Itch is a catalytic ubiquitin ligase that prevents autoimmunity in humans and mice. Here, Moser et al. show that Itch negatively regulates B cell proliferation and metabolic fitness to limit antigen-driven B cell responses and antibody production after immunization.
Abstract
The E3 ubiquitin ligase Itch regulates antibody levels and prevents autoimmune disease in humans and mice, yet how Itch regulates B cell fate or function is unknown. We now show that Itch directly limits B cell activity. While Itch-deficient mice displayed normal numbers of preimmune B cell populations, they showed elevated numbers of antigen-experienced B cells. Mixed bone marrow chimeras revealed that Itch acts within B cells to limit naive and, to a greater extent, germinal center (GC) B cell numbers. B cells lacking Itch exhibited increased proliferation, glycolytic capacity, and mTORC1 activation. Moreover, stimulation of these cells in vivo by WT T cells resulted in elevated numbers of GC B cells, PCs, and serum IgG. These results support a novel role for Itch in limiting B cell metabolism and proliferation to suppress antigen-driven B cell responses.
Introduction
Antibodies are an important component of both protective immunity and autoimmunity. Antibody production occurs when B cells become activated under conditions that promote their differentiation into plasma cells (PCs), and the quality of antibody produced (e.g., isotype, affinity, longevity) is shaped by micro-environmental cues (Shapiro-Shelef and Calame, 2005; Corcoran and Tarlinton, 2016). Most high-affinity class-switched antibodies are derived from B cells that have received signals from T follicular helper (Tfh) cells within germinal centers (GCs), specialized sites of B cell affinity maturation (Berek et al., 1991; Jacob et al., 1991; Victora and Nussenzweig, 2012). Within GCs, B cell maturation into PCs is tightly regulated to ensure production of robust pathogen-specific antibodies and prevent the generation and secretion of autoreactive antibodies. Despite their importance, few therapeutic strategies exist to modulate the magnitude and quality of antibodies elicited after vaccination or during the development of autoimmune disease. A better understanding of the regulatory circuits that control maturation of GC B cells and antibody responses could result in new therapies for controlling antibody levels.
Itch is a ubiquitin ligase that regulates antibody levels in both humans and mice. Mice with a spontaneous mutation in the Itch promoter lack Itch protein and exhibit elevated serum antibody and autoantibody (Perry et al., 1998; Matesic et al., 2006; Parravicini et al., 2008). Similarly, a loss-of-function mutation in the Itch gene has been identified in humans with severe multi-faceted autoimmune disease, accompanied by the production of autoantibodies (Lohr et al., 2010). Despite the likely role for high antibody levels in driving the pathologies observed in Itch deficiency, the mechanisms by which Itch functions to control B cells and antibody production are largely unexplored. To date, much of what is known about how Itch prevents inflammation and immune dysregulation has focused on T helper (Th) cells.
Studies of Itch-deficient mice revealed that Itch limits T cell activation and Th differentiation. Specifically, Itch-deficient T cells are more resistant to anergy induction, are more likely to differentiate into Th2 cells, and are less likely to become Tfh cells (Fang et al., 2002; Venuprasad et al., 2006; Ramos-Hernández et al., 2013; Xiao et al., 2014). This latter finding is surprising when considering the high class-switched antibody levels in these mice. Additionally, it was shown that B lymphocytes that lacked Itch exhibited defects in antigen-triggered B cell receptor (BCR) trafficking into vesicles associated with antigen processing in vitro (Zhang et al., 2007; Xiao et al., 2014). These data would imply that Itch-deficient B cells would be poor antigen-presenting cells to T cells and would be less likely to differentiate into antibody-producing PCs. Thus, the current description of Itch function cannot explain why Itch deficiency results in increased total serum antibody and the emergence of autoantibodies.
In this study, we investigated how Itch regulates the generation of antibody producing B cells and their production of class-switched antibody. We found that Itch acts within B cells to limit the numbers of GC B cells and PCs. In vitro, Itch functions as a negative regulator of B cell proliferation and metabolic fitness subsequent to activation of cells by diverse stimuli. Itch limited mTORC1 activity within hours after B cell activation, supporting a role for Itch in regulating early activation pathways downstream of both the BCR and TLR9. Finally, we determined that loss of Itch in B cells is sufficient to drive increased B cell responses to immunization in vivo, and that GC B cells lacking Itch exhibited enhanced proliferation and mTORC1 activity, associated with increased persistence, output of PCs, and production of class-switched antibodies. Our data establish Itch as a novel negative regulator of activated B cells.
Results
Itch limits quantity of serum antibody and activated B cells
Itch-deficient mice develop increased levels of serum antibodies and autoantibodies (Matesic et al., 2006; Parravicini et al., 2008). Accordingly, we examined serum IgM, IgG1, and IgG2c levels, as well as IgG anti–double-stranded (ds) DNA in mice lacking Itch (Itch KO). Consistent with published data, we found that serum IgM and IgG1 were markedly elevated in Itch KO mice (Fig. 1 A), and levels of autoantibodies were clearly detectible above age-matched controls (Fig. 1 B), albeit not as high as can be seen in New Zealand black × New Zealand white F1 mice, a commonly used model for antibody-mediated lupus-like autoimmune disease (Dubois et al., 1966; Morel, 2010). Thus, Itch is required to limit antibody production and prevent development of autoantibodies in vivo.
Figure 1.
Itch limits serum antibody and activated B cells. (A) Quantity of total antibody in serum of WT and Itch KO mice was determined by ELISA (mice between 8 and 12 wk old, n = 4–8, compiled from two independent experiments, multiple t tests, Holm–Sidak correction). (B) Anti-dsDNA IgG in serum was determined by ELISA (mice between 12 and 24 wk, n = 9–13, compiled from three independent experiments, two-way ANOVA). Serum from New Zealand black × New Zealand white F1 mice was included as a positive control (n = 2). (C and D) Preimmune and activated B cell subsets in spleen were quantified by flow cytometry, and representative flow cytometry plots are shown. Cells were gated on live singlets, and then transitional B cells were CD19+CD93+, FO B cells were CD19+CD93−CD23+, and marginal zone B cells were CD19+CD23lo/−CD21+. For GC and class-switched non-GC B cells, cells were first gated on IgD−CD4−CD8−F480−GR1−CD19+IgM−, and then GC B cells were GL7+CD38−, and class-switched non-GC B cells were GL7− (n = 6–8, compiled from two independent experiments, Mann–Whitney test, WT mice are C57Bl/6). (E) Total IgM-, IgG1-, and IgG2c-secreting PCs were enumerated by ELISPOT (n = 8, Mann–Whitney test, compiled from two independent experiments; error bars indicate SEM; WT mice are C57Bl/6). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
To determine how Itch might function to limit antibody production, we first assessed the numbers and proportions of B cell subsets in the spleen, with the prediction that one or more B cell populations might be perturbed in mice lacking Itch, providing clues to the etiology of the dysregulated antibody production. The numbers of mature preimmune B cells, i.e., transitional, marginal zone, and follicular (FO) B cells, were comparable in number to control mice (Fig. 1 C), suggesting that Itch plays little role in determining B cell lineage output from bone marrow (BM) or in the maintenance of mature naive B cells. However, we found a striking elevation in the numbers of GC B cells and class-switched non-GC B cells in the spleen, and PCs in the BM (Fig. 1, D and E). Class-switched non-GC B cells are a mixed population of activated cells that are not part of GCs, including memory cells.
