T helper 17 cells (Th17) are important for fighting mucosal infections and in the pathogenesis of autoimmune disease. This work identifies the activating transcription factor 7 interacting protein (ATF7ip) as an important factor controlling Th17 differentiation by inhibiting Il2 gene expression.
Abstract
T helper 17 cells (Th17) are critical for fighting infections at mucosal surfaces; however, they have also been found to contribute to the pathogenesis of multiple autoimmune diseases and have been targeted therapeutically. Due to the role of Th17 cells in autoimmune pathogenesis, it is important to understand the factors that control Th17 development. Here we identify the activating transcription factor 7 interacting protein (ATF7ip) as a critical regulator of Th17 differentiation. Mice with T cell–specific deletion of Atf7ip have impaired Th17 differentiation secondary to the aberrant overproduction of IL-2 with T cell receptor (TCR) stimulation and are resistant to colitis in vivo. ChIP-seq studies identified ATF7ip as an inhibitor of Il2 gene expression through the deposition of the repressive histone mark H3K9me3 in the Il2-Il21 intergenic region. These results demonstrate a new epigenetic pathway by which IL-2 production is constrained, and this may open up new avenues for modulating its production.
Introduction
Naive CD4+ T cells differentiate into effector T cells after they encounter antigen presented by antigen-presenting cells within the LN. There are at least five well-defined effector T cell lineages, including T helper 1 (Th1), Th2, T follicular helper cells, regulatory T cells (T reg cells), and Th17 (Zhu et al., 2010). Th17 cells are unique in their requirement to control pathogens at mucosal surfaces (Gaffen et al., 2014; Naglik et al., 2017). Th17 cells produce the cytokines IL-17A, IL-17F, and IL-22, which in turn act on epithelial cells and innate immune cells to help clear the infection. In addition to their function in the normal immune system, Th17 cells have been found to be critical in the pathogenesis of multiple autoimmune diseases (Liu et al., 2009; Shen et al., 2009,; Jadidi-Niaragh and Mirshafiey, 2011; Langley et al., 2014).
Over the last decade, multiple factors have been implicated in the development and inhibition of Th17 cells. Both in vitro and in vivo, the orphan nuclear receptor Rorγt transcription factor has been found to be critical for the development of Th17 cells (Ivanov et al., 2006). Multiple studies have shown that IL-2 is critical for the induction and maintenance of T reg cells (Fontenot et al., 2005; Setoguchi et al., 2005) while also inhibiting Th17 development (Laurence et al., 2007; Yang et al., 2011). Interestingly, while IL-2 inhibits Th17 development, it does not cause a dramatic decrease in the induction of Rorγt. Due to IL-2’s ability to promote immune tolerance, understanding the factors that control Il2 expression may have clinical relevance. One potential avenue to alter T cell Il2 production and CD4+ effector T cell differentiation would be to modulate the epigenetic state of the Il2-Il21 locus.
There has been a significant body of work characterizing the effect of specific repressive histone modifications on effector T cell development (Wang et al., 2016). While generation of the repressive H3K27me3 histone mark in T cells relies on one protein complex centered on the histone methyltransferase, EZH2, there are multiple protein complexes required for the generation of the repressive H3K9me3 histone mark (Schultz et al., 2002; Kimura, 2013; Bulut-Karslioglu et al., 2014). One open question in the field is whether proteins important in the formation of H3K9me3 histone marks modulate helper T cell differentiation. To this end, we sought to determine the potential role for activating transcription factor 7 interacting protein (ATF7ip) secondary to ATF7ip’s expression in the immune system and ATF7ip’s functional role in H3K9me3 formation.
ATF7ip (also known as MCAF1 or mAM) is an epigenetic regulator involved in gene repression through promoting the formation of the H3K9me3 mark (Wang et al., 2001). Through its interactions with binding partners such as the histone methyltransferase SETDB1/ESET (Wang et al., 2001; Timms et al., 2016), MBD1, and members of the human silencing hub complex (Fujita et al., 2003; Ichimura et al., 2005; Minkovsky et al., 2014; Tchasovnikarova et al., 2015), ATF7ip has been implicated in the regulation of gene expression programs in retroviral silencing, cellular senescence, cancer susceptibility, and immune tolerance (Turnbull et al., 2010; Sasai et al., 2013; Waterfield et al., 2014; Timms et al., 2016). At the molecular level, two different functions have been reported for ATF7ip: (1) as an essential cofactor in SETDB1 enzymatic activity and (2) in SETDB1 nuclear localization (Wang et al., 2001; Timms et al., 2016). To characterize the in vivo function of ATF7ip, we created a conditional KO mouse to allow Cre-mediated deletion of ATF7ip in specific cell types. Interestingly, we found that T cell–specific deletion of ATF7ip resulted in a defect in Th17 differentiation. Furthermore, global gene expression studies revealed that one cause of the Th17 defect in ATF7ip-deficient T cells is secondary to the increased production of IL-2. Chromatin immunoprecipitation sequencing (ChIP-seq) for H3K9me3 in naive T cells further refined the mechanism of increased IL-2 production by showing decreased deposition of H3K9me3 in the Il2-Il21 intergenic region. These studies reveal a new role for ATF7ip in promoting Th17 differentiation by epigenetic silencing of Il2 gene expression.
Results and discussion
ATF7ip inhibits in vitro and in vivo Th17 differentiation
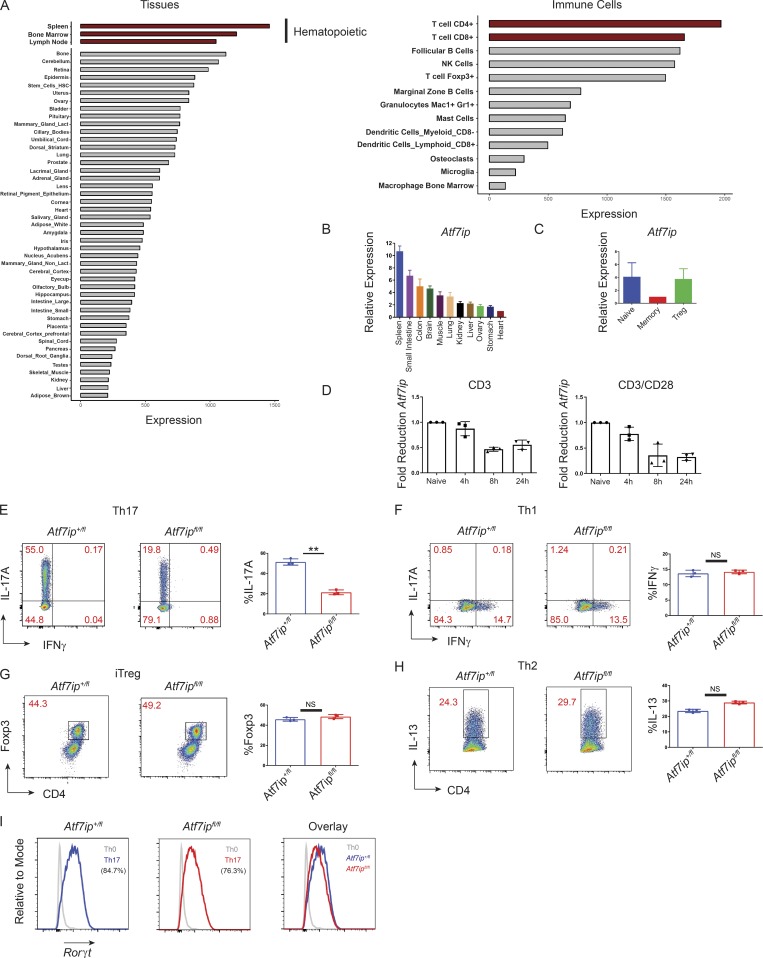
We hypothesized that ATF7ip may have a function in the immune system secondary to the significant expression of Atf7ip mRNA in the cells and tissues of the immune system in public RNA expression databases (BioGPS; Fig. 1 A). Expression profiling for Atf7ip mRNA in multiple tissues demonstrates the highest expression in the spleen (Fig. 1, A and B), with significant expression of Atf7ip in T and B cells (Fig. 1 A). Within CD4+ T cell subsets, Atf7ip was highly expressed in naive T cells (CD62L+CD44−CD25−) and T reg cells with lower expression in effector-memory CD4+ T cells (CD62L−CD44+; Fig. 1 C). Because a cascade of changes in gene expression occur with T cell activation (Liao et al., 2013), we examined whether Atf7ip expression levels change with T cell stimulation. Activation of the TCR of naive T cells (CD62L+CD44−CD25−) with anti-CD3 antibodies, alone or in combination with costimulation of the CD28 coreceptor for the TCR, caused a decrease in Atf7ip mRNA expression (Fig. 1 D), suggesting that the release of negatively regulated targets of ATF7ip may be important in T cell activation and action.
Figure 1.
ATF7ip is required for in vitro Th17 differentiation. (A) Relative expression of Atf7ip mRNA in tissues and immune cells downloaded from the BioGPS expression database. Red bars highlight hematopoietic cells or T cells. (B–D) Relative expression of Atf7ip mRNA in mouse tissues (B), CD4+ T cell subsets (C), or naive CD4+ T cells (D) stimulated with either anti-CD3 (2 µg) or both anti-CD3 (2 µg) and anti-CD28 (2 µg) for the indicated times. (E–H) Naive T cells from CD4-Cre/Atf7ip+/fl (Atf7ip+/fl) and CD4-Cre/Atf7ipfl/fl (Atf7ipfl/fl) mice were in vitro differentiated under Th17 (E), Th1 (F), iT reg cell (G), or Th2 (H) conditions and analyzed for intracellular cytokine or Foxp3 expression. (I) Rorγt protein expression under Th17 conditions. A histogram of Rorγt expression under Th0 conditions is included for reference. Each data point in D–H represents an individual mouse with three mice per genotype. Data are representative of five (E) or two (B–D and F–I) independent experiments. Error bars in B and C show mean with SEM of technical replicates. Error bars (D–H) show mean with SD. **, P < 0.01 by Student’s t test.
