Abstract
Background & Aims
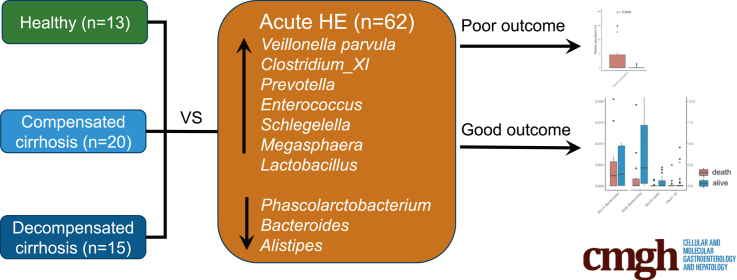
Gut dysbiosis plays a role in hepatic encephalopathy (HE), while its relationship at the acute episode of overt HE (AHE), the disease progression and clinical outcomes remains unclear. We aimed to identify AHE-specific microbiome and its association to patients’ outcomes.
Methods
We profiled fecal microbiome changes for a cohort of 62 patients with cirrhosis and AHE i) before treatment, ii) 2-3 days after medication and iii) 2-3 months after recovery, and three control cohorts i) healthy individuals, patients with ii) compensated or iii) decompensated cirrhosis.
Results
Comparison of the microbiome shift from compensated, decompensated cirrhosis, AHE to recovery revealed the AHE-specific gut-dysbiosis. The gut microbiome diversity was decreased during AHE, further reduced after medication, and only partially reversed during the recovery. The relative abundance of Bacteroidetes phylum in the microbiome decreased, whereas that of Firmicute, Proteobacteria and Actinobacteria increased in patients during AHE compared with those with compensated cirrhosis. A total of 70 operational taxonomic units (OTUs) were significantly different between AHE and decompensated cirrhosis abundances. Of them, the abundance of Veillonella parvula increased the most during AHE via a metagenomics recovery of the genomes. Moreover, the relative abundances of three (Alistipes, Bacteroides, Phascolarctobacterium) and five OTUs (Clostridium-XI, Bacteroides, Bacteroides, Lactobacillus, Clostridium-sedis) at AHE were respectively associated with HE recurrence and overall survival during the subsequent one-year follow-up.
Conclusions
AHE-specific gut OTUs were identified that may be involved in HE development and able to predict clinical outcomes, providing new strategies for the prevention and treatment of HE recurrence in patients with cirrhosis.
Keywords: Gut Microbiota, Gut Dysbiosis, Gut-Liver-Brain Axis, Veillonella parvula
Abbreviations used in this paper: AHE, acute episode of overt hepatic encephalopathy; CTP, Child-Turcotte-Pugh; HE, hepatic encephalopathy; HSD, honest significant difference; KEGG, Kyoto Encyclopedia of Genes and Genomes; NMDS, nonmetric multidimensional scaling; OTU, operational taxonomic unit; rRNA, ribosomal RNA
Graphical abstract
See editorial on page 231.
Summary.
Comparison of the stool microbiome between the decompensated cirrhosis patients with and without acute episode of hepatic encephalopathy (AHE) showed Veillonella parvula and other Firmicutes were associated with the AHE development. Eight taxa at AHE could predict clinical outcomes.
Hepatic encephalopathy (HE) is a severe central neurological dysfunction resulting from the decompensation of liver metabolism function. HE can develop in patients with acute liver disease (type A), portal-systemic shunting without intrinsic liver disease (type B), and cirrhosis (type C or acute on chronic liver failure ). Type C HE represents the majority and a primary cause of mortality in patients with cirrhosis.1 More than one-third of cirrhosis patients will develop HE and approximately 40% of HE patients die within 1 year.2 More importantly, patients with HE often have significantly worse outcomes than those without it.3 The exact underlying mechanisms of HE in patients with cirrhosis remains unclear, but hyperammonemia, systemic inflammation, and the deregulation of glutaminase are critical factors.1
Data accumulated in recent years—including information on efficient treatment of HE by various prebiotics, probiotics, and antibiotics4—suggest that gut microbiota play an important role in HE development in patients with cirrhosis.5, 6, 7 Shen et al8 showed that an engineered low-urease gut microbiome protects mice from developing minimal encephalopathy. More recently, fecal microbiota transplantation from a healthy donor reduced the recurrence of HE and dysbiosis in patients with recurrent HE.9 Apparently, the gut microbiome critically regulates brain functions in patients with decompensated cirrhosis via the gut-liver-brain axis.10, 11 However, how the gut microbiome is involved in HE development, which of its members are involved and whether such dysbiosis can predict clinical outcomes remain unknown.
In this study, we profiled the changes in gut microbiomes of cirrhotic patients with overt HE at the acute episode before treatment, 48–72 hours after active treatment, and the inactive stage (2–3 months after the episode) and compared them with those of healthy individuals and patients with compensated cirrhosis. Accordingly, we identified microbial pathogens associated with the disease activity. We further examined whether their abundance was correlated with patients’ 1-year outcomes, including HE recurrence and overall survival. Our findings provide the gut microbiota dynamics during the disease progression and resolution, disclose the microbial components contributing to the pathogenesis of HE, and offer new targets for the prevention and treatment of HE in patients with cirrhosis.
Results
Patient Population and Amplicon Sequencing
We enrolled a total of 110 patients in this study. There were 62 cirrhosis patients with acute episode of overt HE (AHE) in the emergency unit, 20 outpatients with compensated cirrhosis (C2), 15 patients with advanced stages of cirrhosis (C3), and 13 healthy individuals (C1) (Table 1). Stool samples were collected from patients with AHE within 12 hours on arrival of our emergency unit (n = 62; acute episode state = D1) and 48–72 hours after treatment for AHE in our emergency unit or ward (n = 34; admission stage = D2). Some of the AHE patients went for the collection of their stool samples 2–3 months after recovering from the episode of AHE (n = 18; D3). All the patients were followed up for up to 1 year. The majority of AHE patients were men (n = 49, 79%). In the AHE group, 22 and 42 patients had cirrhosis as Child-Turcotte-Pugh (CTP) class B and C, respectively. The majority of the cirrhotic control group (C2) had CTP class A, except for 2 patients that had class B (CTP score 7). The C3 group had 8 and 7 patients with CTP class B and C, respectively. All patients in the control group have exhibited no symptoms or signs of HE 6 months before sample collection. All individuals in the healthy control group did not have any apparent liver disease. Viral hepatitis B and C and alcoholism represented the primary causes of cirrhosis in both the control (C2 and C3) and AHE groups (D1).
Table 1.
Clinical Characteristics of the Subjects in Study
| Group | C1 | C2 | C3 | D1 | D2 | D3 |
|---|---|---|---|---|---|---|
| Number | 13 | 20 | 15 | 62 | 34 | 18 |
| Women/men | 6/7 | 8/12 | 3/12 | 13/49 | 2/32 | 4/17 |
| Age, y | 47.2 ± 10.2 | 61 ± 8.4 | 58.1 ± 12.3 | 56.7 ± 12.2 | 55.3 ± 11.5 | 55.4 ± 11.5 |
| Antibiotics used (Y/N) | 0/20 | 33/27 | 17/17 | 10/11 | ||
| MELD score | 16.8 ± 5.58 | 18.5 ± 6 | 18.5 ± 6.3 | 14.8 ± 4 | ||
| Liver cirrhosis CTP A/B/C | none | 18/2/0 | 0/8/7 | 0/22/40 | 0/11/23 | 0/10/11 |
| Total bilirubin, mg/dL | 1 ± 0.2 | 1.1 ± 0.6 | 3.5 ± 3.7 | 5.2 ± 5.3 | 5.4 ± 6 | 2.8 ± 1.8 |
| Albumin, g/dL | 4 ± 0.4 | 3 ± 0.6 | 2.8 ± 0.6 | 2.7 ± 0.6 | 3 ± 0.6 | |
| INR | 1.2 ± 0.1 | 1.5 ± 0.6 | 1.6 ± 0.3 | 1.6 ± 0.3 | 1.4 ± 0.2 | |
| Creatine, mg/dL | 1 ± 0.2 | 0.9 ± 0.3 | 1.4 ± 1.4 | 1.4 ± 1.1 | 1.4 ± 1.2 | 1.1 ± 0.5 |
| AST, U/L | 24.6 ± 4.6 | 44.4 ± 16.4 | 68.9 ± 44 | 109 ± 159.8 | 81.8 ± 66 | 67.9 ± 55.6 |
| ALT, U/L | 21.8 ± 7.4 | 37.5 ± 18.9 | 51.7 ± 46.8 | 44 ± 36.4 | 37.5 ± 20.9 | 32.4 ± 15.1 |
| Platelet, 1000/UL | 254.5 ± 42.1 | 112.2 ± 55.5 | 100.7 ± 28.8 | 84.2 ± 44.3 | 88.6 ± 48.4 | 88.4 ± 35.4 |
| PPI used (Y/N) | 3/10 | 6/14 | 4/11 | 22/40 | 10/24 | 4/14 |
| Hepatic encephalopathy history (Y/N) | none | 0/20 | 4/11 | 41/19 | 25/9 | 16/5 |
| Outcome within 1 year: dead/HE recurrence | 19/26 | |||||
| Cirrhosis risk: HBV/HCV/alcoholic/others | 0/0/7/0 | 15/6/3/0 | 6/2/8/0 | 18/19/36/4 | 10/11/21/5 | 7/5/14/1 |
Values are n or mean ± SD. MELD score = 10 * ((0.957 * ln(Creatinine)) + (0.378 * ln(Bilirubin)) + (1.12 * ln(INR))) + 6.43.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; C1, healthy individuals; C2, well-compensated cirrhosis; C3, decompensated cirrhosis without hepatic encephalopathy; CTP, Child-Turcotte-Pugh; D1, acute hepatic encephalopathy at emergency room; D2, after 48–72 h of treatment; D3, recovered 3 months after the hepatic encephalopathy episode; HBV, hepatitis B virus; HCV, hepatitis C virus; HE, hepatic encephalopathy; INR, international normalization ratio of patient's prothrombin time/normalized prothrombin time; MELD, Model for End-Stage Liver Disease; N, no; PPI, proton pump inhibitor; Y, yes.
