Abstract
apoE, a key regulator of plasma lipids, mediates altered functionalities in lipoprotein metabolism and thus affects the risk of coronary artery disease (CAD). The significance of different apoE polymorphisms remains unclear; although the ε4 allele is clearly associated with increased cholesterol levels (which inform CAD risk), direct studies about apoE polymorphisms on CAD risk and development have yielded controversial results. Furthermore, certain species of ceramides—complex lipids abundant in plasma LDL—are markers of increased risk of myocardial infarction and cardiovascular death. Using a high-throughput MS approach, we quantified 30 molecular plasma ceramide species from a cohort of 2,160 apoE-genotyped (rs7412, rs429358) young adults enrolled in the population-based Cardiovascular Risk in Young Finns Study. We then searched this lipidome data set to identify new indications of pathways influenced by apoE polymorphisms and possibly related to CAD risk. This approach revealed a previously unreported association between apoE polymorphism and a consistently documented high-risk CAD marker, Cer(d18:1/16:0). Compared with the apoE ε3/3 reference group, plasma levels of apoE ε4 were elevated and those of apoE ε2 were lowered in all subjects without evidence of apoE-by-sex interactions. apoE associated with seven ceramides that are connected to atherogenically potent macrophages and/or lipoprotein particles; these associations could indicate a plausible linkage between apoE polymorphism and ceramide metabolism, leading to adverse plasma LDL metabolism and atherogenesis. In conclusion, new evidence from plasma ceramides links apoE polymorphism with an increased risk of CAD and extends our understanding of the role of apoE in health and disease.
Keywords: apolipoprotein E, atherosclerosis, dyslipidemias, genes, lipid dysfunction, low-density lipoprotein, metabolism, lipidomics
apoE is a glycoprotein of 299 amino acids with a key role in regulating plasma lipid levels. The three common allelic variants of the apoE gene, ε2, ε3, and ε4, determined by two SNPs at locus 19q13.31, rs429358 and rs7412, code respectively three protein isoforms: E2, E3, and E4. Of the six forming apoE genotypes (ε2/2, ε3/2, ε3/3, ε4/2, ε4/3, and ε4/4), the homozygote parent genotype ε3/3 is the most prevalent, and the protein isoform E3 associates with normal/average plasma lipid levels (1). Despite the early established association of the ε4 allele with elevated serum total cholesterol and LDL cholesterol (LDL-C) (2, 3), as well as the generally recognized causality of high serum total cholesterol and LDL-C with coronary artery disease (CAD) (4), studies investigating the significance of apoE polymorphism on the susceptibility and development of CAD have led to controversial results (5–8). However, it has been shown that genetic components affect atherosclerosis and CAD risk (9, 10). In a Finnish population with higher-than-average incidence of atherosclerosis and nationwide prevalence of the ε4 allele (2), indications that ε4 is linked to the risk of atherosclerosis and CAD have been reported (11). On the other hand, ε2 has been shown to reduce carotid artery intima-media thickness and coronary artery calcification across multiple ethnicities, including Finns (12, 13). Altogether, further research is required to clarify the role of apoE in the metabolic pathways and mechanisms of atherogenesis.
In predicting atherosclerosis and its complications, previous studies have concluded that the traditionally applied standard lipid-panel parameters are inadequate for identifying patients at different risks; therefore, novel, more specific risk markers have been investigated and found with state-of-art lipidomic and metabolomic methods (4). In the current work we concentrate on the effects of apoE polymorphism on plasma ceramides, with an emphasis on predicted CAD risk. Unlike with basic lipids, evidence of the associations of apoE polymorphism with plasma ceramides, or lipidomics in general, is very limited.
To allow for a comprehensive analysis, we quantified 30 molecular plasma ceramide species. Certain ceramides have consistently been reported as high-risk markers of CAD and adverse cardiac outcomes, indicated among CAD patients with an established disease (14–18), as well as at the population level in incident events (19, 20). The aim of our study was to search for novel linkages between apoE polymorphism and risk of CAD by means of plasma ceramides. By implementing a population-based cohort of basically healthy, relatively young adults, we sought to recognize prevailing genotype-related differences prior to adverse manifestations, especially in the markers previously associated with atherogenesis. With our findings we expect to extend our current understanding of the role of apoE in health and disease.
MATERIALS AND METHODS
Study population and data sources
The Cardiovascular Risk in Young Finns Study (YFS) is a Finnish longitudinal general population study on the evolution of cardiovascular risk factors from childhood to adulthood (21). The study began in 1980, when 3,596 children and adolescents aged 3, 6, 9, 12, 15, and 18 years were randomly selected from five university hospital catchment areas in Finland. In 2007, 2,200 participants aged 30–45 years attended the 27-year follow-up. Of these subjects, we included those for whom the apoE genotype data and at least 80% of the lipidomic parameters were available. Therefore, 2,160 participants contributed to the cross-sectional association analyses of apoE genotype and plasma ceramide profile. The YFS was approved by the 1st ethical committee of the Hospital District of Southwest Finland and by local ethical committees (1st Ethical Committee of the Hospital District of Southwest Finland, Regional Ethics Committee of the Expert Responsibility area of Tampere University Hospital, Helsinki University Hospital Ethical Committee of Medicine, The Research Ethics Committee of the Northern Savo Hospital District and Ethics Committee of the Northern Ostrobothnia Hospital District). The study protocol of each study phase corresponded to the proposal by the World Health Organization. All subjects gave written informed consent, and the study was conducted in accordance with the Helsinki declaration. At prior YFS follow-ups, informed consent of every participant under the age of 18 years was obtained from a parent and/or legal guardian.
