The authors report three unrelated children with inherited TLR3 deficiency, impaired TLR3-dependent, IFN-α/β– and/or -λ–mediated, pulmonary epithelial cell–intrinsic immunity to influenza A virus, and life-threatening influenza pneumonitis.
Abstract
Autosomal recessive IRF7 and IRF9 deficiencies impair type I and III IFN immunity and underlie severe influenza pneumonitis. We report three unrelated children with influenza A virus (IAV) infection manifesting as acute respiratory distress syndrome (IAV-ARDS), heterozygous for rare TLR3 variants (P554S in two patients and P680L in the third) causing autosomal dominant (AD) TLR3 deficiency. AD TLR3 deficiency can underlie herpes simplex virus-1 (HSV-1) encephalitis (HSE) by impairing cortical neuron-intrinsic type I IFN immunity to HSV-1. TLR3-mutated leukocytes produce normal levels of IFNs in response to IAV. In contrast, TLR3-mutated fibroblasts produce lower levels of IFN-β and -λ, and display enhanced viral susceptibility, upon IAV infection. Moreover, the patients’ iPSC-derived pulmonary epithelial cells (PECs) are susceptible to IAV. Treatment with IFN-α2b or IFN-λ1 rescues this phenotype. AD TLR3 deficiency may thus underlie IAV-ARDS by impairing TLR3-dependent, type I and/or III IFN–mediated, PEC-intrinsic immunity. Its clinical penetrance is incomplete for both IAV-ARDS and HSE, consistent with their typically sporadic nature.
Introduction
Human influenza is a contagious acute illness caused by influenza viruses, a family of segmented negative-sense single-stranded RNA viruses (Shaw and Palese, 2013). Three types of influenza virus are known to infect humans: influenza A virus (IAV), influenza B virus, and influenza C virus. IAVs are generally the most virulent, and have two highly variable surface glycoproteins: hemagglutinin (HA) and neuraminidase. Two IAV subtypes, H1N1 and H3N2, are currently circulating in humans (Pulendran and Maddur, 2015). Human influenza infection typically causes mild, self-healing clinical manifestations. In rare cases, it may lead to life-threatening pneumonitis, manifesting as acute respiratory distress syndrome (ARDS; Jaber et al., 2010). Encephalitis is another life-threatening form of influenza, which is typically not associated with pneumonia, and is more rare, occurring in only 2–4% of patients hospitalized for severe influenza (Surtees and DeSousa, 2006; Lester-Smith et al., 2009; Glaser et al., 2012). The global prevalence of these two forms of life-threatening seasonal influenza has been estimated at ∼4–6 in 10,000 (World Health Organization, 2018). A significant proportion of ARDS deaths are due to secondary infections with bacteria or other viruses (McCullers, 2014). Known risk factors for influenza ARDS include preexisting comorbid conditions, such as asthma and other chronic pulmonary diseases, cardiovascular diseases, and neurological disabilities (Dawood et al., 2011; McCullers, 2014; FluSurv-NET, 2018). According to recent reports from the Centers for Disease Control and Prevention, such underlying conditions accounted for 92.4% of hospitalizations of adults and 56.7% of hospitalizations of children for influenza during the 2017 and 2018 epidemics (FluSurv-NET, 2018). However, previously healthy patients, normally resistant to other infectious agents, can also develop unexplained life-threatening influenza, manifesting as influenza-associated ARDS or, more rarely, encephalitis (Lester-Smith et al., 2009; Glaser et al., 2012). The pathogenesis of influenza ARDS or encephalitis in such individuals remains largely unknown.
A few genetic etiologies of severe influenza in humans have recently been described. Heterozygous mutations of RANBP2 underlie acute necrotizing encephalopathy, which is not viral but occurs after viral infections, including influenza (Singh et al., 2015). We recently reported the first genetic cause of bona fide influenza encephalitis: autosomal recessive (AR) DBR1 deficiency impairing the metabolism of RNA lariats and underlying viral infections of the brainstem (Zhang et al., 2018). Interestingly, inborn errors of adaptive immunity, such as severe combined immunodeficiency (lack of autologous T cells) and agammaglobulinemia (lack of autologous B cells), do not confer predisposition to severe influenza (either ARDS or encephalitis), despite underlying a very broad range of severe infectious diseases, including many viral diseases of the brains and lungs (Bousfiha et al., 2018; Picard et al., 2018). By contrast, influenza ARDS has been documented in four adults with AD GATA2 deficiency (Pasquet et al., 2013; Donadieu et al., 2018; Sologuren et al., 2018). All but one of these patients had suffered from other infections when struck by influenza (Sologuren et al., 2018). The development of severe influenza in these patients was probably not due to a lack of natural killer (NK) cells, as patients with other forms of NK deficiency are not prone to this disease (Gineau et al., 2012; Hughes et al., 2012; Cottineau et al., 2017; Marcenaro et al., 2017). Instead, it probably involved a lack of development of plasmacytoid dendritic cells (pDCs), the most potent producers of IFN-α/β and -λ, due to their constitutive expression of IRF7 (Kerkmann et al., 2003). We also recently identified AR complete IRF7 deficiency as the first human genetic etiology of influenza ARDS in an otherwise healthy child (Ciancanelli et al., 2015). Upon IAV infection, IRF7 deficiency impairs the production of IFN-α/β and IFN-λ not only by pDCs, but also by fibroblasts and induced pluripotent stem cell (iPSC)–derived pulmonary epithelial cells (PECs; Ciancanelli et al., 2015). Moreover, IRF7 is required for the amplification of both types of antiviral IFNs. Consistently, IRF7-deficient fibroblasts and PECs were found to be highly susceptible to IAV, as shown by viral replication rates in these cells. The relative contributions of human IFN-α/β and -λ, and of pDCs and PECs, to protective immunity to IAV are unknown.
We recently reported a child with AR complete IRF9 deficiency, influenza ARDS, and a history of adverse reaction to the live measles mumps rubella vaccine (Hernandez et al., 2018). The patient’s cells did not activate ISGF3 complexes, which normally consist of STAT1-STAT2-IRF9, in response to IFN-α/β. IRF9 deficiency is also predicted to impair IFN-λ responsiveness, as the receptor for IFN-λ signals through ISGF3 (Hermant and Michiels, 2014). The lack of induction of IRF9- and ISGF3-dependent IFN-stimulated genes (ISGs) accounts for the observed vulnerability of the patient’s fibroblasts to IAV. Interestingly, a small but clearly detectable proportion of ISGs was weakly or normally induced by IFN-α in IRF9-deficient EBV-B cells and fibroblasts. IRF7- and IRF9-independent cellular responses to IFNs may partly account for the very narrow range of infections seen in the IRF7- and IRF9-deficient patients reported to date (Ciancanelli et al., 2015; Hernandez et al., 2018). These residual responses may also contribute to the absence of influenza encephalitis in these patients. It would be premature to draw any firm conclusions from such a small number of patients, but the findings for autosomal dominant (AD) GATA2, AR IRF7, and AR IRF9 deficiencies collectively suggest that the integrity of the human IFN-α/β and -λ circuit is required for protective immunity to influenza viruses in human lungs (Zhang et al., 2019). In this context, we hypothesized that other single-gene inborn errors of immunity related to IFN-α/β and -λ immunity might underlie influenza ARDS in at least some other previously healthy children. We set out to analyze the exomes of 25 unrelated children with influenza ARDS without mutations of GATA2, IRF7, or IRF9. Testing a hypothesis of genetic homogeneity, we searched for genes carrying rare nonsynonymous variants in two or more patients.
Results
Enrichment in TLR3 variants in children with IAV-ARDS
We performed whole-exome sequencing (WES) in 27 unrelated children with IAV-associated ARDS (IAV-ARDS). Following the identification of AR IRF7 and IRF9 deficiencies, each in a single child, we analyzed the exomes of the remaining 25 children, searching for AD genetic etiologies by testing a hypothesis of genetic homogeneity (i.e., two or more affected children heterozygous for disease-causing mutations of the same gene), focusing on genes related to IFN-α/β and -λ immunity. We considered only nonsynonymous variants that (i) were very rare (i.e., with a minor allele frequency [MAF] < 0.001) in 1000 Genomes, the Genome Aggregation Database (gnomAD), which contains WES or whole-genome sequencing data for 141,456 individuals, and in our in-house WES database of 4,186 patients with various infectious diseases; (ii) had a combined annotation-dependent depletion (CADD) score (Kircher et al., 2014) higher than the corresponding mutational significance cutoff (MSC) of the 99% confidence interval (Itan et al., 2016); and (iii) affected a gene closely connected to IRF7 (IRF7 connectivity P value < 0.01), as predicted by the human gene connectome (Itan et al., 2013). We analyzed these data, adjusting for the first three principal components to account for ethnic heterogeneity, as described in a previous study (Belkaya et al., 2017). We compared this group of patients with cohorts of patients with nonviral infections available from our in-house WES database (1,485 individuals in total). This approach identified TLR3 as the best candidate gene (IAV-ARDS cohort versus nonviral cohorts, P value = 0.00052; Table S1), as three unrelated children were carrying a single heterozygous missense mutation of TLR3 (Fig. 1 A). Two children were heterozygous for c.1660C>T p.P554S and the third for c.2039C>T p.P680L. TLR3 acts as a sensor for extracellular double-stranded RNA (dsRNA) and a potent inducer of IFN-α/β and -λ (Alexopoulou et al., 2001). It is also closely connected to IRF7 and IRF9, as predicted by the human gene connectome (Itan et al., 2013). The discovery of these two variants was, nevertheless, surprising, because rare deleterious TLR3 variants, including P554S in particular, were previously shown to underlie HSV-1 encephalitis (HSE; Zhang et al., 2007b; Guo et al., 2011; Lim et al., 2014). Indeed, we also found a significant enrichment in rare TLR3 variants in our HSE cohort (245 WES), serving as a positive control, relative to the same 1,485 control exomes (HSE cohort versus other patient cohorts, P value = 0.0022; Table S2).
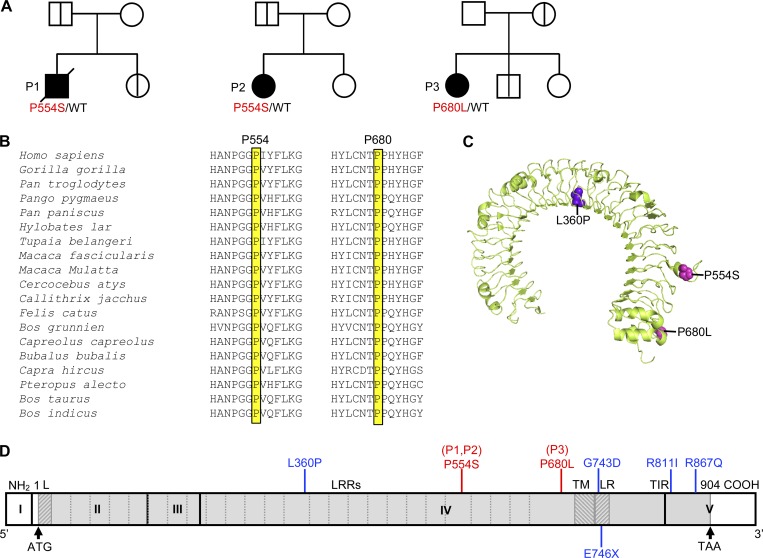
Figure 1.
Heterozygous TLR3 mutations in three unrelated IAV-ARDS patients. (A) Family pedigrees with TLR3 allele segregation. The black symbols indicate patients, and the bold vertical lines indicate healthy carriers of the mutant TLR3 allele. Healthy TLR3 WT relatives are indicated by white symbols. (B) Multiple sequence alignment across 19 vertebrate species. Both residues mutated in P1, P2, and P3 were highly conserved. (C) Predicted three-dimensional structure of the human TLR3 protein ectodomain. The residues affected by the two missense mutations found in IAV-ARDS patients are highlighted in magenta (P554S and P680L). The L360P mutation was previously identified in a patient with HSE (highlighted in violet). (D) Schematic diagram of the structure of the human TLR3 gene and protein, featuring the leader sequence (L), leucine-rich repeats (LRRs) of the ectodomain, transmembrane domain (TM), linker region (LR), and Toll/IL-1 receptor (TIR) domain. Roman numerals indicated the coding exons. Previously reported experimentally validated deleterious mutations found in HSE patients are shown in blue. The mutations found in the three children with severe influenza are shown in red (including the P554S mutation found in three previously reported and one unreported HSE patient).
TLR3 missense variants in three unrelated children with IAV-ARDS
Strikingly, patient 1 (P1) and patient 2 (P2) were both found to be heterozygous for a known, HSE-causing, c.1660C>T p.P554S, TLR3 variant (rs121434431) that is both loss-of-function (LOF) and dominant negative and was already found in four HSE children (Zhang et al., 2007b; Guo et al., 2011; unpublished data). Its global MAF in the gnomAD database is 0.0004, and it has a CADD score of 24.9. P1 was born to nonconsanguineous French parents, and he died from IAV-ARDS in 2008, at the age of 9 yr (see clinical case reports in Human patients section and Fig. S1 A). At the time of death, he was seronegative for HSV-1 (Table S3). P2 was born to nonconsanguineous Belgian parents. She developed IAV-ARDS at 5 wk of age in 2015, and one episode of severe respiratory syncytial virus (RSV) pneumonitis at the age of 2.5 mo. She is now 3 yr 6 mo of age and remains well with monthly intravenous immunoglobulins and yearly influenza vaccination. P3 is heterozygous for the c.2039C>T p.P680L TLR3 variant. The P680L mutation is private to P3, as it has not been reported in any public or in-house databases, indicating that its MAF is <0.000004. Its CADD score is 24.9. P3 was born to nonconsanguineous parents of European descent living in the United States, and she developed three successive episodes of viral ARDS in 2008, 2009, and 2010, at the ages of 5, 6, and 7 yr. Two of these episodes were caused by IAV (2008 and 2010). P3 is now healthy at 14 yr of age and has been well since the introduction of annual vaccination against influenza, as reported for IRF7- and IRF9-deficient patients (Ciancanelli et al., 2015; Hernandez et al., 2018). She is seropositive for IAV H1N1 and H3N2, and for HSV-1 (Fig. S1 A and Table S3). P680L was not found in any other individual in our WES database of 4,186 exomes, whereas P554S was also found in four patients with HSE, one patient with mycobacterial infection, and one patient with pyogenic infection, consistent with its MAF of 0.0004. Finally, in the WES data for these three patients, 238 variants in P1, 407 in P2, and 209 in P3, fulfilled our filtering criteria, affecting 213 genes in P1, 316 genes in P2, and 197 genes in P3 (Table S4), with TLR3 the only gene affected in all three patients. Moreover, there were no homozygous, compound heterozygous, or de novo (trio analysis of P2, P3, and their parents) variants of the 168 genes with an IRF7 connectivity P value < 0.01 (Itan et al., 2013). The P554S and P680L variants of TLR3 were, therefore, considered to be the most plausible candidates warranting further investigation.
