Abstract
Aminoglycosides, a class of antibiotics that includes gentamicin, kanamycin, neomycin, streptomycin, tobramycin and apramycin, are derived from various streptomyces species. Despite the significant increase in the antibacterial resistant pathogens, aminoglycosides remain an important class of antimicrobial drugs due to their unique chemical structure which offers a broad spectrum of activity. The modification of antibiotics and their subsequent use in supramolecular chemistry is rarely reported. Given the importance of aminoglycosides, here we give a brief overview on the modification of 4,5‐ and 4,6‐disubstituted deoxystreptamine classes of aminoglycosides through supramolecular chemistry and their potential for real world applications. We also make the case that the work in this area is gaining momentum, and there are significant opportunities to meet the challenges of modern antibiotics through the modification of aminoglycosides by harnessing the advantages of supramolecular chemistry.
Keywords: supramolecular self-assemblies, aminoglycosides, antibiotics, DNA, superstructures
1. Introduction
Antibiotics are substances that possess bactericidal activity. The most commonly used antibiotics are: β‐Lactams (penicillins, cephalosporins), macrolides, fluoroquinolones, tetracyclines and aminoglycosides. Each class of antibiotic contains multiple drugs that may have unique activity. Among these antibiotics, aminoglycosides are of particular interest due to their broad spectrum of activity, with rapid bacteriostatic and bactericidal action1 and favourable chemical and pharmacokinetic properties. These unique properties make them useful drugs against various types of microbial infections.2 Aminoglycosides are generally most effective against gram‐negative bacteria.3
As the global spread of bacterial resistance to antibiotics increases, aminoglycosides remain an important class of broad spectrum antibiotics despite their age owing to their unique structure and properties. Their structure is composed of aminocyclitol with one or more amino or sugar groups. Aminoglycosides are natural products, synthesized from the fermentation of soil microbes (actinomycetes) or semisynthetic derivatives of those secondary metabolites. The presence of multiple amino and hydroxyl groups make them hydrophilic in nature, quite polar, basic and water soluble.2, 4 It was believed that the problem of bacterial infection was solved when antibiotics were first discovered. However, antibiotics lose their effectiveness as bacteria evolve to gain resistance traits.5
A persistent threat to public health is the spread of multidrug‐resistant infectious diseases. Therefore, it is necessary to develop new antibiotics to replace those that have developed the resistance.6 This can be achieved by either discovery of new classes of antibiotics or by further development of existing antibiotic classes. Aminoglycosides are considered as one of the most effective antibiotics and derived from a range of streptomyces including amikacin, gentamicin, kanamycin, neomycin, streptomycin, tobramycin and apramycin. Streptomycin was first isolated from streptomyces griseus whereas neomycin was derived from streptomyces fradiae. Gentamycin, tobramycin and amikacin were isolated from micromonospora purpurea, streptomyces tenebrarius, and kanamycin, respectively.4a Neomycin is found in many topical medications such as creams, ointments, and eye‐drops.
Aminoglycosides are used as RNA binders as RNA plays a significant role in both bacteriostatic and bacteriocidal action of these aminoglycosides.7 Aminoglycosides contain several rings referred to as cyclitols (saturated 6‐membered ring structures), with five or six amino sugars connected by glycosidic bonds as shown in Figure 1.2 One of another important features of the aminoglycoside based antibiotics is their unique structure, connectivity and stereochemistry of amino and hydroxyl groups which make them unique because they are groups of diverse polyaminated pseudosaccharides.
Figure 1.
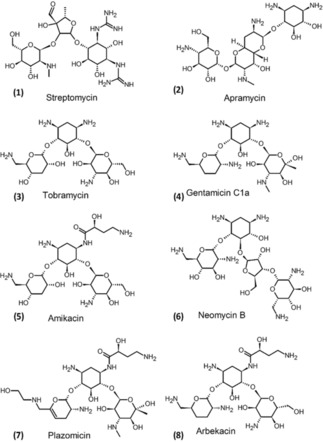
Structures of some aminoglycosides.
Aminoglycosides are important class of antibiotics and widely used for treatment of chronic bacterial diseases of heart, lungs and urinary tract.8 For example, aminoglycosides such as gentamycin and kanamycin are commonly used for the treatment of septicaemia and serious sepsis caused by gram negative bacteria.9 As aminoglycosides are polycationic saccharides at neutral pH, they are expected to interact effectively with RNA through both electrostatic interactions and hydrogen bonding but the functional resistance occurs at acidic pH.9 It should also be noted that aminoglycosides can also be used as an important tool to study the molecular recognition of ribonucleic acid (RNA).10 Based on the aminocyclitol, aminoglycosides are broadly classified into four subclasses:
No deoxystreptamine e. g. streptomycin
Mono‐substituted deoxystreptamine e. g. apramycin
4,5‐di‐substituted deoxystreptamine e. g. neomycin, ribostamycin
4,6‐di‐substituted deoxystreptamine e. g. gentamycin, tobramycin, amikacin, and plazomicin2
As infectious microorganisms have become much more resistant to aminoglycosides, the antibiotic resistance by pathogens is considered as one of the major problems for human health. Therefore, the advanced research on the design of novel but more effective antibiotics is required. Recently, much research is focused on the detailed structural and functional understanding of the interaction between the drug and the target to design these new antibiotics.11 For example, the specific interactions occur between functional groups present on aminoglycosides and nucleotides in RNA causes misreading of genetic code and inhibition of translocation processes.12 These developments triggered a significant attention on the development of modified aminoglycosides such as plazomicin and arbekacin which has some potential to overcome some limitations and are used against multidrug resistant pathogens.2
However, these aminoglycosides suffer from a few drawbacks including several side effects such as nephrotoxicity,4a, 13 and ototoxicity,13b, 14 permanent hearing loss or vestibular damage.15 To reduce these side effects such as nephrotoxicity and ototoxicity in humans, aminoglycosides such as dibekacin, netilmicin, amikacin15 and isepamicin with an improved antibacterial activity were designed.4a, 16
2. Mechanism of Action
Aminoglycoside antibiotics have been shown to be active against various types of gram‐negative and gram‐positive bacteria, particularly the enterobacteriaceae family, including escherichia coli, kebsiella pneumoniae, K. oxytoca, enterobacter cloacae, E. aerogenes, providencia spp., proteus spp., morganella spp., and serratia spp. Although aminoglycosides are used against gram‐negative and gram‐positive bacteria, they are generally most effective against gram‐negative bacteria.3
The mechanism of action of these aminoglycosides involves inhibition of protein synthesis. RNA plays an important role in inhibition of the growth of pathogens.7 Aminoglycosides inhibit protein synthesis by reversible binding with the 16 S ribosomal RNA of the 30 S ribosome, causing miscoding in the translation process, resulting in damage to the cell membrane. Some inhibition involves decoding of the target site by aminoglycosides (Figure 2).10 These antibiotics thus exhibit a post‐antibiotic effect, defined as the continued suppression of bacterial growth following transient antimicrobial exposure.2, 14, 17 Unlike other antibiotics, aminoglycosides break down in the stomach and therefore they cannot be given orally and must be injected.
Figure 2.
