Abstract
Introduction:
Insufficient patient enrollment in clinical trials remains a serious and costly problem and is often considered the most critical issue to solve for the clinical trials community.
In this project, we assessed the feasibility of automatically detecting a patient’s eligibility for a sample of breast cancer clinical trials by mapping coded clinical trial eligibility criteria to the corresponding clinical information automatically extracted from text in the EHR.
Methods:
Three open breast cancer clinical trials were selected by oncologists. Their eligibility criteria were manually abstracted from trial descriptions using the OHDSI ATLAS web application. Patients enrolled or screened for these trials were selected as ‘positive’ or ‘possible’ cases. Other patients diagnosed with breast cancer were selected as ‘negative’ cases. A selection of the clinical data and all clinical notes of these 229 selected patients was extracted from the MUSC clinical data warehouse and stored in a database implementing the OMOP common data model. Eligibility criteria were extracted from clinical notes using either manually crafted pattern matching (regular expressions) or a new natural language processing (NLP) application. These extracted criteria were then compared with reference criteria from trial descriptions. This comparison was realized with three different versions of a new application: rule-based, cosine similarity-based, and machine learning-based.
Results:
For eligibility criteria extraction from clinical notes, the machine learning-based NLP application allowed for the highest accuracy with a micro-averaged recall of 90.9% and precision of 89.7%. For trial eligibility determination, the highest accuracy was reached by the machine learning-based approach with a per-trial AUC between 75.5% and 89.8%.
Conclusion:
NLP can be used to extract eligibility criteria from EHR clinical notes and automatically discover patients possibly eligible for a clinical trial with good accuracy, which could be leveraged to reduce the workload of humans screening patients for trials.
Keywords: Clinical trial, Natural Language Processing, Eligibility criteria, Machine learning
1. INTRODUCTION
Insufficient patient enrollment in clinical trials remains a serious and costly problem and is often considered the most critical issue to solve for the clinical trials community.[1] The majority of patients are never offered an opportunity to enroll in clinical trials (enrollment levels reach only 3% in oncology[2]). The participation of physicians is essential for successful patient enrollment and lack of awareness of trials is often cited as a reason for low enrollment levels.[3] One potential barrier to this awareness is the difficulty in correlating eligibility criteria with patient characteristics in an efficient manner. Eligibility criteria specify the characteristics of study participants and provide a checklist for screening and recruiting participants. They are essential to every clinical research study. Patient eligibility screening is typically a cumbersome and lengthy manual process, ranging from about 10 minutes for criteria with minimal complexity to more than 2 hours for highly complex eligibility criteria.[4]
The adoption of Electronic Health Record (EHR) systems is growing at a fast pace in the U.S. This growth results in very large quantities of patient clinical data becoming available in electronic format. Secondary use of clinical data is essential to fulfill the potential for effective scientific research, high quality healthcare, and improved healthcare management. Using this electronic patient data to automate screening for clinical trials eligibility has demonstrated significant gains in efficiency and accuracy, allowing for a much faster process with higher recall.[4–7] Several barriers remain to fully automate the discovery of patients eligible for clinical trials. Most clinical information required to assess eligibility is recorded in EHR systems, in unstructured narrative text. Clinical trial descriptions of eligibility criteria are also typically only available in narrative text. This format requires computable representations of eligibility criteria and methods based on natural language processing (NLP) to automate the extraction of eligibility criteria from trial descriptions or clinical notes, adding breadth and depth to the limited coded data available in typical EHRs (i.e., diagnostic and procedure codes).
Our hypothesis for this study is that an automated process based on NLP can detect patients eligible for a specific clinical trial by linking the information extracted from the narrative trial description to the corresponding EHR and alerting clinicians caring for the patient.
1.1. Related work:
The automated extraction of eligibility criteria is closely related to clinical information extraction (cf. [8,9] for reviews) and EHR phenotyping (i.e., determining if a patient has a medical condition or a risk for one; cf. [10]) by systems such as DeepPhe [11] or the “learning with Anchors” approach.[12] Significant efforts and success have been reported in the automated extraction and representation of eligibility criteria from trial descriptions [13–16] but only limited research has been reported on the extraction of eligibility criteria from clinical text. Rare examples focused only on pediatric oncology patient pre-screening [17] or finding sentences likely to contain eligibility information.[18]
2. MATERIALS AND METHODS
2.1. Study setting, patient population, and data set:
This study, approved by the Medical University of South Carolina (MUSC) Institutional Review Board (i.e., ethics committee), includes patients treated at the Hollings Cancer Center (Charleston, SC) and a selection of three breast cancer clinical trials open at this institution at the time of the study and involving two investigators in this project (Drs. Aruch and Britten). A cohort of 229 patients diagnosed with breast cancer who were treated at MUSC between 2015 and 2017 were selected in three categories and considered “positive” (i.e., enrolled in a selected trial), “possible” (i.e., screened but failed to be enrolled often for not meeting further screening criteria), or “negative” (i.e., diagnosed with breast cancer, but neither screened nor enrolled) (Table 1).
