Abstract
AIMS:
Skeletal muscle wasting is associated with many chronic diseases. Effective prevention and treatment of muscle wasting remain as a challenging task due to incomplete understanding of mechanisms by which muscle mass is maintained and regulated. This study investigated the functional role of Ubiquitin C-terminal hydrolase L1 (UCHL1) in skeletal muscle.
MAIN METHODS:
Mice with skeletal muscle specific gene knockout of UCHL1 and C2C12 myoblast cells with UCHL1 knockdown were used. Muscle fiber types and size were measured using tissue or cell staining. The mammalian target of rapamycin complex 1 (mTORC1) and mTORC2 activities were assessed with the phosphorylation of their downstream targets.
KEY FINDINGS:
In mouse skeletal muscle, UCHL1 was primarily expressed in slow twitch muscle fibers. Mice with skeletal muscle specific knockout (skmKO) of UCHL1 exhibited enlarged muscle fiber sizes in slow twitch soleus but not fast twitch extensor digitorum longus (EDL) muscle. Meanwhile, UCHL1 skmKO enhanced mTORC1 activity and reduced mTORC2 activity in soleus but not in EDL. Consistently, in C2C12 cells, UCHL1 knockdown increased the myotube size, enhanced mTORC1 activity, and reduced mTORC2 activities as compared with control cells. UCHL1 knockdown did not change the major proteins of mTOR complex but decreased the protein turnover of PRAS40, an inhibitory factor of mTORC1.
SIGNIFICANCE:
These data revealed a novel function of UCHL1 in regulation of mTORC1 activity and skeletal muscle growth in slow twitch skeletal muscle. Given the upregulation of UCHL1 in denervation and spinal muscle atrophy, our finding advances understanding of regulators that are involved in muscle wasting.
Keywords: UCHL1, skeletal muscle, mTOR, mice, C2C12 cell
Introduction
Skeletal muscle is the most abundant tissue in the body, accounting for over 40% of total body mass. Not only responsible for the body locomotion and posture, skeletal muscle is also a critical determinant of metabolism and energy homeostasis in physiological [1, 2] and pathophysiological conditions [3]. Muscle mass loss, or muscle wasting, is associated with a variety of chronic diseases, including denervation, diabetes, heart failure, chronic obstructive pulmonary disease (COPD), and cancer (cachexia) [4]. Effective prevention and treatment of muscle wasting remain a challenge due to the incomplete understanding of the underlying mechanisms [5, 6].
Skeletal muscle growth, mass maintenance, and appropriate function are controlled by multiple factors and signaling pathways. A well-recognized signaling pathway that is involved in muscle growth and metabolism is mammalian target of rapamycin (mTOR) [7]. mTOR is a conserved serine/threonine kinase that is broadly involved in cell survival, growth, metabolism and energy balance [8, 9]. mTOR forms two distinct complexes (mTORC) that possess different functions. Raptor containing mTORC1 promotes protein and lipid synthesis, and inhibits autophage [10], [11, 12]. Rictor-containing mTORC2 receives growth factor stimulation to regulate cell survival and proliferation [10].
Ubiquitin C-terminal hydrolase L1 (UCHL1) was originally identified as a neuronal protein that accounts for nearly 2% of total brain proteins [13]. Functionally, UCHL1 is a deubiquitinating enzyme (DUB) but its functional importance and substrate proteins are still poorly understood [14]. Reduced hydrolytic activity of mutant UCHL1 is implicated in the pathophysiologic process of Parkinson’s [15] and Alzheimer’s disease [16] due to abnormal neurotoxic protein aggregation [17, 18]. Our lab has reported that UCHL1 regulates acetylcholine transporter cytosol-plasma membrane translocation in cholinergic cells [19]. UCHL1 is also found to be highly expressed in some cancer cells to promote cell proliferation and metastasis [20].
Expression of UCHL1 in skeletal muscles was unknown until recently. A few studies have reported the expression of UCHL1 in mouse skeletal muscles in some disease conditions [21, 22]. Our recent study showed that UCHL1 was expressed in C2C12 mouse myoblast cells and was involved in regulation of cell proliferation and differentiation [23]. The function of UCHL1 in skeletal muscles, however, remains to be fully understood. To this end, we have recently developed a mouse line with skeletal muscle specific knockdown (skmKO) of UCHL1. Here we report novel observations of UCHL1 distribution in skeletal muscle and the effects of UCHL1 skmKO on muscle mass and mTORC1 activity.
Methods
Animals and Genotyping
Use of animals in this study were in compliance with NIH guidelines and were reviewed and approved by the University of South Dakota Institutional Animal Care and Use Committee Mouse strain “UCHL1 HEPD0603 _7_h04” generated from EUCOMM/KOMP-CSD ES cells [24–26] was provided by Medical Research Council (MRC)-Harwell, UK, on behalf of the European Mouse Mutant Archive (EMMA) and the International Mouse Phenotyping Consortium (IMPC, www.mousephonogype.org). By crossing this strain with a mouse expressing Flp recombinase, the targeting cassette (LacZ and Neo genes) were removed. Further breeding with wild type (WT) mice separated the Flp transgene out, the resulting strain carrying the exon 2 floxed UCHL1 gene and free of Flp was then crossed to a mouse expressing Cre recombinase driven by the myosin light chain 1 promoter (Myl1tm1(cre)sjb/J, Jackson Lab, stock # 024713) [27] to generate a mouse strain with skeletal muscle specific UCHL1 knockout (skmKO). Genotyping was conducted using PCR with appropriate primers for floxed UCHL1 and Cre. UCHL1 skmKO and WT mouse littermate were used in this study.