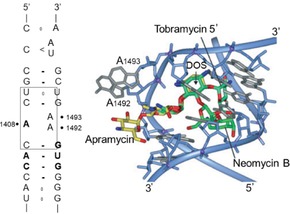
Decoding site in 16 S ribosomal RNA. At left: secondary structure of bacterial decoding sites in which the aminoglycoside binding site is marked by a box. At right: superposition of the crystal structure of three aminoglycosides complexed with the bacterial decoding site RNA. Reprinted by permission from Wiley (Springer Nature), journal: Cellular and Molecular Life Sciences, COPYRIGHT 2007.10
In vitro, there is a decrease in antimicrobial activity of aminoglycosides at acidic pH or under anaerobic conditions.9 The structure of aminoglycosides involves substitution at amino and hydroxyl groups which directly influence the antimicrobial activity.2 In this review we provide a discussion on the 4,5‐ and 4,6‐disubstituted deoxystreptamine classes of aminoglycosides such as neomycin, gentamycin, kanamycin and tobramycin, because they have been in clinical use for many decades as topical, intravenous or nebulizer formulations.
Thus, herein we focus on functionalisation, supramolecular self‐assembly and applications of this important class of aminoglycosides.
3. Self‐Assembly
The rational design of chemical entities which are able to self‐assemble into stable and dynamically ordered superstructures is essential if supramolecular materials are to be implemented in applications such as biomedicine, catalysis, and material and biological chemistry. Stimuli‐triggered self‐assembly plays an important role in the control of structural transformations in supramolecular living entities on the molecular, cellular, and macroscopic domains. Several parameters drive supramolecular self‐assembly of synthetic molecules, including the nature of the solvent, temperature, and molecular design to control electronic and functional properties.18 To achieve controlled supramolecular structures, non‐covalent interactions are used, including hydrogen bonding, hydrophobic interactions, van der Waals, ionic and electrostatic interactions, solvophobic effects, and π‐π stacking between adjacent aromatic planes. Antibiotics are small organic molecules with functional groups that may be used for supramolecular assemblies. However, the rational design of molecules for antibiotics that specifically exploits directional growth in nano‐ or meso‐scale objects are very rare. Due to the aforementioned problems with antibiotic resistance, there is an opportunity to functionalise antibiotics, and create novel supramolecular structures that help overcome shortcomings of the small organic molecule on its own.
In one example, Goekl and co‐workers improved the activity of commercial antibiotics erythromycin, kanamycin, rifampicin, and tetracycline with hydraphiles (see Figure 3 where each of the hydraphile compounds bears three diaza‐18‐crown‐6 (9) residues). The antibiotics were paired with spacers of three different chain lengths: namely dodecyl, tetradecyl, and hexadecyl.19 Their study revealed that the mode of action is different for each antibiotic. Kanamycin interferes in translation (protein synthesis) processes and cell respiration, and causes potassium ion leakage, whereas Erythromycin inhibits protein synthesis by binding to the 50 S ribosome subunit, and rifampicin inhibits RNA synthesis by binding to beta‐subunit of RNA polymerase. Tetracycline, a fused ring antimicrobial antibiotic binds to the 30 S ribosome subunit and prevents peptide chain elongation. Hence, the mechanism of action is different for each antibiotic, and is dependent on their structure. In summary the hydrophilic synthetic ion channels were found to enhance the cytotoxicity to E. coli and B. in co‐administered with erythromycin, kanamycin, rifampicin, and tetracycline.
Figure 3.
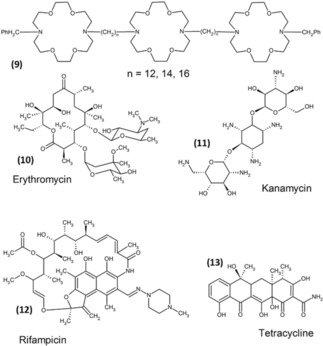
Hydraphile and antimicrobial structure.19
The Fuhrhop group reported the spontaneous formation of rigid fibers on negatively charged surfaces (mica, graphite, DNA) from readily available water‐soluble (>200 mg/mL) antibiotics such as tobramycin, kanamycin and neomycin (Figure 4). Aminoglycosides tobramycin formed up to 15 micrometer long rigid fibers, with a uniform height of 2.4 nm with an apparent helical pitch of 30 nm on the surface of mica at pH 13 (NaOH). Fibers of similar size (length and width) without secondary structure were obtained from aqueous solution at pH 7, which were characterized by AFM.20 Interestingly similar fibers obtained from kanamycin and neomycin. However, λ‐DNA strands (height of 1.0 nm) did not interact with tobramycin coils at pH 7 on mica surface. Interestingly, after treatment with EDTA the height of the λ‐DNA strands grew to 3.8 nm upon only treatment with tobramycin, thus, confirm λ‐DNA a wrapped by the supramolecular fibers. Nevertheless, such effect not observed in the case of kanamycin and neomycin. Authors has concluded that such fibers may interact with F‐actin fibers in biological cells.
Figure 4.
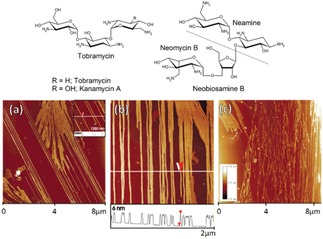
AFM image of (a) tobramycin fibers (NaOH) (profile height is 24 Å), (b) fibrous assemblies of kanamycin, and (c) neomycin all on mica as deposited from aqueous solutions at pH 13. Reprinted from20 with permission of American Chemical Society.
In another report, kanamycin A was functionalised with long alkyl stearic acid chains at one of the amino groups attached to one methylene group (C6′) and other three are at equatorial position of chair form, which makes it highly reactive. Interestingly, functionalised kanamycin formed micelles in aqueous solution, with hydrophilic head regions in contact with the surrounding solvent, and the hydrophobic tail in the micelle centre (Figure 5).21 This new kanamycin with four amino groups is a pseudo‐trisaccharide, and when mono‐amidated with stearic acid the hydrophilic moiety forms −NH3 +….OH hydrogen bonds in aqueous solution, and the hydrophobic moiety allows micelle formation. Intermolecular hydrogen bonds should stabilize the spherical shape of micelles, and larger head groups cover the micelle surface more effectively. Kanamycin inhibits antimicrobial microbial growth by interfering in protein synthesis and strongly interacts with the 30 S subunit of ribosome. Thus, the micelle formation of Kanamycin A 6′‐Stearoylamide may be used to enhance the interaction with ribosome with anionic polyelectrolytes, and may also be used as a drug carrier.
Figure 5.
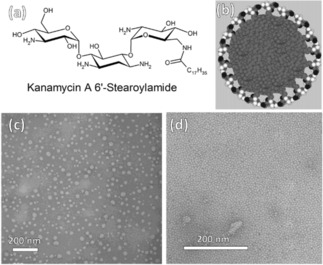
(a) Molecular structure of Kanamycin A 6′‐Stearoylamide, (b) a model of the micelle made of Kanamycin A 6′‐Stearoylamide in solution, and transmission electron micrographs (TEMs) of (c) micelles deposited from a dilute Kanamycin A 6′‐Stearoylamide (10‐5 M) and (c) a 100‐fold concentration on a carbon grid. The diameter of the micelles in (d) is a uniform 6 nm, corresponding to a fluid bilayer of Kanamycin A 6′‐Stearoylamide. Reprinted from21 with permission of American Chemical Society.
