Abstract
Objective
MicroRNA-1271 (miR-1271) has a role in suppressing cell growth, cell cycle and promoting cell apoptosis in many cancers. This research was to explore the great role of miR-1271 in ovarian cancer (OC).
Patients and Methods
RT-qPCR was utilized to evaluate the mRNA levels of miR-1271 and its target gene. The proliferative and invasive abilities were measured using Cell Counting Kit-8 and transwell assays. The overall survival rate of OC patients was assessed by Kaplan–Meier method.
Results
miR-1271 was downregulated in OC tissues, and downregulation of miR-1271 predicted a poor outcome of the OC patients. Zinc finger E-box binding homeobox 1 (ZEB1) was a target gene of miR-1271 and its expression was regulated by miR-1271 in OC. The expression of miR-1271 had a negative connection with the expression of ZEB1 in OC tissues. miR-1271 inhibited cell viability and invasion-mediated epithelial–mesenchymal transition in SKOV3 cells. ZEB1 reversed partial roles of miR-1271 on viability and invasion in OC.
Conclusion
miR-1271 inhibited cell proliferation and invasion-mediated EMT in OC. The newly identified miR-1271/ZEB1 axis provides novel insight into the pathogenesis of OC.
Keywords: miR-1271, proliferation, invasion, epithelial–mesenchymal transition (EMT), ovarian cancer
Introduction
Ovarian cancer (OC), one of the most malignant gynecological cancers, is the most lethal cancer of female.1,2 Therefore, it is urgent to discover new biomarkers for the early diagnosis of OC.
MicroRNAs (miRNAs) could downregulate gene expression by targeting complementary mRNA at post-transcription.3 miRNAs has a huge effect on the growth and metastasis of cancers and are associated with the development and progression of tumor.4 Most reports found that multiple miRNAs including miR-145, miR-23a, miR-16 and miR-30a played an important role in OC.5–8 miR-1271 has been found to be a tumor suppressor in inhibiting the proliferation, migration and invasion and inducing cell apoptosis in hepatocellular carcinoma.9 miR-1271 was downregulated in multiple cancers including osteosarcoma, gastric cancer and non-small cell lung cancer.10–12 miR-1271 inhibited cell growth and reduced cell apoptosis via PDK1 in pancreatic cancer.13 Moreover, miR-1271 suppressed cell proliferation and induced apoptosis in endometrial cancer.14 However, there were few studies that indicated the roles of miR-1271 in OC; therefore, the experiments were carried out to explore the vital roles of miR-1271 in OC.
The zinc finger E-box binding homeobox 1 (ZEB1) encodes a zinc finger transcription factor that may play a role in transcriptional repression of interleukin 2.15 ZEB1 is required for neural differentiation of human embryonic stem cells.16 ZEB1 enhanced tumorigenesis and metastasis by regulating vimentin in hepatocellular carcinoma.17 Xavier et al18 indicated that ZEB1 acted as a potential therapeutic target that promoted the invasiveness and tumorigenicity of canine mammary cancer. Romero et al19 elucidated that was involved in the prognosis of gastric cancer. In our study, we discovered that mIR-1271 mediated the viability, invasion and epithelial–mesenchymal transition (EMT) by directly targeting ZEB1 in SKOV3 cells. ZEB1 reversed partial function of miR-1271 on the viability, invasion and EMT in OCcells.
Patients and methods
Clinical specimens
We screened 50 OC patients who were admitted in the Central Hospital of Shengli Oil Field from June 2016 to June 2018, and we obtained 50 pairs of OC tissues and peritumoral normal tissues. Specimens were immediately frozen in liquid nitrogen and then stored at −80°C after surgery. This study was approved by the Ethics Committee of Central Hospital of Shengli Oil Field. Signed written informed consents were obtained from all participants prior to the study. This study was conducted in accordance with the Declaration of Helsinki.
Cell culture
We obtained the normal ovarian cell line IOSE80 and two human OC cell lines SKOV3 and CAOV3 from American Type Culture Collection (Rockville, MD, USA). All the cells were incubated in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Sigma-Aldrich, Louis, MO, USA) at 37°C in 5% CO2 environmental state.