To interrogate the potential in vivo function of ATF7ip in T cells, a conditional deletion allele of mouse Atf7ip was created using a targeting construct surrounding exon 2 of Atf7ip with loxP Cre-recombination sites (Atf7ipfl/fl mice; Fig. S1, A and B). Deletion of Atf7ip in the germline resulted in embryonic lethality in mice (Fig. S1 C), consistent with previous work in Drosophila (Koch et al., 2009). To examine the function of ATF7ip in T cells, Atf7ipfl/fl mice were crossed to transgenic mice expressing Cre recombinase from the CD4 promoter (CD4-Cre). Quantitative PCR (qPCR) confirmed the deletion of Atf7ip mRNA in T cells (Fig. S1 D), and Western blot confirmed deletion of ATF7ip protein (Fig. S1 E). Extensive immune profiling in both CD4-Cre/Atf7ip+/fl T cells and CD4-Cre/Atf7ipfl/fl T cells indicated no differences in steady-state thymic T cell subsets (Fig. S1 F), T reg cells, or LN (Fig. S1 G)/spleen (not depicted) naive and activated/memory T cells. To interrogate T cell lineage commitment in the absence of ATF7ip, in vitro T cell differentiation assays were performed. Interestingly, naive T cells from CD4-Cre/Atf7ipfl/fl mice were unable to differentiate into IL-17A–producing Th17 cells despite normal induction of INFγ+ Th1 cells, IL-13+ Th2 cells, and Foxp3+ induced T reg cells (iT reg cells; Fig. 1, E–H). The Th17 differentiation defect did not appear to be due to a loss of RORγt (Fig. 1 I) or a defect in cell proliferation (Fig. S2, A and B). Taken together, these data indicated a role for ATF7ip in Th17 effector differentiation.
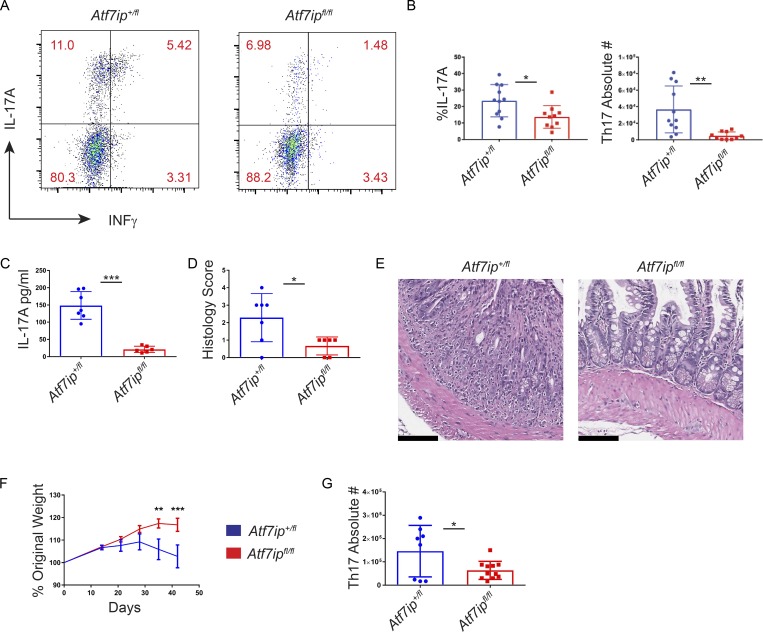
In vivo, Th17 cells have been implicated in the development of multiple autoimmune diseases (Patel and Kuchroo, 2015). In mice, one well-characterized model of Th17-mediated colitis is anti-CD3 colitis (Esplugues et al., 2011; Rutz et al., 2015). After injection of anti-CD3, CD4-Cre/Atf7ipfl/fl mice had a lower percentage and absolute number of IL-17A+ Th17 cells in the small intestine than CD4-Cre/Atf7ip+/fl mice (Fig. 2, A and B), with equal numbers of INFγ+ Th1 cells and Foxp3+ T reg cells in the mesenteric LNs (Fig. S2, C and E) and small intestine (Figs. 2 A and S2, D and F). Furthermore, serum from CD4-Cre/Atf7ipfl/fl mice revealed significantly less IL-17A than serum from CD4-Cre/Atf7ip+/fl mice (Fig. 2 C), and the diminished Th17 response in CD4-Cre/Atf7ipfl/fl mice correlated with less pathological intestinal inflammation (Fig. 2, D and E). The significance of ATF7ip in Th17 pathology was further confirmed in a second model using a T cell transfer model of colitis (Ostanin et al., 2009). Consistent with the results of the CD3-induced colitis model, we found that Rag1−/− mice that received RBhi CD4-Cre/Atf7ipfl/fl T cells did not lose weight, had reduced numbers of Th17 cells in the colon, and had less intestinal pathology (Fig. 2, F and G; and Fig. S2, G and H). Collectively, these data suggest that ATF7ip is required for in vivo Th17-mediated inflammation.
Figure 2.
Atf7ip deletion attenuates colitis in vivo. (A–E) CD4-Cre/Atf7ip+/fl (Atf7ip+/fl) and CD4-Cre/Atf7ipfl/fl (Atf7ipfl/fl) mice were injected intraperitoneal with anti-CD3 antibodies and analyzed 48 h after injection. (A) Flow cytometric analysis of CD4+IL17A+INFγ+ small intestine IELs. (B) Percentage and absolute numbers intraepithelial CD4+ T cells in the small intestine expressing IL-17A. (C) Serum IL-17A levels measured by ELISA. (D) Histological score of the small intestine. (E) H&E staining of the small intestine. Bars, 100 µm. (F) Weight change in Rag1−/− recipients of RBhi naive T cells from either CD4-Cre/Atf7ip+/fl or CD4-Cre/Atf7ipfl/fl mice measured on day 0, 14, 21, 28, 35, and 42. Results are the combination of two experiments with 13 mice per genotype. (G) Absolute number of CD4+IL17A+ T cells in the colonic lamina propria of Rag1−/− mice. Each data point in B–D and G represents an individual mouse. Data are the combination of three (B) or two (C, D, F, and G) independent experiments with three to four mice per group in each experiment. Error bars in B–D and G are mean with SD. Error bars in F are SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; significance by Student’s t test (B, C, and G); Mann–Whitney nonparametric test (D); and two-way ANOVA followed by multiple t tests using the Holm–Sidak method (F).
ATF7ip-deficient T cells produce increased IL-2 under Th17-inducing conditions
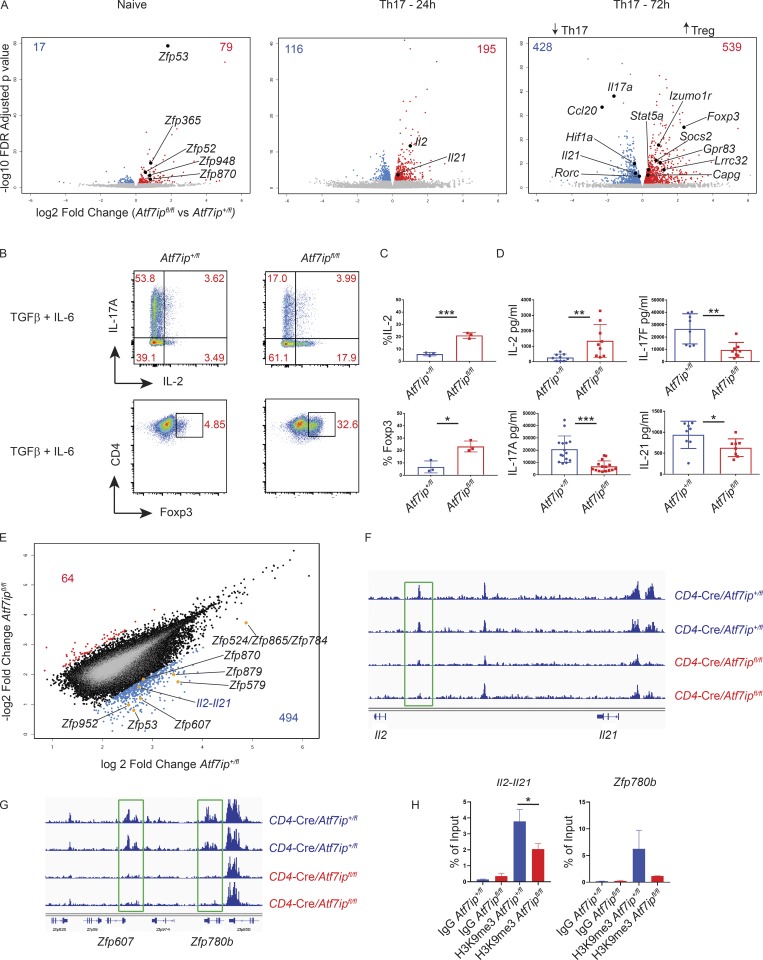
We next turned to the mechanism by which ATF7ip promotes appropriate Th17 differentiation. Secondary to ATF7ip’s role in the formation of the H3K9me3 histone mark, we hypothesized that deletion of Atf7ip in T cells may allow for the aberrant overexpression of an inhibitor of Th17 differentiation. To gain insight into the altered gene regulation in CD4-Cre/Atf7ipfl/fl T cells, global gene expression analysis with RNA sequencing (RNA-seq) was performed in naive T cells and T cells in vitro differentiated under Th17-polarizing conditions for 24 and 72 h to capture early and late changes in gene expression (Fig. S3 A). Consistent with ATF7ip’s function in gene repression, there were more genes up-regulated in CD4-Cre/Atf7ipfl/fl T cells than CD4-Cre/Atf7ip+/fl T cells in all three RNA-seq conditions (Fig. 3 A). A previous study of ATF7ip deletion in HeLa cells implicated ATF7ip in silencing zinc finger proteins (Timms et al., 2016). In line with this study, CD4-Cre/Atf7ipfl/fl naive T cells had several zinc finger protein genes that were significantly overexpressed (false discovery rate [FDR] <0.01) compared with CD4-Cre/Atf7ip+/fl naive T cells (Fig. 3 A). Interestingly, at 24 h of Th17-inducing conditions, CD4-Cre/Atf7ipfl/fl T cells expressed more Il2 mRNA than CD4-Cre/Atf7ip+/fl T cells (Fig. 3 A). Consistent with IL-2’s known function in the maintenance of T reg cells. At 72 h of Th17-inducing conditions, CD4-Cre/Atf7ipfl/fl T cells had a gene signature more in line with T reg cells, with a significant increase in Foxp3 expression along with several genes associated with the T reg cell lineage (Lrrc32, Socs2, Izumo1r, and Capg) and IL-2 signaling (Stat5a; Fig. 3 A; Zemmour et al., 2018). Moreover, CD4-Cre/Atf7ipfl/fl T cells compared with CD4-Cre/Atf7ip+/fl T cells had a significant decrease in many Th17-associated genes (Il17a, Ccl20, Il21, Hif1a, and Rorc; Fig. 3 A). To confirm these findings, we also performed qPCR on differentially expressed genes from the RNA-seq data along with other genes in the Th17 pathway (Fig. S3 B). Consistent with the modest decrease in Rorγt protein expression in CD4-Cre/Atf7ipfl/fl T cells (Fig. 1 I), qPCR did not reveal a decrease in Rorc at early time points.
