Serotonin type 3A receptors are homopentameric ligand-gated ion channels that are thought to assemble via interactions involving the subunits’ extracellular and transmembrane domains. Pandhare et al. reveal that channel assembly is also determined by three arginine residues in the receptor’s intracellular domain.
Abstract
Serotonin type 3 receptors (5-HT3Rs) are cation-conducting pentameric ligand-gated ion channels and members of the Cys-loop superfamily in eukaryotes. 5-HT3Rs are found in the peripheral and central nervous system, and they are targets for drugs used to treat anxiety, drug dependence, and schizophrenia, as well as chemotherapy-induced and postoperative nausea and emesis. Decades of research of Cys-loop receptors have identified motifs in both the extracellular and transmembrane domains that mediate pentameric assembly. Those efforts have largely ignored the most diverse domain of these channels, the intracellular domain (ICD). Here we identify molecular determinants within the ICD of serotonin type 3A (5-HT3A) subunits for pentameric assembly by first identifying the segments contributing to pentamerization using deletion constructs of, and finally by making defined amino acid substitutions within, an isolated soluble ICD. Our work provides direct experimental evidence for the contribution of three intracellular arginines, previously implicated in governing the low conductance of 5-HT3ARs, in structural features such as pentameric assembly.
Introduction
Serotonin type 3 receptors (5-HT3Rs) are cation-conducting pentameric ligand-gated ion channels (pLGICs), also known as the Cys-loop superfamily in eukaryotes. 5-HT3Rs were first discovered in the peripheral nervous system in the gut (Gaddum and Picarelli, 1957). Only much later was it identified that agents developed for the treatment of chemotherapy-induced emesis elicited behavioral effects in rodents indicative of a central nervous system effect. These receptors play a key role in the process of rapid excitatory neurotransmission in the human brain (Yang, 1990; Hargreaves et al., 1994). 5-HT3Rs are targeted by many therapeutic drugs currently prescribed for the management of cancer chemotherapy–induced vomiting (Aapro, 1991) as well as depression (Pandhare et al., 2017). They are also implicated as potential targets for the treatment of some neurological and psychiatric diseases and disorders (Lummis, 2012). The Cys-loop superfamily additionally includes neuronal- and muscle-type nicotinic acetylcholine receptors (nAChRs), glycine receptors, and γ–aminobutyric acid type A receptors (Changeux and Edelstein, 1998; Miller and Smart, 2010). Cys-loop receptors assemble from five homologous subunits to form homo- or heteropentamers. Each subunit contains three domains: (1) an N-terminal extracellular domain (ECD) mainly structured into two antiparallel β-sheets that harbors the ligand-binding site at subunit interfaces, (2) a transmembrane domain (TMD) consisting of four α-helical segments that shape the ion channel, and (3) a variable intracellular domain (ICD; Forman et al., 2015). Recently, there has been remarkable progress in the determination of atomic resolution 3-D structures of a number of eukaryotic anion- (Huang et al., 2015; Phulera et al., 2018; Zhu et al., 2018; Laverty et al., 2019) and cation-conducting (Hassaine et al., 2014; Morales-Perez et al., 2016) members of the Cys-loop superfamily. As a result, our structural understanding of subunit assembly, allosteric regulation, and conformational transitions associated with gating has improved significantly (Nury et al., 2011; Spurny et al., 2013; Du et al., 2015; Huang et al., 2015, 2017; Polovinkin et al., 2018; Zhu et al., 2018; Masiulis et al., 2019).
While significant advances with regard to the structure and function of the ECD and the TMD have been achieved, structure determination of the ICD has remained elusive. The ICD of 5-HT3ARs (5-HT3A–ICD) begins at the C-terminus of M3 as a loop (L1; Fig. 1) and a short α-helix (MX) followed by a stretch of ∼60 amino acids forming an intrinsically disordered region (L2) and a membrane-associated α-helix (MA). The MA helix forms a continuous helix with M4 at the cytoplasmic leaflet of the membrane bilayer. Pioneering EM studies on the cation-conducting Torpedo nAChR reported the presence of the MA helix in these receptors (Unwin, 2005). Similar helical content pre-M4 is predicted for most other cation-conducting Cys-loop receptors but absent from anion-conducting superfamily members (Fig. S1). On the contrary, α-helical content corresponding to the MX helix is predicted for most anion- and cation-conducting channels. Overall, the lengths and sequences of ICDs are remarkably dissimilar between different subunits. The discovery that replacement of the ICD with a short linker for both anion- and cation-conducting Cys-loop receptors led to functional channels (Jansen et al., 2008) inspired a series of studies using the same or similar modifications for functional (Bar-Lev et al., 2011; McKinnon et al., 2011, 2012; Moroni et al., 2011; Unterer et al., 2012) and structural (Hibbs and Gouaux, 2011; Bondarenko et al., 2012, 2014; Mowrey et al., 2013; Hassaine et al., 2014; Miller and Aricescu, 2014; Morales-Perez et al., 2016) studies. Recent cryo-EM studies using full-length 5-HT3ARs resolved L1-MX and the MA helix (Basak et al., 2018; Polovinkin et al., 2018), similar to a study where the ICD had been proteolyzed before crystallization and x-ray structure determination (Hassaine et al., 2014). The inability to resolve L2 likely stems from its inherently “very dynamic” nature (Polovinkin et al., 2018). Therefore, structural, albeit incomplete, insights into the ICD are limited to neuronal and Torpedo nAChRs and 5-HT3ARs, and entirely lacking for anion-conducting Cys-loop receptors (Unwin, 2005; Hassaine et al., 2014; Morales-Perez et al., 2016). Even though full-length γ–aminobutyric acid type A constructs were used for recent cryo-EM structure determination, only a few amino acids of the ICD without defined secondary structure could be resolved (Phulera et al., 2018; Laverty et al., 2019; Masiulis et al., 2019). This provides the first experimental indication for the ICD of anion-conducting pLGICs being significantly different from cation-conducting channels.
Figure 1.
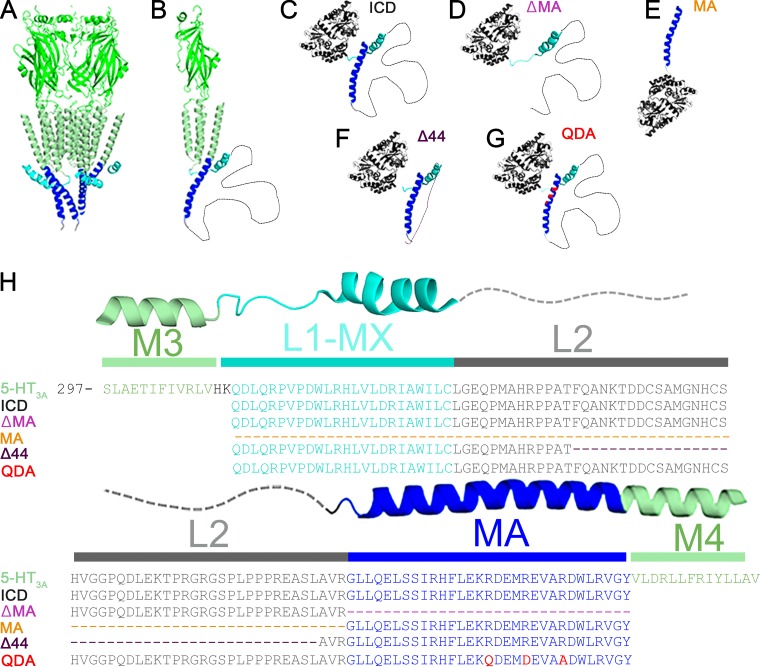
Structural representation and alignments of constructs. (A) Cartoon representation of the pentameric mouse 5-HT3A receptor x-ray structure (PDB ID: 4PIR; Hassaine et al., 2014) viewed parallel to the plane of the membrane. ECD and TMD in green, MA and MX helices in blue and cyan. (B) A single subunit of the 5-HT3A receptor viewed parallel to the membrane. The 62 residues not resolved in L2 are depicted as a dashed line. (C) Cartoon representation of MBP–5-HT3A–ICD–WT. All structural models of the fusion proteins are hypothetical models. The cytoplasmic domain of 5-HT3AR is fused to the C-terminus of MBP and serves as the template for the chimeras. (D) The chimera MBP-ΔMA has the entire MA helix removed. (E) MBP-MA has the MA helix attached to the C-terminus of MBP. (F) MBP-Δ44 contains a 44-amino acid deletion in L2. (G) MBP-QDA contains a triple mutation in its MA helix and no deletions. (H) Multiple sequence alignment of 5-HT3AR and the ICD chimeras highlighting deletions (dashed lines) or mutations (red).