Transfection
The specific the miR-1271 mimic or the miR-1271 inhibitor and negative control plasmids were designed and synthesized from Gene-Pharma (Shanghai, China). SKOV3 cells used for transfection of the vectors were incubated in a 6-well plate. The transfection was performed using Lipofectamine 2000 Reagent (Invitrogen, USA) pursuant to the command of the manufacturer. The stable transfected cells were selected using Geneticin (G418; Thermo Scientific, Shanghai, China), while the transient transfected cells were harvest after transfected 48 hrs.
Quantitative real-time PCR
The total miRNAs were extracted utilizing the miRNeasy Mini Kit (Qiagen, Hilden, Germany) from tissues or cell lines. The TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) was used to synthesize the first cDNA chain. Subsequently, the qPCR was performed using the miRNA-specific TaqMan MiRNA Assay Kit (Applied Biosystems, Foster City, CA, USA). The U6 small nuclear RNA acted as the normalization and the relative levels of miRNA were calculated using 2−ΔΔCt method.
The total RNAs were extracted by using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), and the first cDNA chain was synthesized by Omniscript Reverse Transcription Kit (Qiagen, Hilden, Germany). The RT-qPCR was carried out by the QuantiTect SYBR Green PCR Kit (Qiagen, Hilden, Germany) in a Quantitect SYBR Green PCR System (Qiagen, Hilden, Germany). The quantification of mRNA was used by a 2−ΔΔCt method. The expression of ZEB1 was normalized by glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primers were ZEB1, forward: 5ʹ-TTCACAGTGGAGAGAAGCCA-3ʹ, reverse: 5ʹ-GCCTGGTGATGCTGAAAGAG-3ʹ; GAPDH forward: 5ʹ-TGGCAAAGTGGAGATTGTTG −3ʹ, reverse: 5ʹ-CTTCTGGGTGGCAGTGATG-3ʹ; miR-1271 forward: 5ʹ-CTAGACGTCCAGATTGAATAGAC-3ʹ, reverse: 5ʹ-GTCCGAGCTTGGTCAGAATG-3ʹ; U6 forward: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, reverse: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
Western blot
The total proteins were lysed using radioimmunoprecipitation assay Lysis Buffer (Sigma, St. Louis, MO, USA) containing 10% phenylmethylsulfonyl fluoride (Sigma, St. Louis, MO, USA). Equal proteins were separated by 10% SDS-PAGE and the blots were then electro-transferred the blots onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). After blocking in 5% fat-free milk for 1 hr at room temperature, the membranes were incubated with primary antibodies. The primary antibodies were against ZEB1 (1:1000; Abcam, Cambridge, CA, USA), E-cadherin (1:1000; Abcam, Cambridge, CA, USA) and N-cadherin (1:1000; Abcam, Cambridge, CA, USA). Subsequently, the blots were incubated in secondary horse radish peroxidase-conjugated antibody (Cell Signaling, Danvers, MA, USA) at room temperature for 2 hrs. The protein signals were captured using Enhanced Chemiluminescence Detection Kit (ECL, Pharmacia Biotech, Arlington, MA, USA).
Cell Counting Kit-8 (CCK-8) assay
The cell proliferation ability was measured by CCK-8 assay (Dojindo, Kumamoto, Japan). Briefly, SKOV3 cells that transfected with special vectors were incubated in 96-well plates and cultured for 24 hrs, 48 hrs, 72 hrs and 96 hrs at 37°C under a humidified atmosphere of 5% CO2, followed by addition of 10 μL CCK-8 solution into each well and incubated for 2 hrs. Finally, the absorbance was measured at 450 nm using a microplate reader (Dynex Technologies, Chantilly, VA, USA).
Transwell assay
The cell invasive ability was calculated by the transwell inserts (8-μm membrane, Corning, Cambridge, MA, USA), which were placed in 24-well plates and formed upper and lower two chambers. SKOV3 cell suspension suspended in serum-free medium was added to the upper chamber, whereas 600 μL medium containing 15% FBS was filled into the lower chamber. After the cells were incubated at 37°C for 24 hrs, the noninvaded cells were removed with cotton swabs, while the invaded cells were fixed with 4% paraformaldehyde and then stained with 10% crystal violet. Finally, the cell number was counted by a microscope (Olympus Corporation, Tokyo, Japan).
