Summary
Microbiota from various maternal sites, including the gut, vagina and breast milk, are known to influence colonization in infants. However, emerging evidence suggests that these sites may exert their influence prior to delivery, in turn influencing fetal immune development. The dogma of a sterile womb continues to be challenged. Regardless, there is convincing evidence that the composition of the maternal gut prior to delivery influences neonatal immunity. Therefore, while the presence and function of placental microbiome is not clear, there is consensus that the gut microbiota during pregnancy is a critical determinant of offspring health. Data supporting the notion of bacterial translocation from the maternal gut to extra‐intestinal sites during pregnancy are emerging, and potentially explain the presence of bacteria in breast milk. Much evidence suggests that the maternal gut microbiota during pregnancy potentially determines the development of atopy and autoimmune phenotypes in offspring. Here, we highlight the role of the maternal microbiota prior to delivery on infant immunity and predisposition to diseases. Moreover, we discuss potential mechanisms that underlie this phenomenon.
Keywords: infant immunity, microbiota, pregnancy
Introduction
The human body is home to a range of microorganisms, including viruses, bacteria, fungi, archae and unicellular eukaryotes. The bacterial contingent of this community, the microbiota, are known to affect our health in profound ways, bringing to the forefront the symbiotic relationship that exists between these microbes and their human host. Vertical transmission of microbes from various maternal body sites contribute to the developing infant gut microbiota, including the gut, vagina, skin and breast milk 1, 2. Many of these exert their influences during delivery or postpartum, after exposure to maternal vagina, stool and skin 3, but emerging evidence suggests that their influence may begin in utero. Historically, it has been assumed that the uterine environment is sterile 4. This dogma has since been challenged 5, 6, 7, 8, with unique placental microbiome in some studies 5, 9 but not others 7, 10.
Regardless of whether there is a fetal or placental microbiome, there is evidence of a strong influence of maternal gut microbiota during pregnancy on infant microbiota. Maternal gut strains have been shown to be more persistent in the infant gut and ecologically better adapted compared to those from other sources 11. In mice, genetically labeled bacteria were present in meconium samples that matched those that were orally administered to the mother 8. In addition, germ‐free pregnant dams transiently colonized with a modified Escherichia coli strain and returned to germ‐free status prior to delivery 12 had pups with altered innate lymphoid and mononuclear cells, suggesting that transient changes in maternal microbiota during pregnancy drive fetal immune programming. Therefore, whether or not the fetus is indeed colonized in‐utero, the period during pregnancy is the first point of maternal microbes influencing fetal immunity. The mechanisms through which maternal microbiota prior to delivery program neonatal immunity are yet to be elucidated. Here, we review current literature on the impact of maternal microbiota during pregnancy on infant immunity.
Prenatal gut microbiome and fetal immunity
Antibiotic‐induced shifts in microbiota
A significant body of work implicates exposures and events during gestation as critical determinants of offspring disease predisposition later in life 13, 14, 15, 16. This concept has formed the basis of the developmental origins of health and disease (DOHaD) hypothesis 17, 18. These influences include maternal diet, toxins, stress and smoking and may act through changing metabolism, epigenetics and/or microbiota 19. Support for microbial‐mediated mechanisms are widespread. Tormo‐Badia et al. 20 observed significantly increased proportions of CD8+ T cells in the mesenteric lymph nodes (MLN) of pups born to non‐obese diabetic (NOD) dams treated with antibiotics during pregnancy compared to control pups. Similarly, pups born to mothers treated with antibiotics orally during gestation and postpartum exhibited increased susceptibility to vaccinia virus infection and reduced interferon (IFN)‐γ production by CD8+ T cells when compared to controls 21. Follow‐up experiments revealed that the observed CD8+ T cell impairment was driven by an altered activation and expression of T cell receptor (TCR) critical for sustained cytokine production 22. We recently showed, using mouse models, that altering maternal intestinal microbiota during pregnancy alone impacted inherent immunity in pups at 14 days postpartum 1 (Fig. 1). We intended to only alter the microbiota in maternal gut and not at other body sites by administering oral vancomycin, which has poor oral bioavailability, during gestation. Indeed, we did not detect any vancomycin in maternal plasma 2 days after delivery, indicating that the antibiotic was not systemically absorbed. Although the intervention was targeted to maternal gut in our model, we observed perturbations in breast milk microbiota, and to a lesser degree vaginal microbiota postpartum, suggesting that gut microbiota during pregnancy may also impact that at distal sites 1 and this may, in part, be a mechanism through which maternal gut microbiota influence neonatal immunity.
Figure 1.
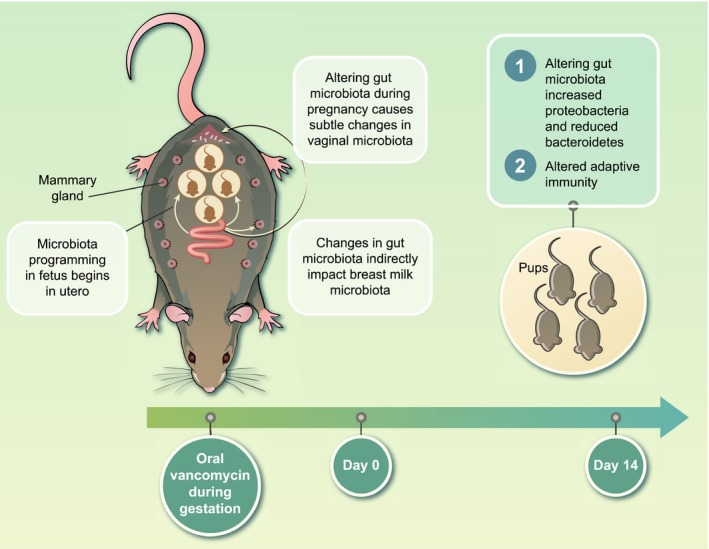
Experimental model of the impact of maternal gut microbiota during gestation on offspring immunity. To test the role of maternal gut microbiota during gestation on offspring immunity, pregnant BALB/c dams were fed vancomycin in drinking water for 5 days prior to delivery. No antibiotics were administered to control dams. All mice in these groups received normal water after delivery. Pups born to dams treated with vancomycin during gestation only had altered intestinal microbiota compared to controls. In addition, pups born to vancomycin breeders had significantly higher splenic cell counts, higher numbers of total B cells as well as follicular B cells versus control pups 1 .