Sciatic Denervation
The sciatic nerve of the hindlimb was severed in 8 to 12 week old male and female mice as described previously [28]. Briefly, mice were anesthetized with 2% isoflurane in oxygen. The right hindlimb was aseptically prepared. A small incision was made in the skin and the muscles of the hindlimb gently retracted to reveal the sciatic nerve. Approximately 3 mm of sciatic nerve was removed using micro-scissors. The skin incision was sealed using surgical glue and Buprenorphine analgesia was given subcutaneously. The contralateral hindlimb was left intact as the control. After two weeks, under isoflurane anesthesia, muscles from denervated and control hindlimb were collected.
Tissue collection
As described previously [28], male mice were anesthetized with isoflurane. Hindlimb muscles were carefully exposed by removing skin. Soleus and extensor digitorum longus (EDL) were collected and frozen on dry ice for Western blot assay. Gastrocnemius muscle were isolated and snap frozen in pre-chilled 2-methylbutane for ~20 seconds [29] and transferred to −70 °C until sectioning. Mid-belly region of muscles was sectioned into 8.5 μm thickness and adhered on the slides. The slides were stored at −70 °C for future staining.
Immunohistochemistry staining
Frozen sections were left at room temperature for 20 min, and then incubated in phosphate buffered saline-Tween 20 (PBST) solution for 5 min. Sections were treated with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for 10 min and rinsed with PBST for 5 min. Sections were microwaved in sodium citrate for 20 min. After 20 min siting at room temperature, sections were rinsed with PBST for 5 min. Sections were blocked with 3% bovine serum albumin (BSA) and 10% AffiniPure Goat Anti-Mouse IgG + IgM Fab (H+L) (Jackson ImmunoResearch, 115-005-044) PBST solution for 1 h at room temperature. After washed with PBST for 5 min, sections were incubated with primary antibodies diluted in 3% BSA and 5% Donkey Serum PBST solution at 4 °C for overnight. Sections were then washed with PBST for 3 times, 5 min each, and then incubated with secondary antibodies conjugated with fluorescent dye Alexa 594 or Alexa 488 diluted in 3% BSA PBST (1:400) for 1 hour at room temperature. Sections were then incubated in PBST containing Hoechst (1:15 000) for 5 min, followed by wash with PBST 2 times, 5 min each. The sections were mounted with coverslips using Fluomount-G solution (Southern Biotech, 0100–01). Immunostaining images were taken using a confocal microscope (Olympus FluoView 500, Center Valley, PA).
Hematoxylin and Eosin (H&E) staining
Muscle sections were dried, recovered at room temperature for 20 min, and subjected to H&E staining using standard procedure [30]. Slides were immersed in hematoxylin solution for 10 min. After thoroughly washing with tap water, slides were stained with Eosin solution for 3 min, and successively transferred in 70% ethanol for 20 sec, 90% ethanol for 20 sec, 100% ethanol for 1 min, and xylene for 3 min. The slides were mounted with coverslips using DPX Mountant for histology media (Sigma, 06522) and imaged.
Measurement of fiber size
Cross-sections of muscles from 3-month-old mice were immunofluorescence-stained for dystrophin or laminin. Four digital images (X 10 or X 20) were taken from non-overlapping areas from each slide, and muscle fiber size was measured from roughly 150 myofibers in three representative fields in each animal of 5 WT mice and 3 UCHL1 skmKO mice using Image J software. The same approach was used to measure myotube width of differentiated C2C12 cells.
Cell culture
As previously described [23], C2C12 myoblasts (1.5×105/ml) were seeded in the growing medium (GM), Dulbecco’s Modified Eagle’s Medium (DMEM, ATCC) supplemented with 10% fetal bovine serum (FBS, GE HyClone, Logan, UT) and 1% penicillin-streptomycin and Hepes. Medium was changed every 24 h until cells grew to 80–90% confluence (~2 days growing in GM). The cells were then transferred to differentiating medium (DM) DMEM (GE HyClone) supplemented with 2% horse serum (GE HyClone) and 1% penicillin-streptomycin and Hepes. Medium were changed in every 24 h.
Knockdown of UCHL1 gene expression
For UCHL1 gene knockdown, 24 h after seeding in GM, cells were treated with siRNA mixture as previously reported [23] with minimal modification. Media were changed to half amount of fresh GM, and siRNA mixed with Lipofectamine RNAiMAX was added into media with a final concentration of 10 nM. After 24 h, medium was changed to fresh DM and cells were grown for desired periods.
Before harvest, cells were rinsed once with warm serum free medium and transferred to serum free medium. Some cells were treated with 100 nM insulin for 10 min.
Total protein extraction and Western blot analysis
As described previously [28], cells or tissues were homogenized in 1X RIPA buffer containing protease inhibitor cocktail (Santa Cruz, Dallas, TX) and 2% phosphatase inhibitor (Research Product International, Mount Prospect, IL). Protein concentration of samples were adjusted to 1μg/μL by a standard BCA assay, and 15 μg of proteins were fractionated in 9–16% gradient gels under 90 V for 3 h. Proteins were transferred onto Nitrocellulose membranes using a Trans-blot apparatus (Bio-Rad, Hercules, CA), and blocked with 3% non-fat milk in PBST solution for 1 h. Proteins were detected by using primary antibodies for overnight incubation at4 °C. The primary antibodies used for Western blot in this study: anti UCHL1 (Abcam, EPR4118, ab108986), anti actin (Santa Cruz Biotech, 2Q1055, sc-58673), and anti GAPDH (Santa Cruz BioTech, 0411, sc-47724), and following antibodies from Cell Signaling Technologies: anti phosphor-AKP473 (D9E, #4060); anti total AKT (40D4, #2920), anti phosphor-S6K389 (1A5, #9206), anti S6K (49D7, #2708), anti mTOR (7C10, #2983), anti raptor (24C12, #2280), anti rictor (53A2, #2114), and anti PRAS40 (#2610) The appropriate secondary antibodies conjugated with Alex-680 or 800 (Invitrogen) were used to recognize primary antibodies and scanned with LI-COR scanner (LI-COR Biosciences, Lincoln, NE). The band density was quantified by subtracting background using LI-COR Image Studio software. The ratio of the band density of the protein of interest vs loading control protein (actin or GAPDH) of each sample was calculated. For protein phosphorylation, the ratio of the band density of the phosphorylated protein vs the total protein of each sample was calculated. To normalize the data from different batches of Western blot, the ratios to the WT control group of the same batch were used for comparison.
