Abstract
The complex, frequently devastating, multi-organ pathophysiology of sickle cell disease has a single root cause: polymerization of deoxygenated sickle hemoglobin. A logical approach to disease modification is, therefore, to interdict this root cause. Ideally, such interdiction would utilize small molecules that are practical and accessible for worldwide application. Two types of such small molecule strategies are actively being evaluated in the clinic. The first strategy intends to shift red blood cell precursor hemoglobin manufacturing away from sickle hemoglobin and towards fetal hemoglobin, which inhibits sickle hemoglobin polymerization by a number of mechanisms. The second strategy intends to chemically modify sickle hemoglobin directly in order to inhibit its polymerization. Important lessons have been learnt from the pre-clinical and clinical evaluations to date. Open questions remain, but this review summarizes the valuable experience and knowledge already gained, which can guide ongoing and future efforts for molecular mechanism-based, practical and accessible disease modification of sickle cell disease.
Introduction
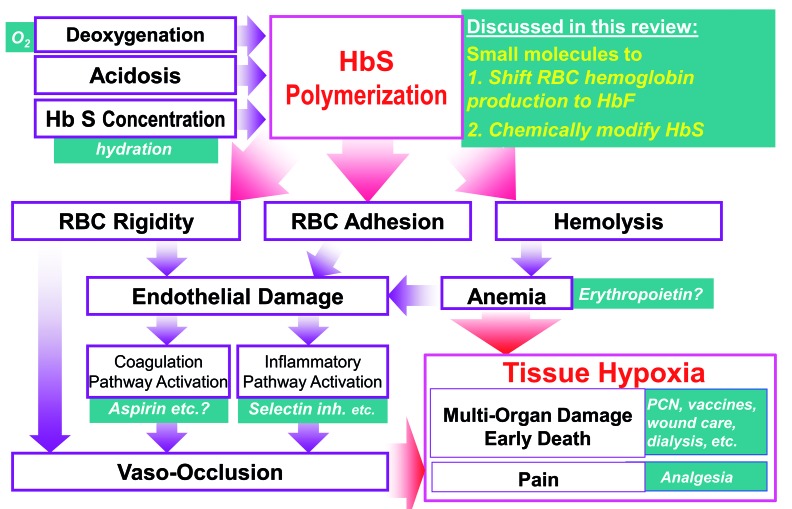
Sickle cell disease (SCD) demands practical, accessible oral therapies, since it is a problem of global scope. It afflicts millions of people worldwide, and has an especially high prevalence in pediatric populations in low-income, malaria-belt countries.1 Such therapies are technically plausible, since despite the complex and potentially devastating multi-organ pathophysiology of SCD, this condition has a single, well-characterized root cause: polymerization of deoxygenated sickle hemoglobin (HbS). The hemoglobin molecule is an assembly of two a-like protein subunits and two β-like protein subunits (a2β2), each with a heme moiety to transport an oxygen molecule. In SCD, the gene for the β sub-unit (HBB) of adult hemoglobin (HbA) contains an ‘A’ to ‘T’ mutation in the seventh codon. The β sub-units (βS) produced by this mutated gene substitute a hydrophilic glutamate with a hydrophobic valine, predisposing deoxygenated HbS (a2βS2) to polymerization and gelation in red blood cells (RBC). This affects RBC viability, rheology and adhesiveness, promoting hemolysis, endothelial damage, occlusion of small blood vessels, and thromboses of large vessels. The hemolytic anemia is frequently severe, and is only partially and non-sustainably compensated by >10-fold increases in erythropoiesis.2 The net consequence of this anemia and vaso-occlusion is decreased oxygen delivery and hypoxic injury to potentially all tissues of the body, manifest clinically as episodic pain, chronic pain, avascular necrosis of bones, infections, overt and silent strokes, renal/respiratory/cardiac/hepatic failure, and early death. In the USA >$1 billion in annual health care costs is attributed to SCD, and even so, the median life expectancy of affected individuals is shortened by two or more decades on average.3,4 Most children with SCD in low-income countries do not even survive to adulthood.1 By way of emphasis, all this morbidity and mortality begins with a single process, polymerization of deoxygenated HbS in RBC, and it is therefore logical to attempt to interdict this root cause. Two major small molecule drug approaches are in active clinical evaluation: (i) small molecules to shift the hemoglobin manufactured by RBC precursors from HbS to fetal hemoglobin (HbF), and (ii) small molecules to chemically modify HbS to impede its polymerization. These active efforts are discussed in turn, with an emphasis on lessons learned so far and remaining open questions.
Small molecule approaches for which there are no active clinical efforts that we are aware of are not discussed here, e.g., small molecules to decrease HbS concentration by increasing RBC hydration.5,6 Methods to interdict HbS polymerization that are not based on small molecule drugs are also not discussed here, because their application in the areas of the world most affected by SCD will be difficult for reasons of infrastructure and costs, e.g., harvesting of autologous hematopoietic stem cells, their engineering ex vivo, then re-infusion after myeloablative bone marrow conditioning by chemotherapy and/or radiation (gene therapy), or use of hematopoietic stem cells from immune-compatible non-SCD donors for transplant – a valuable approach in the West that has been thoroughly and recently reviewed elsewhere.7
Interdicting HbS polymerization by pharmacological induction of HbF
At the fetal stage of life, RBC contain fetal hemoglobin (HbF), an assembly of two a-globin subunits and two γ-globin subunits (a2γ2), with the γ-globin subunits being encoded by duplicated γ-globin genes (HBG2 and HBG1). During human development, the switch from HbF to HbA production begins late in fetal gestation (~ 7 months), and the typical adult pattern of <1% HbF and >90% HbA in RBC is established by ~12 months post-conception.8,9 Several genetic polymorphisms or mutations in humans, some but not all identified, promote persistent, relatively high RBC HbF content beyond infancy. The phenotypes with particularly generous HbF levels (HbF >10%) are referred to as hereditary persistence of fetal hemoglobin (HPFH). SCD patients who co-inherit such genetic variants can, in the best cases, have asymptomatic, normal life-spans.10–12 Notably, HbF has benefits even at lower dynamic ranges than seen in HPFH: HbF levels correlate continuously with fewer vaso-occlusive pain crises, less renal damage, less pulmonary hypertension, fewer strokes and longer survival.4,13–19 In short, nature has demonstrated that HbF is a highly potent modulator of SCD.20
Detailed biochemical studies have demonstrated how: the intracellular concentration of HbS is a major determinant of polymerization kinetics, and HbF substitution for HbS decreases this concentration.20–22 Moreover, HbF does not polymerize with deoxygenated HbS for reasons of molecular structure (the sophisticated biophysics underlying this have recently been reviewed in detail).5 By contrast, HbA can polymerize with deoxygenated HbS.20–22 In short, HbF interdicts the root-cause pathophysiology of SCD. It is logical therefore to attempt to use pharmacology to recapitulate such naturally demonstrated, powerful disease modulation.23
The earliest efforts at HbF induction
The earliest efforts built on the observation that HbF is enriched in RBC produced during the recovery phase of bone marrow from severe insults or stress.24–29 One way of creating such stress is to administer cytotoxic (cell killing) drugs, leading to clinical evaluation in SCD of the oral ribonucleotide reductase inhibitor hydroxyurea.28–30 In the pivotal trial, hydroxyurea (15-35 mg/kg) increased HbF for 2 years in ~50% of the adult SCD patients treated.30,31 As predicted, HbF increases with hydroxyurea correlated strongly with longer RBC half-life,32,33 fewer pain crises,31 and better quality of life34 (the benefits of hydroxyurea therapy in sickle cell mice also depended on HbF induction).35 Trial patients with HbF levels >0.5 g/dL also survived longer15 although a caveat to these analyses was that it was not known whether the higher HbF levels were intrinsic to the patients or a result of the hydroxyurea therapy.
