Abstract
Pheochromocytoma and paraganglioma (PPGL) can be divided into at least four molecular subgroups. Whether such categorizations are independent factors for prognosis or metastatic disease is unknown. We performed a systematic review and individual patient meta-analysis aiming to estimate if driver mutation status can predict metastatic disease and survival. Driver mutations were used to categorize patients accordingly to three different molecular systems: two subgroups (SDHB mutated or wild type), three subgroups (pseudohypoxia, kinase signaling or Wnt/unknown) and four subgroups (tricarboxylic acid cycle, VHL/EPAS1, kinase signaling or Wnt/unknown). Twenty-one studies and 703 patients were analyzed. Multivariate models for association with metastasis showed correlation with SDHB mutation (OR 5.68 [95% CI 1.79–18.06]) as well as norepinephrine (OR 3.01 [95% CI 1.02–8.79]) and dopamine (OR 6.39 [95% CI 1.62–25.24]) but not to PPGL location. Other molecular systems were not associated with metastasis. In multivariate models for association with survival, age (HR 1.04 [95% CI 1.02–1.06]) and metastases (HR 6.13 [95% CI 2.86–13.13]) but neither paraganglioma or SDHB mutation remained significant. Other molecular subgroups did not correlate with survival. We conclude that molecular categorization accordingly to SDHB provided independent information on the risk of metastasis. Driver mutations status did not correlate independently with survival. These data may ultimately be used to guide current and future risk stratification of PPGL.
Introduction
The Cancer Genome Atlas (TCGA) proposed that neuroendocrine tumors of adrenal paraganglia, pheochromocytomas (PCCs) and extra-adrenal paraganglia paragangliomas (PGLs, together denoted PPGL) can be divided into three main molecular subgroups that have been linked to distinct driver genes(Fishbein, et al. 2017): Pseudohypoxia (SDHA, SDHB, SDHC, SDHD, SDHAF2, FH, VHL, EPAS1 and EGLN1), Wnt-altered (CSDE1 or MAML3), and kinase signaling (RET, NF1, TMEM127, MAX, HRAS, FGFR1, and MET)(Bausch, et al. 2017; Castro-Vega, et al. 2015; Fishbein et al. 2017; Letouze, et al. 2013; Toledo, et al. 2015; Welander, et al. 2018). Previous data also support that the pseudohypoxic group can be further divided into two subclusters: tricarboxylic acid (TCA) cycle related (SDHA-SDHD, SDHAF2, or FH), and those with VHL/EPAS1 related (VHL/EPAS1/EGLN1) PPGLs(Burnichon, et al. 2011; Fliedner, et al. 2016; Letouze et al. 2013). Each subgroup is named after their molecular hallmarks and are thought to be associated with distinct biochemical and clinical phenotypes (reviewed in(Crona, et al. 2017; Neumann, et al. 2018)): All pseudohypoxic PPGLs secrete norepinephrine and those related to the TCA-cycle are more predominantly PGLs with relatively high proportion having dopamine secretion. The TCA-cycle subgroup and particularly SDHB carriers are associated with the highest proportion of metastatic disease(Eisenhofer, et al. 2011a; Eisenhofer, et al. 2011b). On the other end of the spectrum is the kinase signaling subgroup that has a more well differentiated phenotype with epinephrine secretion, predominantly adrenal location and rarely develop metastatic disease. PPGLs related to the Wnt-altered subgroup are thought to display intermediate characteristics in terms of catecholamine secretion (mixed noradrenergic and adrenergic) and frequency of metastatic or recurrent disease(Fishbein et al. 2017). It has also been proposed that PPGL with somatic abberations in genes releated to telomere maintenance (inactivation of ATRX or transcriptional activation of TERT) as well as chromatin maintenance (SETD2) could have more aggressive features and may thus be disease modifiers(Fishbein, et al. 2015; Fishbein et al. 2017; Job, et al. 2018).
The predominant cause of death in patients with PPGL is metastasis that occur in about 10–20% of cases(Hamidi, et al. ; Timmers, et al. 2008). Tumor location (PGL versus PCC), germline SDHB mutations (SDHB mutated versus SDHB wild type), ATRX mutation, TERT overexpression, catecholamine secretion (noradrenergic or dopaminergic versus adrenergic) and large size of the primary tumor have all been independently associated with metastasis(Assadipour, et al. 2017; Ayala-Ramirez, et al. 2011; Cho, et al. 2018; Eisenhofer, et al. 2012; Job et al. 2018; Turkova, et al. 2015; Welander, et al. 2011). The disease course of those with metastatic disease is heterogeneous in terms of tumor aggressiveness and overall survival(Hamidi, et al. 2017b). Size of the primary tumor, gender, SDHB mutation, catecholamine profile, ATRX mutation and TERT overexpression are suggested to be prognostic factors for survival(Amar, et al. 2007; Ayala-Ramirez et al. 2011; Hamidi et al. 2017b; Job et al. 2018; Zelinka, et al. 2011).
Although at least 16 common driver genes has been identified in PPGL, the only disease driver that showed a robust correlation to metastatic disease and outcome has been SDHB. We and others have proposed that the improved characterization of PPGL driver mutations provide additional information beyond the dichotomous categorization based on SDHB. However, due to disease rarity and extensive genetic heterogeneity, interpretation of findings are currently restricted due to low statistical power. We hypothesized that a systematic review and individual patient meta-analysis could overcome these challenges and provide information on correlation between driver mutation status and clinical parameters. We particularly focused on predictive factors for metastatic disease and prognostic factors for survival.
Methods
This study followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) workflow(Liberati, et al. 2009). The study reviewed and analyzed published data, these activities fall under an approval by the Regional Ethics Committee in Uppsala, Sweden (Dnr 2015/544).
Search strategy
One investigator (JC) performed a systematic search of PubMed to identify relevant reports published between 2007-01-01 and 2017-12-01. The following search terms were used: “pheochromocytoma” and “paraganglioma”. Reports were initially screened by title for relevance and potentially relevant reports had its abstract reviewed. Case reports, review articles and editorials as well as those publications in other languages than English were not considered. Potentially relevant studies were assessed for eligibility through review of the full-text article.
Study Selection and Data Extraction
Studies fulfilling the following eligibility criteria were included. Criteria (1) genetic sequencing and reporting of PPGL disease drivers: germline mutations: SDHA, SDHB, SDHD, TMEM127, germline and somatic mutations; VHL, RET, NF1, MAX, and somatic mutations; HRAS. Criteria (2), shared data on genetic mutations and clinical characteristics on the individual patient level for both mutation positive and mutation negative cases. Criteria (3), patient identification numbers for cross-validation between different studies from the same study site. Publications were grouped into cohorts based on the study site to allow for reconstruction of each study cohort. Two investigators (JC and SG) reviewed the papers independently and transferred the data into one study database. Values that did not overlap between the two investigators were re-assessed to reach a common conclusion.