On a similar path, the Liang group developed novel aminoglycoside complexes with DNA via electrostatic interactions. These complexes were hydrophobic, leading to the formation of capsules which had a hierarchical shell structure, with smooth, rigid outer layer and a viscoelastic inner layer. This group has also shown that cationic reagents with multiple hydroxyl groups can form kinetically stable structures with DNA in aqueous solution, leading to kanamycin‐DNA capsules. They used kanamycin as the aminoglycoside and studied the interaction with genomic double stranded DNA in aqueous solution as shown in Figure 6.22 They have successfully prepared a class of capsule (containing a shell with a hierarchical structure) made up of DNA and aminoglycosides, with its morphology depending on the shape of the DNA droplet before being immersed in kanamycin solution. Using only electrostatic interactions, these capsules with hierarchical shell structures can be useful in wound dressing and drug delivery. They have described a mechanism for capsule formation, where a droplet of DNA solution is added into kanamycin, the kanamycin molecules quickly interact with DNA at outer layer of the droplet. As kanamycin has four positive charges, it can bind with more than one DNA molecule. The dehydration of the DNA‐kanamycin complex enhances the toughness of the formed shell.
Figure 6.
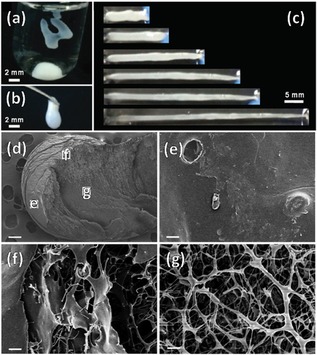
(a & b) DNA‐kanamycin capsules of various morphologies, (c) shape elongations after stretching. CDNA=1.0×10–2 g mL−1;Ckanamycin=1.0×10–1 g mL−1. SEM images of DNA‐kanamycin capsules at selected regions; (e, f and g) the areas indicated in the white boxes in (d). CDNA=1.0×10−2 g mL−1; C kanamycin=1.0×10−1 g mL−1. The scale bar is 200 μm in (d), and 4 μm in (e, f and g). Reprinted from22 with permission of Royal Chemical Society.
RNA is also a major constituent of ribosome, where protein synthesis takes place and plays an important role in the replication of bacteria and viruses. Inhibition of protein synthesis using specific targeted RNA binders can inhibit protein synthesis and subsequently bacterial growth. New RNA binders to inhibit bacterial growth are thus an important target. RNA molecules are good molecular hosts due to their characteristic design, having secondary and tertiary structures.23 Ideally, one would like to identify recognition rules and well defined binding motifs, and utilize that knowledge for the design of specific RNA binders. Aminoglycosides show a preference for RNA binding by electrostatic, non‐intercalative interactions, as aminoglycosides containing amino and sugar are known to bind with ribosome in prokaryotes. They bind with the decoding region of 16 S rRNA of the 30 S subunit, and thus interfere with translation and translocation processes. This ultimately disrupts bacterial protein biosynthesis. The reported SAR for natural aminoglycosides suggests that the charge density present on aminoglycoside is important for RNA binding. Hence aminoglycosides containing four amino groups (kanamycin) show less RNA binding than five or six amino groups (neomycin B and tobramycin). The Tor group7 demonstrated that substituting a hydroxyl with an amino group can convert a poor RNA binder such as kanamycin A to reasonably good binder.
Kanamycin contamination in food is undesirable, therefore a new biosensor for the detection of small molecules such as kanamycin is required, and can be achieved using supramolecular principles. Bansal et al. reported a new, highly sensitive ‘turn‐off/turn‐on’ biosensing approach for detection of kanamycin by using the intrinsic peroxidase‐like activity of gold nanoparticles,24which oxidise peroxidase substrate in the presence of H2O2 to form a colored reaction product. This activity is independent of gold aggregation behavior, which makes it similar to traditional biochemical assays such as ELISA. The peroxidase like activity of gold nanoparticle can be turned‐off by blocking its surface through the adsorption of ssDNA aptamer molecules, but this activity can be turned‐on as the high affinity of the aptamer for the target will induce aptamer desorption from the gold nanoparticle surface.
Aminoglycoside antibiotics lack spectroscopic and electrochemical properties, making their detection challenging. Interestingly, the aptamer also possesses functional characteristics in many cases similar to antibodies. Electrochemical sensors are specific, selective, sensitive and widely applicable for the detection of variety of target analytes. The binding properties of an electrode‐bound RNA or DNA aptamer will give specificity. White and co‐workers presented a systematic study of several approaches to develop a sensor for the detection of aminoglycoside antibiotics (Figure 7).25 They demonstrated several strategies for an electrochemical aptamer, capable of detecting tobramycin, and utilized several strategies to create larger target induced conformation changes in the aptamer structure, which improve the binding affinity, sensitivity, and the detection abilities of an RNA based electrochemical sensor. These electrochemical aptamer based sensors are capable of detecting tobramycin from micro‐ to nano‐molar level. However, the magnitude of these changes ultimately affects the magnitude of the signal gain achieved by the respective sensors. Below is a schematic illustration of two E‐AB sensor architectures: a low signal gain sensor and a high signal gain sensor. The graphical illustration shows a larger change in conformation upon target binding thus leading to a high signal change (see Figure 7).25
Figure 7.
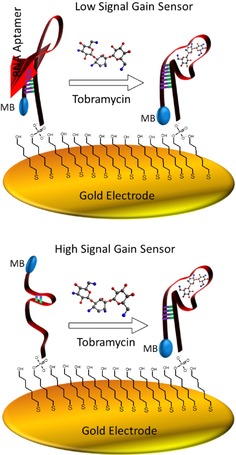
Electrochemical, aptamer‐based sensors employing structure‐switching aptamers rely on specific target‐induced changes in the conformation and/or flexibility of the aptamer. All sensor architectures investigated in this study appear to demonstrate an increase in conformation/flexibility upon target binding.
Several methods are available for kanamycin analysis, including HPLC, chromatography combined with mass spectrometry, capillary electrophoresis and enzyme‐linked immunosorbent assay. But these methods involve costly instruments, complicated procedures and poor sensitivity. Sun et al. developed an electrochemical aptamer based sensor/aptasensor based aptamer/target/aptamer sandwich structure for kanamycin detection which was achieved by reaction of H2O2 and HQ catalysed by horseradish peroxidase (HRP).26 However, the sandwich assay required two aptamers binding to two different sites, making this method several steps, which made the assay time‐consuming.27 Song et al. reported a new kanamycin aptamer as the sensor and HRP as a biocatalyst for signal amplification using a complementary DNA strand.28 Liu et al. developed an aptasensor using complementary strands labeled Fc as the signalling probe, which was found to be more sensitive that the signalling probe using aptamers labeled Fc.29 Recently, scientists have attempted to develop label‐free aptasensors based on target‐induced strand displacement. Jingyi et al. developed a label free and highly sensitive electrochemical aptasensor using graphene for the detection of kanamycin, in which graphene‐gold nanoparticles were used as modified electrodes with double‐stranded DNA(ds‐DNA) duplex probes, improving the sensitivity of the electrochemical sensor (Figure 8).30 Upon addition of kanamycin, the aptamer was displaced from the ds‐DNA duplex into solution, in which methylene blue (MB) was used as a redox indicator. The electrochemical aptasensor for kanamycin displayed a large linear range from 0.1 pmol/L to 10 pmol/L with a low detection limit of 0.03 pmol/L. When this strategy was used for direct analysis of milk samples, the results showed a high sensitivity and a good selectivity.
