ATRA treatment | miRNAs upregulated | miRNAs downregulated |
|---|---|---|---|---|
|
Chen and Stallings (2007) [31] United States |
SK-N-BE | 5 μmol/L for 5 days | 17; miR-330, miR-187, miR-331, miR-200c, miR-216, miR-150, miR-141, miR-200a, miR-326, miR-186, miR-30b, miR-137, miR-184, miR-129, let-7b, let-7a and miR-181a; most significant: miR-184 | 4; miR-323, miR-302a, miR-181b and miR-92; most significant miR-192 |
|
Laneve et al. (2007) [20] Italy |
SK-N-BE | 10 μmol/L for 10 days | 14; miR-9, miR-124a, miR-125a, miR-125b, let-7a, let-7b, miR-7, miR-22, miR-23a, miR-24, miR-26a, miR-30a-5p, miR-100 and miR-103; most significant: miR-124a | – |
|
Evangelisti et al. (2009) [21] Italy |
SH-SY5Y | 10 μmol/L for 6 days | miR-128 | – |
|
Le et al. (2009) [32] United States |
SH-SY5Y | 10 μmol/L for 5 days | 6; miR-7, miR-124a, miR-125b, miR-199a, miR-199a*, and miR-214; most significant: miR-199a* | – |
|
Beverigde et al. (2009) [33] Australia |
SH-SY5Y | 10 μmol/L for 6 days | 12; miR-10a, miR-128a, miR-331, miR-124a, miR-409-5p, miR-210, miR-149, miR-9, miR-423, miR-483, miR-208 and miR-184; most significant: miR-10a. | 32; miR-301, miR-19a*, miR-520d*, miR-18a*, miR-106a*, miR-218, miR-20a, miR-137, miR-15b, miR-20b*, miR-25*, miR-130a, miR-29b, miR-19b*, miR-34a, let-7a, miR-195, miR-106b*, miR-199a, miR-92*, miR-138, miR-93*, miR-432, miR-519e, miR-101, miR-518f-526a, miR-21, miR-134, miR-98, miR-370, miR-525 and miR-91 (aka miR-17-5p); most significant: miR-301. |
|
Ragusa et al. (2010) [34] Italy |
SK-N-BE | 10 μmol/L for 10 days | 3; miR-152, miR-200b and miR-338; most significant: miR-200b | – |
|
Meseguer et al. (2010) [35] Spain |
SH-SY5Y | 1 μmol/L for 4 days | 26; miR-10a, miR-10b, miR-10b, miR-615-5p, miR-211, miR-212, miR-132, miR-22, miR-766, miR-135a, miR-148b*, miR-877, miR-184, miR-135b, miR-30e, miR-214, miR-92b, mir-99b, miR-628-3p, miR-27b, miR-335, miR-25, miR-30a, miR-610, miR-331, 3p, miR-149; most significant: miR-10a and miR-10b | 16; miR-490-3p, miR-154, miR-26a-2, miR-296-3p, miR-422a, miR-486-3p, miR-378, miR-411, miR-543, miR-801, miR-873, miR-107, miR-744, miR-139-3p, miR-487b and miR-106a; most significant: miR-490-3p |
|
Das et al. (2010) [36] Ireland |
SK-N-BE | 5 μmol/L for 7 days | 17; miR-132, miR-10a, miR-10b, miR-210, miR-192, miR-184, miR-146b, miR-152, miR-126, miR-615, miR-21, miR-203, miR-214, miR-24, miR-196a, miR-191 and let-7e; most significant: miR-132 | 17; miR-409-5p, miR-656, miR-299-5p, miR-485-5p, miR-424, miR-127, miR-323, miR-339, miR-576, miR-432, miR-433, miR-7, miR-30e-5p, miR-485-3p, miR-379, miR-134 and miR-487b; most significant: miR-432 |
|
Das et al. (2012) [37] Ireland |
SK-N-BE and SHSY-5Y | 5 μmol/L for 7 days | miR-340 | – |
|
Chen et al. (2010) [38] United Kingdom |
SH-SY5Y | 10 μmol/L for 48 h | 6; miR-132, miR-16, miR-27a, miR-27b, miR-214, and miR-197; most significant: miR-132. | 6; miR-133a, miR-508-3p, miR-7, miR-1, miR-205, and miR-20; most significant: miR-20b |
|
Foley et al. (2011) [39] Ireland |
SK-N-BE, SHSY-5Y and LAN-5 | 5 μmol/L for 7 days | 30; miR-132, miR-10a, miR-10b, miR-210, miR-192, miR-184, miR-146b, miR-152, miR-126, miR-615, miR-21, miR-203, miR-214, miR-24, miR-196a, miR-191, miR-7e, miR-28, miR-95, miR-26b, miR-26a, miR-199a, miR-374, miR-340, miR-125b, miR-125a, miR-7 g, miR-425-5p, miR-190, miR- 361; most significant: miR-132, miR-10a, and miR-10b | 23; miR-409-5p, miR-656, miR-299-5p, miR-485-5p, miR-424, miR-127, miR-323, miR-339, miR-576, miR-432, miR-433, miR-7, miR-30e-5p, miR-485-3p, miR-379, miR-134, miR-487b, miR-382, miR-486, miR-565, miR-149, miR-550 and miR-503; most significant: miR-409-5p |
|
Das and Bhattacharyya (2014) [22] (India) |
SH-SY5Y | 10 μmol/L for 7 days | 31; miR-9, miR-9*, miR-15b, miR-16, miR-34a, miR-100, miR-124, miR-125b, miR-126, miR-132, miR-134, miR-137, let-7a, miR-432, miR-15a, miR-21, miR-23a, miR-27b, miR-29c, miR-103, miR-125a, miR-128a, miR-140, miR-141, miR-185, miR-326, miR-1320, let-b, let-7e, miR-127 and miR-10a; most significant: miR-432 | 33; miR-17, miR-19a, miR-138, miR-145, miR-146a, miR-148a, miR-150, miR-154, miR-190 miR-194, miR-200a, miR-205, miR-221, miR-299, miR-323, miR-335, miR-30e, miR-133a, miR-133b, miR-142-5p, miR-144, miR-152, miR-185, miR-189, miR-191, miR-193, miR-200c, miR-213, miR-219, miR-296, miR-302a, miR-331 and miR-342; most significant: miR-17 |
*Cluster 17
Table 2.