Clinical and biochemical measurements and their use in statistical standardization
To eliminate the effects of the most probable error sources, a comprehensive set of clinical background information was analyzed as confounding candidates. The effect of BMI was considered by including it in the final regression models as a covariate. Based on questionnaires, daily smoking (yes/no), hormonal birth control of women (yes/no), cholesterol-lowering medication (yes/no), and socioeconomic status were all tested as covariates to have negligible or zero effect on the apoE β-values. The distributions of alcohol consumption (daily portions based on a questionnaire, with one portion equaling 12 g of pure alcohol) and physical activity index [graded 5–15, where higher values indicate greater physical activity; the evaluation method has been described elsewhere (22)] were well comparable in every analyzed apoE subgroup and therefore not confounding, which was also confirmed with covariate analyses. In addition, there were only a few pregnant women, and persons with diagnosed diabetes (evaluated with questionnaires) and their distributions did not differ in the analyzed subgroups. Parameters describing the cardiovascular status, such as systolic blood pressure (defined as an average of three consecutive measurements with a random-zero mercury sphygmomanometer), hypertension (based on a questionnaire for medically diagnosed hypertension), and high-sensitivity C-reactive protein (CRP) (measured with an Olympus AU400 automatic analyzer) might well reflect variations in lipidomics, that is, could be considered more as consequential rather than confounding factors. Nevertheless, differences in these measures between the subgroups were also found to be minor, a significant majority of the study population being basically healthy. Based on the described background analyses and to avoid unnecessary selection error, additional exclusions were not made in the final analyses. The effect of possible information bias may well be neglected in our results when comparing the subgroups to each other, with the bias (if any) being presumably distributed comparably.
Serum concentrations of standard lipids
Levels of total cholesterol, LDL-C, HDL cholesterol (HDL-C), triglycerides, and apoA-I and apoB were measured as background descriptive data with methods described elsewhere (23).
apoE genotyping
apoE alleles (ε2, ε3, ε4) were determined on the basis of SNP rs7412 and rs429358 haplotypes. Genomic DNA was extracted from peripheral blood leukocytes by using the QIAamp DNA Blood Minikit and automated biorobot M48 extraction (Qiagen, Hilden, Germany). Genotyping was performed by using Taqman SNP genotyping assays and the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). As a quality control, water controls, random duplicates, and known control samples were run in parallel with unknown DNA samples.
Plasma lipidomic profiling
Ceramide quantification for the stored plasma samples was performed at Zora Biosciences Oy (Espoo, Finland). Lipid extraction was based on a previously described method (24). In brief, 10 µl 10 mM 2,6-di-tert-butyl-4-methylphenol in methanol was added to 10 μl of sample, followed by 20 µl of internal standards (Avanti Polar Lipids Inc., Alabaster, AL) and 300 µl chloroform-methanol (2:1; v:v) (Sigma-Aldrich GmbH, Steinheim, Germany). Samples were mixed and sonicated in a water bath for 10 min, followed by a 40 min incubation and centrifugation (15 min at 5,700 g). The upper phase was transferred and evaporated under nitrogen. Extracted lipids were resuspended in 100 µl water-saturated butanol and sonicated in a water bath for 5 min. Methanol (100 µl) was added to the samples before the extracts were centrifuged for 5 min at 3,500 g, and finally the supernatants were transferred to the analysis plate for mass spectrometric analysis. Details of mass spectrometric analyses have also been described in detail previously (25). The analyses were performed on a hybrid triple-quadrupole/linear ion trap mass spectrometer (QTRAP 5500; AB Sciex, Concord, Canada) equipped with ultra-high-performance liquid chromatography (Nexera-X2; Shimadzu, Kyoto, Japan). Chromatographic separation of the lipidomic screening platform was performed on an Acquity BEH C18, 2.1 × 50 mm i.d. 1.7 µm column (Waters Corporation, Milford, MA). The data were collected using a scheduled multiple-reaction monitoring algorithm, and the data were processed using Analyst and MultiQuant 3.0 software (AB Sciex).
Statistical methods
All statistical analyses were conducted with R version 3.1.2 (https://www.r-project.org). To facilitate comparisons across all lipidomic measures, association magnitudes (β-values) are reported in scaled SD fractions (units) of normalized, ln-transformed lipid concentrations. In sex-stratified cross-sectional analyses, separate sex-specific scaling was applied to lipidomic measures.
Cross-sectional associations of apoE genotype and serum ceramide profile were analyzed using linear multivariable regression models, with each lipidomic measure as the outcome and apoE genotype as the main explanatory value. Regression models were adjusted for age and BMI. The effect of sex was analyzed in both ways, with models stratified by sex and models with sex as a covariate. The apoE × BMI interaction was tested to have a negligible or zero effect on the apoE β-values and was therefore excluded from the final models. Prior to testing the interaction effects of BMI, measures of BMI were mean-centered. Similarly, apoE × sex and apoE × age interactions were excluded from the final models for not showing an effect within any lipidomic measure (P > 0.05). apoE genotype was included in the model as a categorical variable. First, the overall effect of apoE genotype on the linear fit was F-tested by implementing the Benjamini-Hochberg procedure (26) in false discovery rate (FDR) correction and setting the limit at P < 0.05. After that, apoE genotypes were compared with each other post hoc with t-tests (implementing FDR correction), the values of which were inherited from the linear regression function (lm) in R. To allow for a better comparison of the apoE groups, 95% CIs were also calculated.
Data availability statement
The data sets generated and/or analyzed during this study are not publicly available due to restrictions imposed by Finnish legislation, as the contained information could compromise research participant privacy and consent. However, data sharing is possible upon reasonable request, and all decisions are made by the YFS Publication and Data Sharing Board.
RESULTS
Characteristics of cross-sectional analyses
The frequencies of different apoE genotypes, as well as the summary of the descriptive data in the YFS study population in the 2007 follow-up, are presented in Table 1. The P values across apoE genotypes are given in supplemental Table S1.
TABLE 1.
