Abstract
Background
Previous studies have reported associations between ADHD symptoms and DNA methylation in children. We report the first epigenome-wide association study (EWAS) meta-analysis of adult ADHD symptoms, based on peripheral blood DNA methylation (HM450 array) in three population-based adult cohorts.
Methods
An EWAS was performed in the Netherlands Twin Register (NTR, N=2258, mean age 37 years), the Dunedin Study (N=800, age 38 years), and the Environmental Risk Longitudinal Twin Study (E-risk, N=1631, age 18 years), and results were combined through meta-analysis (total sample size= 4689). Second, region-based analyses accounting for the correlation between nearby methylation sites were performed.
Results
One epigenome-wide significant differentially methylated position (DMP) was detected in the Dunedin Study, but meta-analysis did not detect DMPs that were robustly associated across cohorts. In region-based analyses, six significant differentially methylation regions (DMRs) were identified in NTR, 19 in the Dunedin Study and none in E-risk. Of these DMRs, 92% were associated with methylation QTLs, and 68% showed moderate to large blood-brain correlations for DNA methylation levels. DMRs included six non-overlapping DMRs (three in NTR, three in Dunedin) in the Major Histocompatibility Complex (MHC), which were associated with expression of genes in the MHC, including C4A and C4B, previously implicated in schizophrenia.
Conclusions
Our findings point at new candidate loci involved in immune and neuronal functions that await further replication. Our work also illustrates the need for further research to examine to what extent epigenetic associations with psychiatric traits depend on characteristics such as age, comorbidities, exposures, and genetic background.
Keywords: ADHD, meta-analysis, DNA methylation, EWAS, CAARS, epigenetic
Introduction
Attention-deficit/hyperactivity disorder (ADHD) is among the top-ranking psychiatric diagnoses in children and adults(1), and appears to reflect the extreme end of a continuous distribution of ADHD symptoms in the population(2–4). Genetic association studies have identified common and rare variants associated with ADHD(5–7). Numerous environmental risk factors have been reported, including pre- and perinatal factors(8) (birth weight(9), prenatal exposure to maternal smoking(10), toxins(8), maternal fever and infections(11)) and postnatal factors (childhood maltreatment(12), current stress(12), and infections(13)). Recent studies have investigated if DNA methylation levels, which regulate gene expression, are associated with ADHD(14–26). DNA methylation might represent a marker that captures the cumulative effects of genetic variants, stochastic effects, and environmental exposures(27, 28) associated with a trait. Several prenatal exposures, including maternal famine(29), maternal folate(30), and maternal smoking(31), and later life exposures, for example, smoking(32), have been associated with stable long-term changes in DNA methylation in blood and other peripheral tissues. Importantly, DNA methylation patterns are largely tissue-specific. ADHD symptom level-associated DNA methylation differences in peripheral tissues such as blood are likely to reveal epigenetic consequences of differential life conditions that correlate with ADHD (biomarker of exposures). The extent to which DNA methylation in peripheral tissues are informative about epigenetic mechanisms that contribute to inter-individual differences in ADHD symptoms or that correlate with causal epigenetic mechanisms in other tissues is unknown. It has been reported that DNA methylation levels in blood correlate to a limited extent with methylation levels in other tissues, including the brain(33–36). One explanation for such correlations is that methylation Quantitative Trait Locus (mQTL) effects correlate to some extent across tissues, as was recently demonstrated for cis mQTL effects in blood and brain(37).
Several candidate gene studies, most of limited sample size (mean=192, range=82–426), have reported associations between ADHD symptoms in children and DNA methylation in cord blood(38), peripheral blood(19, 21, 39), buccal samples(23), and saliva(21). Other candidate approaches in small samples have reported relationships between DNA methylation, ADHD symptoms, and environmental risk factors(16, 22, 24). To date, three epigenome-wide association studies (EWASs) of ADHD (symptoms) in children have been published(20, 25, 40). The first study measured DNA methylation in saliva in a small group of boys with ADHD and controls (age 7–12, sample size=112), and reported genes with suggestive evidence for association(20). The second study measured DNA methylation in 384 cord blood samples and found no significant differences between children later diagnosed with ADHD and controls(41). The largest EWAS (817 children from a UK population-based study) identified 13 loci, where DNA methylation level in cord blood was significantly predictive of ADHD symptom trajectories between age 7 and 15 years(40); among those was ST3GAL3; one of the significant loci from the Genome-wide Association Study (GWAS) of ADHD(7). Since DNA methylation and ADHD symptoms both change with age, it is unknown if findings in children persist in adulthood. Moreover, the robustness of associations between ADHD symptoms and DNA methylation remains to be investigated in larger studies of multiple cohorts.
Here, we report the first EWAS meta-analysis of ADHD symptoms in adults; this is also the largest EWAS of ADHD symptoms to date. We assessed the association between whole blood DNA methylation and ADHD symptoms in three population-based cohorts: the Netherlands Twin Register (NTR)(42), the Dunedin Multidisciplinary Health and Development Study from New Zealand, and the Environmental risk (E-risk) twin study from the UK. In secondary analyses, we 1) tested if CpGs with a lower, albeit non-significant, p-value showed enrichment for loci previously identified in GWASs of psychiatric disease or in EWASs of psychiatric phenotypes or exposures, 2) analysed differentially methylated regions (DMRs), to examine the evidence for small methylation differences at multiple nearby CpGs, and 3) examined the relationship between DNA methylation and RNA transcript levels in blood, effects of mQTLs, and the correlation between DNA methylation in blood and brain, to facilitate the biological interpretation of findings.
Methods and Materials
Overview
The EWAS was performed in three cohorts (Figure 1): NTR(42) (N=2232 individuals from twin families), the Dunedin study(43) (N=800 unrelated individuals) and E-risk(44) (N=1631 twins). Results were combined in a meta-analysis to identify DMPs and to examine heterogeneity of effects across cohorts. Next, we performed secondary analyses. Genome-wide meta-analysis test statistics were used to test for enrichment of ADHD EWAS signals in loci detected in previous GWA and EWA studies of relevant traits. We performed DMR analyses in each cohort, compared results across cohorts, and performed a meta-analysis-based DMR analysis. Functional follow-up analyses were performed on top-DMPs from the meta-analysis (nominal p-value<1.0×10−5) and on significant DMRs. Further details about the analyses are provided in Appendix 1.
Figure 1.
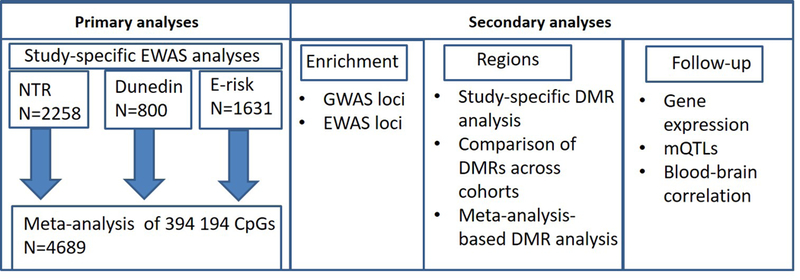
Summary of analyses of epigenome-wide association study (EWAS) in three cohorts: Netherlands Twin Register (NTR), Dunedin Multidisciplinary Health and Development Study from New Zealand, and Environmental Risk (E-Risk) Longitudinal Twin Study. DMR, differentially methylated region; GWAS, genome-wide association study; mQTLs, methylation quantitative trait loci.