Data analysis
Data calculation, graphing and descriptive statistics were performed using Microsoft Excel Data Analysis Package or GraphPad Prism 7.03. Data were presented as Mean ± Standard Error of the Mean (SEM). Statistical significance was compared by one-factor analysis of variance (ANOVA) or two-way ANOVA followed by two-tailed Student’s t-test. Statistical significances were defined as p value less than 0.05.
Results
1. UCHL1 is primarily expressed in oxidative muscle fibers
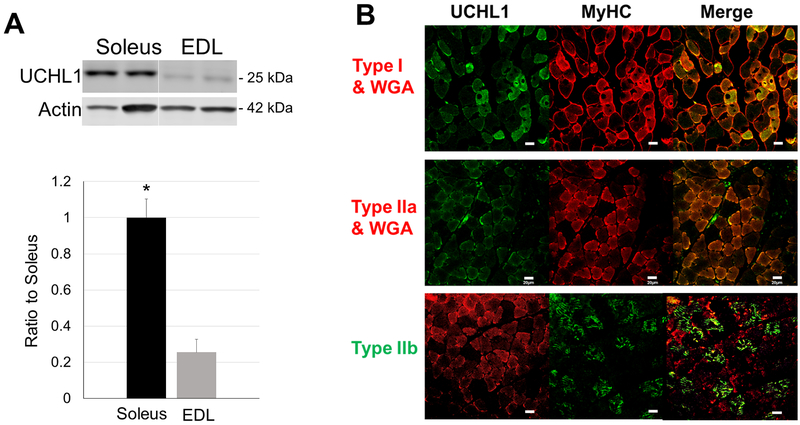
Examining the expression of UCHL1 in mouse skeletal muscles, we observed that UCHL1 expression was muscle type specific. Western blot analyses showed a much higher level of UCHL1 proteins in slow oxidative muscle soleus than in the fast-glycolytic muscle extender digitorum longus (EDL) (Fig. 1A). Immunofluorescent staining further identified that UCHL1 was highly expressed in type I (slow oxidative) muscle fibers, moderately expressed in type IIa (fast oxidative) fibers, but undetectable in type IIb (fast glycolytic) fibers (Fig. 1B). These data indicate that UCHL1 is primarily expressed in oxidative muscle fibers.
Figure 1: UCHL1 is selectively expressed in oxidative muscle fibers.
(A) Representative images of Western blot and quantifications of UCHL1 signal from soleus and EDL normalized with loading control Actin. “*” indicates p < 0.05 as compared to Soleus, n = 8 for Soleus, n = 6 for EDL. (B) Fluorescent staining of gastrocnemius from wild type mice. Immunohistochemistry tissue sections were fluorescently stained with primary antibodies against type I, IIa and IIb myosin heavy chain and UCHL1 followed by appropriate fluorescently conjugated secondary antibodies. The scale bar is 20 μm.
2. UCHL1 skmKO increases muscle fiber size
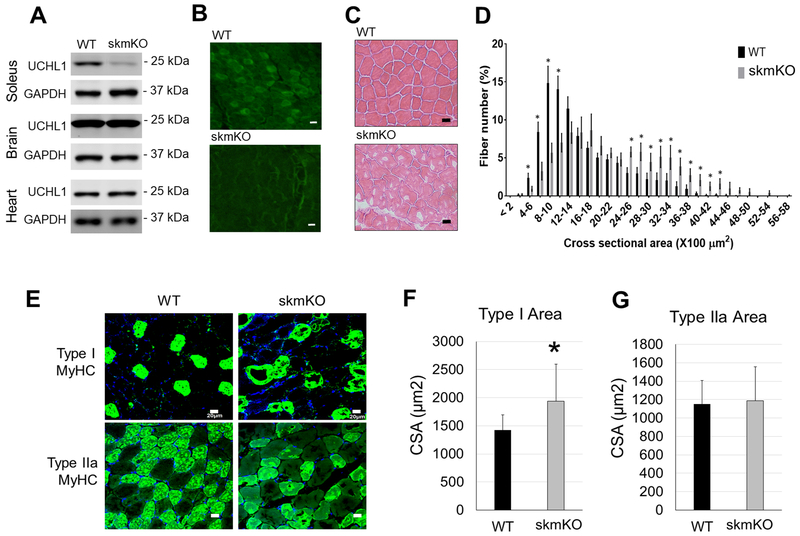
To investigate the functional role of UCHL1 in skeletal muscle, we have generated a mouse line with UCHL1 skmKO. Western blotting and immunofluorescent staining confirmation showed that UCHL1 expression was specifically knocked out in skeletal muscles but not in the brain and the heart (Fig. 2A and 2B). At 2 to 3-months of age, mice with UCHL1 skmKO developed normally without any observable abnormalities. However, H&E staining of gastrocnemius muscle sections revealed enlarged muscle fibers in muscle from skmKO mice as compared to the WT control muscle (Fig. 2C). The distributions of muscle fiber size (cross sectional areas, CSA) of UCHL1 skmKO muscles was shifted rightward. The number of smaller muscle fibers was significantly reduced, and the number of larger fibers was significantly increased in skmKO as compared with WT muscles (Fig. 2D). This indicates that loss of UCHL1 in skeletal myocytes leads to muscle hypertrophy. Noticeably, the hypertrophic fibers were primarily type I fibers (Fig. 2D, F and G), consistent with the UCHL1 expression pattern. Interestingly, low tinctorial areas of H&E staining as well as immunofluorescent staining were often observed in the skmKO muscle. Whether those represent alterations of muscle fiber composition or deposition remain to be further investigated.