Table 1.
Population categories characteristics
| Positive cases | Possible cases | Negative cases | |
|---|---|---|---|
| Patient count | 25 | 4 | 200 |
| Note count | 2470 | 511 | 12143 |
| Average age (y) | 60.01 | 62.68 | 64.58 |
| Gender (% female) | 100% | 100% | 100% |
All clinical text notes (15,124) and a selection of structured clinical data were examined for each patient. A subset of eligibility criteria was selected for each trial and included demographics, cancer staging, biomarkers (e.g., estrogen receptors, progesterone receptors), functional status and menopausal status (Table 2).
Table 2:
Eligibility criteria selected for each clinical trial
| [19] | [20] | [21] |
|---|---|---|
| Female | Female or Male | Female |
| Age 18 years or older | Age 18 years or older | Age 18 years or older |
| ECOG performance status 0–1 | ECOG performance status 0–1 | ECOG performance status 0–2 |
| Breast cancer | Breast cancer | Postmenopausal |
| AJCC clinical stage T1-T3 | AJCC clinical stage T1-T3 | Breast cancer |
| AJCC clinical stage N1 | AJCC clinical stage N1 | AJCC clinical stage T2-T4 |
| AJCC clinical stage M0 | AJCC clinical stage M0 | AJCC clinical stage M0 |
| Estrogen receptor positive | ||
| Allred score 6–8 | ||
| Human Epidermal Growth Factor Receptor 2 (HER2) negative |
2.2. Reference standard development:
2.2.1. Reference standard for eligibility criteria extraction
To train and then evaluate the accuracy of the NLP-based extraction of eligibility criteria, a random selection of 138 clinical notes was extracted from the aforementioned collection of text notes to be manually annotated by an oncology attending physician and a medical resident. The sample size was based on a power analysis allowing for descriptive statistics (averaging 0.9) with confidence intervals of ±0.05.
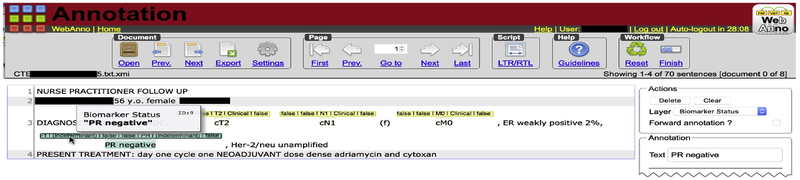
We developed the guideline for the text annotation task over a series of interviews with the oncologist (included as Appendix A). The WebAnno annotation tool [22] was used for the annotation task (Fig. 1). WebAnno allows a domain expert to digitally annotate spans of text within a document (i.e., clinical notes in our case) with attributes and concepts from terminologies. For instance, the string ‘cT1N0M0’ would be annotated with the concept “Clinical Tumor Stage T1 [UMLS CUI C0475372]”.
Figure 1:
WebAnno annotation tool (partial screenshot).
Both oncologist and resident annotated six of these notes for a joint accuracy of 0.61 and an F1-measure of 0.76. The medical resident had an accuracy of 0.88 and an F1-measure of 0.94 when compared with the final reference of these six double-annotated documents after annotation disagreements adjudication. The remaining notes were annotated by the medical resident only. All 138 documents were treated as a reference standard for evaluating the concept extraction of the NLP systems. The remaining 14,986 notes were used for training and development of the NLP systems.
2.2.2. Reference standard for patient-trial classification
Three breast cancer clinical trials open at the MUSC Hollings Cancer Center (HCC) were selected (i.e., [19], [20], [21]) in close collaboration with the oncology experts team. The reference standard for patient-trial classification was derived from the original clinical enrollment status of the patients. As mentioned above (2.1), each patient was associated with one of three possible eligibility statuses for each trial. If enrolled in a selected trial, the patient was considered ‘positive’; if screened but not enrolled, the patient was considered ‘possible’; and if diagnosed with breast cancer but neither screened nor enrolled, the patient was considered ‘negative’ (cf. resulting counts in Table 3). Three patients were positive or possible for more than one trial because of eligibility criteria shared between trials. All patients in Table 3 were negative for at least one trial; 200 patients were negative for all three trials.
