Abstract
The combination of daunorubicin (dnr) and cytarabine (Ara-C) is a cornerstone of treatment for acute myelogenous leukemia (AML); resistance to these drugs is a major cause of treatment failure. Ceramide, a sphingolipid (SL), plays a critical role in cancer cell apoptosis in response to chemotherapy. Here, we investigated the effects of chemotherapy selection pressure with Ara-C and dnr on SL composition and enzyme activity in the AML cell line HL-60. Resistant cells, those selected for growth in Ara-C- and dnr-containing medium (HL-60/Ara-C and HL-60/dnr, respectively), demonstrated upregulated expression and activity of glucosylceramide synthase, acid ceramidase (AC), and sphingosine kinase 1 (SPHK1); were more resistant to ceramide than parental cells; and displayed sensitivity to inhibitors of SL metabolism. Lipidomic analysis revealed a general ceramide deficit and a profound upswing in levels of sphingosine 1-phosphate (S1P) and ceramide 1-phosphate (C1P) in HL-60/dnr cells versus parental and HL-60/Ara-C cells. Both chemotherapy-selected cells also exhibited comprehensive upregulations in mitochondrial biogenesis consistent with heightened reliance on oxidative phosphorylation, a property that was partially reversed by exposure to AC and SPHK1 inhibitors and that supports a role for the phosphorylation system in resistance. In summary, dnr and Ara-C selection pressure induces acute reductions in ceramide levels and large increases in S1P and C1P, concomitant with cell resilience bolstered by enhanced mitochondrial remodeling. Thus, strategic control of ceramide metabolism and further research to define mitochondrial perturbations that accompany the drug-resistant phenotype offer new opportunities for developing therapies that regulate cancer growth.
Keywords: sphingolipids, ceramide, cancer, drug resistance
A chemotherapy regimen consisting of daunorubicin (dnr) and cytarabine (Ara-C) is the cornerstone of induction therapy in acute myelogenous leukemia (AML) (1), a hematological malignancy marked by the accumulation of large numbers of immature myeloblasts in bone marrow. The overall prognosis of AML is poor. In older patients, unfavorable karyotypes are common with antecedent myelodysplastic syndromes (2). In patients receiving therapy with curative intent, less than one half will achieve long-term survival. Chemotherapy resistance, which can be intrinsic or acquired, is the major cause of treatment failure (1), as most patients subsequently relapse with a chemoresistant disease that is difficult to treat and manage.
Resistance to drugs with diverse chemical structures and mechanisms of action is known as multidrug resistance (3). In leukemia, resistance mechanisms that involve membrane-resident proteins belonging to the ABC transporter protein family are of interest (4, 5). In this scenario, enhanced expression of these proteins is often associated with poor prognosis and frequent relapsed or refractory disease (6). ABC transporters act as ATP-dependent efflux pumps and thus reduce the intracellular concentrations of anticancer agents, greatly diminishing clinical efficacy. In addition to drug efflux, other factors contribute to drug resistance, including dysfunctional cell-death inhibition signaling, DNA damage repair, drug inactivation, and mitochondrial alterations (7). In this study, we used chemotherapy selection pressure with Ara-C and dnr, specific agents for treating of AML, to determine whether alterations in sphingolipids (SLs) are affiliated with AML cell survival under conditions of prolonged chemotherapy exposure wherein drug resistance is likely to develop. Because ceramide plays a principle role in initiating apoptosis in cancer cells in response to a myriad of anticancer agents (8–11), we hypothesized that changes in SLs would accompany dnr and Ara-C selection pressure and contribute to cell survival.
Ceramide, a proapoptotic tumor-suppressing SL, is the hydrophobic backbone of most SLs and glycosphingolipids (12). Historically, some of the first evidence of chemotherapy interaction with SL metabolism was reported by Bose et al. (13) and Jaffrezou et al. (14). Using leukemia cell models, these investigators observed that dnr stimulated ceramide formation and elicited apoptosis via the de novo pathway by activating ceramide synthase (CerS) and via sphingomyelin hydrolysis. These studies emphasized the importance of ceramide in dnr-induced cell death. With ceramide playing a central role in the induction of apoptosis (8), it is easy to envision that cancer cell-defense mechanisms might use the conversion and/or destruction of ceramide for survival, and indeed upregulated ceramide glycosylation has been identified as a marker of drug resistance in numerous cancer types, including leukemia (15–17).
Beyond the ability of chemotherapy drugs and ionizing radiation to promote ceramide production (13, 14, 18, 19) and the role of ceramide glycosylation in dampening apoptotic responses (15, 20), little is known regarding the effects of chemotherapy on the more global aspects of SLs. In this work we demonstrate that selection pressure with chemotherapy drugs used to treat AML elicited profound alterations in SL composition and enhanced the expression of SL enzymes that catalyze ceramide glycosylation, hydrolysis, and formation of sphingosine 1-phosphate (S1P). These enzymatic steps limit intracellular ceramide levels as well as underpin mitogenicity via S1P production. These findings could foster innovative approaches for overcoming drug resistance. Of note, we found that HL-60/dnr (dnr-resistant) and HL-60/Ara-C (Ara-C resistant) cells were partially refractory to ceramide, a finding that implicates ceramide in drug resistance biology. Moreover, profound alterations in mitochondrial quality and quantity were accompanied by dnr and Ara-C selection pressure, lending further credence to the notion that increased reliance/capacitance within the oxidative phosphorylation (OXPHOS) system may be a hallmark of AML drug resistance (7).
MATERIALS AND METHODS
Materials
Ara-C and dnr (hydrochloride) were obtained from R&D Systems (Minneapolis, MN); 1.0 and 10 mM stock solutions were made in water, filter-sterilized, and stored at −20°C. Propidium iodide (PI) was obtained from Thermo Fisher Scientific (Waltham, MA). The acid ceramidase (AC) inhibitors DM102 and 2-chloro-N-(2S,3R)-1,3-dihydroxyoctadecan-2-yl acetamide (SACLAC) were obtained from the Research Unit on Bioactive Molecules, Institute for Advanced Chemistry of Catalonia (Barcelona, Spain). The sphingosine kinase 1 (SPHK1) inhibitor SK1-i was a gift from Enzo Life Sciences (Farmingdale, NY). FTY720 hydrochloride (fingolimod), an S1P receptor modulator and SPHK1 inhibitor that also induces SPHK1 degradation (21), and NBD-C6-ceramide [N-[7-(nitrobenzo-2-oxa-1,3-diazole)]6-aminocaproyl-d-erythro-sphingosine] were obtained from Cayman Chemical Co. (Ann Arbor, MI). NBD-C6-ceramide complexed to BSA was from Thermo Fisher Scientific. The glucosylceramide (GCS) inhibitor d,l-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol (PDMP) was purchased from Matreya (State College, PA). N-Hexanoyl-d-erythro-sphingosine (C6-ceramide) and N-stearoyl-d-erythro-sphingosine (C18-ceramide) were purchased from Avanti Polar Lipids (Alabaster, AL). For use in experiments, C18-ceramide was dissolved in 98% methanol and 2% dodecane (5.0 mM) by vortex mixing in 1 dram glass vials and warming at 37°C. The solvent was evaporated under a stream of nitrogen, and the resultant C18-ceramide film was dissolved in 100% ethanol, briefly warmed at 40°C, and injected and vortex mixed into 37°C culture medium containing 10% FBS. Eliglustat was a gift from Dr. James A. Shayman (University of Michigan, Ann Arbor, MI).
Cell culture and establishment of drug-resistant cells
The human AML cell line HL-60 (designated wt for wild-type) was obtained from ATCC (Manassas, VA). The cell line was not tested or authenticated over and above documentation provided by ATCC, which included antigen expression, DNA profile, short tandem repeat profiling, and cytogenetic analysis. Cells were maintained in RPMI-1640 medium (Thermo Fisher Scientific) supplemented with 10% FBS (Peak Serum, Inc., Wellington, CO) and 100 units/ml penicillin and 100 µg/ml streptomycin. Cells were selected for Ara-C and dnr resistance via continuous, long-term exposure to stepwise increments in drug concentrations ranging from 12.5 to 500 nM. Ara-C-selected cells (HL-60/Ara-C) seeded at 0.25 × 106 to 0.5 × 106 cells/ml were first cultured in medium containing 25 nM Ara-C. The drug concentration was doubled every 3–4 weeks over approximately 6–8 passages to a maximum of 500 nM. Cells resistant to dnr (HL-60/dnr) were developed in a similar manner except that the initial concentration was 12.5 nM. HL-60/dnr and HL-60/Ara-C cells were maintained in a medium containing a final concentration of 400 nM dnr and 500 nM Ara-C, respectively. Individual experiments were conducted in the absence of these chemotherapy drugs.