miRNAs regulated by ATRA in AML cell lines
| Study (reference and country) | Cell type | ATRA treatment | miRNAs upregulated | miRNAs downregulated |
|---|---|---|---|---|
|
Garzon et al. (2007) [23] United States |
NB4 | 10−1 μmol /L for 4 days | 9; miR-15a, miR-15b, miR-16-1, let-7a-3, let-7c, let-7d, miR-223, miR-342, and miR-107; most significant: miR-15b | miR-181b |
|
Marchis et al. (2009) [40] Italy |
NB4 | 1 μmol/L for 72 h | miR-342 | – |
| Gao et al. (2011) [41] China | U937, HL60, NB4 and leukemic fresh cell lines | 1 μmol/L for 72 h | 2; miR-15a and miR-16-1; most significant: miR-15a | – |
| Lin et al. (2011) [42] China | NB4, HL60 and K562 | 1 μmol/L for 72 h | – | miR-125b |
| Morris et al. (2013) [43] United States | HL60, NB4, PL21, and THP-1 | 10−3 μmol/L to 1 μmol/L for 21 days | – | miR-150 |
| Zhuang et al. (2014) [44] China | NB4 | 1 μmol/L for 36 h | – | miR-181a |
|
Lin et al. (2015) [45] China |
HL60 and NB4 | 3 μmol/L ATRA for 72 h (HL60) and 2 μmol/L ATRA for 4 days (NB4) | miR-638 | – |
| Bräuer-Hartmann et al. (2015) [46] Germany | NB4 | 10−1 μmol/L for 24 h | – | miR-181 cluster (miR-181a, miR-181b, miR-181c and miR-181d); most significant: miR-181a and miR-181b |
| Yan et al. (2016) [47] United States | HL60 | 10− 1 and 1 μmol/L for 72 h | – | miR-17–92 cluster (miR-17, miR-18a, miR-19b, miR-20a and miR-92); most significant: miR-20a |
Table 3.
miRNAs regulated by ATRA in breast carcinoma cell lines
| Study (reference and country) | Cell type | ATRA treatment | miRNAs upregulated | miRNAs downregulated |
|---|---|---|---|---|
| Terao et al. (2011) [48] | MCF-7 | 1 μmol/L for 72 h | miR-21 | – |
|
Khan et al. (2015) [49] Ireland |
T47D and SK-BR-3 | 1 and 5 μmol/L for 24 h | miR-10a | – |
|
Fisher et al. (2015) [50] Italy |
SKBR3, MCF-7, MDA-MB-157 | 10−1 μmol/L (SKBR3) and 1 μmol/L ATRA (MCF-7 and MDA-MB-157) for 36 h | 13; mir-1031; miR-1225-5p1, miR-12681, miR-141, miR-188-5p1, miR-19b1, miR-200c1, miR-211, miR-221, miR-513a-5p1, miR-5751, miR-6421 and miR-6641a | 17; miR-15b12, miR-9612, miR-20312, miR-18312, miR-210-3p123, miR-362-5p12, miR-14912, miR-125a-5p123, miR-193a-5p12, miR-18512, miR-532-5p12, miR-342-3p12, miR-148a1, miR-3751, miR-193a-3p1, miR-2051 and miR-6601a |
1SKBR3 cell lines; 2MCF-7 cell lines; 3MDA-MB-157 cell lines; aAuthors did not mention the miRNAs most sensitive to ATRA treatment
Table 4.
miRNAs regulated by ATRA in lung cancer cell lines
| Study (reference and country) | Cell type | ATRA treatment | miRNAs upregulated | miRNAs downregulated |
|---|---|---|---|---|
|
Zhu et al. (2015) [3] China |
A549 and H1299 | 10 and 100 μmol/L for 24 h (A549); and 100 μmol/L for 72 h (H1299) | 8; miR-594, miR-519b, miR-504, miR-512-3p, miR-363, miR-517a, miR-518a and miR95; most significant: 512-3p | 5; mir-223, miR-196a, miR-369-3p, miR-146a and miR-142-3pa |
|
Chu et al. (2016) [4] China |
H1299 and A549 | 10 and 100 μmol/L for 72 h | miR-512-5p | – |
aAuthors did not mention the miRNA most sensitive to ATRA treatment
Table 5.
miRNAs regulated by ATRA in pancreatic cancer, glioma, glioblastoma, embryonal carcinoma, and colorectal cancer
| Study (reference and country) | Cell type | ATRA treatment | miRNAs upregulated | miRNAs downregulated |
|---|---|---|---|---|
|
Weiss et al. (2009) [51] Germany |
PaTu8988-S and PaTu8988- | 1 μmol/L for 72 h | miR-10a | – |
|
Xia et al. (2009) [52] China |
U343 and U251 | 1 μmol/L for 48 h | – | miR-125b |
| Chen et al. [53] (2014) Taiwan | U87 MG | 10, 20, 40, and 60 μmol/L for 72 h | 13; miR-302b, miR-302a, miR-302d, miR-30a, miR-146a, miR-224, miR-135a, miR-137, miR-212, miR-628-3p, miR-200c, miR-146b-5p and miR-136; most significant: miR-302b | 15; miR-19a, miR-19b, miR-551b, miR-101, miR-301a, miR-199b-5p, miR-455-5p, miR-565, miR-107, miR-216a, miR-32, miR-33a, miR-17-5p, miR-497 and miR-210; most significant: miR-19a |
|
Chen et al. [54] (2014) China |
NT2/D1 | 10 μmol/L for 21 days | – | miR-134 |
|
Liu et al. [55] (2018) China |
HCT116 | 10, 20, 40, and 60 μmol/L for 24 h | miR-3666 | – |
Neuroblastoma (NB)
NB originates from the aberrant development of primordial neural crest cells and is the most common extracranial solid tumor during childhood and the most common tumor in infants [35]. Several lines of evidence support a role for miRNAs in NB pathogenesis as well as the utility of miRNA profiling in NB diagnostics, classification, and prognosis [35]. Furthermore, the involvement of miRNAs in ATRA-induced differentiation of NB cells was recently reported [35]. In particular, cell lines derived from NB such as SK-N-BE, SH-SY5Y, and LAN-5 can be induced by ATRA treatment to undergo neural cell differentiation and are thus often used as model systems for studying biochemical pathways involved in differentiation [39].
To test the effects of ATRA on NB differentiation and the development of embryonic cells, Chen and Stallings [31] evaluated the expression of 34 human miRNAs in SK-N-BE cells treated with 5 μmol/L ATRA by replacing the culture medium every 24 h for 5 days. The expression profiles of 21 miRNAs were found altered when compared with that of untreated cells (Table 1). Of these miRNAs, 17 were upregulated in ATRA-treated cells and 4 were downregulated. In particular, miR-184 showed the most significant change, as its expression increased 9-fold following ATRA treatment. The authors also observed that the effects of ATRA treatment on miRNA expression were sustained for at least a short period of time after release; for example, after SK-N-BE cells were treated with ATRA for 5 days and released for 3 days, miR-184 levels were 25-fold higher than that of untreated cells, indicating sustained effects of ATRA on miRNA expression. In addition, their results suggested that treatment with ATRA is associated with apoptosis rather than differentiation induction in this cell line.