Table 3:
Study population characteristics
| Positive cases | Possible cases | Negative cases | |
|---|---|---|---|
| Patients | 13 | 1 | 215 |
| Patients | 5 | 2 | 222 |
| Patients | 9 | 2 | 218 |
2.3. Eligibility criteria extraction:
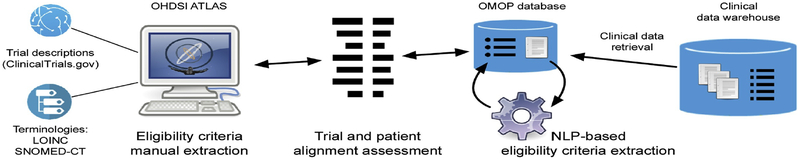
The overall process of automated clinical trial eligibility surveillance consists of three major sub-processes: eligibility criteria extraction from clinical trial descriptions, eligibility criteria extraction from the EHR, and ‘alignment’ of the two sets of eligibility criteria to discover patients eligible for a specific clinical trial (Figure 2).
Figure 2:
Clinical trial eligibility automatic surveillance process
As computable representation of eligibility criteria, we selected a standard data model proposed by the Observational Medical Outcomes Partnership (OMOP) consortium, as experimented by Si and Weng.[23] The OMOP Common Data Model (CDM v5.2) was implemented in a research database, along with query and analysis tools (i.e., OHDSI ATLAS[24]). These tools were then used for manual definition of the selected trial eligibility criteria by an oncology expert. This definition resulted in the creation of sets of criteria defining a “cohort” for each trial. The cohort definitions could be used within ATLAS to query for cohort size and cohort attrition (that is, how much of the population is excluded by each additional criterion). The definition could also be exported as a SQL database query for use with external tools.
On the trial ‘side’, a selection of key eligibility criteria (Table 2) for each selected trial were retrieved from ClinicalTrials.gov [25] trial descriptions and manually abstracted by domain experts with ATLAS allowing us to represent eligibility criteria in a structured and coded form. ATLAS uses the OMOP CDM with a selection of standard terminologies for representing these criteria. The ability to use multiple standard terminologies allowed us to, for example, represent ECOG Performance Status as a LOINC [26] concept (i.e., “ECOG performance status grade Observed”) and Estrogen Receptor positive as a SNOMED-CT [27] concept (i.e., “Estrogen receptor positive tumor”) and yet still reason over these concepts in the same interface. The OMOP CDM database was then loaded with a selection of structured clinical data (patient identifier, gender, date of birth, height, weight, diagnostic codes, procedure codes) and clinical notes from our study population. Clinical notes were stored in the NOTES table.
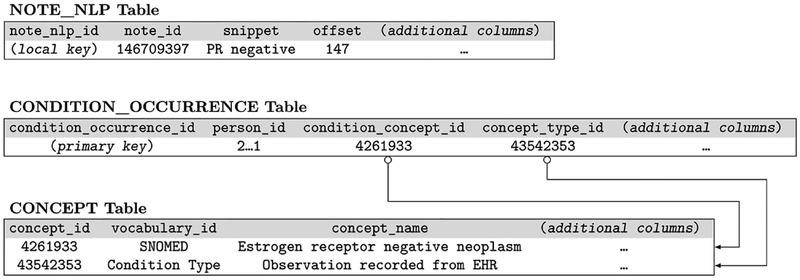
On the EHR ‘side’, a new NLP-based modular software application has been developed within the enterprise-grade Apache UIMA framework.[28] At a very high level, this application 1) retrieves clinical notes from the OMOP CDM database, 2) extracts all mentions of eligibility criteria in said clinical notes, and 3) posts the extracted concepts back into the appropriate structured and coded columns of the OMOP CDM database to create a recent and accurate set of values for these criteria, as per Figure 3. Two variations of this application were developed in tandem to explore the relative performance of a rule-based system and a machine learning system.
Figure 3:
Selected rows for representing a mention of “PR negative” in an OMOP CDM schema.
The rule-based system uses regular expressions to extract mentions of eligibility criteria. The regular expressions (provided in Appendix B with an example) were developed through iterated interviews with an oncology expert about how concepts are mentioned in clinical notes and evaluating output from the training corpus.