Figure 2: UCHL1 skmKO leads to larger fiber size.
(A) Representative images of Western blots for UCHL1 and GAPDH in soleus, brain and heart from WT-Cre and UCHL1 skmKO mice. (B) Representative immunofluorescent staining of UCHL1 in gastrocnemius muscle from WT and UCHL1 skmKO mice. (C) Representative micrographs of H&E staining of gastrocnemius from WT and UCHL1 skmKO mice. (D) Quantification of gastrocnemius muscle fiber cross-section area of 150 fibers in three areas from 5 individual WT mice and 3 individual UCHL1 skmKO mice. “*” indicates p< 0.05. (E) Immunofluorescent staining of Type I or IIa MyHC in gastrocnemius from WT or UCHL1 skmKO mice. (F) and (G) Measurement of cross-section areas of type I (E) and Type IIa (F) MyHC positive muscle fiber, with 6 to 10 fibers per mouse from WT mice (n=4 mice) and UCHL1 skmKO mice (n=3 mice), *p < 0.05 vs. WT, one-way ANOVA followed by t-test. The scale bars in the images indicate 20 μm.
3. UCHL1 skmKO enhances mTORC1 activity in soleus in response to insulin stimulation
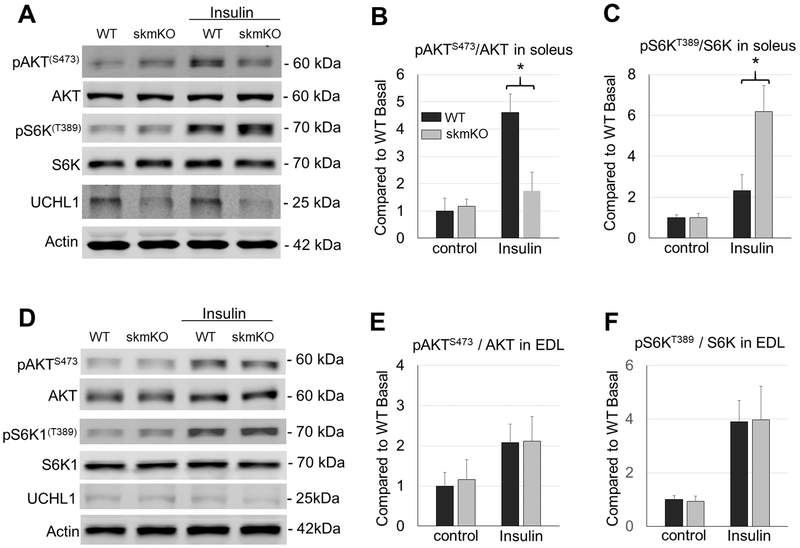
In search for potential signaling pathways that are involved in the UCHL1 skmKO-induced muscle hypertrophy, we found that, although the basal level was unchanged, phosphorylation of p70 ribosomal S6 Kinase at T389 (pS6KT389) by mTORC1 in response to insulin stimulation was significantly elevated in UCHL1 skmKO muscle, (Fig. 3A and B). In contrast, AKT phosphorylation at S473 (pAKTS473) by mTORC2 was significantly inhibited (Fig. 3A and C) in UCHL1 skmKO muscle in response to insulin, compared to WT muscles.
Figure 3: UCHL1 skmKO enhances mTORC1 activity.
(A, D) Representative images of Western blots for pAKTS473, AKT, pS6KT389, S6K, UCHL1, and β-Actin in soleus (A) or EDL (D) from WT and UCHL1 skmKO mice under 4 h of starvation with or without 1.5 IU/kg insulin stimulation for 15 min. Note that some representative protein bands were selected from different gels. (B-C, and E-F) Quantifications of Western blots for pS6KT389/S6K (B, E), and pAKTS473/AKT (C, F) in soleus or EDL. Results were normalized to WT Basal level, black bars are WT mice and grey bars are UCHL1 skmKO mice. “*” indicates p < 0.05, as compared between WT and skmKO group (genotype effect), by two-way ANOVA followed by t-test; n = 6 for each group.
In contrast, phosphorylation of S6KT389 by mTORC1 and pAKTS473 by mTORC2 was comparable in fast twitch muscle EDL from WT and UCHL1 skmKO mice upon insulin stimulation (Fig. 3D, E and F). This result was consistent with the lack of UCHL1 expression in the fast glycolytic muscles.
Overall, these results suggest that UCHL1 elicits a negative effect on mTORC1 and a positive effect on mTORC2 activity in slow twitch skeletal muscle.
