Abstract
Background/Aims
Matrix-assisted laser desorption/ionization time-of-f light mass spectrometry (MALDI-TOF MS) is a new diagnostic tool for microorganism identification. The clinical usefulness of this approach has not been widely examined in Korea. This retrospective pre–post-intervention quasi-experimental study examined the effect of MALDI-TOF MS on patients with multidrug-resistant (MDR) bacteremia in the intensive care unit (ICU).
Methods
All consecutive patients with MDR bacteremia in the ICU of a tertiary care hospital between March 2011 and February 2013 and between March 2014 and February 2016 were enrolled. MALDI-TOF MS was introduced between these periods. In the pre-intervention and intervention groups, microorganisms were identified by conventional means and by MALDI-TOF MS, respectively. The groups were compared in terms of time from venipuncture to microorganism identification and antimicrobial susceptibility test results.
Results
In total, 187 patients (mean age, 61.0 years; 56.7% male) were enrolled. Of these, 97 and 90 were in the pre-intervention and intervention groups, respectively. The intervention group had a significantly shorter time from venipuncture to microorganism identification and antimicrobial susceptibility test results (82.5 ± 21.6 hours vs. 92.3 ± 40.4 hours, p = 0.038). The antibiotics were adjusted in 52 patients (26 each in the pre-intervention and intervention groups) based on these results. These groups did not differ in terms of time from venipuncture to antibiotic adjustment, and multivariate regression analysis showed that MALDI-TOF MS–based microorganism identification was not associated with 28-day mortality.
Conclusions
Our study showed that MALDI-TOF MS accelerated microorganism identification in patients with MDR bacteremia, but did not inf luence 28-day mortality.
Keywords: Drug resistance; Identification; Intensive care units; Spectrometry, mass, matrix-assisted laser desorption-ionization
INTRODUCTION
One of the most important determinants influencing the outcomes of patients in the intensive care unit (ICU), especially those with serious infectious diseases (IDs), is antibiotic resistance. This influence is due largely to the tendency for prevalence of endemic antibiotic resistance in ICU patient populations because of the extensive use of empirical broad-spectrum antimicrobial agents in the ICU, the weakened immune systems of patients, and the frequent use of invasive devices (e.g., venous cannulae) that impact anatomical integrity and protective barriers [1-3]. These high rates of antibiotic resistance in the ICU, which are growing, are associated with high morbidity and mortality rates and costly hospitalization stays [4,5]. Therefore, to improve patient outcomes and lower healthcare costs, a major objective of many hospitals is to reduce the prevalence of multidrug-resistant (MDR) bacterial infections, especially in the ICU [4,6]. One approach to do so is to reduce the duration of empirical broad-spectrum antibiotic therapy by using novel diagnostic tools that are now widely available. These tools can rapidly identify the microorganisms with which patients are infected, thereby promoting the more judicious use of antibiotics [7-10].
One of these new tools used for rapid pathogen identification is matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). It is increasingly being used in hospitals worldwide to identify microorganisms, and several recent studies have shown that it improves the clinical outcomes of patients, especially those with bacteremia [11-14]. However, in Korea, critical care delivery systems are underdeveloped compared with those in Western countries [15-17]. As a result, newly developed rapid pathogen identification tools such as MALDI-TOF MS have not been fully introduced into clinical practice. Therefore, whether MALDI-TOF MS can also be useful for Korean patients with MDR bacteremia remains unclear.
To address this issue, the present study evaluated the clinical efficacy of rapid microorganism identification using MALDI-TOF MS in Korean patients with MDR bacteremia in the ICU by comparing two patient groups: one from before the introduction and one from after the introduction of MALDI-TOF MS in our institution.
METHODS
Study design, ethics, and subject selection
This retrospective quasi-experimental pre–post-intervention study included all consecutive patients who had positive blood culture tests in the ICUs of Pusan National University Hospital, Busan, Korea, between March 2011 and February 2013 and between March 2014 and February 2016. MALDI-TOF MS was introduced in the hospital between these two study periods (i.e., between March 2013 and February 2014). Thus, the patients were divided into the pre-intervention group, in which microorganisms were identified by conventional methods between March 2011 and February 2013, and the intervention group, in which microorganisms were identified by MALDI-TOF MS between March 2014 and February 2016 (all patients in the intervention group underwent microorganism indentification using MALDI-TOF MS only). This study was approved by the Institutional Review Board of Pusan National University Hospital (C-1703-004-052). The need for informed consent from patients was waived due to the observational and retrospective nature of this study. Our study had no impact on the treatment of the patients included.
Pusan National University Hospital is a university-affiliated tertiary referral academic care hospital with 1,100 beds. It has six functionally separate ICUs with 85 beds (medical, 12 beds; surgical, 10 beds; cardio-stroke, 14 beds; neurosurgical, 13 beds; emergency, 20 beds; and trauma, 16 beds). All ICUs have full cardiovascular and close airway monitoring equipment. Each has one full-time ICU specialist. All patients were managed according to therapeutic recommendations based on sepsis survival guidelines and a lung-protective ventilator strategy [18,19].