Gut–breast milk bacterial axis
An early study by Martin et al. reported that select gut bacteria from the maternal gastrointestinal tract can access the mammary glands through an enteromammary pathway 23. Although controversial, some studies have offered a scientific basis for such physiological translocation (reviewed in 24). The mechanisms could involve dendritic cells (DCs) and CD18+ cells 25, 26, which take up non‐pathogenic bacteria from the gut lumen and subsequently transport them to other locations, including lactating mammary glands. Bacterial translocation from the gut to the mammary glands and milk has been observed in mice late in gestation 27. Two lactic acid bacteria strains, Lactococcus lactis and L. salivarius, were orally administered to pregnant dams and later detected in mammary tissue and milk 27. While the breast milk microbiota influences immune development postpartum 28, 29, 30, the composition of these microbiota is partly shaped prior to delivery and is dependent on gut microbiota during pregnancy 1. Therefore, gut composition prior to delivery indirectly drives early offspring immune development via the gut–breast milk axis. However, further research is needed to explore the existence of the bacterial enteromammary pathway. This novel form of maternal–neonatal communication could influence our present understanding of fetal immune development.
Vaginal microbiota during pregnancy and infant immunity
The vagina contains more than 170 species of bacteria, and these communities are less diverse and remarkably stable throughout pregnancy 31, 32, 33. Previous work suggests that the vagina might be a source of microbes that reach the placenta, amniotic fluid and fetus, via translocation across the choriodecidual plate 34, 35). However, microbes in the vagina are probably themselves critical in programming neonatal immunity. A great deal of data exist regarding immunological differences between vaginal versus caesarean‐delivered infants, suggesting a role of vaginal microbiota in immune education in offspring 36, 37, 38. Maternal vaginal microbiota during labor and delivery is probably determined during pregnancy. There is also evidence that maternal vaginal microbiota during pregnancy impacts infant immunity even before passage through the vaginal canal during delivery in utero. Newborns whose mothers were vaginally colonized with Lactobacillus during pregnancy had higher proportions of CD45RO+ cells and reduced IL‐12 in cord blood, indicating that lactobacilli in the maternal vagina impact fetal immune development 39. Benn et al. 40 found that the presence of certain maternal vaginal microbes was associated with risk for wheezing in children at 4–5 years of age. The mechanisms of this interaction remain unknown, but could be due to bacterial metabolites, ascending organisms or simply that women’s vaginal microbiota is largely dependent on that of the gut microbiota.
Maternal microbiota during pregnancy and offspring immune‐related disorders
Allergy and asthma
There is substantial epidemiological evidence that exposure to farming and pets in early life is associated with reduced incidence of asthma and allergies 41, 42, 43. In addition, a large European study revealed that mothers exposed to farm animals during pregnancy were less likely to have children who developed allergies, and the immunological tolerance to allergens was already present in cord blood 44. Similarly, a recent study in a Chinese cohort observed that maternal exposure to farming during pregnancy impacted the quantity and function of neonatal regulatory T cells (Tregs), partly contributing to reduction of incidences of allergies and asthma in offspring 45. Maternal pet exposure has been associated with reduced cord blood levels of immunogloblulin (Ig)E 46 and increased numbers of Tregs which ameliorate the effect of allergy‐mediated T helper type 2 (Th2) cytokines 47, 48. Douwes et al. 49 showed that maternal exposure during pregnancy to animals and/or grain and hay reduced the symptoms of asthma, hay fever and eczema in their children. Prenatal exposure was found to contribute to low prevalence of these atopic diseases, and continued exposure only contributed additional protection in some cases. Importantly, the timing of these exposures is crucial, with the strongest effects observed in utero and during the first year of life 50. The interaction between farm‐derived biological factors and the immune responses and disease susceptibility in the host has also been tested in mouse models. An interesting study by Conrad et al. 51 investigated the asthma‐protective effect of prenatal exposure to farm‐derived microorganisms. Intranasal exposure to Acinetobacter lwoffii F78 (cowshed‐derived bacterium) protected against the development of experimental asthma in the progeny, and this protection was dependent on intact maternal Toll‐like receptor (TLR) signaling 51. While the exact mechanism of allergic protection is unclear in humans, as both prenatal and postnatal pet exposure alters infant gut microbial composition 52 it is likely that modulation of the microbiota plays a role.
Microbiome modulation by probiotics has also been shown to impact pediatric allergy development 53. Combined pre‐ and postnatal probiotic supplementation was shown to be crucial for the preventive effects of probiotics on infant eczema; prenatal or postnatal supplementation alone was ineffective 53, 54. However, others have found the prenatal component to be more influential 55. While probiotic‐induced changes in maternal gut microbiota during pregnancy and lactation may reduce incidence of pediatric allergies, further research is warranted to determine optimal timing and dosage.
