Abstract
The development of cost-effective and rapid assays for the accurate counting of CD4 cells has remained prime focus for disease management. The lack of such assays has severely affected people living in resource-limited disease prevalent areas. CD4 count information plays a vital role in the effective management of HIV disease. There is an unmet need to develop rapid, cost-effective, portable and user-friendly point-of-care (POC) disease diagnostic platform technology for CD4+ T cell counting. Here, we have developed a flow-free magnetic actuation platform that uses antibody-coated magnetic beads to efficiently capture CD4+ T cells from a 30 μL drop of whole blood. On-chip cell lysate electrical impedance spectroscopy has been utilized to quantify the isolated CD4 cells. The developed assay has a limit of detection of 25 cells per μL and provides accurate CD4 counts in the range of 25 – 800 cells per μL. The whole immunoassay along with the enumeration process is very rapid and provides CD4 quantification results within five minutes time frame. The assay does not require off-chip sample preparation steps and minimizes human involvement to a greater extent. The developed impedance-based immunoassay has potential to significantly improve the CD4 enumeration process especially for POC settings.
Keywords: CD4+ T Cells, Human Immunodeficiency Virus, Antiretroviral therapy, Rapid immunoassays, resource-constrained settings
1. Introduction
Human Immunodeficiency Virus (HIV) is one of the deadliest pathogens of present times. It still poses a significant threat to the global health. As per the most recent available statistics around 36.9 million people are living with HIV in 2017 [1]. HIV attacks the CD4+ T cells and can cause acquired immune deficiency syndrome (AIDS). Viral load monitoring and CD4+ T cell enumeration are the two most preferred methods to identify the onset of disease and effectiveness of treatment respectively. The CD4 count provides invaluable information about the overall health of the immune system while plasma viral load determines the level of HIV viremia. Antiretroviral therapy (ART) has been proven to be quite successful in stopping the replication of HIV virus. However, the dilemma is that most of the HIV infected people live in the resource-poor settings lacking basic healthcare facilities, and thus cannot afford the expensive diagnostic tests [2–7].
Although as per recent guidelines of WHO, rapid initiation of ART is highly recommended for all HIV infected patients regardless of their CD4 count [8], CD4 enumeration is still a reliable indicator of the disease staging and can be employed to assess risk of death for patients with advanced level of disease [9, 10]. CD4 count can also be utilized to monitor the effectiveness of treatment. The current gold standards for both viral load count and CD4 enumeration are quite expensive and cannot be employed in disease prevalent areas.
Biosensors and microfluidics have emerged as important tools for various diagnostic applications [11–13]. The stringent requirements of fluid flow control in the current diagnostic technologies have resulted in highly specialized and expensive equipment. The associated costs and other requirements like need of skilled operators to run and maintain these diagnostic devices greatly limit their utilization. The current gold standard for CD4 counting is flow cytometry that is a reliable and accurate method [14], but it is not suitable for resource-limited settings due to high operation/maintenance costs and requirement of skilled human resources. Various researchers have developed point-of-care (POC) devices to enumerate CD4+ T cells [3, 4, 15, 16]. Although, these methods have significantly improved CD4 cells counting process, these devices require precise fluidic manipulations. Peripherals required to ensure strict fluid flow control are bulky and expensive, hence not suitable for POC settings.
A highly sensitive cell lysate impedance spectroscopy based method was presented for the enumeration of CD4 cells in HIV infected people [17]. Electrical impedance based counting processes utilize manual operations for sample preparations, washing and lysis. These manual steps are highly user dependent and can cause significant variations in final count results. Recently, micro-a-fluidic ELISA based device was developed to reliably count up to 30 CD4 cells [2]. However, this enzyme based colorimetric detection requires standardization after a few runs and often control samples can give false positives.
We have developed a flow-free automatic immunoassay that incorporates highly sensitive electrical detection to enumerate CD4 cells. The chip design minimizes the human intervention. The developed method utilizes antibody-conjugated magnetic beads to capture the CD4+ T cells from a whole blood drop with high specificity, and a washing step is performed to remove the debris and other unattached entities (fig. 1). Then cell lysate impedance is used to quantify the captured CD4+ T cells. The developed method uses a disposable microfluidic chip and eliminates the need of manual sample preparation, washing, and lysis steps. It is a cost-effective solution to monitor disease progression and effectiveness of treatment. Supplementary table 1 compares the characteristics of the developed technology with already existing methods utilized for CD4+ T cells counting.