Ethical permission
Ethical permission is described in Appendix 1. Written informed consent was obtained from all participants.
ADHD symptoms
The EWAS in NTR was performed on the Conners Adult ADHD Rating Scale (CAARS) index (total ADHD symptoms; Figure S1a)(45). Sensitivity analyses were performed on DSM-IV-based CAARS inattention and hyperactivity subscales, available for 1846 samples. In the Dunedin Study, DSM-5 ADHD symptoms (Figure S1b) were assessed based on private structured interviews as described previously(46) at age 38 years. In the E-Risk Study, DSM-5 ADHD symptoms (Figure S1c) were assessed based on private structured interviews as described previously(47) at age 18 years.
Peripheral blood DNA methylation
DNA methylation was assessed with the Infinium HumanMethylation450 BeadChip Kit (HM450k; Illumina, San Diego, CA, USA). Normalization was performed with functional normalization in NTR(48), with methylumi in the Dunedin Study, and with dasen in E-Risk(49). The following probes were removed from all cohorts: sex chromosomes, probes with a single nucleotide polymorphism (SNP) within the CpG site (at the C or G position), irrespective of minor allele frequency (MAF), based on the genome of the Netherlands (GONL) reference population(50), probes with common (>5% MAF) SNPs within 10 base pairs (bp) of the single base extension site(51), and ambiguous mapping probes with an overlap of at least 47 bases per probe(52). Only methylation sites that were present in all cohorts were kept in the analysis, leaving 394194 sites.
Statistical analyses
Epigenome-wide association study (EWAS)
The association between DNA methylation levels and ADHD symptoms was tested for each site under a linear model (Dunedin) or generalized estimation equation (gee) model accounting for relatedness of twins and other family members (NTR and E-risk); methylation β-value were assessed as outcome and the following predictors were used: ADHD symptoms, sex, smoking status, white blood cell (WBC) percentages, age at blood sampling (only in NTR), cohort-specific technical covariates (i.e. sample plate and array row or Principal Components (PCs) based on control probes), and Principal Components (PCs) based on genome-wide SNPs. The R-package bacon was used to compute the Bayesian inflation factor(53).
Meta-analysis
A p-value-based fixed-effects sample size-weighted meta-analysis was performed in METAL. The sample size–weighted method was chosen because of the differences in measurement scales of ADHD symptoms across studies. Statistical significance was assessed considering Bonferroni correction for the number of methylation sites tested (alpha=0.05/394194=1.3×10−7). False discovery rate (FDR) q-values are presented in supplementary tables. The I2 statistic provided by METAL was evaluated to assess heterogeneity.
Inattention and hyperactivity
For top-DMPs, sensitivity analyses were performed in which the association of these DMPs with inattention and hyperactivity/impulsivity subscales was tested in NTR.
Overlap with EWAS and GWAS loci
Enrichment analysis was performed to examine whether CpGs in or near loci detected by GWASs, or top-ranking CpGs from previous EWA studies, on average, showed a stronger association with ADHD symptoms than other genome-wide HM450 methylation sites. We considered the most recent GWASs for ADHD(7), major depressive disorder (MDD)(54), schizophrenia(55), and autism spectrum disorders (ASD)(56), and the largest available EWA studies on ADHD symptoms in children(40), schizophrenia(57), smoking(32), and maternal smoking(31).
Differentially methylated regions
We used the python module ‘Comb-p’(58) to identify regions where multiple correlated methylation sites show evidence for association with ADHD symptoms. We report significant regions (Šidák p<0.05) with at least two methylation sites within a 500bp window. Comb-p was applied in each of the 3 cohorts, separately, and on the meta-analysis results.
Functional follow-up analyses
In follow-up analyses, previously published datasets were used to test if DNA methylation level at top-DMPs and at CpGs within significant DMRs were associated with whole blood gene expression level in cis(59), whole blood mQTLs(59), and to examine the correlation between DNA methylation in blood and four brain regions (prefrontal cortex, entorhinal cortex, superior temporal gyrus and cerebellum)(35).
Power analysis
Power analyses are described in Appendix 1.
Results
Meta-analysis
Demographic information of the cohorts is provided in Table 1. Genome-wide EWAS test statistics from each cohort showed no inflation (Table S1, Figure S2). One significant DMP (alpha=1.3×10−7) was detected in Dunedin (cg26197679, chromosome 8 intergenic), and none in NTR and E-risk. Meta-analysis of the three cohorts (N=4689) detected no significant DMPs, and meta-analysis test statistics showed no inflation (Table S1, Figure S2). Summary statistics for genome-wide methylation sites are provided in Table S2. Top-DMPs with a nominal p-value (p<1.0×10−5) are presented in Table 2. Two DMPs showed a negative relationship with ADHD symptoms (cg26197679, intergenic and cg23144852, gene body OPA1), and one DMP showed a positive relationship with ADHD symptoms in all cohorts (cg10984962, gene body AGAP1). The effect sizes for these DMPs in the individual cohorts are presented in Table S3. In NTR, an increase of one standard deviation in CAARS ADHD symptoms (3.9 points) was associated with a methylation change of −0.04%(cg26197679), −0.16%(cg23144852), and 0.10%( cg10984962), respectively.
Table 1.
Cohort characteristics
| NTR (N=2258*) | Dunedin Study (N=800) | E-risk study (N=1631) | |||
|---|---|---|---|---|---|
| Characteristic | Mean (SD) or N (%) |
Characteristic | Mean (SD) or N (%) |
Characteristic | Mean (SD) or N (%) |
| CAARS ADHD index | 7.9 (3.9) | DSM-5 Adult ADHD Symptoms | 1.5 (2.3) | DSM-5 Adult ADHD Symptoms | 5.8 (4.3) |
| Age at blood sampling (years) | 37.3 (12.9) | Age at blood sampling (years) | 38 (0) | Age at blood sampling (years) | 18 (0) |
| Sex: female | 1549 (69.0%) | Sex: female | 392 (49.0%) | Sex: female | 817 (50.1%) |
| Current smoker | 438 (19.4%) | Current smoker | 162 (20.3%) | Current smoker** | 372 (22.8%) |
| Former smoker | 532 (23.6%) | Former smoker | 233 (29.1%) | ||
| Never smoked | 1288 (57.0%) | Never smoked | 405 (50.6%) | Non-smoker ** | 1259 (77.2%) |
In NTR, 2258 samples from 2232 individuals were included (for 26 individuals, two longitudinal DNA samples were included).
