Abstract
Newcastle disease (ND), an acute and highly contagious avian disease caused by virulent Newcastle disease virus (NDV), often results in severe economic losses worldwide every year. Although it is clear that microRNAs (miRNAs) are implicated in modulating innate immune response to invading microbial pathogens, their role in host defense against NDV infection remains largely unknown. Our prior study indicates that gga-miR-19b-3p is up-regulated in NDV-infected DF-1 cells (a chicken embryo fibroblast cell line) and functions to suppress NDV replication. Here we report that overexpression of gga-miR-19b-3p promoted the production of NDV-induced inflammatory cytokines and suppressed NDV replication, whereas inhibition of endogenous gga-miR-19b-3p expression had an opposite effect. Dual-luciferase and gene expression array analyses revealed that gga-miR-19b-3p directly targets the mRNAs of ring finger protein 11 (RNF11) and zinc-finger protein, MYND-type containing 11 (ZMYND11), two negative regulators of nuclear factor kappa B (NF-κB) signaling, in DF-1 cells. RNF11 and ZMYND11 silencing by small interfering RNA (siRNA) induced NF-κB activity and inflammatory cytokine production, and suppressed NDV replication; whereas ectopic expression of these two proteins exhibited an opposite effect. Our study provides evidence that gga-miR-19b-3p activates NF-κB signaling by targeting RNF11 and ZMYND11, and that enhanced inflammatory cytokine production is likely responsible for the suppression of NDV replication.
Keywords: NDV, gga-miR-19b-3p, DF-1 cells, RNF11, ZMYND11, inflammatory cytokine, NF-κB signaling
Introduction
Newcastle disease virus (NDV) was first described in the early 1900s as the contagious agent of the fatal avian disease known as chicken pest (Dimitrov et al., 2016, 2017). It is classified as an avian paramyxovirus-1 (APMV-1) in the Avulavirus genus of the family Paramyxoviridae (Cheng et al., 2016). The viral particles have a negative-sense, single-stranded RNA genome of about 15 kb that contains six genes in the order of 3′-NP-P-M-F-HN-L-5′, encoding six proteins: nucleoprotein (NP), phosphor protein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN), and large protein (L), respectively. As one of the most devastating pathogens for poultry industry, NDV has a wide host range and can naturally or experimentally infect more than 250 bird species (Ganar et al., 2014). Although all NDV isolates belong to a single serotype, they have a high genetic diversity and evolve rapidly. According to recent epidemiological investigations, sub-genotype VIId has become the dominant genotype in many Asian and African countries since the late 1990s (Qin et al., 2008; Wu et al., 2011; Munir et al., 2012). Currently, vaccination is the main way to prevent and control NDV prevalence. However, due to the wide range of host and high genetic diversity, the failure of Newcastle disease (ND) vaccine immunization and reduction of vaccine efficiency often occur in cultivation industry worldwide (Miller et al., 2009, 2013).
During NDV infection, a variety of the host pattern recognition receptors (PRRs) are activated such as toll-like receptor 3 (TLR-3) (Cheng et al., 2014), retinoic-acidinducible gene I (RIG-I) (Kato et al., 2006; Sun et al., 2013), melanoma differentiation associated gene 5 (MDA5) (Rue et al., 2011), and protein kinase R (PKR) (Liao et al., 2016) by recognizing virus double-stranded RNA. These PRRs subsequently activate downstream transcription factors such as interferon regulatory factor 7 (IRF7) (Wang et al., 2014) and NF-κB (Rajmani et al., 2016), leading to inflammatory cytokine production. For NF-κB activation, PRRs initiate a signaling cascade resulting in the IκB kinase (IKK) phosphorylation, which subsequently phosphorylates and ubiquitinates IκB-α bound to NF-κB in resting cells, and results in IκB-α proteasomal degradation, thereby allowing NF-κB nuclear translocation to transcriptionally activate its gene targets (Woronicz et al., 1997). However, to maintain cellular homeostasis, activation of NF-κB signaling pathway is tightly controlled with multiple tiers of regulation (Ruland, 2011), such as RNF11 (Shembade et al., 2009; Dalal et al., 2012; Pranski et al., 2012) and ZMYND11 (Ikeda et al., 2009, 2010). These two proteins negatively regulate NK-κB activity and greatly impact the replication of certain viruses such as Hendra virus (HeV) (Stewart et al., 2013), Japanese encephalitis virus (JEV) (Ashraf et al., 2016), and Epstein-Barr virus (EBV) (Ikeda et al., 2009).
miRNAs are small non-coding RNAs that are usually composed of 21–23 nucleotides. miRNAs negatively regulate the expression of half protein-coding genes involving in cell growth, proliferation, apoptosis, and signal transduction through post-transcriptional mechanisms (Bartel, 2004). Accumulating evidence suggests that virus infection profoundly impacts miRNA expression, and that altered miRNA expression can enhance or suppress anti-viral responses (Trobaugh and Klimstra, 2017). Recent studies have shown that NDV infection alters the levels of several miRNAs. For example, NDV infection induces hsa-miR-485 expression in human embryonic kidney (HEK) 293T cells, which in turn suppresses antiviral response and enhances NDV replication by targeting RIG-I mRNA (Ingle et al., 2015). In contrast, NDV infection down-regulates gga-miR-203a expression in chicken embryos, overexpression of gga-miR-203a effectively increases NDV replication both in vivo and in vitro via targeting transglutaminase 2 (TGM2) (Jia et al., 2018). Up-regulated gga-miR-375 in NDV-infected DF-1 cells suppresses NDV replication via targeting viral M gene or cellular embryonic lethal, abnormal vision, Drosophila-like RNA binding protein 4 (ELAVL4) gene (Wang et al., 2019). These observations collectively suggest that miRNAs play a critical role in NDV infection and replication. Identification of host-dysregulated miRNAs and understanding of their roles in virus replication may shed light on NDV pathogenesis and provide better strategies for the control of NDV infection.
Our present study focuses on the effect of gga-miR-19b-3p on NDV replication and the underlying molecular mechanisms. Here we report that RNF11 and ZMYND11 are two targets of gga-miR-19b-3p in DF-1 cells, and that up-regulation of gga-miR-19b-3p and the subsequent down-regulation of RNF11 and ZMYND11 expression enhance NDV-induced NF-κB activation, leading to increased inflammatory cytokine production and suppression of virus replication. Data presented herein improve our understanding of the role of host miRNAs in regulating NDV replication.
Materials and Methods
Cells and Viruses
DF-1 cells obtained from ATCC (CRL-12203) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies, United States) supplemented with 10% fetal bovine serum (FBS) (Life Technologies, United States), 100 U/mL penicillin and 100 μg/mL streptomycin at 37°C under 5% CO2 atmosphere. Velogenic genotype VII NDV strain JS5/05 (Accession Number: JN631747) was propagated in chicken embryos and the biological characteristics of the virus were determined previously (Hu et al., 2011). Velogenic genotype IV Herts/33 (Accession Number: AY741404) was obtained from Dr. D. J. Alexander (Animal Health and Veterinary Laboratories Agency, United Kingdom) and avirulent genotype II NDV strain La Sota (Accession Number: AF077761) was isolated and maintained in our laboratory.
MiRNA Mimic and Inhibitor
Gga-miR-19b-3p mimic was chemically modified double-stranded oligonucleotides for overexpression of gga-miR-19b-3p and gga-miR-19b-3p inhibitor was single-stranded oligonucleotides for inhibition of gga-miR-19b-3p. All RNA oligonucleotides were designed and synthesized by GenePharma, China. For miRNA transfection, DF-1 cells were transfected with gga-miR-19b-3p mimic and inhibitor and mimic negative control (mimic-NC) and inhibitor negative control (inh-NC) at a final concentration of 100 nM using EL Transfection Reagent (Transgen Biotech, China) following the manufacturer’s instructions.
