Abstract
Objective
Phosphatidylethanolamine (PtdEtn) is a major phospholipid in mammals. It is synthesized via two pathways, the CDP-ethanolamine pathway in the endoplasmic reticulum and the phosphatidylserine (PtdSer) decarboxylase (PSD) pathway in the mitochondria. While the CDP-ethanolamine pathway is considered the major route for PtdEtn synthesis in most mammalian tissues, little is known about the importance of the PSD pathway in vivo, especially in tissues enriched with mitochondria such as skeletal muscle. Therefore, we aimed to examine the role of the mitochondrial PSD pathway in regulating PtdEtn homeostasis in skeletal muscle in vivo.
Methods
To determine the functional significance of this pathway in skeletal muscle in vivo, an adeno-associated viral vector approach was employed to knockdown PSD expression in skeletal muscle of adult mice. Muscle lipid and metabolite profiling was performed using mass spectrometry.
Results
PSD knockdown disrupted muscle phospholipid homeostasis leading to an ∼25% reduction in PtdEtn and an ∼45% increase in PtdSer content. This was accompanied by the development of a severe myopathy, evident by a 40% loss in muscle mass as well as extensive myofiber damage as shown by increased DNA synthesis and central nucleation. In addition, PSD knockdown caused marked accumulation of abnormally appearing mitochondria that exhibited severely disrupted inner membrane integrity and reduced OXPHOS protein content.
Conclusions
The PSD pathway has a significant role in maintaining phospholipid homeostasis in adult skeletal muscle. Moreover, PSD is essential for maintenance of mitochondrial integrity and skeletal muscle mass.
Keywords: Phospholipids, Phosphatidylethanolamine, Phosphatidylserine, Mitochondria, Skeletal muscle, Atrophy
Abbreviations: PtdEtn, phosphatidylethanolamine; PtdSer, phosphatidylserine; PSD, phosphatidylserine decarboxylase
Highlights
-
•
The PSD pathway has an important role in regulating muscle phospholipid homeostasis.
-
•
Disrupting the PSD pathway caused marked muscle wasting and myofibre damage.
-
•
Knockdown of PSD caused accumulation of mitochondria with ultrastructural defects.
-
•
PSD is important in regulating mitochondrial integrity and skeletal muscle mass.
1. Introduction
Phosphatidylethanolamine (PtdEtn) is the second most abundant phospholipid in mammals, comprising 20–50% of total phospholipid content [1]. Compared with other organelles, PtdEtn is abundant in mitochondria accounting for ∼30% of mitochondrial phospholipid [1], [2]. In mammals, the majority of PtdEtn is synthesized by two spatially distinct pathways: 1) the CDP-ethanolamine branch of the Kennedy pathway, located in the endoplasmic reticulum; and 2) decarboxylation of phosphatidylserine (PtdSer) by the enzyme PtdSer decarboxylase (PSD), which is localized to the inner mitochondrial membrane [3], [4]. While the CDP-ethanolamine pathway is considered the major route for PtdEtn synthesis in mammals [5], [6], the PSD pathway is critically important. Indeed, the PSD pathway is not only required to maintain mitochondrial structure and function, it is essential for life [7]. Global elimination of PSD in mice leads to embryonic lethality and accumulation of morphologically abnormal mitochondria in the embryo [7]. Furthermore, silencing PSD expression in CHO cells caused severe mitochondrial ultrastructural defects, impaired respiratory capacity and reduced cell growth [2]. However, beyond these studies, there has been little progress in advancing the understanding of the role of the PSD pathway in mammals.
Recently, we made the observation that the PSD pathway could be an important site of PtdEtn synthesis in muscle [8]. This was based on two lines of evidence. Firstly, the molecular species composition of PtdEtn in muscle largely reflects that of PtdSer, the substrate for PSD; and secondly, the PtdEtn molecular species profile of purified muscle mitochondria is remarkably similar to that of whole muscle [8]. This appears to be unlike other tissues where most PtdEtn is produced by the CDP-ethanolamine pathway, such as the liver which exhibits distinct PtdEtn and PtdSer molecular species composition [9]. In addition, we have shown that when the CDP-ethanolamine pathway is eliminated from skeletal muscle in vivo, the PSD pathway compensates to maintain relatively normal PtdEtn content [8]. Interestingly, part of this compensatory mechanism in muscle lacking the CDP-ethanolamine pathway was the stimulation of mitochondrial biogenesis as an adaptive response to enhance the capacity for PtdEtn synthesis via PSD [8]. Together, this suggests that mitochondria may be an important site of PtdEtn synthesis in muscle.
Apart from these observations, little is known about the in vivo importance of the PSD pathway in muscle. Thus, our aim was to study the role of mitochondria in regulating PtdEtn homeostasis in skeletal muscle in vivo. To examine this, we used an adeno-associated viral (AAV) vector approach to knockdown PSD in muscle of adult mice. Based on our previous observations suggesting that the PSD pathway could be an important route of PtdEtn synthesis in muscle [8], we hypothesized that perturbing PSD activity would disrupt muscle phospholipid homeostasis, evident by an imbalance in PtdEtn and PtdSer content. Consistent with this, PSD knockdown caused a reduction in muscle PtdEtn content and accumulation of PtdSer. In addition, knockdown of PSD severely disrupted mitochondrial integrity, caused muscle damage as well as profound atrophy. Our findings demonstrate that the PSD pathway is an important site of PtdEtn synthesis in skeletal muscle and is required for maintenance of mitochondrial integrity and muscle mass. This reveals unique insight into the role of mitochondria in maintaining skeletal muscle homeostasis.
2. Materials and methods
2.1. Animals
All surgical and experimental procedures were approved by the Deakin University Animal Ethics Committee and were in accordance with the National Health and Medical Research Council of Australia's guidelines on animal experimentation. Male and female C57Bl/6J mice (8 weeks old at the start of study; sourced from an in-house colony) were communally housed (3–5 to a cage) in temperature controlled (22 ± 0.5 °C), 12 h light–dark cycle rooms (light cycle, 7:00–19:00). Mice had free access to water and a standard chow diet (Barastoc Rat & Mouse, Ridley AgriProducts, Australia).
2.2. Adeno-associated virus (AAV) administration
AAV serotype 6 vectors encoding a control scrambled short hairpin RNA sequence (Scrambled; catalogue number shAAV-268700) or a short hairpin targeting PSD (PSD shNRA; catalogue number 7043) under the control of a U6 promoter and expressing eGFP (driven by a CMV promoter) were obtained from Vector BioLabs (Malvern, PA, USA). Eight week old mice were anesthetized with isoflurane after which AAV vectors (5 × 1010 viral genomes in 30 μl of sterile saline) were injected locally into the TA muscle. The left TA muscle served as the control and was injected with the Scrambled AAV while the PSD shRNA AAV was injected into the right TA.
2.3. Tissue collection and processing
At the end of the experimental period (4, 8 or 16 weeks post AAV injection), mice were weighed and anesthetized with isoflurane. TA muscles were dissected, weighed, frozen in liquid nitrogen, or prepared for histological analysis including electron microscopy studies. With the exception of the targeted quantitative metabolite profiling, all assays were undertaken using freeze dried muscle. The entire excised TA muscle was freeze dried at −60 °C under constant vacuum (100 mTorr) for 24 h. Freeze dried muscles were dissected free of blood, connective tissue and visible fat, and powdered for subsequent analysis. For the quantitative metabolomics analysis, TA muscles were dissected from anesthetized mice and were rapidly frozen in liquid nitrogen for subsequent metabolite extraction on the entire wet TA muscle.
2.4. Western blotting
TA muscle was lysed, and protein (30 μg) was resolved by SDS-PAGE (Bio-Rad Laboratories). After transfer to PVDF membranes, stain-free images for total proteome visualization were collected, and membranes were blocked in 5% BSA. After incubation with primary antibody (see details supplied in Supplementary Table 1), appropriate secondary antibodies were applied, and the immunoreactive proteins were detected with enhanced chemiluminescence and quantified by densitometry (Li-Cor Image Studio). The immunoreactive signal was normalized to the density of the total protein loading for each sample, which was obtained by visualization of the stain-free blot image. The mean normalized intensity for the scrambled condition was determined and the intensity of every sample was made relative to this value. As such, the scrambled shRNA control condition has a mean-normalized reference value of 1 (±error). Changes caused by PSD knockdown are scaled relative to this scrambled control reference value.
2.5. RNA extraction and gene expression analysis
Total RNA was extracted from ∼2 mg of powdered freeze-dried TA muscle tissue using Tri-Reagent (Ambion Inc, Austin, TX, USA). Samples were treated with DNAse I (Life Technologies, Mulgrave, Victoria, Australia) to remove any contaminating genomic DNA as recommended by the manufacturer. cDNA was synthesized using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) from half a microgram of RNA followed by treatment with RNase H to remove remaining RNA as per the manufacturer's instructions. Semi-quantitative polymerase chain reaction (qPCR) was performed on cDNA diluted 1:20 using the Mx3000 PCR system (Stratagene, La Jolla, CA, USA) with SYBR Green Master Mix (Applied Biosystems) and 300 nM primers. Primer sequences and gene accession numbers are listed in Supplementary Table 2. The qPCR cycling conditions were: 95 °C for 10 min (1 cycle), 30 s at 95 °C and 60 °C for one min (40 cycles). The mRNA expression was calculated using the 2−ΔCt formula following normalization to cDNA input as determined by the Quant-iT™ OliGreen™ ssDNA Assay Kit (Thermo Fisher Scientific, Scoresby, Victoria, Australia). Data were then normalized to mean of the scrambled condition.