The ICD is involved in determining ion conductance (Kelley et al., 2003; Jansen et al., 2008) and modulation by drugs (Moraga-Cid et al., 2011). Additionally, it interacts with various entities of the intracellular apparatus, which facilitate receptor sorting, assembly, trafficking, and anchorage (Thompson et al., 2010; Nishtala et al., 2016). Studies of subunit assembly and oligomerization have investigated the individual contributions of the ECD (Verrall and Hall, 1992; Kouvatsos et al., 2016) as well as the TMD (Bondarenko et al., 2012; Mowrey et al., 2013) at the molecular level. While both the ECD and the TMD can assemble individually into pentamers (Wells et al., 1998; Liu et al., 2008; Bondarenko et al., 2012, 2014), formation of hexamers of the ECD alone for GLIC (Nury et al., 2010) and nonnative stoichiometries for α2β4 nAChR (Bondarenko et al., 2012) exemplify that additional drivers outside these domains may mediate precise subunit stoichiometries and arrangements observed in nature. Intriguingly, this points toward the highly diverse ICD as a mediator for defined oligomerization and pentamerization, in general. Indeed, we showed for the first time that the 5-HT3A-ICD alone can assemble into highly stable pentamers in the absence of the ECD and the TMD (Pandhare et al., 2016).
This remarkable experimental observation led us to investigate and uncover the molecular determinants for pentameric assembly of the 5-HT3A-ICD. In the present study, we introduced defined deletions over the entire span of the ICD, and finally a set of amino acid substitutions, to determine the motif necessary for its pentameric assembly.
Materials and methods
The following materials were used: BL21-CodonPlus-(DE3)-RIPL cells (Agilent Technologies), ampicillin (Fisher Scientific), chloramphenicol (Fisher Scientific), isopropyl β-D-thiogalactoside (Fisher Scientific), leupeptin (AdipoGen Life Sciences), pepstatin (AdipoGen Life Sciences), phenylmethylsulfonyl fluoride (PMSF; Research Products International), tris(2-carboxyethyl)phosphine hydrochloride (TCEP; Oakwood Chemical), lysozyme (MP Biomedicals), protease inhibitor cocktail III (Research Products International), and DNase I (Alfa Aesar).
Molecular biology
The intracellular domain of the mouse 5-HT3AR (NCBI Protein database accession no. Q8K1F4) was generated as a fusion construct with N-terminal maltose-binding protein (MBP) as we described previously (Pandhare et al., 2019) using the pMAL-c2x (New England Biolabs) variant pMALX (Moon et al., 2010). Based on this MBP–5-HT3A–ICD template containing the entire WT ICD, deletions and substitutions were generated using the QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies) and confirmed by DNA sequencing (GENEWIZ). The resulting amino acid sequences for all constructs are displayed in Fig. 1.
Expression and purification of the MBP–5-HT3A–ICD constructs
All plasmids for expression of MBP–5-HT3A–ICD constructs were transformed into Escherichia coli BL21-CodonPlus-(DE3)-RIPL cells (E. coli; Agilent Technologies). Cells were grown in Terrific Broth medium (1.2% [wt/vol] tryptone, 2.4% [wt/vol] yeast extract, and 0.4% [vol/vol] glycerol) supplemented with ampicillin (100 µg/ml), chloramphenicol (34 µg/ml), and 0.2% (wt/vol) sterile glucose, at 37°C and 250 rpm, in a shaking incubator. All concentrations indicated are final concentrations unless otherwise stated. At OD600 of 0.4–0.5, the cultures were induced by adding 0.4 mM isopropyl β-D-thiogalactoside and allowed to continue growth at 18°C for an additional 8 h. The cells were harvested by centrifugation at 4,600 g and 4°C for 15 min and then resuspended (10 ml buffer/g of the cell pellet) in buffer A (20 mM Tris, pH 7.4; 200 mM NaCl, 1 mM TCEP, and 2 mM EDTA) enriched with a freshly prepared protease inhibitor cocktail containing leupeptin (10 µg/ml), pepstatin (10 µg/ml), and 1 mM PMSF. After the cells were disrupted by treatment with lysozyme (100 µg/ml) and a freeze–thaw cycle, the lysate was clarified by ultracentrifugation at 100,000 g and 4°C for 1 h. The supernatant was passed through a 0.2-µm pore-size filter and loaded onto a pre-equilibrated amylose resin column. After a 30–bed volume wash of buffer A, proteins were eluted with 20 mM maltose. Fractions were analyzed after separation in stain-free precast SDS-PAGE gels (4–20% Mini-PROTEIN TGX Stain-Free; Biorad). These stain-free gels contain trihalo compounds that undergo UV-induced covalent modification of tryptophan residues for subsequent fluorescent detection. Fractions containing purified protein were pooled and concentrated (up to ∼5 mg/ml) using a 50- or 100-kD molecular weight cut-off centrifugal filter (Amicon Ultra-15; Merck Millipore Ltd.) for an additional purification step by size exclusion chromatography (SEC).
SEC
The amylose column–purified and concentrated protein samples were passed through an ENrich SEC 650 10 × 300 high-resolution column (Biorad) pre-equilibrated with buffer B (20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5 mM maltose, and 0.01% NaN3, pH 7.4) for additional purification as well as molecular mass determination. For the apparent molecular mass estimation of each 5-HT3A-ICD construct (WT, deletion, and substitution constructs), the SEC column was calibrated using thyroglobulin, 669 kD; ferritin, 440 kD; aldolase, 158 kD; conalbumin, 75 kD; and ovalbumin, 44 kD according to the instruction manual (GE Healthcare). The column void volume (Vo) was established with Blue Dextran 2000. The molecular mass of each construct was determined based on the calibration curve of the gel-phase distribution coefficient (Kav) versus log molecular weight (log Mr). The gel-phase distribution coefficient is calculated by the equation Kav = (Ve − Vo)/(Vc − Vo), where Ve = elution volume, Vc = geometric column volume, and Vo = column void volume. The purified protein samples obtained after the final SEC step were analyzed by stain-free precast SDS-PAGE gels.
SEC coupled with multi-angle light scattering (SEC-MALS)
We configured a Biorad Biological DuoFlow 10 system coupled with a miniDAWN-TREOS static 3-angle laser light-scattering detector and an Optilab-rEX refractive index detector (Wyatt Technology) for conducting the SEC-MALS experiments. 250 µl of purified protein sample (0.1–0.5 mg) was passed, at a constant flow rate of 0.5 ml/min, through an ENrich SEC 650 10 × 300 high-resolution column (Biorad) thoroughly pre-equilibrated with buffer B at room temperature. UV absorbance was measured with the detector at 280 nm. Light scattering and refractive index were monitored at a wavelength of 658 nm. BioLogic DuoFlow software version 5.3 (Biorad) was used to control the chromatography system, and Astra 5.3.4 software (Wyatt Technology) was used for data collection and analysis. The light-scattering (LS) detectors were normalized with monomeric BSA (Sigma). Baseline settings for all laser detectors as well as peak alignment and band-broadening correction between the UV, LS, and refractive index detectors were performed using Astra software algorithms. Data were processed to determine the weight-average molar mass and polydispersity of the protein sample using the Debye model as per the manufacturer’s instructions. A minimum of three repeat runs was conducted for each construct under identical experimental conditions.