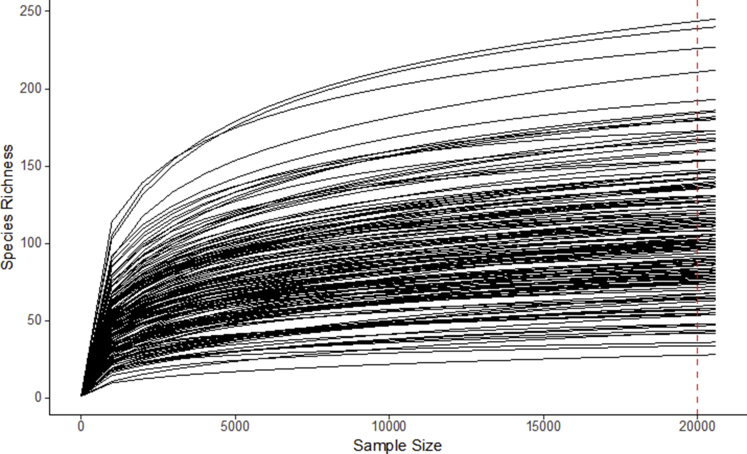
We characterized fecal microbiomes of these 162 samples from 110 subjects using 16S V3-V4 amplicon sequencing (performed by NGS High Throughput Genomics Core at the Biodiversity Research Centre in Academia Sinica, Taipei, Taiwan). A total of 13,505,741 reads were sequenced and passed the quality filters. These reads were further assigned into 1242 operational taxonomy units (OTUs). Of these OTUs, all were successfully assigned at the phylum level and 97.0% at the genus level. On average, each sample had 83,368 reads, which were assigned to 134 OTUs. Rarefaction curve analysis suggested that the microbiome diversity was captured beyond 20,000 reads across the majority of samples as the number of observed species plateaued (Figure 1).
Figure 1.
Rarefaction curves of all 16S amplicon samples from patients.
Dysbiosis of Microbiome Diversity in AHE Patients
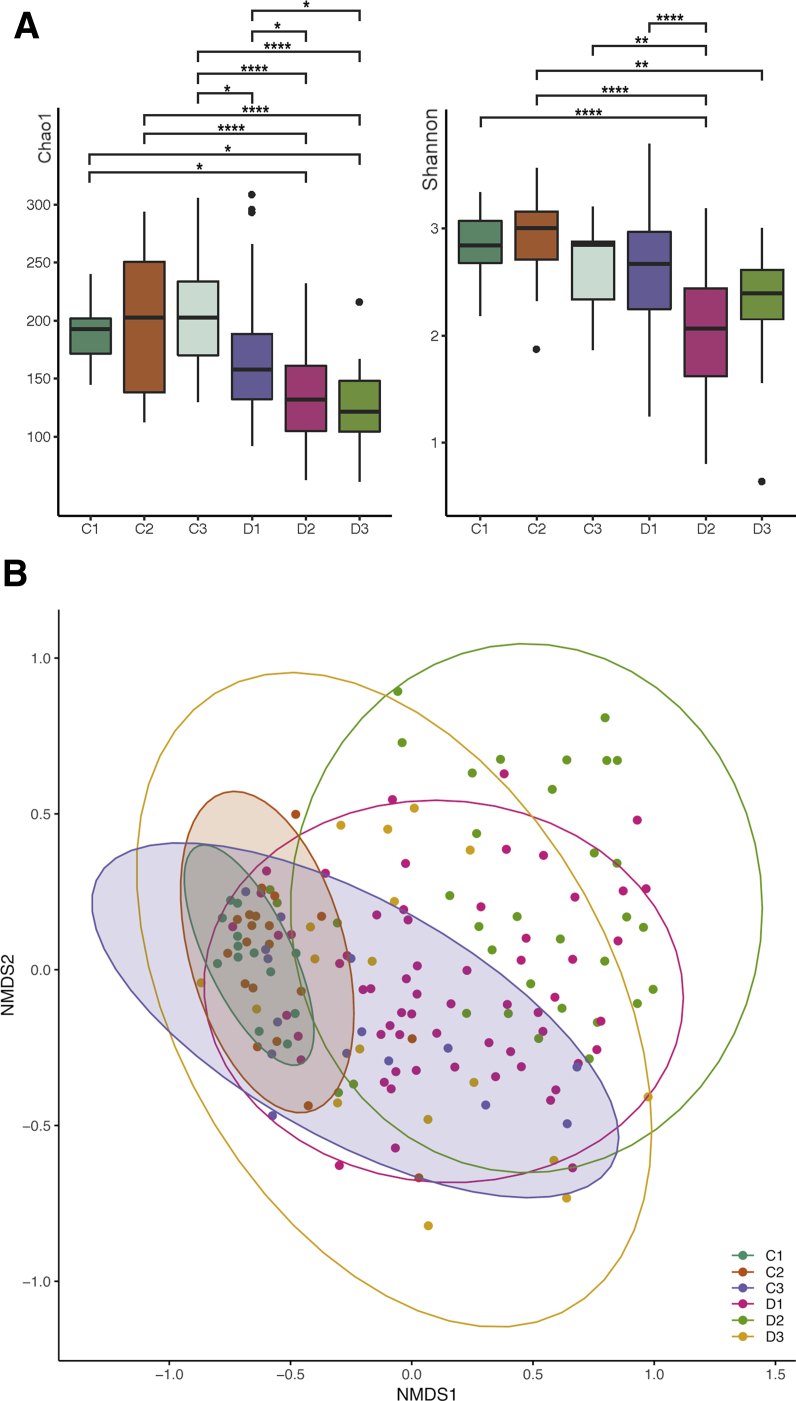
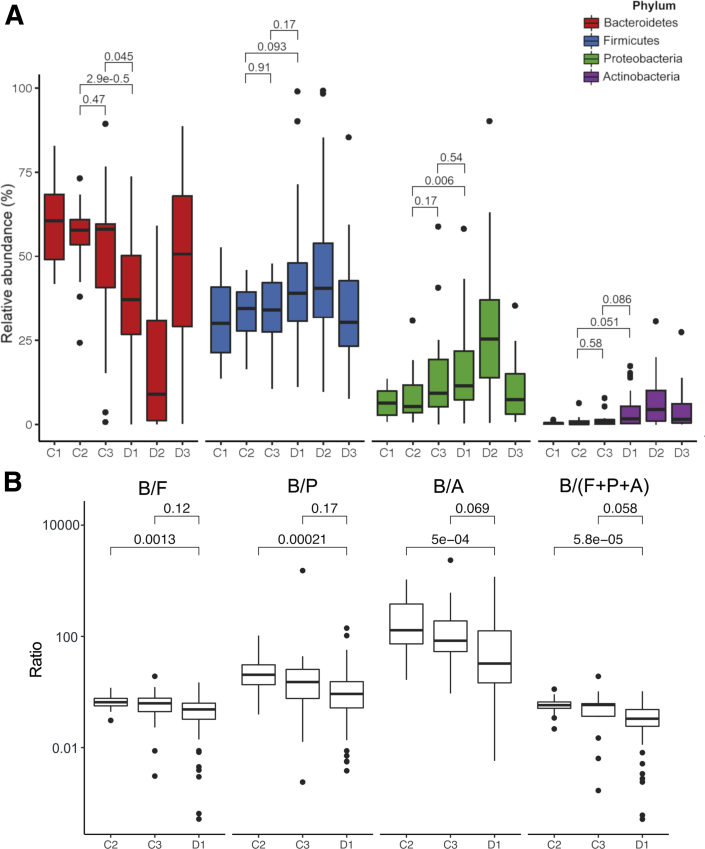
Alpha (within-sample) diversities were calculated using Chao1 (for OTU richness) and Shannon indices (for community diversity) (2A). As multiple conditions were present in this study, we used Tukey’s honest significant difference (HSD) test to assess all combinations of pairwise comparisons. Across both indices, the 3 control groups exhibited no differences in diversity between 3 pairwise comparisons (adjusted P = 1.00). A significant decrease in Chao1 diversity index (HSD; adjusted P = .02) between decompensated cirrhosis patients without (C3) and with AHE (D1) suggests that AHE might contribute to the decrease in microbial richness. Compared with the patients with compensated cirrhosis (C2), Shannon's diversity index shows a decreased trend in the D1 cohort (HSD; adjusted P = .07) but not the Chao1, suggesting some levels of dysbiosis of prevalent OTUs were observed. Gut microbiome diversities from 34 patients after treatment (D2) were significantly decreased further in both indices, suggesting a reduction in both species richness and evenness. A longitudinal follow up of 18 recovered patients (D3) shows an increased trend in Shannon diversity but not in Chao1, suggesting during this period of recovery a higher abundance was found in the remaining species after treatment, without the emergence of new species.
Beta diversities (between sample differences) were assessed using the Bray-Curtis dissimilarity index (Figure 2B). Nonmetric multidimensional scaling (NMDS) using Bray-Curtis dissimilarities showed the microbiomes of the C1 and C2 control groups are clustered together and distinctly separate from AHE patients at different stages. The microbiome profile of C3 did not cluster tightly with other control subjects but partially overlapped with D1. The first NMDS axis separates the control groups (C1 and C2) and patients with different disease status: D1 is shifted to the right of the control groups, D2 is shifted further to the right, and D3 overlaps with the control and C3+D1 groups. Consistent with NMDS positioning, PERMANOVA indicates that the microbiome compositions are significant across different disease statuses (P = .001). Within each pairwise condition of disease status, C1 and C2 control groups share the same microbiome composition (adjusted P = .00) and are somewhat different to C3 (adjusted P value = .945 in C2 vs C3 and P = .33 in C1 vs C3). The gut microbiomes between AHE patients at initial 12 hours (D1) were significantly different to C1+C2 groups (adjusted P = .015) but similar to those with decompensated cirrhosis (C3) or recovered patients (D3) (adjusted P = .21 and .3, respectively). Similar microbiome displayed in C3 to both C1+C2 and D1 groups suggest a gradual change of OTU abundance associated with HE development. Additionally, patients underwent 48–72 hours after treatment (D2) were significantly different to these 3 stages (adjusted P = .015) presumably as a result of the treatments, usually including lactulose, for AHE rather than a microbiome shift associated with disease progression/regression.