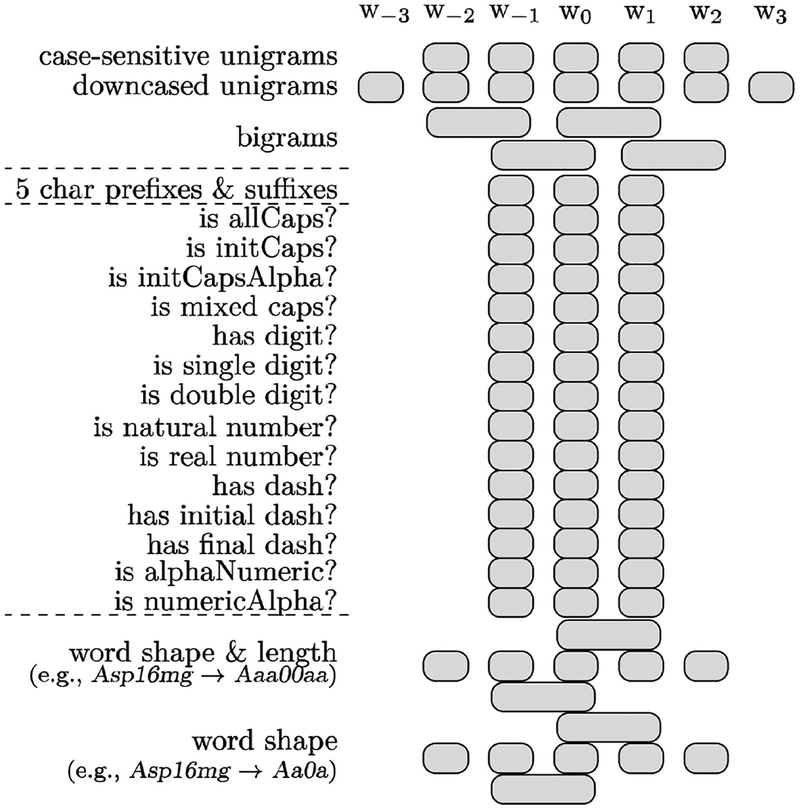
The machine learning-based system has been implemented as a named entity recognition (NER) task based on sequential token-based labeling using a support vector machine (SVM) model trained with lexical features (Fig. 4). The SVM does not require sentence annotations as input and can make independent labeling decisions for each word, which isolates it from sentence boundary errors. In our previous studies involving medical concept extraction,[29] we observed that SVMs allowed for higher recall than other structured learning algorithms including CRF (conditional random fields [30]).
Figure 4:
Features for SVM-based eligibility criteria extraction
Our sequential model found and interpreted mentions of each eligibility criterion and combined them at the patient level. We trained the SVM classifier with a linear kernel for multi-class classification using the LIBLINEAR software package.[31] A tokenizer pre-processed the text by splitting it into word tokens every time a whitespace character was encountered. Then, each word was further divided if a mixture of lowercase letters, uppercase letters, numbers, or other characters were used. We reformatted the training data with BIO tags (B: at the beginning, I: inside, or O: outside of a concept). Figure 4 shows the feature set used with the SVM model. For instance, word features relied on the current word (w0), previous words (w−1, w−2, w−3) and following words (w1, w2, w3). For orthographic features, regular expressions-based features for w0, w−1, w1 were defined. We performed 10-fold cross validation to tune the LIBLINEAR’s parameters. To emphasize recall, we halved the weight of negative examples (i.e., words with O tags).
2.4. Eligibility classification (patient eligibility determination):
To determine the eligibility of a patient for a clinical trial, we assessed the ‘alignment’ of eligibility criteria represented in a structured and coded form on both ‘sides.’ Three different approaches have been implemented and compared: rule-based, cosine similarity-based, and SVM-based.
The first approach uses rules implemented as database queries to align patients with trials. The eligibility criteria used were extracted using the rule-based system described above. The database queries were exported from ATLAS and then applied in a database management tool. They were used to determine how many individual criteria a patient met for a given trial out of all possible criteria. The maximum score (e.g., 8 if there were 8 criteria or 5 if there were only 5 criteria) means all criteria are met. A score of zero meant no criteria were met.
For the cosine and SVM-based classification approaches, all eligibility criteria used were extracted using the SVM-based NER method.
The cosine similarity-based approach was used to compare patient and trial data represented in vectors of eligibility criteria. Each criterion was represented as a component of the vector. The similarity between the two vectors was calculated by the cosine between instance vectors. The more concepts shared between a patient’s concept vector and a trial description vector, the closer they would be in our multi-dimensional space.
