Abstract
Ko143, a potent ABCG2 inhibitor that reverses multidrug resistance, cannot be used clinically due to its unsuitable metabolic stability. We identified benzoyl indoles as reversal agents that reversed ABCG2-mediated multidrug resistance (MDR), with synthetic tractability and enhanced metabolic stability compared to Ko143. Bisbenzoyl indole 2 and monobenzoyl indole 8 significantly increased the accumulation of mitoxantrone (MX) in ABCG2-overexpressing NCI-H460/MX20 cells, and sensitized NCI-H460/MX20 cells to mitoxantrone. Mechanistic studies were conducted by [3H]-MX accumulation assay, Western blot analysis, immunofluorescence analysis and ABCG2 ATPase assay. The results revealed that the reversal efficacies of compounds 2 and 8 were not due to an alteration in the expression level or localization of ABCG2 in ABCG2-overexpressing cell lines. Instead, compounds 2 and 8 significantly stimulated the ATP hydrolysis of ABCG2 transporter, suggesting that these compounds could be competitive substrates of ABCG2 transporter. Overall, the results of our study indicated that compounds 2 and 8 significantly reversed ABCG2-mediated MDR by blocking the efflux of anticancer drugs.
Keywords: ABCG2 transporter, Multidrug resistance, Benzoyl indoles, Metabolic stability, Reversal agents
Graphical Abstract

Benzoyl indoles as ABCG2 reversal compounds have greater synthetic tractability and metabolic stability than the potent ABCG2 inhibitor, Ko143.
Introduction
Multidrug resistance (MDR) is characterized by the resistance of cancer cells to drugs that are structurally and mechanistically distinct.1–3 There are several different factors that contribute to MDR, including enhanced drug efflux, decreased cellular uptake, drug detoxification, altered drug target, enhanced DNA repair, apoptosis inhibition, and altered cell cycle check points.4–6 ATP-binding cassette transporters (ABC transporters) are a superfamily of proteins that transport various substrates by utilizing the energy of ATP hydrolysis.7 ABCG2, as a G-subfamily member of ABC transporter, is a ‘half-transporter’, with one nucleotide binding domains (NBD) and one transmembrane domain (TMD) fused to a single polypeptide chain.8 The functional form of the ABCG2 transporter is a homodimer.9 The ABCG2 transporter is localized in the blood-brain barrier (BBB), blood-testis barrier (BTB), blood-placental barriers (BPB), blood-retinal barriers (BRB), and the luminal surface of liver canaliculi and renal proximal convoluted tubules.10 The ABCG2 transporter, due to its localization on the surfaces of excretory organs can alter the absorption, distribution, metabolism, and elimination (ADME) of certain drugs.11, 12 The substrates of the ABCG2 transporter are primarily hydrophobic and are either negatively or positively charged molecules, e.g., mitoxantrone (MX), methotrexate, topotecan, flavopiridol and SN-38.13 It has been shown that the overexpression of ABCG2 transporter in certain cancers significantly decreases or abrogates the efficacy of various chemotherapeutic drugs.14, 15
Ko143 (Figure 1) was found to be the most potent inhibitor of the ABCG2 transporter.16 A major problem associated with Ko143 was the low total yield (5%) as its synthesis required a multi-step synthesis method.17, 18 Furthermore, Ko143 was rapidly metabolized to a compound that lacked efficacy as an inhibitor of the ABCG2 transporter.19 Thus, in order to develop efficacious ABCG2 inhibitors, it is essential to find compounds that can be easily synthesized that have a suitable pharmacokinetic profile.
Figure 1.
Simplifying Ko143 to benzoyl indole
Jackson et al. determined the cryo-EM structure of ABCG2 protein (PDB: 6ETI) bound to a Ko143 analog, MZ29, and two Fabs.9 The result indicated that the −NH group of the indole ring in the Ko143 analog formed a hydrogen bond with Asn436 and the methoxy group formed a hydrogen bond with Thr435. Thus, the indole and methoxy group may be important in the interaction of the Ko143 analogs and ABCG2. In this study, we simplified the structure of Ko143 to a methoxy benzoyl indole (Figure 1), which has a molecular size similar to Ko143 but with higher conformational flexibility. In addition, a benzoyl naphthalene compound (Figure 1) was synthesized to elucidate the importance of the indole group in reversing ABCG2-mediated drug resistance.
Since Ko143 is rapidly metabolized to an inactive compound in the plasma, the metabolic stability of the indole derivatives would be a major concern. Previously, it has been shown that the benzoyl indole, BPR0L075 (Figure 2), has a half-life of 2.8 h in plasma following the oral administration of 20 mg/kg in rats.20 Therefore, we hypothesized that benzoyl indoles could be developed as a new series of ABCG2 reversal compounds with an improved pharmacokinetic profile.
Figure 2.
The structure of BPR0L075
Results and discussion
Chemistry.
The general method used to synthesize the indole derivatives in this study is shown in Scheme 1. The synthesis involves a straightforward reaction sequence. Initially, the acylation reaction was performed with the use of methoxy indoles and methoxy benzoyl chloride in the presence of POCl3 and AlCl3. Secondly, the hydroxy-substituted compounds were afforded by the deprotection of the methoxy groups. The synthesis of 3-acylindoles has recently gained considerable attention as 3-substituted indoles are not only significant therapeutic compounds but are also used as synthetic intermediates for alkaloid synthesis.21, 22 Several methods for the synthesis of 3-acylindoles have been reported, including Friedel–Crafts or Vilsmeier–Haack acylations, the acylation of indole Grignard reagents or 3-indolylzinc chloride, and the use of iminium ions, dialkoxycarbenium, or N-(2-haloacyl)-pyridinium salts.23–31
Scheme 1.
Synthesis of indole derivatives
The synthetic route of the indole derivatives is shown in Scheme 1. The indoles reacted with acyl chloride in the presence of AlCl3 and POCl3 to obtain compounds 1–12 (Scheme 1). Interestingly, when we synthesized benzoyl indole 1, we obtained 4-OMe bisbenzoyl indole 2 in the same reaction. Similar results occurred in the synthesis of 3, where 2,4-OMe bisbenzoyl indole 4 was obtained. The corresponding hydroxy compounds 13–23 were synthesized by demethylation of compounds 1–12 with boron tribromide. In addition, the benzoyl naphthalene compound 24 (Table 1) was synthesized as a control to elucidate the importance of the indole group in reversing ABCG2-mediated multidrug resistance. Compound 24 was synthesized by the acylation of methoxy naphthalene in the presence of AlCl3 and POCl3. The structures of all the synthesized compounds were confirmed by HRMS (high resolution mass spectrometry), IR (infrared radiation), 1H, 13C NMR and HMBC (heteronuclear multiple bond correlation) data.
Table 1.
The structure and the selected concentration of monobenzoyl indoles and benzoyl naphthalene
 | |||
|---|---|---|---|
| Compd. | R1 | R2 | *Selected concentration (μM) |
| 1 | 4-OMe | 4-OMe | 10 |
| 3 | 4-OMe | 2,4-OMe | 25 |
| 5 | 4-OMe | 3,4,5-OMe | 1 |
| 6 | 5-OMe | 2-OMe | 1 |
| 7 | 5-OMe | 3-OMe | 10 |
| 8 | 5-OMe | 4-OMe | 10 |
| 9 | 5-OMe | 2,4-OMe | 25 |
| 10 | 5-OMe | 3,4,5-OMe | 1 |
| 11 | 5-OMe | 2, 6-OMe | 1 |
| 12 | 7-OMe | 2, 4-OMe | 10 |
| 13 | 4-OH | 4-OH | 25 |
| 15 | 4-OH | 2,4-OH | 25 |
| 16 | 4-OH | 3,4,5-OH | 25 |
| 17 | 5-OH | 2-OH | 25 |
| 18 | 5-OH | 3-OH | 25 |
| 19 | 5-OH | 4-OH | 25 |
| 20 | 5-OH | 2,4-OH | 25 |
| 21 | 5-OH | 3,4,5-OH | 25 |
| 22 | 5-OH | 2,6-OH | 10 |
| 23 | 7-OH | 2,4-OH | 10 |
| 24 | / | / | 10 |
The concentrations that did not decrease the viability of NCI-H460 and NCI-H460/MX20 cells by more than 20%.
The determination of the ABCG2 reversal activity of the indole derivatives at selected (non-toxic) concentrations
The structure of indole derivatives and the selective non-toxic concentration is shown in Table 1 and Table 2. The selective concentration is the concentration that did not decrease the viability of the parental NCI-H460 and drug-resistant NCI-H460/MX20 cell lines by more than 20%. NCI-H460/MX20 cell line overexpressing ABCG2 was selected by MX.32 The reversal efficacies of the indole derivatives were determined (Figure 3) at the selected concentrations by determining their effect on the intracellular levels of mitoxantrone (2.5 μM), a substrate of the ABCG2 transporter.5 As shown in Figure 3, 2.5 μM of MX alone produced a 24±3% inhibition of the proliferation of NCI-H460/MX20 cells and the efficacy of MX increased when combined with the different indole derivatives. For example, MX (2.5 μM) inhibited the proliferation of NCI-H460/MX20 cells by more than 87% when combined with the monobenzoyl indoles 7, 8, 13, 15 and 17, or the bisbenzoyl indoles 2 and 4 at the selected concentrations. However, the benzoyl naphthalene 24 (25 μM) did not significantly increase the efficacy of 2.5 μM of MX. The reversal efficacy of 2,4-OMe benzoyl naphthalene 24 was lower than that of 2,4-OMe benzoyl indoles 3, 9 and 12. Most of the methoxy indoles have greater reversal efficacies than the corresponding hydroxy indoles except methoxy indoles 1, 3 and 6. Interestingly, 5-OMe indole compounds 8 (4-OMe phenyl), 9 (2,4-OMe phenyl), and 10 (3,4,5-OMe phenyl) showed greater reversal efficacies than those corresponding 4-OMe indole compounds 1, 3 and 5, respectively. However, the 4-OH indole compounds 13 and 15 showed better reversal efficacies than those 5-OH indole compounds 19 and 20, respectively. Among the 5-OMe indole compounds 6~11, compounds 7 and 8 with 3-OMe and 4-OMe benzoyl group, respectively, had better reversal efficacies than the other indoles. The above results indicated that the reversal efficacies of the indole compounds are influenced by the position and hydrophilicity of the substituents at the indole ring and the benzoyl group. However, the benzoyl naphthalene 24 (25 μM) did not significantly increase the efficacy of 2.5 μM of MX. The reversal efficacy of 2,4-OMe benzoyl naphthalene 24 was lower than that of 2,4-OMe benzoyl indoles 3, 9 and 12, suggesting that the indole group was critical for the reversal of ABCG2-mediated multidrug resistance.
Table 2.
