Abstract
Carboxypeptidase X, M14 family member 2 (CPXM2), has been associated with several human disorders such as developmental diseases. However, whether CPXM2 is involved in oncogenesis or tumor progression remains unclear. In the present study, we used clinical samples from gastric cancer (GC) patients to investigate potential roles of CPXM2 in GC. We also analyzed datasets from the Oncomine database, The Cancer Genome Atlas (TCGA), and the Kaplan-Meier Plotter to validate these results. We found that CPXM2 was overexpressed in GC and that the overexpression was associated with an unfavorable prognosis, regardless of the Lauren classification and tumor node metastasis staging. In addition, knockdown of CPXM2 in cultured GC cells significantly impeded cell proliferation and migration, as indicated by the cholecystokinin octapeptide, colony formation assay, scratch wound healing assay, and Transwell® migration assay. Furthermore, gene set enrichment analysis using RNA-seq data from TCGA indicated that high CPXM2 expression in GC patients was positively correlated with the HALLMARK_APICAL_JUNCTION and HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION gene sets. Finally, western blotting results revealed that several key molecules involved in the epithelial mesenchymal transition were regulated by CPXM2. Taken together, these results imply an active role for CPXM2 in promoting tumor aggressiveness via epithelial to mesenchymal transition (EMT) modulation in GCs.
Keywords: CPXM2, gastric cancer, prognosis, proliferation, migration, EMT
Introduction
Gastric cancer is one of the major malignancies in the world (1,2). Although its incidence and mortality rates have declined dramatically in Western countries for decades (1,2), GC still constitutes a huge health threat in some parts of the world, such as in China and Japan (3,4). Surgery with radical intention is suitable for only a limited percentage of patients, while many patients are often diagnosed at later stages, or experience postsurgical disease relapse or metastasis, which make their prognoses even worse (5). Comprehensive treatment of advanced GC remains unsatisfactory, so it is of vital importance to elucidate the molecular mechanisms that lead to disease progression of GC.
Metallocarboxypeptidases (MCPs) are zinc-dependent peptidases that cleave off C-terminal amino acid residues from their substrates by catalysis of peptide bond hydrolysis. More than 26 MCPs have been discovered and are categorized into four major subgroups of the M14 family of peptidases based on their structural and functional relevance, namely M14A subfamily or digestive carboxypeptidases, M14B subfamily or regulatory carboxypeptidases, M14C subfamily or bacterial peptidoglycan hydrolyzing enzymes, and M14D subfamily or cytosolic carboxypeptidases. MCPs induce substrate degradation and participate in a variety of physiological and pathological processes such as digestion, development, inflammation, and type 2 diabetes, as reviewed by several authors (6–8). Furthermore, dysregulation of MCPs has been associated with oncogenesis. For example, carboxypeptidase A4 (CPA4) is elevated in GCs and is associated with an aggressive phenotype and unfavorable prognosis (9), and carboxypeptidase E (CPE) and its splice isoform are correlated with metastases in a variety of malignancies (10), while carboxypeptidase M (CPM) is dysregulated in several tumor types (11), highlighting the importance of some carboxypeptidases in modulating tumor behavior. Although the underlying mechanisms of their oncogenic roles are largely unknown, their interaction with growth factors could be of some interest (11). In addition, successful isolation of endogenous inhibitors and synthesis of exogenous inhibitors make MCPs ideal targets for anticancer therapy (8).
Carboxypeptidase X, M14 family member 2 (CPXM2), one of the less characterized MCPs, has been associated with developmental diseases (12,13), late-onset Alzheimer's disease, and cognitive decline in schizophrenia (14,15). However, whether CPXM2 is involved in oncogenesis or tumor progression remains unclear. In the present study, we determined the expression of CPXM2 and its clinical and prognostic relevance in GCs. We also investigated its in vitro activities in cultured GC cells and characterized the potential underlying mechanisms of action. We aimed to shed light on the oncogenic roles of CPXM2 in GC, one of the most fatal malignancies in the world.
Materials and methods
Patients and specimens
Fifteen paired human GC samples and their matched gastric non-cancerous tissues (NTs) were collected at the time of surgical resection at Shanghai Fifth People's Hospital (Shanghai, China) from February 2017 to February 2018. There were 10 males and 5 females, with a median age of 63 (range, 52–77 years). Samples were snap-frozen in liquid nitrogen and stored at −80°C. Paraffin-embedded tissues were retrieved from the Tissue Bank of the Shanghai Fifth People's Hospital, and 4-µm tissue sections were prepared by the Department of Pathology at the same hospital. Tissue microarrays (TMAs) of GCs and adjacent NTs were prepared by Shanghai Outdo Biotech (Shanghai, China). The TMA sections contained paired GCs and NTs from 90 patients with a median follow-up of 30 months. The clinicopathological characteristics of these patients are summarized in Table I. This study was approved by the institutional Ethics Committee of Shanghai Fifth People's Hospital (Ethical approval form no. 2017–097) and adhered to the principles of the Declaration of Helsinki. Informed consent was obtained from each patient before collection of tissues.
Table I.
Clinical and pathological features of the gastric cancer patientsa (n=90).
| Variable | No. of patients (%) |
|---|---|
| Sex | |
| Male | 69 (76.7) |
| Female | 21 (23.3) |
| Age (years) | |
| <70 | 45 (50) |
| ≥70 | 45 (50) |
| Differentiation | |
| G2 | 28 (31.1) |
| G3 | 62 (68.9) |
| Tumor size (cm) | |
| <5 | 35 (38.9) |
| ≥5 | 54 (60.0) |
| NA | 1 (1.1) |
| TNM stage | |
| I | 12 (13.3) |
| II | 27 (30.0) |
| III | 49 (54.5) |
| IV | 2 (2.2) |
| Tumor stage | |
| T1 | 5 (5.6) |
| T2 | 15 (16.7) |
| T3 | 46 (51.1) |
| T4 | 24 (26.6) |
| Nodal stage | |
| N0 | 24 (26.7) |
| N1 | 15 (16.7) |
| N2 | 23 (25.5) |
| N3 | 28 (31.1) |
| Vessel invasion | |
| No | 74 (82.2) |
| Yes | 16 (17.8) |
| Nerve invasion | |
| No | 74 (82.2) |
| Yes | 15 (16.7) |
| NA | 1 (1.1) |
| Expression of CPXM2 | |
| Low | 41 (45.6) |
| High | 49 (54.4) |
Data shown here may be duplicated with data from other published resources that are based on the same cohorts. NA, information not available; CPXM2, carboxypeptidase X, M14 family member 2; TNM, tumor node metastasis.
