Abstract
The dysregulation of microRNAs (miRNAs) is associated with the development and progression of a variety of cancers, including liver cancer. Aberrant expression of miRNA (miR)-124 has been demonstrated in liver cancer, but its functional mechanism in liver cancer is still largely unknown. Metastasis of liver cancer is one of the most common causes of mortality. The present study showed that miR-124 inhibited the proliferation, migration and invasion of liver cancer cells. Furthermore, chloride intracellular channel 1 (CLIC1) was identified as a novel target of miR-124 in liver cancer cells. Overexpression of miR-124 reduced CLIC1 expression at both the protein and mRNA levels in liver cancer cells. Downregulation of CLIC1 decreased the migration and invasion of liver cancer cells without affecting cell proliferation. Taken together, these results showed that CLIC1 is a critical target for miR-124-mediated inhibitory effects on cell migration and invasion. Thus, miR-124 or suppression of CLIC1 may have diagnostic value and therapeutic potential for the treatment of human liver cancer.
Keywords: liver cancer, microRNA-124, migration, invasion, chloride intracellular protein 1
Introduction
Liver cancer is the fifth most frequently diagnosed cancer and the second most common cause of cancer mortality in males worldwide (1). In females, the numbers are as high as ninth and sixth worldwide for diagnosed cancer and cause of mortality, respectively (1). Liver cancer constitutes a major global health problem, with the highest incidence rates in East Asia (1). The major causes of liver cancer include chronic hepatitis B and C virus infection, aflatoxin exposure, alcohol abuse, smoking and obesity (2). Most liver cancer patients are diagnosed at advanced stages with a poor prognosis; surgical resection and chemotherapy may prolong the overall survival time of liver cancer patients (3). Therefore, there is an urgent need to identify effective biomarkers and therapeutic targets for the treatment of liver cancer.
MicroRNAs (miRNAs) are small noncoding RNAs composed of ~22 nt. They usually bind to the 3′-untranslated region (UTR) of mRNAs and suppress gene expression by either degrading the mRNA or suppressing mRNA translation into protein, depending on pairing complementarity (4,5). Growing evidence indicates that miRNAs play important roles in many biological processes, including development, proliferation, differentiation, apoptosis and oncogenesis (5). It has been shown that miRNAs are abnormally expressed in hepatocellular carcinoma (HCC), one of the most common primary liver cancers representing 75–85% of cases (1), and act as tumor oncogenes or suppressors by affecting various targets to regulate tumorigenesis, invasion and metastasis (6). Furthermore, many miRNAs show promise as prognostic markers or therapeutic targets for the treatment of different cancer types (7,8).
miRNA-124 (miR-124) was first identified to be highly expressed in the central nervous system, and it regulates many neuronal activities (9–11). Subsequent studies revealed that miR-124 is significantly downregulated in several types of human cancer and functions as a tumor suppressor (9,12,13). miR-124 is able to recognize cell proliferation-related genes such as EPH receptor A2 (14), Smad4 (15), and AKT-solute carrier family 2 member 1/HKII (16). Several studies have shown aberrant expression of miR-124 in liver cancer, and decreased expression of miR-124 is correlated with shorter overall survival and poor prognosis in patients with liver cancer, suggesting that miR-124 might be a potential biomarker for the early diagnosis of liver cancer (17,18).
Chloride channels are membrane proteins with Cl− permeation pores. The mammalian chloride intracellular ion channel (CLIC) family consists of 6 subfamilies: CLIC1, CLIC-2, CLIC-3, CLIC-4, CLIC-5 and CLIC-6 (19). CLIC1 was initially identified in the human myelomonocytic cell line U937 in 1997 (20). Studies have shown that the CLIC1 protein level is increased in different types of cancer, including epithelial ovarian cancer (21), gastric cancer (22), pancreatic cancer (23) and HCC (24). All of these findings suggest that CLIC1 is an oncogene or a tumor marker in the progression of cancer. The present study identified CLIC1 as a miR-124 target gene by mass-spectrometry, luciferase assay and western blot analysis. Using further knockdown and functional assays, it was confirmed that miR-124 functions as a tumor suppressor through the downregulation of CLIC1. Thus, miR-124 and CLIC1 may serve as a biomarkers or have therapeutic potential for the treatment of human liver cancer.
Materials and methods
Hepatic cell culture and transfection
Human liver cancer HepG2 cells and human fetal hepatic HL-7702 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C and 5% CO2. The cell lines were characterized by Shanghai Yihe Applied Biotechnology Co., Ltd. using short tandem repeat markers. miR-124 precursor oligonucleotide (miR-124) and mismatched sequence as a negative control (cat. no. AM17111; pre-scrambled miRNA control; NC) were synthesized by Ambion; Thermo Fisher Scientific, Inc. The sequence of miR-124 was as follows: Sense, 5′-GGCAUUCACCGCGUGCCUUATT-3′ and antisense, 5′-UAAGGCACGCGGUGAAUGCCAA-3′. HepG2 cells were seeded in 12-well plates (1×105 cells/well) and incubated for 20 h before transfection. The transfection of miRNA (100 nm) or plasmid (2 µg) was carried out with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. For rescue experiments, the miRNA (100 nm) and plasmids (2 µg) were transfected into HepG2 cells using Lipofectamine® 2000.
Plasmid construction
Total RNA was extracted from HepG2 cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.) and was reverse-transcribed using a murine Moloney Leukemia Virus (M-MLV) kit (Takara Biotechnology Co., Ltd.). The PCR was performed using Ex Taq (Takara Biotechnology Co., Ltd.). The 3′-UTR of CLIC1 was amplified by PCR and constructed into the SpeI/HindIII sites downstream of the luciferase gene in the pMIR-REPORT Luciferase miRNA Expression Reporter Vector (Ambion; Thermo Fisher Scientific, Inc.). The primers for the 3′-UTR of CLIC1 were as follows: Forward, 5′-gactagtGCCCCTCCTGGGACTCCCT-3′ and reverse primer, 5′-atgcaagcttTTTTGCGTAAAAACACTTG-3′. A CLIC1 expression vector, pEGFP-N1-CLIC1, was constructed in our laboratory. Primers for CLIC1 mRNA were as follows: Forward, 5′-atcgctcgagATGGCTGAAGAACAACCGCAGG-3′ and reverse, 5′-agtcgacTTATTTGAGGGCCTTTGCCAC-3′. The PCR reaction was carried out under the following conditions: 95°C for 5 min; 30 cycles of 95°C for 30 sec, 56°C for 30 sec and 72°C for 1 min; and 72°C for 5 min. All constructs were confirmed by SpeI/HindIII or XhoI/BamHI restriction digestion and DNA sequencing. The restriction digestion was performed at 37°C for 1 h and the DNA sequencing was performed by BGI.
