Abstract
Background
MiR-221, acting as onco-miR or oncosuppressor-miR, plays an important role in tumor progression; however, the prognostic value of miR-221 in human carcinomas is controversial and inconclusive. The objective of our study was to conducted a systematic review and meta-analysis of miR-221 in various types of human cancers.
Methods
An online search of up-to-date electronic databases, including PubMed and Embase, was conducted to identify as many relevant papers as possible. 32 papers involving 3041 patients with different carcinomas were included in the analysis. Hazard ratios (HRs) of miR-221 were used to evaluate prognostic values.
Results
Thirty-two papers involving 15 cancers were included. MiR-221 was associated with a worse overall survival (OS) in patients, and a combined HR was 1.93 (95% CI of 1.43–2.60, 2080 patients, 22 studies, I-squared = 80.4%, P = 0.000); however, the combined HR for relapse-free survival (RFS) was 1.37 (95% CI of 0.75–2.48, 625 patients, 7 studies, I-squared = 78.8%, P = 0.000), and disease-free survival (DFS) was 1.24 (95% CI of 0.60–2.56, 539 patients, 5 studies, I-squared = 81.8%, P = 0.000).
Conclusion
MiR-221 was shown to be associated with a poor OS in human carcinomas, and thus may serve as a useful predictor of clinical outcomes.
Electronic supplementary material
The online version of this article (10.1186/s12885-019-6079-1) contains supplementary material, which is available to authorized users.
Keywords: MiR-221, Human carcinoma, Prognosis, Meta-analysis
Background
MicroRNAs (miRNAs), small noncoding single-stranded RNAs, play a pivotal role in diverse cellular processes through post-transcriptional regulation of gene expression [1]. MiRNAs are now known to play an essential role in malignancy, functioning as tumor suppressors and oncogenes [2]. The expression of miRNAs is abnormal in different carcinomas and miRNAs are involved in the development and progression of disease [3]. As favorable or unfavorable prognostic biomarkers, many miRNAs are associated with patients’ survival in different cancers [4–6].
MiR-221, located on human chromosome X, is upregulated in many different cancers. As an onco or oncosuppressor-miR, miR-221 plays an important role in tumor progression [7]. High expression of miR-221 is associated with a worse survival in patients with different cancers, such as liver, laryngeal, and lung cancers [8–10]. It has also been reported that miR-221 is a favorable factor in predicting the prognosis of ovarian and renal cancers [11, 12]. Because of the controversy involving the association between miR-221 and survival among patients with different carcinomas, we conducted a systematic review and meta-analysis of the prognostic value of miR-221 in human cancers.
Methods
Search strategy
The objective of our study was to summarize the prognostic value of miR-221 in human carcinomas. We conducted an online search of up-to-date electronic databases, including PubMed and Embase, to identify as many relevant papers as possible. The search was performed by professional literature librarian. The key words, “miR-221” and “cancer” were used. The details of the search strategy in two databases were shown (Additional file 6: Table S1). The reference lists of review papers were hand-searched for further relevant studies. The last search update was performed on July 20, 2019.
Risk of bias assessment
We systematically assessed the quality of all included studies according to the guidelines that we previously described [13]. The assessment details of the studies are as follows: 1) clear information about populations and nations of included participants; 2) clear information about the type of carcinomas involved; 3) clear information about study design (prospective or retrospective); 4) clear information about the arrays used to measure the expression of miR-221; 5) clear information about the type of outcome assessment; and 6) clear information about follow-up. Studies that met the above criteria were included.
The evaluation process was performed by two independent authors, all HRs were extracted directly from papers. The risk of bias was assessed by Cochrane B or Newcastle-Ottawa Scale (NOS).
Statistical analysis
All statistical analyses were conducted using Stata12.0. The HRs with corresponding 95% CIs were used to estimate the strength of the relationship between miR-221 and prognosis. The results were displayed by forest plots. The heterogeneity assumption of the pooled HR was verified by a χ2-based Cochran Q test and Higgins I2 statistic, with an I-squared> 50% and/or a P < 0.1 indicating heterogeneity. Considering the heterogeneity of different articles, a random effect was performed in this meta-analysis. Potential publication bias was determined by Begg’s test with a funnel plot.
Results
Characters of included studies
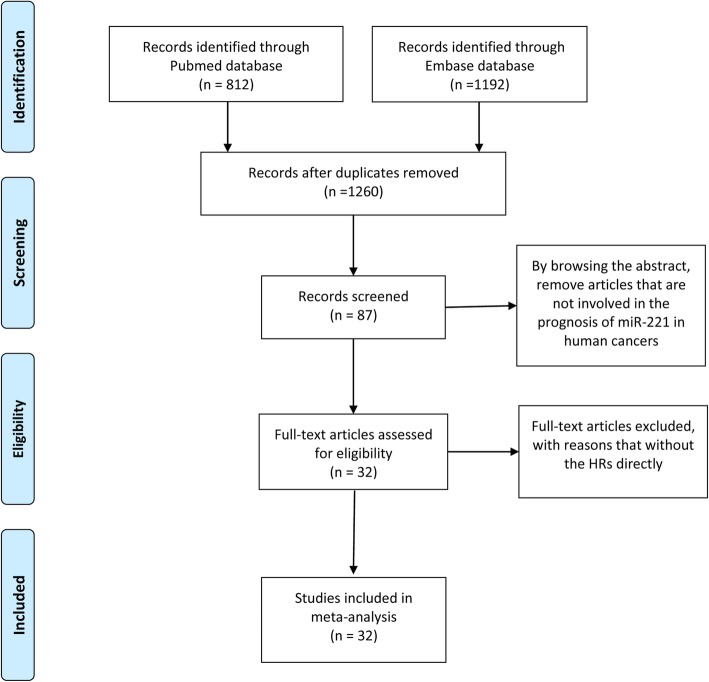
812 and 1192 papers were identified from the databases of Pubmed and Embase respectively. By browsing the titles and abstracts, the articles that were duplicates or not involved in the prognostic value of miR-221 in human cancers were excluded. Then, according to full-text assessment, 32 papers researching the prognostic value of miR-221 in human carcinomas with sufficient data were included in our study (Fig. 1). All included papers have relatively high-quality assessment of the risk of bias, which met the requirements for inclusion.
Fig. 1.
