Abstract
Toxoplasma gondii is a protozoan parasite that has a tremendous impact on human health and livestock. High seroprevalence among humans and other animals is facilitated by the conversion of rapidly proliferating tachyzoites into latent bradyzoites that are housed in tissue cysts, which allow transmission through predation. Epigenetic mechanisms contribute to the regulation of gene expression events that are crucial in both tachyzoites as well as their development into bradyzoites. Acetylation of histones is one of the critical histone modifications that is linked to active gene transcription. Unlike most early-branching eukaryotes, Toxoplasma possesses two GCN5 homologues, one of which, GCN5b, is essential for parasite viability. Surprisingly, GCN5b does not associate with most of the well-conserved proteins found in the GCN5 complexes of other eukaryotes. Of particular note is that GCN5b interacts with multiple putative transcription factors that have plant-like DNA-binding domains denoted as AP2. To understand the function of GCN5b and its role(s) in epigenetic gene regulation of stage switching, we performed co-immunoprecipitation of GCN5b under normal and bradyzoite induction conditions. We report the greatest resolution of the GCN5b complex to date under these various culture conditions. Moreover, reciprocal co-IPs were performed with distinct GCN5b-interacting AP2 factors (AP2IX-7 and AP2XII-4) to delineate the interactomes of each putative transcription factor. Our findings suggest that GCN5b is associated with at least two distinct complexes that are characterized by two different pairs of AP2 factors, and implicate up to four AP2 proteins to be involved with GCN5b-mediated gene regulation.
Keywords: Acetylation, Apicomplexan, Epigenetics, Proteomics, Toxoplasma
Graphical Abstract
Introduction
General control non-derepressible 5 (GCN5) was originally described as a transcriptional activating protein in the yeast Saccharomyces cerevisiae [1]. The discovery that GCN5 possesses lysine acetyltransferase (KAT) activity provided the first direct evidence that histone acetylation modulates gene expression [2]. This finding inaugurated a new field of research that aims to identify and functionally characterize histone post-translational modifications (PTMs), along with the team of proteins that deliver, recognize, and remove these PTMs. This extensive network of proteins collaborates to regulate gene expression by modifying the chromatin associated with DNA.
GCN5 is a highly conserved protein found in all eukaryotes. Most species, including Arabidopsis thaliana, S. cerevisiae, and Drosophila melanogaster, contain a single GCN5, but mammalian species harbor a second GCN5 known as PCAF (p300/CBP Associating Factor). GCN5 functions in a large multi-protein complex called SAGA (Spt-Ada-Gcn5 Acetyltransferase) that is also well-conserved among eukaryotic organisms [3]. In yeast, the SAGA complex has a total mass of ~2 MDa and is composed of at least 19 subunits. Clusters of associating proteins comprising the SAGA complex give it a modular structure that carries out distinct functions including lysine acetylation, chromatin recognition, histone deubiquitination, and interaction with the TFIID complex via TBP (TATA-Binding Protein) [4]. The core acetylation module is comprised of GCN5, ADA2, ADA3, and SGF29. SPT and TAF proteins cluster with TRA1/TRRAP to form a scaffold that bridges transcription factors to TFIID. The histone deubiquitination module is formed by subunit proteins USP8, SGF11, SGF73, and SUS1. Several SAGA subunits, including GCN5, harbor chromatin “reader” domains such as the bromodomain, PHD domain, Tudor domain, or SANT domain.
Far less is known about the GCN5 complexes operating in protozoa, such as parasitesin the phylum Apicomplexa. GCN5 homologues have been identified in key representatives from this phylum, including Plasmodium spp., the causative agent of malaria [5]. Curiously, some apicomplexan parasites, like Toxoplasma gondii, possess two GCN5 KATs (GCN5a and GCN5b) and two ADA2 co-adaptor proteins (ADA2a and ADA2b) [6]. Transmitted by felines or through contaminated food and water, Toxoplasma is capable of infecting virtually all warm-blooded vertebrates and causes opportunistic illness in susceptible hosts. The parasite’s ability to convert from a rapidly replicating form (tachyzoite) into a latent tissue cyst (bradyzoite) is central to transmission and pathogenesis [7]. Histone acetylation and GCN5 KATs in particular are required to regulate genes involved in bradyzoite differentiation [8]
Both GCN5a and GCN5b are present in the parasite nucleus and contain a bromodomain downstream of the KAT catalytic domain [8–10]. In contrast to most other species, however, Toxoplasma GCN5s contain lengthy N-terminal extensions that have little homology to each other or the one found on Plasmodium GCN5 [9–11]. We hypothesized that these unique sequences suggest that apicomplexan SAGA composition would differ from other eukaryotes. Consistent with this hypothesis, aside from an ADA2 homologue (ADA2a), the initial GCN5b complex that we isolated from tachyzoites [10] showed no resemblance to the evolutionarily conserved SAGA complex, or other GCN5-containing complexes like SLIK/SALSA[12] or ATAC [13].