Immunohistochemistry (IHC)
Sections were stained with an anti-CPXM2 polyclonal antibody using IHC in the Department of Pathology at our hospital. In brief, after deparaffination, rehydration in graded ethanol, antigen retrieval with citrate buffer pH 6.0 (1:300 dilution; ZLI-9065; ZSGB-Bio, Beijing, China) and blocking, slides were stained with a rabbit anti-CPXM2 polyclonal antibody (1:50 dilution; cat. no. abs127533a; Absin Bioscience, Shanghai, China) at 4°C overnight. Normal rat IgG (1:50 dilution; cat. no. D110504; Sangon Biotech, Shanghai, China) instead of the primary antibody was used as a control. Subsequently, after washing with PBS, a horseradish peroxidase (HRP)-conjugated secondary antibody (1:2,000; goat anti-rat, cat. no. A0192; Beyotime Institute of Biotechnology, Haimen, China) was added and incubated at room temperature for 1 h. Then, these sections were stained using 3,3′-diaminobenzidine (DAB) (cat. no. GK500705; Shanghai GeneTech Co., Ltd., Shanghai, China) and counterstained with hematoxylin. A modified H score system was used to semi-quantitate CPXM2 expression, as previously described (16). Briefly, the maximal intensity of staining (0, negative; 1, weak; 2, moderate; and 3, strong) was multiplied by the percentage of positive tumor cells (0–100%) to generate the modified H score (range, 0–300). CPXM2 expression was categorized as high or low based on the median H score.
Access to public datasets
The mRNA expression of CPXM2 in GC tissues and normal mucosae was acquired from Oncomine (http://www.oncomine.org) (17,18). The original data for prognostic analysis of CPXM2 were downloaded from the Kaplan-Meier Plotter (http://www.kmplot.com) (19) and UCSC Xena (https://xenabrowser.net/heatmap/).
Cell lines and culture conditions
A gastric epithelial cell line (GES-1) and five GC cell lines (AGS, HGC27, MGC803, NCI-N87 and SNU-1) were obtained from the Type Culture Collection of the Chinese Academy of Science (Shanghai, China) and were validated by short tandem repeat DNA profiling. Cells were cultured in RPMI-1640 (BBI Life Sciences, Shanghai, China) or F12K medium (Zhong Qiao Xin Zhou Biotechnology, Shanghai, China) supplemented with 10% fetal bovine serum (FBS), 100 µg/ml penicillin, and 100 mg/ml streptomycin at 37°C with 5% CO2 in a humidified incubator (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Construction of CPXM2-targeting shRNAs and packaging of lentiviruses
Four targeting shRNAs and a nontargeting scrambled RNA (scramble) were subcloned into the GV248 lentivirus vector by Shanghai GeneChem Co., Ltd., (Shanghai, China). The shCPXM2 target sequences were AGGTTCATCGTGGCATTAA (shCPXM2-1), ACGATGGAATTGACATCAA (shCPXM2-2), TCCCAATATCACCAGAATT (shCPXM2-3) and CTCAGTCCTGGTTTGATAA (shCPXM2-4). Lentiviral stocks were prepared and purified as previously described (20).
Infection of GC cells with lentiviruses
Cells were seeded in 6-well plates at a density of 2×105/ml and cultivated for 24 h. Then, 20 µl of lentivirus solution and 1 ml fresh medium containing 10 µg/ml polybrene (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) were added to each well. The medium was changed after 24 h and an efficient lentiviral transduction was confirmed by a fluorescence microscope at 72 h after infection.
RNA extraction and the quantitative polymerase chain reaction (qPCR)
Total RNA was isolated from cell cultures or from snap-frozen tissues from GC patients using RNAiso Plus (Takara Bio, Kusatsu, Japan) according to the manufacturer's instructions, reverse transcribed with HiScript Q Select RT SuperMix (R132-01; Vazyme, Jiangsu, China) according to the manufacturer's protocol, and subjected to real-time reverse transcription (RT)-PCR using the 2−ΔΔCq method (21). The thermocycling conditions were as follows: 95°C for 30 sec, followed by 40 cycles of 95°C for 10 sec, 60°C for 32 sec, 95°C for 15 sec, 60°C for 60 sec and 95°C for 15 sec. Each sample was determined in duplicate. All PCR products were confirmed by 2.0% agarose gel electrophoresis. The sequences for RT-PCR primers were: CPXM2 forward primer, 5′-GTGCGCGGGAAGAAATGAC-3′ and reverse primer, 5′-CCTCCCTTGAGTGATGACACC-3′. The specificity of primers was validated by sequencing. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (forward primer, 5′-GTCAAGGCTGAGAACGGGAA-3′ and reverse primer 5′-AAATGAGCCCCAGCCTTCTC-3′) served as an internal control. Experiments were repeated three times in duplicate.
Western blot analysis
Total protein was extracted from cell cultures or homogenized tissues from GC patients using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing phenylmethylsulfonyl fluoride (Beyotime Institute of Biotechnology) and proteinase inhibitor cocktail solution (Roche, Basel, Switzerland), and quantitated using the bicinchoninic acid protein assay (Beyotime Institute of Biotechnology) as recommended by the manufacturers. Western blotting was performed according to standard methods as previously described (21). Briefly, the proteins (30 µg) were separated by 10% SDS-PAGE and then transferred to a polyvinylidene fluoride (PVDF) membrane. Subsequently, the membranes were blocked with 5% fat-free dry milk at room temperature for 1 h. Then, the blots were incubated with a rabbit anti-CPXM2 polyclonal antibody (1:1,000 dilution; cat. no. abs127533a; Absin Bioscience, Shanghai, China), and rabbit monoclonal antibodies against E-cadherin, N-cadherin, vimentin and ZEB1 (1:1,000 dilution; cat. nos. 3195, 13116, 5741 and 3396; Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. GAPDH (1:2,000; cat. no. AF1186, rabbit anti-human; Beyotime Institute of Biotechnology) or β-actin (1:2,000; cat. no. AF0003, mouse anti-human; Beyotime Institute of Biotechnology) was detected as a loading control. The membranes were again washed with TBST and incubated with respective horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2,000 dilution; goat anti-rat cat. no. A0192, goat anti-mouse cat. no. A0216 and goat anti-rabbit cat. no. A0239; Beyotime Institute of Biotechnology) at room temperature for 1 h. The proteins were finally examined by an enhanced chemiluminescence system (ECL) (P0018AS; Beyotime Institute of Biotechnology). The grayscale values of protein bands were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Cell proliferation assay
Stably transfected AGS and HGC-27 cells (2×103 cells/well) were seeded in 96-well plates and cultivated for 24, 48, 72 or 96 h. Then, 10 µl cholecystokinin octapeptide (CCK-8) reagent [10% (v/v) in serum-free RPMI-1640 medium; Beyotime Biotechnology] was added to each well and incubated at 37°C for 1 h. The absorbance at 450 nm was measured using a microplate reader (BioTek Synergy 2; BioTek Instruments Inc., Winooski, VT, USA).