In E-risk, the smoking variable that was included as covariate in the analyses and that is presented in this table is current daily smoking at age 18.
Table 2.
Top DMPs from the EWAS meta-analysis.
| cgID | CHR | Position* | Gene | Location | Nearest Gene | Weight | Z score | P | Direction |
|---|---|---|---|---|---|---|---|---|---|
| cg26197679 | 8 | 142310085 | Intergenic | LINC01300 | 4689 | −4.9 | 1.1 × 10−6 | --- | |
| cg23144852 | 3 | 193405999 | OPA1 | Gene body | OPA1 | 4684 | −4.8 | 1.7 × 10−6 | --- |
| cg10984962 | 2 | 236462202 | AGAP1 | Gene body | AGAP1 | 4688 | 4.5 | 9.7 × 10−6 | +++ |
note: CpGs with a p-value lower than 1.0 × 10−5 are shown. CHR=chromosome. P=p-value. Weight=Total sample size in the meta-analysis. Z score=Meta-analysis Z score.
Genome build Hg19 (build 37).
Heterogeneity of top-DMPs
One top-DMP (cg26197679) displayed large between-study heterogeneity (I2=93.6%, heterogeneity p=1.6×10−7). This DMP showed the strongest association with ADHD symptoms and was epigenome-wide significant in the Dunedin study, where an increase of one standard deviation in DSM-5 ADHD symptoms was associated with a methylation change of −0.62%. Although the effect size was weaker in the other cohorts, the direction of association was the same in all cohorts (Table 2). Inspection of all top-ranking sites from the individual cohorts (p<1.0×10−5: 14 sites) revealed that all sites were characterized by substantial between-study heterogeneity (mean I2=87.2%, range=71.3%−93.8%, Table S4). Nine sites (64.3%) showed the same direction of effect across all cohorts.
Inattention and hyperactivity
In sensitivity analyses, the association with inattention and hyperactivity/impulsivity subscales, separately, was tested in NTR for the three top-DMPs from the meta-analysis. This analysis showed that the direction of effect and the strength of the association with each of the subscales was highly similar (Table S5).
Enrichment of EWAS and GWAS loci
In testing for overlap of our EWAS meta-analysis results with findings from previous EWASs and GWASs, we failed to observe enrichment of CpGs previously associated with longitudinal ADHD trajectories in children(40) or schizophrenia in adults(57), CpGs associated with individual(32) or maternal smoking(31), and CpGs near GWAS loci for ADHD, ASD, MDD, or schizophrenia (Table S6). Methylation sites previously associated with schizophrenia(57) showed a small but significant depletion of signal for ADHD symptoms.
Differentially methylated regions
In NTR, six significant DMRs were identified, which spanned from 164 bp to 848 bp, and included three to 32 CpGs (Table 3 and Table S7). One example is illustrated in Figure S3. In the Dunedin study, 19 significant DMRs were identified, spanning from two to 51 CpGs within regions of 18 bp to 1818 bp (Table 3 and Table S7). In E-risk, no significant DMRs were identified. Six distinct DMRs were detected in the Major Histocompatibility Complex (MHC) region (chromosome 6): three in NTR, and three in the Dunedin Study. In line with the heterogeneity of DMP results across cohorts, none of the DMRs detected in NTR and Dunedin overlapped, and DMR analysis on the meta-analysis of the three cohorts did not detect significant DMRs.
Table 3.
Significant DMRs associated with ADHD symptoms
| chr | start | end | N probes | region size (bp) |
Region Šidák p- value |
gene (s) | Gene expression associated with CpGs |
|---|---|---|---|---|---|---|---|
| Netherlands Twin Register (NTR) | |||||||
| 6 | 31650735 | 31651363 | 19 | 628 | 4.3×10−10 | LY6G5C, DDAH2, AIF1, C6orf48, LST1, VWA7, HSPA1L, PRRC2A, MICB | |
| 11 | 67417958 | 67418406 | 11 | 448 | 7.6×10−9 | ACY3 | ACY3 |
| 6 | 31583915 | 31584224 | 4 | 309 | 1.2×10−6 | AIF1 | AIF1, LY6G5C, C6orf48, PRRC2A, LST1, LY6G5C, VWA7, BAG6, HSPA1L |
| 6 | 33245460 | 33246308 | 32 | 848 | 3.0×10−5 | B3GALT4 | B3GALT4, RPS18 |
| 13 | 113613574 | 113613738 | 3 | 164 | 4.8×10−5 | - | |
| 3 | 48632484 | 48633145 | 11 | 661 | 2.1×10−4 | COL7A1 | COL7A1 |
| Dunedin Study | |||||||
| 6 | 32063394 | 32065212 | 51 | 1818 | 2.3 ×10−17 | TNXB | SKIV2L, C4B, C4A, TNXB, TNXA |
| 5 | 191127 | 192104 | 12 | 977 | 4.4 ×10−11 | LRRC14B | - |
| 19 | 17931932 | 17932370 | 8 | 438 | 2.3 ×10−8 | INSL3 | - |
| 20 | 748992 | 749621 | 8 | 629 | 4.9 ×10−8 | C20orf54 | - |
| 6 | 32847441 | 32847846 | 16 | 405 | 1.3 ×10−6 | PPP1R2P1 | HLA-DQA |
| 15 | 75019070 | 75019377 | 10 | 307 | 4.8 ×10−6 | CYP1A1 | - |
| 4 | 56458183 | 56458623 | 7 | 440 | 5.1 ×10−6 | PDCL2 | - |
| 11 | 1769152 | 1769646 | 9 | 494 | 6.8 ×10−6 |
HCCA2; LOC402778; LOC402778 |
- |
| 4 | 186732837 | 186733332 | 8 | 495 | 8.9 ×10−6 | SORBS2 | - |
| 15 | 90727560 | 90728141 | 9 | 581 | 5.5 ×10−5 | SEMA4B | SEMA4B |
| 7 | 1250038 | 1250757 | 10 | 719 | 6.4 ×10−5 | - | |
| 7 | 2728344 | 2728913 | 4 | 569 | 8.8 ×10−5 | AMZ1 | - |
| 8 | 41655673 | 41656080 | 6 | 407 | 1.5×10−4 | ANK1 | - |
| 12 | 25801455 | 25801622 | 4 | 167 | 3.5×10−4 | IFLTD1 | - |
| 6 | 3247958 | 3248099 | 2 | 141 | 2.3×10−3 | - | |
| 14 | 96670628 | 96671170 | 11 | 542 | 3.9×10−3 | BDKRB2 | - |
| 20 | 22795685 | 22795775 | 2 | 90 | 8.7×10−3 | - | |
| 4 | 187985377 | 187985473 | 2 | 96 | 0.02 | - | |
| 8 | 1993528 | 1993546 | 2 | 18 | 0.03 | MYOM2 | - |
chr=chromosome, N probes= total number of methylation sites in the window, bp= base pairs, gene(s)= gene(s) mapping to CpGs in the window. More detailed information on the regions is provided in Table S7. More detailed results for the association between DNA methylation levels and gene expression in cis is provided in Table S8 (NTR) and Table S9 (Dunedin Longitudinal Study). No significant DMRs were found in E-risk.