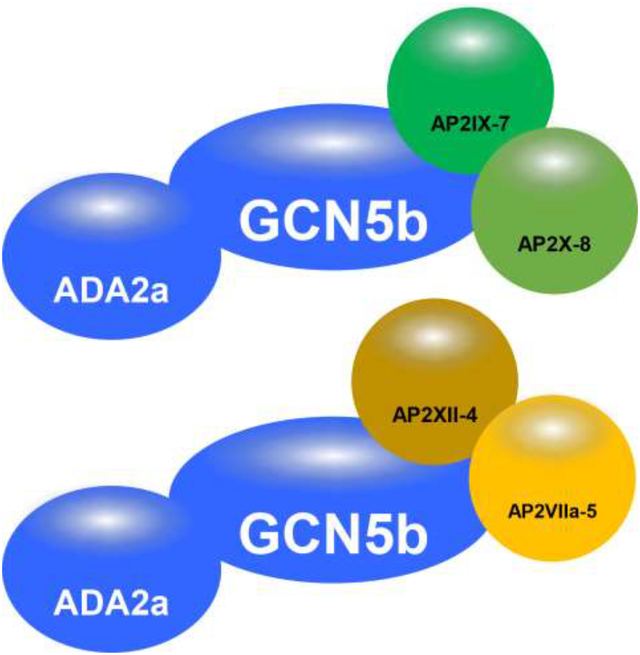
For this study, we conducted a more rigorous investigation of the GCN5b complex in intracellular tachyzoites and examined how the GCN5b complex may alter its composition in tachyzoites subjected to alkaline pH, a stress that induces differentiation into latent bradyzoites. Our findings lend further support for a striking divergent GCN5b-ADA2 core complex that associates with multiple ApiAP2 proteins, which contain a plant-like DNA-binding domain [10]. ApiAP2 proteins are the largest family of transcription factors in apicomplexans (68 in Toxoplasma) and appear to play a central role in the regulation apicomplexan gene expression[14]. Recent studies indicate that ApiAP2 proteins play a central role in gene expression changes during bradyzoite development in Toxoplasma [15–17]. We also elucidated the interactomes of two of these GCN5b-associating ApiAP2 factors, AP2IX-7 and AP2XII-4. Together, the findings are consistent with a model that suggests GCN5b is the catalytic engine for multiple distinct lysine acetyltransferase complexes in Toxoplasma.
Materials and Methods
Parasite strains and cell culture
This study used the RH HAGCN5b and RHΔku80 AP2IX-7-HA strains generated for previous studies [10]. The RH HAGCN5b line overexpresses HAGCN5b exogenously while in the RHΔku80 AP2IX-73xHA line a triple HA tag has been added to the endogenous AP2IX-7 gene. All parasites were maintained in confluent monolayers of human foreskin fibroblasts (HFFs). For unstressed parasites pH 7.4 media was made with Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 1% heat-inactivated fetal bovine serum (FBS). Unstressed parasites were growing in humidified incubators at 37°C with 5% CO2. Alkaline pH 8.3 media was made with RPMI medium supplemented with 1% heat-inactivated FBS. Alkaline stressed parasites were grown in a 37°C incubator with ambient CO2
Endogenous tagging of AP2XII-4
The portion of the ap2xii-4 coding region corresponding to the coding sequence of the C-terminus of the AP2XII-4 protein was amplified using the primers pLIC XII-4 PacI F 5’-ttccaatccaatttaattaaCGAGACAGAAACGGAGACGTC-3’ and pLIC XII-4 PacI R 5’-ccacttccaattttaattaaGACATATCTCGGGCGCGGCCTG-3’ and cloned into the PacI site in pLIC_3HA_DHFR (from Dr. Vern Carruthers, [18]) using InFusion ligase-independent cloning (Takara). The plasmid was linearized with XcmI and transfected into RHΔku80Ahxgprt tachyzoites using Buffer P3 and program FI115 on the Lonza Nucleofector II system.
Nuclear Extraction & Immunoprecipitation
Due to the high level of expression of TgGCN5bHA and relatively low levels of the endogenously tagged AP2s, we used 10 and 20 T–175 cm2 flasks of intracellular tachyzoites, respectively, for the co-immunoprecipitations. To harvest parasites, media was aspirated off the HFF monolayer and 10 mL of 4°C PBS was added to the culture flasks before the flasks were scraped to remove the monolayer. The cell suspensions were passed through a 25 gauge needle five times to lyse parasites from host cells and host cell debris were removed by passing the suspension through a 0.4 μm filter. The purified parasite pellets were stored at −20C.
Cell pellets were resuspended with 0.5 ml of low salt extraction buffer (50mM HEPESNaOH pH7.5, 20% glycerol, 10 mM NaCl, 0.1% NP−140, and one c0mplete protease inhibitor tablet (Millipore-Sigma) per 10 mL of buffer) and incubated on ice for 5 minutes to lyse cells and release nuclei. The nuclei were then pelleted and the pellet was resuspended with 0.5 ml of high salt extraction buffer (low salt buffer with 420 mM NaCl). The suspension was vortexed for 1 min at 4°C before snap freezing in liquid nitrogen and thawing it on ice three times to break open the nuclei. Membrane debris was then removed by centrifugation. The supernatant was kept and used for immuno-precipitation.
Nuclear extract was added to 50 μL of prewashed anti-mouse IgG beads (5873, CST) and incubated at 4°C for 1 hour with mixing. The supernatant was then removed from the beads and added to 50 μL of prewashed Pierce Anti-HA Magnetic Beads (88836, ThermoFisher) and incubated at 4°C with mixing (1 hour for exogenously expressed proteins, overnight for endogenously tagged proteins). After incubation, both sets of beads were washed five times with Pierce IP Lysis Buffer (87787, ThermoFisher) and twice with PBS. Beads were stored in 50 μL of PBS at −80°C until they could be submitted for mass spectrometry.