Itch KO mice display marked Th2 cell–dependent inflammation, which is largely alleviated in mice that also lack IL-4 (Itch/IL4 double KO [DKO] mice; Fang et al., 2002; Moser et al., 2018). Because IL-4 is a potent regulator of B cell class switching and survival (Schultz and Coffman, 1991; Wurster et al., 2002), we next determined whether the increased number of GC B cells, class-switched non-GC B cells, and antibodies were due to B cell exposure to increased IL-4, or another mechanism. We examined serum antibodies and splenic B cells in IL4 KO and Itch/IL4 DKO mice. We found that serum IgM was restored to normal levels after ablation of IL-4. However, IgG remained elevated in Itch/IL4 DKO mice. Interestingly, instead of increased IgG1 (an IL-4–dependent isotype), we saw increased IgG2c (Fig. S1 A). We next examined splenic B cell subsets and found that total numbers of FO, GC, and class-switched non-GC B cells were elevated in Itch/IL4 DKO mice compared with IL4 KO mice (Fig. S1 B). These data support that Itch likely functions to limit numbers of activated B cells and class-switched antibody production by a mechanism that is distinct from its role in controlling IL-4 production. Furthermore, the elevated serum antibody in Itch KO mice likely arose from T-dependent interactions because it was class-switched, and the isotype was dependent upon cytokines likely produced by T cells.
Itch acts within B cells to negatively regulate GC B cell numbers
GC B cells require close interactions with multiple cell types (Vinuesa et al., 2010). Itch is ubiquitously expressed in many cell types; thus, the observed increase in GC B cells in Itch KO mice could be a result of loss of Itch function in B cells, or other cell types. Notably, Itch controls several Th cell subsets that could impact GC B cell function, including Tfh cells, regulatory T cells, Th2 cells, and Th17 cells (Fang et al., 2002; Jin et al., 2013; Xiao et al., 2014; Kathania et al., 2016). Importantly, we found that Itch was expressed in both FO and GC B cells (Fig. S2), supporting that it could also function within B cells. To determine if Itch limits GC B cell numbers through an intrinsic function within B cells, or indirectly by acting in another cell type, we generated mixed BM chimeras. To do this, we injected congenically marked BM from Itch KO and WT mice into lethally irradiated WT mice as illustrated in Fig. 2 A. Following repopulation of the immune compartment, we evaluated peripheral B cell populations. We found that the proportion of CD45+ immune cells that arose from CD45.1 and CD45.2 BM cells was approximately equal in the chimeras (Fig. 2 B and Fig. S3 A). However, Itch KO B cells made up a moderately higher proportion of the FO B cell population in the spleen (60% from Itch KO compared with 40% from WT), and a significantly more disproportionate fraction of the GC pool (80% from Itch KO compared with 20% from WT; Fig. 2 B and Fig. S3, B and C). This enhanced skewing in the GC B cell population was observed in all chimeras. Supporting this, the proportion of Itch KO GC B cells relative to Itch KO FO B cells was threefold higher than the same ratio of the WT populations (Fig. 2 C). To ensure that the increase in CD45.2+ B cells was a consequence of Itch function within B cells and not due to CD45.1/CD45.2 differences, we compared WT–WT mixed chimeras to WT–Itch KO mixed chimeras. We found that the frequency of GC B cells within the CD45.2 compartment was significantly elevated relative to CD45.1 cells only when the CD45.2 cells lacked Itch (Fig. S3 D). These data indicate that Itch functions within B cells to limit B cell numbers, and this effect is amplified within in the GC compartment.
Figure 2.
Itch acts within B cells to negatively regulate GC B cell numbers. (A) Experimental design. (B) Percent of WT (B6.SJL [CD45.1]) and Itch KO (CD45.2) cells within immune populations was enumerated by flow cytometry (n = 6, compiled from two independent experiments, two-way ANOVA with Sidak post-test). (C) Percent of GC B cells of each genotype was divided by percent of FO B cells from the same genotype (n = 6 compiled from two independent experiments, paired t test). (D) NP+ GC B cells were analyzed on day 14 after immunization. Percent of NP+ GC B cells from each genotype was divided by percent FO B cells from the same genotype (n = 4, compiled from two independent experiments, paired t test). (E) Equal numbers of flow cytometry–sorted CD45.1+ and CD45.2+ cells were plated on an NP-BSA–coated ELISPOT plate, and IgG1-secreting PCs were enumerated (n = 5, compiled from two independent experiments, paired t test). For A–E, two sets of BM donors were used to generate the chimeras. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Spontaneous GCs occur continuously in response to undefined environmental and self-antigens. To determine whether Itch limits acutely induced B cell responses to a specific foreign antigen, we immunized mixed BM chimeras with nitrophenyl acetyl (NP)–conjugated OVA in alum. After different time points following immunization, we determined the relative contribution of WT and Itch KO B cells to the population of NP-specific GC B cells and NP-specific PCs in the spleen. Similar to what we observed with the steady state GC B cell populations, NP-specific GC B cells lacking Itch were found in much higher frequency that their WT counterparts (Fig. 2 D). Interestingly, NP+ WT GCs appear to exhibit defective expansion, which may indicate that Itch KO B cells out-compete WT GC B cells for T cell help, excluding them from the GC. Importantly, NP-specific class-switched IgG1 PCs were more frequent among the Itch KO CD45.2 population compared with the CD45.1 WT population (Fig. 2 E), supporting that the function of Itch within B cells has significant consequences for class-switched antigen-specific antibody production.
Proteomic profiling revealed Itch limits cell cycle and mTORC1 in activated B cells
Itch is a ubiquitin ligase that mediates covalent addition of ubiquitin molecules to substrate proteins in order to govern their function and stability (Perry et al., 1998; Aki et al., 2015). To explore the cellular processes regulated by Itch in activated B cells, we used quantitative mass spectrometry to profile the proteomes of WT and Itch-deficient B cells that were activated in vitro. To avoid quantifying proteins that were altered due to exposure of cells to IL-4 before harvest, we used Itch/IL4 DKO and IL4 KO mice for these experiments. We isolated FO B cells from spleens of IL4 KO and Itch/IL4 DKO mice, stimulated the cells with the TLR9 agonist CpG, and harvested cell lysates after 24 h, before cell division, at a time when WT cells express Itch (Fig. S4 A). The proteomes were quantified using multidimensional protein and peptide fractionation followed by tandem mass spectrometry (Layman et al., 2017). More than 6,000 proteins were quantified, with 163 proteins displaying significantly increased or decreased abundance in Itch-deficient B cells based on significant P values (Fig. 3 A). Significantly up- and down-regulated proteins were entered into the Broad Institute Molecular Signatures Database (Liberzon et al., 2015) and searched within the hallmark gene sets. Several hallmark gene sets were enriched within the proteomics data. Two of the most significantly enriched hallmarks were “genes upregulated through activation of MTORC1 complex,” and “cell cycle–related targets of E2F transcription factors” (Fig. 3 B). Importantly, the enriched proteins in these two hallmark gene sets were almost all up-regulated in the Itch-deficient samples. For the mTORC1-related hallmark, 9 out of 11 proteins were up-regulated in Itch/IL4 DKO B cells (Fig. 3 C), and for the cell cycle–related hallmark, six out of eight proteins were up-regulated in Itch/IL4 DKO B cells (Fig. 3 D). These results suggested that Itch negatively regulates mTORC1 activity and cell cycle progression in B cells.
