Abstract
Liquid biomarkers for the early detection of resistance to chemotherapy are important for improving prognosis. This study investigated the usefulness of plasma exosomal microRNA-125b (ex-miRNA-125b) for the early detection of resistance to modified fluorouracil, leucovorin and oxaliplatin (mFOLFOX6)-based first-line chemotherapy in patients with advanced or recurrent (advanced/recurrent) colorectal cancer (CRC). First, ex-miRNAs associated with resistance to mFOLFOX6-based chemotherapy were profiled via miRNA microarray analysis. In this analysis, ex-miR-125b exhibited the greatest upregulation in patients with progressive disease (PD) compared with the findings for patients with stable disease (SD) and healthy controls. Next, another 55 patients with advanced/recurrent CRC who received mFOLFOX6-based first-line chemotherapy underwent a validation study of ex-miR-125b. Blood samples were collected before and during treatment until tumor progression. Ex-miRNA levels were measured via TaqMan microRNA assays. Patients with CRC had significantly higher ex-miR-125b levels than healthy controls. In patients with partial responses, ex-miR-125b levels at the Response Evaluation Criteria in Solid Tumors (RECIST) judgment point were significantly lower than those measured before treatment. In patients with SD, ex-miR-125b levels did not differ before and during treatment. In patients with PD, ex-miR-125b levels at the RECIST judgment point were significantly higher than those measured before treatment. These changes in ex-miR-125b levels were significantly different between groups even 1 month after the initiation of chemotherapy. Progression-free survival (PFS) was significantly worse in patients with high baseline ex-miR-125b levels than in those with low levels. In the Cox analysis, baseline ex-miR-125b levels and KRAS mutation were indicated to be independent prognostic factors for PFS. The present results suggest that plasma ex-miR-125b levels may be useful for the early detection of resistance to mFOLFOX6-based first-line chemotherapy. Furthermore, ex-miR-125b before chemotherapy is a predictive biomarker for PFS in patients with advanced/recurrent CRC.
Keywords: colorectal cancer, exosome, chemoresistance, monitoring biomarker, microRNA-125b, mFOLFOX6
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related deaths in both men and women globally (1). Chemotherapy in combination with molecular targeted compounds is the most widespread treatment for CRC, particularly metastatic CRC. As the first-line treatment for metastatic CRC, modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6)-based chemotherapy is one of the most common regimens used in Japan. However, it was reported that the response rate to first-line chemotherapy is approximately 50%. In addition, patients with chemoresistant tumors experience adverse effects following chemotherapy. There is a need for a minimally invasive biomarker that can predict the therapeutic effect as early as possible. Clinically, the therapeutic effects of chemotherapy can be monitored using imaging modalities, such as CT, and tumor markers, such as CEA and CA19-9. Because of the inherent radiation exposure, CT examination cannot be performed frequently. Furthermore, CEA and CA19-9 have unsatisfactory sensitivity as monitoring biomarkers. Unfortunately, we have no strategy for predicting the response to chemotherapy. Therefore, there is an urgent need to identify effective biomarkers that can provide some indication of the likely response or resistance to chemotherapy.
One candidate monitoring marker for chemotherapy is microRNA (miRNA) (2–10). MiRNAs are short (20–24 nucleotides) non-coding RNAs that are involved in the post-transcriptional regulation of gene expression in multicellular organisms by affecting both the stability and translation of mRNAs. MiRNAs target protein-coding mRNAs at the post-transcriptional level via direct cleavage of mRNAs or inhibition of protein synthesis (11–13). Recent studies identified these miRNAs in plasma and serum (plasma/serum), and their importance as minimally invasive liquid biomarkers for patients with cancer has been reported (2–10).
Several papers reported that circulating miRNA levels could reflect the treatment response to chemotherapy in patients with CRC (14–16). Furthermore, it is noteworthy that these miRNAs have also been identified in plasma/serum exosomes in a remarkably stable form that is protected against endogenous RNase activity (17). Exosomes are nano-sized vesicles (40–150 nm) derived from the luminal membranes of multivesicular bodies that are released via fusion with the cell membrane (18–20). Exosomes can contain tissue-specific signature proteins and intact miRNAs and transfer these components to other cells (18–20). Therefore, exosomal miRNAs (ex-miRNAs) may include potential biomarkers (21–23).
In the present study, we aimed to clarify the potential of plasma ex-miRNAs as minimally invasive liquid biomarkers for the early detection of resistance to mFOLFOX6-based chemotherapy in patients with advanced or recurrent (advanced/recurrent) CRC.
Patients and methods
Study design
First, we selected the exosomal miRNAs associated with resistance to mFOLFOX6 chemotherapy using a miRNA microarray. In this microarray analysis, samples were collected from patients with advanced CRC and stable disease (SD, n=3) or progressive disease (PD, n=3), as well as healthy volunteers (n=3). Next, we validated the usefulness of the selected miRNA (miR-125b) in an additional 55 patients with advanced/recurrent CRC. These patients were treated with mFOLFOX6-based chemotherapy in the first-line setting. These patients were observed from August 2007 to October 2017 at Teikyo University Hospital. The median follow-up period for first-line chemotherapy was 7 months (range, 2–30 months). Peripheral blood samples for ex-miR-125b measurement were collected from each patient before treatment and at various points during first-line mFOLFOX6-based chemotherapy until treatment were halted. In addition, 40 patients with stage I (10 cases), II (10 cases), III (10 cases), and IV CRC (10 cases) underwent examination of the correlation between plasma ex-miR-125b levels and miR-125b expression in tumor tissues. Blood samples were collected before treatment, and primary CRC tissues were collected during surgery. Healthy controls (n=30) consisted of healthy Japanese subjects with a similar average age (63 years) and gender ratio (male: Female =5:4) as patients with CRC.