Mass Spectrometry
Beads were pelleted by centrifugation and resuspended in 8M urea in 100 mM Tris pH 8.5 prior to digestion by endoproteinase LysC and Trypsin Gold (Promega) at 37°C with shaking in a Thermomixer for 12 hours. Digested peptides were separated from the beads using a spin column (Pierce). Approximately 1/3 of the total elution was injected onto a C18 3 uM reversed phase trap column for clean-up prior to separation on an Easy-spray 2 uM 15 cm reversed phase analytical column. Peptides were separated over a 90 minute gradient from 2–25% acetonitrile in front of a Velos Pro Orbitrap or Q-Exactive Plus mass spectrometer in data-dependent acquisition mode.
Bioinformatics Analysis
The MS/MS data obtained was searched against a custom Toxoplasma database which also contained common lab contaminants such as IgGs and human keratins (FASTA database available upon request). Database search was performed using Sequest HT within the Proteome Discoverer 2.2 software package (Thermo) with a precursor mass tolerance of 10 ppm, methionine oxidation considered as a differential modification, and up to two missed tryptic cleavage sites allowed. The data was further analyzed using Scaffold 4 software (Proteome Software) and filtered to a false discovery rate of equal to or less than 1%. The peptide-spectrum matches (PSMs) from Scaffold were used for Statistical Analysis of InNTeractome (SAINT) express analysis launched through the user upload options in the CRAPome as previously described [19–21]. An equal number of user controls was used for the analysis considering that Toxoplasma affinity purification controls are not currently available in the CRAPome.
Immunofluorescence assays.
Localization of tagged AP2XII-4 was determined by immunofluorescence staining, as described previously [22]. Confluent HFF monolayers grown on coverslips were infected with tachyzoites for 18 h, and then infected monolayers were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS, and blocked in 3% bovine serum albumin (BSA). Fixed monolayers were incubated with primary antibody (anti-HA, 1:2,000; Roche) diluted in 3% BSA-PBS overnight at 4C, washed in PBS (three 15-min washes), and then stained with fluorophore-linked secondary antibody (anti-rat-488, 1:2,000; Molecular Probes) for 1 h, followed by washing in PBS (three 15-min washes). Cells were stained with 4’,6-diamidino-2-phenylindole (DAPI) prior to mounting in Vectashield antifade solution.
Western blotting.
Parasite lysates were prepared from infected HFF monolayers that were scraped, washed in PBS, resuspended in parasite lysis buffer (150 mM NaCl, 50 mM Tris-Cl, pH 7.5, 0.1% NP-40) supplemented with mammalian protease inhibitor cocktail (Roche), and sonicated. The insoluble fraction was cleared by centrifugation, and protein concentration was quantified using the Bio-Rad DC assay. Samples were suspended in SDS-PAGE buffer with beta-mercaptoethanol before being loaded on a 4 to 12% Bis-Tris Novex gradient gel and run in morpholinepropanesulfonic acid (MOPS) running buffer. Separated protein was transferred to nitrocellulose membrane and blocked in 4% milk-Tris-buffered saline-Tween (TBST) for 1 h at room temperature followed by incubation with anti-HA (1:2,000; Roche) in 4% milk-TBST, washing in TBST, and anti-rat-horseradish peroxidase (1:2,000; GE Healthcare) incubation in 4% milk-TBST. Chemiluminescent imaging of membranes was performed using Pierce enhanced chemiluminescence (ECL) western blotting substrate and a Protein Simple chemiluminescent imager.
Results and Discussion
Analysis of the Toxoplasma GCN5b complex with SAINT
Our previous study isolated a GCN5b lysine acetyltransferase complex from tachyzoites; likely due to its highly divergent N-terminal extension, we found that this GCN5b complex lacks most subunits that are well-conserved in other eukaryotic SAGA complexes [10]. The aim of the present study was to carry out a more rigorous analysis of the GCN5b interacting proteins and to determine how dynamic this complex is under stress conditions. We employed the same transgenic parasite clone (type I RH strain) engineered to ectopically express recombinant full-length GCN5b tagged with an HA tag at its N-terminus as described in [10]. Using stringent co-immunoprecipitation, followed by mass spectrometry peptide identification and Significance Analysis of INTeractome (SAINT) analysis [21], we have achieved greater resolution of the factors that are associated with GCN5b (SAINT probability score >0.75) (Table 1). SAINT analysis compares peptide counts between positive and negative controls (anti-HA resin vs mouse IgG resin), allowing us to reduce the number of false positives from non-specific interactions. Our data shows that ADA2a and three AP2s (AP2X-7, AP2X-8 and AP2XII-4) are present in the GCN5b complex, but we did not identify other well-conserved eukaryotic SAGA complex members with statistical confidence (Table 1). Consistent with our previous observations [10], two PHD domain containing proteins (TGGT1_224260, TGGT1_234900) and seven hypothetical proteins were also observed. Notably, a significant association between GCN5b and the non-essential paralogue GCN5a was identified in two of the three experimental replicates (Table 1), suggesting that both Toxoplasma GCN5 KATs may carry out a cellular function in the same complex. Two homologues of the Facilitates Chromatin Transcription (FACT) complex (FACT80 and FACT140) were also detected, underlining the role of GCN5b and its associated factors in transcriptional elongation. The involvement of members of the FACT complex with GCN5b is particularly intriguing, given the results of our previous analysis using chromatin immunoprecipitation, followed by microarrays to identify the genomic loci where GCN5b is associated [10]. This determined that GCN5b is predominantly located within gene bodies, further supporting a link between GCN5b and transcriptional elongation. Overall, our findings support that GCN5 KAT complexes in apicomplexan parasites are unusually divergent, consisting a multiple novel, parasite-specific components.
