GTP cyclohydrolase I from Listeria monocytogenes, a putative anti-infective drug target, has been crystallized and the crystal structure was solved at 2.4 Å resolution.
Keywords: GTP cyclohydrolase I, Listeria monocytogenes, crystal structure, high-throughput screening, tetrahydrofolate biosynthesis, listeriosis
Abstract
A putative open reading frame encoding GTP cyclohydrolase I from Listeria monocytogenes was expressed in a recombinant Escherichia coli strain. The recombinant protein was purified and was confirmed to convert GTP to dihydroneopterin triphosphate (K m = 53 µM; v max = 180 nmol mg−1 min−1). The protein was crystallized from 1.3 M sodium citrate pH 7.3 and the crystal structure was solved at a resolution of 2.4 Å (R free = 0.226) by molecular replacement using human GTP cyclohydrolase I as a template. The protein is a D 5-symmetric decamer with ten topologically equivalent active sites. Screening a small library of about 9000 compounds afforded several inhibitors with IC50 values in the low-micromolar range. Several inhibitors had significant selectivity with regard to human GTP cyclohydrolase I. Hence, GTP cyclohydrolase I may be a potential target for novel drugs directed at microbial infections, including listeriosis, a rare disease with high mortality.
1. Introduction
GTP cyclohydrolase I (EC 3.5.4.16) catalyzes a mechanistically complex ring expansion whereby GTP is converted to dihydroneopterin triphosphate (CAS Registry No. 20574-65-6; Supplementary Fig. S1; Yim & Brown, 1976 ▸; Burg & Brown, 1968 ▸; Fukushima et al., 1977 ▸; for a review, see Gräwert et al., 2013 ▸). More specifically, the imidazole ring of the substrate is hydrolytically opened with the assistance of an essential zinc ion (Bracher et al., 2001 ▸), and the resulting pyrimidine intermediate undergoes an Amadori rearrangement of the carbohydrate side chain followed by ring closure (Bracher et al., 1998 ▸; Rebelo et al., 2003 ▸; Tanaka et al., 2005 ▸). The enzyme product, dihydroneopterin triphosphate, serves as the first committed intermediate in the biosynthesis of tetrahydrofolate by many microorganisms and as the first committed intermediate in the biosynthesis of tetrahydrobiopterin by animals (Gräwert et al., 2013 ▸).
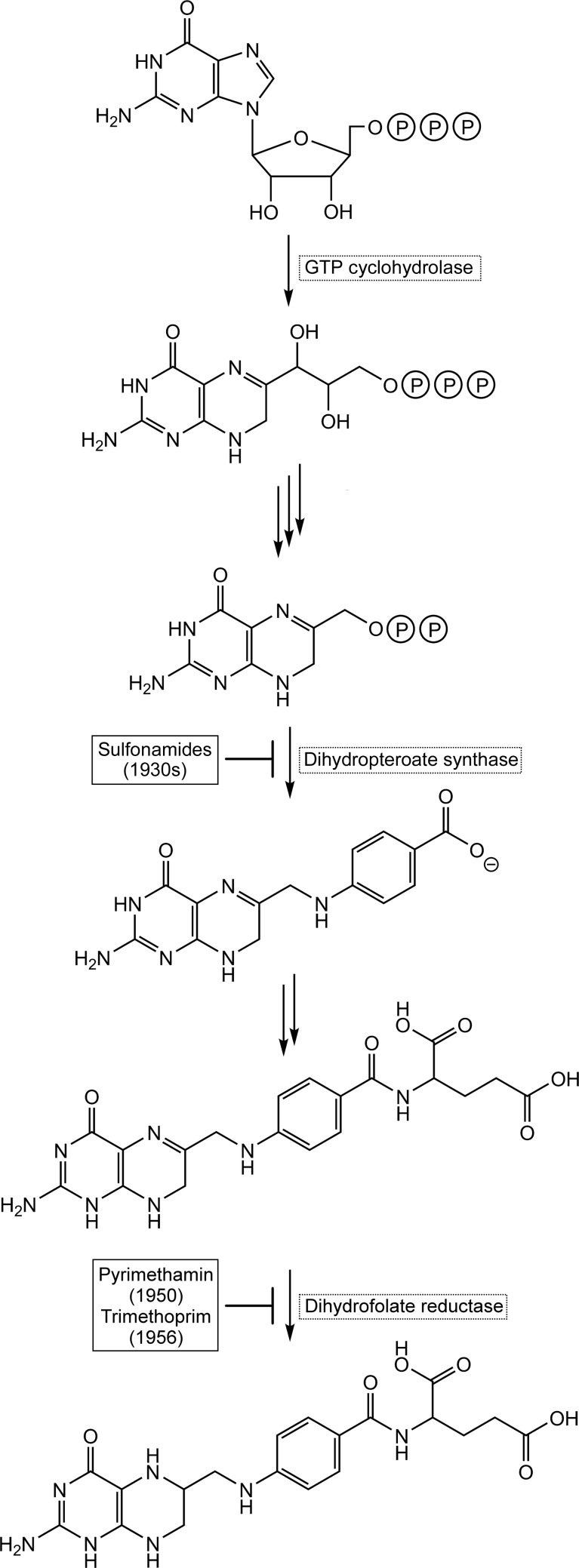
Inhibitors of tetrahydrofolate biosynthesis have played important roles in the treatment of a wide variety of microbial and protozoan infections (Fig. 1 ▸; Brown, 1962 ▸; for a review, see Swarbrick et al., 2008 ▸). Specifically, sulfonamide drugs inhibiting dihydropteroate synthase, the penultimate enzyme of the dihydrofolate pathway, came to the market in the 1930s as the first synthetic agents with broad antimicrobial activity (Nobel Prize awarded to Gerhard Domagk in 1939; Nobel Media AB, 2014a ▸). Whereas sulfonamides have been superseded by more recently developed drugs in the treatment of bacterial infections, they continue to be relevant for certain protozoan diseases such as toxoplasmosis and drug-resistant malaria.