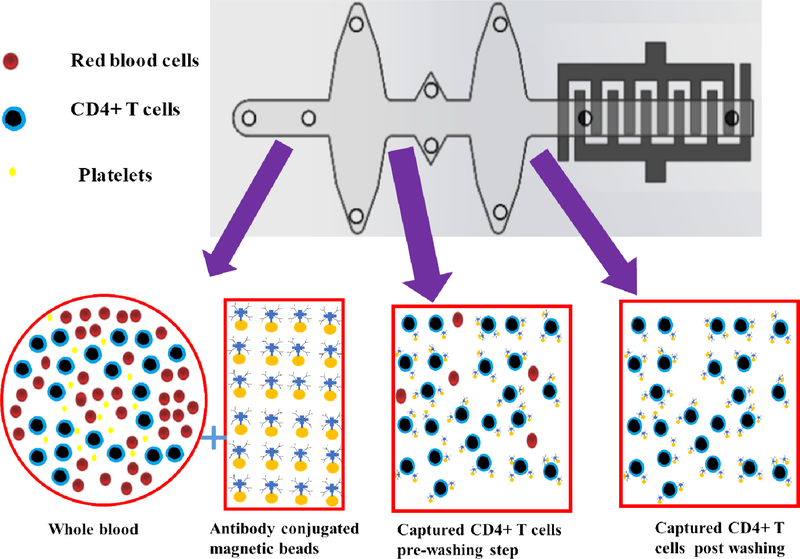
Figure 1.
Pictorial representation of specific capture of CD4 cells from whole blood with antibody conjugated magnetic beads and their subsequent washing steps.
2. Materials and Methods
2.1. Reagents and materials
The details of reagents are provided in the supplementary section.
2.2. Electrode fabrication
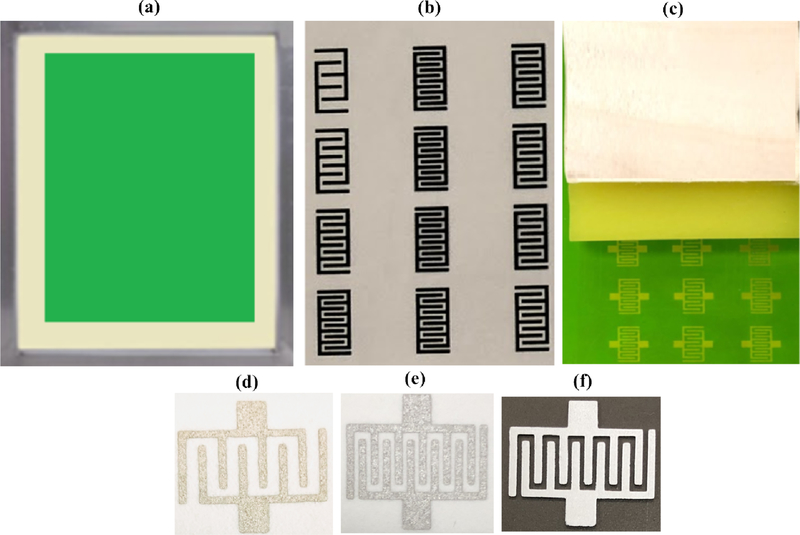
Screen printing is a well-established process to fabricate biosensors for various applications. The end goal was to come up with disposable and cost-effective sensors. Screen printing was utilized for the mass production of interdigitated electrodes. First, a light sensitive emulsion was applied to the surface of an aluminum mesh screen (fig. 2a). Specific patterns of electrodes were designed in AutoCAD software and printed on a transparency sheet using Epson printer (fig. 2b). The transparency sheet containing the designs of electrodes was placed on the top surface of an emulsion coated screen. The screen along with attached transparency sheet was exposed to ultraviolet (UV) light. When UV light passed through the specific areas of the transparency sheet, emulsion was hardened at those locations of screen. Photo emulsion remained water-soluble at all other areas that did not allow UV light to pass (having patterns of electrodes). After this light exposure process, transparency sheet was detached from the screen. The next step was development. Water was sprayed with pressure on the surface of screen. Emulsion was removed from the areas of electrodes. The screen was dried in sun light and it was ready to print electrodes on any substrate including paper, plastic, and fabric etc. (fig. 2c). In order to print the sensors, 20% (w/w) graphene modified silver paste was spread uniformly with the help of a squeegee. The ink penetrated from areas of mesh screen that did not contain photosensitive emulsion, to the surface of substrate and the desired pattern was printed. The sensor was placed in an electric convection oven at 1100C for ten minutes. Visual inspection was carried out to check the uniformity of printed electrodes. It is also equally important to ensure that there are no defective electrodes. Electrically short-circuited electrodes or broken ones are not suitable for experimentation purposes. Continuity testing was also performed to assess the quality of printed electrodes. Only visual/microscopic inspection and continuity tests passed electrodes were utilized for the subsequent biosensing applications.
Figure 2.
Overview of the screen-printing process. (a) Emulsion coated screen. (b) Printing of sensors on transparency sheet. (c) Final version of screen ready for printing various biosensors. (d) 5 IDE based microfluidic device printed on paper. (e) 6 IDE based microfluidic device printed on paper. (f) 6 IDE printed on PMMA.