CLIC1 knockdown
CLIC1 small interfering RNA (siR-CLIC1) and scramble siRNA control (siR-NC) was purchased from Shanghai GeneChem Co., Ltd. The siR-CLIC1 sequence was 5′-GGACCGAGACAGTGCAGAA-3′ and the siR-NC sequence was 5′-TTCTCCGAACGTGTCACGT-3′. HepG2 cells were seeded 80% confluent and transfected with 50 nm siRNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The knockdown effect was evaluated by reverse transcription-quantitative PCR (RT-qPCR) and western blotting at 24, 48 and 72 h post-transfection.
RT-qPCR
Total RNA from HepG2 and HL-7702 cells was extracted with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The expression of miR-124 was measured using a TaqMan MicroRNA assay kit (Ambion; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions (25). Relative expression was calculated using the 2−ΔΔCq method and normalized to the expression of RNU6B (Ambion; Thermo Fisher Scientific, Inc.) (26). For the CLIC1 mRNA analysis, the total RNA was extracted from cells transfected with miR-124 or control with TRIzol® reagent. cDNA was synthesized using a M-MLV (Takara Biotechnology, Co., Ltd.) under the following program: 70°C for 10 min, then cooling on ice, 42°C for 1 h, 70°C for 15 min and held at 4°C. qPCR was performed in triplicate in a 20 µl reaction volume with SYBR-Green (Bio-Rad Laboratories, Inc.) as follows: 95°C for 3 min; and 40 cycles of 95°C for 20 sec, 58°C for 30 sec and 72°C for 20 sec. The primers used for RT-qPCR were as follows: CLIC1 forward, 5′-AATTCAAACCCAGCACTCAATG-3′ and reverse primer, 5′-CAGCACTGGTTTCATCCACTT-3′; and GAPDH forward, 5′-CCACTCCTCCACCTTTGAC-3′ and reverse primer, 5′-ACCCTGTTGCTGTAGCCA-3′. The expression level of GAPDH was used as a control. Relative expression was calculated via the comparative Cq method. The specificity of the PCR products was confirmed by melting curve analysis.
Immunoblotting
Total protein was isolated from cell lines transfected with RNA oligonucleotide and/or plasmid DNA in cell lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mm NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM PMSF and 1% sodium deoxycholate]. Protein concentration was measured using a Bio-Rad protein assay kit (cat. no. 5000002; Bio-Rad Laboratories, Inc.). Protein samples were separated by 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Amersham; GE Healthcare). Primary antibody against CLIC1 (1:1,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-81873) and tubulin (1:5,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-365791) was applied after blocking the membrane with 5% nonfat milk in PBS + 0.1% Tween-20, at room temperature for 1 h, followed by the appropriate horseradish peroxidase-conjugated secondary antibody (1:5,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-516102). The membranes were detected using an ECL western blotting detection system (EMD Millipore). Signals were calculated using ImageJ 1.50 (National Institutes of Health).
Proteomic analysis
HepG2 cells were transfected with miR-124 or pre-scrambled miRNA control (NC). After 48 h of transfection, cellular proteins were extracted using the ProteoPrep Total Extraction Sample kit (Sigma-Aldrich; Merck KGaA) and the protein concentration was determined by Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc.). Proteins were used for two-dimensional gel electrophoresis (2-DGE) as described in the manufacturer's instructions (Bio-Rad Laboratories, Inc.). The first-dimension isoelectric-focusing of 2-DGE was carried out using pH 3–10 immobilized pH gradient ReadyStrips, and in the second dimension, the protein was separated by 8–14% gradient SDS-PAGE. The gels were stained with Coomassie brilliant blue and scanned with a flatbed scanner. The analysis included picture merge and protein spot detection performed by PDQuest 2-DE analysis software (BioRad Laboratories, Inc., version 8.0). Gel comparison was performed using the ‘Automated Detection and Matching’ function in PDQuest software, combined with manual pair correction. The yellow-labeled spots showed at least a 2-fold decrease between the miR-124 sample and control, with statistical significance (P<0.05). Selected protein spots were excised from the gels and identified by mass spectrometry. Mass spectrometry analysis was performed at the Teaching Center of Biology Experiment, School of Life Sciences, Sun Yat-Sen University.
Luciferase assay
HepG2 cells with 80% confluence were transfected with luciferase constructs containing the 3′-UTR of CLIC1 (2 µg) and/or miR-124 or pre-scrambled miRNA control (NC) (100 nm) using Lipofectamine® 2000. After 48 h of transfection, cells were harvested for luciferase assays using the Luciferase Assay System (Promega Corporation), according to the manufacturer's protocol. pMIR-REPORT-β-gal was used for normalization. TargetScan (targetscan.org) was used to search for complementary sites of miR-124 in the 3′-UTR of CLIC1 mRNA.
Cell growth and viability
Cells were counted and plated at a density of 3×103 cells/well in 96-well plates in triplicate. Cell viability was determined at 24, 48 and 72 h post-transfection. Spectrophotometry was performed at λ=450 nm and λref =630 nm after incubation with 10 µl water-soluble tetrazolium salt (WST)-1 in 100 µl medium (Roche Molecular Diagnostics) for 2 h. For the colony formation assays, cells were seeded in a 6-well plate (0.8×103 cells/well) and cultured for 2 weeks. Colonies were fixed with cooled methanol for 10 min at 4°C and stained using 0.1% crystal violet for 10 min at room temperature. The images were taken using a camera. Visible colonies were manually counted. Each group was measured in triplicate.
Scratch wound-healing motility assays
A total of 1.0×106 cells were seeded and grown to confluence in 60-mm dishes with 10% FBS DMEM media. A scratch was made on the cell monolayer using a pipette tip at 48 h after miR-124 or pre-scrambled miRNA control transfection. The cells was washed in PBS twice and cultured in DMEM without FBS. The wound areas were imaged at 24 and 48 h after the scratch was made. The distance between the two edges of the scratch was measured using the ImageJ 1.50 (National Institutes of Health). The images were taken with a Leica Microsystems, Inc. light microscope at a magnification of ×100.