The third approach used an SVM classifier to determine the strength of association between a patient (again, represented as a vector of binary features with “true” values meaning the patient meets a particular criterion) and a clinical trial (likewise represented as a vector of binary features). Similar to eligibility criteria extraction, we used the LIBLINEAR implementation of SVMs, with default parameter values (except negative examples weight set to 0.1). The stronger the association, the more likely a patient was eligible for a particular trial. Unlike the cosine method, which uses only the corresponding criteria on each side, the SVM model considers all possible criteria combinations. When the patient side has m criteria and the clinical trial has n criteria, (m times n) more features are additionally defined to store their co-occurrence information.
3. RESULTS
3.1. Evaluation metrics:
For eligibility criteria extraction, accuracy metrics were based on counts of each annotation as true positive (system output matches the reference standard), false positive (output without match), and false negative (reference standard annotation not in the output). Comparisons were done as partial matches (reference standard annotation and system output overlap with the same information category). We then computed recall (i.e., sensitivity), precision (i.e., positive predictive value), and the F1-measure (a harmonic mean of recall and precision).[32] Each metric was micro-averaged across each mention in clinical notes (i.e., calculated from a confusion matrix combining all mentions in the corpus).
For trial eligibility classification, we used the mean average precision (MAP) and area under the receiver operating characteristic curve (AUC, which approximates the likelihood a model ranks a random patient with the correct trial with a higher score than incorrect trial).
3.2. Eligibility criteria extraction:
The accuracy of each version of the NLP application was evaluated using the reference standard of 138 manually annotated clinical notes. As seen in Table 4, the rule-based version reached 84.6% recall and 64.4% precision. The SVM-based version reached 90.9% recall and 89.7% precision. Table 5 includes a finer-grained evaluation of the NLP application accuracy.
Table 4:
NLP application accuracy for eligibility criteria information extraction
| Rule-based version | SVM-based version | |||
|---|---|---|---|---|
| Criterion | Recall (%) | Precision (%) | Recall (%) | Precision (%) |
| ECOG status | 94.2 | 94.2 | 92.8 | 100.0 |
| Estrogen receptor | 91.5 | 53.4 | 86.9 | 90.9 |
| Progesterone receptor | 95.6 | 69.7 | 94.1 | 86.7 |
| Human Epidermal Growth Factor Receptor 2 | 63.0 | 36.4 | 87.8 | 82.0 |
| Postmenopausal | 88.0 | 72.1 | 86.9 | 84.1 |
| AJCC tumor stage (T) | 84.7 | 83.8 | 95.5 | 97.9 |
| AJCC nodes stage (N) | 79.6 | 82.6 | 72.7 | 95.7 |
| AJCC metastasis stage (M) | 94.5 | 37.7 | 87.5 | 87.4 |
| Overall (micro-average) | 84.6 | 64.4 | 90.9 | 89.7 |
Table 5:
Fine-grained NLP application accuracy for eligibility criteria information extraction
| Rule-based version | SVM-based version | |||
|---|---|---|---|---|
| Criterion | Recall (%) | Precision (%) | Recall (%) | Precision (%) |
| ECOG 0 | 96.9 | 96.9 | 100.0 | 100.0 |
| ECOG 1* | 85.7 | 85.7 | 85.7 | 100.0 |
| ER+ | 61.7 | 91.0 | 91.3 | 86.8 |
| ER− | 32.7 | 93.8 | 82.6 | 95.0 |
| PR+ | 80.0 | 96.8 | 96.0 | 86.6 |
| PR− | 57.5 | 90.2 | 92.1 | 86.8 |
| HER2+ | 24.0 | 100.0 | 85.0 | 89.5 |
| HER− | 31.0 | 98.0 | 90.5 | 74.5 |
| Postmenopausal | 72.1 | 88.0 | 86.89 | 84.1 |
| Not Postmenopausal** | - | - | 46.2 | 100.0 |
| T0 | 100.0 | 100.0 | 100.0 | 100.0 |
| T1 | 89.1 | 87.5 | 96.4 | 93.0 |
| T2 | 79.5 | 81.5 | 97.6 | 98.8 |
| T3* | 82.4 | 90.3 | 88.2 | 100.0 |
| N0 | 79.5 | 85.3 | 91.8 | 97.1 |
| N1 | 84.8 | 78.5 | 99.0 | 100.0 |
| N2 | 77.8 | 58.3 | 100.0 | 90.0 |
| N3 | 100.0 | 75.0 | 0.00 | 0.00 |
| M0 | 100.0 | 98.0 | 100.0 | 96.2 |
| M1 | 2.3 | 50.0 | 90.5 | 78.6 |
No positive instances occurred for ECOG 2–5 or T4
The rule-based system did not extract non-postmenopausal mentions.