The protocol of our study conformed according to the guidelines of the institutional ethics committee, and it was approved by the review board of Teikyo University (09-081-3). Written informed consent was obtained from all patients included in the present study.
Follow-up of patients after chemotherapy
Blood samples were collected every 2–3 weeks according to the chemotherapy protocol. Response to chemotherapy was evaluated according to Response Evaluation Criteria in Solid Tumors (RECIST) every 3 months. The therapeutic response criteria of the target lesion were classified into four groups: Complete response (CR), partial response (PR), SD, and PD. CR was denoted by the disappearance of the entire lesion. In this case, any pathological lymph nodes (whether target or non-target) must have a reduction in the short axis to <10 mm. PR was denoted by an at least 30% decrease in the sum of the diameters of the target lesions compared with the baseline sum diameters. PD was denoted by an at least 20% increase in the sum of the diameters of the target lesions compared with the smallest sum in the study (this includes the baseline sum if it were the smallest in the study). In addition to the relative increase of 20%, the sum must also display an absolute increase of at least 5 mm. The appearance of one or more new lesions was also considered progression. SD was indicated by the presence of insufficient shrinkage to qualify as a PR or a sufficient increase to qualify as PD compared with the smallest sum diameters in the study. Patients with PR after chemotherapy at any time point during first-line chemotherapy were included in the PR group. Patients who did not exhibit a PR or PD during first-line chemotherapy were classified into the SD group. Patients who did not achieve a PR at any time point but exhibited PD during first-line chemotherapy were classified into the PD group. The time point when the classification of patients was performed was defined as the ‘judgment point.’ The judgment point for patients in the PR or PD group was the time when the PR or PD was first observed. The judgment point for patients in the SD group was the time at which the final evaluation of RECIST was performed during first-line chemotherapy.
Purification of exosomes, electron microscopic imaging, and flow cytometry analysis
Peripheral blood was separated via centrifugation at 1,200 × g for 10 min at 4°C to collect plasma. One milliliter of plasma was used for microarray analysis and quantitative real-time reverse transcription-PCR (qRT-PCR). The exosomes of plasma were purified using a previously described ultracentrifugation method (100,000 × g for 70 min at 4°C) and stored at −80°C (24). The morphology of the isolated exosomes was confirmed via electron microscopy (HITACHI H-7600, Japan) as described previously (24). As a marker of exosomes, CD9 expression was analyzed by flow cytometry analysis using an Exo-FACS (Hansa Biomed Life-Sciences, Estonia) according to the manufacturer's protocol. For flow cytometry, FACSCanto™ II and FACSDiva™ software (v 8.0) (Becton, Dickinson and Company, NewJersey, USA) were used. After the gating to not aggregated fraction, fluorescence intensity of the exosome capture beads was detected. As negative control, fluorescence labeled mouse IgG isotype control was used.
Extraction of total RNAs from plasma exosomes and tissues
The total RNAs of exosomes were extracted using a miRNeasy serum/plasma kit (Qiagen, Hilden, Germany). Exosomes purified from specific volumes of plasma were diluted with 1 ml of Qiazol Lysis Reagent. After incubation for 5 min, 3.5 µl of a spike-in control (cel-miR-39 mimic) were added to each sample. Subsequent extraction and cartridge work were then performed according to the manufacturer's protocol. The total RNAs of the frozen tumor tissues and normal tissues were extracted using a miRNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA).
MiRNA microarray analysis
The miRNA expression profiles of the exosomes were examined using a 3D-Gene Human miRNA Oligo chip ver. 20 (TORAY Co., Ltd., Tokyo, Japan), according to the manufacturer's protocol. In total, 2578 genes were mounted onto the chip. The raw data from each spot were normalized by subtracting the background signal mean intensity, which was determined using the 95% confidence intervals of the signal intensities of all blank spots. Valid measurements were considered those in which the signal intensity of both duplicate spots was >2 standard deviations (SDs) of the background signal intensity.
qRT-PCR for miRNAs of plasma exosomes and tissues
MiRNA expression in the plasma exosomes and tissues was measured via qRT-PCR. In the exosomes, cDNA was synthesized from the total RNA using TaqMan MicroRNA primers specific for miR-125b (Thermo Fisher Scientific Inc., Waltham, MA, USA) and a TaqMan Micro-RNA Reverse Transcription Kit (Thermo Fisher Scientific Inc.). MiR-16a was used as an internal control. In the tissues, cDNA was synthesized from the total RNA using TaqMan MicroRNA primers specific for miR-125b (ID 00049) and RNU6B (ID 001093; Thermo Fisher Scientific Inc.) and a TaqMan Micro-RNA Reverse Transcription Kit. RNU6B was used as an internal control. qRT-PCR was performed using a Lightcycler-480 (Roche Applied Science, Basel, Switzerland) and TaqMan Universal PCR Master Mix (Life Technologies) according to the manufacturers' protocols. PCR reaction mixtures were incubated at 95°C for 10 min for denaturation, followed by 45 amplification cycles of 95°C for 15 sec and 60°C 1 min, and 40°C for 30 sec for extension. Each sample was analyzed in duplicate. The relative miRNA expression was calculated using the 2−ΔΔCT method as described previously (24,25).