Transwell assay
Cell migration was detected using a 6-well plate Transwell system without Matrigel coating (BD Biosciences). Cell invasion assays were performed using Matrigel Invasion Chambers (BD Biosciences) precoated with ECM gel (Sigma-Aldrich; Merck KGaA). A total of 1.0×106 cells at 48 h post-transfection were seeded into the upper chamber with serum-free medium. The lower chamber was prepared with medium containing 10% FBS, which served as a chemoattractant. After incubating for 24 h, the non-invasive cells were mechanically removed. The invasive cells on the lower surface of the membrane were then washed, fixed with methanol for 20 min at 4°C and stained with 0.1% crystal violet for 10 min at room temperature. Cells were counted in 10 random optical fields under a Leica Miscrosystems, Inc. light microscope at ×400 magnification. The images were taken by a Leica Miscrosystems, Inc. light microscope at a magnification of ×100.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software, Inc.). All numerical data are displayed as the mean ± SEM. Statistical comparisons among two or more groups were conducted using one-way ANOVA test followed by the Newman-Keuls multiple comparison test, or Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-124 functions as a tumor suppressor in liver cancer cells
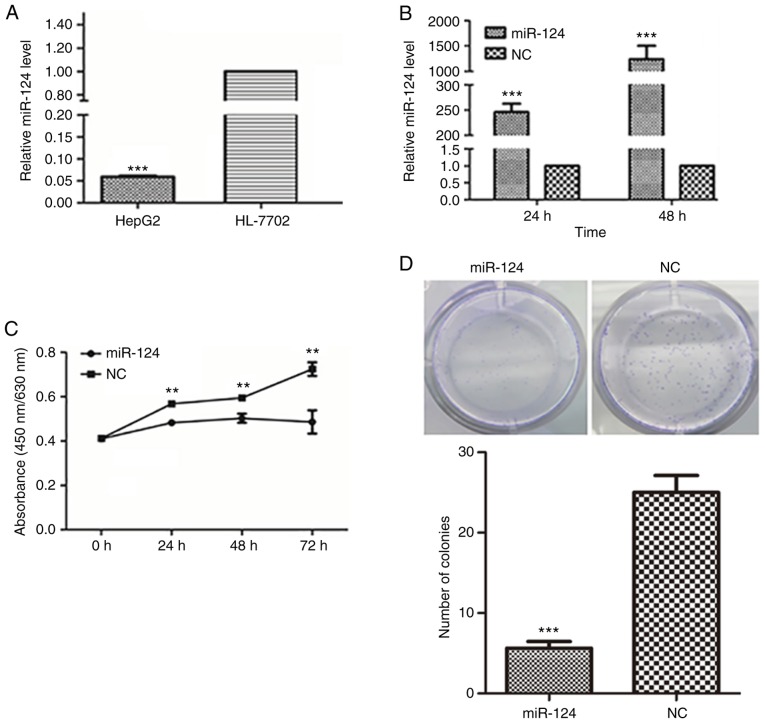
miR-124 levels were compared between a human liver cancer cell line and a normal liver epithelial cell line using RT-qPCR. In HepG2 liver cancer cells, the expression of miR-124 was significantly decreased compared with that in the normal liver epithelial cells (Fig. 1A), suggesting that miR-124 may function as a tumor suppressor in liver cancer cells.
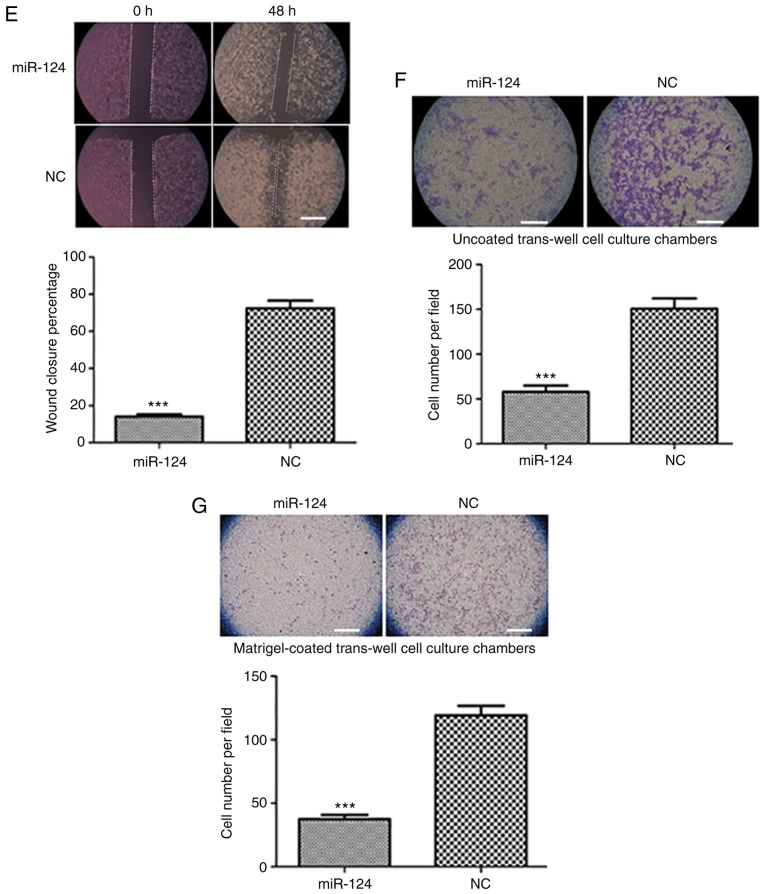
Figure 1.
Ectopic expression of miR-124 suppresses liver cancer cell proliferation, migration and invasion. (A) Detection of the miR-124 expression level in human liver cancer cells and a normal liver epithelial cell line by RT-qPCR with normalization to U6 ***P<0.001 vs. HL-7702. (B) Evaluation of the efficiency of the miR-124 precursor at 24 and 48 h after transfection using RT-qPCR. (C) Summary of the effect of miR-124 on the viability of liver cancer cells, according to the water soluble tetrazolium salt-1 assay. (D) Colony formation assay showing the effect of miR-124 on the cell growth of liver cancer cells. Scale bar, 200 µm. **P<0.01, ***P<0.001 vs. respective NC. RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA; NC, negative control. (E) Analysis of the function of miR-124 on the migration of liver cancer cells by scratch wound-healing motility assay. Scale bar, 200 µm. (F) The effect of miR-124 on the migration of liver cancer cells in the Transwell assay without Matrigel coating. Scale bar, 200 µm. (G) A Matrigel-coated Transwell assay was used to analyze the effect of miR-124 on cell invasion in liver cancer cells. Scale bar, 200 µm. ***P<0.001 vs. respective NC. RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA; NC, negative control.
