Abstract
There are conflicted experimental results on the role of bone morphogenetic proteins (BMPs) in cancer. Some results suggest that BMPs act as tumor suppressors while other findings indicate that BMPs are oncogenic factors. In the present study, we aimed to investigate the association of BMP expression and the survival of breast cancer patients utilizing The Cancer Genome Atlas (TCGA). High expression levels of BMP1 (P<0.001), BMP3 (P=0.002), BMP5 (P=0.002), BMP7 (P<0.001) and BMPR1A (P<0.001) were associated with better overall survival (OS). On the other hand, high expression levels of BMP6 (P<0.001), BMP8A (P=0.031), BMP8B (P<0.001) and BMPR1B (P=0.005) were associated with worse OS. Most of the BMPs demonstrated differential expression levels between normal and tumor tissues. High expression of BMP7 was found to be significantly associated with better prognosis in both estrogen receptor (ER)-positive (ER+) (P<0.001) and ER-negative (ER−) tumors (P<0.001), whereas high expression of BMP6 was associated with better prognosis only in ER+ tumors (P=0.004); it was associated with worse prognosis in ER− tumors (P=0.006). In the ER+ tumors, high expression of BMP7 as well as BMP6 were associated with higher infiltration of anticancer immune cells and cytolytic activity. These results suggest that high expression levels of BMP7 as well as BMP6 possess higher anticancer immunity which results in better prognosis in ER+ breast cancer. In conclusion, BMP expression profiles may be useful for prognostication. Intervention in this pathway may serve to improve outcomes, manage metastatic disease and assist in clinical decision making on optimal therapy based on the risk of recurrence or metastasis.
Keywords: bone morphogenetic protein, BMP, breast cancer, estrogen receptor, immune, macrophage, Treg, cytolytic, immunity, T cell
Introduction
Bone morphogenetic proteins (BMPs) are members of the TGF-β superfamily and are known for their diverse function in cancer progression (1). BMPs were originally believed to play a role solely in the development of bone and cartilage as they were isolated from bone extracts (2); however, the various functions of BMPs have since been discovered. BMPs display aberrant expression in a variety of malignancies (3). In breast cancer, BMP4 and BMP7 are reported to be the most frequently overexpressed BMPs (4). However, their biological functions in breast cancer remain controversial. Some studies have reported that BMPs suppress breast cancer growth, whereas others have reported that BMPs promote it. BMP4 was reported to suppress breast cancer metastasis by upregulating immune responses and inhibition of myeloid-derived suppressor cells (5), whereas it was also reported that high expression of BMP4 promoted cancer cell migration, invasion and metastasis (6). BMP7 was also shown to both facilitate and inhibit cancer growth, migration, invasiveness and metastasis in different breast cancer cell lines (7). BMP receptors (BMPRs) have also reported to play a role in the carcinogenesis and progression of breast cancer (8). As such, there are multiple conflicting reports concerning the role of BMPs in breast cancer.
TGF-β regulates immune cell infiltration in the tumor microenvironment. TGF-β suppresses anticancer immune cell infiltration such as cytotoxic T cells and natural killer cells, whereas it promotes the function of pro-cancer immune cells, such as regulatory T cells (Tregs) and M2 macrophages (9). BMP6 was also found to induce hormone therapy resistance through macrophage-derived cytokines in prostate cancer (10). Thus, we hypothesized that BMPs regulate immune cell infiltration, resulting in differential survival of breast cancer patients based on the expression levels of BMPs.
Here, we investigated the association of BMP expression and prognosis in breast cancer utilizing The Cancer Genome Atlas (TCGA) (11,12). Furthermore, we explored the difference in the association between BMP expression and survival by estrogen receptor (ER) status and sought possible underlying mechanisms using a bioinformatical approach.
Materials and methods
Data acquisition
Clinical data and gene expression from RNA sequences in TCGA breast cancer cohort were downloaded through cBioPortal (13,14) and UCSC Genome Browser (http://genome.ucsc.edu/) and processed, as previously described (15–20). As a validation cohort, we utilized GSE1456, in which there are 62 Luminal A/B patients with gene expression and survival information from Gene Enrichment Omnibus (21). TCGA and GSE1456 are de-identified and publicly available cohorts, thus institutional review board approval was waived.
Gene set enrichment analysis (GSEA)
GSEA was conducted comparing BMP7 high and low as well as BMP6 high and low in both ER-positive (ER+) and ER-negative (ER−) cohorts with the same cutoffs of survival analyses among 50 hallmark sets (23) using software provided by the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp) as described previously (24).
CIBERSORT
Tumor infiltrating immune cell composition was estimated from the gene expression profiles by CIBERSORT algorism (25). Immune cell fraction data were downloaded through TCIA (https://tcia.at/home) (26).
Cytolytic activity
Cytolytic activity was estimated utilizing GZMA and PRF1 expression (27) as described previously (28).
Statistical analysis
Overall survival (OS) was estimated by Kaplan-Meier curve with log-rank test, comparing high and low expression of each gene of interest. The cutoffs were determined by analyzing multiple cutoff points within the range of values and the optimal cutoff points were chosen based on the statistical significance of the varying points as previously reported (22).
Statistical comparisons of gene expression were performed using the Student t-test, and the clinical demographics were compared using the Fisher exact test. Pearson correlations were calculated based on the expression levels of the genes of interest for correlation matrix (Fig. 2C). All statistical analyses were performed using R software (http://www.r-project.org/) and Bioconductor (http://bioconductor.org/).
