Abstract
Fatty acids have various physiological effects on melanoma. For example, palmitic acid (PA) increases melanin levels; linoleic acid and DHA decrease melanin levels; and DHA suppresses tumor growth. In this study, we focused on the relationship between the structure of fatty acids and their physiological effects in melanoma to examine the likely mechanisms of action. We showed that saturated fatty acids and PUFAs display opposing effects on melanin content in melanoma cells. Likewise, PA and EPA have opposing effects in terms of actin polymerization. Our findings suggest that PA and EPA change melanin content in melanoma to alter melanosome trafficking by modulating actin polymerization. Here, we also examined the mechanism of the anti-tumor effect of DHA. We found that DHA interacts with receptor for activated C kinase 1 and represses melanoma cell proliferation by suppressing protein kinase C signaling. Our results suggest a new mechanism to explain the physiological effects of fatty acids.
Keywords: palmitic acid, eicosapentaenoic acid, docosahexaenoic acid, tumor cell proliferation, F-actin, receptor for activated C kinase 1
Fatty acids are major components of triacylglycerol and phospholipids. They are classified in terms of carbon chain length and degree of unsaturation. Lipids in mammals almost always comprise long chain fatty acids with more than 16 carbon atoms. Mammals can synthesize palmitic acid (PA), palmitoleic acid, stearic acid, and oleic acid. However, because mammals lack the enzymes required to produce linoleic acid and α-linoleic acid, these essential fatty acids must be derived from plants. There is a relationship between fatty acid structure and biological function. For example, EPA and DHA are well-studied fatty acids that are known to have a beneficial effect on human health. Epidemiological studies involving Inuit first showed that intake of EPA and DHA reduced the risk of coronary heart disease (1, 2). Subsequently, the association of EPA and DHA intake and reduced risk of coronary heart disease was shown in other countries (3–6). It has been reported that EPA and DHA elicit many physiological effects, including an anti-tumor activity (7–10) and an inhibition of melanogenesis (11). However, the mechanisms of the anti-tumor and the anti-melanogenesis action of EPA and DHA remain unclear. In addition to EPA and DHA, the physiological effects of fatty acids were also reported. For example, in pancreatic β-cells, PA induces apoptosis (12, 13), and oleic acid reduces the deleterious effects of PA and high levels of glucose (14). Diabetic mice that were deficient in Elovl6 displayed improved glycemic control (15). However, the physiological effects and the mechanisms of action of free fatty acid are still a matter of intense debate.
Melanin is a biopolymer that is synthesized from tyrosine by tyrosinase (Tyr), a Tyr-related protein 1 (Tyrp1), and 3,4-dihydroxyphenylalanine (DOPA) chrome tautomerase (16, 17). The biosynthesis of melanin (melanogenesis) occurs within melanosomes, which are discrete membrane-bound organelles of melanocytes. Melanosomes mature within the melanocyte through four morphologically distinct stages. The internal structure of melanosomes is formed during stages I to II. Melanin biosynthesis begins during stage III. In stage IV, all internal structures are masked by the accumulated melanin. In epidermal melanocytes, stage IV melanosomes are translocated along microtubules from the cell center to actin-rich dendritic tips, and then transferred to neighboring keratinocytes (18, 19). Previous studies have demonstrated the opposing effect of linoleic acid and PA on melanogenesis. Linoleic acid increases Tyr protein degradation and suppresses melanogenesis in melanoma (11, 20, 21). By contrast, PA increases Tyr protein and melanin in melanoma (20). Ando and colleagues showed that linoleic acid induces degradation of Tyr via a ubiquitin-proteasome pathway (21, 22).
DHA also suppresses tumor growth in melanoma (8, 9, 23). Recently, it was shown that dietary intake of EPA and DHA reduces the risk of melanoma in humans (24). Previous studies proposed that enhanced nuclear-translocation of β-catenin (25, 26) and downregulation of cyclooxygenase-2 expression (26, 27) are mechanisms for the anti-tumor effect of DHA in melanoma. However, in pancreatic tumors, it was reported that accumulation of β-catenin in the cytoplasm and nucleus could be inhibited by DHA (28). The proposed mechanisms for the anti-tumor effect of DHA in pancreatic tumors involve a buildup of reactive oxygen species and induction of caspase-8-dependent cell death (28). However, the precise mechanisms for the anti-tumor effect of DHA are still disputed.
In this study, we examined the relationship between the structure of the fatty acid and its physiological effect on melanoma, together with the mechanisms of these effects.
MATERIALS AND METHODS
Materials
Myristic acid, PA, stearic acid, oleic acid, and EPA were purchased from Nakarai Tesque (Kyoto, Japan). α-Linoleic acid, stearidonic acid, and DHA were purchased from Cayman Chemical Co. (Ann Arbor, MI). Melanoma cell lines (B16F10, Colo679, G361, HOMM, and HTMM) were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT/AMED, Japan. Melanin and DOPA were purchased from FUJIFILM Wako Pure Chemical Corp. (Osaka, Japan). L-tyrosine (2-13C) was purchased from Otsuka Pharmaceutical Co., Ltd. (Tokushima, Japan). Anti-α-tubulin antibody (clone B-5-1-2) was purchased from Sigma-Aldrich (St. Louis, MO). Anti-α1 Na/K ATPase, anti-Tyrp1, anti-cofilin, anti-phospho-cofilin (Ser 3), anti-Rab27a, and anti-receptor for activated C kinase 1 (Rack1) antibodies were purchased from Abcam Japan (Tokyo, Japan). Anti-Tyr antibody (clone C-19) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Rab38 antibody was purchased from GeneTex (Irvine, CA).
Cell culture and quantitative analysis of melanin
Melanoma cell lines were cultured with RPMI1640 medium (Sigma-Aldrich) containing 10% FBS (Thermo Fisher Scientific, Waltham, MA) and antibiotic-antimycotic solution. Normal human melanocytes (MEMa-LP; Thermo Fisher Scientific) were cultured with medium 254 (Thermo Fisher Scientific) containing 10% FBS, human melanocyte growth supplement 2 (HMGS-2; Thermo Fisher Scientific), and antibiotic-antimycotic solution. Fatty acid dissolved in methanol (MeOH) was added directly to medium containing 10% FBS. The medium for control cells was supplemented with an equivalent amount of MeOH alone. The proteins were extracted from cells by incubation with RIPA lysis buffer [20 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1% NP40, 0.1% sodium deoxycholate, 0.1% SDS] for 30 min at 4°C. The lysate was then clarified by centrifugation and melanin was extracted from the resulting pellet with 1 M NaOH solution. The concentration of melanin and protein was measured by absorbance using a VarioSkan LUX instrument (Thermo Fisher Scientific).
LC/hybrid quadrupole TOFMS
The cells were sonicated in water:MeOH (50:50) solution for 30 s and the mixture was then centrifuged at 20,000 g for 10 min at 4°C. The concentrations of DOPA and tyrosine in supernatants were measured by LC/hybrid quadrupole TOFMS (LC/QTOFMS) (Agilent Technologies, Tokyo, Japan). DOPA and tyrosine were separated on an Accucore C30 column (2.0 mm diameter × 250 mm; Thermo Fisher Scientific) by gradient elution (water containing 0.1% formic acid/acetonitrile containing 0.1% formic acid, 98/2 to 0/100 over 25 min) at a flow rate of 0.2 ml/min. The column temperature was maintained at 40°C. The compounds were identified and quantified by QTOFMS (Agilent Technologies) using Agilent Mass Hunter Workstation software (Agilent Technologies). The drying gas had a flow of 10 l/min and the temperature was maintained 325°C. The Vcap, fragmentor, and skimmer voltages were 3,500, 125, and 65 V, respectively. The pressure of the nebulizer was 30 psig.
Western blot analysis
Total protein was extracted from B16F10 cells using RIPA buffer containing protease phosphatase inhibitor cocktail (Thermo Fisher Scientific). Cytoplasmic proteins were extracted by sonication from B16F10 cells in 100 mM of Tris-HCl (pH 7.6) buffer containing protease phosphatase inhibitors. Membrane proteins were extracted using RIPA buffer from the pellet after cytosol protein extraction. Can Get Signal immunoreaction enhancer solution (TOYOBO, Osaka, Japan) was used for the antibody reaction to proteins fixed on a PVDF membrane. The secondary antibody was labeled with horseradish peroxidase. Immunoreactive bands were visualized by SuperSignal West Dura solution (Thermo Fisher Scientific) and analyzed using ImageQuant LAS 4000 (GE Healthcare, Piscataway, NJ). The intensities of protein bands were analyzed using ImageJ Fiji software (http://imagej.net/Fiji).