3.3. Eligibility classification:
With the mentions extracted from text notes, we applied the three classification methods to determine the eligibility of a patient for each clinical trial. We treated “possible” cases as “positive” cases, which enabled our methods to assign binary labels to each patient/trial combination. Table 6 shows the classification performance results. For each evaluation metric, the better result appears in boldface. To calculate MAP scores, for each clinical trial, we sorted the test cases (patients) by the total number of matching criteria in the query classification, the cosine similarity score in the cosine classification method, and the probability score in the binary SVM classification. The SVM-based classification method outperformed both other classification methods. The MAP of the SVM classification method was 35.2%, which roughly means that we can obtain the relevant samples if less than three times the number of patients is selected. The highest AUC was obtained with the SVM-based eligibility classification version.
Table 6:
Patient trial eligibility classification accuracy
| Query (terms, rules) (%) | Cosine similarity (%) | SVM (%) | |
|---|---|---|---|
| Recall | 100.0, 100.0, 35.7 (78.6) | 57.1, 54.6, 100.0 (70.6) | 100.0, 72.7, 64.3 (79.0) |
| MAP | 10.4, 16.9. 29.8 (19.0) | 6.8, 22.5, 24.6 (18.0) | 37.6, 41.7, 26.2 (35.2) |
| AUC | 83.7 | 75.5 | 89.8 |
4. DISCUSSION
4.1. Results discussion:
The automated detection of patients eligible for the three selected clinical trials allowed for moderate to very good accuracy, depending on the criteria complexity and methods used. In general, SVM and cosine similarity approaches allowed for lower recall than direct database query with “simple” sets of eligibility criteria but allowed for 2–3 times higher recall with more complex sets of criteria. Among patients detected, 2–3 times more were correct with SVM classification than with direct database queries. When balancing recall and precision as in the AUC, the SVM allowed for the highest average accuracy.
Concepts were not all equally covered by the regular expression patterns in the rule-based version of the eligibility criteria extraction. For instance, ‘M1’ (distant metastases) had more ambiguous false positives in simple regular expression patterns than ‘M0’ (no distant metastases). However, the positive presence of M1 was not a criterion important for our three trials while M0 was important. Thus, many of our AJCC metastasis stage (M) precision related errors can be attributed to prioritizing M0 extraction (with a final precision of 98.0%) over M1 extraction (with a final precision of 50%). For the clinical notes corpus as a whole, this strategy reduced performance. For the task at hand, this strategy improved performance. Further regular expression refinement will need to include balancing the performance of these sub-concepts within the larger set of concepts. Our regular expression patterns targeting HER2- suffered from poor recall (31%) despite having high precision (97.6%). Interestingly, this concept appears in clinical notes in a much wider range of formats than any of the other biomarker related concepts, including HER2+. The attempted extraction of M1 and HER2- accounted for approximately a third of all concept extraction errors. Most other concepts extracted by the rule-based system had an F1-measure above 70%.
Among false positive errors produced by the SVM-based NER model, many were caused by mismatches of category types because the mentions in text overlapped with the reference standard. More than half of the errors fell into this case. If ignoring categories, recall would increase to 95.3% and precision to 94.1%. We observed that many false negative errors were due to the use of abbreviations, especially when rarely mentioned in the training data (e.g., “PS” (performance status), “TN” (triple negative), and “LMP” (postmenopausal1)).
4.2. Study limitations:
Limitations to consider include the small sample size, for both the patient eligibility determination and the extraction for eligibility criteria from clinical text. This small sample size probably did not allow for demonstrating the higher potential accuracy machine learning could offer. Another important limitation is the selection of only a subset of the eligibility criteria listed for each trial selected. This partial selection probably prevented higher MAP (approximating positive predictive value).
No temporal information related to eligibility criteria was used in this pilot study. This simplification probably resulted in a higher false positive rate on patients that were not eligible for a trial because of a mismatch between their cancer onset and the trial dates.