Flow chart of study selection process
A total of 3041 patients with different cancers were involved, including ovarian cancer [11, 14], bladder cancer [15], osteosarcoma [16, 17], liver cancer [8, 18–22], laryngeal cancer [9], thyroid cancer [23], lung cancer [10, 24], gastric-colon cancer [25–28], renal cancer [12], breast cancer [29–32], prostate cancer [33–36], cutaneous malignant melanoma (CMM) [37], acute lymphoid leukemia (ALL) [38, 39], and NK/T-cell lymphoma [40]. 20 of them were conducted in Asia (18 in China and 2 in Korea), the remaining studies were conducted in other countries, including Greece, the USA, Egypt, Germany, Italy, and Brazil. The origins of miR-221 in most studies were derived from tumor tissues; 7 were from serum/plasma and 2 were from bone marrow. Quantitative real time PCR (qRT-PCR) was performed in most of the studies for quantification of miR-221; two studies used in-situ hybridization (ISH) to detect miR-221 expression. The details of these papers were summarized in Table 1.
Table 1.
Characters of included 32 papers about the prognostic value of miR-221 in human carcinomas
| Study | Nation | Patients | cancer | Survival | HR(95% CI) | P value | Origins | Measure | Cutoff | Analysis |
|---|---|---|---|---|---|---|---|---|---|---|
| Wu Q (2017) | China | 74 | Ovarian cancer | OS | 0.395 (0.196–0.796) | 0.009 | Tumor | qRT-PCR | Median | Univariate |
| Tsikrika (2017) | Greece | 159 | Bladder cancer |
DFS PFS |
0.712(0.380–1.335) 1.396(0.539–3.620) |
0.290 0.492 |
Tumor | qRT-PCR | ROC | Univariate |
| Deng (2017) | China | 125 | Breast cancer | DFS | 0.480 (0.263–0.879) | 0.017 | Tumor | qRT-PCR | Median | Multivariate |
| Nakka (2017) | USA | 32 | Osteosarcoma | OS | 0.733(0.486–1.1055) | 0.139 | Tumor | qRT-PCR/ISH | Quartation | Multivariate |
| Xie D (2017) | China | 70 | Live cancer | OS | 1.743 (1.004–3.772) | 0.012 | Tumor | qRT-PCR | Median | Multivariate |
| Hussein (2017) | Egypt | 50 | Laryngeal cancer | OS | 6.5 (1.8–22.5) | 0.003 | Tumor | qRT-PCR | ROC | Univariate |
| Dai L (2017) | China | 78 | Thyroid cancer | RFS | 1.41(1.14–1.95) | 0.007 | Tumor | qRT-PCR | Median | Multivariate |
| Chen F (2017) | China | 135 | HCC |
DFS OS |
2.846 (1.564–5.181) 2.969 (1.629–5.408) |
0.001 < 0.001 |
Tumor | qRT-PCR | Median | Multivariate |
| Zhang Y (2016) | China | 104 | Lung cancer | OS | 1.873(1.267–2.768) | 0.002 | Tumor | qRT-PCR | Median | Multivariate |
| Yang Z (2015) | China | 108 | Osteosarcoma |
OS RFS |
7.66(1.83–15.92) 6.82(1.33–13.69) |
0.01 0.01 |
Serum | qRT-PCR | Median | Multivariate |
| Cai K (2015) | China | 182 | Colon cancer | OS | 2.394 (1.210–4.910) | 0.006 | Tumor | qRT-PCR | Median | Multivariate |
| Tao K (2014) | China | 90 | Colon cancer | OS | 2.043 (1.095–3.812) | 0.025 | Tumor | qRT-PCR | Median | Multivariate |
| Vergho (2014) | Germany | 74 | Renal cancer | CSS | 0.47 (0.22–1.00) | 0.0527 | Tumor | qRT-PCR | ROC | Multivariate |
| Lv J (2014) | China | 117 | Lung cancer | OS | 2.425 (1.314–4.475) | 0.005 | Tumor | qRT-PCR | Median | Multivariate |
| Li P (2014) | China | 72 | CMM |
OS DFS |
3.189(1.782–6.777) 2.119(1.962–8.552) |
0.007 0.01 |
Serum | qRT-PCR | Median | Multivariate |
| Gyongyosi B (2014) | Italy | 20 | HCC |
OS PFS |
1.92(0.61–6.10) 1.32(0.47–3.66) |
0.29 0.58 |
Tumor | qRT-PCR | Median | Univariate |
| Falkenberg (2013) | Germany | 86 | Breast cancer | MFS | 2.57(1.1073–5.9647) | 0.028 | Tumor | qRT-PCR | ROC | Multivariate |
| Hong (2013) | China | 96 | Ovarian cancer | OS | 2.243(1.1357–4.4300) | 0.020 | Serum | qRT-PCR | Mean | Multivariate |