Immunofluorescent staining
B16F10 cells were cultured with 50 μM of PA or EPA on culture cover glass (Matsunami Glass, Osaka, Japan). The cells were then washed in PBS, fixed with 10% formalin solution (WAKO) for 10 min, and then washed again in PBS. The cells were incubated with PBS containing 0.2% Triton X-100 (WAKO) for 15 min and then washed with PBS. After blocking with PBS containing 0.5% BSA and 0.1% Triton X-100, the cells were incubated with primary antibody diluted in Can Get Signal immunoreaction enhancer solution for 1 h, and then washed twice with PBS containing 0.1% Triton X-100. The cells were incubated with secondary antibodies labeled with Alexa Fluor 488 or Alexa Fluor 568 (Abcam, Cambridge, UK) for 30 min, and then washed twice with PBS containing 0.1% Triton X-100. Slides were mounted using SlowFade Diamond Antifade Mountant (Thermo Fisher Scientific) and viewed with a laser scanning confocal microscope [FLUOVIEW FV1000-D (OLYMPUS, Tokyo, Japan)]. The images were analyzed using ImageJ Fiji software.
Analysis of actin
B16F10 cells were cultured with 50 μM of PA or EPA on culture cover glass (Matsunami Glass) for 1 day. The cells were then washed in PBS, fixed with 10% formalin solution (WAKO) for 10 min, and then washed again in PBS. The cells were incubated with PBS containing 0.2% Triton X-100 (WAKO) for 15 min and then washed with PBS. The cells were incubated with Phalloidin-iFluor 488 reagent for 1 h and washed twice with PBS. Slides were mounted using SlowFade Diamond Antifade Mountant (Thermo Fisher Scientific) and viewed with a laser scanning confocal microscope [FLUOVIEW FV1000-D (OLYMPUS)]. The images were analyzed using ImageJ Fiji software. The polymerization of actin was analyzed using with Actin Polymerization Biochem kit (Cytoskeleton, Denver, CO). Assays were performed by following the manufacture’s protocol.
BRET analysis of RhoA-Rtkn
Total RNA was purified from B16F10 cells using with NucleoSpin RNA (TaKaRa, Shiga, Japan). The cDNA was synthesized using a PrimeScript RT reagent kit (TaKaRa). The RhoA and Rtkn genes were PCR amplified with KOD Plus (TOYOBO). The primers used to amplify RhoA were 5′-TGAGCAATCGTGGCTGAACT-3′ and 5′-ATGAGGCTGCGTTCACAAGG-3′. The primers used to amplify Rtkn were 5′-AGACTGATCCCAGGAGCGTAT-3′ and 5′-TGGACCTAGAGCCCAGTTGT-3′. The protein coding region of RhoA was PCR amplified from the first PCR product and cloned using NEBuilder (NEB, Ipswich, MA) into pNLF1C vector (Promega, Madison, WI). The protein coding region of Rtkn was also RCP amplified from the first PCR product and cloned using NEBuilder into pHTC vector (Promega). The NanoBRET assay (Promega) was carried out according to the manufacturer’s instructions.
Analysis of melanoma cell proliferation
Melanoma cells (1 × 106 per 6 cm dish) cultured in 25 μM of DHA for 48 h were collected using trypsin-EDTA solution (Thermo Fisher Scientific) and counted with an auto cell counter (Countess 2 FL; Thermo Fisher Scientific). B16F10 cells cultured in 25 μM of DHA for 24 h were incubated in medium containing 10 μM of bromodeoxyuridine (WAKO) for 1 h. The cells were then fixed in ice-cold water:MeOH (30:70) solution. The fixed cells were subsequently incubated in 2 N HCl solution containing 0.5% Triton X-100 for 30 min at room temperature. The cells were then washed in 0.1 M BRAX (WAKO) solution and PBS containing 1% BSA and 0.5% Tween 20 (WAKO). The cells were incubated in anti-bromodeoxyuridine antibody (Abcam) diluted 1 in 50 using PBS containing 1% BSA and 0.5% Tween 20 for 10 min at room temperature. The cells were washed twice in PBS containing 1% BSA and 0.5% Tween 20. The cells were resuspended in PBS containing 5 μg/ml PI, and the cell cycle was analyzed by flow cytometry (Becton Dickinson Japan, Fukushima, Japan). The proliferation of B16F10 cells was analyzed using CytoTrack green (GE Healthcare). B16F10 cells were labeled with CytoTrack green for 15 min, and then the cells were cultured with 25 μM of DHA. The fluorescent levels were measured by flow cytometry every 24 h for 3 days.
Pull-down analysis of DHA binding proteins and their identification
Pull-down assay of DHA-fixed magnetic FG beads (NH2 beads; Tamgawa Seiki, Nagano Japan) was performed according to the manufacturer’s protocol (https://fgb.tamagawa-seiki.com/en/data/pdf/protocolE005.pdf). Briefly, 5 μmol of DHA were activated by incubation with an equal molarity of succinimide and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide in 500 μl of N,N′-dimethylformamide for 2 h at room temperature using a microtube mixer. Activated DHA was fixed on NH2 beads (TAS8848N1130) by incubation with 2.5 mg of FG beads using a microtube mixer for 16 h at room temperature. Unreacted residues were masked using 20% carbonic anhydride in N,N-dimethylformamide. Whole cell lysate solution was prepared from B16F10 cells using extraction buffer [10 mM HEPES-NaOH (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 1% NP-40]. The lysate was centrifuged at 20,000 g for 30 min at 4°C, and the supernatant was collected in a new microtube as protein solution. The protein solution was diluted to 3 mg/ml in dilution buffer [20 mM HEPES-NaOH (pH 7.9), 100 mM KCl, 1 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 1 mM DTT, 0.2 mM PMSF]. The protein solution was preincubated with DMSO, 1 mM DHA, or 1 mM PA for 4 h at 4°C. DHA-labeled FG-beads (0.5 mg) were incubated with 200 μl of the preincubated protein solution for 16 h at 4°C. Incubated DHA-labeled FG-beads were washed three times using a magnetic stand in wash buffer [20 mM HEPES-NaOH (pH 7.9), 100 mM KCl, 1 mM MgCl2, 0.4 mM EDTA, 15% glycerol, 5% NP-40,1 mM DTT, 0.2 mM PMSF]. Proteins bound to the DHA-labeled FG-beads were eluted by incubation at 98°C for 5 min in SDS-PAGE sample buffer [250 mM Tris-HCl (pH 6.8), 0.02% BPB, 8% SDS, 40% glycerol, 20% 2-mercaptoethanol]. The boiled elution samples were separated by SDS-PAGE using a 4–15% NuPAGE gradient gel (Thermo Fisher Scientific). The gel was stained with a Silver staining kit (SilverQuest; Thermo Fisher Scientific). The protein band was excised by an in-gel protein digestion procedure, which was performed using an In-Gel Tryptic Digestion kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, excised gels were destained, reduced with Tris[2-carboxyethyl]phosphine, alkylated with iodoacetamide, and dehydrated with acetonitrile. The proteins were digested with 10 ng/ml trypsin overnight. The solution was purified using Pierce C-18 SpinColumns (Thermo Fisher Scientific) and subjected to nanoLC-MS/MS analysis. Digested peptides were separated using the ADVANCE UHPLC system (MICHROM Bioresources, Billerica, MA) using a Magic C18 nano column (0.1 × 150 mm; MICHROM Bioresources) gradient elution (water containing 0.1% formic acid/acetonitrile to 5/45 in 15 min) at a flow rate of 500 nl/min and analyzed on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) operated with Xcalibur software (version 2.0.7; Thermo Fisher Scientific). Peptides were identified using a MASCOT MS/MS ion search (http://www.matrixscience.com/home.html).
Statistical analysis
Statistically significant differences between the experimental groups were identified using one-way ANOVA and Tukey’s post hoc tests.