2.6. Lipid analysis
For total phospholipid analysis, lipids were extracted from an aliquot of powdered muscle (2–3 mg) according to the methods of Folch [10]. Phospholipid (PtDEtn, PtdSer and PtdCho) content was determined by HPLC (Agilent Technologies 1100 series, Santa Clara, CA, USA) using a mobile phase consisting of acetonitrile:methanol:85% phosphoric acid (90:3:1 v/v/v) as previously described [11]. A standard curve for each phospholipid was generated from commercially available standards (Sigma Chemical, St. Louis, MO, USA). The dried lipid extracts were re-suspended in 100 μL of mobile phase:chloroform (80:20 v/v) and 20 μL was injected for HPLC analysis using an isocratic elution. The mobile phase flow rate was 0.8 mL/min and a 3.9 × 150 mm μPorasil silica column was used (10 μm particle size; Waters Corporation). Phospholipid retention was detected by a UV detector at a wavelength of 203 nm. Data integration and analysis was undertaken using Openlab CDS software (Agilent Technologies).
PtdEtn, PtdSer, and cardiolipin molecular species composition analysis was performed by liquid chromatography, electrospray ionisation-tandem mass spectrometry (LC ESI-MS/MS). Lipids were extracted from an aliquot of powdered freeze dried (1–2 mg) muscle using the Folch extraction [10]. At the time of extraction, samples were spiked with stable isotope labeled internal standards (15:0–18:1 (d7) PtdEtn; 15:0–18:1 (d7) PtdSer; 15:0 (3)–16:1 cardiolipin; Avanti Polar Lipids, AL). Dried extracts were resuspended in 50 μL mobile phase A (100% water with 10 mM Ammonium Formate) and 50 μL mobile phase B (isopropanol:acetonitrile:water; 75:20:5 v/v/v with 10 mM ammonium formate) for subsequent analysis. Mass spectrometry was performed with a Shimadzu 8040 LC/MS/MS triple quadrupole with electrospray ionization (Shimadzu, Melbourne, Australia). Samples (1 μl) were injected onto an SDB column (100 mm × 0.25 mM, 1.8 μm pore size; Agilent), and lipids were eluted using a mobile phase gradient at a flow rate of 0.4 mL/min. The gradient started at 50% mobile phase B and linearly increased to 100% B over 15 min which was then held for 5 min. The gradient was then adjusted back to starting conditions and held for 5 min. Positive ion neutral loss scanning was used to identify PtdEtn (neutral loss of 141 m/z), PtdSer (neutral loss of 185 m/z) and cardiolipin (neutral loss of 867.7 m/z) lipid species. From the identified lipid species, a targeted multiple reaction monitoring (MRM) method was used to analyze each lipid species. Peak areas were determined using LabSolutions software (Shimadzu). The concentration of individual lipid species was determined by relating the integrated peak areas to the appropriate internal standards.
2.7. Histological analysis
TA muscles were frozen in thawing isopentane and stored at −80 °C. Transverse sections (10 μm) were cryosectioned and stained with hematoxylin and eosin (H&E). Images were taken with an Olympus Provis AX70 microscope and Olympus DP70 digital camera (Olympus, Tokyo, Japan). H&E stained sections were used to determine Feret's minimal diameter for all fibers within the field (×40 magnification) from 10 sections per muscle so that on average the diameter was determined on >400 fibers per section. Cross-sections were also stained for oxidative enzyme activity including succinate dehydrogenase, cytochrome oxidase and NADH-tetrazolium reductase as previously detailed [12]. To determine myofiber number and centrally located nuclei, cross sections were stained for laminin and DAPI. Whole mount images were acquired on a Dotslide with BX51 microscope (Olympus, Tokyo, Japan). Image analysis was performed using ImageJ. Immunofluorescence analysis of myosin heavy chain (MHC) isoform expression in the TA muscle was performed according to Bloemberg and Quadrilatero [13]. Briefly, 8 μm sections of TA muscle were affixed to StarFrost Silane coated Slides (ProScitech, Kirwan, Queensland, Australia) and frozen at −80 °C until staining. On the day of staining, slides were thawed at room temperature for 10 min and were then blocked in 10% goat serum in PBS for 1 h at room temperature. Slides were then incubated in a cocktail of primary antibodies for 2 h at room temperature (BA-F8 (1:20; MHCI), SC-71 (1:50; MHCIIa) and BF–F3 (1:20; MHCIIb), Developmental Studies Hybridoma Bank, University of Iowa, Iowa, IA, USA; laminin (1:100) (L9393), Sigma Aldrich). After washing (3 × 5 min in PBS), sections were exposed to fluorescent conjugated secondary antibodies (Alexa Fluor goat anti mouse IgG2b 647, Alexa Fluor goat anti mouse IgG1 488, Alexa Fluor goat anti mouse IgM 555, Alexa Fluor goat anti rabbit IgG H + L 405, all 1:500 (Life Technologies)) diluted in 10% goat serum in PBS for 1 h at room temperature in the dark. Following an additional 3 washes (5 min in PBS), sections were mounted in Prolong Gold (Life Technologies, Australia) and cover-slipped. Slides were visualized using an Olympus Fluoview FV10i confocal microscope (Olympus, Australia). All images were taken using the same laser and sensitivity settings. Images of the entire muscle were taken at 10× magnification (7–12 images total stitched). Using this approach, any unstained fibers were classified as expressing MHCIIx. Individual fiber CSA, number of fibers per cross section and fiber type analysis were determined using SMASH software (MATLAB, University of Pennsylvania, Philadelphia, PA, USA).
2.8. Electron microscopy
TA muscles were dissected into 1 mm × 1 mm pieces and fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) at room temperature for 2 h and postfixed in 2% osmium tetroxide solution for 1 h. Samples were dehydrated in a graded series of ethanol and embedded in epoxy resin before undergoing thin sectioning. The sections were stained with uranyl acetate and lead citrate and viewed on a Hitachi H7500 TEM.
2.9. Heavy water (2H2O) labeling
Heavy water labeling studies were performed on mice studied 4 and 8 weeks after AAV administration. Mice were administered with 2H2O (25 ml/kg i.p. 99% enriched) and were maintained on 8% 2H2O in the drinking water for the specified labeling duration (i.e. between 1 and 4 week). Blood samples were obtained at weekly intervals. Plasma was obtained by centrifugation (3,000 rpm for 5 min at 4 °C) and was used to determine [2H] enrichment of total body water by GC–MS (Agilent 6890N GC coupled to an Agilent 5975C MS) using the acetone exchange method [14].
2.10. Mitochondrial protein synthesis
At the end of the 2 week heavy water labeling period and at 8 weeks after AAV administration, TA muscles were collected and mitochondria were isolated according to previously described methods [8], [15]. Protein was hydrolyzed by addition of 500 μl of 6 N HCl followed by overnight incubation at 110 °C. Samples were ion-exchanged, dried and resuspended in 100 μL of pyridine and were derivatized by the addition of 100 μL MTBSTFA containing 1% TBDMCS. After incubation at 60 °C for 30 min, 2H incorporation into protein-bound alanine was determined by electron ionization (EI) GC–MS using helium as a carrier gas. The deuterium enrichment of the tBDMS derivative of alanine was measured by monitoring the major fragment ion [M-57], 260–262 m/z using selective ion monitoring. Data were corrected for natural isotopic background abundance by using unlabeled biological samples and chemical standards. The fraction of newly synthesized mitochondrial protein was calculated by Equation 1:
| (1) |
where EAla represents the fraction of 2H-labeling (in excess of natural background) in the total protein-bound alanine pool; “n” represents the maximum number of exchangeable carbon bound hydrogens in alanine (3.7 in mammals) [16]; and EBW represents fraction of body water labeling.
2.11. Mixed muscle protein synthesis
Muscle protein synthesis was determined in mice 4 and 8 weeks after AAV administration following either 1 or 4 weeks of heavy water labeling. Protein was hydrolyzed from freeze dried TA muscle (2–3 mg) by the addition of 500 μl of 6 N HCl containing 300 μM U–13C, D4, 15N alanine labeled internal standard followed by overnight incubation at 110 °C. Following acid hydrolysis, samples were evaporated in a speed vacuum (Labconco, Kansas, MO, USA). The dried sample was resuspended in 2:1 chloroform:methanol (v/v) followed by addition of 1 part water to separate the hydrolyzed alanine into the upper aqueous phase from the non-polar metabolites. The aqueous phase was collected and dried in a speed vacuum. Sample derivatization and mass spectrometry were performed as described above in Mitochondrial protein synthesis with the addition of monitoring the internal standard ion (268 m/z). The fraction of newly synthesized protein was calculated from Equation 1 (above). The protein-bound alanine concentration (i.e. protein pool size) in the sample was determined using the isotope dilution technique following correction for natural isotopic background abundance. The absolute amount of newly synthesized protein was then determined by Equation 2:
| (2) |
where fraction newly synthesized is derived from Equation 1; protein-bound alanine is the alanine concentration (nmol/mg); and TA mass is the mass of the entire TA muscle (mg).