Figure 3.
CD4-Cre/Atf7ipfl/fl T cells have increased Il2 and a decreased Th17 gene signature secondary to less H3K9me3 at the Il2-Il21 intergenic region. (A) Volcano plots from RNA-seq data comparing global gene expression analysis of T cells from CD4-Cre/Atf7ipfl/fl mice and CD4-Cre/Atf7ip+/fl mice. Left plot represents naive T cells, middle plot is T cells differentiated for 24 h under Th17-inducing conditions, and right plot is 72 h of Th17-inducing conditions. Red numbers indicate the number of genes that are significantly increased (FDR <0.01) in CD4-Cre/Atf7ipfl/fl T cells, and blue numbers indicate the number of genes that are significantly increased in CD4-Cre/Atf7ip+/fl T cells. RNA-seq was performed in triplicate for each condition. (B) Naive T cells from CD4-Cre/Atf7ip+/fl (Atf7ip+/fl) and CD4-Cre/Atf7ipfl/fl (Atf7ipfl/fl) mice were in vitro differentiated for 4 d under Th17-inducing conditions (IL-6 + TGFβ) and analyzed by flow cytometric analysis for intracellular cytokines (IL-17A, IL-2) or Foxp3. (C) Summary of flow cytometric data in B. (D) ELISA of secreted cytokines (IL-17A, IL-2, IL-17F, and IL-21) from Th17 culture supernatant. (E–G) ChIP-seq data for H3K9me3 from CD4-Cre/Atf7ip+/fl (Atf7ip+/fl) and CD4-Cre/Atf7ipfl/fl (Atf7ipfl/fl) naive T cells performed in duplicate. (E) Scatterplot comparing log2 fold change of H3K9me3 in CD4-Cre/Atf7ip+/fl (Atf7ip+/fl) and CD4-Cre/Atf7ipfl/fl (Atf7ipfl/fl) naive T cells. Blue numbers represent the number of loci with twofold increased H3K9me3 in CD4-Cre/Atf7ip+/fl naive T cells compared with CD4-Cre/Atf7ipfl/fl naive T cells. Red numbers indicate the number of loci with twofold increased H3K9me3 in CD4-Cre/Atf7ipfl/fl naive T cells. (F) Integrated genome viewer H3K9me3 ChIP-seq tracings for the Il2-Il21 intergenic region. (G) Integrated genome viewer H3K9me3 ChIP-seq tracings for the indicated zinc finger proteins (Zfp). Green boxes show sites of twofold decreased H3K9me3 deposition. (H) H3K9me3 ChIP qPCR targeting the site of H3K9me3 deposition within the Il2-Il21 intergenic region and within the Zfp780b gene. Each data point represents an individual mouse. Data in B and C are one representative experiment of three experiments with three mice per group. Data in D are the combination of three experiments with two to three mice per group. Error bars are mean with SD (C and D) and mean and SD of technical replicates (H). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student’s t test.
We next sought to determine if these transcriptional changes also correlated with altered protein production. First, we performed intracellular cytokine staining (ICS) for IL-2 in differentiating T cells. As shown in Fig. 3 B, we found that under Th17-inducing conditions, CD4-Cre/Atf7ipfl/fl T cells produce more IL-2 and less IL-17A than CD4-Cre/Atf7ip+/fl T cells. Furthermore, CD4-Cre/Atf7ipfl/fl T cells have increased intracellular Foxp3 and secrete increased IL-2 and less IL-17A, IL-17F, and IL-21 as measured by ELISA (Fig. 3, B–D). Alternative induction of Th17 cells can be also achieved by the combination of IL-6, IL-23, and IL-1 to produce pathogenic Th17 cells, so named due to their role in neuroinflammation (Ghoreschi et al., 2010). CD4-Cre/Atf7ipfl/fl T cells cultured with IL-6, IL-1, and IL-23 produce less IL-17A/IL-17F and increased IL-2 by ICS and ELISA (Fig. S3, C–E). Altogether, these data indicate that under a variety of Th17-inducing conditions, CD4-Cre/Atf7ipfl/fl T cells produce increased IL-2 and decreased IL-17A.
ATF7ip is required for H3K9me3 deposition at the Il2-Il21 intergenic region
To gain further insight into the mechanism of increased IL-2 production in CD4-Cre/Atf7ipfl/fl T cells, we next explored the potential epigenetic effect of ATF7ip broadly and on the Il2 locus by performing ChIP-seq on T cells using an antibody directed to the H3K9me3 mark. H3K9me3 ChIP-seq was performed in duplicate on naive T cells from both CD4-Cre/Atf7ip+/fl mice and CD4-Cre/Atf7ipfl/fl mice and yielded ∼67,000 H3K9me3 active sites in both genotypes, indicating that there was not a profound reduction of H3K9me3 in naive CD4-Cre/Atf7ipfl/fl T cells (Fig. S3 F). In agreement with the ChIP-seq data, Western blot did not reveal a reduction of H3K9me3 in CD4-Cre/Atf7ipfl/fl T cells compared with CD4-Cre/Atf7ip+/fl T cells (Fig. S3 G).
Although there was not a global difference in H3K9me3, hierarchical clustering showed that the samples clustered by genotype (Fig. S3 H). Interestingly, there were 494 sites with a twofold increase in H3K9me3 deposition in CD4-Cre/Atf7ip+/fl naive T cells compared with CD4-Cre/Atf7ipfl/fl naive T cells, while there were only 64 sites with a twofold increase in CD4-Cre/Atf7ipfl/fl naive T cells (Fig. 3 E). These results support a role for ATF7ip in promoting the formation of the H3K9me3 mark at specific genomic locations in naive T cells. Consistent with prior work in HeLa cells, a large number of zinc finger genes had a significant decrease in H3K9me3 in CD4-Cre/Atf7ipfl/fl naive T cells relative to CD4-Cre/Atf7ip+/fl T cells (Fig. 3, E and G). Moreover, there was also a site of H3K9me3 deposition of ∼1,200 bp in length located on chromosome 3 with a twofold decrease of H3K9me3 (Fig. 3 E). This site is within the Il2-Il21 intergenic region, and ChIP-seq tracings showed a reduction of the H3K9me3 histone mark in this region (Fig. 3 F). H3K9me3 ChIP qPCR confirmed that there was less H3K9me3 within this region (Fig. 3 H). Consistent with increased IL-2 signaling, RNA-seq showed increased Stat5 gene expression in CD4-Cre/Atf7ipfl/fl T cells differentiated for 72 h in Th17-inducing conditions (Fig. 3 A). The increased Stat5 gene expression is unlikely secondary to direct regulation of Stat5 because H3K9me3 ChIP-seq did not show a reduction of H3K9me3 in the Stat5a or Stat5b locus (Fig. S3 I). Together, these results suggest that naive CD4-Cre/Atf7ipfl/fl T cells may be primed to produce more IL-2 secondary to decreased repression by H3K9me3.