Figure 3.
Proteomic profile of Itch KO B cells. (A) Itch-deficient (Itch/IL4 DKO) and control (IL4 KO) FO B cells were stimulated for 24 h with 0.1 μM CpG. Cells were lysed, fractionated, trypsin-digested, and subjected to liquid chromatography and tandem mass spectrometry analysis followed by protein assignation and quantification by MaxQuant. Normalized intensity-based absolute quantification values (for protein abundance) were compared between genotypes to generate fold change. Red dots indicate 163 proteins that were statistically significantly different (n = 3, t test corrected for repeated measures, P < 0.05, IL4 KO mice were on a B6.SJL background [CD45.1]). (B) Top Molecular Signatures Database hallmarks indicated by the 163 proteins that were different between Itch KO and control cells. FDR, false discovery rate. (C) Gene names for the proteins within the top hallmark (genes up-regulated through activation of mTORC1). (D) Gene names for the proteins within the second-ranked hallmark (cell cycle–related targets of E2F transcription factors).
Itch limits B cell proliferation
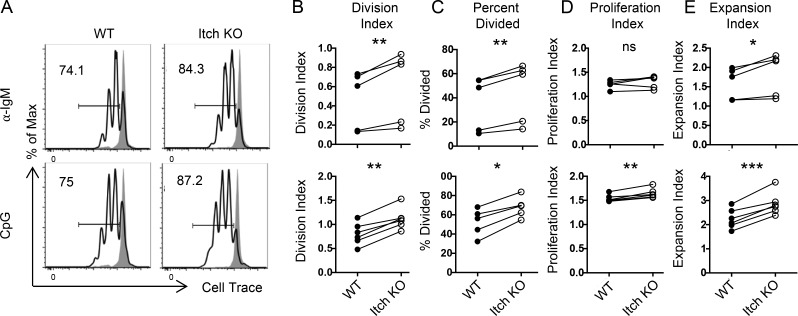
We next wanted to determine (1) if Itch indeed regulated B cell proliferation, as suggested by the proteomics data; and (2) if this regulation was unique to TLR9-driven cell division, or whether Itch would also regulate other mitogenic stimuli. To this end, we isolated FO B cells from WT and Itch KO mice and stimulated them in vitro with anti-IgM or CpG. Importantly, B cells expressed Itch throughout the course of the in vitro stimulation (Fig. S4, A and B). Several indices of the proliferative response were quantified based on analysis of CellTrace dye dilution after 3 d (Parish et al., 2009). First, the division index (a measure of the overall proliferative response) was modestly but consistently increased in Itch KO B cells compared with WT B cells after both BCR and TLR9 stimulation (Fig. 4, A and B). Second, Itch KO B cells stimulated with both anti-IgM and CpG showed an increase in percent divided, supporting that an increased proportion of FO B cells underwent cell division if they lacked Itch (Fig. 4 C). Interestingly, only after CpG stimulation did Itch KO cells show an increase in proliferation index, which measures the number of divisions per dividing cell (Fig. 4 D). This result indicated that the increased proliferation seen in Itch KO B cells after BCR stimulation was mostly due to the increased proportion of cells entering division. In contrast, after TLR9 stimulation, increased division was likely due to increases in both the proportion of cells entering division and the number of divisions per dividing cell. Importantly, in the case of both BCR and TLR9 stimulation, increased proliferation led to increased overall expansion of Itch KO versus WT B cells, as evidenced by the expansion index (Fig. 4 E). These data support that Itch is a negative regulator of B cell proliferation after activation, providing a possible mechanism for increased GC B cell numbers in Itch KO mice.
Figure 4.
Itch limits B cell proliferation. CD23+ FO B cells from WT and Itch KO mice were labeled with CellTrace Violet, then co-cultured and stimulated in vitro with anti-IgM or CpG for 3 d. (A) Representative flow cytometry histograms of CellTrace Violet–labeled FO B cells. Max, maximum. (B–E) Proliferation analysis was performed with FlowJo software. (B–E) Division index (B), percent divided (C), proliferation index (D), and expansion index (E) are shown (n = 5–6, compiled from four independent experiments, paired t test, WT mice were B6.SJL [CD45.1]). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Itch is a negative regulator of mTORC1 activation
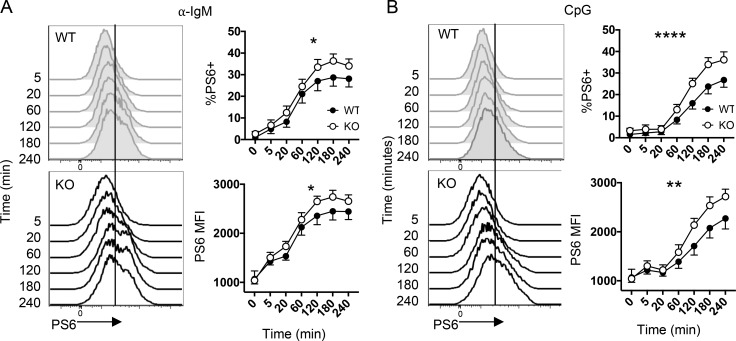
Given that lymphocyte proliferation depends on mTORC1 activity (Zheng et al., 2007; MacIver et al., 2013; McLetchie et al., 2017) and the proteomic profile of Itch KO B cells pointed to increased mTORC1 activation, we next wanted to directly analyze mTORC1 activity. Specifically, we sought to test the kinetics of mTORC1 activity following B cell activation, before cell division. We isolated CD23+ FO B cells and stimulated them in vitro with CpG or anti-IgM, then measured phosphorylation of the direct mTORC1 target, S6, using flow cytometry. We found that WT and Itch KO B cells up-regulated phospho-S6 (P-S6) after activation with both CpG and anti-IgM (Fig. 5, A and B). CpG stimulation induced a slightly delayed P-S6 activation compared with anti-IgM, but in both cases, Itch KO B cells displayed increased levels of P-S6 compared with WT B cells. Increased P-S6 could help explain the increased cell proliferation. Furthermore, because both CpG and anti-IgM stimulation drove higher P-S6 activation within 2 h of stimulation, this suggests that Itch might regulate early signaling events in B cell activation, perhaps through controlling abundance or availability of signaling intermediates in a shared pathway leading to mTORC1 activity downstream of these disparate stimuli. These results support that Itch functions to dampen mTORC1 activation in B cells early after activation in order to limit proliferation.
Figure 5.
