Abstract
Citrullination is a post-translational modification of arginine that is catalyzed by the protein arginine deiminases (PADs). Abnormal citrullination is observed in many autoimmune diseases and cancers. Anti-citrullinated protein antibodies (ACPA) are hallmarks of RA and used as diagnostic markers for disease diagnosis. Even though citrullination is associated with many different pathologies, its role remains unclear due to the challenges associated with the detection of citrullinated proteins since the mass change is only 0.984 Da. Moreover, the functional effects of protein citrullination remain mostly unknown. Herein, we discuss a brief overview of PAD structure and function, recent advances in the detection of citrullinated proteins in complex biological systems and the functional consequences of protein citrullination.
Introduction
Post-translational modifications (PTM) play an important role in regulating complex cellular processes. More than 200 PTMs have been identified with many being linked to human disease. One such modification is citrullination in which an arginine residue is converted into citrulline (Figure 1A). This PTM is generated by the protein arginine deiminases (PAD) [1]. Although the existence of “decitrullinase” has been hypothesized, no such enzyme has been identified and citrullination is generally thought to be an irreversible process. The PAD-catalyzed reaction involves the hydrolysis of the guanidinium group of an arginine residue, which generates a urea and leads to the loss of positive charge and two potential hydrogen bond donors. These changes alter protein activity, protein-protein interactions, and protein-nucleic acid interactions [1,2]. There are five PADs: PADs 1–4 and PAD6.[1] Only PADs 1–4 are active, PAD6 has several mutations that render it inactive [3]. PAD isozymes have unique tissue localizations and substrate specificities. Given that citrullination can irreversibly modify protein function, PAD activity is tightly regulated [4]. Calcium levels are the primary regulator of PAD activity as PADs require high calcium levels (103 to 104 μM) to be active. Thus, in the absence of a signal, they are generally inactive. Once activated, PADs citrullinate a wide range of protein substrates (see below) and this modification regulates numerous cellular processes, including cell signaling, immune responses, and gene regulation.
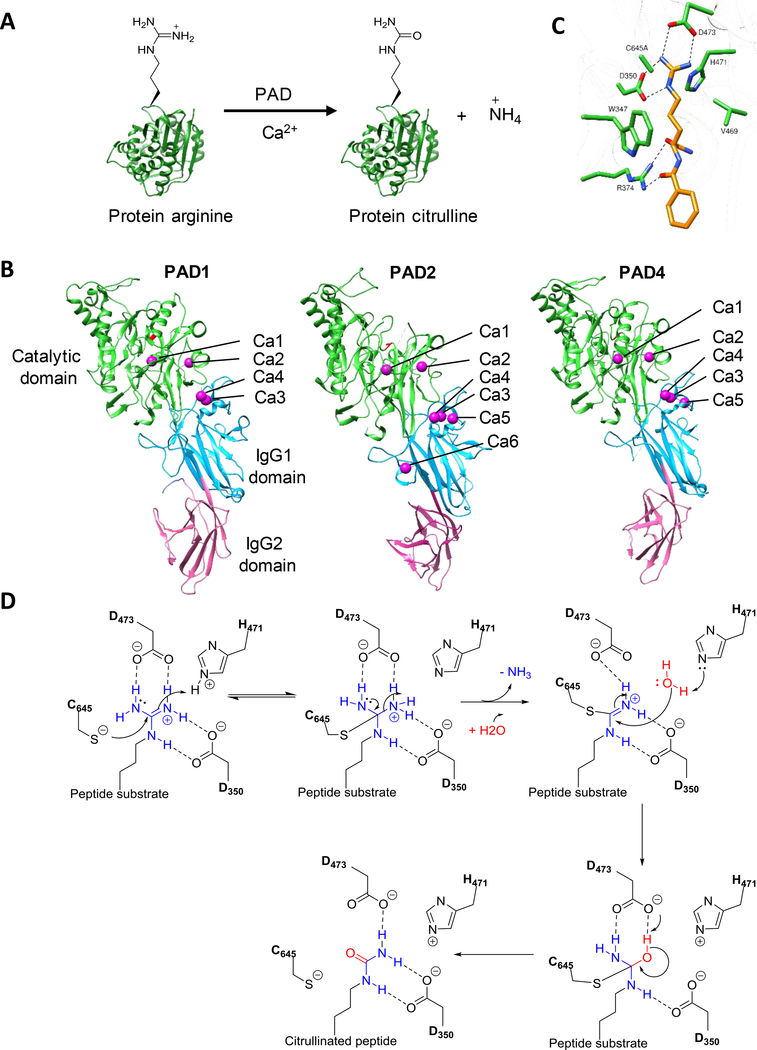
Figure 1.
A. PAD reaction. B. Crystal structure of Ca2+ bound PAD1 (PDB: 5HT5), PAD2 (PDB: 4N2B) and PAD4 (PDB: 1WD9). The catalytic C-terminal, IgG1 and IgG2 domains are colored green, cyan, and pink, respectively. C. Structure of the PAD4-C645A mutant bound to BAA (PDB: 1WDA. D. Proposed catalytic mechanism.
Dysregulated PAD activity is observed in numerous inflammatory diseases [2,5] including rheumatoid arthritis (RA), Parkinson’s disease [6], Alzheimer’s disease [7], atherosclerosis [8], lupus [9], and cancers [10,11]. Notably, PAD inhibitors have shown remarkable efficacy in many of these disease models. RA is the most studied disease amongst this group [12–15]. In the context of protein citrullination, RA is a chronic inflammatory condition that is characterized by damaged bone and cartilage. Studies have established that abnormal protein citrullination is responsible for disease progression and severity. For example, more than 75% of RA patients produce anti-citrullinated protein antibodies (ACPA) [16]. These antibodies recognize specific citrullinated epitopes present on numerous proteins (including vimentin, fibrin, and enolase) and are key drivers of RA. Notably, higher ACPA titers correlate with a more severe disease course [17]. Therefore, the detection of ACPA, through an anti-CCP test is used to diagnose RA [18]. ACPA are generally restricted to RA and thus it is not clear how the PADs contribute to so many other pathologies. Potential explanations include the role of PADs in promoting NET (Neutrophil Extracellular Trap) formation or NETosis [9,19,20]. NETosis is a pro-inflammatory form of programmed cell death, in which a chemokine or pathogen stimulates a subset of neutrophils to decondense their chromatin to form web-like structures that can capture pathogens. Abnormal NET formation is observed in many autoimmune disorders including RA [21], lupus [22], and Alzheimer’s disease [23].