2.12. DNA synthesis
DNA synthesis was determined in TA muscle from mice studied 4 and 8 weeks after AAV administration following either 1 or 4 weeks of heavy water labeling. DNA was extracted from 2–3 mg of freeze dried powdered TA muscle by addition of 250 μl of proteinase K tissue lysis solution (0.2 mg/ml) and overnight incubation at 55 °C. The proteinase K solution was made from a 1 mg/mL stock of proteinase K (Sigma) mixed with VIAGEN Direct PCR Tail Lysis Reagent (1:4 v/v proteinase K: lysis buffer). DNA was precipitated by the addition of 250 μl isopropanol and centrifugation (20,000 ×g; 5 min). The supernatant was carefully discarded, the DNA pellet washed with 70% v/v ethanol (500 μL, 2 min) and the supernatant removed after centrifugation at 20,000 ×g for 5 min. The DNA pellet was left to dry for 15 min, then resuspended in H2O (200 μL) and was incubated at 55 °C for 10 min followed by vortex mixing. The resuspended DNA samples were hydrolyzed and dephosphorylated by the addition of S1 nuclease and potato acid phosphatase as previously described [17]. After hydrolysis, the deoxyribose moiety was derivatized in a two-step reaction as we have previously detailed [17]. The perfluorotriacetyl derivative of DNA-bound deoxyribose was analyzed by GC–MS in the negative chemical ionization (NCI) mode (Agilent 7890B GC system and an Agilent 5977B MSD) with helium as the carrier and methane as the reagent gas. The perfluorotriacetyl derivative of deoxyribose was analyzed by monitoring the 435 mz (M0), 436 mz (M1) and 437 mz (M2) ions in SIM mode. The fraction of newly synthesized DNA was calculated by Equation 3:
| (3) |
where EDeoxyribose represents the fraction of 2H-labeling in DNA-bound deoxyribose in excess of natural isotopic background abundance; “n” represents the maximum number of exchangeable carbon bound hydrogens in deoxyribose (5.7 in mice) [18]; and EBW represents fraction of body water labeling. Note, data are not presented from mice studied 4 weeks after AAV administration with 1 week of heavy water labeling as deuterium incorporation into DNA was not detected. This is likely due to the fact that there was no evidence of muscle damage/regeneration at this point and is consistent with the fact that muscle exhibits slow cell turnover, thus the lack of deuterium incorporation over this short labeling period is expected.
2.13. Enzyme activity
TA muscle (5–10 mg) was homogenized in 1:50 dilution (w/v) of a 175 mM potassium buffer solution, and citrate synthase activity was assayed spectrophotometrically at 37 °C [19]. β-HAD activity was assayed spectrophotometrically at 25 °C, measuring the disappearance of NADH [20]
2.14. Respiratory capacity
Respiration analyses were performed as previously described [8], [21]. Briefly, TA muscles were dissected and the belly of the muscle was biopsied cross-sectionally using a 1 mm punch biopsy needle, so that biopsy margins disrupted the sarcolemma of the myofibers within the biopsy to allow for substrate diffusion. The muscle biopsy was placed in a Seahorse Islet capture plate in αMEM media containing 5 mM glucose. Baseline respiration was measured using 2 cycles of 2 min mix, 2 min wait and 2 min measure, before injection of malate and glutamate (to assess Complex I supported respiration) or succinate (to assess Complex II supported respiration) at final concentrations of 5 mM, 1 mM and 5 mM respectively. Respiration was measured for two 2 min periods, separated by a 2 min mix period. This was followed by injection of ADP (pH7.4) at a final concentration of 2.5 mM and two more measurement cycles. The peak oxygen consumption rate throughout each measurement period was recorded in point-to-point mode. Following the assay, Islet capture plate screens were removed and the biopsy sample retrieved for determination of total protein. Biopsies were homogenized in 100 μL of lysis buffer, and protein concentration was determined using the BCA assay. All respiration values were normalized to total protein contained within each sample and state 3 respiration was established as the peak respiration values obtained following ADP injection [21].
2.15. Targeted quantitative metabolite profiling
For targeted metabolite profiling, TA muscles were dissected from anesthetized mice and muscles were immediately flash frozen in liquid nitrogen. The entire (wet) TA muscle was transferred into a ceramic bead containing cryomill tube (Precellys; Bertin Technologies, France) and 1.2 ml of 3:1 methanol:water (v/v) containing stable isotope labeled internal standards was added. Tubes containing no tissue (extraction blanks) also underwent the entire extraction process to assess any background contamination. The internal standards (Cambridge Isotope Laboratory) included U-13C glucose (2.6 nmol), U-13C glucose-6-phosphate (10 nmol), U-13C fructose-6-phosphate (10 nmol), 2,2,4,4-D4 citric acid (3.2 nmol), D4 succinic acid (3.3 nmol), 2,3,3-D3 malic acid (3.4 nmol), D4 fumaric acid (3.1 nmol), a U-13C, U-15N amino acid mix (3.1 nmol each; contains all primary amino acids except for glutamine, asparagine and tryptophan) and 13C5 glutamine (3.1 nmol). Samples were extracted at −15 °C using a Precellys bead-mill with a Cryolys attachment (Bertin Technologies, France). The homogenate was left to stand on ice for 10 min after which it was spun in a centrifuge at 15,000 rpm for 10 min at 0 °C. The supernatant was transferred to a new tube, 600 μL chloroform and 300 μL water added. The extract was centrifuged at 6,000 rpm for 5 min at 0 °C. For determination of free amino acids and TCA cycle metabolite concentrations, the upper aqueous phase (200 μL) was transferred into a fresh 250 μL glass insert (Agilent) in a 2 ml glass GC vial and was dried in a speed vacuum at 37 °C. This step was repeated two more times (i.e. 600 μL aqueous phase dried in total). Dried samples were derivatized by addition of 25 μL pyridine and 25 μL MTBSTFA + 1% TBDMCS (Sigma) followed by incubation at 60 °C for 30 min, forming TBDMS derivatized molecules for determination of free amino acids and TCA cycle metabolites by EI GC–MS (Agilent). For free glucose, glucose-6-phosphate and fructose-6-phosphate concentrations, the 3 × 200 μL dry down of the remaining aqueous phase was repeated in fresh glass inserts. Dried samples were derivatized by addition of 25 μL of methoxyamine hydrochloride (Sigma) in pyridine (20 mg/mL; Sigma) followed by overnight incubation at room temperature. The following day, 25 μL of BSTFA + 1% TMCS (Thermo Fisher Scientific, Waltham, USA) was added prior to incubation at 60 °C for 30 min. Metabolites were analyzed using EI GC–MS (Agilent). Details of the ions monitored can be provided upon request and are based off the NIST library. Chromatograph peaks were integrated using the Quantitative Mass Hunter Workstation (Agilent). Absolute metabolite concentrations (nmol/mg wet weight) were calculated using the internal standard isotope dilution technique.
2.16. Statistical analysis
All data are presented as mean ± SEM. Data were analyzed by paired t-test or factorial ANOVA where appropriate. For the ANOVA procedures, Tukey's post hoc tests were used to establish differences between groups. Statistical significance was set at P < 0.05. Exact N values as well as the statistical test used are detailed in the figure legends.
3. Results and discussion
3.1. PSD knockdown disrupts muscle phospholipid homeostasis
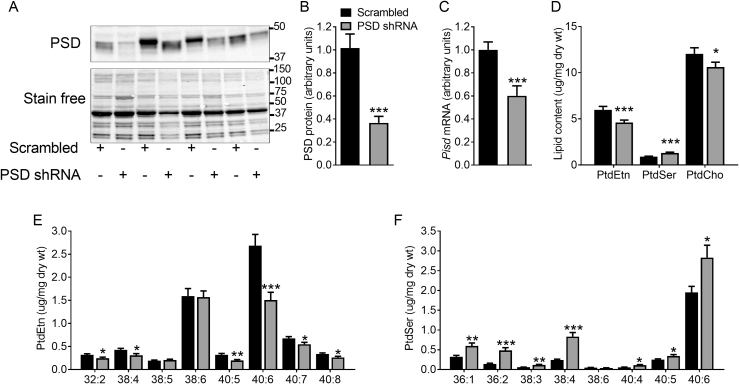
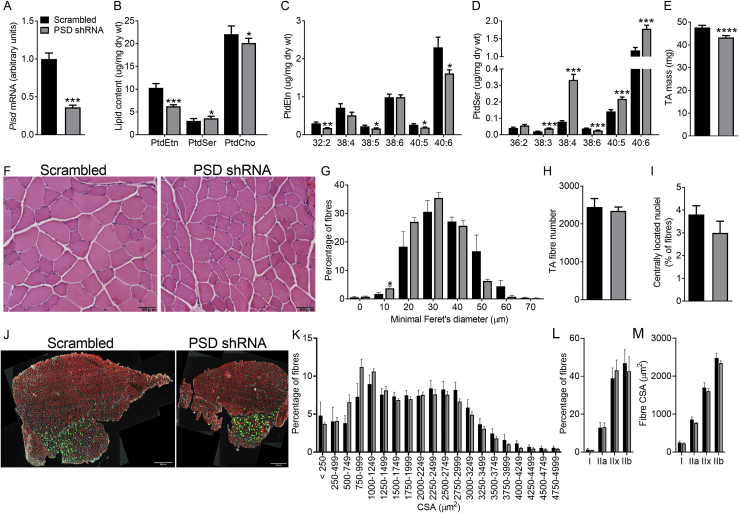
By combining the use of AAV vectors with shRNA, we achieved ∼60% knockdown of PSD protein and ∼40% reduction in Pisd gene expression (which encodes PSD) in the tibialis anterior (TA) muscle of adult mice 8 weeks after AAV injection (Figure 1A–C). This disrupted phospholipid homeostasis, evident by a ∼25% reduction in PtdEtn and a ∼45% increase in PtdSer content (Figure 1D). PSD knockdown also caused a modest reduction in phosphatidylcholine (PtdCho; Figure 1D). As a result, there was an imbalance in the major phospholipids with both the PtdCho:PtdEtn (2.0 ± 0.1 vs. 2.4 ± 0.1 for scrambled vs. PSD shRNA; N = 16; P < 0.01) and PtdEtn:PtdSer ratio (6.6 ± 0.3 vs. 3.6 ± 0.3 for scrambled vs. PSD shRNA; N = 16; P < 0.001) being significantly altered. The reduction in total PtdEtn content was largely mediated by the decrease in (40:6)PtdEtn, the most abundant PtdEtn species in muscle (Figure 1E). Conversely, PSD knockdown resulted in a significant increase in (40:6)PtdSer, the predominant PtdSer species in muscle (Figure 1F). In addition, (38:4)PtdEtn and (40:5)PtdEtn were reduced while there were reciprocal increases in the corresponding PtdSer species. This appears to be consistent with stable isotope labeling studies demonstrating that PSD preferentially produces PtdEtn species containing long-chain polyunsaturated fatty acids [5].