Figure 1.
Bacterial tetrahydrofolate biosynthesis. Enzymes that are existing targets or are potential new targets are shown in black dotted rectangles. Antibiotics inhibiting the enzymes of this pathway are shown near their targets.
The subsequent development of pyrimethamin (in 1950) and trimethoprim (in 1956), which are inhibitors of dihydrofolate reductase, was also honored with Nobel Prizes to Gertrude Elion and George Hitchings in 1988 (Nobel Media AB, 2014 ▸ b); these authors also pioneered the combination of two antifolates directed at two different targets for improved antimicrobial activity.
During the ensuing heyday of the emerging antibiotics era, the emphasis in the field shifted away from synthetic compounds in the direction of natural and semisynthetic agents, while the elucidation of the tetrahydrofolate pathway in the second half of the 20th century has so far not been conducive to the discovery of novel antifolate drugs (for a review, see Burg & Brown, 1968 ▸).
This paper describes the structure elucidation of GTP cyclohydrolase I from Listeria monocytogenes, the foodborne causative agent of listeriosis, which has a mortality rate in the region of 50% (Lorber, 1997 ▸; Schlech & Acheson, 2000 ▸; Granier et al., 2011 ▸). At present, listeriosis infections are usually treated with ampicillin. Cephalosporins are ineffective in Listeria and thus no substitute is available in the case of sensitivity towards β-lactam antibiotics or in the case of emerging resistance. A potential role of the enzyme as an anti-infective drug target is suggested by the discovery of inhibitors which might serve as lead structures, probably also against other pathogens.
2. Materials and methods
2.1. Macromolecule production
2.1.1. Gene cloning and bacterial culture
The putative folE gene from L. monocytogenes (ATCC BAA-679, LGC Standards GmbH, Wesel, Germany) was amplified by two consecutive PCR cycles using the primer pair Fw-LmGTPCHI and Bw-BamHI-LmGTPCHI and the primer pair Fw-EcoRI-LmGTPCHI and Bw-BamHI-LmGTPCHI. The amplificate was digested with EcoRI and BamHI and was ligated into the plasmid pNCO113 that had been treated with the same restriction enzymes. The resulting plasmid pNCO-His6-EK-LmGTP-CHI was transformed into chemically competent XL1-Blue cells (Bullock et al., 1987 ▸; Stratagene, Amsterdam, The Netherlands), affording strain XL1 pNCO-His6-EK-LmGTPCHI. Strain XL1 pNCO-His6-EK-LmGTP-CHI was grown in Terrific Broth containing 170 mg ampicillin per litre for 4 h at 37°C and for a further 36 h at 30°C. The cells were harvested by centrifugation at 4°C and 4000 rev min−1 for 30 min, washed with saline [0.9%(w/v) NaCl] and stored at −20°C.