The TLR3 mutations are predicted to be deleterious and disease-causing
TLR3 is an endosomal receptor of dsRNA (Alexopoulou et al., 2001) and an inducer of IFN-α/β and -λ. Most if not all viral infections result in the production of dsRNA by-products (Jacobs and Langland, 1996), but fewer viruses produce dsRNA viral intermediates stricto sensu (Kumar and Carmichael, 1998; Weber et al., 2006). TLR3 is highly conserved across vertebrate species (Mikami et al., 2012) and has evolved under strong purifying selection in humans (Barreiro et al., 2009). Upon ligand binding, TLR3 recruits a single adaptor, TIR domain–containing adapter protein inducing INF-β, inducing the production of antiviral cytokines, including IFN-β, IFN-λ, and inflammatory cytokines, such as TNF-α and IL-6, through the activation of transcription factors, including IRF3 and NF-κB (Kawai and Akira, 2006). Variants of TLR3 and of six other genes controlling the TLR3 pathway (Casrouge et al., 2006; Zhang et al., 2007a; Pérez de Diego et al., 2010; Audry et al., 2011; Guo et al., 2011; Sancho-Shimizu et al., 2011; Herman et al., 2012; Andersen et al., 2015) confer a predisposition to HSE by impairing neuron- and oligodendrocyte-intrinsic immunity to HSV-1 (Lafaille et al., 2012; Zimmer et al., 2018). Sanger sequencing confirmed the P554S mutation in P1 and P2, and the P680L mutation in P3 (Fig. 1 A and Fig. S1 B). The segregation of these variants within the three families suggested incomplete clinical penetrance for influenza ARDS, as previously shown for families with HSE (Casrouge et al., 2006; Zhang et al., 2007b; Sancho-Shimizu et al., 2011; Herman et al., 2012; Lim et al., 2014; Andersen et al., 2015), as five relatives of the three children with IAV-ARDS were heterozygous for the TLR3 mutation carried by the index case but had remained healthy despite seropositivity for IAV (Fig. 1 A and Table S3). Both variants affect residues strictly conserved across all species for which TLR3 has been sequenced (Fig. 1 B). TLR3 is a type I transmembrane receptor with an N-terminal (N-ter) extracellular domain (ECD), a single transmembrane domain, a linker region, and a cytoplasmic Toll/IL-1 receptor domain (Fig. 1, C and D). Both missense variants are located in the ECD of TLR3, which forms an M-shaped dimer upon ligand binding (Choe et al., 2005; Fig. 1, C and D). The P554S missense mutations have been shown to be both LOF and dominant negative (Zhang et al., 2007b). Its recurrence in two unrelated children with IAV-ARDS further supports its causal role in the pathogenesis. The P680L missense mutation affects residue P680, which has previously been shown to be critical for both TLR3 dimerization and dsRNA binding (Liu et al., 2008; Wang et al., 2010). Moreover, the P680L TLR3 mutant itself has also been previously shown in HEK293 cells to be LOF for NF-κB activation due to a lack of TLR3 dimerization and dsRNA binding (Wang et al., 2010). Collectively, these data suggest that the two TLR3 variants identified in these three children may underlie IAV-ARDS due to AD TLR3 deficiency.
The P680L TLR3 protein is LOF for the induction of anti-viral IFNs
We analyzed the impact of the TLR3 P680L mutation in the TLR3-deficient P2.1 fibrosarcoma cell line, which does not produce detectable amounts of TLR3 protein and does not respond to the dsRNA mimic polyinosinic:polycytidylic acid (poly[I:C]; Sun and Leaman, 2004). We generated cell lines stably transfected with empty plasmid or with plasmids containing C-terminally HA-tagged WT and mutant TLR3 cDNAs. In P2.1 cells expressing WT TLR3, the production of IFNL1 and IFNB mRNAs was induced after 2 and 4 h of poly(I:C) stimulation, respectively, whereas cells expressing the P680L allele displayed no poly(I:C)-stimulated induction of IFNL1 or IFNB mRNA (Fig. 2 A), despite having TLR3 mRNA levels similar to those in the controls (Fig. 2 B). Infection with M51R vesicular stomatitis virus (VSV) mutant, a potent inducer of IFNs via TLR3-independent pathways (Kato et al., 2006), triggered the production of similar amounts of IFNB and IFNL1 mRNAs in all cells, demonstrating that the IFN production machinery was intact. Consistent with a previous report showing that P680L is LOF for NF-κB activation in HEK293 cells (Wang et al., 2010), P680L is, therefore, LOF also in terms of IFNL1 and IFNB induction in P2.1 cells, like the five previously identified HSE-causing TLR3 mutations (L360P, P554S, E746X, G743D+R811I, and R867Q; Zhang et al., 2007b; Guo et al., 2011; Lim et al., 2014). We then tested the other eight very rare (MAF < 0.001) or private missense TLR3 mutations, found in our in-house database of infectious diseases other than influenza and HSE, including mostly fungal, mycobacterial, or pyogenic diseases (Table S5 and Fig. S2 A). Only one of these eight variants, found in two relatives in the fungal disease cohort (a patient and her healthy mother), and with a MAF of 0.00007 in the gnomAD database, was deleterious upon expression in P2.1 cells (Fig. S2, B and C; and Table S5). There is therefore strong enrichment in experimentally proven LOF TLR3 alleles in both our IAV-ARDS (P value = 0.0002) and HSE cohorts (P value = 0.0005), relative to other cohorts of families with nonviral diseases. These data provide evidence that heterozygosity for P554S or P680L TLR3 LOF variants may underlie IAV-ARDS in the three unrelated children under study.
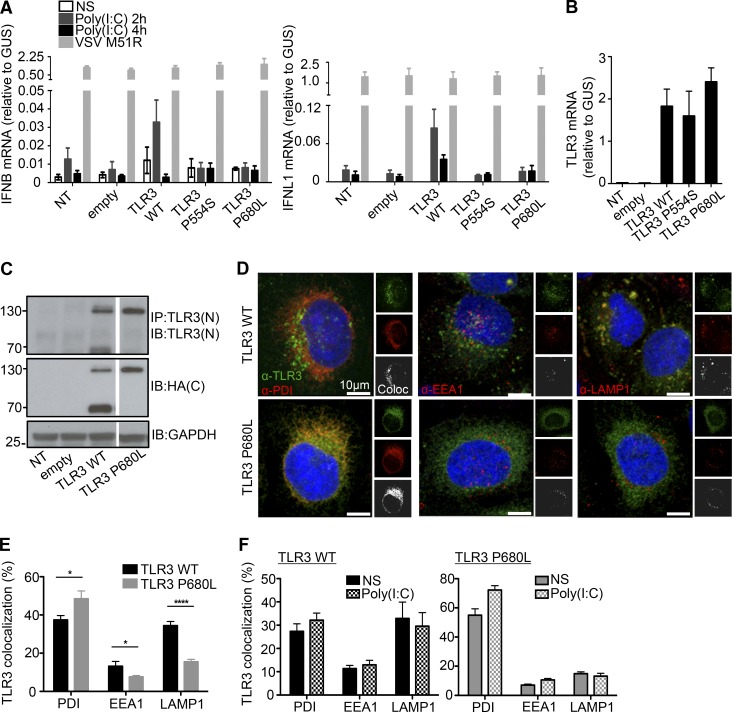
Figure 2.
Expression and function of the P680L mutant TLR3 allele. (A) RT-qPCR for IFNB and IFNL1 mRNA expression without stimulation (NS) or after 2 and 4 h of stimulation with 25 µg/ml poly(I:C) in P2.1 cells not transfected (NT) or stably transfected with empty vector, HA-tagged TLR3 WT, P554S, or P680L. VSV M51R, at a MOI of 1, was used as a positive stimulus for IFN induction via the TLR3-independent pathways. Mean values ± SD were calculated from two independent experiments, with biological duplicates in each experiment. (B) TLR3 mRNA levels were determined by RT-qPCR in P2.1 TLR3-deficient fibrosarcoma cells with or without transfection with empty vector, HA-tagged TLR3 WT, P554S, or P680L. Mean values ± SD were calculated from two independent experiments, with biological duplicates in each experiment. (C) TLR3 was detected on immunoblots. P2.1 cells not transfected or stably transfected with empty vector, HA-tagged TLR3 WT, or P680L were subjected to immunoprecipitation (IP) with anti-TLR3 antibody, and the immunoprecipitated protein was then immunoblotted (IB) with C-ter (C) HA antibody or N-ter (N) TLR3 antibody. GAPDH was used as a loading control for immunoblotting. Reproducible result from six independent experiments is shown. (D–F) Immunofluorescence imaging of P2.1 cells stably expressing HA-tagged WT or P680L. Intracellular distribution was assessed by colocalization (Coloc) with a subcellular marker: anti-PDI antibody for the ER, anti-EEA1 antibody for early endosomes, and anti-LAMP1 antibody for lysosomes. Cells were let un-treated (E) or incubated with 25 µg/ml poly(I:C) for 30 min (F). The images were analyzed with Imaris Coloc software, and plots were generated. About 200 cells were used for each analysis. Mean values ± SD were calculated from two independent experiments. *, P < 0.05; ****, P < 0.0001.
The P680L mutation impairs TLR3 trafficking and cleavage
The P554S variant was characterized in previous studies (Zhang et al., 2007b; Guo et al., 2011). We thus studied the mechanisms of P680L LOF, first by immunoblotting lysates from P2.1 cells stably transfected with the WT or P680L TLR3 allele. Cell lysates were subjected to immunoprecipitation with anti-TLR3 antibody, and the immunoprecipitated was probed with anti-HA and anti-TLR3 antibodies. TLR3 is normally processed to produce the active form by cathepsin-mediated cleavage in the lysosome at residue 323–356 (Toscano et al., 2013). WT TLR3 therefore migrates as two products of ∼130 kD and 70 kD in size, corresponding to the full-length and C-terminal (C-ter) cleaved forms, respectively (Fig. 2 C). Intriguingly, only the full-length P680L product was detected, suggesting that only the uncleaved form was present. This was unexpected, because residue 680 is far from the putative cleavage site (residues 323–356; Garcia-Cattaneo et al., 2012; Toscano et al., 2013). We investigated the reasons for this lack of cleavage by characterizing the subcellular distributions of the WT and P680L TLR3 proteins. Immunofluorescence microscopy showed that WT TLR3 was mostly colocalized with protein disulfide isomerase (PDI; an ER marker: 37.44 ± 2.272%) and LAMP1 (an endolysosome marker: 34.44 ± 2.166%), although some was also found with EEA1 (an early endosome marker: 13.21 ± 2.491%; Fig. 2, D and E). By contrast, TLR3 P680L mostly colocalized with PDI (48.45 ± 4.144%), very little being found with EEA1 (7.635 ± 0.6655%) or LAMP1 (15.54 ± 1.290%), suggesting that P680L TLR3 is largely retained within the ER, with only low rates of transit to the lysosome, where the WT TLR3 is cleaved by cathepsins B and H (Garcia-Cattaneo et al., 2012). Poly(I:C) stimulation of TLR3 WT- and P680L-expressing P2.1 cells had no effect on the subcellular distribution of TLR3 (Fig. 2 F). WT TLR3 cleavage is, thus, constitutive, and occurs independently of poly(I:C) stimulation in lysosomes. Moreover, when lysates from P2.1 cells stably transfected with C-ter HA-tagged WT or P680L TLR3 were subjected to native gels, the P680L protein migrated faster as assessed by immunoblotting with an antibody against N-ter of TLR3 or an anti-HA (C-ter; Fig. S3 A). In comparison, the full-length P680L TLR3 protein migrated at the same speed as that of the WT TLR3 in SDS gels (Fig. S3 B). These findings suggested that the P680L TLR3 protein was misfolded, which may explain why it was retained within the ER and was not cleaved in the lysosome. Treatment with kifunensine, an ER-associated degradation inhibitor (Wang et al., 2011) partially rescued the cleavage of P680L TLR3 (Fig. S3, C and D). Finally, transient cotransfection of C-ter HA- or FLAG-tagged WT and P680L TLR3 in HeLa cells showed an increased proportion of WT TLR3 localized in the ER, and lower being in endolysosomes or lysosomes (Fig. S3 E), despite normal cleavage of WT TLR3 (Fig. S3 F), upon cotransfection with P680L TLR3. The P680L TLR3 might therefore perhaps have a modest dominant negative effect on the WT TLR3 in vivo. Overall, in this experimental setting, P680L is LOF because it is retained in the ER, preventing its proteolytic cleavage in the lysosome. Previous studies showed that P680L cannot dimerize and bind dsRNA (Liu et al., 2008; Wang et al., 2010). Our findings suggest that P680L also accumulates in the ER. Collectively, these findings provide at least three mechanisms by which the P680L allele is LOF (lack of TLR3 dimerization and dsRNA binding, and lack of proteolytic cleavage due to ER retention).