Statistical analysis
The data were expressed as the mean ± SD. The relationship between RECIST and patient characteristics were analyzed using Student's t-test, the chi-squared test, and ANOVA. The Tukey-Kramer method was used as the post hoc test following ANOVA. Progression-free survival (PFS) curves were analyzed using the Kaplan-Meier survival curve method, and the differences were examined using log-rank tests. The cut-off value of relative quantification for miR-125b was set as 0.42, which was the median. Cox proportional-hazard regression analysis was used to estimate the univariate and multivariate hazard ratios for PFS. Univariate analysis was conducted for each factor, and multivariate analysis was performed for factors displaying significance in univariate analysis. All p-values were two-sided, and P<0.05 was considered statistically significant. Statistical analyses were performed using JMP 9.0 software (SAS Inst. Inc. NC, Japan).
Results
Identification of exosomes in plasma
To verify the isolation of exosomes from the ultracentrifuged samples of patients with CRC, morphological identification using transmission electron microscopy and surface marker analysis using FACS were performed (Fig. 1). Using electron microscopy, images of the round microvesicles with diameters of approximately 50–100 nm were captured, as shown in Fig. 1A. In the FACS analysis, CD9-positive exosomes were confirmed (Fig. 1B).
Figure 1.
Morphology and surface marker expression of exosomes. (A) Exosome images produced via transmission electron microscopy. (B) CD9-positive exosomes were analyzed using FACS analysis.
Exosomal miRNA profiling via miRNA array analysis
To identify the ex-miRNA associated with resistance to mFOLFOX6, we conducted miRNA array analysis of plasma exosome samples collected from patients with SD or PD as well as healthy controls (each n=3). The clinical data and background of the patients with CRC and healthy volunteers are described in Table SI and SII. No significant difference in race, sex, or average age was noted between patients with CRC and healthy volunteers. Table I shows the five most highly upregulated ex-miRNAs in patients with PD compared with those in patients with SD and healthy controls. In these comparisons, miR-125b (MIMAT0000423) displayed the highest fold changes as in patients with PD compared with the findings in patients with SD (3.56-fold) and healthy controls (4.41-fold). Based on these results, we selected ex-miR-125b as a predictive marker for resistance to mFOLFOX6-based chemotherapy in patients with advanced/recurrent CRC.
Table I.
Five most highly upregulated plasma ex-miRNAs of patients with PD in comparison with those with SD and healthy volunteers
| Fold changes | ||||
|---|---|---|---|---|
| Ranks | miRNA | MirBase no. | PD patients vs. healthy volunteersa | PD patients vs. SD patientsb |
| 1 | miR-125b | MIMAT0000423 | 4.41 | 3.56 |
| 2 | miR-19a | MIMAT0000073 | 4.02 | 3.32 |
| 3 | miR-21 | MIMAT0000076 | 3.98 | 2.91 |
| 4 | miR-106a | MIMAT0000103 | 3.87 | 2.85 |
| 5 | miR-155 | MIMAT0000046 | 3.43 | 2.61 |
MicroRNA levels of patients with PD (n=3) were compared with those of healthy volunteers (n=3).
MicroRNA levels of patients with PD (n=3) were compared with those of patients with SD (n=3). PD, progressive disease; SD, stable disease; miRNA, microRNA.
Patient characteristics and RECIST
In our study, the patients were divided into two groups, namely the PR or SD (PR/SD) (n=8) and PD groups (n=47). Table II provides a comparison of patient characteristics between these groups. There were no significant differences in age, gender, tumor location, metastatic site, KRAS mutation status, performance status, and serum CA19-9 and CEA levels before chemotherapy between the two groups.
Table II.
Comparison of patient characteristics in the PR/SD and PD groups.
| Variables | PR/SD (n=8) number (%) | PD (n=47) number (%) | P-value |
|---|---|---|---|
| Age (year) | 64.5±4.8 | 62.4±11.3 | 0.60 |
| Sex | |||
| Male | 5 (62.5) | 26 (55.3) | 0.71 |
| Female | 3 (37.5) | 21 (44.7) | |
| Location | |||
| Colon | 3 (37.5) | 31 (66.0) | 0.13 |
| Rectum | 5 (62.5) | 16 (34.0) | |
| Metastatic sites | |||
| 1 | 4 (50.0) | 17 (36.2) | 0.46 |
| >1 | 4 (50.0) | 30 (63.8) | |
| KRAS | |||
| Mutation | 2 (25.0) | 26 (55.3) | 0.08 |
| Wild-type | 6 (75.0) | 21 (46.7) | |
| Performance status (ECOG) | |||
| 0 | 1 (12.5) | 5 (10.6) | 0.10 |
| 1 | 5 (62.5) | 40 (85.1) | |
| 2 | 2 (25.0) | 2 (4.3) | |
| Serum CA19-9a | |||
| ≤37.0 u/ml | 5 (62.5) | 21 (44.7) | 0.35 |
| >37.0 u/ml | 3 (37.5) | 26 (55.3) | |
| Serum CEAa | |||
| ≤5.0 ng/ml | 4 (50.0) | 16 (34.0) | 0.39 |
| >5.0 ng/ml | 4 (50.0) | 31 (66.0) |
Serum CA19-9 and CEA levels were measured before chemotherapy. PD, progressive disease; SD, stable disease; PR, partial response; ECOG, Eastern Cooperative Oncology Group; CEA, carcinoembryonic antigen.
Comparison of ex-miR-125b levels between patients with CRC and healthy controls
A comparison of ex-miR-125b levels between patients with CRC and healthy controls is shown in Fig. 2. Patients with CRC had significantly higher ex-miR-125b levels than healthy controls.