Luciferase reporter assay
TargetScan was used to predict the target genes of miR-1271 and we found that ZEB1 was a candidate gene. To validate whether miR-1271 directly binded to the 3ʹ-UTR of ZEB1 mRNA in OC cells, the binding sequences were mutated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). Luciferase reporter constructs harboring the 3ʹ-UTR of the ZEB1 gene spanning the binding sites for miR-1271 were generated downstream of Renilla luciferase in a psiCheck2 vector (Promega Corporation, Madison, WI). Subsequently, the wild type and the mutant the 3ʹ-UTR of ZEB1 mRNA were inserted into the pmirGlo dual luciferase reporter vectors, which were designated as WT or MUT. The SKOV3 cells were co-transfected with the miR-1271 mimic and the WT or MUT plasmid using Lipofectamine 3000 Reagent (Invitrogen, Carlsbad, CA, USA). The luciferase activity was then measured using a dual luciferase reporter assay system (Promega, Madison, WI, USA).
Statistical analysis
The statistical analysis was performed using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). The independent t-test was used for analyzing the measurement data. Differences between the two groups were analyzed by using the Student’s t-test. Comparisons between multiple groups were performed using a one-way ANOVA test followed by a post hoc test (least significant difference). Kaplan–Meier curve and log-rank test were utilized to assess the association between the expression of miR-1271 or ZEB1 and the overall survival for OC patients. P<0.05 was considered to be statistically significant.
Results
Downregulation of miR-1271 predicted poor prognosis of OC
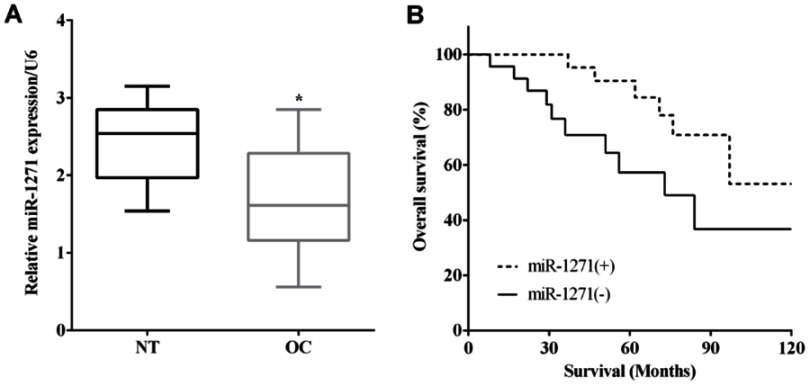
RT-qPCR was applied to evaluate the mRNA levels of miR-1271 in 48 pairs of OC and matched nontumor tissues. miR-1271 was expressed less in OC tissues versus corresponding nontumor tissues (P<0.05) (Figure 1A). Kaplan–Meier method indicated that the expression of miR-223 was associated with a worse outcome of OC patients (P<0.05) (Figure 1B).
Figure 1.
Downregulation of miR-1271 predicted poor prognosis of ovarian cancer. (A) miR-1271 was low expressed in ovarian cancer tissues. (B) Kaplan–Meier method indicated the expression of miR-223 was associated with worse outcome of ovarian cancer patients. *P<0.05.
miR-1271 suppressed cell viability and invasion in SKOV3 cells
The expression of miR-1271 was calculated in two OC cell lines (SKOV3 and CAOV3) and normal ovarian cell line IOSE80. The expression of miR-1271 was lower in SKOV3 (P<0.01) and CAOV3 (P<0.05) cells than that in HET-1A cells (Figure 2A). To explore the roles of miR-1271, SKOV3 cells were transfected with the miR-1271 mimic or the miR-1271 inhibitor to upregulate (P<0.05) or downregulate (P<0.05) miR-1271, and the transfection efficiency was measured by RT-qPCR (Figure 2B).