Patients in the ICU were included in the study if they were ≥ 18 years of age, had positive blood culture tests during their ICU care, and were shown by conventional methods or MALDI-TOF MS to have one of the following six MDR bacteria: methicillin-resistant Staphylococcus aureus (MRSA), extended-spectrum β-lactamase (ESBL)-producing Gram-negative bacteria (Escherichia coli and Klebsiella pneumoniae), carbapenem-resistant Gram-negative rods (Acinetobacter baumannii [CRAB] and Pseudomonas aeruginosa [CRPA]), and vancomycin-resistant Enterococcus faecium (VRE). Patients with polymicrobial bacteremia or a subsequent bacteremia episode after the first (index) bacteremia event were excluded in accordance with the study design of Lockwood et al. [13].
All investigators contributed to the design of the study and confirmed that the study objectives and procedures were honestly disclosed. Between September 2016 and February 2017, the electronic medical records of all enrolled patients were reviewed retrospectively. The relevant medical, laboratory, and radiological data were extracted and used to complete a case report form for each patient, and the data were analyzed.
Microbiology workflow
Blood samples were sent to the microbiology laboratory and were used to inoculate plates containing the appropriate solid agar media as soon as they were received. At the same time, all specimens were Gram stained. Positive blood cultures from the pre-intervention group underwent microorganism identification using conventional and automated biochemical methods (VITEK-2, bioMérieux, Marcy l’Etoile, France). Positive blood cultures from the intervention group underwent microorganism identification using MALDI-TOF MS (Bruker Daltonics, Bremen, Germany). In both groups, antimicrobial susceptibility testing was performed using the VITEK-2 and E-test methods (bioMérieux). The laboratory identified microorganisms and performed antimicrobial susceptibility testing once a day at 9:00 AM for both groups.
Data collection
The following demographic and clinical data were gathered from the medical records of each patient: age, sex, comorbidities before ICU admission, and lengths of ICU stay, hospital stay, and ventilator care. The severity of illness was measured using Acute Physiology and Chronic Health Evaluation (APACHE) II scoring, and accompanying organ failure was measured using Sequential Organ Failure Assessment (SOFA) scoring [20,21]. The APACHE II and SOFA scores were calculated based on laboratory and clinical data recorded on the day of venipuncture for blood culture.
Microbiological information, including microorganism identification and antimicrobial susceptibility test results, were also extracted from the medical records. The time from venipuncture to microorganism identification and antibiotic susceptibility testing was noted. A subset of patients underwent antibiotic adjustment based on the antibiotic susceptibility test results. In these cases, we also determined the time from venipuncture to antibiotic adjustment. Whether an ID specialist was consulted was also evaluated. The antibiotics used for more than 3 days in the 3 months before blood was drawn in the ICU for microorganism culture were also recorded for all patients.
The primary source of infection at the time of ICU admission was also recorded. Moreover, whether the patient needed hemodialysis (defined as use of any form of renal replacement therapy), neuromuscular blocking agents, vasopressors, and/or ventilator care on the day of venipuncture for microorganism culture was documented. Survivors were defined as patients who survived for 28 days after blood was drawn.
Statistical analysis
Continuous variables are expressed as mean ± standard deviation and categorical variable are expressed as number (%). Continuous variables in the pre-intervention and intervention groups were compared using Student t test, and categorical variables were compared using the chi-square test or Fisher exact test (for small samples). Receiver operating characteristic curves were constructed to determine cut-off values. Identification of an optimal cut-off value was based on the maximum Youden’s index [22]. To determine whether MALDI-TOF MS–based microorganism identification served as an independent prognostic factor for 28-day mortality, logistic regression analyses were performed with the inclusion of variables with p < 0.05 in univariate analyses. All statistical analyses were performed using the SPSS version 22.0 (IBM Co., Armonk, NY, USA) and MedCalc Statistical Software version 18.5 (MedCalc Software, Ostend, Belgium). Two-tailed p values < 0.05 were considered to indicate statistical significance.
RESULTS
Baseline characteristics
In total, 1,283 patients in the ICU had positive blood cultures during the two study periods. After applying the eligibility criteria, 187 patients were enrolled in the study (Fig. 1). Blood for microorganism culture was collected 28.2 ± 117.3 days after admission to the ICU. Respiratory infections were the most common source of bacteremia (39.0%), and MRSA was the most commonly identified microorganism (39.6%). During the study period, the total mortality rate in the 28 days after venipuncture for microorganism culture was 38.0%. Table 1 shows the clinical characteristics of all patients, survivors, and non-survivors. Compared with survivors, non-survivors were more likely to require ventilator care, vasopressors, renal replacement therapy, and neuromuscular blocking agents on the day of blood collection for microorganism culture.
