Abstract
Background & Aims
GPBAR1, also known as TGR5, is a G protein–coupled receptor activated by bile acids. Hepatic innate immune cells are involved in the immunopathogenesis of human liver diseases and in several murine hepatitis models. Here, by using genetic and pharmacological approaches, we provide evidence that GPBAR1 ligation attenuates the inflammation in rodent models of hepatitis.
Material and methods
Hepatitis was induced by concanavalin A (Con A) or α-galactosyl-ceramide (α-GalCer). 6b-Ethyl-3a,7b-dihydroxy-5b-cholan-24-ol (BAR501), a selective agonist of GPBAR1, was administrated by o.s.
Results
In the mouse models of hepatitis, the genetic ablation of Gpabar1 worsened the severity of liver injury and resulted in a type I NKT cells phenotype that was biased toward a NKT1, a proinflammatory, IFN-γ producing, NKT cells subtype. Further on, NKT cells from GPBAR1–/– mice were sufficient to cause a severe hepatitis when transferred to naïve mice. In contrast, GPBAR1 agonism rescued wild-type mice from acute liver damage and redirects the NKT cells polarization toward a NKT10, a regulatory, IL-10 secreting, type I NKT cell subset. In addition, GPBAR1 agonism significantly expanded the subset of IL-10 secreting type II NKT cells. RNAseq analysis of both NKT cells type confirmed that IL-10 is a major target for GPABR1. Accordingly, IL-10 gene ablation abrogated protection afforded by GPBAR1 agonism in the Con A model.
Conclusion
Present results illustrate a role for GPBAR1 in regulating liver NKT ecology. Because NKT cells are an essential component of liver immune system, our data provide a compelling evidence for a GPBAR1-IL-10 axis in regulating of liver immunity.
Keywords: Bile Acids, GPBAR1, Autoimmune Hepatitis, Natural Killer T Cells, NKT, Interleukin-10, IL-10
Abbreviations used in this paper: α-GalCer, α-galactosylceramide; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CCL4, carbon tetrachloride; Con A, concanavalin A; CREB, cAMP response element binding protein; FasL, Fas ligand; GAPDH, glyceraldehyde-3-phosphate dehydrogenase (phosphorylating); GPBAR1, G-protein–coupled bile acid receptor 1; IC-FACS, fluorescence-activated cell sorter with intracellular staining; IFN, interferon; NKT10, interleukin-10–biased natural killer T cells; IL, interleukin; LFA-1, lymphocyte function–associated 1; mRNA, messenger RNA; NK, natural killer; NKT, natural killer T; OPN, osteopontin; PBS, phosphate-buffered saline; PE, phycoerythrin; RT-PCR, reverse-transcription polymerase chain reaction; TCR, T cell receptor; TNF-α, tumor necrosis factor alpha
Graphical abstract
Summary.
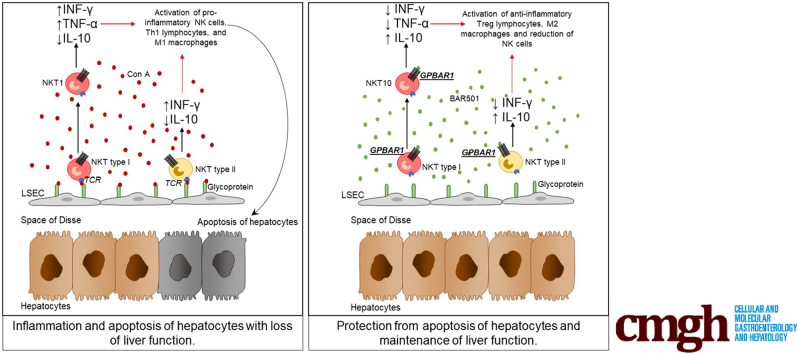
G-protein–coupled bile acid receptor 1 (TGR5) is a receptor for secondary bile acids. Here, we show that G-protein–coupled bile acid receptor 1 is essential for regulation of liver immunity by modulating type I and II natural killer T cells in a interleukin-10 dependent manner.
Natural killer T (NKT) cells are a heterogeneous subset of specialized T cells that exhibit innate cell-like feature of rapid response to antigenic exposure in combination with adaptive cell’s precision of antigenic recognition.1 In contrast to conventional T cells, however, the NKT’s T cell receptor (TCR) recognizes lipid antigens presented by the conserved and nonpolymorphic major histocompatibility complex class 1–like molecule CD1d.2 In addition to the TCRs, and similarly to other cells of innate immunity such as natural killer (NK) and innate lymphoid cells, NKT cells express receptors for cytokines including interleukin-12 (IL-12), IL-18, IL-25, and IL-23, which define functionally polarized subsets of innate-like CD1d-restricted T cells. CD1d-restricted NKT cells can be further divided into 2 main subsets. Type I (classic or invariant) NKT cells, so named because of their limited TCR repertoire, express a semi-invariant TCR alpha chain paired with a heterogeneous Vβ chain repertoire, and type II NKT cells (diverse or nonclassical NKT) which do not use the semi-invariant TCR alpha chain.2 The 2 cell subtypes also differ for their specificity in antigen recognition. Thus, the prototypic antigen recognized by type I NKT cells is the α-galactosylceramide (α-GalCer), a marine sponge cerobroside, which binds to the CD1d receptor and elicits release of large amounts of interferon γ (IFN-γ), while type II NKT cells respond to glycolipids such as sulfatide.2, 3
Several iNKT subtypes have been identified and named as NKT1, NKT2, NKT17, IL17RB, NKTfh, and FOXP3+iNKT, which perform activities and release cytokine patterns that are comparable to their Th counterparts in T lymphocytes (ie, Th1, Th2, and Th17).3, 4 In addition, an IL-10–secreting, regulatory subtype, named NKT10, has been recently identified.5, 6 These NKT10 cells, are regulatory in nature and their activation attenuates disease progression in various experimental settings, including autoimmune encephalomyelitis.5
Previous studies have shown that type I and II NKT cells are enriched in the liver, representing ∼10% of human liver leukocytes but ∼40% in mice,7, 6 and contributing significantly to the liver immunity in both steady state and inflammation.8 Spatially located at the basolateral membrane of liver sinusoids, NKT cells patrol the liver microcirculation functioning as cell sensors for intestinal antigens, representing an essential component of liver innate immune organization, entangling a highly sophisticated signaling network with immune and parenchymal and nonparenchymal cells (liver sinusoidal cells, hepatic stellate cells, and hepatocytes).9, 10, 11, 12, 13
Bile acids are C24-5β-cholanoic acid steroids generated in mammals by the coordinated cooperation of the host and its intestinal microbiota.14, 15 In humans, the 2 main primary bile acids, cholic acid and chenodeoxycholic acid, are synthesized in the liver from the cholesterol breakdown, while secondary bile acids, lithocolic acid and deoxycholic acid, result from the C23 deamidation and 7α-dehydroxylation of cholic acid and chenodeoxycholic acid operated by the intestinal microbiota.15 Both primary and secondary bile acids are steroids that exert regulatory functions by activating a family of evolutionary conserved receptors.14 G-protein–coupled bile acid receptor 1 (GPBAR1), also known as TGR5, is a G protein–coupled receptors, activated by secondary bile acids,16, 17, 18 highly represented in cells of innate immunity contributing to maintenance of state of tolerogenicity by intestinal macrophages in steady state conditions.19
In this report using genetic and pharmacological approaches, we have investigated the role of GPBAR1 in regulating liver NKT cells ecology in murine models acute hepatitis.
Results
GPBAR1 Is Expressed in NKT Cells and Its Activation Reverses the Inflammatory State Induced by Concanavalin A In Vitro
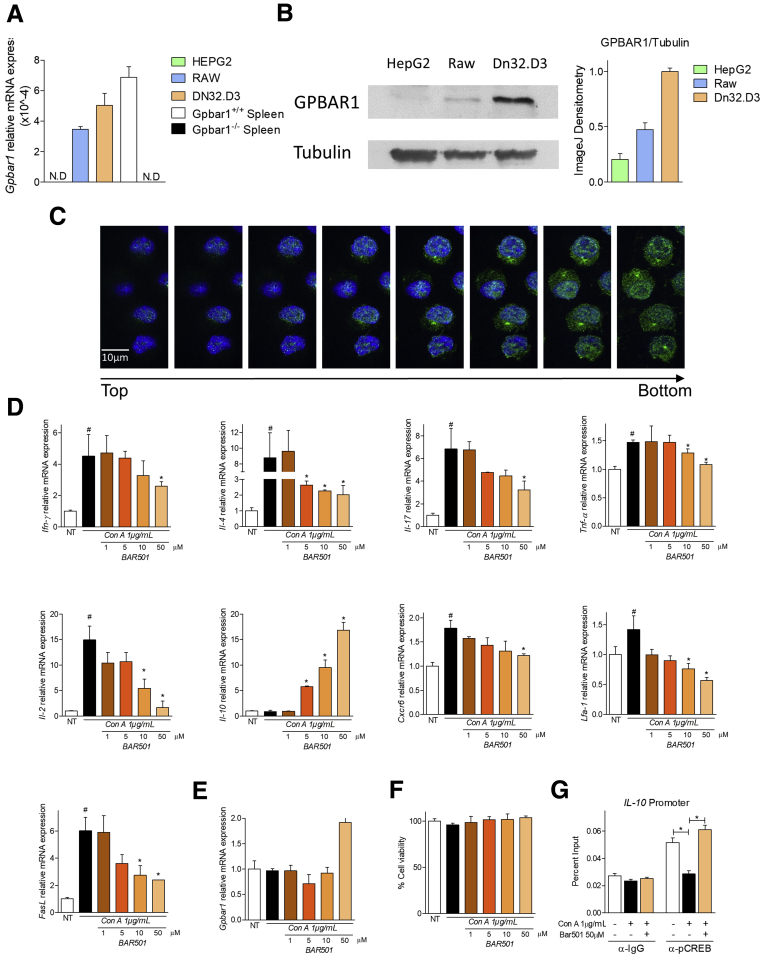
We have first investigated whether NKT express GPBAR1.20, 21, 22 For this purpose, DN32.D3 cells, a murine NKT hybridoma, was used. As shown in Figure 1A, a similar levels of GPBAR1 messenger RNA (mRNA) were detected in DN32.D3 cells and RAW264.7 cells and spleen cells isolated from GPBAR1+/+ mice (2 positive controls). These results were confirmed by Western blot and by IHC analyses (Figure 1B and C). Further on, results shown in Figure 1D, demonstrate that while exposure of DN32.D3 cells to concanavalin A (Con A) increased the expression of proinflammatory genes, this pattern was reversed by co-treating cells with BAR501, a GPBAR1 ligand, which also increased the expression of IL-10 mRNA (Figure 1D, second line). In contrast, none of the treatments modulated the level of GPBAR1 mRNA (Figure 1E). Finally, analysis of cell viability demonstrated that changes in subsets polarization driven by Con A and GPBAR1 agonism did not resulted from death of specific cell subsets (Figure 1F). Because in target cells GPBAR1 activates a signaling pathway that involves the phosphorylation of cAMP response element binding protein (CREB), a transcription factor that binds DNA sequence containing a cAMP response element,23 a ChIP assay was performed on DN32.D3 cells. The data shown in Figure 1G, demonstrated that while exposure to Con A reduced the occupation of cAMP response elements by CREB in the IL-10 promoter, this effect was reversed by BAR501.
Figure 1.
BAR501 counteracts in vitro NKT immune activation. (A) Quantitative real-time PCR analysis of GPBAR1 expression in HepG2 (hepatocytes cell line), RAW264.7 (macrophages cell line), DN32.D3 (NKT cell line) and total spleen of GPBAR1+/+ and GPBAR1–/– mice. The data are normalized to glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) (GAPDH) mRNA. Data are the mean ± SEM of 3 experiments. (B) Western blot analysis for GPBAR1 expression from the cell lysates isolated from HepG2, Raw364.7 and DN32.D3 cells. Tubulin was used as the loading control. (C) Immunohistochemistry of DN32.D3 cells with anti-GPBAR1 antibody in green. (D–G) DN32.D3 cells were incubated for 16 hours with Con A and BAR501. Quantitative real-time PCR analysis of (D) proinflammatory genes IFN-γ, IL-4, IL-17, TNF-α, and IL-2, anti-inflammatory gene IL-10, adhesion molecules CXCR6 and LFA-1, FasL, and (E) GPBAR1. (F) Analysis of cell viability through MTS assay kit. (G) ChIP assay of the CREB binding on IL-10 promoter in DN32.D3 cells. The data are normalized to GAPDH mRNA. Results are the mean ± SEM of 3 experiments. *P < .05.