4. UCHL1 knockdown increases myotube hypertrophy and mTORC1 activity in C2C12 cells
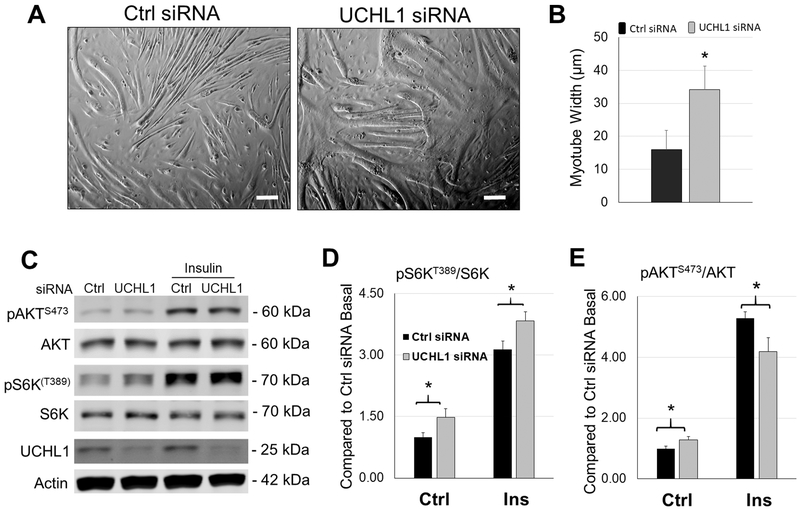
To confirm the in vivo findings in UCHL1 skmKO mouse muscle, we tested the effect of UCHL1 in differentiated C2C12 myotubes. Consistent with the in vivo findings, in differentiated C2C12 cells, UCHL1 gene knockdown (KD) using siRNA significantly increased the width of myotubes as compared to the cells treated with control siRNA (Fig. 4A and B). Furthermore, the mTORC1 activity, measured by pS6KT389 was significantly enhanced, whereas the mTORC2 activity, measured by pAKTS473, was attenuated in the myotubes with UCHL1 KD in response to insulin stimulation, as compared to the cells with control siRNA (Fig. 4C, D and E).
Figure 4: UCHL1 gene knockdown leads to increased mTORC1 activity in C2C12 cells.
(A) Phase-contrast images of 6-day differentiated C2C12 myotubes treated with control (Ctrl) or UCHL1 siRNA. The scale bar = 50 μm, (B) Quantification of myotube width of control (black bar) and UCHL1 KD cells (gray bar). “*” indicates p<0.05; For control group, n=56 fibers from 4 independent dishes;-For UCHL1 KD group, n=64 fibers from 4 independent dishes. (C) Representative images of Western blots of pAKTS473, AKT, pS6KT389, S6K, UCHL1 and β-Actin from C2C12 cells treated with Ctrl or UCHL1 siRNA exposing to two days of DM with or without 100 nM insulin stimulation for 10 min. (D-E) Quantifications of Western blots for pS6KT389/S6K (D), and pAKTS473/AKT (E). Results were normalized to Ctrl siRNA basal level, black bars represent control siRNA and grey bars represent UCHL1 siRNA group. “*” indicates p < 0.05, as compared between control group and UCHL1 knockdown group (genotype effect), by two-way ANOVA followed by t-test; n = 6 for each group.
Altogether, these in vitro data confirm that UCHL1 indeed has an inhibitory effect on mTORC1 activity in muscle. Thus deletion of UCHL1 leads to the enhanced mTORC1 function, which contributes to muscle fiber enlargement by promoting protein synthesis [31].
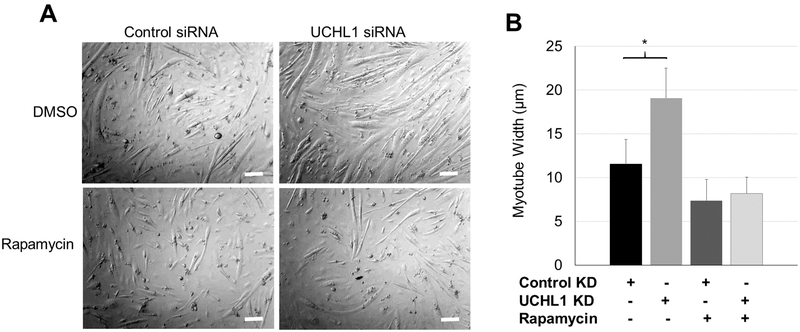
5. Inhibition of mTORC abolished UCHL1 KD-induced myotube enlargement.
To further test the role of mTORC1 in UCHL1 KD-induced myotube enlargement, C2C12 cells were treated with control siRNA or siRNA for UCHL1, and then treated with mTOC1 inhibitory rapamycin (10 nM, 24 hrs) [32] or DMSO. As shown in Fig. 5, rapamycin treatment abolished myotube hypertrophy, confirming that UCHL1 KD-induced increase in mTORC1 activity mediates the myotube enlargement.
Figure 5: Rapamycin abolishes UCHL1-induced myotube hypertrophy.
(A) Phase-contrast images of 3-day differentiated C2C12 myotubes treated with control or UCHL1 siRNA with DMSO or rapamycin. The scale bar = 50 μm (B) Quantification of myotube width of each group. “*” indicates p<0.05 as compared between the control and UCHL1 KD group (genotype effect), by two-way ANOVA followed by t-test; n=40, 37, 32, and 30 for control+DMSO, UCHL1 KD+DMSO, control+rapamycin, and UCHL1 KO+rapamycin from 3 independent dishes per group, respectively.