Figure 1.
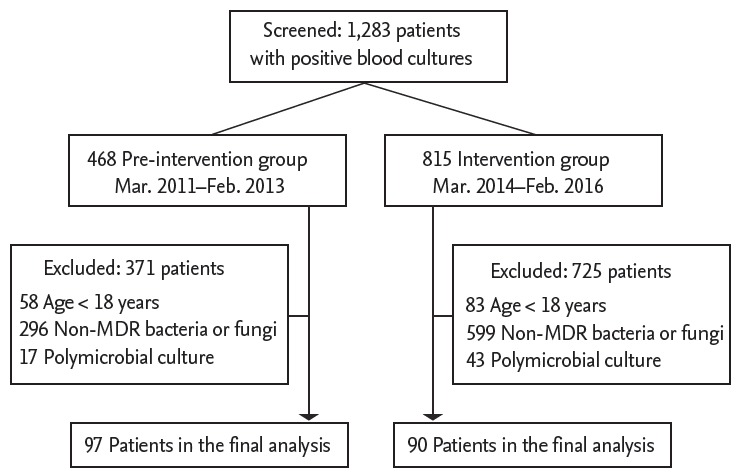
Flowchart of the study participants. MDR, multidrug-resistant.
Table 1.
Demographic and clinical characteristics of all enrolled patients and the survivors and non-survivors for 28 days after blood was drawn
| Characteristic | Total (n = 187) | Survivorsa (n = 116) | Non-survivors (n = 71) | p value |
|---|---|---|---|---|
| Age, yr | 61.0 ± 16.6 | 59.5 ± 15.8 | 63.3 ± 17.6 | 0.130 |
| Male sex | 106 (56.7) | 70 (60.3) | 36 (50.7) | 0.197 |
| Comorbidities | ||||
| Diabetes mellitus | 53 (28.3) | 35 (30.2) | 18 (25.4) | 0.478 |
| Cardiovascular disease | 46 (24.6) | 28 (24.1) | 18 (25.4) | 0.852 |
| Hemato-oncological disease | 42 (22.5) | 12 (10.3) | 30 (42.3) | < 0.001 |
| Cerebrovascular disease | 33 (17.6) | 24 (20.7) | 9 (12.7) | 0.163 |
| Neuromuscular disease | 20 (10.7) | 12 (10.3) | 8 (11.3) | 0.843 |
| Chronic renal disease | 18 (9.6) | 13 (11.2) | 5 (7.0) | 0.349 |
| Chronic liver disease | 15 (8.0) | 10 (8.6) | 5 (7.0) | 0.700 |
| Chronic lung disease | 15 (8.0) | 8 (6.9) | 7 (9.9) | 0.469 |
| Rheumatological disease | 8 (4.3) | 4 (3.4) | 4 (5.6) | 0.481 |
| Biliary disease | 5 (2.7) | 3 (2.6) | 2 (2.8) | > 0.999 |
| Gastrointestinal disease | 5 (2.7) | 3 (2.6) | 2 (2.8) | > 0.999 |
| APACHE II scoreb | 17.5 ± 6.7 | 15.7 ± 6.3 | 20.4 ± 6.3 | < 0.001 |
| SOFA scoreb | 7.5 ± 4.1 | 6.0 ± 3.3 | 9.8 ± 4.3 | < 0.001 |
| Requirement for vasopressorb | 75 (40.1) | 31 (26.7) | 44 (62.0) | < 0.001 |
| Requirement for RRTb | 38 (20.3) | 15 (12.9) | 23 (32.4) | 0.001 |
| Requirement for NMBb | 27 (14.4) | 11 (9.5) | 16 (22.5) | 0.014 |
| Requirement for MVb | 124 (66.3) | 68 (58.6) | 56 (78.9) | 0.004 |
| Duration of MV, day | 30.9 ± 55.4 | 34.0 ± 53.7 | 27.1 ± 57.7 | 0.494 |
| Hospital LOS, day | 71.6 ± 77.6 | 92.2 ± 82.3 | 37.4 ± 54.5 | < 0.001 |
| ICU LOS, day | 35.3 ± 50.0 | 40.1 ± 48.5 | 27.2 ± 51.7 | 0.087 |
| Infection source | ||||
| Respiratory | 73 (39.0) | 46 (39.7) | 27 (38.0) | 0.825 |
| Vascular catheter-related | 30 (16.0) | 18 (15.5) | 12 (16.9) | 0.802 |
| Musculoskeletal | 16 (8.6) | 13 (11.2) | 3 (4.2) | 0.098 |
| Intraabdominal | 14 (7.5) | 10 (8.6) | 4 (5.6) | 0.451 |
| Urinary | 12 (6.4) | 10 (8.6) | 2 (2.8) | 0.137 |
| Skin and soft tissue | 12 (6.4) | 7 (6.0) | 5 (7.0) | 0.768 |
| Unidentified | 30 (16.0) | 12 (10.3) | 18 (25.4) | 0.007 |
| Microorganism identified | ||||
| Methicillin-resistant Staphylococcus aureus | 74 (39.6) | 59 (50.9) | 15 (21.1) | < 0.001 |
| Carbapenem-resistant Acinetobacter baumannii | 54 (28.9) | 29 (25.0) | 25 (35.2) | 0.135 |
| ESBL(+) Klebsiella pneumoniae | 22 (11.8) | 15 (12.9) | 7 (9.9) | 0.527 |
| Vancomycin-resistant Enterococcus faecium | 20 (10.7) | 3 (2.6) | 17 (23.9) | < 0.001 |
| ESBL(+) Escherichia coli | 11 (5.9) | 7 (6.0) | 4 (5.6) | > 0.999 |
| Carbapenem-resistant Pseudomonas aeruginosa | 6 (3.2) | 3 (2.6) | 3 (4.2) | 0.675 |
Values are presented as mean ± SD or number (%).