Cell colony formation assay
A plate colony formation assay was performed as previously described (22). Briefly, stably transfected AGS or HGC-27 cells (5×102 cells/well) were seeded in 6-well plates and cultivated in F12K or RPMI-1640 complete medium at 37°C for 14 days. The cell colonies were washed with phosphate-buffered saline (PBS) twice, fixed with methanol for 20 min, and stained with 0.1% crystal violet in PBS (Beyotime Institute of Biotechnology) for 15 min. The plates were scanned and colonies containing >50 cells were counted manually, and the experiments were performed in triplicate.
Cell migration assay
A Transwell® migration assay was performed as previously reported (22). In brief, cells (4×105 cells/ml) were seeded in serum-free RPMI-1640 or F12K medium in the top chamber of a Transwell® insert. The medium containing 20% FBS in the lower chamber served as a chemoattractant. After incubation for 24 h at 37°C, the cells on the top side of the membrane were removed with a cotton swab, and those on the bottom side were fixed with methanol for 20 min and then stained with crystal violet (0.1% in PBS) for 15 min. Six randomly selected fields per well were photographed under a microscope using the ImageScope software (Leica Biosystems Nussloch GmbH, Nussloch, Germany) with a magnification of ×200, and the numbers of migrated cells were counted manually.
Scratch wound healing assay
A monolayer scratch wound assay was employed as previously described (23). Briefly, cells (4×105 cells/well) were seeded in 12-well plates and grown to nearly 100% confluence. A scratch wound was generated with a 200-µl pipette tip. Wound closure was photographed at 0 and 48 h under a microscope using the ImageScope software (Leica Biosystems Nussloch GmbH) with a magnification of ×40.
Statistical analysis
Statistical analyses were performed using Microsoft Excel 2010 (Microsoft, Redmond, WA, USA), GraphPad Prism7 (GraphPad Software Inc., San Diego, CA, USA), and SPSS statistical software for Windows, version 22 (IBM Corp., Armonk, NY, USA). Paired Student's t-tests were performed for continuous variables between two groups and Wilcoxon signed-rank tests were used if the differences between pairs of data did not follow a Gaussian distribution. One-way analysis of variance (ANOVA) was performed for statistical comparisons among multiple groups along with Bonferroni's post hoc test. Pearson's χ2 test and Fisher's exact test were used for categorical comparisons. Pearson's R correlation test was used to describe correlations between continuous variables. Survival analyses were conducted using the Kaplan-Meier method and differences in survival were examined using the log-rank test. Univariate and multivariate survival analyses were conducted using the Cox proportional hazards regression model. Statistical significance was defined as a value of P<0.05. All statistical tests were two-sided.
Results
CPXM2 is upregulated in GCs
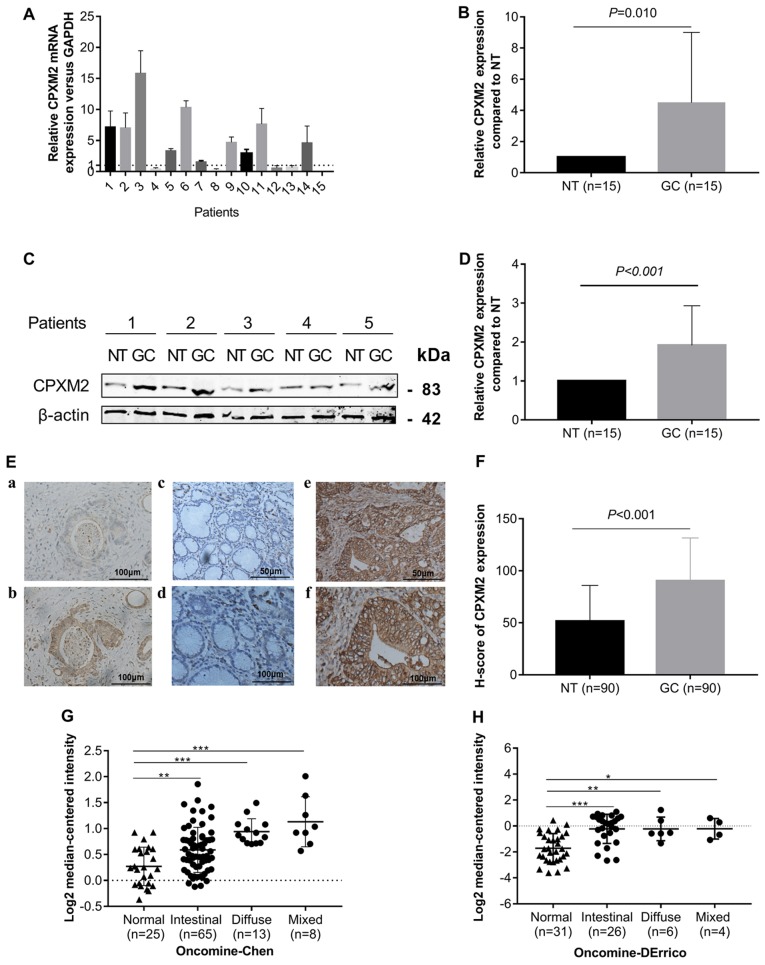
To explore whether CPXM2 is dysregulated in GCs, we first measured CPXM2 expression in 15 GCs and their matched (normal tissues) NTs by qPCR and western blotting. The results indicated that CPXM2 mRNA and protein were elevated in the GC cases compared with the NTs (Fig. 1A-D). We then examined CPXM2 expression in primary GC tissues by IHC. As shown in Fig. 1E and F, CPXM2 was expressed mainly in the cytoplasm and on the cytosolic side of the membrane, while its expression was stronger in GCs than in NTs, as quantified by H scores. To validate these observations further, we analyzed CPXM2 expression in two GC datasets [Chen Gastric, n=111 (17); Derrico Gastric, n=69 (18)] with Lauren's classification in the Oncomine database and found that CPXM2 was unanimously overexpressed in each tumor subtype compared with NTs (Fig. 1G and H). Taken together, these results clearly showed that CPXM2 is upregulated in GCs.