Significant DMRs did not overlap with CpGs from the previous EWAS of ADHD trajectories in children(17) or with schizophrenia in adults(57). One of the six DMRs identified in NTR (chromosome 6: 33245460–3324630) contained CpGs previously associated with smoking (12 of the 32 CpGs) and maternal smoking (3 CpGs). Five of the 19 DMRs identified in the Dunedin Study contained CpGs associated with smoking or maternal smoking (Table S7).
Several DMRs were located in proximity of SNPs associated with schizophrenia(55): of the DMRs in NTR, two were located within 100 kb and one was located within 1 Mb of schizophrenia-associated SNPs, respectively (all in the MHC region). Five DMRs in the Dunedin study were located within 1 Mb of schizophrenia-associated SNPs (two on chromosome 6, two on chromosome 7, and one on chromosome 15). None of the DMRs was located within 1 Mb of significant GWAS loci for ADHD, MDD, or ASD.
Gene expression in cis
To examine potential functional consequences top-DMPs and DMRs, we used previously published data on whole blood DNA methylation and RNA-sequencing (n=2,101 samples). While DNA methylation levels at the ADHD symptom level top-DMPs were not associated with RNA levels of genes in cis, methylation level at CpGs within five of the six significant DMRs detected in NTR were associated with expression levels of 14 genes(Table 3 and Table S8): At one DMR, higher methylation level correlated with lower expression, at another DMR, higher methylation level correlated with higher expression, and at three DMRs, expression of some genes correlated positively and others negatively with methylation level. Of the 19 DMRs identified in the Dunedin Longitudinal Study, the methylation levels at CpGs within three DMRs were associated expression levels of seven genes(Table 3 and Table S9): at two DMRs, a higher methylation level correlated with lower expression, and at one DMR, a higher methylation level correlated with higher expression.
mQTLs
To gain insight into genetic causes of variation underlying top-DMPs and DMRs, we obtained whole blood mQTL data (n=3841 samples) (59). One of the three top-DMPs from the ADHD symptom meta-analysis was associated with six independent SNPs (mQTLs) in cis(Table S10). The majority of DMRs (92.0%; 23) was associated with mQTLs. For ADHD-associated DMRs in NTR, on average 68% of the CpGs within a DMR (range=36.4–92.3%) was associated with at least one mQTL SNP. In total, 164 mQTL associations were identified for NTR DMRs (76.8% were cis mQTLs and 23.2% were trans mQTLs; Table S11), involving 59 CpGs and 55 SNPs. For ADHD-associated DMRs in Dunedin, 323 mQTL associations were identified, involving 126 CpGs and 154 SNPs (88% cis and 13% trans; Table S12). On average, 64.9% (range=0–100%) of CpGs within DMRs identified in the Dunedin study was associated with one or more mQTLs. We highlight one example; a DMR on chromosome 11, detected in NTR and associated with the expression of ACY3, was associated with cis and trans mQTLs, and the correlation structure of DNA methylation within this DMR(Figure S2) mirrored the sharing across CpGs of trans-mQTLs on chromosome 6(Figure S3). Comparing the overlap of mQTLs across cohorts revealed that two DMRs were associated with a common set of trans mQTLs. The DMR on chromosome 11 in NTR (associated with expression of ACY3), and the DMR on chromosome 15 in Dunedin (associated with expression of SEMA4B) were both associated in trans with SNPs on chromosome 6:109626965–109616420 (rs9374080, rs1008084, and rs9386791).
Correlation between DNA methylation level in blood and brain
While DNA methylation levels at the ADHD symptom top-DMPs did not correlate significantly between blood and brain, CpGs within 4 of the 6 DMRs (66.7%) detected in NTR (Table S13), and 13 of the 19 DMRs (68.4%) detected in the Dunedin Study (Table S14) showed significantly correlated DNA methylation levels between blood and one or multiple brain regions. For all CpGs, except for one, the correlation was positive (mean r=0.50, range=−0.42–0.70). The number of CpGs per DMR that showed correlated methylation levels between blood and brain ranged from 1 to 31 (mean=5.5). The DMR with the largest number of CpGs with significant blood-brain correlations (31 CpGs), was a DMR in the MHC region, detected in the Dunedin study and associated with the expression level of multiple genes, including C4A and C4B. An exemplary plot of methylation levels in blood and brain for one CpG in this region (cg01337207) is provided in Figure S5.
Discussion
We performed an EWAS of ADHD symptoms in three population-based adult cohorts. Our hypothesis was that DNA methylation in blood may provide insight into epigenetic consequences of life conditions that correlate with ADHD symptoms, and potentially into epigenetic mechanisms that contribute to ADHD symptoms or that correlate with causal epigenetic mechanisms in the brain. In the Dunedin Study, we identified one significant DMP, where a higher methylation level correlated with fewer ADHD symptoms (cg26197679, chr8 intergenic). This CpG, as well as other top-ranking CpGs from individual cohorts showed considerable heterogeneity, and no significant DMPs were detected in a meta-analysis of the three cohorts. In secondary, region-based analyses, we tested if ADHD symptoms were associated with methylation differences at multiple nearby CpGs that individually failed to reach epigenome-wide significance. We identified six significant DMRs in NTR, 19 in the Dunedin study, and none in E-risk. In line with the heterogeneity of DMP results, none of the DMRs overlapped across cohorts, and the meta-analysis-based DMR analysis did not detect significant DMRs. With respect to effects of differential exposures, although the EWAS signal showed no significant enrichment for CpGs previously associated with smoking, some DMRs contained CpGs previously associated with smoking(32) or maternal smoking(31), even after we adjusted for smoking. There are several possible explanations for this finding(60): 1) residual confounding effects of smoking, 2) second-hand smoking exposure, including maternal prenatal smoking, 3) exposures other than smoking; for instance, ADHD is associated with general substance use(61), and 4) these CpGs are connected to a shared underlying biology of ADHD symptoms and smoking. Importantly, the current study shows that effects of mQTLs and correlated methylation levels between blood and brain exceed effects of smoking: 92% of DMRs was associated with genetic variants, 68% of DMRs showed correlated methylation levels between blood and brain regions, and 24% of DMRs were associated with smoking. These observations suggest that inter-individual differences in DNA methylation at these DMRs are not merely driven by lifestyle differences associated with ADHD symptoms such as smoking, and that some of the methylation differences in whole blood associated with ADHD symptoms might be a marker for methylation variation in the brain.