RESULTS
Opposing effects between saturated fatty acids and PUFAs during melanogenesis
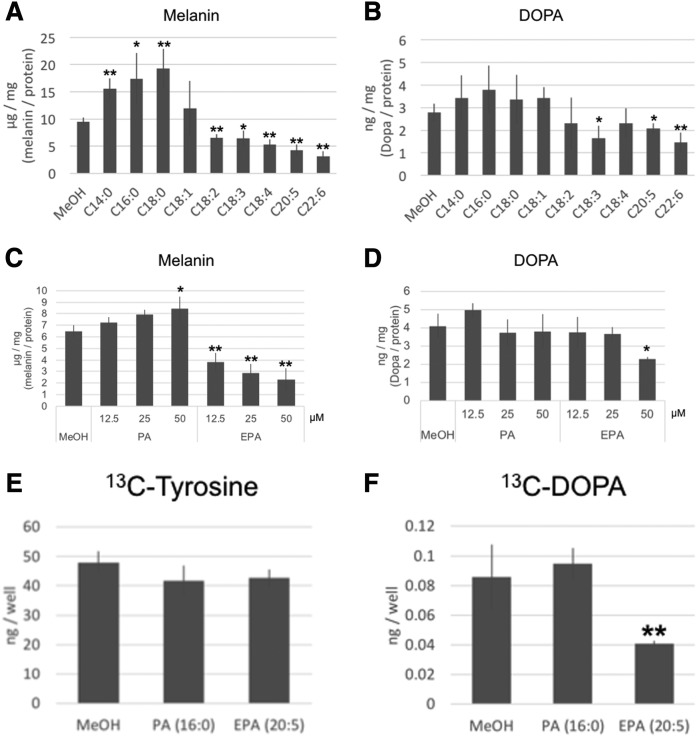
To examine the relationship between the structures of fatty acids and their effect on melanogenesis, we cultured a mouse melanoma cell line (B16F10 cells) in the presence of saturated fatty acids (SFAs) [myristic acid (C14:0), PA (C16:0), and stearic acid (C18:0)], monounsaturated fatty acid [oleic acid (C18:1)], or PUFAs [linoleic acid (C18:2), α-linoleic acid (C18:3), stearidonic acid (C18:4), EPA (C20:5), and DHA (C22:6)] for 3 days. The melanin content of the B16F10 cells was determined following extraction. At a concentration of 50 μM, a cytotoxic effect was only observed in B16F10 cells cultured with DHA (supplemental Table S1). SFAs significantly increased the melanin content of B16F10 cells, whereas PUFAs significantly decreased the melanin content. There was no significant difference of melanin content in the presence of monounsaturated fatty acid (Fig. 1A). To examine Tyr activity of B16F10 cells cultured in the presence of fatty acids, we measured the DOPA contents of B16F10 cells with LC/QTOFMS. The DOPA contents of B16F10 cells cultured with α-linoleic acid, EPA, and DHA were significantly decreased (Fig. 1B). PA increased the melanin level at a concentration of 50 μM. EPA decreased the melanin content at a concentration of >12.5 μM (Fig. 1C). B16F10 cells cultured in the presence of 50 μM of EPA had a decreased level of DOPA (Fig. 1D). These results indicate that oxidation of tyrosine by Tyr was suppressed by PUFAs. To examine this phenomenon in more detail, we analyzed tyrosine metabolism using 13C-labeled tyrosine in B16F10 cells cultured with EPA for 3 days. There was no significant difference of 13C-tyrosine content in the cells, although 13C-DOPA was significantly decreased in B16F10 cells cultured in the presence of EPA. These results indicated that EPA suppressed the conversion of tyrosine to DOPA. There were no significant differences of DOPA content between B16F10 cells cultured in the absence (control) or presence of SFAs (Fig. 1E, F).
Fig. 1.
Quantitative analysis of melanin and DOPA in B16F10 cells. Melanin (A) and DOPA (B) levels were measured in B16F10 cells cultured in the presence of 50 μM of fatty acids for 3 days. C, D: Concentration-dependent effect of PA and EPA on the level of melanin and DOPA in B16F10 cells. The values were normalized by protein concentration. The uptake of tyrosine (E) and production of DOPA from tyrosine (F) in B16F10 cells cultured with 50 μM of PA or EPA were measured by LC/QTOFMS using 13C-tyrosine. The values indicate the relative amount of 13C-tyrosine or 13C-DOPA compared with the control. A–D: Values indicate the mean ± SD of five individual samples. Significant difference: *P < 0.05; **P < 0.01.
EPA suppresses membrane-associated Tyr and Tyrp1
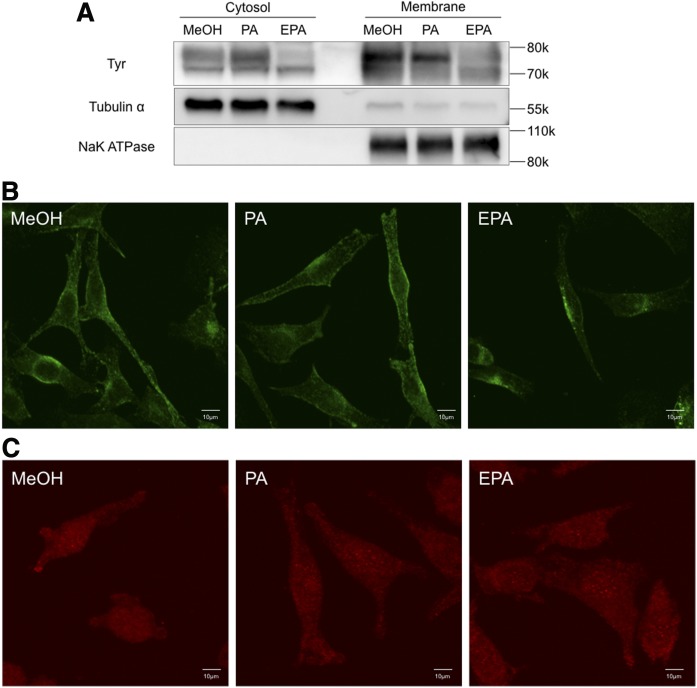
The enzymes that perform melanogenesis are membrane-associated proteins. These melanogenic enzymes are translated in the ER and trafficked to the melanosome by a membrane trafficking system (19). Previous studies demonstrated that Tyr is degraded by the ubiquitin-proteasome pathway in melanoma cells after 3 days of culture in the presence of linoleic acid or DHA (11, 21). To examine whether PA and EPA affect the level and location of Tyr, we quantified this protein by Western blot analysis. According to its amino acid sequence, the molecular mass of Tyr is less than 60 kDa. However, Tyr was detected by Western blot analysis as multiple bands ranging between 70 and 80 kDa owing to extensive levels of glycosylation (29). Indeed, previous reports suggested that mature fully active Tyr exists as a high molecular mass protein of 70–80 kDa (22, 30). We found that B16F10 cells cultured in the presence of EPA had reduced levels of membrane-associated Tyr. Cytosolic levels of glycosylated-Tyr protein also decreased in B16F10 cells cultured in the presence of EPA (Fig. 2A). We also examined Tyrp1 localization in B16F10 cells cultured with PA or EPA by immunostaining. Tyrp1 (also known as Trp1 or gp75) is a melanogenic enzyme. Tyrp1 translated in the ER is stabilized by trafficking during stages III and IV in the melanosome. Tyrp1 was detected around the nuclei and cell membrane of B16F10 cells cultured in the absence or presence of PA. By contrast, the level of Tyrp1 located close to the cell membrane was decreased in B16F10 cells cultured in the presence of EPA (Fig. 2B). Rab38, which is a Rab-type small GTPase, plays an important role during Tyr and Tyrp1 trafficking from the ER to the melanosome. To examine the possible effect of PA and EPA on Rab38, we analyzed Rab38 localization by immunostaining. However, Rab38 localization was unaffected by PA and EPA (Fig. 2C).
Fig. 2.
Tyr, Tyrp1, and Rab38 protein expression in B16F10 cells cultured with fatty acids. A: Western blot analysis of Tyr in B16F10 cells cultured in the presence of 50 μM of PA or EPA for 3 days. Immunofluorescent staining of Tyrp1 (B) and Rab38 (C) in B16F10 cells cultured in the presence of 50 μM of PA or EPA for 3 days.