Figure 2.
miR-1271 suppressed cell viability and invasion in SKOV3 cells. (A) The expression of miR-1271 was lower in SKOV3 and CAOV3 cells than that in HET-1A cells. (B) The transfection efficiency of transfecting the miR-1271 mimic or the miR-1271 inhibitor was measured by RT-qPCR. (C) CCK-8 assay revealed miR-1271 inhibited the ability of viability in SKOV3 cells. (D) Transwell assay validated miR-1271 inhibited the invasive ability. *P<0.05, **P<0.01.
CCK-8 assay revealed that the ability of viability was inhibited by the miR-1271 mimic, whereas it was enhanced by miR-1271 inhibitor in SKOV3 cells (Figure 2C). Transwell assay validated that the invasive ability was suppressed by miR-1271 mimic (P<0.05) while it was improved by miR-1271 inhibitor (P<0.05) (Figure 2D). All the findings indicated miR-1271 suppressed the abilities of viability and invasion in SKOV3 cells.
miR-1271 directly binding to the 3ʹ-UTR of ZEB1 mRNA and regulated the expression of ZEB1
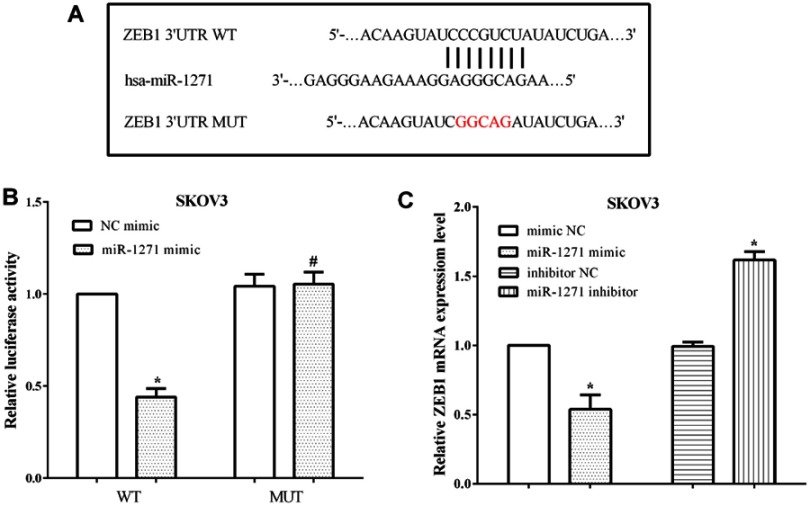
ZEB1 was predicted to be a target gene of miR-1271 by TargetScan, and the binding site was located at 1801–1807 on the 3ʹ-UTR of ZEB1 mRNA. The binding sequences were mutated from GUGCCAA to CACGGUU and then evaluated the luciferase activity (Figure 3A). The luciferase reporter assay proved that miR-1271 mimic reduced the luciferase activity of SKOV3 cells that transfected wild-type ZEB1 3ʹ-UTR (P<0.05); however, it makes no difference on the luciferase activity of cells transfected mutant ZEB1 3ʹ-UTR (P<0.05) (Figure 3B). Moreover, the mRNA levels of ZEB1 were assessed after transfection with the miR-1271 mimic or the miR-1271 inhibitor in SKOV3 cells. Not unexpectedly, overexpression of miR-1271 inhibited the mRNA level of ZEB1 (P<0.05), while knockdown of miR-1271 promoted the expression of ZEB1 in SKOV3 cells (P<0.05) (Figure 3C). All the results indicated that miR-1271 regulated the expression of ZEB1 in OC cells SKOV3.
Figure 3.
miR-1271 directly binded to the 3ʹ-UTR of ZEB1 mRNA and regulated the expression of ZEB1. (A) TargetScan predicted ZEB1 was a target of miR-1271. (B) The luciferase activity revealed miR-1271 targeted the 3ʹ-UTR of ZEB1 mRNA. (C) miR-1271 regulated the expression of ZEB1 in OC cells SKOV3. *P<0.05, #P>0.05.