Laneve et al. [20] analyzed the expression pattern of 70 miRNAs in SK-N-BE cells treated with 10 μmol/L ATRA for 3, 6 and 10 days. They found that 14 miRNAs were upregulated (Table 1), 33 did not exhibit any changes in expression, and 23 could not be detected. Expression levels of the upregulated miRNAs were mostly induced after 3 days upon ATRA treatment and progressively increased after terminal differentiation (10 days). Moreover, the authors observed that the expression levels of miR-9, miR-125b, and miR-125a increased 1.7, 2.2, and 2.6-fold, respectively, compared with that of control cells, and that this increase led to a marked decrease in NB cell proliferation in vitro.
Evangelisti et al. [21] used the same ATRA concentration mentioned above and measured miR-128 expression in SH-SY5Y cell lines after ATRA treatment. The cells were fed every 48 h with ATRA and then treatment was stopped after 6 days. miR-128 expression was found upregulated by approximately 3-fold in treated cells compared with untreated ones.
Similarly, Le et al. [32] treated SH-SY5Y cells with 10 μmol/L ATRA but over the course of 5 days. They analyzed the expression profiles of 175 human miRNAs and found that 12 miRNAs were significantly upregulated (miR-106, let-7b, miR-199a*, miR-124a, miR-143, miR-125b, miR-7, miR-189, miR-199a, miR-27a, miR-21, and miR-214) during treatment; however, after validation with Northern blot analysis, only six were found upregulated during differentiation (Table 1).
Beverigde et al. [33] treated SH-SY5Y cells with 10 μmol/L ATRA for an additional 1 day longer than Le et al. [32]. They performed microarrays and RT-qPCR to examine miRNA expression profiles. Microarray assays identified 44 miRNAs with altered expression after treatment, of which 12 were significantly upregulated and 32 downregulated (Table 1); however, only 10 miRNAs were confirmed altered by RT-qPCR, of which 3 were upregulated (miR-128a, miR-10a, and miR-124a) and 7 downregulated (miR-301, miR-20a, miR-106a, miR-19a, miR-29b, miR-134, and miR-15b). Interestingly, miR-9 exhibited a different expression pattern depending on the method of analysis. It was found upregulated via microarray analysis and downregulated via RT-qPCR. ATRA also induced the downregulation of the entire miR-17 cluster (microarray results). Moreover, five miRNAs that exhibited increased expression (miR-9, miR-124a, miR-128a, miR-208, miR-210, and miR-423) were previously demonstrated to be brain-specific or brain-enriched miRNAs and are considered to play important roles in brain development, neuronal maturation, and neuronal differentiation.
Ragusa et al. [34] analyzed the expression profiles of three miRNAs (miR-152, miR-200b, and miR-338) after ATRA treatment. SK-N-BE cells were treated with ATRA 10 μmol/L and observed on day 5 and 10 after treatment. After 10 days of treatment, all three miRNAs analyzed were upregulated, with the most significant increase observed for miR-200b.
In keeping with the above findings, Meseguer et al. [35] showed that ATRA treatment of SH-SY5Y cells resulted in profound changes in miRNA expression patterns. The authors treated SH-SY5Y cells with 1 μmol/L ATRA and assessed changes in miRNA expression at 0, 24, 48, and 96 h. The expression levels of 42 miRNAs were significantly changed [26 upregulated and 16 downregulated (Table 1)]; in particular, miR-10a and 10b showed the most prominent changes in expression. Furthermore, these changes induced by ATRA contributed to the regulation of SH-SY5Y NB cell differentiation and the associated changes in migratory and invasive activities.
Das et al. [36] treated SK-N-BE NB cells with 5 μmol/L ATRA by replacing the culture medium every 24 h for 7 days to determine changes in methylation patterns and gene expression. In addition, they investigated whether upregulated miRNAs are causally associated with the downregulation of a gene known to cause genome-wide demethylation events. They performed expression analysis of 368 miRNAs using low-density TaqMan arrays and found 17 upregulated miRNAs (≥ 2-fold increase) and 17 downregulated miRNAs (≥ 2-fold decrease; Table 1). Among the upregulated miRNAs possibly involved in controlling DNA methylation, ectopic overexpression of miR-152 significantly decreased cell invasiveness and anchorage independent growth, contributing in part to ATRA-induced differentiation. miR-152 expression was also analyzed in three other NB cell lines (SH-SY5Y, LAN-5 and SK-N-AS) that received the same treatment. As observed in SK-N-BE cells, ATRA treatment induced miR-152 upregulation in SH-SY5Y and LAN-5 cell lines; however, this miRNA was downregulated in SK-N-AS cells.
In a subsequent study with the same treatment regimen, Das et al. [37] observed a correlation between changes in miRNA expression profiles and methylation following ATRA treatment. In SK-N-BE cells, 20 miRNAs were found upregulated and 24 downregulated following ATRA treatment, whereas 13 miRNAs were upregulated in SH-SY5Y cells (the authors did not specify which miRNAs are altered). Only miR-340 was upregulated in both cell lines and showed the highest expression levels. In addition, overexpression of miR-340 was associated with decreased cell viability and limited colony-forming ability.
Chen et al. [38] observed significant changes in the expression profiles of 12 miRNAs, of which half were upregulated and half downregulated, when SH-SY5Y cells were treated with 10 μmol/L ATRA for 48 h (Table 1). Foley et al. [39] treated NB cell lines (SK-N-BE, SH-SY5Y, and LAN-5) with 5 μmol/L ATRA by exchanging the culture medium every 24 h for 7 days, and then analyzed the expression profiles of 364 miRNAs. In SK-N-BE cells, several miRNAs exhibited altered expression levels, of which 53 miRNAs showed significant changes—30 were positively regulated and 23 negatively regulated (Table 1). miRNAs with the highest changes in expression (410-fold increase by day 7) included miR-132, miR-10a, and miR-10b. Similarly, these miRNAs were significantly upregulated in response to ATRA in SK-N-BE, SHSY-5Y, and LAN-5 NB cells.