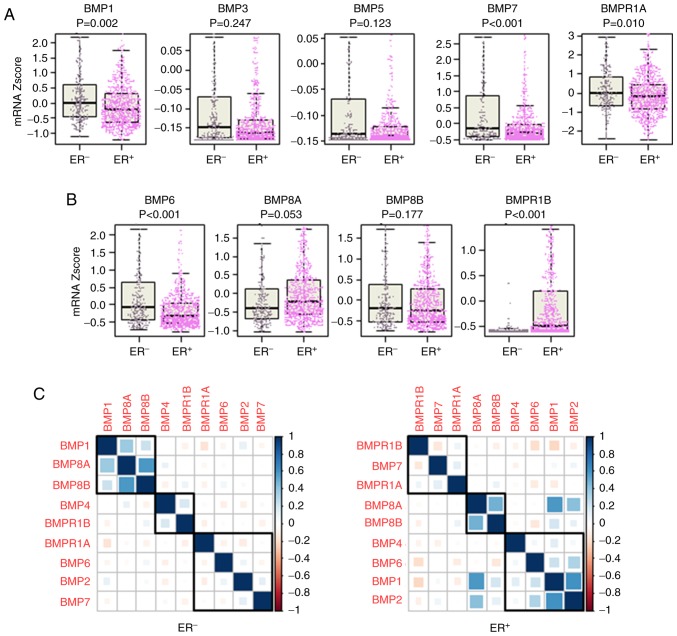
Figure 2.
Comparison of the expression of BMPs with ER status. (A) Tumor-suppressive BMPs in the whole cohort. (B) Oncogenic BMPs in the whole cohort. (C) Correlation of BMPs and BMPRs in the ER+ and ER− tumors. BMP, bone morphogenetic protein; BMPR, BMP receptor; ER, estrogen receptor; ER+, ER-positive; ER−, ER-negative.
Results
Various BMPs and BMPRs are associated with better OS, whereas others are associated with worse OS in breast cancer
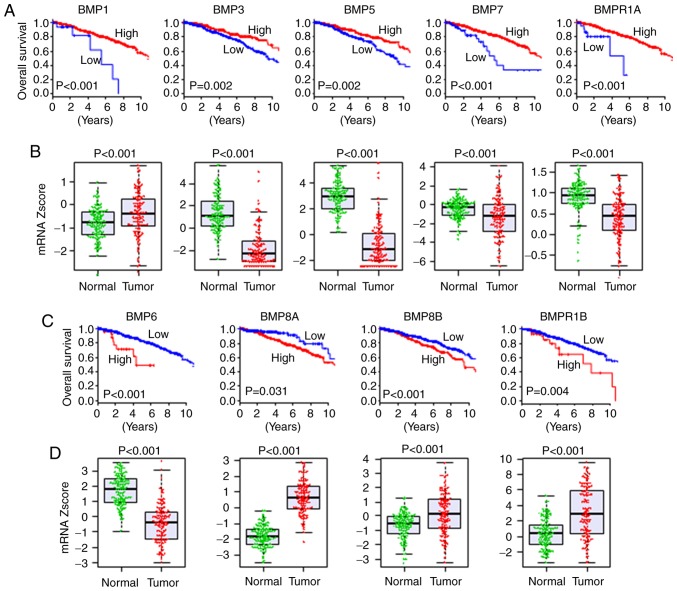
There have been conflicting reports on the association between the expression levels of BMPs and patient survival (29). Thus, we analyzed the association between the expression of each BMP and OS of the breast cancer patients. High expression of BMP1 (P<0.001), BMP3 (P=0.002), BMP5 (P=0.002), BMP7 (P<0.001) and BMPR1A (P<0.001) was indicative of a significantly better OS of the breast cancer patients (Fig. 1A). Conversely, high expression of BMP6 (P<0.001), BMP8A (P=0.031), BMP8B (P<0.001) and BMPR1B (P=0.004) was indicative of a worse OS (Fig. 1C). These findings indicate that the former may act as tumor suppressors, and the latter as oncogenic factors. On the other hand, the expression levels of BMP2 (P=0.083), BMP4 (P=0.071), BMP10 (P=0.441), BMP13 (P=0.112) and BMP15 (P=0.050) were not associated with OS of breast cancer patients (Fig. S1). Of interest, cutoff points of BMP expression with the highest impact on OS were extremely low in some tumor-suppressive BMPs, and hence a poorer prognosis was shown in <10% of patients for BMP1, BMP7 and BMPR1A (Fig. 1A). Similar trends were shown in some oncogenic BMPs, BMP6 and BMPR1B, in which cutoff points with the highest impact of OS were extremely high and <10% of patients were classified as high expression with worse prognosis (Fig. 1C). In summary, we found that extremely low expression of tumor-suppressive BMPs and extremely high expression of oncogenic BMPs were associated with worse OS in breast cancer.
Figure 1.
Association of BMP expression with OS and expression comparison of tumor and adjacent normal tissues in TCGA breast cancer patients. (A) OS in regards to expression of tumor-suppressive BMPs. (B) Gene expression comparison of tumor-suppressive BMPs between tumor and adjacent normal tissues in matched pairs of TCGA breast cancer. (C) OS in regards to expression of oncogenic BMPs. (D) Gene expression comparison of oncogenic BMPs between tumor and adjacent normal tissues in matched pairs of TCGA breast cancer. BMP, bone morphogenetic protein; TCGA, The Cancer Genome Atlas; OS, overall survival.