2.1.2. Protein purification
All steps were carried out at 4°C unless stated otherwise. The cells were thawed on ice, resuspended in buffer A (50 mM Tris–HCl pH 8.0 containing 250 mM NaCl) and disrupted by sonication. The mixture was centrifuged and the supernatant was loaded onto a nickel–NTA agarose column (30 ml; Macherey-Nagel, Düren, Germany) that had been equilibrated with buffer A. The column was washed with 300 ml buffer A followed by 100 and 500 mM imidazole in buffer A. Fractions were combined and concentrated by ultrafiltration. The solution was applied onto a Superdex 200 prep-grade column (5.3 cm2 × 60 cm), which was developed with buffer A. Fractions were combined and concentrated by ultrafiltration. Dithiothreitol was added to a final concentration of 2 mM and the protein was stored at −80°C. Human GTP cyclohydrolase I protein was prepared as reported previously (Auerbach et al., 2000 ▸). Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
Restriction sites are underlined. The first triplet of the folE gene and the stop codon are shown in bold. The His6 tag, His6 tag-coding sequences and enteropeptidase- and enteropeptidase-coding sequences are shown in italics.
| Source organism | L. monocytogenes |
| Forward primer | CACCATCATGGTTCCGATGATGACGATAAGGAGCAAATAGACAAACAAAAGATTGCTGATGCG |
| Forward primer II | ATAATAATAGAATTCATTAAAGAGGAGAAATTAACCATGCATCATCACCACCATCATGGTTCCGATGATGACGATAAG |
| Reverse primer | TATTATTATGGATCC TTAATTATGCTTAATTAAAGCCAAAACTTCACTTCTAAGCTT |
| Cloning vector | pNCO113 |
| Expression vector | pNCO113 |
| Expression host | E. coli |
| Complete amino-acid sequence of the construct produced | MHHHHHHGSDDDDKEQIDKQKIADAVKVILEAVGENPDREGLIDTPMRVARMYEEVFAGLKKDPSVHFDTIFEEQHEELVLVKDIRFSSMCEHHLVPFFGVAHVAYLPQNGRVAGLSKLARVVDDVSRRPQLQERITTTVAEIMMEKLKPLGVMVIMEAEHMCMTIRGVNKPGTKTITSAVRGAFKNDDKLRSEVLALIKHN |
2.2. Crystallization
Crystals were grown by the sitting-drop vapor-diffusion method at 20°C. In initial screening, the Index, Crystal Screen and Crystal Screen 2 kits (Hampton Research, Aliso Viejo, California, USA) were used with a final protein concentration of 5 mg ml−1. The protein was supplied in 10 mM Tris–HCl pH 7.0 containing 75 mM NaCl. Aliquots (1 µl) of protein solution were mixed with 1 µl reservoir buffer solution to obtain a sitting drop. The mother-liquor reservoir contained 60 µl reservoir buffer solution. The protein crystallized from 1.33 M sodium citrate pH 7.3 containing 0.1 M HEPES. Orthorhombic crystals (space group P21, unit-cell parameters a = 78, b = 142, c = 91 Å, 47% solvent content) grew to final dimensions of about 400 × 500 × 300 µm. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Sitting drop |
| Plate type | 96-well |
| Temperature (K) | 283 |
| Protein concentration (mg ml−1) | 5 |
| Buffer composition of protein solution | 10 mM Tris–HCl pH 7.0, 75 mM NaCl |
| Composition of reservoir solution | 0.1 M HEPES pH 7.3, 1.33 M sodium citrate |
| Volume and ratio of drop | 2 µl (1:1) |
| Volume of reservoir (µl) | 60 |
2.3. Data collection and processing
Prior to data collection, crystals were soaked in glycerol for 10 min and were subsequently shock-frozen in a nitrogen stream at 100 K (Oxford Cryosystems). A native data set was collected on the P14 beamline at DESY, Hamburg, Germany. The XDS program package (Kabsch, 2010 ▸) was used to process the collected data. The crystals diffracted to a resolution of 2.4 Å. Data-collection and processing statistics are summarized in Table 3 ▸.