The P680L TLR3 allele underlies AD TLR3 deficiency in fibroblasts
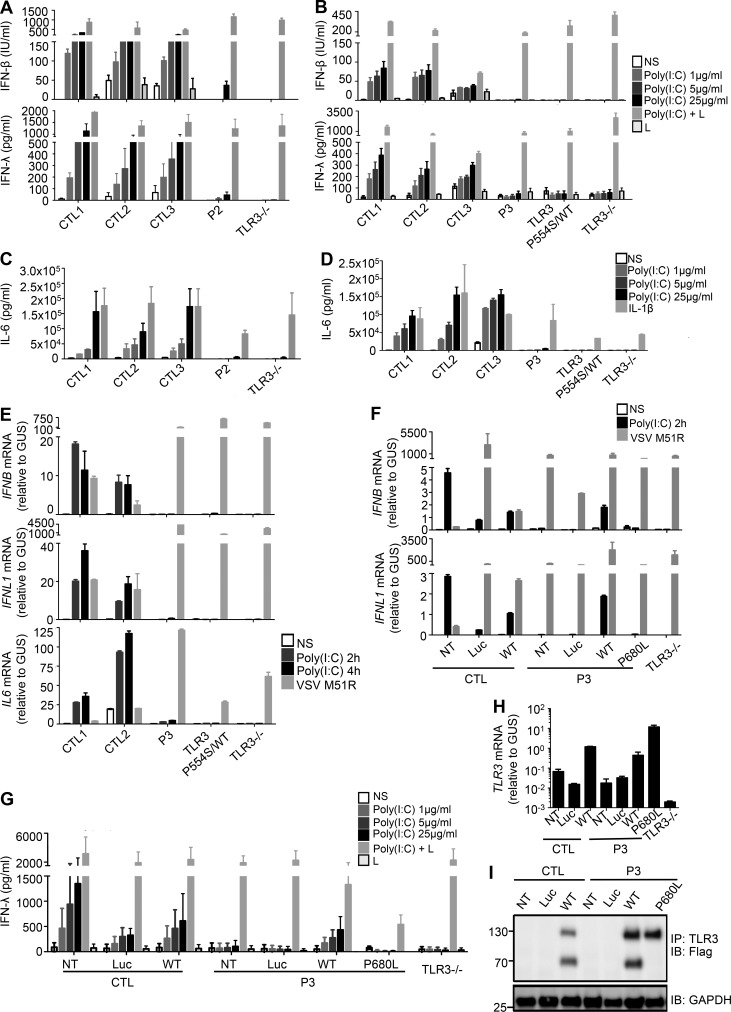
We previously reported that heterozygosity for P554S underlies AD TLR3 deficiency in fibroblasts, in which the LOF P554S allele causes AD TLR3 deficiency due to negative dominance (Zhang et al., 2007b). We also reported that heterozygosity for the HSE-causing LOF L360P or G743D+R811I allele causes AD TLR3 deficiency in fibroblasts due to negative dominance and haploinsufficiency, respectively (Lim et al., 2014). We tested whether heterozygosity for the P680L mutation also caused AD TLR3 deficiency at the cellular level. Human dermal fibroblasts respond to extracellular poly(I:C) stimulation in a TLR3-dependent manner (Zhang et al., 2007b; Guo et al., 2011; Lim et al., 2014). We studied the response to poly(I:C) in SV40-immortalized fibroblasts (SV40-fibroblasts) from P2 (P554S/WT), P3 (P680L/WT), three healthy individuals, a patient with HSE and AR complete TLR3 deficiency (TLR3−/−, due to compound heterozygous P554S and E746X mutations), and a patient with HSE and AD partial TLR3 deficiency (P554S/WT HSE; Zhang et al., 2007b; Guo et al., 2011; Fig. 3, A–D). Fibroblasts from P1 were not available. The fibroblasts from the three healthy controls produced increasing levels of IFN-β, IFN-λ, and IL-6 after 24 h of stimulation with increasing concentrations of poly(I:C), whereas the production of all three cytokines was severely impaired in P2, P3, P554S/WT HSE, and TLR3−/− fibroblasts. Serving as a control, the TLR3-independent induction of IFN-β and IFN-λ was normal upon transfection with poly(I:C) in the presence of lipofectamine, exposing the cytosolic retinoic acid–inducible gene 1 protein (RIG-I)–like receptors RIG-I and MDA5 to poly(I:C) (Yoneyama et al., 2004, 2005). The production of IL-6 in response to IL-1β stimulation was also normal in the patients’ cells. Consistently, 2 h or 4 h of stimulation with extracellular poly(I:C) induced the production of the IFNB, IFNL1, and IL6 mRNAs in healthy control fibroblasts but not in P3, P554S/WT, and TLR3−/− cells, whereas all fibroblasts responded well to infection with VSV M51R (Fig. 3 E). Moreover, SV40-fibroblasts from two relatives of P1 who are heterozygous for the P554S mutation also displayed severely impaired IFN-β, IFN-λ, and IL-6 production after 24 h of stimulation with increasing concentrations of poly(I:C) (Fig. S4, A and B). In this family, heterozygosity for P554S thus confers AD nonresponsiveness to poly(I:C) in fibroblasts with complete penetrance. Finally, the lack of response of P3’s fibroblasts to poly(I:C) was rescued by the stable overexpression of WT, but not P680L, TLR3 (Fig. 3, F–I). Serving as another control, the stable overexpression of TLR3 also rescued poly(I:C) responsiveness in fibroblasts from other HSE patients with AD or AR TLR3 deficiency (Zhang et al., 2007b; Lim et al., 2014), but not in fibroblasts from another HSE patient with AR UNC-93B deficiency (Casrouge et al., 2006; Fig. S4 C). Collectively, these results established that heterozygosity for the recurrent P554S or the newly discovered P680L TLR3 allele underlies AD TLR3 deficiency in human dermal fibroblasts. The P680L TLR3 protein is mostly retained in the ER (Fig. 2, D–F), does not dimerize, and does not bind dsRNA (Wang et al., 2010). Overall, these findings suggest that P680L causes AD TLR3 deficiency by haploinsufficiency, unlike dominant negative P554S (Zhang et al., 2007b), although a modest negative dominance of P680L was not definitively excluded (Fig. S3 E).
Figure 3.
Impaired poly(I:C) responses in SV40-fibroblasts from patients heterozygous for TLR3 P554S or P680L, and rescue by WT TLR3. (A–D) Production of IFN-β, IFN-λ (A and B), and IL-6 (C and D) in SV40-fibroblasts from three healthy controls (CTL1/2/3), P2 (A and C), P3 (B and D), a TLR3 P554S/WT HSE patient, and a TLR3−/− HSE patient, 24 h after stimulation with 1, 5, or 25 µg/ml poly(I:C), or with 25 µg/ml poly(I:C) in the presence of lipofectamine (poly[I:C]+L; A and B), lipofectamine alone (L; A and B), or IL-1β (C and D), as assessed by ELISA. (E) IFNB, IFNL1, and IL6 mRNA levels in SV40-fibroblasts from two CTLs, P3, P554S/WT, and TLR3−/− patients, not stimulated (NS), or stimulated for 2 and 4 h with 25 µg/ml poly(I:C), or infected with VSV M51R at a MOI of 1 for 16 h. GUS was included for normalization. (F–I) Complementation of the impaired poly(I:C) response by introducing WT TLR3 into the patient’s fibroblasts. (F) IFNB, IFNL1 mRNA levels in SV40-fibroblasts from a healthy control (CTL) and P3, without plasmid transfection (NT) or after transfection with luciferase (Luc), Flag-tagged WT or P680L TLR3, and in fibroblasts from a TLR3−/− patient, not stimulated (NS), or stimulated for 2 h with 25 µg/ml poly(I:C), or infected with VSV M51R at a MOI of 1 for 16 h. GUS was included for normalization. (G) Production of IFN-λ, in the absence of stimulation, after 24 h of stimulation with 1, 5, or 25 µg/ml poly(I:C), and after stimulation with 25 µg/ml poly(I:C) in the presence of lipofectamine, or lipofectamine alone, as assessed by ELISA, in SV40-fibroblasts from a CTL and P3, without plasmid transfection or after transfection with Luc, Flag-tagged WT or P680L TLR3, and in fibroblasts from a TLR3−/− patient. (H) TLR3 mRNA levels were assessed by RT-qPCR. (I) TLR3 was detected on immunoblots following IP. Mean values ± SD were calculated from four (A–D and H) or two (E and F) independent experiments, with technical duplicates in each experiment. (G) Results from a single experiment with biological duplicates, representing three independent experiments. (I) Reproducible results from three independent experiments.
Normal responses of TLR3-mutated peripheral blood mononuclear cells to poly(I:C) and IAV
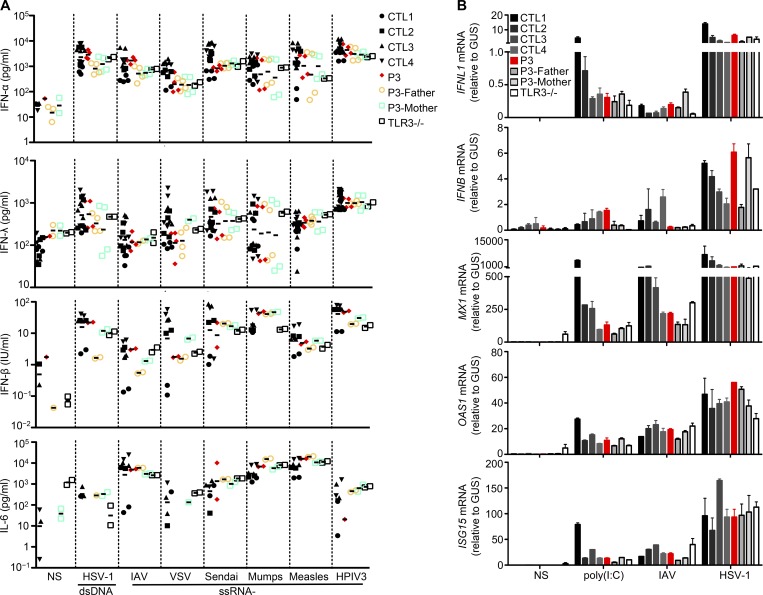
IRF7 and IRF9 deficiencies impair the production of and response to IFN-α/β and -λ in leukocytes and fibroblasts (Ciancanelli et al., 2015; Hernandez et al., 2018). Impaired IFN-α/β and -λ immunity to IAV has also been demonstrated in IRF7-deficient fibroblasts and PECs (Ciancanelli et al., 2015). For both deficits, the respective contributions of leukocytes, including pDCs in particular, and PECs to the pathogenesis of IAV-ARDS remain unknown. We previously showed that TLR3 is largely redundant for the induction of IFNs in leukocytes in response to poly(I:C) and various viruses (Zhang et al., 2007b; Guo et al., 2011). Upon stimulation with 11 viruses, including HSV-1, BK virus, VSV, measles virus, mumps virus, human parainfluenza virus 3 (HPIV3), Newcastle disease virus, Sendai virus, Sindbis virus, encephalomyocarditis virus, and Coxsackievirus B1, peripheral blood mononuclear cells (PBMCs) from patients with AD TLR3 deficiency produce normal amounts of IFN-α, IFN-β, and IFN-λ (Zhang et al., 2007b). Moreover, pDCs from HSE patients with AR or AD TLR3 deficiency produce normal amounts of IFN-α upon stimulation with HSV-1 (Zhang et al., 2007b; Guo et al., 2011), whereas pDCs from an IAV-ARDS patient with AR IRF7 deficiency have impaired IFN-α production following stimulation with HSV-1 or IAV (Ciancanelli et al., 2015). These findings are consistent with constitutive IRF7 expression levels being highest in pDCs (Izaguirre et al., 2003), which do not express TLR3, even upon activation (Kadowaki et al., 2001). Like AR TLR3–deficient PBMCs, P3’s PBMCs produced normal levels of IFN-α, IFN-β, IFN-λ, and IL-6 upon infection with seven different viruses, including HSV-1, IAV, VSV, measles virus, mumps virus, HPIV3, and Sendai virus (Fig. 4 A), relative to PBMCs from controls, and P3’s mother (P680L/WT) and father (WT/WT; Fig. 4 A). The normal response to IAV is especially relevant here. PBMCs from P3 and the AR TLR3–deficient patient also responded normally to poly(I:C) stimulation, in terms of IFNL1, IFNB, MX1, OAS1, and ISG15 mRNA induction (Fig. 4 B). The redundant role of TLR3 in leukocytes may explain the absence of other viral infections in TLR3-deficient patients, and of systemic dissemination of the virus during HSE or IAV-ARDS. These results also suggest that the seemingly normal IFN-α/β and -λ immunity conferred by leukocytes, including PDCs, was insufficient, or arrived too late, to protect the lung against IAV infection in patients with AD TLR3 deficiency. Impaired TLR3-IRF7-IRF9–mediated PEC-intrinsic immunity to IAV may therefore account for susceptibility to IAV-ARDS in patients, mirroring the neuron- and oligodendrocyte-intrinsic immunity accounting for HSE in patients with TLR3 pathway deficiencies (Lafaille et al., 2012).
Figure 4.
Normal IFN response to poly(I:C) and viruses in TLR3-mutated PBMCs. (A) PBMCs from four CTLs, P3, P3’s parents, and a TLR3−/− patient were infected with various types of viruses: double-stranded DNA (dsDNA; HSV-1) and single-stranded RNA (ssRNA−; IAV strain pH1N1, VSV, Sendai virus, mumps virus, measles virus, HPIV3) viruses. The levels of IFN-α, -β, and -λ and IL-6 were measured by ELISA 24 h after infection. (B) The induction of IFNL1, IFNB, MX1, OAS1, and ISG15 mRNA was assessed by RT-qPCR in PBMCs from four CTLs, P3, P3’s parents, and a TLR3−/− patient. RNA was isolated from these cells after 8 h of poly(I:C) stimulation and 24 h of IAV pH1N1 and HSV-1 infection. Mean values ± SD were calculated from biological duplicates from two independent experiments.