Cell viability assays
Cell viability was determined by fluorescence measurement as previously described (22). Briefly, cells were seeded in black-wall 96-well plates in RPMI-1640 medium containing 5% FBS (cell numbers given in figure legends). After the addition of agents (0.2 ml final well volume), cells were incubated at 37°C, 5% CO2, for the times indicated. Viability was determined using PI as follows. Positive control cells were permeabilized by the addition of 10 µl 1.0 mg/ml digitonin and incubated at 37°C, 5% CO2, for 20 min. Plates were then centrifuged at 120 g for 20 min, and after dumping the media, 0.1 ml of a 5.0 µM PI solution in PBS was added. The plate was again incubated for 20 min, and viability was calculated as the mean (n = 6) fluorescence (minus permeabilized vehicle control) at 530 nm excitation and 620 nm emission, using a BIO-TEK Synergy H1 microplate reader (BIO-TEK Instruments, Winooski, VT).
Cell viability was also measured by trypan blue exclusion. For this procedure a Countess II automated cell counter was used (Thermo Fisher Scientific), with disposable hemocytometers, following the manufacturer’s instructions.
Evaluation of apoptosis by Annexin V FITC/PI
Cells were seeded in 6-well plates (1 × 106 cells/ml RPMI-1640 medium containing 10% FBS) and treated with SK1-i (10 µM) for 48 h. Cells were then harvested by centrifugation and mixed with the Annexin V-FITC kit (Trevigen, Gaithersburg, MD) according to the manufacturer’s instructions. The stained cells were examined by flow cytometry on an LSRII flow cytometer (BD Biosciences, San Jose, CA). PI was used to discriminate early apoptosis (Annexin V+/PI− cells) and late apoptosis (Annexin V+/PI+ cells) according to the manufacturer’s instructions. Flow cytometry data were analyzed by FCSalyzer 0.9.17-α.
Hematoxylin and eosin staining
Cytospin preparations (23) of the leukemia cells were stained with hematoxylin and eosin for morphological evaluation. Each microscopic field was captured with 200× magnification. More than three fields were required for review.
GCS, AC, and SPHK1 enzyme activity assays
GCS activity was measured in intact HL-60 wt and in drug-resistant counterparts using C6-NBD-ceramide complexed to BSA as previously described (22, 24). The GCS assays were conducted in the absence of the chemotherapy drugs. Briefly, 100,000 viable cells in 45 µl serum-free RPMI-1640 medium containing 1% BSA were seeded into 96-well plates. The assay was initiated by the addition of 5 µl NBD-C6-ceramide complexed to BSA (25 µM final C6-ceramide substrate concentration) and placed in a tissue-culture incubator for 1 h (the reaction was linear to 90 min). Samples were then placed on ice, and the cells were transferred to 1 dram glass vials for lipid extraction (25). The lower, lipid-containing phase was evaporated to dryness under a stream of nitrogen. Total lipids were dissolved by the addition of 40 μl chloroform-methanol (5:1; v/v) and vortex mixed, and 5 µl was applied to the origin of an HPTLC plate (silica gel 60 F254; Sigma-Aldrich). C6-NBD-ceramide standard was spotted in lateral lanes. Lipids were resolved in a solvent system containing chloroform-methanol-ammonium hydroxide (80:20:2; v/v/v). Products were analyzed directly on the HPTLC plates on a BioRad ChemiDoc Touch and quantified with Image Lab software by BioRad (Hercules, CA). AC activity was evaluated in intact cells using a cell-permeable fluorogenic substrate, RBM14-12 (26, 27), as follows. First, 100,000 cells were seeded in 96-well plates in serum-free RPMI-1640 medium containing 1% BSA, and fluorogenic substrate was added to a final concentration of 16 µM (125 µl final well volume). Plates were then placed in a tissue culture incubator for 2 h. Finally, 50 µl methanol and 100 µl NaIO4 (2.5 mg/ml) in 0.1 M glycine buffer, pH 10.6, was added, and the plates were incubated in the dark for 2 h at 37°C. Fluorescence was measured at 365 nm excitation/410–460 nm emission. SPHK1 activity was measured using a K-3500 kit (Echelon, Salt Lake City, UT) following the manufacturer’s instructions.
Mass spectrometry
Lipidomic analysis, inclusive of C1P, was conducted by LC/ESI/MS/MS as previously described (28). Briefly, total lipids were extracted from cells using ethyl acetate-isopropanol-water (60:30:10; v/v/v) without phase partitioning, and solvents were evaporated (azeotrope) under a stream of nitrogen. Internal standards were added, and separation was achieved using Waters I-class Acquity LC and Waters Xevo TQ-S instruments.
Lactate dehydrogenase determination
Lactate dehydrogenase (LDH) activity was detected in cells using an LDH activity assay kit according to the manufacturer’s instructions (Sigma-Aldrich). Activity is represented as the variation in optical density at 340 nm, which is proportional to the quantity of NADH oxidized. Working reagent (0.1 ml) was added to the wells of a 96-well plate, followed by the addition of 10 µl of the cell lysates (equivalent to 100,000 cells). The plate was shaken well and incubated at 37°C for 20 min. Activity was calculated according to the standard curve for the concentration as follows: pyruvate + NADH + H yields lactate + NAD+.
Cellular respirometry
High-resolution O2 consumption measurements were conducted using the Oroboros Oxygraph-2K (Oroboros Instruments, Innsbruck, Austria) in intact HL-60, HL-60/Ara-C, and HL-60/dnr cells. For each experiment, HL-60 wt and resistant cells were centrifuged at 1,000 rpm for 7 min at room temperature and then suspended in bicarbonate-free RPMI-1640 medium, supplemented with 20 mM HEPES (pH 7.4) and 5% FBS, without Ara-C or dnr, at a final concentration of 2 × 106 viable cells/ml. All experiments were carried out at 37°C in a 2 ml reaction volume (4–6 × 106 viable cells/chamber). Following the assessment of basal respiration, 2.0 µM carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), a mitochondrial uncoupling agent, was added to induce maximal respiratory flux. For experiments involving SACLAC, basal respiration was assessed, followed by the sequential additions of oligomycin (2.5 µM) and increasing concentrations of FCCP (0.5, 1.0, and 2.0 µM).
Immunoblotting
Immunoblotting was conducted as previously described (28, 29) with modifications. SPHK1 (47 kDa), used at 1:1,000 dilution, and β-actin (45 kDa), used at 1:3,000 dilution, were from Cell Signaling Technology (Danvers, MA). P-glycoprotein (P-gp; 180 kDa), used at 1:1,000 dilution, was from Novus Biologicals (Centennial, CO). pSPHK1 (47 kDa), used at 1:1,000 dilution, was from ECM Biosciences (Versailles, KY). GCS (40 kDa), used at 1:1,000 dilution, was from Thermo Fisher Scientific. AC (13 kDa), used at 1:1,000 dilution, was from BD Biosciences. Briefly, cells were lysed in 1× RIPA buffer (Cell Signaling Technology) containing phosphatase inhibitor cocktail 2, protease inhibitor, and PMSF (1.0 mM) according to the manufacturer’s protocol (Sigma-Aldrich). The BCA method was used for determining protein concentrations. Lysates were centrifuged to remove debris and then heated in 1× NuPage LDS sample buffer at 95°C for 5 min (GCS lysate was heated at 55°C for 5 min). Equal amounts of protein were loaded onto 4%–12% NuPAGE gels for electrophoresis and subsequently transferred onto PVDF in Odyssey Blocking Buffer (LI-COR Biotechnology, Lincoln, NE) for 1 h. After incubating with primary antibodies, the membranes were washed three times with PBS containing 0.1% Tween 20. Membranes were then incubated for 45 min with IRDye 680RD-conjugated goat anti-rabbit IgG and IRDye 680RD-conjugated goat anti-mouse IgG secondary antibodies (LI-COR Biotechnology) diluted in Odyssey Blocking Buffer. The blots were then washed three times with PBS-Tween 20 and rinsed with PBS. Proteins were visualized by scanning the membrane on an Odyssey infrared imaging system (LI-COR Biotechnology) with a 700 nm channel.
Quantitative RT-PCR
An RNA extraction kit (Qiagen, Waltham, MA) was used to extract total RNA, which was then reverse-transcribed into cDNA using a ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs, Ipswich, MA). The synthesized cDNAs were amplified with SYBR Premix (Bimake, Houston, TX) using the ABI VII7 RT-PCR system (Thermo Fisher Scientific). The PCR cycling parameters were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Relative mRNA levels were calculated using the comparative Ct method and presented as ratios to their biological controls. The fold change in the expression of each target mRNA relative to β-actin or GAPDH was calculated as 2Δ(ΔCt), where ΔCt = ΔCtAcin − Ctgenes. β-Actin transcript levels were confirmed to correlate well with total RNA levels and therefore were used for normalization throughout the experiments (30, 31). The primers used for RT-PCR were designed by primerbank (http://pga.mgh.harvard.edu/primerbank): ABCB1: F:TTGCTGCTTACATTCAGGTTTCA, R:AGCCTATCTCCTGTCGCATTA; ALDH1A2: F:TTGCAGGGCGTCATCAAAAC, R:ACACTCCAATGGGTTCATGTC; and β-actin: F:GCTGTGCTACGTCGCCCTG, R:GGAGGAGCTGGAAGCAGCC.
