Abstract
Sodium/glucose cotransporter 1 (SGLT1), an essential active glucose transport protein that helps maintain high intracellular glucose levels, was previously shown to interact with epidermal growth factor receptor (EGFR); the SGLT1–EGFR interaction maintains intracellular glucose levels to promote survival of cancer cells. Here, we explore the role of SGLT1 in triple‐negative breast cancer (TNBC), which is the most aggressive type of breast cancer. We performed TCGA analysis coupled to in vitro experiments in TNBC cell lines as well as in vivo xenografts established in the mammary fat pad of female nude mice. Tissue microarrays of TNBC patients with information of clinical–pathological parameters were also used to investigate the expression and function of SGLT1 in TNBC. We show that high levels of SGLT1 are associated with greater tumour size in TNBC. Knockdown of SGLT1 compromises cell growth in vitro and in vivo. We further demonstrate that SGLT1 depletion results in decreased levels of phospho‐EGFR, and as a result, the activity of downstream signalling pathways (such as AKT and ERK) is inhibited. Hence, targeting SGLT1 itself or the EGFR–SGLT1 interaction may provide novel therapeutics against TNBC.
Keywords: EGFR, SGLT1, triple‐negative breast cancer
Abbreviations
- ATCC
American Type Culture Collection
- DAVID
database for annotation, visualization and integration discovery
- EGFR
epidermal growth factor receptor
- HER2
human epidermal growth factor 2
- IgG
immunoglobulin G
- MTT
3‐(4,5‐dimethyl‐thiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide
- PD‐L1
programmed death‐ligand 1
- SGLT1
sodium/glucose cotransporter 1
- shRNA
short hairpin RNA
- siRNA
short interfering RNA
- SPF
specific pathogen‐free
- TNBC
triple‐negative breast cancer
1. Introduction
Breast cancer is the most frequent cancer type in women with 1.38 million new cases (23%) each year throughout the world. It is the fifth leading cause of cancer death (458 000 deaths worldwide) and, globally, the most frequent female cancer deaths in developing and developed countries (Ferlay et al., 2010). Triple‐negative breast cancer (TNBC), which lacks expressions of oestrogen, progesterone and human epidermal growth factor 2 (HER2) receptors, accounts for 10‐20% breast cancers with unsatisfactory therapeutic efficacy (O'Reilly et al., 2015; Reis‐Filho and Tutt, 2008). TNBC is the most aggressive, high‐grade breast cancer type with high risk of metastasis and poor survival rate compared with the other breast cancer subtypes. Chemotherapy has been the only accepted systemic treatment option for TNBC for several years to increase the overall survival rate (Khosravi‐Shahi et al., 2018). However, drug resistance occurs due to its heterogeneity (Du et al., 2015), and this contributes to the recurrence of the metastatic disease (Lee and Djamgoz, 2018). Therefore, it is crucial to identify targeted therapies for TNBC.
Novel targeted therapies, such as endocrine therapies and targeting HER2, were discovered by mapping the genomic landscapes of breast cancer tumours. Genomic analysis has led to insights into the classification of TNBC such as the separation of TNBC into different subtypes based on gene expression (Criscitiello et al., 2012; Garrido‐Castro et al., 2019; Network, 2012). However, due to its complexity and heterogeneity, it has been challenging to identify targeted therapies for TNBC (Lehmann et al., 2011). Atezolizumab (TECENTRIQ®, Roche, Basel, Switzerland), an antiprogrammed death‐ligand 1 (PD‐L1) monoclonal antibody, was approved recently as a first targeted therapy for TNBC with combination of chemotherapy (Abraxane; nab®‐Paclitaxel, Summit, NJ, USA) (Cyprian et al., 2019). As atezolizumab plus chemotherapy combination is only successful for PD‐L1 positive TNBC and TNBC is a heterogeneous disease, further molecular studies are required to identify novel therapeutic targets for TNBC. Over the past decades, potential targeted therapies were discovered for TNBC by targeting different signalling pathways such as epidermal growth factor receptor (EGFR) (Crozier et al., 2016; Nabholtz et al., 2016; Schuler et al., 2012; Shao et al., 2017; Trédan et al., 2015). However, targeting EGFR pathway was not very successful in part due to the resistance mechanisms or activation of different signalling pathway(s) (Baselga et al., 2005; Baselga et al., 2013; Carey et al., 2012; Spector et al., 2005). A previous study showed that inhibition of sodium/glucose cotransporter 1 (SGLT1) sensitized prostate cancer cells to EGFR inhibitors (gefitinib and erlotinib) (Wright et al., 2011), although the precise mechanisms have not been elucidated. Knowing that SGLT1 mainly interacts with the autophosphorylation domain of EGFR (Ren et al., 2013), we hypothesized that SGLT1 may regulate EGFR activity in TNBC.
Sodium/glucose cotransporter 1, encoded by the SLC5A1 gene in humans, is an active glucose transporter, which utilizes sodium gradients to transport glucose into cells independent of extracellular glucose concentration (Rieg and Vallon, 2018; Wright et al., 2011). Various studies have discovered that SGLT1 is overexpressed in different cancer types: prostate cancer (Blessing et al., 2012), ovarian carcinoma (Lai et al., 2012), oral squamous cell carcinoma (Hanabata et al., 2012), head and neck carcinoma (Wright et al., 2011), pancreatic cancer (Casneuf et al., 2008) and colorectal cancer (Guo et al., 2011). In ovarian carcinoma, tumour development and poor prognosis of the disease is associated with the overexpression of SGLT1 (Lai et al., 2012). Overexpression of SGLT1 also has linked with higher clinical stages of colorectal cancer (Guo et al., 2011). Despite of all these observations, the role of SGLT1 in TNBC was not known. In this study, we report that SGLT1 is essential for the survival of TNBC cells in vitro and in vivo. This is achieved, at least in part, via potentiating EGFR activity.
2. Materials and methods
2.1. Cell culture and transfection
BT549, MDA‐MB‐468 and MDA‐MB‐436 cell lines, which all belong to triple‐negative breast carcinoma cells, were purchased from Cell Bank of the Institute of Basic medicine, Chinese Academy of Medical Sciences (Beijing, China), or obtained as NCI‐ICBP45 kit procured through American Type Culture Collection (ATCC; ATCC Breast Cancer Cell Panel, Manassas, VA, USA). BT549 cells were cultured in RPMI 1640 medium (Boster, Wuhan, China) supplemented with 10% FBS (Gibco, Carlsbad, USA), 1% antibiotics and 0.023 IU·mL−1 bovine insulin (Sigma‐Aldrich, Saint Louis, MO, USA). MDA‐MB‐436 and MDA‐MB‐468 cells were cultured in Dulbecco's modified Eagle's medium (Boster) with 20% or 10% FBS (Gibco) and 1% antibiotics. All cells were kept at 37 °C and 5% CO2. The short hairpin RNA (shRNA) product of SGLT1 (sh‐SGLT1) with a targeted sequence of ATCTTTCTCTTATTGGCAA and its negative control (sh‐NC) were purchased from GeneChem (Shanghai, China) and transfected into cells according to manufacturer’s instructions. Short interfering RNA (siRNA) oligos against SGLT1 (MU‐007589‐01‐0002) were purchased from Dharmacon (Lafayette, CO, USA). Sequences are available from Dharmacon or upon request. As a negative control, we used siGENOME RISC‐Free siRNA (Dharmacon). Cells were transfected with the indicated siRNA oligos at a final concentration of 35 nm using Dharmafect 2 reagent (Dharmacon).