ATF7ip-deficient T cells produce increased IL-2 with TCR stimulation
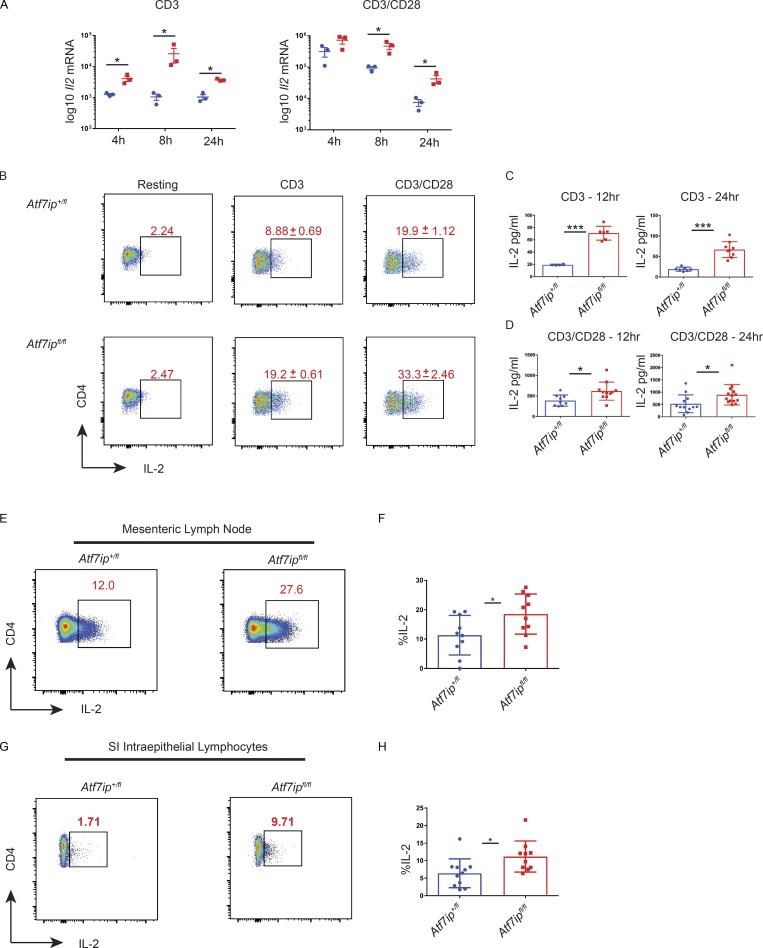
Our ChIP-seq data suggested that ATF7ip might inhibit Il2 gene transcription through the deposition of H3K9me3 in the Il2-Il21 intergenic region. The data shown indicate increased IL-2 production in CD4-Cre/Atf7ipfl/fl T cells under Th17-inducing conditions; however, we hypothesized that CD4-Cre/Atf7ipfl/fl T cells would produce increased IL-2 solely with TCR/CD28 engagement, as naive CD4-Cre/Atf7ipfl/fl T cells have less H3K9me3 in the Il2-Il21 intergenic region. Treatment of CD4-Cre/Atf7ipfl/fl T cells with anti-CD3 or anti-CD3/anti-CD28 resulted in increased levels of Il2 mRNA and IL-2 protein as measured by ICS and ELISA (Fig. 4, A–D). Furthermore, in vivo treatment of CD4-Cre/Atf7ipfl/fl mice with anti-CD3 resulted in an increased percentage and number of IL-2–producing T cells in the mesenteric LN and small intestine intraepithelial lymphocytes (IELs; Fig. 4, E–H).
Figure 4.
CD4-Cre/Atf7ipfl/fl T cells produce increased IL-2 with TCR stimulation. Naive T cells were stimulated for 12–24 h in the presence of TCR stimulation (2 µg anti-CD3) or with both TCR stimulation (2 µg anti-CD3) and costimulation (2 µg anti-CD28). (A) qPCR for Il2 mRNA. (B) Flow cytometric data for IL-2 with ± SD shown. (C and D) IL-2 ELISA from culture supernatant. (E and G) Representative flow cytometry of IL-2 expression in the mesenteric LN (E) or small intestine IELs (G) 48 h after in vivo anti-CD3 treatment. (F and H) Summary of flow cytometry data from the mesenteric LN (E) and small intestine (SI) IELs (G). Each data point represents an individual mouse. Data in A and B are representative of two experiments with three mice per genotype. Data in C, D, F, and H are the combination of three experiments with three to four mice per genotype. Error bars are SEM (A) and mean with SD (B–D, F, and H). *, P < 0.05; ***, P < 0.001 by Student’s t test.
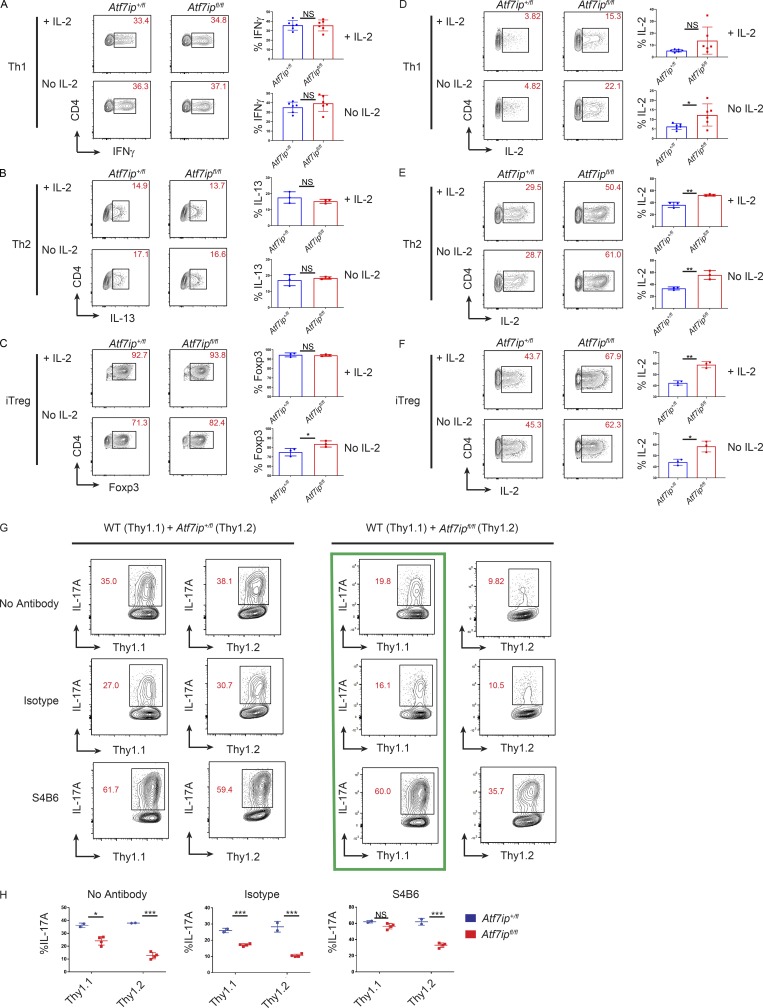
To determine if the increased IL-2 production by CD4-Cre/Atf7ipfl/fl T cells would affect other CD4+ T h cell lineages, we performed in vitro T cell differentiation assays in the presence or absence of IL-2. Interestingly, there was equally effective induction of INFγ+ Th1 cells (Fig. 5 A) and IL-13+ Th2 cells (Fig. 5 B) with or without the addition of IL-2. These results indicate that both CD4-Cre/Atf7ip+/fl T cells and CD4-Cre/Atf7ipfl/fl T cells produce sufficient IL-2 for effective Th1 and Th2 differentiation. In the absence of IL-2, iT reg cell induction was increased in CD4-Cre/Atf7ipfl/fl T cells compared with CD4-Cre/Atf7ip+/fl T cells, consistent with IL-2’s known role in T reg cell function (Fig. 5 C). Prior studies have shown that the cytokines IL-2, IL-4, and IL-12 can negatively regulate IL-2 production, and this inhibition is dependent on Stat5 (Villarino et al., 2006, 2007). As human IL-2 was added to Th1, Th2, and iT reg cell culture conditions, we expected to see an inhibition of endogenous IL-2 production during differentiation; however, CD4-Cre/Atf7ipfl/fl T cells produced increased Il2 mRNA (Fig. S3 J) and IL-2 protein (Fig. 5, D–F) compared with CD4-Cre/Atf7ip+/fl T cells during in vitro differentiation into the Th1, Th2, and iT reg cell lineages. These results indicate that CD4-Cre/Atf7ipfl/fl T cells are able to at least partially overcome cytokine repression of Il2 gene expression and further support our data that the Il2 locus in CD4-Cre/Atf7ipfl/fl T cells is in a less repressed conformation.
Figure 5.
iT reg cell induction is augmented in Atf7ipfl/fl T cells and Atf7ipfl/fl T cells suppress IL-17A production in trans. (A–C) Naive T cells from CD4-Cre/Atf7ip+/fl (Atf7ip+/fl) and CD4-Cre/Atf7ipfl/fl (Atf7ipfl/fl) mice were in vitro differentiated in the presence (+IL-2) or the absence (no IL-2) of IL-2 under Th1 (A), Th2 (B), or iT reg cell (C) conditions and analyzed for intracellular cytokine or Foxp3 expression. (D–F) Naive T cells from CD4-Cre/Atf7ip+/fl (Atf7ip+/fl) and CD4-Cre/Atf7ipfl/fl (Atf7ipfl/fl) mice were in vitro differentiated in the presence (+IL-2) or the absence (no IL-2) of IL-2 under Th1 (D), Th2 (E), or iT reg cell (F) conditions and analyzed for IL-2 expression. (G and H) WT (Thy1.1) naive T cells were mixed at a 50:50 ratio with either CD4-Cre/Atf7ip+/fl (Thy1.2) or CD4-Cre/Atf7ipfl/fl (Thy1.2) naive T cells and cultured for 96 h under Th17-inducing conditions with the addition of no IL-2 blocking antibody (No Antibody), 5 µg isotype control antibody (Isotype), or 5 µg S4B6. (G) Flow cytometric analysis of T cells expressing Thy1.1, Thy1.2, and IL-17A. Green box shows that S4B6 is able to rescue the trans defect in IL-17A production. (H) Summary of flow cytometric data. Each data point represents an individual mouse. Each data point in A–F and H represents an individual mouse. Data are representative of two independent experiments (A and D) or one of two experiments (B, C, E, and F) with three mice per group; data in G and H are representative of two experiments with two to four mice per genotype. Error bars (A–F and H) show mean with SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student’s t test.