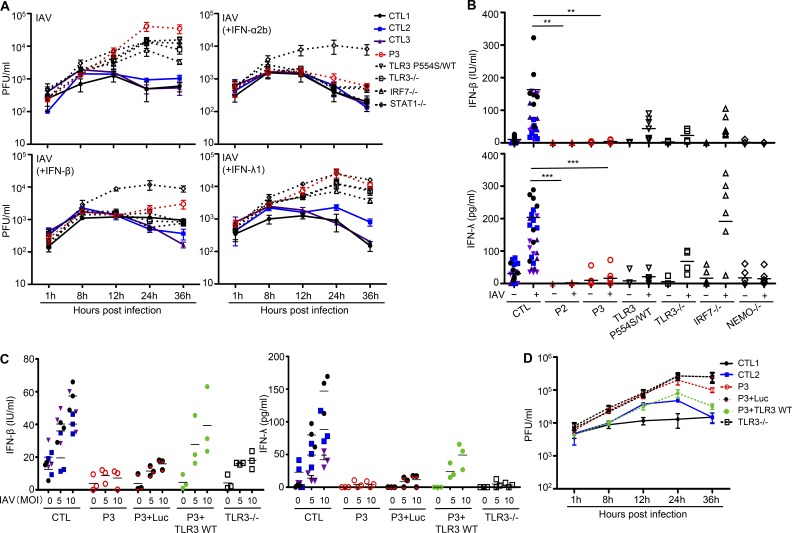
TLR3-mutated fibroblasts are vulnerable to IAV: Rescue by IFN-α2b but not IFN-λ1
We previously reported high levels of IAV replication in human IRF7-deficient (IRF7−/−) and IRF9-deficient (IRF9−/−) dermal fibroblasts, and in IRF7−/− iPSC-derived PECs (Ciancanelli et al., 2015; Hernandez et al., 2018). We also showed that TLR3-deficient fibroblasts and iPSC-derived central nervous system (CNS)–resident neurons and oligodendrocytes were vulnerable to HSV-1 infection (Zhang et al., 2007b; Guo et al., 2011; Lafaille et al., 2012; Lim et al., 2014). We thus hypothesized that TLR3 deficiency might also impair fibroblast- and PEC-intrinsic immunity to IAV. We first infected SV40-fibroblasts from P2, P3, a TLR3 P554S/WT HSE patient, and a TLR3−/− HSE patient, with IAV. We also infected cells from healthy controls, an IRF7−/− patient (Ciancanelli et al., 2015), and a STAT1−/− patient (Chapgier et al., 2006), serving as controls. Virus titers in the P2, P3, TLR3 P554S/WT HSE, and TLR3−/− SV40-fibroblasts were 10 to 100 times higher than in three healthy controls, 24 and 36 h after infection (Fig. 5 A and Fig. S5 A). IAV titers in TLR3-deficient SV40-fibroblasts were higher than those in IRF7−/− cells, and as high as those in STAT1−/− cells (Fig. 5 A). We also found that the production of IFN-β and IFN-λ 24 h after IAV infection was impaired in fibroblasts from P2, P3, TLR3 P554S/WT HSE, and TLR3−/− patients (Fig. 5 B), as well as in SV40-fibroblasts from two relatives of P1 who are heterozygous for the P554S mutation (Fig. S5 B). The viral phenotype of TLR3- and IRF7-deficient fibroblasts was rescued by exogenous IFN-α2b or IFN-β pretreatment (starting 16 h before infection; Fig. 5 A). By contrast, IFN-λ1 did not protect the patients’ fibroblasts, possibly due to the absence of IFN-λ receptor (IFNLR) expression in human dermal fibroblasts (Lazear et al., 2015). The phenotype of STAT1−/− fibroblasts was not rescued in any of the conditions tested. Finally, stable overexpression of WT TLR3 in P3 fibroblasts rescued their IAV-induced IFN-β and IFN-λ production (Fig. 5 C) and IAV susceptibility (Fig. 5 D). Overall, impaired IFN-β production in AD TLR3–deficient fibroblasts may account for their vulnerability to IAV infection. This fibroblast phenotype, although a surrogate phenotype not directly relevant to PECs, suggested that cell-intrinsic immunity was disrupted in nonhematopoietic cells, providing a plausible molecular and cellular basis for IAV-ARDS in the three TLR3-deficient children studied here. These findings mirrored our previous observation that TLR3 deficiency impairs fibroblast immunity to HSV-1 (Zhang et al., 2007b; Guo et al., 2011; Lim et al., 2014), which paved the way for the study of iPSC-derived CNS-resident cells (Lafaille et al., 2012; Zimmer et al., 2018).
Figure 5.
Enhanced susceptibility to IAV in TLR3-mutated fibroblasts, and rescue by WT TLR3. (A) IAV replication, quantified by plaque assays, in SV40-fibroblasts from three CTLs, P3, P554S/WT, TLR3−/−, IRF7−/−, and STAT1−/− patients, 1, 8, 12, 24, and 36 h after infection at a MOI of 10. Cells were untreated or subjected to pretreatment with IFN-α2b, IFN-β, or IFN-λ for 16 h before infection. The data shown are representative of three independent experiments, with biological duplicates in each experiment. (B) Production of IFN-β and IFN-λ in the absence of infection or after 24 h of infection with IAV at a MOI of 1, in SV40-fibroblasts from seven CTLs, P2, P3, a TLR3 P554S/WT HSE patient, TLR3−/−, IRF7−/−, and NF-κB essential modulator (NEMO)–deficient patients, as assessed by ELISA. (C) Production of IFN-β and IFN-λ in the absence of infection or after 24 h of infection with IAV at a MOI of 5 or 10, in SV40-fibroblasts from a CTL and P3, without plasmid transfection or after transfection with Luc, Flag-tagged WT TLR3, and in fibroblasts from a TLR3−/− patient. (D) IAV replication, quantified by plaque assays, in SV40-fibroblasts from two CTLs and P3, without plasmid transfection or after transfection with Luc, Flag-tagged WT TLR3, and in fibroblasts from a TLR3−/− patient, 1, 8, 12, 24, and 36 h after infection at a MOI of 5. Mean values ± SD from five (B) or three (C and D) independent experiments are shown. Biological duplicates were tested in each experiment. **, P < 0.01; ***, P < 0.001.
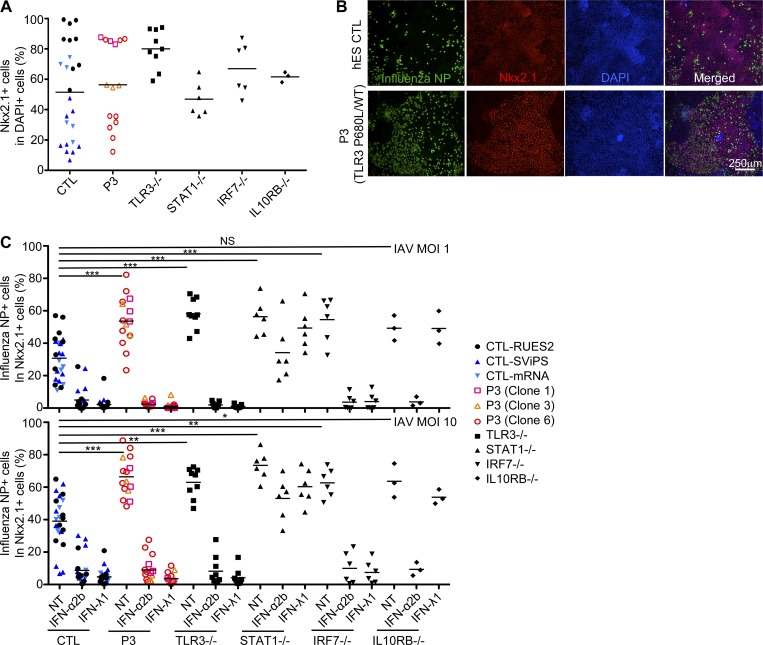
TLR3-deficient iPSC-derived PECs are vulnerable to IAV: Rescue by IFN-α2b or IFN-λ1
PECs are the primary targets of influenza virus in natural conditions (Shieh et al., 2010). These cells express TLR3 and produce IFN-β and IFN-λ upon stimulation with poly(I:C) and influenza virus (Guillot et al., 2005; Le Goffic et al., 2006; Ioannidis et al., 2013; Lazear et al., 2015). We tested the hypothesis that the disruption of PEC-intrinsic TLR3-dependent immunity underlies the pathogenesis of IAV-ARDS in AD TLR3–deficient children, by differentiating PECs from healthy control embryonic stem cells (CTL-RUES2), healthy control iPSCs (Sendai virus–reprogrammed iPSC CTL-SViPS, mRNA-reprogrammed iPSC CTL-mRNA), and iPSCs from P3 (TLR3 P680L/WT), and a patient with AR complete TLR3 deficiency and HSE (TLR3−/−; Guo et al., 2011). We included the IRF7−/− iPSC-derived PECs as a control, as these cells have been shown to be vulnerable to IAV infection (Ciancanelli et al., 2015). We also differentiated PECs from iPSCs from a STAT1−/− patient (Chapgier et al., 2006), and an IL10RB−/− patient (IL-10RB being the second chain of the IFNLR) (Glocker et al., 2009), to distinguish between the roles of IFN-α/β and -λ in PEC-specific antiviral immunity. The patients’ dermal fibroblasts were reprogrammed into iPSCs with Sendai viruses expressing Klf4, c-myc, Sox2, and Oct4, and their karyotype and genotype were confirmed to be correct (Fig. S5, C and D). The iPSCs were then allowed to differentiate into mature PECs for 55 d (Huang et al., 2014). The purity of the iPSC-derived PEC cultures varied between lines and experiments (10.625 to 98.174% NK2 homeobox 1 (Nkx2.1)+ cells in DAPI+ 55-d differentiation culture; Fig. 6 A). The presence of the virus was assessed by immunostaining in the IAV-infected PECs for influenza nucleoprotein (NP) and Nkx2.1 (pulmonary epithelial marker; Fig. 6 B). As for PECs derived from IRF7−/− iPSCs, the proportion of PECs positive for IAV NP was higher for PECs derived from TLR3 P680L/WT and TLR3−/− iPSCs than for those derived from healthy control hESC or iPSC lines (Fig. 6 C). Interestingly, PECs derived from the iPSCs of both STAT1−/− and IL10RB−/− patients also had higher proportions of IAV NP-positive cells, like IRF7−/− and TLR3-mutated patient iPSC-derived PECs (Fig. 6 C). Remarkably, the higher vulnerability to IAV of TLR3 P680L/WT, TLR3−/−, and IRF7−/− PECs was rescued by pretreatment not only with IFN-α2b, but also with IFN-λ1, whereas neither IFN-α2b nor IFN-λ1 protected STAT1−/− cells, and only IFN-α2b protected IL10RB−/− cells (Fig. 6 C). These data suggest that both IFN-α/β and IFN-λ contribute to PEC-intrinsic anti-IAV immunity. The protection conferred by IFN-λ1 in PECs is of particular interest, because IFN-λ1 pretreatment did not have a protective effect in dermal fibroblasts (Fig. 5 A), iPSC-derived neurons, or oligodendrocytes (Lafaille et al., 2012; Zimmer et al., 2018). Thus, IFN-λ1 may be specifically important for protective anti-viral immunity in the lungs, where IFNLR is expressed by PECs (Fig. S5, E–G; Jewell et al., 2010). TLR3 deficiency may, therefore, lead to influenza ARDS due to the impairment of TLR3-dependent, IFN-α/β– and/or IFN-λ–mediated, PEC-intrinsic immunity to IAV.
Figure 6.
Enhanced susceptibility to IAV in TLR3-mutated iPSC-derived lung epithelial cells. Lung epithelial cells were derived from the ES cells of a healthy control (CTL-RUES2, shown as black filled circles in the figure), iPSCs from two healthy controls (Sendai virus–reprogrammed iPSC CTL-SViPS, and mRNA-reprogrammed iPSC CTL-mRNA, shown as dark or light blue triangles, respectively), P3 (three iPSC clones from the same patient, shown with pink squares, yellow triangles, or red circles, respectively), TLR3−/−, IRF7−/−, STAT1−/−, and IL10RB−/− patients. The cells were untreated or subjected to 16 h of pretreatment with IFN-α2b or IFN-λ1, then infected with IAV at a MOI of 1 or 10 for 24 h. The cells were immunostained for influenza NP (green) and Nkx2.1 (red), and their nuclei were stained with DAPI (blue). (A) The percentage of Nkx2.1-positive cells was determined for the DAPI-positive cells. (B) Representative images of CTL and patient PECs, showing the immunostaining of NP, Nkx2.1, and DAPI, 24 h after infection with IAV. hES, human embryonic stem. (C) The percentage of influenza NP-positive cells was then determined for the Nkx2.1-positive cells. We analyzed ∼60,000 Nkx2.1 cells per cell line. Without IFN pretreatment, higher proportions of PECs derived from the iPSCs of TLR3 P680L/WT, TLR3−/−, IRF7−/−, STAT1−/−, and IL10RB−/− patients were positive for IAV NP, than of PECs from CTL-RUES2, iPSC CTL-SViPS, and iPSC CTL-mRNA. The data shown represent the mean values ± SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
This study identifies AD TLR3 deficiency as a novel human genetic etiology of life-threatening childhood pulmonary influenza. Two heterozygous missense LOF mutations were found to underlie AD TLR3 deficiency in three unrelated children with IAV-ARDS. After AR IRF7 deficiency (Ciancanelli et al., 2015) and AR IRF9 deficiency (Hernandez et al., 2018), AD TLR3 deficiency is the third genetic etiology of IAV-ARDS striking otherwise healthy children to be described. Nevertheless, the IRF9-deficient patient may have suffered from an ill-defined adverse effect of measles mumps rubella vaccination (Hernandez et al., 2018), and one of the AD TLR3–deficient patients from severe RSV pulmonary infection. This observation provides additional evidence that life-threatening pulmonary influenza in children may result from single-gene inborn errors of immunity (Alcaïs et al., 2010; Casanova, 2015a,b; Ciancanelli et al., 2016). Together with the observation of severe pulmonary influenza in four adults with AD GATA2 deficiency (Pasquet et al., 2013; Donadieu et al., 2018; Sologuren et al., 2018), these findings suggest that various inborn errors of IFN-α/β and -λ immunity may underlie influenza ARDS in humans. The identification of AD TLR3 deficiency also helps to untangle further the respective contributions of leukocytes and epithelial cells, especially pDCs and PECs, to protective immunity to IAV. In mice and humans, the influenza virus first invades the respiratory tract, replicating in the upper and lower respiratory epithelium and then spreading to other cell types (Perrone et al., 2008; Manicassamy et al., 2010). Studies of human IRF7, IRF9, and GATA2 deficiencies suggested that leukocytes (especially pDCs), or pulmonary cells (especially PECs), or both, were involved in immunity to IAV (Dai et al., 2004; Honda et al., 2005; Ciancanelli et al., 2015; Collin et al., 2015; Onodera et al., 2016; Hernandez et al., 2018). By contrast, TLR3 is constitutively expressed in epithelial cells (Guillot et al., 2005; Le Goffic et al., 2006; Ioannidis et al., 2013) but absent or redundant in most leukocytes, including pDCs (Kadowaki et al., 2001). Previous ex vivo studies have suggested that TLR3 is involved in immunity to IAV in human PECs (Guillot et al., 2005; Le Goffic et al., 2007; Ioannidis et al., 2013). We further demonstrated a severe defect of TLR3-dependent, cell-intrinsic, anti-IAV immunity in patient-specific iPSC-derived PECs, which was rescued by IFN-α2b or IFN-λ1. This finding suggests that PEC-intrinsic immunity is essential for host defense against IAV, and is both TLR3-dependent and IFN-α/β– or -λ–mediated.