Statistical evaluation
All data are expressed as the mean ± SEM from triplicate runs, and all statistical analyses were performed using an unpaired two-tailed Student’s t-test or one- or two-way ANOVA with post hoc analysis as appropriate. Statistical significance in the figures is indicated as follows: *p < 0.05; **p < 0.01; and ***p < 0.001.
RESULTS
Characteristics of HL-60 cells grown under Ara-C and dnr selection pressure
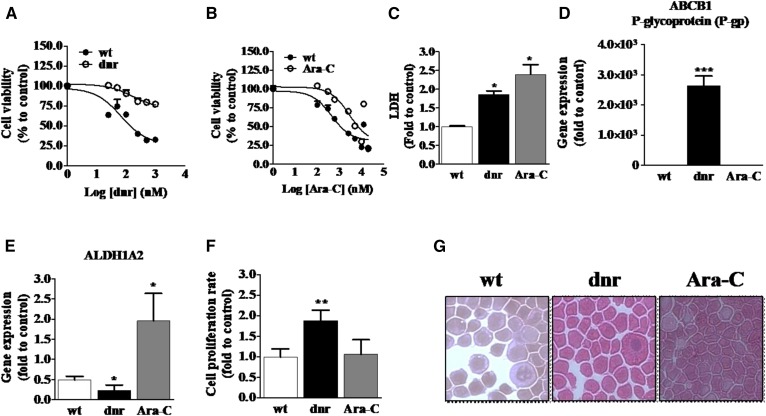
Culturing HL-60 cells for extended periods in medium containing Ara-C and in medium containing dnr yielded cells that were more refractory to these AML drugs compared with chemotherapy-naive HL-60 wt cells. For example, HL-60/dnr cells were approximately 6-fold more resistant to dnr than wt cells (Fig. 1A), whereas HL-60/Ara-C cells were 2.5-fold more refractory to Ara-C compared with wt (Fig. 1B). LDH activity, which can be elevated in drug resistance (32–35), was significantly higher in both HL-60/dnr and HL-60/Ara-C cells compared with wt (Fig. 1C). In addition, as shown by gene expression (Fig. 1D), only dnr-selected cells exhibited upregulated expression of ABCB1 (P-gp), a classic marker of anthracycline resistance (32, 33). Moreover, acquired resistance of leukemia cells to Ara-C has been shown to be associated with the upregulation of aldehyde dehydrogenase 1 (ALDH1A2) (36), a characteristic demonstrated in the HL-60/Ara-C model but not in HL-60/dnr or wt cells (Fig. 1E). These data establish the phenotypes of our cell models as drug-sensitive (wt) and drug-resistant. Experiments to determine growth rates revealed that doubling times for HL-60/dnr cells were 1.8-fold higher than wt and Ara-C counterparts (Fig. 1F), and last, a morphological evaluation revealed no illuminating alterations between wt and drug-resistant variants (Fig. 1G).
Fig. 1.
Characteristics of chemotherapy-naive HL-60 cells and HL-60 cells grown under dnr and Ara-C selection pressure. A: dnr sensitivity in wt and HL-60/dnr cells. B: Ara-C sensitivity in wt and HL-60/Ara-C cells. Cells (50,000) were seeded in black-walled 96-well plates and exposed to either dnr or Ara-C at the concentrations shown for 72 h. Viability was determined using PI staining. C: LDH levels in HL-60 (wt) and dnr- and Ara-C-resistant counterparts. The assay was conducted in dnr- and Ara-C-free media. D: ABCB1 expression. E: ALDH1A2 expression in wt and dnr- and Ara-C-resistant cells. Gene expression was quantitated by RT-PCR as detailed in Materials and Methods. F: Cellular proliferation rates. Cells (400,000) were seeded in 12-well plates and cultured for 72 h, after which the cell number was determined using automated enumeration and disposable hemocytometers. Dnr and Ara-C were present during the proliferation experiment. G: Cell morphology. Cytospin preparations were stained as detailed in Materials and Methods (200× magnification). HL-60 wt (control, chemotherapy-naive), HL-60/dnr (resistant to 400 nM dnr), and HL-60/Ara-C (resistant to 500 nM Ara-C) cells were used in all experiments.
Chemotherapy selection pressure with dnr and Ara-C enhances GCS, AC, and SPHK1 enzyme expression and activity
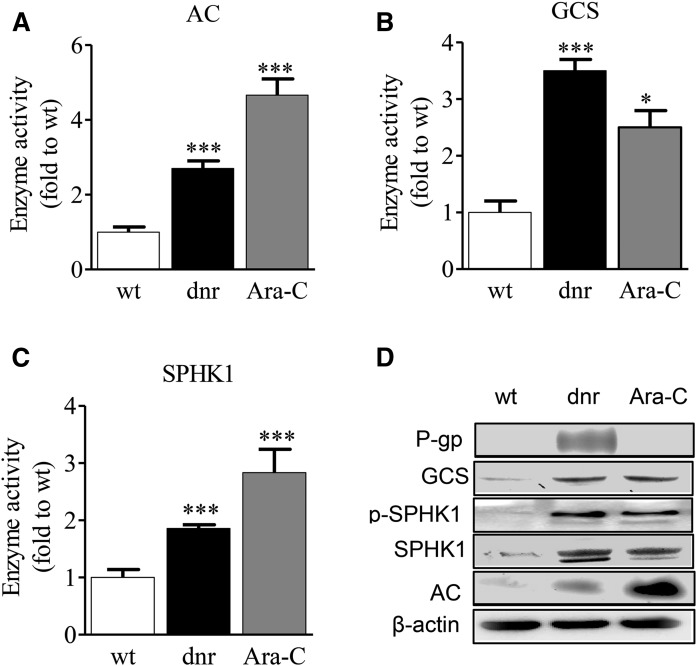
GCS, AC, and SPHK1 are enzymes involved in the regulation of cancer cell growth (37). Subsequent experiments were therefore aimed at determining the impact of dnr and Ara-C selection pressure on these relevant catalysts. GCS, AC, and SPHK1 activities were all augmented by drug selection pressure. The data in Fig. 2A demonstrate that Ara-C selection pressure had a more pronounced effect on AC activity compared with dnr selection pressure, 4.7-fold and 2.7-fold, respectively, compared with wt cells. GCS activity also increased in cells cultured to accommodate dnr and Ara-C. For example, GCS activity in HL-60/dnr and HL-60/Ara-C cells was 3.5- and 2.3-fold over wt, respectively (Fig. 2B). SPHK1 activity in HL-60/dnr and HL-60/Ara-C cells was 1.9- and 2.8-fold over the control, respectively, compared with wt (Fig. 2C). Immunoblot analysis showed that the expression of these enzymes also increased in chemotherapy-selected cells, inclusive of the phosphorylated form of SPHK1, pSPHK1 (Fig. 2D). Last, consistent with mRNA data (Fig. 1D), high P-gp expression was demonstrated only in HL-60/dnr cells (Fig. 2D).
Fig. 2.
Chemotherapy selection pressure with dnr and Ara-C enhances SL enzyme activity in HL-60 cells. A–C: Effect of dnr and Ara-C selection pressure on AC, GCS, and SPHK activity. Note that dnr and Ara-C were not present in the enzyme activity assays. D: Western blot depicting the expression of P-gp, GCS, pSPHK1, SPHK1, and AC in wt and dnr- and Ara-C-selected cells. β-Actin was used as a housekeeping gene. AC and GCS enzymatic activity were determined in intact cells as described in Materials and Methods. SPHK activity was determined by MS as described.
dnr and Ara-C selection pressure contribute to ceramide insensitivity
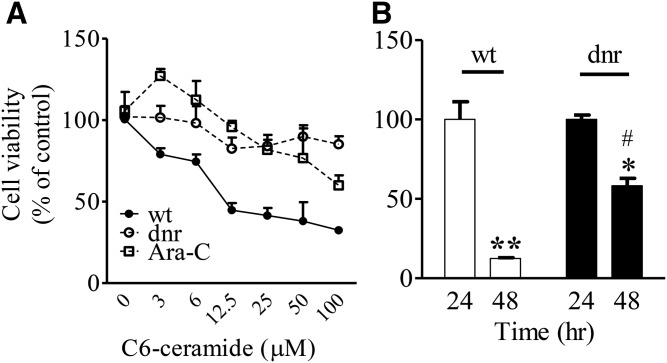
Ceramide is a potent tumor suppressor (8) whose effects can be diminished by catabolic and anabolic steps (15–17). Because chemotherapy-selected cells overexpressed AC and GCS, we sought to determine whether ceramide cytotoxicity would be altered in these cells. As shown in Fig. 3A, HL-60/dnr and HL-60/Ara-C cells were more refractory to C6-ceramide as opposed to wt cells; thus, partial ceramide resistance appears to accompany chemotherapy resistance in our models. In addition to C6-ceramide, we evaluated cellular response to C18-ceramide, a biologically relevant molecular species. The data in Fig. 3B, using wt (white bars) and HL-60/dnr cells (black bars), demonstrate that wt cells were sensitive to C18-ceramide after 48 h of exposure compared with HL-60/dnr cells. Specifically, after 48 h of exposure to C18-ceramide, 12% and 58% of wt and HL-60/dnr cells remained viable, respectively.