Knockdown of RNF11 and ZMYND11
siRNAs (double-stranded RNA oligonucleotides) (Genepharma, China) were used to knockdown the expressions of RNF11 and ZMYND11 in DF-1 cells and the sequences were shown in Table 1. siRNA duplexes were transfected into DF-1 cells at a final concentration of 50 nM using EL Transfection Reagent (Transgen Biotech, China). The interference efficiency was detected by qRT-PCR and Western blot as described below.
TABLE 1.
siRNA sequences.
| siRNAs | Sense (5′-3′) | Anti-sense (5′-3′) |
| SiRNA-ZMYND 11-1 | GCACAUACCUGCGGUUCAUTT | AUGAACCGCAGGUAUGUGCTT |
| SiRNA-ZMYND 11-2 | GCUAUCCUUGUAUACCUAATT | UUAGGUAUACAAGGAUAGCTT |
| SiRNA-ZMYND 11-3 | GCUGUUGAUGUUCCUACUATT | UAGUAGGAACAUCAACAGCTT |
| SiRNA-RNF11-1 | GCCAGAAAUUAGGAAUUAATT | UUAAUUCCUAAUUUCUGGCTT |
| SiRNA-RNF11-2 | CCUACCGAAGGGAGUCUAUTT | AUAGACUCCCUUCGGUAGGTT |
| SiRNA-RNF11-3 | CCACCUGGACUGUAUAGAUTT | AUCUAUACAGUCCAGGUGGTT |
| SiRNA-NC | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
Determination of Viral Titers
DF-1 cells (2 × 105 cells/mL) were seeded on 12-well plates and cultured for 24 h before transfection with indicated RNA oligonucleotides or plasmids. Eighteen hours after transfection, DF-1 cells were infected with JS5/05 strain at a multiplicity of infection (MOI) of 0.1. The culture supernatants were collected and replaced with an equal volume of fresh media at different time points (12, 24, 36, 48, and 60 h) after infection. The viral contents in the supernatants were quantified by 50% tissue culture infective doses (TCID50) on DF-1 cells (Reed and Muench, 1938).
Plasmids Construction
DNA sequences of gga-miR-19b-3p promoter were cloned from genomic DNA of DF-1 cells using specific primers and sub-cloned into KpnI/XhoI restriction sites in the pGL-6 reporter vector (Beyotime, China) to generate the pGL-6-miR-19b-3p reporter vector. Target sequences of gga-miR-19b-3p in RNF11 and ZMYND11 were amplified by polymerase chain reaction (PCR) using a cDNA template synthesized from total RNA of DF-1 cells. Subsequently, both PCR products were sub-cloned into XhoI/NotI restriction sites in the pmiR-RB-reporterTM vector (Ribobio, China), which can express firefly luciferase and renilla luciferase in one plasmid, to generate pmiR-RNF11-3′UTR-wild and pmiR-ZMYND11-3′UTR-wild plasmids. To generate gga-miR-19b-3p target-mutated reporter vector, pmiR-RNF11-3′UTR-mut and pmiR-ZMYND11-3′UTR-mut were achieved by changing the gga-miR-19b-3p binding seed sequences using Fast Mutagenesis System kit (Transgen Biotech, China) according to manufacturer’s instructions. The coding sequences of RNF11 (Accession No. NM001006540) and ZMYND11 (Accession No. XM025148320) were amplified and subsequently cloned into pCMV-blank overexpression vector (Beyotime, China) to generate pCMV-RNF11 and pCMV-ZMYND11 plasmids. All the primers for plasmids construction (shown in Table 2) were synthesized by Sangon Company, China. All the fragments were amplified by PCR using TransStart Fast Pfu Fly DNA Polymerase (Transgen Biotech, China).
TABLE 2.
Primers used for plasmids construction.
| Gene | Forward primers (5′-3′) | Reverse primer (5′-3′) |
| 19b-3p promoter | GGTACCGCCGGGAGG GTCTGCC | CTCGAGTTCAATTTAAC AACAAATTGCAAA |
| RNF11-3′UTR-wild | CTCGAGGCCAGGGTTTCTC CGTGAAC | GCGGCCGCCAAGAGATG CGGTATCCTTTGC |
| ZMYND11-3′UTR-wild | CTCGAGGAGCCATTCCTCCG CTAGAACA | GCGGCCGCCTCGGTCTA AGACCAGCCTCTG |
| RNF11-3′UTR-mut | ATAGTTTTAAGCTTGTGCAC GTAGTTCTTTT | ACGTGCACAAGCTTAAAAC TATTTCTTTGT |
| ZMYND11-3′UTR-mut | TTCCCATTAAGGTGCAGCCT TTAAACA | GCACCTTAATGGGAAGC AAATTCTAAACA |
| RNF11 CDS | ACTAGTATGGGGAACTGCCTC AAGTCC | GGGCCCTCAGTTAGTCTCG TATGATGAAAG |
| ZMYND11 CDS | ACTAGTATGGCACGCTTA ACGAAGAGACG | GGGCCCTCATCTTTTTCTG CGGCAGGTACG |
miRNA Target Prediction and Dual-Luciferase Reporter Gene Assays
Gga-miR-19b-3p targets in host cells were predicted by two online softwares: TargetScan1 and miRDB2. GO Ontology (GO) analysis was conducted to screen the genes on the Database for Annotation, Visualization and Integrated Discovery (DAVID) platform. Luciferase activity was measured using Dual-GLO® Luciferase Assay System Kits (Promega, United States) following the manufacturer’s instructions. For targets verification, DF-1 cells were seeded on 24-well plates with a density of 1 × 105 cells/mL per well. After 24 h, the cells were co-transfected with indicated plasmids and RNA oligonucleotides using EL Transfection Reagent. Forty-eight hours after transfection, luciferase assays were performed with a Fluorescence/Multi-Detection Microplate Reader (Biotek, United States). Renilla luciferase activities were normalized on the basis of firefly luciferase activities. For the detection of promoter activity, DF-1 cells were transfected with pGL-6 or pGL-6-miR-19b-3p plasmid using EL Transfection Reagent. To normalize transfection efficiency, pRL-TK renilla luciferase reporter plasmid (Promega, United States) was also transfected into the cells as an internal control. Then the cells were infected with JS 5/05 at different MOIs at 48h post transfection. Twelve hours post infection (hpi), the dual-luciferase activity was detected and firefly luciferase activities were normalized to renilla luciferase activities. To measure the activity of NF-κB, pNF-κB-luc (Beyotime, China) plasmid and indicated siRNAs or plasmids were transfected into DF-1 cells together with the internal control plasmid pRL-TK (Promega, United States). Eighteen hours after transfection, the cells were infected with JS 5/05 strain at an MOI of 0.1 and dual-luciferase assays were performed as described above at 18 hpi.
miRNA and Total RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
To measure the dynamic expression pattern of gga-miR-19b-3p after NDV infection, DF-1 cells seeded in 12-well plates were infected with JS5/05 strain at different MOIs for 12 h and then miRNAs were extracted using a miRNA Extraction Kit according to manufacturer’s instructions. cDNA synthesis was carried out with a tagged polyT primer according to the protocol of One Step miRNA cDNA Synthesis Kit. Quantification of gga-miR-19b-3p was performed by HG miRNA SYBR Green PCR Kit with a pair of specific primers. Gga-miR-19b-3p expression level was normalized to 5s rRNA levels using the 2–Δ Δ Ct model. All kits used for miRNA extraction, cDNA synthesis and qRT-PCR were purchased from HaiGene Corporation, China.