To further confirm the function of miR-124 in liver cancer cells, HepG2 cells with low miR-124 expression were used as a cell model, and miR-124 was overexpressed in HepG2 cells by transfecting miR-124 precursor; scrambled miRNA precursor was used as the NC. The increased level of miR-124 was confirmed by RT-qPCR (Fig. 1B). Overexpression of miR-124 significantly inhibited the viability and proliferation of HepG2 cells (Fig. 1C and D). In the scratch wound-healing motility assay, transfecting miR-124 into HepG2 cells resulted in slower closure of the wound compared with that in control cells (Fig. 1E). In the Transwell assay without Matrigel coating, HepG2 cells expressing miR-124 migrated to a lesser extent to the lower chamber, compared with the control cells (Fig. 1F). Similarly, cell invasion was also largely inhibited by transfecting miR-124, as demonstrated by the Transwell assay with Matrigel (Fig. 1G). All these observations suggested that miR-124 functions as a tumor suppressor in liver cancer.
miR-124 targets CLIC1 and negatively regulates its expression
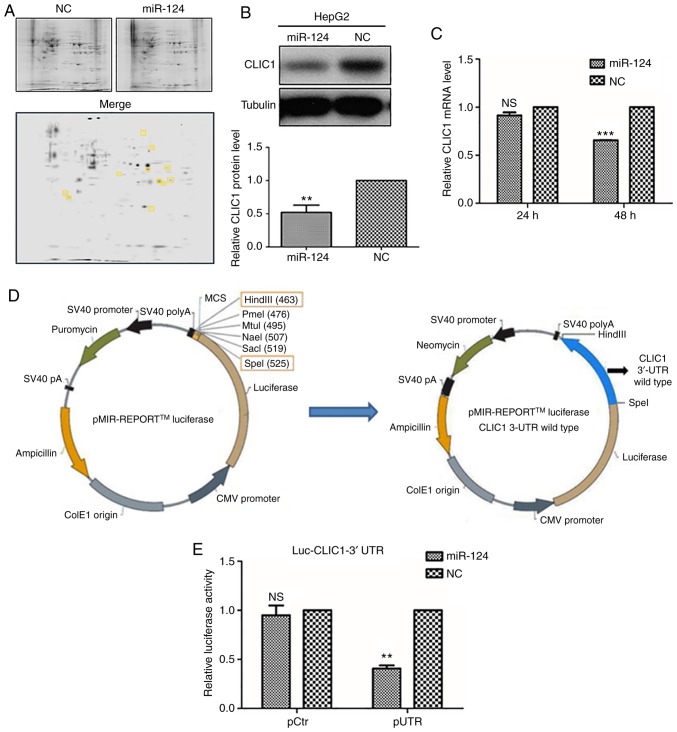
To explore the targets of miR-124 in liver cancer cells, mass spectrometry was used to analyze the differential expression of proteins between miR-124-overexpressing HepG2 cells and control cells (Fig. 2A). Among the proteins detected by two-dimensional gel electrophoresis, CLIC1 expression was decreased in miR-124-overexpressing cells, as compared with control cells (Fig. 2A), suggesting that CLIC1 is one of the potential targets of miR-124. To confirm this finding, HepG2 cells were further transfected with either miR-124 or NC, and the endogenous expression of CLIC1 was detected at both the protein and mRNA levels. Overexpression of miR-124 caused a significant decrease in CLIC1 protein expression (Fig. 2B). miRNAs are able to regulate gene expression by either degrading the mRNA of a target gene or preventing its translation. To better understand the mechanism underlying the downregulation of CLIC1 by miR-124, the mRNA level of CLIC1 was examined in miR-124-overexpressing cells. The RT-qPCR results revealed that the mRNA level of CLIC1 was reduced at 48 h after transfection with miR-124, compared with the NC (Fig. 2C). These data indicated that CLIC1 is a target gene of miR-124, which is likely to downregulate CLIC1 through mRNA degradation.
Figure 2.
CLIC1 is directly regulated by miR-124. (A) Proteomic analysis of differentially expressed proteins between miR-124-overexpressing and negative control cells; The images of the gels were analyzed using PDQuest two-dimensional electrophoresis analysis software (BioRad Laboratories, Inc.; version 8.0). The 11 yellow squares indicate spots with at least a 2-fold decrease in miR-124 expression. Samples were identified by mass spectrometry. (B) Protein levels of CLIC1 were tested after miR-124 transfection in liver cancer cells by western blotting. (C) CLIC1 mRNA levels were tested after miR-124 transfection in liver cancer cells by reverse transcription-quantitative PCR. (D) Diagram of the CLIC1 3′-UTR-containing reporter constructs. (E) Luciferase reporter assay in liver cancer cells **P<0.01, ***P<0.001 vs. respective NC. CLIC1, chloride intracellular protein 1; NC, negative control; miR, microRNA; UTR, untranslated region; pCtr, control plasmid.
miR-124 targets the 3′-UTR of CLIC1 mRNA
To further identify the target region of CLIC1 for miR-124, the 3′-UTR of CLIC1 mRNA was cloned into a luciferase vector (Fig. 2D). Co-transfection of miR-124 significantly inhibited the luciferase activity of the CLIC1 3′-UTR construct (Fig. 2E), suggesting that miR-124 targets the 3′-UTR of CLIC1 mRNA and prevents subsequent translation. Usually, the regulatory effect of miRNAs on target genes depends on the seed region of the miRNA. Bioinformatics analysis using TargetScan was used to search for complementary sites in the 3′-UTR of CLIC1 mRNA for the seed region of miR-124 (data not shown). Unexpectedly, there is no complementary site in the 3′-UTR of CLIC1 for the seed region of miR-124.