Fig. 3.
Dnr and Ara-C selection pressure elicits ceramide resistance in HL-60 cells. A: Sensitivity to C6-ceramide. Cells (50,000/0.2 ml media per well/96-well plate) were exposed to C6-ceramide at the concentrations shown for 72 h. B: Sensitivity to C18-ceramide. HL-60 wt and HL-60/dnr cells (300,000/1.2 ml media per well/24-sell plate) were exposed to C18:0 (5.0 μM) ceramide for 24 and 48 h. Experiments were conducted in dnr- and Ara-C-free media. Viability was measured by PI staining (A) and assessed by automated cell counting using trypan blue (B). C6-ceramide was delivered in DMSO. DMSO at all concentrations had no effect on cell viability. Controls contained vehicle. C18:0 ceramide was delivered to cells in ethanol vehicle (0.1% final concentration) as detailed in Materials and Methods. Ethanol had no effect on cell viability. *Statistical significance versus 24 h treatment HL-60/dnr cells; **statistical significance versus 24 h treatment wt HL-60 cells; #statistical significance (p <0.05) versus wt HL-60 cells at 48 h.
Effect of inhibitors of SL metabolism on HL-60, HL-60/dnr, and HL-60/Ara-C cell viability
Aberrant SL metabolism is an exploitable target in cancer (38, 39); this is primarily because dysfunctional SL metabolism in many instances underlies accelerated growth characteristics associated with cancer. HL-60/dnr- and Ara-C-selected cells displayed elevated GCS, AC, and SPHK1 activity (Fig. 2) and therefore comprise a useful model for studying the effects of SL metabolism inhibitors on the drug-resistant phenotype. The data in Fig. 4A demonstrate that drug-resistant models as well as wt models were nearly equally sensitive to GCS (PDMP, eliglustat), AC (DM102, SACLAC), and SPHK1 (SK1-i, FTY720) inhibitors. These data suggest that targeting SL metabolism would be a useful therapeutic strategy in a wide spectrum of clinical situations, including new as well as relapse populations. Using SK1-i, we determined that the loss of cell viability was attributed to apoptosis, as shown in Fig. 4B. After 48 h of exposure, early apoptosis doubled in wt cells and was manifold higher in Ara-C and dnr-resistant cells in response to SK1-i exposure. Late apoptosis was likewise elevated in wt and drug-resistant counterparts after exposure to SK1-i (Fig. 4B).
Fig. 4.
Effect of inhibitors of SL metabolism on viability in HL-60 wt and chemotherapy-resistant counterparts. A: Cell viability assays. Cells (50,000/well, black-walled 96 well plates) were exposed to the GCS (PDMP, eliglustat), AC (DM102, SACLAC), and SPHK1 (SK1-i, FTY720) inhibitors at the concentrations shown for 72 h in media containing 5% FBS. Viability was measured by PI staining as detailed in Materials and Methods. B: Effect of SK1-i on apoptosis in HL-60 wt, HL-60/dnr, and HL-60/Ara-C cells. Cells were treated with SK1-i (10 μM) for 48 h in dnr- and Ara-C-free media and analyzed by flow cytometry using Annexin-V FITC/PI staining. Early-apoptosis populations are represented by FITC+/PI−. Late apoptosis is represented by FITC+/PI+. Data are the mean ± SEM (n = 6) for each experimental point. **,***Statistical significance versus untreated control; ##,###statistical significance versus wt cells treated with SK1-i. Experiments were conducted in dnr- and Ara-C-free media.
Lipidomics: influence of chemotherapy selection pressure on SL composition
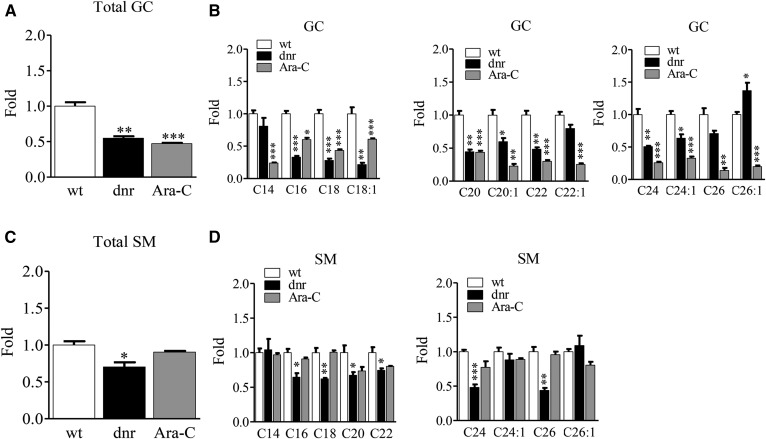
Lipidomic analyses demonstrated a profound increase in S1P levels in dnr-resistant cells that was not seen with Ara-C resistance (Fig. 5A), even though SPHK1 enzymatic activity was elevated in HL-60/Ara-C cells (Fig. 2C). Levels of dh-S1P showed a similar pattern in control, dnr, and Ara-C-resistant cells. Also prominent in HL-60/dnr cells was an increase in the levels of C1P (Fig. 5B), which by mass predominantly consisted of C16-C1P. Drug-resistant cells demonstrated a significant reduction in levels of total ceramides (Fig. 5C) that was reflected in measurements of individual ceramide molecular species, except for increases in several longer-chain, unsaturated species in HL-60/dnr cells. Interestingly, the more notably bioactive ceramide molecular species, C16 and C18, were significantly lower in HL-60/dnr cells. Total levels of glucosylceramides (GCs) in dnr- and Ara-C-selected HL-60 cells were also lower compared with chemotherapy-naive wt cells (Fig. 6A). An analysis of GC molecular species again revealed widespread reduction, except for C26:1 GC, which increased in dnr-selected cells (Fig. 6B). A significant reduction in total SM was also observed in HL-60/dnr cells, as opposed to the Ara-C-selected counterpart (Fig. 6C). Changes in the levels of individual molecular species of SM are shown in Fig. 6D, where reductions were primarily seen with dnr resistance.
Fig. 5.
Effect of dnr and Ara-C selection pressure on S1P, C1P, and ceramide levels in HL-60 cells. A: Effect of selection pressure on levels of S1P and dhS1P. The S1P mass in wt cells was 1.77 ± 0.03 pmol/mg protein. B: Total C1P and C16:0-C1P levels. The total C1P mass in wt cells was 32.75 ± 1.35 pmol/mg protein. C: Total ceramides and ceramide molecular species. wt, HL-60/dnr, and HL-60/Ara-C cells were harvested, pelleted by centrifugation, and washed three times in cold PBS. Lipids were extracted as indicated, and MS was conducted as detailed in Materials and Methods. Results are from experiments run in triplicate. Data are expressed as fold changes compared with wt control cells.
Fig. 6.
Impact of dnr and Ara-C selection pressure in HL-60 cells on levels of higher sphingolipids. A: Total GC levels. B: GC molecular species. C: Total SM levels. D: Levels of SM molecular species. Cell preparations are the same as those in Fig. 5. Data expressed as fold changes compared with wt cells.
Effect of SACLAC on SL levels in HL-60/dnr cells: alterations in lipid composition support cytotoxic response
We chose to study an AC inhibitor because its use affords the inhibition of ceramide hydrolysis as well as subsequent diminution of S1P generated from the ceramide hydrolysis product, sphingosine, by SPHK. We anticipated that SL measurement data in response to SACLAC would allow understanding how SL changes support the observed cytotoxic responses (see Fig. 4A). As shown in Fig. 7, SACLAC exposure had a striking effect on levels of S1P, sphingosine, ceramides, and dh-ceramides, reflective of a proapoptotic, antiproliferative SL profile. For example, SACLAC-treated cells showed a 75% and 70% reduction in levels of sphingosine and S1P, respectively, whereas levels of dh-sphingosine and dh-S1P increased manifold with treatment. In addition, nearly all molecular species of ceramide increased dramatically (Fig. 7B); these increases related to an overall 5-fold increase in total ceramides (Fig. 7B). Likewise, except for C26, all dh-ceramide molecular species increased markedly, translating to an overall 10-fold increase in total dh-ceramides in SACLAC-treated HL-60/dnr cells (Fig. 7C). Decreases in sphingosine and S1P are reflective of the inhibition of ceramide hydrolysis, as are the increases in ceramides. Compensatory increases in GC were also seen in SACLAC-treated HL-60/dnr cells (Fig. 7D), a possible response to reduce ceramide cytotoxicity via glycosylation. Although levels of SM molecular species were altered by SACLAC treatment, total SM levels did not differ in control and treated cells (Fig. 7E), which may be reflective of the need to maintain membrane integrity.