Figure 1.
CPXM2 is upregulated in gastric cancers. (A and B) CPXM2 mRNA expression levels in 15 paired GCs and their adjacent NTs. (C and D) Representative blots and quantification of CPXM2 protein expression levels from samples in (A and B). The average CPXM2 expression was normalized to the expression of β-actin. Three replicates were conducted for each experiment. (E) Immunohistochemical staining of CPXM2 in GCs and NTs. (a and b) IgG and CPXM2 staining in GCs (×400 magnification). (c-f) CPXM2 staining in NTs (c, ×200; d, ×400 magnification) and GCs (e, ×200; f, ×400 magnification). (F) H scores of CPXM2 staining in NTs and GCs. (G and H) A logarithmic 2−ΔΔCq scale was used to represent the fold-changes in CPXM2 mRNA expression in microarray datasets from the Oncomine database: Chen gastric and Derrico gastric, grouped by Lauren classification. *P<0.05, **P<0.01, ***P<0.001 vs. the control group. CPXM2, carboxypeptidase X, M14 family member 2; GC, gastric cancer; NT, normal tissue.
Overexpression of CPXM2 in GCs correlates with an unfavorable prognosis
To test whether CPXM2 overexpression in GCs contributes to increased invasiveness, we analyzed the relationships between CPXM2 expression and clinicopathological variables. As shown in Table II, high CPXM2 expression was significantly associated with larger tumor size and later pTNM stages.
Table II.
Association between CPXM2 expression and clinicopathological variables in the gastric cancer cases (n=90).
| CPXM2 expression | ||||
|---|---|---|---|---|
| Clinicopathological features | N | Low (n=45) | High (n=45) | P-value |
| Sex | ||||
| Male | 68 | 32 (47.1) | 36 (52.9) | |
| Female | 21 | 13 (61.9) | 8 (38.1) | 0.319 |
| Age (years) | ||||
| <70 | 45 | 26 (57.8) | 19 (42.2) | |
| ≥70 | 45 | 19 (42.2) | 26 (57.8) | 0.206 |
| Histological grade | ||||
| G2 | 28 | 14 (50.0) | 14 (50.0) | |
| G3 | 62 | 31 (50.0) | 31 (50.0) | >0.999 |
| Tumor size (cm) | ||||
| <5 | 34 | 22 (64.7) | 12 (35.3) | |
| ≥5 | 54 | 22 (40.7) | 32 (59.3) | 0.048 |
| pT stage | ||||
| T1/T2 | 29 | 12 (41.4) | 17 (58.6) | |
| T3/T4 | 60 | 32 (53.3) | 28 (46.7) | 0.367 |
| pN stage | ||||
| N0 | 25 | 15 (60.0) | 10 (40.0) | |
| N1-N3 | 65 | 30 (46.2) | 35 (53.8) | 0.347 |
| pStage | ||||
| I/II | 42 | 27 (64.3) | 15 (35.7) | |
| III/IV | 48 | 18 (37.5) | 30 (62.5) | 0.020 |
| Vessel invasion | ||||
| No | 74 | 40 (54.1) | 34(45.9) | |
| Yes | 16 | 5 (31.3) | 11 (68.7) | 0.167 |
| Nerve invasion | ||||
| No | 75 | 38 (50.7) | 37 (49.3) | |
| Yes | 15 | 7 (46.7) | 8 (53.3) | >0.999 |
P-values in bold print indicate a statistically significant result. CPXM2, carboxypeptidase X, M14 family member 2.
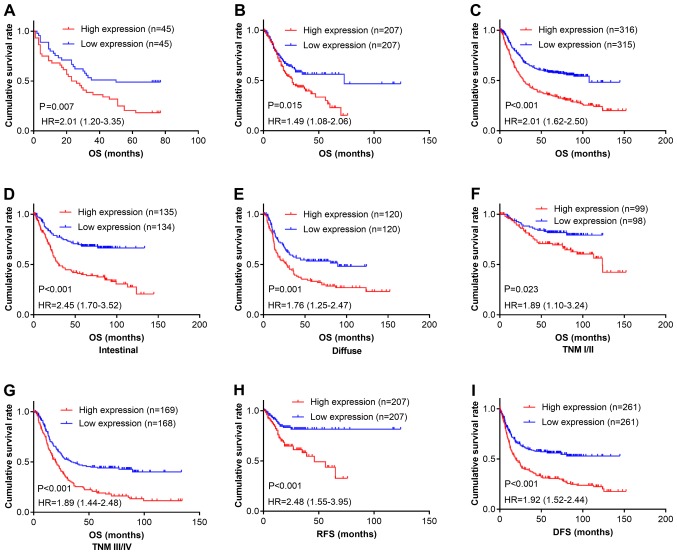
We next determined the relationship between CPXM2 expression and patient outcomes. Kaplan-Meier survival analysis revealed that patients with high CPXM2 expression had shorter overall survival (OS) times than did those with low expression [estimated mean OS, 30.7 months; 95% confidence interval (CI), 23.0–38.5 months vs. OS, 48.0 months; 95% CI, 39.0–56.9 months; log-rank test, P=0.007; Fig. 2A]. In multivariate analysis with a Cox proportional hazards model, CPXM2 overexpression was significantly associated with a shorter OS [hazard ratio (HR), 1.92; 95% CI, 1.08–3.40; P=0.026], after adjustment for age, tumor size, T stage, and N stage (Table III). To confirm the adverse prognostic roles of CPXM2 in patients with GC, we downloaded and analyzed CPXM2 transcription data from TCGA and the Kaplan-Meier Plotter. The results showed that high CPXM2 expression was correlated with worse OS in both datasets (Fig. 2B and C). To eliminate potential influences from confounding factors, we stratified patients in the Kaplan-Meier Plotter dataset by Lauren classification and TNM staging. As indicated by Fig. 2D-G, high CPXM2 expression was consistently associated with unfavorable OS in GC patients, regardless of the Lauren classification and TNM staging. In addition, we explored the potential effects of CPXM2 expression on recurrence-free survival (RFS) or disease-free survival (DFS) in GC patients. The results demonstrated that high CPXM2 expression was also associated with tumor relapse (Fig. 2H and I). Taken together, these findings indicate that CPXM2 is a prognostic marker in GCs and correlates with an unfavorable prognosis.
Figure 2.