The expression of ZEB1 in OC
The expression of ZEB1 in tissues and cell lines was calculated by RT-qPCR. As expected, the expression of ZEB1 in OC was higher than that in corresponding nontumor tissues (P<0.05) (Figure 4A). The correlation analysis validated that it had an inversed correlation between miR-1271 expression and ZEB1 in OC tissues (P<0.05) (Figure 4B). Similarly, the expression of ZEB1 was lower in OC cell lines SKOV3 (P<0.01) and CAOV3 (P<0.05) than normal ovarian cell IOSE80 cells (Figure 4C).
Figure 4.
The expression of ZEB1 in ovarian cancer. (A) The expression of ZEB1 in ovarian cancer was lower than that in nontumor tissues. (B) It had inversed correlation between the expression of miR-1271 and ZEB1 in ovarian cancer tissues. (C) The expression of ZEB1 was lower in ovarian cancer cells SKOV3 and CAOV3 than normal ovarian cell IOSE80 cells. *P<0.05, **P<0.01.
ZEB1 reversed partial function of miR-1271
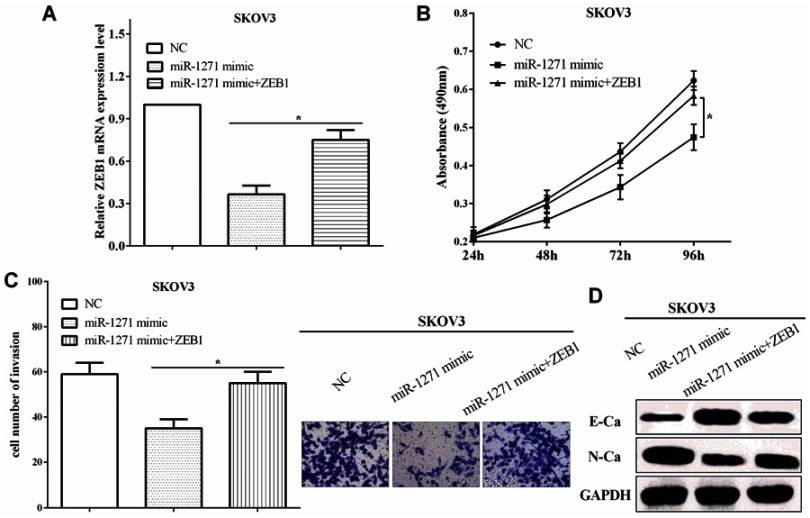
To explore the function of ZEB1 of miR-1271, the expression of ZEB1 was restored in miR-1271-transfected SKOV3 cells, and the transfection efficiency was assessed by RT-qPCR (P<0.05) (Figure 5A). CCK-8 assay indicated that ZEB1 re-expression enhanced the viability of miR-1271-transfected SKOV3 cells (P<0.05) (Figure 5B). Transwell assay validated that ZEB1 re-transfection increased the invasive ability in miR-1271 mimic-transfected SKOV3 cells (P<0.05) (Figure 5C), which suggests that ZEB1 reversed partial function of miR-1271. All the results demonstrate that miR-1271 inhibited the viability and invasion through directly binding to ZEB1. The expression of EMT associated proteins was calculated by Western blot in SKOV3 cells. miR-1271 mimic suppressed the expression of ZEB1 and E-cadherin, while improved the expression of N-cadherin in SKOV3 cells (Figure 5D), which suggested that miR-1271 inhibited the EMT ability through ZEB1.
Figure 5.
ZEB1 reversed partial function of miR-1271. (A) The expression of ZEB1 was restored in miR-1271-transfected SKOV3 cells. (B) CCK-8 assay indicated that re-expression of ZEB1 enhanced the viability of miR-1271-transfected SKOV3 cells. (C) Transwell assay validated that ZEB1 reversed partial function of miR-1271 on invasion. (D) ZEB1 reversed partial function of miR-1271 on the EMT. *P<0.05.
