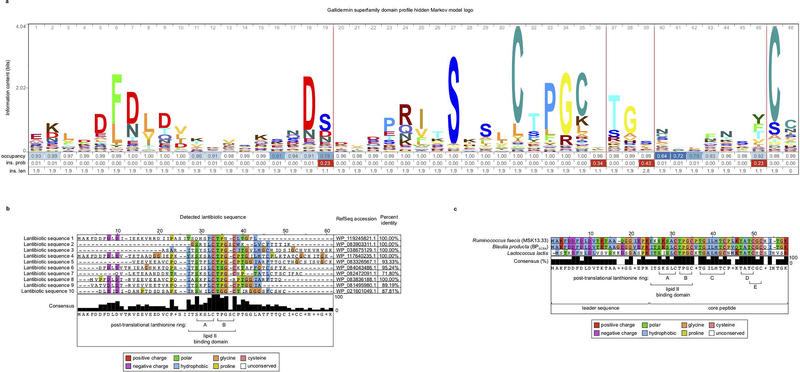
Extended Data Figure 9 |. Identification of lantibiotic sequences from metagenomic sequencing of healthy human fecal samples.
a, the profile hidden Markov model used to identify the gallidermin superfamily domain, illustrated as a logo. b, Multiple sequence alignment of lantibiotic precursor sequences identified from shotgun sequencing of healthy-donor fecal samples. Detected lantibiotic sequences are the assembly of lantibiotic reads from shotgun metagenomic fecal samples. c, 421 species were individually isolated from healthy human fecal samples, whole genome sequenced, assembled, annotated, and mined for lantibiotic precursor sequences to identify a strain of Ruminococcus faecis encoding a homologous lantibiotic. The precursor lantibiotic sequence is compared to the sequences of BPSCSK LanA1–4 lantibiotic and nisin-A by multiple alignment.