Abstract
Background
This is an update of the original review published in the Cochrane Database of Systematic Reviews Issue 10, 2015.
Invasive aspergillosis (IA) is the most common life‐threatening opportunistic invasive mould infection in immunocompromised people. Early diagnosis of IA and prompt administration of appropriate antifungal treatment are critical to the survival of people with IA. Antifungal drugs can be given as prophylaxis or empirical therapy, instigated on the basis of a diagnostic strategy (the pre‐emptive approach) or for treating established disease. Consequently, there is an urgent need for research into both new diagnostic tools and drug treatment strategies. Increasingly, newer methods such as polymerase chain reaction (PCR) to detect fungal nucleic acids are being investigated.
Objectives
To provide an overall summary of the diagnostic accuracy of PCR‐based tests on blood specimens for the diagnosis of IA in immunocompromised people.
Search methods
We searched MEDLINE (1946 to June 2015) and Embase (1980 to June 2015). We also searched LILACS, DARE, Health Technology Assessment, Web of Science and Scopus to June 2015. We checked the reference lists of all the studies identified by the above methods and contacted relevant authors and researchers in the field. For this review update we updated electronic searches of the Cochrane Central Register of Controlled Trials (CENTRAL; 2018, Issue 3) in the Cochrane Library; MEDLINE via Ovid (June 2015 to March week 2 2018); and Embase via Ovid (June 2015 to 2018 week 12).
Selection criteria
We included studies that: i) compared the results of blood PCR tests with the reference standard published by the European Organisation for Research and Treatment of Cancer/Mycoses Study Group (EORTC/MSG); ii) reported data on false‐positive, true‐positive, false‐negative and true‐negative results of the diagnostic tests under investigation separately; and iii) evaluated the test(s) prospectively in cohorts of people from a relevant clinical population, defined as a group of individuals at high risk for invasive aspergillosis. Case‐control and retrospective studies were excluded from the analysis.
Data collection and analysis
Authors independently assessed quality and extracted data. For PCR assays, we evaluated the requirement for either one or two consecutive samples to be positive for diagnostic accuracy. We investigated heterogeneity by subgroup analyses. We plotted estimates of sensitivity and specificity from each study in receiver operating characteristics (ROC) space and constructed forest plots for visual examination of variation in test accuracy. We performed meta‐analyses using the bivariate model to produce summary estimates of sensitivity and specificity.
Main results
We included 29 primary studies (18 from the original review and 11 from this update), corresponding to 34 data sets, published between 2000 and 2018 in the meta‐analyses, with a mean prevalence of proven or probable IA of 16.3 (median prevalence 11.1% , range 2.5% to 57.1%). Most patients had received chemotherapy for haematological malignancy or had undergone hematopoietic stem cell transplantation. Several PCR techniques were used among the included studies. The sensitivity and specificity of PCR for the diagnosis of IA varied according to the interpretative criteria used to define a test as positive. The summary estimates of sensitivity and specificity were 79.2% (95% confidence interval (CI) 71.0 to 85.5) and 79.6% (95% CI 69.9 to 86.6) for a single positive test result, and 59.6% (95% CI 40.7 to 76.0) and 95.1% (95% CI 87.0 to 98.2) for two consecutive positive test results.
Authors' conclusions
PCR shows moderate diagnostic accuracy when used as screening tests for IA in high‐risk patient groups. Importantly the sensitivity of the test confers a high negative predictive value (NPV) such that a negative test allows the diagnosis to be excluded. Consecutive positives show good specificity in diagnosis of IA and could be used to trigger radiological and other investigations or for pre‐emptive therapy in the absence of specific radiological signs when the clinical suspicion of infection is high. When a single PCR positive test is used as the diagnostic criterion for IA in a population of 100 people with a disease prevalence of 16.3% (overall mean prevalence), three people with IA would be missed (sensitivity 79.2%, 20.8% false negatives), and 17 people would be unnecessarily treated or referred for further tests (specificity of 79.6%, 21.4% false positives). If we use the two positive test requirement in a population with the same disease prevalence, it would mean that nine IA people would be missed (sensitivity 59.6%, 40.4% false negatives) and four people would be unnecessarily treated or referred for further tests (specificity of 95.1%, 4.9% false positives). Like galactomannan, PCR has good NPV for excluding disease, but the low prevalence of disease limits the ability to rule in a diagnosis. As these biomarkers detect different markers of disease, combining them is likely to prove more useful.
Plain language summary
A new, non‐invasive diagnostic blood test — polymerase chain reaction — for people at risk of an invasive mould infection (aspergillosis)
Review question We reviewed the evidence about the accuracy of polymerase chain reaction (PCR) tests for diagnosing invasive aspergillosis (IA) among people with defective immune systems from medical treatment such as chemotherapy or following organ or bone marrow transplant.
Background IA is a fungal disease caused by the widespread mould Aspergillus, with Aspergillus fumigatus being the most common species. Most people breathe in Aspergillus spores every day without becoming ill. However people with weakened immune systems or lung diseases are at a higher risk of developing respiratory problems of the lungs and sinuses due to Aspergillus, ranging from allergic complications to IA, which is the most common life‐threatening, invasive fungal infection of people whose immune systems are compromised. Without antifungal treatment, most people with IA will die as a direct result of IA, so early diagnosis and prompt administration of appropriate antifungal treatment are both critical to the survival of these people. The ideal specimen for diagnosing IA would be lung tissue but obtaining this carries a significant risk to the patient so there is a clear need for new, non‐invasive methods such as PCR to demonstrate the fungus’s presence in blood by detecting its nucleic acids.
Study characteristics We conducted our most recent search for studies in March 2018 and combined with an earlier search selected 29 clinical studies reporting the evaluation of PCR tests prospectively in cohorts of people at high risk of IA.
Study funding sources None of the companies involved in the diagnosis of invasive fungal diseases funded any of the studies included in the review.
Quality of the evidence Most studies were at low risk of bias and low concern regarding applicability. However, differences in the reference standard may have contributed to differences we found in the distribution of cases as being classified as IA or not.
Key results Several PCR techniques were used in the studies. Pooling the data from the studies showed that sensitivity and specificity of PCR for the diagnosis of IA varied (from 59% to 79.2% and from 79% to 95.2%, respectively) depending on the interpretative criteria used to define a test as positive. When used as a diagnostic criterion for IA in a population of 100 people with a disease prevalence of 16.3% (overall mean prevalence), a single PCR positive test would have missed three people with the disease, and falsely classified 17 people as having the disease, who would be treated unnecessarily or referred for further tests. A requirement of two positive tests as a diagnostic criterion in a population with the same disease prevalence would miss nine people with the disease and falsely classify four people as having the disease. These numbers should be interpreted with caution because the reference standard is based on the degree of certainty of diagnosis and is rarely proven so cannot provide consistent assessment of cases as being IA or not.
Overall, PCR shows moderate diagnostic accuracy when used as a screening test for IA in high‐risk patient groups. Importantly, when the rate of sensitivity is low, the sensitivity of the tests means that a negative result allows the diagnosis to be excluded with confidence except when the patient is receiving certain antifungal drugs. With the low prevalence of the disease, a high negative predictive value such that a negative test allows the diagnosis to be excluded.
Summary of findings
Summary of findings'. 'Summary of findings table. PCR for the diagnosis of invasive aspergillosis.
|
Review question: what is the diagnostic accuracy of aspergillus PCR blood test for detection of invasive aspergillosis (IA)? Patients/population: patients at risk of IA, including neutropenic cancer patients and HSCT or solid organ transplant recipients Index test: PCR on blood specimens (whole blood or serum). We considered different DNA extraction methods and PCR methods (e.g. nested, ELISA, qPCR) Reference standard: EORTC/MSG criteria for invasive aspergillosis Studies: cohort studies | ||||
| Index Test: interpretative criteria to define a test as positive | Effect (95% CI) | No. of studies | Mean prevalence (range) | What do these results mean? |
| 1 Single PCR specimen | sensitivity: 79.2% (71.0% to 85.5%) specificity: 79.6% (69.9% to 86.6%) |
27 studies | 16.3% (2.5% to 57.1%) | With a prevalence of 16%, 16 out of 100 patients will develop IA. Of these, 3 will be missed by a single PCR test (20.8% of 16); of the 84 patients without IA, 17 will have a false positive result of the PCR test; repeating the test will reduced significantly rates of false positive results. |
| ≥ 2 PCR specimens | sensitivity: 59.6% (40.7% to 76.0%) specificity: 95.1% (87.0% to 98.2%) |
9 studies | 16.3% (2.5% to 57.1%) | With a prevalence of 16%, 16 out of 100 patients will develop IA. Of these, 9 will be missed using the 2 positive PCR test (40.4% of 16); of the 84 patients without IA, 4 will have a false positive result of the PCR test. |
The PCR methods varied notably across studies. Several covariates (in particular, the adoption of antifungal prophylaxis and blinding to the reference test or index test) were found to substantively affect the measures of diagnostic accuracy under evaluation, mainly sensitivity and specificity. CI: confidence interval IA: invasive aspergillosis PCR: polymerase chain reaction
Background
Target condition being diagnosed
Invasive aspergillosis (IA) is a disease resulting from opportunistic fungal infection and mainly affects immunocompromised hosts, particularly neutropenic patients such as those undergoing cancer treatment and hematopoietic stem cell transplantation (HCT) and solid organ transplant recipients (Flückiger 2006; Marr 2002). The highest incidence (10% to 20%) and mortality rates (60% to 90%) of IA have been reported following allogeneic HCT and heart, lung or heart/lung transplantation. The principal reason for such people developing IA is that the underlying disease and its subsequent treatment with chemotherapy induces bone marrow failure resulting in profound leucopenia and impaired innate and cell‐mediated immunity. The leucopenia is marked by a lack of functioning polymorphonuclear leucocytes, resulting in the patient lacking the phagocytic white blood cells needed to fight infections, including aspergillosis. Innate immunity is further impaired by iatrogenic damage to the local defences of the oral cavity, gastrointestinal tract and respiratory tract. Damage to the respiratory tract is poorly understood but hampers effective clearance of fungal spores, especially those of Aspergillus fumigatus which are ubiquitous in the environment, readily airborne and small enough to lodge in the alveolar spaces. The lack of local and systemic immune defences means that any spores that germinate can infect lung tissue and progress to a full‐blown infection. The disease that follows is characterised by invasion of the capillaries (angioinvasion) which can lead to further dissemination to other parts of the lung and indeed other organs, particularly the brain.
Early diagnosis of IA and prompt administration of appropriate antifungal treatment have been recognised as crucial to the survival of people with IA (Marr 2002; Walsh 2008). Antifungal drugs can be given for prevention of infection (prophylaxis), treatment of unexplained fever (empirical therapy), treatment of non‐specific clinical features or mycological evidence (pre‐emptive therapy) and treatment of possible, probable and proven invasive fungal disease (IFD) (directed therapy). Clearly, the earlier that treatment is started the better the outcome. Consequently there is an urgent need for new diagnostic tools to detect infection before disease becomes manifest, thereby allowing effective treatment strategies to be developed. The polymerase chain reaction (PCR) is becoming increasingly popular (Arvanitis 2014; Donnelly 2006; Hope 2005; Mengoli 2009; Tuon 2007; White 2015). However it was not considered robust enough to be included in the international consensus definitions of the European Organisation for Research and Treatment of Cancer/Mycoses Study Group (EORTC/MSG); (Ascioglou 2002; De Pauw 2008).
The prevalence of IA varies from 1 in 100 to about 1 in 6 depending upon the level of compromised immunity, the environmental exposure and preventative measures taken, which can include protected isolation with filtered air and antifungal prophylaxis. The outcome depends upon the extent of infection, whether diagnosis is made and treatment with an effective drug is initiated early; and, importantly, whether or not an individual's immune system begins to recover (Marr 2002; Walsh 2008).
Demonstration of fungi in diseased tissue is still required for a proven diagnosis of IFD. Unlike other infectious diseases, direct demonstration of Aspergillus infection is rarely possible by culture of sterile body fluids, and obtaining tissue from a live patient is seldom feasible because of the risks posed to the patient.by biopsy.
Recently, advances have been made on several fronts. The EORTC/MSG's published definitions of invasive fungal disease (IFD) allow for degrees of certainty of diagnosis: possible, probable and proven (Ascioglou 2002; De Pauw 2008). Definitions of invasive fungal infection were devised in 2002 and revised in 2008 to focus on fungal disease (Table 2). These are based on host factors, radiological features and mycological evidence. Probable and possible cases have to satisfy the same host and radiological criteria and they are only distinguished by the presence or absence of mycological evidence. Biomarkers have potential to detect infection before development of overt disease, allowing treatment to be initiated at an earlier stage. These definitions were only made possible by other contemporaneous developments in the field. Computer‐assisted tomography (CT scan) became more widely available, allowing lesions consistent with pulmonary IA to be detected at an early stage of disease. This offered the possibility of performing bronchoscopy to obtain bronchoalveolar fluid in which the fungus could be detected by microscopy and culture as well as galactomannan (GM). However the technique is not without risk and cannot always be performed when required. By contrast, blood is readily available which opens up the possibility of looking for fungi in an indirect fashion by detecting fungal cell components including the galactomannan of the cell wall of Aspergillus species (Leeflang 2008).
1. European Organisation for Research and Treatment of Cancer/Mycoses Study Group definitions of invasive aspergillosis.
| Original definitions ofAscioglou 2002 | Revised definitions ofDe Pauw 2008 | |
| PROVEN IA | Specimen obtained by needle aspiration or biopsy from a normally sterile and clinically or radiologically abnormal site consistent with an infectious disease processand either histopathological, cytopathological, or direct microscopic examination of the specimen in which hyphae are seen accompanied by evidence of associated tissue damage or recovery of Aspergillus species by culture from the specimen obtained by a sterile procedure excluding bronchoalveolar lavage, cranial sinus cavity, and urine |
|
| PROBABLE IA | At least 1 host factor criterion plus 1 major (or 2 minor) clinical criteria from abnormal site consistent with infectionplus 1 microbiological criterion | At least 1 host factor plus 1 clinical feature plus 1 microbiological criterion |
| POSSIBLE IA | At least 1 host factor criterion plus either 1 major (or 2 minor) clinical criterion from abnormal site consistent with infection or 1 microbiological criterion |
At least 1 host factor plus 1 clinical feature |
Host factor criteria will include the temporal relationship between the onset of fungal disease and the receipt of an allogeneic stem cell transplant.
Clinical features include for example neutropenia, persistent fever, predisposing conditions, prolonged use of corticosteroids; in the case of lower respiratory tract infection, the presence of 1 of the following signs on CT: dense well circumscribed lesions(s) with or without a halo sign or an air crescent sign, cavity.
Microbiological criteria consist of a positive culture including the presence of fungal elements indicating a mould on microscopy or recovery by culture of Aspergillus species from sputum, bronchoalveolar lavage (BAL) fluid, bronchial brush or sinus aspirate samples; positive result for Aspergillus detection of galactomannan antigen in specimens of plasma, serum, BAL, cerebrospinal fluid or 2 or more blood samples. Major clinical criteria are, for example, new infiltrates on computerized tomography imaging (e.g. halo sign) or suggestive radiological findings.
Minor clinical criteria are suggestive symptoms and signs.
The exact definitions of the European Organisation for Research and Treatment of Cancer/Mycoses Study Group criteria and their host factor, microbiological or clinical criteria can be found in Ascioglou 2002 and De Pauw 2008.
The EORTC/MSG definitions help integrate all the clinical and laboratory information available. Combining of host factor (such as neutropenia) with clinical features (such as pulmonary nodules) and mycological evidence (such as detection of GM) allows a high level of certainty of diagnosis to be assigned. These definitions have been adopted widely by regulatory agencies, such as the European Medicines Agency and the US Food and Drug Administration, for evaluating antifungal drug products and diagnostic tests, as well as by the scientific and medical community at large for investigating epidemiology and auditing antifungal stewardship.
Whilst the range of potential drugs currently available allows prophylaxis, pre‐emptive therapy, as well as directed therapy for possible, probable and proven IFD, the ability to identify 'who needs treatment, when, and with what' is sufficiently unreliable that many physicians continue to treat empirically. Not only does this lead to unnecessary costs but it is also not clear how many people are helped or harmed by this approach. There are circumstances when a host factor is present (for instance receipt of an allogeneic hematopoietic stem cell transplant (HSCT)) and mycological evidence exists (such as Aspergillus being recovered from pulmonary secretions) without evidence of active disease. This may represent infection before disease becomes manifest and provides the opportunity for therapy to pre‐empt disease. Consequently there is an urgent need for new diagnostic tools and an assessment of their utility in the clinic. Biomarkers have the potential to detect infection before development of overt disease, allowing treatment to be initiated at an earlier stage.
Index test(s)
There are few direct diagnostic tests and those that are available are limited by the difficulties in obtaining tissue specimens to allow culture, microscopy and histology (Chamilos 2006). Blood in its various forms — whole blood, plasma and serum — is readily available, but only tests for antigens such as GM and beta‐D‐glucan have been deemed acceptable to support a diagnosis (Leeflang 2008; Pfeiffer 2006; Senn 2008). In neutropenic patients, pulmonary abnormalities consistent with invasive aspergillosis, such as nodules, often surrounded by a 'halo sign', can be detected using high‐resolution computed tomography (Greene 2007). However, the 'halo sign' is transient and only detectable during early invasive aspergillosis, after which radiological signs become non‐specific or appear too late to be therapeutically useful (Caillot 2001). Radiological signs also herald established disease so the opportunity to intervene early has been lost.
Molecular methods, such as the PCR, have been investigated in order to improve the diagnosis of IA (Donnelly 2006; Mengoli 2009; White 2010; White 2015). PCR can amplify a single or a few copies of target DNA allowing target detection with great sensitivity and specificity. Moreover it can be quantitative, using the procedural variant called real‐time PCR (qPCR). The sensitivity is based on the enormous potential for exponential amplification of the DNA target (the 'amplicon') due to repeated cycles of the polymerase reaction, where every cycle doubles the quantity of amplicon. Real‐time PCR continuously monitors the amplification of target DNA at every cycle. The threshold cycle number (preferred term Cq) is when the amplicon becomes detectable above the background level, as an exponentially increasing signal, and is proportional to the amount of starting DNA in the reaction. A high initial DNA concentration will require fewer cycles to reach the threshold and has an earlier Cq value. The specificity of PCR resides in the DNA oligonucleotides used as primers, allowing the terminally stable variant of the enzyme DNA polymerase to initiate sequence duplication. These primers join to the DNA target ('annealing') in a very stringent way, allowing only minimal misfit possibility. Moreover, in quantitative real time PCR (q‐RT‐PCR), the use of reporter probes, hydrolysis probes or molecular beacons that bind to the central part of the target sequence increase the assay’s specificity.
PCR has an enormous potential for diagnosing infectious diseases, particularly where traditional culture methods are less effective. The fungal genus Aspergillus is a good example of this kind of approach. The recovery of Aspergillus from blood cultures is rarely achieved even in overwhelming infection. PCR‐based tests on blood specimens have gained popularity as the platforms become more automated and extraction methods and targets become commercially available (White 2010). However, its exclusion from the EORTC/MSG definitions led to the establishment of the European Aspergillus PCR Initiative (EAPCRI), which is a working group of the International Society of Human and Animal Mycology (ISHAM). The EAPCRI has published various studies describing the critical stages in DNA isolation from blood samples (White 2010), and on the critical characteristics of a standardized Aspergillus PCR assay. These studies, allied to the standardization of qPCR assays described in the MIQE (minimum information for publication of quantitative real‐time PCR experiments) guidelines, have helped pave the way for reliable and robust PCR assays for the diagnosis of IA in the clinical setting (Bustin 2009). Progress in the standardization of methodology enabled the development of commercially available Aspergillus PCR assays in the last few years (Rath 2018).
Clinical pathway
As stated above, many physicians still opt for starting antifungal treatment empirically because of diagnostic uncertainty. This approach can lead to unnecessary treatment, which incurs extra costs, and may be harmful to some people. Diagnostic tests can be used to establish (i.e. rule in or rule out) disease. This is particularly useful for people at risk of IA where a highly sensitive test can deliver a high negative predictive value for disease, allowing empirical therapy to be safely withheld even on the basis of a single test result. Conversely, a high positive predictive value is required to rule in the diagnosis. The use of PCR as a screening tool differs fundamentally from its use as confirmation of the diagnosis. Therefore, if prevalence is low (i.e. < 10%), IA can be ruled out during the risk period for as long as any single PCR test result is negative, and there are no clinical signs of disease. Conversely, two or more PCR positive test results could be used for mycological confirmation of clinically suspected disease, also allowing a case of possible IA to be upgraded to probable IA.
Clinical pathways of managing patients can vary according to the risk of IA. Patients at high risk can be screened using GM, PCR (or both), with positive test results being used to trigger an intensive diagnostic workup with CT scanning and bronchoalveolar lavage (BAL) to determine disease (diagnostic driven) or to initiate antifungal treatment to prevent development of disease (pre‐emptive). Screening may occur throughout the period of risk or only when people develop fever. Alternatively, patients may be tested in the presence of symptoms suggestive of disease to confirm diagnosis.