Given that aberrant protein citrullination is a hallmark feature of virtually all inflammatory diseases and some cancers, it is critical that we understand how this PTM contributes to disease pathology. Despite the emerging biology and prominent role of citrullination in many disease pathologies, this PTM is poorly understood in part, because of the technical difficulties associated with detecting protein citrullination. However, tremendous progress has been made in recent years, including the discovery of new tools to detect protein citrullination in complex biological mixtures. Once discovered these citrullinated proteins will help define the role of citrullination in human biology and many could also serve as biomarkers for the detection of a wide array of diseases. Herein, we provide a brief overview of PAD structure and function as well as recent advances in the detection of protein citrullination. Finally, we discuss the functional consequences of protein citrullination.
PAD Structure
Structures of PADs 1, 2 and 4 have been determined thus far.[24–26] PADs 2, 3 and 4 form homodimers, whereas PAD1 is monomeric in solution. Each monomer contains distinct N- and C-terminal domains. The C-terminal catalytic domain adopts an α/β propeller fold whereas the N-terminal domain is further subdivided into two immunoglobulin-like (Ig) subdomains, i.e. IgG1 and IgG2. These N-terminal subdomains are involved in substrate binding and protein-protein interactions [1]. The X-ray crystal structures of PADs 1, 2 and 4 show that these enzymes bind four, six and five calcium ions, respectively (Figure 1B). In all three isozymes, two calcium ions (Ca1 and Ca2) are bound to catalytic domain while the remaining calcium ions are positioned in the IgG2-subdomain. A comparison of the calcium-free form of PAD2 and the calcium-bound forms of PAD2 and PAD4 revealed that calcium binding induces a large conformation change that generates the active site cleft. Upon calcium binding, the catalytic cysteine (C645 and C647 in PAD4 and PAD2, respectively) moves ≥ 5 Å, positioning it for nucleophilic attack on the bound substrate. Calcium titration experiments with PAD2 showed that Ca1 and Ca6 bind first followed by Ca3, Ca4 and Ca5, leading to a conformation change that generates the Ca2 binding site. The binding of calcium in the Ca2 position is crucial for the movement of the active site cysteine into a catalytically competent position.
PAD substrate binding and catalytic mechanism
PADs selectively convert peptidyl-arginine into peptidyl-citrulline; free arginine is not a substrate.[27] The structure of PAD4 bound to benzoyl-L-arginine amide (BAA) identified key catalytic residues (Figure 1C) [28]. These residues include C645, which acts as a nucleophile and H471 which acts as a general acid/base (Figure 1D). Substrate binding is initialized by strong electrostatic interactions between D350 and D473 and the substrate guanidinium. The aliphatic chain of the substrate interacts with side chain of W347 and V469 through hydrophobic interactions. Hydrogen bonding interactions between the side chain of R374 and the carbonyl groups in the peptide backbone ensures that the PADs do not deiminate free arginine. Structures of PAD4 with various histone peptides showed that most of the interactions between PAD4 and its substrates are through the backbone carbonyls of substrate. The lack of specific interactions between the side chain of the substrate peptides and the active site explains the broad specificity of PAD4 and the other PADs.
Mechanistic studies on all four PADs revealed that PADs 1, 2 and 4 proceed through a reverse-protonation mechanism whereas PAD2 utilizes a substrate-assisted mechanism [27–31]. In the former mechanism, the active site nucleophile (C645 in PAD4) is in the thiolate form prior to substrate binding (Figure 1D). Attack of the thioloate on the guanidinium carbon results in the formation of a covalent tetrahedral intermediate. Subsequently, H471 protonates this intermediate, thereby stabilizing it. Collapse of the tetrahedral intermediate and release of ammonia generates an S-alkylthiouronium intermediate. This adduct is hydrolyzed by the attack of a water molecule activated by H471, which leads to the formation of a second tetrahedral intermedium which collapses to regenerate the thiolate and peptidyl-citrulline. Support for this mechanism comes from pH-dependent kinetic inactivation studies which showed that the pKa of the C645 is ~8.3.[27,29–31] Further support for this mechanism comes from the large inverse solvent isotope effect on kcat/KM and the fact that 2-chloroacetamidine, a thiol-reactive compound does not depress the pKa of C645. However, 2-chloroacetamidine does decrease the pKa of C647 in PAD2 by ~1 log unit, suggesting that PAD2 follows a substrate assisted mechanism of thiol deprotonation [29].