There were, however, noteworthy limitations to the induction of HbF by hydroxyurea: (i) average HbF increases at 2 years were modest (3.6%);28–31,36 (ii) HbF increases were particularly unlikely in patients with the lowest baseline HbF levels and thus at highest risk of morbidity and mortality,31,33,37,38 and (iii) HbF increases diminished over time, even in the ~50% of patients with excellent initial HbF induction.31,39
A shared basis for these several limitations was suggested by the correlation between lower HbF increases and fewer reticulocytes (<300,000×109/L) and neutrophils (<7.5×109/L) at baseline: this correlation underscored that HbF induction by cytotoxicity requires sufficient reserves of hematopoietic precursors to mount repeated recoveries from the stress that destroys their counterparts.29,31 Such reserves are circumscribed, subject to attrition via vaso-occlusion in the marrow and kidneys, and decline with aging.31,33,37,38 A declining capacity to compensate for hemolytic anemia is a problem even separate from considerations of sustainable HbF induction via cytotoxicity: SCD patients require erythropoiesis at >10-fold the normal rate simply to sustain hemoglobin levels compatible with life, and dwindling compensatory reticulocytosis is a major cause of early death.2,15,31,40 Therefore, alternative, non-cytotoxic, durable, and more potent methods of inducing HbF are needed.
Directly targeting the enzymes that silence the γ-globin gene
DNA in nuclei is packaged together with RNA and structural proteins – histones - to form chromatin. Chromatin regulates gene transcription by determining accessibility of genes to the massive machinery (~150 proteins) that transcribes genes. Reorganization (‘remodeling’) of chromatin, to facilitate or hinder this machinery, is signaled via post-translational modifications to histones -methylation and acetylation of lysine residues, phosphorylation of threonines and serines – and by modifications to DNA, mainly, methylation of cytosines that precede guanines (CpG). These signals determine whether ATP-dependent chromatin remodelers shift histones towards or away from gene transcription start sites, repositioning these physical barriers to either welcome or obstruct the gene transcribing basal transcription factor machinery.
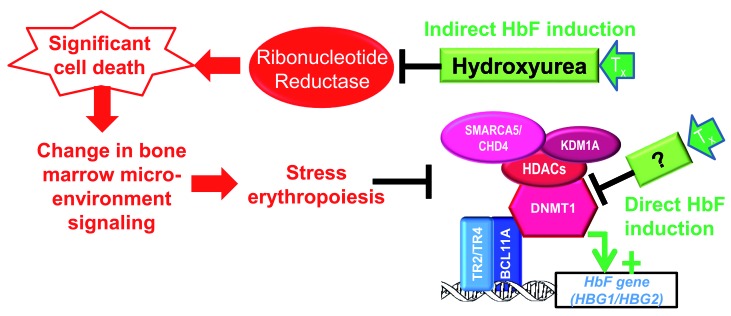
Thus, induction of HbF, even when it is indirectly via bone marrow stress, implies remodeling γ-globin and β-globin gene loci, to activate one and not the other.41 Specifically, persistent HbF expression requires: (i) decreased operation at HBG2/HBG1 of epigenetic enzymes that create ‘off’ marks and that reposition histones to obstruct transcription start sites, and (ii) increased function of the epigenetic enzymes that create epigenetic ‘on’ marks and that reposition histones away from transcription start sites, with vice versa at HBB. Cytotoxic methods of inducing HbF achieve such chromatin remodeling crudely and indirectly, via bone marrow stress29,41,42 (Figure 2).
Figure 2.
Induction of fetal hemoglobin (HbF) requires chromatin remodeling, including DNA hypomethylation, of the HbF gene locus. Bone marrow stress, e.g., from cytotoxic drugs such as hydroxyurea, can create chromatin remodeling during the recovery phase of surviving erythroid precursors. An alternative approach is to remodel the hemoglobin F (HbF) gene locus (HBG) directly, e.g., by directly inhibiting/repressing epigenetic enzymes. Enzymes shown are those known to be recruited by BCL11A, TR2 or TR4 (EHMT2 and PRMT5 are not reported participants in the BCL11A/TR2/TR4 hub). The relative efficiencies of these approaches are illustrated by the greater HbF increases produced in the same non-human primates or patients by decitabine ~0.2 mg/kg twice weekly versus hydroxyurea ~20 mg/kg daily. That is, the molar amount of decitabine administered per week is <1/1000th the amount of hydroxyurea administered per week. We published variations of this figure in Molokie et al.95 and Lavelle et al.23
Figure 1.
Polymerization of sickle hemoglobin drives the multi-organ cascade of sickle cell disease pathophysiology. This review examines the strategies to interdict the multi-organ cascade of sickle cell disease at its inception using small molecules that shift red blood cell precursor production from sickle hemoglobin (HbS) toward fetal hemoglobin (HbF), and small molecules that chemically modify HbS to decrease its polymerization. We published variations of this figure in Molokie et al.95 and Lavelle et al.23
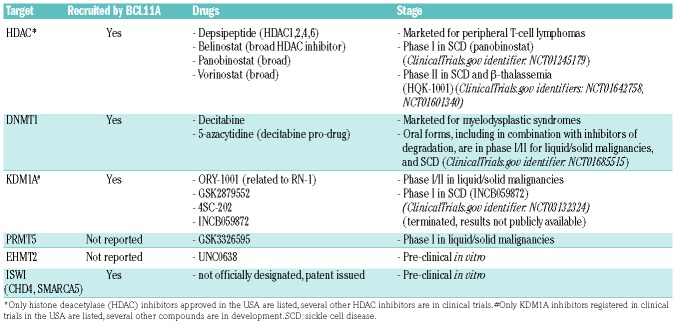
So why not identify repressing epigenetic enzymes and inhibit them directly43,44 (Figure 2)? Cells contain dozens of epigenetic enzymes mediating gene activation and repression, and not all repressing epigenetic enzymes (corepressor protein complexes) are logical molecular targets for therapy. Sequence-specific DNA-binding factors are particular in their epigenetic co-regulator usage, e.g., even distinguishing between closely similar BAF and PBAF coactivator complexes.45–48 Logically, the epigenetic enzymes to target for HbF induction are those that have been directly implicated in silencing of HBG2/HBG1. Multi-protein corepressor complexes directed to the HBG loci by the DNA-binding factors DRED and BCL11A have been characterized in great detail.49–53 Druggable epigenetic silencing enzymes contained in these recruited corepressor complexes include DNA methyltransferase 1 (DNMT1), various histone deacetylases (HDAC), lysine demethylase 1 (LSD1, KDM1A), and chromodomain helicase DNA binding protein 4 (CHD4) and other members of the ISWI family of ATP-dependent chromatin remodelers52–55 (Table 1). Other types of biochemical studies have implicated euchromatic histone lysine methyltransferase 2 (EHMT2, G9a),56 and protein arginine methyltransferase 5 (PRMT5) in the silencing of HBG2/HBG157,58 (Table 1). Yet another approach to identifying candidate targets has been chemical screens for HbF inducers. This approach has identified histone methyltransferases EHMT1 and EHMT2 as candidates for inhibition59,60 (Table 1). Notably not identified by studies thus far, given that there are clinically available inhibitors for these targets, are epigenetic enzymes in polycomb repressor complex 2 (e.g., EZH2).61
Table 1.
Scientifically validated molecular targets for HbF induction and candidate drugs
Since the natural genetic experiment of HPFH provides a fundamental rationale for pursuing pharmacological induction of HbF, by extension, can the genetic variants underlying HPFH help to identify or prioritize molecular targets for manipulation? HPFH-linked point mutations cluster in two regions 115 and 200 base-pairs upstream of the HBG2 start site, suggesting these are sites at which key repressors of HBG2/HBG1 bind.62 BCL11A and ZBTB7A have been shown to bind at these locations, and HPFH mutations have been shown to abrogate such binding.63 Moreover, some HPFH mutations occur at BCL11A rather than β-globin gene loci.51,64 In short, the natural genetic experiment of HPFH also seems to support drugging of the corepressors recruited by BCL11A (and ZBTB7A and DRED).49–53
The candidate targets are discussed below in turn. Histone deacetylases (HDAC)
HDAC were among the first candidate targets identified for HbF induction.65 Moreover, a number of HDAC inhibitors have already been approved by the United States Food and Drug Administration (FDA) to treat peripheral T-cell malignancies (Table 1). Unfortunately, despite exciting pre-clinical results, clinical application of marketed HDAC inhibitors for HbF induction is limited by the pleiotropic roles of HDAC outside of chromatin. That is, clinical side-effects, arising from HDAC participation in multiple cellular and physiological functions, limit the achievement of an epigenetic pharmacodynamic effect in the target compartment, and thus of HbF induction in vivo.66–71 There are efforts to develop HDAC inhibitors that are more selective to specific HDAC than the broad HDAC inhibiting activity of the currently marketed drugs (Table 1), and perhaps these more selective agents will have a more suitable safety profile for HbF induction. The caution remains that even an on-target, specific drug action can generate toxicities if the molecular target of that action has pleiotropic physiological roles.