The structure and the selected concentration of bisbenzoyl indoles
 | ||||
|---|---|---|---|---|
| Compd. | R1 | R2 | R3 | *Selected concentration (μM) |
| 2 | 4-OMe | 4-OMe | OMe | 25 |
| 4 | 2, 4-OMe | 2, 4-OMe | OMe | 25 |
| 14 | 4-OH | 4-OH | OH | 25 |
The concentrations that did not decrease the viability of NCI-H460 and NCI-H460/MX20 cells by more than 20%.
Figure 3.
The reversal efficacy of the indole derivatives in combination with 2.5 μM of MX in NCI-H460/MX20 cells. The inhibition of proliferation indicates the inhibition of the proliferation of NCI-H460/MX20 cells after incubation with MX in the absence or presence of the indole derivatives, compared to the proliferation of cells in the control group. The results were obtained from three independent experiments performed in triplicate. *, P < 0.05, **, P < 0.01, ****, P < 0.0001 versus no indole derivatives group, two-way ANOVA.
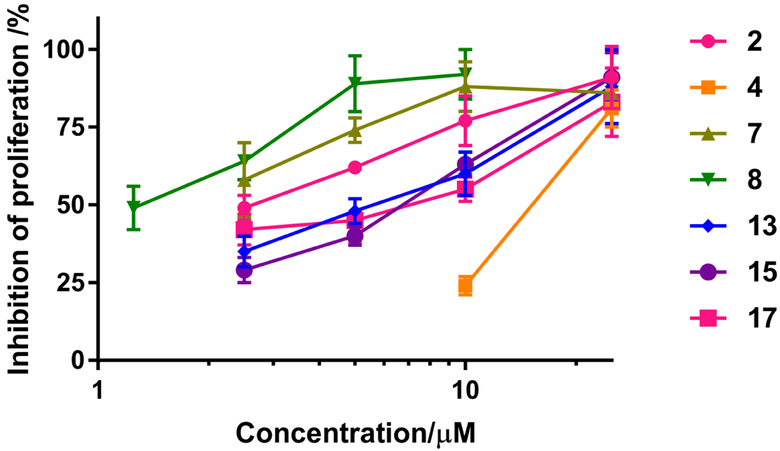
The determination of the ABCG2 reversal efficacy of the indole derivatives 2, 4, 7, 8, 13, 15 and 17
The reversal efficacies of bisbenzoyl indoles 2 and 4, and monobenzol indoles 7, 8, 13, 15 and 17 at different concentrations on the proliferation of NCI-H460/MX20 cells, in combination with 2.5 μM of MX, were determined to ascertain which compound had the greatest efficacy (Figure 4). The results suggested that reversal efficacy increased as the concentration of the indole derivatives increased, which indicated that the reversal efficacies of these indole compounds are dose-dependent. Monobenzoyl indole 8 had the greatest anti-proliferative efficacy among the seven compounds at the same concentration, while bisbenzoyl 2 has greater efficacy than bisbenzoyl 4 (Figure 4). Thus, compounds 2 and 8 exhibited the highest reversal efficacy among the bisbenzoyl and monobenzoyl indoles, respectively. Subsequently, we further investigated the reversal efficacies and inhibition mechanism of compounds 2 and 8 as reversal compounds for ABCG2-mediated drug-resistance.
Figure 4.
The reversal efficacy of the indole derivatives in combination with 2.5 μM of MX. The proliferation of NCI-H460/MX20 cells was inhibited by 25 ± 4 % with 2.5 μM of MX alone. The inhibition of proliferation indicates the inhibition of the proliferation of NCI-H460/MX20 cells after incubation with MX in the absence or presence of the indole derivatives, compared to the proliferation of cells in the control group. The results were obtained from three independent experiments performed in triplicate.
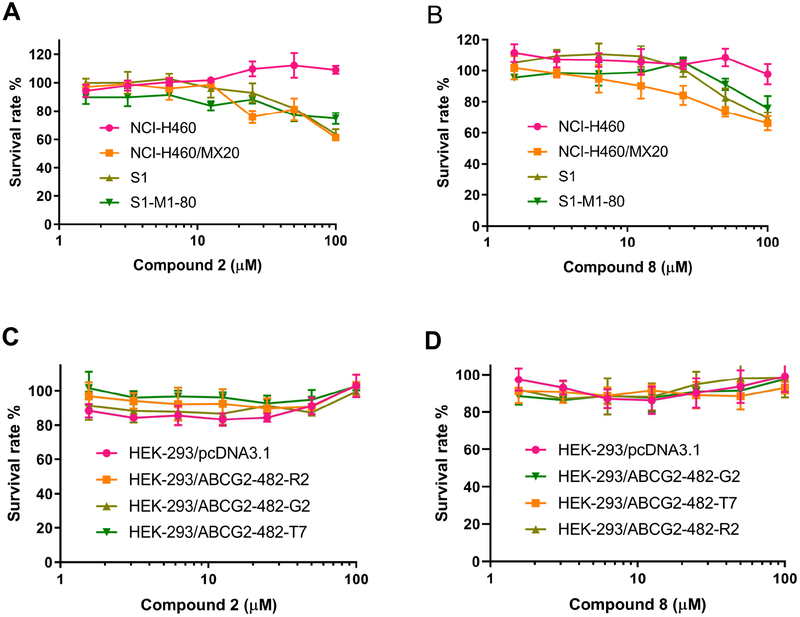
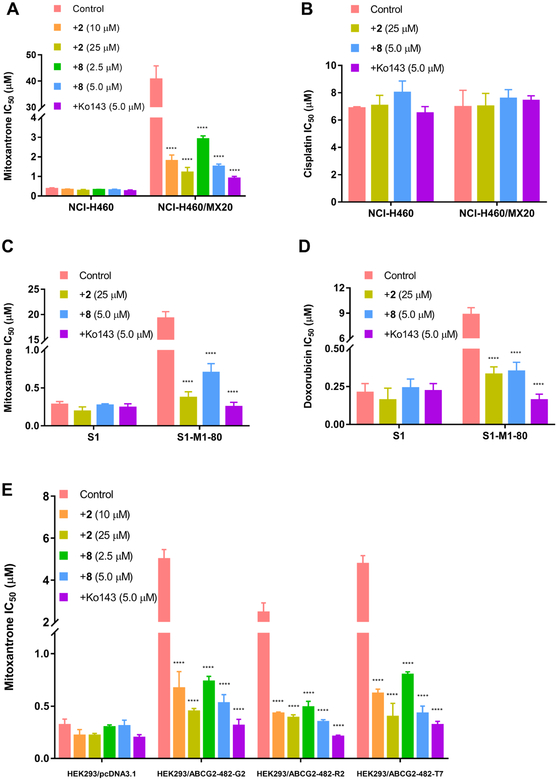
The cytotoxic curves of compounds 2 and 8 on H460, H460/MX20, S1, S1-M1-80, HEK-293/pcDNA3.1, wild-type HEK-293/ABCG2–482-R2, mutant HEK-293/ABCG2–482-G2 and HEK-293/ABCG2–482-T7 cell lines are shown in Figure 5, indicating that compounds 2 and 8 at the concentration of 25 μM and 5 μM exhibited lower than 20% inhibition on all the cell lines we investigated. S1-M1-80 is an ABCG2-mediated multi-drug resistant cell line selected by MX.33 The previous studies revealed that mutations at amino-acid 482 in the ABCG2 gene influence the substrate specificity of the ABCG2 transporter.34 It was also reported that the efficacy of Novobiocin, a ABCG2 inhibitor, would be altered due to the mutations at amino-acid 482. Novobiocin exhibited potent inhibition on wild-type ABCG2, however, almost no inhibition on mutant ABCG2.35 Thus, we used the transfected wild-type HEK293/ABCG2–482-R2, mutant HEK293/ABCG2–482-G2 and HEK293/ABCG2–482-T7 cells to investigate whether the reversal effects of the benzoyl indoles would be different in these transfected HEK293 cell lines. As shown in Figure 6A, compounds 2 and 8 significantly increased the cytotoxicity of MX in ABCG2-overexpressing NCI-H460/MX20 cell line. Compounds 2 and 8 did not significantly sensitize NCI-H460/MX20 cells to cisplatin, which is not a substrate of the ABCG2 transporter (Figure 6B). These results suggested that the sensitizing effects of compounds 2 and 8 is limited to ABCG2 substrates for ABCG2-mediated multidrug resistance cancer cell lines. Furthermore, the reversal efficacy of compounds 2 and 8 were also determined in parental S1 and S1-M1-80 cells, where ABCG2 overexpression was also induced using mitoxantrone.36 Human embryonic kidney (HEK293) cells were transfected with the pcDNA3.1 vector containing a full-length ABCG2, with either coding arginine (R), glycine (G), or threonine (T) at amino acid position 482, to generate wild-type HEK293/ABCG2–482-R2, mutant (HEK293/ABCG2–482-G2 and HEK293/ABCG2–482-T7), respectively. Compounds 2 and 8 significantly increased the cytotoxicity of 1) MX and doxorubicin in S1-M1-80 cells (Figure 6C, 6D) and 2) MX in HEK293/ABCG2–482-R2, HEK293/ABCG2–482-G2 and HEK293/ABCG2–482-T7 cells expressing theABCG2 transporter (Figure 6E).37 Thus, it can be concluded that compounds 2 and 8 might sensitize ABCG2-mediated multidrug resistance cell lines to anticancer drugs by interacting with ABCG2. Even though the potencies of compounds 2 and 8 are not as strong as Ko143, the two compounds are also effective to reverse ABCG2-mediated multi-drug resistance. The representative cytotoxic curves of mitoxantrone, cisplatin and doxorubicin in combination with compounds 2, 8 and Ko143 on different cell lines were also shown in Figure 7 to indicate the effects of compounds 2, 8 and Ko143 on the cytotoxicity of the anti-cancer drugs.
Figure 5.
Cytotoxic curves of compounds 2 and 8 on H460, H460/MX20, S1, S1-M1-80, HEK-293/pcDNA3.1, HEK-293/ABCG2–482-R2, HEK-293/ABCG2–482-G2 and HEK-293/ABCG2–482-T7 cell lines.
Figure 6.
The effect of compounds 2 and 8 on the IC50 values of (A) mitoxantrone and (B) cisplatin in parental NCI-H460 and the drug-resistant cell line, NCI-H460/MX20. The effect of compounds 2 and 8 on the IC50 values of (C) mitoxantrone and (D) doxorubicin in parental S1 and drug-resistant cell line, S1-M1-80. (E) The effect of compounds 2 and 8 on the IC50 values of mitoxantrone in parental HEK293/pcDNA3 and transfected ABCG2-overexpressing HEK293/ABCG2–482-R2, HEK293/ABCG2–482-G2 and HEK293/ABCG2–482-T7 cells. The error bars represent the SD value. Ko143 (5 μM) was used as a positive control. ****, P < 0.0001 versus no indole derivatives group, two-way ANOVA.
Figure 7.
Cytotoxic curves of mitoxantrone, cisplatin and doxorubicin in combination with compounds 2, 8 and Ko143 on H460, H460/MX20, S1, S1-M1-80, HEK-293/pcDNA3.1, HEK-293/ABCG2–482-R2, HEK-293/ABCG2–482-G2 and HEK-293/ABCG2–482-T7 cell lines.