Methods to detect PAD activity
COLDER Assay
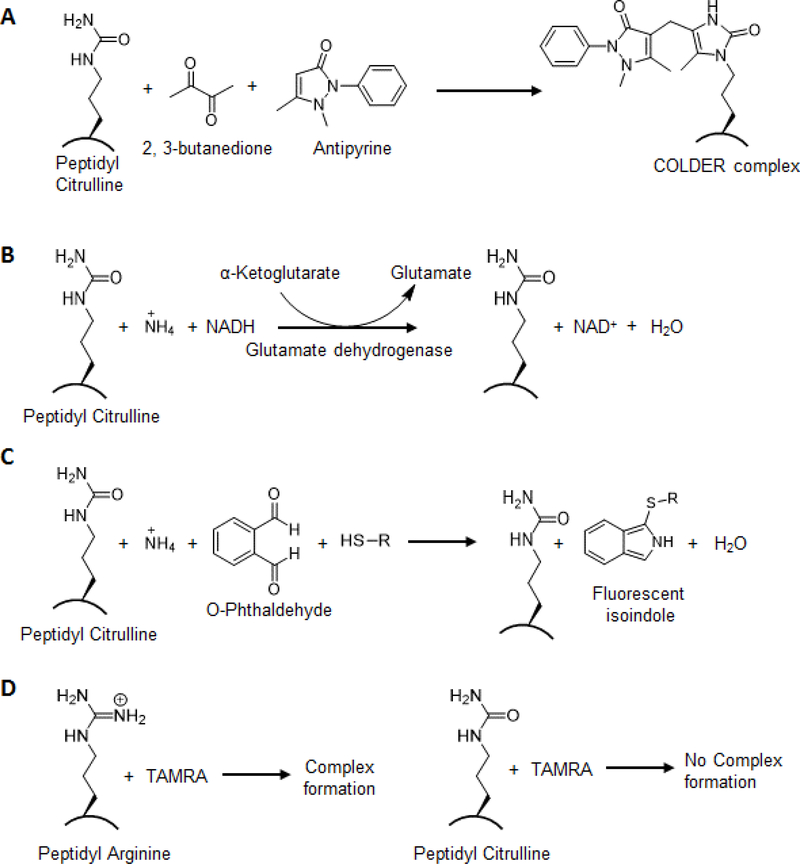
Citrullination is classically detected with the color development reagent (COLDER) assay [32]. In this assay, the ureido group in citrulline is converted into an imidazolone via reaction with 2,3-butanedione in the presence of antipyrine, thiosemicarbazide, and ammonium ferric (III) sulfate under strongly acidic conditions (Figure 2A). This reaction produces rapid color development at 95 °C which is monitored spectrophotometrically.
Figure 2.
In-vitro assays for detection of citrulline. A. COLDER assay. B. and C. Ammonia release assay D. Fluorescence-based detection of PAD activity.
Ammonia Release Assay
This assay measures the release of ammonia, a byproduct of the PAD-catalyzed reaction. To continuously measure the release of ammonia, the assay can be directly coupled to the oxidation of NADH via activating α-ketoglutarate through glutamate dehydrogenase [33]. The rate of reaction is directly proportional to the oxidation of NADH which is measured at 340 nm (Figure 2B). In another assay, the release of ammonia is measured in the presence of o-phthalaldehyde and thiol. The formation of a fluorescence product is measured by excitation at 413 nm and emission at 476 nm (Figure 2C) [34].
HPLC-based Fluorometric Assay
Chikuma and colleagues developed a more sensitive HPLC-based fluorescence assay using N-dansyl-glycyl-arginine as a substrate [35]. The PAD catalyzed conversion of N-dansyl-glycyl-arginine to N-dansyl-glycyl citrulline can be monitored at 533 nm. The reaction product N-dansyl-glycyl-citrulline is further separated by reverse phase HPLC.
Fluorescence-based detection of PAD activity
Several different fluorescence-based substrates have been developed to measure PAD activity. For example, Wang and colleagues non-covalently coupled peptidyl-arginine to tetramethylrhodamine (TAMRA) (Figure 2D) [36]. When complexed, the fluorescence of the dye is quenched by the positive charge on arginine. The amount of citrulline is measured by an increase in fluorescence upon citrullination, which occurs because of the loss of positive charge and the consequent loss in quenching. Wilderman and Pires developed an alternative fluorescence-based assay that takes advantage of fact that trypsin cleaves after arginine but not citrulline [37]. Specifically, they showed trypsin cleavage of Cbz-Arg-AMC leads to an increase in fluorescence, however, trypsin cleavage after PAD treatment blocks the increase in fluorescence.
More recently, Nakagawa and coworkers developed a turn-on fluorescence-based probe that is based on donor-excited photo induced electron transfer (d-PeT) mechanism [38]. This strategy exploits the reaction between a phenylglyoxal and citrulline under acidic conditions. Although most of these assays are easy to perform in vitro with purified enzyme, they are not suitable for detecting the PAD activity in biological mixtures due to either the interference of endogenous urea containing compounds or the low sensitivity of the assay. Furthermore, none of these assays can be used to visualize citrullinated proteins.
Methods to detect PAD substrates
Antibody-Based Detection of Protein Citrullination
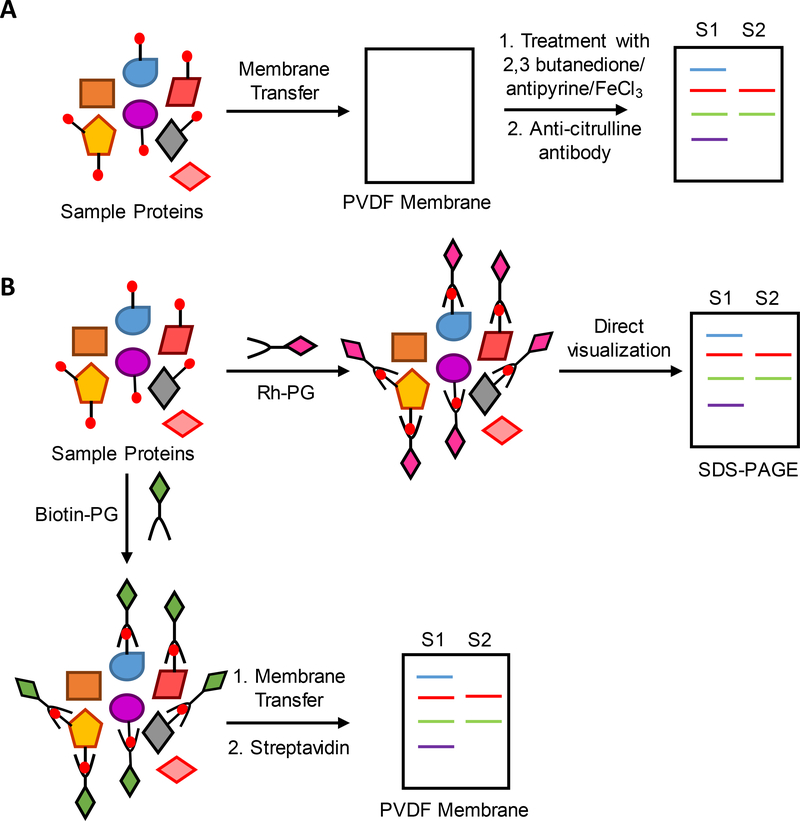
A breakthrough in the field of citrullination came with the development of antibody-based detection systems. These systems are sensitive enough to detect citrullinated proteins of low abundance. There are two types of antibody-based detection systems: 1) the antibody recognizes chemically modified citrullines; and 2) the antibody directly recognizes a citrullinated protein. The former approach was developed by Senshu who raised an antibody against citrullinated histones that had been chemically modified with 2,3-butanedione and antipyrine [39]. In this method, citrullinated proteins are transferred to PVDF and then the membrane is treated with 2,3-butanedione and antipyrine in the presence of FeCl3 under acidic conditions (Figure 3A). The blots are then washed and probed with the antibody that recognizes chemically modified citrullines. Now commercialized by Millipore as the Anti-citrulline modified (ACM) detection kit, this kit is widely used to detect citrullinated proteins.