Table 3. Data collection and processing.
| Diffraction source | Beamline P14 (MX2), PETRA III, EMBL c/o DESY |
| Wavelength (Å) | 1.23953 |
| Temperature (K) | 100 |
| Detector | Dectris PILATUS 6M |
| Crystal-to-detector distance (mm) | 349.213 |
| Rotation range per image (°) | 0.1 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 0.1 |
| Space group | P21 |
| a, b, c (Å) | 78.25, 141.85, 90.78 |
| α, β, γ (°) | 90, 104.61, 90 |
| Mosaicity (°) | 0.251 |
| Resolution range (Å) | 30.00–2.40 (2.53–2.40) |
| Total No. of reflections | 510889 |
| No. of unique reflections | 74531 |
| Completeness (%) | 99.7 (99.8) |
| Multiplicity | 6.9 (6.8) |
| 〈I/σ(I)〉 | 19.2 (3.5) |
| R r.i.m. † | 0.076 (0.617) |
| Overall B factor from Wilson plot (Å2) | 39.9 |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
2.4. Structure solution and refinement
Crystal structure analysis was performed by molecular replacement with Phaser (McCoy et al., 2007 ▸) within the CCP4 package (Winn et al., 2011 ▸) using the atomic coordinates of human GTP cyclohydrolase I (PDB entry 1fb1; Auerbach et al., 2000 ▸) as a template. TLS refinement including noncrystallographic symmetry averaging was performed with the PDB-REDO server (Joosten et al., 2014 ▸). The electron density and model were improved by successive rounds of model building and subsequent refinement using Coot (Emsley et al., 2010 ▸) and REFMAC5 (Murshudov et al., 2011 ▸). The asymmetric unit contains one decamer of GTP cyclohydrolase I. 15 N-terminal residues (MHHHHHHGSDDDDKE) and one C-terminal residue (N) are disordered and thus are missing from the model in every monomer. MolProbity (Chen et al., 2010 ▸) was used for Ramachandran analysis. The atomic coordinates and structure factors of GTP cyclohydrolase I from L. monocytogenes have been deposited in the PDB as entry 4uqf. Refinement statistics are summarized in Table 4 ▸.
Table 4. Structure refinement.
| Resolution range (Å) | 87.84–2.40 (2.462–2.400) |
| Completeness (%) | 99.6 |
| No. of reflections, working set | 70754 (5214) |
| No. of reflections, test set | 3752 (281) |
| Final R cryst | 0.193 (0.309) |
| Final R free | 0.226 (0.314) |
| Cruickshank DPI | 0.25 |
| No. of non-H atoms | |
| Protein | 14475 |
| Water | 0 |
| Total | 14475 |
| R.m.s. deviations | |
| Bonds (Å) | 0.014 |
| Angles (°) | 1.773 |
| Average B factors (Å2) | |
| Protein | 49.6 |
| Ramachandran plot | |
| Favored regions (%) | 97.7 |
| Additionally allowed (%) | 2.1 |
| Outliers (%) | 0.2 |
3. Results and discussion
3.1. Results
A putative folE gene encoding GTP cyclohydrolase I was amplified from L. monocytogenes DNA by PCR (Mullis et al., 1986 ▸) and was cloned into the pNCO113 plasmid (Stüber et al., 1990 ▸), where it was preceded by sequence elements encoding a polyhistidine tag and an enterokinase cleavage site. After transformation into a recombinant Escherichia coli host strain, the plasmid directed the synthesis of a polypeptide with an approximate mass of 22 kDa, as estimated by SDS–PAGE (Laemmli, 1970 ▸), which was in good agreement with the calculated mass of 22.9 kDa. The recombinant protein was isolated by metal-affinity chromatography. The predicted sequence was confirmed by partial Edman degradation (Edman, 1960 ▸) and by mass spectrometry of a tryptic digest (Mann & Wilm, 1994 ▸).
The recombinant protein catalyzes the conversion of GTP to dihydroneopterin triphosphate (v max = 180 nmol mg−1 min−1, K m = 53 µM; Supplementary Fig. S3) as determined by photometric analysis (for details of the enzyme assay, see the supporting information). The formation of the second enzyme product, formate, was also directly detected by NMR spectroscopy using [U-13C10]GTP as substrate. In line with reports on other known GTP cyclohydrolase I orthologs, the addition of divalent cations to the assay mixtures was not required for activity.
Equilibrium sedimentation yielded a molecular weight of 212 ± 30 kDa. Boundary sedimentation resulted in a sedimentation velocity of 9.6 S. Mass spectrometry revealed the presence of 0.9 moles of zinc per mole of polypeptide.