Data collection and cleaning
Patients without available PPGL tissue for analysis were excluded. For patients with multiple primary tumors, the one that occurred at the earliest age was selected, or if the same time point, the row that occurred first in the original data was chosen. In patients with a conflict of data between multiple publications, the most recent value was used. Collected data-points and definitions are provided in section 1 of the supplementary data appendix.
Definition of PPGL driver-gene subgroups
With the TCGA publication as a starting point and taking into account the available literature, we selected three different systems for driver gene categorization: A 2-molecular subgroup system accordingly to SDHB mutation status: SDHB mutated or SDHB wild type. A second system with 3-molecular subgroups categorized according to presence of germline/somatic driver mutations/gene fusions: Pseudohypoxia (SDHA, SDHB, SDHC, SDHD, SDHAF2, FH, VHL, EPAS1 or EGLN1), kinase signaling (NF1, RET, TMEM127, MAX, HRAS, MET or FGFR1) and Wnt/unknown (CSDE1 or MAML3). PPGL with driver mutations associated with different molecular subgroups as well as those without a driver mutation was classified as Wnt/unknown. The cortical admixture subgroup, originally reported by the TCGA project is thought to be defined by non-tumoral cells and was not included(Crona et al. 2017; Fishbein et al. 2017). A third system with 4-molecular subgroups was also used to take into account the distinct features of TCA-cycle related (SDHA, SDHB, SDHC, SDHD, SDHAF2, and FH) and VHL/EPAS1 related (VHL, EPAS1, and EGLN1) PPGLs(Burnichon et al. 2011; Fliedner et al. 2016; Flynn, et al. 2014).
Risk of bias assessment
Risk of bias was determined by two investigators (JC, SG), in cases of discrepant assessement the original papers were re-evaluated to reach a common conclusion. Bias assessment was designed based on a modified Newcastle-Ottawa tool for bias assessment adopted by Hamidi et al.(Hamidi et al. 2017a) that was further modified to this study. Criteria for bias assessment is available in section 2 of the supplementary data appendix.
Statistical Analyses
Nominal data are presented as number of patients and percentages and were analyzed with Chi square test. Scaled data were presented as median and range or 95% confidence interval (CI) and were analyzed with Mann-Whitney U or Kruskal Wallis tests. Logistic regression (univariate/multivariable) was used as appropriate. Survival analysis was performed using Log-Rank, Kaplan-Meier and Cox regression analyses. P-values <0.05 were defined as statistically significant. Variables identified as significant in univariate analysis were included in multivariable analysis (applicable for logistic and for Cox regression). Statistical analyses were performed using SPSS version 22 (IBM, NY, USA) and Stata version 12 (College Station, TX, USA). Figures were drawn with Prism 6.0h (GraphPad Software Inc, USA) and Stata version 12 (College Station, TX, USA).
Results
A PubMed search generated 7689 results and 118 manuscripts were selected for review of eligibility (Supplementary Figure 1). A total of 97 publications did not meet criteria on method for genetic sequencing (n=82) or individual patient data availability (n=13). Two studies were excluded as individual patients could not be matched to previous studies. Twenty one publications matched study criteria and allowed reconstruction of 7 cohorts (Table 1). These 7 cohorts represented 948 individual patients, 32 had data on multiple tumor lesions. Two hundred-forty-five patients were excluded as there was no tumor tissue available. Seven hundred three patients remained, 274 were analyzed with exome sequencing and 429 with a targeted re-sequencing approach.
Table 1.
Study cohorts
| References | Patients, n | Study site | WES, n | Targeted re-sequencing, n |
|---|---|---|---|---|
| (Juhlin, et al. 2015; Stenman, et al. 2016a; Stenman, et al. 2016b; Welander, et al. 2014a; Welander, et al. 2014b; Welander, et al. 2012; Welander et al. 2018) | 137 | Karolinska University Hospital, Stockholm, Sweden; Linköping University Hospital, Linköping, Sweden; Haukeland University Hospital, Bergen, Norway | 19 | 118 |
| (Wilzen, et al. 2016) | 9 | Sahlgrenska University Hospital, Gothenburg, Sweden | 9 | 0 |
| (Dwight, et al. 2018; Flynn et al. 2014; Flynn, et al. 2017; Flynn, et al. 2016) | 48 | The Peter MacCallum Cancer Centre and University of Melbourne, Australia; Kolling Institute and University of Sydney, Australia; Royal Brisbane Hospital, Brisbane, Australia; Royal North Shore Hospital, Sydney, Australia; The Children’s Hospital at Westmead, Sydney, Australia | 44 | 4 |
| (Burnichon, et al. 2012; Burnichon et al. 2011; Castro-Vega, et al. 2014; Castro-Vega et al. 2015; Favier, et al. 2012; Letouze et al. 2013) | 190 | INSERM, Hôpital Europe éen Georges Pompidou and Université Paris Descartes, Sorbonne Paris Cité French, Paris, France. Cortico et Médullosurrénale: les Tumeurs Endocrines (COMETE) Network, France. | 29 | 161 |
| (Fishbein et al. 2017) | 173 | National Institutes of Health, United states and multiple collaborating institutions of the Cancer Genome Atlas, Pheochromocytoma and Paraganglioma project. | 173 | 0 |
| (Curras-Freixes, et al. 2015) | 118 | Centro Nacional de Investigaciones Oncológicas, Madrid, Spain and multiple collaborating institutions throughout Spain. | 0 | 118 |
| (Toledo et al. 2015) | 28 | University of Texas Health Science Center at San Antonio, San Antonio, United States. | 28 | 0 |
WES, Whole Exome Sequencing.
Risk of bias assessment
Risk of bias assessments was performed for each study cohort and it is presented in Supplementary Figure 2. All studies performed retrospective characterization of case series. Assessment of genetic results (7/7 studies low risk) and method coverage (5/5 studies low risk) showed relatively low risk of bias. Clinical data and particularly hormone assessment (6/7 studies high or unclear risk) and follow-up time (7/7 studies high or unclear risk) had a high risk of bias.
Baseline characteristics
Clinical characteristics of the reviewed patients are presented in Table 2. PPGL-related driver mutations were detected in 437 patients (62.6%, 95% CI 58.5–65.7, Supplementary Figure 3, Supplementary Table 1) that were confirmed as germline in 178 (25.3%, 95% CI 22.3–28.7) and somatic in 237 (33.7%, 95% CI 30.3–37.3). The frequency of mutations in the different driver genes are shown in Supplementary Table 1.