The effect of the indole derivatives on the intracellular accumulation of MX in ABCG2-overexpressing cells
The reversal efficacies of compounds 2 and 8 were tested at low concentrations which were not toxic to both parental NCI-H460 and drug-resistant NCI-H460/MX20 cells. Thus, the significant increase in the cytotoxicity of MX in combination of compounds 2 or 8 cannot be considered as additivity of the cytotoxicity. The effect of the compounds on intracellular [3H]-MX accumulation was determined by comparing the accumulation of [3H]-MX in parental and ABCG2-mediated MDR cells with or without the reversal compounds. The intracellular level of [3H]-MX in NCI-H460/MX20 cells in the absence of the reversal compounds was approximately 3.1-fold lower than NCI-H460 cells (which do not overexpress ABCG2) (Figure 8). The incubation of NCI-H460/MX20 cells with compounds 2 (25 μM) and 8 (5.0 μM) significantly increased the accumulation of [3H]-MX by 3.0-fold and 2.7-fold, respectively (Figure 8). Ko143, at 5.0 μM, produced a 3-fold increase in the accumulation of [3H]-MX and there was no statistical difference in the accumulation of [3H]-MX between Ko143 (5.0 μM) and compounds 2 (10 or 25 μM) or compound 8 (5.0 μM). The results revealed that compounds 2 and 8 effectively blocked the efflux of MX by NCI-H460/MX20 cells.
Figure 8.
The effects of 2 and 8 on the intracellular accumulation of [3H]-MX in NCI-H460 and NCI-H460/MX20 cells. The error bars represent the SD value. Ko143 (5.0 μM) was used as positive control for ABCG2-overexpressing cells. ****, P < 0.0001 versus no indole derivatives group, two-way ANOVA.
The expression level and the intracellular localization of ABCG2 in cells overexpressing ABCG2
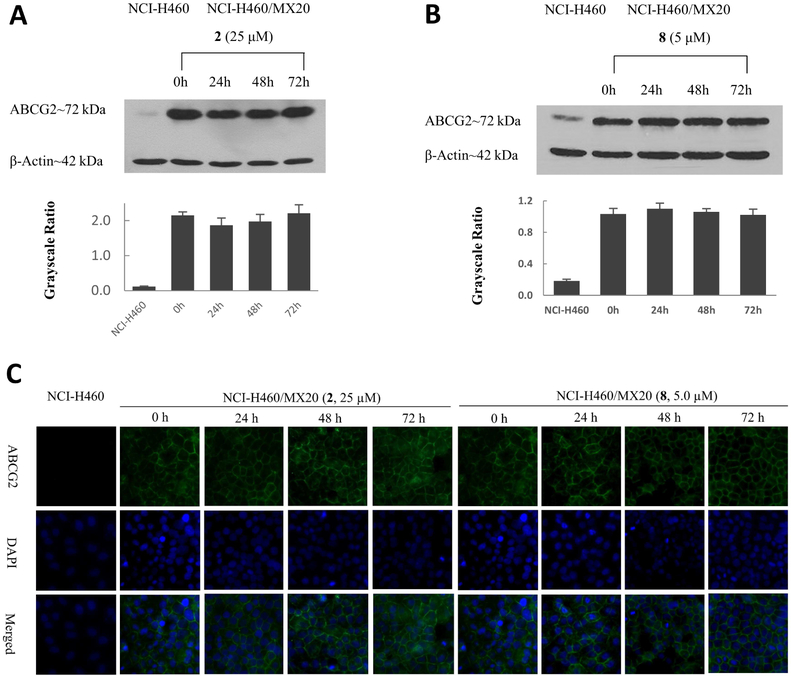
The factors inhibiting the efflux function of ABCG2 transporter could include the down-regulation of expression or the localization alteration of ABCG2 in cells. To determine whether compounds 2 and 8 can cause the down-regulation of ABCG2 protein, the ABCG2 expression levels were determined for cells incubated with 2 (25 μM) or 8 (5.0 μM) for 0, 24, 48, and 72 h. The results showed no significant alteration in the expression level of ABCG2 in the drug-resistant NCI-H460/MX20 cells following incubation with compounds 2 or 8 (Figure 9A, Figure 9B).
Figure 9.
(A) The effect of compound 2, at 25 μM, on the expression levels of ABCG2 protein in NCI-H460/MX20 cells after 24, 48, and 72 h of incubation. Equal amounts of total cell lysate were used for each sample. (B) The effect of compound 8, at 5 μM, on the expression level of ABCG2 in NCI-H460/MX20 cells after 24, 48, and 72 h of incubation. (C)The effect of 2 (25 μM) and 8 (5 μM) on the subcellular localization of ABCG2 in ABCG2-overexpressing NCI-H460/MX20 cells.
Alterations in the localization of the ABCG2 transporter in the membrane has been shown to reverse ABCG2-mediated MDR.38 Therefore, we used an immunofluorescence assay to determine if the localization of ABCG2 transporter in NCI-H460/MX20 cells was altered following incubation with the indole derivatives. As shown in Figure 9C, compounds 2 and 8 did not induce the internalization of cell surface ABCG2 transporter. These results suggest that the reversal mechanism of action for compounds 2 and 8 was not induced by the alteration the expression or localization of ABCG2 transporter.
The effect of the indole compounds on ABCG2 ATPase activity
Since there is no alteration of expression or localization of ABCG2, the inhibition of the efflux function of ABCG2 transporter could be due to the inhibition of the activity of ABCG2 transporter. To determine whether compounds 2 and 8 inhibit or stimulate the activity of ABCG2 ATPase, we performed an ATPase assay using membranes of High Five insect cells overexpressing ABCG2, in the presence of different concentrations of compounds 2 or 8. Both compounds 2 or 8 significantly stimulated the ATPase activity of ABCG2 in a concentration-dependent manner, with a maximal stimulation of 3.01-fold and 2.03-fold greater, than the basal activity (Figure 10), respectively. The concentration of 2 or 8 required to obtain 50% of maximal stimulation was 0.21 and 0.75 μM, respectively. These results suggested that 2 and 8 were the substrates of ABCG2 transporter. Therefore, compounds 2 and 8 could acts as a potential competitive substrate, thus blocking the efflux function of ABCG2 and increasing the accumulation of anticancer drugs.
Figure 10.
The effect of compounds 2 and 8 (0–40 μM) on orthovanadate (Vi)-sensitive ABCG2 ATPase activity. The concentrations of compounds 2 and 8 were plotted at linear in panel (A) and (B), respectively.
Besides the mechanisms we discussed above, inhibition of the phosphorylation of ABCG2 could be another mechanism for reversing ABCG2-mediated multi-drug resistance. As depicted in the previous study, BCRP/ABCG2 can be phosphorylated by Pim-1L and thus the drug resistance would increase.39 We suppose that inhibiting the phosphorylation of ABCG2 could decrease the efflux function of ABCG2, thus increase the accumulation of the anti-cancer drugs in drug-resistant cancer cells. Additional experiments are needed to determine whether the inhibition of phosphorylation of ABCG2 was induced by the benzoyl indoles in our future study.
Docking analysis of indole derivatives with ABCG2
The best-scored docked position of the indole derivatives within the large drug-binding cavity of human ABCG2 is shown in Figure 11. The indole group of 2 forms a hydrogen bond with Thr435 in the B chain of the ABCG2 protein (-NH•••O=C- Thr435). In addition, the phenyl ring of the indole group forms a π-π interaction with Phe439 in the B chain of ABCG2, while the other phenyl ring of 2 forms a π-π interaction with Phe439 in the A chain of ABCG2. Compound 2 was stabilized into a hydrophilic pocket formed by residues Val401, Leu405, Phe432, Val546, Phe547, Met549 of ABCG2. The amine group in compound 8 forms a hydrogen bonding with Asn436 (-NH•••O=C- Asn436). Moreover, the indole group of 8 was stabilized into a hydrophilic pocket formed by nearby residues Phe431, Phe432, Val546, Met549 and Leu555 of the ABCG2 protein. Ko143 formed a hydrogen bond with Asn436 in the B chain of the ABCG2 transporter. Based on the results of the induced-fit docking, compounds 2, 8 and Ko143 had a strong interaction with the human ABCG2 transporter protein, with docking scores of −11.591 and −10.052 and 10.236 kcal/mol, respectively. Interestingly, compound 2 had a higher docking score than 8, but 8 had greater reversal efficacy than 2 in the cytotoxicity and intramolecular accumulation assays. Compound 2 may have a lower reversal efficacy due to its large size, which could impede its entry into the binding site of the human ABCG2 protein.40 As depicted in Figure 11G, even though compounds 2, 8 and Ko143 bind with the same domain of ABCG2 protein, the binding modes of the three compounds are not superimposable, which could contribute to the difference in the reversal efficacies of compounds 2, 8 and Ko143.
Figure 11.
Predicted binding positions of compounds 2 and 8 with the human ABCG2 transporter protein. (A) Docked position of compound 2 within the binding site of the human ABCG2 transporter protein. Compound 2 is shown as a ball and stick model, with the atoms colored as follows: carbon = cyan, hydrogen = white, nitrogen = blue and oxygen = red. The important residues are depicted as sticks with the same color scheme as above except that carbons are indicated by the grey color. Ring centroids were represented as dark-green dots. Dotted yellow lines indicate hydrogen bonds. (B) A two-dimensional ligand−receptor interaction diagram shows the important interactions of compound 2 with the binding site residues of human ABCG2. The amino acids are shown as colored bubbles, cyan indicates polar residues and green indicates hydrophobic residues. Hydrogen bonds are indicated by the purple dotted arrow, and π-π stacking aromatic interactions are indicated by the green lines. (C) Docked position of compound 8 within the binding site of the human ABCG2 transporter protein. (D) A two-dimensional ligand−receptor interaction diagram shows the important interactions of compound 8 with the binding site residues of the human ABCG2 transporter protein. (E) Docked position of Ko143 within the binding site of the human ABCG2 protein. (F) A two-dimensional diagram shows the important contacts of Ko143 with the binding site residues of the human ABCG2 transporter protein. (G) The comparison of the binding mode of 2, 8 and Ko143 with ABCG2 protein. Compounds are shown as a ball and stick model, with the atoms colored as follows: hydrogen = white, nitrogen = blue, oxygen = red. The carbon atoms are labeled in different colors: 2 (grey), 8 (cyan), Ko143 (orange).
Metabolic Stability of the indole derivatives
Approximately 60 % of the drugs are metabolized by CYP 450 in the human liver.41 Human liver microsomes, derived from human liver, contain a wide variety of membrane bound drug metabolizing enzymes. Thus, in vitro metabolic stability testing with human liver microsomes is useful to predict the in vivo hepatic CYP-mediated metabolism of the compounds in the human body. The in vitro metabolic stability testing with human liver microsome has been commonly applied for investigating the metabolism of the drugs prior to in vivo study.42–44 The metabolic stabilities of compounds 2 and 8 were investigated with human liver microsome, and the results indicated that compounds 2 and 8 are more metabolically stable than Ko143 (Table 3). The amount of compounds 2 and 8
Table 3.