Itch limits B cell mTORC1 activity. (A and B) CD23+ FO B cells from WT and Itch KO mice were stimulated in vitro with (A) anti-IgM or (B) CpG for between 5 and 240 min. At the indicated times, P-S6 was detected by flow cytometry. Representative histograms and quantifications are shown. The mean fluorescence intensity (MFI) was quantified from the total population. The vertical line indicates the threshold for a positive P-S6 value (n = 5, compiled from three independent experiments, two-way ANOVA; error bars indicate SEM; WT mice were C57Bl/6 [CD45.2]). *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
We next wanted to ask whether spontaneous GC B cells showed any evidence of increased mTORC1 activation, which could support increased competitive fitness and GC numbers in vivo. We purified total B cells from spleens of WT or Itch KO mice, and we cultured the cells for 1 h with media alone, CpG, or anti-Ig. Next, we briefly surface-stained the cells for GC markers, then immediately fixed the cells and performed intracellular staining for P-S6. We found that GC B cells expressed little P-S6 directly ex vivo, likely due to phosphatase activity during the tissue processing and surface staining. However, after culture ex vivo for 1 h, P-S6 could be detected in GC B cells. GC B cells showed little response to CpG, but a subset responded to anti-Ig, indicating BCR ligation could activate mTORC1 in GC B cells. Itch KO B cells showed increased P-S6 after anti-Ig stimulation compared with WT GC B cells (Fig. S5). These data indicate that Itch limits mTORC1 activity in GC B cells, likely downstream of BCR signals.
Itch limits B cell metabolism
mTORC1 is an important driver of metabolic fitness in B cell development and in GC B cells (Iwata et al., 2016; Ersching et al., 2017). Additionally, mTORC1-dependent changes in metabolism (e.g., up-regulation of glycolysis) support cell division by increasing generation of energy and biomass. Thus, we next wanted to test whether Itch regulated B cell metabolism. We stimulated FO B cells with anti-IgM or CpG for 24 h, then analyzed glycolysis and oxidative phosphorylation using a Seahorse analyzer. We found that after stimulation through either BCR or TLR9, Itch KO B cells displayed increased glycolysis. Itch KO B cells displayed increased maximum glycolytic capacity, as determined by extracellular acidification rate (ECAR) of the media after blockade of the electron transport chain (with oligomycin), supporting that Itch limits the availability or function of the glycolytic pathway (Fig. 6 A). Additionally, we found that Itch exerted an additional negative regulatory influence over the basal level of oxidative phosphorylation, which determines ATP production, but this was observed only after stimulation of TLR9 (Fig. 6 B). These data support that Itch regulates B cell metabolic fitness, and this is consistent with increased mTORC1 activity and proliferation in B cells lacking Itch. Such an increase could underlie the elevated FO and GC B cell competitive fitness of Itch-deficient B cells in vivo.
Figure 6.
Itch limits B cell metabolism. CD23+ FO B cells from WT and Itch KO mice were stimulated in vitro with anti-IgM or CpG for 1 d, and then equal numbers were plated in a Seahorse Extra Cellular Flux Assay plate and subjected to treatment with oligomycin. (A and B) ECAR (A) and OCR (B) are shown. mpH, milli-pH; ETC, electron transport chain (n = 6, six independent experiments, two-way ANOVA with Sidak post-test; error bars indicate SEM; WT mice were C57Bl/6 [CD45.2]). *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant.
Itch functions within B cells to limit antibody production following immunization
The data so far support that Itch regulates mTORC1 activity to limit B cell glycolytic fitness and proliferation. Based on these observations, we would expect that Itch deficiency in B cells would be sufficient to drive enhanced GC B cell and antibody responses after immunization. To test this hypothesis, we employed the B1-8 transgenic mouse model, in which the B1-8 heavy chain is expressed within the endogenous IgH locus, yielding a BCR that recognizes NP when paired with a lambda light chain (Sonoda et al., 1997). Importantly, this BCR can still undergo class switching and somatic hypermutation in the GC (Sonoda et al., 1997). The B1-8 heavy chain is congenically marked; thus, antibodies secreted from transferred B1-8 cells can be identified as IgH[a].
We crossed B1-8 transgenic mice to Itch KO mice, and then isolated 20,000 lambda+ B cells from Itch KO or WT B1-8 mice (CD45.2, IgH[a]). These cells were adoptively transferred into congenically marked WT mice (CD45.1, IgH[b]), and recipients were immunized with NP-OVA in alum (Fig. 7 A). Transferred cells were detected on days 4, 6, 8, 12, and 20 after immunization, and we quantified the numbers of donor NP+ GC B cells at these time points following immunization. We found that WT and Itch KO cells appeared to expand with equal magnitude, with total numbers reaching a peak at day 8. However, while WT B1-8 cell numbers rapidly decreased by day 12 and were barely detectable by day 20, Itch KO B1-8 cells remained elevated at these later time points (Fig. 7 B). The increased persistence of Itch KO GC B cells had important consequences for the emergence of PCs derived from the donor B cells. By day 12, mice that received Itch KO B1-8 cells had more NP-specific IgG1[a] secreting PCs in the spleen than mice that had received WT B1-8 cells (Fig. 7 C). Furthermore, on day 20, the difference in NP-specific IgG1[a] PCs was evident in the BM as well (Fig. 7 D). Importantly, the increase in GC B cells and PCs was accompanied by an increase in high-affinity NP-specific IgG1[a] in the serum (Fig. 7, E–G). The increase in PCs and high-affinity antibody is likely due to the prolonged persistence of GC B cells. With increased numbers of cells persisting longer in the GC, more B cells would achieve higher affinity and produce a larger output of high-affinity PCs. Supporting this idea, we found that NP+ GC B cells isolated on day 12 after adoptive transfer and immunization exhibited increased replacement mutations in the Vh186.2 gene. This indicates that Itch KO GC B cells undergo more rounds of division or survive better in the GCs, allowing accumulation of more mutations (Fig. 7 H). These data support that Itch acts in B cells to limit GC B cell responses and subsequent PC differentiation, thus impacting high-affinity antibody levels.
Figure 7.
B cell Itch limits antibody responses to immunization. (A) Experimental design for adoptive transfer and immunization. (B) Donor NP-binding GC B cells (CD45.2+NP+GL7+) from spleen of recipient mice were enumerated by flow cytometry after immunization (n = 3 for day 4, n = 6 for day 8, n = 10–12 for day 12, and n = 6 or 7 for day 20, two or three independent experiments, two-way ANOVA with Sidak post-test). (C and D) NP-specific IgG1 PCs were enumerated on day 12 and day 20 after immunization in the spleen (C) and BM (D) by ELISPOT (n = 10–12 for day 12 and 6 or 7 for day 20, three independent experiments, two-way ANOVA with Sidak post-test for spleen, Mann–Whitney test for BM). (E–G) Donor-derived NP-specific IgG1[a] was quantified by ELISA. NP-BSA with either a high or low conjugation ratio was used to capture total or high-affinity NP-specific antibody, respectively. (E) Low-affinity antibody = total − high. (F) High-affinity antibody. (G) Ratio of high/low affinity was enumerated within each mouse (for day 12, n = 10–12, for day 20, n = 6 or 7, three independent experiments, two-way ANOVA with Sidak post-test). (H) Chimeras were immunized with NP-OVA, and after 12 d, NP+GL7+ GC B cells were FACS-sorted. Vh186.2 gene was amplified using RT-PCR, and the amplicon was cloned and sequenced. Data are derived from sequences (WT, 18; Itch KO, 34) that were obtained from multiple mice (WT, 4; Itch LP, 10) and two independent experiments (two-way ANOVA and multiple t tests were performed). Error bars indicate SEM. *, P < 0.05; **, P < 0.01.