Figure 3.
Workflow showing the detection of differential levels of protein citrullination from two different biological samples, S1 and S2, using A. ACM kit B. Rhodamine-PG and biotin-PG.
Using the latter approach, Nicholas and colleagues raised a monoclonal antibody against a citrulline-containing decapeptide [40]. This antibody, known as the F95 antibody, is also widely used to detect protein citrullination. Notably, when the ACM and F95 antibodies were compared in a head-to-head test, the F95 antibody showed lower staining suggesting that its sensitivity is poorer than the ACM kit [41]. Abcam latter developed several other commercially available polyclonal antibodies that detect protein citrullination in a sequence independent manner. Abcam have also developed antibodies against specific citrullinated residues on the N-terminal tail of histones H3 (citrulline 2, 8 and 17) and H4 (citrulline 3). Although the use of these antibodies has significantly impacted our understanding of the broader role of citrullination in human biology, they still show bias against amino acid sequences in detecting citrulline, thus making it difficult to detect global citrullination changes in complex biological samples.
Phenylglyoxal-based Probes to Detect Protein Citrullination
Phenylglyoxal has been known to react with arginine under neutral or basic conditions and citrulline under acidic conditions for more than 50 years. Using this principle, Tutturen and colleagues demonstrated that phenylglyoxal forms an adduct with citrulline that is similar to the adduct formed upon reaction with 2,3-butanedione and antipyrine [42]. Taking this concept further, Thompson and colleagues developed rhodamine-PG (Rh-PG) and biotin-PG which link rhodamine and biotin to phenylglyoxal through a triazole linker [43]. Key advantages of Rh-PG include: 1) it labels citrullinated proteins in a sequence independent manner similar to the ACM kit; 2) it has a lower detection limit and could be easily used in a high throughput assay format; and 3) it can visualize protein citrullination in a complex biological proteome after SDS-PAGE gel electrophoresis (Figure 3B). By contrast, the ACM kit requires SDS-PAGE, transfer to PVDF, chemical derivatization of citrulline on the membrane, and then western blotting with the anti-modified citrulline antibody. Using Rh-PG, Bicker et al showed that a PAD inhibitor reduces protein citrullination in the dextran sulfate (DSS)-induced model of ulcerative colitis [43]. Notably, it was possible to correlate the citrullination levels of specific proteins to disease severity. More recently, we used Rh-PG to evaluate RA patient derived serum, synovial fluid and synovial tissue samples [44]. By comparing the data to healthy controls, we showed that RA patients have elevated levels of protein citrullination. Biotin-PG can also visualize protein citrullination, however, this probe has a wider array of applications, including as a surrogate antibody for western blotting (Figure 3B), as a chemical handle to enrich and isolate citrullinated proteins from complex mixtures (see below), and as a detection element to qualitatively and quantitatively analyze citrullinated protein levels [45]. Notably, Lewallen used this probe to develop a sandwich ELISA-based platform to quantify the levels of citrullinated ApoA1. More recently, we used biotin-PG to quantify the levels of citrullinated ApoA1, antithrombin, antiplasmin and antitrypsin in RA patient serum and synovial fluid samples by both western blot and ELISA [44].
MS-based analysis of citrullinated proteins
Mass spectrometry is the only method that can identify the exact site of citrullination. However, the citrullination of an arginine residue only increases protein mass by 0.98 Da. Therefore, it is difficult to precisely identify deimination events because the mass change is difficult to distinguish from a 13C isotope or, more commonly, the deamidation of Asn and Gln residues. Another challenge includes the fact that citrullination results in the loss of a trypsin cleavage site - trypsin does not cleave after citrulline - and this leads to longer peptides that are difficult to detect by tandem MS. These issues are further compounded when analyzing low abundance proteins.
To partially overcome these challenges, Jin et al used collision-induced dissociation (CID) in combination with high energy collision dissociation (HCD) [46]. CID causes the neutral loss of isocyanic acid from the citrullinated peptide and HCD allows detection of lower m/z ions. Using this technique, the authors identified the citrullination sites in several PAD substrates, including glial fibrillary acid protein (GFAP), myelin basic protein, and neurogranin from human brain. More recently, Kuster and colleagues mined the proteomes of 30 human tissues using the above-mentioned strategy and identified 375 citrullination sites on 209 proteins [47]. Despite their tremendous efforts, most of the identified proteins were high abundance proteins suggesting the need for better methods that could enrich for low abundance citrullinated proteins prior to detection.