Online supplemental material
Fig. S1 provides sequence alignments for cation- and anion-conducting pLGICs with predicted α-helical segments highlighted to illustrate predicted structural diversity of these two groups of ICDs. Fig. S2 presents an illustration of charged amino acids of the MA-helix and post-M3 loop using the x-ray structure of 5-HT3A (PDB ID: 4PIR; Hassaine et al., 2014).
Results
Overview of approach
We have shown previously that the intracellular domain of 5-HT3ARs in fusion with MBP self-associates as a pentamer in solution (Pandhare et al., 2016, 2019). To gain further insights into the molecular mechanisms governing pentameric assembly of the 5-HT3A–ICD, we adopted a systematic approach wherein a series of MBP–5-HT3A–ICD constructs, bearing either large deletions or a set of amino acid substitutions within the ICD, were engineered. Our nomenclature for the different constructs is illustrated in detail in Fig. 1. In brief, five constructs were investigated in this study: the initial full-length ICD construct containing the 5-HT3A–ICD consisting of all 115 amino acids of the ICD; three deletion constructs, and one construct based on the full-length ICD with three arginine residues in the MA helix mutated to glutamine, aspartic acid, and alanine (R432Q/R436D/R440A). All constructs were expressed in E. coli and purified to homogeneity. We then used a combination of biophysical and biochemical methods, including denaturing gel electrophoresis, SEC, and SEC-MALS to investigate the oligomeric state of each construct.
Expression and purification of WT and engineered 5-HT3A–ICD constructs
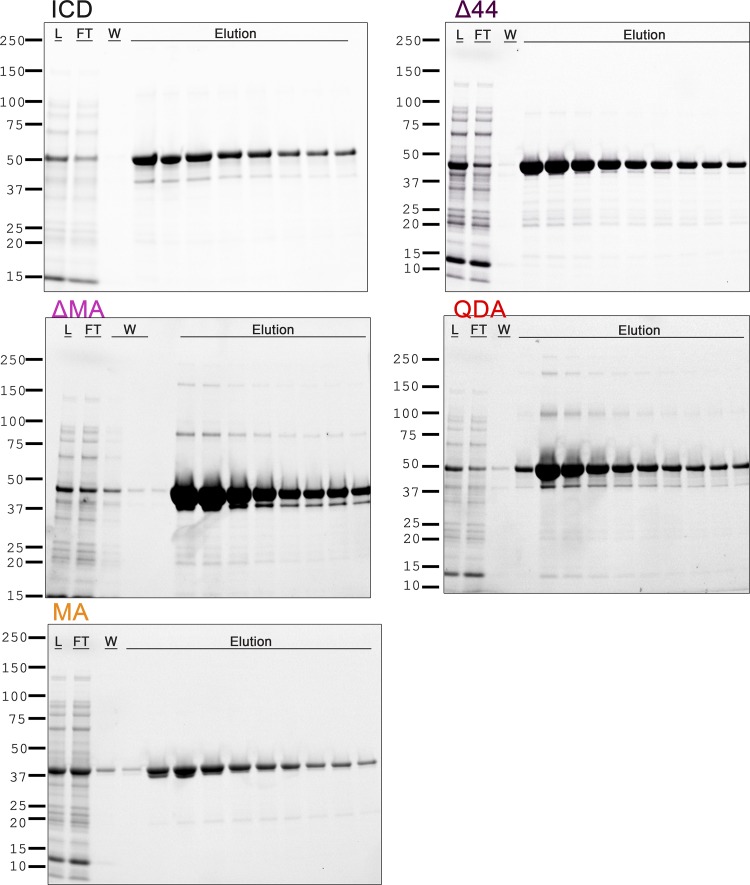
Cytosolic expression in E. coli cells of the 5-HT3A–ICD as a chimeric protein containing an N-terminal MBP was under the control of a Ptac promoter. After careful optimization, ∼20–30 mg of ICD could be purified per liter of E. coli cell culture (Pandhare et al., 2019). Modifications (point or deletion) of this original construct (Fig. 1) were similarly produced with yields of 8–30 mg/liter of E. coli culture. Two-step purification was accomplished on an amylose resin affinity column followed by SEC with SDS-PAGE indicating a purity of >90% and >95%, respectively (Figs. 2 and 3).
Figure 2.
Affinity column purification of the ICD chimeras. The chimeras were purified by amylose resin column chromatography. The soluble protein was loaded onto columns (L, loading material; FT, flow-through), followed by washes (W), and eluted. SDS-PAGE indicates the quality of purified protein and identifies peak fractions.
Figure 3.
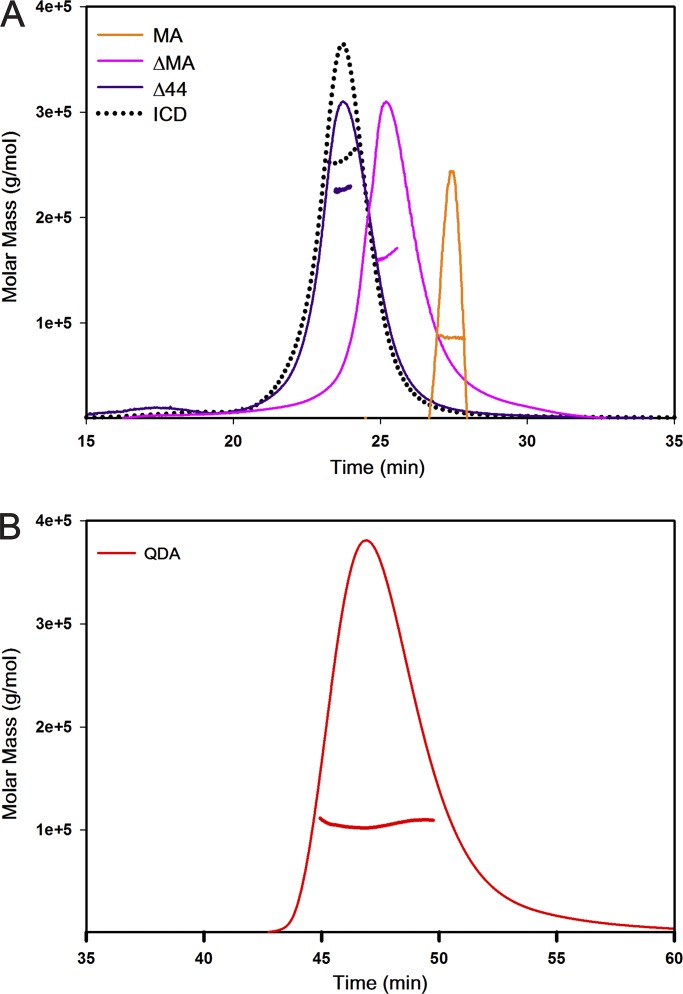
Weight determination of ICD chimeras by SDS-PAGE and SEC. (A) The weight of the ICD chimeras in monomeric state was determined by resolving SEC-purified protein on SDS-gel electrophoresis. Molecular weight standards are shown on top, and ICD (53 kD), ΔMA (49 kD), MA (45 kD), Δ44 (48 kD), and QDA (52 kD) are shown below. (B) The complete SDS-gel electrophoresis image from which the analysis in section A is drawn. A standard protein ladder is shown next to each protein for increased accuracy of the analysis. Lane compositions are as follows: lane 2, ICD; lane 4, ΔMA; lane 6, MA; lane 8, Δ44; and lane 10, QDA. (C) SEC yielded a weight of 267 kD for ICD, 261 kD for Δ44, 180 kD for ΔMA, 141 kD for QDA, and 73 kD for MA. Chromatogram of standard proteins (T, thyroglobulin; F, ferritin; A, aldolase; C, conalbumin; O, ovalbumin) in gray, and chimeras in their respective colors are shown above. A280, UV absorbance at 280 nm.