Rationale
There is no single assay that has been validated for the early diagnosis of IA. Non‐culture‐based methods such as serial GM ELISA screening hold most promise in establishing early diagnosis and may result in improved outcomes, but clinical utility has not been fully established. Moreover, newer methods such as PCR are being investigated (Donnelly 2006; White 2015). As with any diagnostic test, the utility of PCR as either a screening tool or a confirmatory test will depend on the prevalence of disease in the population in which it is used. The use of prophylactic or empirical antifungal agents, availability of protective environments and other diagnostic tests will all influence how the test is used in clinical practice. It is not the aim of this present analysis to establish clinical outcomes but rather to evaluate diagnostic accuracy so that rational use of PCR testing can be applied to different populations.
Objectives
To provide an overall summary of the diagnostic accuracy of PCR‐based tests on blood specimens for the diagnosis of IA in immunocompromised people.
Secondary objectives
When studies included in the analysis also compared the diagnostic performance of PCR techniques and the GM ELISA assay, we comparatively evaluated the diagnostic performance of PCR‐based tests and GM ELISA assays. However, since the objective of this review is not to identify all studies dealing with GM ELISA assays and IA, only those within the study comparison were included in the review.
Methods
Criteria for considering studies for this review
Types of studies
We included studies using PCR techniques on blood specimens for analysis if they:
compared the results of PCR tests with the diagnosis made following the published case definition criteria for invasive fungal disease proposed by the EORTC/MSG; or, for studies published before the publication of these criteria in 2002, used comparable criteria as a reference standard (Ascioglou 2002; De Pauw 2008);
reported data on false‐positive, true‐positive, false‐negative and true‐negative results of the diagnostic tests under investigation separately; and
evaluated the tests prospectively in a cohort of people from a relevant clinical population, defined as a group of individuals at high risk of IA.
We classified studies, on the basis of the sampling method, as being consecutive or non‐consecutive. We regarded studies evaluating specimens from a group of people known to have aspergillosis, and from a separate group of subjects without evidence of disease, as case‐control studies (Lijmer 1999). We included these studies in the systematic review but excluded them from the quantitative analysis.
Aspergillus contamination and false positive PCR results with bronchoalveolar lavage (BAL) and sputum samples can follow inhalation of airborne spores or colonization of the lung (Lewis 2006). Moreover, BAL is an invasive procedure performed only to confirm the aetiology in a subset of cases that already meet the clinical definitions of IA. Thus, to avoid bias related to the patient selection and specimen type, we analysed only studies evaluating PCR on blood (whole blood, serum, and plasma), with exclusion of studies that analyse the accuracy of PCR tests on BAL only.
Participants
Patients at risk of IA, including neutropenic cancer patients and hematopoietic stem cell transplant (HSCT) or solid organ transplant recipients.
Index tests
PCR methods on blood specimens (whole blood or serum). We considered different DNA extraction methods and PCR methods (e.g. nested, ELISA, qPCR).
Target conditions
The target condition of this review is IA (systemic aspergillosis).
Reference standards
Definitions for invasive fungal disease were first published in 2002 by the EORTC/MSG (Ascioglou 2002); they were revised in 2008 (De Pauw 2008; Table 2). These were used as a reference standard and comparable criteria were used for studies published before the publication of the definitions in 2002. The EORTC/MSG definitions divide the patient population into four categories: people with proven IA, people with probable IA, people with possible IA, and people without IA. In accordance with the previous Aspergillus review on Aspergillus GM detection (Leeflang 2008), sensitivity and specificity were assessed in each study considering the proven and probable cases of IA as having the disease, and the cases of possible IA and no IA as not having the disease.
Search methods for identification of studies
The search strategies for MEDLINE, Embase and CENTRAL are listed in Appendix 1.
Electronic searches
We updated searches on the following electronic databases to identify reports of relevant studies for the review update.
Cochrane Central Register of Controlled Trials (CENTRAL; 2018, Issue 3), in the Cochrane Library;
MEDLINE via Ovid (June 2015 to March week 2 2018);
Embase via Ovid (June 2015 to 2018, week 12);
LILACS (June 2015 to September 2018);
Database of Abstracts of Reviews of Effects (DARE) to October 2018;
Health Technology Assessment database to October 2018;
Web of Science to October 2018.
Searching other resources
We also searched for unpublished material on Scopus (www.scopus.com). We checked the reference lists of all the studies identified by the above methods and contacted other authors and trialists in the field.
Data collection and analysis
Selection of studies
Two review authors (PD, RB) independently assessed the abstract (if available) of each reference identified by the search against the inclusion criteria. We resolved any disagreements that arose through discussion and consensus with a third author (MC). For the update review, we screened the search results using Covidence 2014. We retrieved those references that potentially met the inclusion criteria (based on their abstract or title) in full for further independent assessment.
Data extraction and management
We extracted the following data from each included study.
Study design
Study population
Reference standard and performance of the reference standard
Performance of the index test
Technical details of the PCR methods used, including genetic target of PCR and nucleotide probe sequence, and any PCR testing methods; we classified the diagnostic modalities using PCR assays according to the sampling methods and how these relate to the definition of a positive result, namely either positive PCR in at least two consecutive blood samples drawn from the same patient, or a single sample yielding a PCR positive result. When we compared PCR‐based tests to GM, we assessed whether authors explicitly mention the exclusion of the GM ELISA test from the reference test definition (EORTC/MSG criteria). In this case, we performed a direct comparison of the index test and the comparator evaluated in the same study population towards the reference standard.
QUADAS‐2 items
Data for two‐by‐two table (false‐positive, true‐positive, false‐negative and true‐negative results of the diagnostic tests under investigation and reference standard).
Pairs of authors extracted the data; they resolved disagreements by discussion.
Assessment of methodological quality
Assessment of the quality of diagnostic accuracy studies, as recommended in STARD (Standards for Reporting of Diagnostic Accuracy), is of absolute relevance in systematic reviews (Bossuyt 2003; Reitsma 2009; Whiting 2004). For this purpose, we used the Quality Assessment tool for Diagnostic Accuracy Studies (QUADAS‐2) tool , the current version of QUADAS that has been adopted for use by Cochrane and is recommended for use in all Cochrane diagnostic test accuracy reviews to evaluate the risk of bias and applicability of primary diagnostic accuracy studies. Pairs of authors independently assessed the methodological quality of the studies included, and disagreements were resolved by consensus with all of the authors.
QUADAS‐2 consists of the following four key domains.
Patient selection
Index test
Reference standard
Flow and timing
Each is assessed in terms of risk of bias and the first three in terms of concerns regarding applicability. Signalling questions are included to assist in judgements about risk of bias. Risk of bias is judged as 'low', 'high', or 'unclear'. If all signalling questions for a domain are answered 'yes' then risk of bias can be judged 'low'. If any signalling question is answered 'no' this flags the potential for bias. The 'unclear' category is used only when insufficient data are reported to permit a judgment.
Tabular and graphical displays are used to summarise QUADAS‐2 assessments. We did not calculate a summary score estimating the overall quality of an article, since their interpretation is problematic and potentially misleading (Whiting 2005).
The items of the QUADAS‐2 tool and their interpretation are reported in appendix (Appendix 2).
Statistical analysis and data synthesis
The values of sensitivity and specificity are automatically computed in Review Manager 2014. We obtained summary positive (LR+) and negative (LR−) likelihood ratios from the bivariate analysis (see below). We evaluated different interpretive criteria for a PCR‐positive result in the two‐by‐two table, namely a single positive PCR result and two positive PCR results. We have presented individual study results graphically by plotting the estimates of sensitivity and specificity (and their 95% confidence intervals (CIs)) in both forest plots and receiver operating characteristics (ROC) space.
We assessed the operating point sensitivity and specificity of the diagnostic test under scrutiny by a bivariate random‐effects approach (Reitsma 2005). The original method was modified by using a random‐effects bivariate logistic model (Chu 2006). The same procedure permits generation of a hierarchical summary receiver operating characteristic (HSROC) model (Rutter 2001). The bivariate approach examines the influence of covariates on sensitivity and specificity (or both), whilst the HSROC model is focused on threshold and accuracy (Guo 2015; Harbord 2007). In most conditions bivariate and HSROC are equivalent, particularly in the absence of covariates. When there is a considerable degree of between‐study heterogeneity, as is common in meta‐analysis of diagnostic accuracy studies, a prediction region may be preferable to a confidence region(Harbord 2007); this is assured by the bivariate approach. The results of the bivariate model can be used to calculate likelihood ratios. To calculate (negative) predictive values, an estimate of prevalence in addition to values of sensitivity and specificity is required. One can then apply a Bayesian approach to obtain predictive values from these three parameters. We performed bivariate analysis using STATA 11 software. We compared the diagnostic measures of diagnostic accuracy and related 95% CIs by adding binary covariates to the bivariate model.
Investigations of heterogeneity
We assessed heterogeneity by visual inspection of forest plots of sensitivity and specificity, and through visual examination of ROC plot of the raw data. We further investigated heterogeneity by exploring the effects of several study‐level covariates. For this, we performed a multilevel mixed‐effects logistic model using the probability of test positivity as a dependent variable; the group variable was the study, and the disease status was the first explanatory variable. This basic model admitted in turn several additional covariates. When available, we examined the following covariates.
Distinctive groups of patients
Study size (< or ≥ 100 patients)
Children versus adults
Use of antifungal prophylaxis active against Aspergillus species
Variation in PCR techniques (RT‐PCR versus other PCR methods)
We included the interaction between the disease status and the additional covariate into the model as well.
We have analysed the potential influence of risk of bias (e.g. blinding of the index test, blinding of the reference test) by sensitivity analysis.
Results
Results of the search
Of the 2474 references identified, we selected 215 potentially relevant citations (Figure 1). After screening titles and abstracts, we selected 91 articles for full‐text review. Of these, we excluded 62 studies for various reasons (Characteristics of excluded studies): patients were selected retrospectively in 11 studies; 20 studies did not provide sensitivity and/or specificity data for two‐by‐two tables; 13 were case‐control studies; four studies included BAL only or tissue PCR; 3 studies included a subset of previous trials; the index test was inappropriate in 6 studies and the reference standard was inappropriate in 7 studies; 2 studies were in Chinese; 3 studies were duplicates of previously published papers; and, finally, 2 for other reasons. Therefore, 29 studies published between 2000 and 2018 met the inclusion criteria and were included in the meta‐analysis (Aslan 2015; Badiee 2010; Badiee 2017; Barnes 2009; Barnes 2013; Bellanger 2015; Boch 2016; Boluk 2016; Cuenca‐Estrella 2009; da Silva 2010; El Mahallawy 2006; Ferns 2002; Florent 2006; Halliday 2006; Hebart 2000a; Hummel 2009; Imbert 2016; Landlinger 2010; Loeffler 2017; Pini 2015; Ramírez 2009; Rogers 2013; Schwarzinger 2013; Springer 2011; Springer 2016; Suarez 2008; Sugawara 2013; von Lilienfeld‐Toal 2009; White 2006). Three studies reported the diagnostic performance of PCR performed with different methodologies (Aslan 2015; Rogers 2013; Suarez 2008), and one in a different patient setting (Rogers 2013). Therefore data were analysed from 34 data sets.
1.
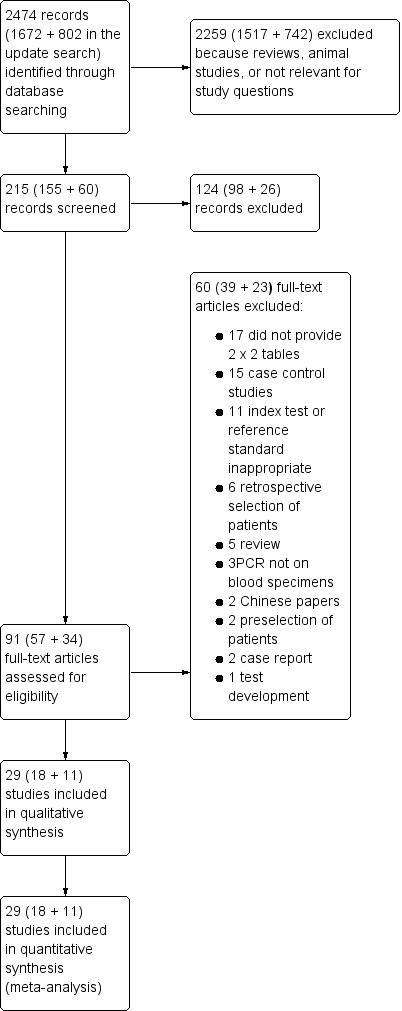
Study flow diagram.
The main characteristics of the studies are summarized in the Characteristics of included studies tables. More than 28,000 clinical blood specimens from 4718 patients at risk of IA were included. Most had received chemotherapy for a haematological malignancy or had been given a hematopoietic stem cell transplant (HSCT). The PCR techniques used are summarized in Table 3. Twenty‐eight of the selected studies (corresponding to 33 data sets) reported the results of a single PCR result, and nine studies (13 data sets) reported using two PCR results. In three studies it was possible to extract the two‐by‐two data in subsets of patients receiving or not receiving anti‐mould prophylaxis (Imbert 2016; Rogers 2013; Springer 2016). Sixteen of the studies included in the analysis also reported results of GM assay (Barnes 2009; Bellanger 2015; Cuenca‐Estrella 2009; da Silva 2010; El Mahallawy 2006; Ferns 2002; Florent 2006; Hummel 2009; Imbert 2016; Loeffler 2017; Rogers 2013; Schwarzinger 2013; Springer 2011; Springer 2016; Suarez 2008; Sugawara 2013). The study by Rogers 2013 presented two cohorts of patients (one from the University Clinic of Wurzburg, and one from Saint James's Hospital, Dublin) according to the PCR test used: Internal Transcribed Spacer (ITS) qPCR and the 28S nested PCR; the study by Suarez 2008 presented data according to the protocols for serum processing (large and small volume); and the study by Aslan 2015 according to two PCR tests used (in‐house and commercially available test).
2. Technical details of the PCR methods used in the studies analysed in this review.
| Study | Sample type | Sample volume | DNA extraction methodsA | PCR methodC | Target gene | Appropriate controls | Requirements for positive by PCR | Methods used (refs) | ||||||
| Cell wall disruptionB | DNA isolation kit/protocol | NegativeD | PositiveE | PCR inhibition | ||||||||||
| Ex | PCR | Ex | PCR | |||||||||||
| Hebart 2000a | Whole blood | 5 ml | Zymolase and NaOH lysis buffer | Protein precipitation and DNA precipitation | PCR‐slot blot | 18S | ‐ | Yes | ‐ | Yes | Yes | Single positive | Einsele 1997 | |
| Ferns 2002 | Whole blood | 2 ml | Lyticase | QIAamp | Nested PCR | mtDNA | Yes | Yes | Yes | Yes | ‐ | Positive on 2 occasions |
Bretagne 1998 Tang 1993 |
|
| Florent 2006 | Serum | 200 μl | ‐ | QIAamp | PCR‐ELISA | mtDNA | ‐ | Yes | ‐ | Yes | Yes | 2 consecutive positives | Bretagne 1998 | |
| Halliday 2006 | Whole blood | 500 μl | Lyticase | GenElute | Nested PCR | 18S | Yes | Yes | ‐ | Yes | Yes | 2 consecutive positives | Skladny 1999 | |
| El Mahallawy 2006 | Serum | ‐ | Lyticase | QIAamp | Standard PCR | 18S | ‐ | Yes | ‐ | Yes | ‐ | Single positive | Williamson 2000 | |
| White 2006 | Whole blood | 2 ml | Glass beads | MagNA Pure | Nested qPCR | 28S | Yes | Yes | Yes | Yes | Yes | Serial positives in single episode | Loeffler 2002; Williamson 2000 | |
| Suarez 2008 | Serum | 1 ml or 200 μl | ‐ | MagNA Pure | qPCR | 28S | ‐ | Yes | ‐ | Yes | ‐ | Single positive | Challier 2004 | |
| Hummel 2009 | Blood | 5 ml | Lyticase | Phenol‐chloroform | Nested PCR | 18S | ‐ | Yes | ‐ | Yes | ‐ | Single positive | Skladny 1999 | |
| Ramírez 2009 | Whole blood | 5 ml | Lyticase and glass beads | QIAamp | qPCR | 18S | ‐ | Yes | ‐ | Yes | ‐ | Single positive | Loeffler 2000 | |
| Barnes 2009 | Whole blood | 2 ml | Glass beads | MagNA Pure | Nested qPCR | 28S | Yes | Yes | Yes | Yes | Yes | Confirmed positiveF | White 2006 | |
| Cuenca‐Estrella 2009 | Whole blood and serum | ‐ | ‐ | QIAamp | qPCR | ITS1 | ‐ | Yes | ‐ | Yes | Yes | 2 consecutive positives | Yoo 2008 | |
| von Lilienfeld‐Toal 2009 | Whole blood | 10 ml | Ceramic beads | Septifast | qPCR | 18S | ‐ | Yes | ‐ | Yes | Yes | ‐ | Lehmann 2008 | |
| Landlinger 2010 | Whole blood | 3 ml | Lyticase | MagNA Pure | qPCR | 28S | ‐ | Yes | ‐ | Yes | Yes | Single positive | Baskova 2007; Watzinger 2004 | |
| Badiee 2010 | Whole blood | 3 to 5 ml | Lyticase | QIAamp | qPCR | 18S | Yes | Yes | ‐ | Yes | ‐ | Single positive | Van Burik 1998; Kami 2001; | |
| da Silva 2010 | Serum | 5 ml Blood | Lyticase | Protein precipitation and DNA precipitation | Standard PCR | 18S | ‐ | Yes | ‐ | Yes | ‐ | 2 consecutive positives | Ribeiro 2006; Van Burik 1998 | |
| Springer 2011G | Whole blood | 3 ml | Glass beads | High Pure PCR Template Preparation Kit (Roche) | qPCR | ITS | ‐ | Yes | ‐ | Yes | ‐ | Single positiveH | ‐ | |
| FastPrep‐24 MP (Biomedicals) | ||||||||||||||
| Whole blood | 5 ml | Glass beads | Standard PCR | ‐ | ‐ | Yes | Yes | Yes | Yes | ‐ | Sachse 2009 | |||
| Rogers 2013G | Whole blood | 3 ml | Glass beads | High Pure PCR Template Preparation Kit (Roche) | Nested qPCR | 28S | Yes | Yes | Yes | Yes | Yes | Single positiveI | White 2006 | |
| Springer 2011 | ||||||||||||||
| qPCR | ITS1 | Yes | Yes | Single positiveI | ||||||||||
| Sugawara 2013 | Whole blood | 1 ml | Beads and lysis buffer | Phenol‐chloroform | Nested PCR and sequencing | 18S | ‐ | Yes | ‐ | Yes | ‐ | Single positive | Nakamura 2010 | |
| Barnes 2013 | Whole blood | 3ml | Glass beads | Various automated extractors – Roche MagNA Pure LC Total NA, BioMerieux EasyMag, Qiagen EZ1 Advance XL tissue kit. | qPCR and nested qPCR | 28S | Yes | Yes | Yes | Yes | Yes | Single and multiple positive thresholds used | White 2006 | |
| Schwarzinger 2013 | Serum | 1 ml | Not required | Roche MagNA Pure LC DNA | qPCR | Mitochonrial | ‐ | Yes | ‐ | Yes | Yes | Single positive | Botterel 2008 | |
| Aslan 2015 | Serum | 0.2 ml | Not required | Qiamp DNA Mini Kit | qPCR | 18S and 28S | Yes | Yes | Yes | Yes | Yes | Single positive | Mycassay Aspergillus and in‐house PCR | |
| Bellanger 2015 | Serum | 1 ml | ‐ | Large Volume MagNa Pure Nucleic acid isolation kit | qPCR | 18S Mitochondrial (L37095) |
‐ | ‐ | ‐ | ‐ | ‐ (no info on controls) |
Single positive |
Millon 2011, Costa 2001 |
|
| Pini 2015 | Serum | 0.5 ml | Not required | High Pure template (Roche) | qPCR | 18S | ‐ | Yes | ‐ | Yes | Yes | Single Positive | Mycassay Aspergillus | |
| Boch 2016 | Whole blood | 3 to 5 ml | Lyticase | Phenol‐chloroform | Nested PCR | 18S | ‐ | Yes | ‐ | Yes | ‐ | Single positive | Skladny 1999 | |
| Boluk 2016 | Serum | ‐ | ‐ | ZR Fungal/Bacterial DNA MiniPrep Kit |
qPCR | Kit (Way2 Gene Fungi) | ‐ | Yes | ‐ | ‐ | Yes | Single Positive | No ref to methods for Asp PCR | |
| Imbert 2016 | Serum | 1 ml | ‐ | MagNA Pure Compact large volume kit on a MagNA Pure device (Roche) |
qPCR | 28S | ‐ | ‐ | Yes | Yes | Yes | Single Positive | Suarez 2008, Challier 2004 | |
| Springer 2016 | Serum | 1 ml | ‐ | Qiaamp UltrasensVirus Kit | qPCR | ITS1‐5.8S | Yes | Yes | Yes, but Bacillus‐DNA was used | Yes | Yes | Single and multiple positive thresholds used | Skladny 1999,Springer 2012 | |
| Badiee 2017 | Serum | 0.2 ml | ‐ | QiaAmp Mini | qPCR | 18S | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | Skladny 1999; Shin 1999 | |
| Loeffler 2017 | Cell‐free blood fraction, mostly serum | 1 ml | ‐ | Qiaamp UltrasensVirus Kit | qPCR | ITS1‐5.8S | Yes | Yes | Yes, but Bacillus‐DNA was used | Yes | Yes | Single positive | Skladny 1999; Springer 2016 | |
‐: not reported; MagNA Pure: an automated DNA isolation system manufactured by Roche; mtDNA: mitochondrial DNA; PCR: polymerase chain reaction; QIAamp: QIAamp DNA isolation kit manufactured by Qiagen; Ex: extraction; ITS: Internal Trascribed Spacer; RCLB: red cell lysis buffer.