Frequencies of different apoE genotypes with summary descriptive data in the YFS cohort in the 2007 follow-up
| All | ε2/2 | ε3/2 | ε3/3 | ε4/2 | ε4/3 | ε4/4 | |
| Subjects | 2,160 | 4 (0.2) | 142 (6.6) | 1,241 (57.5) | 41 (1.9) | 651 (30.1) | 81 (3.8) |
| Males | 974 (45.1) | 2 (0.2) | 56 (5.7) | 568 (58.3) | 17 (1.7) | 296 (30.4) | 35 (3.6) |
| Females | 1,186 (54.9) | 2 (0.2) | 86 (7.3) | 673 (56.7) | 24 (2.0) | 355 (29.9) | 46 (3.9) |
| Age (years) | 37.6 (5.0) | 36.0 (7.4) | 36.9 (5.1) | 37.7 (5.0) | 38.6 (4.0) | 37.5 (5.1) | 38.0 (4.8) |
| BMI (kg/m2) | 26.0 (4.8) | 36.8 (17.2) | 25.5 (4.8) | 26.0 (4.6) | 25.9 (4.5) | 26.0 (4.8) | 26.3 (5.3) |
| Daily smokers | 398 (18.4) | 1 (25) | 21 (14.8) | 229 (18.5) | 6 (14.6) | 127 (19.5) | 14 (17.3) |
| Daily alcohol consumption (g ethanol) | 11.3 (17.0) | 4.7 (6.5) | 12.4 (18.2) | 11.9 (18.8) | 9.8 (13.9) | 10.0 (13.1) | 11.5 (13.7) |
| Cholesterol-lowering medication | 45 (2.1) | 0 (0.0) | 2 (1.4) | 23 (1.9) | 2 (4.9) | 15 (2.3) | 3 (3.7) |
| Diabetes mellitus type 2 | 22 (1.0) | 0 (0.0) | 2 (1.4) | 15 (1.2) | 0 (0.0) | 5 (0.8) | 0 (0.0) |
| Hypertension | 123 (5.7) | 2 (50.0) | 4 (2.8) | 67 (5.4) | 1 (2.4) | 45 (6.9) | 4 (4.9) |
| Total cholesterol (mmol/l) | 5.05 (0.90) | 4.12 (0.76) | 4.49 (0.82) | 5.03 (0.89) | 4.68 (0.82) | 5.22 (0.89) | 5.28 (0.84) |
| LDL-C (mmol/l) | 3.15 (0.81) | 1.94 (0.30) | 2.48 (0.65) | 3.12 (0.79) | 2.76 (0.68) | 3.36 (0.80) | 3.30 (0.76) |
| HDL-C (mmol/l) | 1.34 (0.33) | 1.17 (0.17) | 1.41 (0.37) | 1.35 (0.33) | 1.35 (0.37) | 1.31 (0.31) | 1.35 (0.41) |
| Triglycerides (mmol/l) | 1.40 (0.93) | 2.22 (0.87) | 1.45 (1.00) | 1.38 (0.93) | 1.36 (0.97) | 1.39 (0.82) | 1.70 (1.43) |
| apoB (g/l) | 1.02 (0.26) | 0.79 (0.16) | 0.87 (0.26) | 1.01 (0.26) | 0.91 (0.23) | 1.07 (0.26) | 1.09 (0.25) |
| apoA-I (g/L) | 1.60 (0.26) | 1.60 (0.22) | 1.66 (0.30) | 1.61 (0.26) | 1.59 (0.25) | 1.57 (0.24) | 1.60 (0.28) |
Values are means (SDs) or n (%). The P values across apoE genotypes are given in supplemental Table S1.
Cross-sectional associations of the apoE genotype within measured ceramides
Due to small subgroups of apoE ε2/2, ε4/2, and ε4/4 in our population-based cohort (see Table 1), apoE genotypes were clustered into three subgroups for better statistical comparison: the reference group ε3/3 as well as ε2+ (consisting of ε2/2 and ε3/2) and ε4+ (including all ε4 carriers: ε4/2, ε4/3, and ε4/4). The cross-sectional associations of apoE genotypes within 30 quantified plasma ceramides (and three calculated ceramide ratios) for all subjects are illustrated in Fig. 1. The complete statistics of Fig. 1, including exact P values, are given in supplemental Table S2. As illustrated in supplemental Table S3, the exclusion of the ε4/2 subgroup from the analyses did not have a major effect on the results. In addition, the complete results of apoB, LDL-C, and HDL-C concentration-adjusted analyses are provided in supplemental Tables S4–S6, respectively. Furthermore, the results from inflammatory marker-association analyses (high-sensitivity CRP) are shown in supplemental Table S7. There were no apoE × sex interactions found in relation to any studied measure (P > 0.05), justifying the main analyses being performed with all subjects combined.
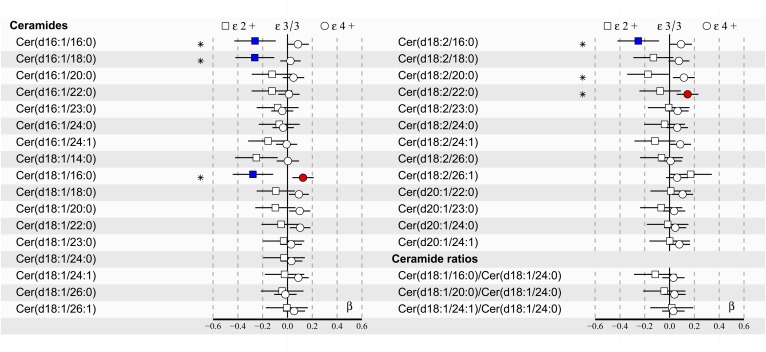
Fig. 1.
apoE effects on 30 plasma ceramides and 3 ceramide ratios in all subjects (n = 2,160) of the YFS cohort in the 2007 follow-up. Regression models were adjusted for age, BMI, and sex. Regression β-coefficients (x-axis) indicate in SD units the change in the lipid measure over apoE genotype subgroups (ε2+, ε3/3, ε4+). The most common ε3/3 subgroup (n = 1,241) is set at the origin (zero SD) and compared post hoc with ε2+ (squares) and ε4+ (circles) subgroups. β-values with 95% CIs are scaled to SD increments from normalized (i.e., ln-transformed) lipid measures. *F-tested effect of apoE with P < 0.05 after FDR correction in the regression model. The post hoc difference with P < 0.05 after FDR correction from ε3/3 is indicated with colors (blue: ε2+; red: ε4+). The apoE ε2+ subgroup (n = 146) includes ε2/2 and ε3/2, and the apoE ε4+ subgroup (n = 773) includes ε4/2, ε4/3, and ε4/4.