APACHE, Acute Physiology and Chronic Health Evaluation; SOFA, Sequential Organ Failure Assessment; RRT, renal replacement therapy; NMB, neuromuscular blockade; MV, mechanical ventilation; LOS, length of stay; ICU, intensive care unit; ESBL, extended-spectrum β-lactamase.
Survivors were defined as patients who survived for 28 days after blood was drawn.
Based on the clinical data that were recorded on the day on which blood was drawn.
Comparison of the pre-intervention and intervention groups
Of the 187 patients enrolled in the study, 97 were in the pre-intervention group and 90 were in the intervention group. The most common microorganisms in the pre-intervention and intervention groups were MRSA and CRAB, respectively (Table 2). However, the two groups did not differ in terms of frequencies of infection with different microorganisms.
Table 2.
Comparison of the pre-intervention and intervention groups in terms of microorganism-related variables
| Variable | Pre-intervention group (n = 97) | Intervention group (n = 90) | p value |
|---|---|---|---|
| Identified microorganisms | |||
| Methicillin-resistant Staphylococcus aureus | 44 (45.5) | 30 (33.3) | 0.093 |
| Carbapenem-resistant Acinetobacter baumannii | 23 (23.7) | 31 (34.4) | 0.106 |
| ESBL(+) Klebsiella pneumoniae | 13 (13.4) | 9 (10.0) | 0.471 |
| Vancomycin-resistant Enterococcus faecium | 7 (7.2) | 13 (14.4) | 0.110 |
| ESBL(+) Escherichia coli | 6 (6.2) | 5 (5.6) | 0.855 |
| Carbapenem-resistant Pseudomonas aeruginosa | 4 (4.1) | 2 (2.2) | 0.684 |
| Time from venipuncture to microorganism identification and antibiotics susceptibility result, hr | |||
| All microorganisms | 92.3 ± 40.4 | 82.5 ± 21.6 | 0.038 |
| Methicillin-resistant Staphylococcus aureus | 91.9 ± 33.9 | 85.4 ± 20.9 | 0.357 |
| Carbapenem-resistant Acinetobacter baumannii | 87.3 ± 54.5 | 77.3 ± 16.4 | 0.338 |
| ESBL(+) Klebsiella pneumoniae | 77.6 ± 24.7 | 75.7 ± 22.2 | 0.854 |
| Vancomycin-resistant Enterococcus faecium | 108.0 ± 48.8 | 96.7 ± 29.5 | 0.522 |
| ESBL(+) Escherichia coli | 113.1 ± 37.2 | 77.6 ± 19.4 | 0.088 |
| Carbapenem-resistant Pseudomonas aeruginosa | 115.5 ± 35.1 | 70.3 ± 17.7 | 0.174 |
Values are presented as number (%) or mean ± SD.
ESBL, extended-spectrum β-lactamase.
The intervention group had a significantly shorter average time from venipuncture to microorganism identification and antimicrobial susceptibility test results than the pre-intervention group (p = 0.038) (Table 2, Fig. 2). However, when the groups were divided according to the causative microorganism, they did not differ in terms of this variable (Table 2).
Figure 2.
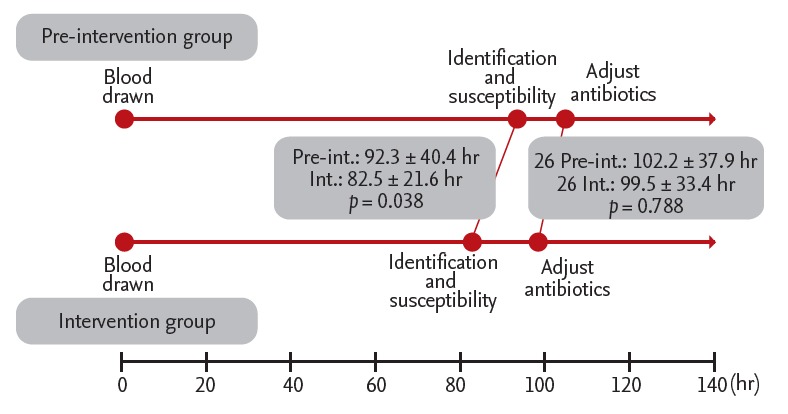
Comparison of the pre-intervention and intervention groups in terms of microbiology workflow. Pre-int., pre-intervention group; Int., intervention group.