Table 1.
SAINT analysis of co-immunoprecipitation of HATgGCN5b (HA) compared 0074o negative control (IgG) under normal growth conditions. PSM – number of peptide spectrum matches
| Total PSMs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Protein | Molecular Weight (kDa) | Fold Change A | Fold Change B | SAINT Probability | HA I | HA II | HA III | IgG I | IgG II | IgG III |
| TGGT1_243440 | histone lysine acetyltransferase GCN5-B | 111 | 7.81 | 4.22 | 1 | 443 | 55 | 181 | 59 | 22 | 46 |
| TGGT1_217050 | ADA2-A transcriptional co-activator SAGA component | 127 | 14.4 | 6.05 | 1 | 63 | 14 | 47 | 9 | 1 | 1 |
| TGGT1_224260 | PHD-finger domain-containing protein | 603 | 81.26 | 30 | 1 | 241 | 21 | 170 | 8 | 1 | 2 |
| TGGT1_291930 | RNA recognition motif-containing protein | 73 | 12.14 | 6.23 | 1 | 33 | 16 | 10 | 0 | 5 | 0 |
| TGGT1_261450 | hypothetical protein | 119 | 10.03 | 8.23 | 1 | 13 | 4 | 4 | 0 | 1 | 0 |
| TGGT1_234900 | PHD-finger domain-containing protein | 475 | 85.19 | 41.31 | 1 | 77 | 4 | 77 | 0 | 0 | 0 |
| TGGT1_221670 | transcriptional elongation factor FACT140 | 135 | 11.36 | 5.89 | 0.96 | 7 | 13 | 0 | 0 | 0 | 0 |
| TGGT1_241850 | hypothetical protein | 479 | 31.03 | 7.33 | 1 | 79 | 1 | 40 | 3 | 0 | 0 |
| TGGT1_205580 | nuclear factor NF4 | 51 | 5.32 | 2.05 | 0.97 | 15 | 14 | 1 | 0 | 3 | 0 |
| TGGT1_244650 | putative eukaryotic initiation factor-5 | 47 | 5.88 | 5.2 | 0.9 | 3 | 5 | 1 | 0 | 0 | 0 |
| TGGT1_247700 | AP2 domain transcription factor AP2XII-4 | 401 | 50.62 | 11.22 | 1 | 64 | 0 | 52 | 0 | 0 | 1 |
| TGGT1_214960 | AP2 domain transcription factor AP2X-8 | 400 | 22.21 | 4.69 | 1 | 38 | 0 | 29 | 2 | 0 | 0 |
| TGGT1_290630 | AP2 domain transcription factor AP2IX-7 | 279 | 11.9 | 2.93 | 0.98 | 21 | 0 | 16 | 2 | 0 | 0 |
| TGGT1_254555 | histone lysine acetyltransferase GCN5-A | 128 | 15.58 | 5.18 | 1 | 101 | 5 | 24 | 7 | 0 | 6 |
| TGGT1_309250 | hypothetical protein | 407 | 42.32 | 6.06 | 1 | 109 | 0 | 77 | 7 | 0 | 1 |
| TGGT1_280590 | hypothetical protein | 288 | 24.02 | 4.87 | 1 | 59 | 0 | 26 | 3 | 0 | 0 |
| TGGT1_226620 | hypothetical protein | 395 | 10.39 | 3.63 | 0.98 | 62 | 3 | 26 | 1 | 2 | 4 |
| TGGT1_274180 | hypothetical protein | 142 | 14.88 | 7.65 | 0.98 | 15 | 0 | 11 | 0 | 0 | 0 |
| TGGT1_212860 | hypothetical protein | 110 | 5.26 | 4.9 | 0.88 | 6 | 1 | 2 | 0 | 0 | 0 |
| TGGT1_261460 | transcriptional elongation factor FACT80 | 61 | 4.13 | 2.94 | 0.84 | 2 | 4 | 0 | 0 | 0 | 0 |
| TGGT1_265330 | putative cell-cycle-associated protein kinase GSK | 44 | 4.38 | 1.73 | 0.75 | 4 | 0 | 5 | 1 | 0 | 0 |
Analysis of the GCN5b complex during bradyzoite induction conditions
Environmental stresses such as alkaline pH induce differentiation from the rapidly replicating tachyzoite to the latent bradyzoite stage [7]. During the initial response to these stresses, tachyzoites undergo a global downregulation of protein translation and preferentially translate a specific subset of transcripts [23]. GCN5b is one of the most significantly preferentially translated transcripts during this stress response, but it is unknown how the GCN5b interactome might be modulated during a stress known to induce transition to the bradyzoite stage [7]. We hypothesized that in response to environmental stress, GCN5b may form an active complex with an alternative set of proteins that are dedicated to upregulating stress response factors and, in the event of persistent stress, bradyzoite developmental genes. Co-IPs and mass spectrometry analysis of GCN5b associated proteins were repeated in tachyzoites that were subjected to alkaline (pH 8.3) stress for 48 hours, conditions under which we have previously shown upregulation of bradyzoite markers such as ldh2 and bag1, even in type I RH strain [17]. Our findings imply that the GCN5b complex is significantly modified during the first 48 hours of the alkaline stress response (Table 2). SAINT analysis of the three co-IP replicates determined that nine proteins remain consistently associated with GCN5b, which likely constitute the core GCN5b functional complex. Interactions between GCN5b and AP2IX-7, AP2X-8, and AP2XII-4 were no longer detectable under alkaline stress conditions. Of interest for future study would be the purification of the GCN5b complex from type II strain Toxoplasma under normal versus differentiating conditions, although this will be more a challenging endeavor due to the substantially reduced amount of parasite material obtained.