2.2. Cell viability assay
Short hairpin RNA‐transfected cells were seeded into 96‐well plates with a density of 1 × 103 cells per well and allowed to grow for 24, 48 and 72 h. When indicated time arrived, 3‐(4,5‐Dimethyl‐thiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) solution (Sangon Biotech, Shanghai, China) was added into the medium with a final concentration of 0.5 mg·mL−1. The solution in each well was carefully sucked after incubation for 4 h at 37 °C, replaced by 100 µL formazan solubilization solution and gently mix for 10 min. OD values at 570 nm were measured using Microplate Reader (Bioteck Instrument, Winooski, VT, USA).
2.3. Western blot analysis
Western blot analysis was performed with lysates from cells or tissues with urea buffer (8 m urea, 1 m thiourea, 0.5% CHAPS, 50 mm DTT and 24 mm spermine). For immunoprecipitations, the cells were lysed for 30 min at 4 °C in pNAS buffer [50 mm Tris/HCl (pH 7.5), 120 mm NaCl, 1 mm EDTA and 0.1% Nonidet P‐40], with protease inhibitors. Indicated antibodies and immunoglobulin G (IgG) agarose were added to the lysate for 16 h at 4 °C. Immunoprecipitates were washed four times with cold PBS followed by the addition of SDS sample buffer. The bound proteins were separated on SDS polyacrylamide gels and subjected to immunoblotting with the indicated antibodies. Primary antibodies were from Abcam (Cambridge, UK) (SGLT1, 1 : 1000, ab14686; β‐actin, 1 : 2000, ab8227; β‐tubulin, 1 : 5000, ab6046; GAPDH, 1 : 2000, ab9385; Phospho‐EGFRTyr1068, 1 : 1000, ab40815), Cell Signaling Technology (Leiden, Netherlands) [SGLT1, 1 : 1000, 5042; Phospho‐EGFRTyr1068, 1 : 1000, 3777, D7A5; EGFR, 1 : 1000, 4267, D38B1; Phospho‐AKT Ser473, 1 : 1000, 9271; Phospho‐AKT Thr308, 1 : 1000, 4056, 244F9; AKT, 1 : 1000, 9272; Phospho‐ERK (Thr202/Tyr204), 1 : 1000, 9101; ERK, 1 : 1000, 9102; Cleaved PARP Asp214, 1 : 1000, 9541] and Millipore (Burlington, MA, USA) (PTEN, 1 : 1000, 04‐409). Signals were detected using an ECL detection system (GE Healthcare) (Chicago, IL, USA) or an Odyssey imaging system (LI‐COR), and evaluated by ImageJ 1.42q software (National Institutes of Health) (Berhesda, MD, USA).
2.4. Immunofluorescence microscopy
Cells were fixed in 4% PBS‐paraformaldehyde for 15 min, incubated in 0.1% Triton X‐100 for 5 min on ice, then incubated in 0.2% Fish Skin Gelatine in PBS for 1 h, and stained for 1 h with an anti‐Phospho‐EGFRTyr1068 (1 : 100; Cell Signaling Technology, 3777). Protein expression was detected using Alexa Fluor (1 : 400; Molecular Probes, Eugene, OR, USA) for 20 min. TO‐PRO‐3 (Invitrogen, Waltham, MA, USA) was used to stain nucleic acids (1 : 2000).
2.5. Animal study
Animal experiments were carried out in accordance with the guidelines and approved protocols of the Ethics Committee of Tongji Hospital (Wuhan, China). Specific pathogen‐free (SPF) nude mice (female, 6 weeks) were purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and housed under SPF condition. After adaptation for 1 week in condition of stable temperature and humidity with 12 h light–dark cycle, mice were randomly divided into sh‐NC group and sh‐SGLT1 group (n = 6 mice per group). ShRNA‐transfected MDA‐MB‐436 cells were harvested, rinsed and resuspended in PBS at a concentration of 2 × 108 cells·mL−1, which were then placed on ice for subsequent operation. Orthotopic injection was performed in sterile condition in clean bench according to protocol described previously (Kocatürk and Versteeg, 2015) with adaptations. Briefly, after anesthetizing the mice intraperitoneally with sterilized 2% Avertin (Sigma‐Aldrich) with a dosage of 0.12 mL/10 g weight, mammary gland was exposed by making a small incision between the fourth nipple and the midline. A total of 1 × 107 cells in 50 µL suspension were completely injected into mammary fat pad located in groin without leaking. Then, the incisions were sutured and mice were attended until they gain consciousness. The lengths and widths of xenografts were measured weekly by vernier calliper, and volume was calculated using following formula: V (mm3) = 1/2*length*widths2. At the end of experiment, mice were sacrificed, while xenografts were completely separated, measured and fixed by 4% paraformaldehyde for histological staining.
2.6. Immunohistochemical and H/E staining and scoring
Tissue microarray of TNBC patients with information of clinical–pathological parameters was purchased from Outdo Biotech (HBreD090Bc01; Shanghai, China). Paraffin‐embedded sections of xenograft tissues were subjected to deparaffinization and rehydration. H/E staining of sections was carried out using H/E staining kit (Beyotime, Shanghai, China) according to manufacturer’s instructions. For immunohistochemical staining of tissue microarray and sections of xenograft and antigen retrieval, blocking of non‐specific binding and incubation of primary antibodies at 4 °C overnight was sequentially conducted. The primary antibodies used were list as follows: anti‐phospho‐EGFR (ab40815; Abcam, 1 : 200) and anti‐SGLT1 (ab14686; Abcam, 1 : 100). After incubation with secondary goat anti‐rabbit immunoglobulin conjugated to peroxidase‐labelled dextran polymer (SV0002; Boster) at 37 °C for 1 h, visualization, counterstaining with haematoxylin and mounting were performed. Semiquantitative evaluations of protein expression were scored on the basis of the intensity and the percentage of phospho‐EGFR‐ or SGLT1‐positive tumour cells as previously described (Wang et al., 2014).
2.7. TCGA data mining and pathway analysis
SLC5A1, mRNA expression z‐score (RNA Seq V2 RSEM) and molecular subtypes of breast invasive carcinoma (TCGA, Provisional) were obtained from cBioPortal for Cancer Genomics website (http://www.cbioportal.org). Molecular subtypes of breast samples were separated based on ER, PR and HER2 status, and mRNA expression of SGLT1 was analysed in each subtype in graphpad prism 8 (La Jolla, CA, USA) by ordinary one‐way ANOVA.
Two different data sets, TCGA_BRCA_RPPA‐2015‐02‐24 and TCGA_BRCA_exp_HiSeqV2‐2015‐02‐ 24, for RPPA protein expression and gene expression RNASeq (IlluminaHiSeq, San Diego, CA, USA), respectively, were extracted from UCSC Cancer Browser (https://genome-cancer.ucsc.edu/) for the analysis of genomic matrix data in TNBC samples. The data of TNBC samples that had SGLT1 mRNA expression from the cBioPortal for Cancer Genomics website were aligned with the samples in genomic matrix of TCGA data for both protein and gene expressions. For both protein and gene expression data sets, top 20% and bottom 20% of samples were chosen for high and low SGLT1 expression, respectively. Then, unpaired t‐test was performed to find the significantly different proteins/genes between high and low SGLT1 groups in r version 3.4.4 (Auckland, New Zealand), P < 0.05. For the protein expression of AKT_pT308 in TCGA protein data set, prism8 was used to plot scatter dot plot between high and low SGLT1 groups.