5. CONCLUSION
Insufficient clinical trial enrollment is a critical issue that has been addressed with various strategies but never with automated processing of unstructured clinical and trial data, as envisioned in this pilot project. Eligibility criteria have been automatically extracted from trial descriptions [13–16] but not from unstructured EHR data. Our objectives after this pilot study will be to integrate the automated extraction of eligibility criteria on both the trial and EHR sides and to eventually notify healthcare providers of patients potentially eligible for clinical trials in a far more timely and comprehensive way than currently possible. The first guiding design principle will be to reduce the search space of a human trial coordinator, in effect reducing the number of patients that need to be reviewed for every successfully enrollable patient. A second guiding principle will be to reduce the onboarding time required to add new trials and new classes of criteria to the system.
SUMMARY TABLE
| What was already known on the topic | What this study added to our knowledge |
|---|---|
|
|
HIGHLIGHTS.
Most eligibility criteria are only mentioned in unstructured clinical text.
Natural language processing (NLP) system allows extracting criteria from text.
NLP system could extract eligibility criteria with up to 95.5% sensitivity.
Patients could be automatically matched with clinical trials with an AUC of 89.8%.
ACKNOWLEDGMENT
We thank Chase Arbra, MD, for clinical text annotations; Abdul AlAbdulsalam for sharing some software focused on cancer TNM staging information extraction; Jean Craig, Erin Quigley and Patricia Rudisill for their technical support acquiring and transforming clinical data.
FUNDING STATEMENT
This work is supported in part by pilot research funding, Hollings Cancer Center’s Cancer Center Support Grant P30 CA138313 at the Medical University of South Carolina, and by NIH/NCATS 5UL1TR001450–03. This work is also supported by UG1 CA189848, and by the South Carolina Centers of Economic Excellence.
Abbreviations:
- AJCC
American Joint Committee on Cancer
- AUC
area under the curve
- CDM
common data model
- ECOG
Eastern Cooperative Oncology Group
- HER
Electronic Health Record
- HER2
Human Epidermal Growth Factor Receptor 2
- MAP
mean average precision
- MUSC
Medical University of South Carolina
- NLP
Natural Language Processing
- OHDSI
Observational Health Data Sciences and Informatics
- OMOP
Observational Medical Outcomes Partnership
- SVM
support vector machine
- UIMA
Unstructured Information Management Architecture
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS STATEMENT
The authors have no competing interests to declare.
A patient whose (l)ast (m)enstrual (p)eriod was over two years ago is considered post-menopausal.
REFERENCES
- 1.Sung NS, Crowley WF, Genel M, Salber P, Sandy L, Sherwood LM, et al. Central challenges facing the national clinical research enterprise. JAMA. 2003. March 12;289(10):1278–1287. [DOI] [PubMed] [Google Scholar]
- 2.Lara PN, Higdon R, Lim N, Kwan K, Tanaka M, Lau DH, et al. Prospective evaluation of cancer clinical trial accrual patterns: identifying potential barriers to enrollment. J Clin Oncol 2001. March 15;19(6):1728–1733. [DOI] [PubMed] [Google Scholar]
- 3.Somkin CP, Ackerson L, Husson G, Gomez V, Kolevska T, Goldstein D, et al. Effect of medical oncologists’ attitudes on accrual to clinical trials in a community setting. J Oncol Pract 2013. November;9(6):e275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penberthy L, Brown R, Puma F, Dahman B. Automated matching software for clinical trials eligibility: measuring efficiency and flexibility. Contemp Clin Trials 2010. May;31(3):207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thadani SR, Weng C, Bigger JT, Ennever JF, Wajngurt D. Electronic screening improves efficiency in clinical trial recruitment. Journal of the American Medical Informatics Association Oxford University Press; 2009. November;16(6):869–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embi PJ, Jain A, Clark J, Harris CM. Development of an electronic health record-based Clinical Trial Alert system to enhance recruitment at the point of care. AMIA Annu Symp Proc 2005;:231–235. [PMC free article] [PubMed] [Google Scholar]
- 7.Heinemann S, Thüring S, Wedeken S, Schäfer T, Scheidt-Nave C, Ketterer M, et al. A clinical trial alert tool to recruit large patient samples and assess selection bias in general practice research. BMC Med Res Methodol BioMed Central; 2011. February 15;11(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–144. [PubMed] [Google Scholar]