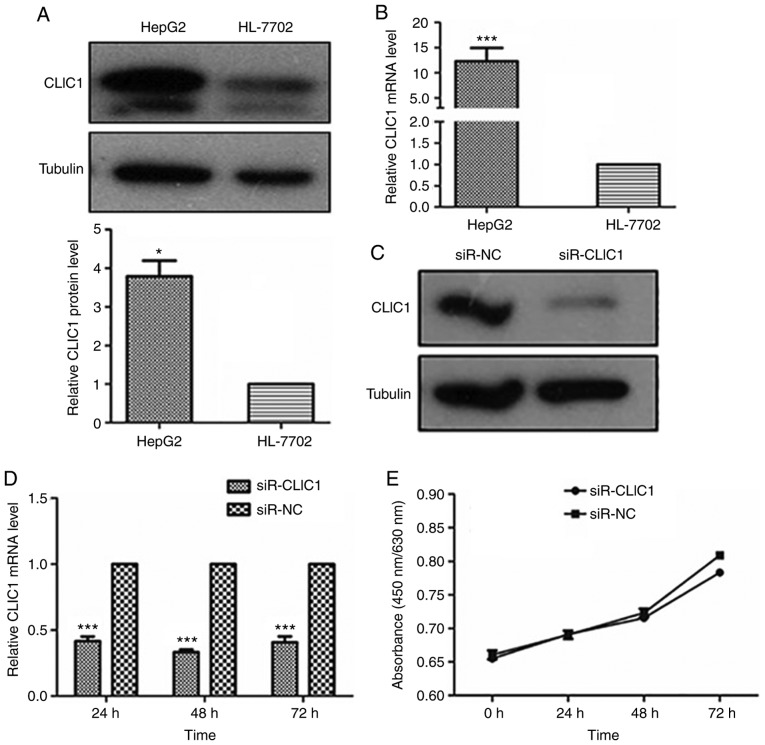
Inhibition of cancer progression in liver cancer cells by CLIC1 knockdown
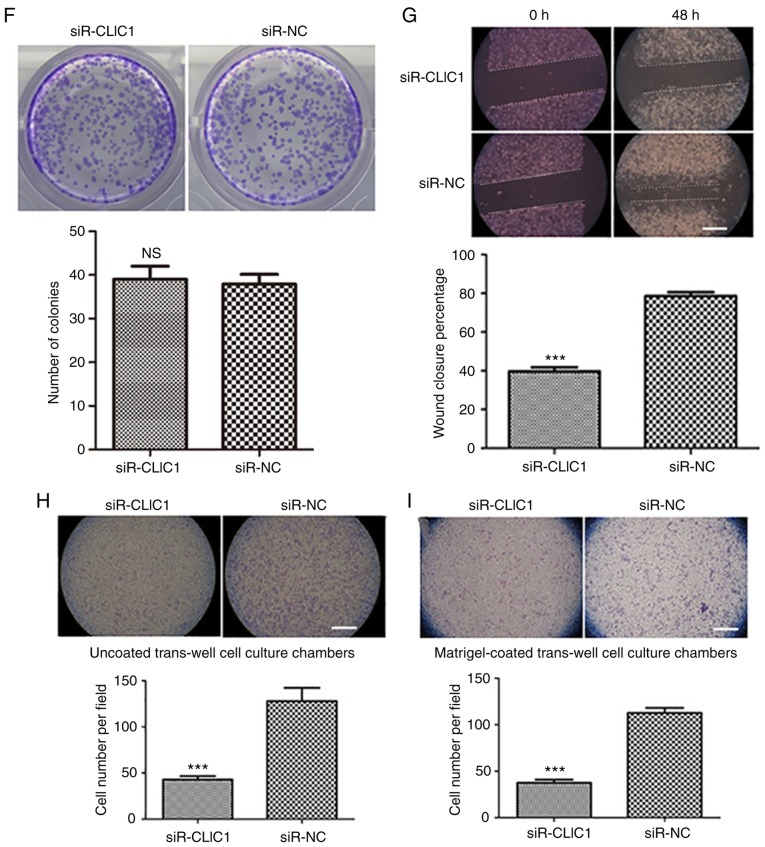
To explore the function of CLIC1 in liver cancer, CLIC1 protein and mRNA levels were measured in a liver cancer cell line (HepG2) and in the normal human hepatocyte cell line HL-7702. Both the protein and mRNA of CLIC1 were highly expressed in HepG2 liver cancer cells, but not in the normal hepatocyte cells (Fig. 3A and B). To confirm that miR-124 functions as a tumor suppressor by downregulating CLIC1, the effect of silencing CLIC1 was assessed in liver cancer cells. As shown in Fig. 3C and D, transfecting CLIC1 siRNA caused significant downregulation of CLIC1 protein and mRNA levels in HepG2 cells compared with control (NC). The CLIC1-deficient cells exhibited a growth rate similar to that of control cells in WST-1 and colony formation assays (Fig. 3E and F), suggesting that CLIC1 had no effects on the viability and proliferation of liver cancer cells. In the scratch wound-healing motility assay, CLIC1-deficient cells showed significantly slower closure of the wound than the control cells (Fig. 3G). In the Transwell assays without Matrigel coating, fewer CLIC1-deficient cells were able to migrate through pores in the membrane to the lower chamber compared with control cells transfected with scramble siRNA (Fig. 3H). In the Matrigel-coated Transwell invasion assay, silencing of CLIC1 inhibited the invasion of liver cancer cells (Fig. 3I). These results demonstrated that suppression of CLIC1 functions as a metastasis inhibitor in liver cancer cells and has an effect similar to that of miR-124 in liver cancer cells.
Figure 3.
CLIC1 is essential for liver cancer cell migration and invasion, but not proliferation. (A) CLIC1 protein is overexpressed in liver cancer cells compared with normal liver epithelial cells. (B) Analysis of the mRNA expression level of CLIC1 by reverse transcription-quantitative PCR. *P<0.05, ***P<0.001 vs. HL-7702. (C) CLIC1 protein and (D) mRNA were reduced by siR-CLIC1 in liver cancer cells. (E) WST1 showed that CLIC1 had no effect on cell viability and cell growth. siR, small interfering RNA; CLIC1, chloride intracellular protein 1; NC, negative control; n.s., not significant. (F) Colony formation assays showed that CLIC1 had no effect on cell viability and cell growth. The cell migration of liver cancer cells after knockdown of CLIC1 was evaluated by (G) scratch wound-healing motility assay and (H) Transwell assay without Matrigel coating. Scale bar, 200 µm. (I) The cell invasion ability of liver cancer cells was evaluated by Matrigel-coated Transwell assay. Scale bar, 200 µm. ***P<0.001 vs. respective siR-NC. siR, small interfering RNA; CLIC1, chloride intracellular protein 1; NC, negative control; n.s., not significant.