Das and Bhattacharyya [22] treated SH-SY5Y cells for 7 days with 10 μmol/L ATRA and investigated the expression levels of 96 miRNAs on day 3 and 7. They found 31 upregulated miRNAs, of which 14 exhibited high expression levels (miR-9, miR-9 *, miR-15b, miR-16, miR-34a, miR-100, miR-124, miR-125b, miR-126, miR-132, miR-134, miR-137, let-7a, and miR-432), and 33 downregulated miRNAs (Table 1). Interestingly, miR-185 presented with two different patterns of expression, downregulation on day 3 and upregulation on day 7.
These results support the notion that miRNA regulation plays a key role in the differentiation of NB cells induced by ATRA and in the phenotypic changes linked to the expression of genes associated with these miRNAs. In addition, multiple lines of evidence indicate that NB treated with ATRA exhibits lower migration and invasion abilities. Changes in miRNA expression profiles in NB cell lines after ATRA treatment are summarized in Table 1.
Acute myeloid leukemia (AML)
AML is characterized by abnormal differentiation and uncontrolled proliferation of immature hematopoietic cells [56]. In hematopoietic malignancies, several miRNAs have been reported to exhibit tumor-suppressive or oncogenic roles in leukemogenesis [44]. Accordingly, it has been suggested that miRNAs are important in the molecular pathogenesis of leukemia by interfering with essential pathways of hematopoietic differentiation [40].
Acute promyelocytic leukemia (APL), the main subtype of acute myeloid leukemia in which positive responses are observed after treatment with ATRA [56], is characterized by chromosomal translocations involving the RA receptor-α (RARA) gene that result in clonal expansion of hematopoietic precursors blocked at the promyelocytic stage of differentiation [40]. Several miRNAs upregulated upon ATRA treatment of APL cell lines have already been identified in different studies [40].
Garzon et al. [23] cultured APL NB4 cells with or without 10− 1 μmol ATRA over 4 days and identified nine upregulated and one downregulated miRNA after comparing expression levels between both groups (Table 2). They also observed that treatment of NB4 cells with ATRA induced granulocytic differentiation, as evidenced by morphological changes and increased expression of the surface antigens CD11b and CD15.
Marchis et al. [40] treated NB4 APL cells with 1 μmol/L ATRA and confirmed that ATRA induced a marked and selective increase in miR-342 expression, starting 24 h after treatment and progressively increasing over 3 days. They also found increased expression of the miR-342 host gene Enah/Vasp-like (EVL) during ATRA treatment. These results indicated that EVL and miR-342 expression is co-regulated, with miR-342 contributing to the ATRA-mediated granulocytic differentiation program of promyelocytic precursors in ATRA-treated NB4 cells.
To uncover the regulation of miR-15a and miR-16-1 induced by ATRA, Gao et al. [41] employed two models, NB4, HL60, and U937 cell lines and fresh leukemic cells extracted from 10 AML patients; both experimental underwent 1 μmol/L ATRA treatment for 72 h. The authors found that treatment with ATRA increased miR-15a and miR-16-1 expression in NB4 cells, which in turn culminated in the inhibition of leukemic cell differentiation. They also observed that miR-15a and miR-16-1 were upregulated in 8 of 10 patient samples. These findings indicated that upregulation of miR-15a and miR-16-1 may be associated with the differentiation induced by ATRA. In addition, they evaluated the time (0 to 72 h) and concentration-dependent (10− 2 to 10 μmol/L ATRA) expression of miR-15a and miR-16-1 in response to ATRA treatment, which induced differentiation in NB4 cells. They found that an increase in time and ATRA concentrations between 10− 1 to 10 μmol/L ATRA were associated with elevated miR-15a and miR-16-1 expression; however, 10− 2 μmol/L ATRA did not induce differentiation and failed to modulate the expression of miR-15a and miR-16-1.
In another study, Lin et al. [42] treated NB4, HL60, and K562 cells with 1 μmol/L ATRA over 72 h to monitor the in vitro expression levels of CDX2 and miR-125b. In NB4 cells, miR-125b expression progressively decreased to 30% of baseline after 72 h following differentiation induction with ATRA. The researchers also demonstrated decreased CDX2 mRNA expression levels that were positively correlated with miR-125b levels after ATRA treatment, suggesting a potential function of CDX2 and miR-125b in inhibiting cell differentiation in AML cells and promoting leukemogenesis.
Morris et al. [43] further examined AML cell lines (NB4, HL60, PL21, and THP-1) that can be induced to differentiate along the granulocytic or monocytic lineage after exposure to ATRA (10− 3 to 1 μmol/L) for 2 (THP-1), 3 (NB4) or 4 (HL60 and PL21) days. The authors found that these AML cells lines express very low levels of endogenous miR-150 in contrast to normal CD34+ progenitors cell lines after exposure to ATRA. However, in AML cell lines, differentiation of miR-150-expressing cells occurs independently of RARA signaling.
Zhuang et al. [44] revealed the downregulation of miR-181a in NB4 cells during treatment with 1 μmol/L ATRA over 36 h. They also observed that ATRA induced NB4 cell differentiation. Their study strongly supported the therapeutic role of ATRA on APL. Furthermore, Lin et al. [45] observed significant upregulation of miR-638 during ATRA-induced myeloid differentiation of HL60 and NB4 cell lines compared with that of untreated control. HL60 cells were treated with 3 μmol/L ATRA for 72 h while NB4 cells were treated with 2 μmol/L ATRA for 4 days. In HL60 cells, upregulation of miR-638 coincided with increased expression of the myeloid-specific surface markers CD14 and CD11b. Moreover, CDK2 overexpression eliminated the inhibitory effect of miR-638 in HL60 cells, which was more significant upon ATRA treatment.
To evaluate changes in miRNA expression mediated by ATRA, Bräuer-Hartmann et al. [46] cultured NB4 cell lines in the presence of 10− 1 μmol/L ATRA and analyzed the expression profiles of miR-181 family members (miR-181a–d) 24 h after treatment. In this study, a significant downregulation of all miR-181 family members was observed. In parallel, they also investigated granulocyte differentiation associated with changes in the expression patterns of the miR-181 family in U937, HL60, and NB4 cells treated with 1 μmol/L ATRA. They found significant downregulation of miR-181a/b in APL cell lines (NB4); however, no significant changes were found in the non-APL cell lines (U937 and HL60). Moreover, inhibition of the miR-181a/b cluster by ATRA treatment effectively repressed cell proliferation and induced apoptosis in APL cells (NB4).
After treatment of AML HL60 cells with 0.1 or 1 μmol/L ATRA for 72 h, Yan et al. [47] found a significant reduction in the expression levels of the miR-17–92 cluster upon ATRA treatment, which decreased by almost 50% in 3 days. In addition, they found that HL60 cell proliferation was reduced after exposure to ATRA, which activated their terminal differentiation into granulocytes.