Majority of oncogenic BMPs are expressed at a high level, and tumor-suppressive BMPs are expressed at a low in the tumors compared to the normal tissues
Given the associations between the expression levels of the BMPs and worse or better survival, we hypothesized that oncogenic BMPs are expressed at a higher level and tumor-suppressive BMPs are expressed at a lower level in tumors compared to adjacent normal tissues. A total of 114 breast cancers with matched tumor and adjacent normal tissues were present in TCGA, where the expression of BMPs and BMPRs was compared. All of the BMPs and BMPRs, which were associated with survival, were expressed at significantly different levels between the normal and tumor tissues. All of the tumor-suppressive BMPs except BMP1; i.e. BMP3, BMP5, BMP7 and BMPR1A, were expressed at a significantly lower level in the tumors, whereas all of the oncogenic BMPs except BMP6; i.e. BMP8A, BMP8B and BMPR1B, were expressed at a higher level in the tumors compared to the normal tissues (Fig. 1B and D). The expression levels of BMP10 and BMP15, which showed no association with OS, were not detectable in the vast majority of samples (106 normal and 108 tumor tissues, and 104 normal and 100 tumor tissues, respectively). BMP2 and BMP4, that did not show an association with OS, were also expressed at a lower level in the tumors (Fig. S1). These results support the notion that oncogenic BMPs are expressed higher, and tumor-suppressive BMPs are expressed lower in the tumors, except BMP1 and BMP6.
Expression of BMPs differs according to ER status
It is well known that certain signaling, such as HER2, crosstalks with ER signaling and the targeted drug sensitivity is enhanced by ER blockade (30). Signaling of BMPs is also known to crosstalk with that of ER (31,32). Therefore, it was of interest to investigate the association of ER positivity and BMP expression. We found that among the tumor-suppressive BMPs, BMP1 (P=0.002), BMP7 (P<0.001) and BMPR1A (P=0.010) were expressed significantly higher in the ER− tumors compared to the ER+ tumors, whereas there was no significant difference in the expression levels of BMP3 (P=0.247) and BMP5 (P=0.123) (Fig. 2A). On the other hand, among the oncogenic BMPs, BMP6 expression was significantly higher in the ER− tumors (P<0.001), whereas BMPR1B expression was significantly higher in the ER+ tumors (P<0.001) (Fig. 2B). Interactions of BMPs and BMPRs were also different between ER+ and ER− tumors (Fig. 2C). In the ER+ tumors, BMP6 was in the same cluster as BMP1, BMP2 and BMP4, where BMP6 exhibited a higher correlation with BMP1 and BMP2, and BMP7 was in the same cluster of BMPRs (BMPR1A and BMPR1B) (Fig. 2C). Whereas in the ER− tumors, BMP6 and BMP7 were in the same cluster with BMP2 and BMPR1A, showing a very weak correlation with each other (Fig. 2C).
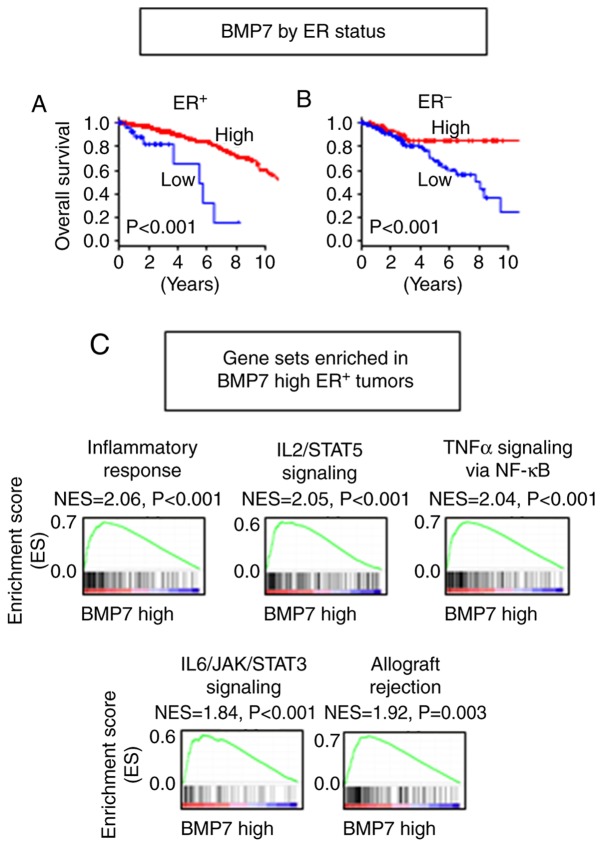
Higher BMP7 expression is associated with better OS in both ER+ and ER− tumors
Since BMP7 was reported to have roles in both promotion and inhibition of cancer growth (4), and showed significant differential expression as well as differential interaction with other BMPs and BMPRs between ER+ and ER− tumors, we focused on BMP7 among the tumor-suppressive BMPs. Higher expression of BMP7 demonstrated better survival in both ER+ (P<0.001) and ER− (P<0.001) cohorts (Fig. 3A and B). Although higher expression of BMP7 was found to be associated with better prognosis in the both ER+ and ER− tumors, GSEA demonstrated that different gene sets were enriched between the ER+ and ER− tumors (Tables SI–SIII). In the ER+ tumors, immune response-related gene sets, such as inflammatory response (P<0.001), IL2/STAT5 signaling (P<0.001), TNFα signaling via NF-κB (P<0.001), IL6/JAK/STAT3 signaling (P<0.001) and allograft rejection (P=0.003), were enriched in the BMP7 high tumors (Fig. 3C, Table SI). As tumor aggressiveness feature-related gene sets, the epithelial-mesenchymal transition (EMT) gene set was also enriched (P<0.001), while cell proliferation, cell cycle-related gene sets were not enriched in the BMP7 high ER+ tumors (Table SI). On the other hand, no gene set was significantly enriched in the BMP7 low tumors. When we compared demographics of the patients with BMP7 high and low ER+ tumors, the patients with BMP7 high ER+ tumors were younger than those with BMP7 low ER+ tumors (Table I). In the ER− tumors, no immune reaction-related gene set was enriched neither in BMP7 high nor low tumors (Tables SII and SIII). These findings suggest that BMP7 plays a tumor-suppressive role in the ER+ tumor by evoking anticancer immunity, which has a more predominant effect than promotion of EMT, whereas it has different mechanisms in the ER− tumors.