More direct evidence for the influence of maternal microbiota during pregnancy on offspring atopic disease is antibiotic use during pregnancy, which can cause prolonged alterations to the microbiota and their metabolites 56. In a Danish birth cohort, the use of prenatal antibiotics was associated with increased odds of atopic dermatitis at 18 months of age in infants of mothers with atopy 57. In addition, in children aged 2–10 years, maternal use of any antibiotics during pregnancy was associated with a 1·3‐fold increased risk of asthma in the offspring [95% confidence interval (CI) = 1·21–1·42] (58). However, in a Swedish study by Örtqvist et al. 59 antibiotic exposure in fetal life was associated with increased risk for asthma in a cohort analyses but not sibling analyses, suggesting that the link between antibiotic use and atopy may be confounded by shared familial risk factors . Furthermore, a recent study showed that maternal antibiotic exposure during pregnancy is associated with a dose‐dependent increase in child asthma risk 60, but so was maternal antibiotic use before pregnancy and in the 9 months postpartum. However, in a well‐designed sibling‐control study, Mulder et al. 61 found prenatal antibiotic use to be predictive of childhood asthma even compared to sibling controls. Although some studies show influence regardless of trimester of maternal antibiotic use 57, this study found the influence to be more profound when antibiotic use occurs during the third trimester 61. Although more research is needed to determine whether timing is crucial, multiple lines of evidence suggest the maternal microbiota during pregnancy plays a key role in preventing an allergy‐prone immune phenotype in infants.
Type 1 diabetes
Apart from allergy and asthma, the prenatal microbiome has also been implicated in other immune disorders, including type 1 diabetes and inflammatory bowel disease. Studies in rodent models of spontaneous type 1 diabetes (T1D) have linked the gut microbiota to disease susceptibility 62, 63. Livanos et al. 64 demonstrated that administering a subtherapeutic low‐dose penicillin (STAT) to mice during the late pregnancy period and postpartum accelerates the incidence of T1D in male offspring compared to untreated controls; however, whether these effects were mediated in the antepartum period is unclear. Either way, there were no differences in the frequencies of the lamina propria Treg or Th17 cells between STAT and control pups. In an elegant murine study conducted by Hu et al., oral administration of a combination of neomycin, polymyxin B and streptomycin to pregnant NOD dams led to a delay in the development of diabetes in these susceptible offspring. The incidence of diabetes was also significantly reduced relative to offspring born to untreated dams 65. Authors observed shifts in microbiota of both dams and pups, suggesting a direct role of the microbiota in modulating the predisposition of T1D in offspring 65. Furthermore, antibiotic treatment of NOD dams during pregnancy led to lower frequencies of both CD4+ and CD8+ T cells production of IFN‐γ in the MLN in pups compared to those born to untreated dams. Interestingly, dams treated with vancomycin during pregnancy had different alterations in gut microbiota and increased susceptibility to T1D via opposing immunological effects, possibly mediated by alterations in antigen‐presenting cells (APCs) 66.
Inflammatory bowel disease
Beyond T1D, the microbiota has also been implicated in inflammatory bowel diseases (IBD) 67, 68, 69. The composition of the gut during pregnancy has been shown to influence IBD outcomes in offspring. For example, antibiotic exposure during pregnancy but not during infancy was associated with an increased risk of very early onset IBD, regardless of whether there was antecedent gastroenteritis 70. In a murine study using an IL‐10 knock‐out colitis model, exposing dams to antibiotics from the third week of gestation until weaning led to increased susceptibility to chemically induced dextran sulfate sodium (DSS) colitis and inflammation in her offspring, lasting into adulthood 71. This suggests that maternal microbiota during pregnancy is a critical determinant of IBD development. Taken together, the T1D and IBD studies discussed demonstrate that maternal dysbiosis during pregnancy has immunological consequences in the offspring and is a determinant of infant predisposition to autoimmune disorders.
Potential mechanism of cross‐talk between maternal microbiota and offspring immunity
Although the exact mechanisms remain to be revealed, microbiota during pregnancy are thought to initiate offspring immune programming in various somewhat interrelated ways which are not necessarily mutually exclusive (Table 1, Fig. 2).
Table 1.
Mechanisms of cross‐talk between maternal microbiota and offspring immunity
| Study theme | Outcome | Species | Refs |
|---|---|---|---|
| Maternal microbiota influence fetal microbiota |
|
Mouse | 73 |
|
Human | 76, 77, 78 | |
|
Human | 5, 9 | |
| Maternal microbiota influence infant microbiota and immunity |
|
Human; | 11 |
|
Mouse | 1, 12, 20 | |
|
Mouse | 99, 100 | |
|
Mouse | 1 | |
| Bacterial metabolites |
|
Mouse; | 86, 87, 89, 90, 101 |
|
Mouse | 12, 91, 93 | |
| Maternal IgG |
|
Mouse | 1 |
|
Human; | 97 | |
|
Mouse | 12 |
Figure 2.