In vitro metabolic stability of compounds 2, 8 and Ko143
| compound | Metabolic stabilitya/% |
|---|---|
| 2 | 56±4.6 |
| 8 | 78±1.2 |
| Ko143 | 23±1.4 |
Metabolic stability was calculated as the % compound remaining after 1 h of incubation with human liver microsomes.
remaining after incubation with human liver microsome for 1 h was 56 and 78%, respectively, compared to 23% for Ko143. Since the low metabolic stability of Ko143 is the major problem for clinical application, we would like to discover metabolically stable reversal agents for ABCG2-mediated multidrug resistance. Inspiringly, the results revealed that compounds 2 and 8 are more metabolically stable than Ko143. Thus, indole compounds 2 and 8 as reversal agents could be valuable and suitable for clinical application in the future.
Conclusions
In this study, we synthesized 23 benzoyl indoles and determined their efficacy and mechanism of action for reversing ABCG2-mediated MDR. Based on the structure-activity relationship data, The reversal efficacies of 2,4-OMe benzoyl indoles 3, 9 and 12 were greater than that of 2,4-OMe benzoyl naphthalene 24, revealing the indole group was more important in enhancing the reversal efficacy compared to naphthalene group. Docking studies showed that the indole moiety of compounds 2 and 8 formed hydrogen bonds with the ABCG2 transporter protein, indicating the importance of the indole group. Furthermore, in most cases, compounds that had methoxy substituents had greater reversal efficacy than those with the hydroxy groups. The increase in reversal efficacy may be due to the stronger interaction between the methoxy group and the hydrophobic pocket residues in the human ABCG2 transporter protein. The methoxy benzoyl indole compounds 2 and 8, had the greatest reversal efficacy among bisbenzoyl and monobenzoyl indole compounds, respectively. Inspiringly, compounds 2 and 8 have better metabolic stability than the potent ABCG2 inhibitor, Ko143, which revealing the value of clinical application of these benzoyl indoles. The reversal efficacy of compound 8 was significantly greater than that of compound 2. The lower reversal efficacy of compound 2 may result from its larger molecule size, thus hindering compound 2 from entering the drug-binding site of the ABCG2 transporter protein. Compounds 2 and 8 could act as competitive substrates of ABCG2 transporter to block the efflux of anticancer drugs, thus sensitizing ABCG2-mediated multidrug resistant cancer cells to the anticancer drugs. Overall, the benzoyl indoles 2 and 8 had significant efficacy as ABCG2-mediated drug resistance reversal agents, with synthetic tractability and enhanced metabolic stability. These results give insight into the rational design of reversal agents targeting ABCG2 transporter.
Experimental Section
General Chemistry.
Chemicals were obtained from Materials and Instrumentation. Chemicals were purchased from Aldrich Chemical Co. (Milwaukee, WI), TCI America (Portland, OR) and Alfa Aesar (Ward Hill, MA), and used without further purification unless otherwise indicated. Purifications by column chromatography were carried out over silica gel (300−400 mesh) and monitored by thin layer chromatography (TLC) performed on GF/UV 254 plates and were visualized using UV light at 254 and 365 nm. Melting points were determined using a SGW X-4 digital melting point apparatus, and the temperatures were not corrected. IR spectra were determined on an EQUINOX 55 Fourier transformation infrared spectrometer. 1H and 13C NMR spectra were recorded on Varian INOVA 500, Bruker AVANCE AV400, or Mercury-Plus 300 NMR spectrometer in CDCl3, DMSO-d6, or Acetone-d6 and TMS as an internal standard. Chemical shifts were expressed as values (ppm) relative to tetramethylsilane as an internal standard, and coupling constants (J values) were given in hertz (Hz). Abbreviations are used as follows: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, dd = doublet of doublet, doublet of doublet of doublets (ddd), br = broad. High resolution (HR) mass spectra were measured on Thermo MAT95XP mass spectrometer.
General procedure for the synthesis of compounds 1–12.
A mixture of methoxy indole (2 mmol) and methoxy benzoylchloride (2 mmol) was stirred in the presence of POCl3 (5 ml) and AlCl3 (0.5 g) in dichloroethane (10 ml) at 60°C for 2.5 h. The reaction was quenched by 30 ml crushed ice and then extracted with dichloromethane (3 × 30 ml). The combined organic layer was washed by saturated sodium carbonate until PH=7, washed with saturated aqueous sodium chloride, dried over with anhydrous Na2SO4, and evaporated to give the crude product. The obtained residue was purified by column chromatography on silica gel using petroleum ether-acetone as the eluent to produce compounds 1–12.
(4-Methoxy-1H-indol-3-yl)(4-methoxyphenyl)methanone (1).
Yield: 36 %, white solid, m.p. 250–251°C; IR (KBr): 3182, 3003, 1620, 1420, 1093 cm−1; 1H NMR (400 MHz, DMSO): δ (ppm) 11.81 (br, 1H), 7.74 (d, J = 8.8 Hz, 2H), 7.64 (d, J = 2.8 Hz, 1H), 7.15 (dd, J =8.0, 7.6 Hz, 1H), 7.11 (dd, J = 8.0, 0.8 Hz, 1H), 7.02 (d, J = 8.8 Hz, 2H), 6.61 (dd, J = 7.6, 0.8 Hz, 1H), 3.85 (s, 3H), 3.59 (s, 3H); 13C NMR (101 MHz, DMSO): δ (ppm) 189.28, 162.57, 153.89, 138.48, 133.27, 131.82, 130.96, 124.00, 116.72, 116.20, 113.61, 105.64, 102.16, 55.86, 55.38; HRMS for C17 H15 N O3 ([M-H]−) calculated: 280.0968, found: 280.0986.
(4-methoxy-1H-indole-3,5-diyl)bis((4-methoxyphenyl)methanone) (2).
Yield: 19 %, yellow solid, m.p. 194–196°C; IR (KBr): 3380, 2935, 1600, 1361, 1075 cm−1; 1H NMR (400 MHz, DMSO): δ (ppm) 12.00 (br, 1H), 7.77 (dd, J = 8.8, 2.4 Hz, 4H), 7.60–7.56 (m, 2H), 7.13 (dd, J = 8.8, 2.0 Hz, 2H), 7.05 (dd, J = 8.8, 2.0 Hz, 2H), 6.78 (d, J = 8.8 Hz, 1H), 3.89 (s, 3H), 3.86 (s, 3H); 13C NMR (101 MHz, DMSO): δ (ppm) 193.86, 189.17, 162.86, 162.59, 158.10, 137.09, 132.84, 131.95, 131.86, 131.58, 131.17, 116.92, 116.89, 115.10, 114.21, 113.82, 101.80, 55.96, 55.94, 55.92; HRMS for C25 H21 N O5 ([M-H]−) calculated: 414.1336, found: 414.1393.
(4-Methoxy-1H-indol-3-yl)(2,4-dimethoxyphenyl)methanone (3).
Yield: 40 %, white solid, m.p. 224–226°C; IR (KBr): 3213, 2937, 1622, 1420, 1088 cm−1; 1H NMR (400 MHz, DMSO): δ (ppm) 11.75 (s, 1H), 7.53 (d, J = 3.2 Hz, 1H), 7.25 (d, J = 8.4 Hz, 1H), 7.10 (t, J = 8.0 Hz, 1H), 7.04 (dd, J = 8.4, 0.8 Hz, 1H), 6.61 (d, J = 2.4 Hz, 1H), 6.58 (dd, J = 7.6, 0.8 Hz, 1H), 6.54 (dd, J = 8.8, 2.0 Hz, 1H), 3.82 (s, 3H), 3.59 (s, 3H), 3.58 (s, 3H); 13C NMR (101 MHz, DMSO): δ (ppm) 188.29, 162.37, 159.32, 154.21, 138.94, 133.35, 131.32, 125.42, 123.90, 119.73, 115.52, 105.65, 104.78, 102.51, 98.88, 55.89, 55.83, 55.53; HRMS for C18 H17 N O4 ([M-H]) calculated: 310.1074, found: 310.1089.
(4-methoxy-1H-indole-3,5-diyl)bis((2,4-dimethoxyphenyl)methanone) (4).
Yield: 8 %, white solid, m.p. 181–184°C; IR (KBr): 3321, 2937, 1620, 1416, 1072 cm−1; 1H NMR (400 MHz, Acetone): δ (ppm) 11.56 (s, 1H), 7.68 – 7.73 (m, 1H), 7.47 (d, J = 8.4 Hz, 1H), 7.43 (d, J = 8.4 Hz, 1H), 7.31 (d, J = 8.4 Hz, 1H), 6.72 (d, J = 2.4 Hz, 1H), 6.70 – 6.65 (m, 2H), 6.64 (d, J = 2.0 Hz, 1H), 6.61 (dd, J = 8.4, 2.4 Hz, 1H), 3.92 (s, 3H), 3.90 (s, 3H), 3.77 (s, 3H), 3.71 (s, 3H), 3.63 (s, 3H); 13C NMR (101 MHz, Acetone): δ (ppm) 194.97, 187.95, 162.96, 162.52, 159.83, 158.90, 158.41, 137.39, 132.00, 131.58, 131.50, 130.13, 125.17, 122.52, 120.82, 115.85, 115.73, 104.84, 104.36, 101.27, 98.55, 98.14, 55.07, 54.98, 54.94; HRMS for C27 H25 N O7 ([M-H]) calculated: 474.1547, found: 474.1560.
(4-Methoxy-1H-indol-3-yl)(3,4,5-trimethoxyphenyl)methanone (5).
Yield: 58 %, white solid, m.p. 239–243°C; IR (KBr): 3114, 2964, 1578, 1422, 1125 cm−1; 1H NMR (400 MHz, DMSO): δ (ppm) 11.87 (br, 1H), 7.76 (d, J = 2.8 Hz, 1H), 7.17 (dd, J = 7.6, 8.0 Hz, 1H), 7.13 – 7.08 (m, 3H), 6.65 (d, J = 7.2 Hz, 1H), 3.78 (s, 6H), 3.76 (s, 3H), 3.67 (s, 3H); 13C NMR (101 MHz, DMSO): δ (ppm) 188.68, 154.02, 152.72, 141.13, 138.71, 135.74, 132.15, 124.22, 116.38, 116.19, 107.38, 105.66, 102.39, 60.60, 56.34, 55.47; HRMS for C19 H19 N O5 ([M-H]) calculated: 340.1179, found: 340.1193.
(5-Methoxy-1H-indol-3-yl)(2-methoxyphenyl)methanone (6).