Figure 3.
Association of BMP7 expression with OS of breast cancer patients by ER status. OS by BMP7 expression in (A) ER+ and (B) ER- tumors. (C) Gene sets enriched in the BMP7 high ER+ tumors. BMP, bone morphogenetic protein; OS, overall survival; ER, estrogen receptor; ER+, ER-positive; ER−, ER-negative.
Table I.
Patient demographics comparing BMP7 high and low as well as BMP6 high and low expression in the ER+ breast cancer patients.
| BMP7 expression | BMP6 expression | |||||
|---|---|---|---|---|---|---|
| Characteristics | High (n=770) n (%) | Low (n=34) n (%) | P-value | High (n=608) n (%) | Low (n=196) n (%) | P-value |
| Age, years | 0.001a | 0.033a | ||||
| <60 | 395 (51.3) | 8 (23.5) | 318 (52.7) | 85 (43.4) | ||
| ≥60 | 375 (48.7) | 26 (76.5) | 290 (47.7) | 111 (56.6) | ||
| Race | 0.658 | 0.167 | ||||
| Asian | 36 (5.2) | 2 (6.7) | 26 (4.6) | 12 (7.5) | ||
| African-American | 106 (15.9) | 3 (10.0) | 81 (14.4) | 28 (17.6) | ||
| Caucasian | 548 (79.4) | 25 (83.3) | 454 (80.9) | 119 (74.8) | ||
| Menopause status | 0.056 | <0.001a | ||||
| Pre | 160 (24.4) | 3 (9.1) | 142 (27.1) | 21 (12.7) | ||
| Post | 497 (75.6) | 30 (90.9) | 382 (72.9) | 145 (87.3) | ||
| pT | 0.343 | 0.663 | ||||
| T1/2 | 640 (83.1) | 25 (75.8) | 501 (82.4) | 164 (84.1) | ||
| T2/3 | 130 (16.9) | 8 (24.2) | 107 (17.6) | 31 (15.9) | ||
| pN | 0.286 | 0.559 | ||||
| N0 | 337 (44.6) | 18 (54.5) | 266 (44.4) | 89 (46.8) | ||
| N1/2/3 | 419 (55.4) | 15 (45.5) | 333 (55.6) | 101 (53.2) | ||
| pM | 0.474 | 0.084 | ||||
| M0 | 628 (97.7) | 25 (96.2) | 494 (98.2) | 159 (95.8) | ||
| M1 | 15 (2.3) | 1 (3.8) | 9 (1.8) | 7 (4.2) | ||
| Stage | 0.842 | 0.572 | ||||
| Stage I/II | 557 (73.9) | 24 (81.8) | 437 (73.2) | 144 (75.4) | ||
| Stage III/IV | 198 (26.1) | 9 (18.2) | 160 (26.8) | 47 (24.6) | ||
| Histology | 0.475 | <0.001a | ||||
| Infiltrating ductal carcinoma | 523 (73.9) | 18 (81.8) | 381 (68.7) | 160 (93.0) | ||
| Infiltrating lobular carcinoma | 185 (26.1) | 4 (18.2) | 177 (31.7) | 12 (7.0) | ||
ER+, estrogen receptor-positive; BMP, bone morphogenetic protein.
P<0.05, significant difference.
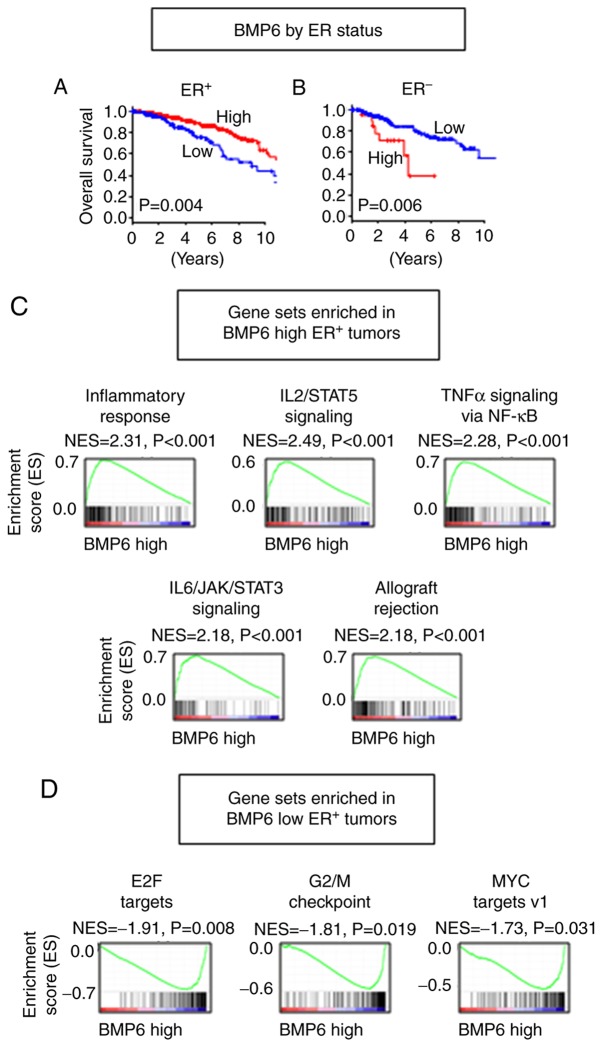
Higher BMP6 expression is associated with better OS in ER+ tumors, whereas it is associated with worse OS in ER− tumors
Since high BMP6 expression was associated with significantly worse survival with lower expression in the tumor tissues, and different expression of BMP6 as well as different correlation with other BMPs and BMPRs between ER+ and ER− tumors were observed, we focused on BMP6 among the oncogenic BMPs. Despite the fact that BMP6 high tumors demonstrated worse OS in the whole cohort, that trend was only apparent in the ER− tumors (P=0.006) (Fig. 4B). Conversely, high expression of BMP6 was found to be associated with better survival in the ER+ tumors (P=0.004) (Fig. 4A). GSEA revealed that immune reaction-related gene sets, including inflammatory response (P<0.001), IL2/STAT5 signaling (P<0.001), TNFα signaling via NF-κB (P<0.001), IL6/JAK/STAT3 signaling (P<0.001) and allograft rejection (P<0.001) were enriched in the BMP6 high ER+ tumors (Fig. 4C and Table SIV). Similar to the BMP7 high ER+ tumors, the EMT gene set was also enriched (P<0.001) in the BMP6 high ER+ tumors (Table SIV). Interestingly, cell proliferation and cell cycle-related gene sets, such as E2F targets (P=0.008), G2/M checkpoint (P=0.019) and MYC targets v1 (33) (P=0.031), were enriched in the BMP6 low ER+ tumors, which suggest that a different mechanism may be in place than BMP7 (Fig. 4D and Table SV). The