For the detection of inflammatory cytokines and IκB-α, DF-1 cells were transfected with indicated RNA oligonucleotides for 18 h before infected with JS 5/05 at an MOI of 0.1. Then the total RNAs were extracted from the cells at 18 hpi using TRIzol (TransGen Biotech, China) following the manufacturer’s instructions. The cDNA synthesis and qRT-PCR reaction were performed using TransScript Green One-Step qRT-PCR Super Mix (TransGen Biotech, China) according to the manufacturer’s instructions. To detect RNF11 and ZMYND11 mRNA expression levels, DF-1 cells were transfected with indicated RNA oligonucleotides or infected with JS 5/05 or Herts/33 or La Sota strain at an MOI of 0.1. Eighteen hours post transfection or infection, qRT-PCR were performed as described above using RNF11 and ZMYND11 specific primers. The relative expressions of inflammatory cytokines, IκB-α, RNF11, and ZMYND11 were all normalized with glyceraldehyde phosphate dehydrogenase (GAPDH) and calculated using the 2–ΔΔCT method. All qRT-PCR experiments were preformed in LightCycler 480 (Roche, Switzerland) and primers used were shown in Table 3.
TABLE 3.
Primers used for qRT-PCR.
| Gene | Forward primers (5′-3′) | Reverse primer (5′-3′) |
| gga-miR-19b-3p | AGTGTGCAAATCCATGCAA | GGTCCAGTTTTTTTTTTTTTTTCAGT |
| IFN-β | TGCACAGCATCCTACTG CTCTTG | GTTGGCATCCTGGTGACGAA |
| TNF-α | GGACAGCCTATGCCAACAAG | ACACGACAGCCAAGTCAACG |
| IL-1β | ACCCGCTTCATCTTCTACCG | TCAGCGCCCACTTAGCTT |
| IL-6 | GGCATTCTCATTTCCTTCT | CTGGCTGCTGGACATTTT |
| IL-8 | CCAAGCACACCTCTCTTCCA | GCAAGGTAGGACGCTGGTAA |
| IκB-α | CTGGACTCCATGAAGGA GGAGG | TCGTGAATGATCGCCA AGTGGAG |
| RNF11 | GCCTTGACCCACCATTGTCTTT | CCTTGACCCAAATGCCAGGAC |
| ZMYND11 | GGCGATCCAACATCTTTGGGC | GCCTTTGACCCTTTACACCCCA |
| 5s RNA | AACGCCCGATCTCGTCTGAT | AGTCTCCCATCCAAGTA CTAACCG |
| GAPDH | GAGGGTAGTGAAGGCTGCTG | CACAACACGGTTGCTGTATC |
Enzyme-Linked Immunosorbent Assay (ELISA)
DF-1 cells in 6-well plates were transfected with indicated RNA oligonucleotides or plasmids and were incubated for 18 h and then either left uninfected or infected with JS 5/05 at an MOI of 0.1 for 18 h. The protein levels of IFN-β, TNF-α, IL-1β, IL-6, and IL-8 in cell cultures were determined by ELISA kits according to the manufacturer’s instructions. The IFN-β (cat. no. CH50022), TNF-α (cat. no. CH50032), IL-1β (cat. no. CH50020), IL-6 (cat. no. CH50016), and IL-8 ELISA kits (cat. no. CH50027) were purchased from Bio-Swamp, China.
For the detection of phosphorylation of IκB-α protein, DF-1 cells seeded on 12-well plates were transfection with indicated RNA oligonucleotides or plasmids for 18 h. Then the cells were infected with JS5/05 strain at an MOI of 0.1 for 18 h. The phosphorylation of IκB-α protein was measured by ELISA kit (SbjBio, China, cat. no. SBJ-C134) according to the manufacturer’s instructions.
Western Blot Analysis
To measure the expression levels of RNF11 and ZMYND11 proteins, DF-1 cells were transfected with indicated RNA oligonucleotides or plasmids or infected with different NDV strains at an MOI of 0.1. Eighteen hours post transfection, the cells were lysed and 20 μg protein samples of each group were used for electrophoresis on 12% sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, and resolved proteins were transferred onto polyvinylidene difluoride (PVDF) membranes using electro-transfer (Bio-Rad, United States). After blocking with 5% skimmed milk for 1 h at room temperature, the membranes were incubated with the appropriate primary antibodies, including rabbit polyclonal anti-RNF11 (LSBio, United States), rabbit polyclonal anti-ZMYND11 (LSBio, United States) and mouse monoclonal anti-β-actin (Transgene, China) followed by incubation with goat anti-rabbit and goat anti-mouse HRP-conjugated secondary antibodies (Beyotime, China). The proteins on the membranes were detected using an ECLTM detection system (Bio-Rad, United States).
To measure the translocation of NF-κB from the cytoplasm to the nucleus, DF-1 cells transfected with indicated RNA oligonucleotides or plasmids were infected with JS 5/05 at an MOI of 0.1 for 18 h. Then cytoplasmic and nuclear proteins of the cells were prepared using nuclear and cytoplasmic protein extraction kit (Beyotime, China) according to the manufacturer’s instructions. Western blot assays were performed as described above using NF-kB p65 rabbit monoclonal antibody (Beyotime, China). Monoclonal rabbit anti-tubulin β and anti-histone H3 antibody (Beyotime, China) were used as control of cytoplasmic and nuclear proteins, respectively. The relative protein quantification was analyzed by Image J software (Bethesda, United States).
Statistical Analysis
All data are the averages of triplicates and are representative of results from at least three independent experiments. The statistical analysis were analyzed using one-way analysis of variance (ANOVA) following Bonferroni’s multiple comparison tests for TCID50, miRNA, Western blot and mRNA detections and two-way ANOVA using a Bonferroni’s multiple comparison tests for viral growth curve using GraphPad Prism 5 (GraphPad Software, San Diego, United States). P < 0.05 was considered as a significant difference.
Results
Gga-miR-19b-3p Is Upregulated During NDV Infection and Inhibits NDV Replication in DF-1 Cells
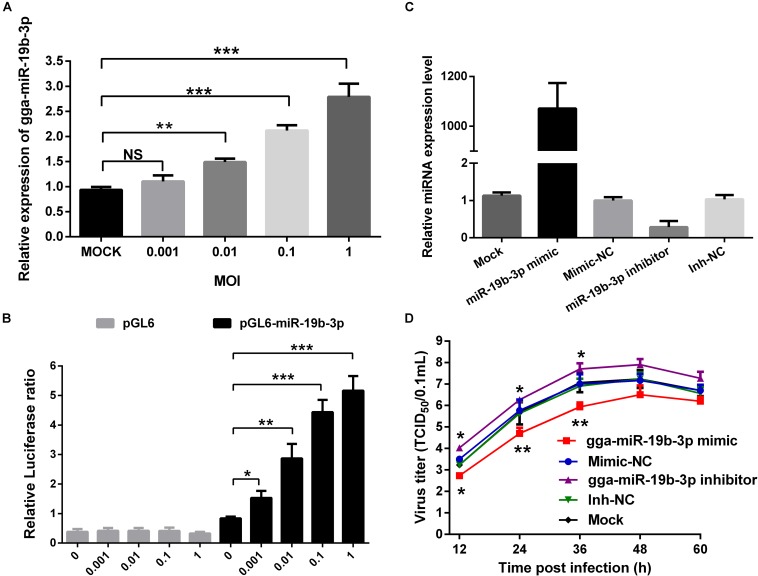
We first conducted qRT-PCR to determine if NDV infection regulated gga-miR-19b-3p expression in DF-1 cells. As shown in Figure 1A, gga-miR-19b-3p was up-regulated in NDV-infected DF-1 cells in a dose-dependent manner. Consistently, NDV infection also increased the gga-miR-19b-3p promoter-driven luciferase reporter gene expression in a dose-dependent manner (Figure 1B).
FIGURE 1.