Oligomeric structure of full-length 5-HT3A–ICD
Affinity- or SEC-purified full-length ICD, when analyzed by SDS-PAGE under reducing conditions, migrated as a major band with a relative molecular weight of 53 kD (Figs. 2 and 3), consistent with its theoretical mass of 53.4 kD. Notably, during SEC ICD eluted much earlier than would be anticipated from its relative molecular weight determined by SDS-PAGE. The elution profile of the protein was similar over the entire range of concentrations examined (0.1–5 mg/ml; data not shown). The hydrodynamic volume (13.01 ml) of a well-resolved peak of ICD predicted an apparent molecular mass of 267 kD when compared with the elution profiles of proteins with known molecular masses (Fig. 3). The elution volume indicates a mass that is five times the monomer mass and therefore a potential pentameric assembly. Asymmetrically shaped molecules containing multiple helical regions with an overall extended conformation, such as human mannose-binding protein, are known to display anomalous behavior on SEC (Lipscombe et al., 1995). As shape-dependent factors can influence the elution volume of macromolecules when analyzed on SEC, the results from this hydrodynamic technique are possibly vulnerable to misinterpretation (Wen et al., 1996). Therefore, we used the shape-independent technique of SEC-MALS to study the self-assembly properties of 5-HT3A–ICD constructs further (Table 1). SEC-MALS determines the absolute molecular mass of macromolecules independent of molecular shape, and the high-resolution separation by SEC facilitates analysis of each resolved protein species in a tandem manner. Examination of the SEC-MALS results revealed that WT 5-HT3A–ICD exists predominantly as pentamers with an absolute molecular mass of 259 kD (Pandhare et al., 2016, 2019). Samples consisting of MBP alone were analyzed by SEC-MALS, indicating that MBP was monomeric in solution (data not shown), consistent with a previous small-angle x-ray scattering study (Rodgers et al., 1996), and that it likely did not influence oligomeric assembly of the ICD.
Table 1. Monomer and oligomer masses.
| Construct | Theoretical molar mass of the monomer (Mw, 103 g/mol) | Calculated molar mass from SEC-MALS (Mw, 103 g/mol) | Dispersity (Ð = Mw/Mn) | Number of monomers (calculated Mw/theoretical Mw) |
|---|---|---|---|---|
| (Mean ± SEM) | (Mean ± SEM) | |||
| ICD | 53.4 | 259 ± 8 | 1.073 ± 0.015 | 4.8 |
| Δ44 | 49.3 | 245 ± 9 | 1.002 ± 0.001 | 4.9 |
| ΔMA | 49.8 | 149 ± 6 | 1.001 ± 0.001 | 2.9 |
| MA | 44.3 | 89 ± 2 | 1.000 ± 0.003 | 2.0 |
| QDA | 53.0 | 106 ± 1 | 1.004 ± 0.003 | 2.0 |
Effect of deletions on ICD self-assembly
Our data so far demonstrate that the pentameric assembly is an intrinsic property of the ICD. Based on a recent cryo-EM structure of the mouse 5-HT3AR with only a partially resolved ICD, the MX and MA helices form a “putative contact” with the MA helix of the neighboring subunit (Polovinkin et al., 2018). Since the involvement of unresolved regions in domain–domain interactions was not apparent, we first sought to understand the effects of systematic deletions within the ICD on their oligomeric states.
We generated three ICD deletion constructs: (1) ICD lacking 44 amino acids from the disordered flexible region between the MX and MA helices (Δ44), (2) ICD lacking the MA helix (ΔMA), and (3) MA helix alone (MA; Fig. 1). The theoretical molecular weights, calculated from their amino acid composition of 49.3 kD, 49.8 kD, and 44.3 kD were comparable to the experimentally determined relative molecular weights of 48 kD, 49 kD, and 45 kD, respectively, when purified protein samples were analyzed by SDS-PAGE (Fig. 3). The faint higher molecular weight bands observed in SDS-PAGE (Fig. 2) are likely comprised of oligomeric assemblies resistant to SDS under the given electrophoresis condition. However, as observed later on for each construct, the purified protein analyzed after SEC (Fig. 3) as well as by multi-angle light scattering (Fig. 4) was homogenous and monodisperse, respectively.
Figure 4.
SEC-MALS determination of oligomeric assembly. (A) Both the ICD (dotted black) and the Δ44 construct (purple line) maintain pentameric assembly, whereas deletions in the ΔMA (pink line) and the MA (orange line) constructs abolish pentamerization of the ICD. (B) The QDA substitutions (red line) disrupt the pentameric assembly of the ICD. A representative SEC-MALS profile showing the Rayleigh ratio. Note that different flow rates were used during the monitoring period for A (0.5 ml/min) and B (0.3 ml/min).
The three deletion constructs showed strikingly different elution profiles in SEC (Fig. 3). Importantly, all constructs eluted in single monodisperse peaks. Based on the elution volumes of 13.04 ml, 13.57 ml, and 14.85 ml for Δ44, ΔMA, and MA, respectively, apparent molecular weights of 261 kD, 171 kD, and 73 kD, respectively, were estimated. These results were inconsistent with a monomeric state for all deletion constructs. While SEC data for Δ44 may indicate a pentamer (49.3 kD × 5 = 247 kD vs. 261 kD), data for ΔMA and MA did not correspond to a defined multimeric state. Therefore, at this juncture, it was imperative to investigate all constructs with SEC-MALS to determine the absolute molecular weight (Table 1). The predominant peak for the Δ44 construct yielded a weight-average molar mass of 245 ± 9 kD (mean ± SEM, n = 5), which corresponds well to a pentameric assembly (theoretical molecular mass: 49.3 kD). Moreover, the pentameric protein is monodisperse because the experimentally determined molar masses were equivalent across the peak, as indicated by the measurements of polydispersity: Mw/Mn of 1.002 ± 0.001, where Mw is the weight-average molar mass, and Mn is the number-average molar mass (Fig. 4; Table 1). Similarly, for the other two deletion constructs, ΔMA and MA, analysis yielded the weight-average molar masses of 149 ± 6 kD (mean ± SEM, n = 3) and 89 ± 2 kD (mean ± SEM, n = 3), respectively, and polydispersity of 1.001 ± 0.001 (mean ± SEM, n = 3) and 1.000 ± 0.003 (mean ± SEM, n = 3), respectively. Therefore, SEC-MALS results established that ΔMA is a trimer (theoretical molecular mass: 49.8 kD) and MA is a dimer (theoretical molecular mass: 44.3 kD) in solution. It is also important to note that we did not observe a plateau in the molar mass profiles or a peak in the elution profiles that corresponded to major quantities of the monomeric form of either of the deletion constructs. Together, the results suggested that we were able to disrupt pentameric assembly of the ICD. Since Δ44 assembled into pentamers, but both ΔMA and MA did not do so, we inferred at this point that the segment removed in Δ44 is not involved in pentamerization, whereas motifs contained in both MA and ΔMA may interact to mediate pentamerization.