BAR501 Protects Against Acute Hepatitis Induced by α-GalCer
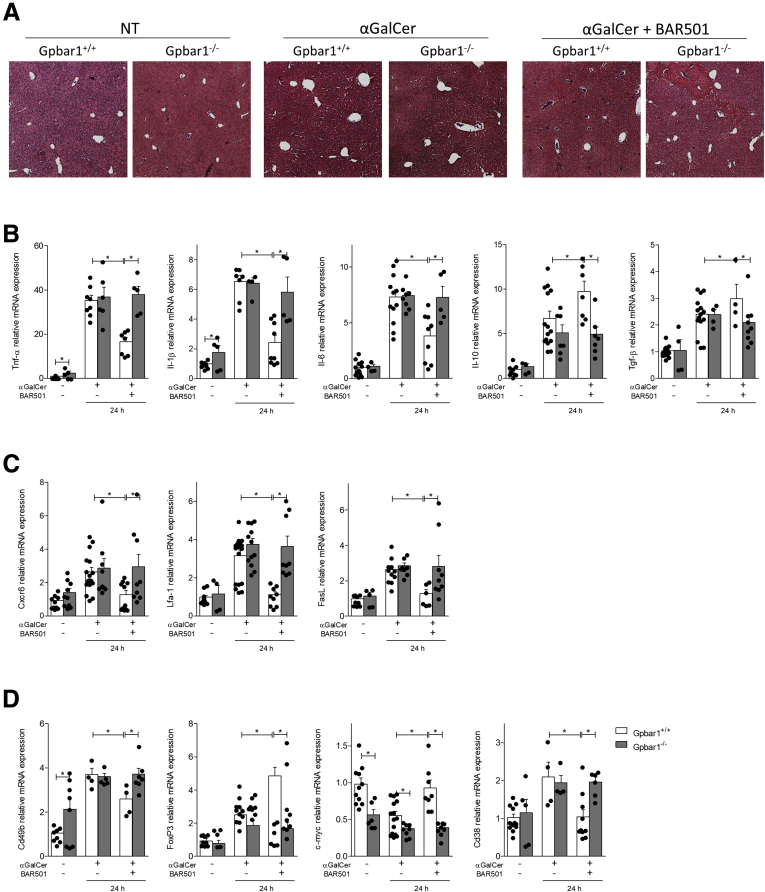
We have then tested whether genetic deletion of GPBAR1 or its activation by BAR501 modulated clinical and biochemical outcomes of acute hepatitis induced in mice by α-GalCer, that causes an immune-mediated hepatitis that is largely contributed by activation of iNKT through the CD1d receptor.20, 21, 24, 25, 26, 27 As shown in Table 1, the peak of the liver injury, measured by assessing aspartate aminotransferase (AST) and alanine aminotransferase (ALT) plasma levels, occurred at 24 hours after α-GalCer administration. The development and severity of hepatitis induced by α-GalCer was exacerbated in GPBAR1–/– mice and, conversely, attenuated by treating wild-type mice with BAR501, while the protective effects of this agent were lost in GPBAR1–/– mice (Figure 2A and Table 1).
Table 1.
Plasmatic Levels of AST and ALT and Liver Index (Obtained From Ratio of Liver Weight and Body Weight × 1000)
| GPBAR1+/+ |
GPBAR1–/– |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NT | 8 h |
24 h |
NT | 8 h |
24 h |
|||||
| α-GalCer | α-GalCer + BAR501 | α-GalCer | α-GalCer + BAR501 | α-GalCer | α-GalCer + BAR501 | α-GalCer | α-GalCer + BAR501 | |||
| AST | 81.1 ± 12.60 | 145.2 ± 122.2 | 107.4 ± 11.01 | 1300 ± 218.0 | 545.8 ± 136.9 | 91 ± 4.000 | 161.3 ± 7.318 | 169.1 ± 15.14 | 2138 ± 964.9 | 2224 ± 423.2 |
| ALT | 38.01 ± 4.371 | 58.54 ± 61.12 | 43.22 ± 4.099 | 1104 ± 253.3 | 445.3 ± 8.803 | 37 ± 3.000 | 62.01 ± 16.3 | 61.1 ± 14.82 | 2356 ± 762.1 | 2321 ± 658.0 |
| Liver Index | 39.47 ± 1.566 | 48.87 ± 2.616 | 43.24 ± 1.217 | 53.8 ± 1.879 | 48.28 ± 0.2271 | 42.53 ± 1.314 | 49.77 ± 1.916 | 48.48 ± 1.056 | 58.37 ± 2.399 | 59.07 ± 2.607 |
Values are mean ± SD.
α-GalCer, α-galactosylceramide; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GPBAR1, G-protein–coupled bile acid receptor 1; NT, not treated.
Figure 2.
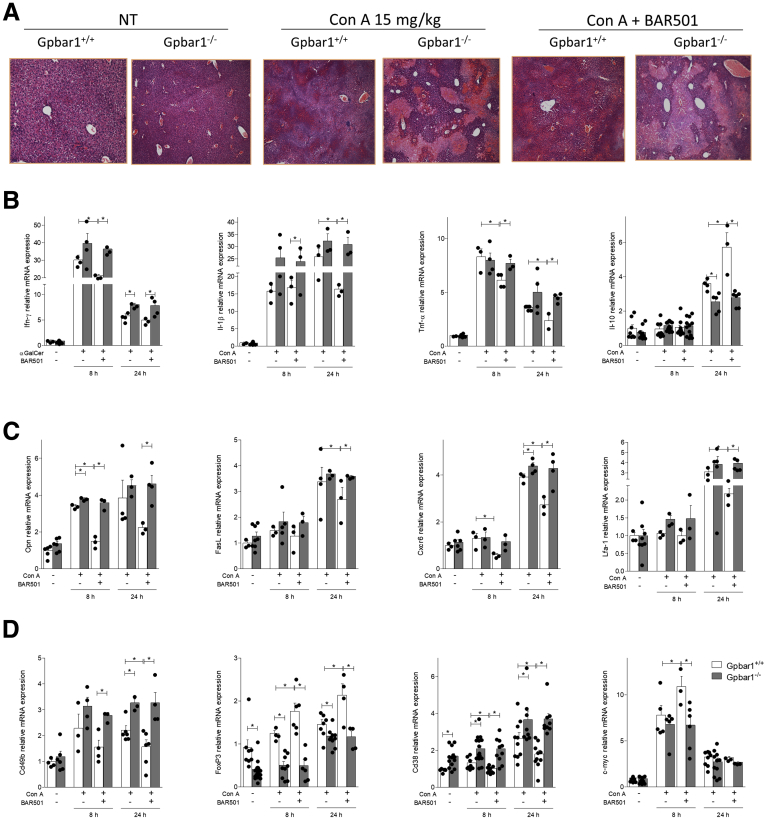
BAR501 reduces the severity of acute hepatitis induced by α-GalCer via GPBAR1. Acute hepatitis was induced in C57BL6 male mice, GPBAR1+/+ and GPBAR1–/–, by α-GalCer (3 μg/300 μL/mouse) intravenous injection for 8 or 24 hours. Mice were randomized to receive α-GalCer alone or in combination with BAR501 (30 mg/kg) daily from 3 days before induction of hepatitis to the time of the sacrifice. (A) Hematoxylin and eosin staining on mice liver tissues. (B–D) Total RNA extracted from liver was used to evaluate by quantitative real-time PCR the relative mRNA expression of (B) proinflammatory genes TNF-α, IL-6, and IL-1β; anti-inflammatory genes IL-10 and TGF-β; (C) adhesion molecules CXCR6 LFA-1, and FasL; and markers specific for different immune cells populations such as (D) Cd49b, FoxP3, c-Myc, and Cd38. The data are normalized to GAPDH mRNA. Each point represents a single mouse. *P < .05.
The GPBAR1 agonist reversed the induction of proinflammatory mediators (tumor necrosis factor alpha [TNF-α], IL-1β IL-6, CXCR6, lymphocyte function–associated 1 [LFA-1], and Fas ligand [FasL]) caused by α-GalCer (Figure 2B and C), while increased the liver expression of anti-inflammatory cytokines (IL-10 and TGF-β) (Figure 2B). In addition, while the expression of Cd49b, a common marker for NK and NKT cells, and Cd38, a marker of M1 macrophages, was increased by α-GalCer administration, this pattern was reverted by the GPBAR1 ligand, which also increased the expression of FoxP3 and c-Myc,21, 24, 25, 26, 27 2 markers for regulatory T cells and M2 macrophages (Figure 2D). These protective effects were lost in GPBAR1–/– mice.
GPBAR1 Protects Against Acute Hepatitis Induced by Con A
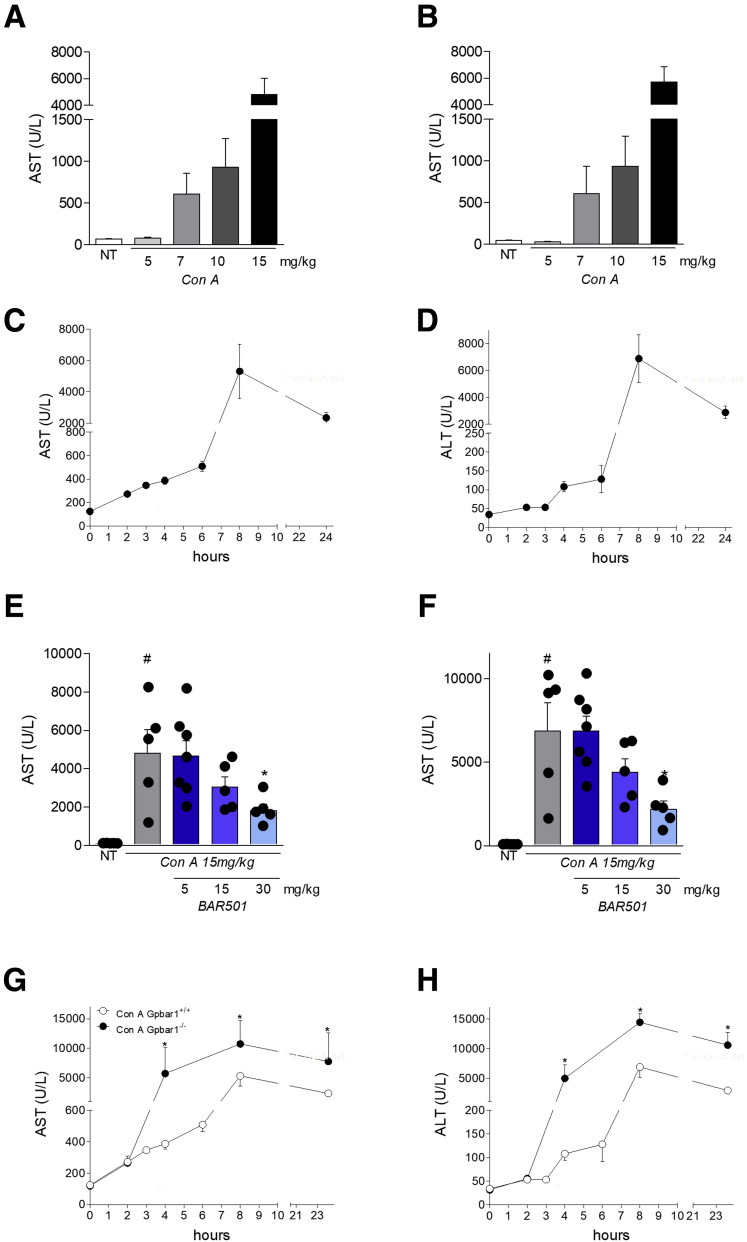
We have then investigated whether GPBAR1 modulates the severity of acute hepatitis induced by Con A, a model of autoimmune liver injury that involves type I and type II NKT cells, and not just the iNKT cells, as it takes place in the α-GalCer model.21, 24, 25, 26, 27, 28, 29 We therefore carried out a Con A dose-finding experiment to obtain a murine model of hepatitis that corresponds as closely as possible to human pathology (Figure 3A and B). We obtained very high values of AST and ALT at 24 hours after the induction of the disease administering 15 mg/kg of Con A. We also tested a higher dose of Con A, 30 mg/kg; however, the higher Con A dosage induced high mortality in mice (about 90%, data not shown) and, on the contrary, doses lower than or equal to 5, 7, and 10 mg/kg did not induce disease (Figure 3A and B). In a preliminary set of experiments shown in Figure 3C and D, we have first determined that peak of illness occurs 8 hours after the administration of the lectin, while at 24 hours the value of AST and ALT start to fall. In these preliminary experiments, a dose-finding investigation, demonstrated that 30-mg/kg BAR501, almost completely reversed the liver injury caused by Con A (Figure 3E and F).
Figure 3.
Characterization of the murine model of acute hepatitis induced by Con A. Acute hepatitis was induced in C57BL6 male mice by intravenous injection of Con A at different dose (5, 7, 10, and 15 mg/kg) 8 or 24 hours. Mice were randomized to receive Con A alone or in combination with BAR501 at different dose (5, 15, and 30 mg/kg) daily from 3 days before induction of hepatitis to the time of the sacrifice. Data shown are plasmatic levels of (A, B) AST and ALT 24 hours after injection of Con A at different doses (5, 7, 10, and 15 mg/kg) or (C, D) at different time points after Con A injection (15 mg/kg). Plasmatic levels of (E, F) AST and ALT in GPBAR1+/+ and GPBAR1–/– mice treated 8 hours with Con A alone or (G, H) in combination with different doses of BAR501 (5, 15, and 30 mg/kg) or at different time points after Con A injection (15 mg/kg). Each point represents a single mouse. *P < .05.
The acute hepatitis were replicated using wild-type and GPBAR1–/– mice challenged with 15 mg/kg Con A. The severity of the liver damage induced by Con A, was exacerbated in GPBAR1–/– mice in comparison with their congenic littermates (Figure 3G and H).
Importantly, while pretreatment with BAR501 effectively attenuated development of liver injury in wild-type mice, the protection was lost in GPBAR1–/– mice (Table 2 and Figure 4A).
Table 2.
Plasmatic Levels of AST and ALT in Each Group and Number of Leukocytes in Liver Tissues
| GPBAR1+/+ |
GPBAR1–/– |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NT | 8 h |
24 h |
NT | 8 h |
24 h |
|||||
| Con A | Con A + BAR501 | Con A | Con A + BAR501 | Con A | Con A + BAR501 | Con A | Con A + BAR501 | |||
| AST | 105.8 ± 16.46 | 5007 ± 1776 | 1271 ± 304.2 | 2812 ± 543.9 | 842 ± 301.1 | 78.92 ± 9.59 | 12183 ± 3696 | 13184 ± 3520 | 9440 ± 4955 | 9011 ± 4829 |
| ALT | 32.44 ± 1.879 | 5066 ± 1509 | 1682 ± 590.9 | 2442 ± 361.4 | 1230 ± 183.3 | 36.31 ± 3.255 | 11043 ± 2322 | 11935 ± 2511 | 9905 ± 3468 | 9170 ± 3595 |
| Leukocytes (106)/ mg of liver | 1.043 ± 0.05937 | 1.386 ± 0.04059 | 1.159 ± 0.08166 | 1.469 ± 0.08061 | 1.697 ± 0.1488 | 1.201 ± 0.05937 | 2.082 ± 0.1462 | 1.834 ± 0.2715 | 2.492 ± 0.5143 | 2.182 ± 0.082 |
Values are mean ± SD.