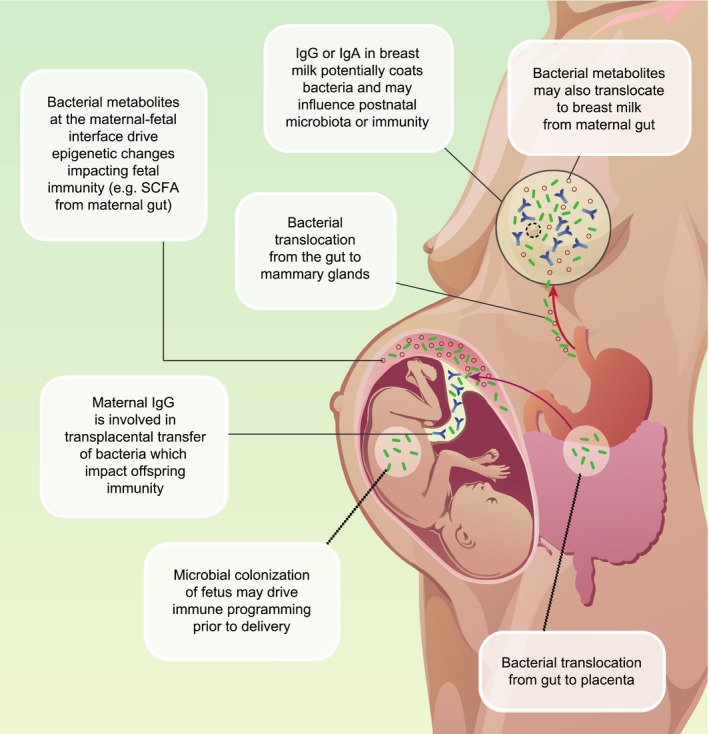
Potential mechanisms of crosstalk between maternal microbiota and offspring immunity. Maternal gut microbiota during pregnancy translocate to the maternal‐fetal interface. Commensals microbes translocate from the maternal gut to the placenta or fetal gut during pregnancy (maternal gut placenta axis) or to mammary glands. These microbes impact developing fetal immunity via various mechanisms including epigenetic changes, release of short chain fatty acids and alteration of the cytokine environment. Bacteria or bacterial metabolites transfer to the mammary glands (gut‐breastmilk axis) impacting infant gut colonization and continued immune development after delivery.
Maternal microbiota influence fetal microbiota
There are data supporting the possibility that the fetal microbiota may develop in utero via the placental barrier or through ingestion of amniotic fluids 72, and therefore may impact the developing fetal immune system. Various studies have indicated that certain bacteria from the maternal gut may translocate to extradigestive sites in healthy hosts 73, 74, 75. Murine and human studies have shown that meconium is colonized with bacteria during pregnancy 76, 77, 78. Jimenez et al. 8 isolated a tagged Enterococcus faecium from the meconium of offspring after orally inoculating the strain to pregnant dams, indicating that maternal gut microbes potentially cross the placenta to offspring gut.
In theory, fetal intestines may be exposed to commensal microbes and their products in swallowed amniotic fluid, which may therefore be an important contributor to early immune development. For example, memory CD4+ and CD8+ T cells can be identified towards the end of the first trimester in human fetal gut 79. Memory CD4 T cells in fetal intestines have been shown to co‐localize with APCs and produce IFN‐γ, IL‐2 or tumor necrosis factor (TNF)‐α, promoting intestinal development 80, 81. Altogether, these suggest that early fetal exposure to microbial antigens may impact immunity. While it is not clear what the relative contribution of maternal versus fetal microbiome is to offspring immunity, it is plausible that both these microbiota are critical in programming fetal immunity prior to delivery.
Maternal microbiota during pregnancy influence early‐life infant microbiota and immunity
Although not a direct effect during pregnancy, maternal microbiota during pregnancy shape the vaginal and breast milk microbiota, which will alter the pioneer infant microbiota during a critical window in immune development. Indeed, we have recently demonstrated that altering maternal gut communities only during gestation indirectly impacts breast milk and, to a lesser degree, vaginal microbiota 1 (Fig. 1). In addition, vancomycin‐induced shifts in maternal gut microbiota profoundly impacted infant gut microbiota 14 days postpartum. Pups born to dams treated with vancomycin during gestation had significantly higher numbers of CD4+ T and B cells compared to controls 1. Together, our findings reveal a multi‐factorial link among maternal gut microbiota during pregnancy, breast milk microbiota, infant intestinal microbiota and postnatal immune development.
Bacterial metabolites
Maternal microbiota may impact infant immunity by the action of bacterial metabolites. Gut bacteria produce numerous metabolites that are critical mediators of various host physiological functions, immune modulation and energy production 82. The immune system senses microbial products (including metabolites) and pathogen‐associated molecular patterns, and the recognition of these molecules can influence host immunity 83, 84, 85. Short‐chain fatty acids (SCFAs) such as acetate, propionate and butyrate are end‐products of bacterial anaerobic fermentation and have been shown to impact intestinal immunity 86, Treg development 87, DC biology 88, 89 and epithelial integrity 90. During pregnancy, SCFAs produced by maternal gut bacteria may indirectly impact the developing fetal immunity. For example, bacterial metabolites potentially translocate from maternal gut during pregnancy to mammary glands, and could influence postnatal immune development during breastfeeding. Furthermore, a reversible maternal colonization model showed that microbial constituents such as aryl hydrocarbon ligands induce transcriptional changes in the fetal gut, enhancing cellularity of the innate immune system 12. Similarly, maternal retinoic acid (RA) induces fetal type 3 innate lymphoid cells and therefore secondary lymphoid organ development 91. Clostridia spp. in the gut can modulate RA concentration by suppressing the expression of retinol dehydrogenase 7 (Rdh7) in intestinal epithelial cells 92.