Figure 8.
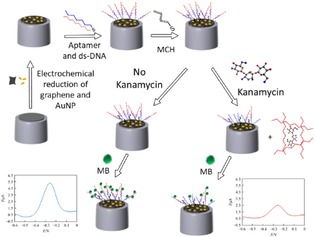
Aptasensor preparation and proposed principle for kanamycin detection. Adopted from Reprinted from30 with permission of Springer.
Alvarez et al. also used aptamers, to selectively recognise antibiotics.31 This group presented an aptamer‐based assay for the detection of neomycin B in milk, and showed that the use of a modified‐RNA sequence does not alter the selectivity of the aptamer, enabling it to distinguish the changes in the functionalities of target molecule.
Supramolecular hydrogels consist of a solid three‐dimensional network with noncovalent interactions such as hydrogen bonds, hydrophobic interaction, and π‐π interactions. Hydrogels of small molecules self‐assemble in water to yield supramolecular nanofibers. Supramolecular hydrogels are synthesized from hydrophilic molecules and stabilised by these interactions, which form macromolecular polymers and a three dimensional gel network. Hydrogen bonding is the main driving force to induce aggregation of small molecules in water to form nanofibers and hydrogels. Successful integration of biologically active molecules in hydrogels has led to rapid development of supramolecular hydrogels as a new class of biomaterial. These hydrogels have recently received much attention due to their potential in medical fields such as drug delivery, tissue culture, and medical treatment.
Yang et al. reported the first hydrogel derived from an aminoglycoside antibiotic (kanamycin), and described how the hydrogel based on kanamycin aminoglycoside preserves the specific interaction with their macromolecular targets, e. g. 16 S rRNA.32 This is a simple approach to explore and identify possible biological targets of supramolecular nanofibers/hydrogels. Figure 9 explains the conversion of functional molecules to hydrogels in a simple way to leverage the self‐assembly of the functional molecule, and the interactions between nanofibers and potential targets. The self‐assembly of nanogels leads to the formation of supramolecular nanofibers, and then supramolecular nanofibers which sequester the potential targets [(e. g. 16 S rRNA (in red)] in the gel phase (Figure 10).
Figure 9.
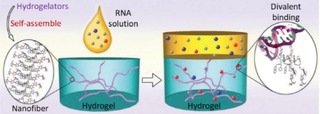
Supramolecular nanofibers sequester the potential targets [(e. g. 16 S rRNA (in red)] in the gel phase. Reprinted from32 with permission of Royal Chemical Society.
Figure 10.
Supramolecular nanofibers sequester the potential targets [(e. g. 16 S rRNA (in red: as shown in Figure 9)] in the gel phase.
Gentamycin is another class of aminoglycoside antibiotic which has a bactericidal effect on both gram positive and gram negative bacteria. It is commonly used against infections caused by Staphylococcus aureus, Staphylococcus epidermidis, Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis and Pseudomonas aeruginosa. Gentamycin is chemically stable and soluble in water. The amino group of gentamycin can protonate, and this positive charge facilitates the drug attachment to negatively charged hosts by electrostatic interactions. The release of this molecules is controlled by encapsulating the same in the porous network. This will allow the slow release of the drug. This was demonstrated by Tamanna et al who used mesoporous silica nanoparticles (MSNs) to control the release of gentamycin via a self‐assembled polyelectrolyte coating.33 They first loaded the gentamycin into the drug carrier by adjusting the pH, and then coated the MSNs with multilayer film of polyelectrolytes such as sodium polystyrene sulfonate (PSS) and poly (allylamine hydrochloride) (PAH). They achieved a high gentamycin loading capacity of 211 micro gm per mg MSNs at pH 7.4. The unique pore structure of the MSNs helps not only to control the release but also offers the space for storing a huge amount of gentamycin.
4. Nano‐Flower Assembly
The unique properties of the nanomaterials are effectively utilized for the delivery of antibiotics. It has been demonstrated that silver nanoparticles are quite effective in replacing conventional antibiotics due to their broad spectrum antimicrobial activity. However, silver nanoflowers oxidise and aggregate easily, and are toxic to mammalian cells. Thus there is a need to develop coatings on antibacterial agents. This was achieved by coating the silver nanoparticles with gold nanostructures. For example, Yan et al. demonstrated that gold‐silver‐gold alloy nanoflowers are highly effective antimicrobial materials with a low toxicity. They demonstrated in vivo inhibition of bacteria by using gold@silver‐gold alloy nanoflowers with a distinct atomic structure (Figure 11).34 Gold@silvergold alloy nanoflowers with rough surface adsorbed firmly on bacterial cell surfaces and damaged the cell membrane. They also used these nanoflowers for treatment of bacterial infection in mouse intestine and observed that there is a significant decrease in bacteria as compared to an untreated mouse which gives similar results to current antibiotics. These nanoflowers bind to the protein surface of bacteria, and leads to damage of bacterial cells and subsequently cell death.
Figure 11.
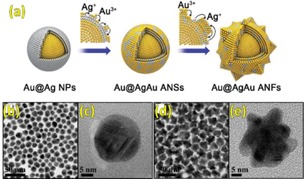
(a) Schematic illustration of the synthesis of Au@AgAu ANFs. The TEM and HRTEM images of (b and c) Au@AgAu ANSs and (d and e) Au@AgAu ANFs. Reprinted from34 with permission of Royal Chemical Society.
Recently, Liu and co‐workers developed a photoelectrochemical (PEC) aptasensor based on a 3D flower‐like TiO2‐MoS2AuNP composite, in which mercapto‐group modified aptamers were immobilised on the photoactive composite as a recognition unit for kanamycin (Figure 12).35 The composite was found to have three important factors: 1) acceleration of electron transfer, 2) increase in the loading of aptamers and 3) improvement in visible light excitation of the sensor. The aptasensor displayed a dynamic range from 0.2 nM to 450 nM for kanamycin, with a detection limit of 0.05 nM under optimal conditions. This is the first example which was demonstrated to be an effective scaffold for the development of photoelectrochemical biosensors.
Figure 12.
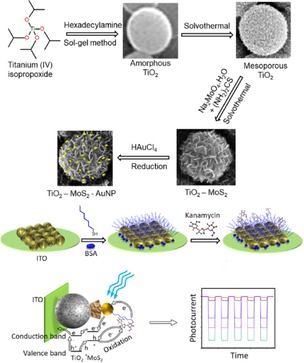
Synthesis of TiO2‐MoS2‐gold nanoparticles, fabrication steps of a kanamycin aptasensor and its PEC mechanism of detection. Reprinted from35 with permission of Elsevier.