Discussion
OC is the most lethal malignant gynecological cancer with high morbidity and mortality rates.20,21 Therefore, it is urgent to explore the novel biomarkers for the early diagnosis and treatment of OC.
miRNAs play great roles on viability and invasion of tumors and are connected to the development and progression of tumor.22 miR-1271 has been reported to act as a tumor suppressor in breast cancer by suppressing cell proliferation via targeting SPIN1.23 miR-1271 was expressed less in hepatocellular carcinoma and it could be associated with oncogenic effects.24 Our findings were consistent with the entire findings, and we discovered that miR-1271 was downregulated in OC tissues and cell lines versus the normal tissues and cell lines. Downregulation of miR-1271 was associated with a worse outcome of OC patients. miR-1271 inhibited cell proliferation and invasion through Wnt signaling of osteosarcoma.25 miR-1271 functioned as a transition inhibitor that inhibited HCC cell metastasis and EMT.26 Previous research has been reported that miR-1271 inhibited cell growth via targeting CCNG1.27 We discovered that miR-1271 impaired the viability, invasion and EMT in OC cells, which were consistent with all the findings mentioned above.
ZEB1 acted as an oncogene and promoted tumorigenesis in several tumors including oral squamous cell carcinoma, glioma and pancreatic cancer.28–30 In hepatocarcinoma cells, ZEB1 regulated the resistance of Dox and mediated the mesenchymal characteristics.31 In glioma, ZEB1 acted as a transcription factor of Prognosis and Predict.32 Consistent with all the findings, ZEB1 was overexpressed in OC tissues and cell lines, and upregulation of ZEB1 predicted poor prognosis. ZEB1 played promoting functions in tumorigenic progression and epigenetic modifications.33 In addition, ZEB1 acted as a potential diagnostic marker in prostate cancer.34 ZEB1 was a target gene of miscellaneous miRNAs including miR-708, miR-205, miR-641 and miR-873.35–38 Consistent with the findings in pancreatic cancer,39 we revealed that miR-1271 mediated viability, invasion and EMT through directly targeting to the 3ʹ-UTR of ZEB1 mRNA in SKOV3 cells. In addition, ZEB1 reversed partial function of miR-1271 on viability, invasion and EMT in OC cells.
Conclusion
miR-1271 was downregulated and ZEB1 was upregulated in OC tissues and cell lines, and downregulation of miR-1271 or upregulation of ZEB1 predicted poor prognosis. miR-1271 targeted ZEB1 and mediated its expression in OC. miR-1271 suppressed cell viability, invasion and EMT in SKOV3 cells by directly binding to the 3ʹ-UTR of ZEB1 mRNA. ZEB1 reversed partial function of miR-1271 in OC cells.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. 2014;384(9951):1376–1388. doi: 10.1016/S0140-6736(13)62146-7 [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 3.Meister G. miRNAs get an early start on translational silencing. Cell. 2007;131(1):25–28. doi: 10.1016/j.cell.2007.09.021 [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- 5.Hua M, Qin Y, Sheng M, et al. miR145 suppresses ovarian cancer progression via modulation of cell growth and invasion by targeting CCND2 and E2F3. Mol Med Rep. 2019;19(5):3575–3583. doi: 10.3892/mmr.2019.10004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhuang RJ, Bai XX, Liu W. MicroRNA-23a depletion promotes apoptosis of ovarian cancer stem cell and inhibits cell migration by targeting DLG2. Cancer Biol Ther. 2019;20(6):897–911. doi: 10.1080/15384047.2019.1579960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li N, Yang L, Sun Y, Wu X. MicroRNA-16 inhibits migration and invasion via regulation of the Wnt/beta-catenin signaling pathway in ovarian cancer. Oncol Lett. 2019;17(3):2631–2638. doi: 10.3892/ol.2019.9923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Zhao S, Yu M. Mechanism of low expression of miR-30a-5p on epithelial-mesenchymal transition and metastasis in Ovarian cancer. Dna Cell Biol. 2019;38(4):341–351. doi: 10.1089/dna.2018.4396 [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Zhao ZX, Huang F, Yuan XW, Deng L, Tang D. MicroRNA-1271 functions as a potential tumor suppressor in hepatitis B virus-associated hepatocellular carcinoma through the AMPK signaling pathway by binding to CCNA1. J Cell Physiol. 