Newcastle disease virus-induced gga-miR-19b-3p inhibits NDV replication in DF-1 cells. (A) DF-1 cells were infected with JS 5/05 strain at indicated MOIs for 12 h, and then the expression of gga-miR-19b-3p was detected by qRT-PCR. (B) The pGL-6-miR-19b-3p or pGL-6 plasmid was transfected into DF-1 cells together with pRL-TK renilla luciferase reporter plasmid as an internal control. Forty-eight hours post transfection, the cells were infected with JS 5/05 strain at different MOIs for 12 h, and then the dual-luciferase activity was detected and firefly luciferase activities were normalized to firefly renilla luciferase activities. (C,D) DF-1 cells were transfected with gga-miR-19b-3p mimic or inhibitor or mimic-NC or inh-NC at a final concentration of 100 nM or left untreated. The gga-miR-19b-3p expression was measured by qRT-PCR at 18 h post transfection (C). Eighteen hours after transfection, DF-1 cells were infected with JS5/05 strain at an MOI of 0.1. The viral road in the supernatants collected at different time points were quantified by TCID50 on DF-1 cells (D). Results are representative of three independent experiments and presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We next evaluated the effect of gga-miR-19b-3p on NDV replication. DF-1 cells were transfected with gga-miR-19b-3p mimic or its inhibitor, followed by infection with NDV (JS 5/05 strain, 0.1 MOI). As shown in Figure 1C, gga-miR-19b-3p mimic increased gga-miR-19b-3p expression about 1000-fold in DF-1 cells, whereas gga-miR-19b-3p antagonist decreased its expression by >70%. Overexpression of gga-miR-19b-3p mimic significantly suppressed NDV replication, whereas gga-miR-19b-3p inhibitor increased NDV replication (Figure 1D).
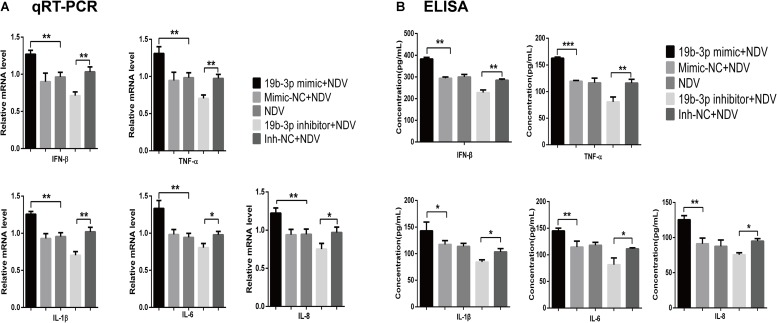
Gga-miR-19b-3p Positively Regulates NDV-Induced Expression of Inflammatory Cytokines
We next determined if gga-miR-19b-3p affected virus replication by regulating the inflammatory cytokine production. As shown in Figures 2A,B, gga-miR-19b-3p mimic significantly increased both mRNA and protein levels of IFN-β, TNF-α, IL-1β, IL-6, and IL-8 in NDV-infected DF-1 cells. In contrast, gga-miR-19b-3p inhibitor significantly decreased the levels of IFN-β, TNF-α, IL-1β, IL-6, and IL-8 mRNA and protein.
FIGURE 2.
Gga-miR-19b-3p promotes NDV-induced expression of inflammatory cytokines. DF-1 cells were transfected with indicated RNA oligonucleotides at a final concentration of 100 nM for 18 h or left untreated before infected with JS 5/05 at an MOI of 0.1. The expression levels of IFN-β, TNF-α, IL-1β, IL-6, and IL-8 were measured by qRT-PCR (A) and ELISA (B) at 18 hpi. Results are representative of three independent experiments and presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Gga-miR-19b-3p Directly Targets RNF11 and ZMYND11 in DF-1 Cells
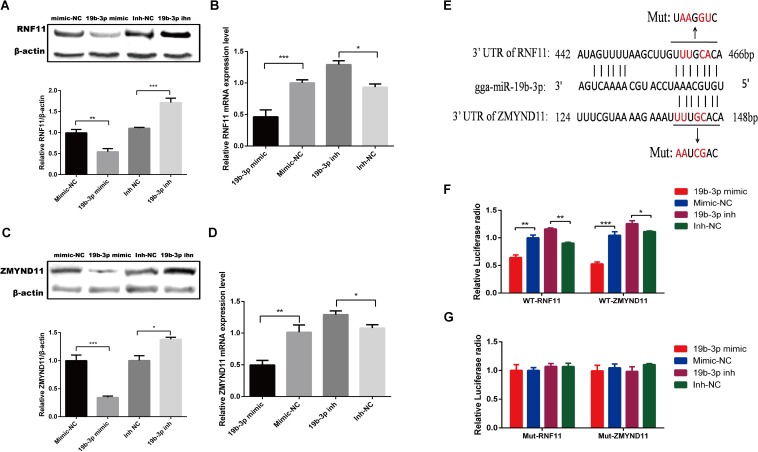
We then investigated the targets of gga-miR-19b-3p that might be involved in regulating NDV-induced inflammatory cytokine production. A total of 571 and 547 targets were predicted by TargetScan and miRDB, respectively. Two hundred and twenty-one common target genes were used to do GO analysis on the DAVID platform. After enrichment by GO analysis, we found that two potential target genes RNF11 and ZMYND11were clustered on immune system process. Moreover, both of them were reported as negatively regulators of NK-κB activity. Finally, RNF11 and ZMYND11 were screened for validation given the GO analysis results and previous study.
To examine whether gga-miR-19b-3p directly targets RNF11 and ZMYND11, We first detected whether gga-miR-19b-3p regulates the RNF11 and ZMYND11 expression. qRT-PCR and Western blot revealed that transfection of DF-1 cells with gga-miR-19b-3p mimic led to decreased RNF11 (Figures 3A,B) and ZMYND11 (Figures 3C,D) expression at both protein and mRNA levels, compared to that transfected with mimic-NC or inh-NC. We then constructed luciferase reporter plasmids containing the wild-type or mutant 3′-UTR of the RNF11 and ZMYND11 genes. Predicted target sequences for gga-miR-19b-3p in the 3′-UTR of RNF11 and ZMYND11 are illustrated in Figure 3E. DF-1 cells were co-transfected with indicated RNA oligonucleotides plus wild-type or mutated-type luciferase reporter plasmids. Forty eight hours after transfection, dual-luciferase reporter assays were performed. gga-miR-19b-3p mimic significantly decreased the ratio of Renilla/Firefly in pmiR-RNF11-3′UTR-wild and pmiR-ZMYND11-3′UTR-wild transfected DF-1 cells, but not in pmiR-RNF11-3′UTR-mut and pmiR-ZMYND11-3′UTR-mut cells (Figures 3F,G). As expected, inhibition of gga-miR-19b-3p exhibited a significant promotion of the luciferase radio in pmiR-RNF11-3′UTR-wild and pmiR-ZMYND11-3′UTR-wild DF-1 cells, whereas no changes in pmiR-RNF11-3′UTR-mut and pmiR-ZMYND11-3′UTR-mut cells (Figures 3F,G). Combined with these results, we demonstrated that RNF11 and ZMYND11 are directly targeted by gga-miR-19b-3p in DF-1 cells.
FIGURE 3.