In both nano‐ & bio‐technology, fluorescent molecules play an important role, for highly sensitive and selective sensing for the detection of cations, anions and neutral molecules, and also for bioimaging and biomolecule detection. Among biomolecules, heparin is widely used for anticoagulation during surgery. Heparin is a highly sulfonated aminoglycoside, and bears a negative charge at physiological pH due to presence of the sulphonate and carboxylic groups. Several methods are available for heparin detection such as use of surface plasmon resonance, colorimetric changes and indicator displacement assays, but they suffer from poor selectivity and specificity. Recently, Nalage et al. have used aminoglycoside antibiotics such as kanamycin A with a pyrenyl (kanamycin‐pyrenyl conjugate) fluorophore, which showed a high selectivity and sensitivity for detection of heparin over other biomolecules such as ATP, AMP, triglycine, aspartic acid, and act‐DNA (Figure 13).36 The kanamycin A‐pyrene conjugate carries polar hydroxyl groups for hydrogen bonding, amino groups for interaction with anionic species and the pyrenyl group for fluorescence. This is the first report where a kanamycin A based fluorescent neutral sensor has been used for specific recognition of heparin. However, the biological assays of the polyvalent kanamycin model showed neither antibacterial activity nor inhibitory activity against rRNA based protein synthesis.37
Figure 13.
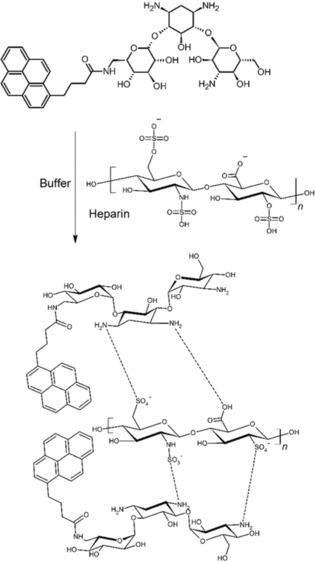
Interaction of heparin with a kanamycin A‐pyrene conjugate.36
RNA and DNA are targets for new detection methods. In recent years, amino modified silica nanoparticles and cationic gold nanoparticles have been used for DNA sensing. Aminoglycoside antibiotics were developed as DNA intercalators in molecular biology, and were also shown to be applicable for detection of DNA, generation of electroactive species and in biotransformation.38 Grace et al. used gold nanoparticles coated with aminoglycosides, which showed an efficient antimicrobial activity against gram‐negative and gram‐positive bacteria.39
We also reported a highly selective and sensitive technique for detection of DNA using kanamycin A‐modified gold nanoparticles, with a high binding affinity towards DNA as shown in Figure 14.40 We also demonstrated the interaction of ethidium bromide, a dye which specifically binds to DNA with kanamycin A‐modified gold nanoparticles. When ethidium bromide‐DNA (EtBr‐DNA) complex interacts with kanamycin A‐modified gold nanoparticles, fluorescence is observed. The EtBr‐DNA complex shows increased fluorescence with kanamycin A‐modified gold nanoparticles when compared to an unmodified gold nanoparticle. Aggregation of gold nanoparticles in the presence of DNA indicates that binding of kanamycin A‐modified gold particles to DNA is occurring.
Figure 14.
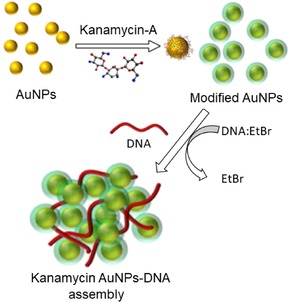
Schematic representation of kanamycin A‐modified gold nanoparticles and their self‐assembly with DNA.
In another report, we demonstrated the synthesis of gold and silver nanoparticles by reducing AuCl4 and AgNO3 using pineapple extract, and studied the interaction of these nanoparticles with aminoglycosides such as kanamycin A and neomycin, in which aminoglycoside antibiotics were used as stabilising agents. This work demonstrated that aminoglycosides play a crucial role, not only in tuning the properties of the Au and Ag nanoparticles, but also formed particles are polydispersed, and are much more stable in solution or in powder form for a longer time. We believe that this protocol is useful for the synthesis of shape controlled gold and silver nanoparticles, with unique structural and spectral properties at room temperature in aqueous medium.41
Belmont et al. reported kanamycin aminoglycoside derived gene delivery systems, in which cationic lipid characterised by an aminoglycoside head group allowed efficient gene transfer into a variety of mammalian cells in vitro, when used either alone or as a liposome formulation with the neutral colipid dioleoylphosphatidylethanolamine (DOPE). Aminoglycosides have six amino groups and several hydroxyl groups, and selective acylation of one of the six amino group provides aminoglycosides with lipophilic properties, which is required for their use as a vector for gene transfer. They also showed that functionalisation of cationic cholesterol conjugate characterized by a kanamycin headgroup and its polyguanidinylated derivative is very efficient for gene transfection into the mouse airways in vivo.42
Excess use of aminoglycoside antibiotics causes damage to kidneys and cranial nerves, which may result in renal dysfunction and irreversible hearing less. Therefore, it is desirable to determine the aminoglycosides level in blood and urine by a simple and efficient method. Yang et al used a copper microparticle‐modified carbon fiber microdisk electrode array for the determination of aminoglycoside antibiotics by capillary electrophoresis.43
5. Applications of Modified Aminoglycosides
The Westermann group used the Ugi multicomponent reaction to functionalise neomycin B and kanamycin B, obtaining multivalent neoglycoconjugates with 2,6‐diamino‐2,6‐dideoxyglucoseby as mono‐, di‐ and tri‐valent carbohydrate clusters as shown in Figure 15.44 These new derivatives are important as Wong et al. earlier reported that incorporation of 2,6‐diamino‐2,6‐dideoxyglucopyranose moieties (ring I of neomycin B) offers very good binding motifs for prokaryotic RNA interaction through the phosphodiester group and the Hoogsteen site of RNA residues (see Figure 15).45
Figure 15.
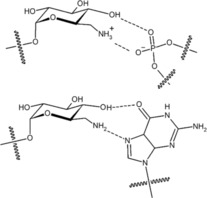
Binding modes of aminoglycosides to RNA.
Cowan and co‐workers46 in 1998 described the synthesis of novel aminoglycosides, and also developed new approaches to the recognition and specific binding of anionic species. Furthermore, they showed how these antibiotics can be used for anion sensors in biological and separation science. Aminoglycosides may provide new approaches to the recognition and specific binding of anionic species due to large number of functional groups they possess and their positive charge density in neutral solution. The coordination chemistry of neomycin B with a variety of negatively charged species was evaluated, with thermodynamics studied by isothermal titration calorimetry. Anion binding typically involves outer sphere coordination. Due to presence of six amino functional groups of neomycin B, a strong affinity to bind a variety of ionic species is expected, making it as a biological sensor and also in ion separation.
Cowan and co‐workers synthesised copper derivatives of aminoglycosides, and showed them to be efficient catalysts for RNA and DNA cleavage at neutral pH (Figure 16).47 They also showed that Cu2+‐kanamycin cleaved plasmid DNA yields residual DNA and larger product fragments along with cytosine (C), guanine (G), thymine (T), and adenine (A), which were confirmed by HPLC profiles. They later found that both the copper aminoglycosides follow a highly specific route mediated by abstraction of the C‐4H's in which opposing strands are separated by two additional base‐paired nucleotides.48 This provides a rational explanation for double‐strand cleavage of duplex DNA. Oxidative degradation of DNA is accomplished a number of ways, including abstraction of proton from C‐1′, C‐2′, C‐3′, C‐4′, and C‐5′ carbons of ribose. The moiety Cu2+‐neomycin or Cu2+‐kanamycin have potential importance in oxidative cleavage of double stranded DNA at C‐4′. This is highly specific at the C‐4′ position, giving a highly efficient route by abstraction of C‐4 H, as compared to all other different oxidative paths.
Figure 16.