Some top-DMPs and top-DMRs mapped to genes that have been previously linked to psychiatric disorders or implicated in brain biology. These are potential candidates for being involved in the underlying biology of ADHD symptoms, provided that these loci also show symptom-associated differences in epigenetic regulation in the brain. For instance, cg10984962; the third-ranking CpG from the meta-analysis, is located in AGAP1, which encodes a protein involved in endosomal trafficking. In neuronal cells, it plays a role in the recycling of muscarinic acetylcholine receptors(62) and was shown to influence dendritic spine morphology(63). Yet, methylation levels in blood did not correlate with methylation levels in the brain at this CpG. Six significant DMRs in distinct sub-regions of the MHC were identified in NTR and the Dunedin Study. The top-DMR in MHC in the Dunedin Study was associated with expression levels of SKIV2L, C4B, C4A, TNXB, and TNXA. CpGs in this DMR were not associated with smoking(32). The C4 genes are of great interest, as they have been previously implicated in functional effects of schizophrenia-associated SNPs in the MHC on postnatal synaptic pruning(64). At many CpGs in this DMR, DNA methylation level in blood showed moderate to strong correlations with DNA methylation levels in multiple brain regions. Yet, since the DMRs did not replicate across cohorts, it remains to be established whether they are relevant to ADHD symptom levels.
The lack of overlap of our findings with CpGs from a previous study, at which methylation level in cord blood significantly predicted ADHD trajectories in childhood(40), could indicate that epigenetic associations relevant to ADHD symptoms are age-specific (which has already been described for genetic contributions to ADHD symptoms(65)). The loci detected in cord blood also did not show association with ADHD symptoms when methylation was assessed at age seven in the previous study(40). Age-specific epigenetic associations may also potentially explain a lack of overlap with GWAS loci. While our EWAS only included adults, the GWAS of ADHD contained mainly children(7).
This is the first EWAS of ADHD symptoms in adults, the largest epigenetic study of ADHD symptoms to date, and the first study of ADHD symptoms to apply a multi-cohort approach. This study also has limitations. Although all cohorts included adults and applied continuous measures of ADHD symptoms reported by the same informant (self-report), there were also differences between cohorts that may have reduced power. First, the CAARS was used in NTR, which is based on DSM-IV, while DSM-5 symptoms were assessed in the E-risk and Dunedin Study. Second, ADHD symptoms were assessed by a self-report scale in NTR and by structured interviews in the Dunedin Study and E-risk. Third, DNA methylation measurements, processing of the data, and quality control were performed separately in each cohort, based on QC and normalization pipelines that were optimized for each cohort. Fourth, cohorts varied in age and reported number of ADHD symptoms. NTR included a broad age range (mean age=37), while participants from the E-risk study were young adults (age=18), and the age of participants from the Dunedin Study was 38. The prevalence of ADHD in these cohorts, based on the instruments used in the current study, has been previously reported: 7% in NTR(66), 3% in the Dunedin Study(46), and 8% in E-risk (47). Fifth, the cohorts were from different countries, and it is possible that epigenetic differences exist between cohorts due to differences in genetic background or in the presence and frequency of environmental exposures.
We performed a power analysis for the three meta-analysis top-DMPs, based on their effect size observed in NTR (Figure S6). The effect sizes of these three DMPs ranged from 0.09%- 0.92% explained variance. For the top-DMP that showed the least heterogeneity (cg10984962; AGAP1; variance explained= 0.28% in NTR), the required sample size to achieve 80% power is 13508. Similar to our population-based EWAS of ADHD symptoms, a recent blood-based EWAS meta-analysis of cognitive abilities identified only a few significant sites for most cognitive measures in >6000 individuals(67).
Several limitations apply to epigenetic epidemiology studies including ours. Firstly, the Illumina 450k array captures only ~1.7% of all CpGs in the genome, and the removal of methylation probes that overlap with genetic polymorphisms, albeit inevitable because these polymorphisms compromise the quality of microarray-based measurements of DNA methylation, may limit the ability to detect DNA methylation-mediated SNP effects on the phenotype. Secondly, blood is unlikely to provide a complete picture of ADHD-related epigenetic processes, because epigenetic marks are largely tissue-specific. Yet, methylation changes in blood have already been associated with ADHD symptoms(40), other psychiatric conditions(57), cognition(67), and hippocampal volume(68) in earlier studies. Furthermore, we found that several of the DMRs detected in blood contain CpGs with correlated methylation levels across blood and brain. Finally, a general constraint in epigenetic studies is that trait-associated variation in DNA methylation may arise secondary to trait development (reverse causality) or may be a marker of trait-associated environmental exposures (such as lifestyle, medication use, infections, and early life environmental factors, such as nutrition, and maternal smoking). These associations nevertheless may provide valuable insight into the underlying biological changes associated with ADHD and its risk factors. The (direction of) causality may be addressed in human studies, for example, with longitudinal study designs or Mendelian Randomization(69).
In conclusion, by performing an EWAS of ADHD symptoms in three population-based adult cohorts, we found no significant sites in an EWAS meta-analysis of 4689 individuals and observed considerable heterogeneity of effects across cohorts. We found several significant cohort-specific DMPs and DMRs, with the MHC region emerging in two cohorts. The significance of these findings is unknown, but they may point at new candidate pathways awaiting further replication. We conclude that larger studies are necessary to identify methylation sites in whole blood that are robustly associated with population-based ADHD symptoms in adults. Our findings also illustrate the need for further research to examine to what extent epigenetic associations with psychiatric traits depend on characteristics such as age, sex, lifetime exposures, genetic background, symptom severity, and comorbidities.
Supplementary Material
Acknowledgements
Netherlands Twin Register: We would like to thank the twins and their family members for their participation. This study was funded by: BBRMI-NL (NWO 184.021.007), NWO 480–15-001/674: Netherlands Twin Registry Repository: researching the interplay between genome and environment, and ACTION. ACTION receives funding from the European Union Seventh Framework Program (FP7/2007–2013) under grant agreement no 602768. Prof. Boomsma gratefully acknowledges the Royal Netherlands Academy of Science Professor Award (PAH/6635). Prof. Franke receives support from a personal grant from the Netherlands Organization for Scientific Research (NWO; Vici grant 016–130-669), from a grant for the Dutch National Science Agenda for the NWANeurolabNL project (grant 400 17 602), from the European Community’s FP7 under grant agreement no 602805 (Aggressotype), and from the European Community’s Horizon 2020 Program (H2020/2014 – 2020) under grant agreement no 728018 (Eat2beNICE).
Dunedin Study: We thank the Dunedin Study members, Unit research staff, and Study founder Phil Silva. The Dunedin Multidisciplinary Health and Development Research Unit is supported by the New Zealand Health Research Council and New Zealand Ministry of Business, Innovation and Employment (MBIE). This research received support from the US-National Institute of Aging (grant number R01AG032282) and the UK Medical Research Council (grant number MR/P005918). Additional support was provided by the Jacobs Foundation. This work used a high-performance computing facility partially supported by grant 2016-IDG-1013 (“HARDAC+: Reproducible HPC for Next-generation Genomics”) from the North Carolina Biotechnology Center.