Table 2:
Clinical Characteristics of the reviewed patients
| Patients (n=703) | Frequency | % | |
|---|---|---|---|
| Gender | Male | 307 | 43.7 |
| Female | 392 | 55.7 | |
| Data not available | 4 | 0.6 | |
| Age at diagnosis | Median (range) | 46 (7–84) | |
| Tumor size mm, | Median (range) | 45 (10–160) | |
| Stage | Non-metastatic | 618 | 87.9 |
| Metastatic | 85 | 12.1 | |
| Catecholamine profile | Epinephrine | 161 | 22.9 |
| Norepinephrine | 139 | 19.8 | |
| Dopamine | 24 | 3.4 | |
| Data not available | 324 | 53.9 | |
| WHO 2004 | Pheochromocytoma | 572 | 81.4 |
| Paraganglioma | 127 | 18.1 | |
| Data not available | 4 | 0.5 | |
| WHO 2017 | PCC | 572 | 81.4 |
| Sympathetic PGL | 96 | 13.7 | |
| Head and Neck PGL | 27 | 3.8 | |
| Data not available | 8 | 1.1 | |
| Time on follow up (months) | Median (range) | 33 (0–316) | |
| Status at the end of follow-up | Alive | 494 | 70.3 |
| Dead | 40 | 5.7 | |
| Data not available | 169 | 24.0 | |
| ATRX mutation status | ATRX mutated | 450 | 63.9 |
| ATRX wild type | 17 | 2.4 | |
| Data not available | 237 | 33.7 | |
| 2-molecular subgroups | SDHB wild type | 645 | 91.7 |
| SDHB mutated | 58 | 8.3 | |
| 3-molecular subgroups | Pseudohypoxia | 177 | 25.2 |
| Kinase signaling | 245 | 34.9 | |
| Wnt/unknown | 281 | 39.9 | |
| 4-molecular subgroups | Pseudohypoxia TCA-cycle | 79 | 11.3 |
| Pseudohypoxia VHL/EPAS1 | 98 | 13.9 | |
| Kinase signaling | 245 | 34.9 | |
| Wnt/unknown | 281 | 39.9 | |
DA, Dopamine; E, Epinephrine; F, Female; HNPGL, Head and Neck PGL; M, Male; NA, Not Available; NE, Norepinephrine; PCC, Pheochromocytoma; PGL, Paraganglioma; sPGLs Sympathetic PGL; TCA, tricarboxylic acid.
Data on age was missing in 3 patients, on tumor size on 291 patients and on follow up length in 167 patients.
Patients were categorized into three different molecular systems. Two-molecular subgroups: SDHB mutated 58 patients (8.3%, 95% CI 6.4–10.5, Table 2) and SDHB wild type 645 patients (91.8% 95% CI 89.5–93.6). Three-molecular subgroups: pseudohypoxia, 177 patients (24.9% 95% CI 21.8–28.2); kinase signaling, 245 patients (34.9%, 95% CI 31.4–38.5); and Wnt/unknown, 281 patients (39.9% 95% CI 36.4–43.6). In the 4-molecular subgroup system, the pseudohypoxia subgroup was further divided into TCA-cycle, 79 patients (11.2%, 95% CI 9.1–13.8) and VHL/EPAS1 related, 98 patients (13.9%, 95% CI 11.6–16.7).
Clinical correlations to molecular subgroups
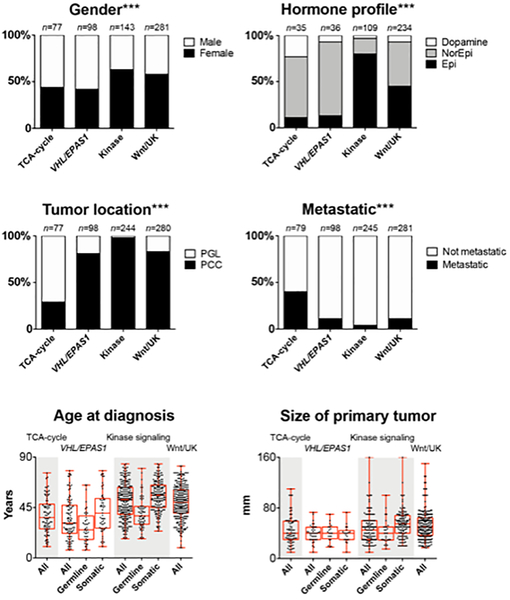
An overview of clinical correlations to the different molecular systems are presented in Figure 1, Supplementary Figure 3 and Supplementary Table 2. Gender, catecholamine profile, WHO classification (PCC versus PGL), metastatic stage, age at diagnosis as well as tumor size were all differently distributed (p-values <0.05) among subgroups of all three molecular systems. Detailed descriptive data is available in section 4 of the supplementary data appendix.
Figure 1:
Clinical correlations to modified molecular subgroups from the Cancer Genome Atlas. Epi, Epinephrine; Kinase; Kinase signaling subgroup; Norepi, Norepinephrine; PCC, Pheochromocytoma; PGL, Paraganglioma; TCA, tricarboxylic acid; UK, unknown; VHL/EPAS1, Pseudohypoxia VHL/EPAS1 related. ***: chi-square test including all four molecular subgroups had a significance level <0.001
Predictive factors of metastatic disease
Frequency of metastatic disease in the cohort was 12.1% (85/703 patients). Categorization accordingly to the 2-,3- and 4-molecular subgroup systems as well as catecholamine profile, WHO classification and ATRX mutation status correlated with metastatic disease in univariate Cox regression analyses (Figure 1, Table 3): Those with SDHB mutated PPGLs had metastatic disease in 46.6% (27/58 patients, OR 8.81 [95% CI 4.92–15.78]; P<0.001) that was higher compared to SDHB wild type 8.9% (58/645 patients) PPGLs. In the 3-molecular subgroup system, metastasis was more common in pseudohypoxia 24.3% (43/177 patients, OR 2.49 [95% CI 1.51–4.13] P<0.001) and less frequent in kinase signaling 4.1% (10/245 patients, OR 0.33 [95% CI 0.16–0.69] P=0.003) compared to Wnt/unknown 11.4% (32/281 patients). In the 4-molecular subgroups classification, metastatic PPGLs occurred more often in TCA-cycle 40.5% (32/79 patients, OR 5.29 [95% CI 2.96–9.47]) but was not different in VHL/EPAS1 related PPGLs 11.2% (11/98 patients, OR 0.98 [95% CI 0.78–2.04]) compared to the Wnt/unknown group.