| Gimenes (2013) | Brazil | 48 | ALL |
OS DFS |
2.31(0.92–5.81) 1.54 (0.57–4.17) |
0.074 0.391 |
Marrow | qRT-PCR | Median | Multivariate |
| Karakatsanis (2013) | Greece | 60 | HCC | OS | 1.72 (1.32–2.50) | 0.002 | Tumor | qRT-PCR | Mean | Multivariate |
| Amankwah (2013) | USA | 65 | Prostate cancer | RFS | 1.79 (0.67–4.76) | 0.25 | Tumor | qRT-PCR | Median | Multivariate |
| Liu K (2012) | China | 92 | Gastric cancer | OS | 2.322 (1.1116–4.8505) | 0.025 | Tumor | qRT-PCR | Mean | Multivariate |
| Hanna (2012) | USA | 377 | Breast cancer | OS | 0.70 (0.51–0.97) | 0.0312 | Tumor | ISH | Quartation | Multivariate |
| Kang (2012) | Korea | 92 | Prostate cancer | RFS | 0.360 (0.171–1.896) | 0.570 | Tumor | qRT-PCR | Median | Univariate |
| Li J (2011) | China | 46 | HCC | OS | 1.903(1.235–2.981) | 0.018 | Serum | qRT-PCR | Mean | Multivariate |
| Yoon (2011) | Korea | 115 | HCC | RFS | 3.07 (1.56–6.07) | 0.001 | Tumor | qRT-PCR | Mean | Multivariate |
| Zhao R (2011) | China | 93 | Breast cancer | OS | 6.871 (1.967–23.997) | 0.003 | plasma | qRT-PCR | Median | Multivariate |
| Schaefer (2010) | Germany | 75 | Prostate cancer | RFS | 0.93 (0.3–2.89) | 0.902 | Tumor | qRT-PCR | Median | Univariate |
| Wang (2010) | China | 32 | ALL | OS | 0.538(0.30–0.9648) | 0.038 | Marrow | qRT-PCR | Median | Multivariate |
| Spahn (2010) | Germany | 92 | Prostate cancer | RFS | 0.525(0.29–0.95) | 0.032 | Tumor | qRT-PCR | ROC | Multivariate |
| Pu (2010) | China | 103 | Colon cancer | OS | 3.478(1.038–11.654) | 0.043 | Plasma | qRT-PCR | Youden | Multivariate |
| Guo (2010) | China | 79 | Lymphoma | OS | 5.714(1.782–18.18) | 0.003 | Plasma | qRT-PCR | Youden | Multivariate |
Note: HCC Hepatocellular carcinoma; ALL Acute lymphoid leukemia; CMM Cutaneous malignant melanoma; ROC Receiver operating characteristic curve; Youden, Youden index; OS Overall survival; RFS Relapse-free survival; DFS Disease-free survival; PFS Progression-free survival; CSS Cancer-special survival; MFS Metastasis-free survival; qRT-PCR Quantitative Real Time PCR; ISH In-situ hybridization
Association of miR-221 with overall survival (OS)
A total of 22 articles researched the association between miR-221 and OS among different carcinomas. Generally, miR-221 was associated with a poor OS, with a pooled HR was 1.93 (95% CI of 1.43–2.60, 2080 patients, 22 studies, I-squared = 80.4%, P = 0.000) (Fig. 2). Due to the heterogeneity, subgroup analyses were performed. Most of the studies originated from China, thus we divided the patients into Asian (Chinese) and non-Asian groups. MiR-221 was significantly related to the OS of Chinese patients (HR = 2.14 (1.53–2.99), 1493 patients, 16 studies, I-squared = 74.2%, P = 0.000), but not non-Asian patients (HR = 1.44 (0.83–2.47), 587 patients, 6 studies, I-squared = 83.3%, P = 0.000) (Table 2 and Additional file 1: Figure S1). Then, we divided studies according to the number of included individuals. The combined HR was 2.28 (95% CI of 1.29–4.41, 1126 patients, 7 studies, I-squared = 85.9%, P = 0.000) in studies with more than 100 participants, and the HR was 1.80 (95% CI of 1.25–2.59, 954 patients, 15 studies, I-squared = 78.4%, P = 0.000) in studies with less than 100 patients (Table 2 and Additional file 2: Figure S2); however, heterogeneity still existed in these subgroups.
Fig. 2.
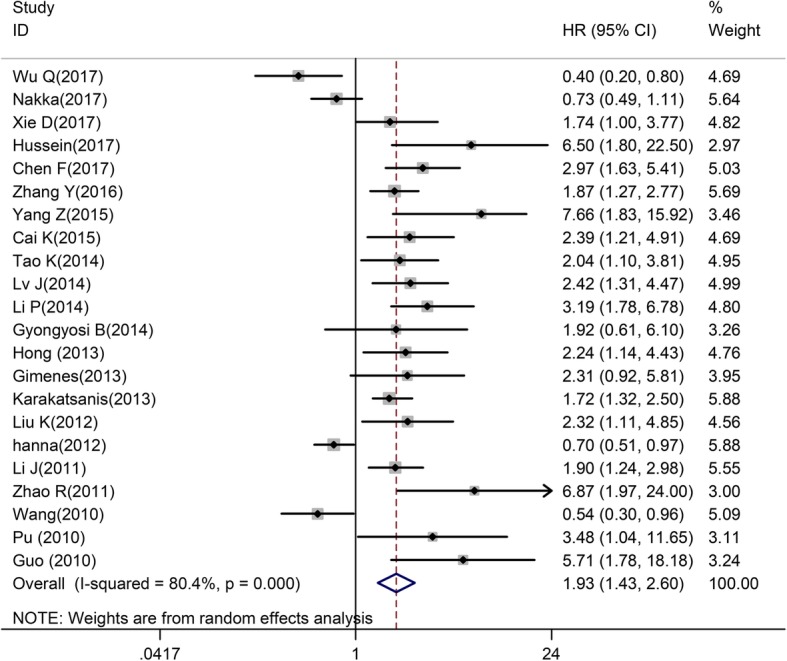
Forrest plots of the studies that evaluated the hazard ratios (HRs) of high miR-221 expression as compared to low expression in OS
Table 2.