Fig. 7.
Effect of SACLAC exposure on SL composition in HL-60/dnr cells. A: Long-chain bases. B: Ceramides. C: dh-ceramides. D: GC molecular species. E: SM molecular species. HL-60/dnr cells (800,000/ml dnr-free, RPMI-1640 medium, 10% FBS) were exposed to 10 μM SACLAC (DMSO vehicle) or DMSO (control) for 28 h. DMSO did not impact cell viability. Cells were then harvested by centrifugation, washed three times in PBS, and frozen at −80°C until evaluation by MS. Viability in SACLAC-treated cells was 80% at harvest. Data are expressed as fold changes compared with untreated controls. Sph, sphingosine.
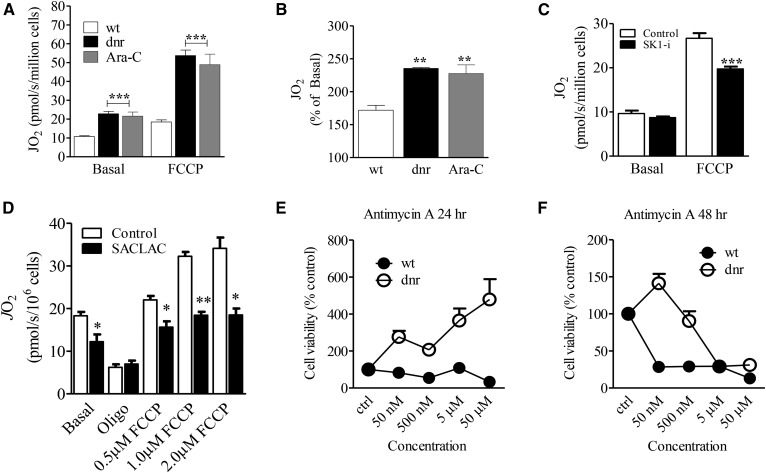
Impact of chemotherapy selection pressure on mitochondrial respiration
Mitochondria are important downstream targets of ceramide (8). We were therefore interested in determining whether mitochondrial metabolism was affected in the setting of dnr and Ara-C resistance, wherein we have demonstrated that changes in SL composition accompany chemotherapy selection pressure and the establishment of the drug-resistant phenotype. Mitochondrial respiratory function was assessed in intact cells under both basal and FCCP-stimulated conditions. Results revealed stark elevations in both basal respiration as well as maximal respiratory capacity in chemotherapy-resistant cells (Fig. 8A). While the increase in absolute respiratory capacity suggests an increase in mitochondrial biogenesis (i.e., altered quantity), the doubling of basal respiration implies that drug resistance in AML may in fact involve substantial bioenergetic remodeling (i.e., altered quality). Consistent with this, when maximal respiratory capacity was normalized to basal JO2 (i.e., FCCP/basal; “spare reserve capacity”), the response to FCCP in both the dnr- and Ara-C-resistant models was nearly 2-fold greater (Fig. 8B). To determine whether there is a link between SLs and mitochondrial metabolism in drug resistance, respirometry experiments were conducted in wt and HL-60/dnr cells following exposure to the SPHK1 inhibitor SK1-i and the AC inhibitor SACLAC. Exposure to SK1-i partially reversed the increase in the OXPHOS spare reserve capacity of HL-60/dnr (Fig. 8C). Similar albeit more prominent responses were seen with AC inhibition by SACLAC (Fig. 8D), which had a profound impact on S1P, ceramide, and dh-ceramide levels in dnr-resistant cells (Fig. 7). These results suggest that mitochondrial remodeling in drug resistance may be downstream of SL alterations, data supportive of a link between SLs and mitochondrial function in the drug-resistant phenotype. Last, increased spare reserve capacity should be associated with resistance to inhibitors of mitochondrial OXPHOS. We therefore tested the effects of Antimycin A on HL-60/dnr cell viability. This agent binds to cytochrome c reductase and inhibits the oxidation of ubiquinol. As shown over a 24–48 h time-course, HL-60 wt cells demonstrated sensitivity, whereas HL-60/dnr cells were more resistant to Antimycin A (Fig. 8E, F).
Fig. 8.
Chemotherapy selection pressure produces comprehensive upregulations in mitochondrial respiration with reversal by inhibitors of SL metabolism. A: Basal respiration and maximal respiratory capacity in chemotherapy-selected and wt HL-60 cells. B: Maximal respiratory capacity (FCCP-stimulated JO2) normalized to basal respiration (FCCP/basal) in wt and dnr- and Ara-C-resistant cells. C: Effect of SK1-i on respiratory capacity (basal and maximal) in HL-60/dnr cells. Cells (800,000/ml dnr-free, RPMI-1640 medium, 10% FBS) were exposed to 10 μM SK1-i for 24 h. D: Effect of SACLAC on respiratory capacity in HL-60/dnr cells. Cells (800,000/ml dnr-free, RPMI-1640 medium, 10% FBS) were exposed to 10 μM SACLAC for 24 h. Viability in treated cells was 81% at harvest. Mitochondrial respiration was conducted as detailed in Materials and Methods. E: Effect of Antimycin A on wt and HL-60/dnr cell viability. Cells (50,000/well) were exposed to the concentrations of Antimycin A indicated (50 nM – 50 μM) for 24 and 48 h, respectively. At termination, cell viability was determined by PI staining. n = 6 experimental points.
DISCUSSION
Prominent mechanisms of drug resistance in AML include diminished dnr intracellular retention directed by transporters such as P-gp, whereas Ara-C resistance is often linked to enhanced ALDH1A2 (36) and reduced capacity of the activating enzyme deoxycytidine kinase and overexpression of the inactivating 5′-nucleotidase, cytoplasmic 5′-nucleotidase II (40–42). Whereas research on chemotherapy-induced apoptosis and downstream signaling has made great strides, the initial signaling events in chemotherapy action remain unclear. Nonetheless, a curious commonality in the actions of dnr and Ara-C is that both agents increase intracellular ceramide levels in leukemia (14, 43–45). This commonality in activity positions SLs at the gateway to the apoptosis initiation process and as such accentuates the importance of SL enzymes as potential therapeutic targets. Previous studies demonstrated an association between SL abnormalities and chemotherapy resistance (38, 39, 46–51); however, relatively few works have focused on chemotherapy selection pressure in AML in response to dnr and Ara-C, the drugs used to treat this disease.
The salient points in this study are i) the upregulation of three key SL branch-point enzymes in AML cells in response to dnr and Ara-C selection pressure; ii) the striking ceramide deficiency and spike in S1P and C1P levels in the dnr-resistant model; iii) mitochondrial remodeling that accompanied dnr and Ara-C resistance that was partially reversed by the inclusion of SPHK1 and AC inhibitors; and iv) the clear effect of AC inhibition by SACLAC on S1P and ceramide levels that accompanied HL-60/dnr cellular demise. These dynamic responses resulting from chemotherapy selection pressure underscore the relevance of SL enzymes in multidrug resistance. Excellent reviews are available on SLs in cancer and drug resistance and on targeting SL metabolism as a therapeutic strategy. Works by Ponnusamy et al. (51) and Selvam and Ogretmen (50) highlight the role of S1P in cancer cell death and drug resistance. A review by Giussani et al. (47) focuses on counteracting the accumulation of ceramide and the role of gangliosides. A review by Lee and Kolesnick (46) focused on the novel role of SM in the sequestration of ABCB1, and summaries by Ogretmen (39) and Morad and Cabot (8) highlight SL metabolism in cancer cell signaling and therapy. Few studies, however, have focused on the impact of prolonged exposure to Ara-C and dnr on SL enzyme expression and SL composition in AML. Of merit here is a study in doxorubicin-resistant HL-60 cells wherein Itoh et al. (52) demonstrated that enhanced GCS and sphingomyelin synthase activity was accompanied by lower ceramide levels compared with HL-60 wt cells. Pertinent work by Bonhoure et al. (53) demonstrated that doxorubicin- and VP-16-resistant HL-60 cells displayed sustained SPHK1 activity and that treatment with an SPHK1 inhibitor led to classic apoptosis. Recent work by Snider et al (54), using short-term exposure of MCF-7 breast cancer cells to doxorubicin (500 nM), showed enhanced S1P levels and decreases in glucosylceramides, similar to our data in HL-60-dnr-selected cells. Paugh et al. (55) also demonstrated therapeutic efficacy of SK1-i in U937 human histiocystic leukemia, Jurkat acute T-cell leukemia cells, and in leukemic blasts from AML patients. These studies underscore the significance of SL enzymes in drug resistance and in therapeutics. Also of note in the current study is the refractory nature of HL-60/dnr and HL-60/Ara-C cells to C6- and C18:0-ceramide that we propose is aligned with upregulated ceramide metabolism. Thus, drug resistance and ceramide resistance appear to be allied.