2019;234(4):3555–3569. doi: 10.1002/jcp.26955 [DOI] [PubMed] [Google Scholar]
- 10.Lu G, Du L, Guo Y, Xing B, Lu J, Wei Y. Expression and role of microRNA-1271 in the pathogenesis of osteosarcoma. Exp Ther Med. 2018;15(2):1934–1940. doi: 10.3892/etm.2017.5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang XJ, Deng J, Liu YW, et al. MiR-1271 inhibits cell proliferation, invasion and EMT in gastric cancer by targeting FOXQ1. Cell Physiol Biochem. 2015;36(4):1382–1394. doi: 10.1159/000430304 [DOI] [PubMed] [Google Scholar]
- 12.Zhou Z, Niu X, Li C, Sheng S, Lu S. Inhibition of the growth of non-small cell lung cancer by miRNA-1271. Am J Transl Res. 2015;7(10):1917–1924. [PMC free article] [PubMed] [Google Scholar]
- 13.Xie F, Huang Q, Liu CH, et al. MiR-1271 negatively regulates AKT/MTOR signaling and promotes apoptosis via targeting PDK1 in pancreatic cancer. Eur Rev Med Pharmacol Sci. 2018;22(3):678–686. doi: 10.26355/eurrev_201802_14293 [DOI] [PubMed] [Google Scholar]
- 14.Li L, Qu YW, Li YP. Over-expression of miR-1271 inhibits endometrial cancer cells proliferation and induces cell apoptosis by targeting CDK1. Eur Rev Med Pharmacol Sci. 2017;21(12):2816–2822. [PubMed] [Google Scholar]
- 15.Wang J, Lee S, Teh CE, Bunting K, Ma L, Shannon MF. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int Immunol. 2009;21(3):227–235. doi: 10.1093/intimm/dxn143 [DOI] [PubMed] [Google Scholar]
- 16.Jiang Y, Yan L, Xia L, et al. Zinc finger E-box-binding homeobox 1 (ZEB1) is required for neural differentiation of human embryonic stem cells. J Biol Chem. 2018;293(50):19317–19329. doi: 10.1074/jbc.RA118.005498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Y, Yu J, Zhang M, Qin F, Lan X. ZEB1 promotes tumorigenesis and metastasis in hepatocellular carcinoma by regulating the expression of vimentin. Mol Med Rep. 2019;19(3):2297–2306. doi: 10.3892/mmr.2019.9866 [DOI] [PubMed] [Google Scholar]
- 18.Xavier P, Cordeiro YG, Rochetti AL, et al. ZEB1 and ZEB2 transcription factors are potential therapeutic targets of canine mammary cancer cells. Vet Comp Oncol. 2018;16(4):596–605. doi: 10.1111/vco.12427 [DOI] [PubMed] [Google Scholar]
- 19.Romero S, Musleh M, Bustamante M, et al. Polymorphisms in TWIST1 and ZEB1 are associated with prognosis of gastric cancer patients. Anticancer Res. 2018;38(7):3871–3877. doi: 10.21873/anticanres.12671 [DOI] [PubMed] [Google Scholar]
- 20.Rooth C. Ovarian cancer: risk factors, treatment and management. Br J Nurs. 2013;22(17):S23–S30. doi: 10.12968/bjon.2013.22.Sup17.S23 [DOI] [PubMed] [Google Scholar]
- 21.Bast RJ, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9(6):415–428. doi: 10.1038/nrc2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du HY, Liu B. MiR-1271 as a tumor suppressor in breast cancer proliferation and progression via targeting SPIN1. Eur Rev Med Pharmacol Sci. 2018;22(9):2697–2706. doi: 10.26355/eurrev_201805_14966 [DOI] [PubMed] [Google Scholar]
- 24.Maurel M, Jalvy S, Ladeiro Y, et al. A functional screening identifies five microRNAs controlling glypican-3: role of miR-1271 down-regulation in hepatocellular carcinoma. Hepatology. 2013;57(1):195–204. doi: 10.1002/hep.25994 [DOI] [PubMed] [Google Scholar]