Yield: 53 %, white solid, m.p. 175–177°C; IR (KBr): 3154, 2957, 1607, 1417, 1067 cm−1; 1H NMR (400 MHz, Acetone): δ (ppm) 10.86 (br, 1H), 7.87 (d, J= 2.4 Hz, 1H),7.55 (d, J = 3.2 Hz, 1H), 7.45 (ddd, J = 8.4, 7.6, 1.6 Hz, 1H), 7.41 (d, J = 8.8 Hz, 1H), 7.34 (dd, J = 7.2, 1.6 Hz, 1H), 7.13 (d, J = 8.4 Hz, 1H), 7.04 (ddd, J =7.6, 7.2, 0.8 Hz, 1H), 6.89 (dd, J = 8.8, 2.8 Hz, 1H), 3.85 (s, 3H), 3.76 (s, 3H); 13C NMR (101 MHz, Acetone): δ (ppm) 190.18, 157.13, 156.76, 136.17, 132.42, 132.35, 130.91, 128.86, 127.43, 120.65, 118.32, 113.75, 113.16, 112.14, 103.84, 55.52, 55.42; HRMS for C17 H15 N O3 ([M-H]) calculated: 280.0968, found: 280.0995.
(5-Methoxy-1H-indol-3-yl)(3-methoxyphenyl)methanone (7).
Yield: 21 %, white solid, m.p. 197–198°C; IR (KBr): 3133, 2919, 1597, 1431, 1025 cm−1; 1H NMR (400 MHz, Acetone): δ (ppm) 10.99 (br, 1H), 7.95 (d, J = 2.4 Hz, 1H), 7.93 – 7.89 (m, 1H), 7.45 (d,J =8.8, Hz, 1H), 7.43 (d, J =8.0, Hz, 1H), 7.40 (m, 1H) 7.35 (dd, J = 2.4, 1.2 Hz, 1H), 7.14 (ddd, J = 8.0, 2.8, 1.6 Hz, 1H), 6.92 (dd, J = 8.8, 2.4 Hz, 1H), 3.88 (s, 3H), 3.87 (s, 3H); 13C NMR (126 MHz, DMSO): δ (ppm) 190.05, 159.59, 156.05, 142.55, 136.44, 132.04, 129.98, 127.54, 121.09, 117.23, 115.27, 113.73, 113.48, 103.68, 55.75, 55.70; HRMS for C17 H15 N O3 ([M-H]) calculated: 280.0968, found: 280.0994.
(5-Methoxy-1H-indol-3-yl)(4-methoxyphenyl)methanone (8).
Yield: 40 %, white solid, m.p. 197–200°C; IR (KBr): 3151, 2956, 1602, 1427, 1026 cm−1; 1H NMR (400 MHz, DMSO): δ (ppm) 11.90 (br, 1H), 7.89 (d, J = 3.2 Hz, 1H), 7.80 (d, J = 8.8 Hz 2H), 7.76 (d, J = 2.4 Hz, 1H), 7.40 (d, J = 8.8 Hz, 1H), 7.07 (d, J = 8.8 Hz 2H), 6.88 (dd, J = 8.8, 2.4 Hz, 1H), 3.85 (s, 3H), 3.80 (s, 3H); 13C NMR (101 MHz, DMSO): δ (ppm) 189.16, 162.08, 155.85, 135.50, 133.53, 131.94, 130.94, 127.68, 115.33, 114.11, 113.35, 113.32, 103.66, 55.84, 55.74; HRMS for C17 H15 N O3 ([M-H]) calculated: 280.0968, found: 280.0992.
(5-Methoxy-1H-indol-3-yl)(2,4-dimethoxyphenyl)methanone (9).
Yield: 20 %, white solid, m.p. °C; IR (KBr): 3189, 2989, 1604, 1422, 1069 cm−1; 1H NMR (500 MHz, DMSO): δ (ppm) 11.79 (br, 1H), 7.69 (d, J = 2.5 Hz, 1H), 7.57 (d, J = 2.8 Hz, 1H), 7.38 (d, J = 7.2 Hz, 1H), 7.26 (d, J = 6.8 Hz, 1H), 6.87 (dd, J = 6.8, 2.5 Hz, 1H), 6.68 (d, J = 1.6 Hz, 1H), 6.61 (dd, J = 6.8, 2.5 Hz, 1H), 3.85 (s, 3H), 3.80 (s, 3H), 3.73 (s, 3H); 13C NMR (101 MHz, DMSO): δ (ppm) 189.38, 161.88, 158.21, 155.91, 136.47, 132.00, 130.13, 126.94, 124.43, 117.38, 113.40, 113.18, 105.08, 103.38, 99.31, 55.92, 55.83, 55.69; HRMS for C18 H17 N O4 ([M-H]) calculated: 310.1074, found: 310.1091.
(5-Methoxy-1H-indol-3-yl)(3,4,5-trimethoxyphenyl)methanone (10).
Yield: 47 %, white solid, m.p. 206–208°C; IR (KBr): 3296, 2973, 1613, 1420, 1122 cm−1; 1H NMR (400 MHz, DMSO): δ (ppm) 11.93 (br, 1H), 8.04 (d, J = 3.2 Hz, 1H), 7.78 (d, J = 2.4 Hz, 1H), 7.41 (d, J = 8.8 Hz, 1H), 7.08 (s, 2H), 6.89 (dd, J = 8.8, 2.4 Hz, 1H), 3.87 (s, 6H), 3.80 (s, 3H), 3.76 (s, 3H); 13C NMR (101 MHz, DMSO) δ: 189.34, 155.95, 153.07, 140.39, 136.38, 136.21, 132.03, 127.70, 115.12, 113.35, 106.41, 103.75, 60.55, 56.43, 55.74; HRMS for C19 H19 N O5 ([M-H]) calculated: 340.1179, found: 340.1200.
(5-methoxy-1H-indol-3-yl)(2,6-dimethoxyphenyl) methanone (11).
Yield: 47 %, white solid, m.p. 205–207°C; IR (KBr): cm-1; 1H NMR (400 MHz, DMSO) δ 11.74 (s, 1H), 7.57 (s, 1H), 7.45 (d, J = 3.1 Hz, 1H), 7.38 (dd, J = 8.6, 7.3 Hz, 2H), 6.86 (dd, J = 8.8, 2.5 Hz, 1H), 6.77 (d, J = 8.4 Hz, 2H), 3.79 (s, 3H), 3.67 (s, 6H); 13C NMR (126 MHz, DMSO) δ 188.06, 157.26, 155.90, 135.90, 132.11, 130.51, 126.46, 120.23, 118.23, 113.52, 113.26, 104.81, 103.02, 56.10, 55.67; HRMS for C18 H17 N O4 ([M-H]) calculated: 310.1074, found:310.1088.
(7-Methoxy-1H-indol-3-yl)(2,4-dimethoxyphenyl)methanone (12).
Yield: 62 %, white solid, m.p. 208–210°C; IR (KBr): 3174, 2994, 1603, 1434, 1117 cm−1; 1H NMR (400 MHz, Acetone): δ (ppm) 11.01 (br, 1H), 7.91 (d, J = 8.0 Hz, 1H), 7.62 – 7.55 (m, 1H), 7.34 (d, J = 8.0 Hz, 1H), 7.16 (dd, J = 7.6, 8.0 Hz, 1H), 6.82 (d, J =8.0 Hz, 1H), 6.68 (d, J = 2.0 Hz, 1H), 6.63 (dd, J = 8.4, 2.4 Hz, 1H), 3.98 (s, 3H), 3.91 (s, 3H), 3.76 (s, 3H); 13C NMR (126 MHz, DMSO): δ (ppm) 189.46, 162.00, 158.26, 146.69, 135.16, 130.25, 127.68, 127.10, 124.33, 123.06, 118.19, 114.26, 105.13, 104.05, 99.31, 55.93, 55.84, 55.79; HRMS for C18 H17 N O4 ([M-H]) calculated: 310.1074, found: 310.1089.
General procedure for the synthesis of compounds 13–23.
The solution of compounds 1–12 in dry CH2Cl2 was added into excess BBr3/CH2Cl2 (1.0 M) at 0°C under the atmosphere of N2. The mixture was warmed to room temperature slowly and stirred for 24 h. The reaction was quenched by 20 ml cold water. The mixture was filtered to obtain the crude product. The obtained crude product was purified by column chromatography on silica gel using chloroform-acetone as eluent to obtain compounds 13–23.
(4-Hydroxy-1H-indol-3-yl)(4-hydroxyphenyl)methanone (13).
Yield: 71 %, white solid, m.p. 293–296°C; IR (KBr): 3284, 3199, 2924, 1611, 1477, 1164 cm−1; 1H NMR (400 MHz, Acetone): δ (ppm) 11.58 (s, 1H), 11.27 (br, 1H), 9.12 (br, 1H), 8.01 (s, 1H), 7.85 (d, J =8.8 Hz, 2H), 7.16 (dd, J = 7.6, 8.0 Hz, 1H), 7.02 (d, J =8.8, 2H), 6.99 (dd, J = 8.0, 0.8 Hz, 1H), 6.60 (dd, J = 8.0, 0.8 Hz, 1H); 13C NMR (101 MHz, Acetone): δ (ppm) 192.03, 161.10, 152.66, 139.18, 136.52, 131.35, 131.00, 125.79, 116.76, 115.64, 115.07, 107.07, 102.75; HRMS for C15 H11 N O3 ([M-H]) calculated: 252.0655, found: 252.0668.
(4-hydroxy-1H-indole-3,5-diyl)bis((4-hydroxyphenyl)methanone) (14)
Yield: 80 %, yellow solid, m.p. >280°C; IR (KBr): 3313, 3138, 2919, 1595, 1394, 1127 cm−1; 1H NMR (400 MHz, DMSO): δ (ppm) 12.69 (s, 1H), 12.43 (br, 1H), 10.34 (br, 2H), 7.89 (s, 1H), 7.81 (dd, J =6.8, 2.0Hz, 2H), 7.67 (dd, J =6.8, 2.0Hz, 2H), 7.57 (d, J = 8.0 Hz, 1H), 6.97 (dd, J =6.8, 2.0Hz, 2H), 6.93 (dd, J =6.8, 2.0Hz, 2H), 6.66 (d, J = 8.4 Hz, 1H); 13C NMR (126 MHz, DMSO): δ (ppm) 193.30, 192.40, 162.11, 161.48, 157.15, 138.53, 138.39, 132.50, 132.21, 131.99, 129.92, 129.68, 116.52, 116.34, 115.81, 115.55, 113.82, 107.31; HRMS for C22 H15 N O5 ([M-H]) calculated: 372.0866, found: 372.0932.
(4-Hydroxy-1H-indol-3-yl)(2,4-dihydroxyphenyl)methanone (15).