In summary, these studies demonstrated that several miRNAs are highly expressed in specific hematopoietic cell lineages and that modulation of their expression induced by ATRA treatment may be correlated with changes in cellular properties or differentiation and may thus represent an effective treatment strategy. miRNAs induced in AML cell lines after ATRA treatment are summarized in Table 2.
Breast carcinoma
Breast cancer is the most common malignancy in women [57] and represents a heterogeneous group of tumors with varying responses to therapeutic agents, including retinoids [48]. Therefore, only a few reports have assessed the effects of retinoids on miRNAs in this carcinoma.
For example, Terao et al. [48] determined the differential profiles of miRNA expression in MCF-7 and MDA-MB-231 cells cultured with or without 1 μmol/L ATRA over 72 h. Although ATRA did not affect the miRNA profiles of MDA-MB-231 cells—as they are estrogen receptor-negative (ERα-)—the retinoid significantly increased expression levels of a single miRNA, miR-21, in MCF-7 cells (estrogen receptor-positive; ERα+) after 2 h, which leveled off at 24 h.
To investigate the potential modulation of miR-10a expression in breast cancer cells, Khan et al. [49] analyzed SKBR3 cells exposed to higher ATRA concentrations (1 and 5 μmol/L)—albeit for shorter duration (24 h). For both concentrations, an increase in miR-10a expression was observed, with higher expression levels induced upon exposure to 5 μmol/L ATRA. The authors also analyzed T47D cells exposed to 1 or 5 μmol/L ATRA for 24 h and found that miR-10a expression was similarly upregulated upon stimulation under both ATRA concentrations.
Fisher et al. [50] treated SKBR3 cells with 10− 1 μmol/L ATRA for up to 36 h and then observed the expression profiles of 57 miRNAs, of which 37 were upregulated and 20 downregulated. The authors chose to only analyze 30 of these miRNAs in detail (Table 3), because they are strongly associated with positive or negative regulation of more than 500 mRNAs involved in different stages of tumor progression (e.g., apoptosis, motility, and cell invasion). This indicates a possible role for ATRA in the control of the metastatic invasiveness of breast cancer. In addition, the authors treated MCF-7, MDA-MB-453, MDA-MB-231, and MDA-MB-157 cells with 1 μmol/L ATRA for up to 36 h. In MCF-7 cells, ATRA negatively regulated 12 miRNAs, which were also downregulated in SKBR3 cells. They also observed a significant ATRA-dependent downregulation of miR-125a-5p and miR-210-3p in MDA-MB-157 cells. On the other hand, none of the miRNAs were found modulated in response to ATRA in MDA-MB-231 and MDA-MB-453 cells. miRNA expression profiles in breast carcinoma cell lines after ATRA treatment are summarized in Table 3.
Lung cancer
Lung cancer is responsible for the largest number of deaths among all cancer types in both men and women [3]. Recently, it has been demonstrated that miRNAs are directly involved in lung cancer development and progression [3].
Zhu et al. [3] treated non-small cell lung cancer A549 cells with 10 or 100 μmol/L ATRA for 12 or 24 h and H1299 cells with 100 μmol/L ATRA for 48 or 72 h. The authors observed changes in the expression profiles of 13 miRNAs in A549 cell lines (Table 4); however, they focused on miR-512-3p because it exhibited a significant response to ATRA treatment and pro-apoptotic tumor suppressor abilities. In A549 cells, miR-512-3p expression increased by approximately 100% at 12 h and reached a peak at 24 h upon 100 μmol/L ATRA treatment, whereas expression in H1299 cells increased by only 7-fold after 72-h treatment with 100 μmol/L ATRA and showed no change at 48 h. In addition, they demonstrated that miR-512-3p overexpression inhibited adhesion, migration, and invasion of A549 and H1299 lung cancer cells.
To explore the effects of RA on the expression profile of miR-512-p, Chu et al. [4] also treated H1299 and A549 cells with 10 or 100 μmol/L ATRA for 24, 48, and 72 h. miRNA expression levels in A549 cells increased 3-fold when exposed to 10 μmol/L ATRA across all time points compared with that of control. When ATRA concentration increased to 100 μmol/L, miR-512-5p expression increased by 5, 17, and 3-fold at 24, 48, and 72 h, respectively. In H1299 cells, miR-512-5p expression increased by approximately 4-fold and 10-fold when treated with 10 μmol/L and 100 μmol/L ATRA, respectively, at all three time points compared with that of control. They also found that apoptosis was stimulated in both lung cancer cell lines. Overexpression of miR-512-5p in A549 and H1299 cells led to a 40 and 46% increase, respectively, in apoptotic cells compared with that of control cells. In addition, ATRA treatment and the consequent increase in miR-512-p expression led to a sharp decrease in glucose uptake by tumor cells and attenuated cell migration capacity by 39% compared to that of control. miRNA expression profiles of lung cancer cell lines after ATRA treatment are summarized in Table 4.
Other neoplasms
Weiss et al. [51] investigated the importance of miR-10a in pancreatic cancer metastasis and the possible interactions between miR-10a expression and RA. The authors employed the pancreatic tumor cell lines PaTu8988-S and PaTu8988-T and administered 1 μmol/L ATRA or a selective RARα antagonist for 72 h. They found that miR-10a expression in nonmetastatic pancreatic cells was positively regulated by ATRA stimulation, whereas miR-10a expression significantly decreased in cells treated with a selective RARα antagonist.
In another study, Xia et al. [52] treated U343 and U251 glioma cell lines with 1 μmol/L ATRA for 48 h and found that ATRA treatment negatively regulates miR-125b expression in both cell lines, although U343 cells were more sensitive to ATRA than were U251 cells. Furthermore, ATRA-mediated miR-125b overexpression led to reduced cell proliferation and increased apoptosis in both cell lines.
Human glioblastoma cells (U87 MG) were administered different doses of ATRA (10, 20, 40, and 60 μmol/L) and analyzed after 24, 48 and 72 h in the study by Chen et al. [53]. ATRA was found to positively regulate different members of the miR-302 cluster (Table 5), especially miR-302b, in a dose-dependent manner. Treatment with 40 μmol/L ATRA more than doubled miR-302b expression 12 h after exposure, and this overexpression was positively associated with cell death, suggesting that an increase in miR-302b expression is associated with suppression of tumorigenesis. Moreover, cell death associated with ATRA treatment may be regulated via miR-320b expression.