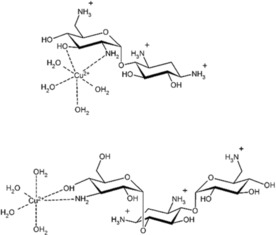
The chemical structure of copper derivatives of aminoglycosides.
Later, they demonstrated in vivo recognition and cleavage of an RNA target sequence with copper aminoglycoside for the first time.49 This suggested that copper aminoglycosides could be a potential source of new metal based drugs that not only recognise but also destroy RNA targets in vivo. To demonstrate in vivo cleavage chemistry, they designed a fluorescence assay based on green fluorescent protein. Copper kanamycin A was used for in vivo cleavage of the R23 RNA target sequence, which resulted in a decrease in translatable mRNA transcript.
The Tor group reported for the first time that neomycin B effectively hydrolyses the phosphodiester group of RNA dinucleoside, as compared to 1,3‐diaminopropane and 3‐aminopropan‐1‐ol at pH 8.0 and reverse phase HPLC confirmed the release of adenosine from loss of adenylyl (3 A–5 A) adenosine (ApA) as shown in Figure 17.50 It has been demonstrated that the DNA‐polyamine conjugate, a sequence‐specific RNA cleaver was able to break the RNA strand most effectively. Furthermore, they also showed that neomycin B is able to enhance the hydrolysis of a ribonucleoside monophosphate. Simple unstructured polyamines and also basic polypeptides were shown to RNA hydrolysers, and aminoglycoside antibiotics exhibited a similar effect. The rate of RNA hydrolysis by neomycin B is pH dependent, and as neomycin has six amino groups it will be protonated when the pH is lowered, and the rate of RNA hydrolysis is expected to decrease. Thus, this method provides a new method of RNA recognition with hydrolysis by using aminoglycosides. These molecules may be useful chemical probes for RNA structure and folding.
Figure 17.
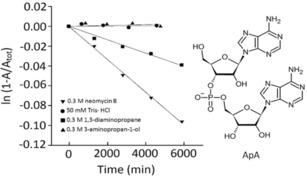
Kinetic analysis of ApA hydrolysis by various organic amines at pH 8.0 and 50 °C. Reprinted from50 with permission of Royal Chemical Society.
Kopaczynska et al.51 studied the interaction of aminoglycoside antibiotics tobramycin, kanamycin and neomycin with calf thymus DNA. Among these three antibiotics, tobramycin was shown to induce the selective condensation of nucleic acid into complex structures, blocking the biological activity of DNA. However, kanamycin and neomycin caused direct damage of genetic material rather than condensation. This different behavior of tobramycin as compared to other two aminoglycosides is due to smaller hydration sphere of tobramycin, indicating the drug may interact with the minor groove of the DNA duplex (see Figure 18). These interactions are important for designing new and efficient drugs, or minimizing their adverse side effects. Figure 18 demonstrates the condensation of tobramycin with DNA in 0.2 : 1, 1 : 1, and 10 : 1 base pair molar ratios. It can be clearly seen that with increasing ratio of tobramycin to DNA, formation of the compact structures such as toroids, spheres, and rods, respectively. Therefore, this recent study on the mechanisms of condensation is important, not only for understanding their functioning in cells, but also to design new drug molecules with minimal adverse side effects. These results also helped in understanding nephrotoxicity issues, and reactions of aminoglycoside antibiotics in the human body.
Figure 18.
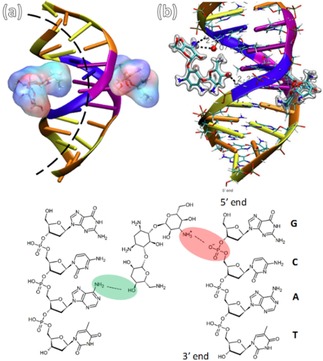
Molecular interaction mechanism of tobramycin with DNA. (a) Two aminoglycosides acting as a clamp induce DNA curvature. (b) Zoom in hydrogen bond between tobramycin molecule and oxygen of deoxyribose 5‐phosphate and the amine groups of tobramycin. Reprinted from51 with permission of Wily‐VCH.
6. Conclusions and Outlook
In summary, this review is a road map to stimulate further modification of the aminoglycoside antibiotics overcome issue in biological sciences. Chemical modification of various aminoglycosides, their self‐assembled nanostructures and interactions with DNA and RNA in real world applications were discussed in detail. We also discussed that aminoglycosides have appropriate binding sites due to the availability of hydroxyl groups which are readily available for hydrogen‐bonding, and can be functionalised through modification. This would pave the way for many more applications in supramolecular chemistry based on aminoglycosides, similar to other earlier reports that described for other molecules. The impending problems of bacterial resistance to antibiotics may well find an answer in the use of supramolecular chemistry with existing antibiotics, and this review offers a lot of possibilities of encouraging further work to this end. To improve the safety and efficacy of the multi drug‐resistant bacteria through optimized dosing regimens has led to a rehabilitated interest of the researcher. However, due to increased resistant power of bacteria against antibiotics make researcher to think to modify the aminoglycosides antibiotics using supramolecular self‐assembled structures. As supramolecular self‐assembly is only a phenomenon in which molecules spontaneously organizes into assembled structures from and such controlled structures of drug molecules into ordered structures has been demonstrated a powerful technique in designing and constructing nano‐ and bio‐techanology.52, 53, 54, 55, 56, 57 Such controls atom and/or molecular‐level in technology are important for the precise placement of components in real world applications.
Nevertheless, there are only few successful designs shown to overcome common aminoglycoside resistance mechanisms against multidrug‐resistant (MDR) pathogens. However, there need to be more study done in future to completely overcome these issues. There is always believe that functionalised aminoglycosides may become critical component of the current antibacterial arsenal due to their broad spectrum of activity, any may take rapid bactericidal action, and also may favorable chemical and pharmacokinetic properties, thus, will make them a clinically useful class of drugs across numerous infection types in future.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Mr. Ratan Wamanrao Jadhav born in Ambagaon, Tq. Deoni, pursuing a PhD under Prof. Sheshanath V. Bhosale. Having UGC‐JRF fellowship at School of Chemical Sciences, Goa University, India. Before joining Bhosale's group, he has completed his MSc in organic chemistry with distinction from Rajarshi Shahu College, Latur, India. He is also qualified various competitive exam's such as NET‐JRF (2 times) with all India rank 51, GATE and SET.

Biographical Information
Dr. Mohammad Al Kobaisi was born in 1971 in Iraq, and received his Ph.D. under the supervision of A/Prof. Colin Rix and Prof. David Mainwaring from RMIT University in 2007, investigating the design of selective adsorbent polymers for sensing applications. Currently, he is a senior researcher at Swinburne University of Technology. His main interest is in materials engineering, especially biopolymer hydrogels in drug delivery and vaccine formulation.

Biographical Information
Dr. Lathe A. Jones obtained his PhD from the University of Queensland (Australia), and has held research positions at the University of Manchester (UK), La Universidad de Zaragoza (Spain), and CIDETEC (Spain). He is currently a senior lecturer at RMIT University, and Deputy Director of CAMIC (Centre for Advanced Materials and Industrial Chemistry). His research interests are in applied chemistry, particularly electrochemistry, industrial chemistry, and supramolecular chemistry.