Figure 1.
PSD knockdown disrupts muscle phospholipid homeostasis. AAV vectors were injected in the tibialis anterior (TA) of 8 week old mice. Studies were conducted 8 weeks after AAV administration (i.e. at 16 weeks of age). A: Representative Western blot of PSD protein levels. B: Quantification of PSD protein levels (N = 9 males). C: Pisd mRNA expression (N = 12; 6 female, 6 male). D: Muscle phosphatidylethanolamine (PtdEtn), phosphatidylserine (PtdSer) and phosphatidylcholine (PtdCho) content (N = 16; 7 female, 9 male). D: PtdEtn molecular species (N = 12; 6 female, 6 male). E: PtdSer molecular species (N = 12; 6 female, 6 male). Data are mean ± SEM. Data were analyzed using paired t-tests. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
3.2. PSD knockdown causes myofiber damage and muscle atrophy
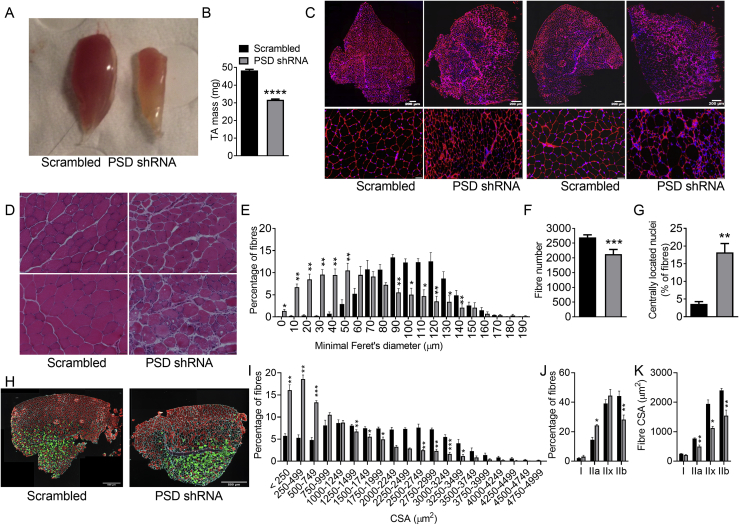
Upon gross visual inspection, it was apparent that 8 weeks after PSD knockdown there was a marked effect on muscle size (Figure 2A). Indeed, PSD-deficient TA muscles weighed ∼35% less than the contralateral TA muscles injected with the scrambled shRNA (Figure 2B). The effect of muscle mass was still evident after the TA muscles had been freeze dried (12.6 ± 0.8 vs. 8.0 ± 0.5 mg dry wet for scrambled vs. PSD shRNA; N = 8; P < 0.001), demonstrating that the difference in mass was not attributable to effects on tissue water content. The effect of PSD knockdown on TA mass affected females (40.9 ± 0.9 vs. 27.9 ± 0.8 mg for scrambled vs. PSD shRNA; N = 58; P < 0.001) and males similarly (52.1 ± 0.5 vs. 33.5 ± 0.5 mg for scrambled vs. PSD shRNA; N = 129; P < 0.001). Histological analysis (Figure 2C–F) revealed the loss of muscle mass was due to both a reduction in fiber size, evident by a left shift in both the minimum Feret's diameter distribution (Figure 2E) and the fiber cross sectional area distribution (Figure 2I), as well as a decrement in fiber number (Figure 2F). PSD-deficient muscle also exhibited greater variability in fiber size, as shown by an increase in the variance coefficient of myofiber diameter (254 ± 15 vs. 535 ± 36 for scrambled vs. PSD shRNA; N = 5; P < 0.01). Additionally, PSD-deficient muscle displayed histological changes consistent with signs of necrosis and muscle damage/regeneration, including increased centrally nucleated fibers (Figure 2G), fibers appearing more rounded in shape, fiber splitting, the presence of cores or vacuoles, pale hematoxylin and eosin (H&E) staining in some fibers, an increase in the thickness of the endomysium and cellular infiltration (Figure 2C,D). Consistent with the histological evidence of increased cellular infiltration, the expression of the pro-inflammatory markers Il1b and Tnfa were elevated in PSD-deficient muscle (Figure 3A). PSD knockdown also resulted in a modest shift in myosin heavy chain (MHC) isoform expression with a reduction in the number of MHCIIb fibers, and an increase in MHCIIa-expressing fibers (Figure 2H,J). In addition, PSD-deficient muscle exhibited a >5-fold increase the mRNA expression Myh3 (Figure 3A), an embryonic isoform of MHC which is re-expressed during muscle regeneration [22]. While the exact cause of damage is not known, we speculate that the alterations in phospholipid content and composition caused by PSD knockdown may have altered the mechanical properties of the sarcolemma thereby making PSD-deficient muscle more prone to contraction-induced damage and subsequent necrosis. From a functional perspective, it would be of interest to determine whether these changes in muscle mass and morphology caused by disrupting the PSD pathway translate to altered contractile function and force generation capacity. One would speculate that with such profound ultrastructural changes, contractile function would be impaired in PSD-deficient muscle. The lack of muscle function data are therefore a limitation of the current study but an area that warrants future investigation.
Figure 2.
PSD knockdown causes muscle atrophy and myofiber damage. AAV vectors were injected in the tibialis anterior (TA) of 8 week old mice. Studies were conducted 8 weeks after AAV administration (i.e. at 16 weeks of age). A: Representative gross appearance of tibialis anterior (TA) muscle 8 weeks after AAV administration. B: TA muscle mass (N = 187; 58 female, 129 male). C: Representative immunofluorescence images of TA muscle cross sections stained with laminin (red) and DAPI (blue). Scale bar is 200 μm for upper panel and 50 μm for the lower panel. D: Representative H&E staining of TA muscle. Scale bar is 50 μm. E: Feret's minimal diameter distribution (N = 4 male). F: Total fiber number (N = 9; 2 female, 7 males). G: Percentage of fibers with centrally located nuclei (N = 5 males). H: Representative image of TA muscle showing laminin (white), myosin heavy chain expression (I, blue; IIa, green; IIx black; IIb red). Scale bar is 500 μm. I: Fiber cross sectional area distribution (N = 4; 2 female, 2 male). J: Quantitative analysis of fiber type distribution (N = 4; 2 female, 2 male). K: Quantitative analysis of fiber type specific cross sectional area (N = 4; 2 female, 2 male). Data are mean ± SEM. Data were analyzed using paired t-tests. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.0001.
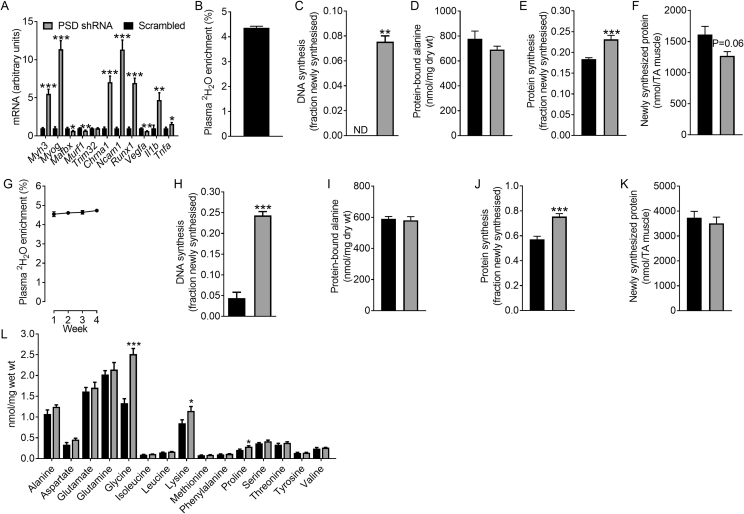
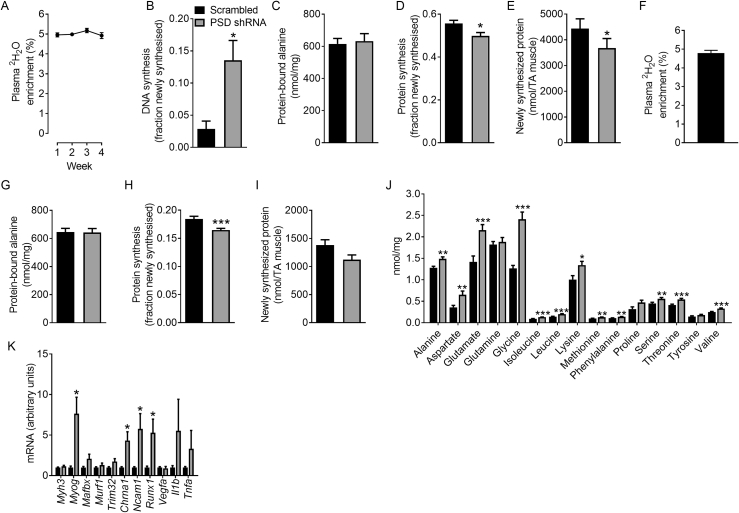
Figure 3.