α-GalCer, α-galactosylceramide; ALT, alanine aminotransferase; AST, aspartate aminotransferase; Con A, concanavalin A; GPBAR1, G-protein–coupled bile acid receptor 1; NT, not treated.
Figure 4.
BAR501 administration protects against acute hepatitis induced by Con A and modulates the expression of pro- and anti-inflammatory markers. Acute hepatitis was induced in C57BL6 male mice, GPBAR1+/+ and GPBAR1–/–, by intravenous injection of Con A (15 mg/kg) at 8 or 24 hours. Mice were randomized to receive Con A alone or in combination with BAR501 (30 mg/kg) daily from 3 days before induction of hepatitis to the time of the sacrifice. (A) Histological evaluation, performed with hematoxylin and eosin staining, of necrosis area on liver sections of GPBAR1+/+ and GPBAR1–/– mice treated with Con A alone or in combination with BAR501 for 8 hours. Total RNA extracted from liver was used to evaluate by quantitative real-time PCR the relative mRNA expression of (B) proinflammatory genes IFN-γ, IL-1β, and TNF-α; anti-inflammatory gene IL-10; (C) markers of NK and NKT cells activation OPN and FasL; adhesion molecules CXCR6 and LFA-1; markers specific for different immune cells populations such as (D) Cd49b for NK and NKT cells; FoxP3 for regulatory T cells; and Cd38 and c-Myc for M1 and M2 macrophages, respectively. The data are normalized to GAPDH mRNA. Each point represents a single mouse. *P < .05.
These changes were confirmed by analysis of the expression of pro and anti-inflammatory biomarkers in the liver. Results shown in Figure 4B, demonstrated that, in comparison with wild-type mice, worsening of liver damage by Con A in GPBAR1–/– mice associated with a robust increase in the expression of inflammatory biomarkers, such as IFN-γ, IL-1β, and TNF-α. Further on, GPBAR1 gene ablation blunted the ability of these mice to produce IL-10 (Figure 4B). Treatment with BAR501 reduced the levels of proinflammatory markers and increased the expression of IL-10 mRNA in the wild-type but not GPBAR1–/– mice (Figure 4B). Furthermore, while exposure to Con A increased the expression of osteopontin (OPN), FasL, CXCR6 and LFA-1 (Figure 4C), this inflammatory pattern was reversed by BAR501 in wild-type but not in GPBAR1–/– mice.
We have then investigated the hepatic expression of markers specific for different immune cells subsets (Figure 4D). At steady state the liver expression of Cd49b and Cd38 was significantly higher in GPBAR1–/– mice, compared with their congenic littermates, and was further enhanced by treating mice with Con A. This pattern, was reversed by dosing the mice with the GPBAR1 ligand, which also increased the liver expression of FoxP3 and c-Myc (Figure 4D). The later effects were observed only in GPBAR1+/+ mice.
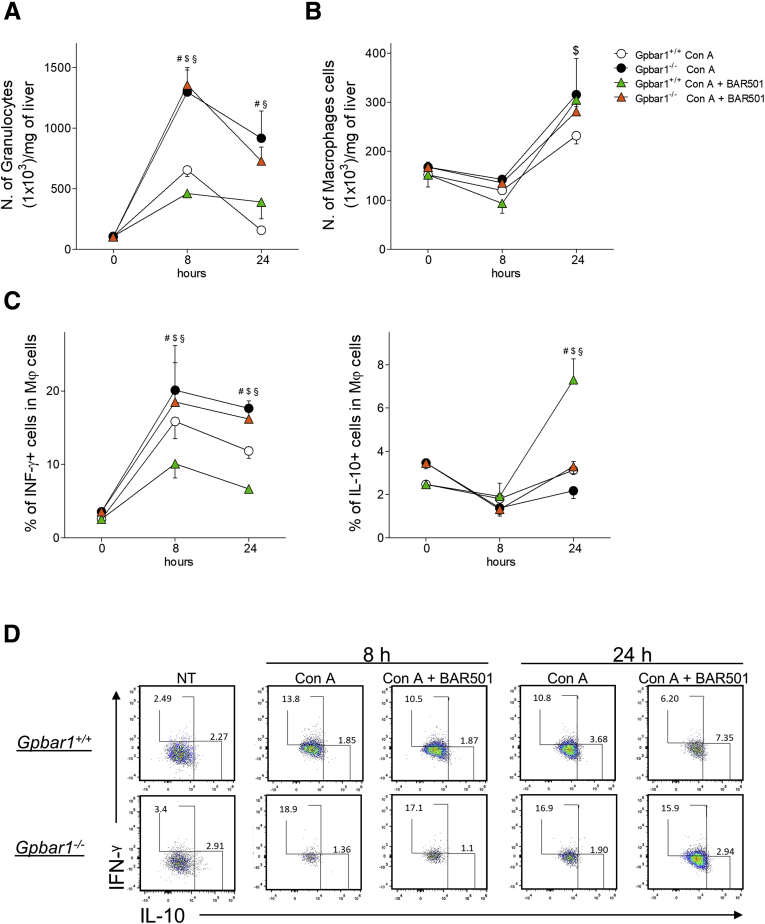
To gain further insights on the role of GPBAR1 in regulating trafficking and functionality of immune cells in this model, we have then carried out a detailed characterization of liver infiltrating cells by fluorescence-activated cell sorter with intracellular flow cytometry staining (IC-FACS) analysis. Exposure to Con A lead to a rapid (8 hours) and massive inflow of granulocytes (CD45+CD11b+Gr1+) into the liver (Figure 5A). This pattern was exacerbated by administering Con A to GPBAR1–/– mice ( ~ 3-fold increase in comparison with wild-type mice) (Figure 5A). As shown in Figure 5A, GPBAR1 agonism had no effect on the relative numbers of infiltrating granulocytes in this model. In contrast to granulocytes, the liver inflow of macrophages, CD45+CD11b+GR1- cells, peaked at 24 hours after Con A injection and was apparently unrelated to the GPBAR1 phenotype (Figure 5B). However, the characterization of proinflammatory (IFN-γ+) and anti-inflammatory (IL-10) demonstrated that BAR501 reduced the inflow of IFN-γ+ macrophages, induced by Con A, while increased the relative percentage of regulatory, IL-10+, macrophages (Figure 5C and D). GPBAR1–/– mice also showed an imbalance in the partition of liver immigrated macrophages, as demonstrated by a higher percentage of proinflammatory macrophages and a lower percentage of anti-inflammatory macrophages found in the liver after the challenge with Con A. Treating GPBAR1–/– mice with BAR501 had no effect on this pattern (Figure 5C and D).
Figure 5.
Effects of BAR501 administration on macrophages cells. Acute hepatitis was induced in C57BL6 male mice, GPBAR1+/+ and GPBAR1–/–, by intravenous injection of Con A (15 mg/kg) a 8 or 24 hours. Mice were randomized to receive Con A alone or in combination with BAR501 (30 mg/kg) daily from 3 days before induction of hepatitis to the time of the sacrifice. Liver tissues were used to perform a detailed characterization of liver infiltrating cells by IC-FACS analysis. Data shown are numbers of (A) granulocytes and (B) macrophages for mg of liver tissue; (C) characterization of proinflammatory (IFN-γ+) and anti-inflammatory (IL-10+) macrophages; (D) flow cytometry analysis of IFN-γ expression and IL-10 expression in CD45+ CD11b+ GR1- cell, recruited into the liver. Results are the mean ± SEM of 7 mice per group. #Wild-type Con A vs knockout Con A; $wild-type Con A vs wild-type Con A + BAR501; §wild-type Con A + BAR501 vs knockout Con A + BAR501. P < .05.
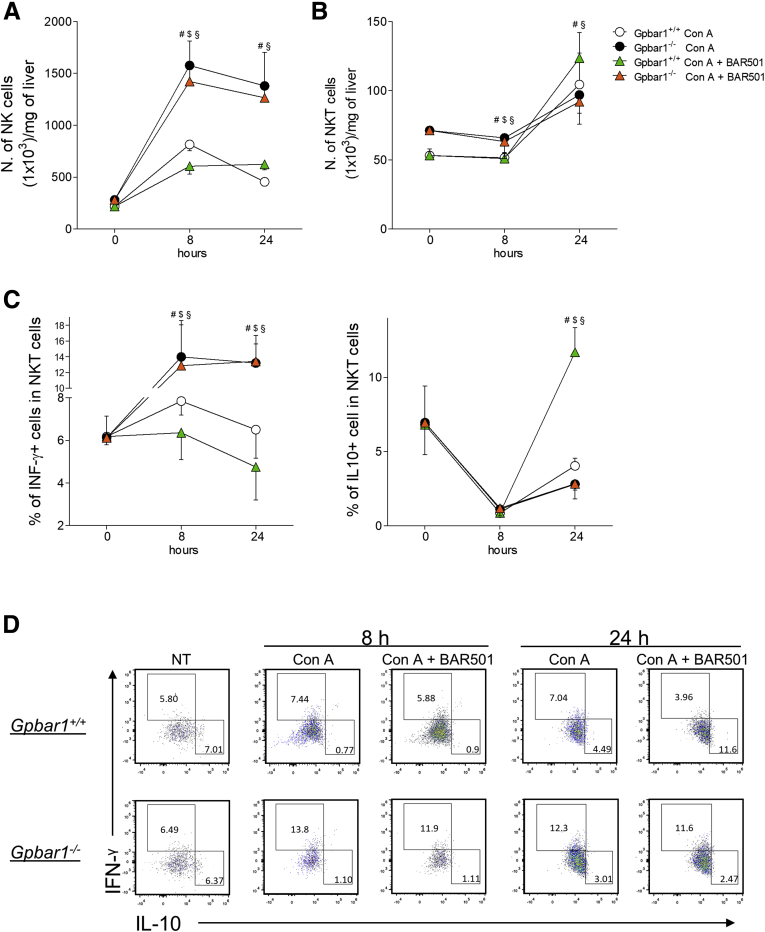
Administration of Con A also increased NK cells number with a peak occurring at 8 hours, and the phenomenon was further exacerbated by GPBAR1 gene ablation (Figure 6A). In contrast to NK cells, NKT cells exhibited a slower kinetics of recruitment (Figure 6B). NKT IFN-γ+ cells were highly predominant in GPBAR1–/– mice, while the percentage of NKT IL-10+ cells was significantly reduced in comparison with their wild-type counterparts (Figure 6C and D). Furthermore, BAR501 reduced the proportion of proinflammatory NKT, both at 8 hours and 24 hours and increased the number of anti-inflammatory NKT cells, at 24 hours, but only in wild-type mice (Figure 6C and D).
Figure 6.
Effects of BAR501 administration on NK and NKT cells. Acute hepatitis was induced in C57BL6 male mice, GPBAR1+/+ and GPBAR1–/–, by intravenous injection of Con A (15 mg/kg) 8 or 24 hours. Mice were randomized to receive Con A alone or in combination with BAR501 (30 mg/kg) daily from 3 days before induction of hepatitis to the time of the sacrifice. Liver tissues were used to perform a detailed characterization of liver infiltrating cells by IC-FACS analysis. Data shown are numbers of (A) NK and (B) NKT cells for milligrams of liver tissue; (C) characterization of proinflammatory (IFN-γ+) and anti-inflammatory (IL-10+) NKT cells; (D) flow cytometry analysis of IFN-γ expression and IL-10 expression in NKT cells recruited into the liver. Results are the mean ± SEM of 7 mice per group. #Wild-type Con A vs knockout Con A; $wild-type Con A vs wild-type Con A + BAR501; §wild-type Con A + BAR501 vs knockout Con A + BAR501. *P < .05.
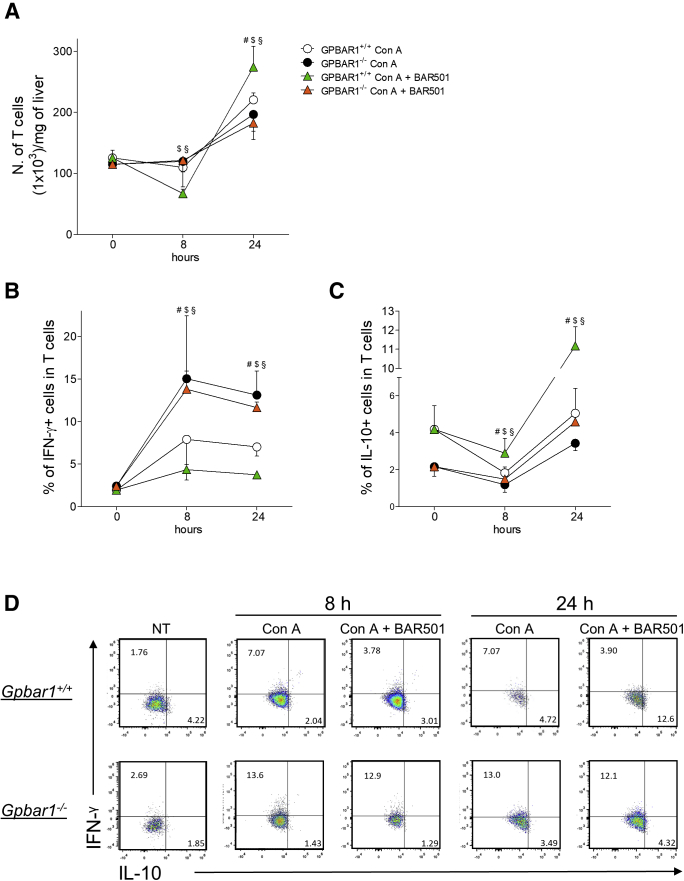
We have then examined the contribution of T lymphocytes to the model and how GPBAR1 regulates this cell subset. The data shown in Figure 7A demonstrated a robust inflow of these cells in the liver, 24 hours after the induction of hepatitis. The number of T cells was further increased by administration of BAR501, although this phenomenon was essentially due to an inflow of regulatory T cells, IL-10+ T lymphocytes, an effect that was not observed in the GPBAR1–/– animals (Figure 7B–D). The effect of the administration of BAR501 on T lymphocytes is to be considered an indirect effect due to the action of the compound on other cells of the immune system, like macrophages and NKT cells, because T lymphocytes do not express the GPBAR1 receptor.23 For the gating strategy, fluorescence minus one strategies were used as described in Materials and Methods.