- 25.Sun X, Zhai H, Chen X, Kong R, Zhang X. MicroRNA-1271 suppresses the proliferation and invasion of colorectal cancer cells by regulating metadherin/Wnt signaling. J Biochem Mol Toxicol. 2018;32:2. doi: 10.1002/jbt.22028 [DOI] [PubMed] [Google Scholar]
- 26.Li C, Jiang Y, Miao R, Qu K, Zhang J, Liu C. MicroRNA-1271 functions as a metastasis and epithelial-mesenchymal transition inhibitor in human HCC by targeting the PTP4A1/c-Src axis. Int J Oncol. 2018;52(2):536–546. doi: 10.3892/ijo.2017.4224 [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Ma L, Rao Q, et al. MiR-1271 inhibits ovarian cancer growth by targeting cyclin G1. Med Sci Monit. 2015;21:3152–3158. doi: 10.12659/msm.895562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren W, Gao L, Qiang C, et al. Kindlin-2-mediated upregulation of ZEB2 facilitates migration and invasion of oral squamous cell carcinoma in a miR-200b-dependent manner. Am J Transl Res. 2018;10(8):2529–2541. [PMC free article] [PubMed] [Google Scholar]
- 29.Liang C, Yang Y, Guan J, et al. LncRNA UCA1 sponges miR-204-5p to promote migration, invasion and epithelial-mesenchymal transition of glioma cells via upregulation of ZEB1. Pathol Res Pract. 2018;214(9):1474–1481. doi: 10.1016/j.prp.2018.07.036 [DOI] [PubMed] [Google Scholar]
- 30.Passacantilli I, Panzeri V, Bielli P, et al. Alternative polyadenylation of ZEB1 promotes its translation during genotoxic stress in pancreatic cancer cells. Cell Death Dis. 2017;8(11):e3168. doi: 10.1038/cddis.2017.562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long L, Xiang H, Liu J, Zhang Z, Sun L. ZEB1 mediates doxorubicin (Dox) resistance and mesenchymal characteristics of hepatocarcinoma cells. Exp Mol Pathol. 2019;106:116–122. doi: 10.1016/j.yexmp.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 32.Edwards LA, Kim S, Madany M, et al. ZEB1 is a transcription factor that is prognostic and predictive in diffuse gliomas. Front Neurol. 2018;9:1199. doi: 10.3389/fneur.2018.01199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Xu L, Li A, Han X. The roles of ZEB1 in tumorigenic progression and epigenetic modifications. Biomed Pharmacother. 2019;110:400–408. doi: 10.1016/j.biopha.2018.11.112 [DOI] [PubMed] [Google Scholar]
- 34.Farfan N, Ocarez N, Castellon EA, Mejia N, de Herreros AG, Contreras HR. The transcriptional factor ZEB1 represses Syndecan 1 expression in prostate cancer. Sci Rep. 2018;8(1):11467. doi: 10.1038/s41598-018-29829-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He J, Xiang D, Lin Y. MicroRNA708 inhibits the proliferation and invasion of osteosarcoma cells by directly targeting ZEB1. Mol Med Rep. 2019;19(5):3948–3954. doi: 10.3892/mmr.2019.10013 [DOI] [PubMed] [Google Scholar]
- 36.El BR, Tinelli S, Tortoreto M, et al. miR-205 enhances radiation sensitivity of prostate cancer cells by impairing DNA damage repair through PKCepsilon and ZEB1 inhibition. J Exp Clin Cancer Res. 2019;38(1):51. doi: 10.1186/s13046-019-1060-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao R, Zheng H, Wu L, Cai P. miRNA-641 inhibits the proliferation, migration, and invasion and induces apoptosis of cervical cancer cells by directly targeting ZEB1. Onco Targets Ther. 2018;11:8965–8976. doi: 10.2147/OTT.S190303 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Li G, Xu Y, Wang S, Yan W, Zhao Q, Guo J. MiR-873-5p inhibits cell migration, invasion and epithelial-mesenchymal transition in colorectal cancer via targeting ZEB1. Pathol Res Pract. 2019;215(1):34–39. doi: 10.1016/j.prp.2018.10.008 [DOI] [PubMed] [Google Scholar]
- 39.Liu H, Wang H, Liu X, Yu T. miR-1271 inhibits migration, invasion and epithelial-mesenchymal transition by targeting ZEB1 and TWIST1 in pancreatic cancer cells. Biochem Biophys Res Commun. 2016;472(2):346–352. doi: 10.1016/j.bbrc.2016.02.096 [DOI] [PubMed] [Google Scholar]