Subgroup analysis of association between the expression of miR-221 and OS
| Categories | Subgroups | No of studies | Pool HR | 95% CI | Result | I2 | P |
|---|---|---|---|---|---|---|---|
| All | OS | 22 | 1.93 | 1.43–2.60 | S | 80.4% | 0.000 |
| Countries | Asian (Chinese) | 16 | 2.14 | 1.53–2.99 | S | 74.2% | 0.000 |
| Non-Asian | 6 | 1.44 | 0.83–2.47 | NS | 83.3% | 0.000 | |
| Samples origins | Tumor tissues | 13 | 1.61 | 1.13–2.29 | S | 81.4% | 0.000 |
| Serum/blood | 7 | 3.25 | 2.15–4.92 | S | 42.2% | 0.109 | |
| Marrow | 2 | 1.07 | 0.26–4.43 | NS | 85.4% | 0.009 | |
| Sample sizes | > 100 | 7 | 2.28 | 1.29–4.11 | S | 85.9% | 0.000 |
| ≤100 | 15 | 1.80 | 1.25–2.59 | S | 78.4% | 0.000 | |
| Cancer types | Colon cancer | 4 | 2.33 | 1.60–3.38 | S | 0.0% | 0.897 |
| Liver cancer | 5 | 1.91 | 1.53–2.38 | S | 0.0% | 0.633 | |
| Lung cancer | 2 | 2.02 | 1.45–2.81 | S | 0.0% | 0.486 | |
| ALL | 2 | 1.07 | 0.26–4.43 | NS | 85.4% | 0.009 | |
| Breast cancer | 2 | 2.02 | 0.22–18.81 | NS | 91.7% | 0.001 | |
| Osteosarcoma | 2 | 2.24 | 0.23–22.29 | NS | 93.7% | 0.000 | |
| Ovarian cancer | 2 | 0.94 | 0.17–5.17 | NS | 91.8% | 0.000 |
Note: ALL Acute lymphoid leukemia; S Significant; NS Non-significant
We further analyzed the subgroups divided based on different cancers. The cancers investigated in more than one paper were included. The results showed that the HR was 2.33 (95% CI of 1.60–3.38, 467 patients, 4 studies, I-squared = 0.0%, P = 0.897) in colon cancer, 1.91 (95% CI of 1.53–2.38, 331patients, 5 studies, I-squared = 0.0%, P = 0.633) in liver cancer, and 2.02 (95% CI of 1.45–2.81, 221 patients, 2 studies, I-squared = 0.0%, P = 0.486) in lung cancer (Table 2 and Additional file 3: Fig. S3). However, the combined HR was 1.07 (95% CI of 0.26–4.43, 80 patients, 2 studies, I-squared = 85.4%, P = 0.009) in ALL, 2.02 (95% CI of 0.22–18.81, 470 patients, 2 studies, I-squared = 91.7%, P = 0.001) in breast cancer, 2.24(95% CI of 0.23–22.29, 140 patients, 2 studies, I-squared = 93.7%, P = 0.000) in osteosarcoma and 0.94 (95% CI of 0.17–5.17, 170 patients, 2 studies, I-squared = 91.8%, P = 0.000) in ovarian cancer (Table 2 and Additional file 4: Figure S4).
Interestingly, the origins of miR-221 in 7 of the papers were derived from serum or plasma. The combined HR was 3.25 (95% CI of 2.15–4.92, 597 patients, 7 studies, I-squared = 42.2%, P = 0.109), which suggested that the expression of miR-221 in serum/plasma was associated with a worse OS. The results differed from a previous study [41]. In addition, the combined HR of the studies from tumor tissues was 1.61 (95% CI of 1.13–2.29, 1403 patients, 13 studies, I-squared = 81.4%, P = 0.000), and the combined HR from marrow was 1.07 (95% CI of 0.26–4.43, 80 patients, 2 studies, I-squared = 85.4%, P = 0.009) (Table 2 and Additional file 5: Figure S5).
Association of miR-221 with relapse-free survival (RFS)/ disease-free survival (DFS)
Seven of papers reported an association between miR-221 and RFS. The combined HR was 1.37 (95% CI of 0.75–2.48, 625 patients, 7 studies, I-squared = 78.8%, P = 0.000) (Fig. 3a). Notably, 4 of them focused on prostate cancer. The combined HR was 0.74 (95% CI of 0.38–1.42, 324 patients, 4 studies, I-squared = 48.6%, P = 0.120), which demonstrated that miR-221 tended to be a favorable predictor of RFS in prostate cancer patients (Fig. 3b). Besides, 5 of studies focused on the DFS of patients, the combined HR was 1.24 (95% CI of 0.60–2.56, 539 patients, 5 studies, I-squared = 81.8%, P = 0.000) (Fig. 3c). Due to the limited number of papers mentioned about RFS/DFS of patients, we were not able to analyze the causes of heterogeneity.
Fig. 3.
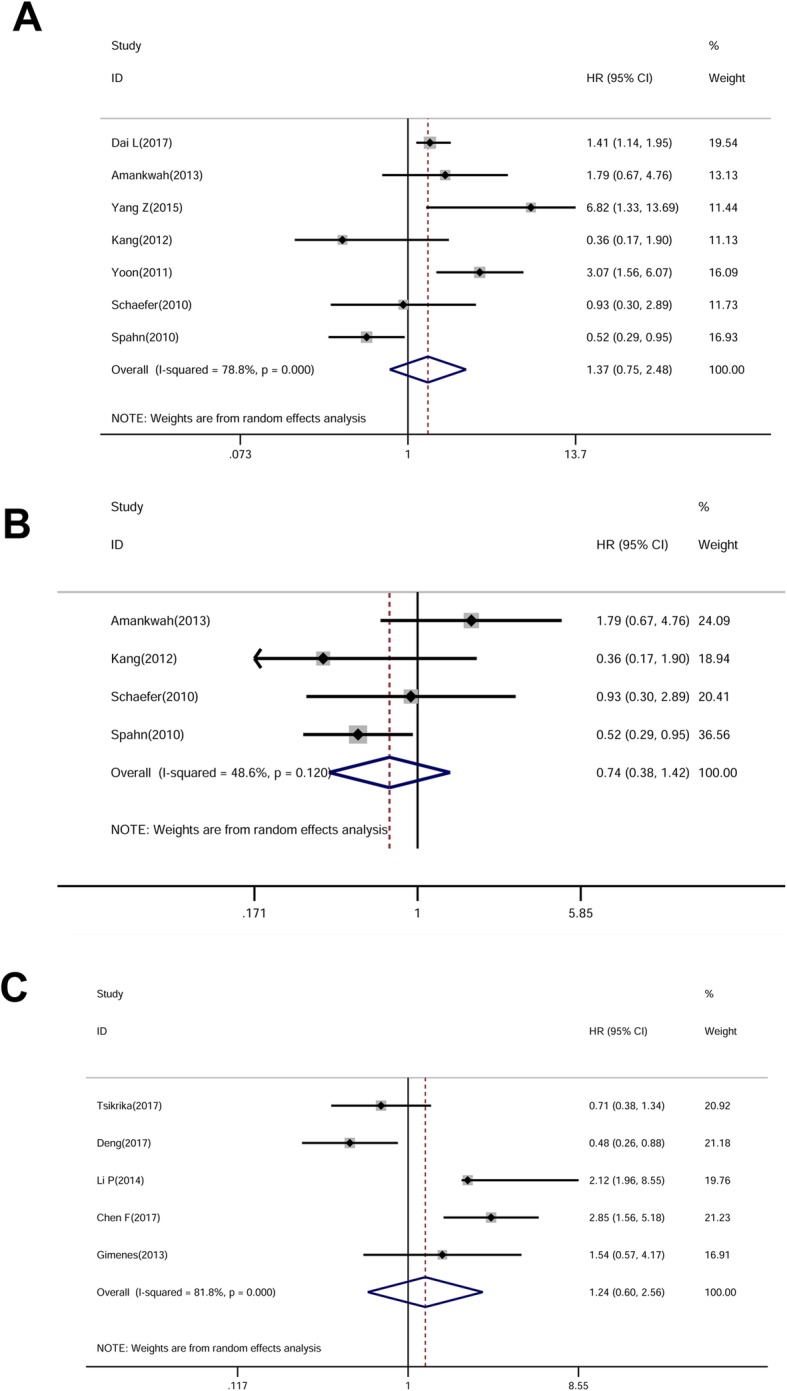
Forrest plots of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in RFS/DFS (a) RFS (b) RFS of prostate cancer (c) DFS