Enrichment of citrullinated proteins using phenylglyoxal-based probes followed by MS analysis
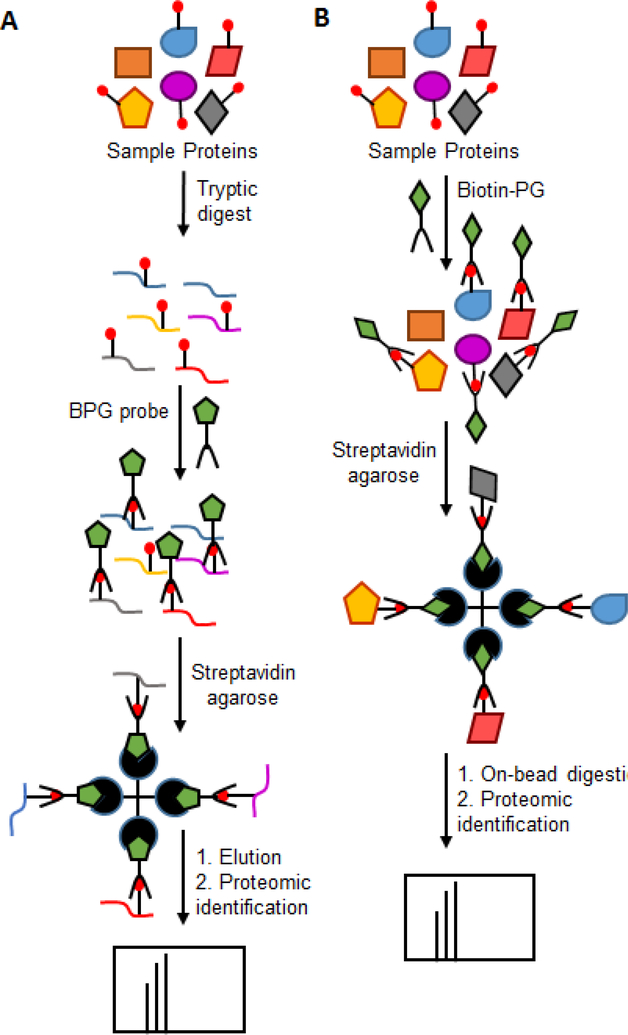
Several chemical modification strategies have been developed for the MS-based detection of citrullinated peptides and proteins. For example, Holm et al used 2,3-butanedione and antipyrine to obtain a modified citrulline adduct that shows a mass increase of 238 Da, thereby facilitating the ready differentiation between citrulline- and arginine-containing peptides [48]. In another approach, Choi labeled citrullinated peptides/proteins with 4-bromophenylglyoxal and detected these sites of citrullination by tandem MALDI MS [49]. Taking this approach further, Tutturen and coworkers developed a method for the enrichment of citrullinated peptides by treating them with biotin-labeled 4-glyoxalbenzoic acid and enriching them on streptavidin beads [50]. The samples were further analyzed by MALDI-TOF MS (Figure 4A). This strategy was used to identify citrullinated proteins from RA synovial fluid samples and the authors reported a 20-fold increase in the detection of citrullinated proteins [51]. However, this methodology involves treating digested samples with the PG-based probe followed by mass spectrometry analysis. Therefore, the number of identified proteins is much lower as this workflow will not detect peptides with other PTMs or are those of low abundance.
Figure 4.
Proteomics approaches for enrichment of citrullinated proteins in RA patient samples taken by A. Tutturen et al [50]. This strategy involves treating digested samples with the PG-based probe followed by mass spectrometry analysis. B. Tilvawala et al [44]. In this method, the citrullinated proteins are enriched before tryptic digestion followed by mass spectrometry analysis.
Along similar lines, Thompson and colleagues developed a more robust enrichment technique using biotin-PG [45]. In this method, the citrullinated proteins are enriched before tryptic digestion, resulting in enhanced coverage and higher confidence that isolated proteins are indeed citrullinated (Figure 4B). Furthermore, the labeling is performed under denaturing conditions using 20% TCA, enabling access to even buried citrullines. Using this approach, the authors identified more than 50 citrullinated proteins from HEK293T PAD2 expressing cells and showed that many of these proteins are involved in mRNA splicing and processing, suggesting for the first time that PAD activity modulates RNA biology [45]. Recently, we used this probe to identify more than 150 novel citrullinated proteins from RA patient serum, synovial fluid and synovial tissue samples [44]. Apart from known citrullinated substrates such vimentin, enolase and keratin, serine protease inhibitors (SERPINs), serine proteases and metabolic enzymes were identified as major classes of proteins that showed elevated levels of citrullination in RA patient samples compared to healthy controls. To date, this is the most comprehensive study of citrullinated proteins present in RA [44].
Functional aspect of protein citrullination
Vimentin plays a significant role in supporting and anchoring the position of organelles in the cytosol. Citrullination of vimentin leads its destabilization, resulting in structural abnormalities that promote apoptosis [52]. Histones are also citrullinated by PAD2 and PAD4 and histone citrullination by PAD4 increases the expression of growth-promoting genes and decreases the expression of growth-inhibiting genes. PAD2 citrullinates histone H3 at R26. This modification is associated with the increased expression of HER2 and more than 200 other genes under the control of the estrogen receptor (ER) [53]. Mechanistically citrullination of this site promotes an open chromatin state that aids the recruitment of ER to its target genes. Citrullination also plays critical role in cells undergoing terminal epidermal differentiation [54]. For example, citrullination of filaggrin results in the partial unfolding of this protein, thereby making it more prone to degradation by the proteasome.
As described above, PAD4 plays a key role in NETosis; PAD4 knockout mice do not form NETs. Here, citrullination aids chromatin decondensation and externalization to form web-like structures that can capture pathogens [19,20]. Given our finding that 33% of the citrullinated proteins in RA synovial tissues are extracellular proteins, we suggested that these proteins are generated by the release of active PAD isozymes from neutrophils during NETosis [44]. Indeed, active PADs are found in the synovial fluid of RA patients [55]. Notably, we identified several novel classes of citrullinated proteins including SERPINs, serine proteases and metabolic enzymes [44]. In fact, 9 of 11 glycolytic enzymes were citrullinated in our study. Citrullination of pyruvate kinase isoform M2 (PKM2) and α-enolase activated and repressed their activity, respectively. NNMT and several SERPINs were also citrullinated. NNMT catalyzes the N-methylation of nicotinamide (NAM) to form N-methylnicotinamide (MeNAM) using S-adenosyl methionine (SAM) as the cofactor. We showed that citrullination abolishes NNMT activity. Interestingly, this effect was only observed when NNMT was citrullinated by either PAD1 or PAD2; PAD3 and PAD4 treated NNMT only show moderate reductions in activity. More recently, Nemmara showed that citrullination of R132 is responsible for the loss in NNMT activity. Using CD spectroscopy, they showed that citrullination of this residue destabilizes the overall structure of NNMT, which leads to its inactivation [56].