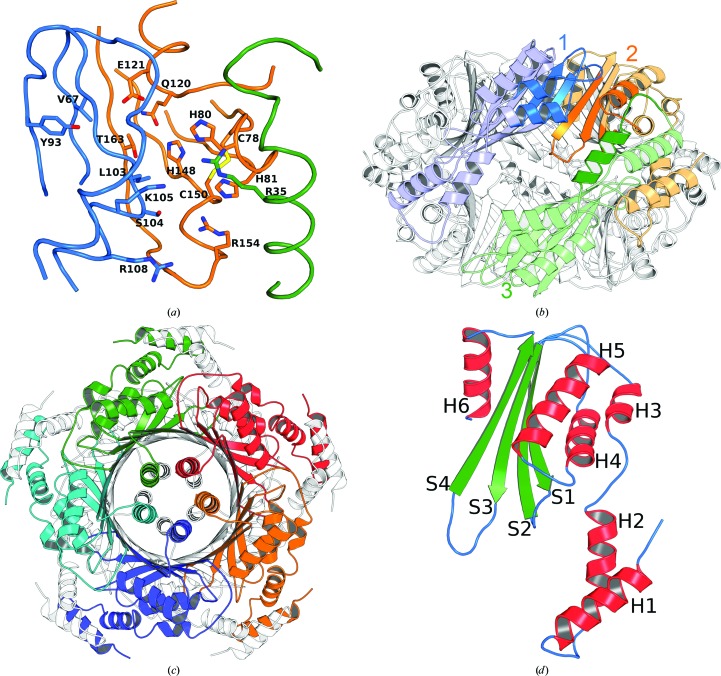
The protein is a D 5-symmetric decamer with close similarity to several orthologous proteins from eubacteria and animals (Fig. 2 ▸, Supplementary Table S1; Nar et al., 1995 ▸). The r.m.s.d. values to previously reported GTP cyclohydrolase I structures were in the range 0.76–1.3 Å for the monomers, whereas r.m.s.d. values in the range 0.12–0.34 Å were found on comparing individual L. monocytogenes GTP cyclohydrolase I monomers. However, no appreciable density representing the essential zinc ions was present, and the two canonical cysteine residues Cys78 and Cys150 that form part of the putative zinc-binding site (for a sequence alignment, see Supplementary Fig. S2) were oxidized to the disulfide level; it is most probable that the zinc had been lost owing to the chelating activity of the citrate that was used as a precipitant, and loss of the metal cation had been followed by oxidation of the zinc-chelating cysteine residues. A similar observation had previously been made for crystals of GTP cyclohydrolase I from E. coli that had been exposed to EDTA. In comparison, the Listeria protein appears to be even more sensitive to chelator-induced zinc-ion loss, but a search for other (nonchelating) crystallization conditions has been unsuccessful to date.
Figure 2.
Structural features of L. monocytogenes GTP cyclohydrolase I. (a) Close-up of the active site. Conserved amino acids are shown in stick representation. (b) Active-site composition; the view is along one C 2 axis. Monomer 1 (blue) and monomer 2 (orange) belong to the ‘top’ pentamer; monomer 3 (green) belongs to the ‘bottom’ pentamer. Intense coloring indicates the active site (one of ten) formed by these monomers. (c) View along the C 5 axis. Monomers of the ‘top’ pentamer are shown in distinct colors and the ‘bottom’ pentamer is in black and white. (d) Monomer architecture. H, helix; S, strand. These figures were prepared using PyMOL (DeLano, 2004 ▸). Corresponding color coding is used in (a) and (b); arbitrary color coding is used in (c) and (d).
In parallel to the findings for other decameric GTP cyclohydrolases (Auerbach et al., 2000 ▸; Nar et al., 1995 ▸; Tanaka et al., 2005 ▸; Maita et al., 2002 ▸), each of the ten active sites of the protein is located at the interface of three subunits (two in one C 5-symmetric unit and a third in the adjacent C 5-symmetric pentamer unit). The residues lining the active-site cavity of the L. monocytogenes protein have high temperature factors, possibly owing to the absence of a divalent cation.