Table 3:
Factors related with increased risk of metastatic disease
| Logistic Regression (Univariate analysis) | Logistic regression (Multivariable analysis) MODEL 1 |
Logistic regression (Multivariable analysis) MODEL 2 |
Logistic regression (Multivariable analysis) MODEL 3 |
|||
|---|---|---|---|---|---|---|
| % of patients with metastatic disease | OR (95% CI); p value | OR (95% CI); p value | OR (95% CI); p value | OR (95% CI); p value | ||
| Gender | Female | 11.2% | 1 (Ref) | - | - | - |
| Male | 13.4% | 1.22 (0.77–1.92); 0.393 | - | - | - | |
| Age at diagnosis | Continuous variable | NA | 0.99 (0.98–1.01); 0.307 | - | - | - |
| Tumor size mm | ≤50 mm | 10.9% | 1 (Ref) | - | - | - |
| >50 mm | 15.7% | 1.52 (0.76–3.05); 0.236 | - | - | - | |
| Hormone | Epinephrine | 3.1% | 1 (Ref) | 1 (Ref) | 1 (Ref) | 1 (Ref) |
| Norepinephrine | 12.9% | 4.64 (1.68–12.86); 0.003 | 3.01 (1.02–8.79); 0.045 | 2.88 (0.94–8.82); 0.065 | 3.12 (1.02–9.56); 0.046 | |
| Dopamine | 29.2% | 12.85 (3.67–44.93); <0.001 | 6.39 (1.62–25.24); 0.008 | 7.86 (2.03–30.4); 0.003 | 6.32 (1.58–25.30); 0.009 | |
| WHO 2004 | PCC | 7.9% | 1 (Ref) | 1 (Ref) | 1 (Ref) | 1 (Ref) |
| PGL | 29.1% | 4.81 (2.95–7.85); <0.001 | 1.52 (0.52–4.43); 0.436 | 3.09 (1.20–7.97); 0.019 | 1.76 (0.57–5.42); 0.324 | |
| WHO 2017 | PCC | 7.9% | 1 (Ref) | * | * | * |
| Sympathetic PGL | 29.2% | 4.82 (2.82–8.23); <0.001 | * | * | * | |
| Head and Neck PGL | 25.9% | 4.09 (1.65–10.21); 0.002 | * | * | * | |
| ATRX mutation status | ATRX mutated | 58.5% | 13.18 (4.78–36.36); <0.001 | * | * | * |
| ATRX wild type | 9.8% | 1 (Ref) | * | * | * | |
| 2-molecular subgroups | SDHB wild type | 8.9% | 1 (Ref) | 1 (Ref) | £ | & |
| SDHB mutated | 46.6% | 8.81 (4.92–15.78); <0.001 | 5.68 (1.79–18.06); 0.003 | £ | & | |
| 3-molecular subgroups | Wnt/unknown | 11.4% | 1 (Ref) | $ | 1 (Ref) | & |
| Pseudohypoxia | 24.3% | 2.49 (1.51–4.13); <0.001 | $ | 0.92 (0.35–2.43); 0.861 | & | |
| Kinase signaling | 4.1% | 0.33 (0.16–0.69); 0.003 | $ | 0.49 (0.13–1.91); 0.305 | & | |
| 4-molecular subgroups | Wnt/unknown | 11.4% | 1 (Ref) | $ | £ | 1 (Ref) |
| Pseudohypoxia TCA-cycle | 40.5% | 5.29 (2.96–9.47); <0.001 | $ | £ | 2.65 (0.83–8.48); 0.101 | |
| Pseudohypoxia VHL/EPAS1- | 11.2% | 0.98 (0.78–2.04); 0.965 | $ | £ | ! | |
| Kinase signaling | 4.1% | 0.33 (0.19–0.69); 0.003 | $ | £ | 0.45 (0.12–1.74); 0.246 | |
95% CI, 95% confidence interval; DA, Dopamine; E, Epinephrine; HNPGL, Head and Neck PGL; NA, Not Available; NE, Norepinephrine; OR, Odds Ratio; PCC, Pheochromocytoma; PGL, Paraganglioma; Ref, Reference; sPGLs Sympathetic PGL; TCA, tricarboxylic acid.
Not included, WHO 2004 classification included in the multivariable analysis.
3- and 4-molecular subgroup systems not included in Model 1 of the multivariable analysis (2-molecular subgroups included instead).
2- and 4-molecular-subgroups not included in Model 2 of the multivariable analysis (3-molecular subgroups classification included instead).
2- and 3-molecular subgroups not included in Model 3 of the multivariable analysis (4-molecular subgroups classification included instead).
could not be calculated due to lack of observations.
ATRX mutation status correlated with increased frequency of metastasis in univariate analysis. But, it was not included in the multivariate model due to a lack of clinical annotations in cases that had analysis of ATRX mutation status.
The three different molecular systems were analyzed separately for association with metastatic disease in multivariate models. Each model included other significant variables in univariate analyses: catecholamine profile (norepinephrine or dopamine compared to epinephrine) and WHO classification (PGL compared to PCC). While ATRX mutated PPGL showed a positive correlation with metastases in the univariate analysis, information on ATRX mutation status was only available in a subset of patients (467/703) that also lacked complete clinical annotations. As such, ATRX mutation status was not included in the multivariate models. In model 1 (exploring the role of 2-molecular subtype; Table 3, Column B), SDHB mutation (OR 5.68 [95% CI 1.79–18.06]; P=0.003) as well as norepinephrine (OR 3.01 [95% CI 1.02–8.79]; P=0.045) and dopamine (OR 6.39 [95% CI 1.62–25.24]; P=0.008) secretion but not WHO classification were associated with metastatic disease. In model 2 (exploring the role of 3-molecular subtype; Table 3, Column C), dopamine secretion (OR 7.86 [95% CI 2.03–30.4], P=0.003), PGL (OR 3.09 [95% CI 1.20–7.97]; P=0.019) but not the 3-molecular subgroup system were associated with metastatic disease. In model 3 (exploring the role of 4-molecular subtype; Table 3, Column D), norepinephrine (OR 3.12 [95% CI 1.02–9.56] p=0.046), dopamine (OR 6.32 [95% CI 1.58–25.3] P=0.009) but not the 4-molecular classification system nor WHO classification showed association with metastasis. Thus, in the context of clinical characteristics the only relevant molecular biomarker for predicting metastasis was categorization accordingly to SDHB mutation status.
Prognostic information
Median survival time for the entire cohort was 240 months (95% CI 202-not reached). Age at diagnosis (Hazard ratio [HR] 1.04 [95% CI 1.01–1.05]; P=0.019), metastatic stage (HR 6.63 [95% CI 3.46–12.7]; p<0.001), PGL (HR 2.6 [95% CI 1.32–5.15]; P=0.006), SDHB mutation (HR [95% CI 1.32–5.94]; P=0.007), pseudohypoxia TCA-cycle (HR 2.28 [95% CI 1.03–5.08]; p=0.043) and ATRX mutation (HR 9.44 [95% CI 3.29–27.15]; P<0.001) correlated with worse survival in univariate cox regression analyses (Table 4, Supplementary Figure 4). In multivariate model 1 (exploring the role of 2-molecular subtype; Table 4, Column B), age (HR 1.04 [95% CI 1.02–1.06]; P=0.001) and metastases (HR 6.13 [95% CI 2.86–13.13]; P<0.001) but not PGL nor SDHB mutation remained significant for survival. In multivariate model 2 (exploring the role of 4-molecular subtype; Table 4, Column C), age (HR 1.04 [95% CI 1.02–1.06]; P<0.001) and metastases (HR 5.85 [95% CI 2.69–12.71]; P<0.001) but not PGL or categorization accordingly to the 4-molecular subgroup system remained significant for survival.