PA induces formation of F-actin, while EPA decreases F-actin levels in melanoma cells
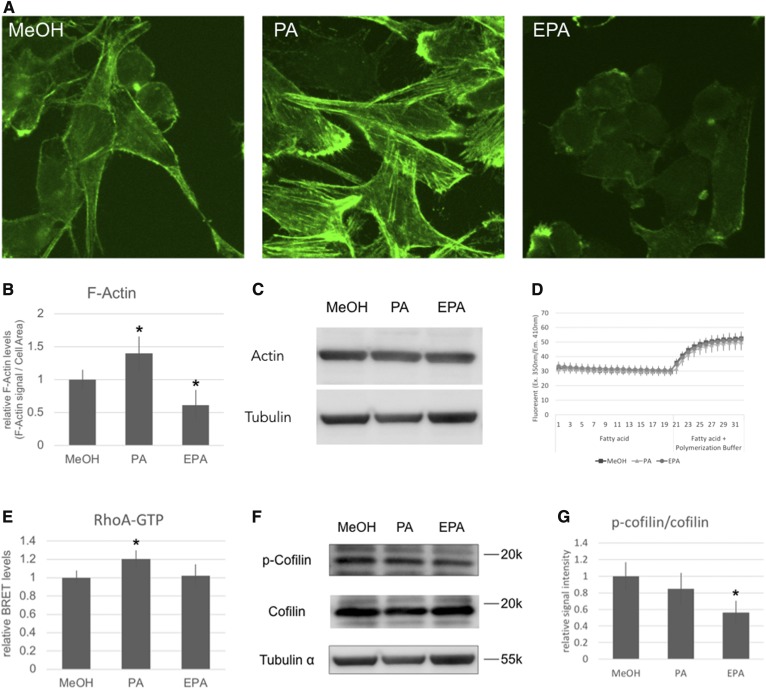
Melanosomes are trafficked to the plasma membrane on microtubules and actin filaments. We examined the effect of PA and EPA on these cytoskeletal structures by β-tubulin and F-actin staining. Although PA and EPA did not significantly perturb the organization of the microtubules, the structure of actin filaments was radically altered. Specifically, PA induced stress fiber formation, whereas EPA decreased the level of F-actin (Fig. 3A, B). While the level of F-actin was dramatically changed by PA and EPA, total actin protein levels were unaltered (Fig. 3C). Next, we examined the direct effect of PA and EPA on actin polymerization by using an in vitro actin polymerization assay. The in vitro experiments confirmed that i) PA does not induce actin polymerization, and ii) EPA does not inhibit actin polymerization (Fig. 3D). These findings indicated that PA and EPA must influence the degree of actin polymerization and depolymerization via an indirect mechanism.
Fig. 3.
Analysis of actin. A: B16F10 cells cultured in the presence of 50 μM of PA or EPA for 24 h were stained by phalloidin. B: Quantitative analysis of the phalloidin fluorescence signal. The fluorescence signals per 100 μm2 of the cell were measured using with ImageJ Fiji software. The values indicate the mean ± SD of four individual fields. C: Western blot analysis of α-actin protein. D: Actin polymerization assay performed in vitro. The values indicate the mean ± SD of eight individual wells. E: BRET assay of RhoA and Rtkn. The values indicate the mean ± SD of eight individual wells. F: Western blot analysis of phosphorylated cofilin and total cofilin. G: The quantitative analysis of the ratio of phosphorylated cofilin to total cofilin. The values indicate the mean ± SD of four individual samples. *Significant difference of P < 0.05.
In the light of these findings, we analyzed RhoA activation and cofilin phosphorylation levels in the B16F10 cells. RhoA, a member of the Rho family of small GTPases, regulates cytoskeletal rearrangements (31–33). Rho GTPase can act as a molecular switch by changing from an inactive GDP-bound conformation to an active GTP-bound conformation. The active GTP-bound RhoA specifically binds Rtkn (34). Moreover, RhoA activation induces the formation of stress fibers (33). We analyzed RhoA activation in B16F10 cells by NanoBRET using RhoA with a HaloTag chimera protein and Rtkn with a NanoLuciferase chimera protein. RhoA was activated in B16F10 cells cultured in the presence of PA (Fig. 3E). The level of activated RhoA in cells cultured in the presence or absence of EPA was found to be equivalent. Cofilin, a key regulator of actin filament dynamics, acts by stimulating the depolymerization and severance of actin filaments (35). Phosphorylated cofilin is enzymatically inactive. Indeed, the activity of cofilin is regulated by a specific kinase and phosphatase. To examine the effect of PA and EPA on cofilin activity, we analyzed the amount of phosphorylated cofilin relative to total cofilin by Western blot analysis. Cells cultured in the presence of EPA had significantly lower levels of phosphorylated cofilin (Fig. 3F) compared with the control (no EPA). No significant difference was observed between the levels of cofilin in cells cultured in the presence and absence of PA.
PA increases and EPA decreases the level of Rab27a in B16F10 cells
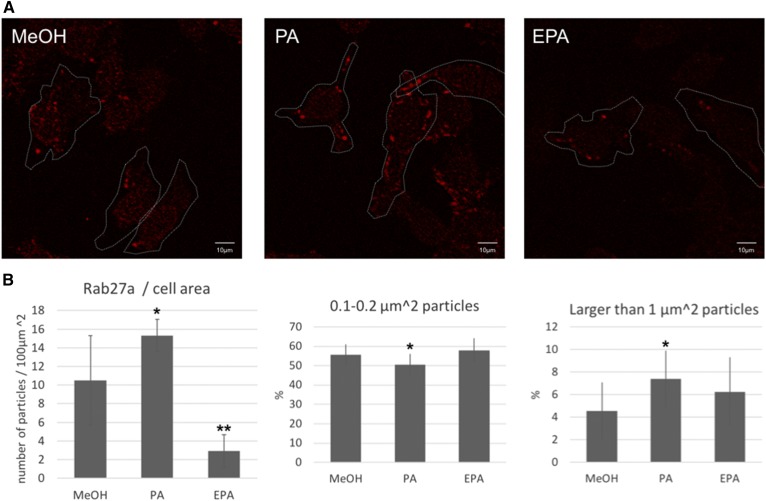
Rab27a regulates actin-based melanosome transport and melanosome anchoring to the plasma membrane (36–41). We used immunofluorescent staining to analyze Rab27a protein in B16F10 cells cultured in the presence of PA or EPA. The number of Rab27a particles increased in cells cultured in the presence of PA for a day. By contrast, the number of Rab27a particles decreased in cells cultured in the presence of EPA for a day (Fig. 4A, B). Intriguingly, the average size of the Rab27a particles found in cells cultured in the presence of PA changed compared with the control. Specifically, B16F10 cells cultured with PA showed a significantly decreased frequency of small Rab27a particles (0.1–0.2 μm2) (Fig. 4C) and significantly increased the frequency of large Rab27a particles (larger than 1 μm2) (Fig. 4D).
Fig. 4.
Immunofluorescent staining of Rab27a. A: B16F10 cells cultured in the presence of 50 μM of PA or EPA for 24 h were stained with a Rab27a-specific antibody. The white broken line indicates the outline of the cell drawn in bright field. B: Quantitative analysis of Rab27a signals. The number of particles and the proportion of small or large sized particles were measured for four individual fields. The values indicate the mean ± SD. Significant difference: *P < 0.05; **P < 0.01.