A DNA isolation protocols may include steps to remove red and white blood cells, fungal cell wall disruption and DNA purification kits.
B Lyticase/Zymolase enzymatically digest fungal cells walls; ceramic or glass beads cause mechanical disruption of the cell wall.
C PCR methods used vary between standard PCR where products are resolved on agarose gels to detect positive or negative reactions or quantitative PCR (qPCR) which allows real time monitoring of the reaction. Nested qPCR involves first round standard PCR and second round qPCR.
D Negative DNA extraction controls feature a sample blank, e.g. blood or sterile solution, that allows detection of any contamination in the DNA isolation protocol.
E Positive DNA extraction controls are a sample blank that is spiked with fungal or specific bacterial spores to ensure that the DNA isolation protocol is working optimally.
F The confirmed positive requires that any single positive sample is confirmed with an additional sample from the same patient. Barnes 2009 also used multiple analyses to determine the effectiveness of single versus multiple positives to yield diagnostic accuracy.
G Studies assessed the effectiveness of more than 1 assay.
H The study analysed the effect of both single and multiple positives.
I The effects of both single and multiple positives were analysed as well as analyses of combined PCR and galactomannan tests.
Methodological quality of included studies
We summarize the quality of studies as assessed by the QUADAS‐2 tool in tables and graphs. Figure 2 shows the overall risk of bias and applicability concerns for the 29 selected studies. Figure 3 presents the quality assessment results for the individual studies. For all QUADAS‐2 domains, most studies were at low risk of bias and low concern regarding applicability. In the patient selection domain, all the studies enrolled a homogenous and representative population of patients at risk of IA; 75% of studies were at low risk of bias because they enrolled participants consecutively and avoided inappropriate exclusions. We graded six studies as being at unclear risk of bias because the manner of patient selection was not stated; and we graded one study at high risk of bias because it included retrospectively a heterogeneous population with various underlying diseases, mostly haematologic and neutropenic, but also patients with a non‐invasive form of aspergillosis.
2.
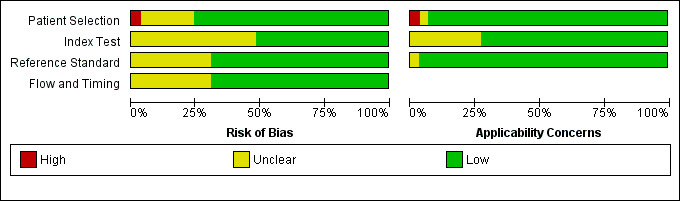
Risk of bias and applicability concerns graph: review authors' judgements about each domain presented as percentages across included studies
3.

Risk of bias and applicability concerns summary: review authors' judgements about each domain for each included study
In the index test domain, we considered 50% of studies to be at low risk of bias and 70% of studies to be at low concern regarding applicability. We judged the remaining studies to be at unclear risk of bias because it was unclear if the index test was performed knowing the results of the reference standard. In the reference standard domain, we judged around 70% of studies to be at low risk of bias because it was stated that the reference standard results were interpreted without knowledge of the results of the index test, while in the remaining studies it was not specified. Applicability was of low concern for almost all studies in the reference standard domain. In the flow and timing domain, we judged 70% of studies to be at low risk of bias because all patients were accounted for in the analysis, the appropriate reference standard was used, and information about uninterpretable results was provided. We had nearly complete information for all studies.
Findings
Results of the meta‐analysis
Based on 29 included studies, the median number of patients per study was 99 (range 17 to 549), and the mean prevalence of proven or probable IA was 16.3% (median 11.1, range 2.5% to 57.1%). The sensitivity and specificity of PCR for the diagnosis of IA varied according to the interpretative criteria used to define a test as positive. For PCR assays, we evaluated the requirement for either one or two consecutive samples to be positive for diagnostic accuracy. With the one positive requirement, the sensitivity reported in the studies ranged from 22% to 100%, and specificity from 2% to 100%. With the two positive requirements the sensitivity reported in the included studies ranged from 0% to 92%, and specificity from 75% to 100%. The summary estimates of sensitivity and specificity were 79.2% (95% CI 71.0% to 85.5%) and 79.6% (95% CI 69.9% to 86.6%) for a single positive result requirement, and 59.6% (95% CI 40.7% to 76.0%) and 95.1% (95% CI 87.0% to 98.2%) for two positive results requirement. LR+/LR− were 3.8 (95% CI 2.6 to 5.7)/0.26 (95% CI 0.18 to 0.36) for a single positive result, and 12.2 (95% CI 4.2 to 35.3)/0.42 (95% CI 0.26 to 0.67) for two positive results. When used in isolation, a single PCR positive test as diagnostic criterion for IA in a population of 100 people with a disease prevalence of 16.3% (overall mean prevalence), three people who have IA would be missed (sensitivity 79.2%, 20.8% false negatives), and 17 people would be unnecessarily treated or referred for further tests (specificity of 79.6%, 21.4% false positive). If we use the 'two positive tests' requirement in a population with the same disease prevalence, it would mean that nine IA people would be missed (sensitivity 59.6%, 40.4% false negatives) and four people would be unnecessarily treated or referred for further tests (specificity of 95.1%, 4.9% false positive).
Heterogeneity
The appearance of the forest plots for PCR show a wide range of diagnostic indices at study level; this was more apparent for specificity using the 'single positive' requirement, and for sensitivity using the 'two positive' requirement. (Figure 4; Figure 5). Visual inspection of the prediction ellipses in the bivariate analysis show a large area occupying most of the full probabilistic space; the degree of eccentricity was more pronounced in the specificity direction for a 'single positive' requirement, and in the sensitivity direction for 'two positives' requirement (Figure 6; Figure 7).
4.
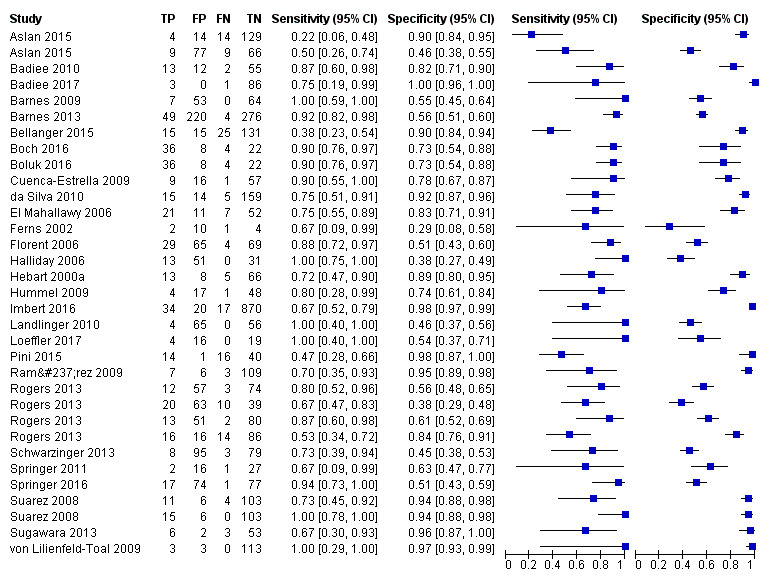
Forest plot of PCR: one (single) positive requirement.
5.
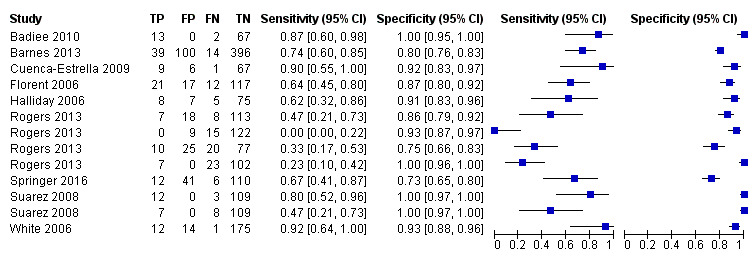
Forest plot of PCR: two positive requirement.
6.
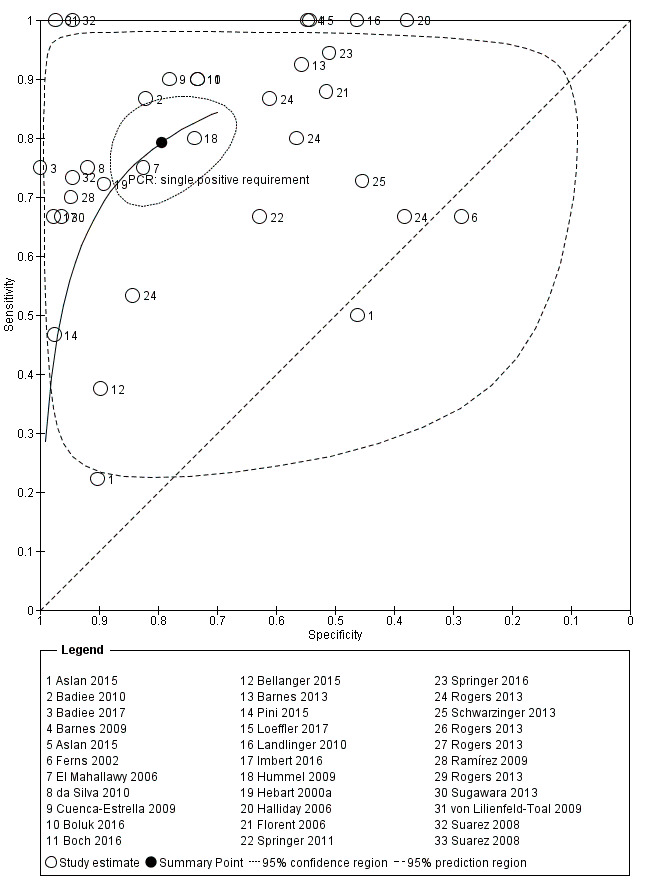
Summary ROC Plot. Bivariate analysis of the sensitivity and specificity of the PCR as a diagnostic tool for Aspergillus invasive infection. One single positive PCR result is required to define the test as positive
7.
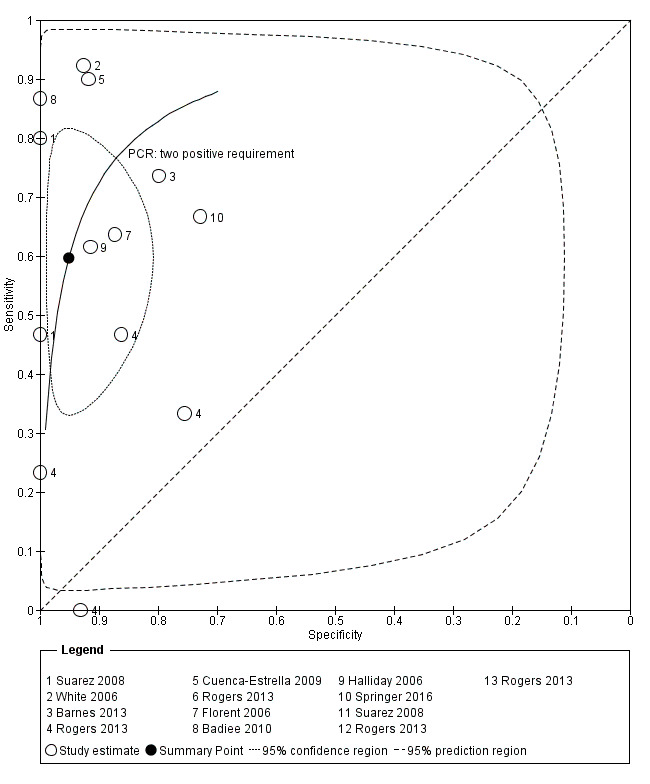
Summary ROC Plot. Bivariate analysis of the sensitivity and specificity of the PCR as a diagnostic tool for Aspergillus invasive infection. Two or more consecutive positive PCR result are required to define the test as positive.
We investigate heterogeneity by subgroups analyses.
Bivariate analysis
Graphs (ellipses) of bivariate models for the two different criteria for PCR positivity are shown in Figure 6 and Figure 7. We excluded unpaired studies for the evaluation of the differential effect of the single positive/two positive criterion. We reduced the number of studies included in the paired analysis to eight, corresponding to 12 comparisons of PCR test (each paired for 'single positive' and 'two positive' criteria; Badiee 2010; Barnes 2013; Cuenca‐Estrella 2009; Florent 2006; Halliday 2006; Rogers 2013; Springer 2011; Suarez 2008). When sensitivity and specificity data from the bivariate model were compared, changing the 'positive results' requirement from one to two increased specificity significantly from 79.5% to 95.1% ( P value < 0.0001). By contrast, the sensitivity decreased significantly from 79.2% to 59.6% (P value < 0.0001). The joint effect on sensitivity and specificity was also significant (P value < 0.0001) (Table 4).
3. Subgroup analyses.
| Covariate | Subgroup | Index | mean | 95% CI | Subgroup Difference: P |
| Anti‐mould prophylaxis | yes | sensitivity | 0.8206 | 0.7536; 0.8725 | not significant |
| no | sensitivity | 0.7577 | 0.6440; 0.8439 | ||
| yes | specificity | 0.6470 | 0.5638; 0.7222 | 0.0387 | |
| no | specificity | 0.7901 | 0.6769; 0.8712 | ||
| EORTC criteria 2008 vs 2002 | 2008 | sensitivity | 0.7311 | 0.6324; 0.8112 | not significant |
| 2002 | sensitivity | 0.7878 | 0.7061; 0.8516 | ||
| 2008 | specificity | 0.7339 | 0.6098; 0.8296 | not significant | |
| 2002 | specificity | 0.8226 | 0.6559; 0.9186 | ||
| Blind reference | yes | sensitivity | 0.7384 | 0.6124; 0.8345 | not significant |
| no | sensitivity | 0.7676 | 0.6652; 0.8460 | ||
| yes | specificity | 0.6284 | 0.5429; 0.7065 | 0.0009 | |
| no | specificity | 0.8553 | 0.7555; 0.9187 | ||
| Blind index | yes | sensitivity | 0.7209 | 0.6402; 0.7895 | not significant |
| no | sensitivity | 0.7584 | 0.6476; 0.8428 | ||
| yes | specificity | 0.6646 | 0.5532; 0.7603 | 0.0161 | |
| no | specificity | 0.8295 | 0.7354; 0.8950 | ||
| In‐house vs commercial kit | In‐house | sensitivity | 0.7489 | 0.6038; 0.8537 | not significant |
| kit | sensitivity | 0.6576 | 0.3274; 0.8835 | ||
| In‐house | specificity | 0.8428 | 0.7263; 0.9155 | not significant | |
| kit | specificity | 0.7674 | 0.4165; 0.9384 | ||
| Whole blood vs serum | WB | sensitivity | 0.8114 | 0.7304; 0.8724 | not significant |
| serum | sensitivity | 0.7130 | 0.5956; 0.8073 | ||
| WB | specificity | 0.7243 | 0.6382; 0.7965 | not significant | |
| serum | specificity | 0.8139 | 0.6661; 0.9056 |
Effects of 6 binary covariates on the sensitivity and specificity of the Aspergillus PCR. Meta‐analytical pooling for proportions (method of logits, DerSimonian‐Laird estimator for tau², inverse variance method), subgroup analysis. Mean values and 95% confidence intervals are reported. “Subgroup Difference: P” reports the comparison between 2 subgroups as difference within the same index for each covariate, as P value. Significant results were found for specificity under prophylaxis (as decrease under prophylaxis), specificity under blind reference (as decrease under blind reference), specificity under blind index (as decrease under blind index). Analysis performed with R version 3.5.3.
Subgroups analysis and bivariate analysis with covariates
We carried out a subgroup analysis of adult and paediatric studies (Boch 2016; El Mahallawy 2006; Halliday 2006; Hummel 2009; Landlinger 2010). The diagnostic yield did not differ significantly between adult and paediatric studies. However, the limited number of paediatric studies does not allow a firm conclusion to be drawn regarding the diagnostic performance of PCR in paediatric patients. We also performed a subgroup analysis according to study size. Studies were defined as small size (15 studies) or large size (14 studies) according to the number of enrolled people (< or ≥ 100). Likewise study size did not have a significant impact on performance of PCR test.
We also performed a subgroup analysis of studies endorsing 2002 EORTC criteria (10 studies: El Mahallawy 2006; Ferns 2002; Florent 2006; Halliday 2006; Hebart 2000a; Hummel 2009; Ramírez 2009; Suarez 2008; von Lilienfeld‐Toal 2009; White 2006) or 2008 criteria (seven studies: Badiee 2010; Barnes 2009; Cuenca‐Estrella 2009; da Silva 2010; Rogers 2013; Springer 2011; Sugawara 2013), using the bivariate method and considering the results of PCR test with the 'single positive' criterion. One study stated the use of EORTC criteria but did not mention which criteria were employed (Landlinger 2010). Lower sensitivity and specificity values were found for studies using 2008 criteria compared to those using 2002 criteria (73.1% (95% CI 63.2 to 81.1) and 73.3% (95% CI 60.9 to 82.9) versus 78.7% (95% CI 70.6 to 85.1) and 82.2% (95% CI 65.5 to 91.8), respectively), but these differences were not statistically significant and probably driven by the low estimates of diagnostic accuracy found in some of the 2008 studies (Rogers 2013; Springer 2011) (Table 4).
Twelve studies used anti‐mould prophylaxis (itraconazole, voriconazole, amphotericins or caspofungin) in the entire population or in a subset of patients under investigation ( Barnes 2009; Barnes 2013; Cuenca‐Estrella 2009; Ferns 2002; Florent 2006; Hummel 2009; Imbert 2016; Loeffler 2017; Rogers 2013; Springer 2016; Sugawara 2013; White 2006) ; ). Thirteen studies did not use antimould prophylaxis at all (Badiee 2010;Badiee 2017; Boch 2016; Boluk 2016; da Silva 2010; El Mahallawy 2006; Halliday 2006Hebart 2000a; Landlinger 2010; Rogers 2013; Schwarzinger 2013; von Lilienfeld‐Toal 2009); or only in a subset of patients (Imbert 2016; Springer 2016). Fluconazole was used as prophylaxis in four studies (Badiee 2010; Halliday 2006; Hebart 2000a; Springer 2011). When examining data under the criterion 'single positive', the anti‐mould prophylaxis produced a significant reduction of specificity (from 0.79 (95% CI 0.67 to 0.87) to 0.64 (95% CI 0.56 to 0.72), coupled with no significant increase of sensitivity (from 0.75 (95% CI 0.64 to 0.84) to 0.82 (95% CI 0.75 to 0.87) (Data table 4).
4. Test.
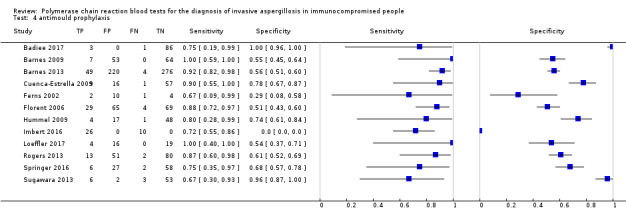
antimould prophylaxis.
The PCR methods varied notably. Some studies were based on gel electrophoretic visualization after proper staining of the amplicons, whereas others were based on automated procedures, such as real‐time PCR, with substantial differences regarding the threshold of detection. We relied on the reported qualitative (positive/negative) test results only, and did not take the possible cut‐point/threshold variation across studies into consideration. Comparison of the three studies — Aslan 2015, Boluk 2016 and Pini 2015 — analyzed in this review that used kit‐based assays to 15 studies (Badiee 2010; Badiee 2017; Bellanger 2015; Cuenca‐Estrella 2009; Imbert 2016; Landlinger 2010; Loeffler 2017; Ramírez 2009; Rogers 2013; Schwarzinger 2013; Springer 2011; Springer 2016; Suarez 2008; von Lilienfeld‐Toal 2009) that used in‐house qPCR assays (excluding end‐point or nested PCR) did not reveal any statistically significant differences between kit and in‐house assays. There was a trend for greater sensitivity and specificity for the in‐house assays compared to commercially available kits (0.74 vs 0.65; 0.84 vs 0.76, respectively), although these differences did not reach statistical significance. Whole blood PCR test had higher sensitivity and lower specificity compared to serum PCR test, but these differences were not statistically significant (Table 4).
Quality items that did have an effect on sensitivity or specificity were blinding of the index test (4% decrease in sensitivity and 17% decrease in specificity) and blinding of the reference standard (5% decrease in sensitivity and 14% decrease in specificity). In other words, failure of blinding produced a spurious increase in overall accuracy.
Predictive values
Positive and negative predictive value (PPV and NPV, respectively) of Aspergillus PCR detection are shown in Figure 8 (Figure 8). The predictive values were calculated by applying the Bayes rule. The use of the two positive criteria produces a significant increase in the PPVs, and only a slight decrease of NPVs. With a mean prevalence of invasive aspergillosis of 16%, the PPV is 42.8% with a 'single positive test' criterion, and 70.3% with 'two positive tests' criterion; for NPV these figures are 95.1% and 92.4%, respectively.