Figure 2.
(A) Alpha diversity pattern with control groups (C1, C2, and C3) and AHE patients at different stages (D1, D2, and D3). Significances of different diversity between samples were assessed by Tukey’s HSD (*P < .05; **P < .01; ***P < .001; ****P < .0001). (B) AHE patients at different stages shift microbial community structure. Samples from both control groups are not distinct from each other (C1 and C2). Samples from AHE patients were shifted away from control groups according to their disease status (D1, D2, and D3). D3 is relatively overlapping with C3 (decompensated cirrhosis without recent HE).
Characteristics of the Fecal Microbiome in AHE Patients at the Phylum Level
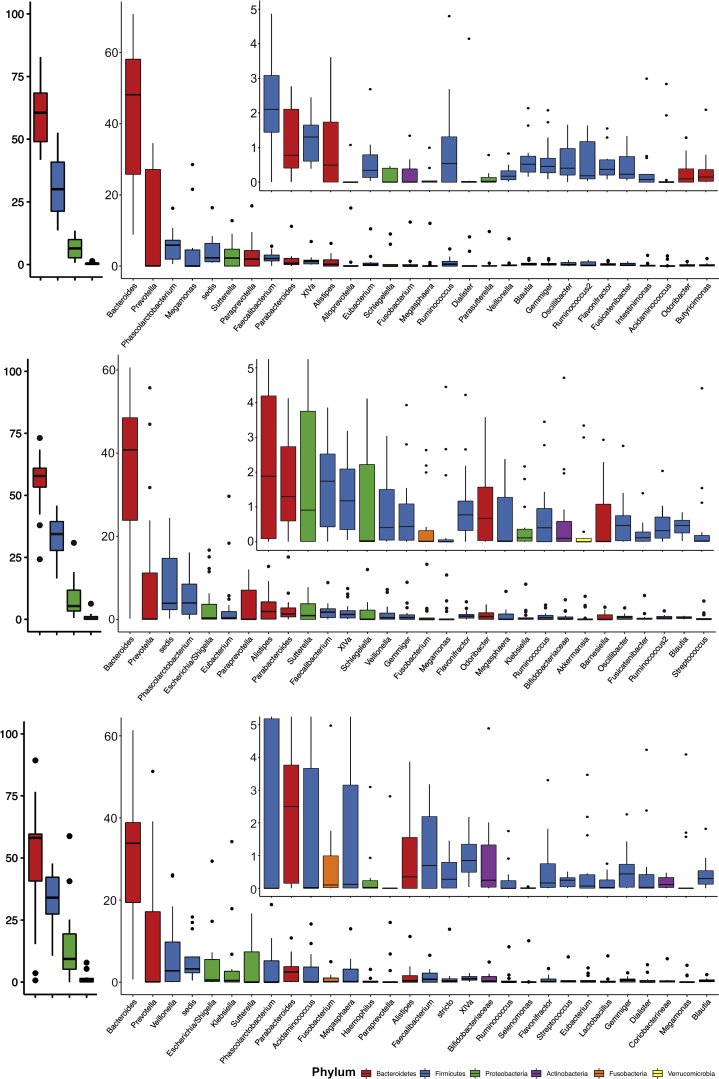
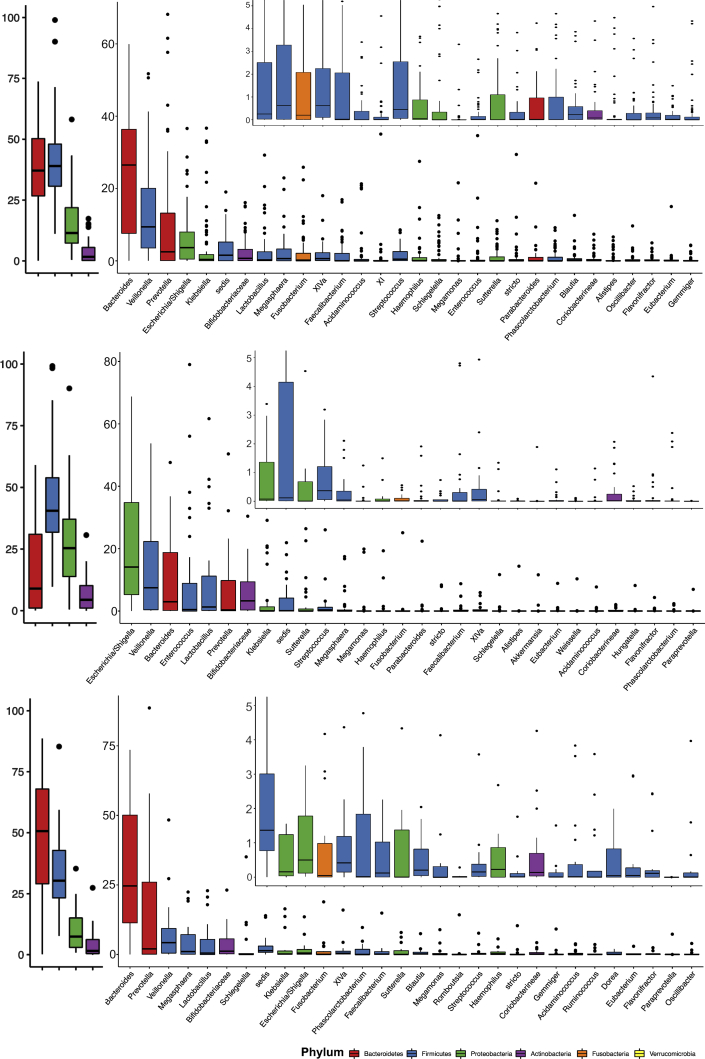
We examined the relative abundance of major taxa amongst AHE patients. The top 4 phyla and 30 genera of the fecal microbiome among groups C1, C2, C3, and D1 are shown in Figures 3 and 4. At the phylum level, the 4 major phyla occupy on average 97.8% of sequences across all samples. The fecal microbiome of AHE patients (D1) is dominated by Firmicutes (on average 41.4%), Bacteroidetes (36.5%), Proteobacteria (15.8%) and Actinobacteria (3.4%). Compared with the control subjects, AHE patients show a definite difference in their fecal microbiome structure with a decreased relative abundance of Bacteroidetes (C1, C2, and C3 vs D1: 60.5%, 55.6% and 47.2% vs 36.5%) and increased relative abundances of 3 other phyla: Firmicutes (31.1%, 33.5%, and 33.2% vs 41.4%), Proteobacteria (6.6%, 8.2%, and 15.5 vs 15.8%), and Actinobacteria (0.4, 0.9%, and 1.4% vs 3.4%) (Figure 5A). Strikingly, the relative abundances of Firmicutes in 5 AHE samples were as high as 90%—compared with a maximum of 52.6% in the control groups. To test if the ratios between Bacteroidetes over combinations of the other 3 major phyla (Firmicutes, Proteobacteria, Actinobacteria, and the sum of these 3 phyla) can be used to differentiate AHE (D1) group against the control subjects, we calculated and found all 4 ratios were all significantly different between compensated patients to either decompensated cirrhosis (C3) or AHE patients (Figure 5B), but no difference was found in the latter 2 groups (Figure 5B).
Figure 3.
Relative abundance of OTUs in healthy patients (top, C1), control cirrhosis patients (middle, C2), and advanced cirrhosis patients (bottom, C3) at the phylum and genus levels. Only the top 30 most abundant OTUs in each group are shown.
Figure 4.
Relative abundance of OTUs in AHE patients (top), AHE patients during treatment (middle), and recovered AHE patients (bottom) at the phylum and genus levels. Only the top 30 most abundant OTUs in each group are shown.
Figure 5.
(A) Relative abundances at the 4 major phylum levels between groups. Boxplots show the median and interquartile range of relative abundances (%). Dots represent the relative abundances of samples. (B) Different combinations of phyla/Bacteroidetes ratios in distinguishing D1 from C2 and C3.
To further pinpoint bacterial OTUs associated with AHE status, counts of the raw sequence of each OTU were transformed and the pairwise comparison of differential abundance among different patient groups was carried out using Wald test of DESeq2.12 A total of 175 unique OTUs were found differentially abundant across all pairwise comparisons (C1 vs C2; C2 vs C3; C3 vs D1; D1 vs D2; D2 vs D3; adjusted P < .05) (Table 2). Consistent with the beta diversity analysis, only 15 OTUs were found to have different abundances between the 2 control groups (C1 vs C2). In contrast, pairwise comparison revealed that 127 OTUs had significantly different abundances between the fecal microbiomes of AHE patients (D1) and those patients with compensated cirrhosis (C2) (Table 2). Of these OTUs, 87 were found reduced in the D1 group, 2.2 times more than the number of increased OTUs. These OTUs made up an overall average decreased relative abundance of 19.4% in Bacteroidetes and increased the abundance of 8.5% in Firmicutes, respectively, confirming the previous observation of the relative abundances for the 4 major phyla (Figure 5A). Compared with the transition stage of decompensated cirrhosis patients (C3), patients with AHE (D1) exhibited 66 OTUs significantly different abundances. A total of 60% of these OTUs were also significantly different in relative abundances compared with C2 suggesting perturbations of these OTUs closely reflect the progression of liver failure and AHE development. Full information including taxa information and P values of all significant abundant OTUs are shown in Supplementary Table S2a.
Table 2.