Biographical Information
Prof. Ajayan Vinu is a Global Innovation Chair and Director at Global Innovative Centre for Advanced Nanomaterials, The University of Newcastle. During these 18 years of research, Prof. Vinu has made a tremendous contribution in the field of nanoporous materials and their application in sensing, energy storage, fuel cells, adsorption and separation, and catalysis. His contribution in nanoporous materials is also clearly reflected by Science Watch international ranking as one of the top 15 researchers in the field and has led to 350 original research papers with ca. 17,600 citations and an H‐index of 68.

Biographical Information
Professor Sheshanath V. Bhosale was born in 1976 in Indral Tq. Deoni, completed his M.Sc. in chemistry from the Maharashtra Udaygiri Mahavidyalaya, Udgir (S.R.T.M.U. Nanded) in 1999. He then worked as a project assistant at NCL, Pune, before moving to the Freie University Berlin, Germany, where he received his Ph.D. (Magna Cum Lauda) in supramolecular chemistry under the supervision of Prof. J. H. Fuhrhop in 2004. Dr. Bhosale pursued his postdoctoral studies with Prof. S. Matile at University of Geneva, Switzerland, under the auspices of a Roche Foundation Fellowship. This was followed by a stay at Monash University, Australia, for five years as an ARC‐APD Fellow. He worked at RMIT University Melbourne for 6 years as ARC‐Future Fellowship followed by Senior researcher. Currently, Prof. Bhosale is now working at the School of Chemical Sciences, Goa University, India as a UGC‐FRP Professor, His research interest in design and synthesis of π‐functional materials, especially small molecules, for sensing, biomaterials and supramolecular chemistry applications.

Acknowledgements
S.V.B. (GU) acknowledges Council of Scientific & Industrial Research (CSIR), India for providing support (Approved No. 02 (0357)/19/EMR‐II) and also acknowledges University Grant Commission (UGC) Faculty Research Program, New Delhi, India for an award of Professorship. R.W.J. acknowledges UGC for providing Junior Research Fellowship.
R. W. Jadhav, M. A. Kobaisi, L. A. Jones, A. Vinu, S. V. Bhosale, ChemistryOpen 2019, 8, 1154.
References
- 1. Xie J., Talaska A. E., Schacht J., Hear. Res. 2011, 281, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krause K. M., Serio A. W., Kane T. R., Connolly L. E., Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen L. F., Kaye D., Infect. Dis. Clin. North Am. 2009, 23, 1053–1075. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Kirst H. A., Allen N. E., Comprehensive Medicinal Chemistry II (Eds.: J. B. Taylor, D. J. Triggle), Elsevier, Oxford, 2007, pp. 629–652; [Google Scholar]
- 4b. Hooper I. R., Amino Acides Pept. Proteines (Eds.: H. Umezawa, I. R. Hooper), Springer Berlin Heidelberg, Berlin, Heidelberg, 1982, pp. 1–35; [Google Scholar]
- 4c. Umezawa S., Advances in Carbohydrate Chemistry and Biochemistry, Vol. 30 (Eds.: R. S. Tipson, D. Horton), Academic Press, 1974, pp. 111–182. [DOI] [PubMed] [Google Scholar]
- 5. Schroeder R., Waldsich C., Wank H., EMBO J. 2000, 19, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Holdren J. P., Lander E. S., President's Council of Advisors on Science and Technology, Washington DC, 2014, p. 1–65; [Google Scholar]
- 6b. Centers for Disease Control and Prevention, Atlanta, GA, 2013, p. 1–114; [Google Scholar]
- 6c. Fischbach M. A., Walsh C. T., Science 2009, 325, 1089–1093; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6d. O′Connell K. M. G., Hodgkinson J. T., Sore H. F., Welch M., Salmond G. P. C., Spring D. R., Angew. Chem. Int. Ed. 2013, 52, 10706–10733; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 10904–10932; [Google Scholar]
- 6e. Gualerzi Claudio O., Brandi Letizia, Fabbretti Attilio, Pon C. L., Antibiotics: Targets, Mechanisms and Resistance, Wiley-VCH, Weinheim, Germany, 2013; [Google Scholar]
- 6f. Wright P. M., Seiple I. B., Myers A. G., Angew. Chem. Int. Ed. 2014, 53, 8840–8869; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 8984–9014; [Google Scholar]
- 6g. Fisher J. F., Mobashery S., MedChemComm 2016, 7, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Michael K., Tor Y., Chem. Eur. J. 1998, 4, 2091–2098. [Google Scholar]
- 8. Hoffman L. R., D′Argenio D. A., MacCoss M. J., Zhang Z., Jones R. A., Miller S. I., Nature 2005, 436, 1171. [DOI] [PubMed] [Google Scholar]
- 9. Daikos G., Jackson G. Gee, Lolans V. T., Livermore D., Adaptive Resistance to Aminoglycoside Antibiotics from First-Exposure Down-Regulation, Vol. 162, 1990. [DOI] [PubMed] [Google Scholar]
- 10. Hermann T., Cell. Mol. Life Sci. 2007, 64, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.
- 11a. Davies J, Smith D. I., Annu. Rev. Microbiol. 1978, 32, 469–508; [DOI] [PubMed] [Google Scholar]
- 11b. Miller G., Sabatelli F., Hare R., Waitz J., Dev. Ind. Microbiol. 1980, 21, 91–104; [Google Scholar]
- 11c. Bryan L. E., Antimicrobial Drug Resistance (Ed.: L. E. Bryan), Academic Press, 1984, pp. 241–277. [Google Scholar]
- 12. Fourmy D., Recht M. I., Blanchard S. C., Puglisi J. D., Science 1996, 274, 1367–1371. [DOI] [PubMed] [Google Scholar]
- 13.
- 13a. Lopez-Novoa J. M., Quiros Y., Vicente L., Morales A. I., Lopez-Hernandez F. J., Kidney Int. 2011, 79, 33–45; [DOI] [PubMed] [Google Scholar]
- 13b. Mingeot-Leclercq M.-P., Tulkens P. M., Antimicrob. Agents Chemother. 1999, 43, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fantin B., Ebert S., Leggett J., Vogelman B., Craig W., J. Antimicrob. Chemother. 1990, 27, 829–836. [DOI] [PubMed] [Google Scholar]
- 15. Forge A., Schacht J., Audiology and Neurotology 2000, 5, 3–22. [DOI] [PubMed] [Google Scholar]
- 16. Lortholary O., Tod M., Petitjean O., Cohen Y., Med. Clin. North Am. 1995, 79, 761–787. [DOI] [PubMed] [Google Scholar]
- 17.