The effect of PSD knockdown on transcriptional markers of regeneration, atrophy, denervation, and inflammation, DNA and protein synthesis as well as free amino acid levels. AAV vectors were injected in the tibialis anterior (TA) of 8 week old mice. Studies to assess gene expression, DNA and protein synthesis and free amino acid levels were conducted 8 weeks after AAV administration (i.e. at 16 weeks of age). A: The effect of PSD knockdown on the expression of atrophy, denervation and inflammation-related genes (N = 12; 6 female, 6 male). B: Plasma 2H2O enrichment after 1 week (i.e. between weeks 7 and 8 after AAV administration) of heavy water labeling (N = 14; 8 female, 6 male). C: DNA synthesis in TA muscles over the 1 week heavy water labeling period (N = 6, 3 female, 3 male). D: Concentration of protein-bound alanine in TA muscle from the 1 week heavy water labeling study (N = 14; 8 female, 6 male). E: Fraction of newly synthesized protein in TA muscle from the 1 week heavy water labeling study (N = 14; 8 female, 6 male). F; The absolute amount of newly synthesized protein in the TA muscle from the 1 week heavy water labeling study (N = 14; 8 female, 6 male). G: Plasma 2H2O enrichment over the 4 week i.e. between weeks 4 and 8 after AAV administration) heavy water labeling study (N = 21; 13 female, 8 male). H: DNA synthesis in TA muscles over the 4 week heavy water labeling period (N = 21; 13 female, 8 male). I: Concentration of protein-bound alanine in TA muscle from the 4 week heavy water labeling study (N = 21; 13 female, 8 male). J: Fraction of newly synthesized protein in the TA muscle from the 4 week heavy water labeling study (N = 21; 13 female, 8 male). K: The absolute amount of newly synthesized protein in the TA muscle from the 4 week heavy water labeling study (N = 21; 13 female, 8 male). L: Concentrations of free amino acids in TA muscle (N = 7; 3 female, 4 male). Data are mean ± SEM. Data were analyzed using paired t-tests. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. ND, not detected.
To begin to understand what may be mediating the loss of muscle mass associated with PSD knockdown, we analyzed the expression of atrophy-related genes. Interestingly, Mafbx and Murf1 expression was reduced in PSD-deficient muscle while Trim32 expression was unchanged (Figure 3A). In contrast, the expression of denervation-related transcripts Myog, Chrna1, Ncam1, and Runx1 were markedly increased in PSD-deficient muscle (Figure 3A), suggesting that denervation could contribute to the atrophy. Since denervation has been shown to reduce muscle capillarization due to down-regulation of vascular endothelial growth factor (VEGF) signalling [23], Vegfa expression was determined and found to be reduced in PSD-deficient muscle (Figure 3A). Based on these observations, we speculate that the alteration in muscle membrane phospholipid content and composition caused by disrupting PSD could affect muscle nerve supply, possibly through alterations in the neuromuscular junction, therefore contributing to the muscle wasting response. Future studies will be required to investigate this mechanism in further detail.
To better characterize the atrophy and regenerative response caused by PSD knockdown we utilized 2H2O labeling to measure DNA and protein synthesis, as determined by 2H incorporation into DNA-bound deoxyribose and protein-bound alanine [16], respectively. These metabolic labeling studies involved the administration of 2H2O to mice for either the final week (Figure 3B,C) or last 4 weeks (Figure 3G,H) of the 8 week study. Regardless of the period of heavy water labeling, there was a dramatic increase in the appearance of newly synthesized DNA in PSD-deficient muscle (Figure 3C,H), suggesting muscle satellite cell proliferation and infiltration of the muscle by newly formed (i.e. bone marrow derived) immune cells as part of the regenerative response. In regard to protein synthesis, while the amount of protein-bound alanine (i.e. protein pool size) was not affected by PSD knockdown (Figure 3D,I), fractional rates of protein synthesis were elevated (25–30%) in PSD-deficient muscle after both the 1 (Figure 3E) and 4 week labeling period (Figure 3J). Given the profound effect of PSD knockdown on muscle mass, we determined the absolute amount of newly synthesized protein across the entire TA muscle. While total protein synthesis was reduced by ∼20% (P = 0.06) in the entire PSD-deficient muscle after 1 week of labeling (Figure 3F), no differences were detected after 4 weeks of labeling (Figure 3K). We also measured the concentration of free amino acids, which revealed that PSD knockdown was associated with an increase in glycine, lysine, and proline (Figure 3L).
3.3. PSD knockdown disrupts mitochondrial integrity
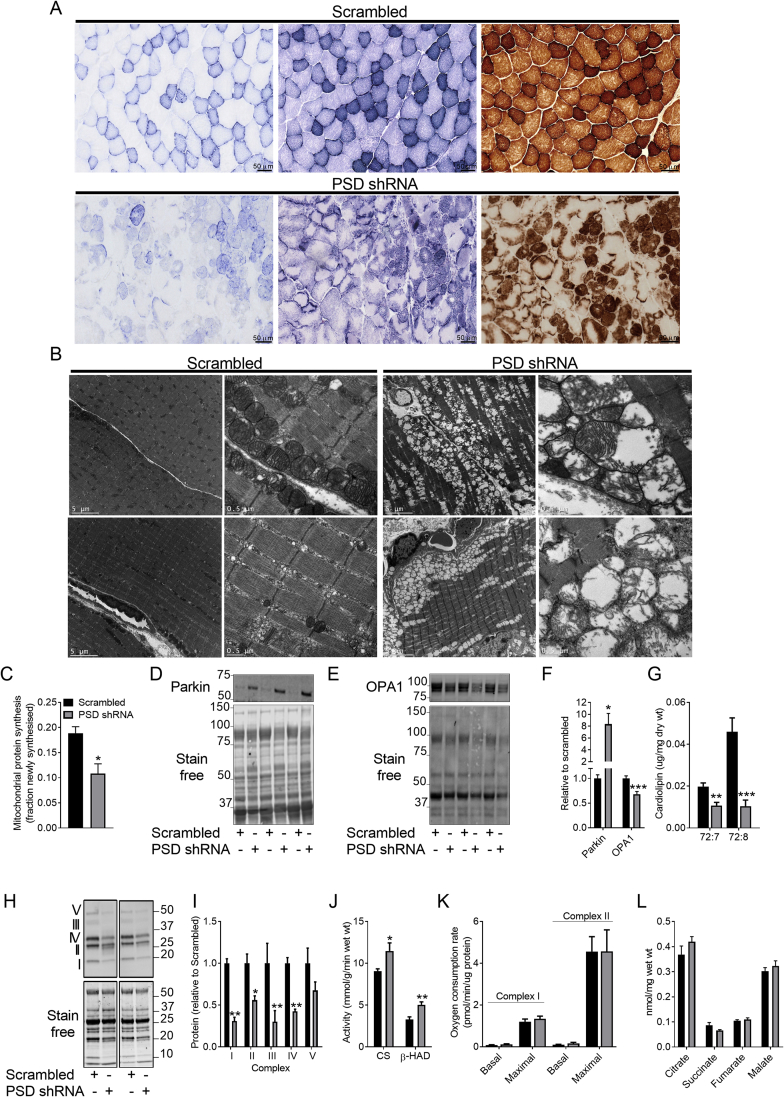
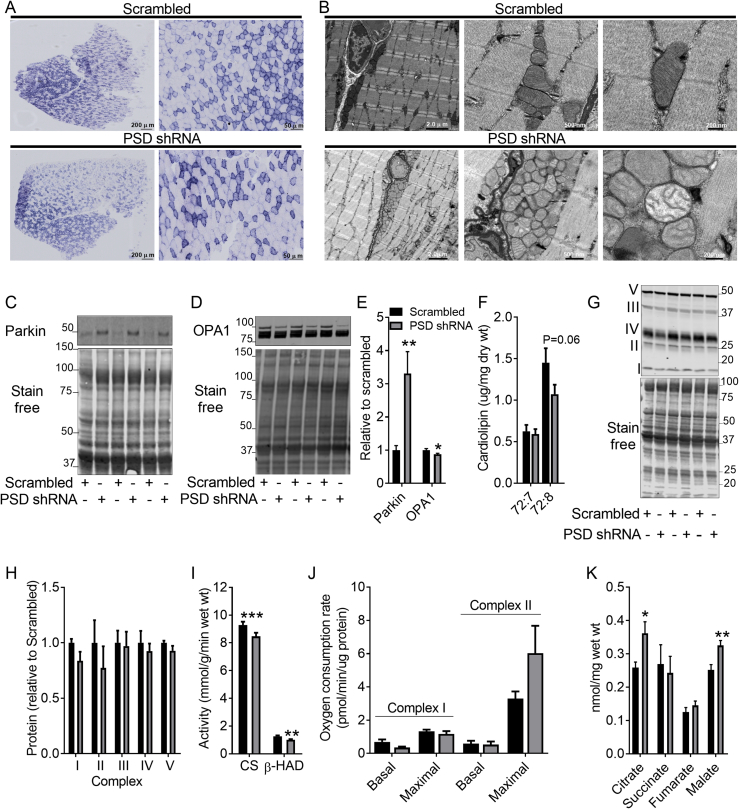
Since PSD-deficiency has been linked to mitochondrial ultrastructural defects [2], [7], we examined whether PSD knockdown affected aspects of mitochondrial structure and function. Histochemical staining of mitochondrial enzymes demonstrated marked effects of PSD knockdown (Figure 4A). Indeed, succinate dehydrogenase staining was weak in fibers from PSD-deficient muscle (Figure 4A; left panel). In addition, the NADH-tetrazolium reductase (Figure 4A; middle panel) and cytochrome oxidase (Figure 4A; right panel) stains revealed that PSD-deficient muscle exhibited fibers with central cores and staining resembling a moth eaten pattern. There was also evidence of excessive cytochrome oxidase staining in some fibers from PSD-deficient muscle (Figure 4A; right panel). Electron microscopy revealed that mitochondrial number was noticeably increased in PSD-deficient muscle (Figure 4B). However, many of these mitochondria exhibited significant ultrastructural changes, appearing swollen with rarefied cristae, demonstrating extensive disruption to the inner mitochondrial membrane and cristae structure. The outer mitochondrial membrane, however, appeared intact. To ascertain whether the increase in mitochondria was due to enhanced mitochondrial biogenesis, 2H2O labeling was used to assess mitochondrial protein synthesis. Interestingly, mitochondrial protein synthesis was reduced in PSD-deficient muscle (Figure 4C), indicating enhanced mitochondrial biogenesis was not likely mediating the increase in mitochondrial content. This raises the possibility that alterations in mitochondrial clearance contribute to mitochondrial accumulation. Interestingly, PSD-deficient muscle exhibited an ∼8-fold increase in parkin (Figure 4D,F), an E3 ubiquitin ligase involved in clearing damaged mitochondria [24]. Despite this, there was still accumulation of mitochondria with ultrastructural abnormalities, suggesting mitophagy was not stimulated. This, is supported by the absence of double-membrane structures engulfing these mitochondria in the electron micrographs (Figure 4B). In addition, PSD-deficient muscle exhibited a ∼30% reduction in OPA1 (Figure 4E,F). Since OPA1 is required for inner mitochondrial membrane fusion and cristae formation [25], [26], this appears to be consistent with the presence of mitochondrial membrane damage and morphological change. Commensurate with the loss of inner mitochondrial membrane integrity, PSD-deficient muscle exhibited a dramatic reduction (∼50–80%) in the major cardiolipin species (Figure 4G), a mitochondrial specific phospholipid almost exclusively located in the inner mitochondrial membrane [27]. Further analysis revealed that while the protein levels of the inner membrane bound OXPHOS complexes were reduced in PSD-deficient muscle (Figure 4H,I), the activity of the matrix enzymes citrate synthase and β-hydroxyacyl CoA dehydrogenase (β-HAD) were elevated (Figure 4J). Together, this provides strong evidence that disrupting the PSD pathway leads to accumulation of mitochondria that exhibit loss of inner membrane integrity.
