Abstract
Loss-of-function mutations in the ATP-binding cassette (ABC) transporter of the inner mitochondrial membrane, ABCB7, cause X-linked sideroblastic anemia with ataxia, a phenotype that remains largely unexplained by the proposed role of ABCB7 in exporting a special sulfur species for use in cytosolic iron-sulfur (Fe-S) cluster biogenesis. Here, we generated inducible ABCB7-knockdown cell lines to examine the time-dependent consequences of loss of ABCB7. We found that knockdown of ABCB7 led to significant loss of mitochondrial Fe-S proteins, which preceded the development of milder defects in cytosolic Fe-S enzymes. In erythroid cells, loss of ABCB7 altered cellular iron distribution and caused mitochondrial iron overload due to activation of iron regulatory proteins 1 and 2 in the cytosol and to upregulation of the mitochondrial iron importer, mitoferrin-1. Despite the exceptionally large amount of iron imported into mitochondria, erythroid cells lacking ABCB7 showed a profound hemoglobinization defect and underwent apoptosis triggered by oxidative stress. In ABCB7-depleted cells, defective heme biosynthesis resulted from translational repression of ALAS2 by iron regulatory proteins and from decreased stability of the terminal enzyme ferrochelatase. By combining chemical crosslinking, tandem mass spectrometry and mutational analyses, we characterized a complex formed of ferrochelatase, ABCB7 and ABCB10, and mapped the interfaces of interactions of its components. A dimeric ferrochelatase physically bridged ABCB7 and ABCB10 homodimers by binding near the nucleotide-binding domains of each ABC transporter. Our studies not only underscore the importance of ABCB7 for mitochondrial Fe-S biogenesis and iron homeostasis, but also provide the biochemical characterization of a multiprotein complex required for heme biosynthesis.
Introduction
ATP-binding cassette (ABC) transporters belong to one of the most abundant families of integral membrane proteins found in all kingdoms of life1,2 and play major roles in several biological processes by mediating the active transport of a variety of molecules across cellular membranes. Three members of the ABC family have thus far been localized to the inner mitochondrial membrane, where they are predicted to act as exporters, since their nucleotide binding domains face the matrix.3,4 These members are ABCB7 (the human ortholog of yeast Atm1), ABCB10 and ABCB8. ABCB6 has been reported to reside either in the outer mitochondrial membrane,5,6 and/or in the Golgi,7 lysosomal,8 and plasma membranes.9
ABCB7 maps to the X-chromosome in mice and humans10 and shows a ubiquitous expression pattern. Knockout studies in mice revealed that expression of ABCB7 was essential for early gestation.11 Mutations in ABCB7 cause X-linked sideroblastic anemia with ataxia (XLSA/A; 301310), which is a recessive disorder characterized by the onset of non-or slowly-progressive cerebellar ataxia and anemia with hypochromia and microcytosis in infancy or early childhood.12–14 Bone marrow examination showed ringed sideroblasts, which give the condition its name. Complementation assays in yeast suggested that each of the human mutations caused a mild partial loss of function.12,13 Conditional gene targeting in mice showed that ABCB7 was essential for hematopoiesis15 and for the development and function of all tissues and organs analyzed.11
Here, we examined the time-dependent consequences of loss of ABCB7 in multiple cell culture models. We found that knockdown (KD) of ABCB7 led to significant loss of mitochondrial iron-sulfur (Fe-S) proteins, which preceded the development of comparatively milder defects in cytosolic Fe-S enzymes. In erythroid cells, loss of Abcb7 caused defective heme biosynthesis and altered cellular iron distribution with mitochondrial iron overload, which triggered oxidative stress and led to apoptosis of erythroid progenitors. By combining chemical crosslinking with tandem mass spectrometry and mutational analyses, we identified a complex formed of ferrochelatase (FECH), ABCB7 and ABCB10 and characterized its overall architecture. Our studies uncovered the importance of ABCB7 for mitochondrial function and iron homeostasis and identified a previously uncharacterized complex that is required for heme biosynthesis.
Methods
Cell lines and cell culture conditions
HEK293T and HeLa cells were purchased from the American Type Culture Collection (ATCC) and propagated in Dulbecco modified Eagle medium with 4.5 g/L glucose, 10% fetal bovine serum, and 2 mM glutamine at 37°C, 5% CO2 in a humidified incubator. G1E-ER4 cells were maintained in Iscove modified Dulbecco medium with 15% fetal bovine serum, 100 U/mL penicillin-streptomycin, 2 U/mL erythropoietin (Sino Biological Inc.), monothioglycerol (1:10,000), and 50 ng/mL Kit-ligand (R&D Systems). GATA1-mediated differentiation was induced by the addition of 100 nmol of β-estradiol to a cell culture at a density of 2×105 cells/mL. All cell lines were tested for mycoplasma.
Short hairpin and small interfering RNA-mediated knockdown of ABCB7, MFRN2 and IRP2 in HEK293T, HeLa or G1E-ER4 cells
The SMARTvector Inducible Lentiviral short hairpin (sh)RNA system (Dharmacon) was used to generate HEK293T and HeLa stable cell clones with tightly controlled expression of three individual shRNA targeting different regions of the ABCB7 transcript and a scrambled shRNA, used as negative control.
Knockdown of Abcb7 or Irp2 in G1E-ER4 cells was achieved with the Accell small interfering RNA delivery system (Dharmacon). Further details of the knockdown procedures, together with information on in vitro and in vivo crosslinking and mass spectrometry, in vitro coupled transcription/translation and pull-down assay of 35S-labeled proteins, the dihydropyrimidine assay, gel electrophoresis, complex I, II and III activity assays, iron and heme measurements, histological staining, flow cytometry studies, assays of superoxide dismutase activity, aconitase and catalase, along with polymerase chain reaction studies and other methods are provided in the Online Supplementary Methods.