In total, 143 of the 187 patients (76.5%) received antibiotics for more than 3 days in the 3 months before blood culture. The two groups did not differ in terms of this variable (p = 0.685). Table 3 shows the antibiotics that were used for more than 3 days in the 3 months before blood culture. In both groups, glycopeptides and carbapenems were the most commonly used antibiotics. The frequency with which patients were given different types of antibiotics in the 3 months before blood culture did not differ between groups (Table 3). Of the 187 study patients, 122 (65.2%) required consultation with an ID specialist (Fig. 3). The intervention group had more consultations with an ID specialist than the pre-intervention group (74.4% vs. 56.7%, p = 0.014).
Table 3.
Comparison of the pre-intervention and intervention groups in terms of antibiotics that were used for more than 3 days in the 3 months before venipuncture for blood culture
| Antibiotic | Pre-intervention group (n = 97) | Intervention group (n = 90) | p value |
|---|---|---|---|
| Glycopeptides | 49 (50.5) | 57 (63.3) | 0.077 |
| Carbapenems | 47 (48.5) | 41 (45.6) | 0.692 |
| Fluoroquinolones | 38 (39.2) | 43 (47.8) | 0.236 |
| Third-generation cephalosporins | 41 (42.3) | 33 (36.7) | 0.434 |
| Anti-pseudomonal penicillins | 25 (25.8) | 27 (30.0) | 0.519 |
| Fourth-generation cephalosporins | 18 (18.6) | 27 (30.0) | 0.067 |
| Colistin | 9 (9.3) | 17 (18.9) | 0.058 |
| Aminoglycosides | 9 (9.3) | 12 (13.3) | 0.380 |
| Minocycline | 4 (4.1) | 10 (11.1) | 0.070 |
| Macrolide | 6 (6.2) | 1 (1.1) | 0.120 |
| Second-generation cephalosporins | 4 (4.1) | 2 (2.2) | 0.684 |
| Linezolid | 3 (3.1) | 3 (3.3) | > 0.999 |
| Trimethoprim-sulfamethoxazole | 3 (3.1) | 2 (2.2) | > 0.999 |
| Tigecycline | 2 (2.1) | 0 | 0.498 |
Values are presented as number (%).
Figure 3.
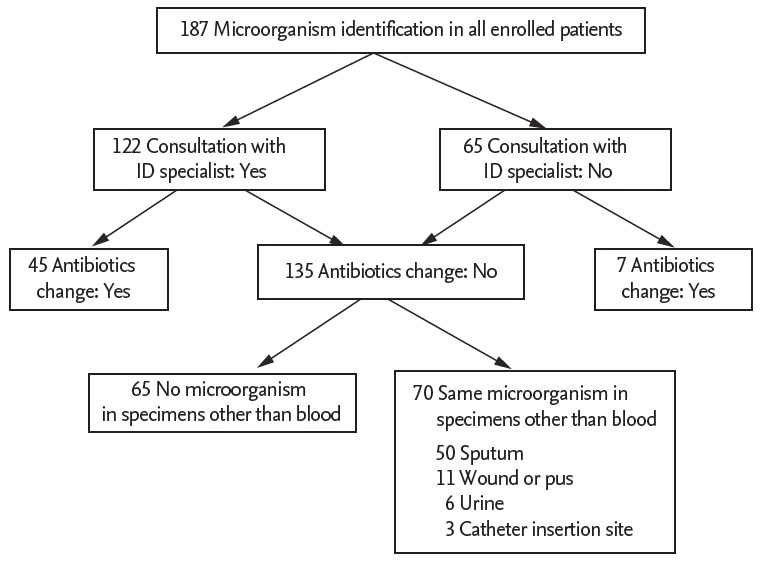
Flowchart of consultation with an infectious diseases specialist and antibiotics adjustment. ID, infectious disease.
The antibiotics used were adjusted in 52 patients (27.8%) based on microorganism identification and antimicrobial susceptibility test results. Of these, 26 were in the pre-intervention group and 26 were in the intervention group. The pre-intervention and intervention groups did not differ in terms of the frequency of antibiotic adjustment (p = 0.751). The most common adjustment in both groups was a change to treat CRAB (32.7%), followed by a change to treat ESBL-producing Gram-negative bacteria (21.2%). However, the two groups did not differ significantly in terms of average time from venipuncture to antibiotic adjustment (Fig. 2). Of the 135 patients who did not undergo adjustment of antibiotics, 70 (51.9%) had the same microorganisms in non-blood specimens that were taken before the blood culture (Fig. 3). No difference was noted in the frequency of this variable between groups (p = 0.490).