2.3. Electrical impedance spectroscopy
Electrical impedance spectroscopy (EIS) is a sensing modality frequently utilized to measure the changes in electrical parameters. EIS is performed at room temperature and does not require any special arrangements. The electrodes play a vital role in this whole process starting from the application of the voltages, to the recording of variations in electrical conductivity. The interdigitated electrodes have been utilized to perform these important tasks in this research. Various number of electrodes were fabricated on two different substrates i.e. paper and PMMA using the screen-printing process in less than 30 minutes shown in (fig. 2(d, e, and f)). PMMA was selected as a final option for the fabrication of microfluidic devices due to its physical properties and improved limit of detection [18–20]. VLS 2.30 laser cutter (VersaLaser, Scottsdale, Arizona) was utilized to cut the PMMA and DSA sheets. The interdigitated electrodes were printed on the base layer PMMA of the last aqueous well of microfluidic device.
2.4. Functionalization of the beads with antibody:
Antibodies have demonstrated excellent detection capabilities [21]. Their characteristics like specificity and higher affinity make them potential candidate for the diagnostic applications.
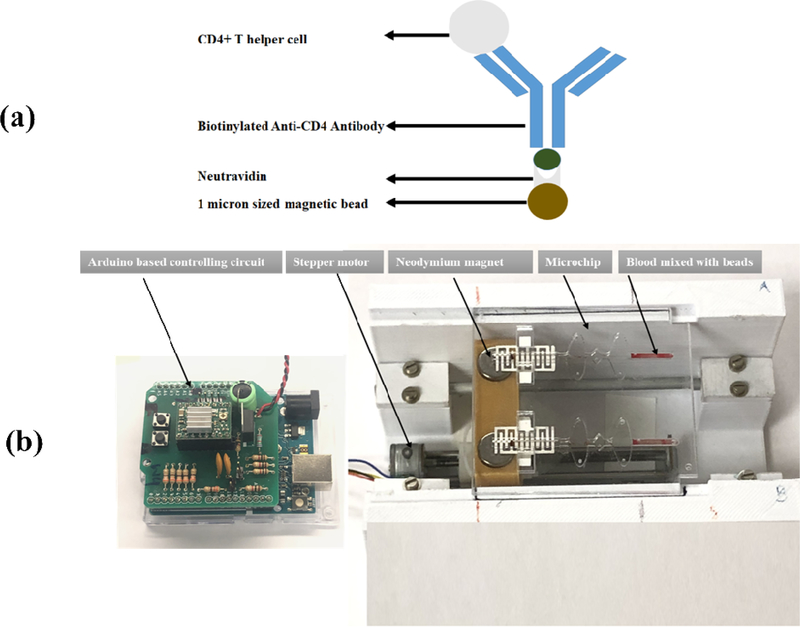
Sera-Mag SpeedBeads Neutravidin™-coated particles having mean diameter of 1 μm were conjugated with anti-CD4 antibody. In order to conjugate the anti-CD4 antibody to the magnetic particles, the strongest non-covalent biological interaction of neutravidin and biotin was utilized. Neutravidin has strong affinity for biotin. A schematic elaborates this concept (fig. 3a).
Figure 3.
(a) Schematic showing the conjugation of antibody to neutravidin coated (b) Flow-free fully automatic magnetic actuation platform loaded with a disposable microfluidic device with screen-printed electrodes.
100 μL of magnetic beads were gently mixed with 900 μL PBS. The clump of magnetic beads was separated with the help of neodymium magnet and a custom-made 3D printed stand. The supernatant was removed with the help of pipette and 1 mL fresh PBS was added to the beads. This process was repeated three times to thoroughly wash magnetic beads. 100 μL of 0.2 mg/mL monoclonal anti-CD4 antibody was added to the magnetic beads. The beads-antibody solution was incubated overnight on a rotator at 40 C. The antibody functionalized magnetic beads were washed three times using the abovementioned washing process. This washing process ensured the removal of any unbound antibody. The antibody conjugated magnetic beads were blocked using 3% bovine serum albumin. This blocking step was performed at room temperature for 1 hour to reduce the non-specific binding. The solution containing the magnetic beads immobilized with antibody and BSA was put on a rotator to ensure proper mixing. After this incubation process, three consecutive washing steps were performed. Magnetic beads were ready for downstream applications. The antibody functionalized magnetic beads were stored at 4 °C for later use.
2.5. Microfluidic chip fabrication and loading of reagents
The chip was fabricated using a non-lithographic process. PMMA and DSA were utilized to fabricate microfluidic device as previously reported [16, 22]. The aim was to come up with a microfluidic setup with static fluids while CD4+ T cells could move with the help of antibody conjugated beads using magnetic actuation. PMMA and DSA sheets were cut using laser cutter as per required dimensions. The microfluidic chip consists of three layers:
Base PMMA layer having thickness of 760 microns
Middle PMMA layer with thickness of 1.5 mm
Top PMMA layer with a thickness of 760 microns
These three layers were assembled together with the help of DSA to form a composite microchip. layer contained the inlets and outlets. The microfluidic chip consists of three types of wells;
(a) rectangular well, (b) elliptical wells, and (c) diamond shaped wells. The first rectangular shaped well was reserved for the capture of CD4+ T cells from whole blood. The whole blood sample was mixed with the antibody conjugated magnetic beads. Low-conductive washing solution consisting of 0.3% dextrose and 8.5% sucrose was filled in the diamond shaped wells. Microchip was designed to have an elliptical well between rectangular and diamond shaped wells. Mineral oil was filled in the elliptical shaped wells to prevent the mixing of aqueous reagents. Interdigitated finger electrodes were printed in the last well using in-house screen-printing technique. The last well containing 6 interdigitated electrodes was filled with 140 μL of nanopure water to ensure the lysis of captured CD4+ T cells. Supplementary table 2 presents the total associated costs of microchip and biological reagents.