Gga-miR-19b-3p directly targets RNF11 and ZMYND11 in DF-1 cells. DF-1 cells were transfected with indicated RNA oligonucleotides at a final concentration of 100 nM for 18 h. Then the expression of RNF11 and ZMYND11 were detected by Western blot (A,C) and qRT-PCR (B,D) assays. The relative levels of RNF11 and ZMYND11 proteins were calculated as follows: band density of RNF11 or ZMYND11/band density of β-actin in the same sample and showed in the below of (A,C). Diagram of the predicted target sites for gga-miR-19b-3p in RNF11 and ZMYND11 were shown in (E). DF-1 cells were co-transfected with indicated RNA oligonucleotides and wild type (F) or mutant luciferase reporter gene vectors (G) before luciferase reporter gene assay was performed. The relative level of renilla luciferase activities was calculated normalized on the basis of firefly luciferase activities. Data are representative of three independent experiments and presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
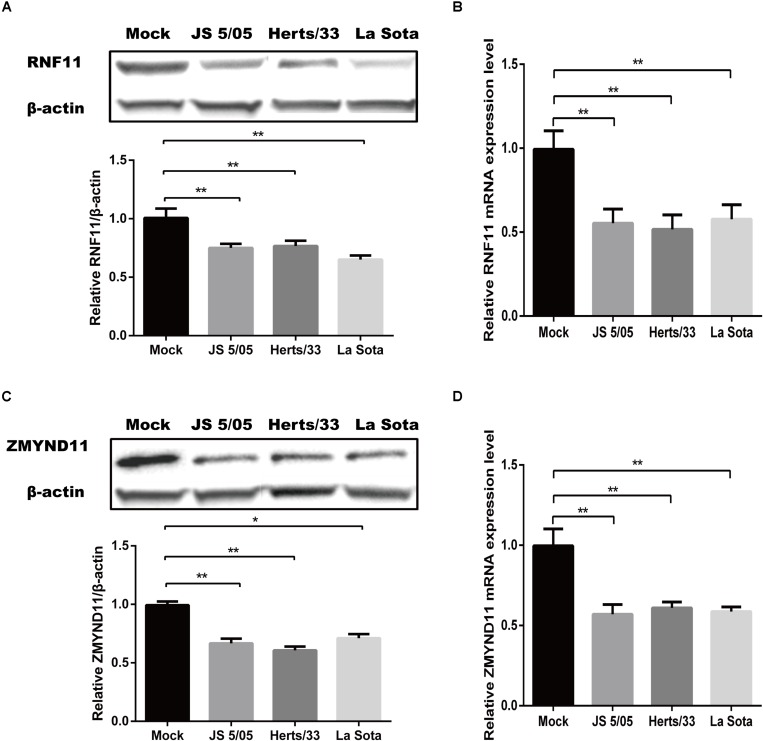
NDV Infection Inhibits the Expressions of RNF11 and ZMYND11
Given that gga-miR-19b-3p directly targeted RNF11 and ZMYND11 mRNAs, we tested if NDV infection indeed affected the expression of these two proteins. qRT-PCR and Western blot revealed that the protein and mRNA levels of RNF11 (Figures 4A,B) and ZMYND11 (Figures 4C,D) were significantly down-regulated in DF-1 cells infected with three different NDV strains, including JS 5/05, Herts/33 and La Sota.
FIGURE 4.
Newcastle disease virus infection inhibits the expressions of RNF11 and ZMYND11. DF-1 cells were infected with different NDV strains (JS 5/05, Herts/33, and La Sota) at an MOI of 0.1 for 18 h. The expressions of RNF11 and ZMYND11 were detected by Western blot (A,C) and qRT-PCR (B,D) assays. The relative levels of RNF11 and ZMYND11 proteins were calculated as described above and showed in the below of (A,C). Data are representative of three independent experiments and presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01.
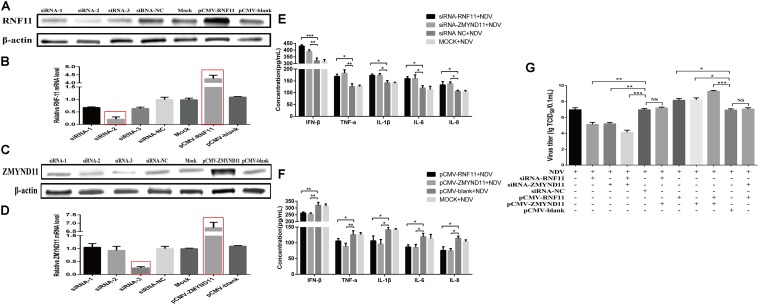
RNF11 and ZMYND11 Modulate NDV Replication via Suppressing NDV-Induced Inflammatory Cytokines
We next determined if RNF11 and ZMYND11 indeed regulated inflammatory production and virus replication. Firstly, we detected the interference and overexpression efficiency of siRNAs and overexpression plasmids by Western blot and qRT-PCR assays. As shown in Figures 5A–D, the levels of RNF11 or ZMYND11 were increased by 4.5 or 7-fold in DF-1 cells transfected with pCMV-RNF11 and pCMV-ZMYND11, respectively. siRNA-RNF11-2 (hereafter called siRNA-RNF11) and siRNA-ZMYND11-3 (hereafter called siRNA-ZMYND11) down-regulated the expression of ZMYND and RNF11 by about 70%. RNF11 or ZMYND11 knockdown in DF-1 cells markedly enhanced the expressions of NDV-induced inflammatory cytokines and suppressed NDV replication, whereas RNF11 or ZMYND11 overexpression blocked NDV-induced inflammatory cytokine expression and promoted NDV replication (Figures 5E–G). Simultaneous knockdown or overexpression of these two proteins showed synergistic effects on viral replication. However, overexpression of ZMYND11 and knockdown of RNF-11, or overexpression of RNF-11 and knockdown of ZMYND11 showed no effect on viral replication.
FIGURE 5.
RNF11 and ZMYND11 inhibit NDV-induced inflammatory cytokines and promote NDV replication in DF-1 cells. DF-1 cells were transfected with indicated siRNAs or siRNA-NC or overexpression plasmids or pCMV-blank. The efficiencies of interference and overexpression in transfected DF-1 cells were measured by Western blot (A,C) and qRT-PCR (B,D) at 18 h post transfection. DF-1 cells transfected with indicated siRNAs or siRNA-NC or overexpression plasmids or pCMV-blank or left untreated were infected with JS 5/05 at an MOI of 0.1 for 18 h. The expression levels of IFN-β, TNF-α, IL-1β, IL-6, and IL-8 were measured by ELISA assay (E,F). DF-1 cells transfected with indicated siRNAs or siRNA-NC or overexpression plasmids or pCMV-blank were infected with JS 5/05 at an MOI of 0.1. After 36 h, viral titers in infected DF-1 cells were measured by TCID50 as described above (G). Data are representative of three independent experiments and presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
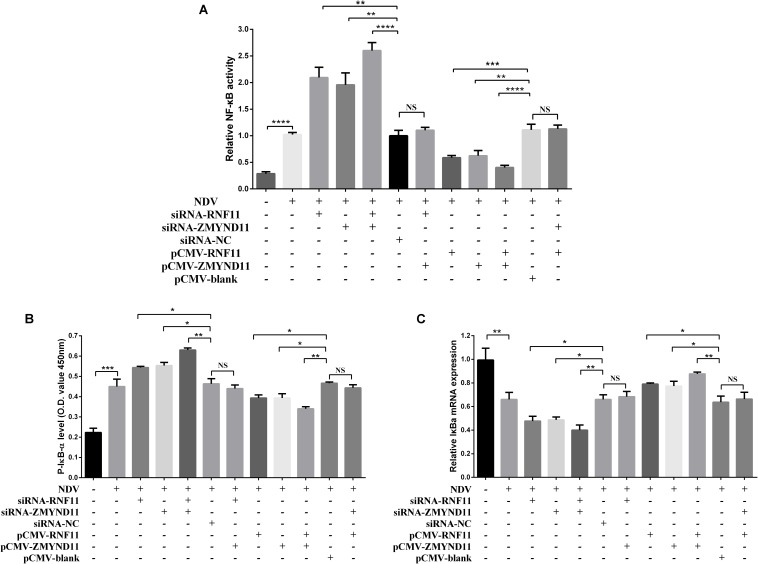
RNF11 and ZMYND11 Suppress NF-κB Activity
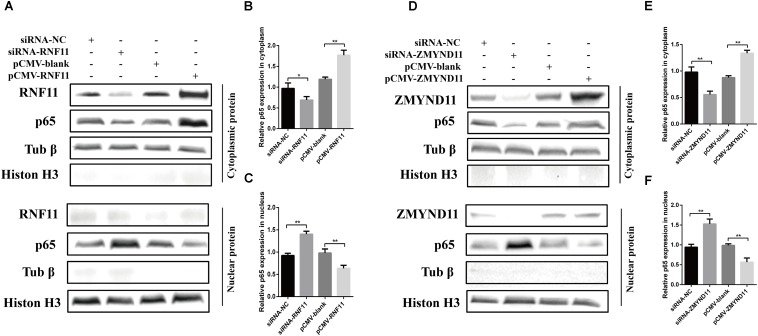
Previous studies demonstrated that RNF11 and ZMYND11 negatively regulate the NF-κB signaling pathway in human cell lines (Ikeda et al., 2010; Dalal et al., 2012). In order to explore the mechanism by which RNF11 and ZMYND11 suppress NDV-induced inflammatory cytokines, we detected NF-κB activity in siRNAs or overexpression plasmids transfected DF-1 cells after NDV infection using a pNF-κB-luc plasmid which containing multiple binding motifs of NF-κB (GGGAATTTCC). We found that knockdown of RNF11 or ZMYND11 in DF-1 cells significantly increased NF-κB activity, while overexpression of them had completely opposite effects (Figure 6A). For further understand the underlying mechanism of the suppression effect on NF-κB activity by RNF11 and ZMYND11, we then detected the changes of p-IκB-α, IκB-α, and p65 nuclear translocation in siRNAs or overexpression plasmids transfected DF-1 cells after NDV infection. As shown in Figures 6B,C, knockdown of RNF11 or ZMYND11 in DF-1 cells significantly promoted p-IκB-α expression and inhibited IκB-α expression, while overexpression of them had completely opposite effects. Similarly, simultaneous knockdown or overexpression of these two proteins also showed synergistic effects on NF-κB activity, p-IκB-α and IκB-α expression. However, overexpression of ZMYND11 and knockdown of RNF-11, or overexpression of RNF-11 and knockdown of ZMYND11 showed no effect. In expectation, transfection of siRNA-RNF11 or siRNA-ZMYND11 increased the translocation of p65, an active subunit of NF-κB with transcriptional activity (Vermeulen et al., 2002), from the cytoplasm to the nucleus (Figures 7A,D). In contrast, treatment of cells with pCMV-RNF11 or pCMV-ZMYND11 significantly inhibited the nuclear translocation of p65 in NDV-infected DF-1 cells (Figures 7A,D). The relative p65 expression in cytoplasm or nucleus was calculated normalized on the basis of tubulin β or histone H3, respectively (Figures 7B,C,E,F).