Epigenetics
In addition, metabolites or other molecules produced by the bacteria potentially impact the developing fetal immune unit through epigenetic modulation. Thorburn et al. 93 observed that exposing mice to acetate in drinking water during pregnancy led to suppression of allergic airway disease in offspring by enhancing Treg cell number and function. Acetate exerted its effects through increased acetylation at the forkhead box protein 3 (FoxP3) promoter, probably by histone deacetylases (HDAC)9 inhibition. In humans, maternal farm exposure has been shown to have an effect on epigenetic regulation of neonatal FoxP3 expression by impacting the Treg‐specific demethylated region (TSDR) conserved element 94. The amount of demethylated TSDR was higher with any single exposure compared with that seen after no exposure, although significantly only for maternal intake of farm milk during pregnancy 95. Michel et al. (2013) observed changes in DNA methylation patterns of asthma‐ and allergy‐related genes in children. Regions in ORMDL sphingolipid biosynthesis regulator 1 (ORMDL1) and signal transducer and activator of transcription (STAT)‐6 were hypomethylated in DNA from farmers’ compared to non‐farmers’ children, while regions in RAD50 double‐strand break repair protein (RAD50) and IL‐13 were hypermethylated 96. Farm exposure possibly mediates epigenetic effects through changes the gut microbiota which, in turn, alters bacterial metabolites 52, 96. Therefore, fetal exposure to maternal bacterial metabolites can impact the developing immune system via induction of epigenetic changes.
Maternal immunoglobulins
Maternal microbiota during pregnancy alters maternal immunity which ultimately impacts passive immunity to the offspring. We recently observed altered levels in breast milk IgG in mothers with vancomycin‐induced alterations in intestinal microbiota during pregnancy 1. While the transferred antibodies in milk are meant to provide immune protection in the neonate, they may also be transferring IgG‐bound bacteria that could impact on the developing immune system. Indeed, the concept of IgA‐ or IgG‐coated bacteria has previously been demonstrated in various body sites 97. Moreover, there is evidence of transplacental IgG‐mediated bacterial transfer. For example, when Gomez de Aguero et al. performed their aforementioned maternal transient colonization experiments 12, they tested the role of IgG in mediating the effects by transferring serum from the transiently colonized dams to unexposed pregnant mice and observed a similar effect on offspring immunity. However, when the serum was depleted of IgG prior to transfer, the impact on offspring immunity was lost. This suggest that fetal programming of immunity in utero is partially dependent on IgG‐mediated transfer of bacterial components. Apart from IgG‐mediated immunomodulation, transplacental immune regulation may also be mediated by cytokines and hormones 98, as well as bacterial components such as lipopolysaccharides 51.
Conclusions
Here, we highlight current literature on the role of maternal microbiota during pregnancy on the developing fetal and infant immunity, including the development of immune‐mediated diseases such as autoimmunity and atopy. We describe potential mechanisms through which maternal gut microbes during pregnancy impact infant immunity. It is clear that immune development in the fetus begins prior to delivery and is probably driven by translocation of microbiota or their metabolites from the maternal gut to the maternal–fetal unit or other mucosal surfaces. While it is appreciated that the largest infusion of microbes occurs at delivery when the neonate comes into contact with external microbiota, data are limited on the role of vaginal microbiota during pregnancy on fetal immunity. Maternal immunoglobulins at these sites augment transfer of these components to the fetus, contributing to microbiota or immune reprogramming. Altogether, the gestational microbiota induce an immune imprint in the fetus that has lasting postnatal immunological consequences.
OTHER ARTICLES PUBLISHED IN THIS REVIEW SERIES
The immunology of the fetal‐placental unit comes of age. Clinical and Experimental Immunology 2019, 198: 11–14.
Embryonic extracellular vesicles as informers to the immune cells at the maternal‐fetal interface. Clinical and Experimental Immunology 2019, 198: 15–23.
The role of neutrophil activation in determining the outcome of pregnancy and modulation by hormones and/or cytokines. Clinical and Experimental Immunology 2019, 198: 24–36.
Overview of procalcitonin in pregnancy and in pre‐eclampsia. Clinical and Experimental Immunology 2019, 198: 37–46.
References
- 1. Nyangahu DD, Lennard KS, Brown BP et al Disruption of maternal gut microbiota during gestation alters offspring microbiota and immunity. Microbiome 2018; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bender JM, Li F, Martelly S et al Maternal HIV infection influences the microbiome of HIV‐uninfected infants. Sci Transl Med 2016; 8:349ra100–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr 1999; 69:1035S–S1045. [DOI] [PubMed] [Google Scholar]
- 4. Funkhouser LJ, Bordenstein SR. Mom knows best: the universality of maternal microbial transmission. PLOS Biol 2013; 11:e1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med 2014; 6:237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stout MJ, Conlon B, Landeau M et al Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol 2013; 208:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leiby JS, Mccormick K, Sherrill‐mix S et al Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome 2018; 6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiménez E, Marín ML, Martín R et al Is meconium from healthy newborns actually sterile? Res Microbiol 2008; 159:187–93. [DOI] [PubMed] [Google Scholar]