DNA methyltransferase 1 (DNMT1)
DNMT1 is well known to maintain methylation marks on DNA through cell division. In addition, DNMT1 is a corepressor that is recruited by sequence-specific DNA-binding factors, e.g., DRED (TR2/TR4) and BCL11A, which direct epigenetic silencing of HBG.43,53,72–82 The deoxycytidine analog decitabine and its pro-drug 5-azacy-tidine, FDA-approved to treat the myeloid malignancy myelodysplastic syndrome, can deplete DNMT1: a nitrogen substituted for a carbon in the decitabine pyrimidine ring covalently binds to DNMT1 and causes its degradation.83 By depleting DNMT1 protein, decitabine disrupts its scaffolding functions for other epigenetic enzymes such as KDM1A.84,85 That is, decitabine does not just inhibit the enzyme function of DNMT1 but produces a broad corepressor disrupting effect. Because the deoxyribose moiety of decitabine is unmodified, it can incorporate into the elongating DNA strand during the S-phase without terminating chain extension or causing cytotoxicity, contrasting with most nucleoside analogs used in the clinic to treat cancer.86,87 High concentrations of decitabine do, however, produce off-target anti-metabolite effects and cytotoxicity, in significant part via its uridine moiety degradation products that can misincorporate into DNA or inhibit thymidylate synthase.88,89 We designed decitabine dose, schedule and route-of-administration regimens to produce non-cytotoxic depletion of DNMT1 in vivo.43,90–93 These regimens increased HbF by >10% in SCD patients who had no HbF response (~0.3%) to hydroxyurea in the pivotal clinical trial.43,81,94 That is, very small, non-cytotoxic doses of ~0.2 mg/kg twice weekly were sufficient to produce large increases in HbF and total hemoglobins, even in patients in whom hydroxyurea ~20 mg/kg/day, >1000-fold the molar amount of decitabine, did not induce HbF (Figure 2).43,44
Marketed decitabine, however, is a parenteral drug with trivial oral bioavailability, undermining potential for worldwide application. We have therefore combined oral decitabine with tetrahydrouridine to inhibit the enzyme that limits its oral bioavailability, cytidine deaminase.95 This combination was well-tolerated and safe in a phase I study in patients with severe SCD. The target decitabine dose of 0.16 mg/kg produced a wide decitabine concentration-time profile (low Cmax, long Tmax) ideal for non-cytotoxic DNMT1 depletion83,96–98 and decreased DNMT1 protein in peripheral blood mononuclear cells by >75% and repetitive element CpG methylation by ~10%. This increased HbF by 4-9%, doubling HbF-enriched RBC (F-cells) up to ~80% of total RBC. Total hemoglobin increased by 1.2-1.9 g/dL (P=0.01) as reticulocytes simultaneously decreased; that is, better quality and efficiency of HbF-enriched erythropoiesis elevated hemoglobin using fewer reticulocytes. Other indications of better RBC quality, biomarkers of hemolysis, thrombophilia and inflammation (lactate dehydrogenase, bilirubin, D-dimer, C-reactive protein) also improved. The side-effects were a concurrent increase in platelets and decrease in neutrophils, expected with non-cytotoxic DNMT1 depletion. In the relatively short treatment duration of 8 weeks, these blood count shifts did not cross thresholds requiring withholding or modification of treatment, that is, neutrophil counts and platelets remained in ranges observed in SCD patients receiving standard-of-care therapies.
The major limitation is the need for longer term studies to demonstrate durable safety and efficacy of the oral tetrahydrouridine/decitabine combination.
Lysine demethylase 1 (LSD1, KDM1A)
KDM1A, like DNMT1, is recruited by the HBG2/HBG1 repressing DNA-binding factors DRED and BCL11A, and KDM1A inhibition with either of two specific inhibitors induced HbF in vitro, in sickle mice and in non-human primates.99–102 Several KDM1A inhibitors are in clinical trials for cancer indications (Table 1). At least two of the compounds in trials (ORY-1001, GSK2879552) are built around a tranylcypropamine warhead that inhibits monoamine oxidases that metabolize catecholamine neurotransmitters in the brain. Although cancer clinical trials are ongoing and unpublished (EudraCT number: 2013-002447-29; ClinicalTrials.gov identifiers: NCT02177812, NCT02034123), there is concern regarding side-effects related to the potential for inhibition of monoamine oxidases other than KDM1A. Thus, there are ongoing efforts to develop and evaluate KDM1A inhibitors with other scaffolds (e.g., ClinicalTrials.gov identifier: NCT01344707). One registered phase I clinical trial evaluated a KDM1A inhibitor for HbF induction in SCD (ClinicalTrials.gov identifier: NCT03132324). This trial has been terminated but results are not publicly available at this time.
Protein arginine methyltransferase 5 (PRMT5)
PRMT5 methylation of histone H4 arginine 3 has been implicated as a signal that recruits additional chromatin-modifying enzymes and represses HBG.57 There is a PRMT5 inhibitor in clinical trials (GSK3326595) for cancer indications. No trials of this molecule for HbF induction in SCD have been registered so far.
Euchromatic histone lysine methyltransferase 2 (EHMT2, G9a)
EHMT2 has been shown to be recruited to the β-globin locus by the sequence-specific DNA binding factor NFE2, and the EHMT2 inhibitor UNC0638 has been shown to induce HbF in vitro.59,60 As of this time, there are no registered clinical trials evaluating EHMT2 inhibition to induce HbF.
Chromodomain helicase DNA binding protein 4 (CHD4) and SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (SMARCA5) (ISWI family of ATP-dependent chromatin remodelers
The culmination of chromatin remodeling for gene repression or activation is nucleosome (histone octamer) repositioning around the transcription start site. This is energetically expensive work executed by SWI/SNF or ISWI family proteins containing the HELICc-DExx ATP-ase domain, with SWI/SNF moving histones away to facilitate basal transcription factor machinery access and ISWI executing the opposite.46,103,104 Since such nucleosome repositioning is the crux of chromatin remodeling, inhibition of this action should in principle offer corresponding potency. CHD4 and SMARCA5 are HELICc-DExx-containing corepressors that are recruited by BCL11A and DRED to repress HBG2/HBG1.54 We have identified a first-in-class drug-like compound series that preliminarily appears to inhibit the HELICc-DExx domains of SMARCA5 and CHD4, and we are actively investigating the potential of this series to induce HbF (US20170253589A1).
Epigenetic targeting - Lessons so far and open questions
Pre-clinical and clinical experience to date provide various lessons and raise some questions regarding epigenetic targeting to induce HbF, as described below.
The consequences of inhibiting an epigenetic enzyme depend on cellular context
The baseline expression pattern of transcription factors is a key determinant of a cell’s fate or function response to epigenetic enzyme inhibition, because sequence-specific DNA-binding factors direct the function of these epigenetic enzymes and are mandatory for gene activation.105 Stated another way, the consequences of inhibiting a particular epigenetic enzyme depend very much on cellular context.105 A corollary of the above is that although inhibiting silencing epigenetic enzymes can produce cell fate or function shifts, these relate to what the cells were to begin with and are not drastic.105 This is of course critical clinically, since a candidate epigenetic therapeutic for SCD will be distributed systemically.
What then about the cellular/transcription factor context of erythropoiesis enables inhibition of DNMT1 etc., to activate HBG2/HBG1? Several groups have found that the developmental switch from HBG2/HBG1 to HBB activation is recapitulated, albeit very rapidly, during erythroid lineage maturation (a ‘maturational switch’ during routine erythropoiesis).106–110 The maturational switch entails removal of activating and acquisition of repressive epigenetic marks at HBG2/HBG1.59,102 with physical migration of the shared enhancer, the locus control region, from the HBG2/HBG1 to the HBB locus.54 These dynamics at HBG2/HBG1 and HBB during erythropoiesis creates an opportunity for pharmacological/biochemical intervention to prevent enhancer migration, to stall the massive gene activating machinery at HBG2/HBG1. That is, HbF induction by inhibiting epigenetic ‘off’ enzymes such as DNMT1 is not predicated on returning the enhancer from HBB back to HBG2/HBG1 (turning a gene that is ‘off’ to ‘on’), but on preventing a switch from HBG2/HBG1 to HBB in the first place (preventing a gene that is ‘on’ from being turned ‘off’). Accordingly, HbF induction by an inhibitor of the silencing epigenetic enzyme EHMT2 (UNC0638) depended on the timing of its addition to cultures of synchronously maturing erythroid progenitors,59 with similar observations in our hands with DNMT1-depleting drugs (personal communication).