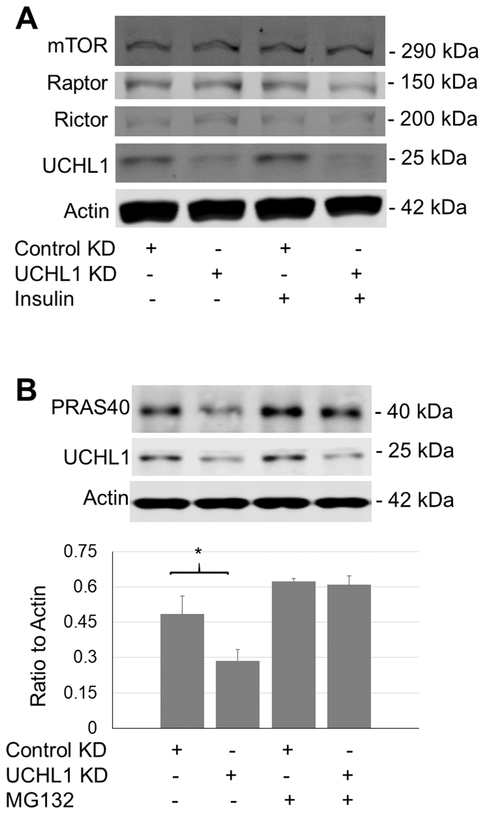
6. UCHL1 KD affects an inhibitory factor of mTORC1
To understand how UCHL1 affects mTORC1 activity, we examined the levels of some key components of the complex and some major proteins that regulate mTORC1 activity. UCHL1KD did not alter the protein levels of mTOR, Raptor, or Riptor, the core components of mTORC1 or mTORC2 (Fig. 6A). Interestingly, UCHL1 KD in C2C12 reduced the protein levels of PRAS40, an mTORC1 inhibitory factor [33], which was reversed by co-treatment with MG132 (Fig. 6B). This result suggests that UCHL1 may prevent PRAS40 from proteasome-mediated degradation, potentially via deubiquitination of this protein.
Figure 6: Effects of UCHL1 knockdown on expression of major regulatory proteins of mTOR complexes.
(A) Western blot image of mTOR, Raptor, Rictor, UCHL1, and actin from C2C12 cells treated with control or UCHL1 siRNA; (B) Western blot image and the quantification of PRAS40 from C2C12 cells treated with control or UCHL1 siRNA with or without rapamycin. “*” indicates p<0.05 as compared between the control and UCHL1 KD group (genotype effect), by two-way ANOVA followed by t-test; n= 4 dishes for each group.
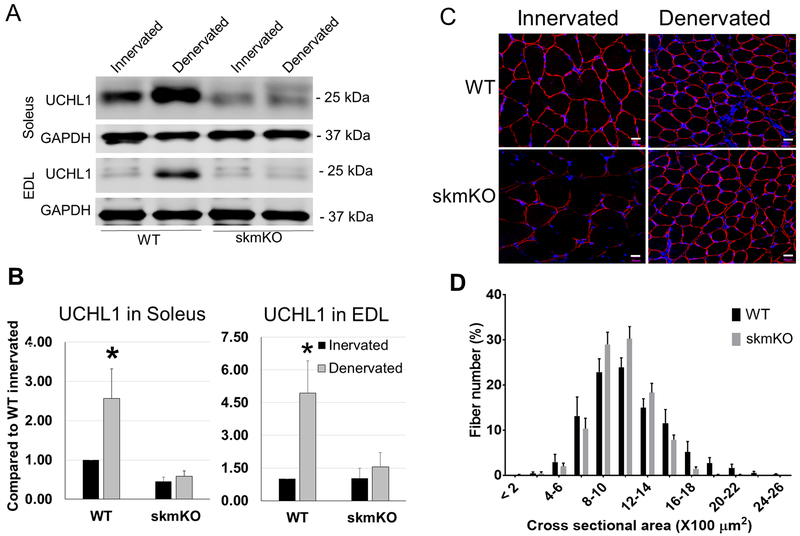
7. UCHL1 skmKO did not compensate denervation-induced muscle atrophy
We found that sciatic nerve denervation significantly increased UCHL1 protein in the denervated muscle, even in the fast glycolytic muscle EDL, which had low basal level of UCHL1. Since UCHL1 skmKO causes muscle hypertrophy, we wanted to test whether UCHL1 skmKO can prevent or alleviate denervation-induced muscle atrophy. As shown in Fig. 7, denervation-induced upregulation of UCHL1 was abolished in the muscle with UCHL1 skmKO. However, denervation-induced reduction of muscle fiber size was similar in UCHL1 skmKO muscle and WT muscle. These data suggest that, the upregulated UCHL1 in denervated muscle is not the key cause of muscle atrophy, rather, likely a compensatory response. Therefore, blockade of UCHL1 activity unlikely can prevent denervation-induced muscle atrophy.
Figure 7: Denervation upregulates UCHL1 in skeletal muscles, but ablation of UCHL1 in skeletal muscles fails to prevent denervation-induced atrophy.
(A) Representative images of Western blots for UCHL1 and GAPDH in soleus and EDL from WT and skmKO mice after 14 days of denervation. (B) Quantifications of Western blots for UCHL1 in soleus and EDL from WT (n=4) and skmKO (n=5). Results were normalized to WT innervated soleus, black bars are samples from innervated muscles, and grey bars are data from denervated muscles, “*” indicates p < 0.05 as compared between WT innervated vs WT denervated group (denervation effect), by two-way ANOVA followed by t-test. (C) Immunohistochemistry staining of cross sections of innervated and denervated gastrocnemius from WT (n=4) and UCHL1 skmKO (n=3) mice, stained with Dystrophin (Red). The scale bars indicate 20 μm. (D) Measurement of denervated gastrocnemius muscle fiber cross-section area of 150 fibers in three areas from WT mice (n=4) and skmKO mice (n=3).