CPXM2 overexpression in GCs is associated with poor patient survival. (A-C) Kaplan-Meier plots for OS of patients in the TMA cohort (A), TCGA cohort (B) and Kaplan-Meier Plotter cohort (C). (D-G) Kaplan-Meier plots for OS of patients with intestinal type (D), diffuse type (E), early stage (F), and late stage (G) GC in the Kaplan-Meier Plotter cohort. (H and I) Kaplan-Meier plots for RFS of patients in the TCGA cohort (H) and Kaplan-Meier Plotter cohort (I). Patients were stratified into low and high CPXM2 expression groups according to CPXM2 mRNA expression (< median vs. ≥ median) in the TCGA and Kaplan-Meier Plotter cohorts, or H scores of CPXM2 staining (< median vs. ≥ median) in the TMA cohort. P-values were obtained using the log-rank test. Censored data are indicated by the + symbol. CPXM2, carboxypeptidase X, M14 family member 2; DFS, disease-free survival; GC, gastric cancer; HR, hazard ratio; NT, adjacent normal tissue; OS, overall survival; RFS, recurrence-free survival; TCGA, The Cancer Genome Atlas; TMA, tissue microarray; TNM, tumor node metastasis.
Table III.
Univariate and multivariate Cox proportional hazard models for overall survival of the GC patients (n=90).
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Clinicopathological features | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex | ||||
| Male | 1 (Reference) | |||
| Female | 0.80 (0.42–1.51) | 0.487 | ||
| Age (years) | ||||
| <70 | 1 (Reference) | 1 (Reference) | ||
| ≥70 | 1.82 (1.08–3.05) | 0.024 | 1.23 (0.69–2.18) | 0.487 |
| Histological grade | ||||
| G2 | 1 (Reference) | |||
| G3 | 1.45 (0.83–2.55) | 0.194 | ||
| Tumor size (cm) | ||||
| <5 | 1 (Reference) | 1 (Reference) | ||
| ≥5 | 3.68 (1.97–6.87) | <0.001 | 2.29 (1.15–4.55) | 0.018 |
| pT stage | ||||
| T1/T2 | 1 (Reference) | 1 (Reference) | ||
| T3/T4 | 1.74 (0.98–3.10) | 0.060 | 1.52 (0.79–2.94) | 0.208 |
| pN stage | ||||
| N0 | 1 (Reference) | 1 (Reference) | ||
| N1-N3 | 2.78 (1.40–5.50) | 0.003 | 2.20 (1.01–4.80) | 0.048 |
| pStage | ||||
| I/II | 1 (Reference) | |||
| III/IV | 3.38 (1.94–5.88) | <0.001 | ||
| Vessel invasion | ||||
| No | 1 (Reference) | |||
| Yes | 1.55 (0.82–2.92) | 0.176 | ||
| Nerve invasion | ||||
| No | 1 (Reference) | |||
| Yes | 1.56 (0.81–3.00) | 0.185 | ||
| CPXM2 expression | ||||
| Low | 1 (Reference) | 1 (Reference) | ||
| High | 2.07 (1.23–3.48) | 0.006 | 1.92 (1.08–3.40) | 0.026 |
P-values in bold print indicate a statistically significant result. CI, confidence interval; HR, hazard ratio; CPXM2, carboxypeptidase X, M14 family member 2.
CPXM2 promotes the proliferation and migration of cultured GC cells
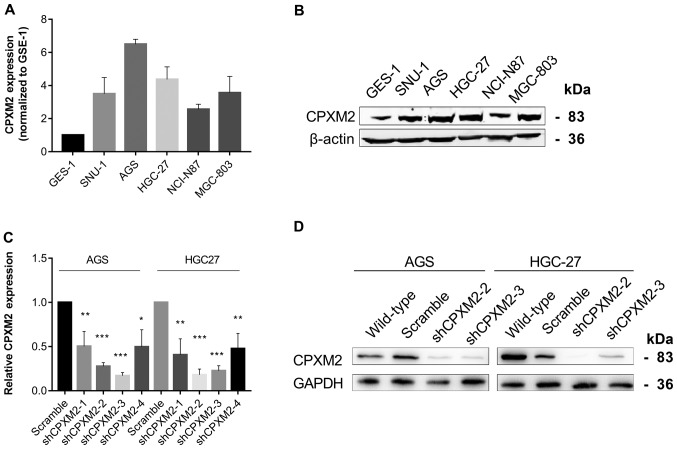
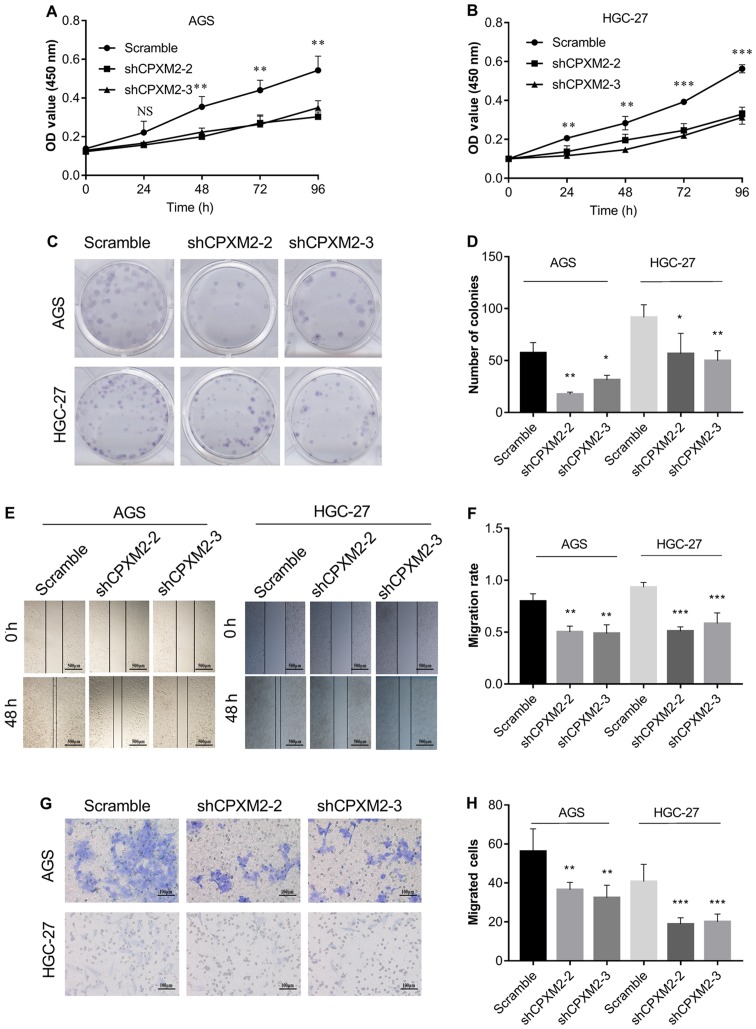
To illustrate further the potential role of CPXM2 in GC progression, we first used qRT-PCR and western blotting to examine the intrinsic expression of CPXM2 in a gastric epithelial cell line, GES-1, and five GC cell lines. The results indicated that CPXM2 expression was significantly increased in GC cells relative to GES-1 cells (Fig. 3A and B). We then knocked down CPXM2 expression in AGS and HGC-27 cells with lentiviruses carrying CPXM2-specific shRNAs (Fig. 3C and D), and selected shCPXM2-2 and shCPXM2-3 for subsequent in vitro experiments. In cultured GC cells, knockdown of CPXM2 expression significantly inhibited cell proliferation, as indicated by the CCK-8 and colony formation assays (Fig. 4A-D). In addition, knockdown of CPXM2 expression significantly decreased cell migration in the scratch wound-healing and Transwell® migration assays (Fig. 4E-H). Collectively, these results clearly indicate that CPXM2 promoted the proliferation and migration of GC cells.