Why are these drugs being evaluated for, or used, to treat cancer?
Some of the most recurrently mutated, deleted or amplified genes in cancers encode for chromatin remodelers. Thus, another concern with epigenetic targeting is whether it might mimic some of these genetic changes and favor activation of oncogenic programs. It is reassuring to some extent, however, that the epigenetic targets and drugs discussed above have been or are being developed to treat and/or prevent cancer. We recently reviewed the biological rationale for this,105 and it is briefly summarized here: cancer cells, including self-replicating cancer cells (cancer or leukemia ‘stem’ cells), contain high amounts of the lineage master transcription factors that normally activate terminal lineage-fates, and depend on specific corepressors (‘addictions’) in order to avoid these terminal fates.111 The pathway of action is activation of the terminal lineage-fates intended by cancer cell lineage master transcription factor content. The same chromatin-‘relaxing’ treatments that trigger terminal lineage-fates of cancer/leukemia stem cells preserve self-renewal of uncommitted tissue stem cells, since these cells express stem cell master transcription factors, not high levels of lineage-specifying transcription factors.92,93 This therapeutic index explains why non-cytotoxic doses and schedules of decitabine can suppress malignant clones and simultaneously improve functional blood counts even in elderly patients with myeloid malignancies.91,105,111–115 Stated simply, several corepressor components (repressing epigenetic enzymes), e.g., DNMT1, HDAC, KDM1A, have been biologically validated as molecular targets for the treatment and prevention of cancer.111
Teratogenic risks
Another concern is the potential for teratogenicity: this should be assumed for individual agents, unless shown otherwise by formal toxicological studies.
Drug metabolism is central to the clinical profile of activity
Drugs, being biologically active, are metabolized, and this too can contribute substantially to their in vivo profile of activity. For example, DNMT1-depleting decitabine is a pyrmidine nucleoside analog pro-drug that depends absolutely for its activity on the pyrimidine metabolism enzyme deoxycytidine kinase: Deoxycytidine kinase executes the initial phosphorylation of decitabine in cells, which rate-limits its conversion into the nucleotide form that actually depletes DNMT1. Serendipitously, deoxycytidine kinase is most highly expressed in the myeloid compartment, especially erythroid precursors. Thus, the clinical profile of decitabine activity is in major part dictated by its metabolically driven tropism for the myeloid compartment.
Baseline HbF levels dictate final HbF levels
There is a wide variation in baseline HbF levels in patients with SCD and even in the general population, reflecting the influence of various genetic polymorphisms on the regulation of this locus.9 Even if a molecular targeted therapy produces similar rates of increase in HbF% (the percentage of total hemoglobin that is HbF) in all patients, the final HbF% will be dictated by the level at which HbF% began. Moreover, in clinical trials we have conducted with DNMT1-depleting decitabine, we have noticed a slightly lower slope to the rate of increase in HbF% in SCD patients with lower HbF% at baseline. Fortunately and importantly, however, HbF induced by this epigenetic strategy was well-distributed among RBC, and the rates of increase of HbF-enriched RBC (F-cells) was actually higher in patients with low F-cells at baseline.43,81,94,95
At some time-point after starting therapy, F-cells entering the circulation are matched by a similar number of F-cells leaving the circulation, producing plateaus in HbF% and F-cell%.
Small molecules to chemically modify HbS to impede polymerization
The scientific foundation for efforts to chemically modify HbS is the two-state allosteric Monod-Wyman-Changeux structural model which characterizes the rapidly reversible equilibrium between the quarternary structure of hemoglobin with low oxygen affinity (fully deoxygenated hemoglobin, ‘‘T’ quarternary structure) and the hemoglobin quarternary structure with high affinity for oxygen (oxygenated hemoglobin, ‘R’ quarternary structure).116,117 The Monod-Wyman-Changeux model demonstrated incompatibility of the R conformation with polymerization, creating a foundation to propose molecules to favor the high oxygen affinity R conformation, as a method to delay HbS polymerization.5
The basic concern with such an approach is that SCD is a disease of decreased oxygen delivery to tissues and, thus, if a chemical modification produces a high oxygen affinity hemoglobin molecule, there is a necessary play off between decreased oxygen supply from increased oxygen affinity of hemoglobin versus increased oxygen supply from less HbS polymerization/higher total hemoglobin. This balancing act is discussed in more detail in the section ‘Lessons learned so far’ below. Ultimately, however, rigorous clinical evaluation is key,5,118 and clinical evaluation has started or is underway for a number of candidate drugs that exploit these principles:
Small molecules to convert hemoglobin to methemoglobin
The earliest clinical effort in this field evaluated the conversion of hemoglobin to methemoglobin following the administration of sodium nitrite or para-amino-proprio-phenone (PAPP) to five patients.119 Both agents were able to increase methemoglobin. Methemoglobin levels of >20% (but not less) produced by sodium nitrite extended RBC survival as measured by chromium-labeling. The methemoglobinemia itself was apparently well-tolerated, but there was no evidence of any clinical benefit. Instead, there were significant side-effects from the administered drugs.119
Interestingly, higher methemoglobin levels produced by PAPP did not extend RBC survival, possibly because PAPP was directly hemolytic.
Small molecules to convert hemoglobin to carboxyhemoglobin
Carbon monoxide can be used to convert hemoglobin to carboxyhemoglobin. Infusion of free pegylated carboxyhe-moglobin (MP4CO), as a hemoglobin-based carbon monoxide carrier, was evaluated in a phase I study.120 In an abstract description of results in 18 patients, the maximum increase in carboxyhemoglobin was to 2%, which returned to pre-dosing levels within 8 h of completion of the MP4CO infusion. There was no significant increase in total hemoglobin. No further studies have been reported.