Figure 4.
PSD knockdown disrupts mitochondrial integrity. AAV vectors were injected in the tibialis anterior (TA) of 8 week old mice. Studies to assess aspects of mitochondrial structure and function were conducted 8 weeks after AAV administration (i.e. at 16 weeks of age). A: Succinate dehydrogenase (left), NADH tetrazolium reductase (middle) and cytochrome oxidase (right) staining. Scale bar is 50 μm. B: Electron micrographs of TA muscles. Scale bar is 5 μm for the left electron micrographs within in each group and 0.5 μm for the right. C: Mitochondrial protein synthesis (N = 7; 5 female, 2 male). D: Representative western blot of parkin protein levels. E: Representative Western blot of OPA1 protein levels. F: Quantification of parkin (N = 4; 2 female, 2 male) and OPA1 (N = 9; 3 female, 6 male) protein content. G: Cardiolipin content (N = 12; 6 female, 6 male). H: Representative western blots of the OXPHOS proteins in Complex I–V of the electron transport chain. I: Quantification of the OXPHOS proteins (N = 5, 2 female, 3 male). J: Citrate synthase and β-HAD activity (N = 10 male). K: Basal and maximal oxygen consumption rates in TA muscles (N = 7, 2 female; 5 male). L: Concentration of TCA cycle metabolites (N = 7; 3 female, 4 male). Data are mean ± SEM. Data were analyzed using paired t-tests except for data in K which was analyzed by two-way ANOVA.∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
Despite the ultrastructural alterations, both basal and maximal rates of oxygen consumption were unaffected in PSD-deficient muscle (Figure 4K). Moreover, the TCA cycle intermediates citrate, succinate, fumarate, and malate were unaffected by PSD knockdown (Figure 4L). While our findings support many of the in vitro effects of PSD knockdown on mitochondrial structure and function, they are also contrasting. Consistent with our data, Tasseva et al. [2] reported that the disruption in mitochondrial structure caused by silencing PSD in CHO cells was also associated with a reduction in OXPHOS complexes. However, unlike our findings, they did in fact report that respiratory capacity was impaired in PSD-deficient cells [2].
3.4. Effects of PSD knockdown on muscle atrophy and mitochondrial ultrastructure precede myofiber damage
To help decipher the temporal relationship between muscle atrophy, myofiber damage and changes in mitochondrial ultrastructure, studies were conducted at an earlier time point (i.e. 4 weeks) after AAV administration. At this early time point, PSD shRNA administration reduced Pisd expression by ∼65% (Figure 5A). Consistent with the 8 week study, muscle phospholipid homeostasis was also altered at this early time point, evident by a 40% reduction in PtdEtn and a 20% increase in PtdSer (Figure 5B). A modest reduction in PtdCho was also noted (Figure 5B). Consequently, there was an imbalance in the major phospholipids in PSD-deficient muscle with significant alterations in the PtdCho:PtdEtn (2.2 ± 0.1 vs.3.3 ± 0.1 for scrambled vs. PSD shRNA; N = 19; P < 0.0001) and PtdEtn:PtdSer ratio (5.0 ± 0.7 vs. 2.3 ± 0.3 for scrambled vs. PSD shRNA; N = 19; P < 0.0001). There were again inverse effects on the predominant PtdEtn and PtdSer species (Figure 5C,D), with a significant reduction in (40:6)PtdEtn while (40:6)PtdSer was increased. At this early time point, a 10% reduction in TA muscle mass was already evident (Figure 5E). Again, this effect was still evident after muscles had been freeze dried (12.3 ± 0.8 vs. 11.2.0 ± 0.6 mg dry wet for scrambled vs. PSD shRNA; N = 8; P < 0.01). Histologically, the myofibers appeared normal with no sign of the damage or remodeling (i.e. absence of centrally located nuclei) that was apparent in PSD-deficient muscle at 8 weeks (Figure 5F). However, fiber size did appear to be somewhat smaller in muscles where the PSD pathway was disrupted (Figure 5F), as demonstrated by the tendency for a left shift in the fiber size distribution curve (Figure 5F,K) and a modest reduction in mean fiber diameter (40.3 ± 2.6 vs. 35.5 ± 0.5 for scrambled vs. PSD shRNA; N = 4; P = 0.16). Since TA fiber number was not affected (Figure 5H), this modest reduction in fiber size likely accounts for the atrophy observed at this time point. There was also no evidence of an increase in the number of central nuclei or immune cell infiltration (Figure 5F,I). No differences were noted in MHC expression or fiber specific cross sectional area (Figure 5J–M).
Figure 5.
Short-term (4 week) disruption of the PSD pathway disturbs phospholipid homeostasis and causes muscle atrophy independent of myofiber damage. AAV vectors were injected in the tibialis anterior (TA) of 8 week old mice. Studies were conducted 4 weeks after AAV administration (i.e. at 12 weeks of age). A: Pisd mRNA expression in TA muscle (N = 6; 3 female, 3 male). B: Muscle phosphatidylethanolamine (PtdEtn), phosphatidylserine (PtdSer) and phosphatidylcholine (PtdCho) content in TA muscle (N = 19; 6 female, 13 male). C: PtdEtn molecular species (N = 7; 4 female, 3 male). D: PtdSer molecular species (N = 7; 4 female, 3 male). E: TA mass (N = 52; 14 female, 38 male). F: Representative H&E staining of TA muscle. Scale bar is 50 μm. G: Feret's minimal diameter distribution (N = 4 male). H: Total TA fiber number (N = 8; 2 female, 4 male). I: Percentage of fibers with centrally located nuclei (N = 4 male). J: Representative image of TA muscle showing laminin (white), myosin heavy chain expression (I, blue; IIa, green; IIx black; IIb red). Scale bar is 500 μm. K: Fiber cross sectional area distribution (N = 4; 2 female, 2 male). L: Quantitative analysis of fiber type distribution (N = 4; 2 female, 2 male). M: Quantitative analysis of fiber type specific cross sectional area (N = 4; 2 female, 2 male). Data are mean ± SEM. Data were analyzed using paired t-tests. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗P < 0.0001.
Interestingly, despite the normal histological appearance, including lack of central nuclei and cellular infiltrate, 2H2O labeling over the entire 4 week post-AAV injection period (Figure 6A) revealed that DNA synthesis was already markedly elevated in PSD-deficient muscle (Figure 6B). This is indicative of satellite cell proliferation occurring prior to migration and differentiation (i.e. the formation of central nuclei) in what appears to be a pre-emptive process in anticipation of the impending damage apparent at the later time point after PSD knockdown. Concurrent measures of protein synthesis were made in order to understand the early mechanisms underlying the atrophy. Since the heavy water labeling studies involved the administration of 2H2O to mice for the entire 4 week period of study (Figure 6A), this allowed us to capture the total cumulative effects on protein synthesis. While disrupting PSD did not affect the concentration of protein-bound alanine (Figure 6C), fractional rates of protein synthesis were lower (Figure 6D). When considered in light of the effects on TA muscle mass, the absolute amount of newly synthesized protein across the entire TA muscle was reduced (Figure 6E). Similar effects were observed when labeling was performed over the final week of the study period (i.e. between week 3 and 4 post AAV injection; Figure 6F–I). These changes in protein synthesis were accompanied by an increase in the levels of free amino acids (Figure 6J), particularly the essential amino acids, which were likely derived from an accelerated rate of protein degradation. Together with the metabolic labeling data, the initial atrophy response therefore appears to be mediated by reduced rates of protein synthesis concomitant with increased protein degradation.
Figure 6.