Figure 2.
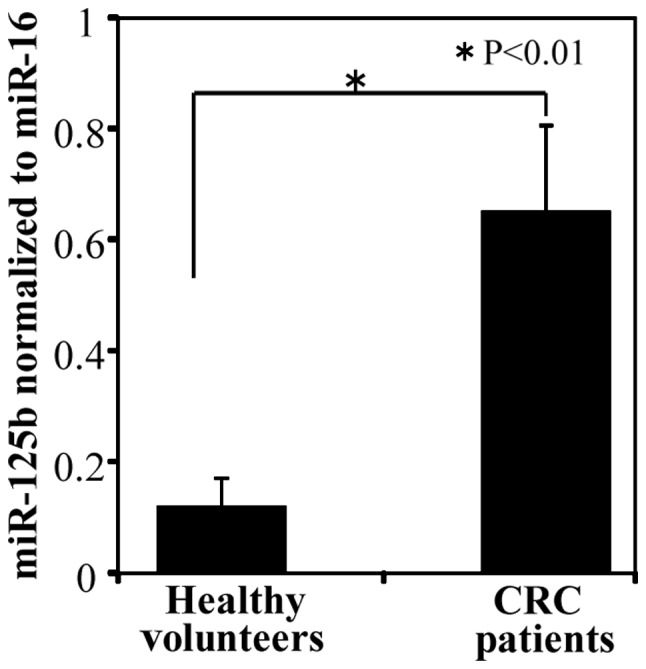
Comparison of ex-miR-125b levels in healthy controls and patients with colorectal cancer (CRC). Ex-miR-125b levels were significantly higher in patients with CRC (n=55) than in healthy controls (n=30). Ex-miR-125b, exosomal microRNA-125b.
Ex-miR-125b levels and RECIST
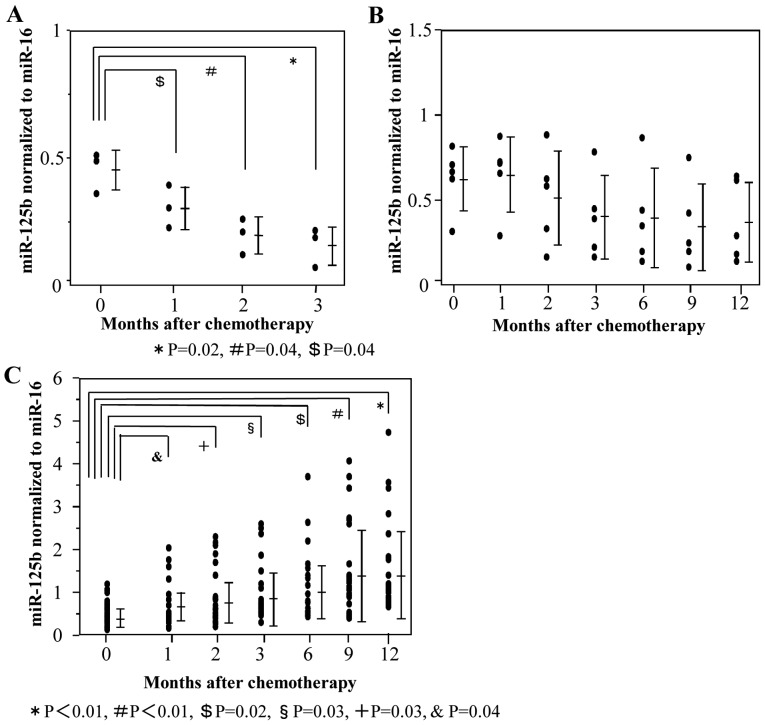
First, plasma ex-miR-125b levels were compared at each point during mFOLFOX6-based treatment (Fig. 3). In patients with PR, ex-miR-125b levels after 1, 2, and 3 months of treatment were significantly lower than those measured before treatment (Fig. 3A). In patients with SD, there were no significant differences of ex-miR-125b levels between before and during treatment (Fig. 3B). In patients with PD, ex-miR-125b levels after 1, 2, 3, 6, 9, and 12 months of treatment were significantly higher than those measured before treatment (Fig. 3C).
Figure 3.
Changes of ex-miR-125b levels in the PR, SD and PD groups during treatment. Ex-miR-125b levels before chemotherapy and after 1, 2, 3, 6, 9 and 12 months of mFOLFOX6-based chemotherapy were compared among patients with (A) PR, (B) SD and (C) PD. Ex-miR-125b, exosomal microRNA-125b; PR, partial response; SD, stable disease; PD, progressive disease.