E-risk Study: The authors are grateful to the study families their participation, and to members of the E-Risk team for their dedication, hard work, and insights. The E-Risk Study is funded by the Medical Research Council (G1002190) and the National Institute of Child Health and Human Development (HD077482). Additional support was provided by a Distinguished Investigator Award from the American Asthma Foundation to Dr. Mill, the Jacobs Foundation and the Avielle Foundation. Dr. Arseneault is the Mental Health Leadership Fellow for the UK ESRC. Dr. Agnew-Blais is an MRC Skills Development Fellow. This work used a high-performance computing facility partially supported by grant 2016-IDG-1013 (“HARDAC+: Reproducible HPC for Next-generation Genomics”) from the North Carolina Biotechnology Center.
Appendix. BIOS Consortium (Biobank-based Integrative Omics Study)
Management Team Bastiaan T. Heijmans (chair)1, Peter A.C. ’t Hoen2, Joyce van Meurs3, Aaron Isaacs4, Rick Jansen5, Lude Franke6.
Cohort collection Dorret I. Boomsma7, René Pool7, Jenny van Dongen7, Jouke J. Hottenga7 (Netherlands Twin Register); Marleen MJ van Greevenbroek8, Coen D.A. Stehouwer8, Carla J.H. van der Kallen8, Casper G. Schalkwijk8 (Cohort study on Diabetes and Atherosclerosis Maastricht); Cisca Wijmenga6, Lude Franke6, Sasha Zhernakova6, Ettje F. Tigchelaar6 (LifeLines Deep); P. Eline Slagboom1, Marian Beekman1, Joris Deelen1, Diana van Heemst9 (Leiden Longevity Study); Jan H. Veldink10, Leonard H. van den Berg10 (Prospective ALS Study Netherlands); Cornelia M. van Duijn4, Bert A. Hofman11, Aaron Isaacs4, André G. Uitterlinden3 (Rotterdam Study).
Data Generation Joyce van Meurs (Chair)3, P. Mila Jhamai3, Michael Verbiest3, H. Eka D. Suchiman1, Marijn Verkerk3, Ruud van der Breggen1, Jeroen van Rooij3, Nico Lakenberg1.
Data management and computational infrastructure Hailiang Mei (Chair)12, Maarten van Iterson1, Michiel van Galen2, Jan Bot13, Dasha V. Zhernakova6, Rick Jansen5, Peter van ’t Hof12, Patrick Deelen6, Irene Nooren13, Peter A.C. ’t Hoen2, Bastiaan T. Heijmans1, Matthijs Moed1.
Data Analysis Group Lude Franke (Co-Chair)6, Martijn Vermaat2, Dasha V. Zhernakova6, René Luijk1, Marc Jan Bonder6, Maarten van Iterson1, Patrick Deelen6, Freerk van Dijk14, Michiel van Galen2, Wibowo Arindrarto12, Szymon M. Kielbasa15, Morris A. Swertz14, Erik. W van Zwet15, Rick Jansen5, Peter-Bram ’t Hoen (Co-Chair)2, Bastiaan T. Heijmans (Co-Chair)1.
1. Molecular Epidemiology Section, Department of Medical Statistics and Bioinformatics, Leiden University Medical Center, Leiden, The Netherlands
2. Department of Human Genetics, Leiden University Medical Center, Leiden, The Netherlands
3. Department of Internal Medicine, ErasmusMC, Rotterdam, The Netherlands
4. Department of Genetic Epidemiology, ErasmusMC, Rotterdam, The Netherlands
5. Department of Psychiatry, VU University Medical Center, Neuroscience Campus Amsterdam, Amsterdam, The Netherlands
6. Department of Genetics, University of Groningen, University Medical Centre Groningen, Groningen, The Netherlands
7. Department of Biological Psychology, VU University Amsterdam, Neuroscience Campus Amsterdam, Amsterdam, The Netherlands
8. Department of Internal Medicine and School for Cardiovascular Diseases (CARIM), Maastricht University Medical Center, Maastricht, The Netherlands
9. Department of Gerontology and Geriatrics, Leiden University Medical Center, Leiden, The Netherlands
10. Department of Neurology, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, The Netherlands
11. Department of Epidemiology, ErasmusMC, Rotterdam, The Netherlands
12. Sequence Analysis Support Core, Leiden University Medical Center, Leiden, The Netherlands
13. SURFsara, Amsterdam, the Netherlands
14. Genomics Coordination Center, University Medical Center Groningen, University of Groningen, Groningen, the Netherlands
15. Medical Statistics Section, Department of Medical Statistics and Bioinformatics, Leiden University Medical Center, Leiden, The Netherlands
Footnotes
Disclosures
Dr. Franke has received educational speaking fees from Shire and Medice. All other authors declare that they have no conflict of interest.
Data availability
The HumanMethylation450 BeadChip and RNA-seq data from the BIOS consortium are available in the European Genome-phenome Archive (EGA), under the accession code EGAD00010000887. Data from the Dunedin Multidisciplinary Health and Development are available via a managed access system (contact: ac115@duke.edu). The HumanMethylation450 BeadChip DNA methylation data from the E-Risk Study are available in GEO under accession number GSE105018.