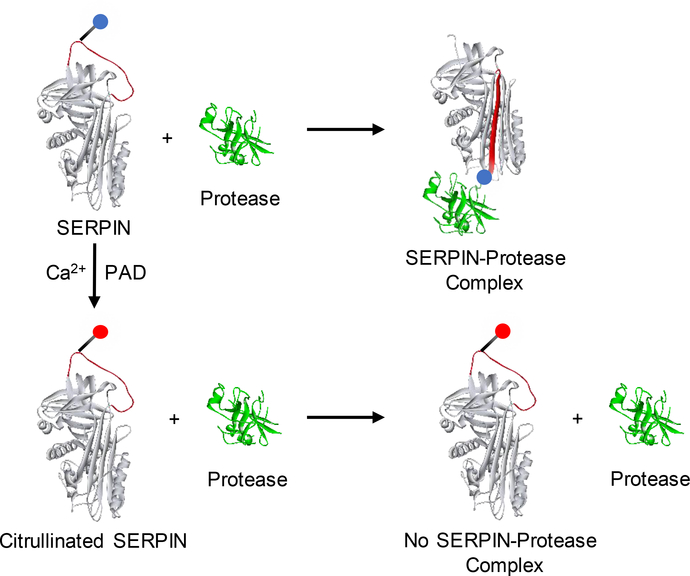
SERPINs are endogenous covalent inhibitors of serine proteases and they regulate many critical protease cascades includes fibrinolysis, blood clotting, complement activation and cell death [57]. SERPINs react with serine proteases through a reactive center loop (RCL), which extends over the body of a SERPIN. The P1 residue in the RCL is responsible for docking SERPINs into the serine protease active site. Once, the RCL is cleaved, SERPINs undergo a radical confirmation change and trap the protease in an inactive acyl-enzyme complex. In many SERPINs, the P1 position is occupied by an arginine residue [58]. We demonstrated that citrullination of a subset of P1-arginine containing SERPINs (including antiplasmin, antithrombin, C1 inhibitor, and tissue plasminogen activator inhibitor (t-PAI)) results in the loss of their inhibitory activity against their cognate proteases (Figure 5) [44]. We confirmed that the loss of inhibition is due to the modification of the P1-arginine by making a mutant version of antiplasmin (R376K) that cannot be citrullinated at the P1-arginine; citrullination does not affect the inhibitory activity of this mutant. Moreover, citrullination of antitrypsin, which contains P1-methionine in place of arginine, had no effect on its inhibitory activity [44]. Thus, citrullination of the P1-arginine is responsible for the loss of SERPIN activity and citrullination of SERPINs is a novel mechanism via which a subset of serine proteases are regulated in human body.
Figure 5.
Reaction of SERPIN and citrullinated SERPIN with protease.
Concluding remarks
Protein citrullination has emerged as an important therapeutic target in many diseases. To understand the role of citrullination in human diseases, there is a need to identify the citrullinated proteins associated with these diseases and link them together. As discussed in this review, many new tools and techniques have recently been developed to detect protein citrullination in complex biological mixtures. Since aberrant citrullination is a hallmark of numerous diseases, including lupus, atherosclerosis and various cancers, we predict that these tools will be widely used to uncover the specific roles of protein citrullination in diverse biological settings as we have recently done to show that citrullination is a novel mechanism to regulate SERPIN activity.
Acknowledgments
Funding
This work was supported in part by NIH grant R35GM118112.
Footnotes
Conflict of interest
P.R.T. founded Padlock Therapeutics and is entitled to payments from Bristol Myers Squibb if certain milestones are met. P.R.T. is a consultant for Celgene and Disarm Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Fuhrmann J, Clancy KW, Thompson PR: Chemical biology of protein arginine modifications in epigenetic regulation. Chem Rev 2015, 115:5413–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ: PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays 2003, 25:1106–1118. [DOI] [PubMed] [Google Scholar]
- 3.Raijmakers R, Zendman AJ, Egberts WV, Vossenaar ER, Raats J, Soede-Huijbregts C, Rutjes FP, van Veelen PA, Drijfhout JW, Pruijn GJ: Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol 2007, 367:1118–1129. [DOI] [PubMed] [Google Scholar]
- 4.Fujisaki M, Sugawara K: Properties of peptidylarginine deiminase from the epidermis of newborn rats. J Biochem 1981, 89:257–263. [DOI] [PubMed] [Google Scholar]
- 5.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR: Protein arginine deiminase 4 (PAD4): Current understanding and future therapeutic potential. Curr Opin Drug Discov Devel 2009, 12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 6.Nicholas AP: Dual immunofluorescence study of citrullinated proteins in Parkinson diseased substantia nigra. Neurosci Lett 2011, 495:26–29. [DOI] [PubMed] [Google Scholar]
- 7.Ishigami A, Ohsawa T, Hiratsuka M, Taguchi H, Kobayashi S, Saito Y, Murayama S, Asaga H,Toda T, Kimura N, et al. : Abnormal accumulation of citrullinated proteins catalyzed by peptidylarginine deiminase in hippocampal extracts from patients with Alzheimer’s disease. J Neurosci Res 2005, 80:120–128. [DOI] [PubMed] [Google Scholar]
- 8.Kinloch A, Lundberg K, Wait R, Wegner N, Lim NH, Zendman AJ, Saxne T, Malmstrom V,Venables PJ: Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis Rheum 2008, 58:2287–2295. [DOI] [PubMed] [Google Scholar]
- 9.Knight JS, Subramanian V, O’Dell AA, Yalavarthi S, Zhao W, Smith CK, Hodgin JB, Thompson PR, Kaplan MJ: Peptidylarginine deiminase inhibition disrupts NET formation and protects against kidney, skin and vascular disease in lupus-prone MRL/lpr mice. Ann Rheum Dis 2015, 74:2199–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang X, Han J: Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol Carcinog 2006, 45:183–196. [DOI] [PubMed] [Google Scholar]