Itch limits proliferation and mTORC1 in B cells in vivo
Because we saw increased numbers of Itch KO GC B1-8 cells after immunization, we wanted to determine if increased proliferation and/or metabolic fitness might underlie increased cell numbers. We posited that Itch might regulate rapid postimmunization responses (as suggested by the in vitro stimulation experiments in Figs. 4, 5, and 6), as well as in sustained GC responses (indicated by the prolonged contraction of Itch KO B1-8 GC B cells shown in Fig. 7 B). To look at early B cell proliferation in vivo, we transferred CellTrace-labeled WT or Itch KO B1-8 cells into recipient mice, immunized the recipient mice, and measured CellTrace dye dilution and P-S6 levels after 3 d (Fig. 8 A). We found that a higher proportion of Itch KO B1-8 cells had proliferated by day 3 after immunization (Fig. 8 B), and Itch KO B1-8 cells expressed higher P-S6 levels after brief (1 h) ex vivo culture (Fig. 8 C), suggesting that Itch limits mTORC1 activity and proliferation in acutely activated B cells in vivo, similar to what is seen in vitro. We next wanted to analyze P-S6 and proliferation at a later stage after immunization that would better reflect a GC B cell. To this end, we analyzed B1-8 cells between days 8 and 12 after immunization, which is the time period when WT B1-8 cells contract, but Itch KO B1-8 cells persist at higher levels (Fig. 7 B). We immunized donor B1-8 and recipient CD45.1 mice. Then, on day 8 after immunization, we isolated B1-8 cells from WT and KO donor mice, labeled them with CellTrace, and injected them into recipient mice (Fig. 8 D). We evaluated proliferation and P-S6 4 d later, on day 12 after immunization. We found that Itch KO B1-8 cells proliferated more than WT (Fig. 8 E), and they expressed higher levels of P-S6 (Fig. 8 F). Together, these data indicate that Itch functions to dampen B cell proliferation and mTORC1 activity in vivo both early and late after immunization, and increased proliferation likely contributes to increased Itch KO B1-8 GC B cell persistence after immunization.
Figure 8.
Itch limits proliferation and mTORC1 in B cells in vivo. (A) Experimental design for B and C. Lambda+ B cells were isolated from WT and Itch KO B1-8 mice (CD45.2). Cells were labeled with CellTrace and transferred into WT recipient mice (B6.SJL CD45.1), which were then immunized with NP-OVA in alum. (B) On day 3, CellTrace dye dilution of spleen NP+CellTrace+CD45.2+ cells was analyzed. (C) P-S6 MFI of NP+CellTrace+CD45.2+ cells after 1 h culture at 37°C in B cell media was determined by flow cytometry. Fold change refers to KO/WT MFI (for B and C: n = 6 or 7, three independent experiments, unpaired t test). (D) Experimental design for E and F. (E and F) Spleen cells were harvested on day 12 after immunization. (E) CellTrace dye dilution of spleen NP+CellTrace+CD45.2+ cells was analyzed. (F) P-S6 MFI of NP+CellTrace+CD45.2+ spleen B cells after 1 h culture at 37°C in B cell media was determined by flow cytometry. Fold change refers to KO/WT MFI (for E and F: n = 5, two independent experiments, unpaired t test). Error bars indicate SEM; *, P < 0.05.
Discussion
In this report, we reveal a B cell–intrinsic role for Itch that aligns with its well-recognized role in preventing autoimmune disease in mice and humans. While loss of Itch had a subtle impact on the formation of preimmune B cell pools, Itch more dramatically limited the numbers of activated B cells. Itch expression restricted the competitive fitness of activated B cells, allowing Itch-deficient B cells to accumulate to high proportions within mixed BM chimeras. Proteomic profiling of Itch-deficient B cells revealed that Itch limited the abundance of (1) proteins that are activated by mTORC1 and (2) proteins that are up-regulated during cell division. Consistent with the proteomics data, Itch-deficient cells displayed increased proliferation, mTORC1 activity, and metabolic fitness after in vitro activation through either BCR (anti-IgM) or TLR9 (CpG). By focusing on early time points after anti-IgM and CpG stimulation, we determined that Itch regulated mTORC1 activity within 2 h of stimulation, supporting the idea that Itch regulates early activation events that are propagated following BCR and TLR engagement. Finally, we demonstrated that loss of Itch in B cells resulted in elevated numbers of antigen-specific GC B cells and PCs, and enhanced antibody production after immunization in vivo, accompanied by increased proliferation and mTORC1 activity. Together these data support a model in which Itch directly limits antigen-driven B cell responses through regulation of mTORC1-dependent metabolic fitness and proliferation, thus dampening PC generation and antibody production.
A key finding from this study is the identification of a novel negative regulatory role for Itch in B cells that aligns with the autoimmune disease phenotype that develops in humans and mice that lack functional Itch. Prior to this study, mechanistic descriptions of Itch function in lymphocytes could not explain the reported elevation in serum IgG and humoral autoimmunity that develops in Itch deficiency (Matesic et al., 2006; Parravicini et al., 2008; Lohr et al., 2010). Tfh cell differentiation, required for efficient IgG responses, was found to be defective in Itch KO lymphocytes in a model of viral infection (Zhang et al., 2007; Xiao et al., 2014), which appears to be at odds with the high levels of class-switched antibody occurring in Itch deficiency. Perhaps the Th2 cell–biased Itch KO T cells may be specifically defective in becoming Tfh cells in response to a Th1 cell/IFN-γ–biased viral infection, but may still support spontaneous GCs (Fang et al., 2002; Oliver et al., 2006). This idea is supported by some evidence of heterogeneity in Tfh cell phonotypes depending on the type of immune response elicited (Yu et al., 2009). Importantly, because Itch KO T cells fail to become Tfh cells, a role for Itch in T cells cannot explain the large increases in class-switched antibody in Itch-deficient mice. On the contrary, a potent gain of function in Itch KO B cells could lead to increased antibody production, with decreased reliance on Tfh cells.
To explore the inhibitory role of Itch in B cells without the confounding effects of Itch function in other cell types, we used both mixed BM chimeras and adoptive transfer of Itch KO B cells into WT mice. In mixed BM chimeras, we observed that Itch KO cells made up a significantly greater proportion of the GC B cell pool than WT cells, in the case of both spontaneous GCs and immunization-induced GCs. The magnitude of GC B cells is a function of differentiation (i.e., GC entry/seeding), proliferation, lifespan, and exit from the GC (e.g., differentiation into PCs; Victora and Nussenzweig, 2012). The results of the adoptive transfer experiments using WT and Itch KO B1-8 cells provide clues as to which determinant of GC B cell numbers is likely regulated by Itch. We found that Itch KO GC B cells accumulated to greater numbers than WT only in the later days of the response, from days 12 to 20 after immunization. Early after transfer and immunization (days 0–3), Itch KO B cells showed increased proliferation and PS-6, but the numbers of GC B cells at the peak of the response (day 8) were similar between WT and Itch KO cells. This indicates that Itch function to limit acute B cell activation may play less of a role in GC entry, but could have lingering effects on the function and/or fitness of GC B cells. Additionally, antigen-specific PCs and antibody derived from Itch KO–transferred cells was greater in magnitude than WT, so exit from the GC by PC differentiation was intact. Thus, it seems likely that Itch limits the proliferation and/or lifespan of GC B cells, leading to the observed increased cell numbers late in the response.