2.6. Automatic magnetic actuation:
Magnetic beads offer great advantages in terms of their ease of actuation with the help of an external magnet. Here, in this project we have developed an automatic platform shown in (fig. 3(b)) to automate the whole process of CD4+ T cells isolation and their subsequent enumeration. The antibody functionalized magnetic beads were mixed with whole blood for a period of 3 minutes. CD4+ T cells were captured by antibody coated magnetic beads (Supplementary fig.1). This 3 minutes’ magnetic actuation was achieved by the back and forth motion of magnets. This actuation was accomplished with the help of magnets mounted on custom made housing underneath the wells. The magnets were very precisely actuated with the help of a stepper motor. This whole automatic process was performed with the help of Arduino Uno microcontroller. As a result of this automation, magnetic actuation process and subsequent CD4+ T cells capture with antibody conjugated magnetic beads process is highly accurate, precise, and reproducible. It also minimizes the human intervention to great extent.
2.7. Washing of magnetic beads cell complex with low conductive solution
To utilize electrical impedance spectroscopy, it is important to wash the isolated CD4+ T cells and beads mixture with a low conductive solution. Washing step helps to get rid of the erythrocytes and other types of non-desired cells and their respective debris present in the whole blood samples. The washing process was accomplished by the back and forth motion of magnets underneath the microfluidic wells. Washing step was of 1.5-minute duration.
2.8. Recording of electrical impedance spectra
The goal of this research is to reliably measure the number of CD4+ T cells using the cell lysate impedance spectroscopy. Nanopure water was selected as a lysing reagent based on its high electrical impedance (Supplementary fig.2). Magnetic beads captured CD4+ T cells were eventually moved to the last rectangular well that contained nanopure water. The interaction of isolated cells with nanopure water resulted in their lysis. The cell lysate changes the electrical conductivity of the solution according to the number of captured CD4+ T cells. Because of cells’ lysis, disintegration of the cell membrane occurred, and ions were released into the solution. This changed the conductivity of the water in accordance with the number of CD4 cells present in the blood sample. So, the corresponding change in the impedance of the solution could easily be measured with the help of E4980AL LCR machine (Agilent Technologies Inc., Palo Alto, CA, USA) (Supplementary fig.3). The LCR machine was connected to the interdigitated electrodes with the help of kelvin probes. The readings for impedance and phase spectra were recorded by automatically sweeping frequency from 20 Hz to 300 kHz. The electrical circuit model of the interdigitated electrodes containing the cell lysate solution and the simulated and experimental results of electrical impedance spectra for 400 cells per microliter are shown in (Supplementary fig. 4).
3. Results and discussion
3.1. Selection of the most sensitive interdigitated electrodes sensor
The sensitivity of 5 and 6 interdigitated electrodes was determined using various concentrations of PBS. The results shown in (Supplementary fig.5) clearly demonstrate that by increasing the interdigitated fingers of the sensors, better sensitivity was achieved [23–25]. It was even possible to reliably detect 0.0001% PBS solution using 6 interdigitated electrodes sensor printed on PMMA only.