Table 2.
SAINT analysis of co-immunoprecipitation of HATgGCN5b (HA) compared to negative controls (IgG) under alkaline (pH 8.2) conditions. PSM – number of peptide spectrum matches.
| Total PSMs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Protein | Molecular Weight (kDa) | Fold Change A | Fold Change B | SAINT Probability | HA I | HA II | HA III | IgG I | IgG II | IgG III |
| TGGT1_243440 | histone lysine acetyltransferase GCN5-B | 111 | 7.65 | 4.26 | 1 | 63 | 67 | 84 | 32 | 25 | 16 |
| TGGT1_217050 | ADA2-A transcriptional co-activator SAGA component | 127 | 23.49 | 8.62 | 1 | 18 | 16 | 33 | 1 | 3 | 0 |
| TGGT1_224260 | PHD-finger domain-containing protein | 603 | 89.04 | 26.96 | 1 | 28 | 20 | 87 | 0 | 0 | 0 |
| TGGT1_291930 | RNA recognition motif-containing protein | 73 | 18.45 | 3.48 | 1 | 24 | 23 | 0 | 2 | 4 | 0 |
| TGGT1_261450 | hypothetical protein | 119 | 9.42 | 4.62 | 1 | 4 | 6 | 0 | 0 | 0 | 0 |
| TGGT1_234900 | PHD-finger domain-containing protein | 475 | 82.93 | 48.74 | 0.96 | 13 | 9 | 28 | 0 | 0 | 0 |
| TGGT1_221670 | transcriptional elongation factor FACT140 | 135 | 18.34 | 8.6 | 0.96 | 12 | 6 | 0 | 0 | 0 | 0 |
| TGGT1_241850 | hypothetical protein | 479 | 11.26 | 4.49 | 0.88 | 4 | 3 | 6 | 0 | 0 | 0 |
| TGGT1_205580 | nuclear factor NF4 | 51 | 7.45 | 1.8 | 0.86 | 9 | 13 | 0 | 3 | 8 | 0 |
| TGGT1_244650 | putative eukaryotic initiation factor-5 | 47 | 7.69 | 4.91 | 0.8 | 4 | 3 | 0 | 0 | 0 | 0 |
The AP2IX-7 interactome
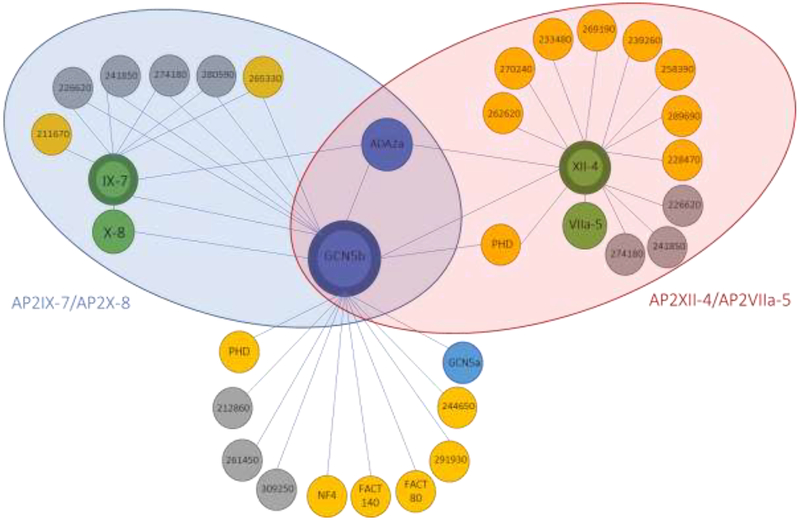
To verify the association of AP2IX-7 with GCN5b, and to determine if this putative DNA-binding protein is associated with other chromatin modifying complexes, we performed reciprocal co-immunoprecipitation and proteomic analysis of the proteins associated with AP2IX-7 using an endogenously HA-tagged AP2IX-7 parasite clone [10]. With this approach, where a hit was classified as having at least a five-fold enrichment in peptides detected in the HA pulldown over the naive IgG control, we confirmed that AP2IX-7 is present in a complex with GCN5b, ADA2a, and AP2X-8 (Table 3). Remarkably, only five other proteins in common with the GCN5b interactome were detected in the AP2IX-7 co-IPs (Table 3, Fig. 2), including four hypothetical proteins and a kinase with homology to the glycogen synthase kinase family (TGGT1_265330). Several proteins with strong confidence SAINT scores in the GCN5b co-IPs were not detected in the AP2IX-7 co-IPs, e.g. AP2XII-4 and the two PHD domain containing proteins. These results suggest that GCN5b is a component of multiple complexes during tachyzoite growth, and that the AP2IX-7/AP2X-8 complex is only associated with a proportion of the total GCN5b in the parasite.
Table 3.