Finally, we attempted to confirm that the defect in Th17 generation in CD4-Cre/Atf7ipfl/fl T cells was secondary to aberrant IL-2 production by performing cell mixing and IL-2 blocking studies. An in vitro Th17 differentiation was performed, mixing WT T cells homozygous for the congenic marker Thy1.1 with equal numbers of either CD4-Cre/Atf7ipfl/fl T cells or CD4-Cre/Atf7ip+/fl T cells homozygous for the congenic marker Thy1.2. In support of a secreted factor causing the Th17 defect, CD4-Cre/Atf7ipfl/fl T cells were able to suppress IL-17A production in trans from WT T cells, whereas CD4-Cre/Atf7ip+/fl T cells had no effect on IL-17A production from WT T cells (Fig. 5, G and H). To confirm that IL-2 is the cause of the Th17 defect, coculture experiments were performed in the presence of the IL-2 blocking antibody S4B6 or an isotype control antibody (Fig. 5, G and H). In agreement with previous studies, IL-2 blockade with S4B6 increased the percentage of IL-17A–producing T cells in all conditions (Laurence et al., 2007). Interestingly, treatment with S4B6 was able to rescue the trans IL-17A defect seen in WT T cells cocultured with CD4-Cre/Atf7ipfl/fl T cells, while the isotype control antibody had no effect (Fig. 5 G, green box). This result further implicates increased secretion of IL-2 as a potential mechanism for the Th17 defect seen in CD4-Cre/Atf7ipfl/fl T cells. While IL-2 blockade was able to rescue the trans defect, S4B6 treatment was unable to completely rescue IL-17A production directly in Thy1.2+CD4-Cre/Atf7ipfl/fl T cells. The partial rescue is likely due to IL-2’s known autocrine function and the inability of the blocking antibody to act before IL-2 binding its receptor. Together, these results implicate increased IL-2 production as one potential mechanism for the Th17 defect seen in CD4-Cre/Atf7ipfl/fl T cells.
This work establishes a new function for ATF7ip in silencing Il2 gene expression in T cells through promoting the deposition of H3K9me3 in the Il2-Il21 intergenic region. Prior work has shown a role for H3K9me3 in silencing II2 in both anergic T cells (Bandyopadhyay et al., 2012) and Th17 cells through the transcription factor Aiolos (Quintana et al., 2012); however, this study is the first to implicate H3K9me3 in silencing Il2 gene expression in naive T cells. As Atf7ip mRNA levels decrease with TCR stimulation, we propose a model by which loss of Atf7ip results in decreased H3K9me3 at the Il2 locus and a more open chromatin conformation allowing the robust IL-2 production necessary for a productive immune response. In ATF7ip-deficient T cells, Il2 gene expression is not effectively silenced, causing the overproduction of IL-2 and the in vitro inhibition of Th17 differentiation. Moreover, the enhanced IL-2 production in ATF7ip-deficient T cells and decreased IL-21 production (Fantini et al., 2007) caused CD4-Cre ATF7ipfl/fl mice to be resistant to Th17-mediated colitis in vivo. IL-2 has previously been reported as an inhibitor of Th17 differentiation through the direct action of STAT5 on the Il17a locus (Laurence et al., 2007; Yang et al., 2011). Interestingly, these studies showed that while IL-2 caused a Th17 defect, there was not a significant decrease in Rorγt protein. These results are consistent with our data showing only a modest defect in Rorγt production in ATF7ip-deficient T cells.
Secondary to the overproduction of IL-2, CD4-Cre ATF7ipfl/fl mice might have been expected to have increased numbers of T reg cells. Under steady-state conditions, we found no difference in the percentage or absolute number of T reg cells in CD4-Cre ATF7ip+/fl mice compared with CD4-Cre ATF7ipfl/fl mice. We saw a trend for increased T reg cells in the small intestine and mesenteric LN of ATF7ip-deficient mice during anti-CD3 colitis; however, this did not reach statistical significance. ATF7ip deficiency may increase T reg cell numbers through IL-2 overproduction in other inflammatory conditions or autoimmune prone mice, and future studies will examine these possibilities.
In CD4+ T cells, ATF7ip is important in regulating the Il2-Il21 intergenic region along with multiple other genomic regions. RNA-seq and ChIP-seq performed in ATF7ip-deficient T cells show that ATF7ip is critical for silencing zinc finger proteins in addition to Il2. A study of ATF7ip-deficient HeLa cells also found that ATF7ip regulated zinc finger proteins, suggesting that ATF7ip has similar functions in diverse cell types (Timms et al., 2016). ATF7ip partners with the histone methyltransferase SETDB1, and the two proteins exert similar effects in HeLa cells with regard to the silencing of zinc finger proteins (Timms et al., 2016). Interestingly, T cell–specific deletion of SETDB1 causes a block in thymic T cell development (Takikita et al., 2016) and augmented Th1 priming (Adoue et al., 2019), while our data show no effect on thymic T cell development or Th1 differentiation. These results imply nonredundant roles for SETDB1 and ATF7ip, and further work will be needed to parse the differential roles of these two epigenetic factors in T cells.
Our work clearly shows that one locus critically regulated by ATF7ip is the Il2-Il21 intergenic region. IL-2 is a pleotropic cytokine with effects in both effector T cell differentiation and the prevention of autoimmunity. Small nucleotide polymorphisms found in the Il2-Il21 intergenic region are linked to multiple autoimmune diseases, including rheumatoid arthritis (Zhernakova et al., 2007), lupus (Sawalha et al., 2008), inflammatory bowel disease (Festen et al., 2009), celiac disease (van Heel et al., 2007), multiple sclerosis (Matesanz et al., 2001), Graves’ disease (Todd et al., 2007), and type 1 diabetes (Cooper et al., 2008). These small nucleotide polymorphisms are hypothesized to lead to attenuated IL-2 production and a susceptibility to autoimmune disease. Due to the possibility that low IL-2 levels play a part in the pathogenesis of autoimmunity, low dose IL-2 therapy has gained momentum as a treatment for multiple autoimmune diseases secondary to its positive effect on T reg cells and its inhibition of Th17 and T follicular helper cells (Klatzmann and Abbas, 2015; Abbas et al., 2018). Furthermore, inhibitors of specific epigenetic pathways are in clinical trials for cancer (Biswas and Rao, 2018), and thus one could envision modulating epigenetic pathways to target autoimmunity. Future work will determine if inhibitors of ATF7ip could be used therapeutically to enable T cells to overproduce IL-2 and potentially alleviate autoimmune disease.
Materials and methods
Mice
Atf7ipfl/fl mice were generated by creating a targeting vector using the pEasyflox-DTA backbone (gift of Dr. Ralf Kuhn, Institute for Developmental Genetics, Helmholtz Center Munich, German Research Center for Environmental Health, Munich, Germany). Homology arms were PCR amplified from bacterial artificial chromosome C57BL/6 ES cell genomic DNA obtained from the Children’s Hospital Oakland Research Institute. The 5′ arm was amplified using primers 5′-GTAGGTGAAATGGTTTGAGAG and 3′-GACTTTTCACGCCCCTGAAG to amplify a 5,026-bp PCR product. The 3′-arm was amplified using primer’s 5′-GTCCATCTGGGGACACGCTGTTG and 3′-GACTCAGCAGTGCTGCTTGC to amplify a 3,294-bp PCR product. The critical exon was amplified using primers 5′-CGGCTCCTTTTTAACTCTTC and 3′-GAAGAACTAATGTGTCGTC to amplify a 2,171-bp product. PCR products were sequenced and then cloned into pEasyflox-DTA using In-Fusion (Takara) one-step cloning. The targeting construct was linearized with Not1 and electroporated into JM8A3.N1 (C57BL/6-derived) ES cell line by the University of California San Francisco (UCSF) ES Cell Targeting Core. ES cell colonies were digested with BamHI, and colonies were screened by Southern blot using a 370-bp probe amplified using primers 5′-GTGATGTTTCTCCCAGCGTG and 3′-TTCCTTTGACTTCCACCGTG. Targeted ES cells were microinjected into blastocysts by the UCSF Gladstone Transgenic Gene-Targeting Core. Male chimeras were bred to C57BL/6 females (the Jackson Laboratory; #000664), and agouti pups were screened by PCR for germline transmission. The neomycin cassette was removed by breeding to B6.Cg-Tg(Pgk1-flpo)10Sykr/J, a mouse strain that expresses flippase recombinase under control of the phosphoglycerate kinase 1 gene (the Jackson Laboratory; #011065). B6.Cg-Tg(Zp3-cre)1Gwh/J were purchased from the Jackson Laboratory (#006888). CD4-Cre and Foxp3-GFP mice were provided by Jeff Bluestone (University of California, San Francisco, San Francisco, CA). B6.Thy1.1 mice were purchased from the Jackson Laboratory (#000406). B6.129S7-Rag1tm1Mom/J were purchased from the Jackson Laboratory (#002216). Mice were maintained in the UCSF specific pathogen–free animal facility in accordance with the guidelines established by the Institutional Animal Care and Use Committee and Laboratory Animal Resource Center, and all experimental procedures were approved by the Laboratory Animal Resource Center at UCSF.
T cell differentiation
T cells were enriched from spleen and LNs using the MagniSort CD4 negative selection kit (Thermo Fisher Scientific). Naive CD4+ T cells were isolated by flow cytometry based on the markers CD4+CD62L+CD44−CD25− or using the Easysep mouse naive T cell isolation kit. 105 naive T cells were cultured for 4 d (Th1, Th17, iT reg cell) or 5 d (Th2) in a 96-well flat-bottom plate coated with 2 µg/ml anti-CD3 (clone 2C11; Tonbo) and 2 µg/ml anti-CD28 (clone 37.51; Tonbo) with the relevant cytokines and blocking antibodies: Th17 (20 ng/ml IL-6, 2 ng/ml TGFβ, 10 µg/ml anti-IL4 [clone 11B11; Tonbo], and 10 µg/ml anti-INFγ [clone XMG1.2; Tonbo]), pathogenic Th17 (20 ng/ml IL-6, 20 ng/ml IL-1β, 20 ng/ml IL-23, 10 µg/ml anti-IL4, and 10 µg/ml anti-INFγ), Th1 (20 ng/ml IL-12, 100 U/ml IL-2, and 10 µg/ml anti-IL4), iT reg cell (20 ng/ml TGFβ and 100 U/ml IL-2), Th2 (500 U/ml IL-4 as supernatant from I3L6 cells [Tepper et al., 1989] or 20 ng/ml mouse IL-4, 100 U/ml IL-2, and 10 µg/ml anti-INFγ), or Th0 (100 U/ml IL-2). After 3 d of culture, T cells cultured under Th2 conditions were removed from stimulation with anti-CD3/anti-CD28 and cultured for an additional 2 d with fresh cytokines. Th17 cultures were performed in Iscove’s medium, Th1/iT reg cell cultures were performed in RPMI, and Th2 cultures were performed in Dulbecco’s medium. All media were supplemented with 10% FBS, penicillin/streptomycin, glucose, pyruvate, β-mercaptoethanol, and Hepes. Cytokines were purchased from R&D Systems (murine IL-4, murine IL-6, and human IL-2), Miltenyi (murine IL-12, murine IL-1β, and murine IL-23), or HumanKine (human TGFβ). For coculture experiments with congenically labeled Thy1.1 and Thy1.2 T cells, 5 µg of isotype control antibody (clone ebioRT IgG2A; Invitrogen) or 5 µg anti-IL2 (Clone eBio S4B6; Invitrogen) was added to cultures.