- 11.Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z: Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer 2009, 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP, Davis T, Matesic LE, Thompson PR, Hofseth LJ: Suppression of colitis in mice by Cl-amidine: a novel peptidylarginine deiminase inhibitor. Am J Physiol Gastrointest Liver Physiol 2011, 300:G929–G938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight JS, Zhao W, Luo W, Subramanian V, O’Dell AA, Yalavarthi S, Hodgin JB, Eitzman DT,Thompson PR, Kaplan MJ: Peptidylarginine deiminase inhibition is immunomodulatory and vasculoprotective in murine lupus. J Clin Invest 2013, 123:2981–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemmara VV, Subramanian V, Muth A, Mondal S, Salinger AJ, Maurais AJ, Tilvawala R, Weerapana E, Thompson PR: The Development of Benzimidazole-Based Clickable Probes for the Efficient Labeling of Cellular Protein Arginine Deiminases (PADs). ACS Chem Biol 2018, 13:712–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemmara VV, Thompson PR: Development of Activity-Based Proteomic Probes for Protein Citrullination. Curr Top Microbiol Immunol 2018, 10.1007/82_2018_132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vossenaar ER, Despres N, Lapointe E, van der Heijden A, Lora M, Senshu T, van Venrooij WJ, Menard HA: Rheumatoid arthritis specific anti-Sa antibodies target citrullinated vimentin. Arthritis Res Ther 2004, 6:R142–R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandra PE, Sokolove J, Hipp BG, Lindstrom TM, Elder JT, Reveille JD, Eberl H, Klause U,Robinson WH: Novel multiplex technology for diagnostic characterization of rheumatoid arthritis. Arthritis Res Ther 2011, 13:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor P, Gartemann J, Hsieh J, Creeden J: A systematic review of serum biomarkers anti-cyclic citrullinated Peptide and rheumatoid factor as tests for rheumatoid arthritis. Autoimmune Dis 2011, 2011:815038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y: PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med 2010, 207:1853–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohrbach AS, Slade DJ, Thompson PR, Mowen KA: Activation of PAD4 in NET formation. Front Immunol 2012, 3:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khandpur R, Carmona-Rivera C, Vivekanandan-Giri A, Gizinski A, Yalavarthi S, Knight JS,Friday S, Li S, Patel RM, Subramanian V, et al. : NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med 2013, 5:178ra140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y, Su K: Neutrophil Extracellular Traps and Systemic Lupus Erythematosus. J Clin Cell Immunol 2013, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietronigro EC, Della Bianca V, Zenaro E, Constantin G: NETosis in Alzheimer’s Disease. Front Immunol 2017, 8:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M: Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol 2004, 11:777–783. [DOI] [PubMed] [Google Scholar]
- 25.Saijo S, Nagai A, Kinjo S, Mashimo R, Akimoto M, Kizawa K, Yabe-Wada T, Shimizu N,Takahara H, Unno M: Monomeric Form of Peptidylarginine Deiminase Type I Revealed by X-ray Crystallography and Small-Angle X-ray Scattering. J Mol Biol 2016, 428:3058–3073.The authors solved structure of PAD1 performed detailed analysis of structural elements of PAD1. Authors further performed detailed structural comparison of PAD1 with PAD2 and PAD4.
- 26.Slade DJ, Fang P, Dreyton CJ, Zhang Y, Fuhrmann J, Rempel D, Bax BD, Coonrod SA, Lewis HD, Guo M, et al. : Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design. ACS Chem Biol 2015, 10:1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H,Thompson PR: Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry 2010, 49:4852–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arita K, Shimizu T, Hashimoto H, Hidaka Y, Yamada M, Sato M: Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4. Proc Natl Acad Sci U S A 2006, 103:5291–5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dreyton CJ, Knuckley B, Jones JE, Lewallen DM, Thompson PR: Mechanistic studies of protein arginine deiminase 2: evidence for a substrate-assisted mechanism. Biochemistry 2014, 53:4426–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knuckley B, Causey CP, Pellechia PJ, Cook PF, Thompson PR: Haloacetamidine-based inactivators of protein arginine deiminase 4 (PAD4): evidence that general acid catalysis promotes efficient inactivation. Chembiochem 2010, 11:161–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knuckley B, Bhatia M, Thompson PR: Protein arginine deiminase 4: evidence for a reverse protonation mechanism. Biochemistry 2007, 46:6578–6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knipp M, Vasak M: A colorimetric 96-well microtiter plate assay for the determination of enzymatically formed citrulline. Anal Biochem 2000, 286:257–264. [DOI] [PubMed] [Google Scholar]
- 33.Liao YF, Hsieh HC, Liu GY, Hung HC: A continuous spectrophotometric assay method for peptidylarginine deiminase type 4 activity. Anal Biochem 2005, 347:176–181. [DOI] [PubMed] [Google Scholar]
- 34.Sugawara K, Oyama F: Fluorogenic reaction and specific microdetermination of ammonia. J Biochem 1981, 89:771–774. [DOI] [PubMed] [Google Scholar]
- 35.Chikuma T, Yamada M, Tsuda A, Yamamoto M, Nakashima K, Yajima R, Kato T: A highly sensitive high-performance liquid chromatography-- fluorometric method for the assay of peptidylarginine deiminase activity. Anal Biochem 2000, 285:230–234. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, Priestman MA, Lawrence DS: Monitoring of protein arginine deiminase activity by using fluorescence quenching: multicolor visualization of citrullination. Angew Chem Int Ed Engl 2013, 52:2323–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wildeman E, Pires MM: Facile fluorescence-based detection of PAD4-mediated citrullination. Chembiochem 2013, 14:963–967. [DOI] [PubMed] [Google Scholar]
- 38.Kunieda K, Yamauchi H, Kawaguchi M, Ieda N, Nakagawa H: Development of a fluorescent probe for detection of citrulline based on photo-induced electron transfer. Bioorg Med Chem Lett 2018, 28:969–973.The authors developed a turn-on fluorescence-based probe that is based on donor-excited photo induced electron transfer (d-PeT) mechanism to detect protein citrullination.