References
- 1.Polanczyk G, Rohde LA (2007): Epidemiology of attention-deficit/hyperactivity disorder across the lifespan. Curr Opin Psychiatry. 20: 386–392. [DOI] [PubMed] [Google Scholar]
- 2.Larsson H, Anckarsater H, Råstam M, Chang Z, Lichtenstein P (2012): Childhood attention-deficit hyperactivity disorder as an extreme of a continuous trait: A quantitative genetic study of 8,500 twin pairs. J Child Psychol Psychiatry Allied Discip. 53: 73–80. [DOI] [PubMed] [Google Scholar]
- 3.Groen-Blokhuis MM, Middeldorp CM, Kan KJ, Abdellaoui A, Van Beijsterveldt CEM, Ehli EA, et al. (2014): Attention-deficit/hyperactivity disorder polygenic risk scores predict attention problems in a population-based sample of children. J Am Acad Child Adolesc Psychiatry. 53: 1123–1129. [DOI] [PubMed] [Google Scholar]
- 4.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. (2017): Discovery Of The First Genome-Wide Significant Risk Loci For ADHD. bioRxiv. . doi: doi: 10.1101/145581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawi Z, Cummins TDR, Tong J, Arcos-Burgos M, Zhao Q, Matthews N, et al. (2017): Rare DNA variants in the brain-derived neurotrophic factor gene increase risk for attention-deficit hyperactivity disorder: A next-generation sequencing study. Mol Psychiatry. 22: 580–584. [DOI] [PubMed] [Google Scholar]
- 6.Demontis D, Lescai F, Børglum A, Glerup S, Østergaard SD, Mors O, et al. (2016): Whole-Exome Sequencing Reveals Increased Burden of Rare Functional and Disruptive Variants in Candidate Risk Genes in Individuals With Persistent Attention-Deficit/Hyperactivity Disorder. J Am Acad Child Adolesc Psychiatry. 55: 521–523. [DOI] [PubMed] [Google Scholar]
- 7.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. (2017): Discovery Of The First Genome-Wide Significant Risk Loci For ADHD. bioRxiv. 145581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mill J, Petronis A (2008): Pre- and peri-natal environmental risks for attention-deficit hyperactivity disorder (ADHD): The potential role of epigenetic processes in mediating susceptibility. J Child Psychol Psychiatry Allied Discip. 49: 1020–1030. [DOI] [PubMed] [Google Scholar]
- 9.Groen-Blokhuis MM, Middeldorp CM, Van Beijsterveldt CEM, Boomsma DI (2011): Evidence for a causal association of low birth weight and attention problems. J Am Acad Child Adolesc Psychiatry. 50. doi: 10.1016/j.jaac.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Dong T, Hu W, Zhou X, Lin H, Lan L, Hang B, et al. (2018): Prenatal exposure to maternal smoking during pregnancy and attention-deficit/hyperactivity disorder in offspring: A meta-analysis. Reprod Toxicol. 76: 63–70. [DOI] [PubMed] [Google Scholar]
- 11.Werenberg Dreier J, Nybo Andersen AM, Hvolby A, Garne E, Kragh Andersen P, Berg-Beckhoff G (2016): Fever and infections in pregnancy and risk of attention deficit/hyperactivity disorder in the offspring. J Child Psychol Psychiatry Allied Discip. 57: 540–548. [DOI] [PubMed] [Google Scholar]
- 12.Thapar A, Cooper M, Eyre O, Langley K (2013): Practitioner review: What have we learnt about the causes of ADHD? J Child Psychol Psychiatry Allied Discip. 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malhotra S, Bhatia NK, Kumar P, Hans C, Bhatia MS (2011): The Potential Role of Infections in Attention-Deficit Hyperactivity Disorder. Delhi Psychiatry J. 14: 361–367. [Google Scholar]
- 14.van Mil NH, Steegers-Theunissen RPM, Bouwland-Both MI, Verbiest MMPJ, Rijlaarsdam J, Hofman A, et al. (2014): DNA methylation profiles at birth and child ADHD symptoms. J Psychiatr Res. 49: 51–59. [DOI] [PubMed] [Google Scholar]
- 15.Park S, Lee J-M, Kim J-W, Cho D-Y, Yun HJ, Han DH, et al. (2015): Associations between serotonin transporter gene (SLC6A4) methylation and clinical characteristics and cortical thickness in children with ADHD. Psychol Med. 45: 3009–3017. [DOI] [PubMed] [Google Scholar]
- 16.Rijlaarsdam J, Cecil CAM, Walton E, Mesirow MSC, Relton CL, Gaunt TR, et al. (2017): Prenatal unhealthy diet, insulin-like growth factor 2 gene (IGF2) methylation, and attention deficit hyperactivity disorder symptoms in youth with early-onset conduct problems. J Child Psychol Psychiatry Allied Discip. 58: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walton E, Pingault J-B, Cecil CAM, Gaunt TR, Relton CL, Mill J, Barker ED (2017): Epigenetic profiling of ADHD symptoms trajectories: a prospective, methylome-wide study. Mol Psychiatry. 22: 250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barker ED, Walton E, Cecil CAM, Rowe R, Jaffee SR, Maughan B, et al. (2017): A Methylome-Wide Association Study of Trajectories of Oppositional Defiant Behaviors and Biological Overlap With Attention Deficit Hyperactivity Disorder. Child Dev. . doi: 10.1111/cdev.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Chen XT, Luo M, Tang Y, Zhang G, Wu D, et al. (2015): Multiple epigenetic factors predict the attention deficit/hyperactivity disorder among the Chinese Han children. J Psychiatr Res. 64: 40–50. [DOI] [PubMed] [Google Scholar]
- 20.Wilmot B, Fry R, Smeester L, Musser ED, Mill J, Nigg T, et al. (2016): Methylomic analysis of salivary DNA in childhood ADHD identifies altered DNA methylation in VIPR2 identifies altered DNA methylation in VIPR2. 57: 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dadds MR, Schollar-Root O, Lenroot R, Moul C, Hawes DJ (2016): Epigenetic regulation of the DRD4 gene and dimensions of attention-deficit/hyperactivity disorder in children. Eur Child Adolesc Psychiatry. 25: 1081–1089. [DOI] [PubMed] [Google Scholar]
- 22.Peter CJ, Fischer LK, Kundakovic M, Garg P, Jakovcevski M, Dincer A, et al. (2016): DNA Methylation Signatures of Early Childhood Malnutrition Associated With Impairments in Attention and Cognition. Biol Psychiatry. 80: 765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinrich H, Grunitz J, Stonawski V, Frey S, Wahl S, Albrecht B, et al. (2017): Attention, cognitive control and motivation in ADHD: Linking event-related brain potentials and DNA methylation patterns in boys at early school age. Sci Rep. 7. doi: 10.1038/s41598-017-03326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sengupta SM, Smith AK, Grizenko N, Joober R (2017): Locus-specific DNA methylation changes and phenotypic variability in children with attention-deficit hyperactivity disorder. Psychiatry Res. 256: 298–304. [DOI] [PubMed] [Google Scholar]
- 25.Gervin K, Nordeng H, Ystrom E, Reichborn-Kjennerud T, Lyle R (2017): Long-term prenatal exposure to paracetamol is associated with DNA methylation differences in children diagnosed with ADHD. Clin Epigenetics. 9. doi: 10.1186/s13148-017-0376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding K, Yang J, Reynolds GP, Chen B, Shao J, Liu R, et al. (2017): DAT1 methylation is associated with methylphenidate response on oppositional and hyperactive-impulsive symptoms in children and adolescents with ADHD. World J Biol Psychiatry. 18: 291–299. [DOI] [PubMed] [Google Scholar]