Results
Knockdown of ABCB7 led to dysfunction of mitochondrial proteins which preceded the development of milder defects in cytosolic Fe-S enzymes
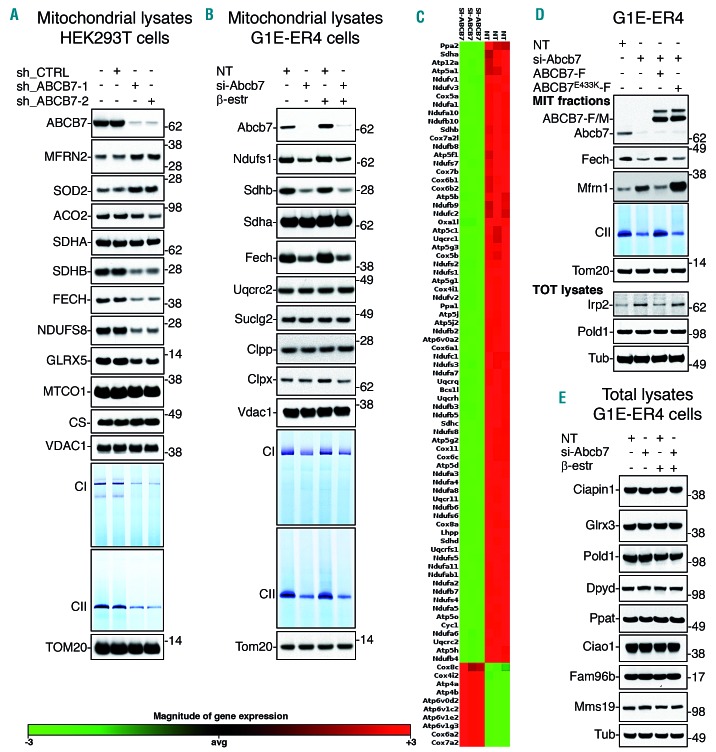
We analyzed the effects of progressive loss of ABCB7 in HEK293T, HeLa and G1E-ER4 cells. To interrogate the role of ABCB7 during erythroid differentiation and address the effect of ABCB7 mutations in XLSA patients,12–14 we studied G1E-ER4 cells, a well-validated murine cell model that recapitulates the early stages of terminal red blood cell development.16 We used an inducible shRNA-mediated KD approach to silence the expression of ABCB7 in HEK293T (Online Supplementary Figure S1A) and HeLa cells (Online Supplementary Figure S1B), which enabled us to discern primary from secondary effects by observing when defects appeared during the time-course after induction of KD. KD of ABCB7 resulted in a time-dependent loss of mitochondrial Fe-S proteins, including FECH, glutaredoxin 5 (GLRX5) and multiple subunits of the respiratory complexes I and II (NDUFS1 and NDUFS8 in complex I and SDHB in complex II) in all the cell lines tested (Figure 1A,B, Online Supplementary Figure S2C-E, and S2H). Loss of ABCB7 also elicited profound transcriptional remodeling that included the downregulation of the vast majority of subunits and assembly factors of the mitochondrial respiratory chain within 48 h after KD (Figure 1C) in G1E-ER4 cells and profoundly affected cell morphology (Online Supplementary Figure S1C). Loss of the catalytic subunits resulted in a significant decrease in the activities of complexes I and II (Figures 1A, B, and D). Mitochondrial aconitase (ACO2) activity was also markedly reduced by 80%, whereas cytosolic aconitase (ACO1) decreased to a lesser extent (40%) (Online Supplementary Figure S2H), likely due to the fact that mitochondrial dysfunction altered cytosolic iron status and activated the RNA-binding activity of iron-regulatory protein-1 (IRP1)17 (see later in the paper), without changing IRP1 protein levels (Online Supplementary Figure S2H). Notably, levels of cytosolic Fe-S proteins, including CIAPIN1, GLRX3, POLD1, DPYD, PPAT, ERCC2, ELP3 and ABCE1, were unchanged 3 days after KD of ABCB7 (Figure 1E and Online Supplementary Figure S2B). Activity of the cytosolic Fe-S enzyme DPYD (Online Supplementary Figure S2F) and radioactive iron incorporation into the cytosolic Fe-S protein NUBP2 were also unchanged (Online Supplementary Figure S2G). Decreased stability of the cytosolic Fe-S proteins DPYD, PPAT and POLD1 was observed 5 days after KD of ABCB7 (Online Supplementary Figure S2C,D); however, the extent of loss of mitochondrial Fe-S proteins at day 5 was much more profound than the decrease in cytosolic Fe-S protein levels (Online Supplementary Figure S2C,D). Consistent with the importance of mitochondria in performing a global regulatory role in numerous cellular processes linked to iron homeostasis, we found that KD of ABCB7 stabilized iron-regulatory protein-2 (IRP2), a master regulator of iron metabolism18 (Figure 1D and Online Supplementary Figure S2C,D). Levels of the erythroid-specific mitochondrial iron transporter mitoferrin-1 (MFRN1) (Figure 1D) and the ubiquitously expressed mitoferrin-2 (MFRN2) in HEK293T and HeLa cells (Figure 1A and Online Supplementary Figure S2D) increased significantly in ABCB7-KD cells. Overall, stabilization of IRP2 in the cytosol and increased MFRN1 or MFRN2 in mitochondria likely increased flux of iron into mitochondria in ABCB7-depleted cells (further investigated later in this study). Re-expression of wildtype ABCB7, but not the pathogenic XLSA E433K mutant13 in the ABCB7-KD cells restored levels of FECH and SDHB, activities of complex II and aconitases and normalized levels of MFRN1 and IRP2 (Figure 1D and Online Supplementary Figure S2H), demonstrating that loss of ABCB7 caused profound early-onset of mitochondrial dysfunction.
Figure 1.
Loss of ABCB7 disrupts mitochondrial function and downregulates genes involved in mitochondrial energy metabolism. (A) Protein blots and respiratory complexes I (CI) and II (CII) in-gel activity assays on mitochondrial lysates from HEK293T cells expressing an inducible control short hairpin (sh)RNA (sh_CTRL) or two shRNA targeting different regions of the ABCB7 transcript (sh_ABCB7-1 and sh_ABCB7-2) for 3 days. Levels of ABCB7, mitoferrin 2 (MTFN2), superoxide dismutase 2 (SOD2), aconitase (ACO2), CII subunits SDHA and SDHB, ferrochelatase (FECH), the CI Fe-S subunit NDUFS8, glutaredoxin 5 (GLRX5), complex IV subunit MTCO1, and citrate synthase (CS) were assayed. Levels of VDAC1 and TOM20 were used as loading controls. (B) Protein blots and CI and CII in-gel activity assays on mitochondrial lysates from G1E-ER4 control cells or from cells depleted of Abcb7 for 3 days. Comparisons between cells at the burst-forming unit erythroid stage (undifferentiated, without β-estradiol) or at the orthochromatophilic stage (differentiated for 72 h with β-estradiol) are shown. Levels of Abcb7, CI Fe-S subunit Ndufs1, CII subunits Sdha and Sdhb, Fech, CIII subunit Uqcrc2, Suclg2, mitochondrial unfoldase Clpx and protease Clpp were assessed by western blot. Levels of Vdac1 and Tom20 were used as loading controls. (C) Log2-fold expression of mitochondrial energy metabolism pathway genes which were differentially expressed in G1E-ER4 cells 48 h after knockdown (KD) of Abcb7 (false discovery rate <0.01, n=3 biological replicates). (D) Complementation assays on G1E-ER4 cells silenced for 3 days to KD the expression of Abcb7 and transfected with wildtype FLAG-tagged ABCB7 or with the X-linked sideroblastic anemia pathogenic mutant ABCB7E433K-F. Levels of Abcb7, Irp2, Pold1, Fech and Mfrn1 are shown, along with CII in-gel activity assay. Tom20 and a-Tubulin (Tub) were used as loading controls for the mitochondrial fractions and total lysates, respectively. (E) Protein blots on total lysates from G1E-ER4 cells. Levels of the cytosolic and nuclear Fe-S proteins Ciapin1, Glrx3, Pold1, Dpyd and Ppat are shown, along with levels of the CIA components Ciao1, Fam96b and Mms19. Tub was used as a loading control. (A, B, D and E, n=6 biological replicates). See also Online Supplementary Figures S13 and S14 for densitometries of immunoblots and statistical analyses. NT: not treated (control).