- 9. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep 2016; 6:23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Theis KR, Romero R, Winters AD et al Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real‐time PCR, 16S rRNA gene sequencing, and metagenomics. Am J Obstet Gynecol 2019; 220:267.e1–267.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferretti P, Pasolli E, Tett A et al Mother‐to‐infant microbial transmission from different body sites shapes the developing Infant gut microbiome. Cell Host Microbe 2018;24:133–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gomez M, de Agüero SC, Ganal‐Vonarburg TF et al The maternal microbiota drives early postnatal innate immune development. Sci Transl Med 2016; 351:35313–9. [DOI] [PubMed] [Google Scholar]
- 13. Cox LM, Yamanishi S, Sohn J et al Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014; 158:705–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vuillermin PJ, Macia L, Nanan R, Tang MLK, Collier F, Brix S. The maternal microbiome during pregnancy and allergic disease in the offspring. Semin Immunopathol 2017; 39:669–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao D, Su H, Cheng J et al Prenatal antibiotic use and risk of childhood wheeze/asthma: a meta‐analysis. Pediatr Allergy Immunol 2015; 26:756–64. [DOI] [PubMed] [Google Scholar]
- 16. Ma J, Prince AL, Bader D et al High‐fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate mode. Nature Commun 2014; 5:3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 2007; 27:363–88. [DOI] [PubMed] [Google Scholar]
- 18. Gluckman PD, Hanson MA, Low FM. Evolutionary and developmental mismatches are consequences of adaptive developmental plasticity in humans and have implications for later disease risk. Phil Trans R Soc B Biol Sci 2019; 374:20180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gollwitzer ES, Marsland BJ. Impact of early‐life exposures on immune maturation and susceptibility to disease. Trends Immunol 2015; 36:684–96. [DOI] [PubMed] [Google Scholar]
- 20. Tormo‐Badia N, Håkansson Å, Vasudevan K, Molin G, Ahrné S, Cilio CM. Antibiotic treatment of pregnant non‐obese diabetic mice leads to altered gut microbiota and intestinal immunological changes in the offspring. Scand J Immunol 2014; 80:250–60. [DOI] [PubMed] [Google Scholar]
- 21. Gonzalez‐Perez G, Hicks AL, Tekieli TM, Radens CM, Williams BL, Lamouse‐Smith ESN. Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. J Immunol 2016; 196:3768–79. [DOI] [PubMed] [Google Scholar]
- 22. Gonzalez‐Perez G, Lamousé‐Smith ESN. Gastrointestinal microbiome dysbiosis in infant mice alters peripheral CD8+T cell receptor signaling. Front Immunol 2017; 8 10.3389/fimmu.2017.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martín R, Langa S, Reviriego C et al The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci Technol 2004; 15:121–7. [Google Scholar]
- 24. Fernandez L, Langa S, Martin V et al The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res 2013; 69:1–10. [DOI] [PubMed] [Google Scholar]
- 25. Rescigno M, Urbano M, Valzasina B et al Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2001; 2:361–7. [DOI] [PubMed] [Google Scholar]
- 26. Vazquez‐Terres A, Jones‐Carson J, Bäumler AJ et al Extraintestinal dissemination of Salmonella by CD18‐expressing phagocytes. Nature 1999; 401:804–8. [DOI] [PubMed] [Google Scholar]
- 27. de Andrés J, Jiménez E, Chico‐Calero I, Fresno M, Fernández L, Rodríguez J. Physiological translocation of lactic acid bacteria during pregnancy contributes to the composition of the milk microbiota in mice. Nutrients 2017; 10:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baldassarre ME, Bellantuono L, Mastromarino P, Miccheli A, Fanelli M, Laforgia N. Gut and breast milk microbiota and their role in the development of the immune function. Curr Pediat Rep 2014; 2:218–26. [Google Scholar]
- 29. Pannaraj PS, Li F, Cerini C et al Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 2017; 171:647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walker WA, Iyengar RS. Breast milk, microbiota, and intestinal immune homeostasis. Pediatr Res 2016; 77:220–8. [DOI] [PubMed] [Google Scholar]
- 31. DiGiulio DB, Callahan BJ, McMurdie PJ et al Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci USA 2015; 112:11060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Romero R, Hassan SS, Gajer P et al The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non‐pregnant women. Microbiome 2014; 136:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aagaard K, Riehle K, Ma J. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS ONE 2012; 7:e36466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet 2008; 371:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hanley J. Child health neonatal infections: group B streptococcus search date March 2007 prophylactic treatment of at‐risk neonates: GBS child health neonatal infections: group B streptococcus. Clin Evid 2008; 1:1–6. [Google Scholar]
- 36. Schlinzig T, Johansson S, Stephansson O et al Surge of immune cell formation at birth differs by mode of delivery and infant characteristics – a population‐based cohort study. PLoS ONE 2017; 12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huurre A, Kalliomaki M, Rautava S, Rinne M, Salminen S, Isolauri E. Mode of delivery‐effects on gut microbiota and humoral immunity. Neonatology 2008; 93:236–40. [DOI] [PubMed] [Google Scholar]
- 38. Sindram‐Trujillo AP, Scherjon SA, Miert PPVHV, Kanhai HHH, Roelen DL, Claas FHJ. Comparison of decidual leukocytes following spontaneous vaginal delivery and elective cesarean section in uncomplicated human term pregnancy. J Reprod Immunol 2004; 62:125–37. [DOI] [PubMed] [Google Scholar]
- 39. Stencel‐Gabriel K, Gabriel I, Wiczkowski A, Paul M, Olejek A. Prenatal priming of cord blood T lymphocytes by microbiota in the maternal vagina. Am J Reprod Immunol 2009; 61:246–52. [DOI] [PubMed] [Google Scholar]
- 40. Benn CS, Thorsen P, Jensen S. Maternal vaginal microflora during pregnancy and the risk of asthma hospitalization and use of antiasthma medication in early childhood. J Allergy Clin Immunol 2002; 110:72–7 . [DOI] [PubMed] [Google Scholar]