FIGURE 6.
RNF11 and ZMYND11 suppress NF-κB activity. pNF-κB-luc plasmid and indicated siRNAs or siRNA-NC or overexpression plasmids or pCMV-blank were co-transfected into DF-1 cells together with the internal control plasmid pRL-TK. Eighteen hours after transfection, the cells were infected with JS 5/05 strain at an MOI of 0.1 or left uninfected, dual-luciferase (A), ELISA (B), and qRT-PCR (C) assays were performed as described above at 18 hpi. NF-κB activities were indicated by the ratio of firefly luciferase activities to renilla luciferase activities. The relative expression of IκB-α was normalized with GAPDH and calculated using the 2–Δ Δ CT method. Data are representative of three independent experiments and presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ****p < 0.0001.
FIGURE 7.
RNF11 and ZMYND11 inhibit the nucleus translocation of p65. DF-1 cells were transfected with indicated siRNAs or siRNA-NC or overexpression plasmids or pCMV-blank for 18 h. Then JS 5/05 strain were used to infect the cells at an MOI of 0.1. Eighteen hours post infection, the cytoplasmic and nuclear proteins of the cells were prepared as described above. Western blot was used to detect the translocation of p65 from cytoplasm to the nucleus (A,D). The relative p65 expression in cytoplasm (B,E) or nucleus (C,F) was analyzed by Image J software and calculated normalized on the basis of tubulin β or histone H3, respectively. Data are representative of three independent experiments and presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01.
Discussion
miRNAs are post-transcriptional regulators that mediate translational repression and/or mRNA degradation by complementary binding to the 3′ UTRs in target mRNAs. Mounting evidence suggest that cellular miRNAs can coordinate host defense against viral infections by direct binding to virus genome or through virus-mediated changes in the host transcriptome (Trobaugh and Klimstra, 2017). For some paramyxoviruses, such as respiratory syncytial virus (RSV) (Bakre et al., 2012; Thornburg et al., 2012), HeV (Stewart et al., 2013), and Nipah virus (NiV) (Foo et al., 2016), critical roles of cellular miRNAs in host defense against virus replication have already been well characterized. Even so, little is known about miRNAs roles in NDV replication until now.
Our previous deep sequencing results showed that the expression of gga-miR-19b-3p is significantly increased in NDV-infected DF-1 cells. Further study suggests that this up-regulated miRNAs might function as an anti-viral factor and play a role in NDV-infected DF-1 cells. Our present study focuses on the mechanism by which gga-miR-19b-3p inhibits NDV replication. We found that NDV infection increased the expression and promoter activity of gga-miR-19b-3p. miR-19b-3p has been shown to play an important role in regulating the replication of other viruses. For example, up-regulated miR-19b enhances CD8+ T cell function via targeting the phosphatase and tensin homolog (PTEN) and inhibits viral production of human immunodeficiency virus (HIV) in T cells (Yin et al., 2018). Our study showed that transfection of gga-miR-19b-3p mimic significantly promoted NDV-induced inflammatory cytokines and inhibited NDV replication in DF-1 cells, suggesting that gga-miR-19b-3p might play an anti-viral role during NDV infection.
Like other paramyxoviruses, NDV infection can be sensed by host innate immune system via PRRs in the cytosol. Activated PRRs lead to activation of the innate immune response and the production of inflammatory cytokines that establishes an antiviral state (Schoggins and Rice, 2011). NF-κB plays a crucial role in the production of inflammatory cytokines (i.e., TNF-α, IL-6), apoptotic factors, and oxidative stress (Deng et al., 2018). A growing body of research suggests that miRNAs can regulate viral replication via influencing NF-κB activity. For example, has-miR-215 directly targets the tripartite motif 22 (TRIM22) to block the NF-κB signaling pathway, and exerts a positively regulatory role on (hepatitis C virus) HCV replication (Tian and He, 2018). Up-regulated miR-221-5p inhibits porcine epidemic diarrhea virus (PEDV) replication by targeting the 3′ UTR of the viral genome and activating the NF-κB-signaling pathway (Zheng et al., 2018). In addition, during HIV infection, the down-regulation of miR-16 results in the activation of the NF-κB signaling pathway, thus inhibiting HIV replication (Li et al., 2010).
RNF11 is an evolutionarily conserved 154 amino acid protein that acts as a negative regulator of NF-κB signaling pathway through its associations with the A20 ubiquitin-editing protein complex (Pranski et al., 2012). Earlier studies have shown that ZMYND11 or BS69, a multidomain cellular protein, can directly interact with tumor necrosis factor receptor associated factor 3 (TRAF3) to negatively regulated Epstein–Barr virus latent membrane protein 1-mediated NF-κB activation and then enhanced IL-6 expression (Ikeda et al., 2010). Moreover, recent studies demonstrated that ZMYND11 could negatively regulate the NF-κB signaling pathway in mycoplasma gallisepticum-infected chicken embryos and DF-1 cells (Hu et al., 2016). In our present study, we found that RNF11 and ZMYND11were directly targets of gga-miR-19b-3p in DF-1 cells. Since gga-miR-19b-3p enhanced NDV-induced inflammatory response, it was imperative to evaluate effect of RNF11 and ZMYND11 on inflammatory cytokines production in NDV-infected DF-1 cells. We found that knockdown of RNF11 and ZMYND11 increased NDV-induced inflammatory cytokines and inhibited NDV replication, similar to the effect of gga-miR-19b-3p. However, overexpression of RNF11 and ZMYND11 showed an opposite effect. These results suggested that gga-miR-19b-3p enhances NDV-induced inflammatory response via targeting RNF11 and ZMYND11. Moreover, different virulence NDV strains infection resulted in a similar suppression of these two proteins expression, indicated that a virulence-independent regulation of RNF11 and ZMYND11 during NDV infection in vitro.