Effect of specific amino acid substitutions in the MA helix on the oligomeric structure of the ICD chimera
The observed involvement of both the proximal and distal segments of the ICD in pentamerization was reminiscent of the salt-bridge networks identified in some of the structural studies (Fig. S2). Three arginine residues of the MA helix (R432, R436, and R440), which have been identified as determinants for the 5-HT3A subunit’s low single-channel conductance, participate in salt bridges with acidic amino acids from neighboring ICDs (Kelley et al., 2003; Hassaine et al., 2014). We replaced these three arginine residues with the aligned residues glutamine, aspartic acid, and alanine of serotonin type 3B (5-HT3B) subunits to obtain the QDA construct (R432Q/R436D/R440A) to investigate their impact on the pentamerization of the ICD. The QDA construct eluted, at a volume of 13.92 ml, as a major symmetrical peak after SEC, which predicted its apparent molecular weight as 137 kD (Fig. 3). The size of the monomer, calculated from its amino acid sequence as well as determined by SDS-PAGE, is 53 kD (Fig. 3). Analogous to the experimental strategy employed for the deletion constructs, we determined an absolute molar mass for the QDA construct by SEC-MALS. The result of the weight-average molar mass of 106 ± 1 kD (mean ± SEM, n = 3) and polydispersity of 1.004 ± 0.003 (mean ± SEM, n = 3) suggested that the QDA construct exists as a dimer in solution (Fig. 4). This result indicates that mutation of the RRR motif and the likely resulting disruption of salt bridges is sufficient to abolish pentameric assembly of the ICD.
Discussion
The ICD is the most diverse and least understood portion of pLGICs across the Cys-loop receptor superfamily. Several studies show that the ICD plays a pivotal role in receptor assembly, trafficking, and targeting (Connolly, 2008; Kracun et al., 2008), functional interactions with cytoplasmic proteins (Lansdell et al., 2005; Nishtala et al., 2016), gating and desensitization (Papke and Grosman, 2014), inward rectification (Baptista-Hon et al., 2013), Ca2+ permeability (Livesey et al., 2008), and single-channel conductance (Kelley et al., 2003; Hales et al., 2006). In the Torpedo nAChR structure, the five MA helices were viewed as an inverted cone protruding into the cytosol and enclosing the intracellular vestibule of the transmembrane pore (Unwin, 2005). The MA helices were thought to frame lateral portals that function as cytosolic ion pathways. In the case of the 5-HT3A–ICD, it has become apparent that several of the experimentally determined functions are, perhaps most critically, contingent on the integrity of its secondary and/or quaternary structure. Mutagenesis studies have demonstrated that the variation in the length and amino acid composition of the ICD can influence the desensitization rate of the receptor channel (McKinnon et al., 2012; Baptista-Hon et al., 2013). Reducing the length of the loop connecting the M3 to the MA helix has been shown to result in loss of inward rectification of macroscopic currents, which has been inferred to be an indicator for the disruption of the portal architecture (Baptista-Hon et al., 2013). Key residues within the MA helix not only profoundly affect the permeability of Ca2+ but also the single-channel conductance uniquely observed in experiments with homomeric 5-HT3ARs. The homomeric 5-HT3AR displays a very low single-channel conductance in the sub-pS range (Brown et al., 1998). This is in stark contrast to the larger currents (16–30 pS) measured from heteromeric serotonin type 3AB receptors as well as other Cys-loop receptors (Davies et al., 1999; Krzywkowski et al., 2008). A seminal study attributed the low conductance to a group of three arginine residues, which are found in the MA helix of the 3A subunit but are absent in the 3B subunit (Kelley et al., 2003). The x-ray structure of mouse 5-HT3AR revealed that some of the conductance-restricting arginine residues as well as other basic residues lining the MA helix are within salt-bridge distance to several acidic residues on the neighboring MA helices and post-M3 loops (Hassaine et al., 2014). Notably, the post-M3 loops thread through the MA portals, leaving no paths for ions to access the channel. In the x-ray structure, the side chain of D312, at the center of the short post-M3 loop, is within salt-bridging distance and straddled by arginine residues R436 and R440 on the adjacent subunit. Interestingly, it has been postulated that inter- and intra-subunit salt bridges confer structural rigidity to the inverted pentacone formed by the five MA helices, and that a controlled removal of these salt bridges increases flexibility in this area (Kozuska et al., 2014). The same study proposed that it was widening of the lateral portals between the MA helices, due to the increased flexibility in the absence of certain salt bridges, and not the abolition of electrostatic repulsion, that relieved the limited conductance of the 5-HT3AR channel. Importantly, the authors based their prediction on a cautious assumption that the mutations they introduced would not destabilize global interhelical interactions that might further widen the portals (Kozuska et al., 2014). Taken together, these findings suggest that the extensive interhelical interactions between the MA helices may be central to holding the intact quaternary structure of the ICD, and thus contribute to the functional landscape of full-length 5-HT3ARs. In this regard, it is intriguing to find that the 5-HT3A–ICD is structured as pentamers in the absence of both the ECD and the TMD (Pandhare et al., 2016). Multiple studies underscore the importance of salt-bridge formation in the stabilization of a protein oligomeric assembly (Binter et al., 2009; Skinner et al., 2017; Pacheco et al., 2018). Therefore, we investigated whether some of the aforementioned function-governing salt bridges additionally play a fundamental role in pentameric assembly.
We previously developed a chimeric strategy, where the 5-HT3A–ICD was linked N-terminally to MBP (Pandhare et al., 2016, 2019). In the present work, we systematically introduced deletions or defined amino acid substitutions within this construct. First, in order to obtain insights into the role of the flexible unresolved region of the ICD, we removed 44 amino acids from this region (Δ44). A similar construct was generated in full-length channels in a study that investigated the significance of the lateral portals within MA helices for inward rectification observed in 5-HT3ARs (Baptista-Hon et al., 2013). Using a series of truncation mutants, the authors determined the minimum number of residues necessary to link M3 with the MA helix without abolishing function. While truncations of 44 and 24 amino acids from the ICD produced functional receptors, the construct with 44 amino acids deleted exhibited loss of rectification. This indicated reduced impediment to ion conduction that was inferred to be caused by deconstruction of the lateral portals. Interestingly, our results showed that truncation of the exact 44 amino acids from the soluble ICD maintained pentamers in solution. The study additionally reported a truncation of 55 amino acids in L2, or removal of 10 amino acids from the proximal part of the ICD that did not produce functional homomeric 5-HT3ARs (Baptista-Hon et al., 2013). We infer that the large 55-residue truncation, as well as truncations made in the proximal ICD, can herald a global disruption and lead to the formation of a global nonnative ICD conformation, which may further impose unfavorable constraints for channel function. Our assumption is supported by the previously reported data in which an engineered 5-HT3AR containing a heptapeptide replacing the entire ICD was not only fully functional but also exhibited enhanced single-channel conductance (Jansen et al., 2008). Within this context, given our experimental evidence of intact pentamers formed by Δ44, we inferred that this shortened ICD plausibly underwent merely a local perturbation likely limited only to the lateral portals without disturbing the key interactions holding together the oligomeric assembly. This might have resulted in a minimally aberrant native-like ICD architecture responsible for the observed retention of function in correspondingly engineered 5-HT3ARs (Baptista-Hon et al., 2013). In the structures there is an ∼54-Å gap between the end of MX and the start of the MA α-helices (M3-WILC to VRG-M4). At least 18 amino acids are needed for a linker to span this distance. Δ44 and Δ55 contain 14 and 3 amino acids in our constructs, respectively. Since Baptista-Hon et al. (2013) used the long variant with 6 additional amino acids in this segment, their Δ44 and Δ55 contain 20 and 9 amino acids, respectively. Consequently, it would be expected that Δ44 has a sufficient remaining L2 loop length to not disrupt the relative arrangement of MX and MA, and indeed it pentamerizes. Δ55, on the other hand, has a too-short L2-linker, and a drastic distortion is expected. In preliminary experiments using dynamic light scattering, we observed disruption of pentamers for Δ55. Collectively, we postulated that the underlying molecular interactions driving the ICD pentamerization must lie within the proximal (L1-MX) as well as the distal (MA) regions of the ICD.