- 41. Fall T, Lundholm C, Örtqvist AK et al Early exposure to dogs and farm animals and the risk of childhood asthma. JAMA Pediatr 2015; 169:e153219. [DOI] [PubMed] [Google Scholar]
- 42. De Meer G, Janssen NAH, Brunekreef B. Early childhood environment related to microbial exposure and the occurrence of atopic disease at school age. Eur J Allergy Clin Immunol 2005; 60:619–25. [DOI] [PubMed] [Google Scholar]
- 43. Jhun I, Phipatanakul W. Early exposure to dogs and farm animals reduces risk of childhood asthma. Evid Based Med 2016; 21:2015–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lluis A, Ballenberger N, Illi S et al Regulation of TH17 markers early in life through maternal farm exposure. J Allergy Clin Immunol 2014; 133:864–71. [DOI] [PubMed] [Google Scholar]
- 45. Yu J, Liu X, Li Y et al Maternal exposure to farming environment protects offspring against allergic diseases by modulating the neonatal TLR–Tregs–Th axis. Clin Trans Allergy 2018; 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Niladri A, Zoratti EM, Strickler R et al Prenatal exposure to household pets influences fetal IgE production. Clin Exp Allergy 2008; 72:181–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bergmann RL, Edenharter G, Bergmann KE et al Predictability of early atopy by cord blood‐IgE and parental history. Clin Exp Allergy 1997; 27:752–60. Available at: http://www.ncbi.nlm.nih.gov/pubmed/9249267 (accessed 30 April 2019). [PubMed] [Google Scholar]
- 48. Wegienka G, Havstad S, Zoratti EM et al Regulatory T cells in prenatal blood samples: variability with pet exposure and sensitization. J Reprod Immunol 2009; 81:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Douwes J, Cheng S, Travier N et al Farm exposure in utero may protect against asthma, hay fever and eczema. Eur Respir J 2008; 32:603–11. [DOI] [PubMed] [Google Scholar]
- 50. Ege MJ, Bieli C, Frei R et al Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school‐age children. J Allergy Clin Immunol 2006; 117:817–23. [DOI] [PubMed] [Google Scholar]
- 51. Conrad ML, Ferstl R, Teich R et al Maternal TLR signaling is required for prenatal asthma protection by the nonpathogenic microbe Acinetobacter lwoffii F78. J Exp Med 2009; 206:2869–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tun HM, Konya T, Takaro TK et al Exposure to household furry pets influences the gut microbiota of infants at 3–4 months following various birth scenarios. Microbiome 2017; 5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jenmalm MC. The mother–offspring dyad: microbial transmission, immune interactions and allergy development. J Intern Med 2017; 38:42–9. [DOI] [PubMed] [Google Scholar]
- 54. Abrahamsson TR, Wu RY, Jenmalm MC. Gut microbiota and allergy: the importance of the pregnancy period. Pediatr Res 2015; 77:214–9. [DOI] [PubMed] [Google Scholar]
- 55. Ismail IH, Licciardi PV, Tang MLK. Probiotic effects in allergic disease. J Paediatr Child Health 2013; 49:709–15. [DOI] [PubMed] [Google Scholar]
- 56. Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host–microbiota mutualism. Nat Rev Microbiol 2011; 9:233–43. [DOI] [PubMed] [Google Scholar]
- 57. Timm S, Schlünssen V, Olsen J, Ramlau‐Hansen CH. Prenatal antibiotics and atopic dermatitis among 18‐month‐old children in the Danish National Birth Cohort. Clin Exp Allergy 2017; 47:929–36. [DOI] [PubMed] [Google Scholar]
- 58. Metsälä J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Prenatal and post‐natal exposure to antibiotics and risk of asthma in childhood. Clin Exp Allergy 2015; 45:137–45. [DOI] [PubMed] [Google Scholar]
- 59. Örtqvist AK, Lundholm C, Kieler H et al Antibiotics in fetal and early life and subsequent childhood asthma: nationwide population‐based study with sibling analysis. Obstet Gynecol Surv 2015; 70:229–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Loewen K, Monchka B, Mahmud SM, Jong G, Azad MB. Prenatal antibiotic exposure and childhood asthma: a population‐based study. Eur Respir J 2018; 52:1702070. [DOI] [PubMed] [Google Scholar]
- 61. Mulder B, Pouwels KB, Schuiling‐Veninga CCM et al Antibiotic use during pregnancy and asthma in preschool children: the influence of confounding. Clin Exp Allergy 2016; 46:1214–26. [DOI] [PubMed] [Google Scholar]
- 62. De Riva A, Wållberg M, Ronchi F et al Regulation of type 1 diabetes development and B‐cell activation in nonobese diabetic mice by early life exposure to a diabetogenic environment. PLoS ONE 2017; 12:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Patrick C, Wang GS, Lefebvre DE et al Promotion of autoimmune diabetes by cereal diet in the presence or absence of microbes associated with gut immune activation, regulatory imbalance, and altered cathelicidin antimicrobial peptide. Diabetes 2013; 62:2036–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Livanos AE, Greiner TU, Vangay P et al Antibiotic‐mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol 2016; 1:16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hu Y, Peng J, Tai N et al Maternal antibiotic treatment protects offspring from diabetes development in nonobese diabetic mice by generation of tolerogenic APCs. J Immunol 2015; 195:4176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hu Y, Jin P, Peng J, Zhang X, Wong FS, Wen L. Different immunological responses to early‐life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J Autoimmun 2016; 72:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Morgan XC, Tickle TL, Sokol H et al Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ott SJ, Musfeldt M, Wenderoth DF et al Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004; 53:685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Frank DN, Robertson CE, Hamm CM et al Disease phenotype and genotype are associated with shifts in inflamm bowel diseases. Inflamm Bowel Dis 2011; 17:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Örtqvist AK, Lundholm C, Halfvarson J, Ludvigsson JF, Almqvist C. Fetal and early life antibiotics exposure and very early onset inflammatory bowel disease: a population‐based study. Gut 2018; 68:218–25. [DOI] [PubMed] [Google Scholar]