Based on multivariate regression analyses of all subjects, six ceramide species (see Fig. 1) showed an association over apoE genotypes with F-tested P < 0.05 after FDR correction. In post hoc comparisons of all subjects against the reference group ε3/3, with FDR corrected P < 0.05, the high-risk CAD ceramide Cer(d18:1/16:0) differed both in apoE ε2+ and ε4+ subgroups, indicating lower levels in the ε2+ and elevated levels in the ε4+ subgroup compared with the reference group ε3/3. In addition, three ceramides, Cer(d16:1/16:0), Cer(d16:1/18:0), and Cer(d18:2/16:0), differed in the ε2+ subgroup, showing lower plasma levels than in the ε3/3 and ε4+ subgroups. Moreover, one ceramide, Cer(d18:2/22:0), had an elevated plasma level in the ε4+ subgroup compared with the most common ε3/3 group.
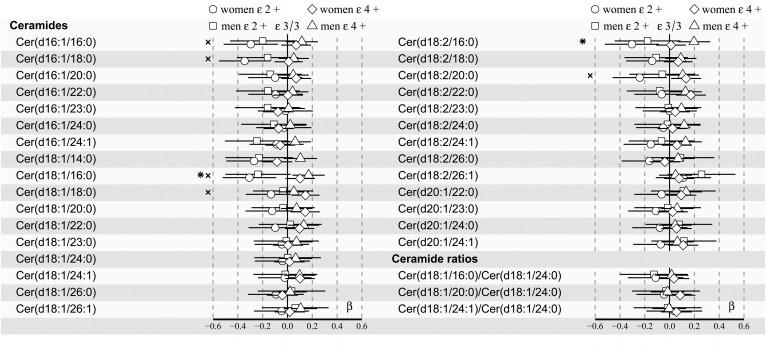
In sex-stratified analyses, the high-risk CAD ceramide Cer(d18:1/16:0) showed an association with apoE genotypes both in women and men (see Fig. 2). In addition, the post hoc analysis compared with ε3/3 showed differences of Cer(d18:1/16:0) in ε2+ women as well as ε4+ men (P = 0.005 and P = 0.01 prior to FDR correction, respectively), as also indicated by the 95% CI in Fig. 2 (see exact FDR-corrected P values in supplemental Table S8). Moreover, the F-tested association (P < 0.05 after FDR correction) of another well-established high-risk CAD ceramide, Cer(d18:1/18:0), was found in women only (Fig. 2), not in all subjects (Fig. 1) or men.
Fig. 2.
apoE effects on 30 plasma ceramides and 3 ceramide ratios in men (n = 974) and women (n = 1,186) of the YFS cohort in the 2007 follow-up. Regression models are adjusted for age and BMI. Regression β-coefficients (x-axis) indicate in SD units the change in the lipid measure over apoE genotype subgroups (ε2+, ε3/3, ε4+). The most common ε3/3 subgroup (n = 568 men; n = 673 women) is set at the origin (zero SD) and compared post hoc with ε2+ (squares: men; circles: women) and ε4+ (triangles: men; diamonds: women) subgroups. β-values with 95% CIs are scaled to SD increments from normalized (i.e., ln-transformed) lipid measures. The F-tested effect of apoE with P < 0.05 after FDR correction in the regression model is indicated with an asterisk for men and an “x” for women. The apoE ε2+ subgroup (n = 58 men; n = 88 women) includes ε2/2 and ε3/2, and the apoE ε4+ subgroup (n = 348 men; n = 425 women) includes ε4/2, ε4/3, and ε4/4.
Cross-sectional associations of the apoE genotype within measured sphingomyelins
The cross-sectional associations of 41 plasma sphingomyelin species over apoE genotypes in all subjects were also analyzed. The complete statistics of these analyses are presented in supplemental Table S9. Twenty-one sphingomyelin species associated with apoE genotypes, with F-tested P < 0.05 after FDR correction; of the species tested, SM(34:1) showed the strongest association. In the post hoc tests compared with ε3/3 and with FDR-corrected P < 0.05, the levels of 14 sphingomyelin species were lower in the ε2 carriers; again, SM(34:1) was the top hit. None of the sphingomyelins quite reached the same statistical limit after FDR correction in the ε4+ subgroup.
DISCUSSION
We investigated the associations of apoE polymorphism with plasma ceramide species with an emphasis on previously reported CAD-risk-related ceramide species. The results of this study show for the first time an association between apoE genotypes and a high-risk CAD ceramide, Cer(d18:1/16:0). Cer(d18:1/16:0) has consistently been connected to an elevated risk of cardiovascular death (16–18), major adverse cardiovascular events (14, 15), and vulnerable plaque characteristics (15) among patients with established CAD, as well as to incident major adverse cardiovascular events among healthy populations (19, 20). Moreover, elevated plasma levels of Cer(d18:1/16:0) have previously been reported among patients with stable CAD compared with healthy controls (27). Reported data on subclinical atherosclerotic associations of Cer(d18:1/16:0), or ceramides in general, is very limited. Nevertheless, Cer(d18:1/16:0) has been linked to an increased risk of carotid artery plaques (28), and SM(34:1), a precursor molecule of Cer(d18:1/16:0), has been linked to increased carotid artery intima-media thickness (29). Our analyses (supplemental Table S7) also indicate an association of Cer(d18:1/16:0) with elevated high-sensitivity CRP levels. In addition to Cer(d18:1/16:0), associations with apoE genotypes were indicated in six other ceramide species as well. These results suggest a novel linkage between apoE polymorphism, lipid metabolism, and CAD, as discussed briefly in the following paragraphs. A more detailed review of previously presented, relevant pathophysiological mechanisms is provided in the supplemental Discussion.