patients with BMP6 high ER+ tumors showed a higher proportion of younger age, pre-menopausal status, and infiltrating lobular carcinoma in histology compared to those with BMP6 low ER+ tumors (Table I). In the ER− tumors, MYC targets v2 (34) gene set was enriched in the BMP6 high tumors (P=0.041), whereas no immune reaction or cell proliferation gene set was enriched (Tables SVI and SVII). These findings suggest that the BMP6 high tumors have more anticancer immunity and the BMP6 low tumors have accelerated cell cycle; thus, high expression of BMP6 is associated with better survival in the ER+ tumors, whereas higher expression of MYC target genes may play a role in the BMP6 high ER− tumors, resulting in worse survival.
Figure 4.
Association of BMP6 expression with OS of breast cancer patients by ER status. OS by BMP6 expression in (A) ER+ and (B) ER− tumors. Gene sets enriched in (C) BMP6 high ER+ tumors, and (D) BMP6 low ER+ tumors. BMP, bone morphogenetic protein; OS, overall survival; ER, estrogen receptor; ER+, ER-positive; ER−, ER-negative.
BMP7 high as well as BMP6 high ER+ tumors have higher anticancer immune cell infiltration and cytolytic activity
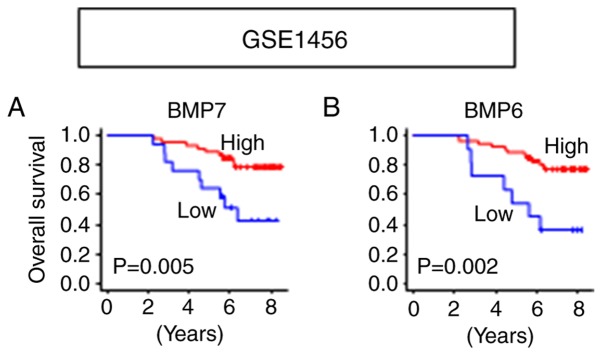
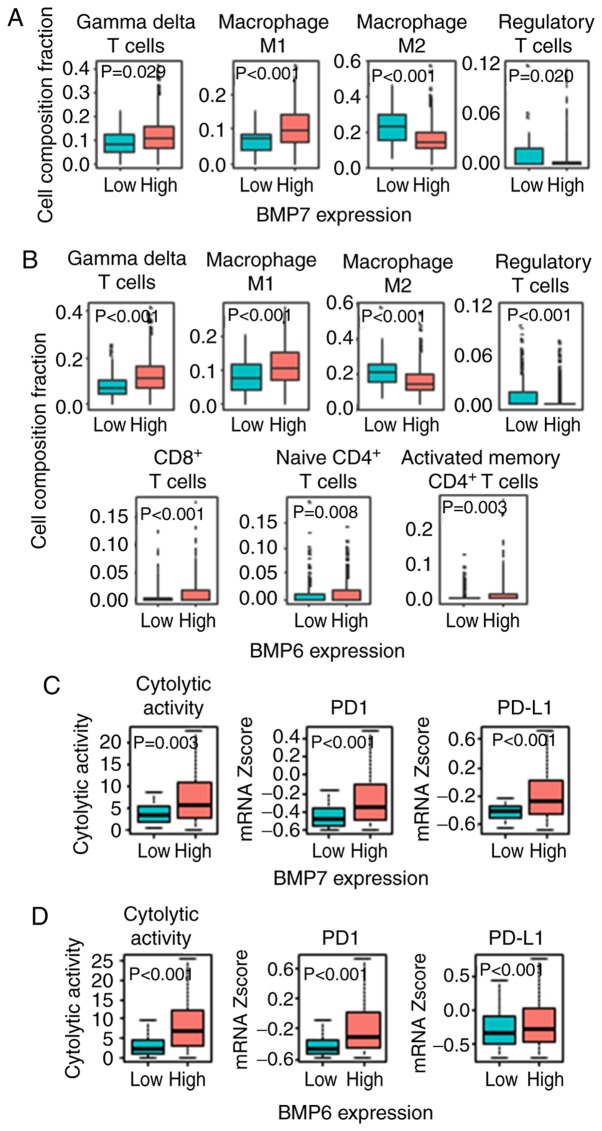
The results that both BMP7 and BMP6 function as tumor suppressors in ER+ tumors were validated in another cohort, GSE1456 (21), in which the patients with high expression of BMP7 (P=0.005) as well as BMP6 (P=0.002) showed significantly better OS in Luminal A/B tumors (Fig. 5). Given these results and the BMP7 and BMP6 high expressing tumor enriched immune-related gene sets, we further hypothesized that the ER+ tumors with high BMP7 as well as high BMP6 expressions have higher anticancer immune cell infiltration. Utilizing CIBERSORT algorithm (25), we found higher anticancer immune cell infiltration, such as gamma-delta (γδ) T cells (P=0.029), M1 macrophages (P<0.001), and less pro-cancer immune cell infiltrations, such as M2 macrophages (P<0.001) and Tregs (P=0.020) in the BMP7 high ER+ tumors (Figs. 6A and S2). A similar trend was found in the BMP6 high ER+ tumors, where higher infiltration of anticancer immune cells, not only γδ T cells (P<0.001) and M1 macrophages (P<0.001), but also CD8+ T cells (P<0.001), CD4+ naïve T cells (P=0.008) and CD4+ activated memory T cells (P=0.003), and less infiltration of pro-cancer immune cells, M2 macrophages (P<0.001) and Tregs (P<0.001) in the BMP6 high ER+ tumors were observed (Figs. 6B and S3). Since higher anticancer immune cell infiltration was observed in both BMP7 and BMP6 high expressing ER+ tumors, we further analyzed the cytolytic activity of those tumors. As shown in Fig. 6C and D, high BMP7 as well as high BMP6 in the ER+ tumors demonstrated higher cytolytic activity compared to those with low expression tumors. These findings imply that ER+ tumors with high BMP7 as well as high BMP6 expression have not only higher immune cell infiltration, but also higher cytolytic activity, resulting in better prognosis.
Figure 5.
Validation of survival analyses utilizing Luminal A/B tumors in GSE1456. (A) OS by BMP7 expression. (B) OS by BMP6 expression. BMP, bone morphogenetic protein; OS, overall survival.
Figure 6.
Immune activity by BMP7 and BMP6 expression in the ER+ tumors. (A) Infiltrating immune cell comparison between the BMP7 high and low ER+ tumors. (B) Infiltrating immune cell comparison between the BMP6 high and low ER+ tumors. (C) Cytolytic activity, PD1 and PD-L1 expression between the BMP7 high and low ER+ tumors. (D) Cytolytic activity, PD1 and PD-L1 expression between the BMP6 high and low ER+ tumors. BMP, bone morphogenetic protein; ER+, ER-positive; PD1, programmed cell death protein 1; PD-L1, programmed death-ligand 1.