8.
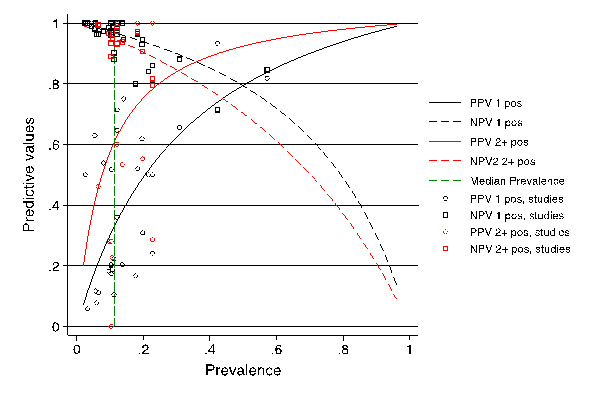
Predictive values. Positive and negative predictive value (PPV and NPV, respectively) of the Aspergillus PCR detection test (y‐axis) as a function of the prevalence of the disease, invasive aspergillosis (x‐axis). The curves are related to the diagnostic criterion (a single positive result or two consecutive positive PCR results). The PVs were calculated by applying the Bayes rule. The mean prevalence of invasive aspergillosis (16.3%) is indicated by the vertical dashed line. It corresponds to PPV1 = 42%, NPV1 = 95%, PPV2 = 70%, NPV2 = 92%.
Comparison between PCR techniques and GM assay
Sixteen studies also evaluated GM assay (Barnes 2009; Bellanger 2015; Cuenca‐Estrella 2009; da Silva 2010; El Mahallawy 2006; Ferns 2002; Florent 2006; Hummel 2009; Imbert 2016; Loeffler 2017; Rogers 2013; Schwarzinger 2013; Springer 2011; Springer 2016; Suarez 2008; Sugawara 2013), but in all studies but one GM was part of the reference standard (Suarez 2008). Thus to avoid incorporation bias, we did not compare data of GM assay to PCR, and did not include them in the current review.
In the study by Suarez 2008, sensitivity and specificity were 100% and 96.7% for qPCR using large sample volume (LSV), and 88.2% and 95.8% for GM. Thus the overall performance of qPCR using LSV was consistently higher than that of GM.
Discussion
Summary of main results
We included 29 primary studies, corresponding to 34 data sets, in the meta‐analyses: 18 RCTs were included in the original review, and we identified 11 additional trials for this update. The mean prevalence of IA (proven or probable) in the included studies was 16.3%. The majority of patients had received chemotherapy for a haematological malignancy or had been given a hematopoietic stem cell transplant (HSCT). Several PCR techniques were used among the included studies. The sensitivity and specificity of PCR for the diagnosis of IA varied according to the interpretative criteria used to define a test as positive. For PCR assays, we evaluated for diagnostic accuracy the requirement for either one or two consecutive samples to be positive. The summary estimate of sensitivity and specificity were 79.2% (95% CI 71.0% to 85.5%) and 79.6% (69.9% to 86.6%) for a single positive test result, and 59.6% (40.7% to 76.0%) and 95.1% (87.0% to 98.2%) for two positive test results. The findings indicate that PCR shows moderate diagnostic accuracy when used as a screening test for invasive aspergillosis in high‐risk patient groups. We found several covariates (in particular, the adoption of antifungal prophylaxis and blinding to the reference test or index test) to substantially affect the measures of diagnostic accuracy under evaluation, particularly sensitivity and specificity. The uneven distribution of these covariates may explain, at least partly, the large heterogeneity found in this analysis. The subgroup analyses suggest that antifungal prophylaxis might impair performance and these conclusions may not be applicable to patients on concurrent antifungal therapy.
Strengths and weaknesses of the review
The findings of this review are based on comprehensive searching, strict inclusion criteria, and standardized data extraction. The strength of our review is that it enables an assessment of the diagnostic accuracy of PCR for detection of IA in a homogenous population of patients at risk of IA. We used the strict inclusion criteria (cohort of consecutive patients, including neutropenic cancer patients and hematopoietic stem cell or solid organ transplant recipients) to cover the spectrum of diseases likely to be encountered in the current or future use of this diagnostic test.
We only included studies that used the EORTC/MSG criteria or a similar reference standard. Differences in the reference standard may have contributed to differences we found in the distribution of patients with probable, possible and no invasive aspergillosis, but not 'proven disease' as this relies on demonstration of the fungus in tissue. For instance, the clinical features in the revised definitions are based solely on radiological evidence of IA whereas the original 2002 definitions also included minor signs such as fever and cough as evidence of disease. Consequently, employing the revised definitions to cases classified as possible IA by the 2002 definitions would only be retained as such if there was radiological evidence. Applying the 2008 definitions would have a similar effect on probable IA for the same reasons.
Anti‐mould prophylaxis reduces the proportion of proven/probable cases of IA (according to EORTC/MSG criteria) which is associated with a lower specificity of the Aspergillus PCR testing of blood. It is likely that PCR can detect infection before overt disease is radiologically detectable. Consequently, people with positive results who did not meet the criteria for proven or probable disease could have had early infection that resolved either with empirical or pre‐emptive antifungal treatment or as a result of resolution of the underlying immunosuppression.
The antifungal administration could mask a proportion of invasive infections, thus lowering the diagnostic recognition of a proportion of them. A raw calculation indicates a prevalence of 17.4% without prophylaxis, 10.4% with prophylaxis. Meanwhile, the PCR could maintain its ability to detect the Aspergillus DNA in the blood of the patients. Alternatively, the prophylaxis could maintain the fungal growth in a pre‐invasive stage, though not impeding the shedding of genomic material into the circulation, possibly enhancing its release through damage to the fungal cell wall or membrane.
The lack of direct comparisons with other biomarkers including GM and beta‐D‐glucan could be a further shortcoming. Looking at our findings and at those of other reviews, the performance of the PCR test is comparable to that reported for GM and superior to beta‐D‐glucan. It is likely that combinations of different biomarkers will provide the optimal diagnostic performance. Also it was difficult to distinguish between using PCR for screening purposes and for confirming the diagnosis as these are associated with low and high a priori likelihood respectively. Furthermore, screening requires testing at regular intervals during the period of risk (typically every 3 to 4 days) whereas tests for confirming the diagnosis of IFD will only be done once.
The molecular basis for azole resistance has been described, and the ability to detect Aspergillus DNA also raises the possibility of rapid detection of antifungal resistance using the same specimen. This could optimise patient management further and should be explored in future studies.
Applicability of findings to the review question
We noted that most studies performed PCR in high‐level reference laboratories, but it is not clear whether intermediate/peripheral laboratories might be settings that match the review question. An important step towards the standardisation and widespread uptake of PCR‐based diagnosis for aspergillosis will be the adoption of effective kit‐based assays. Much has been done by the EAPCRI to establish a standard for PCR that should help laboratories offering the test (www.eapcri.eu). However incorporating PCR into routine practice also requires an explicit protocol indicating who should be tested, when and how frequently, as well as what action should be taken in the event of a given result (Barnes 2018). Moreover the process needs to be completed within a frame so that the results can be used to best advantage by the clinician. This requires an explicit care plan or pathway, a multidisciplinary approach and a clear understanding between the clinic and laboratory to ensure a smooth turnaround.
Authors' conclusions
Implications for practice.
The findings indicate that PCR screening tests show moderately good diagnostic accuracy when used as screening tests for IA in high‐risk patient groups. For a screening strategy, however, with the low prevalence of IA in the observed population and a low pre‐test probability of disease, the moderate sensitivity of the PCR is sufficient to ensure a good negative predictive value, such that disease can be confidently excluded and the need for empiric therapy avoided. As such, screening strategies could replace empirical antifungal therapy in selected high‐risk patients. Consecutive positive test results show excellent specificity in the diagnosis of IA and could be used to trigger radiological and other investigations or for pre‐emptive therapy in the absence of specific radiological signs when the clinical suspicion of infection is high. The subgroup analyses suggest that antifungal prophylaxis could impair performance and these conclusions may not be applicable to people on concurrent antifungal therapy. With the observed prevalence of disease (16.3%), repetition of the PCR test increase considerably the positive predictive values, with a modest decline of the negative predictive values. Therefore we recommend the repetition of the PCR assay in order to increase the diagnostic accuracy.
Implications for research.
It is clear that PCR holds a lot of promise as a useful test for detecting Aspergillus infection although the diagnostic accuracy might be improved further by combining the test with other biomarkers such as GM, and this should be explored in future studies. Further validation is also needed to determine whether using PCR for screening high‐risk patients, not on anti‐mould prophylaxis, could become the standard of care. Future studies that validate PCR for aspergillosis clearly need to distinguish between use of the test to screen for the presence or absence of IA in high‐risk patients if there are no signs of illness, and its use to confirm or exclude the disease when it becomes manifest. IA can be ruled out during the risk period for as long as any single PCR test is negative and there are no clinical signs of disease. Conversely when prevalence of aspergillosis is around 10%, two or more PCR positive results can be used for mycological confirmation to allow a case of possible IA to be upgraded to probable.
The tests need to be incorporated into patient care pathways that compare prophylactic, empirical, pre‐emptive and targeted antifungal drug use looking at impacts on patient management.
It was not possible to investigate the diagnostic utility of combinations of biomarkers (e.g. PCR and GM) because the GM is incorporated into the EORTC/MSG definitions and would introduce incorporation bias. Hence, cases would have to be classified by omitting GM. Further studies are needed to assess clinical utility and cost effectiveness.
What's new
| Date | Event | Description |
|---|---|---|
| 27 August 2019 | Amended | Author name amended. |
| 20 August 2019 | New search has been performed | Search updated. |
| 19 March 2018 | New citation required but conclusions have not changed | Search updated to 19 March 2018. 11 new studies included. |
History
Protocol first published: Issue 1, 2012 Review first published: Issue 9, 2015
| Date | Event | Description |
|---|---|---|
| 14 September 2015 | Amended | Errors in text corrected |
| 14 September 2015 | New citation required but conclusions have not changed | Errors in text corrected |
Acknowledgements
We would like to thank Gail Quinn, Managing Editor of the Cochrane Gynaecological, Neuro‐oncology and Orphan Cancer Group (CGNOG) for continuous support throughout the editorial phase. We would also like to thank Jane Hayes and Joanne Platt, Information Managers of CGNOG, for bibliographical searches. We thank all authors of the included studies who answered our questions and provided additional data.
This project was supported by the National Institute for Health Research (NIHR), via Cochrane Infrastructure funding to the CGNOG. The views and opinions expressed herein are those of the authors and do not necessarily reflect those of the Systematic Reviews Programme, NIHR, National Health Service or the Department of Health.
The project was also supported by the Fungal PCR Initiative (FPCRI), a branch of the International Society for Human and Animal Mycology (ISHAM). Members of the FPCRI Aspergillus clinical translational group are: Rosemary Barnes; Dieter Buchheidt; Catherine Cordonnier; Mario Cruciani; Werner Heinz; Brian Jones; Lena Klingspor; Deborah Lockhart; Johan Maertens; Tom Rogers; Adilia Warris; and Lewis White.
Appendices
Appendix 1. Search strategies
MEDLINE
1 exp Aspergillosis/ 2 exp Pulmonary Aspergillosis/ 3 exp Aspergillus/ 4 (aspergillosis or aspergillus or aspergilloma or "A.fumigatus" or "A. flavus" or "A. clavatus" or "A. terreus" or "A. niger").ti,ab. 5 or/1‐4 6 exp Nucleic Acid Amplification Techniques/ 7 pcr.ti,ab. 8 "polymerase chain reaction*".ti,ab. 9 or/6‐8 10 5 and 9 11 exp Animals/ not Humans/ 12 10 not 11 key: ti,ab. = title,abstract
Embase
1 Aspergillosis/ 2 Lung Aspergillosis/ 3 exp Aspergillus/ 4 (aspergillosis or aspergillus or aspergilloma or "A.fumigatus" or "A. flavus" or "A. clavatus" or "A. terreus" or "A. niger").ti,ab. 5 or/1‐4 6 nucleic acid amplification/ 7 Polymerase Chain Reaction/ 8 pcr.ti,ab. 9 "polymerase chain reaction*".ti,ab. 10 or/6‐9 11 5 and 10 12 (exp Animal/ or Nonhuman/ or exp Animal Experiment/) not Human/ 13 11 not 12 key: ti,ab =title,abstract
CENTRAL
#1 MeSH descriptor: [Aspergillosis] explode all trees #2 MeSH descriptor: [Pulmonary Aspergillosis] explode all trees #3 MeSH descriptor: [Aspergillus] explode all trees #4 aspergillosis or aspergillus or aspergilloma or "A.fumigatus" or "A. flavus" or "A. clavatus" or "A. terreus" or "A. niger" #5 #1 or #2 or #3 or #4 #6 MeSH descriptor: [Nucleic Acid Amplification Techniques] explode all trees #7 MeSH descriptor: [Polymerase Chain Reaction] explode all trees #8 pcr or "polymerase chain reaction*" #9 #6 or #7 or #8 #10 #5 and #9
WEB of Science, LILACS, Database of Abstracts of Reviews of Effects, Health Technology Assessment, Scopus
(Aspergillus or Aspergillosis) AND (Polymerase Chain Reaction or Nucleic Acid Amplification) in title, abstracts and keywords
Appendix 2. QUADAS‐2 Items
| DOMAIN | PATIENT SELECTION | INDEX TEST | REFERENCE STANDARD | FLOW AND TIMING |
| Description | Describe methods of patient selection: Describe included patients (prior testing, presentation, intended use of index test and setting): | Describe the index test and how it was conducted and interpreted: | Describe the reference standard and how it was conducted and interpreted: | Describe any patients who did not receive the index test(s) and/or reference standard or who were excluded from the 2 x 2 table (refer to flow diagram): Describe the time interval and any interventions between index test(s) and reference standard: |
| Signalling questions (yes/no/unclear) | Was a consecutive or random sample of patients enrolled? | Were the index test results interpreted without knowledge of the results of the reference standard? | Is the reference standard likely to correctly classify the target condition? | Was there an appropriate interval between index test(s) and reference standard? |
| Was a case‐control design avoided? | If a threshold was used, was it pre‐specified? | Were the reference standard results interpreted without knowledge of the results of the index test? | Did all patients receive a reference standard? | |
| Did the study avoid inappropriate exclusions? | Did all patients receive the same reference standard? | |||
| Were all patients included in the analysis? | ||||
| Risk of bias: high/low/unclear | Could the selection of patients have introduced bias? | Could the conduct or interpretation of the index test have introduced bias? | Could the reference standard, its conduct, or its interpretation have introduced bias? | Could the patient flow have introduced bias? |
| Concerns regarding applicability: high/low/unclear | Are there concerns that the included patients do not match the review question? | Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Are there concerns that the target condition as defined by the reference standard does not match the review question? |
Item Patient selection. Code this item: Yes. If the characteristics of the spectrum of patients fulfilled the pre‐stated requirements and the method of recruitment was consecutive, or random samples were taken from consecutive series. No. If the sample does not fit with what was pre‐specified as acceptable or if groups with and without the target disorder were recruited separately, particularly with healthy controls. Unclear. If there is insufficient information available to make a judgment. Independent index and reference test (incorporation). Yes.If the index test did not form part of the reference standard. No. If the reference standard formally included the result of the index test. Unclear If it is unclear whether the results of the index test were used in the final diagnosis. .Index test blind for reference test results and vice versa. Yes. If test results (index or reference standard) were interpreted blind to the results of the other test, or blinding is dictated by the test order, or meets the pre‐stated assumptions. No.If it is clear that one set of test results was interpreted with knowledge of the other. Unclear. If it is unclear whether blinding took place. Item Reference Standard Yes. All reference standards used meet the pre‐stated criteria. No. One or more reference standards used do not meet the pre‐stated criteria. Unclear. It is unclear exactly what reference standard was used. Were partial verification and differential verification prevented? Yes. If all patients, or a random selection of patients, who received the index test went on to receive verification of their disease status using a reference standard, even if the reference standard was not the same for all patients. No. If some of the patients who received the index test did not receive verification of their true disease state, and the selection of patients to receive the reference standard was not random. Unclear. If this information is not reported by the study. Item Flow and timing. Yes.If the time between tests was shorter than that required, at least for an acceptably high proportion of patients. No. If the time between tests was longer than that required for an unacceptably high proportion of patients. Unclear. If information on timing of tests is not provided.
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
| Test | No. of studies | No. of participants |
|---|---|---|
| 1 PCR: single positive requirement | 28 | 4989 |
| 2 PCR: two positive requirement | 9 | 2151 |
| 3 no anti‐mould prophylaxis | 13 | 1464 |
| 4 antimould prophylaxis | 12 | 1478 |
| 5 in‐house qPCR | 15 | 2661 |
| 6 qPCR kit | 3 | 302 |
| 7 PCR on whole blood | 15 | 2217 |
| 8 PCR on serum | 13 | 2481 |
1. Test.
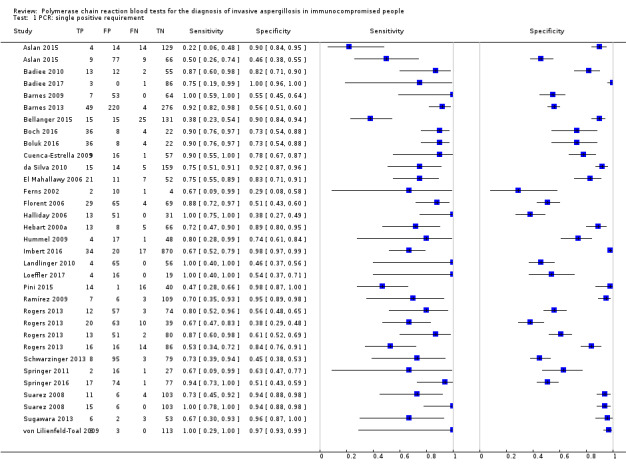
PCR: single positive requirement.
2. Test.
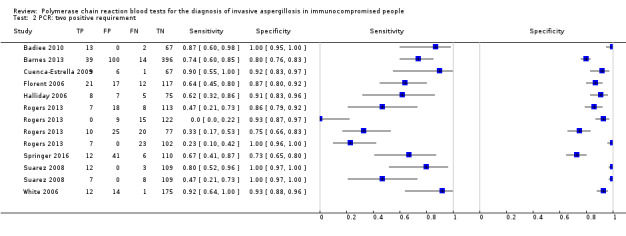
PCR: two positive requirement.
3. Test.
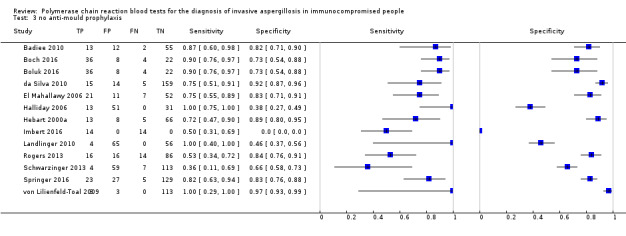
no anti‐mould prophylaxis.
5. Test.
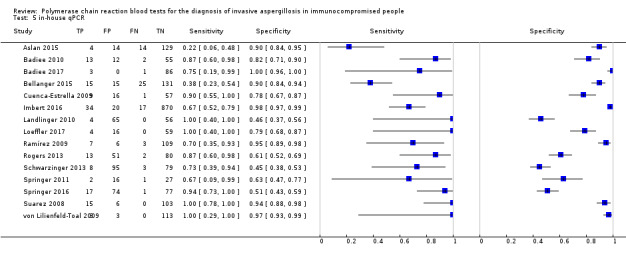
in‐house qPCR.
6. Test.
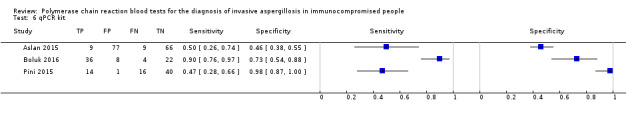
qPCR kit.
7. Test.
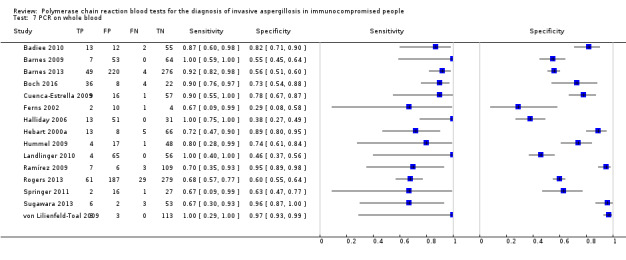
PCR on whole blood.
8. Test.
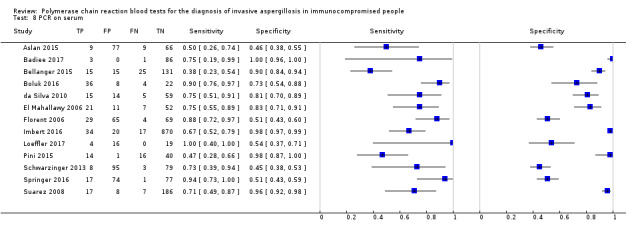
PCR on serum.