Total peptide counts from co-immunoprecipitation replicates of P2IX-73xHA (HA) compared to the negative control (IgG) in normal (7.5) and alkaline stress (8.2) conditions.
| Total Peptide Count | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Protein | Molecular Weight (kDa) | 7.5 HA I | 7.5 HA II |
7.5 IgG I | 7.5 IgG II | 8.2 HA I | 8.2 HA II | 8.2 IgG I | 8.2 IgG II |
| TGGT1_290630 | AP2 domain transcription factor AP2IX-7 | 279 | 31 | 38 | 0 | 0 | 92 | 38 | 1 | 2 |
| TGGT1_214960 | AP2 domain transcription factor AP2X-8 | 400 | 36 | 45 | 0 | 0 | 126 | 35 | 0 | 1 |
| TGGT1_243440 | histone lysine acetyltransferase GCN5-B | 111 | 17 | 24 | 0 | 4 | 38 | 32 | 0 | 0 |
| TGGT1_217050 | ADA2-A transcriptional co-activator SAGA component | 127 | 13 | 25 | 0 | 0 | 50 | 35 | 0 | 0 |
| TGGT1_211670 | S1 RNA binding domain-containing protein | 42 | 9 | 9 | 0 | 0 | 11 | 8 | 0 | 0 |
| TGGT1_265330 | putative cell-cycle-associated protein kinase GSK | 44 | 7 | 9 | 0 | 0 | 19 | 9 | 0 | 0 |
| TGGT1_241850 | hypothetical protein | 479 | 63 | 88 | 0 | 0 | 210 | 92 | 0 | 0 |
| TGGT1_226620 | hypothetical protein | 395 | 56 | 68 | 0 | 0 | 183 | 59 | 0 | 0 |
| TGGT1_280590 | hypothetical protein | 288 | 39 | 55 | 0 | 0 | 124 | 51 | 0 | 0 |
| TGGT1_274180 | (hypothetical protein | 142 | 6 | 13 | 0 | 0 | 37 | 15 | 0 | 0 |
Figure 2. Schematic representation of the GCN5b interactome.
Overview of interacting proteins of GCN5b, AP2IX-7, and AP2XII-4 in tachyzoites under normal culture conditions. Target proteins that were immunoprecipitated are signified with thick borders. Interactors conserved in human and yeast SAGA complexes are colored blue. AP2 factors are green, proteins with an annotated function are in yellow, and hypothetical proteins are in grey. Blue shaded oval signifies the members of the AP2IX-7/AP2X-8 complex and the red shaded oval signifies the members of the AP2XII-4/AP2VIIa-5 complex.
Co-IPs of HAGCN5b under alkaline stress conditions implied that its association with AP2 factors is lost (Table 2). Reciprocal co-IPs with AP2IX-7 were repeated on lysates harvested from parasites subjected to pH 8.3 for 48 hours to confirm this finding and to determine what, if any, other complexes the putative DNA-binding protein might associate with. Unexpectedly, we found no difference in the AP2IX-7 interactome during alkaline stress, including its association with GCN5b (Table 3). This discrepancy between the HAGCN5b and AP2IX-7 co-IPs may be explained by differences in protein expression level between GCN5b and AP2IX-7. The co-IPs of GCN5b were performed on a parasite line over-expressing an ectopic, epitope-tagged copy of GCN5b, while the AP2IX-7 co-IPs were performed using a parasite line in which the endogenous coding locus was tagged. In other words, the levels of GCN5b are enhanced relative to AP2IX-7. Furthermore, since GCN5b appears to inhabit multiple complexes, the relative proportion of these complexes that GCN5b occupies may be dynamic, and the stoichiometry of the GCN5b and AP2IX-7 interaction changes in response to conditions such as alkaline stress. These findings highlight the potential caveats that can arise when analyzing interacting partners of over-expressed proteins.
Purification of the AP2XII-4 complex indicates that GCN5b is present in at least two independent complexes with different pairs of AP2 factors
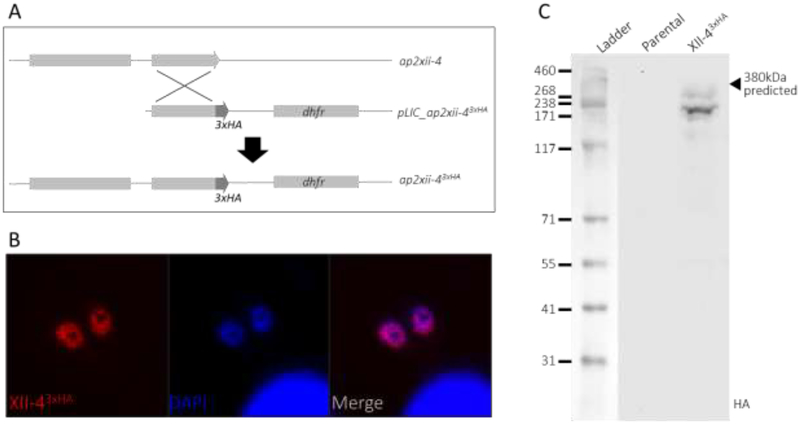
Three AP2 factors were identified to interact with GCN5b (Table 1). Two of these, AP2IX-7 and AP2X-8, constitute a distinct complex that does not contain AP2XII-4. To determine the composition of the complex containing AP2XII-4, we generated an endogenously tagged AP2XII-4 parasite line for proteomic analysis. A C-terminal 3xHA epitope tag was incorporated in-frame with the endogenous ap2xii-4 coding sequence. The tagged AP2XII-4 protein localizes to the parasite nucleus, and western blotting shows that the protein is present as two bands, both migrating slightly faster than the expected 380kDa annotated molecular weight (www.toxodb.org)(Fig. 1).