Publication bias
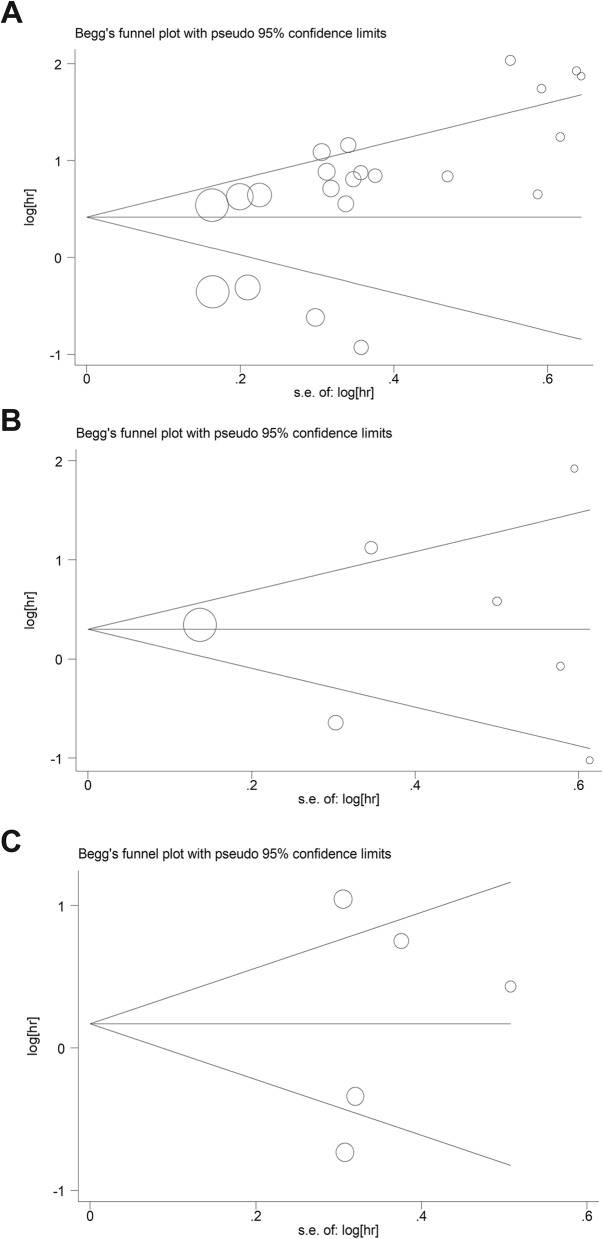
Both Begg’s and Egger’s tests were performed to estimate the potential publication bias in our study. A P < 0.05 indicated the existence of publication bias. There was no apparent publication bias in papers with respect to RFS and DFS; however, we evaluated the potential publication bias with respect to OS (P = 0.015). The funnel plots of Begg’s test were shown in Fig. 4.
Fig. 4.
Funnel plots of Begg’s test included in the meta-analysis of (a) OS (b) RFS (c) DFS
Discussion
MicroRNAs have been reported to be aberrantly expressed and play an important role in predicting the prognosis of human carcinomas [42]. It has been recommended to classify miRNAs into two categories (oncogenes and onco-suppressor miRNAs), which regulate tumor oncogenes or suppressor genes, respectively [43]. MiR-221, either as an oncogene or tumor suppressor gene, is involved in tumor progression in many different cancers. MiR-221 promotes the tumor progression of cancers, such as bladder, prostate, and breast cancers, by targeting downstream molecules, including PTEN, E-cadherin, and suppressors of cytokine signaling 1, 3 (SOCS1, 3) [44–48]. In contrast, miR-221 inhibits tumor progression in diseases such as pancreatic and ovarian cancers, by targeting factors, such as SOCS3 and ADP-ribosylation factor-4 (ARF4) [11, 49]. Interestingly, the dual role of miR-221 has been found in some cancers, such as pancreatic cancer [49–51]. It has been reported that some miRNAs (miR-21 and miR-155) are related to unfavorable clinical outcomes of pancreatic cancer [52]; however, there is still a lack of studies on the prognostic value of miR-221, which will be worth further exploration. Taken together, miR-221 has a dual role in tumor progression of different cancers. Future studies are necessary to elucidate the mechanisms underlying miR-221 in oncology.
The prognostic values of miR-221 have been investigated in different kinds of cancers; however, the roles of miR-221 in different studies have been controversial and inconclusive. A meta-analysis involving miR-221 in human cancers was conducted by Yang et al.in 2014 [53]. Subsequently, additional studies researching the prognostic value of miR-221 have been published in recent years and the results of those studies are inconsistent. Thus, an updated systematic review and meta-analysis was necessary to ascertain the prognostic value of miR-221.
Recently, Zhang et al. conducted a review and meta-analysis of the prognostic value of miR-221/miR-222 in human malignancy [54]; however, we noted that many relevant articles were omitted and some of the results should be re-summarized. Therefore, in the current study we systematically summarized the prognostic value of miR-221 according to the papers published in recent years. The results showed that miR-221 was associated with a worse OS of various of cancers. In agreement with our results, miR-222 (miR-221 highly homologous miRNA) was reported to be associated with poor survival [55]. Subgroup analysis showed that miR-221 was related to the OS of Chinese, but not non-Asians. Regarding different cancers, we found that miR-221 was significantly related to the OS of colon, liver, and lung cancers, but not associated with ALL, osteosarcoma, breast cancer, and ovarian cancer. With respect to the methods of miR-221 quantification, because there were limited articles using the ISH method, subgroup analysis using methods of miR-221 quantification did not produce meaningful results. In addition, the relationship between miR-221 and RFS/DFS of patients was indefinite, but high expression of miR-221 tended to be associated with a favorable RFS in prostate cancer patients. With the limited number of relevant papers, more studies are needed to confirm these conclusions.