Small molecules that delay HbS polymerization by unclear mechanisms
Niprisan (Nix-0699) and related small molecules (SCD-101) are plant-derived molecules that have been found to delay polymerization of deoxygenated HbS, but by unclear mechanisms.121 SCD-101 has been evaluated in a phase IB clinical trial in 26 SCD patients. There were no major adverse events attributed to the drug taken for 28 days, and it appeared to decrease chronic pain and fatigue at higher doses. However, there were no laboratory data providing evidence of decreased hemolysis or increased total hemoglobin, although analysis of peripheral smears suggested improvements in RBC shape.122
Small molecules to increase hemoglobin oxygen affinity
Specific small molecule aldehydes have been found to form reversible Schiff base linkages with the N-terminal amino group of hemoglobin a chains to lock in the high oxygen affinity R conformation, and the polyaromatic adldehyde GBT440 (voxeletor) has been developed through to phase III clinical trial evaluation. In phase I/II randomized, double-blind, placebo-controlled evaluation in SCD patients, some of whom were receiving concurrent therapy with hydroxyurea, there were increases in total hemoglobin of ≥1 g/dL in six of 12 patients who received the drug for 90 days or more.123 There were concurrent decreases in markers of hemolysis (lactate dehy- drogenase, total bilirubin). There were no significant adverse events attributed to study drug. Oxygen delivery was evaluated by measurement of oxygen consumption during cardiopulmonary exercise testing, erythropoietin levels, resting heart rate and heart rate during peak exercise, and these parameters did not suggest decreased oxygen delivery to tissues.123 A subsequent double-blind, randomized, placebo-controlled phase III clinical trial evaluated two different doses of the study drug (900 and 1500 mg per day) in 274 SCD patients, two-thirds of whom remained on stable doses of hydroxyurea initiated well before study enrollment.124 A hemoglobin response, defined as an increase from baseline of >1 g/dL at week 24, occurred in 51% of the patients on the 1500 mg dose, 33% on the 900 mg dose, and 7% on placebo, in intention-to-treat analyses. There were also improvements in biomarkers of hemolysis. The frequency of vaso-occlusive crises did not differ between the treatment arms. Breakdown of vaso-occlusive crisis frequency according to whether or not the patients were taking hydroxyurea was not reported. Erythropoietin levels (as a surrogate for oxygen delivery) as well as grade 3 and serious adverse events were similar between the treatment arms.124
Chemical modification of HbS – lessons learned so far and open questions
Balancing acts
The clinical trial results with GBT440 thus illustrate that chemical modification of hemoglobin to increase its oxygen affinity (promote the hemoglobin R conformation) can indeed significantly decrease hemolysis and significantly increase total hemoglobin. The hope and goal is that higher hemoglobin increases oxygen supply by amounts that exceed any decrease in oxygen supply from the higher oxygen affinity of the modified hemoglobin molecule,5,118 as per the equation:
Thus, increasing total hemoglobin increases oxygen supply, but chemical modification of some of these hemoglobin molecules to increase oxygen affinity decreases effective arterial oxygen saturation and oxygen supply. Some tissues, e.g., the brain, have limited capacity to increase the ‘blood flow’ component in the equation, and hence, are particularly dependent on the ‘arterial oxygen saturation’ x ‘total hemoglobin’ components, as extensively modeled recently.118,125 Underscoring this point, most silent cerebral infarctions in SCD children have been found to be caused by disruption to oxygen supply that is not caused by large vessel vasculopathy, implying anemia and/or blood oxygen saturation are critical drivers of this hypoxic damage.126–128
Even the ‘blood flow’ component of the equation is a balancing act in SCD patients: whole blood viscosity is a key determinant of blood flow; less HbS polymerization, by increasing (improving) RBC deformability, can decrease whole blood viscosity and thus increase blood flow. On the other hand, higher total hemoglobin/hematocrit can increase blood viscosity which can decrease blood flow, even with hematocrits in an anemia range, because of the contribution of baseline low RBC deformability of SCD to viscosity. This blood flow calculus needs to be considered with small molecules aiming to chemically modify HbS, and with small molecules aiming to substitute HbS with HbF.
Ultimately, the risk/benefit calculus for any therapeutic approach requires careful clinical trial determination.
Combinatorial approaches
In oncology, combinations of drugs are almost mandatory, because the target cell population is evolving, and will select to evade the effects of drugs. Although target cells in SCD are not evolving, other biological realities compel consideration of combination therapies. One reality is that most SCD patients will already have tissue/organ damage that can undermine the potential benefits of novel small molecule therapeutics. For example, diminished bone marrow reserve from vaso-occlusive damage and/or replication-mediated exhaustion, which decreases compensatory reticulocytosis, and which contributes to early death,2,15,31,33,37,38,40 could limit the scope of potential benefit that can be produced by HbF inducers or HbS modifiers. Another biological reality, but potentially positive, is demonstrated by the approval by the FDA of the amino acid glutamine as a treatment to reduce the frequency of vaso-occlusive crises in SCD patients.129 Natural substances, which in most humans can be assumed to be satisfactorily maintained by a normal diet, might actually be important pharmaceuticals for SCD patients. By way of bringing such negative and potentially positive biological realities together, it is noteworthy that the natural substance nicotinamide (vitamin B3) markedly expands hematopoietic stem cells in vitro at concentrations that can be readily and safely produced in vivo with oral supplementation.130–132 Moreover, nicotinamide is a direct precursor for the vital energy currency nicotinamide adenine dinucleotide (NAD) which is depleted in SCD RBC, increasing their susceptibility to oxidative damage. In fact, replenishing NAD is one of the rationales for glutamine administration to SCD patients.129 In short, in considering combination therapy, there could be important, highly feasible, but unexplored opportunities around relatively non-toxic natural substances (glutamine, nicotinamide, vitamin D, etc.). Other under-evaluated natural molecules include the kidney hormone erythropoietin, since declining kidney erythropoietin production also contributes to declining compensatory reticulocytosis.2
Combining small molecules to inhibit more than one co-repressing enzyme in the BCL11A hub, each used at doses low enough to avoid side-effects from off-target actions, and with non-overlapping side-effects from on-target actions, might produce greater HbF induction than achieved with a single target. Such molecules should have non-cytotoxic mechanisms of action, to avoid potential injury to needed bone marrow capacity. Unfortunately, there are few non-cytotoxic small molecule drugs targeting rational epigenetic targets, and even fewer for which optimal single molecule application has been characterized (Table 1). That is, more non-cytotoxic epigenetic drugs, and more information on their profiles of side-effects from on-target and off-target actions, are needed to guide any consideration of combination therapy.
What about combining HbF inducers with HbS modifiers? This has in effect been evaluated in the clinical trials of GBT440, since this drug was added to stable doses of hydroxyurea in >60% of clinical trial participants. Hemoglobin increases of >1 g/dL occurred in ~40% of patients taking GBT440 1500 mg alone versus ~55% of patients taking GBT440 1500 mg + hydroxyurea in the phase III trial, but whether vaso-occlusive crisis frequency and other adverse events varied between these two groups was not described.124 The efficacy calculus and hope is that increases in oxygen delivery from better RBC deformability and higher total hemoglobin will exceed decreases in oxygen delivery caused by greater blood viscosity and chemical modification of HbS.
Conclusions
Clinical proof-of-principle that substantial total hemoglobin increases can be produced by non-cytotoxic inhibition of specific epigenetic enzymes, to shift RBC precursor hemoglobin manufacturing from HbS to HbF, and by chemical modification of hemoglobin to promote the high oxygen affinity ‘R’ quarternary structure of the hemoglobin molecule, has already been generated in SCD patients. Clinical evaluation to determine the long-term safety, the impact on symptoms and multi-organ pathophysiology, and the durability of any benefits, is ongoing. There is hope that one or more of the small molecules being evaluated will pass rigorous scrutiny and culminate in practical, accessible, cost-effective, safe and potent disease-modifying therapy for SCD patients worldwide.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/9/1720
Funding source: National Heart, Lung and Blood Institute UO1 HL117658.