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Aslan 2015.
| Study characteristics | |||
| Patient sampling | Neutropenic patients at risk of IA were prospectively included in the trial between January 2011 and January 2012 | ||
| Patient characteristics and setting | 161 febrile neutropenic episodes of 99 patients. Haematology and SCT patients with fever. University Hospital in Turkey | ||
| Index tests | 2 PCR tests were used: an In‐house real‐time PCR and a commercially available test (MAP‐Myconostica Ltd, Manchester, UK). GM also performed | ||
| Target condition and reference standard(s) | Patients were evaluated for IA; cases of IA were defined according to the EORTC/MSG revised criteria (incorrectly used). | ||
| Flow and timing | January 2011 to January 2012 | ||
| Comparative | |||
| Notes | A control group of patients not at risk of IA was also included, but it was possible to extract sensitivity and specificity data just from the relevant clinical population | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Badiee 2010.
| Study characteristics | |||
| Patient sampling | Prospective study, samples collected September 2004 to June 2006. Patients with haematological malignancies (who had received chemotherapy) | ||
| Patient characteristics and setting | Sample size: 194 Males/females: 133/61 Mean age: 33.7 years (range 14 to 80) Presentation: patients with haematological malignancies and solid organ transplantation at risk for IFD Setting: Nemazi Hospital, Shiraz, Iran | ||
| Index tests | DNA extracted through lysis of blood and fungal cells (Van Burik 1998) followed by purification using the QIAamp DNA Mini Kit. Standard PCR was used as well as PCR‐ELISA. . Presence or absence of bands indicated a positive result; positive results were retested with species‐specific probes | ||
| Target condition and reference standard(s) | Patients were evaluated for IA; patient samples (urine, cerebrospinal fluid, pleural and abdominal tap, BAL and sputum) were examined for signs of infection. Cases of IA were defined according to the EORTC/MSG 2002 criteria | ||
| Flow and timing | Samples were collected from 209 patients between September 2004 and June 2006; 985 samples collected from 194 patients were analysed. Blood samples (EDTA) were collected once per week and frozen prior to analysis. Patients were excluded if they did not attend follow‐up for more than 2 weeks. No indication that patients with possible IA were excluded from 2 × 2 analysis | ||
| Comparative | |||
| Notes | This study describes the performance of standard PCR and PCR‐ELISA | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Unclear | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | No | ||
| Unclear | |||
Badiee 2017.
| Study characteristics | |||
| Patient sampling | Consecutive patients | ||
| Patient characteristics and setting | 86 haematologic paediatric patients. Shiraz University of Medical Science, Iran | ||
| Index tests | real time PCR for candidiasis and aspergillosis | ||
| Target condition and reference standard(s) | IFI; EORTC/MSG revised criteria | ||
| Flow and timing | from January 2014 to February 2015 | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Barnes 2009.
| Study characteristics | |||
| Patient sampling | Prospective study between October 2005 and March 2006; at risk febrile patients or SCT patients with graft‐versus‐host disease were tested |
||
| Patient characteristics and setting | Sample size: 125 patients Males/females: 1.4/1 Mean age: 56.2 years (range 16 to 83) Presentation: haematology patients at risk for IFD including SCT, acute myeloid leukemia Setting: University Hospital of Wales | ||
| Index tests | DNA extracted from 2 ml blood, red cell lysis, white cell lysis, bead beating and Magna Pure (Roche) DNA purification (White 2006). Nested PCR with second round on LightCycler (Roche) targeting 28S, 60 cycles all together. All positive samples were repeated | ||
| Target condition and reference standard(s) | IFD was the target condition for PCR assays; GM antigen testing was performed on patient samples, EORTC/MSG 2008 criteria (including GM) were used to define cases of IFD | ||
| Flow and timing | 1028 specimens collected from 125 patients over a 6‐month period. 130 patients were screened but 125 were evaluable. No indication that patients were excluded from 2 × 2 analysis; this analysis was performed for "single non‐reproducible positive PCR", "Single reproducible positive PCR" and "multiple positive PCR" results | ||
| Comparative | |||
| Notes | Report examines diagnostic‐driven care pathway, limited empirical treatment. Data provided for interpretation of single and reproducible results. Very relevant to this review | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Low | |||
Barnes 2013.
| Study characteristics | |||
| Patient sampling | Prospective, consecutive | ||
| Patient characteristics and setting | Sample size: 612 patients, excluded 27 children, 36 due to sample size (> 2) males/females: ? Mean age: ? Presentation: febrile or history of fungal infection orSCT with GVHD Setting: hospital in Cardiff | ||
| Index tests | Aspergillus PCR (Barnes 2009; Lewis 2006) | ||
| Target condition and reference standard(s) | Invasive aspergillosis. EORTC/MSG criteria | ||
| Flow and timing | Between Oct 2005 and June 2009 all adult patients entered into the pathway were audited. Fungal diagnostic test (antigen and PCR) were performed twice weekly in SCT patients and during fever in other patients | ||
| Comparative | |||
| Notes | comparison of single vs double PCR positives; EORTC classification with/without GM/serum. Patients received itraconazole prophylaxis | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Unclear | ||
| Unclear | |||
Bellanger 2015.
| Study characteristics | |||
| Patient sampling | consecutive sample of patients | ||
| Patient characteristics and setting | Sample size: 185 patients Males/females: not stated Mean age: not stated Presentation: inclusion based on risk factors for IA including prolonged neutropenia and aplasia. Setting: haematology ICU, University Hospital Besancon, France | ||
| Index tests | 18S and Mito | ||
| Target condition and reference standard(s) | Invasive aspergillosis. 2008 EORTC/MSG criteria | ||
| Flow and timing | Twice weekly serum samples | ||
| Comparative | |||
| Notes | GM and beta‐glucan also performed | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Boch 2016.
| Study characteristics | |||
| Patient sampling | Prospective, consecutive patients | ||
| Patient characteristics and setting | 99 haematologic patients at risk of IA. Patients with CT signs suggestive of lung infiltrates underwent BAL. University hospitals of Mannheim, Cologne, Essen, Wuerzburg, Regensburg, Erlangen, Heidelberg, Prosper‐Hospital Reckling‐hausen and the General Hospital of Frankfurt/Oder | ||
| Index tests | Diagnostic performance of a galactomannan (GM) enzyme immune assay (EIA), a 1,3‐β‐D‐glucan assay (BDG), an Aspergillus PCR, and a multifungal DNA‐microarray (Chip) alone or in combination were calculated. | ||
| Target condition and reference standard(s) | Invasive aspergillosis. EORTC/MSG criteria | ||
| Flow and timing | Patients were treated from 2012 to 2015 | ||
| Comparative | |||
| Notes | Calculation of diagnostic performance of GM and/or BDG was additionally carried out with the exclusion of these test from defining probable IFD | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Boluk 2016.
| Study characteristics | |||
| Patient sampling | Prospective study with consecutive enrolment of patients, although not clearly stated | ||
| Patient characteristics and setting | Sample size: 70 patients Males/females: not stated Mean age: not stated Presentation: Inclusion based on risk factors for IA including neutropenia, recent use of immunosuppressive drugs including corticosteroids and persistent fever under broad spectrum antibiotic therapy. Setting: Hospital Haematology Clinic, Uludag University, Turkey | ||
| Index tests | They used a commercial PCR kit (Way2 Gene Fungi Kit() on a LightCycler 480 Probes Master. An internal control was used for PCR. | ||
| Target condition and reference standard(s) | Invasive aspergillosis. Patients classified by the 2008 EORTC/MSG criteria | ||
| Flow and timing | Twice weekly serum samples, stored and analysed retrospectively | ||
| Comparative | |||
| Notes | There was no antifungal prophylaxis | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Cuenca‐Estrella 2009.
| Study characteristics | |||
| Patient sampling | Patients with febrile neutropenia considered at risk from IA were studied prospectively between October 2004 and November 2005 | ||
| Patient characteristics and setting | Sample size: 83 patients Males/females: 48/35 Mean age: 52 years Presentation: patients with haematological malignancies and febrile neutropenia at risk for IA Setting: Hospital Universitario 12 de Octubre in Madrid, Spain | ||
| Index tests | DNA extraction: DNA was extracted from the samples using the QiampDNA Mini Kit (Qiagen, Izasa, Madrid, Spain) DNA detection: 2 µl of DNA from each sample were used for each RT‐PCR, which contained a final volume of 20 µl with 3 mM of Cl2Mg, 0.5 µM from each primer, and 0.4 µM of molecular beacon probe. Preincubation was at 95 °C, followed by 45 denaturation cycles (15 s at 95 °C), annealing (30 s at 56 °C), and extension (5 s at 72°C). Each experiment was run twice Definition of positive assay: the results were considered positive when an exponential increase in fluorescence was detected compared with that of the negative controls before cycle 40 of amplification. The detection limit was 10 fg of DNA per µl of sample (cycle 42 of amplification). Aspergillus‐specific: analyses for at least 1, 2 or 3 positive PCR tests retesting. 2244 specimens tested |
||
| Target condition and reference standard(s) | The definitions of proven, probable and possible IA were set according to the definitions of the EORTC/MSG. HRCT and GM testing were also performed as a part of reference standard | ||
| Flow and timing | 4 weekly samples (2 blood and 2 serum) were taken during episodes of febrile neutropenia Time interval sampling: 2004 to 05 Selection/exclusion for analysis: excluding patient 10, for whom the PCR result was negative, it was possible to calculate the time gain in diagnosis for the PCR technique compared to that for HRCT and GM for the other 11 patients with IA Sampling/storage: years (range) Analysis type: at least 2 consecutive positive PCR results missing/uninterpretable results: N |
||
| Comparative | |||
| Notes | Prophylaxis: itraconazole; proven/probable/possible/no IA: 1/9/2; PCR effectiveness (replica/eluat into PCR volume): 2 × 2 of 200 µl. The information collected on each patient, as well as the PCR results, were entered in a database | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
da Silva 2010.
| Study characteristics | |||
| Patient sampling | From October 2000 to August 2003, 172 patients with haematologic malignancies and 27 patients receiving high‐dose chemotherapy in an autologous haematopoietic stem cell transplantation setting were studied prospectively. All patients were screened by PCR twice a week since admitted in the ward | ||
| Patient characteristics and setting | Patients with haematological malignancies and febrile neutropenia at risk for IA Median age 50 years Male/female: 102/70 Setting: Hospital dos Capuchos, Lisbon, Portugal |
||
| Index tests | Blood samples, BAL samples, fungal DNA extraction and PCR conditions were performed as described in Van Burik 1998. The whole process of amplification was done using Taq polymerase (Gibco BRL) and pan‐fungal primers that bind to the conserved regions of the fungal 18S rRNA gene sequence. Established PCR negative and positive controls were used in every assay. 1311 blood specimens tested | ||
| Target condition and reference standard(s) | Fungal infections were classified according to EORTC/MSG revised consensus | ||
| Flow and timing | Peripheral blood samples from patients were screened twice weekly for both methods since admission to the ward. If a positive value was obtained the patient would be screened every day for 3 consecutive days in the first week and then twice weekly again | ||
| Comparative | |||
| Notes | The study also evaluated GM assay, but due to incorporation bias (GM is part of the reference standard), these data were not included in the current review | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
El Mahallawy 2006.
| Study characteristics | |||
| Patient sampling | Febrile, neutropenic paediatric cancer patients were prospectively sampled between April 2003 and April 2004. Patients were included if they had antibiotic‐resistant fever. Patients were given full diagnostic work‐ups for any signs of IFD | ||
| Patient characteristics and setting | Sample size: 91 patients Males/females: 37:25 Mean age: 8 (range 2 to 18) Presentation: "at risk" for IA including febrile neutropenic cancer patients and fever not responding to antibiotics Setting: National Cancer Institute, Cairo University | ||
| Index tests | Serum samples (unknown volume) were treated with Lyticase, then extracted using QIAamp DNA Mini Kit (Qiagen), PCR amplified 420 bp products from 18S gene (universal fungal assay). Single round conventional PCR with 30 cycles. Products detected on agarose gel | ||
| Target condition and reference standard(s) | Target condition was IFD; CT scan, blood culture and Aspergillus antigen detection were used to aid in defining cases of IFD according to the EORTC/MSG (2002) criteria | ||
| Flow and timing | 91 patients tested, unknown sample numbers during 1 year period. All patients were included in 2 × 2 analysis to calculate sensitivity, etc. | ||
| Comparative | |||
| Notes | Pan‐fungal conventional PCR used with low cycles, lack of specific IA information may be a problem for inclusion | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Ferns 2002.
| Study characteristics | |||
| Patient sampling | 94 blood samples from 17 patients at high risk of IA undergoing chemotherapy for acute leukaemia (10) or undergoing allogenic BMT (7) on the haematology unit at the University College London Hospital Trust were screened. | ||
| Patient characteristics and setting | Gender and age: not specified Setting: University College London Hospital Trust |
||
| Index tests | Aspergillus DNA, from whole blood samples, was amplified by nested PCR to detect a 135 bp fragment in the mitochondrial region of Aspergillus fumigatus or Aspergillus flavus (121 bp) | ||
| Target condition and reference standard(s) | IA in haematologic patients. The diagnosis of aspergillosis was classified into proven, probable or possible on the basis of EORTC/MSG criteria | ||
| Flow and timing | PCR results were retrospectively compared with clinical data and antifungal treatment | ||
| Comparative | |||
| Notes | None of the 94 samples from the 17 patients were above the cut‐off value when tested as serum in the Platelia™ Aspergillus antigen ELISA | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Unclear | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Florent 2006.
| Study characteristics | |||
| Patient sampling | From April 2001 through November 2002, all patients (> 15 years) with haematological malignancies who were routinely screened for GM detection were included in the study. Gender and age were not specified. Setting was Hopital Saint‐Louis and Hotel‐Dieu, Paris | ||
| Patient characteristics and setting | A total of 201 patients were enrolled in the study and had 256 consecutive episodes of neutropenia (neutrophil count fewer than 500 cells/mL). During the high‐risk periods for infection and until absolute neutrophil counts increased to greater than 500 cells/mL, all patients were hospitalised in protected facilities with high‐efficiency particulate air filtration associated with laminar air flow for patients undergoing allogeneic stem cell transplantation | ||
| Index tests | DNA was extracted from both serum and fungal cultures by use of the QIAamp DNA Mini Kit (Qiagen), in accordance with the manufacturer’s recommendations. 2 negative controls were used in each DNA extraction experiment. The PCR‐ELISA was performed using the serum sample that was collected for GM detection, which was stored at −20 °C until processing. 1205 specimens tested | ||
| Target condition and reference standard(s) | The criteria proposed by the EORTC/MSG were used. To evaluate the performance of the GM assay either alone or in combination with the PCR‐ELISA, the results of the GM assay were not included in the microbiological criteria for the diagnosis of probable IA | ||
| Flow and timing | Single‐positive results were defined as at least a single positive result, and consecutive positive results were defined as at least 2 positive results obtained consecutively within 1 week. 34 patients did not have consecutive serum samples that were collected within 1 week, and they were excluded from the final analysis. Because of the uncertainty of the diagnosis in patients with possible IA, 3 separate analyses were performed: the first included only proven and probable IA cases; the second included proven and probable IA cases, and possible cases were considered to be proven IA cases; and the third included proven and probable IA cases, and possible cases were not considered to be IA. Inhibitors were detected in 18 serum samples, and these samples were excluded from the analysis. | ||
| Comparative | |||
| Notes | PCR‐ELISA precocity in diagnosing IA was assessed in comparison with the timing of the clinical suspicion of IA, the results of CT, and histological and microbiological criteria as defined by the EORTC/MSG. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Unclear | |||
Halliday 2006.
| Study characteristics | |||
| Patient sampling | Prospective collection of samples from patients undergoing chemotherapy or HSCT who had developed febrile neutropenia between August 2002 and July 2003. Blood samples collected from consecutive patients twice weekly; only patients from whom 3 samples were obtained per febrile episode were analysed | ||
| Patient characteristics and setting | Sample size: 65 patients Males/females: 23:6 Mean age: 37 (range 16 to 62) Presentation: episodes of febrile neutropenia in patients undergoing chemotherapy or HSCT Setting: Westmead Hospital, NSW, Australia | ||
| Index tests | Blood collected twice weekly; DNA extracted from 500 µl EDTA blood using the GenElute Mammalian Genomic DNA Kit (Sigma‐Aldrich) with modified protocol that included RCLB, followed by lyticase treatment; no bead beating. Conventional nested PCR no qPCR assay modified from (Skladny 1999). Aspergillus specific targeting 18S. Sensitivity of 10 CFU/ml | ||
| Target condition and reference standard(s) | Target condition was IA, classified according to the EORTC/MSG criteria (2002). IA defined at the end of "at risk" episodes | ||
| Flow and timing | 998 blood samples from 65 patients (29 adults and 36 children) were collected between August 2002 and July 2003. Separate 2 × 2 analyses were carried out to calculate sensitivity, etc, with possible cases excluded, or with possible cases included as true negatives or true positives. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Hebart 2000a.
| Study characteristics | |||
| Patient sampling | Prospective sample collection from patients who had undergone allogenic SCT between 1996 and 1997. 5 ml EDTA was collected 2 to 4 times weekly from the time of admission until discharge or death. Samples from multiple centres were analysed in Tübingen. | ||
| Patient characteristics and setting | Sample size: 84 patients Males/females: not specified Mean age: 35 years (range 17 to 57) Presentation: patients had undergone allogeneic SCT Setting: University Hospital Würzburg | ||
| Index tests | DNA extracted from 5 ml blood as described by Einsele et al 1997 (JCM); PCR targeting 18S with Aspergillus specific probe (Aspergillus fumigatus, flavus and versicolour) for slot blot testing (not qPCR) | ||
| Target condition and reference standard(s) | IA was the target condition; cases of proven IA were defined as recovery of Aspergillus from normally sterile sites, positive culture or demonstration of hyphae from deep tissue biopsy and autopsy specimens along with clinical symptoms. Probable IA was defined as the presence of clinical signs and symptoms together with radiographic evidence compatible with IA and isolation of Aspergillus from respiratory specimens. | ||
| Flow and timing | 1193 samples from 84 patients collected twice weekly and processed twice weekly. 2 × 2 analysis to calculate sensitivity, etc. Included all patients (possible was not defined). Parameters were calculated for both early and late onset IA. | ||
| Comparative | |||
| Notes | This study utilises definitions of IA that are pre‐EORTC/MSG. Generally seems a compatible study | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Hummel 2009.
| Study characteristics | |||
| Patient sampling | PCR results from all consecutive patients from 3 university children’s hospitals investigated between November 2000 and January 2007 were evaluated in this study | ||
| Patient characteristics and setting | The majority of patients had malignant haematological diseases. Patients from 3 university children’s hospitals | ||
| Index tests | Aspergillus DNA was detected in clinical samples by an experimentally and clinically validated nested PCR assay as described previously (Bucheidt 2001; Bucheidt 2004; Skladny 1999). | ||
| Target condition and reference standard(s) | Invasive aspergillosis; EORTC/MSG criteria | ||
| Flow and timing | between November 2000 and January 2007 | ||
| Comparative | |||
| Notes | Results of serological diagnostic techniques (GM assay, Platelia™ Aspergillus enzyme immunoassay; Bio‐Rad) and post‐mortem histological examination were included for clinical classifications | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Imbert 2016.
| Study characteristics | |||
| Patient sampling | Retrospective single‐centre analysis of all patients at risk of IA | ||
| Patient characteristics and setting | Patients with various underlying diseases, mostly haematologic and neutropenic, but also patients with non‐invasive form of aspergillosis. Hôpital Pitié Salpêtrière, Paris | ||
| Index tests | In‐house A fumigatus real‐time PCR | ||
| Target condition and reference standard(s) | Invasive aspergillosis and a subset of patients with non‐invasive aspergillosis. EORTC/MSG criteria | ||
| Flow and timing | GM and PCR performed in 970 patients, but clinical data available from 941 (5146 serum samples). Retrospective analysis of all patients at risk of IA from February 2012 and October 2014 | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Unclear | |||
Landlinger 2010.
| Study characteristics | |||
| Patient sampling | Clinical specimens from consecutive patients were prospectively collected | ||
| Patient characteristics and setting | 125 paediatric haemato‐oncological patients undergoing intensive chemotherapy (65) or allogeneic stem cell transplantation (60) were analysed during 150 episodes of febrile neutropenia | ||
| Index tests | Pan‐fungal RT‐PCR | ||
| Target condition and reference standard(s) | IA; EORTC/MSG criteria | ||
| Flow and timing | Whenever possible, specimens were collected at first onset of fever, within 48 hours thereafter, and at subsequent time points in the course of the febrile episode, upon availability. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Loeffler 2017.
| Study characteristics | |||
| Patient sampling | Prospective biomarkers screening for IA in haematologic children (alloHSCT). | ||
| Patient characteristics and setting | Haematologic children at risk of IA. University Children's hospital , Wurzburg | ||
| Index tests | PCR conducted according to the EAPCRI criteria. GM assay also performed | ||
| Target condition and reference standard(s) | Invasive aspergillosis. EORTC/MSG criteria | ||
| Flow and timing | Twice weekly systematic screening of high‐risk children by GM and PCR. Patients screened from 2012 to 2015 were all selected for retrospective analysis | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Pini 2015.