Figure 7.
Effects of BAR501 administration on T lymphocytes. Acute hepatitis was induced in C57BL6 male mice, GPBAR1+/+ and GPBAR1–/–, by intravenous injection of Con A (15 mg/kg) 8 or 24 hours. Mice were randomized to receive Con A alone or in combination with BAR501 (30 mg/kg) daily from 3 days before induction of hepatitis to the time of the sacrifice. Liver tissues were used to perform a detailed characterization of liver infiltrating cells by IC-FACS analysis. Data shown are numbers of (A) T cells for mg of liver tissue; characterization of (B) proinflammatory (IFN-γ+) and (C) anti-inflammatory (IL-10+) T cells; (D) Flow cytometry analysis of IFN-γ expression and IL-10 expression in T cells recruited into the liver. Results are the mean ± SEM of 7 mice per group. #Wild-type Con A vs knockout Con A; $wild-type Con A vs wild-type Con A + BAR501; §wild-type Con A + BAR501 vs knockout Con A + BAR501. P < .05.
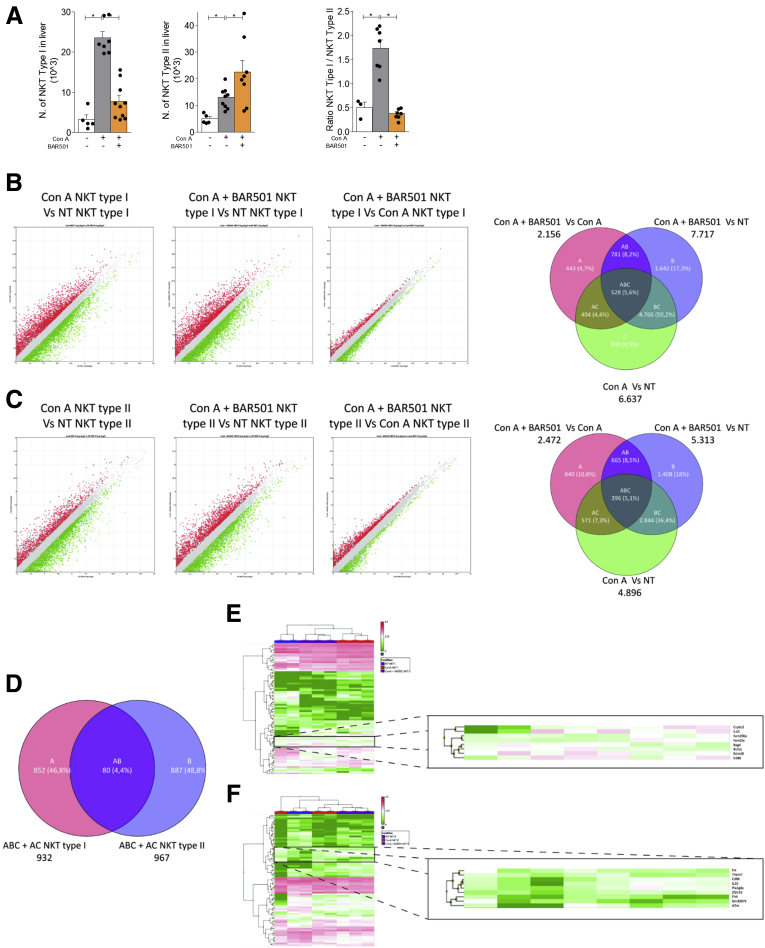
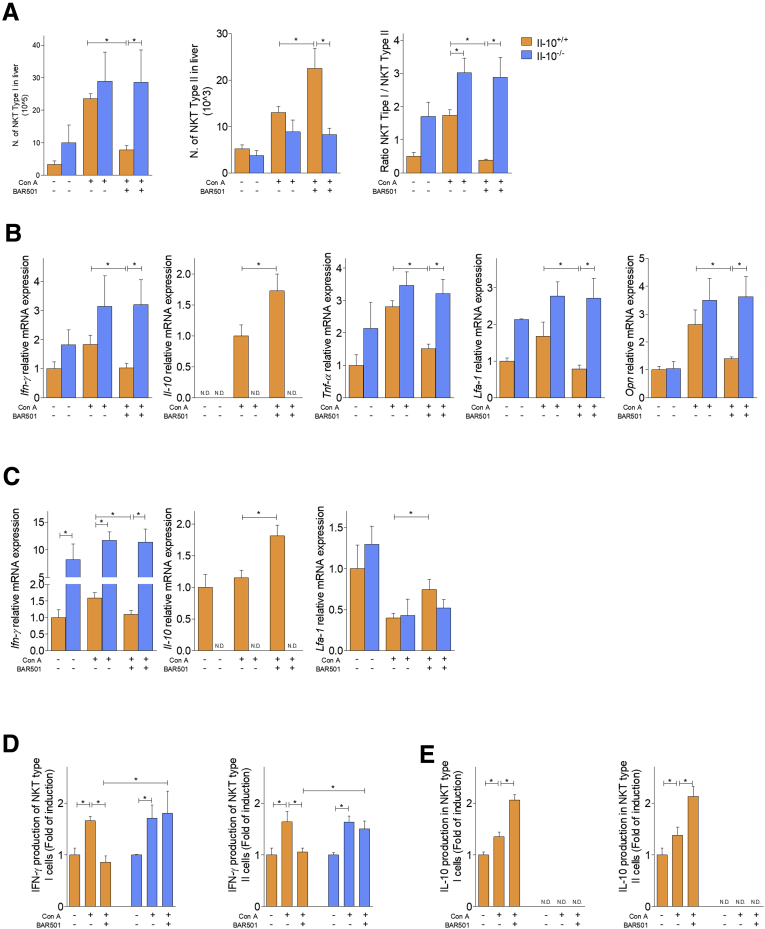
To gain insights on the role of GPBAR1, we have then characterized liver type I and type II NKT cells in wild-type and GPBAR1–/– mice at steady state and in response to Con A (Figure 8).21, 24, 25, 26, 27, 28 Treating mice with Con A increased the number of both type I and II NKT cells, while BAR501 modulated the number of these subpopulations in a opposite way (Figure 8A, left and middle panels). Indeed, while BAR501 reduced the number of IFN-γ+, type I, NKT cells, it increased the number of IL-10+, type II, NKT cells. Accordingly, while the relative ratio of type I/type II NKT cells (a ratio between pro- and anti-inflammatory NKT subsets) increased in response to Con A, this proinflammatory polarization was reversed by GPBAR1 agonism (Figure 8A, right panel).
Figure 8.
RNA sequencing of type I and type II NKT cells. Acute hepatitis was induced in C57BL6 male mice GPBAR1+/+ by intravenous injection of Con A (15 mg/kg) and they were sacrificed 24 hours later. Mice were randomized to receive Con A alone or in combination with BAR501 (30 mg/kg) daily from 3 days before induction of hepatitis to the time of the sacrifice. type I and type II NKT cells were purified from the liver of mice with Con A–induced hepatitis, treated or untreated with BAR501. (A) Number of type I NKT (left panel), type II NKT cells (middle panel), and ratio between the number of NKT types I and type II NKT cells (right panel). RNA-seq analysis results in (B) NKT Type I and (C) NKT Type II cells purified from livers of NT, Con A and Con A + BAR501 mice. (B, C) Scatter plots of average gene expression, showing the comparison results of the RNA-seq data with a TAC software analysis; Venn diagram of the number of the differentially expressed genes showing the overlapping regions (identified as ABC, AC, AB and BC sets) between the 3 experimental groups of purified NKT Type I and Type II cells, respectively (Fold Change <-1.5 or >+1.5, p value <0.05). (D) Venn diagrams obtained combining the intersections ABC + AC of NKT Type I and NKT Type II cells, obtained from previous diagrams; from this combination comes out a list of 80 genes (AB), differentially expressed both in NKT Type I and in NKT Type II cells. Clustered heat map of 80 genes (AB) in (E) NKT Type I and (F) NKT Type II cells respectively, with a magnification that highlights a subset of genes including IL-10.
To further elucidate the effect of GPBAR1 on regulation of liver NKT cells, type I and type II NKT cells isolated from the livers of wild-type mice were subjected to RNAtype I and type II NKT analysis. As illustrated in Figure 8, and Supplementary Table 1, exposure to Con A modulated a wide array of genes in both type I and II NKT cells. Up to 6637 gene transcripts were found to be differentially expressed by type I NKT cell isolated from Con A–treated mice in comparison with naïve mice. Treating Con A mice with BAR501 partially reversed this pattern, resulting in 2156 differentially expressed transcripts, thought that the treatment failed to restore the baseline setting, and up to 7717 genes were found to be differentially expressed by type I NKT cells isolated from Con A mice treated with BAR501 in comparison with naïve type I NKT cells (Figure 8B). Similarly, exposure to Con A resulted in a differential expression of 4896 genes in type II NKT cells in comparison with naïve cells; and, while exposure to the GPBAR1 agonist lead to 2472 differentially expressed transcripts in comparison with Con A alone, up to 5313 genes were still differentially expressed in comparison with naïve mice (Figure 8C). By Venn diagram analysis we isolated 2 gene subsets indicated as “AC” and “ABC” in Figure 8B and C, that were differentially modulated by GPBAR1 agonism in comparison with either naïve or Con A treated mice (Supplementary Table 1). Because the functional effects of GPBAR1 agonism on liver NKT cells lie in this intersection, we have then compared the transcriptome recorded in the “AC” and “ABC” areas in the 2 NKT cell subpopulations. As illustrated in Figure 8D–F and Supplementary Table 1, this comparison gives rise to cluster of 80 genes that were either up- or –downregulated by GPBAR1 agonism in the type I and type II NKT cells. This cluster is bona fide the best representation of the pharmacological effects of GPBAR1 agonism on the transcriptome of Con A–treated cells. As shown by hierarchical cluster analysis (Figure 8E and F), several genes relevant for NKT cells regulation were included in this cluster, including IL-10 and Cd86 among others, but the IL-10 gene was the one with the highest level of regulation in both type I and type II NKT cells (Figure 8E and F and Supplementary Table 1 for the complete list).
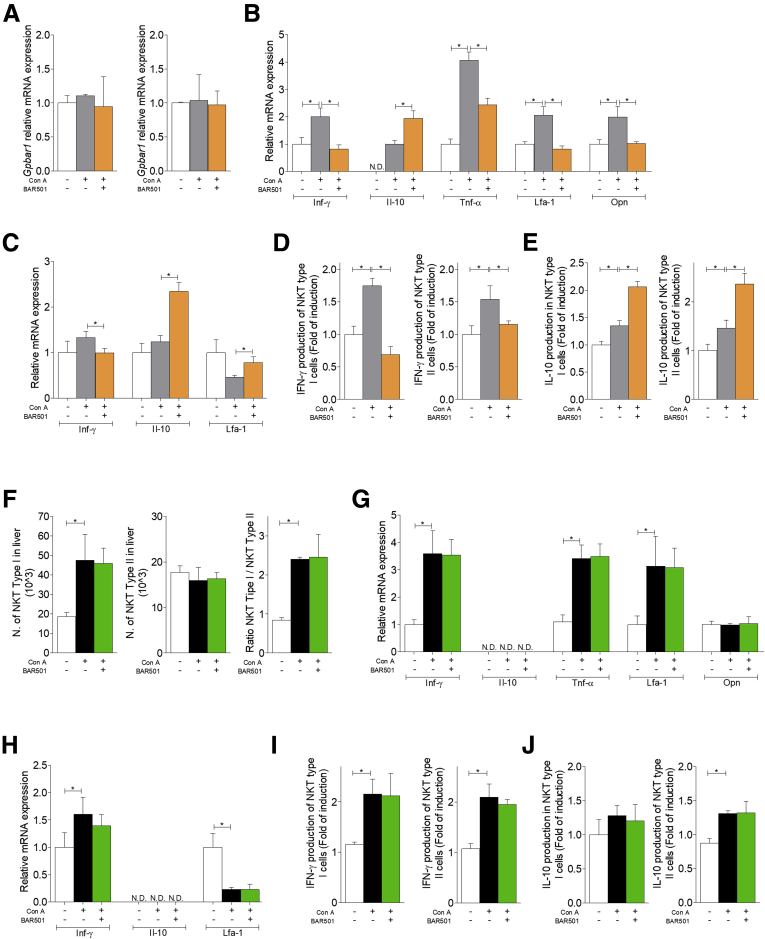
We have therefore confirmed these data by reverse-transcription polymerase chain reaction (RT-PCR) (Figure 9). First, we have confirmed that both type I NKT and type II cells express GPBAR1 (Figure 9A). The subsequent analysis demonstrated that exposure to Con A induced a dramatic polarization of type I NKT cells toward an IFN-γ biased, proinflammatory phenotype. This inflammatory phenotype was reversed by BAR501, which also promoted a robust up-regulation of IL-10 gene expression (Figure 9B). In contrast, we have been unable to detect any expression of IL-4, IL-13, IL-17, and FoxP3 genes indicating that type I NKT subtypes, NKT2, NKT17, and FoxP3+NK regulatory T cells, were not represented in the liver.3, 4, 5, 6, 29 Additionally, while signature markers for type I NKT cells (ie, TNF-α, LFA-1, and OPN) were increased by Con A, BAR501 reversed this pattern (Figure 9B).