Recent studies have identified the two main determinants for GC B cell proliferation and survival: (1) the ability to solicit external survival signals through T cell help and FO dendritic cell–derived integrin interactions, and (2) BCR signaling (Vinuesa et al., 2010). In mixed BM chimeras, increased proportions of Itch KO GC B cells could indicate a competitive advantage in acquiring survival signals or increased responsiveness to positive stimuli (e.g., BCR). Our in vitro studies demonstrated that Itch could regulate B cell proliferation and metabolism without the presence of Tfh cells or FO dendritic cells, supporting the idea that Itch regulates the way B cells respond to activating stimuli (e.g., BCR and TLR9). In the context of GCs, increased BCR signaling would be expected to lead to increased size and duration of GCs (Su et al., 1997; Clayton et al., 2002; Huntington et al., 2006). This is supported by the increased P-S6 levels in spontaneous GC B cells stimulated through the BCR ex vivo with anti-Ig (Fig. S5). Increased BCR signaling in the absence of Itch would lead to enhanced proliferation and survival of GC B cells and could explain the increased numbers of GC B cells observed in Itch KO mice. Although we have not ruled out the possibility that Itch might also regulate interactions with Tfh cells or FO dendritic cells, our data support an important role for Itch in negative regulation of GC B cells through suppression of responses to BCR and/or TLR9 stimulation.
The molecular mechanisms underlying increased metabolic fitness and proliferation in GC B cells lacking Itch are yet to be defined. Itch has been linked to ubiquitylation and degradation of Fox-o1 in T cells (Xiao et al., 2014) and ubiquitylation and endo-lysosomal trafficking of Igβ in naive B cells (Zhang et al., 2007). BCR signaling, mTORC1 activity, and Fox-o1 are key contributors to GC maintenance. BCR signals activate mTORC1, which promotes survival and proliferation through increased glycolysis and generation of biomass (Zhang et al., 2011). BCR-dependent inactivation of Fox-o1 promotes dark zone to light zone transition (Dominguez-Sola et al., 2015; Sander et al., 2015). It was recently shown that the BCR mediates these effects in GC B cells by triggering Akt signals. Importantly, Akt activation in GC B cells required BCR signals and could not be triggered by ligation of CD40 (Luo et al., 2018). Thus, GC BCR signals promote metabolic fitness, light zone transition, and maintenance of GC B cell numbers, and Itch regulation of this axis, possibly through limiting BCR signals, Fox-o1 levels, or anther mechanism, could impact GC B cell numbers and antibody responses. Future studies should explore how Itch enzymatic activity regulates these pathways in GC B cells.
It is important to note that Itch exerted modest negative regulation of FO B cells in addition to the more dramatic effect on GC B cells in vivo. In mixed BM chimeras, Itch KO B cells made up a greater proportion of the FO B cell pool than WT B cells, although we did not observe increased numbers of FO B cells in Itch KO mice. Numbers of mature B cells are determined largely by their capacity for acquiring survival signals (e.g., B lymphocyte stimulator) in the periphery, which regulates both rate of mature B cell generation (i.e., survival during maturation from transitional to FO), and lifespan of FO B cells (Harless et al., 2001; Cancro and Smith, 2003). Because we only observed an increase in FO B cells in the context of a competitive environment, it remains possible that Itch KO FO B cells are able to compete more effectively for survival cytokines than WT B cells. Importantly, these observations indicate that Itch is active within FO B cells. Thus, although Itch appears to have only small effects on FO cell numbers, even in a competitive setting, Itch may have other impacts on FO B cell biology, e.g., to limit cell activation. This idea aligns with the in vitro and in vivo data showing enhanced proliferation and mTORC1 activity in acutely activated FO B cells, and lack of Itch may poise FO B cells to exhibit enhanced responses to activating stimuli. Although we did not see evidence of enhanced GC entry in the adoptive transfer experiments, it is possible that Itch function in FO B cells leads to enhanced spontaneous GC seeding or primes GC B cells to persist longer.
An important remaining question is whether the suppressive role of Itch in B cells is linked to the autoimmune phenotype observed in Itch-null patients and mice. While our results demonstrate that Itch limits B cell proliferation, metabolic fitness, and GC B cell numbers, it has yet to be determined whether loss of this negative regulator would be sufficient to drive autoimmunity. Survival in the GC depends on accumulation of survival signals (e.g., CD40, BCR) to counteract death signals through Fas (Hao et al., 2008). Apoptosis is the default pathway for GC B cells, and Fas is required to prevent emergence of autoreactive B cells that arise during somatic hypermutation in GCs (Victora and Nussenzweig, 2012). It is conceivable that Itch-deficient GC B cells, which exhibit exaggerated proliferation and mTORC1 in response to BCR ligation, may be less sensitive to Fas-mediated apoptosis, allowing emergence of autoreactive cells. Future studies using B cell conditional Itch KO mice in a model of autoimmunity could explore the extent to which Itch activity in B cells opposes autoimmunity.
Using proteomics, we identified pathways regulated by Itch using CpG-stimulated B cells. We chose to screen protein expression rather than gene expression because Itch, a ubiquitin ligase, can directly regulate protein levels. Indeed, we were able to make accurate predictions as to the cellular processes that were regulated by Itch by proteomic profiling of activated B cells. Even more importantly, we found the same pathways were elevated in Itch KO cells activated in vitro by anti-IgM and in vivo during a response to immunization. We found that both proliferation and glycolysis were elevated in Itch KO B cells over WT B cells after in vitro activation of either the B cell or TLR9 receptors. We can attribute this in part to early regulation of mTORC1 activity within hours of activation by either stimulus. Mechanistically, Itch-mediated ubiquitylation likely controls signaling pathways downstream of the BCR and TLR9 that converge on mTORC1 activation to limit metabolic fitness and proliferation. Other B cell activation pathways require mTORC1 as well, including CD40 signaling and TLR4 stimulation (Zhang et al., 2011). Determining if Itch regulates S6 phosphorylation downstream of other activating stimuli will help determine whether Itch regulates specific pathways leading to mTORC1 versus regulating general metabolic reprograming in response to any activation signal. It is important to note that although mTORC1 is a major activator of S6 kinase leading to phosphorylation of S6 and enhanced protein translation, mTORC1-independent S6 phosphorylation has been described (Panner et al., 2006). Future studies should explore the mechanistic details by which Itch controls mTORC1 activation and cell proliferation.
It was recently shown that Itch plays a role in B cell development. Supporting this, Itch KO B cells had modestly decreased expression of RAG and IL7R during the pro–B cell stage, although overall numbers of B cells in the spleen were not changed (Liu et al., 2019). A subtle change in B cell development could impact the B cell repertoire emerging from the BM, possibly contributing to autoreactive cells. CD93 is a marker of both transitional B cells and anergic cells, autoreactive cells that are held in an unresponsive state (Merrell et al., 2006). We did not see an increase in CD93+ cells in Itch KO mice, but it would be useful to quantify anergic cells either in Itch KO mice or in a mixed chimera setting. If Itch regulation of B cell development modifies the BCR repertoire, Itch deficiency could lead to changes in the numbers of autoreactive cells in the periphery. Additionally, if Itch regulates responses to BCR stimulation, Itch could impact the balance of IgM versus IgD signaling, which could alter the numbers and/or responsiveness of anergic cells (Sabouri et al., 2016). A closer look at the role of Itch in B cell anergy could help define the link between Itch function in B cells and autoimmunity.