| Study characteristics | |||
| Patient sampling | Prospective evaluation of patients at risk of IA. Of the 71 eligible patients, 64 were prospectively enroled, while 7 were excluded for incomplete data collection | ||
| Patient characteristics and setting | Haematologic and other patients at risk of IA (COPD, SOT, cancer receiving chemotherapy, cirrhosis) | ||
| Index tests | Qualitative real‐time PCR. GM also performed | ||
| Target condition and reference standard(s) | Invasive aspergillosis. EORTC/MSG criteria for haematologic patients. For the other patients, the criteria proposed by Meersseman 2004 | ||
| Flow and timing | From December 2011 to December 2013. 141 serum samples from 64 evaluable patients | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Unclear | Unclear | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Unclear | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Unclear | |||
Ramírez 2009.
| Study characteristics | |||
| Patient sampling | Prospective sampling of "at risk" patients for IFD between June 2004 and July 2006. Samples also taken from patients for whom confirmation of IFD before, during and after treatment was required | ||
| Patient characteristics and setting | Sample size: 127 patients
Males/females: 64/63 Mean age: 45 years (range 30 to 58) Presentation: patients at risk for IA and those requiring confirmation of IFD Setting: Hospital Universitario de Valme, Seville, Spain |
||
| Index tests | DNA extracted from 5 ml blood (EDTA); used RCLB, glass bead disruption and QiaAmp DNA Mini Kit. LightCycler assay as described by Loeffler 2000. 20 µl PCR included 10 µl template DNA; 50 cycles; followed by melt‐curve analysis. DNA extraction control included, no internal control | ||
| Target condition and reference standard(s) | IA was the target condition; cases were defined according to the EORTC/MSG criteria (2002) | ||
| Flow and timing | 948 clinical samples from 127 patients collected between June 2004 and July 2006. Samples processed immediately or stored prior to processing. 2 × 2 analysis was not conducted. Study focused on analytical sensitivity (60 fg Aspergillus DNA, or 5 to 20 conidia); 1% of the samples were PCR positive | ||
| Comparative | |||
| Notes | This study had 5 proven/probable cases, 17 possible | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | No | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Rogers 2013.
| Study characteristics | |||
| Patient sampling | Consecutive patients at risk of IA. Age not specified | ||
| Patient characteristics and setting | Patients undergoing remission‐induction chemotherapy for acute leukaemia, lymphoma, or myeloma, autologous or allogeneic bone marrow or stem cell transplant were eligible for inclusion. Over the course of the study 146 patients were recruited from Trinity College Dublin & St. James’s Hospital, Dublin, and 132 from the Department of Internal Medicine, University of Würzburg Medical Centre, Würzburg, Germany | ||
| Index tests | ITS qPCR assay targeting the ITS 1/5.8S ribosomal operon was performed as previously described (Springer 2011) | ||
| Target condition and reference standard(s) | The EORTC/MSG definitions were used for categorization of patients with IFD including IA | ||
| Flow and timing | Patient blood samples were collected twice weekly; in UKW the EDTA blood samples were logged and processed prospectively while, in SJH, they were frozen at 80 °C and processed in retrospective batches. DNA extracts were stored at 20 °C until they were processed by the second PCR assay. | ||
| Comparative | |||
| Notes | GM was part of the EORTC/MSG criteria for IFD | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Schwarzinger 2013.
| Study characteristics | |||
| Patient sampling | Consecutive patients at risk of IA (185 patients with AML) with 2214 serum samples prospectively included | ||
| Patient characteristics and setting | Setting: the study was conducted in 13 French teaching hospitals | ||
| Index tests | In‐house R‐T PCR not according to EAPCRI recommendations | ||
| Target condition and reference standard(s) | IA was the target condition; cases were defined according to the EORTC/MSG criteria (2002) | ||
| Flow and timing | GM and R‐T PCR was taken twice‐weekly. The entire set of samples comprised 2214 sera collected from 185 patients. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Springer 2011.
| Study characteristics | |||
| Patient sampling | Consecutive patients at high risk of IA. 536 specimens from 46 patients at high risk for invasive fungal infection were collected | ||
| Patient characteristics and setting | Patients at risk of IA after allogeneic SCT and patients receiving myeloablative chemotherapy with an expected duration of neutropenia (leucocyte count of 1,000/L) of at least 10 days. 19 males (mean age 51 years), 17 females (mean age 58 years) | ||
| Index tests | Quantitative PCR and ITS semi quantitative RT‐PCR assay | ||
| Target condition and reference standard(s) | EORTC/MSG criteria | ||
| Flow and timing | Between January and August 2009, blood samples from patients with a high risk of IFD, together with clinical data, were collected | ||
| Comparative | |||
| Notes | GM performed as a part of EORTC/MSG criteria for IA | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Springer 2016.
| Study characteristics | |||
| Patient sampling | Prospective study with consecutive enrolment of patients. | ||
| Patient characteristics and setting | Sample size: 213 patients with 2128 sera. Males/females: 132/81. Mean age: mean age for women: 53,5 years (range 22 to 80); men: 52,7 years (range 19 to 77). Presentation: in total 213 mostly HM patients with 2128 sera were prospectively included during a 2‐year period. Twice‐weeekly serum samples (GM and PCR) were taken from 203 Allogeneic HSCT patients and patients receiving myelosuppressive chemotherapy for AML (n = 99) ALL (18) CLL (6),MDS (26) Lymphoma (21) multiple myeloma (38) solid tumors (5). Setting: university hospitals in Germany and Austria. | ||
| Index tests | In‐house R‐T PCR according to EAPCRI recommendations | ||
| Target condition and reference standard(s) | IA was the target condition; cases were defined according to the EORTC/MSG criteria (2002) | ||
| Flow and timing | |||
| Comparative | |||
| Notes | GM and R‐T PCR was taken twice‐weekly. The entire set of samples comprised 2259 sera collected from 235 patients. 12 patients with fewer samples than 3, and samples showing any failure/inhibition of the internal controle or the entire PCR reaction (n=56) were excluded | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Suarez 2008.
| Study characteristics | |||
| Patient sampling | All adult patients receiving allogeneic or autologous haematopoietic SCT, or intensive (induction, consolidation, or salvage) chemotherapy for haematological malignancies were included in the study | ||
| Patient characteristics and setting | 124 patients (138 treatment episodes) at risk of IA in the adult Haematology and Bone Marrow Transplant Unit at Necker‐Enfants Malades hospital, a tertiary‐care university hospital (Paris, France) | ||
| Index tests | RT‐PCR on 1342 specimens | ||
| Target condition and reference standard(s) | EORTC/MSG‐documented IA. The diagnosis of IA (proven, probable, or possible) was defined for a given patient as the day on which the first clinical, radiological and/or microbiological EORTC/MSG criteria, other than a GM‐positive result, appeared | ||
| Flow and timing | This study was conducted prospectively from February 2006 to March 2007. The dates of diagnosis and the dates on which the first positive test results for Aspergillus fumigatus DNA and GM were recorded. | ||
| Comparative | |||
| Notes | For GM, incorporation bias avoided | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Sugawara 2013.
| Study characteristics | |||
| Patient sampling | Prospective analysis of consecutive blood samples from patients at risk for IFD | ||
| Patient characteristics and setting | 51 patients with haematologic disorders at high risk for IFD who were treated at Mie University Hospital, Japan. Median age in years (range) 57.5 (17 to 78). Sex (male/female) 37/14 | ||
| Index tests | Pan‐fungal PCR assay on 273 specimens | ||
| Target condition and reference standard(s) | Revised criteria of the EORTC/MSG | ||
| Flow and timing | The study was conducted between April 2007 and October 2010. 273 consecutive blood samples from 64 risk episodes in 51 patients with haematologic disorders were analysed. | ||
| Comparative | |||
| Notes | IFD was documented in 14 episodes (21.9%, 9 probable IFDs and 5 possible IFDs). PCR was positive in all of these 14 episodes, and in 4 of the 50 episodes with no IFD category. In this study, a considerable number of fungi (44.4%) other than major ones such as Aspergillus and Candida species were positive by PCR. Non‐major fungi identified were Cunninghamella species, Fusarium species, Scedosporium apiospermum, Rhodotorula species, Rhizopus species, Paecilomyces lilacinus, and Penicillium sclerotiorum. In 10 of the 18 PCR‐positive episodes, continued PCR screenings disclosed the clearance of the fungal DNA during antifungal therapy. The study also evaluated the diagnostic performance of GM, but GM was also part of the reference standard. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
von Lilienfeld‐Toal 2009.
| Study characteristics | |||
| Patient sampling | 70 patients with febrile neutropenia (median leucocyte count 420/mm³) after chemotherapy | ||
| Patient characteristics and setting | Patients treated between September 2001 and February 2002 and between April 2003 and January 2004 on the Haematology ward of the University Hospital Bonn, Germany. Median age in years (IQR) was 60 (49 to 66). Nunber of males (%) was 38 (54). | ||
| Index tests | Commercial PCR‐based kit to detect the DNA of 20 different pathogens (SeptiFast), including IFD. PCR testing was performed retrospectively. | ||
| Target condition and reference standard(s) | IFD according to the standards of the EORTC/MSG | ||
| Flow and timing | 784 serum samples of 119 febrile neutropenic episodes in 70 patients with haematological malignancies were analysed | ||
| Comparative | |||
| Notes | The only patient with proven IFD (Candida glabrata in 1 blood culture which also grew Klebsiella pneumoniae and Enterococcus faecium) yielded a negative result for fungus in the PCR, although the PCR did detect Enterococcus faecium. All of the patients with probable IFDs had positive results for Aspergillus in the PCR | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
White 2006.
| Study characteristics | |||
| Patient sampling | A group of patients at risk of IA | ||
| Patient characteristics and setting | A group of 203 patients at risk of IFD were tested by RT‐PCR over a 13‐month period (November 2003 to December 2004). The majority (176) were haematology patients, with 133 receiving remission‐induction therapy for acute leukaemia (68 patients) or undergoing SCT (65 patients). The mean age of patients was 48 years. | ||
| Index tests | RT‐PCR | ||
| Target condition and reference standard(s) | IA. The EORTC‐MSG criteria | ||
| Flow and timing | Patients at risk of IFD were tested by RT‐PCR over a 13‐month period (November 2003 to December 2004) | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test All tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | No | ||
| Did all patients receive the same reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
BAL: broncho‐alveolar lavage EDTA: ethylenediaminetetraacetic acid EORTC/MSG: European Organisation for Research and Treatment of Cancer/Mycoses Study Group GM: galactomannan HRCT: high‐resolution computed tomography IA: invasive aspergillosis IFD: invasive fungal disease ITS: internal transcribed spacer PCR: polymerase chain reaction ELISA: enzyme‐linked immunosorbent assay RCLB: red cell lysis buffer RT‐PCR: real time polymerase chain reaction SCT: stem cell transplant
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Adhurti 2011 | no 2x2 data provided |
| Aguado 2015 | no 2x2 data available. a RCT of PCR +GM vs GM only as a screening for directing further diagnostic strategy in pts at risk of IA |
| Armenian 2009 | no 2x2 data provided |
| Auberger 2011 | Retrospective study |
| Badiee 2008 | no 2x2 data provided |
| Badiee 2009 | no 2x2 data provided |
| Badiee 2016a | subset of patients included in another report |
| Badiee 2016b | not invasive aspergillosis, but fungal rhinosinusitis |
| Bernal‐Martinez 2011 | only sensitivity data provided |
| Blennow 2010 | no 2x2 data provided |
| Boch 2015 | retrospective evaluation of a non‐consecutive cohort of patients |
| Bolehovska 2006 | Include several materials and at risk patients (not only haematologic) |
| Bretagne 1998 | retrospective selection of patients at risk of IA from a cohort of haematologic patients |
| Bu Rong 2005 | Case control, not consecutive pts |
| Bucheidt 2001 | case control (control group healthy control) |
| Bucheidt 2004 | no 2x2 data provided |
| Capoor 2017 | case‐control |
| Cesaro 2008 | no 2x2 data provided |
| Challier 2004 | retrospective selection |
| Chryssanthou 1999 | Candida PCR |
| da Silva 2014 | results of PCR and GM not available according to reference standard |
| Danylo 2014 | Index test only in a subset of stored samples |
| Drogari‐Apiranthitou 2016 | PCR on tissue |
| Du 2016 | case control |
| Gupta 2017 | there was no identification to genus level so we could not identify positive aspergillus PCR results |
| Hadrich 2011 | case control |
| Halliday 2005 | Methodological, assay procedure |
| Hasseine 2010 | no 2x2 data provided (published only as abstract) |
| Hebart 2000 | no 2x2 data provided |
| Hummel 2010 | no 2x2 data provided; preliminary selection of patients |
| Idelevich 2015 | not EORTC/MSG criteria as reference standard |
| Johnson 2012 | gold standard different from EORTC; 3 cases only |
| Jones 1998 | BAL only |
| Jordanides 2005 | doesn't distinguish Aspergillus from Candida |
| Kalkank 2010 | no 2x2 data provided (published only as abstract) |
| Kami 2001 | This study has combined patient samples from both a non‐random sampling strategy and from prospective sampling. The authors suggest a case‐control approach. The study does not follow EORTC/MSG criteria for defining IA. |
| Kawazu 2004 | no 2x2 data provided |
| Khalid 2017 | test development, not diagnostic study |
| Klingspor 2006 | only sensitivity data provided |
| Lass‐Florl 2001 | only sensitivity data provided |
| Lehrnbecher 2016 | review |
| Li 2013 | case‐control |
| Liu 2005 | Chinese |
| MacEsic 2017 | cost‐effectivness study |
| Mandhanija 2010 | terms not according EORTC criteria (e.g., suspected cases) |
| Millon 2011 | case control (retrospective selection of patients GM‐posiitive from a cohort of haematologic patients) |
| Morrissey 2013 | no 2x2 data available. a RCT comparing standard diagnostic strategy vs rapid biomarkers diagnostic strategy (PCR +GM) for directing the use of antifungal agents |
| Nakamura 2010 | PCR for bacteria and fungi, one positive case |
| Oz 2016 | duplicate |
| Paholcseck 2015 | retrospective evaluation of a cohort of non‐consecutive patients |
| Paolucci 2013 | not EORTC/MSG criteria as reference standard |
| Reinwald 2014 | retrospective evaluations of patients |
| Scotter 2005 | retrospective, case control |
| Skladny 1999 | retrospective, case control |
| Sonmez 2015 | not 2x2 data available according to reference standard (EORTC/MSG criteria) |
| Springer 2013 | retrospective, case control |
| Sun 2010 | Chinese |
| Tang 2016 | retrospective evaluation in a selected population of patients |
| Teifoori 2011 | No reference standard; no 2x2 tables; not clear if pts were consecutive and when PCR was performed |
| White 2013 | retrospective serum testing for Beta‐Glucan, LFD and PCR |
| Yoo 2005 | NASBA |
| Zhang 2016 | case control |
| Zhao 2016 | case control |
Differences between protocol and review
We intended to use QUADAS, as described in the protocol, but switched to QUADAS‐2 for the review.
Contributions of authors
MC, CM, RAB and PD conceived the original idea for the review and wrote the protocol. MC, CM, RAB, PD, OM, LK, JL, BLJ, and LW reviewed articles for inclusion and extracted data. MC and CM analysed the data and drafted the methodological and statistical analysis part of the draft. All authors reviewed, commented on and approved the final version.
Declarations of interest
RAB has been a consultant to Astellas Pharma, Gilead Sciences, Merck, Sharpe & Dohme, Pfizer and Schering‐Plough, has received a research grant from Pfizer, has been a member of a speakers' bureau for Astellas Pharma, Gilead Sciences, Merck, Sharpe & Dohme, Pfizer and Schering‐Plough, and has received travel grants from Astellas Pharma, Gilead Sciences, Merck, Sharpe & Dohme, Pfizer and Schering‐Plough.
JL has received research grants from Novartis and travel grants from Pfizer and Cephalon.
PD has been a consultant to Gilead Sciences, Ipsat Therapies and Pfizer, has received research grants from AM‐Pharma, Basilea Pharmaceutica and Schering‐Plough, has been a member of a speakers' bureau for Gilead Sciences, Janssen Pharmaceuticals, Pfizer, Schering‐Plough, and Xian‐Janssen and has received travel grants from Merck, Sharpe & Dohme and UCB Pharma.
LK has been an adviser to Astellas, Gilead, Schering‐Plough, has received research grants from Gilead, Merck, Sharpe & Dohme, Schering‐Plough and has received honoraria for educational lectures from Gilead, Pfizer, Merck, Sharpe & Dohme, Schering‐Plough and Janssen.
BLJ has been an advisor to Gilead, MSD, Astellas and Pfizer; has received research grants from Gilead, Astellas, Pfizer, Janssen and MSD; has received honoraria for educational lectures from Gilead, MSD and Pfizer; and owns stock in Gilead, MSD and Pfizer.
JM has served as consultant to Schering‐Plough, Gilead Sciences, Merck, Sharp & Dohme, Pfizer, Bio‐Rad, Fujisawa healthcare, Astellas, Nextar and Zeneus (Cephalon), has received research funding from Bio‐Rad, Merck, Sharp & Dohme, and Pfizer, and has been on the speakers' bureau for Schering‐Plough, Gilead Sciences, Merck, Sharp & Dohme, Pfizer, Bio‐Rad, Fujisawa healthcare, Astellas and Zeneus (Cephalon).
MC, CM, OM: no conflicts of interest to declare with regard to the content of the article.
LW has received research funding from Bruker, Luminex and Renishaw Diagnostics, provided consultancy for Gilead Sciences, has been a member of a speakers' bureau for Gilead Sciences, Merck, Sharpe and Dohme, Bruker and Dynamiker Ltd, and received travels funding from Gilead Sciences, Launch Diagnostics, Bruker and Renishaw Diagnostics.
RAB, JL and PD are founder members of the European Aspergillus PCR Initiative Working Group of the International Society for Human and Animal Mycology, and board members of the EAPCRI, which is registered with the Dutch Chamber of Commerce, number 09165918
None of the authors has any interests, financial or otherwise, in any of the companies involved in the diagnosis of IFD. The authors' disclosures are on public record to ensure their independence and integrity and to help avoid potential conflicts of interest.