Yield: 22 %, yellow solid, m.p. 248–252°C; IR (KBr): 3299, 3016, 2921, 1627, 1402, 1134 cm−1; 1H NMR (400 MHz, Acetone): δ (ppm) 11.45 (s, 2H), 10.55 (s, 1H), 9.43 (s, 1H), 8.15 (s, 1H), 7.94 (d, J = 8.8 Hz, 1H), 7.21 (t, J = 8.0 Hz, 1H), 7.05 (dd, J = 8.0, 0.8 Hz, 1H), 6.65 (dd, J =8.0, 0.8, 1H), 6.54 (dd, J = 8.8, 2.4 Hz, 1H), 6.48 (d, J = 2.4 Hz, 1H); 13C NMR (101 MHz, Acetone): δ (ppm) 193.12, 163.93, 152.13, 139.03, 136.61, 134.47, 125.78, 115.73, 115.49, 114.05, 107.85, 107.73, 103.23; HRMS for C15 H11 N O4 ([M-H]) calculated: 268.0604, found: 268.0619.
(4-Hydroxy-1H-indol-3-yl)(3,4,5-trihydroxyphenyl)methanone (16).
Yield: 85 %, yellow solid, m.p. 300–305°C; IR (KBr): 3494, 3265, 2775, 1615, 1414, 1042 cm−1; 1H NMR (400 MHz, Acetone): δ (ppm) 11.57 (s, 1H), 11.27 (br, 1H), 8.34 (br, 1H), 8.04 (s, 1H), 7.15 (dd, J = 7.6, 8.0 Hz, 1H), 7.03 (s, 2H), 6.98 (dd, J = 8.0, 0.8 Hz, 1H), 6.59 (dd, J = 7.6, 0.8 Hz, 1H); 13C NMR (101 MHz, Acetone): δ (ppm) 192.17, 152.62, 145.18, 139.15, 137.03, 136.55, 130.43, 125.73, 116.69, 115.66, 108.88, 107.04, 102.77; HRMS for C15 H11 N O5 ([M-H]) calculated: 284.0553, found: 284.0566.
(5-Hydroxy-1H-indol-3-yl)(2-hydroxyphenyl)methanone (17).
Yield: 43 %, yellow solid, m.p. 214–217°C; IR (KBr): 3339, 3122, 2797, 1596, 1424, 1131 cm−1; 1H NMR (400 MHz, Acetone): δ (ppm) 12.11 (s, 1H), 11.12 (br, 1H), 8.10–8.06 (m, 2H), 8.00 (dd, J = 8.0, 1.6 Hz, 1H), 7.79 (d, J = 2.4 Hz, 1H), 7.50 (ddd, J = 8.4, 7.2, 1.6 Hz, 1H), 7.41 (d, J = 8.8 Hz, 1H), 7.02 – 6.94 (m, 2H), 6.88 (dd, J = 8.4, 2.4 Hz, 1H). 13C NMR (101 MHz, Acetone): δ (ppm) 194.41, 162.61, 154.51, 135.97, 135.10, 132.32, 132.16, 128.92, 122.42, 119.54, 118.47, 115.55, 114.33, 113.49, 107.35; HRMS for C15 H11 N O3 ([M-H]) calculated: 252.0655, found: 252.0669.
(5-Hydroxy-1H-indol-3-yl)(3-hydroxyphenyl)methanone (18).
Yield: 40 %, white solid, m.p. 313–316°C; IR (KBr): 3310, 3086, 2960, 1579, 1443, 1142 cm−1; 1H NMR (400 MHz, DMSO) δ 11.79 (s, 1H), 9.68 (s, 1H), 9.04 (s, 1H), 7.78 (d, J = 3.2 Hz, 1H), 7.65 (d, J = 2.4 Hz, 1H), 7.31 (dd, J = 12.3, 5.1 Hz, 2H), 7.19 – 7.13 (m, 1H), 7.13 – 7.10 (m, 1H), 6.96 (ddd, J = 8.1, 2.5, 0.9 Hz, 1H), 6.74 (dd, J = 8.7, 2.4 Hz, 1H); 13C NMR (101 MHz, DMSO): δ (ppm) 190.13, 157.68, 153.66, 142.61, 135.94, 131.28, 129.81, 127.79, 119.55, 118.29, 115.37, 114.98, 113.44, 113.06, 106.31; HRMS for C15 H11 N O3 ([M-H]) calculated: 252.0655, found: 252.0664.
(5-Hydroxy-1H-indol-3-yl)(4-hydroxyphenyl)methanone (19).
Yield: 66 %, white solid, m.p. 298–302°C; IR (KBr): 3343, 3103, 2977, 1717, 1435, 1137 cm−1; 1H NMR (400 MHz, DMSO): δ (ppm) 11.71 (br, 1H), 10.04 (s, 1H), 8.98 (s, 1H), 7.79 (d, J = 3.2 Hz, 1H), 7.68 (d, J = 8.8 Hz, 2H), 7.62 (d, J = 2.4 Hz, 1H), 7.28 (d, J = 8.8 Hz, 1H), 6.87 (d,J = 8.8 Hz, 2H), 6.72 (dd, J = 8.8, 2.4 Hz, 1H); 13C NMR (126 MHz, DMSO): δ (ppm) 189.15, 160.70, 153.41, 135.03, 132.20, 131.22, 131.18, 128.03, 115.38, 115.04, 113.29, 112.91, 106.28; HRMS for C15 H11 N O3 ([M-H]) calculated: 252.0655, found: 252.0667.
(5-Hydroxy-1H-indol-3-yl)(2,4-dihydroxyphenyl)methanone (20).
Yield: 23 %, yellow solid, m.p. 217–220°C; IR (KBr): 3596, 3192, 2957, 1629, 1442, 1160 cm−1; 1H NMR (500 MHz, Acetone): δ (ppm) 13.00 (s, 1H), 11.02 (br, 1H), 9.28 (s, 1H), 8.03 (s, 2H), 7.93 (d, J = 8.8 Hz, 1H), 7.73 (d, J = 1.6 Hz, 1H), 7.40 (d, J = 6.8 Hz, 1H), 6.87 (dd, J = 6.8, 2.0 Hz, 1H), 6.47 (dd, J = 6.8, 2.0 Hz, 1H), 6.45 (m, 1H); 13C NMR (101 MHz, Acetone): δ (ppm) 192.82, 165.12, 163.32, 153.24, 133.70, 133.57, 131.20, 128.03, 114.53, 114.18, 113.23, 112.43, 107.19, 106.28, 102.99. HRMS for C15 H11 N O4 ([M-H]) calculated: 268.0604, found: 268.0618.
(5-Hydroxy-1H-indol-3-yl)(3,4,5-trihydroxyphenyl)methanone (21).
Yield: 87 %, white solid, m.p. 272–274°C; IR (KBr): 3456, 3124, 2768, 1629, 1431, 1155 cm−1; 1H NMR (400 MHz, DMSO): δ (ppm) 11.68 (br, 1H), 9.16 (s, 2H), 8.97 (s, 1H), 8.69 (s, 1H), 7.77 (d, J = 2.8 Hz, 1H), 7.60 (d, J = 2.4 Hz, 1H), 7.28 (d, J = 8.8 Hz, 1H), 6.79 (s, 2H), 6.71 (dd, J = 8.8, 2.4 Hz, 1H); 13C NMR (126 MHz, DMSO): δ(ppm) 189.35, 153.19, 145.89, 137.10, 134.74, 131.37, 131.18, 128.03, 115.02, 113.24, 112.91, 108.64, 106.27; HRMS for C15 H11 N O5 ([M-H]) calculated: 284.0553, found: 284.0565.
(5-hydroxy-1H-indol-3-yl)(2,6-dihydroxyphenyl)methanone (22).
Yield: 53 %, white solid, m.p. 237–240°C; IR (KBr): cm-1; 1H NMR (400 MHz, Acetone) δ 10.87 (s, 1H), 9.11 (s, 2H), 7.99 (s, 1H), 7.88 (dd, J = 37.2, 2.7 Hz, 3H), 7.35 (d, J = 8.7 Hz, 1H), 7.17 (t, J = 8.2 Hz, 1H), 6.83 (dd, J = 8.7, 2.4 Hz, 1H), 6.49 (d, J = 8.2 Hz, 3H); 13C NMR (126 MHz, Acetone) δ 190.30, 157.33, 153.54, 136.49, 131.90, 131.22, 127.60, 117.39, 114.95, 112.68, 112.40, 107.68, 106.52; HRMS for C15 H10 N O4 ([M-H]−) calculated: 268.0604, found: 268.0617.
(7-Hydroxy-1H-indol-3-yl)(2,4-dihydroxyphenyl)methanone (23)
Yield: 49 %, yellow solid, m.p. 247–250°C; IR (KBr): 3352, 3126, 2926, 1627, 1440, 1143 cm−1; 1H NMR (400 MHz, Acetone): δ (ppm) 13.01 (s, 1H), 11.17 (br, 1H), 9.28 (br, 1H), 8.89 (br, 1H), 8.02 (d, J = 2.8 Hz, 1H), 7.96 (d, J = 8.8 Hz, 1H), 7.73 (d, J = 8.0 Hz, 1H), 7.07 (dd, J = 7.6, 8.0 Hz, 1H), 6.76 (d, J = 7.6 Hz, 1H), 6.49 (dd, J = 8.8, 2.4 Hz, 1H), 6.44 (d, J= 2.4 Hz, 1H); 13C NMR (126 MHz, Acetone): δ (ppm) 193.11, 165.26, 163.48, 143.65, 133.92, 132.48, 128.91, 126.75, 122.54, 115.56, 114.21, 113.36, 107.61, 107.26, 102.99; HRMS for C15 H11 N O4 ([M-H]−) calculated: 268.0604, found: 268.0613.
(2,6-dimethoxy-1-naphthalenyl)(2,4-dimethoxyphenyl)methanone (24).
A mixture of 2,6-dimethoxynaphthalene (2 mmol) and methoxy benzoylchloride (2 mmol) was stirred in the presence of POCl3 (5 ml) and AlCl3 (0.5 g) in dichloroethane (10 ml) at 60°C for 2.5 h. The reaction was quenched by 30 ml crushed ice and then extracted with dichloromethane (3 × 30 ml). The combined organic layer was washed by saturated sodium carbonate until PH=7, washed with saturated aqueous sodium chloride, dried over with anhydrous Na2SO4, and evaporated to give the crude product. The obtained residue was purified by column chromatography on silica gel. Yield: 63 %, yellow solid, m.p. 99–102°C; IR(KBr): 2937, 1598, 1465, 1024 cm−1; 1H NMR (400 MHz, Acetone): δ (ppm) 11.01 (br, 1H), 7.91 (d, J = 8.0 Hz, 1H), 7.62 – 7.55 (m, 1H), 7.34 (d, J = 8.0 Hz, 1H), 7.16 (dd, J = 7.6, 8.0 Hz, 1H), 6.82 (d, J =8.0 Hz, 1H), 6.68 (d, J = 2.0 Hz, 1H), 6.63 (dd,J = 8.4, 2.4 Hz, 1H), 3.98 (s, 3H), 3.91 (s, 3H), 3.76 (s, 3H);13C NMR (126 MHz, DMSO): δ (ppm) 189.46, 162.00, 158.26, 146.69, 135.16, 130.25, 127.68, 127.10, 124.33, 123.06, 118.19, 114.26, 105.13, 104.05, 99.31, 55.93, 55.84, 55.79; HRMS for C21 H20 O5 ([M+H]+) calculated: 353.1384, found: 353.1385.