3.2. CEM-CD4+T cells enumeration
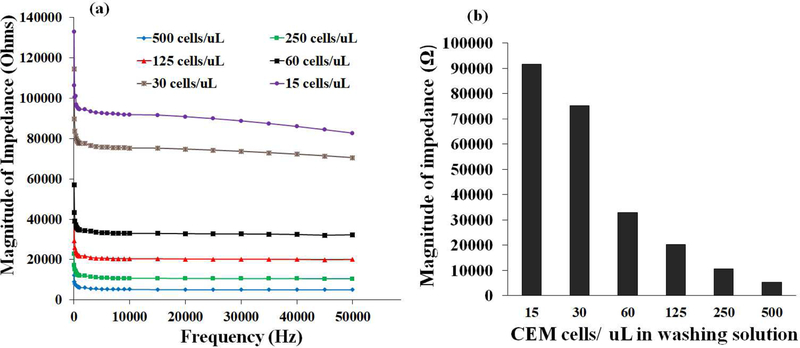
CEM-CD4+ T cells were grown in lab. The supplementary section provides the detailed information about CEM-CD4+ T cell culture. To validate the concept of electrical impedance sensing, we initially spiked the CEM-CD4+ T cells in low-conductive sugar solution. The morphology of cells was observed using an optical microscope (Nikon). Buffer solution was spiked with various CEM-CD4+ T cells to have multiple cell concentrations i.e. (500, 250, 125, 60, 30, and 15 cells per μL). Hemocytometer and Luna automated cell counter L10001 (Logos Biosystems) were used to confirm the number of CEMCD4+ T cells in each dilution. It is utmost important that each dilution must has specific number of CEM-CD4+ T cells. Then, each sample with pre-known cells concentration was loaded into the disposable microfluidic chip. Antibody conjugated magnetic beads were added to cell sample. These beads were mixed with cells using the back and forth motion of a magnet. This process was carried out for 3 minutes to ensure capture of CD4+ T cells from the spiked samples. The captured CEM cells along with magnetic beads were moved to washing well. Washing was performed to get rid of debris and any other unwanted entities. It is utmost important to select a low-conductive washing solution. The readings for magnitude of impedance of various washing solutions were recorded with this method (Supplementary fig.1). A solution consisting of 0.3% dextrose and 8.5% sucrose was selected for washing purposes due to its high magnitude of impedance [17]. Furthermore, it has been experimentally proven that cells remain intact in this washing solution (8.5% sucrose and 0.3% dextrose) for a period of 60 minutes [26]. The washing process was accomplished by the back and forth motion of magnets underneath the microfluidic wells. Washing step was of 1.5-minute duration. The washed CD4 cells were eventually taken to last well that contained nanopure water on the surface of interdigitated electrode sensor. The cell lysate impedance readings were recorded with the help of LCR equipment (fig. 4). It was observed that the different number of cells could be distinguished based on their respective impedance spectra. These initial findings confirmed the feasibility of utilization of electrical impedance spectroscopy along with the antibody conjugated magnetic beads-based cell isolation.
Figure 4.
(a) Magnitude of impedance Vs frequency graphs for various concentrations of CEM cells mixed in buffer solution. (b) Magnitude of electrical impedance for various concentrations of CEM cells at 10 kHz
3.3. CD4+ T cells counting from the whole blood
The next step involved the testing of whole blood to enumerate the CD4+ T cells. To accomplish this, de-identified whole blood from healthy donors was obtained from a local blood bank (Continental Services Group, Inc. Miami, USA). The blood specimens were collected in K2 anticoagulant EDTA tubes. All samples were tested within 3 days of blood withdrawal.
The initial CD4 count in whole blood was determined using two pre-existing well-established methods.
EasySep Direct Human CD4 T cell isolation kit (Stem Cell Technologies, 19662) was utilized as per manufacturer protocol. The isolated CD4+ T cells were stained with DAPI and Alexa Flour 488 (AF488) mouse anti-human CD4 antibody to confirm their identity.
Direct staining of CD4+ T cells from whole blood after lysing RBC. ACK lysis buffer was used to lyse RBC. The solution was centrifuged at 500 g for 5 minutes. After removing supernatant, cell pellet was suspended in the PBS solution. Cells were stained with DAPI for 10 minutes in dark and AF-488 conjugated anti-CD4 antibody for 90 minutes at room temperature. The bright field and fluorescent images of CD4+ T cells isolated from whole blood were recorded (Supplementary fig.6).
After validating the identity of the captured CD4+ T cells from both methods, their counting was done using a hemocytometer under an optical microscope. Three different readings were recorded for each sample and then average was used. LUNA-FL™ dual fluorescence cell counter was also used to validate these counts.
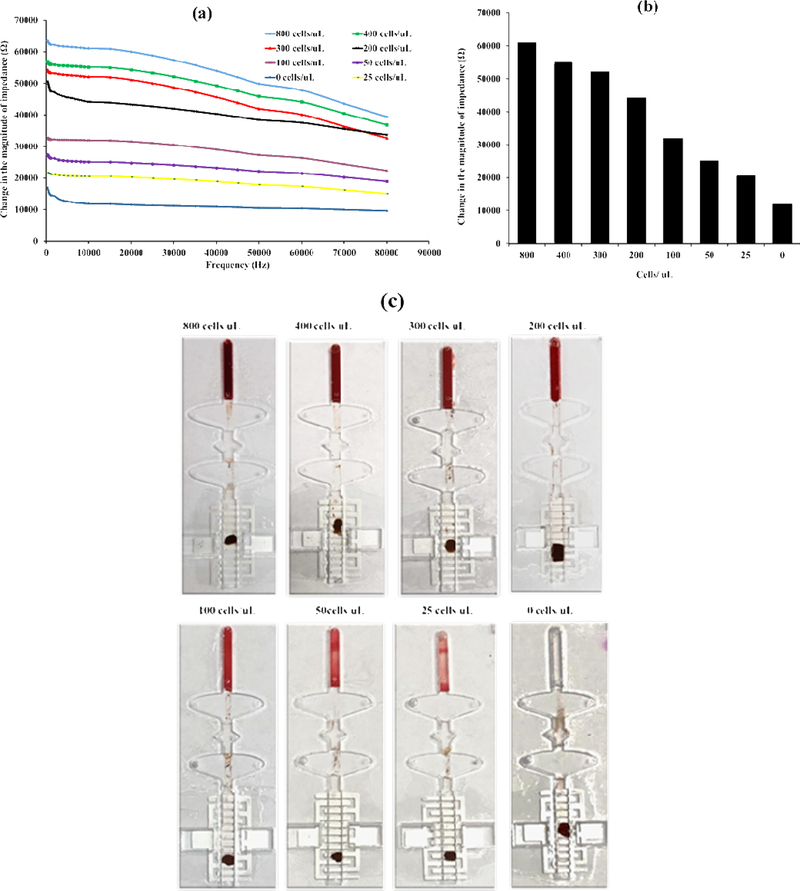