DHA delayed the cell cycle of melanoma
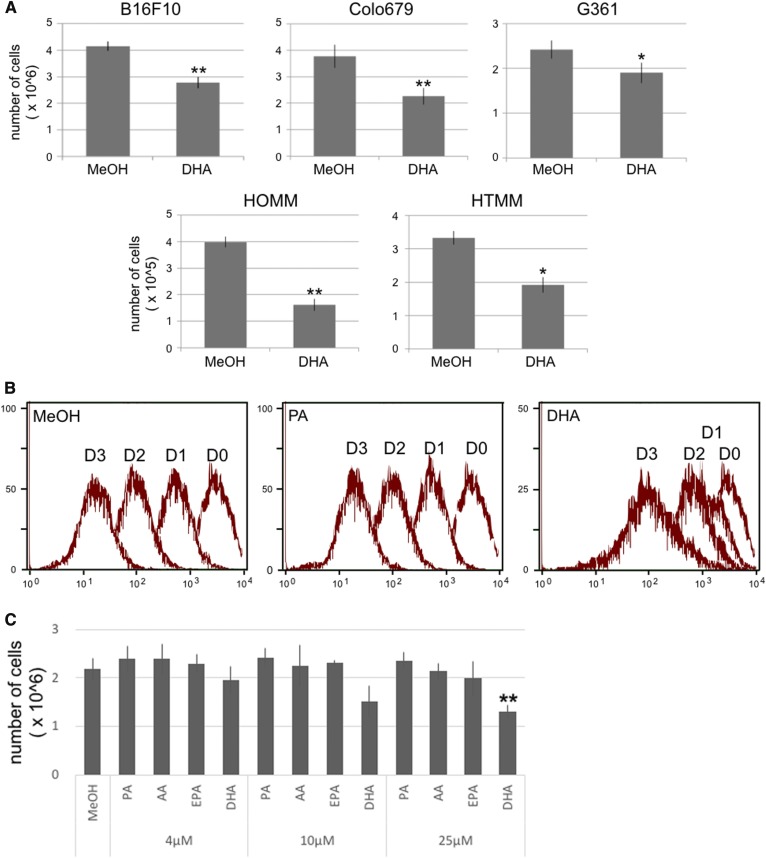
To examine the anti-tumor effect of DHA on melanoma, we analyzed the cell growth and cell cycle of melanoma cell lines cultured in the presence of DHA. First, we analyzed the effect of DHA on the cell growth of mouse (B16F10) and human (Colo679, G361, HOMM, and HTMM) melanoma cell lines. DHA suppressed tumor cell growth in all five melanoma cell lines that we examined (Fig. 5A). Next, we analyzed the effect of DHA on the cell cycle of B16F10 cells by flow cytometry. The proportion of S phase cells decreased in B16F10 cells cultured in the presence of DHA, and the proportion of G1 and G2/M phase cells increased (Table 1). These findings indicated that DHA suppressed cell growth in melanoma. Thus, we analyzed cell proliferation in B16F10 cells cultured in the presence of DHA, and we found that DHA delayed B16F10 proliferation (Fig. 5B). PA, arachidonic acid (AA), and EPA did not suppress B16F10 cell proliferation at a concentration greater than 4–25 μM (Fig. 5C).
Fig. 5.
The proliferation and cell cycle analysis of melanoma. A: The mouse and human melanoma cell lines were cultured in the presence of 25 μM of DHA for 48 h. The values indicate the mean ± SD of three individual samples. B: B16F10 cells were stained with CytoTrack green for 15 min, and then the fluorescence signals in the cells were measured by flow cytometry every 24 h for 3 days. D0, D1, D2, and D3 indicate day 0, day 1, day 2, and day 3, respectively. C: Effect of fatty acids on the proliferation of B16F10 cells. Significant difference: *P < 0.05; **P < 0.01.
TABLE 1.
The effect of DHA on the cell cycle of B16F10 cells by flow cytometry
| Cell Cycle (%) | |||
| G1 | S | G2/M | |
| MeOH | 49.57 ± 0.002 | 44.81 ± 0.015 | 0.57 ± 0.001 |
| 25 μM PA | 52.75 ± 0.060a | 43.25 ± 0.090 | 0.51 ± 0.010 |
| 25 μM DHA | 52.36 ± 0.080a | 38.84 ± 0.012a | 2.09 ± 0.007a |
The values indicate the mean ± SD of three individual samples.
A significant difference compared with MeOH of P < 0.01.
DHA interacts with Rack1
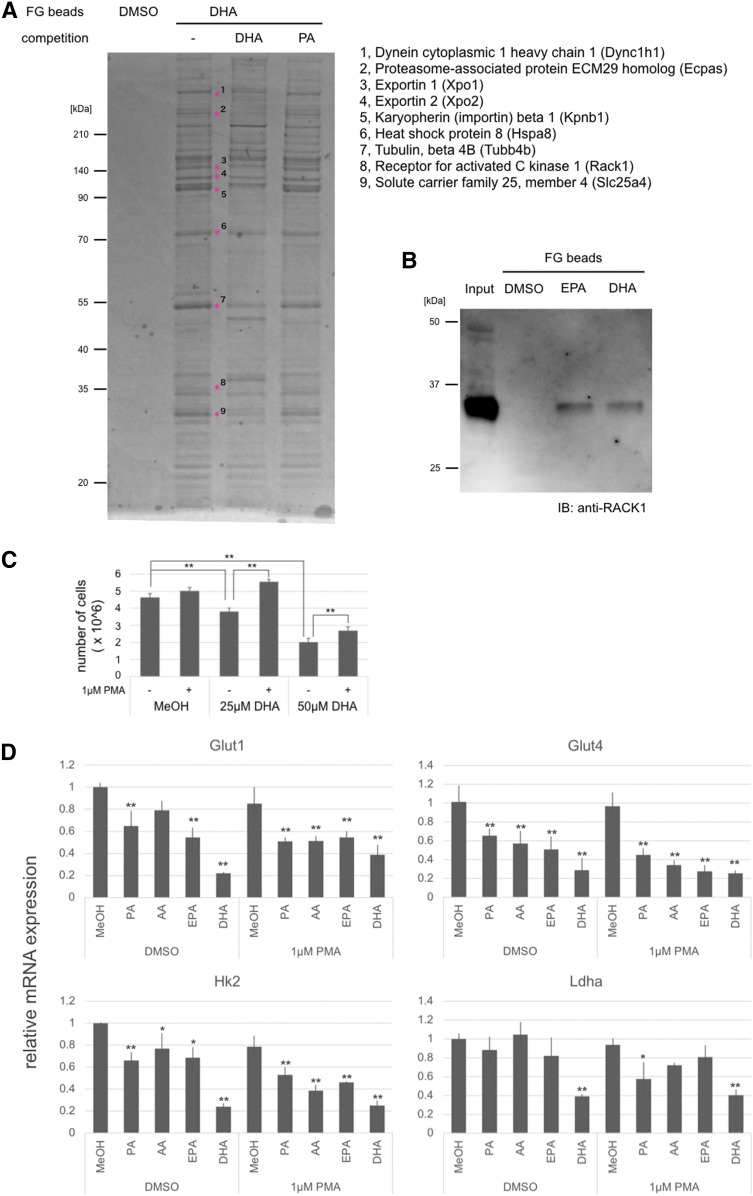
To reveal the anti-tumor mechanism of DHA, we examined proteins that directly interact with DHA by pull-down assay using magnetic beads. Numerous candidates were detected as potential DHA binding proteins. To narrow down the number of DHA binding proteins according to specificity, we preincubated proteins extracted from B16F10 with 1 mM of DHA or PA. We identified nine proteins that decreased interaction with DHA-fixed beads after preincubation with DHA: dynein cytoplasmic 1 heavy chain 1 (Dync1h1), proteasome-associated protein ECM29 homolog (Ecpas), exportin 1 (Xpo1), exportin 2 (Xpo2), karyopherin (importin) β1 (Kpnb1), heat shock protein 8 (Hspa8), tubulin β 4B (Tubb4b), Rack1, and solute carrier family 25 member 4 (Slc25a4) (Fig. 6A). In particular, we focused on Rack1, which interacted only with activated protein kinase C (PKC) to serve both as an anchor protein for PKC and as a scaffold protein for the recruitment PKCs (and other proteins) into a signaling complex (42). Multiple studies suggest that PKC-mediated signaling plays an important role in a variety of cancers (42). We confirmed interaction of DHA and Rack1 using a Rack1-specific antibody (Fig. 6B). EPA was also found to interact with Rack1.
Fig. 6.
Affinity purification and protein identification of DHA binding protein. A: Silver-stained polyacrylamide gel. Bands highlighted with an asterisk (*) indicate proteins that interact specifically with DHA. B: Immunoblotting analysis indicating the binding of Rack1 to DHA. C: Proliferation assay of B16F10 cells cultured with DHA and PMA. D: Gene expression of glucose transporters and glycolysis enzymes. The values indicate the mean ± SD of three individual samples. Significant difference: *P < 0.05; **P < 0.01.