Paired Comparison (With DAOTU P < .05)
| Group1 | Group2 | DAOTUs with P < .05 | More Abundant in Group 1 | More Abundant in Group 2 |
|---|---|---|---|---|
| C1 | C2 | 15 | 5 | 10 |
| C2 | C3 | 26 | 21 | 5 |
| C2 | D1 | 127 | 87 | 40 |
| C3 | D1 | 70 | 26 | 44 |
| D1 | D2 | 84 | 71 | 13 |
| D2 | D3 | 94 | 17 | 77 |
| D1 (n = 62) | D3 (n = 18) | 71 | 24 | 47 |
| D1 (n = 18) | D3 (n = 18) | 42 | 17 | 25 |
C1, healthy individuals; C2, well-compensated cirrhosis; C3, decompensated cirrhosis without hepatic encephalopathy; CTP, Child-Turcotte-Pugh; D1, acute hepatic encephalopathy at emergency room; D2, after 48–72 h of treatment; D3, recovered 3 months after the hepatic encephalopathy episode; DAOTU, differentially abundant operational taxonomic unit between Group 1 and Group2.
Abundance of Signature OTUs in Longitudinal Follow-Up
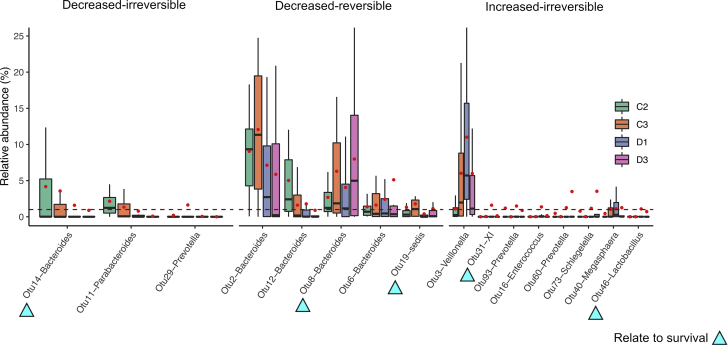
The changes in the gut microbiome from the acute episode to recovery of HE are shown in Figure 4. We hypothesized that those with a less than 1% of total fecal microbiome abundance were unlikely clinically significant. We thus report the relative mean abundance of 16 signature AHE-associated OTUs in Table 3, defined here as OTUs that 1) have a mean relative abundance of at least 1% in the patients either with decompensated cirrhosis (C3) or with AHE (D1) and 2) were significantly different in abundance between the 2 stages. The trend of perturbations of these 16 signature-OTUs imply their involvement during AHE development. We sought to further characterize these 16 signature-OTUs depending on their relative abundances in a longitudinal follow-up group of 18 patients (D3) using Wald test of DESeq2.12 From the signature OTUs formed 3 groups: 1) 3 OTUs with significantly lower abundances in D1 than those in C3 and persistently low abundance in D3, which we termed decreased-irreversible; 2) 5 OTUs with significantly lower abundances in D1 than those in C3 but with higher abundance in D3 than in D1, which we termed decreased-reversible; and 3) 8 OTUs with significantly higher abundances in D1 than in C2 that remain high in D3, which we termed increased-irreversible. The relative abundances of the signature OTUs during these stages (C2, C3, D1, and D3) are shown in Figure 6.
Table 3.
The 16 OTUs With Significantly Different Abundance Between C3 and D1 and With the Mean Abundance at Least 1%
| OTU_ID | C1 |
C2 |
C3 |
D1 |
D2 |
D3 |
Adjusted P Value | Recovered in D3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||||
| Increased OTUs | |||||||||||||||
| Proteobacteria | |||||||||||||||
| Schlegelella | Otu73 | 0.288 | 0.710 | 0.780 | 2.714 | 0.029 | 0.085 | 1.172 | 2.924 | 0.202 | 1.097 | 3.526 | 8.696 | .000 | No |
| Firmicutes | |||||||||||||||
| Megasphaera | Otu40 | 0.037 | 0.112 | 0.452 | 1.551 | 1.031 | 1.861 | 1.105 | 1.518 | 1.105 | 3.317 | 1.258 | 4.720 | .022 | No |
| Veillonella | Otu3 | 0.736 | 1.916 | 1.085 | 1.758 | 6.015 | 7.910 | 11.007 | 12.473 | 10.549 | 14.133 | 5.976 | 11.785 | .017 | No |
| Enterococcus | Otu16 | 0.006 | 0.008 | 0.012 | 0.024 | 0.020 | 0.031 | 1.369 | 5.038 | 9.645 | 18.166 | 0.160 | 0.484 | .039 | No |
| Lactobacillus | Otu46 | 0.001 | 0.004 | 0.003 | 0.004 | 0.011 | 0.017 | 1.086 | 3.184 | 1.855 | 9.842 | 0.714 | 2.613 | .003 | No |
| XI | Otu31 | 0.007 | 0.019 | 0.008 | 0.028 | 0.039 | 0.081 | 1.603 | 7.644 | 0.034 | 0.132 | 0.132 | 0.345 | .028 | No |
| Bacteroidetes | |||||||||||||||
| Prevotella | Otu60 | 0.004 | 0.009 | 0.476 | 1.616 | 0.022 | 0.084 | 1.254 | 4.101 | 0.510 | 1.543 | 3.492 | 11.627 | .000 | No |
| Prevotella | Otu93 | 0.001 | 0.002 | 1.192 | 5.317 | 0.002 | 0.005 | 1.487 | 4.098 | 0.526 | 2.298 | 0.880 | 3.121 | .000 | No |
| Decreased OTUs | |||||||||||||||
| Firmicutes | |||||||||||||||
| sedis | Otu19 | 1.871 | 2.922 | 1.311 | 2.721 | 1.788 | 2.986 | 0.401 | 0.923 | 0.155 | 0.533 | 1.134 | 2.826 | .001 | Yes |
| Bacteroidetes | |||||||||||||||
| Prevotella | Otu29 | 0.000 | 0.000 | 0.205 | 0.916 | 1.635 | 6.154 | 0.050 | 0.390 | 0.000 | 0.002 | 0.000 | 0.001 | .027 | No |
| Bacteroides | Otu12 | 5.443 | 6.557 | 5.021 | 5.913 | 1.607 | 2.290 | 1.780 | 4.062 | 1.070 | 4.475 | 0.899 | 2.285 | .001 | Yes |
| Bacteroides | Otu6 | 2.423 | 3.383 | 1.381 | 1.718 | 1.623 | 2.030 | 2.417 | 4.898 | 1.017 | 4.020 | 5.117 | 12.504 | .040 | Yes |
| Bacteroides | Otu8 | 6.020 | 5.370 | 2.687 | 3.227 | 6.290 | 7.899 | 4.051 | 6.931 | 0.982 | 2.411 | 7.990 | 9.865 | .000 | Yes |
| Bacteroides | Otu2 | 15.265 | 12.722 | 9.060 | 6.392 | 12.069 | 8.806 | 7.128 | 10.163 | 3.470 | 6.150 | 5.869 | 9.434 | .000 | Yes |
| Bacteroides | Otu14 | 1.889 | 3.663 | 4.176 | 8.492 | 3.571 | 8.650 | 1.591 | 4.525 | 0.771 | 2.862 | 0.892 | 3.775 | .005 | No |
| Parabacteroides | Otu11 | 1.419 | 2.389 | 2.151 | 3.227 | 1.334 | 2.269 | 0.788 | 2.946 | 0.693 | 3.835 | 0.083 | 0.201 | .016 | No |
OTU, operational taxonomic unit.
Figure 6.
Distribution of relative abundances for 16 signature OTUs at the C2, C3, D1, and D3 stages. Boxplots show the median and interquartile range of relative abundance (%). Red dots denote mean relative abundances of each category, the raw values of which are listed in Table 3. Arrowheads indicate those whose abundances related to clinical outcomes.
Eight of the 16 signature-OTUs have significantly lower relative abundances in patients with AHE (D1) compared with those with decompensated cirrhosis (C3). The majority of them belong to the genus Bacteroides (Table 3), which explains the general decreasing trend in the Bacteroidetes phylum of AHE patients (Figure 3). OTU8 and OTU19 with reduced in abundance in AHE cohort and show significantly higher relative abundances in the recovered patients (D3, the decreased-reversible subgroup) (6), suggesting these are originally autochthonous OTUs that were present in relatively higher abundance in both the recovered patients and control subjects. They might play a protective role in the development of HE.
In contrast, we hypothesized the 8 OTUs in the increased-irreversible subgroup might play a pathogenic role in AHE (D1 vs D3). Although all the increased OTUs were not apparent different between AHE and recovered patients (D1 vs D3; adjusted P > 0.05), a decreased trend was observed in all these OTUs except OTU40 (Figure 6). The relative abundances of these taxa in the control groups were usually low, ranging from 0 to 6.0% and they belonged to 7 different genera, suggesting that AHE is influenced by a diverse microbiome (Table 3).