Figure 9.
Effects of BAR501 on NKT cells in GPBAR1+/+and GPBAR1–/–mice. Acute hepatitis was induced in C57BL6 male mice GPBAR1+/+ and GPBAR1–/– by intravenous injection of Con A (15 mg/kg) and they were sacrificed 24 hours later. Mice were randomized to receive Con A alone or in combination with BAR501 (30 mg/kg) daily from 3 days before induction of hepatitis to the time of the sacrifice. type I and type II NKT cells were purified from the liver of mice with Con A–induced hepatitis, treated or untreated with BAR501. In GPBAR1+/+ mice real-time PCR analysis of (A) GPBAR1 expression in type I (left panel) and type II (right panel) NKT cells. Real-time PCR analysis of (B) IFN-γ, IL-10, TNF-α, LFA-1, and OPN expression in type I NKT cells. Real-time PCR analysis of (C) IFN-γ, IL-10, and LFA-1 expression in type II NKT cells. ELISA test to evaluate IFN-γ and IL-10 production by NKT cells, were performed on culture supernatants. Data shown are (D) IFN-γ production by type I and type II NKT cells; (E) IL-10 production by type I and type II NKT cells. The data of GPBAR1–/– mice shown (F) number of type I (left panel) and type II cells (middle panel) NKT cells, and ratio between the number of type I and type II NKT cells (right panel). Real-time PCR analysis of (G) IFN-γ, IL-10, TNF-α, LFA-1, and OPN expression in type I NKT cells. Real-time PCR analysis of (H) IFN-γ, IL-10, and LFA-1 expression in type II NKT cells. ELISA test to evaluate IFN-γ and IL-10 production by NKT cells, were performed on culture supernatants. Data shown are (I) IFN-γ production by type I and type II NKT cells; (J) IL-10 production by type I and type II NKT cells. Results are the mean ± SEM of 10 points per group. *P < .05.
Characterization of type II NKT cells confirmed that in the mice livers this NKT subset is relatively poorly represented,21, 24, 25, 26, 27, 29 but is regulatory in nature. Accordingly, naïve type II NKT cells expressed IL-10 gene and reacted to Con A by a minor induction of expression of IFN-γ mRNA (Figure 9C). Furthermore, exposure to BAR501 induced a large increase in IL-10 gene expression (Figure 9C). These findings were confirmed at protein level (Figure 9D and E). Finally, analysis of the phenotype of type I and II NKT cells isolated from GPBAR1–/– mice (Figure 9F–J), demonstrated that GPBAR1 gene ablation, abrogated protection exerted by BAR501 and resulted in a phenotype of type I and type II NKT cells that was dramatically biased toward IFN-γ, while IL-10 mRNA was not detectable.
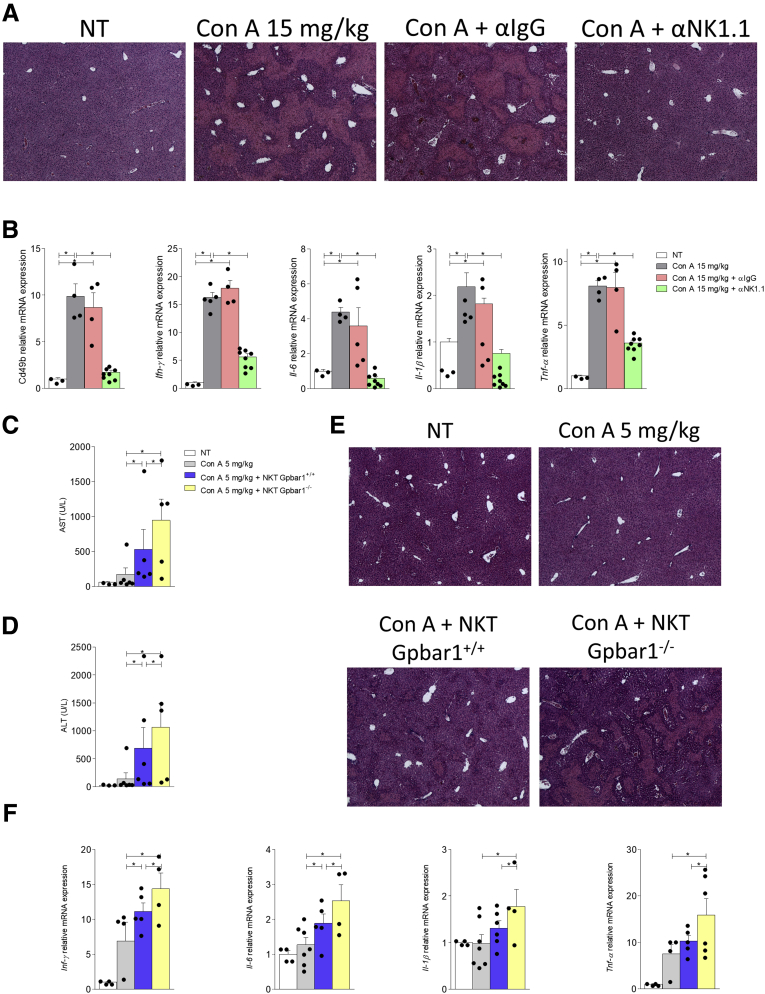
To further confirm the involvement of NKT cells in this model, we have administered the Con A to mice that were depleted of their NK and NKT cells by preliminary treatment with α-NK1.1 antibody. The results of these investigations, shown in Table 3 and Figure 10A and B, demonstrated that pretreatment with α-NK1.1 antibody abrogated the development of liver injury caused by Con A.
Table 3.
Plasmatic Levels of AST and ALT in Each Group
| NT | 24 h |
|||
|---|---|---|---|---|
| Con A | Con A + α-IgG | Con A + α-NK1.1 | ||
| AST | 46.67 ± 4.005 | 3154 ± 432.5 | 2734 ± 773.3 | 76.25 ± 3.899 |
| ALT | 26.00 ± 2.00 | 3410 ± 230.9 | 2797 ± 732.5 | 36.75 ± 5.331 |
Values are mean ± SD.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; Con A, concanavalin A; NT, not treated.
Figure 10.
The pathogenetic role of NKT cells in concanavalin A-acute hepatitis. C57BL6 male mice have been pretreated with α-NK1.1 or α-IgG antibody (as a negative control) 200 μg/topo intraperitoneally the day before hepatitis induction. Then hepatitis was induced by the administration of Con A at the concentration of 15 mg/kg intravenously and the mice were sacrificed 24 hours later. (A) Histological evaluation, performed with hematoxylin and eosin staining, of necrosis area on liver sections of mice treated with Con A alone or in combination with α-NK1.1 or α-IgG antibody. Total RNA extracted from liver was used to evaluate by quantitative real-time PCR the relative mRNA expression of (B) proinflammatory genes Cd49b, IFN-γ, IL-6, IL-1β, and TNF-α. The data are normalized to GAPDH mRNA. To determine whether the difference in the severity of the hepatitis developed after the administration of Con A in GPBAR1+/+and GPBAR1–/– mice could depend on the absence of the GPBAR1 receptor in NKT cells, we performed a cell transfer experiment of liver-purified NKT cells obtained from GPBAR1+/+ and GPBAR1–/– 24 hours after the treatment with Con A 15 mg/kg. Subsequently, we transferred 4 × 106 NKT cells resuspended in a saline solution with Con A, administered at a dose of 5 mg/kg. Data shown are (C, D) plasmatic levels of AST and ALT for each experimental group. (E) Histological evaluation, performed with hematoxylin and eosin staining, of necrosis area on liver sections. Total RNA extracted from liver was used to evaluate by quantitative real-time PCR the relative mRNA expression of (F) proinflammatory genes IFN-γ, IL-6, IL-1β, and TNF-α. Each point represents a single mouse. *P < .05.
The pretreatment with α-NK1.1 antibody, as expected, reduced the expression of Cd49b in the liver, a markers of NK and NKT cells, and consequently reduced the expression of proinflammatory cytokines IFN-γ, IL-6, IL-1β, and TNF-α.
To tight the role of GPBAR1 to the NKT population, we have carried on a cell transfer experiment using liver-purified NKT cells obtained from GPBAR1+/+ and GPBAR1–/– 24h after the treatment with 15 mg/kg Con A. For these purposes liver-derived 4 × 106 NKT cells were resuspended in a saline solution with Con A and administered intravenously in combination with 5 mg/kg Con A (a dose of Con A that does not elicit acute liver injury as shown in Figure 3A and B), into wild-type C57BL/6NCrl recipient mice. The data of AST and ALT (Figure 10C and D) and histology analysis of livers of the recipient mice, confirmed that while 5 mg/kg Con A did not cause any liver damage the transfer of NKT cell from Con A challenged mice resulted in a severe liver injury that was significantly exacerbated by GPBAR1 gene ablation (Figure 10 C–F). The RT-PCR analysis of liver proinflammatory biomarkers (Figure 10F) IFN-γ, IL-6, IL-1β, and TNF-α confirmed that mice that were transplanted with GPBAR1–/– NKT cells expressed the highest levels of these cytokines. Together, these data support a regulatory role for GPBAR1 in NKT cells (Figure 4).
Regulatory Effects of GPBAR1 Agonism Are IL-10 Dependent
Because the previously mentioned data strongly indicate that beneficial effects exerted by GPBAR1 activation correlates with induction of a subtype of IL-10–biased NKT cells (NKT10), we have investigated whether IL-10 was required by GPBAR1 in order to modulate the response of NKT cells. For this purpose, we challenged IL-10–/– mice with Con A with or without administration of BAR501. Analysis of AST and ALT plasma levels shown in Table 4, demonstrated that development of liver injury was dramatically exacerbated in IL-10–/– mice and the protective effect of BAR501 was lost in these mice.
Table 4.
Plasmatic Levels of AST and ALT in IL-10+/+ and IL-10–/– Mice
| IL-10+/+ |
IL-10–/– |
|||||
|---|---|---|---|---|---|---|
| NT | Con A | Con A + BAR501 | NT | Con A | Con A + BAR501 | |
| AST | 62 ± 11.34 | 3000 ± 578.8 | 1570 ± 240.8 | 55 ± 3.109 | 5909 ± 1193 | 5732 ± 1216 |
| ALT | 30.67 ± 3.676 | 3521 ± 719.1 | 1910 ± 265.7 | 28.5 ± 1.500 | 6793 ± 1229 | 6628 ± 981.5 |
Values are mean ± SD.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; Con A, concanavalin A; IL, interleukin; NT, not treated.
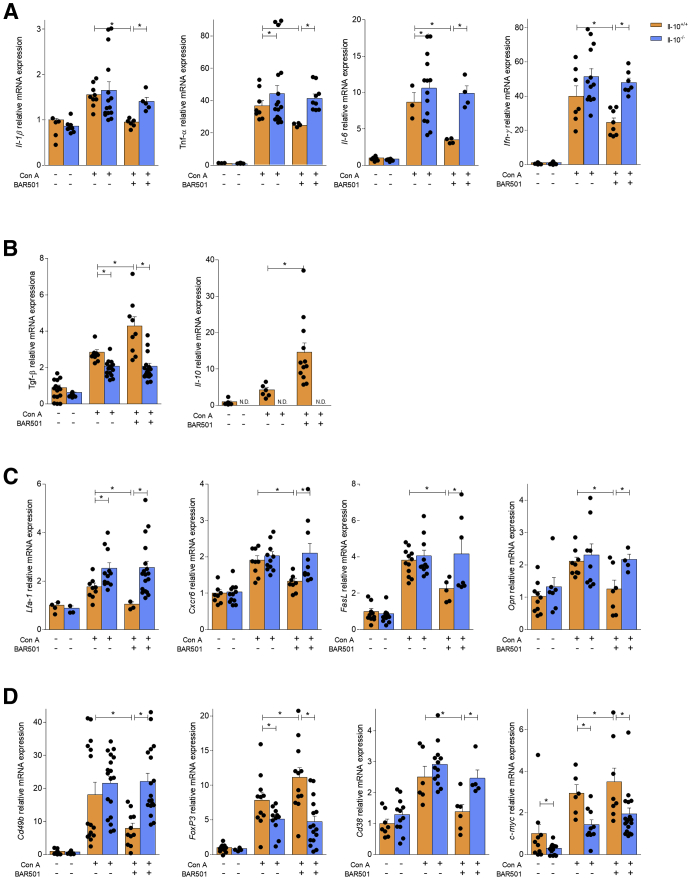
At histopathology analysis, we found that, in comparison with wild-type mice, IL-10–/– mice had a severe disease that was not reversed by BAR501 (data not shown). These findings were confirmed by analysis of pro- and anti-inflammatory biomarkers by RT-PCR (Figure 11A–D).
Figure 11.
IL-10 is required by GPBAR1 to modulate the response of NKT cells. Acute hepatitis was induced in C57BL6 male mice, IL-10+/+ and IL-10–/–, by intravenous injection of Con A (15 mg/kg) at 8 hours. Mice were randomized to receive Con A alone or in combination with BAR501 (30 mg/kg) daily from 3 days before induction of hepatitis to the time of the sacrifice. Total RNA extracted from liver was used to evaluate by quantitative real-time PCR the relative mRNA expression of (A) proinflammatory genes IL-1β, TNF-α, IL-6, and IFN-γ; (B) anti-inflammatory genes TGF-β and IL-10; (C) adhesion molecules CXCR6 and LFA-1, FasL, and OPN; and markers specific for different immune cells populations such as (D) Cd49b, FoxP3, Cd38, and c-Myc. The data are normalized to GAPDH mRNA. Each point represents a single mouse. *P < .05.