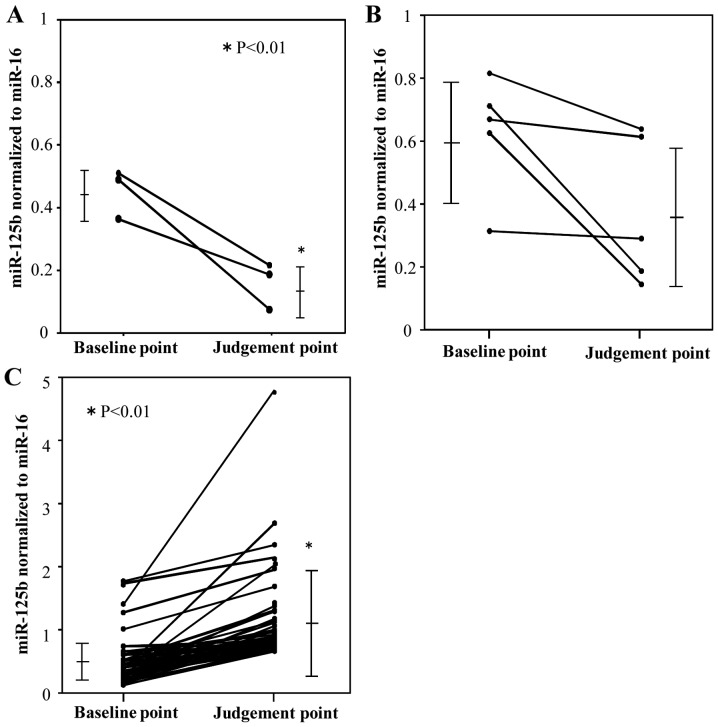
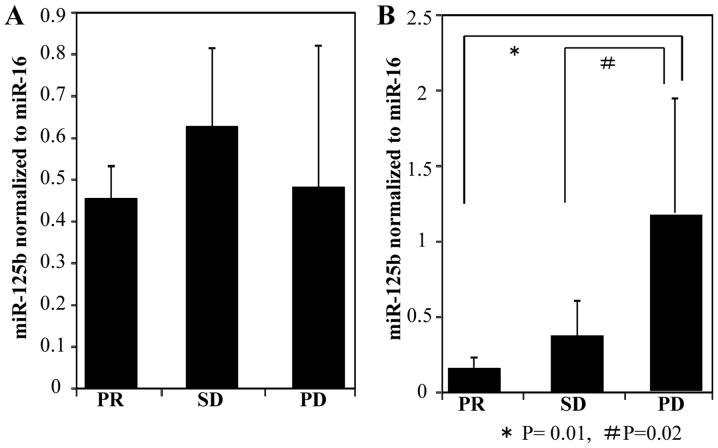
Next, plasma ex-miR-125b levels were compared at two time points, namely before treatment initiation (baseline point) and the judgment point (Fig. 4). The judgment point was 3 months after treatment initiation for patients with PR (n=3). The judgment point was 6 (n=1), 9 (n=1), or 12 months (n=3) for patients with SD, whereas that for patients with was 3 (n=7), 6 (n=13), 9 (n=8), or 12 months (n=19). Among patients with PR, ex-miR-125b levels were significantly lower at the judgment point than at the baseline point (Fig. 4A). Among patients with SD, ex-miR-125b levels did not significantly differ between these two points (Fig. 4B). Among patients with PD, ex-miR-125b levels were significantly higher at the judgment point than at the baseline point (Fig. 4C). In addition, ex-miR-125b levels were compared among the PR, SD, and PD groups at the baseline and judgment points (Fig. 5). At the baseline point, ex-miR-125b levels did not differ among the three groups (Fig. 5A). At the judgment point, ex-miR-125b levels were significantly higher in the PD group than in the PR and SD groups (Fig. 5B).
Figure 4.
Comparison of ex-miR-125b levels in the PR, SD and PD groups at the baseline and RECIST judgment points. Ex-miR-125b levels at the baseline and judgment points were compared among the (A) PR, (B) SD and (C) PD groups. Ex-miR-125b, exosomal microRNA-125b; PR, partial response; SD, stable disease; PD, progressive disease; RECIST, Response Evaluation Criteria in Solid Tumors.
Figure 5.
Comparison of ex-miR-125b levels among the PR, SD and PD groups. (A) Ex-miR-125b levels at the treatment initiation point (baseline point) were compared among the groups. (B) Ex-miR-125b levels at the Response Evaluation Criteria in Solid Tumors judgment point after treatment (judgment point) were compared among the groups. Ex-miR-125b, exosomal microRNA-125b; PR, partial response; SD, stable disease; PD, progressive disease.
These results support the potential of ex-miR-125b as a biomarker for the early selection of resistance to mFOLFOX6-based treatment.
Kaplan-Meier PFS curves based on ex-miR-125b levels
The Kaplan-Meier PFS curves of patients with advanced/recurrent CRC (n=55) were compared according to the presence of low or high ex-miR-125b levels. In this analysis, patients with high ex-miR-125b levels exhibited significantly worse PFS than those with low levels (Fig. 6).
Figure 6.
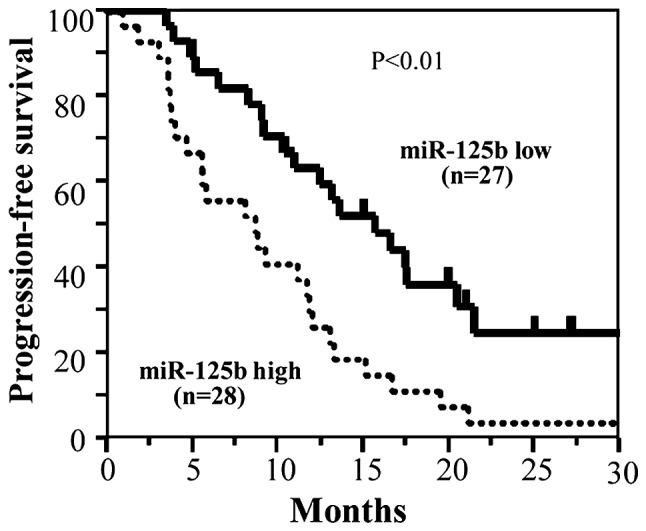
Kaplan-Meier progression-free survival rates based on ex-miR-125b levels in patients with advanced and recurrent colorectal cancer. Ex-miR-125b, exosomal microRNA-125b.