CLIC1 overexpression reverses the miR-124-mediated inhibition of cancer cell migration and invasion
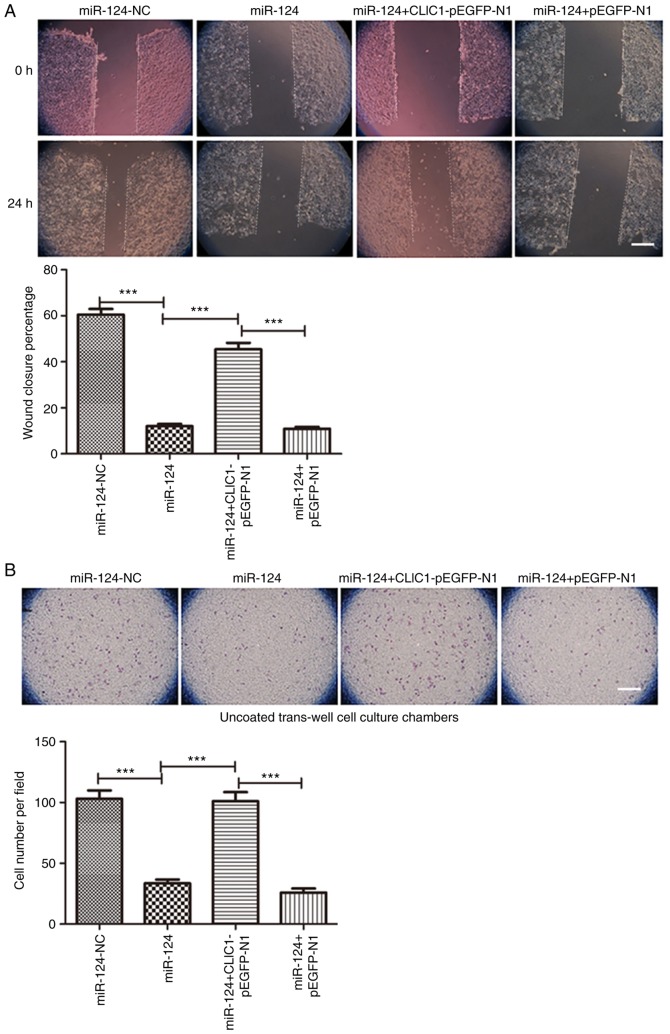
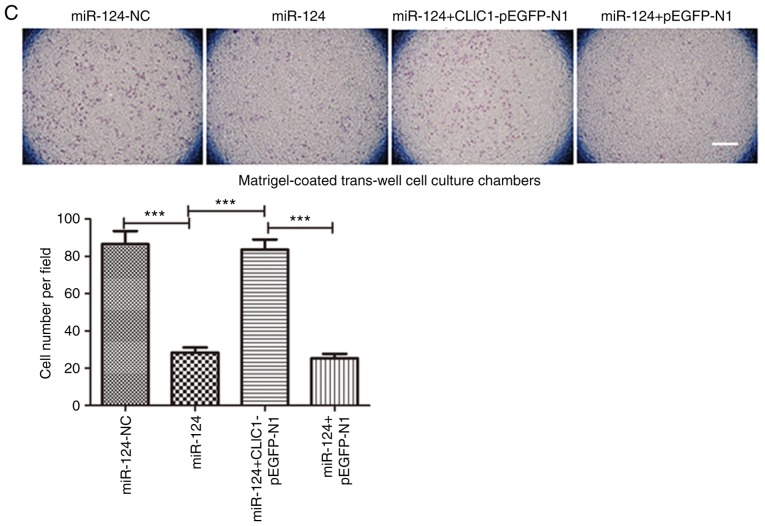
Since CLIC1 is one of the targets of miR-124, CLIC1 was overexpressed in HepG2 cells transfected with an expression plasmid together with miR-124. As expected, miR-124 itself and miR-124 with the pEGFP-N1 vector both inhibited cell migration in the scratch wound-healing motility assay (Fig. 4A). Similar to the miR-124 negative control, cells expressing CLIC1 and miR-124 migrated faster and had faster closure of wound healing than those transfected with miR-124 alone or miR-124 with the vehicle vector pEGFP-N1. In the Transwell assay without Matrigel coating, there were fewer cells able to migrate through the membrane in both the miR-124 and miR-124 + pEGFP-N1 vector groups, but cells expressing CLIC1 and miR-124 migrated faster than cells transfected with miR-124 or miR-124 + pEGFP-N1 vector (Fig. 4B). Similar to cell migration, CLIC1 also reversed the miR-124-induced inhibition of liver cancer cell invasion in the Matrigel-coated Transwell invasion assay (Fig. 4C). These results further support the hypothesis that CLIC1 is target of miR-124 and is involved in the inhibition of liver cancer cell migration and invasion.
Figure 4.
CLIC1 is involved in the miR-124-induced inhibition of liver cancer cell migration and invasion. (A) Scratch wound-healing motility assay of liver cancer cells transfected with miR-124, miR-124-NC, miR-124 + CLIC1 and miR-124 + vector control. Scale bar, 200 µm. (B) The Transwell assay without Matrigel coating showed the cell migration ability of liver cancer cells co-transfected with miR-124 and CLIC1 expression vector compared with the controls. Scale bar, 200 µm. (C) Cell invasion ability of liver cancer cells co-transfected with miR-124 and CLIC1, evaluated by Matrigel-coated Transwell assay. Scale bar, 200 µm. ***P<0.001. NC, negative control; CLIC1, chloride intracellular protein 1; miR, microRNA.
Discussion
The goal of the present study was to investigate the role of miR-124 in hepatocarcinogenesis. miR-124 is highly expressed in the brain and was previously demonstrated to be a ‘brain-enriched’ miRNA (10,11). In recent years, researchers have demonstrated that the expression of miR-124 is dysregulated in almost all tumors, such as colorectal cancer (27), gastric cancer (28), lung cancer (16) and hepatocellular carcinoma (29). In the progression of cancer, miR-124 not only acts as a suppressor of tumor growth but also decreases cell invasion and metastasis. These previous results suggest that miR-124 plays an important role in tumorigenesis and tumor progression. The present study found that miR-124 was downregulated in liver cancer cells and that miR-124 could suppress liver cancer cell proliferation, migration and invasion, indicating its role as a tumor suppressor in liver cancer.
In an earlier study, it was shown that overexpression of miR-124 suppressed tumor growth by targeting STAT3 (30), but the mechanism of action of miR-124 in liver cancer metastasis remained elusive. Invasion, as one of the hallmarks of malignant tumors, renders liver cancer difficult to cure. To elucidate the mechanism of metastasis, miR-124 targets were investigated using 2-DGE and mass spectrometry to analyze the differentially expressed proteins between miR-124-overexpressing and control cells. The results of the mass spectrometry led to a focus on CLIC1 as a potential target. Furthermore, the results revealed that miR-124 directly targeted CLIC1 to inhibit cell migration and invasion in liver cancer cells. Moreover, CLIC1 rescued the miR-124-mediated repression of cell migration and invasion. For the first time, to the best of our knowledge, CLIC1 was identified as a functional target of miR-124 in the inhibition of cell migration and invasion. These findings suggested that miR-124 and CLIC1 play critical roles in liver cancer metastasis.
From the 2-DGE and mass spectrometry results, several proteins with decreased expression were identified. Based on literature research, CLIC1 is highly expressed in several human malignant tumors, such as nasopharyngeal carcinoma, ovarian cancer and glioma, and was considered a promising diagnostic biomarker (31–33). According to functional research, CLIC1 plays an important role in neoplastic transformation (34,35), and promotes the cell motility and invasion of gallbladder carcinoma (36) and prostate cancer (37). However, the function of CLIC1 in liver cancer is still elusive. The present results identified CLIC1 to be a functional target of miR-124; thus, this was a focus of further investigation.