As of now initial CD4 counts were known, so, various dilutions of blood were made, and samples were tested using our developed setup. Initially, the impedance of sensor having nanopure water was measured. After this the CD4 capture experiment was conducted. The recording of the developed immunoassay is presented in supplementary video 1. The cells of interest were isolated with antibody conjugated magnetic beads, then cells-beads complexes were washed with low-conductive washing solution. Eventually, the washed sample was brought to the last well. This chamber contained 6 interdigitated finger electrode sensor immersed with nanopure water. As a result of the exposure of captured cells to this nanopure water, cell lysis occurred, and the impedance of the solution was changed. The electrical impedance readings were recorded. As we already knew the initial impedance of the nanopure water, and now we determined the impedance after the introduction of captured lysed cells. The difference of these two readings gave the impedance of the isolated cells shown in (Fig. 5(a)). The change in the magnitude of electrical impedance for various concentrations of cells at one specific frequency i.e. 10 kHz are plotted in (Fig. 5(b)). It was observed that the change in the magnitude of impedance relates to the to the number of CD4+ T cells present in the test sample. Images of the microchip after the successful completion of immunoassay are presented in (Fig. 5(c)). The utilization of highly specific monoclonal antibody has resulted in capture efficiency of 95% and 90% for spiked CEM-CD4+ T cells and CD4+ T helper cells obtained from whole blood respectively.
Figure 5.
(a) Change in the magnitude of impedance vs frequency graphs for various concentrations of CD4+ T cells. (b) Change in the magnitude of electrical impedance for various concentrations of CD4+ T cells at 10 kHz. (c) Images of the microfluidic chips after the successful completion of the immunoassay.
The performance of developed assay was also evaluated using de-identified clinical whole blood samples. Three different whole blood samples from healthy individuals were used to perform CD4 testing and validation. CD4 counts were obtained by our developed method and the measured readings showed good match with the CD4 counts obtained by the conventional hemocytometer based fluorescently tagged CD4 cells counting method. Table 1 enlists the results. To check the specificity of the immunoassay, the developed assay was also tested with Human Umbilical Vein Endothelial Cells (HUVECs) as a negative control. None of the HUVECs cells were captured. The magnitude of impedance resembled closely with the one already recorded for 0 cells per μL. This indicated high specificity of the developed assay. We have also performed the marker staining of the HUVEC cells using AF488 conjugated anti-CD4 antibody. It was found that none of these cells provided green fluorescent emissions. It is a matter of fact that HUVECs cells do not express CD4 glycoprotein. Our results obtained by the developed cell lysate impedance spectroscopy method also validated this concept.
Table 1:
Comparison of CD4 + T cells count results from whole blood using the developed and conventional methods.
| Sample ID | Conventional method to count fluorescently tagged CD4+ T cells using hemocytometer under microscope. (cells /μL) (Source= whole blood) |
Standard deviation of conventional method to count fluorescently tagged CD4+ T cells using hemocytometer under microscope. | Developed method (cells /μL) (Source= whole blood) |
Standard deviation of developed method |
|---|---|---|---|---|
| Blood Sample 1 | 792 | 5.42 | 794 | 7.31 |
| Blood Sample 2 | 250 | 6.97 | 247 | 12.72 |
| Blood Sample 3 | 572 | 19.61 | 588 | 17.2 |
To test the repeatability of the developed CD4 counting method, a de-identified blood sample was tested. Both channels of the microfluidic chip were loaded with the blood from the same source. The immunoassay was carried out in exactly similar conditions. The cell lysate impedance readings were recorded. The change in the magnitude of electrical impedance measured using both sensors, and the readings were consistent (47376.16 Ω and 45589.4303 Ω), and it was compared with the standard readings of cell lysate impedance (Fig. 5b). The developed method gave CD4+ T count of 250 cells per μL. The results were validated by performing an independent CD4 count using the EasySep Direct Human CD4 T cell isolation kit and manual cell counting. The second method provided 245 cells per μL.
The developed fully automatic CD4+ T cells counting immunoassay platform requires only 12V DC to operate. Hence, there is no need of continuous electric supply and portable batteries can be utilized for successfully running the assay. This flexible requirement makes it more suitable for field testing in resource-constrained settings.