Figure 1. Epitope tagging of TgAP2XII-4 at its endogenous locus.
A. Plasmid integration at the ap2xii-4 locus by single crossover recombination to insert sequence coding for a C-terminal 3xHA tag in frame with the ap2xii-4 coding sequence. B. Immunofluorescence staining of HA-tagged AP2XII-43xHA tachyzoites (red), co-localizing with DAPI DNA stain (blue). C. Western blotting of parental RHΔHXΔku80 and AP2XII-43xHA parasite lysates with anti-HA antibody.
Co-immunoprecipitation followed by proteomic analysis was performed on lysate from AP2XII-43xHA intracellular tachyzoites (Table 4). Results confirmed the interaction of AP2XII-4 and GCN5b, as well as with ADA2a. Of note, a fourth AP2 factor was identified, AP2VIIa-5, which likely functions in conjunction with AP2XII-4 in a manner analogous to the association of AP2IX-7 and AP2X-8. There was no evidence of either AP2IX-7 or AP2X-8 interaction with AP2XII-4, reinforcing the model that GCN5b forms different complexes with different sets of AP2 factors. Although there were no PHD domain containing proteins associated with the AP2IX-7/AP2X-8 complex, one PHD domain protein (TGGT1_ 234900) was identified in the AP2XII-4 complex, hinting at the different functionalities of these distinct GCN5b complexes. It is likely that the core GCN5b complex is recruited by distinct AP2 factors (or pairs of AP2 factors) to regulate different gene networks or target different substrates. To date, the subsets of genes that are regulated by the four AP2 factors identified in this study are unknown. Both AP2IX-7 and AP2XII-4 scored as highly essential (−4.22 and −5.12, respectively) in a genome-wide CRISPR screen [24], suggesting that each pair of AP2s that partner with GCN5b are recruitedto genes that carry out critical cellular processes. Future studies will attempt to identify these parasite genes through ChIP-seq analysis of the individual AP2s. Our findings here will help steer future studies to discern the precise functions of the distinct GCN5b complexes.
Table 4.
Total peptide counts from co-immunoprecipitation replicates of AP2XII-43xHA (HA) compared to negative controls (IgG).
| Total peptide count | ||||||
|---|---|---|---|---|---|---|
| Gene ID | Protein | Molecular Weight (kDa) | HA I | HA II | IgG I | IgG II |
| TGGT1_247700 | AP2 domain transcription factor AP2XII-4 | 401 | 37 | 38 | 0 | 0 |
| TGGT1_243440 | histone lysine acetyltransferase GCN5-B | 111 | 10 | 22 | 0 | 0 |
| TGGT1_217050 | ADA2-A transcriptional co-activator SAGA component | 127 | 7 | 22 | 0 | 0 |
| TGGT1_234900 | PHD-finger domain-containing protein | 475 | 25 | 47 | 0 | 0 |
| TGGT1_203690 | AP2 domain transcription factor AP2VIIa-5 | 266 | 4 | 9 | 0 | 0 |
| TGGT1_262620 | RNA recognition motif-containing protein | 32 | 8 | 12 | 0 | 0 |
| TGGT1_270240 | MAG1 protein | 49 | 19 | 13 | 0 | 0 |
| TGGT1_233480 | SAG-related sequence SRS29C | 39 | 15 | 3 | 0 | 0 |
| TGGT1_269190 | glyceraldehyde-3-phosphate dehydrogenase GAPDH2 | 105 | 8 | 10 | 0 | 0 |
| TGGT1_239260 | histone H4 | 11 | 7 | 17 | 0 | 2 |
| TGGT1_258390 | putative DnaJ protein | 45 | 5 | 7 | 0 | 0 |
| TGGT1_289690 | glyceraldehyde-3-phosphate dehydrogenase GAPDH1 | 53 | 3 | 8 | 0 | 0 |
| TGGT1_258870A | hypothetical protein | 61 | 6 | 7 | 0 | 0 |
| TGGT1_268580 | hypothetical protein | 54 | 5 | 17 | 0 | 0 |
| TGGT1_215980 | hypothetical protein | 24 | 5 | 5 | 0 | 0 |
| TGGT1_228470 | ribosomal protein RPL15 | 24 | 6 | 4 | 0 | 0 |
Supplementary Material
Highlights.
The SAGA lysine acetyltransferase complex is unusually divergent in Toxoplasma gondii
TgGCN5b forms multiple complexes with different ApiAP2 factors.
ApiAP2 factors that interact with TgGCN5b appear to do so as distinct pairs.