Loss of Abcb7 altered cellular iron distribution and caused oxidative stress and apoptosis of erythroid progenitors
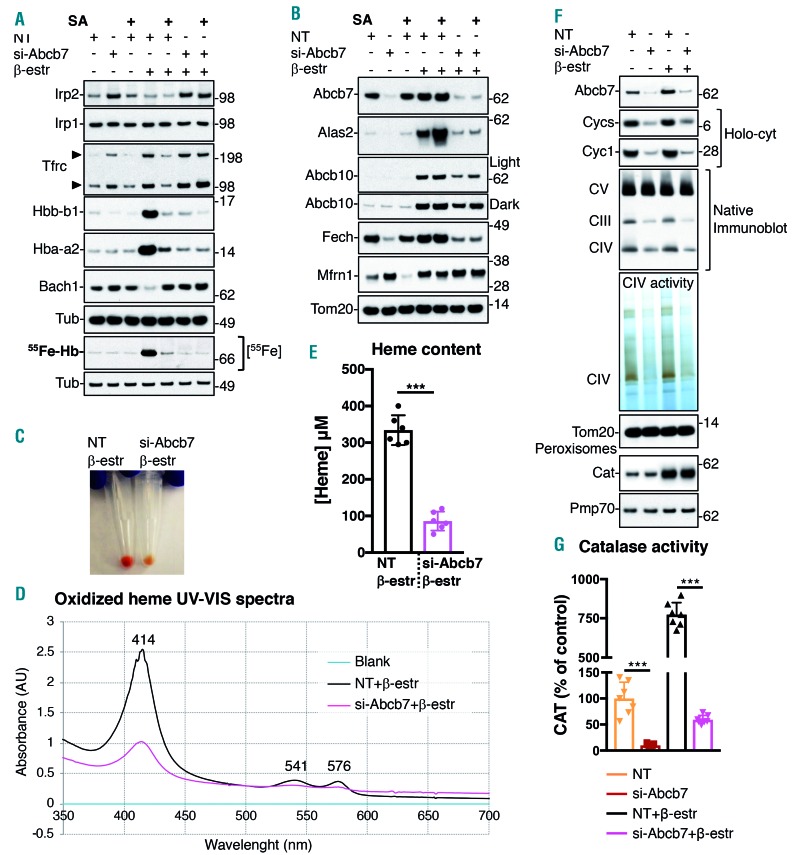
We investigated the consequences of loss of Abcb7 in developing G1-ER4 cells, which require large amounts of iron inside mitochondria to support heme synthesis. We first analyzed changes in the activation of the iron responsive element (IRE)-binding activities of Irp1 and Irp2, which post-transcriptionally shape the expression of the mammalian iron metabolism proteome by binding to IRE in the messenger RNA of transcripts involved in iron homeostasis.18 G1E-ER4 cells robustly activated the IRE-binding activities of Irp1 and Irp2 within 10 h after induction of differentiation, with maximal activation at 24 h and 48 h (Figure 2A). By 72 h in differentiation medium, when cells had reached the orthochromatophilic erythroid stage and fulfilled their extracellular iron demands, the IRE-binding activities of Irp1 and Irp2 diminished (Figure 2A). In contrast, in Abcb7-KD G1E-ER4 cells, Irp2 activation was elicited well before induction of differentiation and its levels remained undiminished throughout the time course (Figure 2A). Control cells differentiated for 72 h showed increased sequestration of radiolabeled iron into ferritin (55Fe-autoradiogram) (Figure 2B), as already reported.19 In control cells, iron was also successfully incorporated into protoporphyrin IX (PPIX) by Fech, as radiolabeled hemoglobin (55Fe-Hb) increased significantly (Figure 2B). Compared to control (NT), cells lacking Abcb7 exhibited prolonged activation of the cytosolic iron starvation response with stabilization of Irp2, decreased cytosolic aconitase (Aco1) activity and increased IRE-binding activities of IRP (Figure 2A-C). Hyperactivation of IRP caused translational repression of ferritin (Figure 2B) and decreased radiolabeled-iron sequestration into the heteropolymeric storage form of the protein (Figure 2B). Under the transcriptional control of Gata-1,20 differentiating cells upregulated hemoglobin subunits (Hba-a2 and Hbb-b1) and Alas2 (Figure 2B,D, Online Supplementary Figure S3). Mfrn1 increased during differentiation to fulfill the exceptional iron demand for heme synthesis (Figure 2B and Online Supplementary Figure S3). Abcb7-depleted cells failed to increase levels of Alas2, due to translational repression by IRP at the 5’-IRE present in its transcript21,22 (Figure 2D), even though Alas2 transcript levels were over 50-fold higher than those in undifferentiated cells (Online Supplementary Figure S3). Importantly, levels of Bach1, an erythroid transcriptional repressor that is rapidly degraded under conditions of heme sufficiency,23 increased upon KD of Abcb7 (Figure 2D), and levels of Hbb-b1 and Hba-a2, which are Bach1 targets,24 were repressed (Figure 2B and Online Supplementary Figure S3). Notably, Tfrc and Mfrn1 levels were elevated in Abcb7-KD cells prior to differentiation (Figure 2B and Online Supplementary Figure S4A,B). Total cellular iron content in early erythroid progenitors depleted of Abcb7 was about 7-fold higher than that in controls (Online Supplementary Figure S5A) and iron accumulated in mitochondria (Figure 2E), whereas the pool of available cytosolic labile iron was significantly reduced (Figure 2F), indicating a condition of cytosolic functional iron deficiency. Mitochondrial iron overload increased the production of reactive oxygen species by 30% (Mitosox+ cells) (Figure 2G,H), which damaged plasma membranes, as shown by the increased percentage of annexin V+ cells (Figure 2G,H and Online Supplementary Figure S5B), even though both mitochondrial and cytosolic superoxide dismutase enzymes (Sod2 and Sod1) were activated (Figure 2I), and were transcriptionally upregulated about 2- and 5-fold, respectively (Online Supplementary Figure S6A-C). Unexpectedly, most of the genes involved in the antioxidant response were downregulated in Abcb7-KD cells (Online Supplementary Figure S6A). Manganese accumulated in mitochondria of Abcb7-KD cells (Online Supplementary Figure S5C), raising the possibility that metalation of Sod2 accounted for the increase in man ganese. KD of ABCB7 in HEK293T cells also caused mitochondrial iron accumulation (Online Supplementary Figure S5D), and activation of SOD enzymes (Online Supplementary Figure S5E,F). As MFRN2 was significantly upregulated upon silencing of ABCB7 in the non-erythroid HEK293T cells (Figure 1A), we knocked down its expression and found that cells depleted of MFRN2 maintained intact mitochondrial function (Online Supplementary Figure S5D,G). KD of MFRN2 in ABCB7-depleted cells lowered mitochondrial iron accumulation (Online Supplementary Figure S5D), but levels and activities of Fe-S-dependent enzymes did not return to normal (Online Supplementary Figure S5H), suggesting that mitochondrial iron overload was not the primary cause of the compromise of Fe-S proteins in the matrix.
Figure 2.
Loss of Abcb7 alters cellular iron distribution and leads to oxidative damage and apoptosis of erythroid progenitors. (A) Iron responsive elemebt (IRE)-binding activities of Irp1 and Irp2 in control and Abcb7-knockdown (KD) G1E-ER4 cells before differentiation (-β-estradiol) and 10, 24, 48 or 72 h after differentiation in the presence of β-estradiol. (B) In-gel aconitase activity assay on control or Abcb7-KD G1E-ER4 cells before and after 72 h of differentiation, showing the activities of mitochondrial and cytosolic aconitase (Aco2 and Aco1, respectively). Immunoblots to Irp2, Irp1, Aco2, Tfrc, ferritin (Ft), Hba-a2, Hbb-b1 and Mfrn1 on the same set of samples analyzed in the aconitase activity assays are shown. Tubulin (Tub) was used as a loading control. 55Fe autoradiogram on G1E-ER4 cells treated as for the aconitase assays, showing levels of radiolabeled iron incorporated into the iron storage protein Ft or into hemoglobin (Hb). (C) IRE-binding activities of Irp1 and Irp2 in G1-ER4 cells silenced for Abcb7 for 3 days and differentiated in the presence of β-estradiol for 72 h, and immunoblots to Irp2 and to Tub. (D) Protein levels of Irp2, Alas2 and Bach1 in G1E-ER4 cells before and 30 h after differentiation. (E) Iron content in mitochondria was significantly increased in Abcb7-KD (day 3) cells before (left lanes) differentiation. (F) The cytosolic labile iron pool (LIP) was decreased in cells treated as in (E). The mean fluorescence intensity (MFI) is shown in arbitrary units (a.u.). (G) Representative flow cytometry analysis to sort G1E-ER4 control or Abcb7-KD cells that were double-positive for the apoptotic marker (annexin V) and for mitochondrial reactive oxygen species production (Mitosox) (top, right square). (H) Quantification of G1E-ER4 cells double-positive for annexin V and Mitosox from data shown in (G). (I) In-gel activity assay of Sod1 and Sod2 in G1E-ER4 cells treated as in (E) showed significantly higher activity and protein levels in cells depleted of Abcb7 for 3 days. Data in (E) (F) and (H) are expressed as mean ± standard deviation. (A-I, n=5). See also Online Supplementary Figure S14 for densitometries of immunoblots and statistical analyses. NT: not treated (control). ***P<0.001
Heme biosynthesis defect in cells lacking ABCB7
Loss of Abcb7 during erythroid differentiation led to an 80% reduction in the levels of 55Fe-Hb, to the IRP-mediated translational repression of Alas2 and to Bach1-mediated blockage of Hba-a2 and Hbb-b1 transcription, pointing to a potential defect in heme biosynthesis. Treatment with succinylacetone, a potent inhibitor of Alad, confirmed that the defect of 55Fe-Hb synthesis caused by KD of Abcb7 during differentiation was comparable to the inhibition of the heme biosynthetic pathway (Figure 3A, 55Fe-Hb autoradiogram, lanes 5 and 6). Alas2, Fech and Alad protein levels decreased in cells lacking Abcb7 (Figure 3B and Online Supplementary Figure S7A), whereas levels of the heme biosynthetic enzyme Cpox,25 of Sucla2 and Suclg2, which provide succinyl-CoA to the first rate-limiting step of the pathway.26 and of the unfoldase Clpx, required for the pyridoxal-phosphate-dependent activation of Alas2,27 did not change (Online Supplementary Figure S7A). Abcb7-KD cells exhibited impaired hemoglobinization (Figure 3C). Heme content decreased by 75% (Figure 3D,E) and activities of the heme-containing enzymes cytochrome c oxidase (Figure 3F) and peroxisomal catalase (Figure 3G and Online Supplementary Figure S7C-E) were reduced by more than 80%. Levels of respiratory complexes III and IV, which require three and two heme centers, respectively, were significantly reduced in Abcb7-KD cells (Figure 3F). Heme-bound cytochromes c (Cyc) and c1 (Cyc1) decreased (Holocyt) (Figure 3F), without changes in the levels of total apocytochromes (Online Supplementary Figure S7B). The heme-regulated eIF2a kinase (HRI), which inhibits the general translation initiation factor eIF2a under conditions of heme deficiency,28 may mediate the translational repression of the globin chains, which are also transcriptionally repressed by Bach1 (Online Supplementary Figure S3), in Abcb7-depleted erythroid progenitors to prevent accumulation of globins in excess of heme, and may also repress translation of the heme biosynthetic enzymes Alas2 and Alad.28 A defect in heme biosynthesis was also observed in HEK293T and HeLa cells upon KD of ABCB7 (Online Supplementary Figure S7F-K).