Table 4:
Survival analysis
| Median Survival (months) (95% CI) |
Cox regression (Univariate analysis) HR (95% CI); p value |
Cox regression (Multivariable analysis) HR (95% CI); p value MODEL 1 |
Cox regression (Multivariable analysis) HR (95% CI); p value MODEL 2 |
||
|---|---|---|---|---|---|
| Gender | Female | 240 (202-nr) | 1 (Ref) | - | - |
| Male | Nr (!) | 1.14 (0.59–2.19); 0.686 | - | - | |
| Age at diagnosis | Continuous variable | NA | 1.02 (1.01–1.05); 0.019 | 1.04 (1.02–1.06); 0.001 | 1.04 (1.02–1.06); <0.001 |
| Tumor size mm | ≤50 mm | 240 (nr-nr) | 1 (Ref) | - | - |
| >50 mm | Nr (!) | 0.82 (0.34–1.99); 0.654 | - | - | |
| Stage | Non-metastatic | 240 (202-nr) | 1 (Ref) | 1 (Ref) | 1 (Ref) |
| Metastatic | 156 (84-nr) | 6.63 (3.46–12.70); <0.001 | 6.13 (2.86–13.13); <0.001 | 5.85 (2.69–12.71); <0.001 | |
| Catecholamine | Epinephrine | 240 (nr-nr) | 1 (Ref) | - | - |
| Norepinephrine | Nr (192-nr) | 1.15 (0.39–3.35); 0.800 | - | - | |
| Dopamine | 168 (156-nr) | 3.06 (0.85–11.04); 0.088 | - | - | |
| WHO 2004 | PCC | 240 (202-nr) | 1 (Ref) | 1 (Ref) | 1 (Ref) |
| PGL | 192 (156-nr) | 2.60 (1.32–5.15); 0.006 | 1.56 (0.64–3.81); 0.332 | 1.47 (0.56–3.87); 0.440 | |
| WHO 2017 | PCC | 240 (202-nr) | 1 (Ref) | * | * |
| Sympathetic PGL | 192 (117-nr) | 2.76 (1.37–5.58); 0.005 | * | * | |
| Head and Neck PGL | Nr (!) | 1.54 (0.21–11.53); 0.672 | * | * | |
| ATRX mutation status | ATRX mutated | 100 (3-nr) | 9.44 (3.29–27.15); <0.001 | * | * |
| ATRX wild type | Nr (!) | 1 (ref) | * | * | |
| 2-molecular subgroups | SDHB wild type | 240 (202-nr) | 1 (Ref) | 1 (Ref) | £ |
| SDHB mutated | 168 (117-nr) | 2.80 (1.32–5.94); 0.007 | 1.45 (0.47–4.44); 0.514 | £ | |
| 3-molecular subgroups | Wnt/unknown | Nr (!) | 1 (Ref) | - | - |
| Pseudohypoxia | 202 (156-nr) | 1.66 (0.79–3.48); 0.180 | - | - | |
| Kinase signaling | 240 (nr-nr) | 0.66 (0.28–1.55); 0.335 | - | - | |
| 4-molecular subgroups | Wnt/unknown | Nr (!) | 1 (Ref) | $ | 1 (Ref) |
| Pseudohypoxia TCA-cycle | 168 (117-nr) | 2.28 (1.03–5.08); 0.043 | $ | 1.43 (0.45–4.55); 0.543 | |
| Pseudohypoxia VHL/EPAS1 | 202 (192-nr) | 0.86 (0.25–2.98); 0.814 | $ | 0.88 (0.24–3.24); 0.851 | |
| Kinase signaling | 240 (nr-nr) | 0.66 (0.28–1.56); 0.341 | $ | 0.79 (0.33–1.95); 0.616 |
95% CI, 95% confidence interval; DA, Dopamine; E, Epinephrine; HNPGL, Head and Neck PGL; HR, Hazard Ratio; NA, Not available; NE, Norepinephrine; Nr, not reached; PCC, Pheochromocytoma; PGL, Paraganglioma; Ref, Reference; sPGLs Sympathetic PGL; TCA, tricarboxylic acid.
could not be calculated due to lack of observations.
Not included, WHO 2004 classification included in the multivariable analysis.
4-molecular-subgroups system not included in Model 1 of the multivariable analysis (2-molecular subgroups included instead).
2-molecualr subgroups system not included in Model 2 of the multivariable analysis (4-molecular subgroups classification included instead).
ATRX mutation status correlated to survival in univariate analysis. But, it was not included in the multivariate model due to a lack of clinical annotations in cases that had analysis of ATRX mutation status.
A subgroup analysis of patients with metastatic disease (n=57) did not show any clinical or molecular factors associated with survival in univariate Cox regression analysis (Supplementary Table 3). Even though there was a trend towads worse overall survival on patients with PPGLs classified as pseudohypoxia TCA-cycle related and Wnt/unknown identified in Kaplan-Meier curves (Supplementary Figure 5), such differences did not reach statistical significance due to limited power and number of events (log-rank test P=0.1620).
Discussion
We performed a meta-analysis on data from a systematic review of 703 PPGL patients published by 21 genome sequencing studies, this is to our knowledge, the largest review in the literature. We focused on identifying predictive factors of metastatic disease, the major determinant of outcome from PPGL disease. While tumor location, biochemical phenotype and the driver gene classifications all showed different frequencies of metastatic disease in the univariate analyses, the only categorization accordingly to a driver gene that remained significant in the multivariate models was SDHB mutation status. In univariate analysis age, tumor location, metastatic disease, SDHB and TCA-cycle related PPGL showed difference in survival. But, no molecular information remained significant for survival in the multivariate model.
The aggregated frequency of driver mutations presented in our review was 62.2%, 24.6% in germline and 32.9% on the somatic level. This number is slightly lower than the frequencies observed in the included TCGA study (27% germline and 39% somatic driver mutations) that used the most comprehensive genetic analysis of all included studies(Fishbein et al. 2017). Major driver genes in the reviewed studies were NF1, VHL, RET, SDHB, and HRAS that were mutated in 45.2% in of PPGL. A second group of driver genes, EPAS1, SDHD, SDHA, MAML3, MAX, and TMEM127 occurred less frequently and had a cumulative frequency of 8.8%. A third group of genes were only found to be mutated in a minority of patients, cumulative frequency 2.8%; CSDE1, FGFR1, MET, SDHC, SDHAF2, FH, and EGLN1. It should be noted that MAML3, CSDE1, FGFR1, and MET were recently discovered in this disease and were therefore only partially included in the sequencing analyses of the reviewed studies.