PMA decreased the anti-tumor effect of DHA
To examine the relationship between the anti-tumor effect of DHA and the PKC signaling pathway, we cultured B16F10 cells in the presence of DHA and PMA. PMA decreased the anti-tumor effect of DHA (Fig. 6C). However, PMA did not significantly change the proliferation of B16F10 cells. Previous reports showed that PKC affected glucose metabolism by enhancing expression of glucose transporter genes and glycolysis-related genes (43, 44). Moreover, inhibition of glycolysis has been shown to suppress B16F10 cell proliferation (45). We examined changes in the expression level of glucose transporter genes and glycolysis-related kinase genes. PMA did not alter the level of expression of these genes. However, expression of Glut4 and Hk2 was suppressed by four fatty acids (PA, AA, EPA, and DHA) examined in this study. The gene expression of Glut1 was suppressed by EPA and DHA. The gene expression of Ldha was suppressed by DHA. Interestingly, DHA suppressed the expression of glucose transporters and glycolysis-related genes to a greater degree than PA, AA, and EPA (Fig. 6D). Inhibition or activation of the PKC signal did not change the anti-melanogenic effect of EPA (supplemental Fig. S1).
Effect of fatty acids on normal human melanocytes
To examine the effect of fatty acids on normal human melanocytes, we cultured these cells with PA, oleic acid, EPA, or DHA. PA, oleic acid, EPA, and DHA did not alter the proliferation of normal human melanocytes (supplemental Table S2). Moreover, the melanin and Tyr protein expression levels in human melanocytes were not changed during culture with PA or EPA (supplemental Fig. S2).
DISCUSSION
Several studies have examined the physiological effects of fatty acids on melanoma. However, the relationship between fatty acid structures and their physiological effects on melanoma, as well as the corresponding mechanism of action, had not previously been studied. Here, we show that cells cultured in the presence of SFAs had increased levels of melanin (Fig. 1), whereas PUFAs decreased melanin levels (Fig. 1). Moreover, DHA was found to suppress tumor proliferation in melanoma (Fig. 5, Table 1).
EPA decreased the level of Tyr and DOPA in B16F10 cells but did not suppress import of tyrosine into the cells (Figs. 1, 2). These findings indicate that EPA suppresses melanogenesis by decreasing the activity of Tyr. Thus, the results for EPA are in good agreement with previous studies that investigated linoleic acid (20) and DHA (11). In B16F10 cells cultured in the presence of PA, we observed increased levels of melanin (Fig. 1). This result is in agreement with a previous study (20). Ando et al. (22) suggested that PA and linoleic acid affect trafficking of Tyr. In this study, we found that EPA decreased the levels of Tyrp1 located near the plasma membrane (Fig. 2D). Therefore, we reasoned that PA and EPA might alter the localization of both Tyr and Tyrp1. First, we examined the expression and location of Rab38 by immunostaining because this protein is required for Tyr and Tyrp1 transport to the melanosome. However, incubation of the cells in the presence of PA and EPA did not alter the level and localization of Rab38 protein (Fig. 2E). Next, analysis of the cytoskeletal structure of B16F10 cells revealed that PA and EPA radically changed the formation of F-actin. Specifically, PA increased the active form of RhoA (Fig. 3E) and induced stress fibers (Fig. 3A) as well as the accumulation of Rab27a (Fig. 4) in B16F10 cells. EPA decreased the level of phosphorylated cofilin (inactive form) (Fig. 3F, G), F-actin (Fig. 3A), and Rab27a (Fig. 4). Rab27a is known to recruit myosin Va for melanosomes (36, 38) and regulate actin-based movement of melanosomes (46). Dysfunction of Rab27a causes perinuclear localization of melanosomes and the suppression of coat color in mice (36, 47). Our results suggest that PA accumulated melanosome in the cell by stress fiber formation and EPA destabilized melanosome by decreasing F-actin. The opposing effects of PA and EPA on melanogenesis may result from their modulation of actin polymerization. Thus, the regulation of F-actin formation by fatty acids may be a significant phenomenon in the cell. In addition to this study, at least two other reports conclude that PUFAs suppress F-actin formation (48, 49). However, there is a paucity of studies that examine the relationship between F-actin formation and SFAs. Further investigation is needed to reveal the overall physiological effect of these fatty acids and to correlate the diseases that lead to an imbalance in F-actin formation.
DHA suppressed the proliferation of human and mouse melanoma cell lines to delay cell cycle progression (Fig. 5, Table 1). We identified nine proteins that specifically bind DHA (Fig. 6). These were: i) Dync1h1, which acts as a motor for the intracellular retrograde motility of vesicles and organelles along microtubules (50); ii) Ecaps, an adaptor/scaffolding protein that may play a role in ERAD and other enhanced proteolysis pathways (51); iii) Xpo1 and Xpo2, which mediate nuclear export of cellular proteins (cargos) bearing a leucine-rich nuclear export signal (NES) and of RNAs; iv) Kpnb1, which binds to nuclear localization signals (NLS) and functions in nuclear protein import (52); v) Hspa8, a molecular chaperone that plays a pivotal role in the protein quality control system; vi) Tubb4b, a major constituent of microtubules; vii) Rack1, a scaffolding protein that binds activated PKCs; and viii) Slc25a4, which functions in mitochondrial ADP/ATP transport and catalyzes the exchange of cytoplasmic ADP with mitochondrial ATP across the mitochondrial inner membrane (53). The interactions of DHA and these nine proteins are interesting new insights, but the physiological significance of each interaction remains to be determined. The results from this study indicate the importance of Rack1 for the anti-tumor effect mediated by DHA. Indeed, previous studies showed that Rack1 activates PKC and oncogenic signaling as well as promoting tumor cell proliferation (54–57). PKC signaling also regulates the gene expression of glucose transporters and glycolysis enzymes (43, 44). DHA suppressed the expression of glucose transporter gene and glycolysis-related genes in B16F10 cells (Fig. 5D). Inhibition of glucose transport and glycolysis appears to be a cause of suppressed melanoma cell proliferation brought about by DHA. The DHA-mediated suppression of melanoma cell proliferation was reduced by PMA treatment (Fig. 6C). This observation suggests that DHA inhibits melanoma proliferation by suppression of PKC signaling. Several studies have shown that DHA suppresses PKC signaling in T cells (58–60). Thus, DHA may modulate PKC signaling by interacting with Rack1 in several cell types. With respect to the anti-melanogenic activity, PMA did not change the melanin content of EPA-treated B16F10 cells (supplemental Fig. S1). These results indicated that the anti-melanogenic effect of PUFA might involve another signaling pathway.
Proliferation and melanogenesis in human melanocytes were not influenced by PA, oleic acid, EPA, or DHA (supplemental Table S2, supplemental Fig. S2). Melanin accumulation by PA, suppression of melanogenesis by EPA, and anti-proliferation by DHA were observed in melanoma cells.
In this study, we demonstrate that fatty acids elicit physiological functions by modulating actin polymerization and PKC signaling by interacting with Rack1. These findings represent newly determined effects mediated by free fatty acids. Fatty acids, actin, and Rack1 are ubiquitous molecules in cells. Therefore, PA-induced stress fiber formation and EPA-induced depolymerization of F-actin as well as interaction between DHA and Rack1 might be widespread phenomena.