To ascertain whether these tumors are good candidates for immune check point inhibition, PD1 and PD-L1 expressions were compared between BMP7 high and low as well as BMP6 high and low expression in the ER+ tumors. PD1 and PD-L1 expression levels were significantly higher in the BMP7 high expressing as well as BMP6 high expressing ER+ tumors (Fig. 6C and D). These findings suggest that high expression of BMP7 and BMP6 in the ER+ tumors may be good candidates for immune check point inhibition.
Discussion
In the present study, we found that each BMP was associated differentially with OS of breast cancer patients. High expression levels of BMP1, BMP3, BMP5, BMP7 and BMPR1A were associated with better prognosis in the whole cohort of breast cancer patients, whereas those of BMP6, BMP8A, BMP8B and BMPR1B were associated with worse prognosis. BMP7 was associated with better prognosis in both the ER+ and ER− tumors. In contrast, higher BMP6 expression was found to be associated with better prognosis in the ER+ tumors, but was associated with worse prognosis in the ER− tumors. High expression of BMP7 as well as BMP6 was associated with higher immune response in the ER+ tumors, but not in the ER− tumors.
BMPs play complex roles in cancer progression, specifically in breast cancer (34). Our findings, that high expression levels of BMP7 and BMP6 are associated with better prognosis in ER+ breast cancer, is consistent with previous reports. Utilizing breast cancer cell lines, Takahashi et al demonstrated that BMP6 and BMP7 inhibit estrogen-induced proliferation (35). It was reported that higher BMP7 expression is associated with higher rates of bone metastasis, however, lower BMP7 expression is associated with local relapse in breast cancer (36). In our analysis BMP7 expression was found to be associated with a better overall survival, irrespective of ER status. In the same token, BMP6 expression has been demonstrated to suppress breast cancer metastasis in both ER+ and ER− tumors (37,38). It was also found that BMP6 silencing increased chemo-resistance to doxorubicin by upregulating multiple drug resistance glycoproteins in breast cancer (39). In our analysis, BMP6 expression was found to be associated with worse survival in the whole patient cohort. Notably, the reverse finding was observed in the ER+ tumors, where BMP6 plays a tumor-suppressive role. This suggests a complex interplay with the hormonal pathway.
There have been few reports that link BMP7 or BMP6 to immune response. During the wound healing process, BMP7 plays an anti-fibrotic role by causing inflammation (40). BMP6 expression was found to be elevated in the ovary of polycystic ovarian syndrome which is known to be associated with inflammation, when compared to normal ovarian tissues (41). Overall, the role of BMPs in breast cancer remains poorly understood and is associated with complex interactions with a multitude of pathways and effectors that may act to completely alter the effector function of BMP signaling.
Although novel findings were discovered, there are limitations to the present study. First, our results were confirmed in a few cohorts due to availability. Second, all of our findings were based on gene expression of the primary tumor, thus experimental approaches are needed to elucidate further molecular mechanisms.