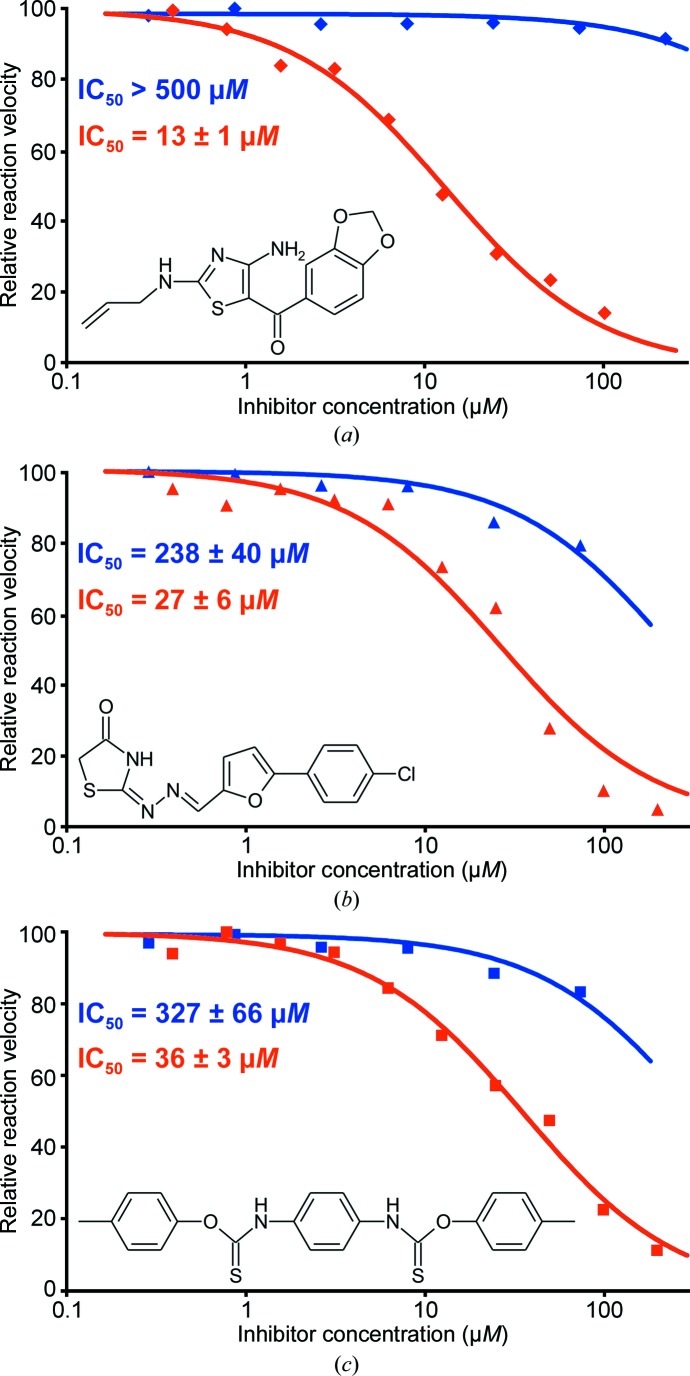
In order to explore the druggability of the protein, we screened a small library of about 9000 highly diverse compounds (Bracher et al., 1998 ▸). Assays were run in 384-well plates and were monitored photometrically at 330 nm. Hits were rescreened against GTP cyclohydrolase I from L. monocytogenes and human GTP cyclohydrolase I, and IC50 values were determined using the photometric assay. Semilogarithmic dose–response curves showed symmetrical, S-shaped features, and the overall Z factor (Zhang et al., 1999 ▸) of the screen was 0.87. Three heterocyclic compounds inhibited the L. monocytogenes enzyme with IC50 values in the low double-digit micromolar range while not acting as strong inhibitors of human GTP cyclohydrolase I (Fig. 3 ▸; for details of the high-throughput assay, see the supporting information). Although stronger inhibitors have been reported down to a K i of 5.4 nM (Tanaka et al., 2005 ▸), all of the inhibitors reported so far are substrate or product analogs.
Figure 3.
Dose–response curves. Red, GTP cyclohydrolase I from L. monocytogenes; blue, human GTP cyclohydrolase I. Inhibition of the Listeria enzyme is significantly stronger compared with the human enzyme. IC50 values were calculated with DynaFit (BioKin, Watertown, Massachusetts, USA; Kuzmič, 1996 ▸).
3.2. Discussion
Animals and many microorganisms feature homodecameric, D 5-symmetric GTP cyclohydrolases I with molecular masses of about 200 kDa. The enzyme from L. monocytogenes is similar in terms of sequence and three-dimensional structure to orthologs from eubacteria (E. coli and Yersinia pestis). The structure is also similar to mammalian cyclohydrolase I (rat and mouse), although the mammalian protomers are somewhat longer.
Our data confirm that GTP cyclohydrolase I from L. monocytogenes is well suited for high-throughput screening. Importantly, screening does not require any supplementary enzymes, since the enzyme product can be monitored directly by photometry.
Screening a small library of about 9000 structurally diverse compounds retrieved an inhibitor with an IC50 of 13 µM; importantly, the compound was much less active against human GTP cyclohydrolase I (IC50 > 500 µM).