- 27.van Dongen J, Nivard MG, Willemsen G, Hottenga J-J, Helmer Q, Dolan C V, et al. (2016): Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun. 7: 11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McRae AF, Powell JE, Henders AK, Bowdler L, Hemani G, Shah S, et al. (2014): Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol. 15: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. (2008): Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci. 105: 17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joubert BR, den Dekker HT, Felix JF, Bohlin J, Ligthart S, Beckett E, et al. (2016): Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun. 7: 10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. (2016): DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am J Hum Genet. 98: 680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, et al. (2016): Epigenetic Signatures of Cigarette Smoking. Circ Cardiovasc Genet. 9: 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slieker RC, Bos SD, Goeman JJ, Bovée JV, Talens RP, Van Der Breggen R, et al. (2013): Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics and Chromatin. 6. doi: 10.1186/1756-8935-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walton E, Hass J, Liu J, Roffman JL, Bernardoni F, Roessner V, et al. (2016): Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophr Bull. 42: 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hannon E, Lunnon K, Schalkwyk L, Mill J (2015): Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 10: 1024–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RD, Jones MJ, Meaney MJ, Turecki G, Kobor MS (2017): BECon: A tool for interpreting DNA methylation findings from blood in the context of brain. Transl Psychiatry. 7. doi: 10.1038/tp.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi T, Wu Y, Zeng J, Zhang F, Xue A, Jiang L, et al. (2018): Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun.. doi: 10.1038/s41467-018-04558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Mil NH, Steegers-Theunissen RPM, Bouwland-Both MI, Verbiest MMPJ, Rijlaarsdam J, Hofman A, et al. (2014): DNA methylation profiles at birth and child ADHD symptoms. J Psychiatr Res. 49: 51–59. [DOI] [PubMed] [Google Scholar]
- 39.Park S, Lee J-M, Kim J-W, Cho D-Y, Yun HJ, Han DH, et al. (2015): Associations between serotonin transporter gene (SLC6A4) methylation and clinical characteristics and cortical thickness in children with ADHD. Psychol Med. 45: 3009–3017. [DOI] [PubMed] [Google Scholar]
- 40.Walton E, Pingault J-B, Cecil CAM, Gaunt TR, Relton CL, Mill J, Barker ED (2017): Epigenetic profiling of ADHD symptoms trajectories: a prospective, methylome-wide study. Mol Psychiatry. 22: 250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gervin K, Hammerø M, Akselsen HE, Moe R, Nygard H, Brandt I, et al. (2011): Extensive variation and low heritability of DNA methylation identified in a twin study. Genome Res. 21: 1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willemsen G, Vink JM, Abdellaoui A, den Braber A, van Beek JHDA, Draisma HHM, et al. (2013): The Adult Netherlands Twin Register: twenty-five years of survey and biological data collection. Twin Res Hum Genet. 16: 271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poulton R, Moffitt TE, Silva PA (2015): The Dunedin Multidisciplinary Health and Development Study: overview of the first 40 years, with an eye to the future. Soc Psychiatry Psychiatr Epidemiol. 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moffitt TE, Adlam A, Affleck G, Andreou P, Aquan-Assee J, Arseneault L, et al. (2002): Teen-aged mothers in contemporary Britain. J Child Psychol Psychiatry Allied Discip. 43: 727–742. [DOI] [PubMed] [Google Scholar]
- 45.Conners CK; Erhardt D; Sparrow E (1999): Conners’ Adult ADHD Rating Scales (CAARS) technical manual. North Tonawanda, NY; Multi-Health Systems. [Google Scholar]
- 46.Moffitt TE, Houts R, Asherson P, Belsky DW, Corcoran DL, Hammerle M, et al. (2015): Is adult ADHD a childhood-onset neurodevelopmental disorder? Evidence from a four-decade longitudinal cohort study. Am J Psychiatry. 172: 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agnew-Blais JC, Polanczyk G V., Danese A, Wertz J, Moffitt TE, Arseneault L (2016): Evaluation of the persistence, remission, and emergence of Attention-deficit/hyperactivity disorder in young adulthood. JAMA Psychiatry. 73: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fortin J, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, et al. (2014): Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pidsley R Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC (2013): A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 14: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Francioli LC, Menelaou A, Pulit SL, Van Dijk F, Palamara PF, Elbers CC, et al. (2014): Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. . doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 51.Price ME, Cotton AM, Lam LL, Farré P, Emberly E, Brown CJ, et al. (2013): Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Lemire M, Choufani S, Butcher DT, Zanke BW, Gallinger S, et al. (2013): Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2294. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Iterson M, Van Zwet EW, Heijmans BT (2017): Controlling bias and inflation in association studies using the empirical null distribution. Genome Biol. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wray NR, Sullivan PF (2017): Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. bioRxiv. . doi: 10.1101/167577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, et al. (2014): Biological insights from 108 schizophrenia-associated genetic loci. Nature. 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grove J, Ripke S, Als TD, Mattheisen M, Walters R, Won H (2017): Common risk variants identified in autism spectrum disorder. bioRxiv. . doi: 10.1101/224774doi. [DOI] [Google Scholar]
- 57.Hannon E, Dempster E, Viana J, Burrage J, Smith AR, Macdonald R, et al. (2016): An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 17: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pedersen BS, Schwartz DA, Yang I V, Kechris KJ (2012): Comb-p : software for combining, analyzing, grouping and correcting spatially correlated P -values. 28: 2986–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonder MJ, Luijk R, Zhernakova D V., Moed M, Deelen P, Vermaat M, et al. (2017): Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 49: 131–138. [DOI] [PubMed] [Google Scholar]
- 60.van Dongen J, Bonder MJ, Dekkers KF, Nivard MG, van Iterson M, Willemsen G, et al. (2018): DNA methylation signatures of educational attainment. npj Sci Learn. 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fayyad J, Sampson NA, Hwang I, Adamowski T, Aguilar-Gaxiola S, Al-Hamzawi A, et al. (2017): The descriptive epidemiology of DSM-IV Adult ADHD in the World Health Organization World Mental Health Surveys. ADHD Atten Deficit Hyperact Disord. 9: 47–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bendor J, Lizardi-Ortiz JE, Westphalen RI, Brandstetter M, Hemmings HC, Sulzer D, et al. (2010): AGAP1/AP-3-dependent endocytic recycling of M5 muscarinic receptors promotes dopamine release. EMBO J. . doi: 10.1038/emboj.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnold M, Cross R, Singleton KS, Zlatic S, Chapleau C, Mullin AP, et al. (2016): The Endosome Localized Arf-GAP AGAP1 Modulates Dendritic Spine Morphology Downstream of the Neurodevelopmental Disorder Factor Dysbindin. Front Cell Neurosci. . doi: 10.3389/fncel.2016.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. (2016): Schizophrenia risk from complex variation of complement component 4. Nature. 530: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang Z, Lichtenstein P, Asherson PJ, Larsson H (2013): Developmental twin study of attention problems: High heritabilities throughout development. JAMA Psychiatry. 70: 311–318. [DOI] [PubMed] [Google Scholar]
- 66.Boomsma DI, Saviouk V, Hottenga JJ, Distel MA, de Moor MHM, Vink JM, et al. (2010): Genetic epidemiology of attention deficit hyperactivity disorder (ADHD index) in adults. PLoS One. 5. doi: 10.1371/journal.pone.0010621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marioni RE, McRae AF, Bressler J, Colicino E, Hannon E, Li S, et al. (2018): Meta-analysis of epigenome-wide association studies of cognitive abilities. Mol Psychiatry. . doi: 10.1038/s41380-017-0008-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jia T, Chu C, Liu Y, Van Dongen J, Armstrong NJ, Bastin ME, et al. (2018): Epigenome-wide meta-analysis of blood DNA methylation and its association with subcortical volumes : findings from the ENIGMA Epigenetics Working. bioRchiv. . doi: 10.1101/460444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dekkers KF, van Iterson M, Slieker RC, Moed MH, Bonder MJ, van Galen M, et al. (2016): Blood lipids influence DNA methylation in circulating cells. Genome Biol. 17: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