Flow cytometry and intracellular staining
LN and spleen were isolated by dissection from mice and then mashed through a 70-µm filter. Spleen cells were lysed in ammonium-chloride-potassium lysis buffer to remove red blood cells. Cells isolated from spleen and LNs were counted, and 1–5 × 106 cells were first stained in PBS and Ghost Live/Dead (Tonbo), followed by blocking in 24G2 before staining with the appropriate antibodies for flow cytometry. For transcription factor staining, cells were fixed overnight in the eBioscience Foxp3/Transcription Factor/Fixation-Concentrate kit (Thermo Fisher Scientific). After fixation, cells were permeabilized and stained with the appropriate antibodies. For ICS, cells were stimulated for 4 h in Brefeldin A (eBioscience) and eBioscience Cell Stimulation Cocktail (500×). Cells were then fixed and permeabilized using the BD cytofix/cytoperm kit before staining with the appropriate antibodies.
Antibodies used for flow-cytometry were as follows: PE-Cy7-conjugated CD4 (clone RM4-5; Tonbo), PE-Cy7-conjugated B220 (clone RA3-6B2; BioLegend), PE-Cy7-conjugated CD62L (clone MEL-14; Tonbo), PE-Cy7-conjugated CD45 (clone 30-F11; Invitrogen), PE-conjugated IL-17A (clone eBio17B7; Invitrogen), PE-conjugated CD62L (clone MEL-14; Tonbo), PE-conjugated CD44 (clone IM7; BioLegend), PE-conjugated CD8 (clone 53-6.7; Tonbo), PE-conjugated RORγ (Q31-378; BD Biosciences), PE-conjugated IL-13 (clone eBio13A; Invitrogen), FITC-conjugated Thy1.2 (clone 53-2.1; BD Biosciences), FITC-conjugated IFNγ (clone xMG1.2; Invitrogen), FITC-conjugated Foxp3 (clone FJK-16s; Invitrogen), FITC-conjugated CD44 (clone IM7; BioLegend), FITC-conjugated CD8 (clone 53-6.7; Tonbo), allophycocyanin (APC)-conjugated Thy1.1 (clone OX-7; BioLegend), APC-conjugated CD25 (clone PC61.5; Tonbo), APC-conjugated CD4 (clone GK1.5; BioLegend), APC-conjugated IL-2 (clone JES6-5H4; Invitrogen), APC-conjugated CD62L (clone MEL-14; Tonbo), PerCP-conjugated CD45 (clone 30-F11; Invitrogen), eFluor 450-conjugated TCRβ (clone H57-597; Tonbo), eFluor 450-conjugated CD8 (clone 53-6.7; Tonbo), PerCP-Cy5.5-conjugated CD4 (clone GK1.5; BioLegend), Ghost UV 450 (Tonbo), and Ghost 510 (Tonbo).
Colitis
Anti-CD3 colitis was performed by injecting 50 µg of anti-CD3 (clone 145-2C11; Tonbo) intraperitoneally. 48 h after injection, mice were euthanized, and blood was collected by cardiac puncture, tissue was collected for H&E histology, and IELs were isolated for flow cytometry. Sample sizes of three to four mice per genotype were used for each experimental replicate. No mice were excluded from the study. Rag colitis was performed by injecting CD4+RBhiCD25−CD44− T cells into Rag1−/− hosts (4 × 105/200 µl PBS). Weight was monitored weekly for 6 wk. Lamina propria lymphocytes were isolated for flow cytometry, and tissue was collected for histology. Histology for colitis was scored blinded using the following scoring system (Rutz et al., 2015): 0, no immune cell infiltration; 1, minimal inflammation with <10% infiltration of the mucosa; 2, mild inflammation, 10–25% of the mucosa infiltrated; 3, moderate inflammation, 26–50% infiltration; 4, extensive infiltration, 51–75% affected; and 5, diffuse infiltration, >75% of the mucosa infiltrated.
Isolation of small intestine IELs
Peyer’s patches and fat were removed from the small intestine, and the intestine was cut longitudinally and then into pieces ∼1.0 cm in length. After washing in PBS, the pieces of small intestine were incubated with vigorous shaking (230 rpm) in HBSS containing 5% FBS, 10 mM Hepes, and 1 mM dithiothreitol for 30 min at 37°C. After extensive vortexing and filtering through a 70-µm filter, the flowthrough was spun down at 1,500 rpm for 5 min. The cell pellet was resuspended in 40% Percoll and then underlaid with 80% Percoll and spun at 2,500 rpm for 20 min with no brake. The interphase containing the IELs was collected for flow cytometry. Isolated IEL cells were resuspended in complete RPMI containing Brefeldin A (eBioscience) and eBioscience Cell Stimulation Cocktail (500×) containing PMA/ionomycin and incubated for 4 h before flow cytometry for intracellular cytokines. For T reg cell analysis, isolated IELs were analyzed directly for flow cytometry of Foxp3 expression.
Isolation of colonic lamina propria lymphocytes
Lamina propria lymphocytes were isolated using magnetic-activated cell sorting with the MACS Lamina Propria Dissociation Kit (Miltenyi Biotec) following the manufacturer’s protocol. Briefly, Peyer’s patches and fat were removed from the small intestine, and the intestine was cut longitudinally and then into pieces ∼1.0 cm in length. After extensive washing and enzymatic digestion, tissue was homogenized with the gentle MACS Dissociator. Homogenized tissue was filtered through a 100-µm filter, and flowthrough was spun down at 300 g for 10 min. The cell pellet containing epithelial cells and lymphocytes was further enriched using the MagniSort CD4 negative selection kit (Thermo Fisher Scientific). Isolated T cells were resuspended in complete RPMI containing Brefeldin A and Cell Stimulation Cocktail containing PMA/ionomycin and incubated for 4 h before flow cytometry for intracellular cytokines.
ELISA
Naive T cells were cultured under Th17-inducing conditions for 4 d or with anti-CD3/anti-CD28 for 12 or 24 h, and supernatant was collected. IL-2, IL-17A, IL-17F, and IL-21 ELISAs were performed using Ready-Set-Go ELISA Kits (eBioscience).
RNA isolation and qPCR
RNA was isolated from T cell cultures using the RNeasy micro kit (Qiagen). Mouse tissue RNA was isolated by homogenization in Trizol reagent (Invitrogen). RNA was isolated from T cell subsets by sorting cells from a Foxp3-GFP mouse based on the following markers: naive (CD62L+CD44−CD25−), memory-activated (CD62L−CD44+), and T reg cells (GFP+). Isolated RNA was reverse transcribed using superscript IV reverse transcription (Invitrogen) with Oligodt20 primers (Invitrogen). qPCR was performed on an ABI 7500 fast machine using Quantabio PerfeCTa qPCR Tough Mix. All reactions were normalized to Actin (ActB), and fold induction was calculated using the ddCT method. All primer-probes were purchased from ABI: Il17a (Mm00439618_m1), Il17f (Mm00521423_m1), Il2 (Mm99999222_m1), Atf7ip (Mm00479827_m1), Rorc (Mm01261022_m1), and ActB (Mm00607939_s1).
RNA-seq
Total RNA was extracted using Trizol and purified using a Qiagen microcolumn. Illumina libraries were generated from total RNA using the Illumina TruSeq Library Prep Kit and sequenced on the Illumina HiSeq 4000 to a depth of ≥20 million reads. Sequence alignment was performed using STAR (version 2.4.2a; Dobin et al., 2013). Mappings were restricted to those that were uniquely assigned to the mouse genome, and unique read alignments were used to quantify expression and aggregated on a per-gene basis using the Ensembl (GRCm38.78) annotation. Differentially expressed genes between experimental groups were then determined using DESeq2 (v1.16.1; Love et al., 2014).
ChIP
ChIP was performed by Active Motif. Primary naive T cells were fixed with 1% formaldehyde for 15 min and quenched with 0.125 M glycine. Chromatin was isolated by the addition of lysis buffer, followed by disruption with a Dounce homogenizer. Lysates were sonicated, and the DNA was sheared to an average length of 300–500 bp. Genomic DNA (input) was prepared by treating aliquots of chromatin with RNase, proteinase K, and heat for de-cross-linking, followed by ethanol precipitation. Pellets were resuspended, and the resulting DNA was quantified on a NanoDrop spectrophotometer. Extrapolation to the original chromatin volume allowed quantitation of the total chromatin yield. An aliquot of chromatin (15 µg) was precleared with protein A agarose beads (Invitrogen). Genomic DNA regions of interest were isolated using 5 µg of antibody against H3K9me3 (Active Motif; #39161, Lot #2). Complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Cross-links were reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation. The quality of ChIP-isolated DNA was tested by qPCR. qPCR reactions were performed in triplicate on specific genomic regions using SYBR Green Supermix (Bio-Rad). The resulting signals were normalized for primer efficiency by carrying out qPCR for each primer pair using input DNA.