To further dissect the GPBAR1/IL-10 axis, we have then investigated how BAR501 modulates type I and type II NKT cells in IL-10–/– mice challenged with Con A (Figure 12). As expected and similarly to GPBAR1–/– mice, IL-10–/– mice at steady state had a higher number of type I NKT cells and relatively low number of type II NKT cells in comparison with their wild-type counterparts (Figure 12A) and reacted to Con A administration by a dramatic exacerbation of this pattern. Furthermore, similarly to GPBAR1–/– mice, also in IL-10–/– mice, BAR501 failed to reverse this pattern, neither the GPBAR1 agonism modulated the expression of IFN-γ, IL-10, TNF-α, LFA-1, and OPN in type I and type II NKT cells (Figure 12B and C). These data were confirmed by the analysis of cytokines secretion in cell supernatants (Figure 12D and E).
Figure 12.
Effects of BAR501 on NKT cells in IL-10+/+and IL-10–/–mice. Acute hepatitis was induced in C57BL6 male mice, IL-10+/+ and IL-10–/–, by intravenous injection of Con A (15 mg/kg) at 8 hours. Mice were randomized to receive Con A alone or in combination with BAR501 (30 mg/kg) daily from 3 days before induction of hepatitis to the time of the sacrifice. type I and type II NKT cells were purified from the liver of mice with Con A–induced hepatitis, treated or untreated with BAR501. (A) Number of type I (left panel) and type II (middle panel) NKT cells and ratio between the number of type I and type II NKT cells (right panel). Real-time PCR analysis of (B) IFN-γ, IL-10, TNF-α, LFA-1, and OPN expression in type I NKT cells. Real-time PCR analysis of (C) IFN-γ, IL-10, and LFA-1 expression in type II NKT cells. Enzyme-linked immunosorbent assay test to evaluate IFN-γ and IL-10 production by NKT cells, were performed on culture supernatants. Data shown are (D) IFN-γ production by type I and type II NKT cells; (E) IL-10 production by type I and type II NKT cells. Results are the mean ± SEM of 6 points per group. *P < .05.
GPBAR1 Protects Against Chronic Hepatitis Induced by Carbon Tetrachloride
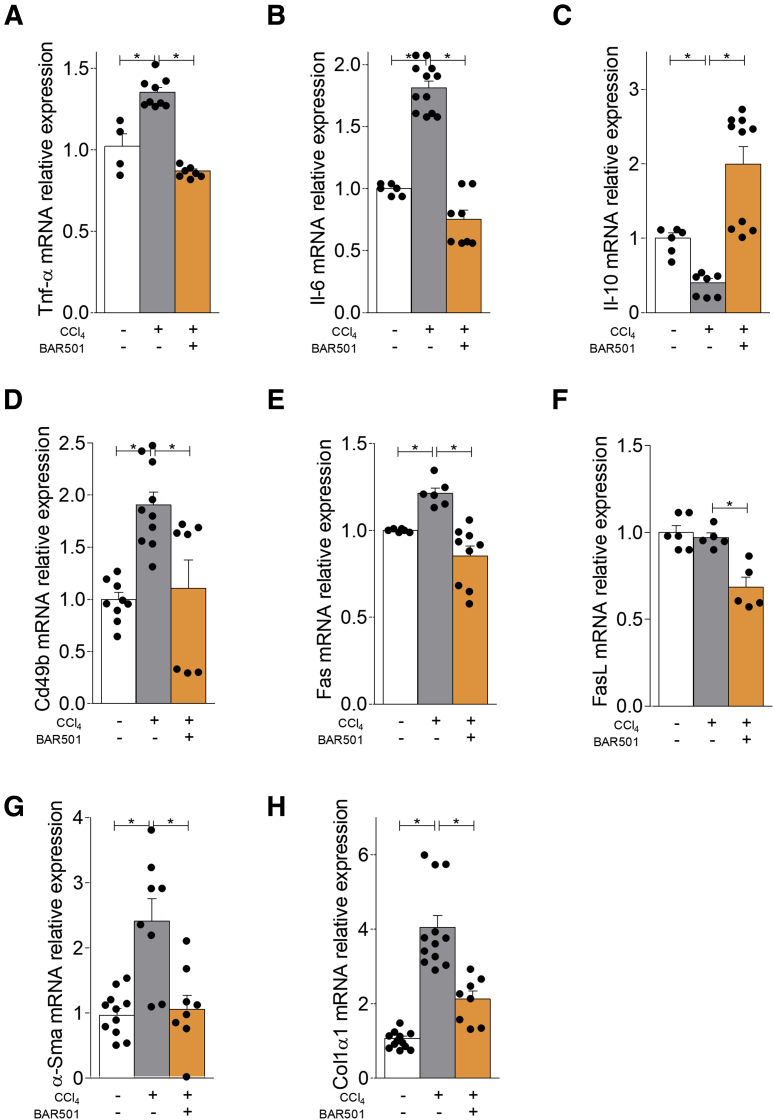
NKT cells are thought to contribute to initiation and maintenance of liver inflammation and progression to fibrosis. To investigate the role of GPBAR1 in a liver model of chronic inflammation, we investigated the effects exerted by BAR501 in mice rendered cirrhotic by carbon tetrachloride (CCl4).30 Results of this investigation demonstrated that while exposure to CCl4 increased the liver expression of TNF-α (Figure 13A) and IL-6 (Figure 13B), and reduced the expression of the IL-10 mRNA (Figure 13C), this pattern was reversed by treating mice with BAR501 (Figure 13C). Furthermore, the activation of GPBAR1 reduced the liver expression of Cd49b (Figure 13D) and Fas and FasL (Figure 13E and F) indicating a lower influx of activated NK and NKT cells. Additionally, while exposure to CCl4 increased the expression of α-SMA and Col1α1 mRNA (Figure 13G and H), this profibrotic pattern was completely reversed by BAR501 (Figure 13G and H).
Figure 13.
BAR501 administration protects against chronic liver injury induced by CCl4modulating the expression of pro- and anti-inflammatory markers. Chronic hepatitis was induced in C57BL6 male mice by intraperitoneal injection of 500 μL/kg body weight of CCl4 in an equal volume of paraffin oil twice a week for 9 weeks. Mice were randomized to receive CCl4 alone or in combination with BAR501 (30 mg/kg) daily. Total RNA extracted from liver was used to evaluate by quantitative real-time PCR the relative mRNA expression of (A, B) proinflammatory genes TNF-α and IL-6; (C) anti-inflammatory gene IL-10, markers for NK and NKT cells; (D–F) Cd49b, Fas, and FasL, and markers of fibrosis; and (G, H) α-SMA and col1α1. The data are normalized to GAPDH mRNA. Each point represents a single mouse. *P < .05.
Discussion
Primary bile acids are steroid hormones generated by cholesterol breakdown in the liver and further metabolized by the intestinal microbiota to generate secondary bile acids.31 Both primary and secondary bile acids exert homeostatic functions by activating a group of cell membrane and nuclear transcription factors, collectively known as bile acid activated receptors.14, 32
Here, we report that similarly to other cells of innate immunity (specifically monocytes and macrophages), NKT cells express GPBAR1 a receptor for secondary bile acids. Thus, in a broad manner, GPBAR1 could be considered a member of the relatively small family of G protein–coupled receptors functioning as extranuclear receptors for steroids, whose prototype is the G protein–coupled estrogen receptor 1 (also known as GPR30).33 These receptors mediate the nongenomic effects exerted by their steroidal ligands, thought that GPBAR1 also exerts genomic effects.14
Over the years evidence accumulated that GPBAR1 is expressed by nonparenchymal liver cells, sinusoidal endothelial cells and resident macrophages, and by both circulating monocytes and intestinal and liver resident macrophages,34 and its ablation in mice results in an inflammation-prone phenotype, characterized by increased susceptibility to develop severe inflammation in response to experimental colitis.35, 36 Although loss of function single-nucleotide polymorphisms of GPBAR1, have been identified in patients with primary sclerosing cholangitis and ulcerative colitis, the role of this receptor in regulating liver immunity is poorly understood.
NKT cells are placed at the interface between the innate and adaptive immunity and represent the most abundant lymphocyte subset in the mice (40%) and human (10%) livers.7 NKT cells are known for exerting both proinflammatory and regulatory roles and are considered pathogenic in liver immune-mediated disorders such autoimmune hepatitis, primary sclerosing cholangitis, and primary biliary cholangitis, and in rodent models of nonalcoholic steatohepatitis.37, 38, 39, 40 However, akin to cells of the innate immune system (eg, neutrophils, macrophages, dendritic cells, NK cells), NKT cells become rapidly activated after agonist recognition or binding by regulatory mediators in a TCR-independent manner, releasing robust amounts of effector cytokines and chemokines. Here, we have provided evidence that GPBAR1 is expressed by and modulate the polarization of both type I and type II NKT cells toward a regulatory phenotype which is dominated by the expansion of NKT10 cells and increased production of IL-10 by type II NKT cells. However, it should be considered that the relevance of NKT cells in developing of liver autoimmune disorders in clinical settings, is only partially demonstrated and there is evidence that mucosal-associated invariant T cells (ie, innate-like T cells characterized by an evolutionarily conserved semi-invariant TCR consisting of an invariant α chain [Vα7.2-Jα33/Ja20/Jα12 in humans] and varying β chains) could contribute to the development of autoimmune diorders.41
The models of acute hepatitis induced by α-GalCer and Con A are driven by an acute expansion of IFN-γ producing cells, that are characterized as NKT1 cells. Here, we have confirmed these finding by demonstrating that, not only isolating type I NKT cells in Con A treated mice by α-GalCer:CD1d complex resulted in a cell population that was highly biased toward production of IFN-γ, but analysis of markers for other NKT subsets (ie, IL-4 [NKT2], IL17 [NKT17], and FoxP3 [NKTFoxp3]) give raise to negative results. The effects exerted by Con A on the NKT1 subset was significantly enhanced by GPBAR1 gene ablation. Thus, not only was the number of NKT1 cells recovered in the liver of GPBAR1–/– mice challenged with Con A significantly higher than that of their wild-type counterparts, but these cells were more activated and secreted higher amount of IFN-γ, their signature cytokine. The fact that BAR501, a GPBAR1 ligand, effectively curtailed the NKT1 response, further confirmed a role for GPBAR1 in balancing pro- and anti-inflammatory signals that control the differentiation of type I NKT cells.
Several mechanisms might account for the beneficial effects exerted by GPBAR1 in regulating type I NKT cell differentiation toward a NKT10 phenotype. First of all, we have observed that exposure to BAR501 markedly reduced the liver and cells expression of CXCR6 and LFA-1, 2 adhesion molecules that have been shown essential for invariant NKT cells (by large the prevalent phenotype of type I NKT), to reach the liver, crawl within hepatic sinusoids and to stop upon T cell antigen receptor activation. In addition, GPBAR1 agonism might function as a negative regulator of effector mechanisms in NKT1 cells by inhibiting effector mechanisms as demonstrated in macrophages23 and as suggested in the present report by the reduced production of IFN-γ and TNF-α by type I NKT cells in ex vivo assay (Figure 6). Finally, GPBAR1 might act by driving type I NKT differentiation toward an alternative, regulatory phenotype, that, in its turn, redirects differentiation and function of type I NKT cells. Supporting the later explanation, we have shown that GPBAR1 is essential to produce a significant number of NKT10 cells in the Con A model, and that GPBAR1 activation increases the number of NKT10 cells in the liver along with the expression of IL-10.5
NKT10 cells are found at low frequency in naïve mice and in human peripheral blood mononuclear cells, but expand greatly in response to immune activation with α-GalCer.5 The origin of NKT10 cells is controversial. Thought that were originally considered to represent a sort of late, anergic phenotype, of α-GalCer–primed type I NKT cells. However, a growing body evidence, including the present report, demonstrate that appearance of NKT10 cells in mice could be elicited in models of inflammation that are α-GalCer independent. Further on, we have shown that the number of NKT10 cells in the liver increases dramatically in response to GPBAR1 agonism. Together with the fact that this response could not be elicited in GPBAR1–/– mice, these results strongly support a role for this receptor in the development of NKT10 cells in the liver.
The mechanism that mediates the beneficial effects of GPBAR1, is essentially due to its ability to regulate IL-10.23 Thus, GPBAR1 agonism, increases dramatically IL-10 production from DN32.D3 cells, a NKT cell line. In these cells, BAR501 elicited a potent regulatory response characterized a genomic regulation of IL-10 generation, as demonstrated by ChIP assay,23 along with a downregulation of IFN-γ, IL-4, IL-17, and TNF-α and CXCR6, LFA-1, and FasL. In addition, in vivo experiments and RNAtype I and type II NKT analysis demonstrated that GPBAR1 activation by BAR501, resulted in a robust regulation of IL-10 gene expression. Additionally, in sharp contrast with their wild-type counterparts, type I NKT cells isolated from GPBAR1–/– mice challenged with Con A, were characterized by undetectable amount of IL-10 mRNA. Finally, IL-10 deficient mice, not only developed a severe disease in response to Con A, but were resistant to treatment with BAR501, and type I NKT cells obtained from these mice were severely biased toward an IFN-γ–prone NKT1 phenotype. Importantly, these effects could not be reversed by GPBAR1 agonism.
The essential role of IL-10, in mediating the beneficial effects of GPBAR1 agonism, was further confirmed by analysis of type II NKT cells. This cell type, is significantly less characterized than their type I counterparts, and are also significantly less represented in the liver and systemic circulation. We have found that in wild-type mice challenged with Con A, GPBAR1 agonism increased the number of type II NKT cells and that this effect was lost in the GPBAR1–/– mice. Additionally, functional characterization of these cells demonstrated that BAR501 effectively redirect their polarization toward an IL-10 biased phenotype, as indicated by increased production of IL-10 in ex vivo assay. Finally, IL-10–/– mice were characterized by reduced number of type II NKT cells that were also severely biased toward IFN-γ generation.