Figure 3.
Heme biosynthesis defect in cells lacking Abcb7. (A) Protein blots to Irp2, Irp1, Tfrc, Hbb-b1, Hba-a2, and Bach1 in control (NT) and Abcb7-KD G1E-ER4 cells before (-β-estradiol) and after (+β-estradiol) 72 h of differentiation. The effect of the Alad inhibitor succinylacetone (SA) during cell differentiation was also tested. 55Fe incorporated into hemoglobin (Hb) in cells silenced for Abcb7 was profoundly decreased. (B) Mitochondrial fractions from G1E-ER4 cells treated as in (A) were probed with antibodies against Abcb7, Alas2, Abcb10, Fech, and Mfrn1. Tom20 was used as a loading control. (C) Cell pellets of control (NT β-estr) or Abcb7-knockdown (KD) (si-Abcb7 β-estr) G1E-ER4 cells differentiated for 72 h showed decreased hemoglobinization in cells depleted of Abcb7. (D) Representative oxidized UV-VIS spectra of heme, with the characteristic Soret band at 414 nm and additional peaks at 541 and 576 nm, in control and Abcb7-KD G1E-ER4 cells 72 h after differentiation showed significantly lower heme levels in cells depleted of Abcb7. (E) Heme levels in control and GIE-ER4 cells depleted of Abcb7 for 3 days. Values are means ± standard deviation. (F) Levels of heme-bound cytochromes c (Cyc) and c1 (Cyc1) were significantly decreased in Abcb7-depleted G1E-ER4 cells. Native immunoblots to Atp5a (CV subunit), Uqcrc2 (CIII subunit) and Mtco1 (CIV subunit) showed that levels of heme-containing complexes CIII and CIV were significantly decreased in Abcb7-KD cells. CIV activity, which requires two heme centers, was also decreased. Tom20 was used as a loading control. Peroxisomal fractions showed comparable levels of catalase protein in control and Abcb7-KD G1E-ER4 cells. Catalase (Cat) protein levels increased to the same extent in control and Abcb7-KD cells during differentiation. Pmp70 (also known as Abcd3) is a marker of peroxisomes and was used as a loading control. (G) Catalase activity in control and Abcb7-KD cells before and after differentiation showed profound defects in heme-dependent catalase activity in cells depleted of Abcb7. Values are expressed as % of control. Values are means ± standard error of mean. (A-C, E-G, n=6; D, n=3). See also Online Supplementary Figure S15 for densitometries of immunoblots and statistical analyses. ***P<0.001
Irp2 activation in Abcb7-depleted cells sustained mitochondrial iron overload mediated by mitoferrin-1 upregulation
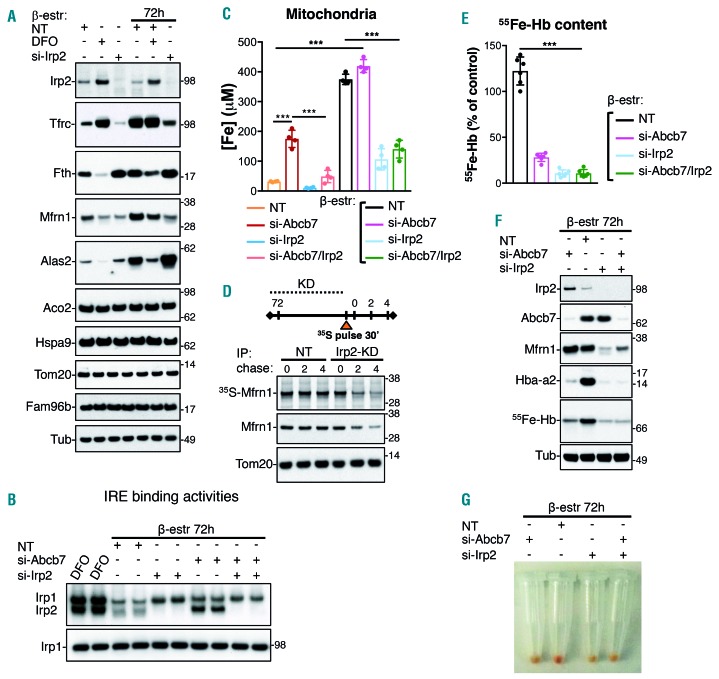
We investigated the correlation between loss of Abcb7 and activation of IRP, by analyzing the effect of double KD of Abcb7 and Irp2 in developing erythroid cells. KD of Irp2 significantly diminished the stability of Tfrc (Figure 4A) and decreased Mfrn1 to levels comparable to those of cells treated with desferrioxamine (Figure 4A). A detectable compensatory activation of the IRE-binding activity of Irp1 in Irp2-KD cells (Figure 4B) was insufficient to stabilize Tfrc to levels comparable to those in controls. Mitochondrial iron levels decreased by more than 50% (Figure 4C), due to the decreased half-life of MFRN1 under the iron-limiting conditions generated by the lack of Irp2 (Figure 4D). Levels of radiolabeled hemoglobin dropped by more than 80% (Figure 4E,F), causing an evident hemoglobinization defect in cells lacking Irp2 (Figure 4G). Double KD of Abcb7 and Irp2 reduced iron overload in early erythroid progenitors (Figure 4C). However, it also severely impaired iron delivery to mitochondria during differentiation (Figure 4C) and impaired hemoglobinization (Figure 4G), confirming the essential role of Irp2 in erythropoiesis.29
Figure 4.