Effect of short-term (4 week) disruption of the PSD pathway on transcriptional markers of regeneration, atrophy, denervation, and inflammation, DNA and protein synthesis as well as free amino acid levels. AAV vectors were injected in the tibialis anterior (TA) of 8 week old mice. Studies to assess DNA and protein synthesis and free amino acid levels were conducted 4 weeks after AAV administration (i.e. at 12 weeks of age). A: Plasma 2H2O enrichment over the entire 4 week heavy water labeling period in mice studied 4 weeks after AAV administration (N = 8; 1 female, 7 male). B: DNA synthesis in TA muscles over the 4 week heavy water labeling period in mice studied 4 weeks after AAV administration (N = 8; 1 female, 7 male). C: Concentration of protein-bound alanine in TA muscle obtained from mice studied 4 weeks after AAV administration and labeled with heavy water for 4 weeks (N = 8; 1 female, 7 male). D: Fraction of newly synthesized protein in TA muscle obtained from mice studied 4 weeks after AAV administration and labeled with heavy water for 4 weeks (N = 8; 1 female, 7 male). E: The absolute amount of newly synthesized protein in the entire TA muscle obtained from mice studied 4 weeks after AAV administration and labeled with heavy water for 4 weeks (N = 8; 1 female, 7 male). F: Plasma 2H2O enrichment after 1 week (i.e. between weeks 3 and 4 after AAV administration) of heavy water labeling period in mice studied 4 weeks after AAV administration (N = 11; 5 female, 6 male). G: Concentration of protein-bound alanine in TA muscle obtained from mice studied 4 weeks after AAV administration and labeled with heavy water for 1 week (N = 11; 5 female, 6 male). H: Fraction of newly synthesized protein in TA muscle obtained from mice studied 4 weeks after AAV administration and labeled with heavy water for 1 week (N = 11; 5 female, 6 male). I: Absolute amount of newly synthesized protein in the entire TA muscle obtained from mice studied 4 weeks after AAV administration and labeled with heavy water for 1 week (N = 11; 5 female, 6 male). J: Concentrations of free amino acids in TA muscle (N = 11; 6 female, 6 male). K: The effect of PSD knockdown on the expression of atrophy, denervation, and inflammation-related genes (N = 6; 3 female, 3 male). Data are mean ± SEM. Data were analyzed using paired t-tests. ∗P < 0.05; ∗∗P < 0.01;∗∗∗P < 0.001.
To further investigate potential mechanisms underlying this early atrophy response associated with disrupting PSD, atrophy and denervation-related transcripts were examined. While there no change in atrophy-related genes (Figure 6K), the expression of denervation-related transcripts (Myog, Chrna1, Ncam1, and Runx1) were upregulated in PSD-deficient muscle (Supplementary Fig. 1D). In addition PSD knockdown had no effect on Vegfa, Il1b or Tnfa mRNA (Figure 6K). These findings therefore suggest that some degree of denervation may contribute to the modest atrophy caused by PSD knockdown at this early time point.
Aspects of mitochondrial structure and function were also assessed at the 4 week time point. While there was no apparent difference in the staining pattern or intensity for succinate dehydrogenase (Figure 7A), there were modest mitochondrial ultrastructural changes typified by a less dense matrix and more rarefied cristae (Figure 7B). Notably, these effects were markedly less severe than when PSD was disrupted for a longer duration (i.e. 8 weeks; Figure 4B). Similar to the observations at 8 weeks (Figure 4D–F), parkin was increased (∼3-fold) while OPA1 expression was reduced (Figure 7C–E). In addition, there was a non-significant reduction (26%; P = 0.06) in tetralinoleoyl-cardiolipin (72:8) levels (Figure 7F). Taken together, this is consistent with the presence of some degree of mitochondrial damage and morphological change. While there were no alterations in the OXPHOS complexes (Figure 7G,H), there modest reductions in citrate synthase and β-HAD activity (Figure 7I). Consistent with the 8 week data, substrate-driven oxygen consumption in intact muscle biopsy specimens was not affected (Figure 7J) nor were succinate and fumarate levels (Figure 7K). In contrast, citrate and malate were increased (Figure 7K). Together, these data demonstrate that the initiation of muscle wasting is associated with modest alterations in mitochondrial ultrastructure and marked satellite cell proliferation, responses that precede the onset of severe myofiber damage.
Figure 7.
Effect of short-term (4 week) disruption of the PSD pathway on mitochondrial structure and function. AAV vectors were injected in the tibialis anterior (TA) of 8 week old mice. Studies to assess aspects of mitochondrial structure and function were conducted 4 weeks after AAV administration (i.e. at 12 weeks of age). A: Succinate dehydrogenase staining. Scale bar is 200 μm for the panel (whole mount) and 50 μm for the right panel. B: Representative electron micrograph of TA muscle. Scale bar is 2 μm for the left electron micrograph, 500 nm for the middle and 200 nm for the right. C: Representative western blot of parkin protein levels. D: Representative western blot of OPA1 protein levels. E: Quantification of parkin and OPA1 protein content (N = 7; 4 female, 3 male). F: Cardiolipin content in TA muscles (N = 7; 4 female, 3 male). G: Representative western blots of the OXPHOS proteins in Complex I–V of the electron transport chain. H: Quantification of the OXPHOS proteins (N = 7; 4 female, 3 male). I: Citrate synthase and β-HAD activity in TA muscle (N = 6 male). J: Basal and maximal oxygen consumption rates in TA muscles (N = 7; 2 female, 5 male). K: Concentration of TCA cycle metabolites (N = 11; 5 female, 6 male). Data are mean ± SEM. Data were analyzed using paired t-tests.∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
In addition to examining effects at this early time point, studies were conducted at 16 weeks post-AAV to ascertain whether the atrophy associated with disrupting PSD was progressive. At 16 weeks, Pisd expression was reduced by ∼30% (Supplementary Fig. 1A), which was somewhat less than the level of knockdown observed at both 4 and 8 weeks. We believe this can be explained by the fact that AAVs do not incorporate into the host genome. Thus, the myofiber regeneration and satellite cell proliferation caused by PSD knockdown dilutes transgene expression since the AAV is not expressed in new myonuclei [28]. Despite this, the loss of muscle mass (∼40%) was similar to that observed at 8 weeks (Supplementary Fig. 1B) Thus, there appears to be a progressive loss of TA mass within 8 weeks after PSD knockdown, after which muscle mass stabilized and an apparent new steady state is reached. The kinetics of the atrophy response are in line with the protein turnover dynamics and free amino acid data. Specifically, the atrophy observed within the initial 4 weeks was associated with reduced protein synthesis and presumably increased proteolysis, evident by a uniform increase in muscle free amino acids. However, consistent with the stabilization of muscle mass loss at 8 weeks, free amino acid levels and absolute protein synthesis was similar to the control muscle, suggesting normalization of proteolysis. The loss of muscle mass was again associated with an upregulation of the denervation-related transcripts Myog, Chrna1, Ncam1, and Runx1 while no differences in atrophy and inflammation-related genes were noted (Supplementary Figure 1C). Furthermore, a number of the histological alterations observed at 8 weeks were also evident at 16 weeks (Supplementary Fig. 1D), with a reduction in fiber size, increased variation in fiber size, as well as the persistence of centrally located nuclei (Supplementary Figs. 1D–F).
4. Summary and conclusions
A number of interesting questions in relation to the regulation of muscle phospholipid metabolism and muscle mass emerge from our findings. Firstly, since there are two pathways for PtdEtn synthesis, why is the CDP-ethanolamine pathway unable to fully compensate for partial loss (i.e. ∼60% knockdown) of PSD? This may relate to the observation that each pathway exhibits distinct substrate specificity, with the CDP-ethanolamine pathway preferentially producing PtdEtn with mono- or di-unsaturated fatty acids, while PSD favors generation of PtdEtn species containing long-chain polyunsaturated fatty acids [5]. Moreover, while PtdEtn produced via the CDP-ethanolamine pathway in the endoplasmic reticulum can be exported to other organelles, it appears that PtdEtn produced by PSD is retained within mitochondria [5]. A threshold of PSD activity therefore appears to be essential in order to produce adequate amounts of the full complement of mitochondrial PtdEtn species required for mitochondrial membrane integrity and ultrastructure. Alternatively, the physiological effects caused by loss of PSD activity may not necessarily be mediated by the reduction in PtdEtn but rather by mitochondrial accumulation of PtdSer. In contrast to PtdEtn which is highly enriched in mitochondria, PtdSer is a relatively minor constituent [29]. Upon transport to the mitochondria, PtdSer is rapidly decarboxylated to PtdEtn by PSD, the primary route for PtdSer degradation in mammals and the only pathway for mitochondrial PtdSer catabolism [30]. The low abundance of PtdSer is outweighed by its physiological importance, a feature attributed to its unique chemical properties, including its negative charge [30]. Indeed, PtdSer is the most abundant negatively charged phospholipid in eukaryotic membranes, thus subtle alterations in mitochondrial PtdSer could confer significant changes in membrane electrostatic charge which may lead to a range of disturbances including alterations in membrane integrity and membrane-protein interactions [30].