Previous genetic-wide association studies have mapped the apoE locus to the sphingolipid metabolism pathway (30), which is also the major hub for ceramide metabolism (31). Inflammatory conditions such as atherosclerosis may increase the production of ceramides (32). Ceramides are mainly generated through the SMase pathway, which breaks downs sphingomyelin in the cell membrane and releases ceramides (32). Alternatively, ceramides can be generated via a de novo pathway from palmitate, serine, and a covalently linked fatty acid that is added to the molecule by ceramide synthase in all cell types, with the liver being an active site of circulating ceramide production (33, 34). A variety of cell types present in atherosclerotic lesions, including macrophages and endothelial cells, secretes SMase (35). In plasma, ceramides are bound to lipoproteins and are particularly abundant in LDL (36). Moreover, LDL isolated from human plasma has also been reported to possess SMase activity (37) that is capable of catalyzing the formation of ceramides in the adjacent lipoproteins as well (38).
Ceramides have been found to enrich the LDL of atherosclerotic lesions compared with plasma LDL (39). Elevated sphingolipid content (including certain-risk ceramides) of LDL particles (40), as well as macrophage-derived SMase (32, 41), have been shown to promote the aggregation of LDLs and VLDLs. Aggregated lipoprotein particles are prone to accumulate within the arterial intima, increase their uptake by macrophages, and thus promote atherosclerotic plaque formation, as well as future CAD death (32, 40). In addition, macrophages loaded with oxidized LDLs are demonstrated to contain more intracellular ceramides, with Cer(d18:1/16:0) and Cer(d18:1/18:0) being among the reported species (42). On the other hand, the deficiency and/or inhibition of the LDL receptor-degrading PCSK9 enzyme has been found to decrease certain plasma ceramide levels, including Cer(d18:1/16:0) and Cer(d18:1/18:0) (43, 44), as well as lower certain ceramide contents in LDL and VLDL particles, including all of the seven ceramide species we found to be associated with apoE genotypes (43). Supplemental Tables S4 and S5 further indicate a strong connection between these ceramides and apoB-containing LDL, as well as VLDL (and IDL), particles, whereas associations with HDL are weak to none (supplemental Table S6). A consistent association of apoE polymorphism with all of the apoB-containing lipoproteins along with their subfractions have been shown recently in the same YFS cohort (45). Altogether, ceramides would seem to be a plausible and novel common nominator in the abundantly investigated chain of apoE polymorphism, plasma LDL metabolism, and the risk of CAD.
In addition to the SMase pathway, it may be possible that the altered concentrations of certain ceramide species could also result from changes in the de novo pathway due to altered ceramide synthase activities. For example, the de novo pathway has been reported to be influenced by high-fat-diet administration (especially saturated fatty acids) and/or palmitate treatment, which in several reference studies has been shown to increase molecular ceramide contents, most consistently C16:0 and C18:0 species, independent of tissue or cell type (46).
To our knowledge, previous studies of apoE interactions with sphingolipid metabolism in atherogenesis have not investigated the effect of apoE polymorphism. However, apoE has been observed to increase the macrophage uptake of ceramides (47). Moreover, apoE has been reported to prevent SMase-induced hydrophobic interaction and aggregation of lipoproteins (48). In addition, it has been demonstrated that the majority of apoE in atherosclerotic lesions is synthesized locally by lesion macrophages, and the local apoE expression by macrophages is atheroprotective (49). On the other hand, increased cellular ceramides have been reported to reduce macrophage-derived apoE secretion (demonstrated with the E3 phenotype) without increased cell retention of nascent apoE (50). Furthermore, apoE has been demonstrated to prefer binding on oxidized LDL loading-induced, ceramide-enriched microdomain surfaces of human macrophages, which could contribute to atherogenesis (42). apoE has also been shown to prefer binding to ceramides compared with sphingomyelins (48), and SMase-stimulated ceramide formation can therefore induce the membrane remodeling activity of apoE and free cholesterol. If hypothesized, all of these reported findings would be related to the apoE parent isoform E3, and E2 or E4 might, therefore, present altered interactions worth investigating, possibly affecting atherogenic processes via, for example, lipoprotein particle aggregation or adjustment of sphingolipid transfer balance in macrophages. In any case, apoE4 (but not E3) in macrophages has already been demonstrated to enhance atherosclerotic plaque development in an LDL receptor-dependent manner (51).
Our study has several important strengths. Our study cohort has a long and systematic follow-up, offering a representative cross-sectional sample of relatively young Finnish adults. In addition, the comprehensive background information gathered from our cohort allowed for a thorough analysis and consideration of the most expected confounding factors. Furthermore, our analyses reflect genotype-related differences in a basically healthy general population, avoiding confounding/confusing expressions of illnesses expected when using morbid study subjects. In addition, the modern and sophisticated mass spectrometry technology enabled the measurement of circulating ceramides with good accuracy. We also acknowledge certain limitations in our cross-sectional study. Our present scope did not allow direct analyses of subclinical atherosclerosis measures. As previously reported in the Finnish general population (2, 52), subgroups of apoE ε2/2, ε4/2, and ε4/4 were also small in our population-based setting and did not allow for complete analyses of all six genotypes separately. In addition, a completely comprehensive comparison between (sex-stratified) men and women might require an even larger sample size for increased statistical power; however, the absence of apoE × sex interactions was confirmed in the analyses of all subjects. Nevertheless, based on reference studies (e.g., 2, 3), these recognized limitations are not expected to alter the effect directions related to different apoE alleles and are thus less likely to have any major impact on the main discoveries presented and discussed here.
In conclusion, based on quantified molecular ceramide species from plasma, apoE polymorphism is associated with a risk of developing CAD, especially by indicating altered levels of the previously reported clinical high-risk marker Cer(d18:1/16:0) in all subjects. Furthermore, the concentration of every apoE-associating ceramide species in our study has been reported to be reduced in LDL and VLDL particles by PCSK9 inhibition (44), whereas excessive sphingolipid loading has been reported to promote CAD death (40). These findings suggest a plausible linkage between ceramide metabolism, apoE polymorphism, plasma LDL metabolism, and atherogenesis. Changes have already been discovered in a basically healthy general population and extend our understanding of the role of apoE in health and disease. For further work, similar analyses could be performed with patients with incident CAD and established CAD to discover whether the observed associations of apoE polymorphism would change. Associations with subclinical atherosclerosis measures (e.g., carotid artery intima-media thickness or plaques) should also be investigated for further indications. In addition, longitudinal follow-up studies could be performed with older test subjects. Furthermore, the effects of apoE polymorphism on the sphingolipid/ceramide metabolism pathway might be worth investigating, such as whether the macrophage functions or lipoprotein aggregation tendency would become different depending on the apoE isoform. Additional inflammatory marker correlations might also be worth investigating, and the ceramide levels could also be correlated with sphingomyelinase activities.