Effect of MALDI-TOF MS–based microorganism identification on clinical outcomes
The two groups did not differ in terms of the mortality rate within 28 days of venipuncture (40.2% in the pre-intervention group and 35.6% in the intervention group, p = 0.549). In addition, the in-hospital mortality rate did not differ between groups (48.5% in the pre-intervention group and 46.7% in the intervention group, p = 0.807). To evaluate the prognostic utility of MALDI-TOF MS, we assessed the mortality rates of the 52 patients who underwent antibiotic adjustment. MALDI-TOF MS was not significantly associated with this mortality rate (47.1% vs. 52.9%, p > 0.999).
A multivariate logistic regression analysis of data from the 52 patients who underwent antibiotic adjustment showed that diabetes and malignant disease were comorbidities independently associated with 28-day mortality. However, MALDI-TOF MS–based microorganism identification was not associated with 28-day mortality (Table 4).
Table 4.
| Factor | Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
| OR | 95% CI | p value | OR | 95% CI | p value | |
| APACHE II score ≥ 18c | 3.556 | 1.006–12.562 | 0.049 | |||
| SOFA score ≥ 9c | 2.182 | 0.598–7.961 | 0.237 | |||
| Diabetes as a comorbidity | 4.296 | 1.176–15.702 | 0.027 | 5.608 | 1.306–24.081 | 0.020 |
| Malignant disease as a comorbidity | 4.227 | 1.002–17.843 | 0.050 | 6.383 | 1.262–32.283 | 0.025 |
| Vancomycin-resistant Enterococcus faecium bacteremia | 9.000 | 1.580–51.261 | 0.013 | |||
| Consultation with an infectious diseases specialist | 0.145 | 0.025–0.852 | 0.033 | |||
| Microorganism identification via MALDI-TOF MS | 1.191 | 0.373–3.801 | 0.768 | |||
OR, odds ratio; CI, confidence interval; APACHE, Acute Physiology and Chronic Health Evaluation; SOFA, Sequential Organ Failure Assessment; MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
Defined as death within 28 days after venipuncture for blood culture.
In total, 52 patients (26 in the pre-intervention group and 26 in the intervention group) who underwent antibiotics adjustment were included in this analysis.
Based on the clinical data that were recorded on the day of venipuncture. Identification of an optimal cut-off value was based on the maximum Youden’s index.
DISCUSSION
This study is the first to examine the clinical efficacy of MALDI-TOF MS–based microorganism identification for patients with MDR bacteremia in ICUs in Korea. The use of MALDI-TOF MS for microorganism identification significantly reduced the time from venipuncture to microorganism identification and antimicrobial susceptibility test results, but did not significantly impact survival, in our cohort of patients with MDR bacteremia in the ICU.
Our finding that MALDI-TOF MS reduces the time from venipuncture to microorganism identification and antimicrobial susceptibility test results is consistent with the findings of several studies [12-14,23-25]. However, one marked difference between previous studies and our findings is that both groups, particularly the intervention group, in our study had longer times from venipuncture to microorganism identification and antimicrobial susceptibility test results (92.3 and 82.5 hours in the pre-intervention and intervention groups, respectively) than did equivalent groups in previous studies (48.0 to 84.0 and 23.0 to 55.9 hours, respectively) [13,24]. This difference may reflect our hospital’s policy of performing microorganism identification tests once per day due to limited resources (e.g., excessive workload and lack of manpower). To improve the clinical usefulness of MALDI-TOF MS, universal protocols must be established.
Our study demonstrated that the use of MALDI-TOF MS to identify bacteremia-causing microorganisms in patients in the ICU was not associated with 28-day mortality. This finding may reflect the fact that most patients had received empirical broad-spectrum antibiotics before blood microorganism identification and antimicrobial susceptibility testing. Moreover, in 37% of the patients, the same microorganisms had been identified in non-blood specimens (e.g., sputum and urine) taken before the bacteremia arose. Thus, in approximately one-third of patients, the antibiotics administered had already been adjusted before bacteremia developed and blood was taken for culture. These factors may have limited the ability of our study to detect a significant effect of MALDI-TOF MS on mortality rates.