References
- 1.Piel FB, Hay SI, Gupta S, Weatherall DJ, Williams TN. Global burden of sickle cell anaemia in children under five, 2010-2050: modelling based on demographics, excess mortality, and interventions. PLoS Med. 2013;10(7):e1001484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saraf S, Farooqui M, Infusino G, et al. Standard clinical practice underestimates the role and significance of erythropoietin deficiency in sickle cell disease. Br J Haematol. 2011;153(3):386–392. [DOI] [PubMed] [Google Scholar]
- 3.Kauf TL, Coates TD, Huazhi L, Mody-Patel N, Hartzema AG. The cost of health care for children and adults with sickle cell disease. Am J Hematol. 2009;84(6):323–327. [DOI] [PubMed] [Google Scholar]
- 4.Platt OS, Brambilla DJ, Rosse WF, et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994;330(23):1639–1644. [DOI] [PubMed] [Google Scholar]
- 5.Eaton WA, Bunn HF. Treating sickle cell disease by targeting HbS polymerization. Blood. 2017;129(20):2719–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ataga KI, Smith WR, De Castro LM, et al. Efficacy and safety of the Gardos channel blocker, senicapoc (ICA-17043), in patients with sickle cell anemia. Blood. 2008;111(8):3991–3997. [DOI] [PubMed] [Google Scholar]
- 7.Tisdale J. Improvements in haploidentical transplantation for sickle cell disease and beta-thalassaemia. Lancet Haematol. 2019;6(4):e168–e169. [DOI] [PubMed] [Google Scholar]
- 8.Sankaran VG, Xu J, Orkin SH. Advances in the understanding of haemoglobin switching. Br J Haematol. 2010;149(2):181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rochette J, Craig JE, Thein SL. Fetal hemoglobin levels in adults. Blood Rev. 1994;8(4):213–224. [DOI] [PubMed] [Google Scholar]
- 10.Conley CL, Weatherall DJ, Richardson SN, Shepard MK, Charache S. Hereditary persistence of fetal hemoglobin: a study of 79 affected persons in 15 Negro families in Baltimore. Blood. 1963;21:261–281. [PubMed] [Google Scholar]
- 11.Perrine RP, Pembrey ME, John P, Perrine S, Shoup F. Natural history of sickle cell anemia in Saudi Arabs. A study of 270 subjects. Ann Intern Med. 1978;88(1):1–6. [DOI] [PubMed] [Google Scholar]
- 12.Ngo DA, Aygun B, Akinsheye I, et al. Fetal haemoglobin levels and haematological characteristics of compound heterozygotes for haemoglobin S and deletional hereditary persistence of fetal haemoglobin. Br J Haematol. 2012;156(2):259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platt OS, Thorington BD, Brambilla DJ, et al. Pain in sickle cell disease. Rates and risk factors. N Engl J Med. 1991;325(1):11–16. [DOI] [PubMed] [Google Scholar]
- 14.Powars DR, Weiss JN, Chan LS, Schroeder WA. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood. 1984;63(4):921–926. [PubMed] [Google Scholar]
- 15.Steinberg MH, Barton F, Castro O, et al. Effect of hydroxyurea on mortality and morbidity in adult sickle cell anemia: risks and benefits up to 9 years of treatment. JAMA. 2003;289(13):1645–1651. [DOI] [PubMed] [Google Scholar]
- 16.Atweh GF, Schechter AN. Pharmacologic induction of fetal hemoglobin: raising the therapeutic bar in sickle cell disease. Curr Opin Hematol. 2001;8(2):123–130. [DOI] [PubMed] [Google Scholar]
- 17.Powars DR, Elliott-Mills DD, Chan L, et al. Chronic renal failure in sickle cell disease: risk factors, clinical course, and mortality. Ann Intern Med. 1991;115(8):614–620. [DOI] [PubMed] [Google Scholar]
- 18.Lebensburger J, Johnson SM, Askenazi DJ, et al. Protective role of hemoglobin and fetal hemoglobin in early kidney disease for children with sickle cell anemia. Am J Hematol. 2011;86(5):430–432. [DOI] [PubMed] [Google Scholar]
- 19.Aban I, Baddam S, Hilliard LM, et al. Severe anemia early in life as a risk factor for sickle-cell kidney disease. Blood. 2017;129(3):385–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akinsheye I, Alsultan A, Solovieff N, et al. Fetal hemoglobin in sickle cell anemia. Blood. 2011;118(1):19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg MA, Husson MA, Bunn HF. Participation of hemoglobins A and F in polymerization of sickle hemoglobin. J Biol Chem. 1977;252(10):3414–3421. [PubMed] [Google Scholar]
- 22.Nagel RL, Bookchin RM, Johnson J, et al. Structural bases of the inhibitory effects of hemoglobin F and hemoglobin A2 on the polymerization of hemoglobin S. Proc Natl Acad Sci U S A. 1979;76(2):670–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavelle D, Engel JD, Saunthararajah Y. Fetal hemoglobin induction by epigenetic drugs. Semin Hematol. 2018;55(2):60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeSimone J, Biel SI, Heller P. Stimulation of fetal hemoglobin synthesis in baboons by hemolysis and hypoxia. Proc Natl Acad Sci U S A. 1978;75(6):2937–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeSimone J, Heller P, Adams JG. Hemopoietic stress and fetal hemoglobin synthesis: comparative studies in vivo and in vitro. Blood. 1979;54(5):1176–1181. [PubMed] [Google Scholar]
- 26.DeSimone J, Heller P, Amsel J, Usman M. Magnitude of the fetal hemoglobin response to acute hemolytic anemia in baboons is controlled by genetic factors. J Clin Invest. 1980;65(1):224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeSimone J, Heller P, Biel M, Zwiers D. Genetic relationship between fetal Hb levels in normal and erythropoietically stressed baboons. Br J Haematol. 1981;49(2):175–183. [DOI] [PubMed] [Google Scholar]
- 28.DeSimone J, Biel M, Heller P. Maintenance of fetal hemoglobin (HbF) elevations in the baboon by prolonged erythropoietic stress. Blood. 1982;60(2):519–523. [PubMed] [Google Scholar]
- 29.Lavelle D, DeSimone J, Heller P, Zwiers D, Hall L. On the mechanism of Hb F elevations in the baboon by erythropoietic stress and pharmacologic manipulation. Blood. 1986;67(4):1083–1089. [PubMed] [Google Scholar]
- 30.Charache S, Terrin ML, Moore RD, et al. Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell Anemia. N Engl J Med. 1995;332(20):1317–1322. [DOI] [PubMed] [Google Scholar]
- 31.Steinberg MH, Lu ZH, Barton FB, et al. Fetal hemoglobin in sickle cell anemia: determinants of response to hydroxyurea. Multicenter Study of Hydroxyurea. Blood. 1997;89(3):1078–1088. [PubMed] [Google Scholar]
- 32.Ballas SK, Marcolina MJ, Dover GJ, Barton FB. Erythropoietic activity in patients with sickle cell anaemia before and after treatment with hydroxyurea. Br J Haematol. 1999;105(2):491–496. [PubMed] [Google Scholar]
- 33.Charache S, Dover GJ, Moore RD, et al. Hydroxyurea: effects on hemoglobin F production in patients with sickle cell anemia. Blood. 1992;79(10):2555–2565. [PubMed] [Google Scholar]
- 34.Ballas SK, Barton FB, Waclawiw MA, et al. Hydroxyurea and sickle cell anemia: effect on quality of life. Health Qual Life Outcomes. 2006;4:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebensburger JD, Pestina TI, Ware RE, Boyd KL, Persons DA. Hydroxyurea therapy requires HbF induction for clinical benefit in a sickle cell mouse model. Haematologica. 2010;95(9):1599–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal JB, Strouse JJ, Beach MC, et al. Hydroxyurea for the treatment of sickle cell disease. Evid Rep Technol Assess. 2008;(165):1–95. [PMC free article] [PubMed] [Google Scholar]
- 37.Machado RF, Anthi A, Steinberg MH, et al. N-terminal pro-brain natriuretic peptide levels and risk of death in sickle cell disease. JAMA. 2006;296(3):310–318. [DOI] [PubMed] [Google Scholar]
- 38.Ware RE, Eggleston B, Redding-Lallinger R, et al. Predictors of fetal hemoglobin response in children with sickle cell anemia receiving hydroxyurea therapy. Blood. 2002;99(1):10–14. [DOI] [PubMed] [Google Scholar]
- 39.Green NS, Manwani D, Qureshi M, et al. Decreased fetal hemoglobin over time among youth with sickle cell disease on hydroxyurea is associated with higher urgent hospital use. Pediatr Blood Cancer. 2016;63(12):2146–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West MS, Wethers D, Smith J, Steinberg M. Laboratory profile of sickle cell disease: a cross-sectional analysis. The Cooperative Study of Sickle Cell Disease. J Clin Epidemiol. 1992;45(8):893–909. [DOI] [PubMed] [Google Scholar]
- 41.Goren A, Simchen G, Fibach E, et al. Fine tuning of globin gene expression by DNA methylation. PLoS One. 2006;1:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platt OS, Orkin SH, Dover G, et al. Hydroxyurea enhances fetal hemoglobin production in sickle cell anemia. J Clin Invest. 1984;74(2):652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saunthararajah Y, Hillery CA, Lavelle D, et al. Effects of 5-aza-2 ′-deoxycytidine on fetal hemoglobin levels, red cell adhesion, and hematopoietic differentiation in patients with sickle cell disease. Blood. 2003;102(12):3865–3870. [DOI] [PubMed] [Google Scholar]