Discussion
Originally discovered as a neuronal protein, UCHL1 in skeletal muscle has not been recognized until recently [21, 22] and its functional roles in muscle are unknown. We recently reported that UCHL1 has an inhibitory effect on myoblast C2C12 differentiation [23]. This study reveals for the first time that UCHL1 is primarily expressed in type I fibers of mouse muscle, suggesting that UCHL1 may be specifically involved in regulation of growth and function of slow twitch muscle. Indeed, skeletal muscle specific gene knockout of UCHL1 leads to muscle fiber hypertrophy, specifically in type I fiber; Furthermore, in vivo and in vitro gene deletion of UCHL1 resulted in enhanced mTORC1 activity, suggesting that the muscle hypertrophy induced by UCHL1 deletion is at least partially through regulation of mTORC1 activity.
mTOR is a conserved serine/threonine kinase that is broadly involved in cell survival, growth, metabolism and energy balance [8–10] and is a key signaling pathway in regulation of skeletal muscle growth and mass [7, 34]. Raptor containing mTORC1 promotes protein synthesis by phosphorylation and activation of S6K [7], [11, 12]. Our data show that both in vivo and in vitro UCHL1 deletion leaded to increased phosphorylation of S6K and muscle hypertrophy. In C2C12 cells, inhibition of mTORC1 by rapamycin prevented UCHL1 knockdown-induced myotube enlargement. There is evidence that mTORC1 positively regulates C2C12 differentiation and myotube formation [35, 36]. Therefore, our data suggest that UCHL1 functions as a negative regulator of mTOC1. Of note, in vivo, UCHL1 deletion does not lead to a significant and constitutive activation of mTORC1 at rest, but enhances the mTORC1 activity in response to insulin stimulation. This suggests that UCHL1 may not have a tonic inhibition but impose a negative effect on mTORC1 activation upon stimulation.
It remains to be fully addressed as how UCHL1 regulates mTORC1 activity. UCHL1 KD in C2C12 cells did not alter protein levels of raptor, rictor, or mTOR, suggesting that UCHL1 may not directly act on these proteins. On the other hand, mTORC1 activity is controlled by several proteins and signaling pathways. For example, the heterodimer consisting of tuberous sclerosis (TSC) 1 and 2 is a key negative regulator of mTORC1 [37]. PRAS40 is another inhibitor of mTORC1 [33]. We were not able to detect TSC protein in mouse skeletal muscle and C2C12 cells. However, our in vitro data showed that UCHL1 KD reduced PRAS40 protein level, which was prevented by MG132 treatment. Given a DUB function of UCHL1, this result suggests that UCHL1 may deubiquitinate PRAS40 to attenuate the proteasomal degradation of this protein. This provides a potential mechanism by which UCHL1 can restrain mTORC1 activity in slow muscle.
UCHL1 level was upregulated in denervated muscle. Given the negative effect of UCHL1 on muscle mass and mTORC1 activity, it was possible that the upregulation of UCHL1 contribute to the denervation-induced muscle atrophy. However, in UCHL1 skmKO muscle, although the denervation-induced upregulation of UCHL1 was completely abolished, the muscle atrophy was not alleviated. This result suggest that upregulation of UCHL1 is not a major cause of muscle atrophy. This is consistent with a previous report that pharmacologically inhibiting UCHL1 was not beneficial but detrimental in spinal muscle atrophy [22]. The functional significance of the upregulated UCHL1 in muscle remains to be further addressed.
This is the first study to examine UCHL1 function in skeletal muscle using a tissue specific gene knockout approach. The results reveal for the first time that UCHL1 is functionally expressed in oxidative muscle. Still, many critical questions remain to be fully addressed. First, we need to understand the physiological functions of UCHL1 in muscle. The inhibitory effect of UCHL1 on muscle growth and hypertrophy could be physiologically important. Muscle hypertrophy can be physiological and beneficial, such as exercise-induced muscle hypertrophy. On the other hand, pathological muscle hypertrophy is detrimental, such as those present in Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy (BMD) [38]. In the future, whether UCHL1 skmKO-induced muscle hypertrophy is beneficial or detrimental will be verified by testing the strength and endurance of muscles with UCHL1 skmKO. Second, it is necessary to know whether muscle UCHL1 is involved in other physiological and pathological changes. Our preliminary data (not shown here) indicate that muscle UCHL1 protein level is significantly upregulated in denervated muscle and aged muscle. Our future work is to determine whether this upregulation of UCHL1 in muscle plays a beneficial or detrimental role in this condition. In addition, selective expression in oxidative muscle suggests that UCHL1 may be functionally associated with oxidative metabolism, which is also worth further investigation. Moreover, the interaction of UCHL1 and mTOR complexes and regulatory proteins remains to be further investigated.
In summary, this study reveals for the first time the selective expression of UCHL1 in skeletal muscle and its effects on muscle mass and mTORC1 activity. Taken together, these data suggests that UCHL1 is a novel regulator of muscle growth, function, and metabolism.
Acknowledgement
Funding: This study is supported in part by National Institutes of Health grants R15HL118696, R03AG051926 (to YL), R01HL072166, and R01HL131667 (to XW), the USD BBS graduate program, a USD CBBRe pilot grant, and a USD BBS PQCD pilot grant (to YL). USD Graduate Research and Creative Scholarship Grant and USD-N3 Student Travel Award (to HG).