In order to correlate these findings to patient phenotype, we categorized PPGLs into subgroups accordingly to the biological hallmarks of the tumor as per driver mutation status. A novel category, Wnt/unknown, was created to allow for groups with adequate patient numbers for the statistical analyses. We recognize that Wnt/unknown represent a diverse group of PPGLs that is likely to be dissected as investigators employ more comprehensive methods for genome sequencing in near future. Such improved categorization could include additional data on newly discovered PPGL driver genes, such as EGLN2(Yang, et al. 2015), SLC25A11(Buffet, et al. 2018), MDH2(Cascon, et al. 2015), DNMT3A(Remacha, et al. 2018), H3F3A(Toledo et al. 2015) as well as information on disease modifying genes related to telomere maintenance as well as chromatin modification(Fishbein et al. 2015; Fishbein et al. 2017; Job et al. 2018).
Tumor location, biochemical phenotype and molecular subgroup are three interconnected factors that are all known to be associated with PPGL metastasis (reviewed in(Crona et al. 2017)). Welander et al. reviewed the frequency of metastatic disease in patients with hereditary PPGL: RET, 2.9%; VHL, 3.4%; SDHD, 3.5%; and SDHB, 30.7%(Welander et al. 2011). A systematic review later showed that metastasis occurred in 17% of SDHB and 8% of SDHD carriers(van Hulsteijn, et al. 2012). The findings in our review and meta-analysis corroborate these studies that define SDHB (46.6%), pseudohypoxia (24.3%) and pseudohypoxia TCA-cycle related (40.5%) PPGL as having a relatively high risk of metastatic disease. Different from previous studies, PPGLs related to either VHL/EPAS1 (11.2%) or Wnt/unknown (11.4%) subgroups had an intermediate frequency of metastasis whereas the kinase signaling subgroup was validated as having a relatively low frequency of metastatic disease (4.1%). However, only molecular categorization accordingly to SDHB mutation status, but not other molecular systems or mutations, was associated with metastasis in the multivariate models.
SDHB is a validated negative prognostic factor for survival in metastatic PPGL(Amar et al. 2007; Assadipour et al. 2017; Turkova et al. 2015) but it was previously not established whether SDHB show an independent association in a multivariate model that includes other relevant clinical parameters. Although our survival analysis did not show significant results for the molecular subgroups, Kaplan-Meier curves clearly indicate trend towards worse outcome on both TCA-cycle and Wnt/unknown PPGL. Remarkably, no deaths occurred in patients with pseudohypoxia VHL/EPAS1 as well as kinase signaling PPGLs. This information must be considered with caution since the number of events was very low.
Our review and analysis has a number of limitations: clinical annotations in general and hormone evaluations in particular showed a high risk of bias. Lack of data on ATRX inactivation or TERT expression is also a relevant limitation as it has been associated with higher frequency of metastasis as well as poor survival(Fishbein et al. 2015; Fishbein et al. 2017; Job et al. 2018). Selection bias is also likely as a majority of reviewed manuscripts comes from well recognized groups at tertiary centers. Another bias may have been incorporated from our exclusion of patients without available tumor tissue, which could include a selection bias that exclude a relavant subgroup of patients(Roman-Gonzalez, et al. 2018). The analysis of survival in the whole study cohort is likely skewed by the higher age in patients with sporadic PPGLs, that are less likely to have metastasis, compared to the pseudohypoxia group that develop disease earlier mainly due to genetic predisposition. Disease-related survival would be a preferred measurement, even though it could not be explored due to lack of data. Finally, there was a significant loss of patients for the multivariate analysis due to incomplete clinical annotations, which the subsequent limited statistical power that this implies.
Our findings demonstrated SDHB as independently associated with PPGL metastasis and do not favor the use of information on other driver genes as it was not independently correlated to metastatic disease. Due to relatively low patient number and various risks of bias, we predict that the observed trends for both metastasis and survival still indicate that there is a potential of molecular information to yield relevant information on PPGL outcome in future. To test this hypothesis large, preferably prospective, series with very complete clinical and genetic annotation will be required(Kimura, et al. 2014; Koh, et al. 2017; Turkova et al. 2015).
Conclusion
Our review and individual-patient meta-analysis validated previous phenotype correlations including different frequencies of metastasis in-between PPGL driver genes. However, only SDHB mutation status remained significant in the multivariate model. Instead, the biochemical profile including dopamine secretion emerged as a more useful predictor of metastatic disease. Categorization accordingly to a driver gene mutation was not an independent factor associated with survival in this study.
Supplementary Material
Acknowledgements
This work was supported by grants from Akademiska Sjukhuset, Uppsala, the Paradifference foundation (http://www.paradifference.org) and Lions Cancerforskningsfond, Uppsala and by the National Cancer Institute and the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Angela Lamarca was partially-funded by the ASCO Conquer Cancer Foundation Young Investigator Award.
Footnotes
Disclosures
JC received lecture honoraria from Novartis.
References
- Amar L, Baudin E, Burnichon N, Peyrard S, Silvera S, Bertherat J, Bertagna X, Schlumberger M, Jeunemaitre X, Gimenez-Roqueplo AP, et al. 2007. Succinate dehydrogenase B gene mutations predict survival in patients with malignant pheochromocytomas or paragangliomas. J Clin Endocrinol Metab. 92 3822–3828. Epub 2007 July 3824. [DOI] [PubMed] [Google Scholar]
- Assadipour Y, Sadowski SM, Alimchandani M, Quezado M, Steinberg SM, Nilubol N, Patel D, Prodanov T, Pacak K & Kebebew E 2017. SDHB mutation status and tumor size but not tumor grade are important predictors of clinical outcome in pheochromocytoma and abdominal paraganglioma. Surgery 161 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala-Ramirez M, Feng L, Johnson MM, Ejaz S, Habra MA, Rich T, Busaidy N, Cote GJ, Perrier N, Phan A, et al. 2011. Clinical risk factors for malignancy and overall survival in patients with pheochromocytomas and sympathetic paragangliomas: primary tumor size and primary tumor location as prognostic indicators. J Clin Endocrinol Metab. 96 717–725. Epub 2010 Dec 2029. [DOI] [PubMed] [Google Scholar]