To explore the pathway analysis of the TCGA gene data analysis, the database for annotation, visualization and integration discovery (DAVID) functional annotation web tool was conducted (version 6.8; https://david.ncifcrf.gov (Huang et al., 2009)). A total of 1325 genes, which were positively correlated with SGLT1 in TCGA gene data, were analysed with DAVID web tool and obtained the lists of enriched Kyoto Encyclopedia of Genes and Genomes pathway, REACTOME pathway, BIOCARTA, BBID and EC number. A P value ≤ 0.05 was considered significant. The pathways were sorted from lowest P value, and top 34 pathways were chosen which then were sorted with highest number of shared genes. Subsequently, we then plotted that histogram plot with the top 15 pathways in graphpad prism 8.
2.8. Statistical analysis
Comparison of two groups was statistically calculated by Student's t‐test. Chi‐square test or Fisher exact test were used to evaluate the relationship of SGLT1 expression and clinical parameters of TNBC. Correlation between expressions of SGLT1 and phospho‐EGFR was analysed using Pearson's correlation. Data were shown as mean ± SD. Statistical analysis was conducted using spss version 19.0 (Endicott, NY, USA). Unpaired t‐test was performed for TCGA data analysis in r version 3.4.4. To identify the statistical difference of AKT_pT308 between high and low SGLT1 group in TCGA protein data, unpaired t‐test was performed in graphpad prism 8 software.
3. Results
3.1. SGLT1 expression is higher in TNBC, and this associates with a larger tumour size
The expression of SGLT1 was upregulated in various cancer types (Blessing et al., 2012; Casneuf et al., 2008; Guo et al., 2011; Hanabata et al., 2012; Lai et al., 2012; Wright et al., 2011). To investigate whether the expression of SGLT1 is different between each molecular subtype of breast cancer, TCGA breast invasive carcinoma (Provisional) data were analysed. The mRNA levels of SGLT1 (SLC5A) were significantly higher in TNBC and HER2‐positive subtypes than luminal A and luminal B (Fig. S1). No significant difference between TNBC‐ and HER2‐positive subtypes was observed (Fig. S1).
In order to examine whether SGLT1 expression levels correlate with clinical–pathological features in TNBC, we performed immunohistochemistry staining of SGLT1 in TNBC tissue microarrays. Table 1 shows clinical and pathological characteristics of the 90 TNBC patient samples. We found only tumour size was significantly affected by SGLT1 expression (Table 1). Representative images of high and low expression of SGLT1 in TNBC are shown in Fig. 1A. Tumour size was significantly larger in high SGLT1 TNBC samples compared with the low SGLT1 TNBC samples (P = 0.006; Fig. 1B).
Table 1.
The relationship between patients’ clinical–pathological characteristics and SGLT1 expression in TNBC. The other P values were obtained by chi‐square test.
| Characteristics | N | SGLT1 | P value | |
|---|---|---|---|---|
| Low expression | High expression | |||
| Age | 90 | |||
| ≤ 50 | 46 | 28 | 18 | 0.290 |
| > 50 | 44 | 21 | 23 | |
| Location | 90 | |||
| Left breast | 46 | 23 | 23 | 0.406 |
| Right breast | 44 | 26 | 18 | |
| Grade | 90 | |||
| I–II | 36 | 19 | 17 | 0.832 |
| III | 54 | 30 | 24 | |
| Size | 86 | |||
| ≤ 2 cm | 37 | 26 | 11 | 0.016* |
| > 2 cm | 49 | 21 | 28 | |
| Positive LN | 35 | |||
| ≤ 2 | 21 | 10 | 11 | 0.728a |
| > 2 | 14 | 5 | 9 | |
P value of Fisher’s exact test. The other P values were obtained by chi‐square test. *P < 0.05.
Figure 1.
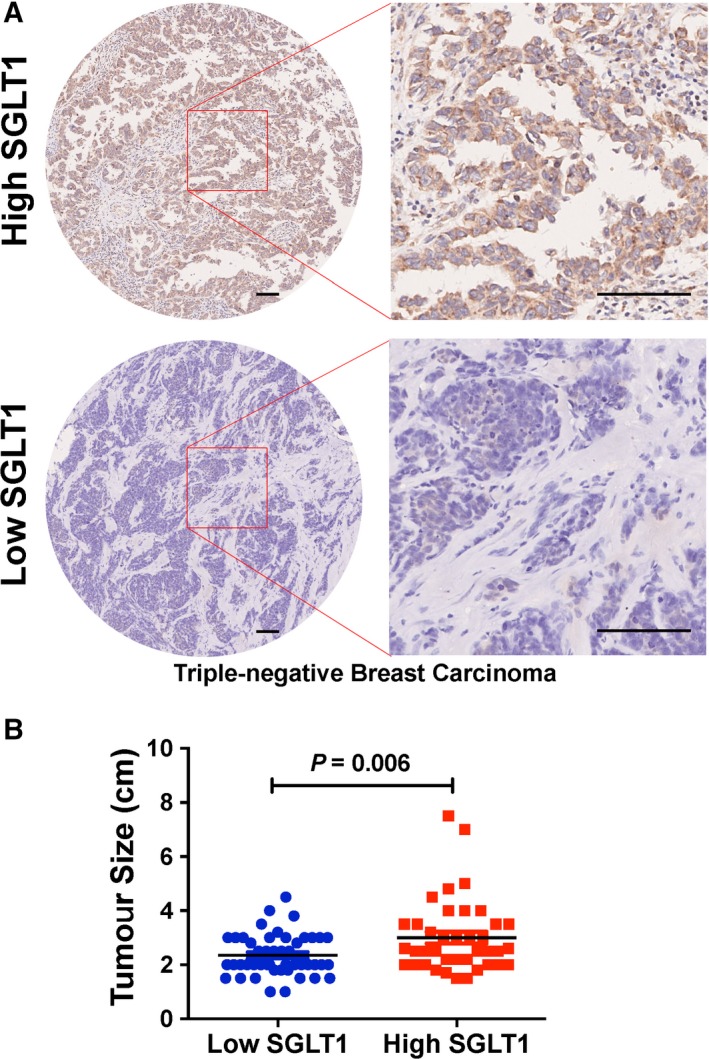
SGLT1 expression levels associate with tumour size in triple‐negative breast carcinoma (TNBC). (A) Representative SGLT1 staining pattern (high or low SGLT1) in 90 TNBC tissue microarray cores. Scale bar: 100 μm. (B) The relationship between SGLT1 expression and tumour size in TNBC samples was analysed. Chi‐square test was performed (P = 0.006).
3.2. SGLT1 depletion impairs cell viability of TNBC
Given the fact that SGLT1 levels associate with tumour size, we next checked whether SGLT1 depletion impairs cell viability. MTT assay was performed in control or SGLT1‐depleted MDA‐MB‐436, MDA‐MB‐468 and BT549 cell lines (Fig. S2). The OD values in MDA‐MB‐436, MDA‐MB‐468 and BT549 cells were significantly decreased in SGLT1‐depleted ones compared with the controls in various time points (Fig. 2A). We subsequently performed an in vivo study to determine the effect of SGLT1 knockdown on tumour growth. MDA‐MB‐436 tumours were established in the mammary fat pad of female nude mice. As shown in Fig. 2B, tumours were found in four of six mice where control MDA‐MB‐436 cells were injected, whereas only two of six mice had tumours where SGLT1‐depleted MDA‐MB‐436 cells were injected. The volume and weight of tumours present in the mice injected with SGLT1‐depleted MDA‐MB‐436 cells were significantly lower than that in the control group injected with control cells (Fig. 2B). Hence, a reduction in SGLT1 expression is able to inhibit TNBC cell growth in vitro and in vivo.