- 71. Miyoshi J, Bobe AM, Miyoshi S et al Peripartum exposure to antibiotics promotes persistent gut dysbiosis, immune imbalance, and inflammatory bowel disease in genetically prone offspring. Cell Rep 2017; 20:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Walker RW, Clemente JC, Peter I, Loos RJ. The prenatal gut microbiome: are we colonized with bacteria in utero? Pediatr Obes 2017; 12(Suppl 1):3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jimenez E, Fernandez L, Marin ML et al Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol 2005; 51:270–4. [DOI] [PubMed] [Google Scholar]
- 74. Abrahamsson TR, Sinkiewicz G, Jakobsson T, Fredrikson M, Björkstén B. Probiotic lactobacilli in breast milk and infant stool in relation to oral intake during the first year of life. J Pediatr Gastroenterol Nutr 2009; 49:349–54. [DOI] [PubMed] [Google Scholar]
- 75. Arroyo R, Martín V, Maldonado A, Jiménez E, Fernández L, Rodríguez JM. Treatment of infectious mastitis during lactation: antibiotics versus oral administration of Lactobacilli isolated from breast milk. Clin Infect Dis 2010; 50:1551–8. [DOI] [PubMed] [Google Scholar]
- 76. Hu J, Nomura Y, Bashir A et al Diversified microbiota of meconium is affected by maternal diabetes status. PLoS ONE 2013; 8:e78257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Madan JC, Salari RC, Saxena D et al Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonat 2012; 97:456–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ardissone AN, De La Cruz DM, Davis‐Richardson AG et al Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS ONE 2014; 9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Spencer J, Dillon SB, Isaacson PG, Macdonaldt TT. T cell subclasses in fetal human ileum. Clin Exp Immunol 1986; 65:553–8. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1542495/pdf/clinexpimmunol00120-0083.pdf (accessed 30 April 2019). [PMC free article] [PubMed] [Google Scholar]
- 80. Li N, van Unen V, Abdelaal T et al Memory CD4 + T cells are generated in the human fetal intestine. Nat Immunol 2019; 20:301–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schreurs RRCE, Baumdick ME, Sagebiel AF et al Human fetal TNF‐α‐cytokine‐producing CD4+ effector memory T cells promote intestinal development and mediate inflammation early in life. Immunity 2019; 50:462–476.e8. [DOI] [PubMed] [Google Scholar]
- 82. Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab 2012; 16:559–64. [DOI] [PubMed] [Google Scholar]
- 83. Vinolo MAR, Rodrigues HG, Hatanaka E, Sato FT, Sampaio SC, Curi R. Suppressive effect of short‐chain fatty acids on production of proinflammatory mediators by neutrophils. J Nutr Biochem 2011; 22:849–55. [DOI] [PubMed] [Google Scholar]
- 84. Arpaia N, Campbell C, Fan X et al Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature 2013; 504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kato K, Uetake C, Kato T et al Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504:446–50. [DOI] [PubMed] [Google Scholar]
- 86. Macfarlane S, Macfarlane GT. Regulation of short‐chain fatty acid production. Proc Nutr Soc 2003; 62:67–72. [DOI] [PubMed] [Google Scholar]
- 87. Smith PM, Howitt MR, Panikov N et al The microbial metabolites, short chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Trompette A, Gollwitzer ES, Yadava K et al Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014; 20:159–66. [DOI] [PubMed] [Google Scholar]
- 89. Tan J, McKenzie C, Vuillermin PJ et al Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep 2016; 15:2809–24. [DOI] [PubMed] [Google Scholar]
- 90. Fukuda S, Toh H, Hase K et al Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011; 469:543–9. [DOI] [PubMed] [Google Scholar]
- 91. Van De Pavert SA, Ferreira M, Domingues RG et al Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature 2014; 508:123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Grizotte‐Lake M, Zhong G, Duncan K et al Commensals suppress intestinal epithelial cell retinoic acid synthesis to regulate interleukin‐22 activity and prevent microbial dysbiosis. Immunity 2018; 49:1103–15.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Thorburn AN, McKenzie CI, Shen S et al Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun 2015; 6:7320. [DOI] [PubMed] [Google Scholar]
- 94. Schaub B, Liu J, Höppler S et al Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol 2009; 123:774–82.e5. [DOI] [PubMed] [Google Scholar]
- 95. Michel S, Busato F, Genuneit J et al Farm exposure and time trends in early childhood may influence DNA methylation in genes related to asthma and allergy. Eur J Allergy Clin Immunol 2013; 68:355–64. [DOI] [PubMed] [Google Scholar]
- 96. Vuitton DA, Dalphin JC. From farming to engineering: the microbiota and allergic diseases. Engineering 2017; 3:98–109. [Google Scholar]
- 97. Simón‐Soro Á, D’Auria G, Collado MC, Džunková M, Culshaw S, Mira A. Revealing microbial recognition by specific antibodies. BMC Microbiol 2015; 15:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. PrabhuDas M, Bonney E, Caron K et al Immune mechanisms at the maternal‐fetal interface: perspectives and challenges. Nat Immunol 2015; 16:237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gonzalez-Perez G, Hicks AL, Tekieli TM, Radens CM, Williams BL, Lamousé-Smith ESN. Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. J Immunol 2016;196:3768–79. [DOI] [PubMed] [Google Scholar]
- 100. Myles IA, Fontecilla NM, Janelsins BM, Vithayathil PJ, Segre JA, Datta SK. Parental dietary fat intake alters offspring microbiome and immunity. J Immunol 2013; 191:3200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Atarashi K, Nishimura J, Shima T et al ATP drives lamina propria TH17 cell differentiation. Nature 2008; 455:808–12. [DOI] [PubMed] [Google Scholar]