- Bausch B, Schiavi F, Ni Y, Welander J, Patocs A, Ngeow J, Wellner U, Malinoc A, Taschin E, Barbon G, et al. 2017. Clinical Characterization of the Pheochromocytoma and Paraganglioma Susceptibility Genes SDHA, TMEM127, MAX, and SDHAF2 for Gene-Informed Prevention. JAMA Oncol 3 1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffet A, Morin A, Castro-Vega LJ, Habarou F, Lussey-Lepoutre C, Letouze E, Lefebvre H, Guilhem I, Haissaguerre M, Raingeard I, et al. 2018. Germline Mutations in the Mitochondrial 2-Oxoglutarate/Malate Carrier SLC25A11 Gene Confer a Predisposition to Metastatic Paragangliomas. Cancer Res 78 1914–1922. [DOI] [PubMed] [Google Scholar]
- Burnichon N, Buffet A, Parfait B, Letouze E, Laurendeau I, Loriot C, Pasmant E, Abermil N, Valeyrie-Allanore L, Bertherat J, et al. 2012. Somatic NF1 Inactivation is a Frequent Event in Sporadic Pheochromocytoma. Hum Mol Genet 6 6. [DOI] [PubMed] [Google Scholar]
- Burnichon N, Vescovo L, Amar L, Libe R, de Reynies A, Venisse A, Jouanno E, Laurendeau I, Parfait B, Bertherat J, et al. 2011. Integrative genomic analysis reveals somatic mutations in pheochromocytoma and paraganglioma. Hum Mol Genet 20 3974–3985. [DOI] [PubMed] [Google Scholar]
- Cascon A, Comino-Mendez I, Curras-Freixes M, de Cubas AA, Contreras L, Richter S, Peitzsch M, Mancikova V, Inglada-Perez L, Perez-Barrios A, et al. 2015. Whole-Exome Sequencing Identifies MDH2 as a New Familial Paraganglioma Gene. J Natl Cancer Inst 107. [DOI] [PubMed] [Google Scholar]
- Castro-Vega LJ, Buffet A, De Cubas AA, Cascon A, Menara M, Khalifa E, Amar L, Azriel S, Bourdeau I, Chabre O, et al. 2014. Germline mutations in FH confer predisposition to malignant pheochromocytomas and paragangliomas. Hum Mol Genet 23 2440–2446. [DOI] [PubMed] [Google Scholar]
- Castro-Vega LJ, Letouze E, Burnichon N, Buffet A, Disderot PH, Khalifa E, Loriot C, Elarouci N, Morin A, Menara M, et al. 2015. Multi-omics analysis defines core genomic alterations in pheochromocytomas and paragangliomas. Nat Commun 6 6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YY, Kwak MK, Lee SE, Ahn SH, Kim H, Suh S, Kim BJ, Song KH, Koh JM, Kim JH, et al. 2018. A clinical prediction model to estimate the metastatic potential of pheochromocytoma/paraganglioma: ASES score. Surgery 164 511–517. [DOI] [PubMed] [Google Scholar]
- Crona J, Taieb D & Pacak K 2017. New Perspectives on Pheochromocytoma and Paraganglioma: Toward a Molecular Classification. Endocr Rev 38 489–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curras-Freixes M, Inglada-Perez L, Mancikova V, Montero-Conde C, Leton R, Comino-Mendez I, Apellaniz-Ruiz M, Sanchez-Barroso L, Aguirre Sanchez-Covisa M, Alcazar V, et al. 2015. Recommendations for somatic and germline genetic testing of single pheochromocytoma and paraganglioma based on findings from a series of 329 patients. J Med Genet 52 647–656. [DOI] [PubMed] [Google Scholar]
- Dwight T, Flynn A, Amarasinghe K, Benn DE, Lupat R, Li J, Cameron DL, Hogg A, Balachander S, Candiloro ILM, et al. 2018. TERT structural rearrangements in metastatic pheochromocytomas. Endocr Relat Cancer 25 1–9. [DOI] [PubMed] [Google Scholar]
- Eisenhofer G, Lenders JW, Siegert G, Bornstein SR, Friberg P, Milosevic D, Mannelli M, Linehan WM, Adams K, Timmers HJ, et al. 2012. Plasma methoxytyramine: a novel biomarker of metastatic pheochromocytoma and paraganglioma in relation to established risk factors of tumour size, location and SDHB mutation status. Eur J Cancer 48 1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhofer G, Lenders JW, Timmers H, Mannelli M, Grebe SK, Hofbauer LC, Bornstein SR, Tiebel O, Adams K, Bratslavsky G, et al. 2011a. Measurements of plasma methoxytyramine, normetanephrine, and metanephrine as discriminators of different hereditary forms of pheochromocytoma. Clin Chem. 57 411–420. doi: 410.1373/clinchem.2010.153320 Epub 152011 January 153324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhofer G, Pacak K, Huynh TT, Qin N, Bratslavsky G, Linehan WM, Mannelli M, Friberg P, Grebe SK, Timmers HJ, et al. 2011b. Catecholamine metabolomic and secretory phenotypes in phaeochromocytoma. Endocr Relat Cancer 18 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier J, Buffet A & Gimenez-Roqueplo AP 2012. HIF2A mutations in paraganglioma with polycythemia. N Engl J Med. 367 2161; author reply 2161–2162. doi: 2110.1056/NEJMc1211953#SA1211951. [DOI] [PubMed] [Google Scholar]
- Fishbein L, Khare S, Wubbenhorst B, DeSloover D, D’Andrea K, Merrill S, Cho NW, Greenberg RA, Else T, Montone K, et al. 2015. Whole-exome sequencing identifies somatic ATRX mutations in pheochromocytomas and paragangliomas. Nat Commun 6 6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbein L, Leshchiner I, Walter V, Danilova L, Robertson G, Johnson AR, Lichtenberg TM, Murray BA, Ghayee HK, Else T, et al. 2017. Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma. Cancer Cell 31 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliedner SM, Shankavaram U, Marzouca G, Elkahloun A, Jochmanova I, Daerr R, Linehan WM, Timmers H, Tischler AS, Papaspyrou K, et al. 2016. Hypoxia-Inducible Factor 2alpha Mutation-Related Paragangliomas Classify as Discrete Pseudohypoxic Subcluster. Neoplasia 18 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn A, Benn D, Clifton-Bligh R, Robinson B, Trainer AH, James P, Hogg A, Waldeck K, George J, Li J, et al. 2014. The genomic landscape of phaeochromocytoma. J Pathol 236 78–89. [DOI] [PubMed] [Google Scholar]
- Flynn A, Dwight T, Benn D, Deb S, Colebatch AJ, Fox S, Harris J, Duncan EL, Robinson B, Hogg A, et al. 2017. Cousins not twins: intratumoural and intertumoural heterogeneity in syndromic neuroendocrine tumours. J Pathol 242 273–283. [DOI] [PubMed] [Google Scholar]
- Flynn A, Dwight T, Harris J, Benn D, Zhou L, Hogg A, Catchpoole D, James P, Duncan EL, Trainer A, et al. 2016. Pheo-Type: A Diagnostic Gene-expression Assay for the Classification of Pheochromocytoma and Paraganglioma. J Clin Endocrinol Metab 101 1034–1043. [DOI] [PubMed] [Google Scholar]