Further research of mechanism by which RNF11 and ZMYND11 influence NDV-induced inflammatory response, we found that silencing of these two proteins by siRNAs promotes NDV-induced NF-κB activity indicated that these two proteins negatively regulate NF-κB signaling pathway in avian cells. It has been well established that phosphorylation and subsequent proteasomal degradation of IκB-α and translocation of NF-κB from the cytoplasm to the nucleus are key determinants of NF-κB activation (Woronicz et al., 1997; Zhu et al., 2015). Therefore, we also determined the effect of RNF11 and ZMYND11 on the expression of p-IκB-α and IκB-α and nucleus translocation of NF-κB after NDV infection in DF-1 cells. We found that treatment of DF-1 cells with siRNA-RNF11 and siRNA-ZMYND11 enhanced the expression of p-IκB-α and nucleus translocation of p65 and inhibited IκB-α expression, whereas its overexpression prevented the expression of p-IκB-α and p65 nuclear translocation and increased IκB-α expression. All these results strongly indicate that RNF11 and ZMYND11can negatively regulate NF-κB activity in DF-1 cells after NDV infection and their down-regulation after NDV infection may be responsible for gga-miR-19b-3p stimulative role on NDV-induced inflammatory cytokines expression. However, the molecular mechanism by which RNF11 and ZMYND11 negatively regulate NF-κB activity in DF-1 cells remains to be further investigated.
To further elucidate the association of the regulated roles of RNF11 and ZMYND11, we knocked down or overexpressed of both proteins concurrently or knocked down one of them while overexpressed the other one before NDV infection. We found that simultaneous knockdown or overexpression of both proteins showed synergistic effects on viral replication and NF-κB activity. However, overexpression of ZMYND11 and knockdown of RNF-11, or overexpression of RNF-11 and knockdown of ZMYND11 showed no effect on viral replication and NF-κB activity. However, we believe that these results do not indicate that RNF11 and ZMYND11 have similar abilities in regulating NDV replication and NF-κB activity. Since the interference efficiency of both genes is approximately 30%, while the overexpression efficiency of both genes is approximately five to sevenfold. Moreover, the expression levels of endogenous RNF11 and ZMYND11 are also different in DF-1 cells. Therefore, further precise experiments are needed to confirm which protein plays more critical role on NDV replication.
In summary, our results indicate that NDV-induced gga-miR-19b-3p inhibits the expressions of RNF11 and ZMYND11 by directly binding them, thereby enhancing NF-κB activity and increasing inflammatory cytokines production, which finally leads to the inhibition of NDV replication in DF-1 cells. However, the study of gga-miR-19b-3p in vivo is more complicated than that in DF-1 cells, further studies are needed to elucidate the role of gga-miR-19b-3p in host response to NDV infection.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files.
Author Contributions
YC was responsible for experiment design, data analysis, and writing the manuscript. WL, HX, YD, HC, TZ, XLL, TL, LG, SZ, and YP were responsible for performing the experiments. JL, JH, XW, MG, and XWL were responsible for suggestion during the experiments performance. ZH, SH, and XFL were responsible for revising the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by grants from the National Key Research and Development Program of China (2017YFD0500101), the National Key Technology R&D Program of China (2015BAD12B03). The Earmarked Fund For China Agriculture Research System (CARS-40), the National Key Research and Development Program of China (2017YFD500101 and 2017YFD0500702), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
- Ashraf U., Zhu B., Ye J., Wan S., Nie Y., Chen Z., et al. (2016). MicroRNA-19b-3p modulates japanese encephalitis virus-mediated inflammation via targeting RNF11. J. Virol. 90 4780–4795. 10.1128/jvi.02586-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakre A., Mitchell P., Coleman J. K., Jones L. P., Saavedra G., Teng M., et al. (2012). Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J. Gen. Virol. 93(Pt 11), 2346–2356. 10.1099/vir.0.044255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Cheng J., Sun Y., Zhang X., Zhang F., Zhang S., Yu S., et al. (2014). Toll-like receptor 3 inhibits newcastle disease virus replication through activation of pro-inflammatory cytokines and the type-1 interferon pathway. Arch. Virol. 159 2937–2948. 10.1007/s00705-014-2148-6 [DOI] [PubMed] [Google Scholar]
- Cheng X., Wang W., Xu Q., Harper J., Carroll D., Galinski M. S., et al. (2016). Genetic modification of oncolytic newcastle disease virus for cancer therapy. J. Virol. 90 5343–5352. 10.1128/jvi.00136-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal N. V., Pranski E. L., Tansey M. G., Lah J. J., Levey A. I., Betarbet R. S. (2012). RNF11 modulates microglia activation through NF-kappaB signalling cascade. Neurosci. Lett. 528 174–179. 10.1016/j.neulet.2012.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Zeng Q., Wang M., Cheng A., Jia R., Chen S., et al. (2018). Suppression of NF-kappaB activity: a viral immune evasion mechanism. Viruses 10:409. 10.3390/v10080409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov K. M., Afonso C. L., Yu Q., Miller P. J. (2017). Newcastle disease vaccines-A solved problem or a continuous challenge? Vet. Microbiol. 206 126–136. 10.1016/j.vetmic.2016.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov K. M., Ramey A. M., Qiu X., Bahl J., Afonso C. L. (2016). Temporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus). Infect. Genet. Evol. 39 22–34. 10.1016/j.meegid.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Foo C. H., Rootes C. L., Cowley K., Marsh G. A., Gould C. M., Deffrasnes C., et al. (2016). Dual microRNA Screens reveal that the immune-responsive miR-181 Promotes henipavirus entry and cell-cell fusion. PLoS Pathog. 12:e1005974. 10.1371/journal.ppat.1005974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganar K., Das M., Sinha S., Kumar S. (2014). Newcastle disease virus: current status and our understanding. Virus Res. 184 71–81. 10.1016/j.virusres.2014.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Zhao Y., Wang Z., Hou Y., Bi D., Sun J., et al. (2016). Chicken gga-miR-19a targets ZMYND11 and plays an important role in host defense against Mycoplasma gallisepticum (HS Strain) infection. Front. Cell Infect. Microbiol. 6:102. 10.3389/fcimb.2016.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Hu S., Meng C., Wang X., Zhu J., Liu X. (2011). Generation of a genotype VII Newcastle disease virus vaccine candidate with high yield in embryonated chicken eggs. Avian. Dis. 55 391–397. 10.1637/9633-122410-Reg.1 [DOI] [PubMed] [Google Scholar]
- Ikeda O., Miyasaka Y., Yoshida R., Mizushima A., Oritani K., Sekine Y., et al. (2010). BS69 cooperates with TRAF3 in the regulation of Epstein-Barr virus-derived LMP1/CTAR1-induced NF-kappaB activation. FEBS Lett. 584 865–872. 10.1016/j.febslet.2010.01.060 [DOI] [PubMed] [Google Scholar]
- Ikeda O., Sekine Y., Mizushima A., Oritani K., Yasui T., Fujimuro M., et al. (2009). BS69 negatively regulates the canonical NF-kappaB activation induced by Epstein-Barr virus-derived LMP1. FEBS Lett. 583 1567–1574. 10.1016/j.febslet.2009.04.022 [DOI] [PubMed] [Google Scholar]