Finally, how is cellular respiration not compromised despite the marked alterations in mitochondrial ultrastructure? This is not a trivial question to address and likely involves complex compensatory mechanisms in an attempt to maintain homeostasis. While speculative, it is possible that the loss of inner mitochondrial membrane integrity, and presumably impaired energy generation on an individual mitochondrion level, is compensated by an increase in mitochondrial number, which appears to occur through accumulation rather than biogenesis since mitochondrial protein synthesis was reduced. This resembles observations made in muscle-specific mitochondrial transcription factor A (Tfam) knockout mice, which exhibit a myopathy characterized by accumulation of abnormal mitochondria with distorted cristae yet preserved ATP production [31]. Together with our data, this supports the concept that an increase in mitochondrial content could compensate for damage to individual mitochondria in order to maintain energy homeostasis [31]. Moreover, it is possible that long-term energy homeostasis is maintained via a reduction in cell volume (i.e. atrophy) and attainment of a new smaller steady-state mass (i.e. plateau in TA mass between 8 and 16 weeks). Indeed, a recent model supports such a phenomenon whereby mitochondrial energetics can be preserved in the face of potentially pathological mitochondrial DNA mutations through reductions in cell volume until available functioning power sources can match energy demand [32].
In conclusion, our findings provide important insight into the biological function of mitochondria in muscle by ascribing a key role of PSD in regulating skeletal muscle phospholipid homeostasis. In addition, we demonstrate that this pathway is critically important for the maintenance of mitochondrial integrity and skeletal muscle mass.
Author contributions
Conceptualization: A.S., G.M.K. and C.R.B.; Methodology: A.S., G.M.K., D.L.C., S.L.M and C.R.B.; Investigation: A.S., G.M.K., S.A.M., D.L.C., V.C.F, P.A.D.G., A.L., S.H., G.K., A.R.C., M.L.B., T.A., S.L.M. and C.R.B.; Writing – Original Draft: A.S., G.M.K., C.R.B, Writing – Review and Editing: A.S., G.M.K., S.A.M., D.L.C., V.C.F, P.A.D.G., A.L., S.H., G.K., A.R.C., M.L.B., T.A., S.L.M. and C.R.B.; Funding Acquisition: A.S., G.M.K, G.K. and C.R.B., Supervision: A.S., G.M.K. and C.R.B.
Data statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgements
G.M.K. (DE180100859) and C.R.B. (FT160100017) are supported by Australian Research Council fellowships. A.L. is supported by an Alfred Deakin Postdoctoral Fellowship. T.A is supported by an Australian Government Research Training Program Scholarship. This work was supported in part by a Diabetes Australia Research Program grant and funding from the Faculty of Health, Deakin University. The funding bodies had no involvement in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication. The authors acknowledge the facilities and scientific and technical assistance of Monash Micro Imaging, Monash University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.06.020.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.van der Veen J.N., Kennelly J.P., Wan S., Vance J.E., Vance D.E., Jacobs R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochimica et Biophysica Acta. 2017;1859:1558–1572. doi: 10.1016/j.bbamem.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Tasseva G., Bai H.D., Davidescu M., Haromy A., Michelakis E., Vance J.E. Phosphatidylethanolamine deficiency in Mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. Journal of Biological Chemistry. 2013;288:4158–4173. doi: 10.1074/jbc.M112.434183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Percy A.K., Moore J.F., Carson M.A., Waechter C.J. Characterization of brain phosphatidylserine decarboxylase: localization in the mitochondrial inner membrane. Archives of Biochemistry and Biophysics. 1983;223:484–494. doi: 10.1016/0003-9861(83)90613-6. [DOI] [PubMed] [Google Scholar]
- 4.Zborowski J., Dygas A., Wojtczak L. Phosphatidylserine decarboxylase is located on the external side of the inner mitochondrial membrane. FEBS Letters. 1983;157:179–182. doi: 10.1016/0014-5793(83)81141-7. [DOI] [PubMed] [Google Scholar]
- 5.Bleijerveld O.B., Brouwers J.F., Vaandrager A.B., Helms J.B., Houweling M. The CDP-ethanolamine pathway and phosphatidylserine decarboxylation generate different phosphatidylethanolamine molecular species. Journal of Biological Chemistry. 2007;282:28362–28372. doi: 10.1074/jbc.M703786200. [DOI] [PubMed] [Google Scholar]
- 6.Sundler R., Akesson B., Nilsson A. Quantitative role of base exchange in phosphatidylethanolamine synthesis in isolated rat hepatocytes. FEBS Letters. 1974;43:303–307. doi: 10.1016/0014-5793(74)80667-8. [DOI] [PubMed] [Google Scholar]
- 7.Steenbergen R., Nanowski T.S., Beigneux A., Kulinski A., Young S.G., Vance J.E. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. Journal of Biological Chemistry. 2005;280:40032–40040. doi: 10.1074/jbc.M506510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selathurai A., Kowalski G.M., Burch M.L., Sepulveda P., Risis S., Lee-Young R.S. The CDP-ethanolamine pathway regulates skeletal muscle diacylglycerol content and mitochondrial biogenesis without altering insulin sensitivity. Cell Metabolism. 2015;21:718–730. doi: 10.1016/j.cmet.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Leonardi R., Frank M.W., Jackson P.D., Rock C.O., Jackowski S. Elimination of the CDP-ethanolamine pathway disrupts hepatic lipid homeostasis. Journal of Biological Chemistry. 2009;284:27077–27089. doi: 10.1074/jbc.M109.031336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. Journal of Biological Chemistry. 1957;226:497–509. [PubMed] [Google Scholar]
- 11.Wang F., Zhao Y., Wang P. Separation and determination of phospholipids in biological samples by high-performance liquid chromatography. Journal of Chromatographic Science. 2003;41:142–144. doi: 10.1093/chromsci/41.3.142. [DOI] [PubMed] [Google Scholar]
- 12.Dubowitz V., Sewry C.A., Oldfors A. 4th ed. Sanders; Oxford: 2013. Muscle biopsy: a practical approach. [Google Scholar]
- 13.Bloemberg D., Quadrilatero J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS One. 2012;7:e35273. doi: 10.1371/journal.pone.0035273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalski G.M., Kloehn J., Burch M.L., Selathurai A., Hamley S., Bayol S.A. Overexpression of sphingosine kinase 1 in liver reduces triglyceride content in mice fed a low but not high-fat diet. Biochimica et Biophysica Acta. 2015;1851:210–219. doi: 10.1016/j.bbalip.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Bruce C.R., Thrush A.B., Mertz V.A., Bezaire V., Chabowski A., Heigenhauser G.J. Endurance training in obese humans improves glucose tolerance and mitochondrial fatty acid oxidation and alters muscle lipid content. American Journal of Physiology. Endocrinology and Metabolism. 2006;291:E99–E107. doi: 10.1152/ajpendo.00587.2005. [DOI] [PubMed] [Google Scholar]
- 16.Gasier H.G., Fluckey J.D., Previs S.F. The application of 2H2O to measure skeletal muscle protein synthesis. Nutrition and Metabolism. 2010;7:31. doi: 10.1186/1743-7075-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foletta V.C., Palmieri M., Kloehn J., Mason S., Previs S.F., McConville M.J. Analysis of mammalian cell proliferation and macromolecule synthesis using deuterated water and gas chromatography–mass spectrometry. Metabolites. 2016;6 doi: 10.3390/metabo6040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busch R., Neese R.A., Awada M., Hayes G.M., Hellerstein M.K. Measurement of cell proliferation by heavy water labeling. Nature Protocols. 2007;2:3045–3057. doi: 10.1038/nprot.2007.420. [DOI] [PubMed] [Google Scholar]
- 19.Srere P.A. Citrate synthase. Methods in Enzymology. 1969;13:3–11. [Google Scholar]
- 20.Lowry O.H., Passonneau J.V. Academic Press; New York: 1972. A flexible system of enzymatic analysis. [Google Scholar]
- 21.Gaur V., Connor T., Sanigorski A., Martin S.D., Bruce C.R., Henstridge D.C. Disruption of the class IIa HDAC corepressor complex increases energy expenditure and lipid oxidation. Cell Reports. 2016;16:2802–2810. doi: 10.1016/j.celrep.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Schiaffino S., Rossi A.C., Smerdu V., Leinwand L.A., Reggiani C. Developmental myosins: expression patterns and functional significance. Skeletal Muscle. 2015;5:22. doi: 10.1186/s13395-015-0046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagatsuma A., Osawa T. Time course of changes in angiogenesis-related factors in denervated muscle. Acta Physiologica. 2006;187:503–509. doi: 10.1111/j.1748-1716.2006.01594.x. [DOI] [PubMed] [Google Scholar]
- 24.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of Cell Biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahn J.J., Stackley K.D., Chan S.S. Opa1 is required for proper mitochondrial metabolism in early development. PLoS One. 2013;8:e59218. doi: 10.1371/journal.pone.0059218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tezze C., Romanello V., Desbats M.A., Fadini G.P., Albiero M., Favaro G. Age-associated loss of OPA1 in muscle impacts muscle mass, metabolic homeostasis, systemic inflammation, and epithelial senescence. Cell Metabolism. 2017;25:1374–1389. doi: 10.1016/j.cmet.2017.04.021. e1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mejia E.M., Hatch G.M. Mitochondrial phospholipids: role in mitochondrial function. Journal of Bioenergetics and Biomembranes. 2016;48:99–112. doi: 10.1007/s10863-015-9601-4. [DOI] [PubMed] [Google Scholar]
- 28.Naso M.F., Tomkowicz B., Perry W.L., 3rd, Strohl W.R. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs. 2017;31:317–334. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath S.E., Daum G. Lipids of mitochondria. Progress in Lipid Research. 2013;52:590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Leventis P.A., Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annual Review of Biophysics. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 31.Wredenberg A., Wibom R., Wilhelmsson H., Graff C., Wiener H.H., Burden S.J. Increased mitochondrial mass in mitochondrial myopathy mice. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aryaman J., Johnston I.G., Jones N.S. Mitochondrial DNA density homeostasis accounts for a threshold effect in a cybrid model of a human mitochondrial disease. Biochemical Journal. 2017;474:4019–4034. doi: 10.1042/BCJ20170651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.