One of the most important findings of our study is that the time from blood culture to antibiotic adjustment did not differ between the pre-intervention and intervention groups, meaning that the advantage of rapid microorganism identification by MALDI-TOF MS did not result in rapid antibiotic adjustment. Although this result may be attributable to the small number of patients who underwent antibiotic adjustment in our study, our findings suggest that optimal interventions are needed not only for real-time notification to attending physicians regarding pathogen identification, but also for reinforcement of antimicrobial stewardship programs. Despite the severity of MDR bacteremia, only One of the most important findings of our study is that the time from blood culture to antibiotic adjustment did not differ between the pre-intervention and intervention groups, meaning that the advantage of rapid microorganism identification by MALDI-TOF MS did not result in rapid antibiotic adjustment. Although this result may be attributable to the small number of patients who underwent antibiotic adjustment in our study, our findings suggest that optimal interventions are needed not only for real-time notification to attending physicians regarding pathogen identification, but also for reinforcement of antimicrobial stewardship programs. Despite the severity of MDR bacteremia, only 65% of our patients had consultations with an ID specialist. Moreover, only 24% of these patients had changes to their antimicrobial treatment that were suggested by an ID specialist. These findings may be typical of the situation in Korea, where few antimicrobial stewardship programs have been established. This factor may be responsible for the recent increase in the consumption of empirical broad-spectrum antibiotics in Korea [26]. Well-regulated antimicrobial stewardship is known to be essential for limiting multidrug resistance worldwide [27,28]. Indeed, the treatment of patients with MDR bacteremia significantly improves when rapid diagnostics is coupled with antibiotic stewardship [11-13,29]. In addition, a study showed that the combination of MALDI-TOF MS and real-time antimicrobial stewardship could achieve optimal treatment outcomes as well as a faster time to optimal antimicrobial therapy compared with MALDI-TOF MS alone [30]. Therefore, the development of a well-regulated national antibiotic stewardship program combined with rapid diagnostic tests (i.e., those based on MALDI-TOF MS) is necessary in our country. Antimicrobial stewardship programs should have a multidisciplinary approach, including an attending physician, ID physician(s), and ID pharmacist(s), as well as real-time notifications to the attending physician(s) regarding pathogen identification [12,30].
Our study has several limitations. First, it had a non-randomized design and was retrospective in nature, which may have resulted in selection bias. Second, our before-after design may not have taken into account changes in the standard of care over the study period that may have improved mortality. However, no significant change in standard sepsis management occurred during the study period. Third, our data are from a single center, and thus the results may not be representative of the general situation in Korea. Finally, we had a small sample, which prevented us from assessing the prognostic utility of MALDI-TOF MS in patients with specific bacteria or sources of infection.
In conclusion, the present study showed that MALDI-TOF MS reduced the time from venipuncture to microorganism identification and antimicrobial susceptibility testing. However, it did not serve as a prognostic indicator in patients with MDR bacteremia. This result may reflect the limitations of this study. Prospective large-scale studies of the optimal use and prognostic utility of MALDI-TOF MS–based microorganism identification in Korean ICUs are warranted.
KEY MESSAGE
1. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) accelerated microorganism identification in patients with multidrug-resistant bacteremia in the intensive care unit.
2. However, MALDI-TOF MS–based microorganism identification did not influence 28-day mortality.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and ICT) (2016R1C1B1008529).
Footnotes
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Brusselaers N, Vogelaers D, Blot S. The rising problem of antimicrobial resistance in the intensive care unit. Ann Intensive Care. 2011;1:47. doi: 10.1186/2110-5820-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu J, Duan X, Wu H, Zhou Q. Surveillance and correlation of antimicrobial usage and resistance of Pseudomonas aeruginosa: a hospital population-based study. PLoS One. 2013;8:e78604. doi: 10.1371/journal.pone.0078604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLaughlin M, Advincula MR, Malczynski M, Qi C, Bolon M, Scheetz MH. Correlations of antibiotic use and carbapenem resistance in enterobacteriaceae. Antimicrob Agents Chemother. 2013;57:5131–5133. doi: 10.1128/AAC.00607-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 5.Vardakas KZ, Rafailidis PI, Konstantelias AA, Falagas ME. Predictors of mortality in patients with infections due to multi-drug resistant Gram negative bacteria: the study, the patient, the bug or the drug? J Infect. 2013;66:401–414. doi: 10.1016/j.jinf.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 6.Magill SS, Edwards JR, Bamberg W, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makristathis A, Riss S, Hirschl AM. A novel fluorescence in situ hybridization test for rapid pathogen identification in positive blood cultures. Clin Microbiol Infect. 2014;20:O760–O763. doi: 10.1111/1469-0691.12561. [DOI] [PubMed] [Google Scholar]