4. Discussions
As per recent guidelines of WHO, ART should be initiated immediately after the diagnosis of HIV. HIV virus attacks CD4 cells. So, as a result CD4 cells start decreasing. The range of CD4+ T lymphocytes in healthy individuals is 500–1600 cells per μL [15]. When the CD4 count drops below a certain threshold number i.e. 200 cells per μL, a patient progresses to advanced stage of disease called Acquired Immunodeficiency Syndrome (AIDS). This depletion of CD4 cells can also lead to other opportunistic infections and eventually cause death. Antiretrovirals (ARVs) are prescribed for the prevention and treatment of HIV. CD4 count is an important tool to measure the effectiveness of the ART in HIV infected people. There is a dire need to develop immunoassays to accurately count CD4 cells, especially for the disease prevalent resource-constrained settings. The desired technology must fulfill some basic requirements like cost-effectiveness, rapid, reliability and accuracy. The overall goal of this research is to achieve these requirements using the integration of the printed electronics with microfluidic technology. The interdigitated electrodes are fabricated on the bottom layer of microfluidic chip and a fully enclosed and disposable device is developed for the counting of CD4 cells. These microfluidic devices provide an inexpensive, portable, and environment friendly solution for CD4 enumeration. The whole idea was to come up with a system comparable with the conventional CD4 cell counting methods. The features like cost-effectiveness, portability, and ease of use make it an ideal candidate for the POC settings, and other rural areas clinics that lack basic facilities. This technology can also be utilized to capture other cells from a drop of whole blood. The only modification required would be the change of antibody.
It is a platform technology that can be applied to other diagnostic applications like the spread of infectious diseases. Time is a very crucial element in such scenarios to curtail the spread of diseases and developed technology can significantly help in developing rapid diagnostics devices.
We presented a highly sensitive method for the quantification of CD4+ T cells from whole blood at the resource-limited settings. The CD4+ T cells were captured from a 30 μL of whole blood. It is convenient to obtain such a small volume of blood from patients. Electrical impedance spectroscopy was used to enumerate the captured CD4+ T cells. The electrical impedance changes according to the number of cells present in blood. Captured cells release ions in their lysed state. The developed assay is fully automatic, and thus minimizes the role of skilled technicians. It also reduces the chances of any possible contamination. Anyone with some basic knowledge of pipetting can perform the assay. The user or healthcare workers just need to fill the wells in the presented microfluidic chip. These wells can be prefilled and stored at 4 °C for later use. The developed assay is fully automatic and flow-free. It does not require any human assistance during the test. The whole process is accomplished within five minutes. It is a matter of fact that flow-based assays require expensive pumps to maintain a constant flow conditions, and the resulting high shear stress also adversely affects the capture efficiency. Here, we have designed and developed a cost-effective magnetic actuation platform integrated with disposable microfluidic devices. To ensure the validity of the assay, each microfluidic device has two exactly similar microchannels; one is reserved for sample and other is used as a control. This ensures that control is subjected to exactly similar conditions and processes. The provision of control ensures the validity of immunoassay. The developed device can accurately count CD4+ cells up to 25 cells per μL. The developed system provides a cost-effective and rapid (5-minute sample-in-answer-out) solution for POC CD4 counting as compared to the various existing technologies.
5. Conclusions
We developed a flow-free microfluidic device for the capture and enumeration of CD4 cells from whole blood. The automatic magnetic actuation is performed for the isolation, washing, and later quantification of CD+ T cells. It is for the first time as per the best of our knowledge that a fully automatic system is developed where on chip electrical impedance sensing is used along with the magnetic bead-based cell manipulations. Electrical impedance spectroscopy was utilized to rapidly quantify CD4+ T cells. The experimental findings demonstrated that CD4+ T cells can be specifically isolated from a mixed population of cells using antibody conjugated magnetic beads and subsequently quantified using cell lysate impedance spectroscopy. This method is well suited for resource-limited settings with very short turnaround time (five minutes) for the detection of CD4+ T cells from 30 μL whole blood. The developed proof of concept holds great potential to effectively monitor the HIV treatment in an efficient and reliable manner especially in resource-constrained areas. It is envisioned that this presented microfluidic platform technology would enable rapid and cost-effective CD4 counting in POC settings.
Supplementary Material
Cost-effective and rapid assays for the accurate counting of CD4 cells.
CD4 enumeration is still a reliable indicator of the disease staging and can be employed to assess risk of death for patients with advanced level of disease
A fully automatic and highly sensitive electrical detection method to enumerate CD4 cells.
A field deployable platform technology for CD4+ T cells counting in the resource-limited settings.
Acknowledgements
We acknowledge research support from NIH R15AI127214, Institute for Sensing and Embedded Networking Systems Engineering (I-SENSE) Research Initiative Award, FAU Faculty Mentoring Award, Humanity in Science Award, and a start-up research support from College of Engineering and Computer Science, Florida Atlantic University, Boca Raton, FL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References:
- 1.UNAIDS. Global HIV & AIDS statistics — 2018 fact sheet. 2018. [cited 2019 04/01/2019]; Available from: http://www.unaids.org/en/resources/fact-sheet.