In another study by Chen et al. [54], pluripotent human embryonal carcinoma cells (NT2/D1) were treated with 10 μmol/L ATRA for 21 days. A decrease in the expression levels of the transcription factor FOXM1 was observed, which was mediated by the ATRA-induced overexpression of miR-134. Moreover, miR-134 overexpression resulted in decreased NT2/D1 cell pluripotency. ATRA was found to function by binding to the ligand-inducible transcription factors that activate or repress the transcription of downstream target genes, controlling cell growth and differentiation in both embryonic and adult cells.
Liu et al. [55] further investigated the association between miR-3666 expression and the effects of ATRA treatment on human colorectal cancer (CRC) cells. After HCT116 cells were treated with increasing doses of ATRA (10, 20, 40, and 60 μmol/L) for 24 h, ATRA concentrations between 20 and 60 μmol/L were found to increase miR-3666 expression in a dose-dependent manner. Moreover, ATRA-induced positive modulation of miR-3666 in HCT116 cells was associated with regulation of cancer cell viability, apoptosis, migration, and invasiveness. The authors also reported that 40 μmol/L ATRA treatment can decrease CRC cell viability by 50%, which was mediated by miR-3666 expression. Thus, ATRA may be a potential preventative agent against CRC development by regulating miR-3666 expression.
Discussion
The studies included in this review demonstrated that more than 300 miRNAs are induced upon ATRA treatment in neoplastic cell lines. The ATRA concentrations used in these studies varied greatly (10− 3 μmol/L to 102 μmol/L). In addition, the duration of ATRA treatment ranged from 1 to 21 days, with the most common duration being 3 days. Overall, the findings indicated that the optimum ATRA treatment parameters to determine changes in miRNA expression in neoplastic cell lines are 10 μmol/L and 3 days.
The findings demonstrated that miR-10a is possibly the most sensitive to ATRA treatment, considering that this miRNA showed altered expression patterns in response to ATRA in seven different studies [33, 35, 36, 39, 49, 51], which addressed three different types of cancers—neuroblastoma, breast cancer, and pancreatic cancer. In addition, treatment of NB cell lines with 5 μmol/L ATRA for 7 days led to a 410-fold increase in miR-10a expression. Moreover, miR-10a was the most common miRNA among all upregulated miRNAs, as it was found upregulated in all studies that measured its expression. Meanwhile, miR-134 was the most common downregulated miRNA [33, 36, 39, 54] and was associated with two different types of cancer, neuroblastoma and embryonal carcinoma.
We observed a large variation in the number of miRNAs evaluated in each study, with some studies analyzing more than 300 miRNAs and others only a single miRNA. In addition, some authors chose not to elucidate in depth all miRNAs that were sensitive to ATRA treatment or the significant findings associated with ATRA exposure, which may have led to imprecise conclusions. Finally, several studies demonstrated that ATRA largely inhibits cell growth and proliferation and promotes apoptosis of neoplastic cells.
Conclusion
Overall, our systematic review supports the use of ATRA as a potential strategy for the prevention and treatment of various neoplasms. However, it should be noted that these studies may not accurately represent changes occurring in vivo, which highlights the need for additional studies to fully elucidate the modulation of miRNAs induced by ATRA in the treatment of neoplasms.
Acknowledgements
This work was based on master’s dissertation of Lara Lima entitled “Associação entre os hábitos alimentares e as concentrações séricas das vitaminas A e E com os preditores de prognóstico histopatológico do carcinoma papilífero da tireoide” from the Postgraduate Program in Nutrition of Federal University of Rio Grande do Norte, Natal, Brazil, in 2018.
Abbreviations
- AML
acute myeloid leukemia
- APL
acute promyelocytic leukemia
- ATRA
all-trans retinoic acid
- CRC
colorectal cancer
- ERα-
estrogen receptor-negative
- ERα+
estrogen receptor-positive
- EVL
Enah/Vasp-like
- miRNAs
microRNAS
- NB
neuroblastoma
- RA
retinoic acid
- RARA
retinoic acid receptor-α
Authors’ contributions
LL and DM conceived and drafted the study design and independently screened the titles and abstracts of the identified studies. LL and DM designed and executed the search strategy. LL assessed the full texts for eligibility and extracted data independently from the included studies. LL described the selection process and prepared the initial draft of the manuscript. DM elaborated on the figures. TCTM, JNGA, ISFL, CXA, JG, and VNS provided supervision as well as revised and commented on subsequent drafts of the manuscript. All authors read and agreed to the contents of this article.
Funding
This work was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001. JNGA and DM are recipients of fellowships from CAPES.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lara Lima, Email: laraclarisse_lima@hotmail.com.
Thaísa Cristina Tavares de Melo, Email: thaiimeelo@gmail.com.
Diego Marques, Email: diegomarquescs@outlook.com.
Jéssica Nayara Góes de Araújo, Email: jessnaysub@hotmail.com.
Isabela Samária Fernandes Leite, Email: isamariafl@outlook.com.
Camila Xavier Alves, Email: camila_xavieralves@yahoo.com.br.
Julieta Genre, Email: jgenre@gmail.com.
Vivian Nogueira Silbiger, Phone: +55 84 3342 9807, Email: viviansilbiger@hotmail.com.