Recovery From a Metagenomics Dataset Shows Veillonella parvula as the Most Prevalent Species in AHE Patients
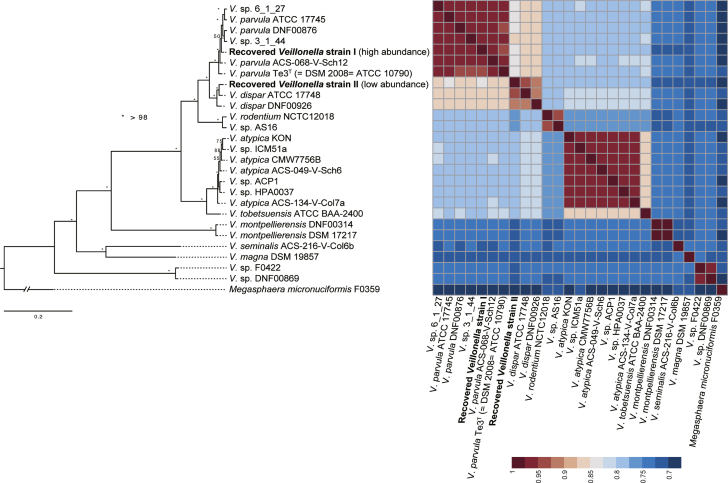
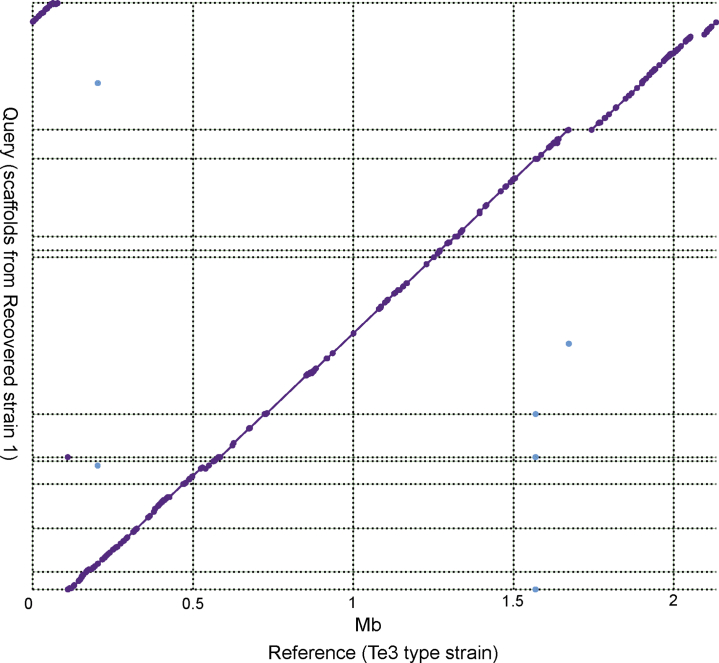
The taxon with most striking difference in relative abundance between AHE and control cirrhosis patients is Veillonella (OTU3); its abundance in AHE patients is 10 times that of control cirrhosis patients (Figure 6 and Table 3) (C2 vs D1 = 1.1% vs 11.0%; adjusted P < .001) and was found to make up 50.1% of 1 patient with AHE (Supplementary Table S2). As 16S ribosomal RNA (rRNA) sequence similarity between Veillonella species can be as high as 99.9%,13 we sought to resolve this OTU at the species level by sequencing the metagenomes of patients with the highest relative abundance of OTU3 (Supplementary Table S2). Two Veillonella genomes with different depths of relative abundance (average relative abundance across the assembly: 29% and 1%) were reconstructed (see Materials and Methods). The more highly abundant Veillonella species OTU3 was assembled into 12 scaffolds of 1.9 Mb and N50 of 253 kb. To characterize OTU3 into a species level, a maximum likelihood phylogeny from a concatenated protein alignment of 318 single copy orthologs revealed that OTU3 was grouped with Veillonella parvula (Figure 7). A whole-genome alignment shows that OTU3 is mostly in synteny with the V. parvula Te3T (Figure 8), with an average nucleotide identity (ANI) of 94.8%. In contrast, the other recovered Veillonella strain with much lower abundance (at 1%) was grouped with the V. dispar (Figure 7).
Figure 7.
Phylogenetic tree of Veillonella species.
Figure 8.
Whole genome alignment between the reconstructed OTU3 genome vs the Te3 Veillonella parvula type strain. Dashed line indicates scaffold boundaries and purple dots/line denote matched alignment.
Phascolarctobacterium Was Able to Predict Recurrence Outcome
We sought to identify specific OTUs associated with the outcomes of HE recurrences by correlating the relative abundance of the 16 AHE-specific OTUs to the clinical outcomes—overall survival and recurrence of HE—of the 62 AHE patients during the subsequent 1-year follow-up. Initially, at the genus level, there were a total of 7 genera that show differential abundance between episodic recurrence of HE (mean abundance >1%; adjusted P < .05) (Table 4). The Fusobacterium (no recurrence vs recurrence: 3.5% vs 1.17%) and Veillonella (11.6% vs 17.2%) were the most reduced and increased genera in the episodic recurrence of HE, respectively. Interrogating this approach further at the OTU level, a total of 5 of 70 C3-D1 differentially abundant OTUs were identified between AHE patients with vs without recurrences (Supplementary Table S2a) (adjusted P < .05). Of these, none were part of the 8 signature OTUs that had decreased abundance in AHE patients (Figure 6), suggesting abundance changes to recurrence OTUs may have already taken place prior to the development of AHE.
Table 4.
Differential Abundant Genera Between Episodic Recurrence of Hepatic Encephalopathy
| Mean Relative Abundance (%) |
Abundance in Recurrent Patients | Adjusted P Value | ||
|---|---|---|---|---|
| No Recurrence | Recurrence | |||
| Fusobacterium | 3.501 | 1.170 | Reduced | .0070 |
| Haemophilus | 1.256 | 1.257 | Increased | .0070 |
| Clostridium_sensu_stricto | 0.564 | 1.781 | Increased | .0033 |
| Phascolarctobacterium | 1.681 | 0.290 | Reduced | .0042 |
| Veillonella | 11.661 | 17.216 | Increased | .0076 |
| Megamonas | 2.028 | 0.876 | Reduced | .0090 |
| Faecalibacterium | 2.529 | 1.151 | Reduced | .0086 |
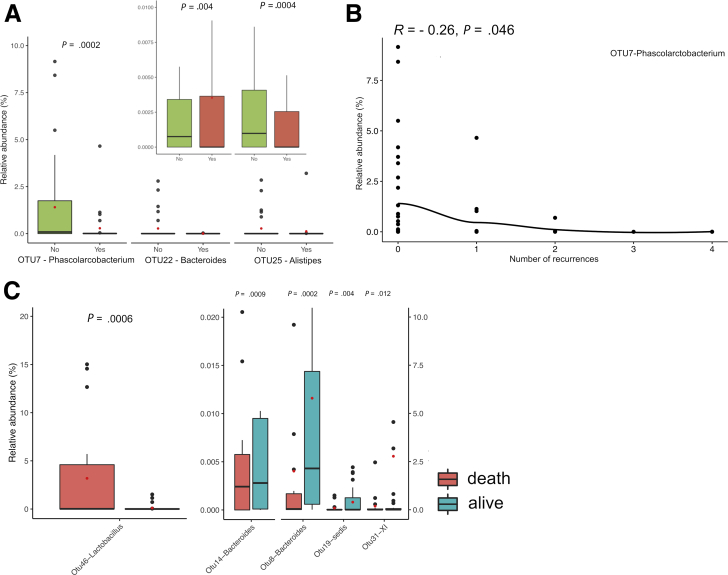
Hypothesizing that abundances of OTUs may have already perturbed in decompensated HE patients, we broadened the search to C2-D1 comparisons and identified a further 10 differentially abundant OTUs. Further abundance changes in these OTUs were not observed in decompensated cirrhosis patients (C3), but as demonstrated in Figure 9A, it is apparent that Phascolarctobacterium (OTU7), which was most decreased in decompensated patients without and with AHE, was even lower in patients who underwent HE recurrences (no recurrence vs recurrence: 1.68% vs 0.29%; adjusted P = .03). Further investigation revealed that there is a negative correlation between the number of HE recurrences and relative abundance of OTU7 at AHE stage (Pearson’s r = –0.26, P = .046) (Figure 9B).
Figure 9.
(A) Boxplots of relative abundances for 3 OTUs at the AHE stage from patients with or without recurrences. (B) Scatterplot of the relative abundances of OTU7 at AHE stage against the number of recurrence (Pearson’s r = –0.26, P = .046). (C) Boxplots of relative abundances for 5 OTUs that were with significantly different abundance between patients with different mortality outcome.
Five OTUs Associated With Mortality
We found that abundances of 5 of the 16 signature OTUs were significantly associated with patients’ mortality (adjusted P < .05) (Figure 9C). OTU46 (Lactobacillus; 1.0% vs 0.01%, D1 vs C3), which increased in abundance in AHE, and was found to be significantly higher in patients that passed away (Figure 9C, left panel), indicative of a pathogenic role in the development of AHE. By contrast, OTUs14 (Bacteroides), 8 (Bacteroides), 19 (Clostridium_incertae_sedis), and 31 (Clostridium XI) had low abundances but were significantly higher in patients that survived (Figure 9C, right panel), suggesting that they might be protective gut germs from the development of HE.
Microbial Functional Prediction in AHE Patients
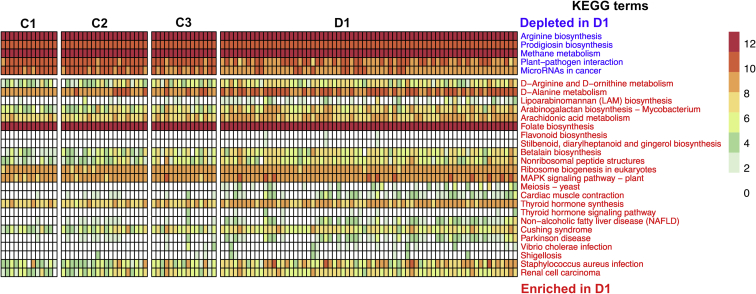
To investigate the functional mechanisms of gut microbiota in the development of HE, we inferred abundance of Kyoto Encyclopedia of Genes and Genomes (KEGG) (release 67.1) orthologues from genomes directly inferred from the 16S rRNA OTU abundance table using Piphillin.14 Differential abundances of 28 KEGG terms between patients with compensated cirrhosis (C3) and patients with AHE (D1) were identified using the negative binomial Wald test implemented in DESeq2 (Figure 10). Many of the AHE-enriched KEGG terms are involved in pathogen-induced diseases or ammonia and amino acid metabolisms such as Arginine biosynthesis and D-arginine, D-ornithine, and D-alanine metabolism (Figure 10). Interestingly, there were 2 pathways—1 related to nonalcoholic fatty liver disease and the other arachidonic acid metabolism that were found to be both increased in the gut microbiome of patients with inflammatory bowel disease15, 16 and associated with neurodegenerative disorders.17
Figure 10.
The abundances of control cirrhosis–enriched and AHE-enriched KEGG functional terms. Abundances were plotted as transformed values from counts of inferred KEGG orthologues. For visualization comparison, abundances from healthy individuals were also plotted.