Cox analysis of PFS based on ex-miR-125b levels
The results of Cox univariate and multivariate regression analyses for PFS are shown in Table III. As variables, gender, location, metastatic site, KRAS mutation, performance status, serum CEA, serum CA19-9 and ex-miR-125b were examined. In the univariate analysis, KRAS mutation and ex-miR-125b were identified as significant variables for PFS, and these variables remained significance in multivariate analysis.
Table III.
Cox univariate and multivariate analyses of prognostic factors for PFS in patients with colorectal cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | Regression coefficient | HR (95% CI) | P-value | Regression coefficient | HR (95% CI) | P-value |
| Sex | 0.02 | 1.02 (0.56–1.83) | 0.937 | |||
| Location | −0.17 | 0.84 (0.44–1.54) | 0.577 | |||
| Metastatic sites | 0.03 | 1.06 (0.51–2.19) | 0.867 | |||
| KRAS | 0.56 | 0.78 (0.46–0.89) | 0.041 | 0.79 | 0.85 (0.56–0.97) | 0.047 |
| Performance Status (ECOG) | −0.45 | 0.41 (0.10–1.69) | 0.215 | |||
| Serum CA19-9a | −0.01 | 0.99 (0.54–1.78) | 0.973 | |||
| Serum CEAa | 0.02 | 1.02 (0.55–1.45) | 0.159 | |||
| miR-125b baseline | −0.66 | 0.52 (0.27–0.95) | 0.035 | 1.04 | 0.71 (0.36–0.94) | 0.041 |
Serum CA19-9 and CEA levels were measured before chemotherapy. PFS, progression-free survival; HR, hazard ratio; CI, confidence interval; miR, microRNA; ECOG, Eastern Cooperative Oncology Group; CEA, carcinoembryonic antigen.
These results suggest that ex-miR-125b has independent prognostic value for PFS in patients with advanced/recurrent CRC.
MiR-125b expression in plasma exosomes and primary tumor tissues
Fig. 7 shows the correlation of miR-125b levels between plasma exosomes and primary tumor tissues in the same patients. A positive correlation was observed between the levels.
Figure 7.
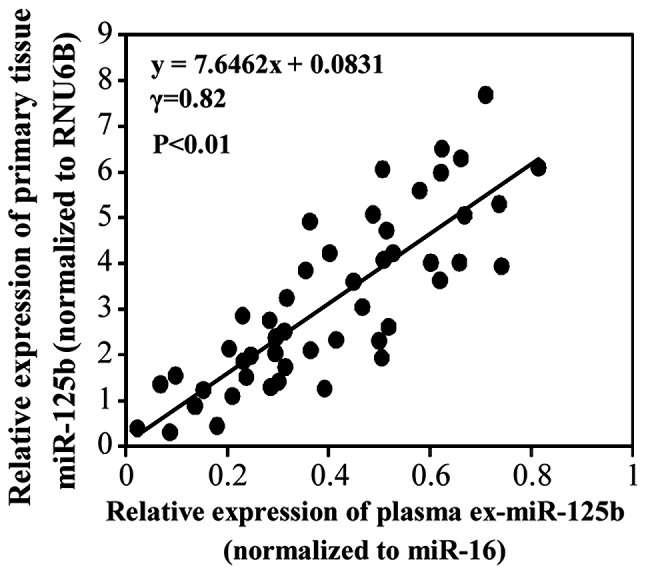
Correlation of miR-125b expression between plasma exosomes and primary CRC tissues. The relative expression of miR-125b in plasma exosomes and cancer tissues displayed a significant positive correlation in patients with CRC. This study included paired samples (plasma and tissue) obtained from 40 patients with TNM stage I (n=10), II (n=10), III (n=10) and stage IV CRC (n=10). miR-125b, microRNA-125b; CRC, colorectal cancer.
Discussion
In the present study, we demonstrated the potential of ex-miR-125b as a minimally invasive liquid biomarker for the early selection of patients with CRC with resistance to mFOLFOX6-based first-line chemotherapy. In addition, our data demonstrated that ex-miR-125b is an independent predictive factor for PFS.
Chemotherapy is the first-line treatment for unresectable advanced/recurrent CRC, with mFOLFOX6-based chemotherapy being the most common regimen. Despite an improvement in the overall prognosis of patients with CRC, these chemotherapeutics fail to work in the majority of patients because of intrinsic or acquired drug resistance. Unfortunately, no effective biomarkers that can monitor resistance to mFOLFOX6-based chemotherapy are available, and thus, the identification of a new biomarker is highly desired.
Recently, circulating plasma/serum miRNAs have attracted interest as novel biomarkers for diagnosis and the prediction of prognosis for various cancers including CRC. Previously, we reported the usefulness of ex-miR-21 in the prediction of recurrence and prognosis in patients with stage II, III, or IV CRC (24). However, few studies examined the potential of plasma ex-miRNA for the early detection of resistance to chemotherapy in patients with CRC. Initially, we searched for plasma ex-miRNAs predicting resistance to mFOLFOX6-based treatment via a genome-wide miRNA profiling approach using microarray analysis. Our research indicated that ex-miR-125b was remarkably upregulated in patients with PD compared with its levels in patients with SD and healthy controls. Based on these results, we selected miR-125b as a biomarker for resistance to mFOLFOX6-based chemotherapy in patients with advanced/recurrent CRC.