The effects of Ara-C and dnr selection pressure on AC and SPHK1 activity merit extra mention. AC lowers intracellular ceramide levels via hydrolysis, a mechanism of ceramide clearance. However, the more impending scenario is that AC activity produces sphingosine that is phosphorylated by SPHK1 to generate S1P, a mitogenic entity that enhances cancer cell growth (56, 57). AC plays a prominent role in therapeutic response in prostate cancer (58, 59); moreover, AC is upregulated in AML (29), wherein targeting the downstream metabolic enzyme, SPHK1, has been shown to be of therapeutic efficacy (60–62). GCS, AC, and SPHK1, as indicators of tumor cell resistance, thus play key roles in regulating cancer cell fate by pitting cell survival against apoptosis. For example, in the dnr model, chemotherapy selection pressure altered SL composition in a manner that drove down ceramide levels and increased S1P; this tilts the SL rheostat in favor of S1P and proliferation. Of note, HL-60/dnr cells grew at nearly twice the rate of wt and HL-60/Ara-C cells, and in this scenario, we suggest that the elevated levels of S1P in dnr resistance contribute to this mitogenic phenotype. Interestingly, both dnr- and Ara-C-resistant models displayed comparable upregulation of SPHK1, yet S1P levels were higher in dnr-resistant cells. Although not pursued, it is possible that enhanced S1P lyase and phosphatase activities accompany Ara-C resistance, as these enzymes tightly regulate S1P levels (63). This idea is supported by data showing that Ara-C and wt cells have similar proliferation rates (Fig. 1F).
Interestingly, drug-resistant cells demonstrated a ceramide deficit, most notably in the dnr model. With this, the family of CerSs becomes an attractive candidate for future investigation. The decreases in antiproliferative C16- and C18-ceramides in HL-60/dnr cells may offer protection from these apoptosis- and autophagy-inducing molecular species and suggests that specific CerSs may be involved. However, little is known regarding the effects of dnr and Ara-C resistance on CerS. Fekry et al. (64) have shown that the CerS6 protein is markedly elevated in cancer cells exposed to methotrexate, and its product, C16-ceramide, mediated antiproliferative effects. However, that study did not encompass methotrexate resistance. Also noteworthy in our lipidomic analyses were the decreases in GC levels that accompanied chemotherapy selection pressure, similar to work in MCF-7 cells after 24 h exposure to 500 nM doxorubicin (54) but discordant with previous works demonstrating that GC levels are elevated in chemotherapy-resistant cancer cells (15–17). It is possible that low GC levels reflect deficits in ceramide levels, even though GCS expression and activity were elevated in the drug-resistant model. It is also important to note that C6-NBD-ceramide was used as a substrate in the GCS activity assays. This short-chain, fluorescent ceramide analogue may behave differently compared with natural long-chain species. In addition, this discrepancy in GC levels and GCS activity may be the result of metabolic flux not picked up by our snapshot SL analyses. Last, C1P, which was first shown by Dressler and Kolesnick (65) to be synthesized in an HL-60 cell model from SM-derived ceramide, was elevated 4-fold in HL-60/dnr cells. This is an interesting finding with perhaps profound implications, as C1P plays essential roles in a myriad of biological functions (66), including cancer cell survival and dissemination (67). The C1P findings merit further exploration, specifically regarding dnr resistance.
Ara-C-resistant, dnr-resistant, and wt cells demonstrated similar sensitivities to SL enzyme inhibitors, indicative of pan-efficacy. Thus, SL inhibitors could serve as therapeutics in both sensitive and resistant clinical settings. Figure 2D demonstrates the presence, albeit slight, of GCS, SPHK1, and AC in wt cells, and therefore these enzymes are present and vulnerable to inhibition. Along these lines, SPHK1 inhibitors have been shown to be effective in wt leukemia cells, including K562, U937, and HL-60 (55, 68), and Bonhoure et al. (53) have shown that F-12509a, a SPHK1 inhibitor, led to ceramide accumulation and decreased S1P content and elicited apoptosis equally in chemosensitive and chemoresistant cell lines. These data reinforce the idea that SL enzymes are exploitable targets and suggest that cancer cells engage SL enzymes to maintain tumorigenicity. The likely impact of GCS, AC, and SPHK in our model of drug resistance converges on the SL rheostat wherein S1P levels increased and levels of total ceramides decreased. Apropos in this instance are the data demonstrating the role of SLs in cytotoxic response to the AC inhibitor SACLAC. Here, the data are crystal clear (see Fig. 7). As depicted in Fig. 9, SACLAC treatment elicited S1P and sphingosine decreases and increases in dh-sphingosine (sphinganine) and tumor-suppressing ceramides and dh-ceramides. It is not known whether SACLAC inhibits dihydroceramide desaturase 1 (Des1); however, we suggest that SACLAC elicits product inhibition of Des1 that is caused by the accumulation of ceramide. The marked increases in dh-ceramides could likewise elicit product inhibition of the dh-CerSs, resulting in increases in sphinganine and dh-S1P. Last, and interestingly, SACLAC-treated cells appeared to offset ceramide increases via conversion to GC (see Fig. 7) and several molecular species of SM; however, this was not adequate to stave off cell death.
Fig. 9.

Schematic depicting the effect of SACLAC on sphingolipid pathway interactions for the promotion of cell death. The schematic is based on the MS results shown in Fig. 7 (HL-60/dnr cells exposed to SACLAC) and SACLAC cytotoxicity (shown in Fig. 4A). SACLAC inhibition of AC produces decreases in the AC product, sphingosine, and decreases in downstream S1P (down arrows). Due to AC inhibition, ceramide levels increase (up arrow). Excess ceramides elicit product inhibition of Des1 (dh-ceramide desaturase) (block sign), which in turn promotes a buildup of dh-ceramides (up arrow) resulting in product inhibition of dh-CerSs (block sign), promoting a sphinganine surplus (up arrow).
The data in mitochondria are of keen relevance. Increased reliance on mitochondrial OXPHOS has recently been described as a common biochemical phenotype in chemoresistant human AML (7). In support of these data, we observed clear upregulations in mitochondria respiratory capacity in drug-resistant AML, consistent with increased mitochondrial biogenesis. Interestingly, in addition to higher FCCP-supported JO2, both drug-resistant cell lines also presented with a near doubling of basal respiration. In the intact cell, basal respiration is governed by the cellular demand for ATP regeneration (69), meaning a two-fold increase in mitochondrial content would not be anticipated to double basal respiration unless the increase in mitochondrial biogenesis is equally accompanied by a near doubling of ATP turnover. Such a doubling of ATP turnover could stem from accelerated ATP hydrolysis throughout the cell or a change in OXPHOS efficiency. Taken together, the biochemical evidence presented herein suggests that AML drug resistance is accompanied by both quantitative and qualitative alterations in mitochondrial bioenergetics. Of note, the increased respiratory activity that was characteristic in drug selection pressure was shown to be allied with reduced sensitivity to the inhibition of mitochondrial OXPHOS. Of importance regarding SLs and mitochondrial function in drug resistance are data showing that inhibitors of SL metabolism, SK1-i and SACLAC, partially reversed the increase in spare reserve capacity that accompanied dnr selection pressure. This evidence of SL-bioenergetic crosstalk implies that mitochondrial adaptations inherent to the drug-resistant phenotype may require a ceramide-restricted SL milieu. Based on these findings, simultaneous targeting of both SL metabolism and OXPHOS may represent a novel treatment regime to combat drug resistance. Future research aimed at the identification of the precise mechanistic signatures that demarcate normal leukemic mitochondria from those in drug-resistant populations may in fact pave the way for the development of novel mitochondrial-targeted chemotherapeutics.