- 17a. Puglisi J. D., Blanchard S. C., Green R., Nat. Struct. Mol. Biol. 2000, 7, 855–861; [DOI] [PubMed] [Google Scholar]
- 17b. Avent M. L., Rogers B. A., Cheng A. C., Paterson D. L., Intern. Med. 2011, 41, 441–449. [DOI] [PubMed] [Google Scholar]
- 18. Al Kobaisi M., Bhosale S. V., Latham K., Raynor A. M., Bhosale S. V., Chem. Rev. 2016, 116, 11685–11796. [DOI] [PubMed] [Google Scholar]
- 19. Atkins J. L., Patel M. B., Cusumano Z., Gokel G. W., Chem. Commun. 2010, 46, 8166–8167. [DOI] [PubMed] [Google Scholar]
- 20. Kopaczynska M., Lauer M., Schulz A., Wang T., Schaefer A., Fuhrhop J.-H., Langmuir 2004, 20, 9270–9275. [DOI] [PubMed] [Google Scholar]
- 21. Gouzy M.-F., Lauer M., Gonzaga F., Schulz A., Fuhrhop J.-H., Langmuir 2002, 18, 10091–10094. [Google Scholar]
- 22. Zhou J., Wen H., Ke F., Shi D., Brisky A. A., Wang N., Zhu L., Qiu X., Liang D., Chem. Commun. 2014, 50, 9525–9528. [DOI] [PubMed] [Google Scholar]
- 23. Hermann T., Westhof E., Curr. Opin. Biotechnol. 1998, 9, 66–73. [DOI] [PubMed] [Google Scholar]
- 24. Sharma T. K., Ramanathan R., Weerathunge P., Mohammadtaheri M., Daima H. K., Shukla R., Bansal V., Chem. Commun. 2014, 50, 15856–15859. [DOI] [PubMed] [Google Scholar]
- 25. Schoukroun-Barnes L. R., Wagan S., White R. J., Anal. Chem. 2014, 86, 1131–1137. [DOI] [PubMed] [Google Scholar]
- 26. Sun X., Li F., Shen G., Huang J., Wang X., Analyst 2014, 139, 299–308. [DOI] [PubMed] [Google Scholar]
- 27. Palchetti I., Mascini M., Anal. Bioanal. Chem. 2012, 402, 3103–3114. [DOI] [PubMed] [Google Scholar]
- 28. Song H.-Y., Kang T.-F., Li N.-N., Lu L.-P., Cheng S.-Y., Anal. Methods 2016, 8, 3366–3372. [Google Scholar]
- 29. Liu R., Yang Z., Guo Q., Zhao J., Ma J., Kang Q., Tang Y., Xue Y., Lou X., He M., Electrochim. Acta 2015, 182, 516–523. [Google Scholar]
- 30. Zheng J., Feng R., He C., Li X., Chem. Res. Chin. Univ. 2018, 34, 952–958. [Google Scholar]
- 31. de-los-Santos-Álvarez N., Lobo-Castañón M. J., Miranda-Ordieres A. J., Tuñón-Blanco P., J. Am. Chem. Soc. 2007, 129, 3808–3809. [DOI] [PubMed] [Google Scholar]
- 32. Yang Z., Kuang Y., Li X., Zhou N., Zhang Y., Xu B., Chem. Commun. 2012, 48, 9257–9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamanna T., Bulitta J. B., Yu A., J. Mater. Sci. Mater. Med. 2015, 26, 117. [DOI] [PubMed] [Google Scholar]
- 34. Yan W., Yang L., Wang H., Zhang J., Shen W., Nanoscale 2018, 10, 15661–15668. [DOI] [PubMed] [Google Scholar]
- 35. Liu X., Liu P., Tang Y., Yang L., Li L., Qi Z., Li D., Wong D. K. Y., Biosens. Bioelectron. 2018, 112, 193–201. [DOI] [PubMed] [Google Scholar]
- 36. Nalage S. V., Bhosale S. V., Bhargava S. K., Bhosale S. V., Tetrahedron Lett. 2012, 53, 2864–2867. [Google Scholar]
- 37. Tanaka H., Nishida Y., Furuta Y., Kobayashi K., Bioorg. Med. Chem. Lett. 2002, 12, 1723–1726. [DOI] [PubMed] [Google Scholar]
- 38.
- 38a. Angelini T. E., Liang H., Wriggers W., Wong G. C. L., PNAS 2003, 100, 8634–8637; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38b. Sanders L. K., Guáqueta C., Angelini T. E., Lee J.-W., Slimmer S. C., Luijten E., Wong G. C. L., Phys. Rev. Lett. 2005, 95, 108302; [DOI] [PubMed] [Google Scholar]
- 38c. Sanders L. K., Xian W., Guáqueta C., Strohman M. J., Vrasich C. R., Luijten E., Wong G. C. L., PNAS 2007, 104, 15994–15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grace A. N., Kannaiyan P., Quinolone Antibiotic-Capped Gold Nanoparticles and Their Antibacterial Efficacy Against Gram Positive and Gram Negative Organisms, Vol. 1, 2007. [Google Scholar]
- 40. Nalage S. V., Bhosale S. V., Bhosale S. V., Nanosci. Nanotech. Let. 2012, 4, 400–404. [Google Scholar]
- 41. Nalage S., Bhosale S. V., Bhosale S., The Open Nanoscience Journal 2011, 5, 78–82. [Google Scholar]
- 42. Belmont P., Aissaoui A., Hauchecorne M., Oudrhiri N., Petit L., Vigneron J.-P., Lehn J.-M., Lehn P., J. Gene Med. 2002, 4, 517–526. [DOI] [PubMed] [Google Scholar]
- 43. Yang W.-C., Yu A.-M., Chen H.-Y., J. Chromatogr. A 2001, 905, 309–318. [DOI] [PubMed] [Google Scholar]
- 44. Westermann B., Dörner S., Chem. Commun. 2005, 2116–2118. [DOI] [PubMed] [Google Scholar]
- 45. Wong C.-H., Hendrix M., Manning D. D., Rosenbohm C., Greenberg W. A., J. Am. Chem. Soc. 1998, 120, 8319–8327. [Google Scholar]
- 46. Ohyama T., Cowan J. A., Chem. Commun. 1998, 467–468. [Google Scholar]
- 47.
- 47a. Sreedhara A., Freed J. D., Cowan J. A., J. Am. Chem. Soc. 2000, 122, 8814–8824; [Google Scholar]
- 47b. Sreedhara A., Patwardhan A., Cowan J. A., Chem. Commun. 1999, 1147–1148. [Google Scholar]
- 48. Patwardhan A., Cowan J. A., Chem. Commun. 2001, 1490–1491. [Google Scholar]
- 49. Chen C.-A., Cowan J. A., Chem. Commun. 2002, 196–197. [DOI] [PubMed] [Google Scholar]
- 50. Kirk S. R., Tor Y., Chem. Commun. 1998, 147–148. [Google Scholar]
- 51. Kopaczynska M., Schulz A., Fraczkowska K., Kraszewski S., Podbielska H., Fuhrhop J. H., Eur. Biophys. J. 2016, 45, 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sawada T., Serizawa T., Bull. Chem. Soc. Jpn. 2018, 91, 455–466. [Google Scholar]
- 53. Li B. L., Setyawati M. I., Chen L., Xie J., Ariga K., Lim C.-T., Garaj S., Leong D. T., ACS Appl. Mater. Interfaces 2017, 9, 15286–15296. [DOI] [PubMed] [Google Scholar]
- 54. Komiyama M., Yoshimoto K., Sisido M., Ariga K., Bull. Chem. Soc. Jpn. 2017, 90, 967–1004. [Google Scholar]
- 55. He H., Xu B., Bull. Chem. Soc. Jpn. 2018, 91, 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ariga K., Leong D. T., Mori T., Adv. Funct. Mater. 2018, 28, 1702905. [Google Scholar]
- 57. Kobaisi M. Al, Bhosale S. V., Latham K., S. V. Bhosale 2016. ,16, 11685–11796 [DOI] [PubMed] [Google Scholar]