- 9.Demner-Fushman D, Elhadad N. Aspiring to Unintended Consequences of Natural Language Processing: A Review of Recent Developments in Clinical and Consumer-Generated Text Processing. Yearb Med Inform 2016:224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richesson RL, Sun J, Pathak J, Kho A. A survey of clinical phenotyping in selected national networks: demonstrating the need for high-throughput, portable, and computational methods. Artificial Intelligence in Medicine 2016;71:57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savova GK, Tseytlin E, Finan S, Castine M, Miller T, Medvedeva O, et al. DeepPhe: A Natural Language Processing System for Extracting Cancer Phenotypes from Clinical Records. Cancer Res. 2017. November 1;77(21):e115–e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halpern Y, Horng S, Choi Y, Sontag D. Electronic medical record phenotyping using the anchor and learn framework. J Am Med Inform Assoc 2016; 23(4):731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc 2011;18 Suppl 1(Supplement 1):i116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boland MR, Tu SW, Carini S, Sim I, Weng C. EliXR-TIME: A Temporal Knowledge Representation for Clinical Research Eligibility Criteria. AMIA Summits Transl Sci Proc 2012;2012:71–80. [PMC free article] [PubMed] [Google Scholar]
- 15.Duryea R, Danese MD. Human Readable Expression of Structured Algorithms for Describing and Storing Clinical Study Criteria and for Generating and Visualizing Queries. AMIA Summits Transl Sci Proc 2015:1–5. [Google Scholar]
- 16.Levy-fix G, Yaman A, Weng C. Structuring Clinical Trial Eligibility Criteria with the Common Data Model. AMIA Summits Transl Sci Proc 2015. [Google Scholar]
- 17.Ni Y, Wright J, Perentesis J, Lingren T, Deleger L, Kaiser M, et al. Increasing the efficiency of trial-patient matching: automated clinical trial eligibility Pre-screening for pediatric oncology patients. BMC Med Inform Decis Mak; 2015;15(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shivade C, Hebert C, Lopetegui M, de Marneffe M-C, Fosler-Lussier E, Lai AM. Textual inference for eligibility criteria resolution in clinical trials. J Biomed Inform 2015. December;58 Suppl:S211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.NLM. Standard or Comprehensive Radiation Therapy in Treating Patients With Early-Stage Breast Cancer Previously Treated With Chemotherapy and Surgery. 2013. Available from: https://clinicaltrials.gov/ct2/show/NCT01872975
- 20.NLM. Comparison of Axillary Lymph Node Dissection With Axillary Radiation for Patients With Node-Positive Breast Cancer Treated With Chemotherapy. 2013. Available from: https://clinicaltrials.gov/ct2/show/NCT01901094
- 21.NLM. Fulvestrant and/or Anastrozole in Treating Postmenopausal Patients With Stage II-III Breast Cancer Undergoing Surgery. 2013. Available from: https://clinicaltrials.gov/ct2/show/NCT01953588
- 22.de Castilho RE, Mujdricza-Maydt E. A web-based tool for the integrated annotation of semantic and syntactic structures. Proceedings of the Workshop on Language Technology Resources and Tools for Digital Humanities; Osaka, Japan; 2016:76–84. [Google Scholar]
- 23.Si Y, Weng C. An OMOP CDM-Based Relational Database of Clinical Research Eligibility Criteria. Stud Health Technol Inform 2017;245:950–954. [PMC free article] [PubMed] [Google Scholar]
- 24.Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational Health Data Sciences and Informatics (OHDSI) - Opportunities for Observational Researchers. Stud Health Technol Inform 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 25.ClinicalTrials.gov. Available from: https://clinicaltrials.gov
- 26.Regenstrief Institute. Logical observation identifier names and codes (LOINC). Available from: http://loinc.org/
- 27.NLM. SNOMED Clinical Terms® (SNOMED-CT®). Available from: http://www.nlm.nih.gov/research/umls/Snomed/snomed_main.html
- 28.Apache. UIMA (Unstructured Information Management Architecture). Available from: http://uima.apache.org/
- 29.Kim Y, Riloff E, Hurdle JF. A Study of Concept Extraction Across Different Types of Clinical Notes. AMIA Annu Symp Proc 2015:737–746. [PMC free article] [PubMed] [Google Scholar]
- 30.Lafferty JD, McCallum A, Pereira F. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proc. 18th International Conf. on Machine Learning 2001;:282–289. [Google Scholar]
- 31.Fan R-E, Chang K-W, Hsieh C-J, Wang X-R, Lin C-J. LIBLINEAR: A Library for Large Linear Classification. JMLR 2008;9(Aug):1871–1874. [Google Scholar]
- 32.van Rijsbergen CJ. Information Retrieval. Butterworth; 1979. [Google Scholar]