Figure 2.
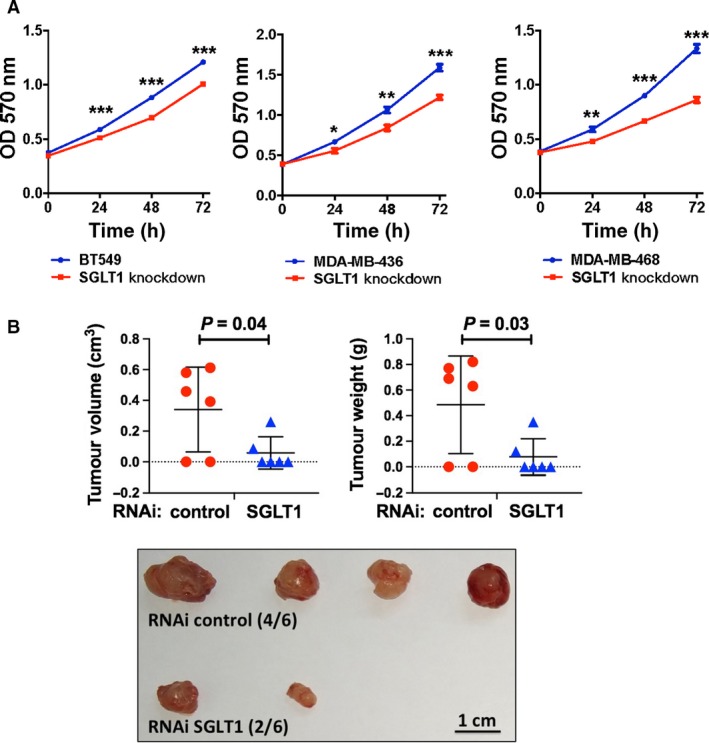
SGLT1 depletion impairs cell viability of TNBC in vitro and in vivo. (A) Knockdown of SGLT1 in BT549, MDA‐MB‐436 and MDA‐MB‐468 cells resulted in cell growth inhibition as performed by MTT assay. Data are represented as a mean value ± standard deviation of three independent experiments. Student’s t‐test was conducted to identify statistical differences in each time point. *P < 0.05, **P < 0.01 and ***P < 0.001. (B) Graphs showing tumour volume (P = 0.04) or weight (P = 0.03) present in the mice injected with control or SGLT1‐depleted MDA‐MB‐436 cells. Pictures of the tumours formed were also shown. Scale bar: 1 cm. Student’s t‐test was performed between control and SGLT1 RNAi groups to identify statistical difference of tumour volume and weight.
3.3. TCGA analysis reveals a link between SGLT1 status and AKT signalling in TNBC samples
In order to find how SGLT1 regulates tumour growth, we analysed data from the TCGA project. Protein (RPPA) TCGA breast invasive carcinoma data and Gene, RNASeq (IlluminaHiSeq) TCGA breast invasive carcinoma data sets were obtained from UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/). Protein expression data included 410 breast invasive carcinoma samples and 142 proteins; and gene expression data contained 1215 samples and 20530 genes. Both protein (RPPA) and gene expression, RNASeq (IlluminaHiSeq), breast invasive carcinoma samples from Cancer Genome Browser were aligned with TNBC samples, as shown in Fig. S1. The alignment showed 59 and 160 TNBC samples in protein (RPPA) and gene, RNASeq (IlluminaHiSeq) data, respectively. In both protein and gene expression data sets, the top 20% and bottom 20% samples were separated into two groups: high and low SGLT1‐expressing samples, respectively, and unpaired t‐test was performed to find the significantly different proteins/genes between the groups. A heat map of protein expression for 19 proteins, including phosphorylated AKT (AKT_pT308), that were significantly different between high and low SGLT1‐expressing samples (P < 0.05) is shown in Fig. 3A. The scatter dot plot showed the protein expression of phosphorylated AKT (AKT_pT308) for each sample in high and low SGLT1 groups (Fig. 3B). A total of 2279 significantly different genes between high and low SGLT1‐expressing samples are shown by the heat map (Fig. 3C).
Figure 3.
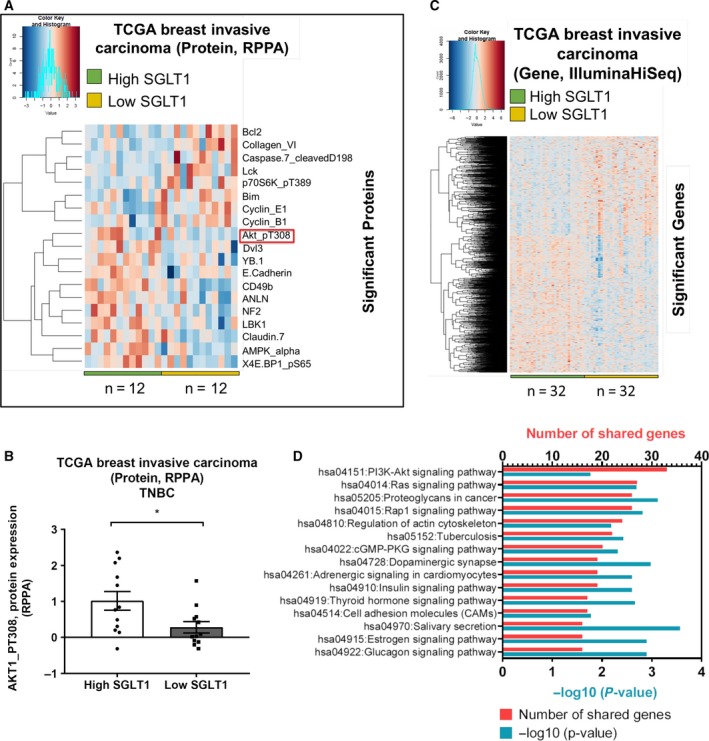
TCGA analysis reveals a link between SGLT1 status and AKT signalling in TNBC samples. Heat maps of the columns indicate each individual sample in high and low SGLT1 groups across each protein (A) and gene (C) in TCGA breast invasive carcinoma data, which obtained from Cancer Genome Browser. Rows indicate protein expressions (RPPA) (A) and gene expressions (IlluminaHiSeq) (C). Dark blue colour indicates low expression of proteins/genes, and dark pink colour illustrates highly expressed proteins/genes. Unpaired t‐test was analysed to find the significantly different proteins/genes in TCGA breast invasive carcinoma data sets. n represents the number of samples in each group. (B) Box plot shows the protein expression of AKT_pT308 between high and low SGLT1 groups in TNBC, TCGA breast invasive carcinoma. *P < 0.05. (D) The genes that were positively regulated with SGLT1 were analysed in DAVID website to show which pathways are regulated. Histogram shows top 15 pathways based on lowest P value. Y‐axis shows the pathways, and top x‐axis and bottom x‐axis show the number of shared genes in each pathway and −log10 (P value), respectively.
To demonstrate whether the significantly positive genes with SGLT1 in TCGA data set are involved in the same pathway, DAVID, online website (https://david.ncifcrf.gov) was used to perform a pathway analysis. We found PI3K–AKT signalling pathway is the top pathway that has the highest hit genes, followed by Ras signalling and other pathways (Fig. 3D). These analyses suggested that SGLT1 may interact with AKT signalling pathway.