To test the above stated hypothesis, we decided to explore the effect of deletion of a proximal (MA construct) or a distal (ΔMA construct) portion of the ICD on its oligomeric state. SEC-MALS indicated disruption of pentameric assembly in both constructs, and MA was determined as a dimer and ΔMA as a trimer. The fact that the MA construct did not pentamerize was somewhat surprising since the MA helices are tightly associated in the crystal structure, where rings of hydrophobic residues L402, L406, I409, and L413 shape a 17-Å-long narrow channel (Hassaine et al., 2014). Given the observation that ΔMA also disrupts pentamerization, while Δ44 retains the pentameric state, we hypothesized that the elements of both MA and ΔMA are required for driving the pentamerization. In particular, the results pointed toward both the proximal and distal segments of the ICD as being determinants for pentameric assembly. Interestingly, when we previously made chimeras by using the prokaryotic pLGIC from GLIC that naturally lacks an ICD and inserted ICDs from different eukaryotic anion- or cation-conducting channels, we found that the linking amino acids, on both the proximal and distal insertion sites, were crucial for functional 5-HT3A–ICD insertion (Goyal et al., 2011). Remarkably, for other ICDs that we investigated (nAChα7, Glyα1, GABAρ1), the linking amino acids for the ICDs had little impact on function, indicating a unique characteristic for the 5-HT3A–ICD (Mnatsakanyan et al., 2015).
We next sought to examine if the conductance-limiting arginines are additionally involved in maintaining the quaternary pentameric structure of the ICD. Interestingly, the location of the RRR coincides with a kink in the paths of the long continuous MA-M4 helices that collectively leads to a significantly tighter packing of MA helices as they traverse away from the membrane. The bottom aperture of the MA bundle has a diameter of 4.2 Å, too small for conduction of hydrated cations, but wider than the remaining opening of the lateral windows through which the post-M3 loop is threaded (Hassaine et al., 2014). Combined replacement of RRR with QDA substantially augments single-channel conductance (Kelley et al., 2003), a phenomenon originally attributed to the removal of repulsive forces of the positive charges of the arginines upon conducted cations. On the other hand, constrained geometric simulations, upon abolishing certain salt bridges, have predicted enhanced structural flexibility within the MA helices and led to postulation that a rigid structural framework of the inverted cone formed by the MA helices might be crucial for the limited conductance of the 5-HT3AR channel (Kozuska et al., 2014). Interestingly, the QDA substitutions in our soluble construct disrupted the pentameric state of the full-length ICD and reduced it to dimers. Clearly, this finding does not appear to support the “charge-repulsion theory,” but rather provides the first experimental evidence suggesting a structural role of the triple arginines that are critical to sustaining tight packing of the MA bundle as well as pentameric assembly of the ICD. We infer that the disjunction of interactions due to the QDA substitutions is related to breakage of salt bridges observed in the x-ray structure.
Sequence alignment between all available 5-HT3A sequences indicates that the triple arginines as well as the aspartate in the post-M3 loop are absolutely conserved. Among 5-HT3B subunit sequences, neither motif is observed. Many 5-HT3B sequences have negatively charged side chains one or two residues before 5-HT3AD312, with or without a positive side chain at the D312-equivalent position. Only very few 5-HT3B sequences have one of the three MA helix arginines. However, the 5-HT3B sequences, similar to the 5-HT3A sequences, have many other charged residues within the MA helix that may contribute to salt-bridge networks (Hassaine et al., 2014; Kozuska et al., 2014). However, as our MA construct shows, those are not sufficient to drive pentamerization of the ICD alone, and therefore we do not expect homopentameric 5-HT3B or heteropentameric serotonin type 3AB ICDs to be as tightly interdigitated as homopentameric 5-HT3A subunits alone. We infer that the heteropentameric ICDs are therefore more loosely packed and that this contributes to their higher single-channel conductance.
We do not know exactly why the ΔMA construct forms trimers under the given experimental conditions; nor do we know the exact driving forces behind dimerization of the MA and QDA constructs in solution. It is a possibility, as documented elsewhere (Skinner et al., 2017), that the breakage of preexisting salt bridges can induce protein remodeling via initiation of partial unfolding and reorganization of electrostatic interactions leading to the formation of nonnative oligomers. Nevertheless, it is important to emphasize that we did not observe pentamers formed by any of the engineered proteins during our investigation, with the notable exceptions of the ICD and Δ44.
Finally, to the best of our knowledge, this is the first experimental evidence that extends our understanding beyond the known functional contributions of the three arginine residues to limiting conductance, and posits that their existence within the MA helix of the 5-HT3AR is critical for preserving a pentameric state of the ICD.
Supplementary Material
Acknowledgments
Merritt C. Maduke served as editor.
We thank the Texas Tech University Health Sciences Center Core Facilities; some of the images and or data were generated in the Image Analysis Core Facility & Molecular Biology Core Facility supported by the Texas Tech University Health Sciences Center, Lubbock, TX.
Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number R01NS077114 (to M. Jansen). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: M. Jansen designed the research. A. Pandhare, E. Pirayesh, A.G. Stuebler and M. Jansen conceived the WT and engineered constructs, performed the experiments, analyzed the data, and wrote the paper.