Cell lines and cell culture
The human lung cancer cell line NCI-H460, and its MX-selected derivative ABCG2-overexpressing cell line, NCI-H460/MX20, were used in this study. 32 The human colon cancer cell line S1, and its MX-selected derivative ABCG2-overexpressing cell line, S1-M1-80, were also used in this study.33 HEK293/pcDNA3.1, wild-type HEK293/ABCG2–482-R2, mutant HEK293/ABCG2–482-G2 and HEK293/ABCG2–482-T7 cells were established by transfecting HEK293 cell with either the empty pcDNA3.1 vector or pcDNA3.1 vector containing a full-length ABCG2, with coding arginine (R), threonine (T), or glycine (G) at amino acid position 482, following selection with G418 and maintenance in medium with 2 mg/ml of G418.34 All the cell lines were maintained in Dulbecco’s Modified Eagle Medium (Corning Inc.) containing 10% fetal bovine serum (Gibco Inc.) and 1% penicillin/streptomycin (Gibco Inc.) at 37°C with 5% CO2.
Cytotoxicity and MDR reversal assay
Cytotoxicity tests and reversal experiments were performed using the MTT colorimetric assay. Cells were harvested and re-suspended in a final concentration of 5 × 103 cells/well for NCI-H460, NCI-H460/MX20, S1, S1-M1-80, HEK293/pcDNA3.1, HEK293/ABCG2–482-R2, HEK293/ABCG2–482-G2 and HEK293/ABCG2–482-T7 cells. Cells were seeded evenly into 96-well plates. To determine the cytotoxicity of the indole derivatives, incremental concentrations of each drug were added into the well after 24 h of incubation. To determine the MDR reversal efficacy of the indole derivatives, the chemotherapeutic drugs (mitoxantrone, doxorubicin and cisplatin) were added into the designated wells after 2 h pre-incubation with the indole derivatives or Ko143 at non-toxic concentrations. After 44 h of drug incubation, the MTT reagent (4 mg/mL) was added into the wells, and then the plates were incubated for an additional 4 h. Subsequently, the supernatant was discarded and 100 μL of DMSO was added to dissolve the formazan crystals. Cell viability was determined by measuring the absorbance at a wavelength of 570 nm. All the experiments were repeated three independent experiments in triplicate, and the mean and standard deviation (SD) values were calculated.
[3H]-MX accumulation assay
To determine the effect of compounds 2 and 8 on the intracellular accumulation of [3H]-MX on NCI-H460 and NCI-H460/MX20 cells, the cells (105 cells / well) were seeded in the 24-well plates and incubated at 37°C with 5 % CO2. After 12 h of incubation, compounds 2 (10 or 25 μM), 8 (2.5 or 5.0 μM), or Ko143 (5.0 μM) were added and the plate was incubated at 37°C for 2 h. Cells were then incubated with 0.01 μM of the [3H]-MX - containing medium for an additional 2 h at 37°C, with or without 2 (10 or 25 μM), 8 (2.5 or 5.0 μM), and Ko143 (5.0 μM). The cells were washed twice with ice-cold PBS, trypsinized and lysed at the end of incubation. The radioactivity was measured using the Packard TRI-CARB1 190À liquid scintillation analyzer.
Western blot analysis
Cell lysates were prepared as previous reports45. Equal amounts of total cell lysates (15 μg protein) were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred onto polyvinylidene fluoride (PVDF) membranes. After incubation in a blocking solution (5% milk) for 2 h at room temperature, the membranes were incubated overnight with primary monoclonal antibodies against actin at 1:1000 dilution of ABCG2 (BXP 21) (Gene Tex Inc.) at 4°C, and were further incubated with horseradish peroxide (HRP)-conjugated secondary antibody (Thermo Fisher Scientific Inc.) at 1:1000 dilution for 2 h at room temperature. The protein–antibody complex was detected using an enhanced chemiluminescence detection system. The Grayscale ratio was analyzed by ImageJ and normalized by the grayscale of the ABCG2 protein divided by that of β-actin.
Immunofluorescence analysis
For immunofluorescence analysis, NCI-H460 and NCI-H460/MX20 cells were seeded in 24-well plates at 10000 cells/well and incubated for 24 h. The cells were incubated with or without 2 (25 μM) or 8 (5.0 μM) for 24 h, 48 h, and 72 h. Thereafter, cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature and then rinsed with PBS twice, followed by permeabilization with 1% triton X-100 for 10 min at 4°C. The cells were again washed twice with PBS, and then blocked with 6% BSA for 1 h at 37°C. Fixed cells were incubated with monoclonal antibody against the ABCG2 protein (BXP 21) (Gene Tex Inc.) (1:50) overnight at 4°C, followed by two washes with PBS. The cells were then further incubated with Alexa flour 488 goat anti-mouse IgG (1:50) (Abcam plc.) for 1 h at 37°C. After the cells were washed twice with PBS, 4’,6-diamidino-2-phenylindole (DAPI) (2 μg/mL) was used for nuclear counterstaining. The immunofluorescence images were generated using a Nikon TE-2000S fluorescence microscope (Nikon Instruments Inc, Melville, NY)
ABCG2 ATPase assay
The Vi-sensitive ATPase activity of ABCG2 in the membrane vesicles of High Five insect cells was determined as previously described, using the PREDEASY ATPase assay kit46.
Molecular modeling
Molecular modeling was performed in Maestro v11.1 on a Mac Pro 6-core Intel Xenon X5 processor with a Macintosh Operating System (OS X El Capitan), using the Maestro v11.1 (Schrödinger, LLC, New York, NY, 2017) software as described previously47. The protein preparation of the wild-type human ABCG2 (PDB ID: 6ETI)9 was performed and the grid was generated by selecting residues in the substrate-binding pocket in the TMD. The structure of ligands was built and prepared. The ligands with the best-scored conformation were obtained through Glide XP docking and were used to generate the receptor grid for induced-fit docking (IFD) at the binding site. The IFD protocol with default parameters was performed and the docking scores (kcal/mol) were obtained. The conformation with the highest docking score for each compound was used for graphical analysis.
In vitro metabolic stability testing
The mixture contained microsomal proteins (0.5 mg/mL), NADPH (3 mmol), MgCl2 (3 mmol), and the test compound (100 μM) in 74 mmol potassium phosphate buffer (pH 7.4). The final incubation volume was 250 μL. The incubation was carried out aerobically at 37°C. The mixture was pre-incubated without NADPH for 10 min at 37°C and NADPH was added to start the reaction. At 0, 10, 30 min and 1 h after the start of reaction, an aliquot (50 μL) of the incubation mixture was taken from each incubation and mixed with 500 μL of ice-cold acetonitrile to terminate the reaction. Subsequently, the sample was precipitated by centrifugation (12000 rpms) at room temperature. The resulting supernatant was analyzed using HPLC (LC, Agilent 1260 Infinity; column, Eclipse Plus C18; column temperature, 25°C; mobile phase, solvent A, acetonitrile, solvent B, water, gradient elution, 25–99% solvent A; flow rate, 1 mL/min; UV signals were recorded at 254 nm) for each compound to determine the metabolic rate20.
Statistical analysis
All experiments were repeated at least three times. All data are expressed as the mean ± SD and the data were analyzed by using a two-way ANOVA. Differences were considered significant when P < 0.05.
Supplementary Material
Highlights.
We synthesized benzoyl indoles with synthetic tractability as reversal agents that reversed ABCG2-mediated MDR.
Benzoyl indoles could interact with the substrate-binding site of ABCG2.
Benzoyl indoles have enhanced metabolic stability compared to Ko143.
The combination of benzoyl indoles with anticancer drug could be a novel treatment strategy to evade ABCG2-mediated MDR in cancer.
Acknowledgments
We are thankful to Dr. Susan E. Bates and Dr. Robert W. Robey for generously providing us ABCG2-transfected cell lines and mitoxantrone selected NCI-NCI-H460/MX20 cell line. We are thankful to Dr. Tanaji T. Talele providing us the computational source for the docking study. We thank Dr. Charles R. Ashby, Jr. for carefully reviewing and revising our manuscript. We thank Juanjuan Huang for drawing the figure of graphic abstract. This work was supported by the National Natural Science Foundation of China (21272290, J1103305), the Fund for Innovative Chemical Experiment and Research of School of Chemistry in Sun Yat-sen University, the National Institute of Health-USA (1R15GM116043–01), the St. John’s University Research Seed Grant (No. 579-1110-7002). Dr. Yangmin Chen (MediMedia Managed Markets, ICON plc) provided editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
There are no conflicts to declare.