ChIP-seq
ChIP-seq and subsequent data analysis was performed by Active Motif. Illumina sequencing libraries were prepared from the ChIP and input DNAs by the standard consecutive enzymatic steps of end-polishing, dA-addition, and adaptor ligation. After a final PCR amplification step, the resulting DNA libraries were quantified and sequenced on Illumina’s NextSeq 500 (75-nt reads, single end). Reads were aligned to the mouse genome (mm10) using the Burrows-Wheeler Aligner algorithm (default settings; Li and Durbin, 2009). Duplicate reads were removed, and only uniquely mapped reads (mapping quality ≥25) were used for further analysis. Alignments were extended in silico at their 3′ ends to a length of 200 bp, which is the average genomic fragment length in the size-selected library, and assigned to 32-nt bins along the genome. The resulting histograms (genomic “signal maps”) were stored in bigWig files. H3K9me3-enriched regions were identified using the Spatial Clustering for Identification of ChIP-Enriched Regions algorithm (spatial clustering for identification of ChIP-enriched regions; Zang et al., 2009) at a cutoff of FDR 1 × 10−10 and a max gap parameter of 600 bp. Peaks that were on the ENCODE (Encyclopedia of DNA Elements) blacklist of known false ChIP-seq peaks were removed. Signal maps and peak locations were used as input data to the Active Motifs proprietary analysis program, which creates Excel tables containing detailed information on sample comparison, peak metrics, peak locations, and gene annotations.
RNA expression data
The expression levels of Atf7ip were assessed in mouse cells and tissues using the BioGPS dataset (http://biogps.org/dataset/GSE10246/), GeneAtlas MOE430, gcrma. A subset of the data from probeset 1454973, containing only the baseline expression profiles of mouse cells and tissues, was used to visualize the gene expression profile.
Western blot
Extracts for Western blotting were isolated as previously performed (Timms et al., 2016). Five million CD4+ T cells were lysed in cytoplasmic buffer containing 10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, EDTA-free protease inhibitor cocktail, and 0.1% IGEPAL. The nuclear pellet was isolated by centrifugation at 1,500 rpm for 5 min. The total nuclear extract was isolated by incubation of the nuclear pellet with 1% SDS and 1:100 benzonase. The nuclear extract was heated in sample buffer containing SDS for 10 min at 70°C before separation with SDS-PAGE and transfer to a polyvinylidene fluoride membrane. The membrane was then blocked with 5% milk in TBST (Tris-buffered saline and Tween 20) followed by 1-h incubation with the primary antibody. The membrane was washed 4× for 5 min in Tris-buffered saline and Tween 20 before incubation in HRP-conjugated secondary antibody. The membrane was visualized with ECL reagent (Thermo Fisher Scientific). Antibodies used for Western blot were as follows: anti-MACF1 (ab84497; Abcam); rabbit anti-mouse anti-histone H3 antibody, Nuclear Loading Control and ChIP Grade (Ab1791; Abcam); rabbit anti-mouse anti-histone H3 (trimethyl K9) antibody, ChIP Grade (Ab8898; Abcam), peroxidase affinipure goat anti-rabbit IgG (H+L; 111-035-144; Jackson ImmunoResearch).
ChIP qPCR
5 × 106 naive T cells were used for each ChIP reaction following the protocol from the Diagenode LowCell# Chip Kit protein G. Briefly, cells were lysed and then sonicated for 8 min using a Covaris s220 sonicator. ChIP was performed using an H3K9me3 antibody from Abcam (ab8898) and an irrelevant IgG control (Diagenode C15410206). qPCR was performed using SYBR green with the following primer pairs: Il2/Il21: forward, 5′-GGGGCAGTAACTTCGACTTG-3′, and reverse, 5′-AGTCGAGCTGAGATGTGGAA-3′; and Zfp780b: forward, 5′-AACTGTGTTTGCCGGAGATG-3′, and reverse, 5′-GTCCTGTGCTGGCCATTAAC-3′. The primer concentration per 25-µl qPCR reaction was 200 nM. 5 µl of ChIP-isolated DNA was used per reaction. Relative Ct values obtained were normalized against the total input to obtain the percentage of amplified ChIP DNA per input.
Statistical analysis
All experiments were performed using randomly assigned mice without investigator blinding. No data were excluded. Statistical significance between two groups was calculated using an unpaired, parametric, two-tailed Student’s t test or the nonparametric Mann–Whitney test. For T cell transfer colitis experiments, the groups were first compared with two-way ANOVA followed by multiple comparisons between groups using multiple t tests (Holm–Sidak method). Experimental groups included a minimum of three biological replicates. Intragroup variation was not assessed. All statistical analysis was performed using Prism 7 (GraphPad Software). Figures display means ± SD unless noted otherwise. A P value <0.05 was considered statistically significant. No statistical methods were used to predetermine sample size.
Data availability
RNA-seq and ChIP-seq data are deposited in the Gene Expression Omnibus under accession nos. GSE131511 and GSE131533.
Online supplemental material
Fig. S1 details creation of the Atf7ip conditional KO mouse. Fig. S2 shows data on T cell proliferation in Atf7ipfl/fl mice and provides supplementary data for colitis experiments. Fig. S3 provides qPCR data on genes identified by RNA-seq, data on pathogenic Th17 differentiation, and supplementary ChIP-seq data.
Supplementary Material
Acknowledgments
We thank Jeff Bluestone, Alex Marson, Mark Ansel, and the Anderson and Bluestone laboratories for helpful discussions. Atf7ipfl/fl mice were made with the assistance of the UCSF ES Cell Targeting Core and the Gladstone Transgenic Gene Targeting Core. Biostatistics support was provided by the UCSF Functional Genomics Core and Diabetes Center Biostatics Core.
This work was supported by National Institutes of Health (NIH) grant R37 AI097457 (M.S. Anderson); NIH grant UO1 DK107383 (M.S. Anderson); NIH Diabetes Research Center grant P30 DK063720 (M.S. Anderson, Single Cell Analysis Center); NIH Shared Instrument Grant 1S10OD021822-01 (Single Cell Analysis Center), Lupus Research Alliance Grant A132045 (M.R. Waterfield), Arthritis National Research Foundation Grant A131146 (M.R. Waterfield), Rheumatology Research Foundation grant A131906 (M.R. Waterfield), and NIH grant K08 A1121513 (M.R. Waterfield).
The authors declare no competing financial interests.
Author contributions: M.R. Waterfield conceived the study, designed and performed experiments, analyzed data. J.H. Sin conceived and designed experiments with M.R. Waterfield, and analyzed data. C. Zuckerman assisted with flow cytometry and performed Western blot. J.T. Cortez helped to create Atf7ipfl/fl mice. W.L. Eckalbar designed and performed bio statistical analysis of RNA-seq data and analyzed data with D.J. Erle. M.R. Waterfield and M.S. Anderson directed the study and wrote the manuscript.
References
- Abbas A.K., Trotta E., Simeonov D.R., Marson A., and Bluestone J.A.. 2018. Revisiting IL-2: Biology and therapeutic prospects. Sci. Immunol. 3:eaa1482 10.1126/sciimmunol.aat1482 [DOI] [PubMed] [Google Scholar]
- Adoue V., Binet B., Malbec A., Fourquet J., Romagnoli P., van Meerwijk J.P.M., Amigorena S., and Joffre O.P.. 2019. The Histone Methyltransferase SETDB1 Controls T Helper Cell Lineage Integrity by Repressing Endogenous Retroviruses. Immunity. 50:629–644.e8. 10.1016/j.immuni.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Montagna C., and Macian F.. 2012. Silencing of the Il2 gene transcription is regulated by epigenetic changes in anergic T cells. Eur. J. Immunol. 42:2471–2483. 10.1002/eji.201142307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., and Rao C.M.. 2018. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur. J. Pharmacol. 837:8–24. 10.1016/j.ejphar.2018.08.021 [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu A., De La Rosa-Velázquez I.A., Ramirez F., Barenboim M., Onishi-Seebacher M., Arand J., Galán C., Winter G.E., Engist B., Gerle B., et al. . 2014. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 55:277–290. 10.1016/j.molcel.2014.05.029 [DOI] [PubMed] [Google Scholar]
- Cooper J.D., Smyth D.J., Smiles A.M., Plagnol V., Walker N.M., Allen J.E., Downes K., Barrett J.C., Healy B.C., Mychaleckyj J.C., et al. . 2008. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 40:1399–1401. 10.1038/ng.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M, and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplugues E., Huber S., Gagliani N., Hauser A.E., Town T., Wan Y.Y., O’Connor W. Jr., Rongvaux A., Van Rooijen N., Haberman A.M., et al. . 2011. Control of TH17 cells occurs in the small intestine. Nature. 475:514–518. 10.1038/nature10228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini M.C., Rizzo A., Fina D., Caruso R., Becker C., Neurath M.F., Macdonald T.T., Pallone F., and Monteleone G.. 2007. IL-21 regulates experimental colitis by modulating the balance between Treg and Th17 cells. Eur. J. Immunol. 37:3155–3163. 10.1002/eji.200737766 [DOI] [PubMed] [Google Scholar]
- Festen E.A., Goyette P., Scott R., Annese V., Zhernakova A., Lian J., Lefèbvre C., Brant S.R., Cho J.H., Silverberg M.S., et al. . 2009. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut. 58:799–804. 10.1136/gut.2008.166918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Rasmussen J.P., Gavin M.A., and Rudensky A.Y.. 2005. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 6:1142–1151. 10.1038/ni1263 [DOI] [PubMed] [Google Scholar]