A 3′-UTR luciferase assay was performed, and it was observed that co-transfection of miR-124 and the 3′-UTR of CLIC1 in the pMIR-REPORT vector resulted in decreased luciferase activity. Notably, in the 3′-UTR of CLIC1 mRNA, there was no complementary sequence of miR-124 found using the target prediction software (TargetScan). The best-known mechanism of miRNA regulation is direct binding with the seed sequence located in the 3′-UTR of the mRNA. Studies have demonstrated that miRNAs have some other regulatory methods, such as interactions with regulatory proteins, miRNAs and long noncoding RNAs to regulate protein expression. Eiring et al (38) showed that miR-328 interacts with poly(rC) binding protein 2 to modulate mRNA translation, and the interaction is independent of the miRNA seed sequence. Tang et al (39) reported that miRNA-709 regulates cell apoptosis through the miRNA-15a/16-1 pathway. miRNAs also regulate gene expression by targeting long noncoding RNAs. Du et al (40) reported that miR-124 regulates ERK/MAPK by targeting MALAT1. Altogether, these data suggest that miRNAs function in cells both through base pairing with seed sequences and though interference with other regulatory proteins, miRNAs, and long non coding RNAs. However, many miRNA regulatory mechanisms remain unknown. There may exist some unknown regulatory interaction between miR-124 and CLIC1. A future study look into the mechanism of how miR-124 regulates CLIC1 expression. CLIC1 protein expression was also markedly decreased in HepG2 cells transfected with miR-124. Subsequently, it was demonstrated that CLIC1 is a functional target of miR-124. The knockdown of CLIC1 affected cancer cell migration and invasion, but not cell proliferation. Similarly, overexpressed CLIC1 reversed the tumor suppressor function of miR-124 in terms of cell migration and invasion, but not cell viability. This suggested that the oncogenic function of CLIC1 in liver cancer cells correlates with cell motility and related pathways, but not pathways related to the cell cycle. It has been reported that CLIC1 regulates the migration and invasion of colon cancer cells by decreasing the regulatory volume decrease capacity (41). A study by Wang et al (42) reported that the inhibition of CLIC1 channel activity leads to decreased cell migration through the ROS/ERK pathway. Further studies may be undertaken to examine the regulatory function of CLIC1 in cell motility-related signaling pathways.
In conclusion, the present study demonstrated that miR-124 is downregulated in liver cancer cells, and identified a new functional target gene of miR-124. Further in vitro studies showed that miR-124 repressed the migration and invasion of liver cancer cells by reducing the expression of CLIC1. Moreover, the knockdown of CLIC1 in miR-124 ectopically-expressing liver cancer cells reversed the effects of miR-124. Combined with all aforementioned studies, the present data contribute to the understanding of the biological function of miR-124 in tumor metastasis.
Acknowledgements
The authors would like to thank Mr. Yang Yang and Ms. Manhui Li for their insight and technical support. The authors would also like to thank Ms. Li Li for the correction of the English in the manuscript.
Funding
The present study was supported by research grants from the Scientific Cooperation Planning Project of Guizhou Province [grant. no. Qian Ke He LH Zi (2015)7530]; the National Natural Scientific Foundation of China (grant. no. 81171447); Program of Science and Technology Department of Guizhou Province (grant. no. QKHPTRC-201905612); and the ZMU Startup Fund for Doctors (grant. no. F-696).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JZ and YL designed the research. XY and YL performed the research and prepared all the figures. YC and QY assisted with the data analysis. YL, JZ and YC wrote the main manuscript text. All authors reviewed the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Marengo A, Rosso C, Bugianesi E. Liver cancer: Connections with obesity, fatty liver, and cirrhosis. Annu Rev Med. 2016;67:103–117. doi: 10.1146/annurev-med-090514-013832. [DOI] [PubMed] [Google Scholar]
- 3.Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 6.Hu Y, Yang C, Yang S, Cheng F, Rao J, Wang X. MiR-665 promotes hepatocellular carcinoma cell migration, invasion, and proliferation by decreasing Hippo signaling through targeting PTPRB. Cell Death Dis. 2018;9:954. doi: 10.1038/s41419-018-0978-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Sarvizadeh M, Malekshahi ZV, Razi E, Sharifi H, Moussavi N, Taghizadeh M. MicroRNA: A new player in response to therapy for colorectal cancer. J Cell Physiol. 2018;234:8533–8540. doi: 10.1002/jcp.27806. [DOI] [PubMed] [Google Scholar]
- 8.Imani S, Wu RC, Fu J. MicroRNA-34 family in breast cancer: From research to therapeutic potential. J Cancer. 2018;9:3765–3775. doi: 10.7150/jca.25576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrasekar V, Dreyer JL. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci. 2009;42:350–362. doi: 10.1016/j.mcn.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. MiR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dal Moro F, Valotto C, Guttilla A, Zattoni F. Urinary markers in the everyday diagnosis of bladder cancer. Urologia. 2013;80:265–275. doi: 10.5301/urologia.5000041. [DOI] [PubMed] [Google Scholar]
- 13.Shi XB, Xue L, Ma AH, Tepper CG, Gandour-Edwards R, Kung HJ, deVere White RW. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene. 2013;32:4130–4138. doi: 10.1038/onc.2012.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Q, Xu L, Wang C, Fan W, Yan H, Li Q. MicroRNA-124-3p represses cell growth and cell motility by targeting EphA2 in glioma. Biochem Biophys Res Commun. 2018;503:2436–2442. doi: 10.1016/j.bbrc.2018.06.173. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Gong Q, Li M, Xu J, Zheng Y, Ge P, Chi G. MicroRNA-124 inhibits the proliferation of C6 glioma cells by targeting Smad4. Int J Mol Med. 2017;40:1226–1234. doi: 10.3892/ijmm.2017.3088. [DOI] [PubMed] [Google Scholar]
- 16.Zhao X, Lu C, Chu W, Zhang B, Zhen Q, Wang R, Zhang Y, Li Z, Lv B, Li H, Liu J. MicroRNA-124 suppresses proliferation and glycolysis in non-small cell lung cancer cells by targeting AKT-GLUT1/HKII. Tumour Biol. 2017;39:1010428317706215. doi: 10.1177/1010428317706215. [DOI] [PubMed] [Google Scholar]
- 17.Long HD, Ma YS, Yang HQ, Xue SB, Liu JB, Yu F, Lv ZW, Li JY, Xie RT, Chang ZY, et al. Reduced hsa-miR-124-3p levels are associated with the poor survival of patients with hepatocellular carcinoma. Mol Biol Rep. 2018;45:2615–2623. doi: 10.1007/s11033-018-4431-1. [DOI] [PubMed] [Google Scholar]