This study also sheds light on the relative contributions of the two types of anti-viral IFNs in pulmonary immunity to IAV. The few patients with biallelic mutations of IFNAR1, IFNAR2, JAK1, TYK2, STAT1, and STAT2 impairing cellular responses to IFN-α/β, and those with biallelic mutations of IL10RB, JAK1, STAT1, and STAT2 impairing cellular responses to IFN-λ, do not appear to be particularly prone to influenza ARDS (Minegishi et al., 2006; Glocker et al., 2009; Hambleton et al., 2013; Duncan et al., 2015; Kreins et al., 2015; Eletto et al., 2016; Hoyos-Bachiloglu et al., 2017; Moens et al., 2017). The small number of patients with influenza ARDS may reflect incomplete penetrance, as suggested by the rarity of influenza ARDS among the much larger number of GATA2-deficient patients reported (Hsu et al., 2015; Donadieu et al., 2018). For example, other infections may have caused the premature deaths of some of these patients, preventing their exposure to influenza viruses (STAT1). Some of these defects may be partial, as opposed to complete (JAK1 and perhaps IFNAR2 and STAT2). It is also possible that some genes involved in IFN-α/β and -λ immunity are entirely redundant for host defense against influenza viruses in the lungs, whereas others are essential (Deeg et al., 2017; Casanova and Abel, 2018). Indeed, some cells can respond to specific individual IFNs (IFN-α/β or IFN-λ) in an IFNAR2-, JAK1-, and TYK2-independent manner (de Weerd et al., 2013; Kreins et al., 2015; Fuchs et al., 2016). In this context, we found that TLR3−/−, TLR3 P680L/WT, IRF7−/−, STAT1−/−, and IL10RB−/− patient iPSC-derived PECs were highly vulnerable to IAV infection, highlighting the critical role of TLR3- and type I and III IFN-mediated immunity to IAV infection in PECs. It is not yet possible to attribute specific roles to individual IFNs or their ISGs in the lungs. Nevertheless, in this study, the susceptibility of TLR3- and IRF7-deficient PECs to IAV was rescued by prior treatment with IFN-α2b or IFN-λ1. This finding is of particular interest, because the joint expression of IFNLR and IL10RB, which form the receptor for IFN-λ, is restricted to epithelial cells, such as PECs, in particular (Ank et al., 2008; Sommereyns et al., 2008), whereas that of IFNAR1 and 2, which form the receptor for IFN-α/β, is ubiquitous (Navarro et al., 1996). Our findings are consistent with recent studies of human and mouse PECs ex vivo, and of studies in mice in vivo (Jewell et al., 2010; Ioannidis et al., 2013; Hermant and Michiels, 2014; Mahlakõiv et al., 2015; Klinkhammer et al., 2018; Werder et al., 2018; Wieland and Heim, 2019). Recombinant IFN-λ may have specific therapeutic benefits in patients with pulmonary influenza, avoiding the side effects of IFN-α inherent to the systemic responsiveness to this cytokine (Davidson et al., 2016).
The identification of AD TLR3 deficiency as a genetic etiology of influenza ARDS is intriguing in at least two ways. It has been suggested that TLR3 acts as a sentinel for almost all viruses generating extracellular or endosomal dsRNA intermediates or by-products (Alexopoulou et al., 2001), but TLR3-deficient mice have been shown to be susceptible to some viruses but normally resistant or even more resistant than WT mice to other viruses (Zhang et al., 2013). In particular, TLR3-deficient mice infected with IAV survive longer than control mice despite having higher lung virus titers (Le Goffic et al., 2006). The discrepancy between TLR3-deficient mice and humans, in terms of the outcome of pulmonary IAV infection, may reflect differences between experimental infections in inbred mice and natural infections in outbred humans (Casanova and Abel, 2004; Quintana-Murci et al., 2007; Casanova et al., 2013; Davis and Brodin, 2018). Our report is also surprising in light of the six previously reported TLR3-deficient HSE patients, with AD or AR, partial or complete TLR3 deficiency (Zhang et al., 2007b; Guo et al., 2011; Lim et al., 2014). Moreover, nine other HSE patients have been shown to carry mutations of five other genes controlling the TLR3 pathway (Casrouge et al., 2006; Pérez de Diego et al., 2010; Audry et al., 2011; Sancho-Shimizu et al., 2011; Herman et al., 2012; Andersen et al., 2015). Impaired CNS-intrinsic, TLR3-dependent, IFN-mediated, anti–HSV-1 immunity underlies HSE in those patients, as suggested by the high susceptibility of the patients’ iPSC-derived CNS neurons and oligodendrocytes to HSV-1 (Lafaille et al., 2012). All the reported TLR3 pathway–deficient patients with HSE seem to be normally resistant to other common viruses, including IAV (Casrouge et al., 2006; Zhang et al., 2007b; Pérez de Diego et al., 2010; Sancho-Shimizu et al., 2011; Herman et al., 2012; Lim et al., 2014; Andersen et al., 2015). Moreover, the three patients with AD TLR3 deficiency and IAV-ARDS did not develop HSE, even though one of them was seropositive for HSV-1. Nevertheless, we have shown that these disorders display incomplete penetrance for HSE in affected families (Casrouge et al., 2006; Zhang et al., 2007b; Sancho-Shimizu et al., 2011; Herman et al., 2012; Lim et al., 2014; Andersen et al., 2015). The clinical penetrance of AD TLR3 deficiency for severe influenza is also incomplete, as five relatives of the three AD TLR3–deficient children with severe influenza are heterozygous and healthy despite seropositivity for IAV. The development of HSE in some TLR3 heterozygotes and IAV-ARDS in others may thus reflect the incomplete penetrance of AD TLR3 deficiency for both HSE and IAV-ARDS.
In genetic terms, our study establishes not only that HSE and IAV-ARDS can be allelic at the TLR3 locus, but also that they can be caused by the same type of TLR3 LOF variations (AD and LOF through negative dominance or haploinsufficiency) and even by the same TLR3 variant (P554S). The observation that AD TLR3 deficiency can underlie HSE or IAV-ARDS in otherwise healthy children normally resistant to IAV and HSV-1, respectively, is not only consistent with the incomplete clinical penetrance seen for both viral infections in TLR3 heterozygotes, but also with both infectious diseases being typically sporadic and mutually exclusive in the general population. Various factors may affect clinical penetrance, including pathogen-related factors (viral load and strain) and host-related factors (age at infection and modifier genes). Most individuals with AD TLR3 deficiency in the general population are probably asymptomatic, a minority suffering from HSE, another from IAV-ARDS, and very few, if any, having both HSE and IAV-ARDS. Intriguingly, one adult with AD TLR3 deficiency (due to P554S) developed CVB3 myocarditis (Gorbea et al., 2010). This may be coincidental, as opposed to causal, given the lack of documented mechanism (Casanova et al., 2014) and the fact that our own study of patients with viral myocarditis detected no significant enrichment in TLR3 or TLR3-pathway gene mutations (although only a small proportion had proven CVB3 myocarditis; Belkaya et al., 2017). Nevertheless, AD TLR3 deficiency is a “non-Mendelian monogenic disorder” (Casanova, 2015a,b), accounting for at least two sporadic infectious diseases, HSE and IAV-ARDS, in otherwise healthy children. This study supports the role of TLR3 in PEC-intrinsic immunity to IAV, and highlights the potential clinical value of IFN-λ for treating patients with influenza ARDS. TLR3 deficiency may underlie viral infectious diseases affecting isolated organs of the human body, not only the CNS, but also the lung. Mutations in other genes controlling TLR3 and IFN-λ immunity may be present in other children with influenza ARDS. Further human genetics and immunological studies of diverse isolated infectious diseases are required to clarify the roles of TLR3, IFN-α/β and -λ, and other host defense pathways in organ-intrinsic immunity to infections.
Materials and methods
Human patients
All patients were living in and followed up in their countries of origin (France for P1, Belgium for P2, United States for P3). Informed consent was obtained in the home country of each patient, in accordance with local regulations and with institutional review board (IRB) approval. Experiments were conducted in the United States and France in accordance with local regulations and with the approval of the IRB of The Rockefeller University and Institut National de la Santé et de la Recherche Médicale, respectively. Detailed clinical case reports are provided below.
P1 was a 9-yr-old boy with nonconsanguineous parents of French origin. There was no family history of infectious diseases. P1 was admitted to hospital with ARDS in February 2008. Nasopharyngeal and tracheal aspirate secretions tested positive for IAV by the immunofluorescence method. Despite intensive care and extracorporeal membrane oxygenation support, P1 developed multiple organ failure and died 1 wk later. Serological tests showed that P1 had been infected with RSV in the past without severe complications (Table S3).
P2 was a girl born to nonconsanguineous parents of Belgian origin. She was 5 wk old when she was first admitted to a pediatric intensive care unit (PICU) with ARDS. IAV was positive in nasopharyngeal secretions by the immunofluorescence method. Non-invasive ventilation was needed. The patient was also treated with oseltamivir (Tamiflu) and recovered. At 2.5 mo old, she was again admitted to a PICU, due to severe RSV bronchiolitis complicated by pneumonitis. Non-invasive ventilation was used again. The patient was treated with broad-spectrum antibiotics and recovered. Since then, she had some other hospitalizations, at ages of 3 mo old and 4.5 mo old, for respiratory disease (mainly bronchitis; no viral origin was identified) requiring only oxygen supplementation. She also had gastroenteritis at 2 mo old due to rotavirus infection, requiring hospitalization for rehydration, and needing classical pediatric ward but no PICU care. The patient showed hypogamma G at 5 mo old, intravenous immunoglobulin (IVIG) treatment was started when she was 6 mo old, and since then she is fine. She is now 3 yr 6 mo old. Her lymphocyte phenotyping showed an immature B cell phenotype, with low memory B cells (total and switched) and high transitional B cells, compatible with her young age. She is still under IVIG treatment to date, and remains well.
P3 was 5 yr old when she was first seen in hospital for severe influenza infection. She was born to nonconsanguineous parents of European descent who live in the United States. There was no family history of infectious diseases. She developed three separate episodes of viral infection–associated septic shock, when she was 5, 6, and 7 yr old. The first episode of ARDS occurred in April 2008 and required intubation and pressure support. Viral tests were positive for both IAV and RSV. P3 was treated with broad-spectrum antibiotics and oseltamivir (Tamiflu), and gradually improved. She then remained well until October 2009, when she again developed headache, sore throat, fever, and vomiting, which rapidly progressed over 48 h to hypotensive shock requiring pressure support. Polycythemia (hemoglobin, 22 g/dl) and hypogammaglobulinemia (282 mg/dl) were diagnosed, but the viral etiology remained undetermined. Again, her condition gradually improved, and she suffered no sequalae. In November 2010, P3 had a third episode of septic shock, and a rapid influenza test was positive. She recovered from the third episode of influenza ARDS, and has since been taking Tamiflu (oseltamivir phosphate) daily and IgG monthly, and is regularly vaccinated against influenza. She remains otherwise healthy. The most recent serological tests, performed at the age of 10 yr, showed P3 to have antibodies against varicella zoster virus, cytomegalovirus, EBV, hepatitis A virus, measles virus, mumps virus, rubella virus, parvovirus B19, and RSV (Table S3).
Serum samples obtained from both patients after the first hospitalization contained high titers of antibodies against H1N1 but not against H3N2, indicating that both patients were infected with H1N1 viruses (Fig. S1 A).
WES and Sanger sequencing
Genomic DNA was isolated by phenol-chloroform extraction from peripheral blood cells or primary fibroblasts from the patients. DNA (3 µg) was sheared with a Covaris S2 Ultrasonicator (Covaris). An adapter-ligated library was prepared with the TruSeq DNA Sample Prep Kit (Illumina). Exome capture was performed with the SureSelect Human All Exon 50 Mb kit (Agilent Technologies). Paired-end sequencing was performed on an Illumina HiSeq 2000 (Illumina), generating 100-base reads. The sequences were aligned with the human genome reference sequence (hg19 build), with Burrows-Wheeler Aligner (Li and Durbin, 2009). Downstream processing was performed with the Genome Analysis Toolkit (GATK; McKenna et al., 2010), SAMtools (Li et al., 2009), and Picard Tools (http://picard.sourceforge.net). Substitution and indel calls were made with GATK Unified Genotyper and GATK IndelGenotyperV2, respectively. All calls with a Phred-scaled single nucleotide polymorphism quality ≤20 and a read coverage ≤2 were filtered out. All variants were annotated with an annotation software system developed in-house. For the Sanger sequencing of TLR3 variants, the exons of TLR3 were amplified by PCR, purified by ultracentrifugation through Sephadex G-50 Superfine resin (Amersham-Pharmacia-Biotech), and sequenced with the Big Dye Terminator Cycle Sequencing Kit on an ABI Prism 3700 apparatus (Applied Biosystems). WES data of the three patients have been deposited in the Sequence Read Archive website (accession no. PRJNA542116).