- 44.Saunthararajah Y, Lavelle D, DeSimone J. DNA hypo-methylating agents and sickle cell disease. Br J Haematol. 2004;126(5):629–636. [DOI] [PubMed] [Google Scholar]
- 45.Sikorski TW, Joo YJ, Ficarro SB, et al. Proteomic analysis demonstrates activator-and chromatin-specific recruitment to promoters. J Biol Chem. 2012;287(42):35397–35408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–928. [DOI] [PubMed] [Google Scholar]
- 47.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11(2):377–389. [DOI] [PubMed] [Google Scholar]
- 48.Kadam S, McAlpine GS, Phelan ML, et al. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 2000;14(19):2441–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sankaran VG, Menne TF, Xu J, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–1842. [DOI] [PubMed] [Google Scholar]
- 50.Tanabe O, Katsuoka F, Campbell AD, et al. An embryonic/fetal beta-type globin gene repressor contains a nuclear receptor TR2/TR4 heterodimer. EMBO J. 2002;21(13):3434–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menzel S, Garner C, Gut I, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet. 2007;39(10):1197–1199. [DOI] [PubMed] [Google Scholar]
- 52.Cui S, Kolodziej KE, Obara N, et al. Nuclear receptors TR2 and TR4 recruit multiple epigenetic transcriptional corepressors that associate specifically with the embryonic β-type globin promoters in differentiated adult erythroid cells. Mol Cell Biol. 2011;31(16): 3298–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu J, Bauer DE, Kerenyi MA, et al. Corepressor-dependent silencing of fetal hemoglobin expression by BCL11A. Proc Natl Acad Sci U S A. 2013;110(16):6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X, Zhang Y, Chen Y, et al. In situ Capture of chromatin interactions by biotinylated dCas9. Cell. 2017;170(5):1028–1043.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Constantoulakis P, Knitter G, Stamatoyannopoulos G. On the induction of fetal hemoglobin by butyrates: in vivo and in vitro studies with sodium butyrate and comparison of combination treatments with 5-AzaC and AraC. Blood. 1989;74(6):1963–1971. [PubMed] [Google Scholar]
- 56.Chaturvedi CP, Hosey AM, Palii C, et al. Dual role for the methyltransferase G9a in the maintenance of beta-globin gene transcription in adult erythroid cells. Proc Natl Acad Sci U S A. 2009;106(43):18303–18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rank G, Cerruti L, Simpson RJ, et al. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood. 2010;116(9):1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Z, He Y, Ju J, et al. The role of WDR5 in silencing human fetal globin gene expression. Haematologica. 2012;97(11):1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krivega I, Byrnes C, de Vasconcellos JF, et al. Inhibition of G9a methyltransferase stimulates fetal hemoglobin production by facilitating LCR/gamma-globin looping. Blood. 2015;126(5):665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renneville A, Van Galen P, Canver MC, et al. EHMT1 and EHMT2 inhibition induces fetal hemoglobin expression. Blood. 2015;126(16):1930–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Velcheti V, Wong KK, Saunthararajah Y. EZH2 inhibitors: take it EZy, it is all about context. Cancer Discov. 2019;9(4):472–475. [DOI] [PubMed] [Google Scholar]
- 62.Wang X, Thein SL. Switching from fetal to adult hemoglobin. Nat Genet. 2018;50(4): 478–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martyn GE, Wienert B, Yang L, et al. Natural regulatory mutations elevate the fetal globin gene via disruption of BCL11A or ZBTB7A binding. Nat Genet. 2018;50(4):498–503. [DOI] [PubMed] [Google Scholar]
- 64.Uda M, Galanello R, Sanna S, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A. 2008;105(5):1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ginder GD, Whitters MJ, Pohlman JK, Burns LJ. Transcriptional activation of an embryonic globin gene in adult erythroid cells in vivo. Prog Clin Biol Res. 1985;191:463–474. [PubMed] [Google Scholar]
- 66.Scuto A, Kirschbaum M, Kowolik C, et al. The novel histone deacetylase inhibitor, LBH589, induces expression of DNA damage response genes and apoptosis in Ph-acute lymphoblastic leukemia cells. Blood. 2008;111(10):5093–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci U S A. 2010;107(33):14639–14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Conti C, Leo E, Eichler GS, et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70(11):4470–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaymes TJ, Padua RA, Pla M, et al. Histone deacetylase inhibitors (HDI) cause DNA damage in leukemia cells: a mechanism for leukemia-specific HDI-dependent apoptosis? Mol Cancer Res. 2006;4(8):563–573. [DOI] [PubMed] [Google Scholar]
- 70.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6(1):38–51. [DOI] [PubMed] [Google Scholar]
- 71.Patthamalai P, Fuchareon S, Chaneiam N, et al. A phase 2 trial of HQK-1001 in HbE-beta thalassemia demonstrates HbF induction and reduced anemia. Blood. 2014;123(12): 1956–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van der Ploeg LH, Flavell RA. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980;19(4):947–958. [DOI] [PubMed] [Google Scholar]
- 73.Mavilio F, Giampaolo A, Care A, et al. Molecular mechanisms of human hemoglobin switching: selective undermethylation and expression of globin genes in embryonic, fetal, and adult erythroblasts. Proc Natl Acad Sci U S A. 1983;80(22):6907–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tagle DA, Koop BF, Goodman M, et al. Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J Mol Biol. 1988;203(2):439–455. [DOI] [PubMed] [Google Scholar]
- 75.DeSimone J, Heller P, Hall L, Zwiers D. 5-Azacytidine stimulates fetal hemoglobin synthesis in anemic baboons. Proc Natl Acad Sci U S A. 1982;79(14):4428–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ley TJ, DeSimone J, Anagnou NP, et al. 5-azacytidine selectively increases gamma-globin synthesis in a patient with beta+ thalassemia. N Engl J Med. 1982;307(24):1469–1475. [DOI] [PubMed] [Google Scholar]
- 77.Ley TJ, DeSimone J, Noguchi CT, et al. 5-Azacytidine increases gamma-globin synthesis and reduces the proportion of dense cells in patients with sickle cell anemia. Blood. 1983;62(2):370–380. [PubMed] [Google Scholar]
- 78.Charache S, Dover G, Smith K, et al. Treatment of sickle cell anemia with 5-aza-cytidine results in increased fetal hemoglobin production and is associated with nonrandom hypomethylation of DNA around the gamma-delta-beta-globin gene complex. Proc Natl Acad Sci U S A. 1983;80(15):4842–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dover GJ, Charache S, Boyer SH, Vogelsang G, Moyer M. 5-Azacytidine increases HbF production and reduces anemia in sickle cell disease: dose-response analysis of subcutaneous and oral dosage regimens. Blood. 1985;66(3):527–532. [PubMed] [Google Scholar]
- 80.Lowrey CH, Nienhuis AW. Brief report: treatment with azacitidine of patients with end-stage beta-thalassemia. N Engl J Med. 1993;329(12):845–848. [DOI] [PubMed] [Google Scholar]
- 81.Koshy M, DeSimone J, Molokie R, et al. Augmentation of fetal hemoglobin (HbF) levels by low-dose short-duration 5 ′-aza-2-deoxycytidine (decitabine) administration in sickle cell anemia patients who had no HbF elevation following hydroxyurea therapy. Blood. 1998;92(10):30b-b. [Google Scholar]
- 82.Cui S, Kolodziej KE, Obara N, et al. Nuclear receptors TR2 and TR4 recruit multiple epigenetic transcriptional corepressors that associate specifically with the embryonic beta-type globin promoters in differentiated adult erythroid cells. Mol Cell Biol. 2011;31(16):3298–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33(1):9–10. [DOI] [PubMed] [Google Scholar]
- 84.Clements EG, Mohammad HP, Leadem BR, et al. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 2012;40(10):4334–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brenner C, Luciani J, Bizet M, et al. The interplay between the lysine demethylase KDM1A and DNA methyltransferases in cancer cells is cell cycle dependent. Oncotarget. 2016;7(37):58939–58952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Covey JM, D’Incalci M, Tilchen EJ, Zaharko DS, Kohn KW. Differences in DNA damage produced by incorporation of 5-aza-2′-deoxycytidine or 5,6-dihydro-5-azacytidine into DNA of mammalian cells. Cancer Res. 1986;46(11):5511–5517. [PubMed] [Google Scholar]