Discussion
In this study, we profiled the dynamic shift of gut microbiomes from AHE throughout its treatment and recovery process in patients with decompensated cirrhosis and compared it to microbiomes in patients with compensated, decompensated cirrhosis without recent history of HE and healthy individuals. Such longitudinal profiling of fecal dysbiosis enables us not only to identify HE-associated dysbiosis but also to distinguish between those taxa potentially involved in HE pathogenesis from those of neutral bystanders and those that are pathogenic for HE from those that protect the body from HE. Furthermore, the correlation between HE-associated fecal dysbiosis and clinical outcomes not only validates their pathogenic roles in HE development but also makes it possible to predict HE recurrence and overall survival based on the fecal dysbiosis. Our findings that gut dysbiosis in acute episode of HE predicts clinical outcomes are reminiscent of a very recent studies that gut dysbiosis on admission in hospitalized cirrhosis patients could predict the clinical outcomes including extrahepatic organ failure, hepatic failure, and mortality.18
It has been previously observed that astrocytic impartment was associated with elevated serum ammonia levels,10 leading to the hypothesis that excess ammonia was produced in the gut microbiome of HE patients. Our AHE-enriched microbial inferred functional terms support such hypothesis. Besides, these enriched germs also implicated in nonalcoholic fatty liver disease suggest a shared repertoire of core bacterial species are implicated to development of cirrhosis and its associated complications, such as HE.
At the phylum level, we found an apparent trend in global changes in the relative abundances of the 4 most abundant phyla: Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria. In the AHE (D1), there was a downregulation of Bacteroidetes and upregulation of Firmicutes, Proteobacteria, and Actinobacteria, suggesting that many taxa belonging to Bacteroidetes are autochthonous and good (favorable or protective) gut germs for health, but many belonging to Firmicutes, Proteobacteria, and Actinobacteria are unfavorable or pathogenic germs. Chen et al6 reported a reduction in Bacteroidetes levels in patients with cirrhosis but an increase in Proteobacteria and Fusobacteria. We also found decreased Fusobacteria during AHE, but with relatively initial low abundances (Supplementary Table S2a). As it is relatively feasible for clinical applications by assaying gut microbiome at the phylum level, further studies are warranted by including a larger sample size to examine whether fecal dysbiosis measured at the phylum level predicts clinical outcomes of patients with HE.
At the genus level, we found that Veillonella (11.0%), Clostridium_XI (1.6%), Prevotella (1.5%), and Enterococcus (1.4%) were significantly higher in AHE. Although none had statistical significance, all showed decreased relative abundance when the disease recovered (D1 vs D3) (Table 3), suggesting a potential pathogenic role in the development of HE. The relative abundances of Bacteroidetes and Veillonella were found to decrease and increase (respectively) even further with the episodic recurrence of HE. Moreover, we also found that increased Lactobacillus at the AHE stage were significantly associated with patient mortality within 1 year (Figure 9C, left panel). Previously, Bajaj et al7 correlated gut dysbiosis with the severe chronic liver disease and found a negative correlation of Clostridium XIV, Lachnospiraceae, Ruminococcaceae, and Rikenellaceae and a positive correlation of Staphylococcae, Enterococcaceae, and Enterobacteriaceae with the severity of chronic liver disease. Decreased abundance of Clostridium XIV, Lachnospiraceae, Ruminococcaceae, and Rikenellaceae are also found in our AHE group (Supplementary Table S2a) in relatively low abundances (<1% for each individual OTU in healthy and well-compensated cirrhosis individuals), indicating that patients with decompensated cirrhosis in both Asian (present study) and American7 populations share common gut dysbiosis with each other. Our findings are also consistent with a recent report that high abundance of Lactobacillaceae but low abundance of Lachnospiracease in the stool and saliva samples is associated with the development of minimal HE in cirrhotic patients.19
Our findings are also reminiscent of the previous findings that increased fecal Veillonella and Klebsiella in patients with decompensated cirrhosis is associated with poor clinical outcomes.20, 21 In particular, by sequencing the metagenome of 1 patient with a gut microbiome, more than half of which was solely the Veillonella OTU3, we were able to reconstruct the genome of this species with high completeness and pinpoint its phylogenetic position to be V. parvula. This is the first time that increased V. parvula was found to be associated with AHE patients, while only V. atypica, V. dispar, and V. sp. oral taxon were previously observed to be enriched in liver cirrhosis patients.20 Veillonella is gram-negative anaerobic cocci, which comprises part of the healthy flora of the mouth and esophagus. More recently, it was detected in patients with decompensated cirrhosis and, thus, was regarded as specific OTUs of oral microbiomes that translocate to the gut in patients with decompensated cirrhosis.22, 23, 24 We annotated the genome of V. parvula and found no urease gene encoded. Further studies that use animal models to elucidate their roles in HE pathogenesis, particularly in cirrhosis mice, may contribute to the prevention and treatment of HE.25
On the other hand, taxa with decreased relative abundances in AHE—including Bacteroides (OTUs 2, 8, and 12), Clostridium_incertae_sedis (OTU19) and Phascolactobacterium (OTU7)—saw their relative abundance restored during the recovery stage, indicating their potential roles in protecting the body from HE attacks. Interestingly, we found that 22 of the 127 OTUs with significant difference in abundance belongs to family Lachnospiraceae and were all decreased in D1 as compared with the control subjects (Supplementary Table S2a). This finding is consistent with a previous report from the United States that there was an inverse correlation between Lachnospiracea abundance and cirrhosis severity.6
We have broadened the comparison criteria of cohorts for OTUs that correlate with HE recurrence to those with compensated cirrhosis (C2). The reasoning for this is that often the C3 patients suffer an episode of AHE soon after admission, suggesting that the microbiome of the C3 group is transitioning toward if not already alike the microbiome of AHE patients as demonstrated in Figure 2B. This will overlook the changes to the recurrence OTUs otherwise significant with an earlier stage of cirrhosis development. Here, we found that a lower Phascolarctobacterium abundance is correlated with a higher frequency of HE recurrence. Indeed, it has been reported that the succinate producer Phascolarctobacterium was the predominant taxon in healthy young Chinese cohorts,26 and its abundance was observed to be significantly reduced in both patients with ulcerative colitis and Crohn’s diseases.27 Together, these findings suggest that Phascolarctobacterium plays a role in protecting the body from attacks by significant human diseases.
Previously, Bajaj et al28 reported no significant difference in stool microbiome between cirrhosis patients with and without HE. They further found a difference in microbiome between the colonic mucosa and stool and a correlation of the mucosal microbiome with HE.29 Here, we reported the identification of the gut dysbiosis with differential abundance between cirrhotic patients with and without HE. Our explanation of this seeming discrepancy is due to the difference in patients with HE recruited to theirs and our studies. In Bajaj et al’s studies, the HE cohort contained those who were not at acute episode of HE but within 3 months after that,28, 29 which was more equivalent to our D3 (recovered stage) than D1 (acute episode) cohort. Despite this, they also found a higher abundance of Veillonellaceae in the HE group compared with the non-HE group,28 which is consistent with our findings. It is intriguing to speculate that the gut mucosa may maintain a microbiome imprint in the recovered stage, which might reflect that at acute episode of HE. Nevertheless, it is much more feasible for clinical application by assaying the microbiome in stool rather than that in gut mucosa because it is inadequate to perform any unnecessary invasive procedure in patients with AHE.
In sum, our results revealed that specific bacterial taxa are associated with the disease progression of HE. These taxa are useful for predicting disease outcome, and further exploration may reveal that some of them may act as probiotics that suppress HE.
Materials and Methods
Patient Collection
We enrolled all 147 samples from Chang Gung Memorial Hospital, Taiwan from November 2014 to March 2016. The AHE group comprised patients sent to our emergency unit due to an acute episode of HE. Cirrhosis was diagnosed on the basis of clinical and laboratory data, including ultrasonography and symptoms and signs of portal hypertension. The severity of HE was graded using the West Haven Criteria, which are based on the level of impairment to autonomy, changes in consciousness, intellectual function, behavior, and the dependence on therapy.30, 31 All first-time stool samples were collected within 12 hours of the patient entering our emergency unit. Patients were excluded if 1) they had received an antibiotics treatment before stool sample collection, 2) fecal sample collection would interfere with treatment, or 3) they were diagnosed with another disease that could influence conscious level (eg, a cerebral vascular accident or metabolic encephalopathy). A total of 62 fecal samples were collected from 62 cases with cirrhosis and AHE. A CTP score was used to assess the cirrhosis’ severity.32 Of the 62 patients, 22 had CTP class B and 40 had CTP class C (D1). An additional 34 fecal samples were collected from 34 of these patients with AHE 48–72 hours after treatment (D2), routinely including lactulose. Proton pump inhibitors were prescribed for some patients at 20–40 mg per day for period of 3 months. For a longitudinal study of post-AHE changes after AHE, a third fecal sample was further collected from 18 of the patients 2–4 months after their episode of AHE who had no overt HE (D3). We collected 20 stool samples from patients with compensated cirrhosis (without a history of ascites, HE, variceal hemorrhage, and associated severe complication) (C2), 15 samples from patient with advanced stages (CTP score >7) of cirrhosis with no overt HE 6 months before sampling (C3) and 13 from healthy individuals (C1) as control subjects (Table 1).
DNA Extraction and Amplicon Library Construction and Sequencing
Total genomic DNA extraction protocol from stool samples was a modification of the PowerSoil DNA isolation kit (Cat. #12888-100; MoBio, Carlsbad, CA), taken from the Human Microbiome Project (http://hmpdacc.org/doc/HMP_MOP_Version12_0_072910.pdf). In the initial processing of the stool samples, 1.0 g of sample was resuspended in 2.0 mL of MoBio bead solution by vigorous vortexing for 30 seconds in a 15-mL Falcon tube. The sample suspension was centrifuged for 5 minutes at 1500× rcf, and 900 μL of supernatant was transferred into MoBio Garnet bead tube with 750 μL of lysis buffer. The supernatant mixture was heated at 65°C for 10 minutes, and then at 95°C for 10 minutes. The rest of the protocol is as detailed by the manufacturer except for the following: 1) PowerLyser-24 setting during homogenization was set at 4200 rpm for 45 seconds, 2) volume of Solution C4 added was 1040 μL, and 3) spin columns were washed twice with 500 μL of Solution C5.