New search for studies and content updated (no change to conclusions)
References
References to studies included in this review
Aslan 2015 {published data only}
- Aslan M, Oz Y, Aksit F, Akay OM. Potential of polymerase chain reaction and galactomannan for the diagnosis of invasive aspergillosis in patients with febrile neutropenia. Mycoses 2015;58(6):343‐9. [DOI] [PubMed] [Google Scholar]
Badiee 2010 {published data only}
- Badiee P, Alborzi A. Detection of Aspergillus species in bone marrow transplant patients. Journal of Infection in Developing Countries 2010;4(8):511‐6. [DOI] [PubMed] [Google Scholar]
Badiee 2017 {published data only}
- Badiee P, Zareifar S, Haddadi P, Jafarian H. Incidence of fungal infections in pediatric patients with hematologic neoplasms. Archives of Pediatric Infectious Diseases 2017;5(3):e41317. [Google Scholar]
Barnes 2009 {published data only}
- Barnes RA, White PL, Bygrave C, Evans N, Healy B, Kell J. Clinical impact of enhanced diagnosis of invasive fungal disease in high‐risk haematology and stem cell transplant patients. Journal of Clinical Pathology 2009;62(1):64‐9. [DOI] [PubMed] [Google Scholar]
Barnes 2013 {published data only}
- Barnes RA, Stocking K, Bowden S, Poynton MH, White PL. Prevention and diagnosis of invasive fungal disease in high‐risk patients within an integrative care pathway. Journal of Infection 2013;67(3):206‐14. [DOI] [PubMed] [Google Scholar]
Bellanger 2015 {published data only}
- Bellanger AP, Millon L, Berceanu A, Grenouillet F, Grenouillet FE, Larosa F, et al. Combining Aspergillus mitochondrial and ribosomal QPCR, in addition to galactomannan assay, for early diagnosis of invasive aspergillosis in hematology patients. Medical Mycology 2015;53(7):760‐4. [DOI] [PubMed] [Google Scholar]
Boch 2016 {published data only}
- Boch T, Spiess B, Cornely OA, Vehreschild JJ, Rath PM, Steinmann J, et al. Diagnosis of invasive fungal infections in haematological patients by combined use of galactomannan, 1,3‐beta‐D‐glucan, Aspergillus PCR, multifungal DNA‐microarray, and Aspergillus azole resistance PCRs in blood and bronchoalveolar lavage samples: results of a prospective multicentre study. Clinical Microbiology and Infection 2016;22(10):862‐8. [DOI] [PubMed] [Google Scholar]
Boluk 2016 {published data only}
- Boluk G, Kazak E, Ozkalemkas F, Ener B, Akalin H, Agca H. Comparison of galactomannan, beta‐d‐glucan, and aspergillus DNA in sera of high‐risk adult patients with hematological malignancies for the diagnosis of invasive aspergillosis. Turkish Journal of Medical Sciences 2016;46(2):335‐42. [DOI] [PubMed] [Google Scholar]
Cuenca‐Estrella 2009 {published data only}
- Cuenca‐Estrella M, Meije Y, Diaz‐Pedroche C, Gomez‐Lopez A, Buitrago M‐J, Bernal‐Martinez L, et al. Value of serial quantification of fungal DNA by a real‐time PCR‐based technique for early diagnosis of invasive Aspergillosis in patients with febrile neutropenia. Journal of Clinical Microbiology 2009;47(2):379‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
da Silva 2010 {published data only}
- Silva RL, Ribeiro P, Abreu N, Ferreira T, Fernandes T, Monteiro A, et al. Early diagnosis of invasive aspergillosis in neutropenic patients. Comparison between serum galactomannan and polymerase chain reaction. Clinical Medicine Insights. Oncology 2010;4:81‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
El Mahallawy 2006 {published data only}
- El‐Mahallawy HA, Shaker HH, Ali Helmy H, Mostafa T, Abo‐Sedah AR. Evaluation of pan‐fungal PCR assay and Aspergillus antigen detection in the diagnosis of invasive fungal infections in high risk paediatric cancer patients. Medical Mycology 2006;44:733‐9. [DOI] [PubMed] [Google Scholar]
Ferns 2002 {published data only}
- Ferns B, Fletcher H, Bradley S, Mackinnon S, Hunt C, Tedder RS. The prospective evaluation of a nested polymerase chain reaction assay for the early detection of Aspergillus infection in patients with leukaemia or undergoing allograft treatment. British Journal of Haematology 2002;119(3):720‐5. [DOI] [PubMed] [Google Scholar]
Florent 2006 {published data only}
- Florent M, Katsahian S, Vekhoff A, Levy V, Rio B, Marie JP, et al. Prospective evaluation of a polymerase chain reaction‐ELISA targeted to Aspergillus fumigatus and Aspergillus flavus for the early diagnosis of invasive aspergillosis in patients with hematological malignancies. Journal of Infectious Diseases 2006;193(5):741‐7. [DOI] [PubMed] [Google Scholar]
Halliday 2006 {published data only}
- Halliday C, Hoile R, Sorrell T, James G, Yadav S, Shaw P, et al. Role of prospective screening of blood for invasive aspergillosis by polymerase chain reaction in febrile neutropenic recipients of haematopoietic stem cell transplants and patients with acute leukaemia. British Journal of Haematology 2006;132(4):478‐86. [DOI] [PubMed] [Google Scholar]
- Halliday C, Wu QX, James G, Sorrell T. Development of a nested qualitative real‐time PCR assay to detect Aspergillus species DNA in clinical specimens. Journal of Clinical Microbiology 2005;43(10):5366‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Hebart 2000a {published data only}
- Hebart H, Loffler J, Reitze H, Engel A, Schumacher U, Klingebiel T, et al. Prospective screening by a panfungal polymerase chain reaction assay in patients at risk for fungal infections: implications for the management of febrile neutropenia. British Journal of Haematology 2000;111(2):635‐40. [DOI] [PubMed] [Google Scholar]
Hummel 2009 {published data only}
- Hummel M, Spiess B, Roder J, Komorowski G, Durken M, Kentouche K, et al. Detection of Aspergillus DNA by a nested PCR assay is able to improve the diagnosis of invasive aspergillosis in paediatric patients. Journal of Medical Microbiology 2009;58(Pt 10):1291‐7. [DOI] [PubMed] [Google Scholar]
Imbert 2016 {published data only}
- Imbert S, Gauthier L, Joly I, Brossas JY, Uzunov M, Touafek F, et al. Aspergillus PCR in serum for the diagnosis, follow‐up and prognosis of invasive aspergillosis in neutropenic and nonneutropenic patients. Clinical Microbiology and Infection 2016;22(6):562.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Landlinger 2010 {published data only}
- Landlinger C, Preuner S, Baskova L, Grotel M, Hartwig NG, Dworzak M, et al. Diagnosis of invasive fungal infections by a real‐time panfungal PCR assay in immunocompromised pediatric patients. Leukemia 2010;24(12):2032‐8. [DOI] [PubMed] [Google Scholar]
Loeffler 2017 {published data only}
- Loeffler J, Hafner J, Mengoli C, Wirth C, Heussel CP, Loffler C, et al. Prospective biomarker screening for diagnosis of invasive aspergillosis in high‐risk pediatric patients. Journal of Clinical Microbiology 2017;55(1):101‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pini 2015 {published data only}
- Pini P, Bettua C, Orsi CF, Venturelli C, Faglioni L, Forghieri F, et al. Clinical performance of a commercial real‐time PCR assay for Aspergillus DNA detection in serum samples from high‐risk patients: comparison with a galactomannan enzyme immunoassay. European Journal of Clinical Microbiology & Infectious Diseases 2015;34(1):131‐6. [DOI] [PubMed] [Google Scholar]
Ramírez 2009 {published data only}
- Ramírez M, Castro C, Palomares JC, Torres MJ, Aller AI, Ruiz M, et al. Molecular detection and identification of Aspergillus spp. from clinical samples using real‐time PCR. Mycoses 2009;52(2):129‐34. [DOI] [PubMed] [Google Scholar]
Rogers 2013 {published data only}
- Rogers TR, Morton CO, Springer J, Conneally E, Heinz W, Kenny C, et al. Combined real‐time PCR and galactomannan surveillance improves diagnosis of invasive aspergillosis in high risk patients with haematological malignancies. British Journal of Haematology 2013;161(4):517‐24. [DOI] [PubMed] [Google Scholar]
Schwarzinger 2013 {published data only}
- Schwarzinger M, Sagaon‐Teyssier L, Cabaret O, Bretagne S, Cordonnier C, PREVERT Investigators. Performance of serum biomarkers for the early detection of invasive aspergillosis in febrile, neutropenic patients: a multi‐state model. Plos ONE 2013;8(6):e65776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Springer 2011 {published data only}
- Springer J, Loeffler J, Heinz W, Schlossnagel H, Lehmann M, Morton O, et al. Pathogen‐specific DNA enrichment does not increase sensitivity of PCR for diagnosis of invasive aspergillosis in neutropenic patients. Journal of Clinical Microbiology 2011;49(4):1267‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Springer 2016 {published data only}
- Springer J, Lackner M, Nachbaur D, Girschikofsky M, Risslegger B, Mutschlechner W, et al. Prospective multicentre PCR‐based Aspergillus DNA screening in high‐risk patients with and without primary antifungal mould prophylaxis. Clinical microbiology and infection 2016;22:80‐86. [DOI] [PubMed] [Google Scholar]
Suarez 2008 {published data only}
- Suarez F, Lortholary O, Buland S, Rubio MT, Ghez D, Mahe V, et al. Detection of circulating Aspergillus fumigatus DNA by real‐time PCR assay of large serum volumes improves early diagnosis of invasive aspergillosis in high‐risk adult patients under hematologic surveillance. Journal of Clinical Microbiology 2008;46(11):3772‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sugawara 2013 {published data only}
- Sugawara Y, Nakase K, Nakamura A, Ohishi K, Sugimoto Y, Fujieda A, et al. Clinical utility of a panfungal polymerase chain reaction assay for invasive fungal diseases in patients with haematologic disorders. European Journal of Haematology 2013;90(4):331‐9. [DOI] [PubMed] [Google Scholar]
von Lilienfeld‐Toal 2009 {published data only}
- Lilienfeld‐Toal M, Lehmann LE, Raadts AD, Hahn‐Ast C, Orlopp KS, Marklein G, et al. Utility of a commercially available multiplex real‐time PCR assay to detect bacterial and fungal pathogens in febrile neutropenia. Journal of Clinical Microbiology 2009;47(8):2405‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
White 2006 {published data only}
- White PL, Linton CJ, Perry MD, Johnson EM, Barnes RA. The evolution and evaluation of a PCR assay for the detection of invasive aspergillosis in haematology patients. Clinical Infectious Diseases 2006;42(4):479‐86. [DOI] [PubMed] [Google Scholar]
References to studies excluded from this review
Adhurti 2011 {published data only}
- Adurthi S, Sahoo TP, Shafiulla M, Radhika B, Jacob LA, Appaji L, et al. Invasive fungal infections in haematological malignancies at a regional cancer institute‐role of panfungal PCR. European Journal of Cancer Supplements 2011;9(1):18. [Google Scholar]
Aguado 2015 {published data only}
- Aguado JM, Vazquez L, Fernandez‐Ruiz M. Serum galactomannan versus a combination of galactomannan and polymerase chain reaction‐based aspergillus DNA detection for early therapy of invasive aspergillosis in high‐risk hematological patients: A randomized controlled trial. Clinical Infectious Diseases 2015;60(3):405‐14. [DOI] [PubMed] [Google Scholar]
Armenian 2009 {published data only}
- Armenian SH, Nash KA, Kapoor N, Franklin JL, Gaynon PS, Ross LA, et al. Prospective monitoring for invasive aspergillosis using galactomannan and polymerase chain reaction in high risk pediatric patients. Journal of Pediatric Hematology/oncology 2009;31(12):920‐6. [DOI] [PubMed] [Google Scholar]
Auberger 2011 {published data only}
- Auberger J, Clausen J, Mrazek C, Lass‐Florl C, Nachbaur D. Breakthrough fungal infections (bIFI), fungal colonization and emergence of resistant strains during posaconazole (PCZ) prophylaxis in patients at risk. Real‐life data from a single center institutional retrospective observational study. Onkologie 2011;34:232. [DOI] [PubMed] [Google Scholar]
Badiee 2008 {published data only}
- Badiee P, Kordbacheh P, Alborzi A, Ramzi M, Shakiba E. Molecular detection of invasive aspergillosis in hematologic malignancies. Infection 2008;36(6):580‐4. [DOI] [PubMed] [Google Scholar]
Badiee 2009 {published data only}
- Badiee P, Kordbacheh P, Alborzi A, Malekhoseini S, Ramzi M, Mirhendi H, et al. Study on invasive fungal infections in immunocompromised patients to present a suitable early diagnostic procedure. International Journal of Infectious Diseases 2009;13(1):97‐102. [DOI] [PubMed] [Google Scholar]
Badiee 2016a {published data only}
- Badiee P, Hashemizadeh Z, Ramzi M, Karimi M, Mohammadi R. Non‐invasive methods to diagnose fungal infections in pediatric patients with hematologic disorders. Jundishapur Journal of Microbiology 2016;9(11):e41573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Badiee 2016b {published data only}
- Badiee P, Moghadami M, Rozbehani H. Comparing immunological and molecular tests with conventional methods in diagnosis of acute invasive fungal rhinosinusitis. Journal of Infection in Developing Countries 2016;10(1):90‐5. [DOI] [PubMed] [Google Scholar]
Bernal‐Martinez 2011 {published data only}
- Bernal‐Martinez L, Gago S, Buitrago MJ, Gomez‐Lopez A, Rodriguez‐Tudela JL, Cuenca‐Estrella M. Analysis of performance of a PCR‐based assay to detect DNA of Aspergillus fumigatus in whole blood and serum: a comparative study with clinical samples. Journal of Clinical Microbiology 2011;49(10):3596‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Blennow 2010 {published data only}
- Blennow O, Remberger M, Klingspor L, Omazic B, Fransson K, Ljungman P, et al. Randomized PCR‐based therapy and risk factors for invasive fungal infection following reduced‐intensity conditioning and hematopoietic SCT. Bone Marrow Transplantation 2010;45(12):1710‐8. [DOI] [PubMed] [Google Scholar]
Boch 2015 {published data only}
- Boch T, Reinwald M, Postina P, Cornely OA, Vehreschild JJ, Heusel CP, et al. Identification of invasive fungal diseases in immunocompromised patients by combining an Aspergillus specific PCR with a multifungal DNA‐microarray from primary clinical samples. Mycoses 2015;58(12):735‐45. [DOI] [PubMed] [Google Scholar]
Bolehovska 2006 {published data only}
- Bolehovska R, Pliskova L, Buchta V, Cerman J, Hamal P. Detection of Aspergillus spp. in biological samples by real‐time PCR. Biomedical Papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia 2006;150(2):245‐8. [DOI] [PubMed] [Google Scholar]
Bretagne 1998 {published data only}
- Bretagne S, Costa JM, Bart‐Delabesse E, Dhedin N, Rieux C, Cordonnier C. Comparison of serum galactomannan antigen detection and competitive polymerase chain reaction for diagnosing invasive aspergillosis. Clinical Infectious Diseases 1998;26(6):1407‐12. [DOI] [PubMed] [Google Scholar]
Bucheidt 2001 {published data only}
- Buchheidt D, Baust C, Skladny H, Ritter J, Suedhoff T, Baldus M, et al. Detection of Aspergillus species in blood and bronchoalveolar lavage samples from immunocompromised patients by means of 2‐step polymerase chain reaction: clinical results. Clinical Infectious Diseases 2001;33(4):428‐35. [DOI] [PubMed] [Google Scholar]
Bucheidt 2004 {published data only}
Bu Rong 2005 {published data only}
- Bu R, Sathiapalan RK, Ibrahim MM, Al‐Mohsen I, Almodavar E, Gutierrez MI, et al. Monochrome LightCycler PCR assay for detection and quantification of five common species of Candida and Aspergillus. Journal of Medical Microbiology 2005;54(Pt 3):243‐8. [DOI] [PubMed] [Google Scholar]
Capoor 2017 {published data only}
- Capoor MR, Puri S, Raheja H, Mohindra R, Gupta DK, Verma PK, et al. Screening of invasive fungal infections by a real‐time panfungal (pan‐ACF) polymerase chain reaction assay in patients with haematological malignancy. Indian Journal of Medical Microbiology 2017;35(1):41‐7. [DOI] [PubMed] [Google Scholar]
Cesaro 2008 {published data only}
- Cesaro S, Stenghele C, Calore E, Franchin E, Cerbaro I, Cusinato R, et al. Assessment of the LightCycler PCR assay for diagnosis of invasive aspergillosis in paediatric patients with onco‐haematological diseases. Mycoses 2008;51(6):497‐504. [DOI] [PubMed] [Google Scholar]
Challier 2004 {published data only}
- Challier S, Boyer S, Abachin E, Berche P. Development of a serum‐based Taqman real‐time PCR assay for diagnosis of invasive aspergillosis. Journal of Clinical Microbiology 2004;42(2):844‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Chryssanthou 1999 {published data only}
- Chryssanthou E, Klingspor L, Tollemar J, Petrini B, Larsson L, Christensson B, et al. PCR and other non‐culture methods for diagnosis of invasive Candida infections in allogeneic bone marrow and solid organ transplant recipients. Mycoses 1999;42(4):239‐47. [DOI] [PubMed] [Google Scholar]
Danylo 2014 {published data only}
- Danylo A, Courtemanche C, Pelletier R, Boudreault AA. Performance of MycAssay Aspergillus DNA real‐time PCR assay compared with the galactomannan detection assay for the diagnosis of invasive aspergillosis from serum samples. Medical Mycology 2014;52(6):577‐83. [DOI] [PubMed] [Google Scholar]
da Silva 2014 {published data only}
- Silva TV, Carneiro LC, Ramos Fdos S. PCR as a screening test for invasive aspergillosis in haematological patients: a pilot study. Mycopathologia 2014;177(1‐2):111‐4. [DOI] [PubMed] [Google Scholar]
Drogari‐Apiranthitou 2016 {published data only}
- Drogari‐Apiranthitou M, Panayiotides I, Galani I, Konstantoudakis S, Arvanitidis G, Spathis, A, et al. Diagnostic value of a semi‐nested PCR for the diagnosis of mucormycosis and aspergillosis from paraffin‐embedded tissue: a single center experience. Pathology, Research and Practice 2016;212(5):393‐7. [DOI] [PubMed] [Google Scholar]
Du 2016 {published data only}
- Du L, Xia Y, He Y, Pu Q, Hua R, Wu W. Development and evaluation of enzyme‐linked immunosorbent assay of nucleic acid sequence‐based amplification for diagnosis of invasive aspergillosis. AMB Express 2016;6(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Gupta 2017 {published data only}
- Gupta P, Ahmad A, Khare V, Kumar A, Banerjee G, Verma N, et al. Comparative evaluation of pan‐fungal real‐time PCR, galactomannan and (1‐3)‐beta‐D‐glucan assay for invasive fungal infection in paediatric cancer patients. Mycoses 2017;60(4):234‐40. [DOI] [PubMed] [Google Scholar]
Hadrich 2011 {published data only}
- Hadrich I, Mary C, Makni F, Elloumi M, Dumon H, Ayadi A, et al. Comparison of PCR‐ELISA and Real‐Time PCR for invasive aspergillosis diagnosis in patients with hematological malignancies.. Medical Mycology 2011;49:489‐94. [DOI] [PubMed] [Google Scholar]
Halliday 2005 {published data only}
- Halliday C, Wu QX, Gregory J, Sorrell T. Development of a nested qualitative real‐time PCR assay to detect Aspergillus species DNA in clinical specimens. Journal of Clinical Microbiology 2005;43(10):5366‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Hasseine 2010 {published data only}
- Hasseine L, Marty P, Cassaing S, Robert‐Gangneux F, Gaudron L, Knecht R, et al. Multicentre prospective monitoring for empirical treatment of invasive fungal infection using high negative predictive value diagnostic strategies. Clinical Microbiology and Infection 2010;16:S207‐8. [Google Scholar]
Hebart 2000 {published data only}
- Hebart H, Löffler J, Meisner C, Serey F, Schmidt D, Böhme A, et al. Early detection of aspergillus infection after allogeneic stem cell transplantation by polymerase chain reaction screening. Journal of Infectious Diseases 2000;181(5):1713‐9. [DOI] [PubMed] [Google Scholar]
Hummel 2010 {published data only}
- Hummel M, Spiess B, Cornely OA, Dittmer M, Mörz H, Buchheidt D. Aspergillus PCR testing: results from a prospective PCR study within the AmBiLoad trial. European Journal of Haematology 2010;85(2):164‐9. [DOI] [PubMed] [Google Scholar]
Idelevich 2015 {published data only}
- Idelevich EA, Silling G, Niederbracht Y, Penner H, Sauerland MC, Tafelski S, et al. Impact of multiplex PCR on antimicrobial treatment in febrile neutropenia: a randomized controlled study. Medical Microbiology and Immunology 2015;204(5):585‐92. [DOI] [PubMed] [Google Scholar]
Johnson 2012 {published data only}
- Johnson GL, Bibby DF, Wong S, Agrawal SG, Bustin SA. A MIQE‐compliant real‐time PCR assay for Aspergillus detection. PLoS One 2012;7(7):e40022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jones 1998 {published data only}
- Jones ME, Fox AJ, Barnes AJ, Oppenheim BA, Balagopal P, Morgenstern GR, et al. PCR‐ELISA for the early diagnosis of invasive pulmonary aspergillus infection in neutropenic patients. Journal of Clinical Pathology 1998;51(9):652‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jordanides 2005 {published data only}
- Jordanides NE, Allan EK, McLintock LA, Copland M, Devaney M, Stewart K, et al. A prospective study of real‐time panfungal PCR for the early diagnosis of invasive fungal infection in haemato‐oncology patients. Bone Marrow Transplant 2005;35(4):389‐95. [DOI] [PubMed] [Google Scholar]
Kalkank 2010 {published data only}
- Kalkanc A, Aki SZ, Yegin ZA, Suyan E, Tuncca O, Koktur N, et al. Molecular approaches to fungal infections in high‐risk haematology patients. Clinical Microbiology and Infection 2010;16:S229. [Google Scholar]
Kami 2001 {published data only}
- Kami M, Fukui T, Ogawa S, Kazuyama Y, Machida U, Tanaka Y, et al. Use of real‐time PCR on blood samples for diagnosis of invasive aspergillosis. Clinical Infectious Diseases 2001;33(9):1504‐12. [DOI] [PubMed] [Google Scholar]
Kawazu 2004 {published data only}
- Kawazu M, Kanda Y, Nannya Y, Aoki K, Kurokawa M, Chiba S, et al. Prospective comparison of the diagnostic potential of real‐time PCR, double‐sandwich enzyme‐linked immunosorbent assay for galactomannan, and a (1‐‐>3)‐beta‐D‐glucan test in weekly screening for invasive aspergillosis in patients with hematological disorders. Journal of Clinical Microbiology 2004;42(6):2733‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Khalid 2017 {published data only}
- Khalid A, Chua AL, Tzar MN, Santhanam J. development of prototype multiplexed lateral flow assay (LFA) for rapid detection of invasive fungal infections. Medical journal of Malaysia. 2017; Vol. 72 (supplement 2):24.