In summary, we have provided evidence that GPBAR1, a receptor for secondary bile acids, is essential to regulate type I and II NKT polarization in the liver and development of tolerogenic phenotype, suggesting that his pathway could be exploited for treating human liver autoimmune disorders.
Materials and Methods
Cell Culture
DN32.D3 cells were cultured in Dulbecco’s modified Eagle medium supplemented with 5% fetal bovine serum, 1% antibiotics (penicillin and streptomycin) and 1% L-glutamine at 37°C in a humidified air atmosphere with 5% CO2. Cells were trypsinized, counted with a Burker chamber and plated (150.000 cells/well). The day of the experiment, the cells were treated with Con A or Con A in combination with 4 different concentrations of BAR501 (1, 5, 10 and 50 μM) and then left in incubation for 18 hours. At the end of the incubation period, the cells were detached and collected in a 1.5 ml microtube.
Cells Proliferation Assay
Briefly, DN32.3D were plated in 96-well tissue culture plate (final cell density: 5 × 104 cells/well) in 100 μL of RPMI 1640 medium supplemented with 10% fetal bovine serum, 1% L-glutamine and 1% penicillin and streptomycin at 37°C and 5% CO2, and incubated 16 hours with Con A 1 μg/mL alone or in combination with 4 different concentrations of BAR501 (1, 5, 10, and 50 μM), or only with vehicle. After the 18-hour incubation period, CellTiter96 Aqueous One Solution was added (20 μL/well) and incubated for another 4 hours at 37°C and 5% CO2. The plate was then read at 490 nm. The background readings in the wells with medium were subtracted from the sample well read-outs.
Immunohistochemistry
DN32.D3 cells were transferred to polarized microscope slides (Bio-Optica, Milano, Italy) using the Cytospin 2 (The Design Council). Cells were air dried for 5 minutes and then fixed for 15 minutes at room temperature with a 4% paraformaldehyde in phosphate-buffered saline (PBS) solution (Chem Cruz, Huissen, Netherlands). Cells were washed twice and incubated for 45 minutes at room temperature with a 5% BSA solution (Sigma, Darmstadt, Germany). Cells were then incubated for 2 hours at room temperature with primary antibody against TGR5 (Novus Biological, Centennial, CO), diluted 1:100 in 5% bovine serum albumin blocking solution (Sigma). Cells were then washed 3 times with PBS and incubated again for 1 hour at room temperature with the secondary antibody goat anti-rabbit f(ab)2 (H+L) AlexaFluor 488 (Thermo Fisher Scientific, Waltham, MA) diluted 1:250 in 5% BSA blocking solution. Furthermore, cells were washed 3 times with PBS and incubated with Hoechst 33342 trihydrochloride trihydrate (Thermo Fisher Scientific) and finally washed twice with PBS. Cells were finally sealed with Prolong Diamond mounting medium (Thermo Fisher Scientific) and a dedicated 1.5-mm microscopy glass coverslip. Images were captured using an Axio Observer.Z1 microscope, equipped with ApoTome 2 and AxioCam MRm camera (Carl Zeiss Microscopy, Zaventem, Belgium). A 63×/0.75 plan NEOFLUAR objective was used to capture images with ApoTome enabled at 15 phases, covering the entire thickness of the cells with Z-axis, 0.30-μM spaced focal planes. Images were later deconvolved using the ZEN software built-in plugin (Carl Zeiss Microscopy) and cleaned stacks of images were finally used to realize extended depth of focus flat projections (maximum projection method).
Western Blotting
DN32.D3, Raw, and HepG2 cells were lysed in E1A lysis buffer (250-mM NaCl, 50-mM HEPES pH 7.0, 0.1% NP40, 50-mM EDTA) containing phosphatase and protease inhibitors cocktail; aliquots from each sample containing equal amounts of protein (80 μg) were run on a 15% sodium dodecyl sulfate–polyacrylamide gel and transferred to a nitrocellulose membrane. The blot was subsequently blocked for 1 hour with 5% milk powder in Tris-buffered saline /Tween 20 at room temperature and then probed overnight (at 4°C) with a rabbit polyclonal antibody recognizing TGR5 (1:2000; Thermo Scientific) and mouse monoclonal anti-αTubulin (1:10000; Sigma-Aldrich). After overnight incubation, appropriate horseradish peroxidase-labeled secondary antibody, at a dilution of 1:10,000, were used. Positive signals were developed by Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore Darmstadt, Germany) and analyzed with ImageJ 1.48v (National Institutes of Health, Bethesda, MD).
Animals and Hepatitis Protocols
C57BL6 mice were from Charles River. GPBAR1 null mice on C57BL/6NCrl background, and their C57BL/6NCrl congenic littermates were originally donated by Dr Galya Vassileva (Schering-Plough Research Institute, Kenilworth, NJ).42 IL-10 null mice (B6.129P2-Il10tm1Cgn/J) were from the Jackson Laboratory. The colonies were maintained in the animal facility of University of Perugia. Mice were housed under controlled temperatures (22°C) and photoperiods (12-hour light/dark cycle), allowed unrestricted access to standard mouse chow and tap water and allowed to acclimate to these conditions for at least 5 days before inclusion in an experiment. The study was conducted in agreement with the Italian law and the protocol was approved by an ethical committee of University of Perugia and by a National Committee of Italian Ministry of Health permit no. 214/2017-PR. The health and body conditions of the animals were monitored daily by the Veterinarian in the animal facility. Only male mice were used in each experiment. Hepatitis was induced in C57BL6 mice, GPBAR1+/+, and GPBAR1–/– mice, and in IL-10–/– mice by intravenous injection of Con A, 15 mg/kg, or α-GalCer (3 μg/300 μL/mice).43 In experiments in which hepatitis was induced by Con A or α-GalCer, BAR501, 6β-ethyl-3a,7b-dihydroxy-5b-cholan-24-ol,44 was administrated orally at different doses (5, 15, or 30 mg/kg), daily for 3 days before induction of hepatitis to the time of the sacrifice.23 A dose finding study is shown in Figure 3. At the end of the experiment, surviving mice were sacrificed, blood samples collected by cardiac puncture and the liver and spleen collected and weighed.
To study the role of NK and NKT cells in the pathogenesis of hepatitis we pretreated C57BL6 male mice with α-NK1.1 or α-IgG antibody (as a negative control) 200 μg/topo intraperitoneally the day before hepatitis induction. Then hepatitis was induced by the administration of Con A at the concentration of 15 mg/kg intravenously and the mice were sacrificed 24 hours later. We also performed an NKT cell transfer experiment. In this experimental set we treated donor GPBAR1+/+ and GPBAR1–/– mice with Con A 15 mg/kg. Donor mice were sacrificed 24 hours after hepatitis induction and liver NKT cells were purified. Subsequently, we transferred 4 × 106 NKT cells resuspended in a saline solution with Con A (5 mg/kg, dose not able to induce acute hepatitis alone as shown in the dose-finding experiment of Con A), into 8-week wild-type C57BL/6NCrl recipient mice. The recipient mice were sacrificed 24 hours after cell transplantation, blood samples collected by cardiac puncture and the liver and spleen collected and weighed.
We have also carried out a model of chronic hepatitis induced by CCl4 administration. For this purpose, C57BL6 male mice were administered intraperitoneal 500 μL/kg body weight of CCl4 in an equal volume of paraffin oil twice a week for 9 weeks. CCl4 mice were randomized to receive BAR501 (30 mg/kg daily by gavage) or vehicle (distilled water).
Histology
Samples of liver were first fixed in buffered formalin, cut into 5-μm-thick sections (∼150 μm between each section, 4–8 per fragment per liver) and then stained with hematoxylin and eosin.
Reverse-Transcription PCR
Liver samples were immediately frozen in liquid nitrogen and stored at –80°C until used, were mechanically homogenated with the aid of a pestle, and the obtained materials were resuspended in 1 mL of Trizol (Thermo Fisher Scientific). The RNA was extracted according to the manufacturer's protocol. After purification from genomic DNA by DNase-I treatment (Thermo Fisher Scientific), 1 μg of RNA from each sample was reverse-transcribed using random hexamer primers with Superscript-II (Thermo Fisher Scientific) in a 20-μL reaction volume; 10-ng complementary DNA were amplified in a 20-μL solution containing 200 nM of each primer and 10 μL of SYBR Select Master Mix (Thermo Fisher Scientific). All reactions were performed in triplicate, and the thermal cycling conditions were as follows: 3 min at 95°C, followed by 40 cycles of 95°C for 15 seconds, 56°C for 20 seconds, and 72°C for 30 seconds, using a Step One Plus machine (Applied Biosystems, Foster City, CA). The relative mRNA expression was calculated accordingly to the ΔCt method. Primers used in this study were designed using the PRIMER3 (http://frodo.wi.mit.edu/primer3/) software using the National Center for Biotechnology Information database. Alternatively, for some genes the TaqMan probes and TaqMan GEX Master Mix (Thermo Fisher Scientific) were used. PRT-PCR primers used in this study were as follow (forward and reverse): GPBAR1 (for GGCCTGGAACTCTGTTATCG; rev GTCCCTCTTGGCTCTTCCTC), IFN-γ (for GCTTTGCAGCTCTTCCTCAT; rev ATCCTTTTGCCAGT), TNF-α (for CCACCACGCTCTTCTGTCTA; rev AGGGTCTGGGCCATAGAACT), IL-4 (for CCTCACAGCAACGAAGAACA; rev ATCGAAAAGCCCGAAAGAGT), IL-17 (for TCCAGAAGGCCCTCAGACTA; rev TGAGCTTCCCAGATCACAGA) IL-6 (for CTTCACAAGTCGGAGGCTTA; rev TTCTGCAAGTGCATCATCGT), IL-2 (for AGCAGGCCACAGAATTGAAA; rev AATGTGTTGTCAGAGCCCTTT) IL-1β (for GCTGAAAGCTCTCCACCTCA; rev AGGCCACAGGTATTTTGTCG), TGF-β (for TTGCTTCAGCTCCACAGAGA; rev TGGTTGTAGAGGGCAAGGAC), IL-10 (for CCCAGAAATCAAGGAGCATT; rev CTCTTCACCTGCTCCACTGC), FoxP3 (for TCTTCGAGGAGCCAGAAGAG; rev AGCTCCCAGCTTCTCCTTTT), CXCR6 (for GTCCATGCTCACTCTCACCT; rev GGCCCCAGATCTTCCACTTA), LFA-1 (for GCAGAACAAGAACCCCGATG; rev TCTTTGAACACGTGTGCCAC), Cd49 (for CCATTCGCACCAAGTACTCC; rev ATAGCCATCCAGGGACCTTC), OPN (for GCTTGGCTTATGGACTGAGG; rev CGCTCTTCATGTGAGAGGTG) and FasL (for ATAGCCAACCCCAGTACACC; rev GCGGTTCCATATGTGTCTTCC). The following TaqMan probes: Cd38 (Mm01220906_m1 Thermo Fisher Scientific), and c-Myc (Mm00487804_m1 Thermo Fisher Scientific) were also used.
Mouse Liver NKT Cells Purification
Nonparenchymal mouse liver cells were isolated from the liver using the “Liver Dissociation Kit” from Miltenyi Biotec (130-105-807) according to the manufacturer's instructions. type I NKT (NK1.1+CD3+ α-GalCer:CD1d complex +) and type II NKT (NK1.1+CD3+ α-GalCer:CD1d complex -) cells were purified from liver using first NK1.1+ iNKT Cell Isolation mouse Kit from Miltenyi Biotec (130-096-513), to obtain a positive selection of all NKT cells, and then a phycoerythrin (PE) anti-mouse α-GalCer:CD1d complex Antibody (BioLegend, L363) and Anti-PE MicroBeads (Miltenyi Biotec, 130-048-801) to separate the type I and type II NKT cells.
AmpliSeq Transcriptome
The analysis of type I and II NKT cells was carried out using an Ion S5 Sequencer with Torrent Suite Software v6. The analyses were performed with a range of <–1.5-fold and >+1.5-fold, using Transcriptome Analysis Console Software (version 4.0.1), certified for AmpliSeq analysis (Thermo Fisher Scientific). High-quality RNA was extracted from type I and type II NKT cells, purified from the liver of mice, using the PureLink RNA Mini Kit, according to the manufacturer's instructions. RNA quality and quantity were assessed with the Qubit RNA HS Assay Kit and a Qubit 3.0 fluorometer (Invitrogen, Carlsbad, CA) followed by agarose gel electrophoresis. Libraries were generated using the Ion AmpliSeq Transcriptome Mouse Gene Expression Core Panel and Chef-Ready Kit (Comprehensive evaluation of AmpliSeq transcriptome, a whole transcriptome RNAtype I and type II NKT methodology). Briefly, 10 ng of RNA was reverse transcribed with SuperScript Vilo complementary DNA Synthesis Kit before library preparation on the Ion Chef instrument. The resulting complementary DNA was amplified to prepare barcoded libraries using the Ion Code PCR Plate, and the Ion AmpliSeq Transcriptome Mouse Gene Expression Core Panel, Chef-Ready Kit, according to the manufacturer's instructions. Barcoded libraries were combined to a final concentration of 100 pM, and used to prepare Template-Positive Ion Sphere Particles to load on Ion 540 Chips, using the Ion 540 Kit-Chef. Sequencing was performed on an Ion S5 Sequencer with Torrent Suite Software v6. The analyses were performed with a range of range of <–1.5-fold and >+1.5-fold, using Transcriptome Analysis Console Software, certified for AmpliSeq analysis (Thermo Fisher Scientific).