In order to discuss the potential role of bacterial GTP cyclohydrolase I as an anti-infective drug target, the role of GTP cyclohydrolase I in bacterial pathogens as well as in the human host must be addressed in some detail. Importantly, in 2006 it was found that certain eubacteria do not carry a folE gene but produce a homotetrameric protein designated GTP cyclohydrolase IB that is under the control of the folE2 gene (El Yacoubi et al., 2006 ▸). In contrast to the decameric GTP cyclohydrolases, the type IB protein is not strictly dependent on zinc ions but can use several different divalent transition-metal ions, including iron, for catalysis. In a remarkable parallel to the decameric type I enzymes, the active sites of the type IB enzymes are also located at the interfaces of three adjacent subunits. Moreover, the protein segments that are located at the surface of the active-site cavity align almost perfectly with the cognate sequence elements of GTP cyclohydrolase I, although they have been reshuffled with respect to their location in the overall protein sequence. The folE2-type GTP cyclohydrolase provides a metabolic advantage under conditions of limited zinc supply.
The L. monocytogenes genome does not contain a folE2 gene. However, certain other pathogenic eubacteria contain folE2 genes, either alone or in combination with folE and folE1 genes; multiple genomic copies of folE and folE1 have also been observed. Whether it would be possible to develop inhibitors that act against the type IB as well as the type I GTP cyclohydrolase is as yet unknown. Inhibitors that act only against one type of GTP cyclohydrolase would be significantly restricted with regard to their therapeutic spectrum.
Yet another caveat needs to be considered with regard to the use of GTP cyclohydrolases (type I and/or type IB) as anti-infective drug targets, namely the structural similarity of bacterial GTP cyclohydrolase I to human GTP cyclohydrolase I. The human enzyme catalyzes the first committed step in the biosynthesis of tetrahydrobiopterin that is required for the catabolism of phenylalanine and for the biosynthesis of nitric oxide (for reviews, see Fischer & Bacher, 2005 ▸; Gräwert et al., 2013 ▸). Ideally, any putative drugs directed at microbial GTP cyclohydrolases should be exempt from inhibition of the human ortholog. Nevertheless, the stringency of this argument may not be absolute. In fact, the human enzyme is considered to be a potential target for pain relief (Tegeder et al., 2006 ▸). Although a genetic deficiency of GTP cyclohydrolase causes severe neurological deficits during fetal and childhood development, its inhibition for the duration of treatment of an acute infectious disease in adults for short periods may be acceptable.
Historically, the success of antifolates as anti-infective drugs has benefited massively from the possibility of the simultaneous inhibition of two folate-pathway enzymes, namely dihydropteroate synthase and dihydrofolate reductase (Fig. 1 ▸). Despite the caveats addressed above, the possibility of adding a third potential anchor point to the arsenal of antifolate strategies may be worth considering. In this context, it is also relevant to note that fungal protozoan pathogens only have the classical GTP cyclohydrolase I, in contrast to the more complex situation in bacterial pathogens.
Supplementary Material
PDB reference: GTP cyclohydrolase I, 4uqf
Supporting Methods, Supplementary Figures and Supplementary Table. DOI: 10.1107/S2053230X19010902/no5161sup1.pdf
Acknowledgments
The compound library was kindly provided by BASF SE, Ludwigshafen, Germany. We thank Ruslan Sanishvili, Argonne National Laboratory for organizing the CCP4 summer school in 2010.
Funding Statement
This work was funded by Deutsche Forschungsgemeinschaft grant . Hans-Fischer-Gesellschaft grant .
References
- Auerbach, G., Herrmann, A., Bracher, A., Bader, G., Gütlich, M., Fischer, M., Neukamm, M., Garrido-Franco, M., Richardson, J., Nar, H., Huber, R. & Bacher, A. (2000). Proc. Natl Acad. Sci. USA, 97, 13567–13572. [DOI] [PMC free article] [PubMed]
- Bracher, A., Eisenreich, W., Schramek, N., Ritz, H., Götze, E., Herrmann, A., Gütlich, M. & Bacher, A. (1998). J. Biol. Chem. 273, 28132–28141. [DOI] [PubMed]
- Bracher, A., Schramek, N. & Bacher, A. (2001). Biochemistry, 40, 7896–7902. [DOI] [PubMed]
- Brown, G. M. (1962). J. Biol. Chem. 237, 536–540. [PubMed]
- Bullock, W. O., Fernandez, J. M. & Short, J. M. (1987). Biotechniques, 5, 376–377.