- Ingle H., Kumar S., Raut A. A., Mishra A., Kulkarni D. D., Kameyama T., et al. (2015). The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 8:ra126. 10.1126/scisignal.aab3183 [DOI] [PubMed] [Google Scholar]
- Jia Y. Q., Wang X. L., Wang X. W., Yan C. Q., Lv C. J., Li X. Q., et al. (2018). Common microRNA(-)mRNA interactions in different newcastle disease virus-infected chicken embryonic visceral tissues. Int. J. Mol. Sci. 19:E1291. 10.3390/ijms19051291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Takeuchi O., Akira S. (2006). [Cell type specific involvement of RIG-I in antiviral responses]. Nihon Rinsho 64 1244–1247. [PubMed] [Google Scholar]
- Li T., Morgan M. J., Choksi S., Zhang Y., Kim Y. S., Liu Z. G. (2010). MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat. Immunol. 11 799–805. 10.1038/ni.1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Gu F., Mao X., Niu Q., Wang H., Sun Y., et al. (2016). Regulation of de novo translation of host cells by manipulation of PERK/PKR and GADD34-PP1 activity during Newcastle disease virus infection. J. Gen. Virol. 97 867–879. 10.1099/jgv.0.000426 [DOI] [PubMed] [Google Scholar]
- Miller P. J., Afonso C. L., El Attrache J., Dorsey K. M., Courtney S. C., Guo Z., et al. (2013). Effects of Newcastle disease virus vaccine antibodies on the shedding and transmission of challenge viruses. Dev. Comp. Immunol. 41 505–513. 10.1016/j.dci.2013.06.007 [DOI] [PubMed] [Google Scholar]
- Miller P. J., Estevez C., Yu Q., Suarez D. L., King D. J. (2009). Comparison of viral shedding following vaccination with inactivated and live Newcastle disease vaccines formulated with wild-type and recombinant viruses. Avian. Dis. 53 39–49. 10.1637/8407-071208-Reg.1 [DOI] [PubMed] [Google Scholar]
- Munir M., Abbas M., Khan M. T., Zohari S., Berg M. (2012). Genomic and biological characterization of a velogenic Newcastle disease virus isolated from a healthy backyard poultry flock in 2010. Virol. J. 9:46. 10.1186/1743-422x-9-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pranski E. L., Dalal N. V., Herskowitz J. H., Orr A. L., Roesch L. A., Fritz J. J., et al. (2012). Neuronal RING finger protein 11 (RNF11) regulates canonical NF-kappaB signaling. J. Neuroinflamm. 9:67. 10.1186/1742-2094-9-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z. M., Tan L. T., Xu H. Y., Ma B. C., Wang Y. L., Yuan X. Y., et al. (2008). Pathotypical characterization and molecular epidemiology of Newcastle disease virus isolates from different hosts in China from 1996 to 2005. J. Clin. Microbiol. 46 601–611. 10.1128/jcm.01356-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajmani R. S., Gupta S. K., Singh P. K., Gandham R. K., Sahoo A. P., Chaturvedi U., et al. (2016). HN protein of Newcastle disease virus sensitizes HeLa cells to TNF-alpha-induced apoptosis by downregulating NF-kappaB expression. Arch. Virol. 161 2395–2405. 10.1007/s00705-016-2923-7 [DOI] [PubMed] [Google Scholar]
- Reed L. J., Muench H. (1938). A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 27 493–497. 10.1093/oxfordjournals.aje.a118408 15955576 [DOI] [Google Scholar]
- Rue C. A., Susta L., Cornax I., Brown C. C., Kapczynski D. R., Suarez D. L., et al. (2011). Virulent Newcastle disease virus elicits a strong innate immune response in chickens. J. Gen. Virol. 92 931–939. 10.1099/vir.0.025486-0 [DOI] [PubMed] [Google Scholar]
- Ruland J. (2011). Return to homeostasis: downregulation of NF-kappaB responses. Nat. Immunol. 12 709–714. 10.1038/ni.2055 [DOI] [PubMed] [Google Scholar]
- Schoggins J. W., Rice C. M. (2011). Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1 519–525. 10.1016/j.coviro.2011.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N., Parvatiyar K., Harhaj N. S., Harhaj E. W. (2009). The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J. 28 513–522. 10.1038/emboj.2008.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. R., Marsh G. A., Jenkins K. A., Gantier M. P., Tizard M. L., Middleton D., et al. (2013). Promotion of Hendra virus replication by microRNA 146a. J. Virol. 87 3782–3791. 10.1128/jvi.01342-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Ding N., Ding S. S., Yu S., Meng C., Chen H., et al. (2013). Goose RIG-I functions in innate immunity against Newcastle disease virus infections. Mol. Immunol. 53 321–327. 10.1016/j.molimm.2012.08.022 [DOI] [PubMed] [Google Scholar]
- Thornburg N. J., Hayward S. L., Crowe J. E., Jr. (2012). Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-kappaB. mBio 3:e00220-12. 10.1128/mBio.00220-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., He Z. (2018). miR-215 enhances HCV replication by targeting TRIM22 and inactivating NF-kappaB signaling. Yonsei Med. J. 59 511–518. 10.3349/ymj.2018.59.4.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobaugh D. W., Klimstra W. B. (2017). MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 23 80–93. 10.1016/j.molmed.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L., De Wilde G., Notebaert S., Vanden Berghe W., Haegeman G. (2002). Regulation of the transcriptional activity of the nuclear factor-kappaB p65 subunit. Biochem. Pharmacol. 64 963–970. 10.1016/s0006-2952(02)01161-9 [DOI] [PubMed] [Google Scholar]
- Wang X., Jia Y., Wang X., Wang C., Lv C., Li X., et al. (2019). MiR-375 has contrasting effects on Newcastle disease virus growth depending on the target gene. Int. J. Biol. Sci. 15 44–57. 10.7150/ijbs.25106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ning Z., Sun M., Gao S., Kang Y., Xie P., et al. (2014). Interferon regulatory factor 7- (IRF7-) mediated immune response affects Newcastle disease virus replication in chicken embryo fibroblasts. Acta Vet. Hung. 62 500–511. 10.1556/AVet.2014.023 [DOI] [PubMed] [Google Scholar]
- Woronicz J. D., Gao X., Cao Z., Rothe M., Goeddel D. V. (1997). IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science 278 866–869. 10.1126/science.278.5339.866 [DOI] [PubMed] [Google Scholar]
- Wu S., Wang W., Yao C., Wang X., Hu S., Cao J., et al. (2011). Genetic diversity of Newcastle disease viruses isolated from domestic poultry species in Eastern China during 2005-2008. Arch. Virol. 156 253–261. 10.1007/s00705-010-0851-5 [DOI] [PubMed] [Google Scholar]
- Yin L. B., Song C. B., Zheng J. F., Fu Y. J., Qian S., Jiang Y. J., et al. (2018). Elevated expression of miR-19b enhances CD8(+) T Cell function by targeting PTEN in HIV infected long term non-progressors with sustained viral suppression. Front. Immunol. 9:3140. 10.3389/fimmu.2018.03140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Xu L., Liu Y., Li C., Zhang L., Wang T., et al. (2018). MicroRNA-221-5p inhibits porcine epidemic diarrhea virus replication by targeting genomic viral RNA and activating the NF-kappaB pathway. Int. J. Mol. Sci. 19:E3381. 10.3390/ijms19113381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Ye J., Nie Y., Ashraf U., Zohaib A., Duan X., et al. (2015). MicroRNA-15b modulates Japanese encephalitis virus-mediated inflammation via targeting RNF125. J. Immunol. 195 2251–2262. 10.4049/jimmunol.1500370 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the manuscript and/or the supplementary files.