References
- 1.Thun MJ, Delancey JO, Jemal A, Ward EM. The global burden of cancer: priorities for prevention. Carcinogenesis. 2010;31:100–110. doi: 10.1093/carcin/bgp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxter E, Windloch K, Gannon F, Lee JS. Epigenetic regulation in cancer progression. Cell Biosci. 2014;4:1–11. doi: 10.1186/2045-3701-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu X, Gao G, Chu K, Yang X, Ren S, Li Y, et al. Inhibition of RAC1-GEF DOCK3 by miR-512-3p contributes to suppression of metastasis in non-small cell lung cancer. Int J Biochem Cell Biol. 2015:1–12. [DOI] [PubMed]
- 4.Chu K, Gao G, Yang X, Ren S. miR-512-5p induces apoptosis and inhibits glycolysis by targeting p21 in non-small cell lung cancer cells. Int J Oncol. 2016;48:577–586. doi: 10.3892/ijo.2015.3279. [DOI] [PubMed] [Google Scholar]
- 5.Chen PH, Shih CM, Chang WC, Cheng CH, Lin CW, Ho KH, et al. MicroRNA-320-inhibited E2F3 transcription factor is related to all trans retinoic acid-induced glioma cell. J Neurochem. 2014:731–42. [DOI] [PubMed]
- 6.Bouriez D, Giraud J, Gronnier C, Varon C. Efficiency of all-trans retinoic acid on gastric cancer: a narrative literature review. Int J Mol Sci. 2018;19:1–21. doi: 10.3390/ijms19113388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyle KM, Dean CA, Thomas ML, Vidovic D, Giacomantonio CA, Helyer L, et al. DNA methylation predicts the response of triple-negative breast cancers to all-trans retinoic acid. Cancers (Basel) 2018;10:1–17. doi: 10.3390/cancers10110397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quiroga PL, Soria EA, Valentich MA, Eynard AR, Quiroga PL, Soria EA, et al. Differential potentiation of retinoic acid effects against human breast cancer cells by unsaturated fatty acids. Nutr Cancer. 2018;70:1137–1144. doi: 10.1080/01635581.2018.1497669. [DOI] [PubMed] [Google Scholar]
- 9.Creutzig U, Dworzak M, Rasche M, Reinhardt D, Hospital C, Medical H. Acute promyelocytic leukemia: new treatment strategies with ATRA and ATO–AML-BFM-recommendations. Klin Padiatr. 2018;230:299–304. doi: 10.1055/a-0750-5963. [DOI] [PubMed] [Google Scholar]
- 10.Sobas M, Montesinos P, Boluda B, Bernal T, Vellenga E, Nomdedeu J, et al. An analysis of the impact of CD56 expression in de novo acute promyelocytic leukemia patients treated with upfront all-trans retinoic acid and anthracycline-based regimens. Leuk Lymphoma. 2018:1–6. [DOI] [PubMed]
- 11.Krestinina O, Fadeev R, Lomovsky A, Baburina Y, Kobyakova M, Akatov V. Melatonin can strengthen the effect of retinoic acid in HL-60 cells. Mol Sci. 2018;19:1–13. doi: 10.3390/ijms19102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walasek A. The new perspectives of targeted therapy in acute myeloid leukemia. Adv Clin Exp Med. 2019;28:271–276. doi: 10.17219/acem/81610. [DOI] [PubMed] [Google Scholar]
- 13.Friesenbichler W, Krizmanich W, Lakatos K, Dworzak M, Amann G, Furtwängler R, et al. Outcome of two patients with bilateral nephroblastomatosis/Wilms tumour treated with an add-on 13-cis retinoic acid therapy – case report. Pediatr Hematol Oncol. 2018;35:218–224. doi: 10.1080/08880018.2018.1515284. [DOI] [PubMed] [Google Scholar]
- 14.Jobani BM, Najafzadeh N, Mazani M, Arzanlou M, Vardin MM. Molecular mechanism and cytotoxicity of allicin and all-trans retinoic acid against CD44+ versus CD117+ melanoma cells. Phytomedicine. 2018;18:30173–30179. doi: 10.1016/j.phymed.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Yu S, Chen H, Chang G, Chen C, Chen H, Singh S, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Waetzig V, Andres C, Frehse S, Herdegen T. Retinoic acid-induced survival effects in SH-SY5Y neuroblastoma cells. J Cell Biochem. 2019;120:5974–5986. doi: 10.1002/jcb.27885. [DOI] [PubMed] [Google Scholar]
- 17.Zage PE. Novel therapies for relapsed and refractory neuroblastoma. Children. 2018;5:1–27. doi: 10.3390/children5110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 20.Laneve P, Di Marcotullio L, Gioia U, Fiori ME, Ferretti E, Gulino A, et al. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Cell Biol. 2007;104:7957–7962. doi: 10.1073/pnas.0700071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evangelisti C, Florian MC, Massimi I, Dominici C, Giannini G, Galardi S, et al. MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. FASEB J. 2009;23:4276–4287. doi: 10.1096/fj.09-134965. [DOI] [PubMed] [Google Scholar]
- 22.Das E, Bhattacharyya NP. MicroRNA-432 contributes to dopamine cocktail and retinoic acid induced differentiation of human neuroblastoma cells by targeting NESTIN and RCOR1 genes. FEBS Lett. 2014;588:1706–1714. doi: 10.1016/j.febslet.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Garzon R, Pichiorri F, Palumbo T, Visentini M, Aqeilan R, Cimmino A, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 24.Biology C, Sanchez-martínez D, Krzywinska E, Rathore MG, Saumet A, Cornillon A, et al. All-trans retinoic acid (ATRA) induces miR-23a expression, decreases CTSC expression and granzyme B activity leading to impaired NK cell cytotoxicity. Int J Biochem Cell Biol. 2014;49:42–52. doi: 10.1016/j.biocel.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Li C, Imai M, Yamasaki M, Hasegawa S, Takahashi N. Effects of pre- and post-administration of vitamin a on the growth of refractory cancers in xenograft mice. Biol Pharm Bull. 2017;40:486–494. doi: 10.1248/bpb.b16-00933. [DOI] [PubMed] [Google Scholar]
- 26.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 27.Khew-goodall Y, Goodall GJ. Myc-modulated miR-9 makes more metastases. Nat Cell Biol. 2010;12:209–211. doi: 10.1038/ncb0310-209. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Roslan S, Johnstone CN, Wright JA, Bracken CP, Anderson M, et al. MiR-200 can repress breast cancer metastasis through ZEB1-independent but moesin-dependent pathways. Oncogene. 2013:1–12. [DOI] [PubMed]
- 29.Roy SS, Gonugunta VK, Bandyopadhyay A, Rao MK, Goodall GJ, Sun L, et al. Significance of PELP1/HDAC2/miR-200 regulatory network in EMT and metastasis of breast cancer. Oncogene. 2013:1–10. [DOI] [PMC free article] [PubMed]
- 30.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Ioannidis JPA, Clarke M, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Stallings RL, Chen Y, Stallings RL. Differential patterns of microrna expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–983. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 32.Le MTN, Xie H, Zhou B, Chia PH, Rizk P, Um M, et al. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29:5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beveridge NJ, Tooney PA, Carroll AP, Tran N, Cairns MJ. Down-regulation of miR-17 family expression in response to retinoic acid induced neuronal differentiation. Cell Signal. 2009;21:1837–1845. doi: 10.1016/j.cellsig.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 34.Ragusa Marco, Majorana Alessandra, Banelli Barbara, Barbagallo Davide, Statello Luisa, Casciano Ida, Guglielmino Maria Rosa, Duro Laura Rita, Scalia Marina, Magro Gaetano, Di Pietro Cinzia, Romani Massimo, Purrello Michele. MIR152, MIR200B, and MIR338, human positional and functional neuroblastoma candidates, are involved in neuroblast differentiation and apoptosis. Journal of Molecular Medicine. 2010;88(10):1041–1053. doi: 10.1007/s00109-010-0643-0. [DOI] [PubMed] [Google Scholar]