- 87.Schermelleh L, Haemmer A, Spada F, et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35(13):4301–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zauri M, Berridge G, Thezenas ML, et al. CDA directs metabolism of epigenetic nucleosides revealing a therapeutic window in cancer. Nature. 2015;524(7563):114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Almqvist H, Axelsson H, Jafari R, et al. CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluo-rouracil. Nat Commun. 2016;7:11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olivieri NF, Saunthararajah Y, Thayalasuthan V, et al. A pilot study of subcutaneous decitabine in beta-thalassemia intermedia. Blood. 2011;118(10):2708–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saunthararajah Y, Sekeres M, Advani A, et al. Evaluation of noncytotoxic DNMT1-depleting therapy in patients with myelodysplastic syndromes. J Clin Invest. 2015;125(3):1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Milhem M, Mahmud N, Lavelle D, et al. Modification of hematopoietic stem cell fate by 5aza 2′ deoxycytidine and trichostatin A. Blood. 2004;103(11):4102–4110. [DOI] [PubMed] [Google Scholar]
- 93.Hu Z, Negrotto S, Gu X, et al. Decitabine maintains hematopoietic precursor self-renewal by preventing repression of stem cell genes by a differentiation-inducing stimulus. Mol Cancer Ther. 2010;9(6):1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.DeSimone J, Koshy M, Dorn L, et al. Maintenance of elevated fetal hemoglobin levels by decitabine during dose interval treatment of sickle cell anemia. Blood. 2002;99(11):3905–3908. [DOI] [PubMed] [Google Scholar]
- 95.Molokie R, Lavelle D, Gowhari M, et al. Oral tetrahydrouridine and decitabine for non-cytotoxic epigenetic gene regulation in sickle cell disease: a randomized phase 1 study. PLoS Med. 2017;14(9):e1002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984; 81(22):6993–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patel K, Dickson J, Din S, et al. Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38(13):4313–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J Biol Chem. 1982;257(4):2041–2048. [PubMed] [Google Scholar]
- 99.Shi L, Cui S, Engel JD, Tanabe O. Lysine-specific demethylase 1 is a therapeutic target for fetal hemoglobin induction. Nat Med. 2013;19(3):291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cui S, Lim KC, Shi L, et al. The LSD1 inhibitor RN-1 induces fetal hemoglobin synthesis and reduces disease pathology in sickle cell mice. Blood. 2015;126(3):386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jagadeeswaran R, Vazquez BA, Thiruppathi M, et al. Pharmacological inhibition of LSD1 and mTOR reduces mitochondrial retention and associated ROS levels in the red blood cells of sickle cell disease. Exp Hematol. 2017;50:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rivers A, Vaitkus K, Ibanez V, et al. The LSD1 inhibitor RN-1 recapitulates the fetal pattern of hemoglobin synthesis in baboons (P. anubis). Haematologica. 2016;101(6):688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137(3):445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2008;27(1):100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Velcheti V, Schrump D, Saunthararajah Y. Ultimate precision: targeting cancer but not normal self-replication. Am Soc Clin Oncol Educ Book. 2018;38:950–963. [DOI] [PubMed] [Google Scholar]
- 106.Kingsley PD, Malik J, Emerson RL, et al. "Maturational" globin switching in primary primitive erythroid cells. Blood. 2006;107(4):1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mabaera R, Richardson CA, Johnson K, et al. Developmental-and differentiation-specific patterns of human gamma-and beta-globin promoter DNA methylation. Blood. 2007;110(4):1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McGrath K, Palis J. Ontogeny of erythropoiesis in the mammalian embryo. Curr Top Dev Biol. 2008;82:1–22. [DOI] [PubMed] [Google Scholar]
- 109.Ni H, Yang XD, Stoeckert CJ., Jr Maturation and developmental stage-related changes in fetal globin gene expression are reproduced in transiently transfected primary adult human erythroblasts. Exp Hematol. 1999;27(1):46–53. [DOI] [PubMed] [Google Scholar]
- 110.Papayannopoulou TH, Brice M, Stamatoyannopoulos G. Stimulation of fetal hemoglobin synthesis in bone marrow cultures from adult individuals. Proc Natl Acad Sci U S A. 1976;73(6):2033–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Velcheti V, Radivoyevitch T, Saunthararajah Y. Higher-level pathway objectives of epigenetic therapy: a solution to the p53 problem in cancer. Am Soc Clin Oncol Educ Book. 2017;37:812–824. [DOI] [PubMed] [Google Scholar]
- 112.Saleh MFM, Saunthararajah Y. Severe pyoderma gangrenosum caused by myelodysplastic syndrome successfully treated with decitabine administered by a noncytotoxic regimen. Clin Case Rep. 2017;5(12):2025–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu Y, Tabarroki A, Billings S, et al. Successful use of very low dose subcutaneous decitabine to treat high-risk myelofibrosis with Sweet syndrome that was refractory to 5-azacitidine. Leuk Lymphoma. 2014;55(2):447–449. [DOI] [PubMed] [Google Scholar]
- 114.Tabarroki A, Saunthararajah Y, Visconte V, et al. Ruxolitinib in combination with DNA methyltransferase inhibitors: clinical responses in patients with symptomatic myelofibrosis with cytopenias and elevated blast(s) counts. Leuk Lymphoma. 2015;56(2):497–499. [DOI] [PubMed] [Google Scholar]
- 115.Saunthararajah Y, Triozzi P, Rini B, et al. p53-independent, normal stem cell sparing epigenetic differentiation therapy for myeloid and other malignancies. Semin Oncol. 2012;39(1):97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. [DOI] [PubMed] [Google Scholar]
- 117.Eaton WA, Henry ER, Hofrichter J, et al. Evolution of allosteric models for hemoglobin. IUBMB Life. 2007;59(8-9):586–599. [DOI] [PubMed] [Google Scholar]
- 118.Hebbel RP, Hedlund BE. Sickle hemoglobin oxygen affinity-shifting strategies have unequal cerebrovascular risks. Am J Hematol. 2018;93(3):321–325. [DOI] [PubMed] [Google Scholar]
- 119.Beutler E. The effect of methemoglobin formation in sickle cell disease. J Clin Invest. 1961;40:1856–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Howard J, Thein SL, Galacteros F, et al. Safety and tolerability of MP4CO: a dose escalation study in stable patients with sick le cell disease. Blood. 2013;122(21):2205.23945154 [Google Scholar]
- 121.Iyamu EW, Turner EA, Asakura T. In vitro effects of NIPRISAN (Nix-0699): a naturally occurring, potent antisickling agent. Br J Haematol. 2002;118(1):337–343. [DOI] [PubMed] [Google Scholar]
- 122.Swift R, Abdulmalik O, Chen QK, et al. SCD-101: a new anti-sickling drug reduces pain and fatigue and improves red blood cell shape in peripheral blood of patients with sickle cell disease. Blood. 2016;128(22):121. [Google Scholar]
- 123.Howard J, Hemmaway CJ, Telfer P, et al. A phase 1/2 ascending dose study and open-label extension study of voxelotor in patients with sickle cell disease. Blood. 2019;133(17):1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vichinsky E, Hoppe CC, Ataga KI, et al. A phase 3 randomized trial of voxelotor in sickle cell disease. N Engl J Med. 2019. June 14 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 125.Watchmaker JM, Juttukonda MR, Davis LT, et al. Hemodynamic mechanisms underlying elevated oxygen extraction fraction (OEF) in moyamoya and sickle cell anemia patients. J Cereb Blood Flow Metab. 2018;38(9):1618–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Choudhury NA, DeBaun MR, Ponisio MR, et al. Intracranial vasculopathy and infarct recurrence in children with sickle cell anaemia, silent cerebral infarcts and normal transcranial Doppler velocities. Br J Haematol. 2018;183(2):324–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ford AL, Ragan DK, Fellah S, et al. Silent infarcts in sickle cell disease occur in the border zone region and are associated with low cerebral blood flow. Blood. 2018;132(16): 1714–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Quinn CT. Silent cerebral infarction: supply and demand. Blood. 2018;132(16):1632–1634. [DOI] [PubMed] [Google Scholar]
- 129.Niihara Y, Miller ST, Kanter J, et al. A Phase 3 trial of L-glutamine in sickle cell disease. N Engl J Med. 2018;379(3):226–235. [DOI] [PubMed] [Google Scholar]
- 130.Peled T, Shoham H, Aschengrau D, et al. Nicotinamide, a SIRT1 inhibitor, inhibits differentiation and facilitates expansion of hematopoietic progenitor cells with enhanced bone marrow homing and engraftment. Exp Hematol. 2012;40(4):342–355.e1. [DOI] [PubMed] [Google Scholar]
- 131.Anand S, Thomas S, Hyslop T, et al. Transplantation of ex vivo expanded umbilical cord blood (NiCord) decreases early infection and hospitalization. Biol Blood Marrow Transplant. 2017;23(7):1151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Horwitz ME, Chao NJ, Rizzieri DA, et al. Umbilical cord blood expansion with nicotinamide provides long-term multilineage engraftment. J Clin Invest. 2014;124(7):3121–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]