- Fujita N., Watanabe S., Ichimura T., Ohkuma Y., Chiba T., Saya H., and Nakao M.. 2003. MCAF mediates MBD1-dependent transcriptional repression. Mol. Cell. Biol. 23:2834–2843. 10.1128/MCB.23.8.2834-2843.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen S.L., Jain R., Garg A.V., and Cua D.J.. 2014. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat. Rev. Immunol. 14:585–600. 10.1038/nri3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Laurence A., Yang X.P., Tato C.M., McGeachy M.J., Konkel J.E., Ramos H.L., Wei L., Davidson T.S., Bouladoux N., et al. . 2010. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 467:967–971. 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T., Watanabe S., Sakamoto Y., Aoto T., Fujita N., and Nakao M.. 2005. Transcriptional repression and heterochromatin formation by MBD1 and MCAF/AM family proteins. J. Biol. Chem. 280:13928–13935. 10.1074/jbc.M413654200 [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., and Littman D.R.. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Jadidi-Niaragh F., and Mirshafiey A.. 2011. Th17 cell, the new player of neuroinflammatory process in multiple sclerosis. Scand. J. Immunol. 74:1–13. 10.1111/j.1365-3083.2011.02536.x [DOI] [PubMed] [Google Scholar]
- Kimura H. 2013. Histone modifications for human epigenome analysis. J. Hum. Genet. 58:439–445. 10.1038/jhg.2013.66 [DOI] [PubMed] [Google Scholar]
- Klatzmann D., and Abbas A.K.. 2015. The promise of low-dose interleukin-2 therapy for autoimmune and inflammatory diseases. Nat. Rev. Immunol. 15:283–294. 10.1038/nri3823 [DOI] [PubMed] [Google Scholar]
- Koch C.M., Honemann-Capito M., Egger-Adam D., and Wodarz A.. 2009. Windei, the Drosophila homolog of mAM/MCAF1, is an essential cofactor of the H3K9 methyl transferase dSETDB1/Eggless in germ line development. PLoS Genet. 5:e1000644 10.1371/journal.pgen.1000644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley R.G., Elewski B.E., Lebwohl M., Reich K., Griffiths C.E., Papp K., Puig L., Nakagawa H., Spelman L., Sigurgeirsson B., et al. FIXTURE Study Group . 2014. Secukinumab in plaque psoriasis--results of two phase 3 trials. N. Engl. J. Med. 371:326–338. 10.1056/NEJMoa1314258 [DOI] [PubMed] [Google Scholar]
- Laurence A., Tato C.M., Davidson T.S., Kanno Y., Chen Z., Yao Z., Blank R.B., Meylan F., Siegel R., Hennighausen L., et al. . 2007. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 26:371–381. 10.1016/j.immuni.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Lin J.X., and Leonard W.J.. 2013. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 38:13–25. 10.1016/j.immuni.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.J., Yadav P.K., Su J.L., Wang J.S., and Fei K.. 2009. Potential role of Th17 cells in the pathogenesis of inflammatory bowel disease. World J. Gastroenterol. 15:5784–5788. 10.3748/wjg.15.5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matesanz F., Fedetz M., Collado-Romero M., Fernández O., Guerrero M., Delgado C., and Alcina A.. 2001. Allelic expression and interleukin-2 polymorphisms in multiple sclerosis. J. Neuroimmunol. 119:101–105. 10.1016/S0165-5728(01)00354-X [DOI] [PubMed] [Google Scholar]
- Minkovsky A., Sahakyan A., Rankin-Gee E., Bonora G., Patel S., and Plath K.. 2014. The Mbd1-Atf7ip-Setdb1 pathway contributes to the maintenance of X chromosome inactivation. Epigenetics Chromatin. 7:12 10.1186/1756-8935-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik J.R., König A., Hube B., and Gaffen S.L.. 2017. Candida albicans-epithelial interactions and induction of mucosal innate immunity. Curr. Opin. Microbiol. 40:104–112. 10.1016/j.mib.2017.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostanin D.V., Bao J., Koboziev I., Gray L., Robinson-Jackson S.A., Kosloski-Davidson M., Price V.H., and Grisham M.B.. 2009. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am. J. Physiol. Gastrointest. Liver Physiol. 296:G135–G146. 10.1152/ajpgi.90462.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D.D., and Kuchroo V.K.. 2015. Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity. 43:1040–1051. 10.1016/j.immuni.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Quintana F.J., Jin H., Burns E.J., Nadeau M., Yeste A., Kumar D., Rangachari M., Zhu C., Xiao S., Seavitt J., et al. . 2012. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat. Immunol. 13:770–777. 10.1038/ni.2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz S., Kayagaki N., Phung Q.T., Eidenschenk C., Noubade R., Wang X., Lesch J., Lu R., Newton K., Huang O.W., et al. . 2015. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature. 518:417–421. 10.1038/nature13979 [DOI] [PubMed] [Google Scholar]
- Sasai N., Saitoh N., Saitoh H., and Nakao M.. 2013. The transcriptional cofactor MCAF1/ATF7IP is involved in histone gene expression and cellular senescence. PLoS One. 8:e68478 10.1371/journal.pone.0068478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawalha A.H., Kaufman K.M., Kelly J.A., Adler A.J., Aberle T., Kilpatrick J., Wakeland E.K., Li Q.Z., Wandstrat A.E., Karp D.R., et al. . 2008. Genetic association of interleukin-21 polymorphisms with systemic lupus erythematosus. Ann. Rheum. Dis. 67:458–461. 10.1136/ard.2007.075424 [DOI] [PubMed] [Google Scholar]
- Schultz D.C., Ayyanathan K., Negorev D., Maul G.G., and Rauscher F.J. III. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16:919–932. 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi R., Hori S., Takahashi T., and Sakaguchi S.. 2005. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J. Exp. Med. 201:723–735. 10.1084/jem.20041982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Goodall J.C., and Hill Gaston J.S.. 2009. Frequency and phenotype of peripheral blood Th17 cells in ankylosing spondylitis and rheumatoid arthritis. Arthritis Rheum. 60:1647–1656. 10.1002/art.24568 [DOI] [PubMed] [Google Scholar]
- Takikita S., Muro R., Takai T., Otsubo T., Kawamura Y.I., Dohi T., Oda H., Kitajima M., Oshima K., Hattori M., et al. . 2016. A Histone Methyltransferase ESET Is Critical for T Cell Development. J. Immunol. 197:2269–2279. 10.4049/jimmunol.1502486 [DOI] [PubMed] [Google Scholar]
- Tchasovnikarova I.A., Timms R.T., Matheson N.J., Wals K., Antrobus R., Göttgens B., Dougan G., Dawson M.A., and Lehner P.J.. 2015. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 348:1481–1485. 10.1126/science.aaa7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper R.I., Pattengale P.K., and Leder P.. 1989. Murine interleukin-4 displays potent anti-tumor activity in vivo. Cell. 57:503–512. 10.1016/0092-8674(89)90925-2 [DOI] [PubMed] [Google Scholar]
- Timms R.T., Tchasovnikarova I.A., Antrobus R., Dougan G., and Lehner P.J.. 2016. ATF7IP-Mediated Stabilization of the Histone Methyltransferase SETDB1 Is Essential for Heterochromatin Formation by the HUSH Complex. Cell Reports. 17:653–659. 10.1016/j.celrep.2016.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J.A., Walker N.M., Cooper J.D., Smyth D.J., Downes K., Plagnol V., Bailey R., Nejentsev S., Field S.F., Payne F., et al. Wellcome Trust Case Control Consortium . 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39:857–864. 10.1038/ng2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C., Rapley E.A., Seal S., Pernet D., Renwick A., Hughes D., Ricketts M., Linger R., Nsengimana J., Deloukas P., et al. UK Testicular Cancer Collaboration . 2010. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat. Genet. 42:604–607. 10.1038/ng.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel D.A., Franke L., Hunt K.A., Gwilliam R., Zhernakova A., Inouye M., Wapenaar M.C., Barnardo M.C., Bethel G., Holmes G.K., et al. . 2007. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat. Genet. 39:827–829. 10.1038/ng2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino A.V., Stumhofer J.S., Saris C.J., Kastelein R.A., de Sauvage F.J., and Hunter C.A.. 2006. IL-27 limits IL-2 production during Th1 differentiation. J. Immunol. 176:237–247. 10.4049/jimmunol.176.1.237 [DOI] [PubMed] [Google Scholar]
- Villarino A.V., Tato C.M., Stumhofer J.S., Yao Z., Cui Y.K., Hennighausen L., O’Shea J.J., and Hunter C.A.. 2007. Helper T cell IL-2 production is limited by negative feedback and STAT-dependent cytokine signals. J. Exp. Med. 204:65–71. 10.1084/jem.20061198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Cao R., Xia L., Erdjument-Bromage H., Borchers C., Tempst P., and Zhang Y.. 2001. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 8:1207–1217. 10.1016/S1097-2765(01)00405-1 [DOI] [PubMed] [Google Scholar]
- Wang Z., Yin H., Lau C.S., and Lu Q.. 2016. Histone Posttranslational Modifications of CD4+ T Cell in Autoimmune Diseases. Int. J. Mol. Sci. 17:1547 10.3390/ijms17101547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield M., Khan I.S., Cortez J.T., Fan U., Metzger T., Greer A., Fasano K., Martinez-Llordella M., Pollack J.L., Erle D.J., et al. . 2014. The transcriptional regulator Aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nat. Immunol. 15:258–265. 10.1038/ni.2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.P., Ghoreschi K., Steward-Tharp S.M., Rodriguez-Canales J., Zhu J., Grainger J.R., Hirahara K., Sun H.W., Wei L., Vahedi G., et al. . 2011. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12:247–254. 10.1038/ni.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C., Schones D.E., Zeng C., Cui K., Zhao K., and Peng W.. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 25:1952–1958. 10.1093/bioinformatics/btp340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour D., Zilionis R., Kiner E., Klein A.M., Mathis D., and Benoist C.. 2018. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 19:291–301. 10.1038/s41590-018-0051-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A., Alizadeh B.Z., Bevova M., van Leeuwen M.A., Coenen M.J., Franke B., Franke L., Posthumus M.D., van Heel D.A., van der Steege G., et al. . 2007. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am. J. Hum. Genet. 81:1284–1288. 10.1086/522037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., and Paul W.E.. 2010. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 28:445–489. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP-seq data are deposited in the Gene Expression Omnibus under accession nos. GSE131511 and GSE131533.