Supplementary Material
Acknowledgments
The authors thank Irina Lisinen for expert data management. The Cardiovascular Risk in Young Finns Study has been financially supported by Academy of Finland Grants 322098, 286284, 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi); the Social Insurance Institution of Finland; Competitive State Research Financing of the Expert Responsibility area of Kuopio, Tampere and Turku Hospitals Grant X51001; the Juho Vainio Foundation; the Paavo Nurmi Foundation; the Finnish Foundation for Cardiovascular Research; the Finnish Cultural Foundation; the Sigrid Juselius Foundation; the Tampere Tuberculosis Foundation; the Emil Aaltonen Foundation; the Yrjö Jahnsson Foundation; the Signe and Ane Gyllenberg Foundation; the Finnish Diabetes Association; the Pirkanmaa Regional Fund of the Finnish Cultural Foundation; Tampere University Hospital; EU Horizon 2020 Grant 755320 for TAXINOMISIS; and European Research Council Grant 742927 for the MULTIEPIGEN project.
Footnotes
Abbreviations:
- CAD
- coronary artery disease
- CRP
- C-reactive protein
- FDR
- false discovery rate
- HDL-C
- HDL cholesterol
- LDL-C
- LDL cholesterol
- YFS
- Cardiovascular Risk in Young Finns Study
This work was supported by the Juhani Aho Foundation for Medical Research, the Päivikki and Sakari Sohlberg Foundation, the Aarne Koskelo Foundation, and the Faculty of Medicine and Health Technology, Tampere University. R. Laaksonen is an employee and shareholder of Zora Biosciences Oy. M. Hilvo and D. Kauhanen are employees of Zora Biosciences Oy. None of the other authors declare a conflict of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Phillips M. C. 2014. Apolipoprotein E isoforms and lipoprotein metabolism. IUBMB Life. 66: 616–623. [DOI] [PubMed] [Google Scholar]
- 2.Lehtimäki T., Moilanen T., Viikari J., Akerblom H. K., Ehnholm C., Rönnemaa T., Marniemi J., Dahlen G., and Nikkari T.. 1990. Apolipoprotein E phenotypes in Finnish youths: a cross-sectional and 6- year follow-up study. J. Lipid Res. 31: 487–495. [PubMed] [Google Scholar]
- 3.Davignon J., Gregg R. E., and Sing C. F.. 1988. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 8: 1–21. [DOI] [PubMed] [Google Scholar]
- 4.Laaksonen R. 2016. Identifying new risk markers and potential targets for coronary artery disease: the value of the lipidome and metabolome. Cardiovasc. Drugs Ther. 30: 19–32. [DOI] [PubMed] [Google Scholar]
- 5.Zhu H., Xue H., Wang H., Ma Y., Liu J., and Chen Y.. 2016. The association of apolipoprotein E (APOE) gene polymorphisms with atherosclerosis susceptibility: a meta-analysis. Minerva Cardioangiol. 64: 47–54. [PubMed] [Google Scholar]
- 6.Zhao Q. R., Lei Y. Y., Li J., Jiang N., and Shi J. P.. 2017. Association between apolipoprotein E polymorphisms and premature coronary artery disease: a meta-analysis. Clin. Chem. Lab. Med. 55: 284–298. [DOI] [PubMed] [Google Scholar]
- 7.Xu M., Zhao J., Zhang Y., Ma X., Dai Q., Zhi H., Wang B., and Wang L.. 2016. Apolipoprotein E gene variants and risk of coronary heart disease: a meta-analysis. BioMed Res. Int. 2016: 3912175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A., Kumar P., Prasad M., Misra S., Kishor Pandit A., and Chakravarty K.. 2016. Association between apolipoprotein ε4 gene polymorphism and risk of ischemic stroke: a meta-analysis. Ann. Neurosci. 23: 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikpay M., Goel A., Won H., Hall L. M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C. P., Hopewell J. C., et al. . 2015. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47: 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson C. P., Goel A., Butterworth A. S., Kanoni S., Webb T. R., Marouli E., Zeng L., Ntalla I., Lai F. Y., Hopewell J. C., et al. . 2017. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 49: 1385–1391. [DOI] [PubMed] [Google Scholar]
- 11.Ilveskoski E., Perola M., Lehtimäki T., Laippala P., Savolainen V., Pajarinen J., Penttilä A., Lalu K. H., Männikkö A., Liesto K. K., et al. . 1999. Age-dependent association of apolipoprotein E genotype with coronary and aortic atherosclerosis in middle-aged men: an autopsy study. Circulation. 100: 608–613. [DOI] [PubMed] [Google Scholar]
- 12.Ilveskoski E., Loimaala A., Mercuri M. F., Lehtimäki T., Pasanen M., Nenonen A., Oja P., Bond M. G., Koivula T., Karhunen P. J., et al. . 2000. Apolipoprotein E polymorphism and carotid artery intima-media thickness in a random sample of middle-aged men. Atherosclerosis. 153: 147–153. [DOI] [PubMed] [Google Scholar]
- 13.Natarajan P., Bis J. C., Bielak L. F., Cox A. J., Dörr M., Feitosa M. F., Franceschini N., Guo X., Hwang S., Isaacs A., et al. . 2016. Multiethnic exome-wide association study of subclinical atherosclerosis. Circ Cardiovasc Genet. 9: 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anroedh S., Hilvo M., Akkerhuis K. M., Kauhanen D., Koistinen K., Oemrawsingh R., Serruys P., van Geuns R., Boersma E., Laaksonen R., et al. . 2018. Plasma concentrations of molecular lipid species predict long-term clinical outcome in coronary artery disease patients. J. Lipid Res. 59: 1729–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng J. M., Suoniemi M., Kardys I., Vihervaara T., de Boer S. P., Akkerhuis K. M., Sysi-Aho M., Ekroos K., Garcia-Garcia H. M., Oemrawsingh R. M., et al. . 2015. Plasma concentrations of molecular lipid species in relation to coronary plaque characteristics and cardiovascular outcome: results of the ATHEROREMO-IVUS study. Atherosclerosis. 243: 560–566. [DOI] [PubMed] [Google Scholar]