3.4. SGLT1 binds EGFR and positively regulates EGFR activity
Search Tool for the Retrieval of Interacting Genes (https://string-db.org) analysis showed the interaction between SGLT1 and EGFR (Fig. S3), which was reported earlier (Ren et al., 2013; Weihua et al., 2008). In fact, we also observed EGFR–SGLT1 interaction in MDA‐MB‐468 human breast cancer cells. Endogenous EGFR and SGLT1 co‐immunoprecipitated together in MDA‐MB‐468 cells (Fig. 4A).
Figure 4.
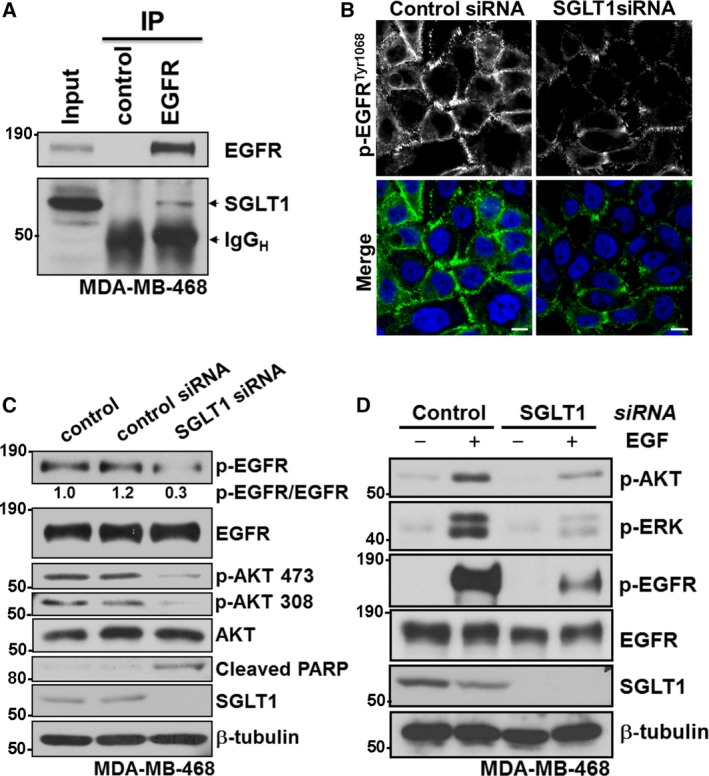
SGLT1 binds EGFR and positively regulates EGFR activity. (A) Total cell lysates from MDA‐MB‐468 cells were immunoprecipitated with an anti‐EGFR antibody or control IgG. EGFR and SGLT1 levels are indicated. IgGH indicates IgG heavy chain. (B) Immunofluorescence staining of Phospho‐EGFRTyr1068 (green) in MDA‐MB‐468 cells transfected with SGLT1 siRNA or control siRNA. TO‐PRO‐3 (blue) was used to stain nuclei. Scale bar: 10 µm. (C) Protein expression of Phospho‐EGFRTyr1068, EGFR, Phospho‐AKT Ser473, Phospho‐AKT Ser308, AKT, Cleaved PARP Asp214 and SGLT1 in MDA‐MB‐468 cells with indicated treatments. β‐tubulin was used as a loading control. (D) Protein expression of Phospho‐EGFRTyr1068, EGFR, Phospho‐AKT Ser473, Phospho‐ERK (Thr202/Tyr204) and SGLT1 in MDA‐MB‐468 cells with indicated treatments. β‐tubulin was used as a loading control.
Given that SGLT1 interacts primarily with the auto‐phosphorylation domain of EGFR (Ren et al., 2013), we asked whether SGLT1 status could also affect EGFR activity on the other hand. Consistently, the immunofluorescence signal of phospho‐EGFRTyr1068 was at the cell membrane in MDA‐MB‐468 cells (Fig. 4B). To test the biological importance of the observed EGFR–SGLT1 interaction, we examined the level of phospho‐EGFRTyr1068 by immunofluorescence staining in MDA‐MB‐468 cells treated with control small interfering RNA (siRNA) or siRNA against SGLT1. Importantly, SGLT1 depletion by RNA interference (RNAi) largely abolished the membrane signal of phospho‐EGFRTyr1068 (Fig. 4B). For most of the analyses described here, we used pools of four siRNA against each gene (‘SMARTpools’ from Dharmacon). However, deconvolution of siRNA pools into their constituent individual oligonucleotides is an important step in minimizing the potential for off‐target effects to compromise the analysis of gene knockdown studies (Echeverri et al., 2006). Figure S4 showed that deconvolution of SGLT1 siRNA SMARTpool, two out of four different oligonucleotides, clearly led to the downregulation of SGLT1 and reduced level of phospho‐EGFRTyr1068.
We next asked whether SGLT1 status could affect the activity of EGFR downstream pathways, which was investigated by examining the EGFR signalling cascade in MDA‐MB‐468 cells transfected with control siRNA or SGLT1 siRNA. The basal phosphorylation levels of EGFR (Tyr1068) and AKT (Ser473 and Thr308) were reduced in SGLT1‐depleted MDA‐MB‐468 cells compared with control cells or control siRNA‐treated cells. Total protein levels of EGFR and AKT, however, were not affected (Fig. 4C). Following SGLT1 gene silencing in MDA‐MB‐468 cells, a rapid and profound loss of cell viability was observed, most likely resulting from a robust induction of apoptosis. This is evident by the detection of poly ADP ribose polymerase (PARP) cleavage, a well‐described indicator of effector caspase activation and consequent cell death (Fig. 4C).
The difference was more notable upon epidermal growth factor (EGF) treatment. A much stronger response induced by addition of EGF was observed in control siRNA‐treated cells compared with SGLT1‐depleted MDA‐MB‐468 cells, reflected by the phosphorylation levels of EGFR (Tyr1068), ERK (Thr202/Tyr204) and AKT (Ser473; Fig. 4D). These data suggest that by binding to EGFR, SGLT1 potentiates EGFR signalling.
To further validate the in vitro findings, the regulation of EGFR phosphorylation by SGLT1 was investigated in vivo using tumour sections derived from control shRNA or SGLT1 shRNA‐transfected MDA‐MB‐436 cells (Fig. 2B). Using adjacent tumour sections derived from MDA‐MB‐436–control shRNA cells, we observed positive SGLT1 and EGFR phosphorylation. In tumours derived from MDA‐MB‐436–SGLT1 shRNA cells, SGLT1‐negative tumour regions showed reduced levels of EGFR phosphorylation (Fig. 5). In addition, the correlation of SGLT1 expression and EGFR phosphorylation levels was analysed in TNBC samples (Fig. 6). High SGLT1 correlated with high EGFR phosphorylation levels in TNBC (P < 0.0001). These observations agree with the finding that SGLT1 potentiates EGFR signalling in TNBC.
Figure 5.