Interestingly, Rong et al. reported that the association between the expression of miR-221 in serum/plasm and prognosis of patients was non-significant (HR = 0.94 (0.47–1.87), I-squared = 84.2%, P = 0.000) [41]; however, in our study, 7 papers focusing on miR-221 from serum/plasma were included. We found that miR-221 was related to a poor OS. The inconsistency of the two studies was possibly due to the different standards used in the articles, and more published studies were added in our study. The Guo et al. study was included in our study and Rong et al. study [40]. In the Guo’s study, the HR of multivariate analysis was calculated using high expression of miR-221 as a baseline. Therefore, the HRs directly obtained from this study should be transformed.
There were limitations in the current study that must be mentioned. First, the numbers of studies with some cancers were limited and it was difficult to conclude that a reliable association existed between miR-221 with those cancers. More studies researching miR-221 in different cancers will be necessary in the future. Second, although subgroup analysis was performed, the heterogeneity still existed in some groups. With the limited number of papers, we could not adequately explore the reasons for heterogeneity. Third, studies with negative results were generally less likely to be published. Therefore, we could not deny the potential existence of publication bias.
Conclusion
By summarizing the results of published papers about the prognostic value of miR-221 in human cancer, we found that high expression of miR-221 was associated with a worse OS; however, the association of miR-221 with RFS/DFS was not significant. Future studies with a larger number of cases are recommended to validate the role of miR-221 in human carcinomas.
Additional files
Figure S1. Subgroup analyses of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in OS by nations, (A) Chinses (B) non-Asian (TIF 24678 kb)
Figure S2. Subgroup analyses of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in OS by the number of individuals, (A) > 100 (B) ≤100 (TIF 24678 kb)
Figure S3. Subgroup analyses of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in OS by cancers, (A) Colon cancer (B) Live cancer (C) Lung cancer (TIF 15382 kb)
Figure S4. Subgroup analyses of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in OS by cancers, (A) ALL (B) Breast cancer (C) Osteosarcoma (D) Ovarian cancer (DOCX 19 kb) (TIF 14956 kb)
Figure S5. Subgroup analyses of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in OS by origins, (A) Tumor tissues (B) Serum/plasm (C) Marrow (TIF 24676 kb)
Table S1. Literature search strategy of PubMed database. The Embase database is searched in a similar way to PubMed. (DOCX 19 kb)
Acknowledgements
The authors sincerely thank all medical staff who participates in this study.
Abbreviations
- ALL
Acute lymphoid leukemia
- CI
Confidence interval
- CMM
Cutaneous malignant melanoma
- DFS
Disease-free survival
- HR
Hazard ratio
- OS
Overall survival
- RFS
Relapse-free survival
Authors’ contributions
Conceived of the study: KL, ES; Literature searching: KL, LW; Data extraction: KL, LW; Data analysis: KL, LW; Draft the manuscript: KL, ES; Approved the final version of the manuscript: KL, LW and ES. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The datasets analyzed during the current study are available in the PubMed and Embase repositories. Persistent web links of each study included in the datasets are provided in the “Reference” part.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kangkang Liu and Lining Wang contributed to the work equally and should be regarded as co-first authors.
Contributor Information
Kangkang Liu, Email: tjykdxlkk@163.com.
Lining Wang, Email: tjykdxwln@163.com.
Erlin Sun, Phone: +86-13032259605, Email: drelsun@163.com.
References
- 1.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. 2016;16(5):279–294. doi: 10.1038/nri.2016.40. [DOI] [PubMed] [Google Scholar]
- 2.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. MicroRNAs in cancer management. Lancet Oncol. 2012;13(6):e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 3.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Lu L, Mao X, Shi P, He B, Xu K, Zhang S, Wang J. MicroRNAs in the prognosis of triple-negative breast cancer: a systematic review and meta-analysis. Medicine. 2017;96(22):e7085. doi: 10.1097/MD.0000000000007085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teoh SL, Das S. The role of MicroRNAs in diagnosis, prognosis, metastasis and resistant cases in breast Cancer. Curr Pharm Des. 2017;23(12):1845–1859. doi: 10.2174/1381612822666161027120043. [DOI] [PubMed] [Google Scholar]
- 6.Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. 2017;172:34–49. doi: 10.1016/j.pharmthera.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 7.Di Martino MT, Rossi M, Caracciolo D, Gulla A, Tagliaferri P, Tassone P. Mir-221/222 are promising targets for innovative anticancer therapy. Expert Opin Ther Targets. 2016;20(9):1099–1108. doi: 10.1517/14728222.2016.1164693. [DOI] [PubMed] [Google Scholar]
- 8.Xie D, Yuan P, Wang D, Jin H, Chen H. Expression and prognostic significance of miR-375 and miR-221 in liver cancer. Oncol Lett. 2017;14(2):2305–2309. doi: 10.3892/ol.2017.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussein S, Mosaad H, Rashed HE, El-Anwar MW. Up-regulated miR-221 expression as a molecular diagnostic marker in laryngeal squamous cell carcinoma and its correlation with Apaf-1 expression. Cancer Biomark. 2017;19(3):279–287. doi: 10.3233/CBM-160444. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Zhao Y, Sun S, Liu Z, Zhang Y, Jiao S. Overexpression of MicroRNA-221 is associated with poor prognosis in non-small cell lung cancer patients. Tumour Biol. 2016;37(8):10155–10160. doi: 10.1007/s13277-015-4662-x. [DOI] [PubMed] [Google Scholar]