Cell culture
Primary cultures of human fibroblasts were obtained from skin biopsy specimens obtained from patients or healthy controls and transformed with an SV40 vector, as previously described (Zhang et al., 2007b), to create immortalized SV40-fibroblast cell lines. Stably transfected SV40-fibroblasts were obtained by transfecting cells with pTRIP-TLR3iresRFP using the Nucleofector X-001 program (Lonza) according to the manufacturer’s protocol, with selection on puromycin (2 µg/ml). The TLR3-deficient P2.1 fibrosarcoma cell line was provided by D.W. Leaman (University of Toledo, Toledo, OH; Sun and Leaman, 2004). Stably transfected P2.1 cells were established by transfection with pUNO-TLR3 WT (Invivogen) or mutants (generated by mutagenesis with a kit from Thermo Fisher Scientific), in the presence of X-tremegene 9, with selection on blasticidin (10 µg/ml). SV40-fibroblasts, P2.1, and Madin-Darby canine kidney cells (American Type Culture Collection) were maintained in DMEM supplemented with 10% FCS. PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation from whole blood collected from patients and healthy controls.
iPSCs were obtained by reprogramming the patients’ primary fibroblasts by infection with a nonintegrating CytoTune Sendai viral vector kit (Life Technologies) in accordance with IRB-approved protocol 16-I-N139. Reprogrammed cells were karyotyped to ensure genomic integrity. PECs were derived from embryonic stem cells or iPSCs as described by Huang et al. (2014).
TLR3 agonists and viral infection
We used the TLR3 agonist poly(I:C) (Amersham), at concentrations of 1, 5, and 25 µg/ml. Cells were stimulated with 25 µg/ml poly(I:C) in the presence of Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. Cells or supernatants were harvested, and their cytokine mRNA or protein production was assessed by quantitative RT-PCR (RT-qPCR) or ELISA, respectively. For the infection of PBMCs with viruses, we plated half a million cells each individual well of 48-well plates. We used HSV-1 (strain KOS-1), at a multiplicity of infection (MOI) of 1, IAV (strain A/H1N1/CA/2009) at a MOI of 1, VSV (Indiana strain) at a MOI of 1, Sendai virus (E92 strain) at a MOI of 1, mumps virus at a MOI of 0.1, measles virus (Edmonston strain) at a MOI of 0.05, and HPIV3 (EA102 strain) at a MOI of 1. After 24 h, cells and supernatants were collected for cytokine determinations by ELISA and the measurement of RNA levels for IFN and ISG genes by RT-qPCR.
RT-qPCR
Total RNA was extracted from SV40-fibroblasts, PBMCs, P2.1, or iPSC-derived PECs with the RNeasy mini kit (QIAGEN). Extracted RNA was treated with DNase I for 1 h at 37°C (Roche). RNA was reverse-transcribed with random hexamers and the Superscript III first-strand cDNA synthesis system (Life Technologies). RT-qPCR was performed with Taqman universal PCR master mix and Taqman gene expression kits from Life Technologies, with an ABI PRISM 7700 Sequence Detection System. We used β-glucuronidase (GUS) for normalization. Results were analyzed by the ΔCt method, in accordance with the kit manufacturer’s instructions.
Cytokine determinations
Levels of IFN-α, -β, and -λ and IL-6 production were assessed by ELISA after 24 h of cell stimulation. Separate ELISAs were performed for each of IFN-α (eBioscience), IFN-β (PBL), IFN-λ (R&D Biosystems), and IL-6 (eBioscience), according to the kit manufacturer’s instructions.
Immunoblots
Total cell extracts were prepared from SV40-fibroblasts and P2.1 cells. Equal amounts of protein from each sample were subjected to immunoprecipitation with a goat anti-human TLR3 antibody directed against the human TLR3 ectodomain (R&D Systems). The immunoprecipitated protein was subjected to SDS-PAGE or native gel, and the proteins were blotted onto polyvinylidene difluoride membrane (Bio-Rad). These polyvinylidene difluoride membranes were then probed with an antibody against human TLR3 (R&D Systems). Anti-HA (Invivogen) antibodies were also used. Membranes were stripped and reprobed with an antibody against GAPDH (Sigma-Aldrich) to control for protein loading. Antibody binding was detected by enhanced chemiluminescence (Amersham-Pharmacia-Biotech). Semi-quantification of the TLR3 immunoblots was performed using the ImageJ in accordance with the program’s instructions.
Kifunensine treatment
6 h after coating, P2.1 cells with and without stable transfection of WT or P680L TLR3 were incubated with 166 µM kifunensine (Toronto Research Chemicals) for 48 h. The cells were then washed twice with cold PBS and lysed in 1% NP-40 lysis buffer, then subjected to SDS-PAGE for immunoblotting.
Influenza virus infection and titration
IAV (A/H1N1/CA/2009) was provided by C. Basler (Georgia State University, Atlanta, GA). For IAV infection, 4 × 104 SV40-fibroblasts were plated in poly-L-lysine–coated individual wells of 48-well plates 1 d before infection. We used IAV at a MOI of 1, 5, or 10 to infect cells in PBS supplemented with 0.3% BSA, 1 mM CaCl2, 1 mM MgCl2, and penicillin/streptomycin. After 1 h, the cells were washed twice with Dulbecco's PBS and transferred to 200 µl of DMEM supplemented with 0.3% BSA, 0.1% FBS, penicillin/streptomycin, and 1 µg/ml of N-tosyl-L-phenylalanine chloromethyl ketone (TPCK)–treated trypsin (Sigma-Aldrich), in which they were maintained until harvesting at 1, 8, 12, 24, or 36 h. Where indicated, cells were pretreated with IFN-α2b (Intron A; Schering-Plough) at a concentration of 104 IU/ml for 18 h before infection. Viral replication was assessed by determining viral titers in plaque assays on Madin-Darby canine kidney cells. 10-fold serial dilutions of viral suspensions were allowed to adsorb onto the cells for 1 h at room temperature. The cells were washed twice with DPBS, and incubated in agar overlay containing MEM, 0.5% dextran, 2.5% NaHCO3, 1 µg/ml TPCK-treated trypsin, and 0.8% agar.
Hemagglutination inhibition assay
The patient’s serum samples were treated with trypsin-heat-periodate to remove nonspecific inhibitors of hemagglutination, as previously described (Casrouge et al., 2006). Briefly, we mixed one volume of serum with half a volume of 8 mg/ml trypsin (Sigma-Aldrich) in 0.1 M phosphate buffer, pH 8.2, and incubated the mixture for 18 h at 37°C to destroy the receptor. The trypsin was inactivated by incubating the mixture at 56°C for 30 min. The samples were allowed to cool to room temperature and were mixed with three volumes of 0.11 M metapotassium periodate and incubated at room temperature for 15 min. Three volumes of 1% glycerol saline were added, and the samples were incubated for a further 15 min at room temperature. The samples were mixed with 2.5 volumes of 0.85% saline, to yield a final testing dilution of 1:10. The viruses used for the hemagglutination inhibition (HI) assay were A/Netherlands/602/2009/H1N1 and A/New York/2008/H3N2. HI assays were performed according to standard protocols (Yu et al., 2008). Two-fold serial dilutions of sera were mixed and incubated in Nunc V-bottom 96-well microtiter plates (Nalge Nunc International) for 30 min at 4°C with 4 HA units of virus per well. Turkey red blood cells were added to a final concentration of 0.5%, and the plate was incubated on ice for 30 min. HI was determined as the inverse of the last serum dilution at which the cells were not agglutinated.
Immunofluorescence
P2.1 cells stably transfected with HA-tagged pUNO-hTLR3 WT or P680L were plated on 8-well microslides (ibiTreat; ibidi). Cells were fixed by incubation in 4% paraformaldehyde for 10 min and washed twice with PBS supplemented with 0.1% glycine, before blocking with PBS supplemented with 3% BSA, 10% goat serum, and 0.3% Triton X-100 for 2 h at room temperature. The cells were incubated with primary antibodies at 4°C overnight and washed three times with blocking buffer. They were then incubated with secondary antibodies (1:1,000 dilution) and DAPI for 30 min and washed three times with PBS. Cells were mounted in ProLong Diamond mounting reagent (Life Technologies). Images were acquired with a TCS SP8 inverted laser scanning confocal microscope (Leica) at the bioimaging resource center of Rockefeller University. Image analysis was performed with Imaris (Bitplane) Coloc, using Manders’ coefficient.
For cotransfection experiments, HeLa cells were subjected to transient transfection with WT or P680L TLR3 (either with an HA or a FLAG tag at C-ter) using X-tremeGENE9 transfection reagent (Sigma-Aldrich). After 48 h of incubation, cells were fixed with 4% paraformaldehyde for 15 min at 37°C, washed three times with PBS, permeabilized with 0.1% Triton, and blocked with 0.1% Triton containing 6% donkey serum for 1 h. The cell cultures were stained overnight with a combination of the following primary antibodies: anti-HA (mouse; Santa Cruz Biotechnology) and anti-EEA1 (rabbit; Thermo Fisher Scientific) or anti-LAMP1 (rabbit; Abcam) or anti-calreticulin (rabbit; Abcam). Cells were washed three times with PBS and incubated with secondary antibodies: anti-rabbit Alexa Fluor 568 (Invitrogen) or anti-mouse Alexa Fluor 633 (Invitrogen). Cells were washed three times with PBS and finally incubated with an anti-FLAG antibody conjugated with Alexa Fluor 488 (R&D Systems). Slides were mounted in DAPI containing Prolong Gold mounting media (Thermo Fisher Scientific). Slides were examined with a confocal laser microscope (Confocal Leica SP8 gSTED). All images were acquired with APO CS2 63×/1.4 oil objectives. Images were exported as TIFF files, and image analysis was performed with ImageJ software. Co-localization studies were done using the JACoP plugin (Bolte and Cordelières, 2006). For each independent experiment, 5–12 cells were analyzed per condition. Each field was selected from DAPI-positive staining, which was used for defining the nuclear region (NR). Only the cells cotransfected with both TLR3-HA–tagged and TLR3-FLAG–tagged plasmids were analyzed, the nontransfected cells used as internal controls, in addition to cells transfected with the empty vector.
Microscopy
Pulmonary epithelial cultures were subjected to positive and negative control staining on day 55, as previously described (Huang et al., 2014). The cells were infected with influenza, fixed with 4% paraformaldehyde, washed with PBS, permeabilized with 0.25% Triton and 5% normal donkey serum (Jackson ImmunoResearch), and blocked by incubation in 5% normal donkey serum. The cell cultures were stained with a combination of the following primary antibodies: TTF-1/Nkx2.1 (rabbit; Seven Hills Bioreagents) and influenza A NP (HT103 mouse antibody) in 5% normal donkey serum in PBS at 4°C overnight. The cultures were then incubated with donkey anti-mouse whole IgG–Alexa Fluor 488 (Jackson ImmunoResearch) and donkey anti-rabbit whole IgG-Cy3 (Jackson ImmunoResearch) at 1:300 dilutions in 5% normal donkey serum at room temperature for 2 h. The cells were washed twice and incubated with DAPI for 5 min at room temperature. The stained cultures were stored in Vectashield Mounting Medium (Vector Laboratories) at 4°C in the dark. Images were obtained with a motorized Leica DMI 6000B fluorescence microscope coupled to a Leica DFC365 FX digital camera and operated by LASAF 6.2 software (Leica Microsystems). All images were acquired with HCX PL S-APO 10×/NA 0.3 or HCX PL FL L 20×/NA 0.4 objectives. The tile scan images were taken with the image tiling module coupled with either the autofocus or z-stack scanning (1.5 µm/stack) module, and autostitched with LAS AF 6.2 software. The images were exported as JPG files and processed (contrast and brightness adjustments) with Photoshop CS5.1 (Adobe Systems).
Statistical analysis
When applicable, results are presented as mean ± SD. Mean values were compared between control cells and cells from the patients by two-tailed Student’s t tests. Statistical analysis was performed in GraphPad Prism 5. Statistical significance was denoted with NS, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 in the figures.
Online supplemental material
Fig. S1 shows heterozygous TLR3 mutations in three unrelated IAV-ARDS patients. Fig. S2 shows the expression and function data of various mutant TLR3 alleles from the Human Genetics of Infectious Diseases (HGID) database. Fig. S3 shows the experimental data that P680L TLR3 is misfolded and retained in the ER. Fig. S4 shows impaired poly(I:C) responses in the TLR3 P554S or P680L heterozygous SV40-fibroblasts from P1 family members or P3. Fig. S5 shows the enhanced susceptibility to IAV in TLR3 P554S heterozygous SV40-fibroblasts, and the characterization of TLR3-mutated patient iPSCs. Table S1 shows the ranking of the top 10 enriched mutated IRF7-connected genes in the IAV-ARDS cohort versus controls. Table S2 shows the comparison of numbers of rare TLR3 nonsynonymous variant carriers in the flu cohort and in other infectious disease cohorts of the HGID laboratory in-house WES database. Table S3 shows the viral serological test results in patients and their family members. Table S4 shows the number of genes harboring rare nonsynonymous or essential splicing variants in the three patients, as revealed by WES. Table S5 shows the list of rare TLR3 variants found in the HGID laboratory in-house WES database for various infectious diseases.
Supplementary Material
Acknowledgments
We thank the patients and their families for participating in this study. We thank the members of both branches of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions; Tatiana Kochetkov for technical assistance; Yoann Seeleuthner, Bertrand Boisson, and Aurélie Cobat for bioinformatic assistance; and Dominick Papandrea, Cécile Patissier, and Yelena Nemirovskaya for administrative assistance.
The work was funded in part by the National Center for Advancing Translational Sciences, National Institutes of Health, Clinical and Translational Science Award program (grant no. UL1TR001866), National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant nos. R01AI088364 and R21AI137371), National Institute of Neurological Disorders and Stroke, National Institutes of Health (grant no. R01NS072381), Cooperative Center on Human Immunology, National Institute of Allergy and Infectious Diseases (grant no. U19AI111825), Rockefeller University, Institut National de la Santé et de la Recherche Médicale, Paris Descartes University, the French National Research Agency under the “Investments for the future” program (grant no. ANR-10-IAHU-01), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (grant no. ANR-10-LABX-62-IBEID), IEIHSEER (grant no. ANR-14-CE14-0008-01), the Center for Research on Influenza Pathogenesis, an National Institute of Allergy and Infectious Diseases-funded Center of Excellence for Influenza Research and Surveillance contract (HHSN272201400008C), the St. Giles Foundation, and the Thrasher Research Fund. N. Hernandez was supported by the Medical Scientist Training Program grant from the National Institute of General Medical Sciences, National Institutes of Health, under award no. T32GM007739. I. Meyts was funded by Fonds voor Wetenschappelijk Onderzoek Vlaanderen project no. G0C8517N. K. Dobbs and L.D. Notarangelo are supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: H.K. Lim, S.X.L. Huang, J. Chen, P. Bastard, K. Dobbs, N. Hernandez, N. Goudin, F.G. Lafaille, L. Lorenzo, P. Luthra, M.L. Hasek, E.J. García, T. Kochetkov, F. Rozenberg, A. García-Sastre, M.J. Ciancanelli, L.D. Notarangelo, and H.-W. Snoeck performed the experiments. Q. Zhang, M.J. Ciancanelli, and S-Y. Zhang coordinated recruitment of the influenza patient cohort. S. Boucherit assisted with patient recruitment. O. Gilliaux, C. Vedrinne, M.D. Keller, M. Celard, J.S. Orange, and I. Meyts contributed patient samples and collected clinical data. H.K. Lim, G. Kerner, O. Gilliaux, P. Bastard, B. Bigio, Y. Itan, I. Meyts, L. Abel, and S-Y. Zhang analyzed the data. J.-L. Casanova and S-Y. Zhang supervised the research. J.-L. Casanova and S.-Y. Zhang wrote the paper with the help of all co-authors.