Supplementary Material
Acknowledgments
The authors thank Ms. Akiko Shiraishi for her technical assistance.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- BRET
- bioluminescence resonance energy transfer
- DOPA
- 3,4-dihydroxyphenylalanine
- LC/QTOFMS
- LC/hybrid quadrupole TOFMS
- MeOH
- methanol
- PA
- palmitic acid
- PKC
- protein kinase C
- Rack1
- receptor for activated C kinase 1
- SFA
- saturated fatty acid
- Tyr
- tyrosinase
- Tyrp1
- tyrosinase-related protein 1
This work was supported by funds from the Basic Biotechnology Project of Iwate Prefecture, Japan and Ministry of Education, Culture, Sports, Science and Technology of Japan Grant-in-Aid for Young Scientists (B) JP16K18775 (to H.Y.). The authors declare that there are no competing interests associated with this work.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Dyerberg J., and Bang H. O.. 1979. Haemostatic function and platelet polyunsaturated fatty acids in Eskimos. Lancet. 2: 433–435. [DOI] [PubMed] [Google Scholar]
- 2.Kromann N., and Green A.. 1980. Epidemiological studies in the Upernavik district, Greenland. Incidence of some chronic diseases 1950-1974. Acta Med. Scand. 208: 401–406. [PubMed] [Google Scholar]
- 3.Kromhout D., Bosschieter E. B., and de Lezenne Coulander C.. 1985. The inverse relation between fish consumption and 20-year mortality from coronary heart disease. N. Engl. J. Med. 312: 1205–1209. [DOI] [PubMed] [Google Scholar]
- 4.Tavani A., Pelucchi C., Negri E., Bertuzzi M., and La Vecchia C.. 2001. n-3 Polyunsaturated fatty acids, fish, and nonfatal acute myocardial infarction. Circulation. 104: 2269–2272. [DOI] [PubMed] [Google Scholar]
- 5.Yuan J. M., Ross R. K., Gao Y. T., and Yu M. C.. 2001. Fish and shellfish consumption in relation to death from myocardial infarction among men in Shanghai, China. Am. J. Epidemiol. 154: 809–816. [DOI] [PubMed] [Google Scholar]
- 6.Iso H., Kobayashi M., Ishihara J., Sasaki S., Okada K., Kita Y., Kokubo Y., and Tsugane S.. 2006. Intake of fish and n3 fatty acids and risk of coronary heart disease among Japanese: the Japan Public Health Center-Based (JPHC) study cohort I. Circulation. 113: 195–202. [DOI] [PubMed] [Google Scholar]
- 7.Stillwell W., Ehringer W., and Jenski L. J.. 1993. Docosahexaenoic acid increases permeability of lipid vesicles and tumor cells. Lipids. 28: 103–108. [DOI] [PubMed] [Google Scholar]
- 8.Albino A. P., Juan G., Traganos F., Reinhart L., Connolly J., Rose D. P., and Darzynkiewicz Z.. 2000. Cell cycle arrest and apoptosis of melanoma cells by docosahexaenoic acid: association with decreased pRb phosphorylation. Cancer Res. 60: 4139–4145. [PubMed] [Google Scholar]
- 9.Zajdel A., Wilczok A., Chodurek E., Gruchlik A., and Dzierzewicz Z.. 2013. Polyunsaturated fatty acids inhibit melanoma cell growth in vitro. Acta Pol. Pharm. 70: 365–369. [PubMed] [Google Scholar]
- 10.So W. W., Liu W. N., and Leung K. N.. 2015. Omega-3 polyunsaturated fatty acids trigger cell cycle arrest and induce apoptosis in human neuroblastoma LA-N-1 cells. Nutrients. 7: 6956–6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balcos M. C., Kim S. Y., Jeong H., Yun H., Baek K. J., Kwon N. S., Park K., and Kim D.. 2014. Docosahexaenoic acid inhibits melanin synthesis in murine melanoma cells in vitro through increasing tyrosinase degradation. Acta Pharmacol. Sin. 35: 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimabukuro M., Zhou Y. T., Levi M., and Unger R. H.. 1998. Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proc. Natl. Acad. Sci. USA. 95: 2498–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maedler K., Spinas G. A., Dyntar D., Moritz W., Kaiser N., and Donath M. Y.. 2001. Distinct effects of saturated and monounsaturated fatty acids on beta-cell turnover and function. Diabetes. 50: 69–76. [DOI] [PubMed] [Google Scholar]
- 14.Maedler K., Oberholzer J., Bucher P., Spinas G. A., and Donath M. Y.. 2003. Monounsaturated fatty acids prevent the deleterious effects of palmitate and high glucose on human pancreatic beta-cell turnover and function. Diabetes. 52: 726–733. [DOI] [PubMed] [Google Scholar]
- 15.Zhao H., Matsuzaka T., Nakano Y., Motomura K., Tang N., Yokoo T., Okajima Y., Han S., Takeuchi Y., Aita Y., et al. 2017. Elovl6 deficiency improves glycemic control in diabetic db/db mice by expanding β-cell mass and increasing insulin secretory capacity. Diabetes. 66: 1833–1846. [DOI] [PubMed] [Google Scholar]
- 16.Hearing V. J., and Jiménez M.. 1987. Mammalian tyrosinase–the critical regulatory control point in melanocyte pigmentation. Int. J. Biochem. 19: 1141–1147. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi T., Urabe K., Winder A., Jiménez-Cervantes C., Imokawa G., Brewington T., Solano F., García-Borrón J. C., and Hearing V. J.. 1994. Tyrosinase related protein 1 (TRP1) functions as a DHICA oxidase in melanin biosynthesis. EMBO J. 13: 5818–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marks M. S., and Seabra M. C.. 2001. The melanosome: membrane dynamics in black and white. Nat. Rev. Mol. Cell Biol. 2: 738–748. [DOI] [PubMed] [Google Scholar]
- 19.Raposo G., and Marks M. S.. 2007. Melanosomes–dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 8: 786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ando H., Funasaka Y., Oka M., Ohashi A., Furumura M., Matsunaga J., Matsunaga N., Hearing V. J., and Ichihashi M.. 1999. Possible involvement of proteolytic degradation of tyrosinase in the regulatory effect of fatty acids on melanogenesis. J. Lipid Res. 40: 1312–1316. [PubMed] [Google Scholar]
- 21.Ando H., Watabe H., Valencia J. C., Yasumoto K. I., Furumura M., Funasaka Y., Oka M., Ichihashi M., and Hearing V. J.. 2004. Fatty acids regulate pigmentation via proteasomal degradation of tyrosinase: a new aspect of ubiquitin-proteasome function. J. Biol. Chem. 279: 15427–15433. [DOI] [PubMed] [Google Scholar]
- 22.Ando H., Wen Z-M., Kim H-Y., Valencia J. C., Costin G-E., Watabe H., Yasumoto K., Niki Y., Kondoh H., Ichihashi M., et al. 2006. Intracellular composition of fatty acid affects the processing and function of tyrosinase through the ubiquitin-proteasome pathway. Biochem. J. 394: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumgartner M., Sturlan S., Roth E., Wessner B., and Bachleitner-Hofmann T.. 2004. Enhancement of arsenic trioxide-mediated apoptosis using docosahexaenoic acid in arsenic trioxide-resistant solid tumor cells. Int. J. Cancer. 112: 707–712. [DOI] [PubMed] [Google Scholar]
- 24.Donat-Vargas C., Berglund M., Glynn A., Wolk A., and Åkesson A.. 2017. Dietary polychlorinated biphenyls, long-chain n-3 polyunsaturated fatty acids and incidence of malignant melanoma. Eur. J. Cancer. 72: 137–143. [DOI] [PubMed] [Google Scholar]
- 25.Serini S., Zinzi A., Ottes Vasconcelos R., Fasano E., Riillo M. G., Celleno L., Trombino S., Cassano R., and Calviello G.. 2016. Role of β-catenin signaling in the anti-invasive effect of the omega-3 fatty acid DHA in human melanoma cells. J. Dermatol. Sci. 84: 149–159. [DOI] [PubMed] [Google Scholar]
- 26.Serini S., Fasano E., Piccioni E., Monego G., Cittadini A. R. M., Celleno L., Ranelletti F. O., and Calviello G.. 2012. DHA induces apoptosis and differentiation in human melanoma cells in vitro: Involvement of hur-mediated COX-2 mRNA stabilization and β-catenin nuclear translocation. Carcinogenesis. 33: 164–173. [DOI] [PubMed] [Google Scholar]
- 27.Denkins Y., Kempf D., Ferniz M., Nileshwar S., and Marchetti D.. 2005. Role of ω-3 polyunsaturated fatty acids on cyclooxygenase-2 metabolism in brain-metastatic melanoma. J. Lipid Res. 46: 1278–1284. [DOI] [PubMed] [Google Scholar]