- 16.Laaksonen R., Ekroos K., Sysi-Aho M., Hilvo M., Vihervaara T., Kauhanen D., Suoniemi M., Hurme R., März W., Scharnagl H., et al. . 2016. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur. Heart J. 37: 1967–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarasov K., Ekroos K., Suoniemi M., Kauhanen D., Sylvänne T., Hurme R., Gouni-Berthold I., Berthold H. K., Kleber M. E., Laaksonen R., et al. . 2014. Molecular lipids identify cardiovascular risk and are efficiently lowered by simvastatin and PCSK9 deficiency. J. Clin. Endocrinol. Metab. 99: E45–E52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sigruener A., Kleber M. E., Heimerl S., Liebisch G., Schmitz G., and Maerz W.. 2014. Glycerophospholipid and sphingolipid species and mortality: the Ludwigshafen Risk and Cardiovascular Health (LURIC) study. PLoS One. 9: e85724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havulinna A. S., Sysi-Aho M., Hilvo M., Kauhanen D., Hurme R., Ekroos K., Salomaa V., and Laaksonen R.. 2016. Circulating ceramides predict cardiovascular outcomes in the population-based FINRISK 2002 cohort. Arterioscler. Thromb. Vasc. Biol. 36: 2424–2430. [DOI] [PubMed] [Google Scholar]
- 20.Wang D. D., Toledo E., Hruby A., Rosner B. A., Willett W. C., Sun Q., Razquin C., Zheng Y., Ruiz-Canela M., Guasch-Ferré M., et al. . 2017. Plasma ceramides, Mediterranean diet, and incident cardiovascular disease in the PREDIMED trial (Prevención con dieta Mediterránea). Circulation. 135: 2028–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raitakari O. T., Juonala M., Rönnemaa T., Keltikangas-Järvinen L., Räsänen L., Pietikäinen M., Hutri-Kähönen N., Taittonen L., Jokinen E., Marniemi J., et al. . 2008. Cohort profile: the cardiovascular risk in Young Finns Study. Int. J. Epidemiol. 37: 1220–1226. [DOI] [PubMed] [Google Scholar]
- 22.Telama R., Yang X., Leskinen E., Kankaanpää A., Hirvensalo M., Tammelin T., Viikari J. S. A., and Raitakari O. T.. 2014. Tracking of physical activity from early childhood through youth into adulthood. Med. Sci. Sports Exerc. 46: 955–962. [DOI] [PubMed] [Google Scholar]
- 23.Nuotio J., Pitkänen N., Magnussen C. G., Buscot M., Venäläinen M. S., Elo L. L., Jokinen E., Laitinen T., Taittonen L., Hutri-Kähönen N., et al. . 2017. Prediction of adult dyslipidemia using genetic and childhood clinical risk factors: the Cardiovascular Risk in Young Finns Study. Circ Cardiovasc Genet. 10: e001604. [DOI] [PubMed] [Google Scholar]
- 24.Mamtani M., Kulkarni H., Wong G., Weir J. M., Barlow C. K., Dyer T. D., Almasy L., Mahaney M. C., Comuzzie A. G., Glahn D. C., et al. . 2016. Lipidomic risk score independently and cost-effectively predicts risk of future type 2 diabetes: results from diverse cohorts. Lipids Health Dis. 15: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braicu E. I., Darb-Esfahani S., Schmitt W. D., Koistinen K. M., Heiskanen L., Pöhö P., Budczies J., Kuhberg M., Dietel M., Frezza C., et al. . 2017. High-grade ovarian serous carcinoma patients exhibit profound alterations in lipid metabolism. Oncotarget. 8: 102912–102922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y., Drai D., Elmer G., Kafkafi N., and Golani I.. 2001. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125: 279–284. [DOI] [PubMed] [Google Scholar]
- 27.Meikle P. J., Wong G., Tsorotes D., Barlow C. K., Weir J. M., Christopher M. J., MacIntosh G. L., Goudey B., Stern L., Kowalczyk A., et al. . 2011. Plasma lipidomic analysis of stable and unstable coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 31: 2723–2732. [DOI] [PubMed] [Google Scholar]
- 28.Zhao W., Wang X., Deik A. A., Hanna D. B., Wang T., Haberlen S. A., Shah S. J., Lazar J. M., Hodis H. N., Landay A. L., et al. . 2019. Elevated plasma ceramides are associated with antiretroviral therapy use and progression of carotid artery atherosclerosis in HIV infection. Circulation. 139: 2003–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saulnier-Blache J. S., Wilson R., Klavins K., Graham D., Alesutan I., Kastenmüller G., Wang-Sattler R., Adamski J., Roden M., Rathmann W., et al. . 2018. Ldlr−/− and ApoE−/− mice better mimic the human metabolite signature of increased carotid intima media thickness compared to other animal models of cardiovascular disease. Atherosclerosis. 276: 140–147. [DOI] [PubMed] [Google Scholar]
- 30.Demirkan A., van Duijn C. M., Ugocsai P., Isaacs A., Pramstaller P. P., Liebisch G., Wilson J. F., Johansson Å, Rudan I., Aulchenko Y. S., et al. . 2012. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 8: e1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hannun Y. A., and Obeid L. M.. 2002. The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 277: 25847–25850. [DOI] [PubMed] [Google Scholar]