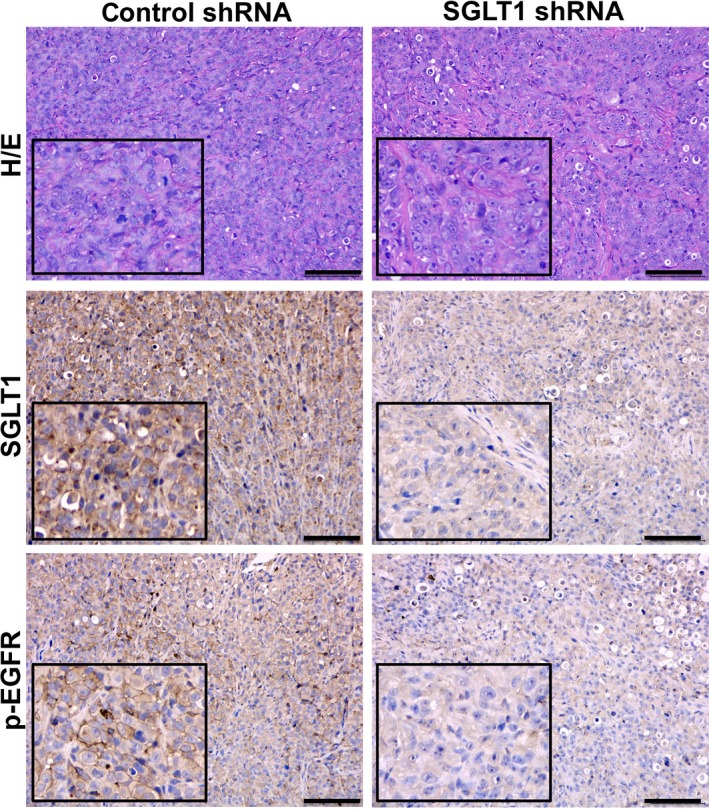
Downregulation of SGLT1 reduces EGFR phosphorylation level in vivo. Adjacent tumour sections from representative cases of tumour xenografts formed by MDA‐MB‐436 cells treated with control or SGLT1 shRNA were stained with H/E. The expression of SGLT1 and p‐EGFR is also indicated. Scale bars: 100 μm.
Figure 6.
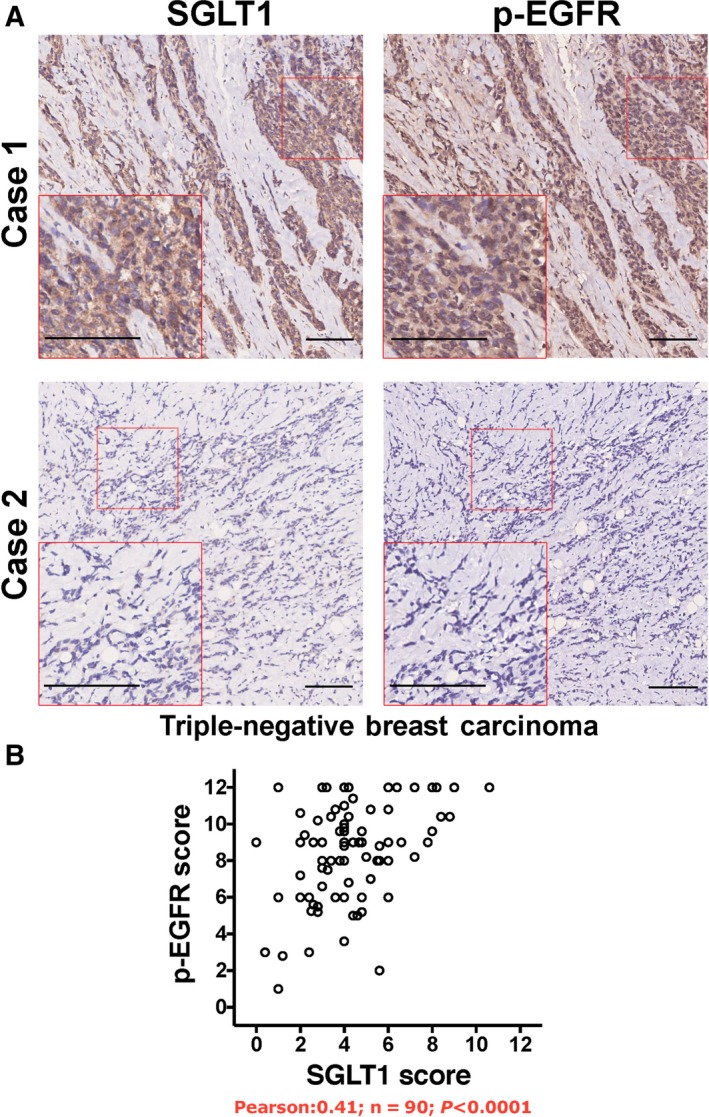
SGLT1 expression levels positively correlate with EGFR phosphorylation in TNBC. (A) Adjacent tumour sections from representative cases show SGLT1 and p‐EGFR expression in TNBC. Scale bar: 100 μm. (B) The relationship between SGLT1 expression and EGFR phosphorylation was analysed by Pearson correlation test (R = 0.41, n = 90, P < 0.0001).
4. Discussion
Chemotherapy has been the only systemic treatment option for TNBCs for several years to increase the overall survival rate of the patients (Khosravi‐Shahi et al., 2018), but chemotherapy resistance is the major challenge for the treatment of the patients with TNBC (Lee and Djamgoz, 2018). Therefore, several studies detected the molecular changes before and after the chemotherapy to identify potential targeted therapy for TNBC (Balko et al., 2014). Identification of targeted therapy for the patients with TNBC could help to prevent the risk of resistance of the treatment option and recurrence of the disease. In this study, we showed that targeting of SGLT1 has the potential to treat TNBC. We found SGLT1 status associates with tumour size in TNBC. Similarly, SGLT1 expression showed significant association with clinical–pathological characteristics and prognosis of ovarian cancer; SGLT1 overexpression significantly correlated with increased pT status and poor prognosis (Lai et al., 2012). Guo et al. (2011) also reported that higher expression of SGLT1 significantly associated with the clinical stage of colorectal cancer (Guo et al., 2011).
SGLT1 is an active glucose transporter and plays a critical role in glucose absorption and retention in the body (Wright et al., 2011). In cancer cells, a high rate of glucose uptake is required to meet the increased energy needs and leads to abnormal growth of cancerous cells (Hanahan and Weinberg, 2011). Depletion or inhibition of SGLT1 may reduce the energy supply to the cancer cells. In addition, we demonstrated that downregulation of SGLT1 dampened EGFR‐AKT or EGFR‐ERK pathways activity, which phosphorylates a plethora of targets to activate the cell cycle, prevent apoptosis and trigger cellular growth (Downward, 2003; Manning and Cantley, 2007). As a result, SGLT1 depletion induced apoptosis in TNBC cells and inhibited their growth in vitro and in vivo. Consistently, overexpression of SGLT1 protected renal epithelial cells (Ikari et al., 2005) and intestinal epithelial cells (Yu et al., 2008) from apoptosis.
Upon activation by its growth factor ligands, EGFR undergoes a transition from an inactive monomeric form to an active homodimer (Yarden and Schlessinger, 1987). EGFR dimerization stimulates its intrinsic intracellular protein–tyrosine kinase activity. As a result, autophosphorylation of several tyrosine residues in the C‐terminal domain of EGFR occurs (Downward et al., 1984; Lemmon et al., 2014). This autophosphorylation elicits downstream activation and signalling. For example, growth factor receptor‐bound protein 2 binds activated EGFR at phospho‐Tyr1068, which is crucial to the EGF‐induced activation of Ras signalling pathway (Rojas et al., 1996). It was demonstrated autophosphorylation of EGFR is taking place primarily by activated EGFR located at the cell membrane (Sousa et al., 2012). Downregulation of SGLT1 by RNAi resulted in the decreased level of phospho‐EGFRTyr1068, and as a result, the activity of downstream signalling pathways (such as AKT and ERK) was inhibited, suggesting that SGLT1 may facilitate the autophosphorylation of EGFR upon ligand binding. This result was also supported by significantly positive correlation between phospho‐EGFR and SGLT1 in TNBC samples.