References
- Alcaïs A., Quintana-Murci L., Thaler D.S., Schurr E., Abel L., and Casanova J.L.. 2010. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann. N. Y. Acad. Sci. 1214:18–33. 10.1111/j.1749-6632.2010.05834.x [DOI] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A.C., Medzhitov R., and Flavell R.A.. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 413:732–738. 10.1038/35099560 [DOI] [PubMed] [Google Scholar]
- Andersen L.L., Mørk N., Reinert L.S., Kofod-Olsen E., Narita R., Jørgensen S.E., Skipper K.A., Höning K., Gad H.H., Østergaard L., et al. 2015. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J. Exp. Med. 212:1371–1379. 10.1084/jem.20142274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ank N., Iversen M.B., Bartholdy C., Staeheli P., Hartmann R., Jensen U.B., Dagnaes-Hansen F., Thomsen A.R., Chen Z., Haugen H., et al. 2008. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J. Immunol. 180:2474–2485. 10.4049/jimmunol.180.4.2474 [DOI] [PubMed] [Google Scholar]
- Audry M., Ciancanelli M., Yang K., Cobat A., Chang H.H., Sancho-Shimizu V., Lorenzo L., Niehues T., Reichenbach J., Li X.X., et al. 2011. NEMO is a key component of NF-κB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J. Allergy Clin. Immunol. 128:610–7.e1: 4. 10.1016/j.jaci.2011.04.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro L.B., Ben-Ali M., Quach H., Laval G., Patin E., Pickrell J.K., Bouchier C., Tichit M., Neyrolles O., Gicquel B., et al. 2009. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 5:e1000562 10.1371/journal.pgen.1000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaya S., Kontorovich A.R., Byun M., Mulero-Navarro S., Bajolle F., Cobat A., Josowitz R., Itan Y., Quint R., Lorenzo L., et al. 2017. Autosomal Recessive Cardiomyopathy Presenting as Acute Myocarditis. J. Am. Coll. Cardiol. 69:1653–1665. 10.1016/j.jacc.2017.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Bousfiha A., Jeddane L., Picard C., Ailal F., Bobby Gaspar H., Al-Herz W., Chatila T., Crow Y.J., Cunningham-Rundles C., Etzioni A., et al. 2018. The 2017 IUIS Phenotypic Classification for Primary Immunodeficiencies. J. Clin. Immunol. 38:129–143. 10.1007/s10875-017-0465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L. 2015a Human genetic basis of interindividual variability in the course of infection. Proc. Natl. Acad. Sci. USA. 112:E7118–E7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L. 2015b Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc. Natl. Acad. Sci. USA. 112:E7128–E7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L., and Abel L.. 2004. The human model: a genetic dissection of immunity to infection in natural conditions. Nat. Rev. Immunol. 4:55–66. 10.1038/nri1264 [DOI] [PubMed] [Google Scholar]
- Casanova J.L., and Abel L.. 2018. Human genetics of infectious diseases: Unique insights into immunological redundancy. Semin. Immunol. 36:1–12. 10.1016/j.smim.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L., Abel L., and Quintana-Murci L.. 2013. Immunology taught by human genetics. Cold Spring Harb. Symp. Quant. Biol. 78:157–172. 10.1101/sqb.2013.78.019968 [DOI] [PubMed] [Google Scholar]
- Casanova J.L., Conley M.E., Seligman S.J., Abel L., and Notarangelo L.D.. 2014. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J. Exp. Med. 211:2137–2149. 10.1084/jem.20140520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casrouge A., Zhang S.Y., Eidenschenk C., Jouanguy E., Puel A., Yang K., Alcais A., Picard C., Mahfoufi N., Nicolas N., et al. 2006. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 314:308–312. 10.1126/science.1128346 [DOI] [PubMed] [Google Scholar]
- Chapgier A., Wynn R.F., Jouanguy E., Filipe-Santos O., Zhang S., Feinberg J., Hawkins K., Casanova J.L., and Arkwright P.D.. 2006. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J. Immunol. 176:5078–5083. 10.4049/jimmunol.176.8.5078 [DOI] [PubMed] [Google Scholar]
- Choe J., Kelker M.S., and Wilson I.A.. 2005. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science. 309:581–585. 10.1126/science.1115253 [DOI] [PubMed] [Google Scholar]
- Ciancanelli M.J., Huang S.X., Luthra P., Garner H., Itan Y., Volpi S., Lafaille F.G., Trouillet C., Schmolke M., Albrecht R.A., et al. 2015. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 348:448–453. 10.1126/science.aaa1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli M.J., Abel L., Zhang S.Y., and Casanova J.L.. 2016. Host genetics of severe influenza: from mouse Mx1 to human IRF7. Curr. Opin. Immunol. 38:109–120. 10.1016/j.coi.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin M., Dickinson R., and Bigley V.. 2015. Haematopoietic and immune defects associated with GATA2 mutation. Br. J. Haematol. 169:173–187. 10.1111/bjh.13317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottineau J., Kottemann M.C., Lach F.P., Kang Y.H., Vély F., Deenick E.K., Lazarov T., Gineau L., Wang Y., Farina A., et al. 2017. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J. Clin. Invest. 127:1991–2006. 10.1172/JCI90727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Megjugorac N.J., Amrute S.B., and Fitzgerald-Bocarsly P.. 2004. Regulation of IFN regulatory factor-7 and IFN-alpha production by enveloped virus and lipopolysaccharide in human plasmacytoid dendritic cells. J. Immunol. 173:1535–1548. 10.4049/jimmunol.173.3.1535 [DOI] [PubMed] [Google Scholar]
- Davidson S., McCabe T.M., Crotta S., Gad H.H., Hessel E.M., Beinke S., Hartmann R., and Wack A.. 2016. IFNλ is a potent anti-influenza therapeutic without the inflammatory side effects of IFNα treatment. EMBO Mol. Med. 8:1099–1112. 10.15252/emmm.201606413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.M., and Brodin P.. 2018. Rebooting Human Immunology. Annu. Rev. Immunol. 36:843–864. 10.1146/annurev-immunol-042617-053206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood F.S., Kamimoto L., D’Mello T.A., Reingold A., Gershman K., Meek J., Arnold K.E., Farley M., Ryan P., Lynfield R., et al. Emerging Infections Program Network . 2011. Children with asthma hospitalized with seasonal or pandemic influenza, 2003-2009. Pediatrics. 128:e27–e32. 10.1542/peds.2010-3343 [DOI] [PubMed] [Google Scholar]
- Deeg C.M., Hassan E., Mutz P., Rheinemann L., Götz V., Magar L., Schilling M., Kallfass C., Nürnberger C., Soubies S., et al. 2017. In vivo evasion of MxA by avian influenza viruses requires human signature in the viral nucleoprotein. J. Exp. Med. 214:1239–1248. 10.1084/jem.20161033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weerd N.A., Vivian J.P., Nguyen T.K., Mangan N.E., Gould J.A., Braniff S.J., Zaker-Tabrizi L., Fung K.Y., Forster S.C., Beddoe T., et al. 2013. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 14:901–907. 10.1038/ni.2667 [DOI] [PubMed] [Google Scholar]
- Donadieu J., Lamant M., Fieschi C., de Fontbrune F.S., Caye A., Ouachee M., Beaupain B., Bustamante J., Poirel H.A., Isidor B., et al. French GATA2 study group . 2018. Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica. 103:1278–1287. 10.3324/haematol.2017.181909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C.J., Mohamad S.M., Young D.F., Skelton A.J., Leahy T.R., Munday D.C., Butler K.M., Morfopoulou S., Brown J.R., Hubank M., et al. 2015. Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci. Transl. Med. 7:307ra154 10.1126/scitranslmed.aac4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletto D., Burns S.O., Angulo I., Plagnol V., Gilmour K.C., Henriquez F., Curtis J., Gaspar M., Nowak K., Daza-Cajigal V., et al. 2016. Biallelic JAK1 mutations in immunodeficient patient with mycobacterial infection. Nat. Commun. 7:13992 10.1038/ncomms13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FluSurv-NET 2018. http://gis.cdc.gov/grasp/fluview/FluHospChars.html.
- Fuchs S., Kaiser-Labusch P., Bank J., Ammann S., Kolb-Kokocinski A., Edelbusch C., Omran H., and Ehl S.. 2016. Tyrosine kinase 2 is not limiting human antiviral type III interferon responses. Eur. J. Immunol. 46:2639–2649. 10.1002/eji.201646519 [DOI] [PubMed] [Google Scholar]
- Garcia-Cattaneo A., Gobert F.X., Müller M., Toscano F., Flores M., Lescure A., Del Nery E., and Benaroch P.. 2012. Cleavage of Toll-like receptor 3 by cathepsins B and H is essential for signaling. Proc. Natl. Acad. Sci. USA. 109:9053–9058. 10.1073/pnas.1115091109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gineau L., Cognet C., Kara N., Lach F.P., Dunne J., Veturi U., Picard C., Trouillet C., Eidenschenk C., Aoufouchi S., et al. 2012. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J. Clin. Invest. 122:821–832. 10.1172/JCI61014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser C.A., Winter K., DuBray K., Harriman K., Uyeki T.M., Sejvar J., Gilliam S., and Louie J.K.. 2012. A population-based study of neurologic manifestations of severe influenza A(H1N1)pdm09 in California. Clin. Infect. Dis. 55:514–520. 10.1093/cid/cis454 [DOI] [PubMed] [Google Scholar]
- Glocker E.O., Kotlarz D., Boztug K., Gertz E.M., Schäffer A.A., Noyan F., Perro M., Diestelhorst J., Allroth A., Murugan D., et al. 2009. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361:2033–2045. 10.1056/NEJMoa0907206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbea C., Makar K.A., Pauschinger M., Pratt G., Bersola J.L., Varela J., David R.M., Banks L., Huang C.H., Li H., et al. 2010. A role for Toll-like receptor 3 variants in host susceptibility to enteroviral myocarditis and dilated cardiomyopathy. J. Biol. Chem. 285:23208–23223. 10.1074/jbc.M109.047464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot L., Le Goffic R., Bloch S., Escriou N., Akira S., Chignard M., and Si-Tahar M.. 2005. Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J. Biol. Chem. 280:5571–5580. 10.1074/jbc.M410592200 [DOI] [PubMed] [Google Scholar]
- Guo Y., Audry M., Ciancanelli M., Alsina L., Azevedo J., Herman M., Anguiano E., Sancho-Shimizu V., Lorenzo L., Pauwels E., et al. 2011. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J. Exp. Med. 208:2083–2098. 10.1084/jem.20101568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambleton S., Goodbourn S., Young D.F., Dickinson P., Mohamad S.M., Valappil M., McGovern N., Cant A.J., Hackett S.J., Ghazal P., et al. 2013. STAT2 deficiency and susceptibility to viral illness in humans. Proc. Natl. Acad. Sci. USA. 110:3053–3058. 10.1073/pnas.1220098110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman M., Ciancanelli M., Ou Y.H., Lorenzo L., Klaudel-Dreszler M., Pauwels E., Sancho-Shimizu V., Pérez de Diego R., Abhyankar A., Israelsson E., et al. 2012. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med. 209:1567–1582. 10.1084/jem.20111316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermant P., and Michiels T.. 2014. Interferon-λ in the context of viral infections: production, response and therapeutic implications. J. Innate Immun. 6:563–574. 10.1159/000360084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Melki I., Jing H., Habib T., Huang S.S.Y., Danielson J., Kula T., Drutman S., Belkaya S., Rattina V., et al. 2018. Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J. Exp. Med. 215:2567–2585. 10.1084/jem.20180628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., and Taniguchi T.. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 434:772–777. 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- Hoyos-Bachiloglu R., Chou J., Sodroski C.N., Beano A., Bainter W., Angelova M., Al Idrissi E., Habazi M.K., Alghamdi H.A., Almanjomi F., et al. 2017. A digenic human immunodeficiency characterized by IFNAR1 and IFNGR2 mutations. J. Clin. Invest. 127:4415–4420. 10.1172/JCI93486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A.P., McReynolds L.J., and Holland S.M.. 2015. GATA2 deficiency. Curr. Opin. Allergy Clin. Immunol. 15:104–109. 10.1097/ACI.0000000000000126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.X., Islam M.N., O’Neill J., Hu Z., Yang Y.G., Chen Y.W., Mumau M., Green M.D., Vunjak-Novakovic G., Bhattacharya J., and Snoeck H.W.. 2014. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 32:84–91. 10.1038/nbt.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C.R., Guasti L., Meimaridou E., Chuang C.H., Schimenti J.C., King P.J., Costigan C., Clark A.J., and Metherell L.A.. 2012. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J. Clin. Invest. 122:814–820. 10.1172/JCI60224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis I., Ye F., McNally B., Willette M., and Flaño E.. 2013. Toll-like receptor expression and induction of type I and type III interferons in primary airway epithelial cells. J. Virol. 87:3261–3270. 10.1128/JVI.01956-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan Y., Zhang S.Y., Vogt G., Abhyankar A., Herman M., Nitschke P., Fried D., Quintana-Murci L., Abel L., and Casanova J.L.. 2013. The human gene connectome as a map of short cuts for morbid allele discovery. Proc. Natl. Acad. Sci. USA. 110:5558–5563. 10.1073/pnas.1218167110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan Y., Shang L., Boisson B., Ciancanelli M.J., Markle J.G., Martinez-Barricarte R., Scott E., Shah I., Stenson P.D., Gleeson J., et al. 2016. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat. Methods. 13:109–110. 10.1038/nmeth.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre A., Barnes B.J., Amrute S., Yeow W.S., Megjugorac N., Dai J., Feng D., Chung E., Pitha P.M., and Fitzgerald-Bocarsly P.. 2003. Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells. J. Leukoc. Biol. 74:1125–1138. 10.1189/jlb.0603255 [DOI] [PubMed] [Google Scholar]
- Jaber S., Conseil M., Coisel Y., Jung B., and Chanques G.. 2010. [ARDS and influenza A (H1N1): patients’ characteristics and management in intensive care unit. A literature review]. Ann. Fr. Anesth. Reanim. 29:117–125. 10.1016/j.annfar.2009.12.026 [DOI] [PubMed] [Google Scholar]