Irp2 activation sustains mitochondrial iron overload mediated by mitoferrin-1 upregulation in erythroid cells depleted of Abcb7. (A) Irp2, Tfrc, ferritin H (Fth), Mfrn1, Alas2, Aco2, Hspa9 and Fam96b levels in G1E-ER4 cells before and during differentiation and either upon treatment with the iron chelator desferrioxamine (DFO) or upon knockdown (KD) of Irp2. Tom20 and tubulin (Tub) were used as loading controls for mitochondrial and cytosolic fractions, respectively. (B) Iron-responsive element (IRE)-binding activities of Irp1 and Irp2 in G1E-ER4 cells differentiated for 72 h and transfected with small interfering (si)-RNA to KD the expression of Irp2, Abcb7 or both for 3 days. DFO-treated samples were run alongside to compare the KD effect with iron deficiency on the activation of iron regulatory proteins (IRP). Irp1 blotting did not show a change in protein levels. (C) Mitochondrial iron in control and Abcb7-, Irp2- or Abcb7/Irp2- double-KD G1E-ER4 cells before and after 72 h of differentiation. Loss of Irp2 reduced iron delivery to mitochondria. (D) A pulse-chase experiment was performed to assess the turnover rate of Mfrn1 under the iron-limiting conditions generated by KD of Irp2. G1E-ER4 cells were silenced for 72 h to KD the expression of Irp2. Cells were then pulsed for 30 min with 35S-Cys/Met, followed by incubation for the indicated time points in differentiation medium. Radiolabeled Mfrn1 was visualized by autoradiography after immunoprecipitation and sodium dodecylsulfate polyacrylamide gel electropheresis (top panel), whereas total protein levels were assessed by immunoblot (lower panels). (E) Levels of radiolabeled iron incorporated into hemoglobin in Abcb7-, Irp2- and Abcb7/Irp2 double-KD cells. (F) Irp2, Abcb7, Mfrn1 and Hba-a2 levels in G1E-ER4 cells differentiated for 72 h in the presence of β-estradiol and silenced for either Abcb7, or Irp2 or simultaneously for Abcb7 and Irp2. A representative 55Fe-autoradiogram shows significantly decreased levels of radioactive iron incorporated into hemoglobin (Hb) in cells depleted of Abcb7, Irp2 or both. (G) Cell pellets of G1E-ER4 cells differentiated for 72 h and transfected with siRNA to KD the expression of Abcb7, Irp2 or both showed defective hemoglobinization in cells lacking Abcb7, Irp2 or both proteins. (A-D, F, G, n=4; E, n=6). See also Online Supplementary Figure S16 for densitometries of immunoblots and statistical analyses. NT: not treated (control). ***P<0.001
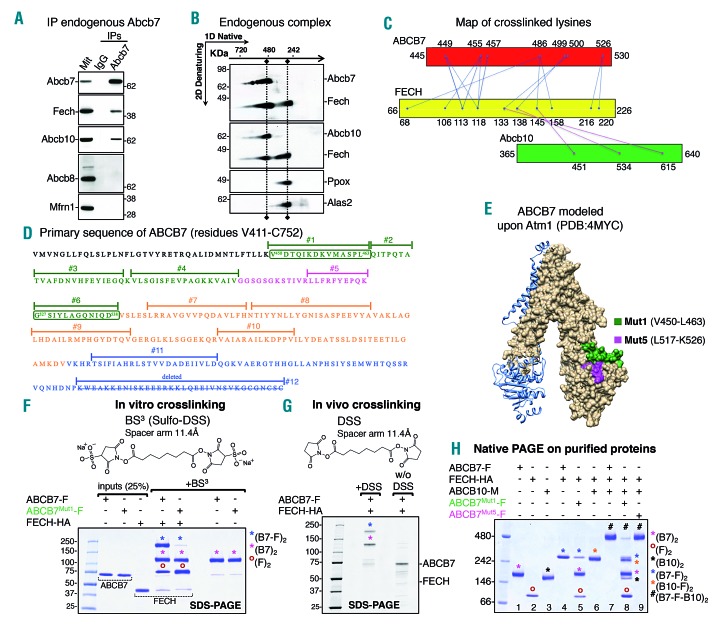
ABCB7 formed a complex with FECH and ABCB10
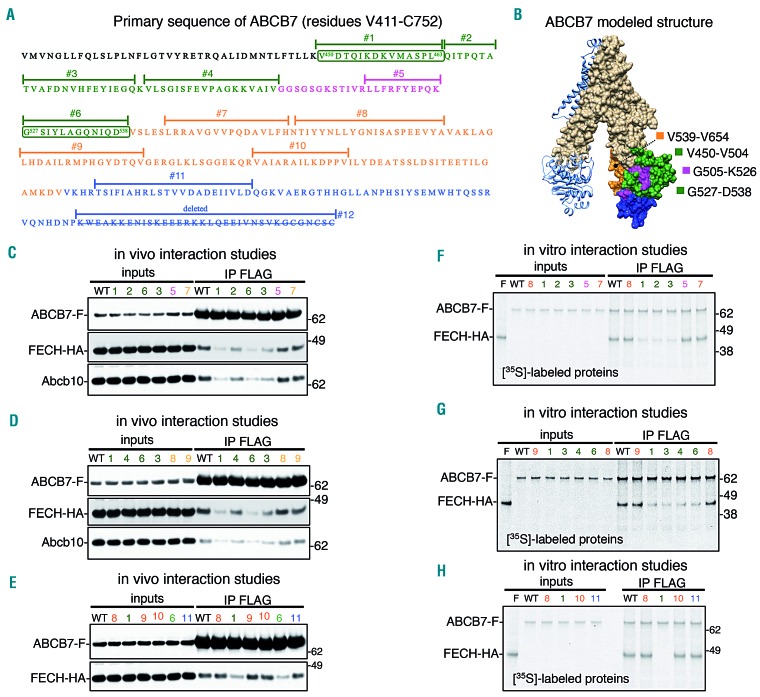
The stability of Fech was reduced by the KD of Abcb7. We found that endogenous Abcb7 interacted with Fech and Abcb10 in G1E-ER4 cells during differentiation (Figure 5A), in agreement with published studies that reported the interaction of Fech with Abcb730–32 or with Abcb10.31,32 Using two-dimensional/Blue-Native(BN)/sodium dodecylsulfate polyacrylamide gel electrophoresis (2D-SDS PAGE) separation, we further demonstrated that Abcb7, Fech and Abcb10 co-migrated in a single multimeric complex of an approximate molecular weight of 480 kDa (Figure 5B). Interestingly, a second distinctive pool of Fech formed a complex of 250 kDa, which also contained the heme-synthesizing enzymes Alas2 and Ppox (Figure 5B), consistent with previously published results that proposed a direct transfer of PPIX from Ppox to Fech for iron insertion,32,33 and that suggested a potential regulatory role for the Fech-Alas2 interaction.32 We performed chemical crosslinking of the ABCB7/FECH complex in vitro on purified proteins and in vivo on mitochondria isolated from G1E-ER4 cells co-expressing human ABCB7-FLAG and FECH-HA, coupled with tandem mass spectrometry (XL-MS) to investigate the architecture of the ABCB7/FECH complex. The analysis yielded 147 MS/MS fragmentation spectra corresponding to 29 high-confidence lysine-lysine crosslinks (Online Supplementary Table S1, Figure 5C and Online Supplementary Figure S8A), among which 12 were intersubunit crosslinks between ABCB7 and FECH and ten were crosslinks between endogenous Abcb10 and FECH (Online Supplementary Table S1, Figure 5C and Online Supplementary Figure S8A,B). No crosslinks were detected between ABCB7 and Abcb10, suggesting that FECH mediated an indirect interaction between ABCB7 and ABCB10, as we subsequently confirmed by coimmunoprecipitation experiments. The analysis also identified a high confidence FECH-FECH intersubunit crosslink (K286-K286) (Online Supplementary Table S1). Identification of this crosslinked species is consistent with the distances derived from the crystal structure of FECH, which shows the two lysines at the FECH dimer interface with a Ca-Ca distance of 10.163Å, in agreement with the length of the DSS crosslinker (11.4 Å) (Online Supplementary Figure S9A). The crystal structure of FECH revealed that the PPIX substrate was deeply bound within a pocket that was enclosed by three movable regions34 (Online Supplementary Figure S9B), one of which (residues 90-115 of FECH) was involved in binding the nucleotide-binding domain of ABCB7, according to our data (Online Supplementary Table S1, Figure 5C and Online Supplementary Figure S9C). We performed alanine scanning mutagenesis on the C terminus of ABCB7 (residues V450-C752) (Figure 5D,E), and tested the mutants in vivo and in vitro for their ability to interact with FECH. SDS-PAGE analysis on crosslinked purified ABCB7 and FECH proteins, showed three major bands in the presence of the crosslinker (BS3) with approximate molecular weights of 80, 120 and 230 kDa (Figure 5F and Online Supplementary Figure S10A), which met the expected molecular weights of a dimer of FECH (84 kDa), a dimer of ABCB7 (138.4 kDa), and a heterotetrameric complex consisting of a dimer of ABCB7 interacting with a dimer of FECH (222.4 kDa). Importantly, the same complexes at 120 and 230 kDa were also detected on SDS-PAGE after in vivo crosslinking on isolated mitochondria, followed by anti-FLAG immunoprecipitation of ABCB7-FLAG (Figure 5G). We also confirmed the previously reported interaction of ABCB10 with FECH31 in vitro with purified proteins (Online Supplementary Figure S10A,B), which demonstrated that binding of FECH to ABCB10 was direct. BN-PAGE analysis on ABCB7, FECH and ABCB10 purified proteins showed that each ABC transporter dimerized when loaded individually (Figure 5H), confirming the results obtained using the crosslinker (Figure 5F and Online Supplementary Figure S10A). Both ABCB7 and ABCB10 were able to interact physically with dimers of FECH and, when combined, ABCB7, FECH and ABCB10 assembled into a complex of approximately 480 kDa (Figure 5H, complex designated with the # symbol), which conformed to the predicted size of a hetero-hexameric complex composed of dimers of ABCB7, FECH, and ABCB10. The ABCB7-FECH interaction was disrupted upon expression of ABCB7Mut1, in which the peptide sequence from Val450 through Leu463 had been replaced by alanines (Figures 