In line with the previous report indicating that loss of EGFR protein but not its tyrosine kinase activity sensitized cancer cells to chemotherapeutic agent (Weihua et al., 2008), EGFR tyrosine kinase inhibitors did not produce therapeutic effects for certain cancers (Cohen et al., 2003; Dancey and Freidlin, 2003; Fukuoka et al., 2003). It has been highlighted EGFR tyrosine kinase inhibitors or monoclonal antibodies against TNBC showed low response rate, and the combination treatment with tyrosine kinase inhibitors or monoclonal antibodies and chemotherapy also showed low response rate and no benefits in the survival rate of patients (Baselga et al., 2013; Carey et al., 2012; Finn et al., 2009; Lee and Djamgoz, 2018; Schuler et al., 2010). Therefore, further works are needed to find out the best way to use EGFR targeted therapy in TNBC. EGFR–SGLT1 interaction stabilizes SGLT1 for the high uptake of glucose by cancerous cells and leads to the progression of cancer (Ren et al., 2013; Weihua et al., 2008). EGFR depletion (Weihua et al., 2008) or deletion of the SGLT1 interacting domain in EGFR promoted the downregulation of SGTL1 via the proteasome machinery (Ren et al., 2013). Together, targeting SGLT1 itself or EGFR–SGLT1 interaction might potentially provide novel therapeutics for TNBC patients.
5. Conclusion
In summary, this study shows that high levels of SGLT1 are associated with greater tumour size in TNBC. SGLT1 depletion compromises cell growth in vitro and in vivo. We further demonstrate that downregulation of SGLT1 results in decreased levels of phospho‐EGFR, and as a result, the activity of downstream signalling pathways (such as AKT and ERK) is inhibited. Hence, SGLT1 is required for the survival of TNBC via potentiation of EGFR activity.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
HL, AE and PP contributed equally in this work.
Supporting information
Fig. S1. TCGA analysis of SGLT1 expression levels in different molecular subtypes of breast invasive carcinoma samples (TCGA, Provisional).
Fig. S2. Knockdown of SGLT1 in TNBC cells via RNAi.
Fig. S3. SGLT1 and its interacting partners.
Fig. S4. SGLT1 positively regulates EGFR activity.
Acknowledgements
This project was supported by the National Natural Science Foundation of China [81772827], the Ministry of Science and Technology of China National Key Research and Development Projects [2016YFC0904701], the Academy of Medical Sciences/the Wellcome Trust Springboard Award [SBF002\1038] and the Wessex Medical Trust. HL was grateful for the scholarship provided by the Whale Education Foundation (Shenzhen, China). AE was supported by the Wessex Medical Trust. YZ was supported by the Natural Science Foundation of Hubei Province [No. 2018CFB611]. WC was supported by China Medical University Hospital [DMR‐108‐100]. JD was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001070), the UK Medical Research Council (FC001070) and the Wellcome Trust (FC001070).
References
- Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, Owens P, Sanders ME, Kuba MG, Sánchez V et al. (2014) Molecular profiling of the residual disease of triple‐negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov 4, 232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Albanell J, Ruiz A, Lluch A, Gascón P, Guillém V, González S, Sauleda S, Marimón I, Tabernero JM et al. (2005) Phase II and tumor pharmacodynamic study of gefitinib in patients with advanced breast cancer. J Clin Oncol 23, 5323–5333. [DOI] [PubMed] [Google Scholar]
- Baselga J, Gómez P, Greil R, Braga S, Climent MA, Wardley AM, Kaufman B, Stemmer SM, Pêgo A, Chan A et al. (2013) Randomized phase II study of the anti‐epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple‐negative breast cancer. J Clin Oncol 31, 2586–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blessing A, Xu L, Gao G, Bollu LR, Ren J, Li H, Wu X, Su F, Huang W‐C, Hung M‐C et al. (2012) Sodium/glucose co‐transporter 1 expression increases in human diseased prostate. J Cancer Sci Ther 4, 306–312. [Google Scholar]
- Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero‐Torres A et al. (2012) TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple‐negative breast cancer. J Clin Oncol 30, 2615–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casneuf VF, Fonteyne P, Van Damme N, Demetter P, Pauwels P, de Hemptinne B, De Vos M, Van de Wiele C and Peeters M (2008) Expression of SGLT1, Bcl‐2 and p53 in primary pancreatic cancer related to survival. Cancer Invest 26, 852–859. [DOI] [PubMed] [Google Scholar]
- Cohen EEW, Rosen F, Stadler WM, Recant W, Stenson K, Huo D and Vokes EE (2003) Phase II trial of ZD1839 in recurrent or metastatic squamous cell carcinoma of the head and neck. J Clin Oncol 21, 1980–1987. [DOI] [PubMed] [Google Scholar]
- Criscitiello C, Azim HA, Schouten PC, Linn SC and Sotiriou C (2012) Understanding the biology of triple‐negative breast cancer. Ann Oncol 23, vi13–vi18. [DOI] [PubMed] [Google Scholar]
- Crozier JA, Advani PP, LaPlant B, Hobday T, Jaslowski AJ, Moreno‐Aspita A and Perez EA (2016) N0436 (Alliance): a phase II trial of irinotecan plus cetuximab in patients with metastatic breast cancer previously exposed to anthracycline and/or taxane‐containing therapy. Clin Breast Cancer 16, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyprian FS, Akhtar S, Gatalica Z and Vranic S (2019) Targeted immunotherapy with a checkpoint inhibitor in combination with chemotherapy: A new clinical paradigm in the treatment of triple‐negative breast cancer. Bosn J Basic Med Sci 1–7. 10.17305/bjbms.2019.4204. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancey JE and Freidlin B (2003) Targeting epidermal growth factor receptor ‐ are we missing the mark? Lancet 362, 62–64. [DOI] [PubMed] [Google Scholar]
- Downward J (2003) Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3, 11–22. [DOI] [PubMed] [Google Scholar]
- Downward J, Parker P and Waterfield M (1984) Autophosphorylation sites on the epidermal growth factor receptor. Nature 311, 483–485. [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N et al. (2006) Minimizing the risk of reporting false positives in large‐scale RNAi screens. Nat Methods 3, 777–779. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C and Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127, 2893–2917. [DOI] [PubMed] [Google Scholar]
- Finn RS, Press MF, Dering J, Arbushites M, Koehler M, Oliva C, Williams LS and Di Leo A (2009) Estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2), and epidermal growth factor receptor expression and benefit from lapatinib in a randomized trial of paclitaxel with lapatinib or placebo as first‐line treatment in HER2‐negative or unknown metastatic breast cancer. J Clin Oncol 27, 3908–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudoh S, Rischin D et al. (2003) Multi‐institutional randomized phase II trial of gefitinib for previously treated patients with advanced non‐small‐cell lung cancer. J Clin Oncol 21, 2237–2246. [DOI] [PubMed] [Google Scholar]