Taken together, our findings demonstrate that Itch is a negative regulator that limits the accumulation of GC B cells, the generation of PCs, and antibody levels before and after immunization. Itch limited proliferation and mTORC1 activity in B cells activated in vitro and in vivo. These data suggest that small increases in proliferation and glycolytic capacity of GC B cells might translate into large changes in antibody quantity and quality in vivo. From a clinical perspective, this could indicate that therapeutic interventions targeting GC B cell proliferation could have large effects on antibody responses. Our data support the idea that therapies designed to boost or suppress Itch function in GC B cells could be used to regulate antibody levels and quality in clinical settings.
Materials and methods
Mice
WT, Itch KO (Perry et al., 1998), IL4 KO, Itch/IL4 DKO, Rag KO, and B1-8 Tg (Sonoda et al., 1997) mice were bred in-house at the Children’s Hospital of Philadelphia (CHOP). Mice were used between 8 and 14 wk of age (unless otherwise noted), and within experiments they were matched for age and sex. Animal housing, care, and experimental procedures were performed in compliance with the CHOP Institutional Animal Care and Use Committee.
Immunization
Mice were immunized by intraperitoneal injection with 200 μl of 0.25 mg/ml NP-OVA with a conjugation ratio of 16NP/OVAL (N-5051; Biosearch Technologies) adsorbed to alum. A 0.5 mg/ml solution of NP-OVA was mixed with 10% aluminum potassium sulfate, and the pH was adjusted to 6.5 using 1 M potassium hydroxide, dropwise. The mixture was incubated at 4°C overnight. Then precipitate was collected by centrifugation and washed with PBS, and precipitant was resuspended in PBS to give a final concentration of 0.25 mg/ml.
BM chimeras
BM chimeras were generated by irradiating recipient mice using an X-Rad Irradiator. For B6.SJL CD45.1 recipients, lethal irradiation was used (1,100 rad), and for Rag KO recipients, sublethal irradiation was used (400 rad). The next day, BM from donor mice was collected, RBCs were lysed, remaining cells were enumerated, and then CD45.1 and CD45.2 cells were mixed at a 1:1 ratio. 2 million mixed BM cells in media were injected into recipient mice via tail vein injections. Following irradiation, mice were kept on Sulfatrim antibiotic water for 3 wk. Mice were analyzed ≥8 wk after injection of BM cells.
Flow cytometry and antibodies
Cells from the spleen were isolated, and single-cell suspensions were stained with a fixable viability dye (Life Technologies), then pretreated with unlabeled anti-CD16/CD32 (Fc Block; BD Pharmingen). Cells were then stained in FACS buffer (2.5% fetal calf serum plus 0.1% sodium azide) with the following mixtures of directly conjugated antibodies, purchased from BioLegend, unless otherwise noted: anti-mouse CD19, IgD, IgM, CD21/35, CD23 (BD Pharmingen), GL-7, CD38, CD4, CD8a, GR-1, F4/80, MHCII, and CD138. Identification of NP-specific B cells was performed by staining with NP-PE, with a conjugation ratio of 23NP/PE (N-5070; Biosearch Technologies). For proliferation studies, cells were labeled with CellTrace violet (Life Technologies).
For P-S6 staining, cells were fixed with 4% paraformaldehyde at 37°C for 10 min, washed in FACS buffer, and fixed in 90% methanol for 20 min on ice. Cells were washed in FACS buffer and stained with anti–P-S6–biotin for 1 h at room temperature. Cells were washed in FACS buffer, and fluorochrome-conjugated streptavidin was added for 15 min at 4°C.
Samples were analyzed using a Fortessa (BD Biosciences) flow cytometer, and flow-cytometric sorting was performed using the MoFlo Astrios (Beckman Coulter) at the CHOP flow cytometry core facility. Data analysis was done using FlowJo (Treestar).
B cell isolation and in vitro stimulation
FO B cells and lambda+ B1-8 cells were isolated by magnetic separation using the Milltenyi anti-PE positive selection kit and anti-mouse CD23-PE (BD Pharmingen) or anti-mouse lambda (Southern Biotech), respectively. FO B cells were stimulated in vitro in B cell media with either 0.1 μg/ml CpG oligodeoxynucleotide (ODN) 1826 (5′-TCCATGACGTTCCTGACGTT-3′ synthesized with phosphorothioate bonds for added stability; Integrated DNA Technologies) or 12.5 μg/ml anti-mouse IgM H+L (Jackson ImmunoResearch). Incubations were performed at 37°C and 10% CO2. B cell media was RPMI 1640 (GE) supplemented with 10% fetal bovine serum (Atlanta), Hepes, nonessential amino acids, sodium pyruvate, and 2-mercaptoethanol.
Total B cells were isolated by negative magnetic separation using the EasySep B cell isolation kit (StemCell). Purified B cells were cultured for 1 h at 37°C with media alone, 1 μM CpG ODN 1826 (5′-TCCATGACGTTCCTGACGTT-3′ synthesized with phosphorothioate bonds; Integrated DNA Technologies), or 10 μg/ml goat anti-mouse Ig (Southern Biotech). After 1 h, cells were washed and surface stained for GC markers for 15 min, washed, then immediately fixed with 4% paraformaldehyde. P-S6 staining was performed as described above in Flow cytometry and antibodies.
GC B cells were isolated using flow-cytometric sorting. Spleens from mice that had received adoptive transfer of either WT or Itch KO B18 cells and immunization with NP-OVA in alum were stained for NP-PE and GC markers, then sorted on NP+GL7+CD23−.
ELISA
Total serum antibody ELISAs were performed by coating ELISA plates with anti-mouse Ig overnight (Southern Biotech). For antigen-specific antibody ELISAs, plates were coated with 100 μl of a 1:1,000 dilution of NP-BSA (Biosearch Technologies). High-affinity antibody was detected by coating with a NP-BSA with a low conjugation ratio (5NP/BSA; Biosearch Technologies), and total antigen-specific antibody was determined by coating with NP-BSA with a high conjugation ratio (30NP/BSA; Biosearch Technologies). Low-affinity antibody was determined by subtracting high-affinity from the total. Plates were blocked, and then standards (Southern Biotech) and serum sample dilutions between 1:5,000 and 1:125,000 were added and incubated for 2 h at room temperature. After washing, isotype-specific detection antibodies conjugated to horseradish peroxidase (Southern Biotech) were added for 1 h at room temperature. 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was added, and the reaction was stopped by adding 1 M phosphoric acid. Absorbance at 450 nm was determined. The standard for NP-specific mouse IgG1 was a gift from Mike Cancro (University of Pennsylvania, Philadelphia, PA).