- 39.Senshu T, Sato T, Inoue T, Akiyama K, Asaga H: Detection of citrulline residues in deiminated proteins on polyvinylidene difluoride membrane. Anal Biochem 1992, 203:94–100. [DOI] [PubMed] [Google Scholar]
- 40.Nicholas AP, Whitaker JN: Preparation of a monoclonal antibody to citrullinated epitopes: its characterization and some applications to immunohistochemistry in human brain. Glia 2002, 37:328–336. [PubMed] [Google Scholar]
- 41.Nicholas AP, King JL, Sambandam T, Echols JD, Gupta KB, McInnis C, Whitaker JN: Immunohistochemical localization of citrullinated proteins in adult rat brain. J Comp Neurol 2003, 459:251–266. [DOI] [PubMed] [Google Scholar]
- 42.Tutturen AE, Holm A, Jorgensen M, Stadtmuller P, Rise F, Fleckenstein B: A technique for the specific enrichment of citrulline-containing peptides. Anal Biochem 2010, 403:43–51. [DOI] [PubMed] [Google Scholar]
- 43.Bicker KL, Subramanian V, Chumanevich AA, Hofseth LJ, Thompson PR: Seeing citrulline: development of a phenylglyoxal-based probe to visualize protein citrullination. J Am Chem Soc 2012, 134:17015–17018.In this work, the authors developed a technique for the visualization of citrullinated proteins in complex protein samples to a lower limit not yet described. This method allows for fast, accurate analysis of citrullination levels in a variety of biological sample types.
- 44.Tilvawala R, Nguyen SH, Maurais AJ, Nemmara VV, Nagar M, Salinger AJ, Nagpal S,Weerapana E, Thompson PR: The Rheumatoid Arthritis-Associated Citrullinome. Cell Chem Biol 2018, 25:691–704 e696.In this article, the authors identified more than 150 novel citrullinated proteins from RA serum, synovial fluid and synovial tissue samples using an optimized enrichment technique that employs biotin-PG. The authors further described the functional consequences of citrullination on SERPIN activity.
- 45.Lewallen DM, Bicker KL, Subramanian V, Clancy KW, Slade DJ, Martell J, Dreyton CJ,Sokolove J, Weerapana E, Thompson PR: Chemical Proteomic Platform To Identify Citrullinated Proteins. ACS Chem Biol 2015, 10:2520–2528.This work uses biotin-conjugated phenylglyoxal to selectively enrich for citrulline-containing proteins. This method was used in cell lysates over expressing PAD2 to show citrullination of multiple RNA binding proteins.
- 46.Jin Z, Fu Z, Yang J, Troncosco J, Everett AD, Van Eyk JE: Identification and characterization of citrulline-modified brain proteins by combining HCD and CID fragmentation. Proteomics 2013, 13:2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee CY, Wang D, Wilhelm M, Zolg DP, Schmidt T, Schnatbaum K, Reimer U, Ponten F, Uhlen M, Hahne H, et al. : Mining the Human Tissue Proteome for Protein Citrullination. Mol Cell Proteomics 2018, 17:1378–1391.The authors used collision-induced dissociation (CID) in combination with high energy collision dissociation (HCD). Using this methodology, the authors mined the proteomes of 30 human tissues and identified 375 citrullination sites on 209 proteins
- 48.Holm A, Rise F, Sessler N, Sollid LM, Undheim K, Fleckenstein B: Specific modification of peptide-bound citrulline residues. Anal Biochem 2006, 352:68–76. [DOI] [PubMed] [Google Scholar]
- 49.Choi M, Song JS, Kim HJ, Cha S, Lee EY: Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of peptide citrullination site using Br signature. Anal Biochem 2013, 437:62–67. [DOI] [PubMed] [Google Scholar]
- 50.Tutturen AE, Holm A, Fleckenstein B: Specific biotinylation and sensitive enrichment of citrullinated peptides. Anal Bioanal Chem 2013, 405:9321–9331.This article describes a method for the detection of citrullinated proteins in synovial fluid and identifies more than 100 novel sites of citrullination. Using this protocol, citrullinated proteins were enriched more than 20-fold compared to direct MS analysis.
- 51.Tutturen AE, Fleckenstein B, de Souza GA: Assessing the citrullinome in rheumatoid arthritis synovial fluid with and without enrichment of citrullinated peptides. J Proteome Res 2014, 13:2867–2873. [DOI] [PubMed] [Google Scholar]
- 52.Inagaki M, Takahara H, Nishi Y, Sugawara K, Sato C: Ca2+-dependent deamination-induced disassembly of intermediate filaments involves specific modification of the amino-terminal head domain. J Biol Chem 1989, 264:18119–18127. [PubMed] [Google Scholar]
- 53.Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD, Slade DJ, Dreyton CJ,Subramanian V, Bicker KL, et al. : Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor alpha target gene activation. Proc Natl Acad Sci U S A 2012, 109:13331–13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Senshu T, Kan S, Ogawa H, Manabe M, Asaga H: Preferential deimination of keratin K1 and filaggrin during the terminal differentiation of human epidermis. Biochem Biophys Res Commun 1996, 225:712–719. [DOI] [PubMed] [Google Scholar]
- 55.Spengler J, Lugonja B, Ytterberg AJ, Zubarev RA, Creese AJ, Pearson MJ, Grant MM,Milward M, Lundberg K, Buckley CD, et al. : Release of Active Peptidyl Arginine Deiminases by Neutrophils Can Explain Production of Extracellular Citrullinated Autoantigens in Rheumatoid Arthritis Synovial Fluid. Arthritis Rheumatol 2015, 67:3135–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nemmara VV, Tilvawala R, Salinger AJ, Miller L, Nguyen SH, Weerapana E, Thompson PR: Citrullination Inactivates Nicotinamide- N-methyltransferase. ACS Chem Biol 2018, 13:2663–2672.This article describes a technique to identify citrullination sites in recombinant protein using phenylglyoxal. The authors further explained the functional consequences of citrullination on NNMT activity.
- 57.Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, et al. : The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem 2001, 276:33293–33296. [DOI] [PubMed] [Google Scholar]
- 58.Khan MS, Singh P, Azhar A, Naseem A, Rashid Q, Kabir MA, Jairajpuri MA: Serpin Inhibition Mechanism: A Delicate Balance between Native Metastable State and Polymerization. J Amino Acids 2011, 2011:606797. [DOI] [PMC free article] [PubMed] [Google Scholar]