5F, H), whereas ABCB7Mut5 (residues L517-K526 replaced by alanines) was able to bind FECH and form a complex with ABCB10 to the same extent as ABCB7-WT (Figure 5H). ABCB7Mut6, in which the peptide sequence between amino acid residues Gly527 and Asp538 were replaced by alanines, also did not form a complex with FECH (Online Supplementary Figure S10A). Immunoprecipitation of recombinantly expressed FLAG-tagged ABCB7 wildtype or its mutants (Figure 6A,B) in vivo in G1E-ER4 cells, which co-expressed FECH-HA and that had been silenced to KD the expression of endogenous Abcb7, demonstrated that two short sequences in the C terminus of ABCB7, residues V450-L463 and G527-D538, were essential molecular mediators of the interaction with FECH (Figure 6C-E and Online Supplementary Figure S10C,D), thereby confirming our results obtained in vitro with purified proteins. Residues between Gln464 and Val504 of ABCB7 were also involved in stabilizing the binding of FECH (mutants 2, 3 and 4) (Figure 6C,D, Online Supplementary Figure S10D), whereas the ABCB7 sequences between G505 and Q525 and downstream of Val539 were not involved in the interaction with FECH (Figure 6C-E and Online Supplementary Figure S10C,D). Endogenous Abcb10 coimmunoprecipitated with the ABCB7/FECH complex (Figure 6C,D), consistent with formation of the native complex of 480 kDa (Figure 5H), and the amount recovered in the eluates after immunoprecipitation of ABCB7 decreased upon expression of the ABCB7 mutants, which were defective in binding FECH, suggesting that formation of a multimeric complex containing ABCB7 and ABCB10 homodimers was bridged by a dimeric FECH, rather than through direct physical contacts between the two ABC transporters. In vitro pull-down assays with 35S-radiolabeled ABCB7 wildtype or mutants in the presence of FECH confirmed direct binding of FECH to ABCB7 (Figure 6F-H). The half-life of FECH was significantly reduced in cells lacking endogenous Abcb7 and transfected with ABCB7Mut1 (Online Supplementary Figure S11A,B), which was unable to interact with Fech, indicating that formation of a functional ABCB7/FECH complex was required for FECH stability and completion of heme synthesis.
Figure 5.
ABCB7 forms a complex with ferrochelatase and Abcb10 through direct interaction revealed by crosslinking. (A) Immunoprecipitation (IP) of endogenous Abcb7 in G1E-ER4 cells 30 h after induction of differentiation showed formation of a complex between Abcb7, Fech and Abcb10 that did not include Abcb8 or Mfrn1. (B) Native/two-dimensional (2D) sodium dodecylsulfate polyacrylamide gel electropheresis (SDS-PAGE) analysis on mitochondrial lysates from G1E-ER4 cells showed co-migration of Abcb7, Fech and Abcb10 in a single complex of an approximate molecular weight of 480 kDa. A second distinctive pool of Fech co-migrated with the heme-synthesizing enzymes Ppox and Alas2, indicating formation of a complex at 250 kDa. (C) Map showing the crosslinked sites in the ABCB7/FECH/Abcb10 complex. Inter-subunit crosslinks between ABCB7 and FECH are in blue, and inter-subunit crosslinks between FECH and Abcb10 are in magenta. The lysine residues crosslinked by DDS in the protein sequences are represented by dots. (D) Primary sequence of the C-terminal domain of ABCB7 between residues Val 411 and Cys752. Peptide sequences that were substituted by alanines to test their involvement in the interaction with Fech are highlighted in different colors. (E) Three-dimensional structure of ABCB7 modeled on the structure of S. cerevisiae Atm1 (PDB:4MYC3) using Swiss-Model.49 The last 44 amino acid residues of ABCB7 are missing from the structure because yeast lacks these terminal residues. One of the two protomers of ABCB7 in the dimeric structure is represented in the surface-mode and the green and magenta sequences in the C terminus of ABCB7 indicate the peptide sequences subjected to alanine scanning mutagenesis in Mut1 and Mut5, respectively, to assess their involvement in the interaction with FECH. (F) Coomassie staining of inputs and in vitro crosslinked products on SDS-PAGE. Magenta asterisks denote dimers of ABCB7 wildtype (WT) or the mutant (Mut1) in which amino acid residues between Val450 and Leu463, involved in binding FECH, were replaced by alanines. Blue asterisks indicate the tetrameric ABCB7-FECH complex. Brown circles indicate FECH dimers. (G) SDS-PAGE analysis after in vivo crosslinking on mitochondria isolated from cells co-transfected with ABCB7-F and FECH-HA, followed by anti-FLAG immunoprecipitation. (H) Native PAGE on purified ABCB7-FLAG, FECH-HA and ABCB10-Myc proteins shows that both ABC transporters dimerized when loaded individually (lanes 1 and 3; magenta and black asterisks correspond to dimers of ABCB7 and ABCB10, respectively). Both ABCB7 and ABCB10 were able to interact physically with dimers of FECH (lanes 4 and 6; blue and orange asterisks denote the hetero-tetrameric ABC transporter-FECH complexes). ABCB7, FECH and ABCB10, when combined together in vitro, formed a multiprotein complex with a 2:2:2 stoichiometry consisting of dimers of each of the components (in lane 7, the complex denoted with the # symbol). ABCB7Mut1, in which amino acid residues between Val450 and Leu463 were replaced by alanines, was unable to interact with FECH (lane 5) and formation of the hexameric complex was disrupted (lane 8), whereas Mut5 showed no defect in interacting with FECH (lane 9). Brown circles indicate FECH dimers (lane 2). (A, B, n=5; F, G and H, n=3).
Figure 6.
Mutational analysis of the C-terminal domain of ABCB7 identified the region V450-D538 as being involved in binding ferrochelatase. (A) Primary sequence of the C-terminal domain of ABCB7. (B) Modeled crystal structure of ABCB7. The numbered peptide sequences in (A) and the corresponding colored regions in (B) refer to the amino acid residues that were subjected to alanine scanning mutagenesis to assess their involvement in interacting with FECH. (C-E) Immunoprecipitation (IP) of FLAG-tagged ABCB7 wildtype (B7) or the mutants, as indicated, expressed in G1E-ER4 cells that had been silenced for 3 days to knockdown the expression of endogenous Abcb7 and that co-expressed HA-tagged FECH. Mutants 1, 2, 3, 4 and 6 of ABCB7 [green peptides and domains in (A) and (B), respectively] showed significantly decreased binding to FECH and to Abcb10. (F-H) In vitro pull-down assays of 35S-labeled FLAG-tagged ABCB7 wildtype (B7) or mutants, as indicated, in the presence of 35S–FECH (F) confirmed the results obtained in vivo (C-E) and demonstrated that binding of FECH to ABCB7 was through direct physical interaction. (C-E, n=6. F-H, n=4). See also Online Supplementary Figure S17 for densitometries of immunoblots and statistical analyses.
Discussion
Loss-of-function mutations in ABCB7 cause XLSA with ataxia, a recessive disorder characterized by the presence in the patients’ bone marrow of nucleated erythroblasts that exhibit granules of iron accumulated in the mitochondria surrounding the nuclei.