Until now, studies of biomarkers using free plasma/serum microRNAs not encapsulated in exosomes have also been reported. In our preliminary experiments, we compared the levels of plasma free miR-125b and ex-miR-125b. We found that the free miR-125b levels were unstable compared to those of ex-miR-125b. In particular, the free plasma miRNA levels markedly decreased in samples that had been frozen for a long period (data not shown). To obtain stable and reliable data, we measured ex-miR-125b, but not free plasma miR-125b. Furthermore, we compared the usefulness of free miR-16 and ex-miR-16 as internal standard, and found that ex-miR-16 is stable and can use as an internal standard in CRC patients (data not shown). Other studies have also used ex-miR-16 as an internal standard of CRC (26).
MiR-125b was one of the first miRNAs identified in C. elegans (27). MiR-125b belongs to the miR-125 family, consisting of miR-125a, miR-125b-1, and miR-125b-2, which result in almost identical products of distinct genes. Several tumors including breast, gastric and pancreatic cancer lesions exhibit upregulated miR-125b expression (27). In the present study, we demonstrated that plasma ex-miR-125b levels were significantly higher in patients with CRC than in healthy controls. Yamada et al also recorded elevated serum miR-125b levels in patients whose total CRC diameters were 5 mm or less (28). These results suggest that miR-125b could discriminate between patients with early CRC and healthy controls.
Next, we examined the relationship between ex-miR-125b levels and resistance to mFOLFOX6-based first-line chemotherapy in patients with advanced/recurrent CRC. In patients with PR, miR-125b levels were significantly lower at the judgment point than at the baseline point. Among patients with SD, no significant differences in miR-125b levels were noted. Among patients with PD, miR-125b levels were significantly higher at the judgment point than at the baseline point. These results indicated a significant association between ex-miR-125b levels and tumor status. Furthermore, our results demonstrated that ex-miR-125b levels significantly changed after 1 month of mFOLFOX6-based first-line treatment in patients with PR and PD. These results suggest that ex-miR-125b can facilitate the early selection of patients exhibiting resistance to mFOLFOX6-based chemotherapy. Prior studies revealed associations of elevated miR-125b expression with chemoresistance and metastasis. Using biopsy samples, Svoboda et al reported that high miR-125b and miR-137 levels are associated with worse response to capecitabine chemoradiotherapy in patients with rectal cancer (29).
D'Angelo et al reported that high miR-125b expression in tissue and serum was associated with a poor treatment response in locally advanced rectal cancer (30). Interestingly, Tanaka et al identified miR-125b-1 expression in cancer tissues as a predictive biomarker for the efficacy of peptide vaccine treatment against CRC (31). However, our study is the first to demonstrate the potential of plasma ex-miR-125b as a biomarker for the early detection of resistance to mFOLFOX6-based chemotherapy in advanced/recurrent CRC.
Furthermore, we demonstrated that PFS was significantly worse in patients with high ex-miR-125b levels than in those with low levels. Cox multivariate analysis identified ex-miR-125b as an independent predictive biomarker for PFS. Regarding plasma miRNAs and PFS, Hansen et al reported that changes in circulating plasma miR-126 levels during treatment are related to PFS in patients with metastatic CRC treated with bevacizumab (32). However, no paper has discussed the relationship between ex-miR-125b and PFS. To the best of our knowledge, this is the first study to clarify the predictive value of plasma ex-miR-125b levels for PFS in patients with advanced/recurrent CRC treated with mFOLFOX6-based chemotherapy.
Numerous studies demonstrated the roles of miR-125b in cell proliferation, apoptosis, and differentiation, suggesting its potentially important role during carcinogenesis. MiR-125b expression has been detected in several tumor types. MiR-125b appears to downregulate anti-apoptotic proteins, resulting in reduced apoptosis and enhanced cellular proliferation, thereby promoting tumor growth. Recent studies found that miR-125b directly targets the tumor suppressor gene p53, which is essential for the maintenance of genome stability and plays a central role in regulating apoptosis. Using CRC cell lines, Lu et al reported that miR-125b and miR-100 mediate cetuximab resistance via negative regulation of Wnt/β-catenin (33).
However, we recognize some limitations of our study. Because of the small number of patients with CR (n=0) and PR (n=3), we compared the PR/SD (n=8) and PD (n=47) groups, but not the CR/PR and SD/PD groups, before analyzing the clinical data. To clarify the usefulness of ex-miR-125b as a monitoring biomarker for mFOLFOX6-based chemotherapy, a larger prospective study comparing the CR/PR and SD/PD groups is required.
In summary, we identified ex-miR-125b as a promising minimally invasive biomarker for the early detection of resistance to mFOLFOX6-based first-line chemotherapy in patients with CRC. The early detection of poor response to mFOLFOX6-based chemotherapy using ex-miR-125b may prevent unnecessary side effects and facilitate more rigorous and safer individualized treatment.
Supplementary Material
Acknowledgements
The authors would like to thank Miss J Tamura (Department of Surgery, Teikyo University School of Medicine, Tokyo, Japan) for her excellent technical assistance and all members of the colorectal group (Department of Surgery, Teikyo University School of Medicine, Tokyo, Japan) for their clinical suggestions.
Glossary
Abbreviations
- RT-PCR
reverse transcription-polymerase chain reaction
- CRC
colorectal cancer
Funding
This study is partly supported by Grants-in-Aid for Scientific Research from the Japanese Society for the Promotion of Science (grant nos. 15K10150, 17K10655 and 18K08716).
Availability of data and materials
All data generated or analyzed during the present study are included in this article.