- Hamidi O, Young WF Jr., Gruber L, Smestad J, Yan Q, Ponce OJ, Prokop L, Murad MH & Bancos I 2017a. Outcomes of patients with metastatic phaeochromocytoma and paraganglioma: A systematic review and meta-analysis. Clin Endocrinol (Oxf) 87 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi O, Young WF Jr., Iniguez-Ariza NM, Kittah NE, Gruber L, Bancos C, Tamhane S & Bancos I 2017b. Malignant Pheochromocytoma and Paraganglioma: 272 Patients Over 55 Years. J Clin Endocrinol Metab 102 3296–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job S, Draskovic I, Burnichon N, Buffet A, Cros J, Lepine C, Venisse A, Robidel E, Verkarre V, Meatchi T, et al. 2018. Telomerase activation and ATRX mutations are independent risk factors for metastatic pheochromocytoma and paraganglioma. Clin Cancer Res. [DOI] [PubMed] [Google Scholar]
- Juhlin CC, Stenman A, Haglund F, Clark VE, Brown TC, Baranoski J, Bilguvar K, Goh G, Welander J, Svahn F, et al. 2015. Whole-exome sequencing defines the mutational landscape of pheochromocytoma and identifies KMT2D as a recurrently mutated gene. Genes Chromosomes Cancer 54 542–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Takayanagi R, Takizawa N, Itagaki E, Katabami T, Kakoi N, Rakugi H, Ikeda Y, Tanabe A, Nigawara T, et al. 2014. Pathological grading for predicting metastasis in phaeochromocytoma and paraganglioma. Endocr Relat Cancer 21 405–414. [DOI] [PubMed] [Google Scholar]
- Koh JM, Ahn SH, Kim H, Kim BJ, Sung TY, Kim YH, Hong SJ, Song DE & Lee SH 2017. Validation of pathological grading systems for predicting metastatic potential in pheochromocytoma and paraganglioma. PLoS One 12 e0187398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, et al. 2013. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell 23 739–752. [DOI] [PubMed] [Google Scholar]
- Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J & Moher D 2009. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med 6 e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann HP, Young WF Jr., Krauss T, Bayley JP, Schiavi F, Opocher G, Boedeker CC, Tirosh A, Castinetti F, Ruf J, et al. 2018. 65 YEARS OF THE DOUBLE HELIX: Genetics informs precision practice in the diagnosis and management of pheochromocytoma. Endocr Relat Cancer 25 T201–t219. [DOI] [PubMed] [Google Scholar]
- Remacha L, Curras-Freixes M, Torres-Ruiz R, Schiavi F, Torres-Perez R, Calsina B, Leton R, Comino-Mendez I, Roldan-Romero JM, Montero-Conde C, et al. 2018. Gain-of-function mutations in DNMT3A in patients with paraganglioma. Genet Med. [DOI] [PubMed] [Google Scholar]
- Roman-Gonzalez A, Zhou S, Ayala-Ramirez M, Shen C, Waguespack SG, Habra MA, Karam JA, Perrier N, Wood CG & Jimenez C 2018. Impact of Surgical Resection of the Primary Tumor on Overall Survival in Patients With Metastatic Pheochromocytoma or Sympathetic Paraganglioma. Ann Surg 268 172–178. [DOI] [PubMed] [Google Scholar]
- Stenman A, Juhlin CC, Haglund F, Brown TC, Clark VE, Svahn F, Bilguvar K, Goh G, Korah R, Lifton RP, et al. 2016a. Absence of KMT2D/MLL2 mutations in abdominal paraganglioma. Clin Endocrinol (Oxf) 84 632–634. [DOI] [PubMed] [Google Scholar]
- Stenman A, Welander J, Gustavsson I, Brunaud L, Backdahl M, Soderkvist P, Gimm O, Juhlin CC & Larsson C 2016b. HRAS mutation prevalence and associated expression patterns in pheochromocytoma. Genes Chromosomes Cancer 55 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers HJ, Brouwers FM, Hermus AR, Sweep FC, Verhofstad AA, Verbeek AL, Pacak K & Lenders JW 2008. Metastases but not cardiovascular mortality reduces life expectancy following surgical resection of apparently benign pheochromocytoma. Endocr Relat Cancer 15 1127–1133. [DOI] [PubMed] [Google Scholar]
- Toledo RA, Qin Y, Cheng ZM, Gao Q, Iwata S, Silva GM, Prasad ML, Ocal IT, Rao S, Aronin N, et al. 2015. Recurrent Mutations of Chromatin-Remodeling Genes and Kinase Receptors in Pheochromocytomas and Paragangliomas. Clin Cancer Res 22 2301–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkova H, Prodanov T, Maly M, Martucci V, Adams K, Widimsky J Jr., Chen CC, Ling A, Kebebew E, Stratakis C, et al. 2015. Characteristics and outcomes of metastatic SDHB and sporadic pheochromocytoma/paraganglioma: An NIH study. Endocr Pract 22 302–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hulsteijn LT, Dekkers OM, Hes FJ, Smit JW & Corssmit EP 2012. Risk of malignant paraganglioma in SDHB-mutation and SDHD-mutation carriers: a systematic review and meta-analysis. J Med Genet 25 25. [DOI] [PubMed] [Google Scholar]
- Welander J, Andreasson A, Brauckhoff M, Backdahl M, Larsson C, Gimm O & Soderkvist P 2014a. Frequent EPAS1/HIF2alpha exons 9 and 12 mutations in non-familial pheochromocytoma. Endocr Relat Cancer 21 495–504. [DOI] [PubMed] [Google Scholar]
- Welander J, Andreasson A, Juhlin CC, Wiseman RW, Backdahl M, Hoog A, Larsson C, Gimm O & Soderkvist P 2014b. Rare germline mutations identified by targeted next-generation sequencing of susceptibility genes in pheochromocytoma and paraganglioma. J Clin Endocrinol Metab 99 E1352–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welander J, Larsson C, Backdahl M, Hareni N, Sivler T, Brauckhoff M, Soderkvist P & Gimm O 2012. Integrative genomics reveals frequent somatic NF1 mutations in sporadic pheochromocytomas. Hum Mol Genet 24 24. [DOI] [PubMed] [Google Scholar]
- Welander J, Lysiak M, Brauckhoff M, Brunaud L, Soderkvist P & Gimm O 2018. Activating FGFR1 Mutations in Sporadic Pheochromocytomas. World Journal of Surgery 42 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welander J, Soderkvist P & Gimm O 2011. Genetics and clinical characteristics of hereditary pheochromocytomas and paragangliomas. Endocr Relat Cancer 18 R253–276. [DOI] [PubMed] [Google Scholar]
- Wilzen A, Rehammar A, Muth A, Nilsson O, Tesan Tomic T, Wangberg B, Kristiansson E & Abel F 2016. Malignant pheochromocytomas/paragangliomas harbor mutations in transport and cell adhesion genes. Int J Cancer 138 2201–2211. [DOI] [PubMed] [Google Scholar]
- Yang C, Zhuang Z, Fliedner SM, Shankavaram U, Sun MG, Bullova P, Zhu R, Elkahloun AG, Kourlas PJ, Merino M, et al. 2015. Germ-line PHD1 and PHD2 mutations detected in patients with pheochromocytoma/paraganglioma-polycythemia. J Mol Med (Berl) 93 93–104. [DOI] [PubMed] [Google Scholar]
- Zelinka T, Musil Z, Duskova J, Burton D, Merino MJ, Milosevic D, Widimsky J Jr. & Pacak K 2011. Metastatic pheochromocytoma: does the size and age matter? Eur J Clin Invest 41 1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.