In the context of ER expression, individualized patient BMP expression profiles may act as a prognostic tool to guide therapy or may even be used as a therapeutic target in itself. This revelation in findings will continue to evolve, and the correlation between the BMP activation in regards to different pathways and the effect on prognosis in the clinical realm remain to be clearly elucidated.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- BMP
bone morphogenetic protein
- BMPR
BMP receptor
- TCGA
The Cancer Genome Atlas
- OS
overall survival
- ER
estrogen receptor
- EMT
epithelial-mesenchymal transition
- Treg
regulatory T cell
- GSEA
Gene set enrichment analysis
Funding
This work was supported by NIH grant R01CA160688 to KT and the National Cancer Institute (NCI) grant P30CA016056 and U24CA232979 involving the use of Roswell Park Comprehensive Cancer Center Shared Resources.
Availability of data and materials
Clinical data and gene expression from RNA sequence in TCGA breast cancer cohort were downloaded through cBioPortal (http://www.cbioportal.org/) and UCSC Genome Browser (http://genome.ucsc.edu/). Clinical data and gene expression of GSE1456 were downloaded from Gene Expression Omnibus.
Author's contributions
EK conceptualized the research design and performed the analysis and prepared the manuscript. AAM conceptualized the research design and prepared the manuscript. LY carried out supervision of the analyses. KT carried out supervision of the analysis and prepared the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
TCGA and GSE1456 include data which have been de-identified and are publicly available cohorts, thus institutional review board approval and patient consent were waived.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19:71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. Novel regulators of bone formation: Molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 3.Langenfeld E, Kong Y, Langenfeld J. Bone morphogenetic protein 2 stimulation of tumor growth involves the activation of Smad-1/5. Oncogene. 2006;25:685–692. doi: 10.1038/sj.onc.1209110. [DOI] [PubMed] [Google Scholar]
- 4.Alarmo EL, Kuukasjärvi T, Karhu R, Kallioniemi A. A comprehensive expression survey of bone morphogenetic proteins in breast cancer highlights the importance of BMP4 and BMP7. Breast Cancer Res Treat. 2007;103:239–246. doi: 10.1007/s10549-006-9362-1. [DOI] [PubMed] [Google Scholar]
- 5.Cao Y, Slaney CY, Bidwell BN, Parker BS, Johnstone CN, Rautela J, Eckhardt BL, Anderson RL. BMP4 inhibits breast cancer metastasis by blocking myeloid-derived suppressor cell activity. Cancer Res. 2014;74:5091–5102. doi: 10.1158/0008-5472.CAN-13-3171. [DOI] [PubMed] [Google Scholar]
- 6.Guo D, Huang J, Gong J. Bone morphogenetic protein 4 (BMP4) is required for migration and invasion of breast cancer. Mol Cell Biochem. 2012;363:179–190. doi: 10.1007/s11010-011-1170-1. [DOI] [PubMed] [Google Scholar]
- 7.Alarmo EL, Pärssinen J, Ketolainen JM, Savinainen K, Karhu R, Kallioniemi A. BMP7 influences proliferation, migration, and invasion of breast cancer cells. Cancer Lett. 2009;275:35–43. doi: 10.1016/j.canlet.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 8.Pickup MW, Hover LD, Guo Y, Gorska AE, Chytil A, Novitskiy SV, Moses HL, Owens P. Deletion of the BMP receptor BMPR1a impairs mammary tumor formation and metastasis. Oncotarget. 2015;6:22890–22904. doi: 10.18632/oncotarget.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, Pang Y, Moses HL. TGF-beta and immune cells: An important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31:220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee GT, Jung YS, Ha YS, Kim JH, Kim WJ, Kim IY. Bone morphogenetic protein-6 induces castration resistance in prostate cancer cells through tumor infiltrating macrophages. Cancer Sci. 2013;104:1027–1032. doi: 10.1111/cas.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green TP, Fennell M, Whittaker R, Curwen J, Jacobs V, Allen J, Logie A, Hargreaves J, Hickinson DM, Wilkinson RW, et al. Preclinical anticancer activity of the potent, oral Src inhibitor AZD0530. Mol Oncol. 2009;3:248–261. doi: 10.1016/j.molonc.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin L, Andersen JN, Futreal PA. Cancer genomics: From discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terakawa T, Katsuta E, Yan L, Turaga N, McDonald KA, Fujisawa M, Guru KA, Takabe K. High expression of SLCO2B1 is associated with prostate cancer recurrence after radical prostatectomy. Oncotarget. 2018;9:14207–14218. doi: 10.18632/oncotarget.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirose Y, Nagahashi M, Katsuta E, Yuza K, Miura K, Sakata J, Kobayashi T, Ichikawa H, Shimada Y, Kameyama H, et al. Generation of sphingosine-1-phosphate is enhanced in biliary tract cancer patients and is associated with lymphatic metastasis. Sci Rep. 2018;8:10814. doi: 10.1038/s41598-018-29144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young J, Kawaguchi T, Yan L, Qi Q, Liu S, Takabe K. Tamoxifen sensitivity-related microRNA-342 is a useful biomarker for breast cancer survival. Oncotarget. 2017;8:99978–99989. doi: 10.18632/oncotarget.