To gain insights into the primary effects of loss of ABCB7, we generated cell lines with inducible silencing of ABCB7. KD of ABCB7 elicited a dramatic loss of multiple mitochondrial Fe-S enzymes after only 3 days, whereas defects in cytosolic Fe-S proteins were not observed until 5 days after KD. Similarly, studies in Atm1-depleted yeast cells found a severe growth defect caused by mitochondrial dysfunction, which included loss of oxidative respiration and defective heme biosynthesis.35–37 A growing list of human diseases, including sideroblastic anemia, manifest severe mitochondrial iron overload,17 and many of these disorders affect the core components of the iron-sulfur cluster (ISC) machinery, including frataxin,38 glutaredoxin 522,39 and HSPA9.40 The molecular mechanism underlying the mitochondrial iron accumulation has not been unveiled. Studies in cell culture models have revealed that a feature of defects manifesting mitochondrial iron overload included the activation of the cytosolic iron starvation response,17,38,41 which increased the IRE-binding activities of IRP1 and IRP2, and upregulation of the ubiquitously expressed mitochondrial iron importer MFRN2.41,42 Our studies show that activation of IRP in the cytosol of cells depleted of ABCB7 and upregulation of the mitochondrial iron importers, MFRN1 and MFRN2, occurred 3 days after KD of ABCB7 and accounted for the increased influx of iron in mitochondria. The decreased cytosolic aconitase activity in ABCB7-depleted cells was therefore likely the result of the conversion of IRP1 from cytosolic aconitase into IRE-binding apoprotein, rather than the result of impaired cytosolic ISC biogenesis, as previously proposed.11 Similarly, yeast cells depleted of Atm1 were reported to activate the iron regulon, which encodes the high affinity iron uptake system.43,44
Despite the large amount of iron imported in mitochondria, cells lacking Abcb7 showed a defect in heme biosynthesis, which resulted not only from translational repression of Alas2 by IRP, but also from the decreased stability of ferrochelatase. Recent studies proposed that in a mouse model of frataxin deficiency, mitochondrial iron overload was mediated by the upregulation of MFRN2, which was driven by defective heme biosynthesis, due to loss of FECH.45 Interestingly, mitochondrial iron accumulation has also been reported in erythroblasts from patients with erythropoietic protoporphyria due to decreased ferrochelatase levels,46 suggesting that loss of FECH and/or its product, heme, in cells with an intact IRE-IRP system may drive mitochondrial iron overload.
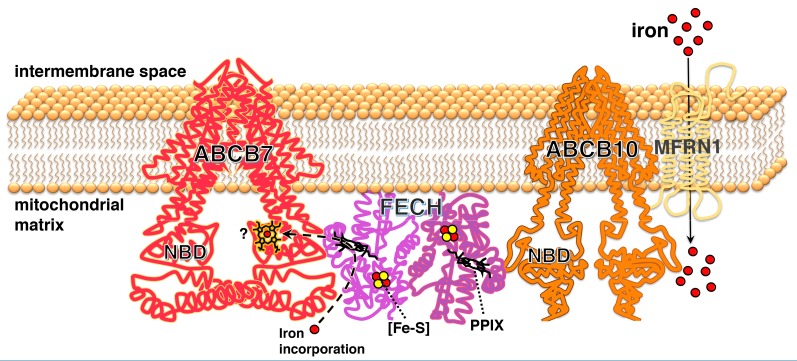
The identification and characterization of protein–protein interactions are fundamental to understanding the mechanisms and regulation of most biological processes, and interacting partners provide insights into biological functions that can be exploited for therapeutic purposes. We found that endogenous Abcb7 formed a complex with Fech and Abcb10 during erythroid differentiation. We performed XL-MS to investigate the architecture of the ABCB7/FECH complex, followed by extensive mutational analyses of ABCB7. Our studies identified two sequences in the C-terminal domain of ABCB7, residues V450-V504 and G527-D538, which were major molecular determinants of the interaction with FECH. Interestingly, these regions are adjacent to the Walker A motif of ABCB7, which is essential for nucleotide binding and transport activity.47 The overall architecture of the ABCB7/FECH/ABCB10 complex is shown in our proposed model (Figure 7), in which a functional dimeric FECH bridges ABCB7 and ABCB10 homo -dimers. Interestingly, residues 90-115 of FECH enclose the enzymatic active site containing PPIX and specifically bind ABCB7 near its nucleotide-binding domain. It is tempting to speculate that hydrolysis of ATP by ABCB7 may drive a conformational rearrangement on the region 90-115 of FECH, which would enable opening of the pocket and release of the newly synthesized proto-heme.
Figure 7.
Dimeric ferrochelatase bridges ABCB7 and ABCB10 homodimers in a molecular complex required for heme biosynthesis. A functional dimeric ferrochelatase (FECH) bridges ABCB7 and ABCB10 by binding near the nucleotide-binding domains (NDB) of each ATB-binding cassette (ABC) transporter. Two sequences in the C terminus of human ABCB7, regions V450-L463 and G527-D538, were essential for binding to FECH. Amino acid residues 90-115 of FECH, which formed the closing gate of the active site of the enzyme containing protoporphyrin IX (PPIX) were specifically involved in the interaction with ABCB7. The region between lysines 133 and 145 of FECH interacted with both ABCB7 and ABCB10. Likely the 133-145 region of one protomer of FECH interacted with ABCB7 and the corresponding region of the other protomer of the dimeric enzyme interacted with ABCB10. Interestingly, FECH interacted directly with regions of ABCB7 and ABCB10 near the NBD which are enriched in histidines, residues that are known heme-ligating amino acids (see also Online Supplementary Figure S12), raising the possibility that one or both ABC transporters may function as a mitochondrial matrix heme exporter.
A previous model was proposed based on the identification of a Fech/Abcb10/Mfrn1 complex,31 in which the interaction of Fech with Abcb10 and Mfrn1 was required to integrate mitochondrial iron import with its utilization for heme synthesis. In the same study,31 a separate interaction of Fech with Abcb7 was also reported. We did not identify Mfrn1 as part of the Abcb7/Fech/Abcb10 complex (Figure 5A and Online Supplementary Table S1). However, since Mfrn1 was reported to be the direct interacting partner of Abcb10,31 it is possible that in our coimmunoprecipitation experiments using an anti-Abcb7 antibody we mainly immunocaptured Abcb7 and its closest interacting partner, FECH, which in turn mediated an interaction with Abcb10; thus, we may not have detected other interactions of Abcb10 with Mfrn1 which are more physically distant from the Abcb7 partners. Nonetheless, Mfrn1 upregulation was essential to meet the exceptionally high iron demand for heme biosynthesis of erythroid cells during differentiation (Figure 4A-G), and decreased Mfrn1 half-life in the iron-deficient conditions generated by loss of Irp2 caused mitochondrial iron deficiency and impaired heme biosynthesis (Figure 4A-G). Our studies offer a potential molecular mechanism for reported cases of erythropoietic protoporphyria in patients harboring mutations between residues 68 and 220,48 which we found were involved in binding ABCB7. Overall, our studies highlight the importance of ABCB7 for mitochondrial function and provide the biochemical characterization of a functional complex formed of ABCB7, FECH and ABCB10, which is required for cellular iron homeostasis, mitochondrial function and heme biosynthesis. Our work suggests that more definitive experiments deploying coordinated activity of the entire complex may aid identification of its physiological substrates.