- 11.Wu Q, Ren X, Zhang Y, Fu X, Li Y, Peng Y, Xiao Q, Li T, Ouyang C, Hu Y, et al. MiR-221-3p targets ARF4 and inhibits the proliferation and migration of epithelial ovarian cancer cells. Biochem Biophys Res Commun. 2018;497(4):1162–1170. doi: 10.1016/j.bbrc.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Vergho DC, Kneitz S, Kalogirou C, Burger M, Krebs M, Rosenwald A, Spahn M, Loser A, Kocot A, Riedmiller H, et al. Impact of miR-21, miR-126 and miR-221 as prognostic factors of clear cell renal cell carcinoma with tumor thrombus of the inferior vena cava. PLoS One. 2014;9(10):e109877. doi: 10.1371/journal.pone.0109877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu K, Zhao K, Wang L, Sun E. Prognostic value of microRNA-155 in human carcinomas: An updated meta-analysis. Clin Chim Acta. 2018;479:171–180. doi: 10.1016/j.cca.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 14.Hong F, Li Y, Xu Y, Zhu L. Prognostic significance of serum microRNA-221 expression in human epithelial ovarian cancer. J Int Med Res. 2013;41(1):64–71. doi: 10.1177/0300060513475759. [DOI] [PubMed] [Google Scholar]
- 15.Tsikrika FD, Avgeris M, Levis PK, Tokas T, Stravodimos K. Scorilas a: miR-221/222 cluster expression improves clinical stratification of non-muscle invasive bladder cancer (TaT1) patients’ risk for short-term relapse and progression. Genes Chromosomes Cancer. 2018;57(3):150–161. doi: 10.1002/gcc.22516. [DOI] [PubMed] [Google Scholar]
- 16.Nakka M, Allen-Rhoades W, Li Y, Kelly AJ, Shen J, Taylor AM, Barkauskas DA, Yustein JT, Andrulis IL, Wunder JS, et al. Biomarker significance of plasma and tumor miR-21, miR-221, and miR-106a in osteosarcoma. Oncotarget. 2017;8(57):96738–96752. doi: 10.18632/oncotarget.18236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, Zhang Y, Zhang X, Zhang M, Liu H, Zhang S, Qi B, Sun X. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed Pharmacother. 2015;75:153–158. doi: 10.1016/j.biopha.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Chen F, Li XF, Fu DS, Huang JG, Yang SE. Clinical potential of miRNA-221 as a novel prognostic biomarker for hepatocellular carcinoma. Cancer Biomark. 2017;18(2):209–214. doi: 10.3233/CBM-161671. [DOI] [PubMed] [Google Scholar]
- 19.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52(4):297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406(1):70–73. doi: 10.1016/j.bbrc.2011.01.111. [DOI] [PubMed] [Google Scholar]
- 21.Yoon SO, Chun SM, Han EH, Choi J, Jang SJ, Koh SA, Hwang S, Yu E. Deregulated expression of microRNA-221 with the potential for prognostic biomarkers in surgically resected hepatocellular carcinoma. Hum Pathol. 2011;42(10):1391–1400. doi: 10.1016/j.humpath.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 22.Gyongyosi B, Vegh E, Jaray B, Szekely E, Fassan M, Bodoky G, Schaff Z, Kiss A. Pretreatment MicroRNA level and outcome in Sorafenib-treated hepatocellular carcinoma. J Histochem Cytochem. 2014;62(8):547–555. doi: 10.1369/0022155414537277. [DOI] [PubMed] [Google Scholar]
- 23.Dai L, Wang Y, Chen L, Zheng J, Li J, Wu X. MiR-221, a potential prognostic biomarker for recurrence in papillary thyroid cancer. World J Surg Oncol. 2017;15(1):11. doi: 10.1186/s12957-016-1086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv J, Xu L, Xu Y, Qiu M, Yang X, Wang J, Yin R, Xu L. Expression of MiRNA-221 in non-small cell lung cancer tissues and correlation with prognosis. Zhongguo fei ai za zhi Chin J Lung Cancer. 2014;17(3):221–225. doi: 10.3779/j.issn.1009-3419.2014.03.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai K, Shen F, Cui JH, Yu Y, Pan HQ. Expression of miR-221 in colon cancer correlates with prognosis. Int J Clin Exp Med. 2015;8(2):2794–2798. [PMC free article] [PubMed] [Google Scholar]
- 26.Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H, Yu H. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6(4):391–401. [PMC free article] [PubMed] [Google Scholar]
- 27.Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25(10):1674–1680. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu K, Li G, Fan C, Diao Y, Wu B, Li J. Increased expression of MicroRNA-221 in gastric cancer and its clinical significance. J Int Med Res. 2012;40(2):467–474. doi: 10.1177/147323001204000208. [DOI] [PubMed] [Google Scholar]
- 29.Deng L, Lei Q, Wang Y, Wang Z, Xie G, Zhong X, Wang Y, Chen N, Qiu Y, Pu T, et al. Downregulation of miR-221-3p and upregulation of its target gene PARP1 are prognostic biomarkers for triple negative breast cancer patients and associated with poor prognosis. Oncotarget. 2017;8(65):108712–108725. doi: 10.18632/oncotarget.21561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falkenberg N, Anastasov N, Rappl K, Braselmann H, Auer G, Walch A, Huber M, Hofig I, Schmitt M, Hofler H, et al. MiR-221/−222 differentiate prognostic groups in advanced breast cancers and influence cell invasion. Br J Cancer. 2013;109(10):2714–2723. doi: 10.1038/bjc.2013.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanna JA, Wimberly H, Kumar S, Slack F, Agarwal S, Rimm DL. Quantitative analysis of microRNAs in tissue microarrays by in situ hybridization. BioTechniques. 2012;52(4):235–245. doi: 10.2144/000113837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao R, Wu J, Jia W, Gong C, Yu F, Ren Z, Chen K, He J, Su F. Plasma miR-221 as a predictive biomarker for chemoresistance in breast cancer patients who previously received neoadjuvant chemotherapy. Onkologie. 2011;34(12):675–680. doi: 10.1159/000334552. [DOI] [PubMed] [Google Scholar]