- 32.Bismuth J., Lin P., Yao Q., and Chen C.. 2008. Ceramide: a common pathway for atherosclerosis? Atherosclerosis. 196: 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen P. J., and Tennagels N.. 2014. On ceramides, other sphingolipids and impaired glucose homeostasis. Mol. Metab. 3: 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wattenberg B. W. 2018. The long and the short of ceramides. J. Biol. Chem. 293: 9922–9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marathe S., Schissel S. L., Yellin M. J., Beatini N., Mintzer R., Williams K. J., and Tabas I.. 1998. Human vascular endothelial cells are a rich and regulatable source of secretory sphingomyelinase. Implications for early atherogenesis and ceramide-mediated cell signaling. J. Biol. Chem. 273: 4081–4088. [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee S. 1998. Sphingolipids in atherosclerosis and vascular biology. Arterioscler. Thromb. Vasc. Biol. 18: 1523–1533. [DOI] [PubMed] [Google Scholar]
- 37.Holopainen J. M., Medina O. P., Metso A. J., and Kinnunen P. K.. 2000. Sphingomyelinase activity associated with human plasma low density lipoprotein. J. Biol. Chem. 275: 16484–16489. [DOI] [PubMed] [Google Scholar]
- 38.Kinnunen P. K. J., and Holopainen J. M.. 2002. Sphingomyelinase activity of LDL: a link between atherosclerosis, ceramide, and apoptosis? Trends Cardiovasc. Med. 12: 37–42. [DOI] [PubMed] [Google Scholar]
- 39.Schissel S. L., Tweedie-Hardman J., Rapp J. H., Graham G., Williams K. J., and Tabas I.. 1996. Rabbit aorta and human atherosclerotic lesions hydrolyze the sphingomyelin of retained low-density lipoprotein. Proposed role for arterial-wall sphingomyelinase in subendothelial retention and aggregation of atherogenic lipoproteins. J. Clin. Invest. 98: 1455–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruuth M., Nguyen S. D., Vihervaara T., Hilvo M., Laajala T. D., Kondadi P. K., Gisterå A., Lähteenmäki H., Kittilä T., Huusko J., et al. . 2018. Susceptibility of low-density lipoprotein particles to aggregate depends on particle lipidome, is modifiable, and associates with future cardiovascular deaths. Eur. Heart J. 39: 2562–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeong T., Schissel S. L., Tabas I., Pownall H. J., Tall A. R., and Jiang X.. 1998. Increased sphingomyelin content of plasma lipoproteins in apolipoprotein E knockout mice reflects combined production and catabolic defects and enhances reactivity with mammalian sphingomyelinase. J. Clin. Invest. 101: 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallner S., Grandl M., Liebisch G., Peer M., Orsó E., Sigrüner A., Sobota A., and Schmitz G.. 2016. oxLDL and eLDL induced membrane microdomains in human macrophages. PLoS One. 11: e0166798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hilvo M., Simolin H., Metso J., Ruuth M., Öörni K., Jauhiainen M., Laaksonen R., and Baruch A.. 2018. PCSK9 inhibition alters the lipidome of plasma and lipoprotein fractions. Atherosclerosis. 269: 159–165. [DOI] [PubMed] [Google Scholar]
- 44.Jänis M. T., Tarasov K., Ta H. X., Suoniemi M., Ekroos K., Hurme R., Lehtimäki T., Päivä H., Kleber M. E., März W., et al. . 2013. Beyond LDL-C lowering: distinct molecular sphingolipids are good indicators of proprotein convertase subtilisin/kexin type 9 (PCSK9) deficiency. Atherosclerosis. 228: 380–385. [DOI] [PubMed] [Google Scholar]
- 45.Karjalainen J. P., Mononen N., Hutri-Kähönen N., Lehtimäki M., Juonala M., Ala-Korpela M., Kähönen M., Raitakari O., and Lehtimäki T.. 2019. The effect of apolipoprotein E polymorphism on serum metabolome - a population-based 10-year follow-up study. Sci. Rep. 9: 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi S., and Snider A. J.. 2015. Sphingolipids in high fat diet and obesity-related diseases. Mediators Inflamm. 2015: 520618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morita S. Y., Kawabe M., Sakurai A., Okuhira K., Vertut-Doï A., Nakano M., and Handa T.. 2004. Ceramide in lipid particles enhances heparan sulfate proteoglycan and low density lipoprotein receptor-related protein-mediated uptake by macrophages. J. Biol. Chem. 279: 24355–24361. [DOI] [PubMed] [Google Scholar]
- 48.Morita S. Y., Nakano M., Sakurai A., Deharu Y., Vertut-Doï A., and Handa T.. 2005. Formation of ceramide-enriched domains in lipid particles enhances the binding of apolipoprotein E. FEBS Lett. 579: 1759–1764. [DOI] [PubMed] [Google Scholar]
- 49.Fazio S., Babaev V. R., Murray A. B., Hasty A. H., Carter K. J., Gleaves L. A., Atkinson J. B., and Linton M. F.. 1997. Increased atherosclerosis in mice reconstituted with apolipoprotein E null macrophages. Proc. Natl. Acad. Sci. USA. 94: 4647–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lucic D., Huang Z. H., Gu D., Subbaiah P. V., and Mazzone T.. 2007. Cellular sphingolipids regulate macrophage apolipoprotein E secretion. Biochemistry. 46: 11196–11204. [DOI] [PubMed] [Google Scholar]
- 51.Altenburg M., Johnson L., Wilder J., and Maeda N.. 2007. Apolipoprotein E4 in macrophages enhances atherogenesis in a low density lipoprotein receptor-dependent manner. J. Biol. Chem. 282: 7817–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehnholm C., Lukka M., Kuusi T., Nikkilä E., and Utermann G.. 1986. Apolipoprotein E polymorphism in the Finnish population: gene frequencies and relation to lipoprotein concentrations. J. Lipid Res. 27: 227–235. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated and/or analyzed during this study are not publicly available due to restrictions imposed by Finnish legislation, as the contained information could compromise research participant privacy and consent. However, data sharing is possible upon reasonable request, and all decisions are made by the YFS Publication and Data Sharing Board.