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoki T, Katsuta E, Yan L, Takabe K, Ito F. Low DMT1 expression associates with increased oxidative phosphorylation and early recurrence in hepatocellular carcinoma. J Surg Res. 2019;234:343–352. doi: 10.1016/j.jss.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sporn JC, Katsuta E, Yan L, Takabe K. Expression of microRNA-9 is associated with overall survival in breast cancer patients. J Surg Res. 2019;233:426–435. doi: 10.1016/j.jss.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Katsuta E, Qi Q, Peng X, Hochwald SN, Yan L, Takabe K. Pancreatic adenocarcinomas with mature blood vessels have better overall survival. Sci Rep. 2019;9:1310. doi: 10.1038/s41598-018-37909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pawitan Y, Bjöhle J, Amler L, Borg AL, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ, Panel members. Strategies for subtypes-dealing with the diversity of breast cancer: Highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katsuta E, Yan L, Nagahashi M, Raza A, Sturgill JL, Lyon DE, Rashid OM, Hait NC, Takabe K. Doxorubicin effect is enhanced by sphingosine-1-phosphate signaling antagonist in breast cancer. J Surg Res. 2017;219:202–213. doi: 10.1016/j.jss.2017.05.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayanan S, Kawaguchi T, Yan L, Peng X, Qi Q, Takabe K. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann Surg Oncol. 2018;25:2323–2331. doi: 10.1245/s10434-018-6506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye L, Bokobza S, Li J, Moazzam M, Chen J, Mansel RE, Jiang WG. Bone morphogenetic protein-10 (BMP-10) inhibits aggressiveness of breast cancer cells and correlates with poor prognosis in breast cancer. Cancer Sci. 2010;101:2137–2144. doi: 10.1111/j.1349-7006.2010.01648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sudhan DR, Schwarz LJ, Guerrero-Zotano A, Formisano L, Nixon MJ, Croessmann S, González Ericsson PI, Sanders M, Balko JM, Avogadri-Connors F, et al. Extended adjuvant therapy with neratinib plus fulvestrant blocks ER/HER2 crosstalk and maintains complete responses of ER+/HER2+ breast cancers: Implications to the ExteNET trial. Clin Cancer Res. 2019;25:771–783. doi: 10.1158/1078-0432.CCR-18-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Athanasios F, Afrodite N, Effstratios P, Demetrios K. Co-expression of bone morphogenetic protein 6 with estrogen receptor a in endometriosis. Arch Gynecol Obstet. 2012;285:1001–1007. doi: 10.1007/s00404-011-2082-8. [DOI] [PubMed] [Google Scholar]
- 32.Shee K, Jiang A, Varn FS, Liu S, Traphagen NA, Owens P, Ma CX, Hoog J, Cheng C, Golub TR, et al. Cytokine sensitivity screening highlights BMP4 pathway signaling as a therapeutic opportunity in ER(+) breast cancer. FASEB J. 2019;33:1644–1657. doi: 10.1096/fj.201801241R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies SR, Watkins G, Douglas-Jones A, Mansel RE, Jiang WG. Bone morphogenetic proteins 1 to 7 in human breast cancer, expression pattern and clinical/prognostic relevance. J Exp Ther Oncol. 2008;7:327–338. [PubMed] [Google Scholar]
- 35.Takahashi M, Otsuka F, Miyoshi T, Otani H, Goto J, Yamashita M, Ogura T, Makino H, Doihara H. Bone morphogenetic protein 6 (BMP6) and BMP7 inhibit estrogen-induced proliferation of breast cancer cells by suppressing p38 mitogen-activated protein kinase activation. J Endocrinol. 2008;199:445–455. doi: 10.1677/JOE-08-0226. [DOI] [PubMed] [Google Scholar]
- 36.Alarmo EL, Korhonen T, Kuukasjärvi T, Huhtala H, Holli K, Kallioniemi A. Bone morphogenetic protein 7 expression associates with bone metastasis in breast carcinomas. Ann Oncol. 2008;19:308–314. doi: 10.1093/annonc/mdm453. [DOI] [PubMed] [Google Scholar]
- 37.Yang S, Du J, Wang Z, Yuan W, Qiao Y, Zhang M, Zhang J, Gao S, Yin J, Sun B, Zhu T. BMP-6 promotes E-cadherin expression through repressing deltaEF1 in breast cancer cells. BMC Cancer. 2007;7:211. doi: 10.1186/1471-2407-7-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du J, Yang S, An D, Hu F, Yuan W, Zhai C, Zhu T. BMP-6 inhibits microRNA-21 expression in breast cancer through repressing deltaEF1 and AP-1. Cell Res. 2009;19:487–496. doi: 10.1038/cr.2009.34. [DOI] [PubMed] [Google Scholar]
- 39.Lian WJ, Liu G, Liu YJ, Zhao ZW, Yi T, Zhou HY. Downregulation of BMP6 enhances cell proliferation and chemoresistance via activation of the ERK signaling pathway in breast cancer. Oncol Rep. 2013;30:193–200. doi: 10.3892/or.2013.2462. [DOI] [PubMed] [Google Scholar]
- 40.Tandon A, Sharma A, Rodier JT, Klibanov AM, Rieger FG, Mohan RR. BMP7 gene transfer via gold nanoparticles into stroma inhibits corneal fibrosis in vivo. PLoS One. 2013;8:e66434. doi: 10.1371/journal.pone.0066434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt J, Weijdegård B, Mikkelsen AL, Lindenberg S, Nilsson L, Brännström M. Differential expression of inflammation-related genes in the ovarian stroma and granulosa cells of PCOS women. Mol Hum Reprod. 2014;20:49–58. doi: 10.1093/molehr/gat051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Clinical data and gene expression from RNA sequence in TCGA breast cancer cohort were downloaded through cBioPortal (http://www.cbioportal.org/) and UCSC Genome Browser (http://genome.ucsc.edu/). Clinical data and gene expression of GSE1456 were downloaded from Gene Expression Omnibus.