Figure 3.
Silencing of CPXM2 in GC cell lines. (A and B) Expression of CPXM2 in a gastric mucosa cell line (GES-1) and five GC cell lines. (C) AGS and HGC-27 cells were infected with lentiviruses carrying shCPXM2-1, shCPXM2-2, shCPXM2-3, shCPXM2-4, or scrambled control shRNA, and then underwent quantitative reverse transcription polymerase chain reaction. (D) AGS and HGC-27 cells were infected with lentiviruses carrying shCPXM2-2, shCPXM2-3, or scrambled control shRNA, and then underwent western blotting validation. *P<0.05, **P<0.01, ***P<0.001 vs. the control group. CPXM2, carboxypeptidase X, M14 family member 2; GC, gastric cancer.
Figure 4.
In vitro activities of CPXM2 in GC cells. AGS and HGC-27 cells were infected with lentiviruses carrying shCPXM2-2, shCPXM2-3, or scrambled control shRNA, and then underwent CCK-8 assay (A and B), colony formation assay (C and D), scratch wound-healing assay (E and F), and Transwell® migration assay (G and H). CPXM2 silencing in these cells significantly inhibited cell proliferation and migration. Three replicates were conducted for each experiment. *P<0.05, **P<0.01, ***P<0.001 vs. the control group. CCK-8, Cell Counting Kit-8; OD, optical density; CPXM2, carboxypeptidase X, M14 family member 2; GC, gastric cancer.
CPXM2 may promote GC progression by modulating EMT
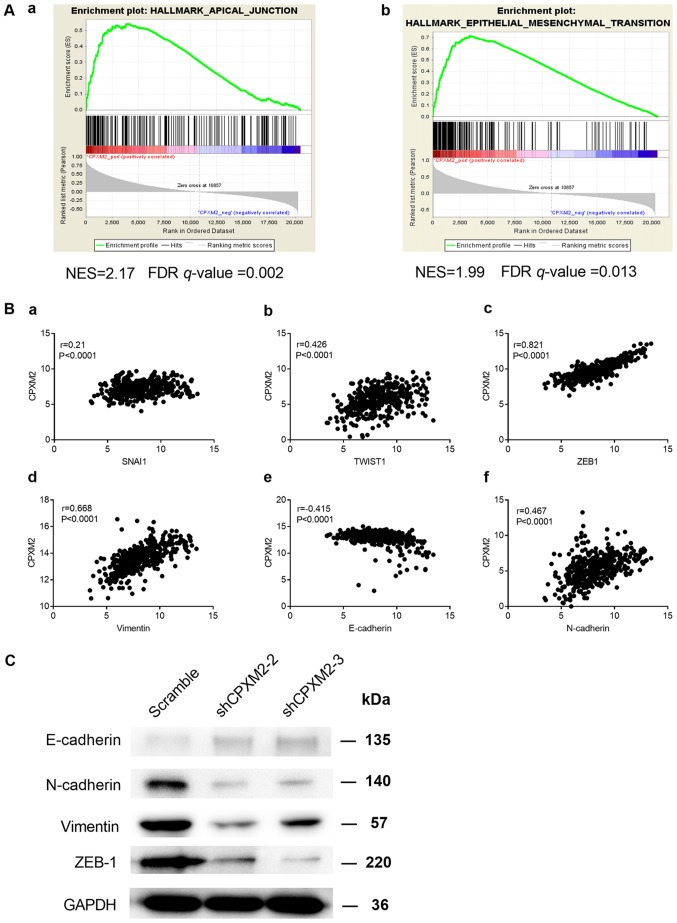
To characterize the potential mechanism of CPXM2 in promoting tumor progression, we used the RNA-seq data for GCs from TCGA to conduct a gene set enrichment analysis (GSEA) (18). We found that CPXM2 expression was positively correlated with the HALLMARK_APICAL_JUNCTION and HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION gene sets (Fig. 5A). In addition, CPXM2 was significantly correlated with key genes involved in EMT (Fig. 5B). Furthermore, the epithelial marker E-cadherin was increased while the mesenchymal markers N-cadherin and vimentin and EMT-related transcription factor ZEB1 were decreased, after CPXM2 was silenced in GC cells (Fig. 5C), Taken together, these results imply that CPXM2 plays an active role in promoting GC tumor aggressiveness via EMT modulation.
Figure 5.
Potential involvement of CPXM2 in the regulation of EMT. (A) GSEA enrichment plots indicate that CPXM2 expression was positively correlated with apical junction and EMT gene signatures. High CPXM2 expression in GC patients was positively correlated with the (a) HALLMARK_APICAL_JUNCTION and (b) HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION gene sets. (B) CPXM2 was significantly correlated with several key genes associated with EMT: (a) SNAI1, (b) TWIST1, (c) ZEB1, (d) vimentin, (e) E-cadherin and (f) N-cadherin. (C) Western blotting analysis was used to compare expression of epithelial and mesenchymal markers between GC cells infected with shCPXM2 or scramble. GAPDH was used as loading control. CPXM2, carboxypeptidase X, M14 family member 2; FDR, false discovery rate; GC, gastric cancer; EMT, epithelial to mesenchymal transition; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GSEA, gene set enrichment analysis; NES, normalized enrichment score.