Amplicons were generated as previously described by Kozich et al,33 targeting the V3 and V4 regions of 16S rRNA. The reaction mix consisted of 17 μL of AccuPrime Pfx Supermix, 2.0 μL of primer each at 10 μM, and 1.0 μL of template genomic DNA. Thermocycling conditions were initial denaturation at 95°C for 2 minutes; 25 cycles of 95°C for 20 seconds, 55°C for 15 seconds, and 72°C for 5 minutes; and a final extension at 72°C for 10 minutes. Polymerase chain reaction products were analyzed using agarose gel electrophoresis to ensure expected products were obtained. The normalization of amplicons was carried out using SequalPrep Normalisation Plate Kit (Cat. #A1051001; Invitrogen, Waltham, MA). After normalization, 5.0 μL of the normalized amplicon was taken from each well to generate a library pool. The concentration of DNA library was measured using Invitrogen Qubit Fluorometer. The DNA library was concentrated using an equal volume of AMPure XP bead (Cat. #A63881; Agencourt AMPure XP; Beckman Coulter, Indianapolis, IN). For the metagenome sequencing from patients with the highest OTU4 relative abundance, genomic DNA from the same source that previously underwent amplicon generation was sheared in microTUBE-130 (Covaris, Woburn, MA) using a 500 bp shearing program (duty factor 10%, treatment for 60 seconds). A genomic library was prepared using TruSeq DNA LT Sample Prep Kit (Illumina, San Diego, CA). The input of 1-μg sheared DNA was used for end-repair, A-tailing, adaptor ligation, and gel size selection. Size range at 600–700 bps range was selected from the gel and amplified by 5 cycles of polymerase chain reaction. The sequencing was performed by the NGS core lab in Academia Sinica, Taiwan. Sequencing of 16S amplicon was carried out using Illumina MiSeq reagent kit v2, 2 × 250bp and metagenomics using Illumina HiSeq rapid mode 2 × 150 bp output format.
Data Analysis
All the clinical data were recorded and expressed as follows. The data of continuous variables were expressed as the mean ± SD, and the data of categorical variables were expressed as fractions. The Kolmogorov–Smirnov test was used to determine the normal distribution for each variable. The Student t test was used to compare between continuous variables which were normally distributed; otherwise, the Mann–Whitney U test was used. Categorical data were examined using the chi-square test or Fisher’s exact test. Data analyses were performed using SPSS 13 (IBM Corporation, Armonk, NY).
Amplicon raw sequencing data were prepared using UParse,34 and meta-taxonomic analysis was carried out using Phyloseq (v1.22.3)35 in an R (v3.4.2; R Foundation for Statistical Computing, Vienna, Austria) environment.
Metagenomic raw sequencing data were first sequence adaptor and sequence quality trimmed with Trimmomatic (ver 0.32) (options LEADING:30 TRAILING:30 SLIDINGWINDOW:4:30 MINLEN:50),36 the host sequences were removed and subsequently assembled using meta-SPAdes (ver 3.11.1),37 and genomes were recovered using MetaBAT2 (v 2.12.1)38 and taxonomically classified using Kraken (v 1.0).39 Genome bins that were classified as Veillonella were used for subsequent analysis. Single copy orthologs of 2 genomes recovered from this sample and 24 published Veillonella strains were collected from National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=29465) and determined by OrthoFinder (v 2.1.2).40 Then, MAFFT (v 7.310)41 was used to align sequences in each of 318 single-copy orthogroups. The alignments were concatenated and used to infer a maximum likelihood phylogeny using RAxML (v 7.7.8)42 with 100 bootstrap replicates. Whole genome alignment was inferred using MUMMER4.43
Footnotes
Conflicts of interest The authors disclose no conflicts.
Author contributions S.Y.H. conceived the concept and designed the studies. S.Y.H. and I.J.T. secured the data and funding. C.M.S., C.F.C., Y.F.L., H.M.K., H.Y.H., Y.N.G., and M.J.L. performed the experiments. S.Y.H., Y.F.L, I.J.T. and C.M.S. wrote the manuscript. K.F.C., and W.S.T. collected patients and clinical information.
Funding Funding was provided by the Chang Gung Medical Foundation, Taiwan, to Sen-Yung Hsieh (CMRPG 3E1411-3; 3H1031-3) and the National Health Research Institute, Taiwan, to Isheng J. Tsai (NHRI-EX106-10619SC).
Contributor Information
Isheng J. Tsai, Email: ijtsai@gate.sinica.edu.tw.
Sen-Yung Hsieh, Email: siming@cgmh.org.tw.
Supplementary Material
Supplementary Table S2.
Relative Abundance of OTU3 Across All Samples
| Disease Status | Relative Abundance of OTU3 (%) |
|---|---|
| C1 | 0.01 |
| C1 | 0.03 |
| C1 | 0.03 |
| C1 | 0.05 |
| C1 | 0.09 |
| C1 | 0.14 |
| C1 | 0.15 |
| C1 | 0.16 |
| C1 | 0.29 |
| C1 | 0.31 |
| C1 | 0.42 |
| C1 | 0.79 |
| C1 | 7.07 |
| C2 | 0.00 |
| C2 | 0.01 |
| C2 | 0.02 |
| C2 | 0.02 |
| C2 | 0.04 |
| C2 | 0.04 |
| C2 | 0.11 |
| C2 | 0.12 |
| C2 | 0.14 |
| C2 | 0.22 |
| C2 | 0.26 |
| C2 | 0.43 |
| C2 | 0.92 |
| C2 | 1.01 |
| C2 | 1.25 |
| C2 | 1.27 |
| C2 | 1.49 |
| C2 | 2.95 |
| C2 | 4.98 |
| C2 | 6.42 |
| C3 | 0.01 |
| C3 | 0.01 |
| C3 | 0.05 |
| C3 | 0.06 |
| C3 | 0.14 |
| C3 | 0.95 |
| C3 | 1.24 |
| C3 | 1.97 |
| C3 | 2.05 |
| C3 | 6.56 |
| C3 | 7.30 |
| C3 | 10.30 |
| C3 | 16.04 |
| C3 | 21.27 |
| C3 | 22.27 |
| D1 | 0.01 |
| D1 | 0.02 |
| D1 | 0.03 |
| D1 | 0.03 |
| D1 | 0.07 |
| D1 | 0.08 |
| D1 | 0.14 |
| D1 | 0.42 |
| D1 | 0.42 |
| D1 | 0.45 |
| D1 | 0.75 |
| D1 | 0.75 |
| D1 | 1.29 |
| D1 | 1.55 |
| D1 | 2.18 |
| D1 | 2.24 |
| D1 | 2.95 |
| D1 | 3.11 |
| D1 | 3.46 |
| D1 | 3.65 |
| D1 | 4.30 |
| D1 | 4.48 |
| D1 | 4.86 |
| D1 | 5.05 |
| D1 | 5.29 |
| D1 | 5.30 |
| D1 | 5.32 |
| D1 | 5.45 |
| D1 | 5.63 |
| D1 | 5.65 |
| D1 | 5.70 |
| D1 | 5.72 |
| D1 | 5.74 |
| D1 | 6.30 |
| D1 | 7.16 |
| D1 | 7.25 |
| D1 | 7.92 |
| D1 | 8.08 |
| D1 | 8.10 |
| D1 | 8.18 |
| D1 | 8.84 |
| D1 | 10.00 |
| D1 | 10.76 |
| D1 | 13.28 |
| D1 | 13.29 |
| D1 | 13.58 |
| D1 | 16.40 |
| D1 | 17.07 |
| D1 | 17.72 |
| D1 | 18.89 |
| D1 | 21.69 |
| D1 | 26.53 |
| D1 | 27.02 |
| D1 | 27.26 |
| D1 | 29.64 |
| D1 | 30.06 |
| D1 | 30.48 |
| D1 | 30.85 |
| D1 | 32.01 |
| D1 | 41.20 |
| D1 | 50.09 |
| D1 | 50.72 |
| D2 | 0.08 |
| D2 | 0.11 |
| D2 | 0.11 |
| D2 | 0.12 |
| D2 | 0.13 |
| D2 | 0.15 |
| D2 | 0.19 |
| D2 | 0.24 |
| D2 | 0.33 |
| D2 | 0.34 |
| D2 | 0.35 |
| D2 | 0.62 |
| D2 | 0.62 |
| D2 | 0.69 |
| D2 | 1.71 |
| D2 | 1.96 |
| D2 | 1.99 |
| D2 | 2.75 |
| D2 | 3.94 |
| D2 | 6.91 |
| D2 | 7.72 |
| D2 | 11.05 |
| D2 | 12.62 |
| D2 | 15.19 |
| D2 | 15.64 |
| D2 | 17.72 |
| D2 | 18.46 |
| D2 | 21.90 |
| D2 | 23.11 |
| D2 | 28.78 |
| D2 | 29.64 |
| D2 | 35.60 |
| D2 | 44.66 |
| D2 | 53.28 |
| D3 | 0.01 |
| D3 | 0.01 |
| D3 | 0.08 |
| D3 | 0.15 |
| D3 | 0.31 |
| D3 | 0.32 |
| D3 | 0.44 |
| D3 | 0.87 |
| D3 | 0.90 |
| D3 | 1.36 |
| D3 | 2.05 |
| D3 | 3.04 |
| D3 | 3.97 |
| D3 | 6.28 |
| D3 | 6.69 |
| D3 | 12.20 |
| D3 | 20.92 |
| D3 | 47.96 |
OTU3, operational taxonomic unit 3.