- Jacobs B.L., and Langland J.O.. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 219:339–349. 10.1006/viro.1996.0259 [DOI] [PubMed] [Google Scholar]
- Jewell N.A., Cline T., Mertz S.E., Smirnov S.V., Flaño E., Schindler C., Grieves J.L., Durbin R.K., Kotenko S.V., and Durbin J.E.. 2010. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J. Virol. 84:11515–11522. 10.1128/JVI.01703-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki N., Ho S., Antonenko S., Malefyt R.W., Kastelein R.A., Bazan F., and Liu Y.J.. 2001. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J. Exp. Med. 194:863–869. 10.1084/jem.194.6.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105. 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- Kawai T., and Akira S.. 2006. TLR signaling. Cell Death Differ. 13:816–825. 10.1038/sj.cdd.4401850 [DOI] [PubMed] [Google Scholar]
- Kerkmann M., Rothenfusser S., Hornung V., Towarowski A., Wagner M., Sarris A., Giese T., Endres S., and Hartmann G.. 2003. Activation with CpG-A and CpG-B oligonucleotides reveals two distinct regulatory pathways of type I IFN synthesis in human plasmacytoid dendritic cells. J. Immunol. 170:4465–4474. 10.4049/jimmunol.170.9.4465 [DOI] [PubMed] [Google Scholar]
- Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkhammer J., Schnepf D., Ye L., Schwaderlapp M., Gad H.H., Hartmann R., Garcin D., Mahlakõiv T., and Staeheli P.. 2018. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife. 7:e33354 10.7554/eLife.33354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreins A.Y., Ciancanelli M.J., Okada S., Kong X.F., Ramírez-Alejo N., Kilic S.S., El Baghdadi J., Nonoyama S., Mahdaviani S.A., Ailal F., et al. 2015. Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 212:1641–1662. 10.1084/jem.20140280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., and Carmichael G.G.. 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62:1415–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaille F.G., Pessach I.M., Zhang S.Y., Ciancanelli M.J., Herman M., Abhyankar A., Ying S.W., Keros S., Goldstein P.A., Mostoslavsky G., et al. 2012. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature. 491:769–773. 10.1038/nature11583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Nice T.J., and Diamond M.S.. 2015. Interferon-λ: Immune Functions at Barrier Surfaces and Beyond. Immunity. 43:15–28. 10.1016/j.immuni.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goffic R., Balloy V., Lagranderie M., Alexopoulou L., Escriou N., Flavell R., Chignard M., and Si-Tahar M.. 2006. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2:e53 10.1371/journal.ppat.0020053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goffic R., Pothlichet J., Vitour D., Fujita T., Meurs E., Chignard M., and Si-Tahar M.. 2007. Cutting Edge: Influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J. Immunol. 178:3368–3372. 10.4049/jimmunol.178.6.3368 [DOI] [PubMed] [Google Scholar]
- Lester-Smith D., Zurynski Y.A., Booy R., Festa M.S., Kesson A.M., and Elliott E.J.. 2009. The burden of childhood influenza in a tertiary paediatric setting. Commun. Dis. Intell. Q. Rep. 33:209–215. [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.K., Seppänen M., Hautala T., Ciancanelli M.J., Itan Y., Lafaille F.G., Dell W., Lorenzo L., Byun M., Pauwels E., et al. 2014. TLR3 deficiency in herpes simplex encephalitis: high allelic heterogeneity and recurrence risk. Neurology. 83:1888–1897. 10.1212/WNL.0000000000000999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Botos I., Wang Y., Leonard J.N., Shiloach J., Segal D.M., and Davies D.R.. 2008. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 320:379–381. 10.1126/science.1155406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlakõiv T., Hernandez P., Gronke K., Diefenbach A., and Staeheli P.. 2015. Leukocyte-derived IFN-α/β and epithelial IFN-λ constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog. 11:e1004782 10.1371/journal.ppat.1004782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B., Manicassamy S., Belicha-Villanueva A., Pisanelli G., Pulendran B., and García-Sastre A.. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. USA. 107:11531–11536. 10.1073/pnas.0914994107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcenaro E., Notarangelo L.D., Orange J.S., and Vivier E.. 2017. Editorial: NK Cell Subsets in Health and Disease: New Developments. Front. Immunol. 8:1363 10.3389/fimmu.2017.01363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J.A. 2014. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 12:252–262. 10.1038/nrmicro3231 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M.A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami T., Miyashita H., Takatsuka S., Kuroki Y., and Matsushima N.. 2012. Molecular evolution of vertebrate Toll-like receptors: evolutionary rate difference between their leucine-rich repeats and their TIR domains. Gene. 503:235–243. 10.1016/j.gene.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Minegishi Y., Saito M., Morio T., Watanabe K., Agematsu K., Tsuchiya S., Takada H., Hara T., Kawamura N., Ariga T., et al. 2006. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 25:745–755. 10.1016/j.immuni.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Moens L., Van Eyck L., Jochmans D., Mitera T., Frans G., Bossuyt X., Matthys P., Neyts J., Ciancanelli M., Zhang S.Y., et al. 2017. A novel kindred with inherited STAT2 deficiency and severe viral illness. J. Allergy Clin. Immunol. 139:1995–1997.e9. 10.1016/j.jaci.2016.10.033 [DOI] [PubMed] [Google Scholar]
- Navarro S., Colamonici O.R., and Llombart-Bosch A.. 1996. Immunohistochemical detection of the type I interferon receptor in human fetal, adult, and neoplastic tissues. Mod. Pathol. 9:150–156. [PubMed] [Google Scholar]
- Onodera K., Fujiwara T., Onishi Y., Itoh-Nakadai A., Okitsu Y., Fukuhara N., Ishizawa K., Shimizu R., Yamamoto M., and Harigae H.. 2016. GATA2 regulates dendritic cell differentiation. Blood. 128:508–518. 10.1182/blood-2016-02-698118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquet M., Bellanné-Chantelot C., Tavitian S., Prade N., Beaupain B., Larochelle O., Petit A., Rohrlich P., Ferrand C., Van Den Neste E., et al. 2013. High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood. 121:822–829. 10.1182/blood-2012-08-447367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez de Diego R., Sancho-Shimizu V., Lorenzo L., Puel A., Plancoulaine S., Picard C., Herman M., Cardon A., Durandy A., Bustamante J., et al. 2010. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 33:400–411. 10.1016/j.immuni.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone L.A., Plowden J.K., García-Sastre A., Katz J.M., and Tumpey T.M.. 2008. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 4:e1000115 10.1371/journal.ppat.1000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard C., Bobby Gaspar H., Al-Herz W., Bousfiha A., Casanova J.L., Chatila T., Crow Y.J., Cunningham-Rundles C., Etzioni A., Franco J.L., et al. 2018. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J. Clin. Immunol. 38:96–128. 10.1007/s10875-017-0464-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B., and Maddur M.S.. 2015. Innate immune sensing and response to influenza. Curr. Top. Microbiol. Immunol. 386:23–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L., Alcaïs A., Abel L., and Casanova J.L.. 2007. Immunology in natura: clinical, epidemiological and evolutionary genetics of infectious diseases. Nat. Immunol. 8:1165–1171. 10.1038/ni1535 [DOI] [PubMed] [Google Scholar]
- Sancho-Shimizu V., Pérez de Diego R., Lorenzo L., Halwani R., Alangari A., Israelsson E., Fabrega S., Cardon A., Maluenda J., Tatematsu M., et al. 2011. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J. Clin. Invest. 121:4889–4902. 10.1172/JCI59259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M.L., and Palese P.. 2013. Orthomyxoviridae. In Fields Virology . Ninth edition. Lippincott Williams & Wilkins, Philadelphia. 1151–1185. [Google Scholar]
- Shieh W.J., Blau D.M., Denison A.M., Deleon-Carnes M., Adem P., Bhatnagar J., Sumner J., Liu L., Patel M., Batten B., et al. 2010. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am. J. Pathol. 177:166–175. 10.2353/ajpath.2010.100115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.R., Sedani S., Lim M., Wassmer E., and Absoud M.. 2015. RANBP2 mutation and acute necrotizing encephalopathy: 2 cases and a literature review of the expanding clinico-radiological phenotype. Eur. J. Paediatr. Neurol. 19:106–113. 10.1016/j.ejpn.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Sologuren I., Martínez-Saavedra M.T., Solé-Violán J., de Borges de Oliveira E. Jr., Betancor E., Casas I., Oleaga-Quintas C., Martínez-Gallo M., Zhang S.Y., Pestano J., et al. 2018. Lethal Influenza in Two Related Adults with Inherited GATA2 Deficiency. J. Clin. Immunol. 38:513–526. 10.1007/s10875-018-0512-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommereyns C., Paul S., Staeheli P., and Michiels T.. 2008. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4:e1000017 10.1371/journal.ppat.1000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., and Leaman D.W.. 2004. Ectopic expression of toll-like receptor-3 (TLR-3) overcomes the double-stranded RNA (dsRNA) signaling defects of P2.1 cells. J. Interferon Cytokine Res. 24:350–361. 10.1089/107999004323142213 [DOI] [PubMed] [Google Scholar]
- Surtees R., and DeSousa C.. 2006. Influenza virus associated encephalopathy. Arch. Dis. Child. 91:455–456. 10.1136/adc.2005.092890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscano F., Estornes Y., Virard F., Garcia-Cattaneo A., Pierrot A., Vanbervliet B., Bonnin M., Ciancanelli M.J., Zhang S.Y., Funami K., et al. 2013. Cleaved/associated TLR3 represents the primary form of the signaling receptor. J. Immunol. 190:764–773. 10.4049/jimmunol.1202173 [DOI] [PubMed] [Google Scholar]
- Wang F., Song W., Brancati G., and Segatori L.. 2011. Inhibition of endoplasmic reticulum-associated degradation rescues native folding in loss of function protein misfolding diseases. J. Biol. Chem. 286:43454–43464. 10.1074/jbc.M111.274332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu L., Davies D.R., and Segal D.M.. 2010. Dimerization of Toll-like receptor 3 (TLR3) is required for ligand binding. J. Biol. Chem. 285:36836–36841. 10.1074/jbc.M110.167973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Wagner V., Rasmussen S.B., Hartmann R., and Paludan S.R.. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059–5064. 10.1128/JVI.80.10.5059-5064.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werder R.B., Lynch J.P., Simpson J.C., Zhang V., Hodge N.H., Poh M., Forbes-Blom E., Kulis C., Smythe M.L., Upham J.W., et al. 2018. PGD2/DP2 receptor activation promotes severe viral bronchiolitis by suppressing IFN-λ production. Sci. Transl. Med. 10:eaao0052 10.1126/scitranslmed.aao0052 [DOI] [PubMed] [Google Scholar]
- Wieland S.F., and Heim M.H.. 2019. The IFN-λ Pony Express. Nat. Immunol. 20:522–524. 10.1038/s41590-019-0362-9 [DOI] [PubMed] [Google Scholar]
- World Health Organization 2018. Influenza (Seasonal) fact sheet. Available at: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed August 2018).
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., and Fujita T.. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737. 10.1038/ni1087 [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y.M., Gale M. Jr., Akira S., et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858. 10.4049/jimmunol.175.5.2851 [DOI] [PubMed] [Google Scholar]
- Yu X., Tsibane T., McGraw P.A., House F.S., Keefer C.J., Hicar M.D., Tumpey T.M., Pappas C., Perrone L.A., Martinez O., et al. 2008. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 455:532–536. 10.1038/nature07231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Y., Jouanguy E., Sancho-Shimizu V., von Bernuth H., Yang K., Abel L., Picard C., Puel A., and Casanova J.L.. 2007a Human Toll-like receptor-dependent induction of interferons in protective immunity to viruses. Immunol. Rev. 220:225–236. 10.1111/j.1600-065X.2007.00564.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., et al. 2007b TLR3 deficiency in patients with herpes simplex encephalitis. Science. 317:1522–1527. 10.1126/science.1139522 [DOI] [PubMed] [Google Scholar]
- Zhang S.Y., Herman M., Ciancanelli M.J., Pérez de Diego R., Sancho-Shimizu V., Abel L., and Casanova J.L.. 2013. TLR3 immunity to infection in mice and humans. Curr. Opin. Immunol. 25:19–33. 10.1016/j.coi.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Y., Clark N.E., Freije C.A., Pauwels E., Taggart A.J., Okada S., Mandel H., Garcia P., Ciancanelli M.J., Biran A., et al. 2018. Inborn Errors of RNA Lariat Metabolism in Humans with Brainstem Viral Infection. Cell. 172:952–965.e18. 10.1016/j.cell.2018.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Y., Jouanguy E., Zhang Q., Abel L., Puel A., and Casanova J.L.. 2019. Human inborn errors of immunity to infection affecting cells other than leukocytes: from the immune system to the whole organism. Curr. Opin. Immunol. 59:88–100. 10.1016/j.coi.2019.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer B., Ewaleifoh O., Harschnitz O., Lee Y.S., Peneau C., McAlpine J.L., Liu B., Tchieu J., Steinbeck J.A., Lafaille F., et al. 2018. Human iPSC-derived trigeminal neurons lack constitutive TLR3-dependent immunity that protects cortical neurons from HSV-1 infection. Proc. Natl. Acad. Sci. USA. 115:E8775–E8782. 10.1073/pnas.1809853115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.