Acknowledgments
The authors thank Dr. Manik Ghosh for technical help, members of the Rouault laboratory for helpful discussion and the Eunice Kennedy Shriver NICHD Intramural Research Program for support.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/9/1756
References
- 1.Gbelska Y, Krijger JJ, Breunig KD. Evolution of gene families: the multidrug resistance transporter genes in five related yeast species. FEMS Yeast Res. 2006;6(3):345–355. [DOI] [PubMed] [Google Scholar]
- 2.Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 2018;18(7):452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srinivasan V, Pierik AJ, Lill R. Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1. Science. 2014;343(6175):1137–1140. [DOI] [PubMed] [Google Scholar]
- 4.Lee JY, Yang JG, Zhitnitsky D, Lewinson O, Rees DC. Structural basis for heavy metal detoxification by an Atm1-type ABC exporter. Science. 2014;343(6175):1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zutz A, Gompf S, Schagger H, Tampe R. Mitochondrial ABC proteins in health and disease. Biochim Biophys Acta. 2009;1787(6):681–690. [DOI] [PubMed] [Google Scholar]
- 6.Krishnamurthy PC, Du G, Fukuda Y, et al. Identification of a mammalian mitochondrial porphyrin transporter. Nature. 2006;443(7111):586–589. [DOI] [PubMed] [Google Scholar]
- 7.Tsuchida M, Emi Y, Kida Y, Sakaguchi M. Human ABC transporter isoform B6 (ABCB6) localizes primarily in the Golgi apparatus. Biochem Biophys Res Commun. 2008;369(2):369–375. [DOI] [PubMed] [Google Scholar]
- 8.Kiss K, Brozik A, Kucsma N, et al. Shifting the paradigm: the putative mitochondrial protein ABCB6 resides in the lysosomes of cells and in the plasma membrane of erythrocytes. PLoS One. 2012;7(5):e37378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paterson JK, Shukla S, Black CM, et al. Human ABCB6 localizes to both the outer mitochondrial membrane and the plasma membrane. Biochemistry. 2007;46(33):9443–9452. [DOI] [PubMed] [Google Scholar]
- 10.Savary S, Allikmets R, Denizot F, et al. Isolation and chromosomal mapping of a novel ATP-binding cassette transporter conserved in mouse and human. Genomics. 1997;41(2):275–278. [DOI] [PubMed] [Google Scholar]
- 11.Pondarre C, Antiochos BB, Campagna DR, et al. The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Hum Mol Genet. 2006;15(6):953–964. [DOI] [PubMed] [Google Scholar]
- 12.Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum Mol Genet. 1999;8(5):743–749. [DOI] [PubMed] [Google Scholar]
- 13.Bekri S, Kispal G, Lange H, et al. Human ABC7 transporter: gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic iron-sulfur protein maturation. Blood. 2000;96(9):3256–3264. [PubMed] [Google Scholar]
- 14.Maguire A, Hellier K, Hammans S, May A. X-linked cerebellar ataxia and sideroblastic anaemia associated with a missense mutation in the ABC7 gene predicting V411L. Br J Haematol. 2001;115(4):910–917. [DOI] [PubMed] [Google Scholar]
- 15.Pondarre C, Campagna DR, Antiochos B, Sikorski L, Mulhern H, Fleming MD. Abcb7, the gene responsible for X-linked sideroblastic anemia with ataxia, is essential for hematopoiesis. Blood. 2007;109(8):3567–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17(3):1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rouault TA. Mitochondrial iron overload: causes and consequences. Curr Opin Genet Dev. 2016;38:31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouault TA, Maio N. Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J Biol Chem. 2017;292(31):12744–12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryu MS, Zhang D, Protchenko O, Shakoury-Elizeh M, Philpott CC. PCBP1 and NCOA4 regulate erythroid iron storage and heme biosynthesis. J Clin Invest. 2017;127(5):1786–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welch JJ, Watts JA, Vakoc CR, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104(10):3136–3147. [DOI] [PubMed] [Google Scholar]
- 21.Melefors O, Goossen B, Johansson HE, Stripecke R, Gray NK, Hentze MW. Translational control of 5-aminolevulinate synthase mRNA by iron-responsive elements in erythroid cells. J Biol Chem. 1993;268(8):5974–5978. [PubMed] [Google Scholar]
- 22.Ye H, Jeong SY, Ghosh MC, et al. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest. 2010;120(5):1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zenke-Kawasaki Y, Dohi Y, Katoh Y, et al. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol Cell Biol. 2007;27(19):6962–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tahara T, Sun J, Igarashi K, Taketani S. Heme-dependent up-regulation of the alpha-globin gene expression by transcriptional repressor Bach1 in erythroid cells. Biochem Biophys Res Commun. 2004;324(1):77–85. [DOI] [PubMed] [Google Scholar]
- 25.Dailey HA, Meissner PN. Erythroid heme biosynthesis and its disorders. Cold Spring Harb Perspect Med. 2013;3(4):a011676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson KD, Laver WG, Neuberger A. Initial stages in the biosynthesis of porphyrins. 2. The formation of delta-aminolaevulic acid from glycine and succinylcoenzyme A by particles from chicken erythrocytes. Biochem J. 1958;70(1):71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitman JC, Paw BH, Chung J. The role of ClpX in erythropoietic protoporphyria. Hematol Transfus Cell Ther. 2018;40(2):182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JJ. Translational control by heme-regulated eIF2alpha kinase during erythropoiesis. Curr Opin Hematol. 2014;21(3):172–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooperman SS, Meyron-Holtz EG, Olivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005;106(3): 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taketani S, Kakimoto K, Ueta H, Masaki R, Furukawa T. Involvement of ABC7 in the biosynthesis of heme in erythroid cells: interaction of ABC7 with ferrochelatase. Blood. 2003;101(8):3274–3280. [DOI] [PubMed] [Google Scholar]
- 31.Chen W, Dailey HA, Paw BH. Ferrochelatase forms an oligomeric complex with mitofer-rin-1 and Abcb10 for erythroid heme biosynthesis. Blood. 2010;116(4):628–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medlock AE, Shiferaw MT, Marcero JR, et al. Identification of the mitochondrial heme metabolism complex. PLoS One. 2015;10(8): e0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medlock AE, Dailey TA, Ross TA, Dailey HA, Lanzilotta WN. A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase. J Mol Biol. 2007;373(4):1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medlock A, Swartz L, Dailey TA, Dailey HA, Lanzilotta WN. Substrate interactions with human ferrochelatase. Proc Natl Acad Sci U S A. 2007;104(6):1789–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kispal G, Csere P, Guiard B, Lill R. The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 1997;418(3):346–350. [DOI] [PubMed] [Google Scholar]
- 36.Leighton J, Schatz G. An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. EMBO J. 1995;14(1):188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao R, Kim H, Koppolu UM, Ellis EA, Scott RA, Lindahl PA. Biophysical characterization of the iron in mitochondria from Atm1p-depleted Saccharomyces cerevisiae. Biochemistry. 2009;48(40):9556–9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puccio H, Simon D, Cossee M, et al. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat Genet. 2001;27(2):181–186. [DOI] [PubMed] [Google Scholar]
- 39.Camaschella C, Campanella A, De Falco L, et al. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood. 2007;110(4):1353–1358. [DOI] [PubMed] [Google Scholar]
- 40.Schmitz-Abe K, Ciesielski SJ, Schmidt PJ, et al. Congenital sideroblastic anemia due to mutations in the mitochondrial HSP70 homologue HSPA9. Blood. 2015;126(25): 2734–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crooks DR, Jeong SY, Tong WH, et al. Tissue specificity of a human mitochondrial disease: differentiation-enhanced mis-splicing of the Fe-S scaffold gene ISCU renders patient cells more sensitive to oxidative stress in ISCU myopathy. J Biol Chem. 2012;287(48):40119–40130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martelli A, Puccio H. Dysregulation of cellular iron metabolism in Friedreich ataxia: from primary iron-sulfur cluster deficit to mitochondrial iron accumulation. Front Pharmacol. 2014;5:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutherford JC, Ojeda L, Balk J, Muhlenhoff U, Lill R, Winge DR. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J Biol Chem. 2005;280(11):10135–10540. [DOI] [PubMed] [Google Scholar]
- 44.Hausmann A, Samans B, Lill R, Muhlenhoff U. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J Biol Chem. 2008;283(13):8318–8330. [DOI] [PubMed] [Google Scholar]
- 45.Martelli A, Schmucker S, Reutenauer L, et al. Iron regulatory protein 1 sustains mitochondrial iron loading and function in frataxin deficiency. Cell Metab. 2015;21(2):311–323. [DOI] [PubMed] [Google Scholar]
- 46.Rademakers LH, Koningsberger JC, Sorber CW, Baart de la Faille H, Van Hattum J, Marx JJ. Accumulation of iron in erythroblasts of patients with erythropoietic protoporphyria. Eur J Clin Invest. 1993;23(2):130–138. [DOI] [PubMed] [Google Scholar]
- 47.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rufenacht UB, Gouya L, Schneider-Yin X, et al. Systematic analysis of molecular defects in the ferrochelatase gene from patients with erythropoietic protoporphyria. Am J Hum Genet. 1998;62(6):1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]