Supplementary Materials
References
- 1.Romero-Gomez M., Montagnese S., Jalan R. Hepatic encephalopathy in patients with acute decompensation of cirrhosis and acute-on-chronic liver failure. J Hepatol. 2015;62:437–447. doi: 10.1016/j.jhep.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Gustot T., Fernandez J., Garcia E., Morando F., Caraceni P., Alessandria C., Laleman W., Trebicka J., Elkrief L., Hopf C., Solis-Munoz P., Saliba F., Zeuzem S., Albillos A., Benten D., Montero-Alvarez J.L., Chivas M.T., Concepcion M., Cordoba J., McCormick A., Stauber R., Vogel W., de Gottardi A., Welzel T.M., Domenicali M., Risso A., Wendon J., Deulofeu C., Angeli P., Durand F., Pavesi M., Gerbes A., Jalan R., Moreau R., Gines P., Bernardi M., Arroyo V. Clinical course of acute-on-chronic liver failure syndrome and effects on prognosis. Hepatology. 2015;62:243–252. doi: 10.1002/hep.27849. [DOI] [PubMed] [Google Scholar]
- 3.Cordoba J., Ventura-Cots M., Simon-Talero M., Amoros A., Pavesi M., Vilstrup H., Angeli P., Domenicali M., Gines P., Bernardi M., Arroyo V. Characteristics, risk factors, and mortality of cirrhotic patients hospitalized for hepatic encephalopathy with and without acute-on-chronic liver failure (ACLF) J Hepatol. 2014;60:275–281. doi: 10.1016/j.jhep.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Tilg H., Cani P.D., Mayer E.A. Gut microbiome and liver diseases. Gut. 2016;65:2035–2044. doi: 10.1136/gutjnl-2016-312729. [DOI] [PubMed] [Google Scholar]
- 5.Victor D.W., 3rd, Quigley E.M. Hepatic encephalopathy involves interactions among the microbiota, gut, brain. Clin Gastroenterol Hepatol. 2014;12:1009–1011. doi: 10.1016/j.cgh.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y., Yang F., Lu H., Wang B., Chen Y., Lei D., Wang Y., Zhu B., Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562–572. doi: 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 7.Bajaj J.S., Heuman D.M., Hylemon P.B., Sanyal A.J., White M.B., Monteith P., Noble N.A., Unser A.B., Daita K., Fisher A.R., Sikaroodi M., Gillevet P.M. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940–947. doi: 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen T.C., Albenberg L., Bittinger K., Chehoud C., Chen Y.Y., Judge C.A., Chau L., Ni J., Sheng M., Lin A., Wilkins B.J., Buza E.L., Lewis J.D., Daikhin Y., Nissim I., Yudkoff M., Bushman F.D., Wu G.D. Engineering the gut microbiota to treat hyperammonemia. J Clin Invest. 2015;125:2841–2850. doi: 10.1172/JCI79214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bajaj J.S., Kassam Z., Fagan A., Gavis E.A., Liu E., Cox I.J., Kheradman R., Heuman D., Wang J., Gurry T., Williams R., Sikaroodi M., Fuchs M., Alm E., John B., Thacker L.R., Riva A., Smith M., Taylor-Robinson S.D., Gillevet P.M. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: a randomized clinical trial. Hepatology. 2017;66:1727–1738. doi: 10.1002/hep.29306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahluwalia V., Betrapally N.S., Hylemon P.B., White M.B., Gillevet P.M., Unser A.B., Fagan A., Daita K., Heuman D.M., Zhou H., Sikaroodi M., Bajaj J.S. Impaired gut-liver-brain axis in patients with cirrhosis. Sci Rep. 2016;6:26800. doi: 10.1038/srep26800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bajaj J.S. The role of microbiota in hepatic encephalopathy. Gut Microbes. 2014;5:397–403. doi: 10.4161/gmic.28684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gronow S., Welnitz S., Lapidus A., Nolan M., Ivanova N., Glavina Del Rio T., Copeland A., Chen F., Tice H., Pitluck S., Cheng J.F., Saunders E., Brettin T., Han C., Detter J.C., Bruce D., Goodwin L., Land M., Hauser L., Chang Y.J., Jeffries C.D., Pati A., Mavromatis K., Mikhailova N., Chen A., Palaniappan K., Chain P., Rohde M., Goker M., Bristow J., Eisen J.A., Markowitz V., Hugenholtz P., Kyrpides N.C., Klenk H.P., Lucas S. Complete genome sequence of Veillonella parvula type strain (Te3) Stand Genomic Sci. 2010;2:57–65. doi: 10.4056/sigs.521107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwai S., Weinmaier T., Schmidt B.L., Albertson D.G., Poloso N.J., Dabbagh K., DeSantis T.Z. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One. 2016;11:e0166104. doi: 10.1371/journal.pone.0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sartini A., Gitto S., Bianchini M., Verga M.C., Di Girolamo M., Bertani A., Del Buono M., Schepis F., Lei B., De Maria N., Villa E. Non-alcoholic fatty liver disease phenotypes in patients with inflammatory bowel disease. Cell Death Dis. 2018;9:87. doi: 10.1038/s41419-017-0124-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenson W.F. The universe of arachidonic acid metabolites in inflammatory bowel disease: can we tell the good from the bad? Curr Opin Gastroenterol. 2014;30:347–351. doi: 10.1097/MOG.0000000000000075. [DOI] [PubMed] [Google Scholar]
- 17.Thomas M.H., Pelleieux S., Vitale N., Olivier J.L. Dietary arachidonic acid as a risk factor for age-associated neurodegenerative diseases: potential mechanisms. Biochimie. 2016;130:168–177. doi: 10.1016/j.biochi.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Bajaj J.S., Vargas H.E., Reddy K.R., Lai J.C., O'Leary J.G., Tandon P., Wong F., Mitrani R., White M.B., Kelly M., Fagan A., Patil R., Sait S., Sikaroodi M., Thacker L.R., Gillevet P.M. Association between intestinal microbiota collected at hospital admission and outcomes of patients with cirrhosis. Clin Gastroenterol Hepatol. 2019;17:756–765.e3. doi: 10.1016/j.cgh.2018.07.022. [DOI] [PubMed] [Google Scholar]
- 19.Bajaj J.S., Fagan A., White M.B., Wade J.B., Hylemon P.B., Heuman D.M., Fuchs M., John B.V., Acharya C., Sikaroodi M., Gillevet P.M. Specific gut and salivary microbiota patterns are linked with different cognitive testing strategies in minimal hepatic encephalopathy. Am J Gastroenterol. 2019 Jan 28 doi: 10.14309/ajg.0000000000000102. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin N., Yang F., Li A., Prifti E., Chen Y., Shao L., Guo J., Le Chatelier E., Yao J., Wu L., Zhou J., Ni S., Liu L., Pons N., Batto J.M., Kennedy S.P., Leonard P., Yuan C., Ding W., Chen Y., Hu X., Zheng B., Qian G., Xu W., Ehrlich S.D., Zheng S., Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 21.Bajaj J.S., Betrapally N.S., Gillevet P.M. Decompensated cirrhosis and microbiome interpretation. Nature. 2015;525:E1–E2. doi: 10.1038/nature14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajaj J.S., Betrapally N.S., Hylemon P.B., Heuman D.M., Daita K., White M.B., Unser A., Thacker L.R., Sanyal A.J., Kang D.J., Sikaroodi M., Gillevet P.M. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. 2015;62:1260–1271. doi: 10.1002/hep.27819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acharya C., Sahingur S.E., Bajaj J.S. Microbiota, cirrhosis, and the emerging oral-gut-liver axis. JCI Insight. 2017;2:94416. doi: 10.1172/jci.insight.94416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y., Ji F., Guo J., Shi D., Fang D., Li L. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci Rep. 2016;6:34055. doi: 10.1038/srep34055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajaj J.S., Idilman R., Mabudian L., Hood M., Fagan A., Turan D., White M.B., Karakaya F., Wang J., Atalay R., Hylemon P.B., Gavis E.A., Brown R., Thacker L.R., Acharya C., Heuman D.M., Sikaroodi M., Gillevet P.M. Diet affects gut microbiota and modulates hospitalization risk differentially in an international cirrhosis cohort. Hepatology. 2018;68:234–247. doi: 10.1002/hep.29791. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Guo Z., Xue Z., Sun Z., Zhang M., Wang L., Wang G., Wang F., Xu J., Cao H., Xu H., Lv Q., Zhong Z., Chen Y., Qimuge S., Menghe B., Zheng Y., Zhao L., Chen W., Zhang H. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J. 2015;9:1979–1990. doi: 10.1038/ismej.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V., Reyes J.A., Shah S.A., LeLeiko N., Snapper S.B., Bousvaros A., Korzenik J., Sands B.E., Xavier R.J., Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bajaj J.S., Ridlon J.M., Hylemon P.B., Thacker L.R., Heuman D.M., Smith S., Sikaroodi M., Gillevet P.M. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168–G175. doi: 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bajaj J.S., Hylemon P.B., Ridlon J.M., Heuman D.M., Daita K., White M.B., Monteith P., Noble N.A., Sikaroodi M., Gillevet P.M. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675–G685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cash W.J., McConville P., McDermott E., McCormick P.A., Callender M.E., McDougall N.I. Current concepts in the assessment and treatment of hepatic encephalopathy. QJM. 2010;103:9–16. doi: 10.1093/qjmed/hcp152. [DOI] [PubMed] [Google Scholar]
- 31.Ferenci P., Lockwood A., Mullen K., Tarter R., Weissenborn K., Blei A.T. Hepatic encephalopathy–definition, nomenclature, diagnosis, and quantification: final report of the working party at the 11th World Congresses of Gastroenterology, Vienna, 1998. Hepatology. 2002;35:716–721. doi: 10.1053/jhep.2002.31250. [DOI] [PubMed] [Google Scholar]
- 32.Pugh R.N., Murray-Lyon I.M., Dawson J.L., Pietroni M.C., Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60:646–649. doi: 10.1002/bjs.1800600817. [DOI] [PubMed] [Google Scholar]
- 33.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 35.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang D.D., Froula J., Egan R., Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emms D.M., Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K., Standley D.M. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 2014;1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- 42.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England) 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 43.Marcais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.