References
- Aapro, M.S. 1991. 5-HT3 receptor antagonists. An overview of their present status and future potential in cancer therapy-induced emesis. Drugs. 42:551–568. 10.2165/00003495-199142040-00002 [DOI] [PubMed] [Google Scholar]
- Baptista-Hon, D.T., Deeb T.Z., Lambert J.J., Peters J.A., and Hales T.G.. 2013. The minimum M3-M4 loop length of neurotransmitter-activated pentameric receptors is critical for the structural integrity of cytoplasmic portals. J. Biol. Chem. 288:21558–21568. 10.1074/jbc.M113.481689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Lev, D.D., Degani-Katzav N., Perelman A., and Paas Y.. 2011. Molecular dissection of Cl−-selective Cys-loop receptor points to components that are dispensable or essential for channel activity. J. Biol. Chem. 286:43830–43841. 10.1074/jbc.M111.282715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak, S., Gicheru Y., Samanta A., Molugu S.K., Huang W., Fuente M., Hughes T., Taylor D.J., Nieman M.T., Moiseenkova-Bell V., et al. 2018. Cryo-EM structure of 5-HT3A receptor in its resting conformation. Nat. Commun. 9:514. 10.1038/s41467-018-02997-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binter, A., Staunig N., Jelesarov I., Lohner K., Palfey B.A., Deller S., Gruber K., and Macheroux P.. 2009. A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase. FEBS J. 276:5263–5274. 10.1111/j.1742-4658.2009.07222.x [DOI] [PubMed] [Google Scholar]
- Bondarenko, V., Mowrey D., Tillman T., Cui T., Liu L.T., Xu Y., and Tang P.. 2012. NMR structures of the transmembrane domains of the α4β2 nAChR. Biochim. Biophys. Acta. 1818:1261–1268. 10.1016/j.bbamem.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko, V., Mowrey D.D., Tillman T.S., Seyoum E., Xu Y., and Tang P.. 2014. NMR structures of the human α7 nAChR transmembrane domain and associated anesthetic binding sites. Biochim. Biophys. Acta. 1838:1389–1395. 10.1016/j.bbamem.2013.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A.M., Hope A.G., Lambert J.J., and Peters J.A.. 1998. Ion permeation and conduction in a human recombinant 5-HT3 receptor subunit (h5-HT3A). J. Physiol. 507:653–665. 10.1111/j.1469-7793.1998.653bs.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changeux, J.P., and Edelstein S.J.. 1998. Allosteric receptors after 30 years. Neuron. 21:959–980. 10.1016/S0896-6273(00)80616-9 [DOI] [PubMed] [Google Scholar]
- Connolly, C.N. 2008. Trafficking of 5-HT3 and GABAA receptors (Review). Mol. Membr. Biol. 25:293–301. 10.1080/09687680801898503 [DOI] [PubMed] [Google Scholar]
- Davies, P.A., Pistis M., Hanna M.C., Peters J.A., Lambert J.J., Hales T.G., and Kirkness E.F.. 1999. The 5-HT3B subunit is a major determinant of serotonin-receptor function. Nature. 397:359–363. 10.1038/16941 [DOI] [PubMed] [Google Scholar]
- Du, J., Lü W., Wu S., Cheng Y., and Gouaux E.. 2015. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. 526:224–229. 10.1038/nature14853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman, S.A., Chiara D.C., and Miller K.W.. 2015. Anesthetics target interfacial transmembrane sites in nicotinic acetylcholine receptors. Neuropharmacology. 96(Pt B):169–177. 10.1016/j.neuropharm.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddum, J.H., and Picarelli Z.P.. 1957. Two kinds of tryptamine receptor. Br. J. Pharmacol. Chemother. 12:323–328. 10.1111/j.1476-5381.1957.tb00142.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal, R., Salahudeen A.A., and Jansen M.. 2011. Engineering a prokaryotic Cys-loop receptor with a third functional domain. J. Biol. Chem. 286:34635–34642. 10.1074/jbc.M111.269647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales, T.G., Dunlop J.I., Deeb T.Z., Carland J.E., Kelley S.P., Lambert J.J., and Peters J.A.. 2006. Common determinants of single channel conductance within the large cytoplasmic loop of 5-hydroxytryptamine type 3 and α4β2 nicotinic acetylcholine receptors. J. Biol. Chem. 281:8062–8071. 10.1074/jbc.M513222200 [DOI] [PubMed] [Google Scholar]
- Hargreaves, A.C., Lummis S.C., and Taylor C.W.. 1994. Ca2+ permeability of cloned and native 5-hydroxytryptamine type 3 receptors. Mol. Pharmacol. 46:1120–1128. [PubMed] [Google Scholar]
- Hassaine, G., Deluz C., Grasso L., Wyss R., Tol M.B., Hovius R., Graff A., Stahlberg H., Tomizaki T., Desmyter A., et al. 2014. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 512:276–281. 10.1038/nature13552 [DOI] [PubMed] [Google Scholar]
- Hibbs, R.E., and Gouaux E.. 2011. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 474:54–60. 10.1038/nature10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., Chen H., Michelsen K., Schneider S., and Shaffer P.L.. 2015. Crystal structure of human glycine receptor-α3 bound to antagonist strychnine. Nature. 526:277–280. 10.1038/nature14972 [DOI] [PubMed] [Google Scholar]
- Huang, X., Chen H., and Shaffer P.L.. 2017. Crystal structures of human GlyRα3 bound to ivermectin. Structure. 25:945–950. [DOI] [PubMed] [Google Scholar]
- Jansen, M., Bali M., and Akabas M.H.. 2008. Modular design of Cys-loop ligand-gated ion channels: functional 5-HT3 and GABA ρ1 receptors lacking the large cytoplasmic M3M4 loop. J. Gen. Physiol. 131:137–146. 10.1085/jgp.200709896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, S.P., Dunlop J.I., Kirkness E.F., Lambert J.J., and Peters J.A.. 2003. A cytoplasmic region determines single-channel conductance in 5-HT3 receptors. Nature. 424:321–324. 10.1038/nature01788 [DOI] [PubMed] [Google Scholar]
- Kouvatsos, N., Giastas P., Chroni-Tzartou D., Poulopoulou C., and Tzartos S.J.. 2016. Crystal structure of a human neuronal nAChR extracellular domain in pentameric assembly: Ligand-bound α2 homopentamer. Proc. Natl. Acad. Sci. USA. 113:9635–9640. 10.1073/pnas.1602619113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuska, J.L., Paulsen I.M., Belfield W.J., Martin I.L., Cole D.J., Holt A., and Dunn S.M.. 2014. Impact of intracellular domain flexibility upon properties of activated human 5-HT3 receptors. Br. J. Pharmacol. 171:1617–1628. 10.1111/bph.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracun, S., Harkness P.C., Gibb A.J., and Millar N.S.. 2008. Influence of the M3-M4 intracellular domain upon nicotinic acetylcholine receptor assembly, targeting and function. Br. J. Pharmacol. 153:1474–1484. 10.1038/sj.bjp.0707676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywkowski, K., Davies P.A., Feinberg-Zadek P.L., Bräuner-Osborne H., and Jensen A.A.. 2008. High-frequency HTR3B variant associated with major depression dramatically augments the signaling of the human 5-HT3AB receptor. Proc. Natl. Acad. Sci. USA. 105:722–727. 10.1073/pnas.0708454105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdell, S.J., Gee V.J., Harkness P.C., Doward A.I., Baker E.R., Gibb A.J., and Millar N.S.. 2005. RIC-3 enhances functional expression of multiple nicotinic acetylcholine receptor subtypes in mammalian cells. Mol. Pharmacol. 68:1431–1438. 10.1124/mol.105.017459 [DOI] [PubMed] [Google Scholar]
- Laverty, D., Desai R., Uchański T., Masiulis S., Stec W.J., Malinauskas T., Zivanov J., Pardon E., Steyaert J., Miller K.W., et al. 2019. Cryo-EM structure of the human α1β3γ2 GABAA receptor in a lipid bilayer. Nature. 565:516–520. 10.1038/s41586-018-0833-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe, R.J., Sumiya M., Summerfield J.A., and Turner M.W.. 1995. Distinct physicochemical characteristics of human mannose binding protein expressed by individuals of differing genotype. Immunology. 85:660–667. [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Ramanoudjame G., Liu D., Fox R.O., Jayaraman V., Kurnikova M., and Cascio M.. 2008. Overexpression and functional characterization of the extracellular domain of the human α1 glycine receptor. Biochemistry. 47:9803–9810. 10.1021/bi800659x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey, M.R., Cooper M.A., Deeb T.Z., Carland J.E., Kozuska J., Hales T.G., Lambert J.J., and Peters J.A.. 2008. Structural determinants of Ca2+ permeability and conduction in the human 5-hydroxytryptamine type 3A receptor. J. Biol. Chem. 283:19301–19313. 10.1074/jbc.M802406200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummis, S.C. 2012. 5-HT(3) receptors. J. Biol. Chem. 287:40239–40245. 10.1074/jbc.R112.406496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiulis, S., Desai R., Uchański T., Serna Martin I., Laverty D., Karia D., Malinauskas T., Zivanov J., Pardon E., Kotecha A., et al. 2019. GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature. 565:454–459. 10.1038/s41586-018-0832-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon, N.K., Reeves D.C., and Akabas M.H.. 2011. 5-HT3 receptor ion size selectivity is a property of the transmembrane channel, not the cytoplasmic vestibule portals. J. Gen. Physiol. 138:453–466. 10.1085/jgp.201110686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon, N.K., Bali M., and Akabas M.H.. 2012. Length and amino acid sequence of peptides substituted for the 5-HT3A receptor M3M4 loop may affect channel expression and desensitization. PLoS One. 7:e35563. 10.1371/journal.pone.0035563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, P.S., and Aricescu A.R.. 2014. Crystal structure of a human GABAA receptor. Nature. 512:270–275. 10.1038/nature13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, P.S., and Smart T.G.. 2010. Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol. Sci. 31:161–174. 10.1016/j.tips.2009.12.005 [DOI] [PubMed] [Google Scholar]