Klingspor 2006 {published data only}
- Klingspor L, Jalal S. Molecular detection and identification of Candida and Aspergillus spp. from clinical samples using real‐time PCR. Clinical Microbiology and Infection 2006;12(8):745‐53. [DOI] [PubMed] [Google Scholar]
Lass‐Florl 2001 {published data only}
- Lass‐Florl C, Aigner J, Gunsilius E, Petzer A, Nachbaur D, Gastl G, et al. Screening for Aspergillus spp. using polymerase chain reaction of whole blood samples from patients with haematological malignancies. British Journal of Haematology 2001;113(1):180‐4. [DOI] [PubMed] [Google Scholar]
Lehrnbecher 2016 {published data only}
- Lehrnbecher T, Robinson PD, Fisher BT, Castagnola E, Groll AH, Steinbach WJ, et al. Galactomannan, beta‐D‐glucan, and polymerase chain reaction‐based assays for the diagnosis of invasive fungal disease in pediatric cancer and hematopoietic stem cell transplantation: a systematic review and meta‐analysis. Clinical Infectious Diseases 2016;63(10):1340‐8. [DOI] [PubMed] [Google Scholar]
Li 2013 {published data only}
- Li Y, Gao L, Ding Y, Xu Y, Zhou M, Huang W, et al. Establishment and application of real‐time quantitative PCR for diagnosing invasive Aspergillosis via the blood in hematological patients: targeting a specific sequence of Aspergillus 28S‐ITS2. BMC Infectious Diseases 2013;13:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Liu 2005 {published data only}
- Liu JQ, Feng LM, Song H, Meng HT, Jin J, Qian WB. [Detection of Aspergillus infection in blood of malignant hematopoietic tumor patients by two‐step polymerase chain reaction]. Zhonghua nei ke za zhi [Chinese Journal of Internal Medicine] 2005;44:726‐9. [PubMed] [Google Scholar]
MacEsic 2017 {published data only}
- MacEsic N, Morrissey CO, Liew D, Bohensky MA, Chen SC‐A, Gilroy NM, et al. Is a biomarker‐based diagnostic strategy for invasive aspergillosis cost effective in high‐risk haematology patients?. Medical Mycology 2017;55(7):705‐12. [DOI] [PubMed] [Google Scholar]
Mandhanija 2010 {published data only}
- Mandhaniya S, Iqbal S, Sharawat SK, Xess I, Bakhshi S. Diagnosis of invasive fungal infections by RQ‐PCR assay in pediatric acute leukemia induction. Journal of Clinical Oncology 2010;28(15_suppl):9579. [DOI: 10.1200/jco.2010.28.15_suppl.9579] [DOI] [PubMed] [Google Scholar]
Millon 2011 {published data only}
- Millon L, Grenouillet F, Legrand F, Loewert S, Bellanger AP, Gbaguidi‐Haore H, et al. Ribosomal and mitochondrial DNA target for real‐time PCR diagnosis of invasive aspergillosis. Journal of Clinical Microbiology 2011;49(3):1058‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Morrissey 2013 {published data only}
- Morrissey CO, Chen SC, Sorrell TC, Milliken S, Bardy PG, Bradstock KF, et al. Australasian Leukaemia Lymphoma Group and the Australia and New Zealand Mycology Interest Group. Galactomannan and PCR versus culture and histology for directing use of antifungal treatment for invasive aspergillosis in high‐risk haematology patients: a randomised controlled trial. Lancet Infectious Diseases 2013;13(6):519‐28. [DOI] [PubMed] [Google Scholar]
Nakamura 2010 {published data only}
- Nakamura A, Sugimoto Y, Ohishi K, Sugawara Y, Fujieda A, Monma F, et al. Diagnostic value of PCR analysis of bacteria and fungi from blood in empiric‐therapy‐resistant febrile neutropenia. Journal of Clinical Microbiology 2010;48(6):2030‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Oz 2016 {published data only}
- Oz Y, Aslan M, Aksit F, Metintas S, Gunduz E. The effect of clinical characteristics on the performance of galactomannan and PCR for the diagnosis of invasive aspergillosis in febrile neutropenic patients. Mycoses 2016;59(2):86‐92. [DOI] [PubMed] [Google Scholar]
Paholcseck 2015 {published data only}
- Paholcsek M, Fidler G, Konya J, Rejto L, Mehes G, Bukta E, et al. Combining standard clinical methods with PCR showed improved diagnosis of invasive pulmonary aspergillosis in patients with hematological malignancies and prolonged neutropenia. BMC Infectious Diseases 2015;15:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Paolucci 2013 {published data only}
- Paolucci M, Stanzani M, Melchionda F, Tolomelli G, Castellani G, Landini MP, et al. Routine use of a real‐time polymerase chain reaction method for detection of bloodstream infections in neutropaenic patients. Diagnostic Microbiology and Infectious Disease 2013;75(2):130‐4. [DOI] [PubMed] [Google Scholar]
Reinwald 2014 {published data only}
- Reinwald M, Konietzka CA, Kolve H. Assessment of Aspergillus‐specific PCR as a screening method for invasive aspergillosis in paediatric cancer patients and allogeneic haematopoietic stem cell recipients with suspected infections. Mycoses 2014;57(9):537‐43. [DOI] [PubMed] [Google Scholar]
Scotter 2005 {published data only}
- Scotter JM, Campbell P, Anderson TP, Murdoch DR, Chambers ST, Patton NW. Comparison of PCR‐ELISA and galactomannan detection for the diagnosis of invasive aspergillosis. Pathology 2005;37(3):246‐53. [DOI] [PubMed] [Google Scholar]
Skladny 1999 {published data only}
- Skladny H, Buchheidt D, Baust C, Krieg‐Schneider F, Seifarth W, Leib‐Mosch C, et al. Specific detection of Aspergillus species in blood and bronchoalveolar lavage samples of immunocompromised patients by two‐step PCR. Journal of Clinical Microbiology 1999;37(12):3865‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sonmez 2015 {published data only}
- Sonmez A, Eksi F, Pehlivan M, Haydaroglu Sahin H. Investigating the presence of fungal agents in febrile neutropenic patients using different microbiological, serological, and molecular methods. Udruzenje Basicnih Mediciniskih Znanosti [Bosnian Journal of Basic Medical Sciences] 2015;15(3):40‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Springer 2013 {published data only}
- Springer J, Morton CO, Perry M, Heinz WJ, Paholcsek M, Alzheimer M, et al. Multicenter comparison of serum and whole‐blood specimens for detection of Aspergillus DNA in high‐risk hematological patients. Journal of Clinical Microbiology 2013;51(5):1445‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sun 2010 {published data only}
- Sun Y, Ji Y, Xu L, Liu D, Liu K, Huang X. [Combination of real‐time polymerase chain reaction assay and serum galactomannan in the diagnosis of invasive aspergillosis in patients with hematological malignancies and recipients of hematopoietic stem cell transplantation]. Zhonghua yi xue za zhi 2010;90:375‐8. [PubMed] [Google Scholar]
Tang 2016 {published data only}
- Tang Q, Tian S, Yu N, Zhang X, Jia X, Zhai H, et al. Development and Evaluation of a Loop‐Mediated Isothermal Amplification Method for Rapid Detection of Aspergillus fumigatusJ. Journal of Clinical Microbiology 2016;54(4):950‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Teifoori 2011 {published data only}
- Teifoori F, Rodbar MS, Sharifi Z, Ghaffari H. Detection of Aspergilus DNA, by a Nested PCR assay and molecula study of Aspergillus fumigatus, by PCR amplification of IGS Region in BMT patients. Mycoses 2011;54:87. [Google Scholar]
White 2013 {published data only}
- White PL, Parr C, Thornton C, Barnes RA. Evaluation of real‐time PCR, galactomannan enzyme‐linked immunosorbent assay (ELISA), and a novel lateral‐flow device for diagnosis of invasive aspergillosis. Journal of Clinical Microbiology 2013;51(5):1510‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Yoo 2005 {published data only}
- Yoo J, Choi J, Choi S, Lee D, Shin W, Min W, et al. Application of nucleic acid sequence‐based amplification for diagnosis of and monitoring the clinical course of invasive aspergillosis in patients with hematologic diseases. Clinical Infectious Diseases 2005;40(3):392‐8. [DOI] [PubMed] [Google Scholar]
Zhang 2016 {published data only}
- Zhang S, Wang S, Wan Z, Que C, Li R, Yu J. Quantitative real‐time PCR and platelia galactomannan assay for the diagnosis of invasive pulmonary aspergillosis: bronchoalveolar lavage fluid performs better than serum in non‐neutropaenic patients. Mycopathologia 2016;181(9‐10):625‐9. [DOI] [PubMed] [Google Scholar]
Zhao 2016 {published data only}
- Zhao Y, Paderu P, Railkar R, Douglas C, Iannone R, Shire N, et al. Blood Aspergillus RNA is a promising alternative biomarker for invasive aspergillosis. Medical Mycology 2016;54(8):801‐7. [DOI] [PubMed] [Google Scholar]
Additional references
Arvanitis 2014
- Arvanitis M, Ziakas PD, Zacharioudakis IM, Zervou FN, Caliendo AM, Mylonakis E. PCR in diagnosis of invasive aspergillosis: a meta‐analysis of diagnostic performance. Journal of Clinical Microbiology 2014;52(10):3731‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ascioglou 2002
- Ascioglu S, Rex JH, Pauw B, Bennett JE, Bille J, Crokaert F, et al. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clinical Infectious Diseases 2002;34(1):7‐14. [DOI] [PubMed] [Google Scholar]
Barnes 2018
- Barnes RA, White PL, Morton CO, Rogers TR, Cruciani M, Loeffler J, et al. Diagnosis of aspergillosis by PCR: Clinical considerations and technical tips. Medical Mycology 2018;56(suppl.1):60‐72. [DOI] [PubMed] [Google Scholar]
Baskova 2007
- Baskova LC, Landlinger S, Preuner T, Lion T. The Pan‐AC assay: a single‐reaction real‐time PCR test for quantitative detection of a broad range of Aspergillus and Candida species. Journal of Medical Microbiology 2007;56(Pt 9):1167‐73. [DOI] [PubMed] [Google Scholar]
Bossuyt 2003
- Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. AJR: American Journal of Roentgenology 2003;181(1):51‐5. [DOI] [PubMed] [Google Scholar]
Botterel 2008
- Botterel F, Farrugia C, Ichai P, Costa JM, Saliba F, Bretagne S. Real‐time PCR on the first galactomannan‐positive serum sample for diagnosing invasive aspergillosis in liver transplant recipients. Transplant Infectious Disease 2008;10(5):333‐8. [DOI] [PubMed] [Google Scholar]
Bustin 2009
- Bustin SA, Benes V, Garson JA. The MIQE guidelines: minimum information for publication of quantitative real‐time PCR experiments. Clinical Chemistry 2009;55(4):611‐22. [DOI] [PubMed] [Google Scholar]
Caillot 2001
- Caillot D, Couaillier JF, Bernard A, Casasnovas O, Denning DW, Mannone L, et al. Increasing volume and changing characteristics of invasive pulmonary aspergillosis on sequential thoracic computed tomography scans in patients with neutropenia. Journal of Clinical Oncology 2001;19(1):253–9. [DOI] [PubMed] [Google Scholar]
Chamilos 2006
- Chamilos G, Kontoyannis DP. Defining the diagnosis of invasive aspergillosis. Medical Mycology 2006;44(Supplement_1):S163‐73. [DOI] [PubMed] [Google Scholar]
Chu 2006
- Chu H, Cole SR. Bivariate meta‐analysis of sensitivity and specificity with sparse data: a generalized linear mixed model approach. Journal of Clinical Epidemiology 2006;59(12):1331‐2. [DOI] [PubMed] [Google Scholar]
Costa 2001
- Costa C, Vidaud D, Olivi M, Bart‐Delabesse E, VidaudM, Bretagne S. Development of two real‐time quantitative TaqMan PCR assays to detect circulating Aspergillus fumigatus DNA in serum. Journal of Microbiological Methods 2001;44(3):263‐9. [DOI] [PubMed] [Google Scholar]
Covidence 2014 [Computer program]
- Veritas Health Innovation. Covidence. Version Accessed prior to 24 July 2019. Melbourne, Australia: Veritas Health Innovation, 2014.
De Pauw 2008
- Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clinical Infectious Diseases 2008;46(12):1813‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Donnelly 2006
- Donnelly JP. Polymerase chain reaction for diagnosing invasive aspergillosis: getting closer but still a ways to go. Clinical Infectious Diseases 2006;42(4):487‐9. [DOI] [PubMed] [Google Scholar]
Einsele 1997
- Einsele H, Hebart H, Roller G. PCR for detection and differentiation of various fungal pathogens in blood samples. Journal of Clinical Microbiology 1997;35:1353‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Flückiger 2006
- Flückiger U, Marchetti O, Bille J, Eggimann P, Zimmerli S, Imhof A, et al. Treatment options of invasive fungal infections in adults. Swiss Medical Weekly 2006;136(29‐30):447‐63. [DOI] [PubMed] [Google Scholar]
Greene 2007
- Greene RE, Schlamm HT, Oestmann JW, Stark P, Durand C, Lortholary O, et al. Imaging findings in acute invasive pulmonary aspergillosis: clinical significance of the halo sign. Clinical Infectious Diseases 2007;44(3):373‐9. [DOI] [PubMed] [Google Scholar]
Guo 2015
- Guo J, Riebler A. meta 4diag : Bayesian bivariate meta‐analysis of diagnostic test studies for routine practice. Journal of Statistical Software 2015;83(1):1‐31. [Google Scholar]
Harbord 2007
- Harbord RM, Deeks JJ, Egger M, Whiting P, Sterne JAC. A unification of models for meta‐analysis of diagnostic accuracy studies. Biostatistics 2007;8(2):239‐51. [DOI] [PubMed] [Google Scholar]
Hope 2005
- Hope WW, Walsh TJ, Denning DW. Laboratory diagnosis of invasive aspergillosis. Lancet Infectious Diseases 2005;5(10):609‐22. [DOI] [PubMed] [Google Scholar]
Leeflang 2008
- Leeflang MM, Debets‐Ossenkopp YJ, Visser CE, Scholten RJ, Hooft L, Bijlmer HA, et al. Galactomannan detection for invasive aspergillosis in immunocompromized patients. Cochrane Database of Systematic Reviews 2008, Issue 4. [DOI: 10.1002/14651858.CD007394] [DOI] [PubMed] [Google Scholar]
Lehmann 2008
- Lehmann LE, Hunfeld KP, Emrich T, Haberhausen G, Wissing H, Hoeft A, et al. A multiplex real‐time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Medical Microbiology and Immunology 2008;197(3):313‐324. [DOI] [PubMed] [Google Scholar]
Lewis 2006
- Lewis PL, Barnes RA. Aspergillus PCR ‐ Platforms, strengths and weaknesses. Medical Mycology 2006;44((Supplement_1)):S191‐8. [DOI] [PubMed] [Google Scholar]
Lijmer 1999
- Lijmer JG, Mol BW, Heisterkamp S, Bonsel GJ, Prins MH, Meulen JH, et al. Empirical evidence of design‐related bias in studies of diagnostic tests. JAMA 1999;282(11):1061‐6. [DOI] [PubMed] [Google Scholar]
Loeffler 2000
- Loeffler J, Henke N, Hebart H, Schmidt D, Hagmeyer L, Schumacher U, et al. Quantification of fungal DNA by using fIuorescence resonance energy transfer and the light cycler system. Journal of Clinical Microbiology 2000;38(2):586‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Loeffler 2002
- Loeffler J, Schmidt K, Hebart H, Schumacher U, Einsele H. Automated extraction of genomic DNA from medically important yeast species and filamentous fungi by using the MagNA Pure LC system. Journal of Clinical Microbiology 2002;40(6):2240‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Marr 2002
- Marr KA, Carter RA, Boeckh M, Martin P, Corey L. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood 2002;100(13):4358‐66. [DOI] [PubMed] [Google Scholar]
Meersseman 2004
- Meersseman W, Vandecasteele SJ, Wilmer A, Verbeken E, Peetermans WE, Wijngaerden E. Invasive aspergillosis in critically ill patients without malignancy. American Journal of Respiratory and Critical Care Medicine 2004;170(6):621‐5. [DOI] [PubMed] [Google Scholar]
Mengoli 2009
- Mengoli C, Cruciani M, Barnes RA, Loeffler J, Donnelly JP. Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta‐analysis. Lancet Infectious Diseases 2009;9(2):89‐96. [DOI] [PubMed] [Google Scholar]
Pfeiffer 2006
- Pfeiffer CD, Fine JP, Safdar N. Diagnosis of invasive aspergillosis using a galactomannan assay: a meta‐analysis. Clinical Infectious Diseases 2006;42(10):1417‐27. [DOI] [PubMed] [Google Scholar]
Rath 2018
- Rath PM, Steinmann J. Overview of commercially available PCR assays for the detection of Aspergillus spp. DNA in patient samples. Frontiers in Microbioogy 2018;9:740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Reitsma 2005
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. Journal of Clinical Epidemiology 2005;58(10):982‐90. [DOI] [PubMed] [Google Scholar]
Reitsma 2009
- Reitsma JB, Rutjes AWS, Whiting P, Vlassov VV, Leeflang MMG, Deeks JJ. Chapter 9: Assessing methodological quality. In: Deeks JJ, Bossuyt PM, Gatsonis C (editors), Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0.0. The Cochrane Collaboration, 2009. Available from srdta.cochrane.org.
Review Manager 2014 [Computer program]
- Nordic Cochrane Centre, The Cochrane Collaboration. Review Manager 5 (RevMan 5). Version 5.3. Copenhagen: Nordic Cochrane Centre, The Cochrane Collaboration, 2012.
Ribeiro 2006
- Ribeiro P, Costa F, Monteiro A, Caldas J, Silva M, Ferreira G, et al. Polymerase chain reaction screening for fungemia and/or invasive fungal infectionsin patients with hematologic malignancies. Supportive Care in Cancer 2006;14(5):469‐74. [DOI] [PubMed] [Google Scholar]
Rutter 2001
- Rutter CM, Gatsonis CA. A hierarchical regression approach to meta‐analysis of diagnostic test accuracy evaluations. Statistics in Medicine 2001;20(19):2865‐84. [DOI] [PubMed] [Google Scholar]
Sachse 2009
- Sachse S, Straube E, Lehmann M, Bauer M, Russwurm S, Schmidt KH. Truncated human cytidylate‐phosphate‐deoxyguanylate‐binding protein for improved nucleic acid amplification technique‐based detection of bacterial species in human samples. Journal of Clinical Microbiology 2009;47(4):1050‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Senn 2008
- Senn L, Robinson JO, Schmidt S, Knaup M, Asahi N, Satomura S, et al. 1,3‐Beta‐D‐glucan antigenemia for early diagnosis of invasive fungal infections in neutropenic patients with acute leukemia. Clinical Infectious Diseases 2008;46(6):878‐85. [DOI] [PubMed] [Google Scholar]
Shin 1999
- Shin JH, Nolte FS, Holloway BP, Morrison CJ. Rapid identification of up to three Candida species in a single reaction tube by a 5' exonuclease assay using fluorescent DNA probes. Journal of Clinical Microbiology 1999;37(1):165‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Springer 2012
- Springer J, Schloßnagel H, Heinz W, et al. A novel extraction method combining plasma with a whole‐blood fraction shows excellent sensitivity and reproducibility for patients at high risk for invasive aspergillosis. Journal of Clinical Microbiology 2012;50(8):2585‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Tang 1993
- Tang CM, Holden DW, Aufauvre‐Brown A, Cohen J. The detection of Aspergillus spp. by the polymerase chain reaction and its evaluation in bronchoalveolar lavage fluid. American Review of Respiratory Disease 1993;148(5):1313‐7. [DOI] [PubMed] [Google Scholar]
Tuon 2007
- Tuon FF. A systematic literature review on the diagnosis of invasive aspergillosis using polymerase chain reaction (PCR) from bronchoalveolar lavage clinical samples. Revista Iberoamericana de Micologia 2007;24(2):89‐94. [PubMed] [Google Scholar]
Van Burik 1998
- Burik JA, Myerson D, Schrekhise RW, Bowden RA. Panfungal PCR assay for detection of fungal infection in human blood specimens. Journal of Clinical Microbiology 1998;36(5):1169‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Walsh 2008
- Walsh TJ, Anaissie EJ, Denning DW, Herbrecht R, Kontoyiannis DP, Marr KA, et al. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clinical Infectious Diseases 2008;46(3):327‐60. [DOI] [PubMed] [Google Scholar]
Watzinger 2004
- Watzinger F, Suda M, Preuner S, Baumgartinger R, Ebner K, Baskova L, et al. Real‐time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. Journal of Clinical Microbiology 2004;42(11):5189‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
White 2010
- White PL, Bretagne S, Klingspor L, Melchers WJ, McCulloch E, Schulz B, et al. Aspergillus PCR: one step closer to standardization. Journal of Clinical Microbiology 2010;48(4):1231‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
White 2015
- White PL, Wingard JR, Bretagne S, Löffler J, Patterson TF, Slavin MA, et al. Aspergillus polymerase chain reaction: systematic review of evidence for clinical use in comparison with antigen testing. Clinical Infectious Diseases 2015;61(8):1293‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Whiting 2004
- Whiting P, Rutjes AW, Dinnes J, Reitsma J, Bossuyt PM, Kleijnen J. Development and validation of methods for assessing the quality of diagnostic accuracy studies. Health Technology Assessment 2004;8:III(25):1‐234. [DOI] [PubMed] [Google Scholar]
Whiting 2005
- Whiting P, Harbord R, Kleijnen J. No role for quality scores in systematic reviews of diagnostic accuracy studies. BMC Medical Research Methodology 2005;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Williamson 2000
- Williamson EC, Leeming JP, Palmer HM, Steward CG, Warnock D, Marks DI, et al. Diagnosis of invasiveaspergillosis in bone marrow transplant recipients by polymerase chain reaction. British Journal of Haematology 2000;108(1):132‐9. [DOI] [PubMed] [Google Scholar]
Yoo 2008
- Yoo JH, Choi SM, Choi JH, Kwon EY, Park C, Shin WS. Construction of internal control for the quantitative assay of Aspergillus fumigatus using real‐time nucleic acid sequence‐based amplification. Diagnostic Microbiology and Infectious Diseases 2008;60(1):121‐4. [DOI] [PubMed] [Google Scholar]