- 2.Wang S, et al. , Micro-a-fluidics ELISA for Rapid CD4 Cell Count at the Point-of-Care. Scientific reports, 2014. 4: p. 3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shafiee H, et al. , Paper and flexible substrates as materials for biosensing platforms to detect multiple biotargets. Scientific reports, 2015. 5: p. 8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asghar W, et al. , Engineering long shelf life multi-layer biologically active surfaces on microfluidic devices for point of care applications. Scientific reports, 2016. 6: p. 21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sher M, et al. , based analytical devices for clinical diagnosis: recent advances in the fabrication techniques and sensing mechanisms. Expert review of molecular diagnostics, 2017. 17(4): p. 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coarsey CT, et al. , Strategies in Ebola virus disease (EVD) diagnostics at the point of care. Critical reviews in microbiology, 2017. 43(6): p. 779–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruhl FJ, Rapp BE, and Länge K, Biosensors for diagnostic applications, in Molecular Diagnostics. 2011, Springer; p. 115–148. [DOI] [PubMed] [Google Scholar]
- 8.Organization, W.H., Guidelines for managing advanced HIV disease and rapid initiation of antiretroviral therapy, July 2017. 2017. [PubMed]
- 9.Ford N, et al. , The future role of CD4 cell count for monitoring antiretroviral therapy. The Lancet Infectious Diseases, 2015. 15(2): p. 241–247. [DOI] [PubMed] [Google Scholar]
- 10.Thorslund S, et al. , Bioactivated PDMS microchannel evaluated as sensor for human CD4+ cells—The concept of a point-of-care method for HIV monitoring. Sensors and Actuators B: Chemical, 2007. 123(2): p. 847–855. [Google Scholar]
- 11.Silveira CM, Monteiro T, and Almeida MG, Biosensing with paper-based miniaturized printed electrodes–a modern trend. Biosensors, 2016. 6(4): p. 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Economou A, Kokkinos C, and Prodromidis M, Flexible plastic, paper and textile lab-on-a chip platforms for electrochemical biosensing. Lab on a Chip, 2018. 18(13): p. 1812–1830. [DOI] [PubMed] [Google Scholar]
- 13.Rapp BE, Gruhl FJ, and Länge K, Biosensors with label-free detection designed for diagnostic applications. Analytical and bioanalytical chemistry, 2010. 398(6): p. 2403–2412. [DOI] [PubMed] [Google Scholar]
- 14.Wasserberg D, et al. , All-printed cell counting chambers with on-chip sample preparation for point-of-care CD4 counting. Biosensors and Bioelectronics, 2018. 117: p. 659–668. [DOI] [PubMed] [Google Scholar]
- 15.Kanakasabapathy MK, et al. , Rapid, label-free CD4 testing using a smartphone compatible device. Lab on a Chip, 2017. 17(17): p. 29100–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coarsey C, et al. , Development of a flow-free magnetic actuation platform for an automated microfluidic ELISA. RSC Advances, 2019. 9(15): p. 8159–8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng X, et al. , Cell detection and counting through cell lysate impedance spectroscopy in microfluidic devices. Lab on a Chip, 2007. 7(6): p. 746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo S, et al. , Microbubbles loaded with nickel nanoparticles: A perspective for carbon sequestration. Analytical chemistry, 2017. 89(20): p. 10827–0833. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, et al. , Integration of cell phone imaging with microchip ELISA to detect ovarian cancer HE4 biomarker in urine at the point-of-care. Lab on a Chip, 2011. 11(20): p. 3411–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rappa K, et al. , Quantitative analysis of sperm rheotaxis using a microfluidic device. Microfluidics and Nanofluidics, 2018. 22(9): p. 100. [Google Scholar]
- 21.Salvador J-P, Vilaplana L, and Marco M-P, Nanobody: outstanding features for diagnostic and therapeutic applications. Analytical and bioanalytical chemistry, 2019: p. 1–11. [DOI] [PubMed] [Google Scholar]
- 22.Asghar W, et al. , Microfluidic Chip for Detection of Fungal Infections. ACS omega, 2019. 4(4): p. 7474–7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Draz MS, et al. , Hybrid Paper–Plastic Microchip for Flexible and High Performance Point of Care Diagnostics. Advanced Functional Materials, 2018: p. 1707161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Safavieh M, et al. , Paper microchip with a graphene-modified silver nano-composite electrode for electrical sensing of microbial pathogens. Nanoscale, 2017. 9(5): p. 1852–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safavieh M, et al. A graphene-modified cellulose paper microchip for HIV detection in Advances in Global Health through Sensing Technologies 2015. 2015. International Society for Optics and Photonics. [Google Scholar]
- 26.Chiou PY, Ohta AT, and Wu MC, Massively parallel manipulation of single cells and microparticles using optical images. Nature, 2005. 436(7049): p. 370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.