Flow Cytometry
Flow cytometry analyses were carried out using a 3-laser standard configuration ATTUNE NxT (Life Technologies, Carlsbad, CA). Data was analyzed using FlowJo software (TreeStar) and the gates set using a fluorescence-minus-one controls strategy. Fluorescence minus one controls are samples that include all conjugated Abs present in the test samples except one. The channel in which the conjugated Ab is missing is the one for which the fluorescence minus one provides a gating control. The following monoclonal antibodies were used: CD45 Brillant Violet 711 (BioLegend, San Diego, CA 30-F11), CD11b PE-Cyanine7 (eBioscience, San Diego, CA M1/70), GR1 Brillant Violet 510 (BioLegend, RB6-8C5), Clec4F Affinity Purified Goat IgG (RD System, Minneapolis, MN), Donkey anti-goat IgG AlexaFlour 488 (Invitrogen), CD49b PE (eBioscience, DX5), CD3 PerCP-Cyanine 5.5 (eBioscience, 145-2C11), IFN-γ eFluor450 (eBioscience, XMG1.2) and IL-10 FITC (eBioscience, JES5-16E3).
Enzyme-Linked Immunosorbent Assay
The concentrations of IFN-γ and IL-10 on liver-derived type I and type II NKT cells supernatants were measured by enzyme-linked immunosorbent assay kits (Mouse IL-10 and IFN gamma ELISA Kit ready Set; Invitrogen; REF:BMS606TW0 and 88-7105-22 Invitrogen) at 24 hours postplating cells.
Statistical Analysis
The analysis of variance followed by nonparametric Mann-Whitney U test or a 2-tailed unpaired Student t test were used for statistical comparisons (P < 0.05) using the Prism 6.0 software (GraphPad Software, San Diego, CA).
Footnotes
Author Contributions S. Fiorucci conceived the study; M. Biagioli, C. Fiorucci, C. Di Giorgio and M. Magro carried out animal studies; A. Carino, S. Marchianò, and R. Roselli performed biochemical and molecular analyses; A. Carino, M. Biagioli, and S. Marchianò performed RNAseq; A. Carino, M. Biagioli, and S. Marchianò performed data analysis; A. Zampella provided BAR501; P. Scarpelli carried out immunohistochemistry; M. Biagioli and O. Bereshchenko performed and analyzed flow cytometry; S. Fiorucci, M. Biagioli, and E. Distrutti wrote the manuscript. All authors have read and approved the final version of the manuscript.
Conflicts of interest The authors declare no conflicts of interest.
Supplementary Material
References
- 1.Makino Y., Kanno R., Ito T., Higashino K., Taniguchi M. Predominant expression of invariant V alpha 14+ TCR alpha chain in NK1.1+ T cell populations. Int Immunol. 1995;7:1157–1161. doi: 10.1093/intimm/7.7.1157. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A., Savage P.B., Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 3.Engel I., Seumois G., Chavez L., Samaniego-Castruita D., White B., Chawla A., Mock D., Vijayanand P., Kronenberg M. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol. 2016;17:728–739. doi: 10.1038/ni.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constantinides M.G., Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol. 2013;25:161–167. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sag D., Krause P., Hedrick C.C., Kronenberg M., Wingender G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest. 2014;124:3725–3740. doi: 10.1172/JCI72308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui K., Yan G., Zheng X., Bai L., Wei H., Sun R., Tian Z. Suppression of natural killer cell activity by regulatory NKT10 cells aggravates alcoholic hepatosteatosis. Front Immunol. 2017;8:1414. doi: 10.3389/fimmu.2017.01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klugewitz K., Adams D.H., Emoto M., Eulenburg K., Hamann A. The composition of intrahepatic lymphocytes: shaped by selective recruitment? Trends Immunol. 2004;25:590–594. doi: 10.1016/j.it.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Zhu S., Zhang H., Bai L. NKT cells in liver diseases. Front Med. 2018;12:249–261. doi: 10.1007/s11684-018-0622-3. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad A., Alvarez F. Role of NK and NKT cells in the immunopathogenesis of HCV-induced hepatitis. J Leukoc Biol. 2004;76:743–759. doi: 10.1189/jlb.0304197. [DOI] [PubMed] [Google Scholar]
- 10.Mattner J. Natural killer T (NKT) cells in autoimmune hepatitis. Curr Opin Immunol. 2013;25:697–703. doi: 10.1016/j.coi.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sepiashvili R.I., Balmasova I.P., Kabanova E.V., Malova E.S., Slavianskaia T.A. Hepatitis B virus: biology, immunopathogenesis, NK/NKT system in viral persistence. Zh Mikrobiol Epidemiol Immunobiol. 2006;6:76–83. [PubMed] [Google Scholar]
- 12.Takeda K., Hayakawa Y., Van Kaer L., Matsuda H., Yagita H., Okumura K. Critical contribution of liver natural killer T cells to a murine model of hepatitis. Proc Natl Acad Sci U S A. 2000;97:5498–5503. doi: 10.1073/pnas.040566697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan X., Ding Y., Zhu P., Dou R., Liang Z., Yang D., Huang Z., Wang W., Wu X., Weng X. Elevated hepatic CD1d levels coincide with invariant NKT cell defects in chronic hepatitis B virus infection. J Immunol. 2018;200:3530–3538. doi: 10.4049/jimmunol.1701801. [DOI] [PubMed] [Google Scholar]
- 14.Fiorucci S., Distrutti E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol Med. 2015;21:702–714. doi: 10.1016/j.molmed.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Chiang J.Y.L. Bile acid metabolism and signaling in liver disease and therapy. Liver Res. 2017;1:3–9. doi: 10.1016/j.livres.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., Nakamura T., Itadani H., Tanaka K. Identification of membrane-type receptor for bile acids (M-BAR) Biochem Biophys Res Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 17.Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., Fukusumi S., Habata Y., Itoh T., Shintani Y., Hinuma S., Fujisawa Y., Fujino M. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 18.Fiorucci S., Cipriani S., Mencarelli A., Renga B., Distrutti E., Baldelli F. Counter-regulatory role of bile acid activated receptors in immunity and inflammation. Curr Mol Med. 2010;10:579–595. doi: 10.2174/1566524011009060579. [DOI] [PubMed] [Google Scholar]
- 19.Fiorucci S., Biagioli M., Zampella A., Distrutti E. Bile acids activated receptors regulate innate immunity. Front Immunol. 2018;9:1853. doi: 10.3389/fimmu.2018.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar V. NKT-cell subsets: promoters and protectors in inflammatory liver disease. J Hepatol. 2013;59:618–620. doi: 10.1016/j.jhep.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumoto H., Kawamura T., Kobayashi T., Kanda Y., Kawamura H., Abo T. Coincidence of autoantibody production with the activation of natural killer T cells in α-galactosylceramide-mediated hepatic injury. Immunology. 2011;133:21–28. doi: 10.1111/j.1365-2567.2011.03405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossjohn J., Pellicci D.G., Patel O., Gapin L., Godfrey D.I. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol. 2012;12:845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biagioli M., Carino A., Cipriani S., Francisci D., Marchianò S., Scarpelli P., Sorcini D., Zampella A., Fiorucci S. The bile acid receptor GPBAR1 regulates the M1/M2 phenotype of intestinal macrophages and activation of GPBAR1 rescues mice from murine colitis. J Immunol. 2017;199:718–733. doi: 10.4049/jimmunol.1700183. [DOI] [PubMed] [Google Scholar]
- 24.Dasgupta S., Kumar V. Type II NKT cells: a distinct CD1d-restricted immune regulatory NKT cell subset. Immunogenetics. 2016;68:665–676. doi: 10.1007/s00251-016-0930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhodapkar M.V., Kumar V. Type II NKT cells and their emerging role in health and disease. J Immunol. 2017;198:1015–1021. doi: 10.4049/jimmunol.1601399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halder R.C., Aguilera C., Maricic I., Kumar V. Type II NKT cell-mediated anergy induction in type I NKT cells prevents inflammatory liver disease. J Clin Invest. 2007;117:2302–2312. doi: 10.1172/JCI31602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monteiro M., Almeida C.F., Caridade M., Ribot J.C., Duarte J., Agua-Doce A., Wollenberg I., Silva-Santos B., Graca L. Identification of regulatory Foxp3+ invariant NKT cells induced by TGF-beta. Identification of regulatory Foxp3+ invariant NKT cells induced by TGF-beta. J Immunol. 2010;185:2157–2163. doi: 10.4049/jimmunol.1000359. [DOI] [PubMed] [Google Scholar]
- 28.Mencarelli A., Renga B., Migliorati M., Cipriani S., Distrutti E., Santucci L., Fiorucci S. The bile acid sensor farnesoid X receptor is a modulator of liver immunity in a rodent model of acute hepatitis. The bile acid sensor farnesoid X receptor is a modulator of liver immunity in a rodent model of acute hepatitis. J Immunol. 2009;183:6657–6666. doi: 10.4049/jimmunol.0901347. [DOI] [PubMed] [Google Scholar]
- 29.Toyabe S., Seki S., Iiai T., Takeda K., Shirai K., Watanabe H., Hiraide H., Uchiyama M., Abo T. Requirement of IL-4 and liver NK1+ T cells for concanavalin A-induced hepatic injury in mice. J Immunol. 1997;159:1537–1542. [PubMed] [Google Scholar]
- 30.Scholten D., Trebicka J., Liedtke C., Weiskirchen R. The carbon tetrachloride model in mice. Lab Anim. 2015;49:4–11. doi: 10.1177/0023677215571192. [DOI] [PubMed] [Google Scholar]
- 31.Ridlon J.M., Kang D.J., Hylemon P.B. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Forman B.M., Goode E., Chen J., Oro A.E., Bradley D.J., Perlmann T., Noonan D.J., Burka L.T., McMorris T., Lamph W.W., Evans R.M., Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 33.Prossnitz E.R., Barton M. The G-protein-coupled estrogen receptor GPER in health and disease. Nat Rev Endocrinol. 2011;7:715–726. doi: 10.1038/nrendo.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keitel V., Reinehr R., Gatsios P., Rupprecht C., Görg B., Selbach O., Häussinger D., Kubitz R. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 2007;45:695–704. doi: 10.1002/hep.21458. [DOI] [PubMed] [Google Scholar]
- 35.Cipriani S., Mencarelli A., Chini M.G., Distrutti E., Renga B., Bifulco G., Baldelli F., Donini A., Fiorucci S. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hov J.R., Keitel V., Laerdahl J.K., Spomer L., Ellinghaus E., ElSharawy A., Melum E., Boberg K.M., Manke T., Balschun T., Schramm C., Bergquist A., Weismüller T., Gotthardt D., Rust C., Henckaerts L., Onnie C.M., Weersma R.K., Sterneck M., Teufel A., Runz H., Stiehl A., Ponsioen C.Y., Wijmenga C., Vatn M.H., IBSEN Study Group. Stokkers P.C., Vermeire S., Mathew C.G., Lie B.A., Beuers U., Manns M.P., Schreiber S., Schrumpf E., Häussinger D., Franke A., Karlsen T.H. Mutational characterization of the bile acid receptor TGR5 in primary sclerosing cholangitis. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geissmann F., Cameron T.O., Sidobre S., Manlongat N., Kronenberg M., Briskin M.J., Dustin M.L., Littman D.R. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 2005;3:e113. doi: 10.1371/journal.pbio.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishikawa S., Ikejima K., Yamagata H., Aoyama T., Kon K., Arai K., Takeda K., Watanabe S. CD1d-restricted natural killer T cells contribute to hepatic inflammation and fibrogenesis in mice. J Hepatol. 2011;54:1195–1204. doi: 10.1016/j.jhep.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Perino A., Pols T.W., Nomura M., Stein S., Pellicciari R., Schoonjans K. TGR5 reduces macrophage migration through mTOR-induced C/EBPβ differential translation. J Clin Invest. 2014;124:5424–5436. doi: 10.1172/JCI76289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S.J., Yang Y.H., Tsuneyama K., Leung P.S., Illarionov P., Gershwin M.E., Chuang Y.H. Innate immunity and primary biliary cirrhosis: activated invariant natural killer T cells exacerbate murine autoimmune cholangitis and fibrosis. Hepatology. 2011;53:915–925. doi: 10.1002/hep.24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Böttcher K., Rombouts K., Saffioti F., Roccarina D., Rosselli M., Hall A., Luong T., Tsochatzis E.A., Thorburn D., Pinzani M. MAIT cells are chronically activated in patients with autoimmune liver disease and promote profibrogenic hepatic stellate cell activation. Hepatology. 2018;68:172–186. doi: 10.1002/hep.29782. [DOI] [PubMed] [Google Scholar]
- 42.Vassileva G., Golovko A., Markowitz L., Abbondanzo S.J., Zeng M., Yang S., Hoos L., Tetzloff G., Levitan D., Murgolo N.J., Keane K., Davis H.R., Jr., Hedrick J., Gustafson E.L. Targeted deletion of GPBAR1 protects mice from cholesterol gallstone formation. Biochem J. 2006;398:423–430. doi: 10.1042/BJ20060537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Küsters S., Gantner F., Künstle G., Tiegs G. Interferon gamma plays a critical role in T cell-dependent liver injury in mice initiated by concanavalin A. Gastroenterology. 1996;111:462–471. doi: 10.1053/gast.1996.v111.pm8690213. [DOI] [PubMed] [Google Scholar]
- 44.Festa C., Renga B., D'Amore C., Sepe V., Finamore C., De Marino S., Carino A., Cipriani S., Monti M.C., Zampella A., Fiorucci S. Exploitation of cholane scaffold for the discovery of potent and selective farnesoid X receptor (FXR) and G-protein coupled bile acid receptor 1 (GP-BAR1) ligands. J Med Chem. 2014;57:8477–8495. doi: 10.1021/jm501273r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.