- 33.Amankwah EK, Anegbe E, Park H, Pow-Sang J, Hakam A, Park JY. MiR-21, miR-221 and miR-222 expression and prostate cancer recurrence among obese and non-obese cases. Asian J Androl. 2013;15(2):226–230. doi: 10.1038/aja.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang SG, Ha YR, Kim SJ, Kang SH, Park HS, Lee JG, Cheon J, Kim CH. Do microRNA 96, 145 and 221 expressions really aid in the prognosis of prostate carcinoma? Asian J Androl. 2012;14(5):752–757. doi: 10.1038/aja.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spahn M, Kneitz S, Scholz CJ, Stenger N, Rudiger T, Strobel P, Riedmiller H, Kneitz B. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int J Cancer. 2010;127(2):394–403. doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- 36.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126(5):1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 37.Li P, He QY, Luo CQ, Qian LY. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med Sci Monit. 2014;20:2472–2477. doi: 10.12659/MSM.891327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gimenes-Teixeira HL, Lucena-Araujo AR, Dos Santos GA, Zanette DL, Scheucher PS, Oliveira LC, Dalmazzo LF, Silva-Junior WA, Falcao RP, Rego EM. Increased expression of miR-221 is associated with shorter overall survival in T-cell acute lymphoid leukemia. Exp Hematol Oncol. 2013;2(1):10. doi: 10.1186/2162-3619-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Li Z, He C, Wang D, Yuan X, Chen J, Jin J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44(3):191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo HQ, Huang GL, Guo CC, Pu XX, Lin TY. Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Dis Markers. 2010;29(5):251–258. doi: 10.1155/2010/474692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rong MH, Dang YW, Chen G. Lack of significant association between plasma/serum miR-221 expression and poor survival of carcinoma: a meta-analysis. TheScientificWorldJournal. 2013;2013:394030. doi: 10.1155/2013/394030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14(13):1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 43.Mavrakis KJ, Leslie CS, Wendel HG. Cooperative control of tumor suppressor genes by a network of oncogenic microRNAs. Cell Cycle. 2011;10(17):2845–2849. doi: 10.4161/cc.10.17.16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao N, Ma G, Zhang J, Zhu W. MiR-221-5p enhances cell proliferation and metastasis through post-transcriptional regulation of SOCS1 in human prostate cancer. BMC Urol. 2018;18(1):14. doi: 10.1186/s12894-018-0325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Lu Y, Yu L, Han X, Wang H, Mao J, Shen J, Wang B, Tang J, Li C, et al. MiR-221/222 promote cancer stem-like cell properties and tumor growth of breast cancer via targeting PTEN and sustained Akt/NF-kappaB/COX-2 activation. Chem Biol Interact. 2017;277:33–42. doi: 10.1016/j.cbi.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Chang JK, Hou JQ, Zhao ZH, Zhang LD. Inhibition of miR-221 influences bladder cancer cell proliferation and apoptosis. Eur Rev Med Pharmacol Sci. 2017;21(14):3193–3199. [PubMed] [Google Scholar]
- 47.Li B, Lu Y, Wang H, Han X, Mao J, Li J, Yu L, Wang B, Fan S, Yu X, et al. MiR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed Pharmacother. 2016;79:93–101. doi: 10.1016/j.biopha.2016.01.045. [DOI] [PubMed] [Google Scholar]
- 48.Pan Y, Li J, Zhang Y, Wang N, Liang H, Liu Y, Zhang CY, Zen K, Gu H. Slug-upregulated miR-221 promotes breast cancer progression through suppressing E-cadherin expression. Sci Rep. 2016;6:25798. doi: 10.1038/srep25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie J, Wen JT, Xue XJ, Zhang KP, Wang XZ, Cheng HH. MiR-221 inhibits proliferation of pancreatic cancer cells via down regulation of SOCS3. Eur Rev Med Pharmacol Sci. 2018;22(7):1914–1921. doi: 10.26355/eurrev_201804_14714. [DOI] [PubMed] [Google Scholar]
- 50.Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S, Philip PA, Li Y. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res. 2013;3(5):465–477. [PMC free article] [PubMed] [Google Scholar]
- 51.Yang W, Yang Y, Xia L, Yang Y, Wang F, Song M, Chen X, Liu J, Song Y, Zhao Y, et al. MiR-221 promotes Capan-2 pancreatic ductal adenocarcinoma cells proliferation by targeting PTEN-Akt. Cell Physiol Biochem. 2016;38(6):2366–2374. doi: 10.1159/000445589. [DOI] [PubMed] [Google Scholar]
- 52.Frampton AE, Krell J, Jamieson NB, Gall TM, Giovannetti E, Funel N, Mato Prado M, Krell D, Habib NA, Castellano L, et al. MicroRNAs with prognostic significance in pancreatic ductal adenocarcinoma: a meta-analysis. Eur J Cancer. 2015;51(11):1389–1404. doi: 10.1016/j.ejca.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 53.Yang J, Zhang JY, Chen J, Xu Y, Song NH, Yin CJ. Prognostic role of microRNA-221 in various human malignant neoplasms: a meta-analysis of 20 related studies. PLoS One. 2014;9(1):e87606. doi: 10.1371/journal.pone.0087606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang P, Zhang M, Han R, Zhang K, Ding H, Liang C, Zhang L. The correlation between microRNA-221/222 cluster overexpression and malignancy: an updated meta-analysis including 2693 patients. Cancer Manag Res. 2018;10:3371–3381. doi: 10.2147/CMAR.S171303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei T, Ye P, Peng X, Wu LL, Yu GY. Prognostic value of miR-222 in various cancers: a systematic review and meta-analysis. Clin Lab. 2016;62(8):1387–1395. doi: 10.7754/Clin.Lab.2016.160102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Subgroup analyses of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in OS by nations, (A) Chinses (B) non-Asian (TIF 24678 kb)
Figure S2. Subgroup analyses of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in OS by the number of individuals, (A) > 100 (B) ≤100 (TIF 24678 kb)
Figure S3. Subgroup analyses of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in OS by cancers, (A) Colon cancer (B) Live cancer (C) Lung cancer (TIF 15382 kb)
Figure S4. Subgroup analyses of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in OS by cancers, (A) ALL (B) Breast cancer (C) Osteosarcoma (D) Ovarian cancer (DOCX 19 kb) (TIF 14956 kb)
Figure S5. Subgroup analyses of the studies that evaluated the HRs of high miR-221 expression as compared to low expression in OS by origins, (A) Tumor tissues (B) Serum/plasm (C) Marrow (TIF 24676 kb)
Table S1. Literature search strategy of PubMed database. The Embase database is searched in a similar way to PubMed. (DOCX 19 kb)
Data Availability Statement
The datasets analyzed during the current study are available in the PubMed and Embase repositories. Persistent web links of each study included in the datasets are provided in the “Reference” part.