- 18.Wu LP, Wu J, Shang A, Yang M, Li LL, Yu J, Xu LR, Wang CB, Wang WW, Zhu JJ, Lu WY. MiR-124 inhibits progression of hepatocarcinoma by targeting KLF4 and promises a novel diagnostic marker. Artif Cells Nanomed Biotechnol. 2018;46(Suppl 1):S159–S167. doi: 10.1080/21691401.2017.1415918. [DOI] [PubMed] [Google Scholar]
- 19.Ulmasov B, Bruno J, Woost PG, Edwards JC. Tissue and subcellular distribution of CLIC1. BMC Cell Biol. 2007;8:8. doi: 10.1186/1471-2121-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valenzuela SM, Martin DK, Por SB, Robbins JM, Warton K, Bootcov MR, Schofield PR, Campbell TJ, Breit SN. Molecular cloning and expression of a chloride ion channel of cell nuclei. J Biol Chem. 1997;272:12575–12582. doi: 10.1074/jbc.272.19.12575. [DOI] [PubMed] [Google Scholar]
- 21.Yu W, Cui R, Qu H, Liu C, Deng H, Zhang Z. Expression and prognostic value of CLIC1 in epithelial ovarian cancer. Exp Ther Med. 2018;15:4943–4949. doi: 10.3892/etm.2018.6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li BP, Mao YT, Wang Z, Chen YY, Wang Y, Zhai CY, Shi B, Liu SY, Liu JL, Chen JQ. CLIC1 Promotes the progression of gastric cancer by regulating the MAPK/AKT pathways. Cell Physiol Biochem. 2018;46:907–924. doi: 10.1159/000488822. [DOI] [PubMed] [Google Scholar]
- 23.Lu J, Dong Q, Zhang B, Wang X, Ye B, Zhang F, Song X, Gao G, Mu J, Wang Z, et al. Chloride intracellular channel 1 (CLIC1) is activated and functions as an oncogene in pancreatic cancer. Med Oncol. 2015;32:616. doi: 10.1007/s12032-015-0616-9. [DOI] [PubMed] [Google Scholar]
- 24.Wei X, Li J, Xie H, Wang H, Wang J, Zhang X, Zhuang R, Lu D, Ling Q, Zhou L, et al. Chloride intracellular channel 1 participates in migration and invasion of hepatocellular carcinoma by targeting maspin. J Gastroenterol Hepatol. 2015;30:208–216. doi: 10.1111/jgh.12668. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Lu Y, Yue X, Li H, Luo X, Wang Y, Wang K, Wan J. MiR-124 suppresses growth of human colorectal cancer by inhibiting STAT3. PLoS One. 2013;8:e70300. doi: 10.1371/journal.pone.0070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Taniguchi K, Sugito N, Kumazaki M, Shinohara H, Yamada N, Nakagawa Y, Ito Y, Otsuki Y, Uno B, Uchiyama K, Akao Y. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett. 2015;363:17–27. doi: 10.1016/j.canlet.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 28.Xiao HJ, Ji Q, Yang L, Li RT, Zhang C, Hou JM. In vivo and in vitro effects of microRNA-124 on human gastric cancer by targeting JAG1 through the Notch signaling pathway. J Cell Biochem. 2018;119:2520–2534. doi: 10.1002/jcb.26413. [DOI] [PubMed] [Google Scholar]
- 29.Cao J, Qiu J, Wang X, Lu Z, Wang D, Feng H, Li X, Liu Q, Pan H, Han X, et al. Identification of microRNA-124 in regulation of hepatocellular carcinoma through BIRC3 and the NF-kB pathway. J Cancer. 2018;9:3006–3015. doi: 10.7150/jca.25956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y, Yue X, Cui Y, Zhang J, Wang K. MicroRNA-124 suppresses growth of human hepatocellular carcinoma by targeting STAT3. Biochem Biophys Res Commun. 2013;441:873–879. doi: 10.1016/j.bbrc.2013.10.157. [DOI] [PubMed] [Google Scholar]
- 31.Chang YH, Wu CC, Chang KP, Yu JS, Chang YC, Liao PC. Cell secretome analysis using hollow fiber culture system leads to the discovery of CLIC1 protein as a novel plasma marker for nasopharyngeal carcinoma. J Proteome Res. 2009;8:5465–5474. doi: 10.1021/pr900454e. [DOI] [PubMed] [Google Scholar]
- 32.Singha B, Harper SL, Goldman AR, Bitler BG, Aird KM, Borowsky ME, Cadungog MG, Liu Q, Zhang R, Jean S, et al. CLIC1 and CLIC4 complement CA125 as a diagnostic biomarker panel for all subtypes of epithelial ovarian cancer. Sci Rep. 2018;8:14725. doi: 10.1038/s41598-018-32885-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, He S, Tu Y, Ji P, Zong J, Zhang J, Feng F, Zhao J, Zhang Y, Gao G. Elevated expression of chloride intracellular channel 1 is correlated with poor prognosis in human gliomas. J Exp Clin Cancer Res. 2012;31:44. doi: 10.1186/1756-9966-31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He YM, Zhang ZL, Liu QY, Xiao YS, Wei L, Xi C, Nan X. Effect of CLIC1 gene silencing on proliferation, migration, invasion and apoptosis of human gallbladder cancer cells. J Cell Mol Med. 2018;22:2569–2579. doi: 10.1111/jcmm.13499. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Peretti M, Angelini M, Savalli N, Florio T, Yuspa SH, Mazzanti M. Chloride channels in cancer: Focus on chloride intracellular channel 1 and 4 (CLIC1 AND CLIC4) proteins in tumor development and as novel therapeutic targets. Biochim Biophys Acta. 2015;1848:2523–2531. doi: 10.1016/j.bbamem.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 36.Wang JW, Peng SY, Li JT, Wang Y, Zhang ZP, Cheng Y, Cheng DQ, Weng WH, Wu XS, Fei XZ, et al. Identification of metastasis-associated proteins involved in gallbladder carcinoma metastasis by proteomic analysis and functional exploration of chloride intracellular channel 1. Cancer Lett. 2009;281:71–81. doi: 10.1016/j.canlet.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 37.Tian Y, Guan Y, Jia Y, Meng Q, Yang J. Chloride intracellular channel 1 regulates prostate cancer cell proliferation and migration through the MAPK/ERK pathway. Cancer Biother Radiopharm. 2014;29:339–344. doi: 10.1089/cbr.2014.1666. [DOI] [PubMed] [Google Scholar]
- 38.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang R, Li L, Zhu D, Hou D, Cao T, Gu H, Zhang J, Chen J, Zhang CY, Zen K. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: Evidence for a microRNA hierarchy system. Cell Res. 2012;22:504–515. doi: 10.1038/cr.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du M, Chen W, Zhang W, Tian XK, Wang T, Wu J, Gu J, Zhang N, Lu ZW, Qian LX, et al. TGF-? regulates the ERK/MAPK pathway independent of the SMAD pathway by repressing miRNA-124 to increase MALAT1 expression in nasopharyngeal carcinoma. Biomed Pharmacother. 2018;99:688–696. doi: 10.1016/j.biopha.2018.01.120. [DOI] [PubMed] [Google Scholar]
- 41.Wang P, Zhang C, Yu P, Tang B, Liu T, Cui H, Xu J. Regulation of colon cancer cell migration and invasion by CLIC1-mediated RVD. Mol Cell Biochem. 2012;365:313–321. doi: 10.1007/s11010-012-1271-5. [DOI] [PubMed] [Google Scholar]
- 42.Wang P, Zeng Y, Liu T, Zhang C, Yu PW, Hao YX, Luo HX, Liu G. Chloride intracellular channel 1 regulates colon cancer cell migration and invasion through ROS/ERK pathway. World J Gastroenterol. 2014;20:2071–2078. doi: 10.3748/wjg.v20.i8.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.