We thank Drs. Ke Ma and Janice M. Huss from City of Hope for their suggestions on the work. We also appreciate Dr. Zhaoqing Zheng from USD Sanford School of Medicine for her suggestions about some experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Rose AJ and Richter EA, Skeletal muscle glucose uptake during exercise: how is it regulated? Physiology (Bethesda), 2005. 20: p. 260–70. [DOI] [PubMed] [Google Scholar]
- 2.Richter EA and Hargreaves M, Exercise, GLUT4, and skeletal muscle glucose uptake. Physiol Rev, 2013. 93(3): p. 993–1017. [DOI] [PubMed] [Google Scholar]
- 3.DeFronzo RA and Tripathy D, Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care, 2009. 32 Suppl 2: p. S157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutt V, et al. , Skeletal muscle atrophy: Potential therapeutic agents and their mechanisms of action. Pharmacol Res, 2015. 99: p. 86–100. [DOI] [PubMed] [Google Scholar]
- 5.Glass D and Roubenoff R, Recent advances in the biology and therapy of muscle wasting. Ann N Y Acad Sci, 2010. 1211: p. 25–36. [DOI] [PubMed] [Google Scholar]
- 6.McKinnell IW and Rudnicki MA, Molecular mechanisms of muscle atrophy. Cell, 2004. 119(7): p. 907–10. [DOI] [PubMed] [Google Scholar]
- 7.Yoon MS, mTOR as a Key Regulator in Maintaining Skeletal Muscle Mass. Front Physiol, 2017. 8: p. 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laplante M and Sabatini DM, mTOR signaling in growth control and disease. Cell, 2012. 149(2): p. 274–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoncu R, Efeyan A, and Sabatini DM, mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol, 2011. 12(1): p. 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxton RA and Sabatini DM, mTOR Signaling in Growth, Metabolism, and Disease. Cell, 2017. 169(2): p. 361–371. [DOI] [PubMed] [Google Scholar]
- 11.Ma XM and Blenis J, Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol, 2009. 10(5): p. 307–18. [DOI] [PubMed] [Google Scholar]
- 12.Bodine SC, et al. , Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol, 2001. 3(11): p. 1014–9. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson KD, et al. , The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science, 1989. 246(4930): p. 670–3. [DOI] [PubMed] [Google Scholar]
- 14.Osaka H, et al. , Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet, 2003. 12. [DOI] [PubMed] [Google Scholar]
- 15.Leroy E, et al. , The ubiquitin pathway in Parkinson’s disease. Nature, 1998. 395(6701): p. 451–2. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, et al. , Overexpression of ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) delays Alzheimer’s progression in vivo. Sci Rep, 2014. 4: p. 7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, et al. , The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell, 2002. 111(2): p. 209–18. [DOI] [PubMed] [Google Scholar]
- 18.Wang YL, et al. , Accumulation of beta- and gamma-synucleins in the ubiquitin carboxyl-terminal hydrolase L1-deficient gad mouse. Brain Res, 2004. 1019(1–2): p. 1–9. [DOI] [PubMed] [Google Scholar]
- 19.Hartnett S, et al. , Ubiquitin C-terminal hydrolase L1 interacts with choline transporter in cholinergic cells. Neurosci Lett, 2014. 564: p. 115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HJ, et al. , Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene, 2009. 28(1): p. 117–27. [DOI] [PubMed] [Google Scholar]
- 21.Vasu VT, et al. , Sarcolipin and ubiquitin carboxy-terminal hydrolase 1 mRNAs are over-expressed in skeletal muscles of alpha-tocopherol deficient mice. Free Radic Res, 2009. 43(2): p. 106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powis RA, et al. , Increased levels of UCHL1 are a compensatory response to disrupted ubiquitin homeostasis in spinal muscular atrophy and do not represent a viable therapeutic target. Neuropathol Appl Neurobiol, 2014. 40(7): p. 873–87. [DOI] [PubMed] [Google Scholar]
- 23.Gao H, Hartnett S, and Li Y, Ubiquitin C-Terminal Hydrolase L1 regulates myoblast proliferation and differentiation. Biochem Biophys Res Commun, 2017: p. file:///C:/Users/Hongbo/AppData/Local/Temp/mmc2.docx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettitt SJ, et al. , Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods, 2009. 6(7): p. 493–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ringwald M, et al. , The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res, 2011. 39(Database issue): p. D849–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skarnes WC, et al. , A conditional knockout resource for the genome-wide study of mouse gene function. Nature, 2011. 474(7351): p. 337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bothe GW, et al. , Selective expression of Cre recombinase in skeletal muscle fibers. Genesis, 2000. 26(2): p. 165–6. [PubMed] [Google Scholar]
- 28.Gao H and Li YF, Distinct signal transductions in fast- and slow-twitch muscles upon denervation. Physiol Rep, 2018. 6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, et al. , Do’s and don’ts in the preparation of muscle cryosections for histological analysis. J Vis Exp, 2015(99): p. e52793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Yue F, and Kuang S, Muscle Histology Characterization Using H&E Staining and Muscle Fiber Type Classification Using Immunofluorescence Staining. Bio Protoc, 2017. 7(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruvinsky I and Meyuhas O, Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci, 2006. 31(6): p. 342–8. [DOI] [PubMed] [Google Scholar]
- 32.Leontieva OV, Demidenko ZN, and Blagosklonny MV, Rapamycin reverses insulin resistance (IR) in high-glucose medium without causing IR in normoglycemic medium. Cell Death Dis, 2014. 5: p. e1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sancak Y, et al. , PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell, 2007. 25(6): p. 903–15. [DOI] [PubMed] [Google Scholar]
- 34.Risson V, et al. , Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J Cell Biol, 2009. 187(6): p. 859–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willett M, et al. , Inhibition of mammalian target of rapamycin (mTOR) signalling in C2C12 myoblasts prevents myogenic differentiation without affecting the hyperphosphorylation of 4EBP1. Cell Signal, 2009. 21(10): p. 1504–12. [DOI] [PubMed] [Google Scholar]
- 36.Yoon MS and Chen J, Distinct amino acid-sensing mTOR pathways regulate skeletal myogenesis. Mol Biol Cell, 2013. 24(23): p. 3754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoki K, et al. , TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol, 2002. 4(9): p. 648–57. [DOI] [PubMed] [Google Scholar]
- 38.Kornegay JN, et al. , The paradox of muscle hypertrophy in muscular dystrophy. Phys Med Rehabil Clin N Am, 2012. 23(1): p. 149–72, xii. [DOI] [PMC free article] [PubMed] [Google Scholar]