Acknowledgements
This research was supported by grants from National Institutes of Health (AI116496 to WJS and T32 AI007637 to MH and T32 AI060519 to JM) and the Indiana Center for AIDS Research Junior Investigator Pilot Grants program (VJ). We are grateful to Dr. Vern Carruthers, University of Michigan for kindly sharing the pLIC_3HA_DHFR plasmid and also the staff of the Proteomics Core at Indiana University School of Medicine for their technical assistance. We also thank members of the Sullivan lab and others in the Biology of Intracellular Pathogens (BIP) group at IUSM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L, Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors, EMBO J 13 (1994) 4807–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD, Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation, Cell 84 (1996) 843–851. [DOI] [PubMed] [Google Scholar]
- [3].Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL, Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex, Genes Dev 11 (1997) 1640–1650. [DOI] [PubMed] [Google Scholar]
- [4].Wu P-YJ, Ruhlmann C, Winston F, Schultz P, Molecular architecture of the S. cerevisiae SAGA complex, Mol. Cell 15 (2004) 199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- [5].Fan Q, An L, Cui L, Plasmodium falciparum histone acetyltransferase, a yeast GCN5 homologue involved in chromatin remodeling, Eukaryotic Cell 3 (2004) 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bhatti MM, Livingston M, Mullapudi N, Sullivan WJ, Pair of unusual GCN5 histone acetyltransferases and ADA2 homologues in the protozoan parasite Toxoplasma gondii, Eukaryotic Cell 5 (2006) 62–76. doi: 10.1128/EC.5.1.62-76.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jeffers V, Tampaki Z, Kim K, Sullivan WJ, A latent ability to persist: differentiation in Toxoplasma gondii, Cell. Mol. Life Sci 75 (2018) 2355–2373. doi: 10.1007/s00018-018-2808-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Naguleswaran A, Elias EV, McClintick J, Edenberg HJ, Sullivan WJ, Toxoplasma gondii lysine acetyltransferase GCN5-A functions in the cellular response to alkaline stress and expression of cyst genes, PLoS Pathog 6 (2010) e1001232. doi: 10.1371/journal.ppat.1001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dixon SE, Bhatti MM, Uversky VN, Dunker AK, Sullivan WJ, Regions of intrinsic disorder help identify a novel nuclear localization signal in Toxoplasma gondii histone acetyltransferase TgGCN5-B, Mol. Biochem. Parasitol 175 (2011) 192–195. doi: 10.1016/j.molbiopara.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang J, Dixon SE, Ting L-M, Liu T-K, Jeffers V, Croken MM, Calloway M, Cannella D, Hakimi MA, Kim K, Sullivan WJ, Lysine acetyltransferase GCN5b interacts with AP2 factors and is required for Toxoplasma gondii proliferation, PLoS Pathog 10 (2014) e1003830. doi: 10.1371/journal.ppat.1003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dixon SE, Stilger KL, Elias EV, Naguleswaran A, Sullivan WJ, A decade of epigenetic research in Toxoplasma gondii, Mol. Biochem. Parasitol 173 (2010) 1–9. doi: 10.1016/j.molbiopara.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, Grant PA, The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway, Mol. Cell. Biol 22 (2002) 8774–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR, Washburn MP, Abmayr SM, Workman JL, Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila, Mol. Cell. Biol 26 (2006) 871–882.doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jeninga MD, Quinn JE, Petter M, ApiAP2 Transcription Factors in Apicomplexan Parasites, Pathogens 8 (2019). doi: 10.3390/pathogens8020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Walker R, Gissot M, Croken MM, Huot L, Hot D, Kim K, Tomavo S, The Toxoplasma nuclear factor TgAP2XI-4 controls bradyzoite gene expression and cyst formation, Mol. Microbiol 87 (2013) 641–655. doi: 10.1111/mmi.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Radke JB, Lucas O, De Silva EK, Ma Y, Sullivan WJ, Weiss LM, Llinas M, White MW, ApiAP2 transcription factor restricts development of the Toxoplasma tissue cyst, Proc. Natl. Acad. Sci. U.S.A 110 (2013) 6871–6876. doi: 10.1073/pnas.1300059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang S, Holmes MJ, Radke JB, Hong D-P, Liu T-K, White MW, Sullivan WJ, Toxoplasma gondii AP2IX-4 Regulates Gene Expression during Bradyzoite Development, MSphere 2 (2017). doi: 10.1128/mSphere.00054-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Huynh M-H, Carruthers VB, Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80, Eukaryotic Cell 8 (2009) 530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Smith-Kinnaman WR, Berna MJ, Hunter GO, True JD, Hsu P, Cabello GI, Fox MJ, Varani G, Mosley AL, The interactome of the atypical phosphatase Rtr1 in Saccharomyces cerevisiae, Mol Biosyst 10 (2014) 1730–1741. doi: 10.1039/c4mb00109e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mellacheruvu D, Wright Z, Couzens AL, Lambert J-P, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, Al-Hakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin Z-Y, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJR, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras A-C, Nesvizhskii AI, The CRAPome: a contaminant repository for affinity purification-mass spectrometry data, Nat. Methods 10 (2013) 730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras A-C, Choi H, SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software, J Proteomics 100 (2014) 37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jeffers V, Kamau ET, Srinivasan AR, Harper J, Sankaran P, Post SE, Varberg JM, Sullivan WJ, Boyle JP, TgPRELID, a Mitochondrial Protein Linked to Multidrug Resistance in the Parasite Toxoplasma gondii, MSphere 2 (2017). doi: 10.1128/mSphere.00229-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Joyce BR, Tampaki Z, Kim K, Wek RC, Sullivan WJ, The unfolded protein response in the protozoan parasite Toxoplasma gondii features translational and transcriptional control, Eukaryotic Cell 12 (2013) 979–989. doi: 10.1128/EC.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sidik SM, Huet D, Ganesan SM, Huynh M-H, Wang T, Nasamu AS, Thiru P, Saeij JPJ, Carruthers VB, Niles JC, Lourido S, A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes, Cell 166 (2016) 1423–1435.e12. doi: 10.1016/j.cell.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.