Footnotes
Abbreviations:
- AML
- acute myelogenous leukemia
- AC
- acid ceramidase
- Ara-C
- cytarabine
- dnr
- daunorubicin
- CerS
- ceramide synthase
- Des1
- dihydroceramide desaturase 1
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- GC
- glucosylceramide
- GCS
- glucosylceramide synthase
- LDH
- lactate dehydrogenase
- OXPHOS
- oxidative phosphorylation
- PDMP
- d,l-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol
- P-gp
- P-glycoprotein
- PI
- propidium iodide
- SACLAC
- 2-chloro-N-(2S,3R)-1,3-dihydroxyoctadecan-2-yl acetamide
- SL
- sphingolipid
- SPHK1
- sphingosine kinase 1
- S1P
- sphingosine 1-phosphate
This work was supported by National Institutes of Health Grant P01 CA171983 and by a grant from the Brody Brothers Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Luger S. M. 2017. How can one optimize induction therapy in AML? Best Pract. Res. Clin. Haematol. 30: 301–305. [DOI] [PubMed] [Google Scholar]
- 2.Rashidi A., Weisdorf D. J., and Bejanyan N.. 2018. Treatment of relapsed/refractory acute myeloid leukaemia in adults. Br. J. Haematol. 181: 27–37. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman M. M., Lavi O., Hall M. D., and Gillet J. P.. 2016. Toward a better understanding of the complexity of cancer drug resistance. Annu. Rev. Pharmacol. Toxicol. 56: 85–102. [DOI] [PubMed] [Google Scholar]
- 4.Fukuda Y., Lian S., and Schuetz J. D.. 2015. Leukemia and ABC transporters. Adv. Cancer Res. 125: 171–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaffer B. C., Gillet J. P., Patel C., Baer M. R., Bates S. E., and Gottesman M. M.. 2012. Drug resistance: still a daunting challenge to the successful treatment of AML. Drug Resist. Updat. 15: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartholomae S., Gruhn B., Debatin K. M., Zimmermann M., Creutzig U., Reinhardt D., and Steinbach D.. 2016. Coexpression of multiple ABC-transporters is strongly associated with treatment response in childhood acute myeloid leukemia. Pediatr. Blood Cancer. 63: 242–247. [DOI] [PubMed] [Google Scholar]
- 7.Farge T., Saland E., de Toni F., Aroua N., Hosseini M., Perry R., Bosc C., Sugita M., Stuani L., Fraisse M., et al. . 2017. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 7: 716–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morad S. A., and Cabot M. C.. 2013. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer. 13: 51–65. [DOI] [PubMed] [Google Scholar]
- 9.Galadari S., Rahman A., Pallichankandy S., and Thayyullathil F.. 2015. Tumor suppressive functions of ceramide: evidence and mechanisms. Apoptosis. 20: 689–711. [DOI] [PubMed] [Google Scholar]
- 10.Huang C., and Freter C.. 2015. Lipid metabolism, apoptosis and cancer therapy. Int. J. Mol. Sci. 16: 924–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tirodkar T. S., and Voelkel-Johnson C.. 2012. Sphingolipids in apoptosis. Exp. Oncol. 34: 231–242. [PubMed] [Google Scholar]
- 12.Zheng W., Kollmeyer J., Symolon H., Momin A., Munter E., Wang E., Kelly S., Allegood J. C., Liu Y., Peng Q., et al. . 2006. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim. Biophys. Acta. 1758: 1864–1884. [DOI] [PubMed] [Google Scholar]
- 13.Bose R., Verheij M., Haimovitz-Friedman A., Scotto K., Fuks Z., and Kolesnick R.. 1995. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell. 82: 405–414. [DOI] [PubMed] [Google Scholar]
- 14.Jaffrézou J. P., Levade T., Bettaieb A., Andrieu N., Bezombes C., Maestre N., Vermeersch S., Rousse A., and Laurent G.. 1996. Daunorubicin-induced apoptosis: triggering of ceramide generation through sphingomyelin hydrolysis. EMBO J. 15: 2417–2424. [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y. Y., Han T. Y., Giuliano A. E., and Cabot M. C.. 1999. Expression of glucosylceramide synthase, converting ceramide to glucosylceramide, confers adriamycin resistance in human breast cancer cells. J. Biol. Chem. 274: 1140–1146. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y. Y., Han T. Y., Giuliano A. E., and Cabot M. C.. 2001. Ceramide glycosylation potentiates cellular multidrug resistance. FASEB J. 15: 719–730. [DOI] [PubMed] [Google Scholar]
- 17.Xie P., Shen Y. F., Shi Y. P., Ge S. M., Gu Z. H., Wang J., Mu H. J., Zhang B., Qiao W. Z., and Xie K. M.. 2008. Overexpression of glucosylceramide synthase in associated with multidrug resistance of leukemia cells. Leuk. Res. 32: 475–480. [DOI] [PubMed] [Google Scholar]
- 18.Senchenkov A., Litvak D. A., and Cabot M. C.. 2001. Targeting ceramide metabolism–a strategy for overcoming drug resistance. J. Natl. Cancer Inst. 93: 347–357. [DOI] [PubMed] [Google Scholar]
- 19.Aureli M., Murdica V., Loberto N., Samarani M., Prinetti A., Bassi R., and Sonnino S.. 2014. Exploring the link between ceramide and ionizing radiation. Glycoconj. J. 31: 449–459. [DOI] [PubMed] [Google Scholar]
- 20.Hajj C., Becker-Flegler K. A., and Haimovitz-Friedman A.. 2015. Novel mechanisms of action of classical chemotherapeutic agents on sphingolipid pathways. Biol. Chem. 396: 669–679. [DOI] [PubMed] [Google Scholar]
- 21.Lim K. G., Tonelli F., Li Z., Lu X., Bittman R., Pyne S., and Pyne N. J.. 2011. FTY720 analogues as sphingosine kinase 1 inhibitors: enzyme inhibition kinetics, allosterism, proteasomal degradation, and actin rearrangement in MCF-7 breast cancer cells. J. Biol. Chem. 286: 18633–18640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morad S. A. F., Davis T. S., MacDougall M. R., Tan S. F., Feith D. J., Desai D. H., Amin S. G., Kester M., Loughran T. P. Jr., and Cabot M. C.. 2017. Role of P-glycoprotein inhibitors in ceramide-based therapeutics for treatment of cancer. Biochem. Pharmacol. 130: 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh C. M. 2013. Preparation of cells for microscopy using cytospin. Methods Enzymol. 533: 235–240. [DOI] [PubMed] [Google Scholar]
- 24.Gupta V., Patwardhan G. A., Zhang Q. J., Cabot M. C., Jazwinski S. M., and Liu Y. Y.. 2010. Direct quantitative determination of ceramide glycosylation in vivo: a new approach to evaluate cellular enzyme activity of glucosylceramide synthase. J. Lipid Res. 51: 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 26.Bedia C., Casas J., Garcia V., Levade T., and Fabrias G.. 2007. Synthesis of a novel ceramide analogue and its use in a high-throughput fluorogenic assay for ceramidases. ChemBioChem. 8: 642–648. [DOI] [PubMed] [Google Scholar]
- 27.Gouazé-Andersson V., Flowers M., Karimi R., Fabrias G., Delgado A., Casas J., and Cabot M. C.. 2011. Inhibition of acid ceramidase by a 2-substituted aminoethanol amide synergistically sensitizes prostate cancer cells to N-(4-hydroxyphenyl) retinamide. Prostate. 71: 1064–1073. [DOI] [PubMed] [Google Scholar]
- 28.Morad S. A., Ryan T. E., Neufer P. D., Zeczycki T. N., Davis T. S., MacDougall M. R., Fox T. E., Tan S. F., Feith D. J., Loughran T. P. Jr., et al. . 2016. Ceramide-tamoxifen regimen targets bioenergetic elements in acute myelogenous leukemia. J. Lipid Res. 57: 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan S. F., Liu X., Fox T. E., Barth B. M., Sharma A., Turner S. D., Awwad A., Dewey A., Doi K., Spitzer B., et al. . 2016. Acid ceramidase is upregulated in AML and represents a novel therapeutic target. Oncotarget. 7: 83208–83222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmittgen T. D., and Livak K. J.. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 31.Nolan T., Hands R. E., and Bustin S. A.. 2006. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1: 1559–1582. [DOI] [PubMed] [Google Scholar]
- 32.McLornan D. P., McMullin M. F., Johnston P., and Longley D. B.. 2007. Molecular mechanisms of drug resistance in acute myeloid leukaemia. Expert Opin. Drug Metab. Toxicol. 3: 363–377. [DOI] [PubMed] [Google Scholar]
- 33.Urasaki Y., Ueda T., Yoshida A., Fukushima T., Takeuchi N., Tsuruo T., and Nakamura T.. 1996. Establishment of a daunorubicin-resistant cell line which shows multi-drug resistance by multifactorial mechanisms. Anticancer Res. 16: 709–714. [PubMed] [Google Scholar]
- 34.Zhou M., Zhao Y., Ding Y., Liu H., Liu Z., Fodstad O., Riker A. I., Kamarajugadda S., Lu J., Owen L. B., et al. . 2010. Warburg effect in chemosensitivity: targeting lactate dehydrogenase-A re-sensitizes taxol-resistant cancer cells to taxol. Mol. Cancer. 9: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimopoulos M. A., Barlogie B., Smith T. L., and Alexanian R.. 1991. High serum lactate dehydrogenase level as a marker for drug resistance and short survival in multiple myeloma. Ann. Intern. Med. 115: 931–935. [DOI] [PubMed] [Google Scholar]