Discussion
Our knowledge concerning carboxypeptidase X, M14 family member 2 (CPXM2) is still quite limited. Most studies have related CPXM2 to developmental diseases, mental disorders, and neurodegenerative diseases (12–15). By analyzing differentially expressed genes in dermal fibroblasts from patients with Apert syndrome and controls, Çetinkaya et al showed CPXM2 to be a gene with Gene Ontology terms associated with extracellular matrix organization, which may regulate early differentiation of connective tissues (12). Using both growth-restricted and normal-term placentas, Sabri et al recognized CPXM2 as one of the most upregulated genes in fetal growth restriction. Using bioinformatic analysis, these authors also suggested a potential connection between fetal growth restriction and gastrointestinal diseases (13). There are at least 26 metallocarboxypeptidases (MCPs), several of which have already been identified as potential tumor biomarkers (6,9–11,24). However, the underlying mechanism to explain the roles of these MCPs in modulating oncogenesis and tumor progression is still lacking.
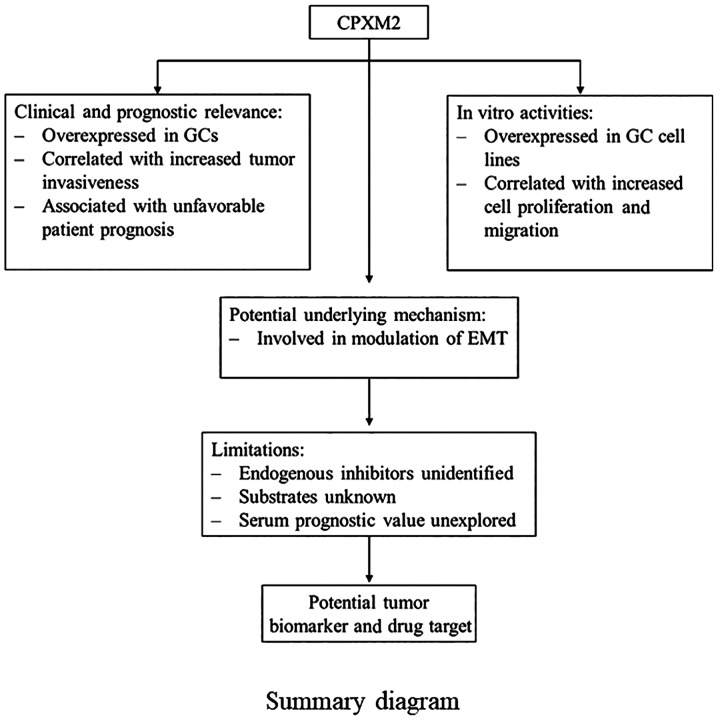
In the present study, we showed that CPXM2 is a candidate tumor biomarker for gastric cancer (GC) (Fig. 6), regardless of the Lauren classification or TNM stage. Therefore, targeting CPXM2 may be a possible solution to impede GC progression. In fact, several endogenous and synthesized carboxypeptidase inhibitors have been reported, although their applications as anticancer reagents are only at a preliminary stage (6,8). Latexin, known for its activity as an endogenous carboxypeptidase inhibitor, is dysregulated and exhibits tumor suppressor potential in several malignancies (25–27). However, analysis of microarray data comparing latexin-overexpressing MGC-803 GC cells with control cells from GEO (GSE15787) (25) found no significant association between latexin and CPXM2 expression (fold change=1.0). Thus, there may be other endogenous inhibitors of CPXM2 that control its enzymatic activity.
Figure 6.
A summary diagram indicating that CPXM2 is a candidate tumor biomarker for GC. CPXM2, carboxypeptidase X, M14 family member 2; GC, gastric cancer; EMT, epithelial to mesenchymal transition.
We also showed that CPXM2 may function by modulating epithelial to mesenchymal transition (EMT). As EMT is a critical process that mediates tumor progression and metastasis (28), the present study provides further evidence that CPXM2 is closely associated with tumor progression in GCs. Considering the catalytical activity of CPXM2, we postulate that CPXM2 may catalyze key molecules that regulate EMT in GCs. Several other MCPs also induce tumor progression and metastasis. For example, CPM is one of the most well-characterized carboxypeptidases. By modulating key molecules such as kinins and chemokines, CPM affects proliferation, angiogenesis, and metastasis in a number of malignancies (11). CPE is another MCP that has been extensively investigated. Both CPE and its splice variant have been proposed as prognostic biomarkers in a variety of tumors, while the latter promotes tumor invasion and metastasis (10). Sun et al demonstrated that CPA4 was associated with tumor invasion and metastasis in a cohort of GC patients. Using correlation analyses, these authors revealed a possible interaction of CPA4 with p53 and Ki-67, which are closely associated with tumor progression (9). Although the tumor-promoting roles of some MCPs have been identified, the underlying mechanisms are largely unknown. We provide a new mechanistic understanding of the effects of these MCPs that centers on their potential modulation of EMT.
To the best of our knowledge, this study is the first to investigate the prognostic value and molecular function of CPXM2 in cancers. However, there are several unanswered questions in this study. First, as mentioned above, we do not know whether there are any endogenous inhibitors of CPXM2. Second, we do not know the substrates of CPXM2. Third, further studies are needed to confirm whether CPXM2 can be used as a serum prognostic biomarker for GC patients.
In summary, we identified CPXM2 as a novel prognostic marker for GC. It was found to accelerate tumor progression by promoting GC cell proliferation and migration via modulation of EMT-associated central pathways, indicating that a detailed study of this putative marker is warranted.
Acknowledgements
We greatly appreciate the technological help from the Department of Pathology of our hospital for the IHC staining and data analysis. We also appreciate the valuable work carried out by Dr Jun Hou at Zhongshan Hospital (Shanghai, China) for her interpretation of the IHC staining. We would like to thank Dr Lijie Ma for his work in packaging the lentiviral particles.
Funding
The present study was supported by the Shanghai Fifth People's Hospital (grant nos. 2016WYRC01 and 2017WYRCSG01); the Medical System of Shanghai Minhang District (grant no. 2017MWDXK01); the Shanghai Minhang District Health and Family Planning Commission (grant no. 2016MW03); and the Shanghai Minhang District Science and Technology Commission (grant no. 2017MHZ02). The funding sources were not involved in the study design; in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Availability of data and materials
The datasets supporting the conclusions of this article are included within this article and its additional images. Raw data are available from the corresponding author on reasonable request.