References
- 1.Dole M; Nunez G; Merchant AK; Maybaum J; Rode CK; Bloch CA; Castle VP Bcl-2 inhibits chemotherapy-induced apoptosis in neuroblastoma. Cancer Res 1994, 54, 3253–9. [PubMed] [Google Scholar]
- 2.Wang Y; Suh YA; Fuller MY; Jackson JG; Xiong S; Terzian T; Quintas-Cardama A; Bankson JA; El-Naggar AK; Lozano G Restoring expression of wild-type p53 suppresses tumor growth but does not cause tumor regression in mice with a p53 missense mutation. J Clin Invest 2011, 121, 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentires-Alj M; Barbu V; Fillet M; Chariot A; Relic B; Jacobs N; Gielen J; Merville MP; Bours V NF-kappaB transcription factor induces drug resistance through MDR1 expression in cancer cells. Oncogene 2003, 22, 90–7. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman MM Mechanisms of cancer drug resistance. Annu Rev Med 2002, 53, 615–27. [DOI] [PubMed] [Google Scholar]
- 5.Gottesman MM; Fojo T; Bates SE Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer 2002, 2, 48–58. [DOI] [PubMed] [Google Scholar]
- 6.Synold TW; Dussault I; Forman BM The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med 2001, 7, 584–90. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YK; Wang YJ; Gupta P; Chen ZS Multidrug Resistance Proteins (MRPs) and Cancer Therapy. Aaps j 2015, 17, 802–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor NMI; Manolaridis I; Jackson SM; Kowal J; Stahlberg H; Locher KP Structure of the human multidrug transporter ABCG2. Nature 2017, 546, 504–509. [DOI] [PubMed] [Google Scholar]
- 9.Jackson SM; Manolaridis I; Kowal J; Zechner M; Taylor NMI; Bause M; Bauer S; Bartholomaeus R; Bernhardt G; Koenig B; Buschauer A; Stahlberg H; Altmann KH; Locher KP Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat Struct Mol Biol 2018, 25, 333–340. [DOI] [PubMed] [Google Scholar]
- 10.Robey RW; To KK; Polgar O; Dohse M; Fetsch P; Dean M; Bates SE ABCG2: a perspective. Adv Drug Deliv Rev 2009, 61, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnamurthy P; Schuetz JD Role of ABCG2/BCRP in biology and medicine. Annu Rev Pharmacol Toxicol 2006, 46, 381–410. [DOI] [PubMed] [Google Scholar]
- 12.Ni Z; Bikadi Z; Rosenberg MF; Mao Q Structure and function of the human breast cancer resistance protein (BCRP/ABCG2). Curr Drug Metab 2010, 11, 603–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volk EL; Farley KM; Wu Y; Li F; Robey RW; Schneider E Overexpression of wild-type breast cancer resistance protein mediates methotrexate resistance. Cancer Res 2002, 62, 5035–40. [PubMed] [Google Scholar]
- 14.Steinbach D; Sell W; Voigt A; Hermann J; Zintl F; Sauerbrey A BCRP gene expression is associated with a poor response to remission induction therapy in childhood acute myeloid leukemia. Leukemia 2002, 16, 1443–7. [DOI] [PubMed] [Google Scholar]
- 15.van den Heuvel-Eibrink MM; Wiemer EA; Prins A; Meijerink JP; Vossebeld PJ; van der Holt B; Pieters R; Sonneveld P Increased expression of the breast cancer resistance protein (BCRP) in relapsed or refractory acute myeloid leukemia (AML). Leukemia 2002, 16, 833–9. [DOI] [PubMed] [Google Scholar]
- 16.Allen JD; van Loevezijn A; Lakhai JM; van der Valk M; van Tellingen O; Reid G; Schellens JH; Koomen GJ; Schinkel AH Potent and specific inhibition of the breast cancer resistance protein multidrug transporter in vitro and in mouse intestine by a novel analogue of fumitremorgin C. Mol Cancer Ther 2002, 1, 417–25. [PubMed] [Google Scholar]
- 17.Li Y; Hayman E; Plesescu M; Prakash SR Synthesis of potent BCRP inhibitor—Ko143. 2008, 49, 1480–1483. [Google Scholar]
- 18.MA Yang-Min LB Progress in the Synthesis of Indolyl Diketopiperazines. Chin. J. Org. Chem 2010, 30, 1624–1639. [Google Scholar]
- 19.Liu K; Zhu J; Huang Y; Li C; Lu J; Sachar M; Li S; Ma X Metabolism of KO143, an ABCG2 inhibitor. Drug Metab Pharmacokinet 2017, 32, 193–200. [DOI] [PubMed] [Google Scholar]
- 20.Tung YS; Coumar MS; Wu YS; Shiao HY; Chang JY; Liou JP; Shukla P; Chang CW; Chang CY; Kuo CC; Yeh TK; Lin CY; Wu JS; Wu SY; Liao CC; Hsieh HP Scaffold-hopping strategy: synthesis and biological evaluation of 5,6-fused bicyclic heteroaromatics to identify orally bioavailable anticancer agents. J Med Chem 2011, 54, 3076–80. [DOI] [PubMed] [Google Scholar]
- 21.La Regina G; Sarkar T; Bai R; Edler MC; Saletti R; Coluccia A; Piscitelli F; Minelli L; Gatti V; Mazzoccoli C; Palermo V; Mazzoni C; Falcone C; Scovassi AI; Giansanti V; Campiglia P; Porta A; Maresca B; Hamel E; Brancale A; Novellino E; Silvestri R New arylthioindoles and related bioisosteres at the sulfur bridging group. 4. Synthesis, tubulin polymerization, cell growth inhibition, and molecular modeling studies. J Med Chem 2009, 52, 7512–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu YS; Coumar MS; Chang JY; Sun HY; Kuo FM; Kuo CC; Chen YJ; Chang CY; Hsiao CL; Liou JP; Chen CP; Yao HT; Chiang YK; Tan UK; Chen CT; Chu CY; Wu SY; Yeh TK; Lin CY; Hsieh HP Synthesis and evaluation of 3-aroylindoles as anticancer agents: metabolite approach. J Med Chem 2009, 52, 4941–5. [DOI] [PubMed] [Google Scholar]
- 23.Ketcha DM; Gribble GW A convenient synthesis of 3-acylindoles via Friedel Crafts acylation of 1-(phenylsulfonyl)indole. A new route to pyridocarbazole-5,11-quinones and ellipticine. The Journal of Organic Chemistry 1985, 50, 5451–5457. [Google Scholar]
- 24.Wenkert E; Moeller PDR; Piettre SR; McPhail AT Yuehchukene analogs. The Journal of Organic Chemistry 1988, 53, 3170–3178. [Google Scholar]
- 25.Katritzky AR; Suzuki K; Singh SK; He H-Y Regiospecific C-Acylation of Pyrroles and Indoles Using N-Acylbenzotriazoles. The Journal of Organic Chemistry 2003, 68, 5720–5723. [DOI] [PubMed] [Google Scholar]
- 26.Anthony W Notes- Novel Synthesis of Heterocyclic Ketones. The Journal of Organic Chemistry 1960, 25, 2049–2053. [Google Scholar]
- 27.Davidsen SK; Summers JB; Albert DH; Holms JH; Heyman HR; Magoc TJ; Conway RG; Rhein DA; Carter GW N-(Acyloxyalkyl)pyridinium Salts as Soluble Prodrugs of a Potent Platelet Activating Factor Antagonist. Journal of Medicinal Chemistry 1994, 37, 4423–4429. [DOI] [PubMed] [Google Scholar]
- 28.Bergman J; Venemalm L Acylation of the zinc salt of indole. Tetrahedron 1990, 46, 6061–6066. [Google Scholar]
- 29.Powers JC Synthesis of Piperidylindoles. The Journal of Organic Chemistry 1965, 30, 2534–2540. [Google Scholar]
- 30.Flo C; Pindur U Reactions of Electron-Rich Heterocycles with Orthocarboxylic Acid Derivatives. Part 10. Formylation and Alkylation of Carbazoles with Ambident Dialkoxycarbenium Tetrafluoroborates. Wiley: 1987. [Google Scholar]
- 31.Bergman J; Bäckvall JE; Lindström JO Synthesis and reactions of some 3-(2-haloacyl)indoles. Tetrahedron 1973, 29, 971–976. [Google Scholar]
- 32.Honjo Y; Hrycyna CA; Yan Q-W; Medina-Pérez WY; Robey RW; van de Laar A; Litman T; Dean M; Bates SE Acquired Mutations in the MXR/BCRP/ABCP Gene Alter Substrate Specificity in MXR/BCRP/ABCP-overexpressing Cells. Cancer Research 2001, 61, 6635–6639. [PubMed] [Google Scholar]
- 33.Robey RW; Honjo Y; van de Laar A; Miyake K; Regis JT; Litman T; Bates SE A functional assay for detection of the mitoxantrone resistance protein, MXR (ABCG2). Biochim Biophys Acta 2001, 1512, 171–82. [DOI] [PubMed] [Google Scholar]
- 34.Robey RW; Honjo Y; Morisaki K; Nadjem TA; Runge S; Risbood M; Poruchynsky MS; Bates SE Mutations at amino-acid 482 in the ABCG2 gene affect substrate and antagonist specificity. Br J Cancer 2003, 89, 1971–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang CH; Chen YC; Kuo ML Novobiocin sensitizes BCRP/MXR/ABCP overexpressing topotecan-resistant human breast carcinoma cells to topotecan and mitoxantrone. Anticancer Res 2003, 23, 2519–23. [PubMed] [Google Scholar]
- 36.Robey RW; Honjo Y; van de Laar A; Miyake K; Regis JT; Litman T; Bates SE A functional assay for detection of the mitoxantrone resistance protein, MXR (ABCG2). Biochimica et biophysica acta 2001, 1512, 171–182. [DOI] [PubMed] [Google Scholar]
- 37.Robey RW; Honjo Y; Morisaki K; Nadjem TA; Runge S; Risbood M; Poruchynsky MS; Bates SE Mutations at amino-acid 482 in the ABCG2 gene affect substrate and antagonist specificity. British journal of cancer 2003, 89, 1971–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chua VY; Larma I; Harvey J; Thomas MA; Bentel JM Activity of ABCG2 Is Regulated by Its Expression and Localization in DHT and Cyclopamine-Treated Breast Cancer Cells. J Cell Biochem 2016, 117, 2249–59. [DOI] [PubMed] [Google Scholar]
- 39.Xie Y; Xu K; Linn DE; Yang X; Guo Z; Shimelis H; Nakanishi T; Ross DD; Chen H; Fazli L; Gleave ME; Qiu Y The 44-kDa Pim-1 kinase phosphorylates BCRP/ABCG2 and thereby promotes its multimerization and drug-resistant activity in human prostate cancer cells. J Biol Chem 2008, 283, 3349–56. [DOI] [PubMed] [Google Scholar]
- 40.Buchwald P Activity-Limiting Role of Molecular Size: Size-Dependency of Maximum Activity for P450 Inhibition as Revealed by qHTS Data. Drug Metabolism and Disposition 2014, 42, 1785. [DOI] [PubMed] [Google Scholar]
- 41.Bertz RJ; Granneman GR Use of in vitro and in vivo data to estimate the likelihood of metabolic pharmacokinetic interactions. Clin Pharmacokinet 1997, 32, 210–58. [DOI] [PubMed] [Google Scholar]
- 42.McGinnity DF; Soars MG; Urbanowicz RA; Riley RJ Evaluation of fresh and cryopreserved hepatocytes as in vitro drug metabolism tools for the prediction of metabolic clearance. Drug Metab Dispos 2004, 32, 1247–53. [DOI] [PubMed] [Google Scholar]
- 43.Baker JA; Altman MD; Martin IJ Interpretation of in Vitro Metabolic Stability Studies for Racemic Mixtures. ACS Med Chem Lett 2018, 9, 843–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masimirembwa CM; Bredberg U; Andersson TB Metabolic stability for drug discovery and development: pharmacokinetic and biochemical challenges. Clin Pharmacokinet 2003, 42, 515–28. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W; Fan YF; Cai CY; Wang JQ; Teng QX; Lei ZN; Zeng L; Gupta P; Chen ZS Olmutinib (BI1482694/HM61713), a Novel Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor, Reverses ABCG2-Mediated Multidrug Resistance in Cancer Cells. Front Pharmacol 2018, 9, 1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta P; Zhang YK; Zhang XY; Wang YJ; Lu KW; Hall T; Peng R; Yang DH; Xie N; Chen ZS Voruciclib, a Potent CDK4/6 Inhibitor, Antagonizes ABCB1 and ABCG2-Mediated Multi-Drug Resistance in Cancer Cells. Cell Physiol Biochem 2018, 45, 1515–1528. [DOI] [PubMed] [Google Scholar]
- 47.Zhang YK; Zhang XY; Zhang GN; Wang YJ; Xu H; Zhang D; Shukla S; Liu L; Yang DH; Ambudkar SV; Chen ZS Selective reversal of BCRP-mediated MDR by VEGFR-2 inhibitor ZM323881. Biochem Pharmacol 2017, 132, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.