- 36.Kawasoe M., Yamamoto Y., Okawa K., Funato T., Takeda M., Hara T., Tsurumi H., Moriwaki H., Arioka Y., Takemura M., Matsunami H., Markey S. P., and Saito K.. 2013. Acquired resistance of leukemic cells to AraC is associated with the upregulation of aldehyde dehydrogenase 1 family member A2. Exp. Hematol. 41: 597–603.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morad S. A. F., and Cabot M. C.. 2018. The onus of sphingolipid enzymes in cancer drug resistance. Adv. Cancer Res. 140: 235–263. [DOI] [PubMed] [Google Scholar]
- 38.Ryland L. K., Fox T. E., Liu X., Loughran T. P., and Kester M.. 2011. Dysregulation of sphingolipid metabolism in cancer. Cancer Biol. Ther. 11: 138–149. [DOI] [PubMed] [Google Scholar]
- 39.Ogretmen B. 2018. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer. 18: 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galmarini C. M., Thomas X., Calvo F., Rousselot P., Rabilloud M., El Jaffari A., Cros E., and Dumontet C.. 2002. In vivo mechanisms of resistance to cytarabine in acute myeloid leukaemia. Br. J. Haematol. 117: 860–868. [DOI] [PubMed] [Google Scholar]
- 41.Ross D. D. 2000. Novel mechanisms of drug resistance in leukemia. Leukemia. 14: 467–473. [DOI] [PubMed] [Google Scholar]
- 42.Negoro E., Yamauchi T., Urasaki Y., Nishi R., Hori H., and Ueda T.. 2011. Characterization of cytarabine-resistant leukemic cell lines established from five different blood cell lineages using gene expression and proteomic analyses. Int. J. Oncol. 38: 911–919. [DOI] [PubMed] [Google Scholar]
- 43.Bradshaw C. D., Ella K. M., Thomas A. L., Qi C., and Meier K. E.. 1996. Effects of Ara-C on neutral sphingomyelinase and mitogen- and stress-activated protein kinases in T-lymphocyte cell lines. Biochem. Mol. Biol. Int. 40: 709–719. [DOI] [PubMed] [Google Scholar]
- 44.Grazide S., Maestre N., Veldman R. J., Bezombes C., Maddens S., Levade T., Laurent G., and Jaffrezou J. P.. 2002. Ara-C- and daunorubicin-induced recruitment of Lyn in sphingomyelinase-enriched membrane rafts. FASEB J. 16: 1685–1687. [DOI] [PubMed] [Google Scholar]
- 45.Terrisse A. D., Bezombes C., Lerouge S., Laurent G., and Jaffrezou J. P.. 2002. Daunorubicin- and Ara-C-induced interphasic apoptosis of human type II leukemia cells is caspase-8-independent. Biochim. Biophys. Acta. 1584: 99–103. [DOI] [PubMed] [Google Scholar]
- 46.Lee W. K., and Kolesnick R. N.. 2017. Sphingolipid abnormalities in cancer multidrug resistance: chicken or egg? Cell. Signal. 38: 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giussani P., Tringali C., Riboni L., Viani P., and Venerando B.. 2014. Sphingolipids: key regulators of apoptosis and pivotal players in cancer drug resistance. Int. J. Mol. Sci. 15: 4356–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dimanche-Boitrel M. T., and Rebillard A.. 2013. Sphingolipids and response to chemotherapy. Handb. Exp. Pharmacol. 216: 73–91. [DOI] [PubMed] [Google Scholar]
- 49.Gouaze-Andersson V., and Cabot M. C.. 2011. Sphingolipid metabolism and drug resistance in hematological malignancies. Anticancer. Agents Med. Chem. 11: 891–903. [DOI] [PubMed] [Google Scholar]
- 50.Selvam S. P., and Ogretmen B.. 2013. Sphingosine kinase/sphingosine 1-phosphate signaling in cancer therapeutics and drug resistance. Handb. Exp. Pharmacol. 216: 3–27. [DOI] [PubMed] [Google Scholar]
- 51.Ponnusamy S., Meyers-Needham M., Senkal C. E., Saddoughi S. A., Sentelle D., Selvam S. P., Salas A., and Ogretmen B.. 2010. Sphingolipids and cancer: ceramide and sphingosine-1-phosphate in the regulation of cell death and drug resistance. Future Oncol. 6: 1603–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Itoh M., Kitano T., Watanabe M., Kondo T., Yabu T., Taguchi Y., Iwai K., Tashima M., Uchiyama T., and Okazaki T.. 2003. Possible role of ceramide as an indicator of chemoresistance: decrease of the ceramide content via activation of glucosylceramide synthase and sphingomyelin synthase in chemoresistant leukemia. Clin. Cancer Res. 9: 415–423. [PubMed] [Google Scholar]
- 53.Bonhoure E., Pchejetski D., Aouali N., Morjani H., Levade T., Kohama T., and Cuvillier O.. 2006. Overcoming MDR-associated chemoresistance in HL-60 acute myeloid leukemia cells by targeting sphingosine kinase-1. Leukemia. 20: 95–102. [DOI] [PubMed] [Google Scholar]
- 54.Snider J. M., Trayssac M., Clarke C. J., Schwartz N., Snider A. J., Obeid L. M., Luberto C., and Hannun Y. A.. 2019. Multiple actions of doxorubicin on the sphingolipid network revealed by flux analysis. J. Lipid Res. 60: 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paugh S. W., Paugh B. S., Rahmani M., Kapitonov D., Almenara J. A., Kordula T., Milstien S., Adams J. K., Zipkin R. E., Grant S., et al. . 2008. A selective sphingosine kinase 1 inhibitor integrates multiple molecular therapeutic targets in human leukemia. Blood. 112: 1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pyne N. J., El Buri A., Adams D. R., and Pyne S.. 2017. Sphingosine 1-phosphate and cancer. Adv. Biol. Regul. 68: 97–106. [DOI] [PubMed] [Google Scholar]
- 57.Olivera A., Kohama T., Edsall L., Nava V., Cuvillier O., Poulton S., and Spiegel S.. 1999. Sphingosine kinase expression increases intracellular sphingosine-1-phosphate and promotes cell growth and survival. J. Cell Biol. 147: 545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saad A. F., Meacham W. D., Bai A., Anelli V., Elojeimy S., Mahdy A. E., Turner L. S., Cheng J., Bielawska A., Bielawski J., et al. . 2007. The functional effects of acid ceramidase overexpression in prostate cancer progression and resistance to chemotherapy. Cancer Biol. Ther. 6: 1455–1460. [DOI] [PubMed] [Google Scholar]
- 59.Cheng J. C., Bai A., Beckham T. H., Marrison S. T., Yount C. L., Young K., Lu P., Bartlett A. M., Wu B. X., Keane B. J., et al. . 2013. Radiation-induced acid ceramidase confers prostate cancer resistance and tumor relapse. J. Clin. Invest. 123: 4344–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hengst J. A., Dick T. E., Sharma A., Doi K., Hegde S., Tan S. F., Geffert L. M., Fox T. E., Sharma A. K., Desai D., et al. . 2017. SKI-178: a multitargeted inhibitor of sphingosine kinase and microtubule dynamics demonstrating therapeutic efficacy in acute myeloid leukemia models. Cancer Transl. Med. 3: 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Powell J. A., Lewis A. C., Zhu W., Toubia J., Pitman M. R., Wallington-Beddoe C. T., Moretti P. A., Iarossi D., Samaraweera S. E., Cummings N., et al. . 2017. Targeting sphingosine kinase 1 induces MCL1-dependent cell death in acute myeloid leukemia. Blood. 129: 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dick T. E., Hengst J. A., Fox T. E., Colledge A. L., Kale V. P., Sung S. S., Sharma A., Amin S., Loughran T. P. Jr., Kester M., et al. . 2015. The apoptotic mechanism of action of the sphingosine kinase 1 selective inhibitor SKI-178 in human acute myeloid leukemia cell lines. J. Pharmacol. Exp. Ther. 352: 494–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leong W. I., and Saba J. D.. 2010. S1P metabolism in cancer and other pathological conditions. Biochimie. 92: 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fekry B., Esmaeilniakooshkghazi A., Krupenko S. A., and Krupenko N. I.. 2016. Ceramide synthase 6 is a novel target of methotrexate mediating its antiproliferative effect in a p53-dependent manner. PLoS One. 11: e0146618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dressler K. A., and Kolesnick R. N.. 1990. Ceramide 1-phosphate, a novel phospholipid in human leukemia (HL-60) cells. Synthesis via ceramide from sphingomyelin. J. Biol. Chem. 265: 14917–14921. [PubMed] [Google Scholar]
- 66.Hoeferlin L. A., Wijesinghe D. S., and Chalfant C. E.. 2013. The role of ceramide-1-phosphate in biological functions. Handb. Exp. Pharmacol. 215: 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomez-Muñoz A. 2018. The role of ceramide 1-phosphate in tumor cell survival and dissemination. Adv. Cancer Res. 140: 217–234. [DOI] [PubMed] [Google Scholar]
- 68.Neviani P., Santhanam R., Oaks J. J., Eiring A. M., Notari M., Blaser B. W., Liu S., Trotta R., Muthusamy N., Gambacorti-Passerini C., et al. . 2007. FTY720, a new alternative for treating blast crisis chronic myelogenous leukemia and Philadelphia chromosome-positive acute lymphocytic leukemia. J. Clin. Invest. 117: 2408–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willis W. T., Jackman M. R., Messer J. I., Kuzmiak-Glancy S., and Glancy B.. 2016. A simple hydraulic analog model of oxidative phosphorylation. Med. Sci. Sports Exerc. 48: 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]