Authors' contributions
CK designed the study. GN, XW, YY, JR, TS, ZH, LC, JX and RH performed the experiments and analyzed the data. XW, YY and GN wrote the manuscript. CK and JR helped to revise the manuscript. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
This study was approved by the Institutional Ethics Committee of the Fifth People's Hospital of Shanghai, Fudan University (Ethical approval form no. 2017-097) and adhered to the principles in the Declaration of Helsinki. Informed consent was obtained from each patient before tissue collection for experimentation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Malvezzi M, Bonifazi M, Bertuccio P, Levi F, La Vecchia C, Decarli A, Negri E. An age-period-cohort analysis of gastric cancer mortality from 1950 to 2007 in Europe. Ann Epidemiol. 2010;20:898–905. doi: 10.1016/j.annepidem.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez D, Pallares I, Vendrell J, Aviles FX. Progress in metallocarboxypeptidases and their small molecular weight inhibitors. Biochimie. 2010;92:1484–1500. doi: 10.1016/j.biochi.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Gomis-Rüth FX. Structure and mechanism of metallocarboxypeptidases. Crit Rev Biochem Mol Biol. 2008;43:319–345. doi: 10.1080/10409230802376375. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez D, Pallares I, Covaleda G, Aviles FX, Vendrell J. Metallocarboxypeptidases and their inhibitors: Recent developments in biomedically relevant protein and organic ligands. Curr Med Chem. 2013;20:1595–1608. doi: 10.2174/0929867311320120009. [DOI] [PubMed] [Google Scholar]
- 9.Sun L, Guo C, Yuan H, Burnett J, Pan J, Yang Z, Ran Y, Myers I, Sun D. Overexpression of carboxypeptidase A4 (CPA4) is associated with poor prognosis in patients with gastric cancer. Am J Transl Res. 2016;8:5071–5075. [PMC free article] [PubMed] [Google Scholar]
- 10.Cawley NX, Wetsel WC, Murthy SR, Park JJ, Pacak K, Loh YP. New roles of carboxypeptidase E in endocrine and neural function and cancer. Endocr Rev. 2012;33:216–253. doi: 10.1210/er.2011-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denis CJ, Lambeir AM. The potential of carboxypeptidase M as a therapeutic target in cancer. Expert Opin Ther Targets. 2013;17:265–279. doi: 10.1517/14728222.2012.741122. [DOI] [PubMed] [Google Scholar]
- 12.Çetinkaya A, Taskiran E, Soyer T, Şimşek-Kiper PÖ, Utine GE, Tuncbilek G, Boduroğlu K, Alikaşifoğlu M. Dermal fibroblast transcriptome indicates contribution of WNT signaling pathways in the pathogenesis of Apert syndrome. Turk J Pediatr. 2017;59:619–624. doi: 10.24953/turkjped.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Sabri A, Lai D, D'Silva A, Seeho S, Kaur J, Ng C, Hyett J. Differential placental gene expression in term pregnancies affected by fetal growth restriction and macrosomia. Fetal Diagn Ther. 2014;36:173–180. doi: 10.1159/000360535. [DOI] [PubMed] [Google Scholar]
- 14.Chen YC, Hsiao CJ, Jung CC, Hu HH, Chen JH, Lee WC, Chiou JM, Chen TF, Sun Y, Wen LL, et al. Performance metrics for selecting single nucleotide polymorphisms in Late-onset Alzheimer's disease. Sci Rep. 2016;6:36155. doi: 10.1038/srep36155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto R, Ikeda M, Ohi K, Yasuda Y, Yamamori H, Fukumoto M, Umeda-Yano S, Dickinson D, Aleksic B, Iwase M, et al. Genome-wide association study of cognitive decline in schizophrenia. Am J Psychiatry. 2013;170:683–684. doi: 10.1176/appi.ajp.2013.12091228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howitt BE, Sun HH, Roemer MG, Kelley A, Chapuy B, Aviki E, Pak C, Connelly C, Gjini E, Shi Y, et al. Genetic basis for PD-L1 expression in squamous cell carcinomas of the cervix and vulva. JAMA Oncol. 2016;2:518–522. doi: 10.1001/jamaoncol.2015.6326. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–3215. doi: 10.1091/mbc.e02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 19.Szasz AM, Lanczky A, Nagy A, Forster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi JY, Ma LJ, Zhang JW, Duan M, Ding ZB, Yang LX, Cao Y, Zhou J, Fan J, Zhang X, et al. FOXP3 Is a HCC suppressor gene and Acts through regulating the TGF-β/Smad2/3 signaling pathway. BMC Cancer. 2017;17:648. doi: 10.1186/s12885-017-3633-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao S, Zhou L, Niu G, Li Y, Zhao D, Zeng H. Differential regulation of orphan nuclear receptor TR3 transcript variants by novel vascular growth factor signaling pathways. FASEB J. 2014;28:4524–4533. doi: 10.1096/fj.13-248401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu HM, Chen Y, Liu L, Zhang CG, Wang W, Gong K, Huang Z, Guo MX, Li WX, Li W. C1orf61 acts as a tumor activator in human hepatocellular carcinoma and is associated with tumorigenesis and metastasis. FASEB J. 2013;27:163–173. doi: 10.1096/fj.12-216622. [DOI] [PubMed] [Google Scholar]
- 23.Niu G, Ye T, Qin L, Bourbon PM, Chang C, Zhao S, Li Y, Zhou L, Cui P, Rabinovitz I, et al. Orphan nuclear receptor TR3/Nur77 improves wound healing by upregulating the expression of integrin β4. FASEB J. 2015;29:131–140. doi: 10.1096/fj.14-257550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kos J, Vižin T, Fonović UP, Pišlar A. Intracellular signaling by cathepsin X: Molecular mechanisms and diagnostic and therapeutic opportunities in cancer. Semin Cancer Biol. 2015;31:76–83. doi: 10.1016/j.semcancer.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Basang Z, Ding H, Lu Z, Ning T, Wei H, Cai H, Ke Y. Latexin expression is downregulated in human gastric carcinomas and exhibits tumor suppressor potential. BMC Cancer. 2011;11:121. doi: 10.1186/1471-2407-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue Z, Zhou Y, Wang C, Zheng J, Zhang P, Zhou L, Wu L, Shan Y, Ye M, He Y, Cai Z. Latexin exhibits tumor-suppressor potential in pancreatic ductal adenocarcinoma. Oncol Rep. 2016;35:50–58. doi: 10.3892/or.2015.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni QF, Tian Y, Kong LL, Lu YT, Ding WZ, Kong LB. Latexin exhibits tumor suppressor potential in hepatocellular carcinoma. Oncol Rep. 2014;31:1364–1372. doi: 10.3892/or.2014.2966. [DOI] [PubMed] [Google Scholar]
- 28.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within this article and its additional images. Raw data are available from the corresponding author on reasonable request.