- Garrido‐Castro AC, Lin NU and Polyak K (2019) Insights into molecular classifications of triple‐negative breast cancer: improving patient selection for treatment. Cancer Discov 9, 176–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo GF, Cai YC, Zhang B, Xu RH, Qiu HJ, Xia LP, Jiang WQ, Hu PL, Chen XX, Zhou FF et al (2011) Overexpression of SGLT1 and EGFR in colorectal cancer showing a correlation with the prognosis. Med Oncol 28, S197–S203. [DOI] [PubMed] [Google Scholar]
- Hanabata Y, Nakajima Y, Morita K, Kayamori K and Omura K (2012) Coexpression of SGLT1 and EGFR is associated with tumor differentiation in oral squamous cell carcinoma. Odontology 100, 156–163. [DOI] [PubMed] [Google Scholar]
- Hanahan D and Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT and Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ikari A, Nagatani Y, Tsukimoto M, Harada H, Miwa M and Takagi K (2005) Sodium‐dependent glucose transporter reduces peroxynitrite and cell injury caused by cisplatin in renal tubular epithelial cells. Biochim Biophys Acta Biomembr 1717, 109–117. [DOI] [PubMed] [Google Scholar]
- Khosravi‐Shahi P, Cabezón‐Gutiérrez L and Custodio‐Cabello S (2018) Metastatic triple negative breast cancer: optimizing treatment options, new and emerging targeted therapies. Asia Pac J Clin Oncol 14, 32–39. [DOI] [PubMed] [Google Scholar]
- Kocatürk B and Versteeg HH (2015) Orthotopic injection of breast cancer cells into the mammary fat pad of mice to study tumor growth. J Vis Exp 96, e51967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B, Xiao Y, Pu H, Cao Q, Jing H and Liu X (2012) Overexpression of SGLT1 is correlated with tumor development and poor prognosis of ovarian carcinoma. Arch Gynecol Obstet 285, 1455–1461. [DOI] [PubMed] [Google Scholar]
- Le Du F, Eckhardt BL, Lim B, Litton JK, Moulder S, Meric‐Bernstam F, Gonzalez‐Angulo AM and Ueno NT (2015) Is the future of personalized therapy in triple‐negative breast cancer based on molecular subtype? Oncotarget 6, 12890–12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A and Djamgoz MBA (2018) Triple negative breast cancer: emerging therapeutic modalities and novel combination therapies. Cancer Treat Rev 62, 110–122. [DOI] [PubMed] [Google Scholar]
- Lehmann BDB, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y and Pietenpol JA (2011) Identification of human triple‐negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121, 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J and Ferguson KM (2014) The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb Perspect Biol 6, a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD and Cantley LC (2007) AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholtz JM, Chalabi N, Radosevic‐Robin N, Dauplat MM, Mouret‐Reynier MA, Van Praagh I, Servent V, Jacquin JP, Benmammar KE, Kullab S et al. (2016) Multicentric neoadjuvant pilot Phase II study of cetuximab combined with docetaxel in operable triple negative breast cancer. Int J Cancer 138, 2274–2280. [DOI] [PubMed] [Google Scholar]
- Network TCGA (2012) Comprehensive molecular portraits of human breast tumors. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly EA, Gubbins L, Sharma S, Tully R, Guang MH, Weiner‐Gorzel K, McCaffrey J, Harrison M, Furlong F, Kell M et al (2015) The fate of chemoresistance in triple negative breast cancer (TNBC). BBA Clin 3, 257–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis‐Filho JS and Tutt ANJ (2008) Triple negative tumours: a critical review. Histopathology 52, 108–118. [DOI] [PubMed] [Google Scholar]
- Ren J, Bollu LR, Su F, Gao G, Xu L, Huang WC, Hung MC and Weihua Z (2013) EGFR‐SGLT1 interaction does not respond to EGFR modulators, but inhibition of SGLT1 sensitizes prostate cancer cells to EGFR tyrosine kinase inhibitors. Prostate 73, 1453–1461. [DOI] [PubMed] [Google Scholar]
- Rieg T and Vallon V (2018) Development of SGLT1 and SGLT2 inhibitors. Diabetologia 61, 2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M, Yao S and Lin YZ (1996) Controlling epidermal growth factor (EGF)‐stimulated ras activation in intact cells by a cell‐permeable peptide mimicking phosphorylated EGF receptor. J Biol Chem 271, 27456–27461. [DOI] [PubMed] [Google Scholar]
- Schuler M, Awada A, Harter P, Canon JL, Possinger K, Schmidt M, De Grève J, Neven P, Dirix L, Jonat W et al. (2012) A phase II trial to assess efficacy and safety of afatinib in extensively pretreated patients with HER2‐negative metastatic breast cancer. Breast Cancer Res Treat 134, 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MH, Uttenreuther‐Fisher MM, Piccart‐Gebhart MJ and Harbeck N (2010) BIBW 2992, a novel irreversible EGFR/HER1 and HER2 tyrosine kinase inhibitor, for the treatment of patients with HER2‐negative metastatic breast cancer after failure of no more than two prior chemotherapies. J Clin Oncol 28, 1065–1065. [Google Scholar]
- Shao F, Sun H and Deng C‐X (2017) Potential therapeutic targets of triple‐negative breast cancer based on its intrinsic subtype. Oncotarget 8, 73329–73344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa LP, Lax I, Shen H, Ferguson SM, Camilli PD and Schlessinger J (2012) Suppression of EGFR endocytosis by dynamin depletion reveals that EGFR signaling occurs primarily at the plasma membrane. Proc Natl Acad Sci USA 109, 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector NL, Xia W, Burris H, Hurwitz H, Dees EC, Dowlati A, O'Neil B, Overmoyer B, Marcom PK, Blackwell KL et al (2005) Study of the biologic effects of lapatinib, a reversible inhibitor of ErbB1 and ErbB2 tyrosine kinases, on tumor growth and survival pathways in patients with advanced malignancies. J Clin Oncol 23, 2502–2512. [DOI] [PubMed] [Google Scholar]
- Trédan O, Campone M, Jassem J, Vyzula R, Coudert B, Pacilio C, Prausova J, Hardy‐Bessard AC, Arance A, Mukhopadhyay P et al. (2015) Ixabepilone alone or with cetuximab as first‐line treatment for advanced/metastatic triple‐negative breast cancer. Clin Breast Cancer 15, 8–15. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bu F, Royer C, Serres S, Larkin JR, Soto MS, Sibson NR, Salter V, Fritzsche F, Turnquist C et al. (2014) ASPP2 controls epithelial plasticity and inhibits metastasis through β 2‐catenin‐dependent regulation of ZEB1. Nat Cell Biol 16, 1092–1104. [DOI] [PubMed] [Google Scholar]
- Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ and Hung MC (2008) Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell 13, 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E, Loo D and Hirayama B (2011) Biology of human sodium glucose transporters. Physiol Rev 91, 733–794. [DOI] [PubMed] [Google Scholar]
- Yarden Y and Schlessinger J (1987) Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry 26, 1443–1451. [DOI] [PubMed] [Google Scholar]
- Yu LCH, Huang CY, Kuo W‐T, Sayer H, Turner JR and Buret AG (2008) SGLT‐1‐mediated glucose uptake protects human intestinal epithelial cells against Giardia duodenalis‐induced apoptosis. Int J Parasitol 38, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. TCGA analysis of SGLT1 expression levels in different molecular subtypes of breast invasive carcinoma samples (TCGA, Provisional).
Fig. S2. Knockdown of SGLT1 in TNBC cells via RNAi.
Fig. S3. SGLT1 and its interacting partners.
Fig. S4. SGLT1 positively regulates EGFR activity.