- 35.Meseguer S, Mudduluru G, Escamilla JM, Allgayer H, Barettino D. MicroRNAs-10a and -10b contribute to retinoic acid-induced differentiation of neuroblastoma cells and target the alternative splicing regulatory factor SFRS1 (SF2/ASF ) J Biol Chem. 2011;286:4150–4164. doi: 10.1074/jbc.M110.167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das S, Foley N, Bryan K, Watters KM, Bray I, Murphy DM, et al. MicroRNA mediates DNA demethylation events triggered by retinoic acid during neuroblastoma cell differentiation. Mol Cell Pathobiol. 2010;70:7874–7882. doi: 10.1158/0008-5472.CAN-10-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das S, Bryan K, Buckley PG, Piskareva O, Bray IM, Foley N, et al. Modulation of neuroblastoma disease pathogenesis by an extensive network of epigenetically regulated microRNAs. Oncogene. 2013;32:2927–2936. doi: 10.1038/onc.2012.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H, Shalom-feuerstein R, Riley J, Zhang S, Tucci P, Agostini M, et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem Biophys Res Commun. 2010;394:921–927. doi: 10.1016/j.bbrc.2010.03.076. [DOI] [PubMed] [Google Scholar]
- 39.Foley NH, Bray I, Watters KM, Das S, Bryan K, Bernas T, et al. MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 2011;18:1089–1098. doi: 10.1038/cdd.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchis ML, De BM, Salvatori B, Puzzolo MC, Bozzoni I, Fatica A. A new molecular network comprising PU.1, interferon regulatory factor proteins and miR-342 stimulates ATRA-mediated granulocytic differentiation of acute promyelocytic leukemia cells. Leukemia. 2009;23:856–862. doi: 10.1038/leu.2008.372. [DOI] [PubMed] [Google Scholar]
- 41.Gao S, Yang J, Chen C, Zhang S, Xing C, Li H. miR-15a/16–1 enhances retinoic acid-mediated diff erentiation of leukemic cells and is up-regulated by retinoic acid. Leuk Limphona. 2011;52:2365–2371. doi: 10.3109/10428194.2011.601476. [DOI] [PubMed] [Google Scholar]
- 42.Lin K, Zhang X, Feng D, Zhang H, Zeng C, Han B, et al. miR-125b, a target of CDX2, regulates cell differentiation through repression of the core binding factor in hematopoietic malignancies. J Biol Chem. 2011;286:38253–38263. doi: 10.1074/jbc.M111.269670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris VA, Zhang A, Yang T, Stirewalt DL, Ramamurthy R, Oehler VG. MicroRNA-150 expression induces myeloid differentiation of human acute leukemia cells and normal hematopoietic progenitors. PLoS One. 2013;8:e75815. doi: 10.1371/journal.pone.0075815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuang LK, Xu GP, Pan XR, Lou YJ, Zou QP, Xia D, et al. MicroRNA-181a-mediated downregulation of AC9 protein decreases intracellular cAMP level and inhibits ATRA-induced APL cell differentiation. Cell Death Dis. 2014;5:e1161. doi: 10.1038/cddis.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y, Li D, Liang Q, Liu S, Zuo X, Li L, et al. miR-638 regulates differentiation and proliferation in leukemic cells by targeting cyclin-dependent kinase 2. J Biol Chem. 2015;290:1818–1828. doi: 10.1074/jbc.M114.599191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bräuer-hartmann D, Hartmann J, Wurm AA, Gerloff D. PML/RARα-regulated miR-181a/b-cluster targets the tumor suppressor RASSF1A in acute promyelocytic leukemia. Cancer Res. 2016;75:3411–3424. doi: 10.1158/0008-5472.CAN-14-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan Y, Hanse EA, Stedman K, Benson JM, Lowman XH, Subramanian S, et al. Transcription factor C/EBP-β induces tumor-suppressor phosphatase PHLPP2 through repression of the miR-17–92 cluster in differentiating AML cells. Cell Death Differ. 2016;23:1232–1242. doi: 10.1038/cdd.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terao M, Fratelli M, Kurosaki M, Zanetti A, Guarnaccia V, Paroni G, et al. Induction of miR-21 by retinoic acid in estrogen receptor-positive breast carcinoma cells: biological correlates and molecular targets. J Biol Chem. 2011;286:4027–4042. doi: 10.1074/jbc.M110.184994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan S, Wall D, Curran C, Newell J, Kerin MJ, Dwyer RM. MicroRNA-10a is reduced in breast cancer and regulated in part through retinoic acid. BMC Cancer. 2015;15:345. doi: 10.1186/s12885-015-1374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisher JN, Terao M, Fratelli M, Kurosaki M, Paroni G, Zanetti A, et al. MicroRNA networks regulated by all-trans retinoic acid and Lapatinib control the growth, survival and motility of breast cancer cells. Oncotarget. 2015;6:13176–13200. doi: 10.18632/oncotarget.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, et al. Retinoic acid receptor antagonists inhibit mir-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137:2136–2145. doi: 10.1053/j.gastro.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 52.Xia H, He T, Liu C, Cui Y, Song P, Jin X, et al. MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem. 2009;23:347–358. doi: 10.1159/000218181. [DOI] [PubMed] [Google Scholar]
- 53.Chen P-H, Shih C-M, Chang W-C, Cheng C-H, Lin C-W, Ho K, et al. MicroRNA-320-inhibited E2F3 transcription factor is related to all trans retinoic acid-induced glioma cell apoptosis. J Neurochem. 2014;131:731–742. doi: 10.1111/jnc.12820. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Meng L, Yu Q, Dong D, Tan G, Huang X, et al. The miR-134 attenuates the expression of transcription factor FOXM1 during pluripotent NT2/D1 embryonal carcinoma cell differentiation. Exp Cell Res. 2015;330:442–450. doi: 10.1016/j.yexcr.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 55.Liu W, Song Y, Zhang C, Gao P, Huang B. The protective role of all-transretinoic acid (ATRA) against colorectal cancer development is achieved via increasing miR-3666 expression and decreasing E2F7 expression. Biomed Pharmacother. 2018;104:94–101. doi: 10.1016/j.biopha.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 56.Su M, Alonso S, Jones JW, Yu J, Kane MA, Jones J, et al. All-trans retinoic acid activity in acute myeloid leukemia: role of cytochrome P450 enzyme expression by the microenvironment. PLoS One. 2015;10:e0127790. doi: 10.1371/journal.pone.0127790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cogan T, Cogan M, Tamil L. RAMS: remote and automatic mammogram screening. Comput Biol Med. 2019;107:18–29. doi: 10.1016/j.compbiomed.2019.01.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.