- 28.Park M., and Kim H.. 2017. Anti-cancer mechanism of docosahexaenoic acid in pancreatic carcinogenesis: a mini-review. J. Cancer Prev. 22: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Újvári A., Aron R., Eisenhaure T., Cheng E., Parag H. A., Smicun Y., Halaban R., and Hebert D. N.. 2001. Translation rate of human tyrosinase determines its N-linked glycosylation level. J. Biol. Chem. 276: 5924–5931. [DOI] [PubMed] [Google Scholar]
- 30.Halaban R., Pomerantz S. H., Marshall S., Lambert D. T., and Lerner A. B.. 1983. Regulation of tyrosinase in human melanocytes grown in culture. J. Cell Biol. 97: 480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridley A. J., and Hall A.. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and stress fibres in response to growth factors. Cell. 70: 389–399. [DOI] [PubMed] [Google Scholar]
- 32.Hall A. 1998. Rho GTPases and the actin cytoskeleton. Science. 279: 509–514. [DOI] [PubMed] [Google Scholar]
- 33.Burridge K., and Wennerberg K.. 2004. Rho and Rac take center stage. Cell. 116: 167–179. [DOI] [PubMed] [Google Scholar]
- 34.Reid T., Furuyashiki T., Ishizaki T., Watanabe G., Watanabe N., Fujisawa K., Morii N., Madaule P., and Narumiya S.. 1996. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the Rho-binding domain. J. Biol. Chem. 271: 13556–13560. [DOI] [PubMed] [Google Scholar]
- 35.Bamburg J. R., and Wiggan O. P.. 2002. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 12: 598–605. [DOI] [PubMed] [Google Scholar]
- 36.Hume A. N., Collinson L. M., Rapak A., Gomes A. Q., Hopkins C. R., and Seabra M. C.. 2001. Rab27a regulates the peripheral distribution of melanosomes in melanocytes. J. Cell Biol. 152: 795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bahadoran P., Aberdam E., Mantoux F., Buscà R., Bille K., Yalman N., De Saint-Basile G., Casaroli-Marano R., Ortonne J. P., and Ballotti R.. 2001. Rab27a: a key to melanosome transport in human melanocytes. J. Cell Biol. 152: 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X., Rao K., Bowers M. B., Copeland N. G., Jenkins N. A., and Hammer J. A. III.. 2001. Rab27a enables myosin Va-dependent melanosome capture by recruiting the myosin to the organelle. J. Cell Sci. 114: 1091–1100. [DOI] [PubMed] [Google Scholar]
- 39.Fukuda M., and Kuroda T. S.. 2002. Slac2-c (synaptotagmin-like protein homologue lacking C2 domains-c), a novel linker protein that interacts with Rab27, myosin Va/VIIa, and actin. J. Biol. Chem. 277: 43096–43103. [DOI] [PubMed] [Google Scholar]
- 40.Wu X. S., Rao K., Zhang H., Wang F., Sellers J. R., Matesic L. E., Copeland N. G., Jenkins N. A., and Hammer J. A.. 2002. Identification of an organelle receptor for myosin-Va. Nat. Cell Biol. 4: 271–278. [DOI] [PubMed] [Google Scholar]
- 41.Kuroda T. S., and Fukuda M.. 2004. Rab27A-binding protein Slp2-a is required for peripheral melanosome distribution and elongated cell shape in melanocytes. Nat. Cell Biol. 6: 1195–1203. [DOI] [PubMed] [Google Scholar]
- 42.Schechtman D., and Mochly-Rosen D.. 2001. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 20: 6339–6347. [DOI] [PubMed] [Google Scholar]
- 43.Cleland P. J., Appleby G. J., Rattigan S., and Clark M. G.. 1989. Exercise-induced translocation of protein kinase C and production of diacylglycerol and phosphatidic acid in rat skeletal muscle in vivo. Relationship to changes in glucose transport. J. Biol. Chem. 264: 17704–17711. [PubMed] [Google Scholar]
- 44.Otake S., Kobayashi M., Narumi K., Sasaki S., Kikutani Y., Furugen A., Watanabe M., Takahashi N., Ogura J., Yamaguchi H., et al. 2013. Regulation of the expression and activity of glucose and lactic acid metabolism-related genes by protein kinase C in skeletal muscle cells. Biol. Pharm. Bull. 36: 1435–1439. [DOI] [PubMed] [Google Scholar]
- 45.Gao C., Yan X., Wang B., Yu L., Han J., Li D., and Zheng Q.. 2016. Jolkinolide B induces apoptosis and inhibits tumor growth in mouse melanoma B16F10 cells by altering glycolysis. Sci. Rep. 6: 36114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chabrillat M. L., Wilhelm C., Wasmeier C., Sviderskaya E. V., Louvard D., and Coudrier E.. 2005. Rab8 regulates the actin-based movement of melanosomes. Mol. Biol. Cell. 16: 1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson S. M., Yip R., Swing D. A., O’Sullivan T. N., Zhang Y., Novak E. K., Swank R. T., Russell L. B., Copeland N. G., and Jenkins N. A.. 2000. A mutation in Rab27a causes the vesicle transport defects observed in ashen mice. Proc. Natl. Acad. Sci. USA. 97: 7933–7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witt W., Büttner P., Jannasch A., Matschke K., and Waldow T.. 2014. Reversal of myofibroblastic activation by polyunsaturated fatty acids in valvular interstitial cells from aortic valves. Role of RhoA/G-actin/MRTF signalling. J. Mol. Cell. Cardiol. 74: 127–138. [DOI] [PubMed] [Google Scholar]
- 49.Bürgin-Maunder C. S., Brooks P. R., and Russell F. D.. 2013. Omega-3 fatty acids modulate Weibel-Palade body degranulation and actin cytoskeleton rearrangement in PMA-stimulated human umbilical vein endothelial cells. Mar. Drugs. 11: 4435–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu X., Chen X., Wan Q., Zheng Z., and Du Q.. 2016. Nuclear mitotic apparatus (numa) interacts with and regulates astrin at the mitotic spindle. J. Biol. Chem. 291: 20055–20067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haratake K., Sato A., Tsuruta F., and Chiba T.. 2016. KIAA0368-deficiency affects disassembly of 26S proteasome under oxidative stress condition. J. Biochem. 159: 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mühlhäusser P., Müller E. C., Otto A., and Kutay U.. 2001. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2: 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson K., Majd H., Dallabona C., Reinson K., King M. S., Alston C. L., He L., Lodi T., Jones S. A., Fattal-Valevski A., et al. 2016. Recurrent de novo dominant mutations in SLC25A4 cause severe early-onset mitochondrial disease and loss of mitochondrial DNA copy number. Am. J. Hum. Genet. 99: 860–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ron D., Chen C. H., Caldwell J., Jamieson L., Orr E., and Mochly-Rosen D.. 1994. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc. Natl. Acad. Sci. USA. 91: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.López-Bergami P., Habelhah H., Bhoumik A., Zhang W., Wang L. H., and Ronai Z.. 2005. Receptor for RACK1 mediates activation of JNK by protein kinase C. Mol. Cell. 19: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang W., Zong C. S., Hermanto U., Lopez-Bergami P., Ronai Z., and Wang L.. 2006. RACK1 recruits STAT3 specifically to insulin and insulin-like growth factor 1 receptors for activation, which is important for regulating anchorage-independent growth. Mol. Cell. Biol. 26: 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang D., Wang Q., Zhu T., Cao J., Zhang X., Wang J., Wang X., Li Y., Shen B., and Zhang J.. 2013. RACK1 promotes the proliferation of THP1 acute myeloid leukemia cells. Mol. Cell. Biochem. 384: 197–202. [DOI] [PubMed] [Google Scholar]
- 58.Denys A., Hichami A., and Khan N. A.. 2001. Eicosapentaenoic acid and docosahexaenoic acid modulate MAP kinase (ERK1/ERK2) signaling in human T cells. J. Lipid Res. 42: 2015–2020. [PubMed] [Google Scholar]
- 59.Denys A., Hichami A., and Khan N. A.. 2005. n-3 PUFAs modulate T-cell activation via protein kinase C-α and -ε and the NF-κB signaling pathway. J. Lipid Res. 46: 752–758. [DOI] [PubMed] [Google Scholar]
- 60.Fan Y. Y., Ly L. H., Barhoumi R., McMurray D. N., and Chapkin R. S.. 2004. Dietary docosahexaenoic acid suppresses T cell protein kinase C theta lipid raft recruitment and IL-2 production. J. Immunol. 173: 6151–6160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.