- Burg, A. W. & Brown, G. M. (1968). J. Biol. Chem. 243, 2349–2358. [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2004). Abstr. Pap. Am. Chem. Soc. 228, U313–U314.
- Edman, P. (1960). Ann. N. Y. Acad. Sci. 88, 602–610. [DOI] [PubMed]
- El Yacoubi, B., Bonnett, S., Anderson, J. N., Swairjo, M. A., Iwata-Reuyl, D. & de Crécy-Lagard, V. (2006). J. Biol. Chem. 281, 37586–37593. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fischer, M. & Bacher, A. (2005). Nat. Prod. Rep. 22, 324–350. [DOI] [PubMed]
- Fukushima, K., Richter, W. E. & Shiota, T. (1977). J. Biol. Chem. 252, 5750–5755. [PubMed]
- Granier, S. A., Moubareck, C., Colaneri, C., Lemire, A., Roussel, S., Dao, T.-T., Courvalin, P. & Brisabois, A. (2011). Appl. Environ. Microbiol. 77, 2788–2790. [DOI] [PMC free article] [PubMed]
- Gräwert, T., Fischer, M. & Bacher, A. (2013). IUBMB Life, 65, 310–322. [DOI] [PubMed]
- Joosten, R. P., Long, F., Murshudov, G. N. & Perrakis, A. (2014). IUCrJ, 1, 213–220. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kuzmič, P. (1996). Anal. Biochem. 237, 260–273. [DOI] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Lorber, B. (1997). Clin. Infect. Dis. 24, 1–11.
- Maita, N., Okada, K., Hatakeyama, K. & Hakoshima, T. (2002). Proc. Natl Acad. Sci. USA, 99, 1212–1217. [DOI] [PMC free article] [PubMed]
- Mann, M. & Wilm, M. (1994). Anal. Chem. 66, 4390–4399. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G. & Erlich, H. (1986). Cold Spring Harb. Symp. Quant. Biol. 51, 263–273. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nar, H., Huber, R., Meining, W., Schmid, C., Weinkauf, S. & Bacher, A. (1995). Structure, 3, 459–466. [DOI] [PubMed]
- Nobel Media AB (2014a). Gerhard Domagk – Biographical. http://www.nobelprize.org/nobel_prizes/medicine/laureates/1939/domagk-bio.html.
- Nobel Media AB (2014b). The Nobel Prize in Physiology or Medicine 1988. https://www.nobelprize.org/nobel_prizes/medicine/laureates/1988/press.html.
- Rebelo, J., Auerbach, G., Bader, G., Bracher, A., Nar, H., Hösl, C., Schramek, N., Kaiser, J., Bacher, A., Huber, R. & Fischer, M. (2003). J. Mol. Biol. 326, 503–516. [DOI] [PubMed]
- Schlech, W. F. III & Acheson, D. (2000). Clin. Infect. Dis. 31, 770–775. [DOI] [PubMed]
- Stüber, D., Matile, H. & Garotta, G. (1990). Immunological Methods, Vol. IV, edited by I. Lefkovits & B. Pernis, pp. 121–152. New York: Academic Press.
- Swarbrick, J., Iliades, P., Simpson, J. S. & Macreadie, I. (2008). Open Enzym. Inhib. J. 1, 12–33.
- Tanaka, Y., Nakagawa, N., Kuramitsu, S., Yokoyama, S. & Masui, R. (2005). J. Biochem. 138, 263–275. [DOI] [PubMed]
- Tegeder, I., Costigan, M., Griffin, R. S., Abele, A., Belfer, I., Schmidt, H., Ehnert, C., Nejim, J., Marian, C., Scholz, J., Wu, T., Allchorne, A., Diatchenko, L., Binshtok, A. M., Goldman, D., Adolph, J., Sama, S., Atlas, S. J., Carlezon, W. A., Parsegian, A., Lötsch, J., Fillingim, R. B., Maixner, W., Geisslinger, G., Max, M. B. & Woolf, C. J. (2006). Nature Med. 12, 1269–1277. [DOI] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Yim, J. J. & Brown, G. M. (1976). J. Biol. Chem. 251, 5087–5094. [PubMed]
- Zhang, J.-H., Chung, T. D. Y. & Oldenburg, K. R. (1999). J. Biomol. Screen. 4, 67–73. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: GTP cyclohydrolase I, 4uqf
Supporting Methods, Supplementary Figures and Supplementary Table. DOI: 10.1107/S2053230X19010902/no5161sup1.pdf