- 8.Seng P, Drancourt M, Gouriet F, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 9.Kerremans JJ, Verboom P, Stijnen T, et al. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J Antimicrob Chemother. 2008;61:428–435. doi: 10.1093/jac/dkm497. [DOI] [PubMed] [Google Scholar]
- 10.Vincent JL, Brealey D, Libert N, et al. Rapid diagnosis of infection in the critically ill, a multicenter study of molecular detection in bloodstream infections, pneumonia, and sterile site infections. Crit Care Med. 2015;43:2283–2291. doi: 10.1097/CCM.0000000000001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verroken A, Defourny L, le Polain de Waroux O, et al. Clinical impact of MALDI-TOF MS identification and rapid susceptibility testing on adequate antimicrobial treatment in sepsis with positive blood cultures. PLoS One. 2016;11:e0156299. doi: 10.1371/journal.pone.0156299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wenzler E, Goff DA, Mangino JE, Reed EE, Wehr A, Bauer KA. Impact of rapid identification of Acinetobacter baumannii via matrix-assisted laser desorption ionization time-of-flight mass spectrometry combined with antimicrobial stewardship in patients with pneumonia and/or bacteremia. Diagn Microbiol Infect Dis. 2016;84:63–68. doi: 10.1016/j.diagmicrobio.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 13.Lockwood AM, Perez KK, Musick WL, et al. Integrating rapid diagnostics and antimicrobial stewardship in two community hospitals improved process measures and antibiotic adjustment time. Infect Control Hosp Epidemiol. 2016;37:425–432. doi: 10.1017/ice.2015.313. [DOI] [PubMed] [Google Scholar]
- 14.Kock R, Wullenweber J, Horn D, Lanckohr C, Becker K, Idelevich EA. Implementation of short incubation MALDI-TOF MS identification from positive blood cultures in routine diagnostics and effects on empiric antimicrobial therapy. Antimicrob Resist Infect Control. 2017;6:12. doi: 10.1186/s13756-017-0173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JH, Hong SK, Kim KC, et al. Influence of full-time intensivist and the nurse-to-patient ratio on the implementation of severe sepsis bundles in Korean intensive care units. J Crit Care. 2012;27:414. doi: 10.1016/j.jcrc.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Phua J, Koh Y, Du B, et al. Management of severe sepsis in patients admitted to Asian intensive care units: prospective cohort study. BMJ. 2011;342:d3245. doi: 10.1136/bmj.d3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwak SH, Jeong CW, Lee SH, Lee HJ, Koh Y. Current status of intensive care units registered as critical care subspecialty training hospitals in Korea. J Korean Med Sci. 2014;29:431–437. doi: 10.3346/jkms.2014.29.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39:165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acute Respiratory Distress Syndrome Network. Brower RG, Matthay MA, et al. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N Engl J Med. 2000;342:1301–1308. doi: 10.1056/NEJM200005043421801. [DOI] [PubMed] [Google Scholar]
- 20.Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13:818–829. [PubMed] [Google Scholar]
- 21.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22:707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 22.Perkins NJ, Schisterman EF. The inconsistency of “optimal” cutpoints obtained using two criteria based on the receiver operating characteristic curve. Am J Epidemiol. 2006;163:670–675. doi: 10.1093/aje/kwj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clerc O, Prod’hom G, Vogne C, Bizzini A, Calandra T, Greub G. Impact of matrix-assisted laser desorption ionization time-of-flight mass spectrometry on the clinical management of patients with Gram-negative bacteremia: a prospective observational study. Clin Infect Dis. 2013;56:1101–1107. doi: 10.1093/cid/cis1204. [DOI] [PubMed] [Google Scholar]
- 24.Huang AM, Newton D, Kunapuli A, et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis. 2013;57:1237–1245. doi: 10.1093/cid/cit498. [DOI] [PubMed] [Google Scholar]
- 25.Martiny D, Debaugnies F, Gateff D, et al. Impact of rapid microbial identification directly from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry on patient management. Clin Microbiol Infect. 2013;19:E568–E581. doi: 10.1111/1469-0691.12282. [DOI] [PubMed] [Google Scholar]
- 26.Yoon YK, Park GC, An H, Chun BC, Sohn JW, Kim MJ. Trends of antibiotic consumption in korea according to national reimbursement data (2008-2012): a population-based epidemiologic study. Medicine (Baltimore) 2015;94:e2100. doi: 10.1097/MD.0000000000002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holloway KA, Rosella L, Henry D. The impact of WHO essential medicines policies on inappropriate use of antibiotics. PLoS One. 2016;11:e0152020. doi: 10.1371/journal.pone.0152020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard P, Pulcini C, Levy Hara G, et al. An international cross-sectional survey of antimicrobial stewardship programmes in hospitals. J Antimicrob Chemother. 2015;70:1245–1255. doi: 10.1093/jac/dku497. [DOI] [PubMed] [Google Scholar]
- 29.Osthoff M, Gurtler N, Bassetti S, et al. Impact of MALDI-TOF-MS-based identification directly from positive blood cultures on patient management: a controlled clinical trial. Clin Microbiol Infect. 2017;23:78–85. doi: 10.1016/j.cmi.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Beganovic M, Costello M, Wieczorkiewicz SM. Effect of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) alone versus maldi-tof ms combined with real-time antimicrobial stewardship interventions on time to optimal antimicrobial therapy in patients with positive blood cultures. J Clin Microbiol. 2017;55:1437–1445. doi: 10.1128/JCM.02245-16. [DOI] [PMC free article] [PubMed] [Google Scholar]