- Mnatsakanyan, N., Nishtala S.N., Pandhare A., Fiori M.C., Goyal R., Pauwels J.E., Navetta A.F., Ahrorov A., and Jansen M.. 2015. Functional chimeras of GLIC obtained by adding the intracellular domain of anion- and cation-conducting Cys-loop receptors. Biochemistry. 54:2670–2682. 10.1021/acs.biochem.5b00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, A.F., Mueller G.A., Zhong X., and Pedersen L.C.. 2010. A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 19:901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga-Cid, G., Yevenes G.E., Schmalzing G., Peoples R.W., and Aguayo L.G.. 2011. A single phenylalanine residue in the main intracellular loop of α1 γ-aminobutyric acid type A and glycine receptors influences their sensitivity to propofol. Anesthesiology. 115:464–473. 10.1097/ALN.0b013e31822550f7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Perez, C.L., Noviello C.M., and Hibbs R.E.. 2016. X-ray structure of the human α4β2 nicotinic receptor. Nature. 538:411–415. 10.1038/nature19785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni, M., Biro I., Giugliano M., Vijayan R., Biggin P.C., Beato M., and Sivilotti L.G.. 2011. Chloride ions in the pore of glycine and GABA channels shape the time course and voltage dependence of agonist currents. J. Neurosci. 31:14095–14106. 10.1523/JNEUROSCI.1985-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowrey, D.D., Cui T., Jia Y., Ma D., Makhov A.M., Zhang P., Tang P., and Xu Y.. 2013. Open-channel structures of the human glycine receptor α1 full-length transmembrane domain. Structure. 21:1897–1904. 10.1016/j.str.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishtala, S.N., Mnatsakanyan N., Pandhare A., Leung C., and Jansen M.. 2016. Direct interaction of the resistance to inhibitors of cholinesterase type 3 protein with the serotonin receptor type 3A intracellular domain. J. Neurochem. 137:528–538. 10.1111/jnc.13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nury, H., Bocquet N., Le Poupon C., Raynal B., Haouz A., Corringer P.J., and Delarue M.. 2010. Crystal structure of the extracellular domain of a bacterial ligand-gated ion channel. J. Mol. Biol. 395:1114–1127. 10.1016/j.jmb.2009.11.024 [DOI] [PubMed] [Google Scholar]
- Nury, H., Van Renterghem C., Weng Y., Tran A., Baaden M., Dufresne V., Changeux J.P., Sonner J.M., Delarue M., and Corringer P.J.. 2011. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 469:428–431. 10.1038/nature09647 [DOI] [PubMed] [Google Scholar]
- Pacheco, S., Gómez I., Sánchez J., García-Gómez B.I., Czajkowsky D.M., Zhang J., Soberón M., and Bravo A.. 2018. Helix α-3 inter-molecular salt bridges and conformational changes are essential for toxicity of Bacillus thuringiensis 3D-Cry toxin family. Sci. Rep. 8:10331. 10.1038/s41598-018-28753-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandhare, A., Grozdanov P.N., and Jansen M.. 2016. Pentameric quaternary structure of the intracellular domain of serotonin type 3A receptors. Sci. Rep. 6:23921. 10.1038/srep23921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandhare, A., Pappu A.S., Wilms H., Blanton M.P., and Jansen M.. 2017. The antidepressant bupropion is a negative allosteric modulator of serotonin type 3A receptors. Neuropharmacology. 113(Pt A):89–99. 10.1016/j.neuropharm.2016.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandhare, A., Stuebler A.G., Pirayesh E., and Jansen M.. 2019. A modified clear-native polyacrylamide gel electrophoresis technique to investigate the oligomeric state of MBP-5-HT3A-intracellular domain chimeras. Protein Expr. Purif. 153:45–52. 10.1016/j.pep.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papke, D., and Grosman C.. 2014. The role of intracellular linkers in gating and desensitization of human pentameric ligand-gated ion channels. J. Neurosci. 34:7238–7252. 10.1523/JNEUROSCI.5105-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phulera, S., Zhu H., Yu J., Claxton D.P., Yoder N., Yoshioka C., and Gouaux E.. 2018. Cryo-EM structure of the benzodiazepine-sensitive α1β1γ2S tri-heteromeric GABAA receptor in complex with GABA. eLife. 7:e39383. 10.7554/eLife.39383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polovinkin, L., Hassaine G., Perot J., Neumann E., Jensen A.A., Lefebvre S.N., Corringer P.J., Neyton J., Chipot C., Dehez F., et al. 2018. Conformational transitions of the serotonin 5-HT3 receptor. Nature. 563:275–279. 10.1038/s41586-018-0672-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers, K.K., Bu Z., Fleming K.G., Schatz D.G., Engelman D.M., and Coleman J.E.. 1996. A zinc-binding domain involved in the dimerization of RAG1. J. Mol. Biol. 260:70–84. 10.1006/jmbi.1996.0382 [DOI] [PubMed] [Google Scholar]
- Skinner, J.J., Wang S., Lee J., Ong C., Sommese R., Sivaramakrishnan S., Koelmel W., Hirschbeck M., Schindelin H., Kisker C., et al. 2017. Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome. Proc. Natl. Acad. Sci. USA. 114:13453–13458. 10.1073/pnas.1711543114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurny, R., Billen B., Howard R.J., Brams M., Debaveye S., Price K.L., Weston D.A., Strelkov S.V., Tytgat J., Bertrand S., et al. 2013. Multisite binding of a general anesthetic to the prokaryotic pentameric Erwinia chrysanthemi ligand-gated ion channel (ELIC). J. Biol. Chem. 288:8355–8364. 10.1074/jbc.M112.424507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, A.J., Lester H.A., and Lummis S.C.. 2010. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 43:449–499. 10.1017/S0033583510000168 [DOI] [PubMed] [Google Scholar]
- Unterer, B., Becker C.M., and Villmann C.. 2012. The importance of TM3-4 loop subdomains for functional reconstitution of glycine receptors by independent domains. J. Biol. Chem. 287:39205–39215. 10.1074/jbc.M112.376053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin, N. 2005. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J. Mol. Biol. 346:967–989. 10.1016/j.jmb.2004.12.031 [DOI] [PubMed] [Google Scholar]
- Verrall, S., and Hall Z.W.. 1992. The N-terminal domains of acetylcholine receptor subunits contain recognition signals for the initial steps of receptor assembly. Cell. 68:23–31. 10.1016/0092-8674(92)90203-O [DOI] [PubMed] [Google Scholar]
- Wells, G.B., Anand R., Wang F., and Lindstrom J.. 1998. Water-soluble nicotinic acetylcholine receptor formed by α7 subunit extracellular domains. J. Biol. Chem. 273:964–973. 10.1074/jbc.273.2.964 [DOI] [PubMed] [Google Scholar]
- Wen, J., Arakawa T., and Philo J.S.. 1996. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal. Biochem. 240:155–166. 10.1006/abio.1996.0345 [DOI] [PubMed] [Google Scholar]
- Yang, J. 1990. Ion permeation through 5-hydroxytryptamine-gated channels in neuroblastoma N18 cells. J. Gen. Physiol. 96:1177–1198. 10.1085/jgp.96.6.1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S., Noviello C.M., Teng J., Walsh R.M. Jr., Kim J.J., and Hibbs R.E.. 2018. Structure of a human synaptic GABAA receptor. Nature. 559:67–72. 10.1038/s41586-018-0255-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.