Authors' contributions
HI conceived and designed the study. TY, HI and YH wrote the manuscript. TY and HI performed the experiment. YH, TH, KM, KN, MT, RS, TA, TT and TO collected the clinical data. HI and YH reviewed and edited the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
The study protocol conformed to the guidelines of the Teikyo University Ethics Committee, and it was approved by the review board of Teikyo University (approval no. 09-081-3). Written informed consent was obtained from all patients.
Patient consent for publication
Written informed consent was obtained from all patients for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T, Alizadeh-Navaei R, Allen C, Alsharif U, Alvis-Guzman N, Amini E, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: A systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4:1553–1568. doi: 10.1001/jamaoncol.2018.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 3.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang L, Huang D, Tan C, Sheng W, Du X. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One. 2012;7:e44398. doi: 10.1371/journal.pone.0044398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S. Plasma miR-21: A potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544–551. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 6.Giráldez MD, Lozano JJ, Ramírez G, Hijona E, Bujanda L, Castells A, Gironella M. Circulating microRNAs as biomarkers of colorectal cancer: Results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol. 2013;11:681–688.e3. doi: 10.1016/j.cgh.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Hofsli E, Sjursen W, Prestvik WS, Johansen J, Rye M, Tranø G, Wasmuth HH, Hatlevoll I, Thommesen L. Identification of serum microRNA profiles in colon cancer. Br J Cancer. 2013;108:1712–1719. doi: 10.1038/bjc.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo X, Stock C, Burwinkel B, Brenner H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One. 2013;8:e62880. doi: 10.1371/journal.pone.0062880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X, Yu L, Wang L, Wang J, Wu Y, et al. A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer. 2015;136:152–161. doi: 10.1002/ijc.28136. [DOI] [PubMed] [Google Scholar]
- 11.Aslam MI, Taylor K, Pringle JH, Jameson JS. microRNAs are novel biomarkers of colorectal cancer. Br J Surg. 2009;96:702–710. doi: 10.1002/bjs.6628. [DOI] [PubMed] [Google Scholar]
- 12.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 13.Croce CM, Calin GA. miRNAs, cancer, and stem cell ivision. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 14.Stiegelbauer V, Perakis S, Deutsch A, Ling H, Gerger A, Pichler M. MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World J Gastroenterol. 2014;20:11727–11735. doi: 10.3748/wjg.v20.i33.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Zhang K, Bi M, Jiao X, Zhang D, Dong Q. Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anticancer Drugs. 2014;25:346–352. doi: 10.1097/CAD.0000000000000049. [DOI] [PubMed] [Google Scholar]
- 16.Molina-Pinelo S, Carnero A, Rivera F, Estevez-Garcia P, Bozada JM, Limon ML, Benavent M, Gomez J, Pastor MD, Chaves M, et al. MiR-107 and miR-99a-3p predict chemotherapy response in patients with advanced colorectal cancer. BMC Cancer Sep. 2014;14:656. doi: 10.1186/1471-2407-14-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z. miRNA in plasma exosome is stable under different storage conditions. Molecules. 2014;19:1567–1575. doi: 10.3390/molecules19021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 19.Simons M, Raposo G. Exosomes-vesicular carriers for intercelluar communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Hannafon BN, Ding WQ. Intercelluar communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu M, Huang Z, Zhu D, Zhou X, Shan X, Qi LW, Wu L, Cheng W, Zhu J, Zhang L, et al. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget. 2017;8:17081–17091. doi: 10.18632/oncotarget.15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo RH, Cheng WF, Wang F, Qi LW, Chen Y, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2017;8:6513–6525. doi: 10.18632/oncotarget.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, Xiang Y, Wu N, Wu L, Bai L, Li Y. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. 2017;8:13048–13058. doi: 10.18632/oncotarget.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukamoto M, Iinuma H, Yagi T, Matsuda K, Hashiguchi Y. Circulating exosomal MicroRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology. 2017;92:360–370. doi: 10.1159/000463387. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, Hirata H, Kuroda Y, Nambara S, Hayashi N, et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017;8:78598–78613. doi: 10.18632/oncotarget.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banzhaf-Strathmann J, Edbauer D. Good guy or bad guy: The opposing roles of microRNA 125b in cancer. Cell Commun Signal. 2014;12:30. doi: 10.1186/1478-811X-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, et al. Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res. 2015;21:4234–4242. doi: 10.1158/1078-0432.CCR-14-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svoboda M, Izakovicova Holla L, Sefr R, Vrtkova I, Kocakova I, Tichy B, Dvorak J. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;33:541–547. [PubMed] [Google Scholar]
- 30.D'Angelo E, Fassan M, Maretto I, Pucciarelli S, Zanon C, Digito M, Rugge M, Nitti D, Agostini M. Serum miR-125b is a non-invasive predictive biomarker of the pre-operative chemoradiotherapy responsiveness in patients with rectal adenocarcinoma. Oncotarget. 2016;7:28647–28657. doi: 10.18632/oncotarget.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka H, Hazama S, Iida M, Tsunedomi R, Takenouchi H, Nakajima M, Tokumitsu Y, Kanekiyo S, Shindo Y, Tomochika S, et al. miR-125b-1 and miR-378a are predictive biomarkers for the efficacy of vaccine treatment against colorectal cancer. Cancer Sci. 2017;108:2229–2238. doi: 10.1111/cas.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen TF, Carlsen AL, Heegaard NH, Sørensen FB, Jakobsen A. Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br J Cancer. 2015;112:624–629. doi: 10.1038/bjc.2014.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, Singh B, Franklin JL, Wang J, Hu H, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat Med. 2017;23:1331–1341. doi: 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the present study are included in this article.