For anti-dsDNA ELISAs, 100 μg/ml salmon sperm DNA in sodium bicarbonate buffer was filtered through a 0.45-µm filter then used to coat an ELISA plate overnight at 37°C. The next day, the plate was washed with deionized water and blocked with 0.2% BSA for 1 h. The plate was washed with wash buffer (PBS + 0.1% Tween) three times, and then samples were diluted in blocking buffer and added in serial dilutions, starting at 1:400. Samples were incubated at 37°C for 2 h. Plates were washed three times, and then secondary antibody for total mouse IgG-HRP (Southern Biotech) was added for 1 h at 37°C. Plates were washed five times, and then TMB substrate was added and the reaction was stopped by adding 1 M phosphoric acid. Absorbance at 450 nm was determined.
ELISPOT
ELISPOT plates (Millipore) were coated overnight with appropriate capture antibody; for total PCs, 100 μl of a 1:1,000 dilution of anti-mouse Ig (Southern Biotech) in sodium bicarbonate buffer was incubated overnight at 4°C, and for antigen specific ELISPOTs, plates were coated with 100 μl of a 1:250 dilution of NP-BSA (23NP/BSA; Biosearch Technologies).
After coating, plates were washed with PBS, then blocked with blocking buffer (PBS containing 2% BSA). Single-cell suspensions of RBC-lysed spleenocytes or BM was prepared in RPMI (GE) supplemented with 10% fetal bovine serum (Atlanta). Serial twofold dilutions of cells were plated, staring with 1 million cells. Cells were incubated in the plates for 4 h at 37°, and plates were washed (PBS containing 0.1% Tween), then incubated with biotin-conjugated isotype-specific detection antibodies diluted in blocking buffer overnight at 4°C (Southern Biotech). Plates were washed, and then 100 μl of 1:10,000 extravadin-alkaline phosphatase was added in blocking buffer. After incubating for 1 h at room temperature in the dark, plates were washed with washing buffer, and 100 μl nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolyphosphate (Thermo Fisher Scientific) substrate was added. Plates were incubated at room temperature in the dark for 10–30 min. Then plates were washed in water and allowed to dry, and spots were enumerated. PCs per BM (one hind limb) were determined by the following formula: no. PCs = no. spots per well/cells per well × total BM counts (determined using a hemacytometer).
For mixed BM chimera ELISPOTs, CD45.1 and CD45.2 cells were sorted from the spleen via flow-cytometric sorting, and then serial dilutions of either CD45.1 or CD45.2 cells were plated starting with 1 million cells.
Proteomics
FO B cells were isolated from spleens of IL4 KO or Itch/IL4 DKO mice by positive selection of CD23+ cells. Cells were stimulated for 24 h with 0.2 μM CpG ODN. Cells were then lysed and run on a 10% polyacrylamide gel (Bio-Rad). The gel was then stained with Coomassie Blue, and each lane, divided into three fractions, was analyzed by liquid chromatography and tandem mass spectrometry by the Proteomics Core at CHOP as previously described (Layman et al., 2017). Protein identification and quantification were performed using MaxQuant. Intensity-based absolute quantification values (Cox et al., 2014) were compared between IL4 KO and Itch/IL4 DKO mice with three biological replicates per genotype. Statistical significance was determined by t test corrected for multiple comparisons. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014243.
Seahorse
ECAR and oxygen consumption rate (OCR) values were measured by the Agilent Seahorse XFe96 instrument at the University of Pennsylvania Diabetes Research Center, Islet Cell Biology Core. Base XF media, extracellular flux assay plates, cell culture plates, and mitochondrial stress test kits were purchased from Agilent. All procedures were performed according to the manufacturer’s instructions and as previously described (Traba et al., 2016). Cell culture plates were coated overnight with CellTak (Corning) and allowed to air-dry. FO B cells were isolated from WT and Itch KO mice and stimulated in vitro overnight, and then 250,000 cells per well were plated onto CellTak-coated plates in mitochondrial stress test media supplemented with stimulation reagents (e.g., anti-IgM and CpG). ECAR and OCR were measured before and after treatment with oligomycin, and data were analyzed by Wave software (Agilent). Basal glycolysis was ECAR before adding oligomycin, and max glycolysis was ECAR after treatment with oligomycin. Non–electron transport chain oxygen consumption was OCR after addition of oligomycin. Basal oxidative respiration was OCR before adding oligomycin.
Real-time PCR
Purified naive or GC B cells were lysed in Trizol (Invitrogen) and stored at −80°C. RNA was extracted with chloroform, precipitated with 70% ethanol, and resuspended in RNase-free water. cDNA was generated using the High Capacity RNA-to-cDNA kit (Thermo Fisher Scientific). Real-time PCR was performed using TaqMan Gene Expression Master Mix (Thermo Fisher Scientific) with Itch-directed primer/probes (Applied Biosystems). Amplification and detection were performed on a 7500 Real-Time PCR System (Applied Biosystems) at the CHOP Nucleic Acid and Protein Core facility.
Ig heavy chain variable region sequencing and mutation analysis
Sequencing of NP-specific B cells was performed as previously described (Goenka et al., 2014). In brief, flow cytometry–sorted WT or Itch KO NP+ GC B1-8 cells were lysed and stored in TRIZOL. RNA was isolated using chloroform extraction, precipitation in 70% ethanol, and recovery in RNase-free water. RNA was converted to cDNA using Superscript III reverse transcription (Invitrogen) and pooled constant region-specific primers for the gamma chain to preferentially amplify IgG transcripts (Rohatgi et al., 2008). cDNA was subjected to two rounds of nested PCR using Expand High Fidelity Polymerase (Roche) with 5′ primers for VH186.2 (Lu et al., 2001) and 3′ primers for all Ig heavy chain junction region genes (Rohatgi et al., 2008). Amplified bands of ∼350 bp were purified using Qiaquick gel extraction kit (Qiagen) and cloned into pCR-TOPO vector using the TOPO TA Cloning kit (Invitrogen). Plasmid was prepared from individual bacterial colonies using a QiaPrep spin miniprep kit (Qiagen), and Ig heavy chain variable gene inserts were sequenced at the CHOP Nucleic Acid and Protein Core Facility. Mutations were analyzed using the International Immunogenetics Information System V-Quest database (Brochet et al., 2008).
Statistics
Statistical methods are stated in each figure legend, and statistics were calculated using Prism6 for Mac OS X software (GraphPad). *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1 shows the spleen B cell, serum antibody, and BM PC phenotype in Itch/IL4 DKO mice and IL4 KO control mice. Fig. S2 shows Itch mRNA expression in naive and GC B cells. Fig. S3 shows the proportions of CD45.1 and CD45.2 B cells in WT-WT and WT-KO mixed BM chimeras. Fig. S4 shows Itch mRNA expression in B cells that have been stimulated for 24 h in vitro. Fig. S5 shows P-S6 levels in spontaneous GC B cells that have been briefly stimulated ex vivo.
Supplementary Material
Acknowledgments
We thank Keisuke Sawada for excellent technical expertise. Additional thanks to these Core facilities: CHOP Flow Cytometry Core, CHOP Nucleic Acid and Protein Core, and Penn Islet Cell Biology Core.
The research was supported by National Institutes of Health grants RO1AI093566 and RO1AI114515.
The authors declare no competing financial interests.
Author contributions: E.K. Moser designed and performed experiments, analyzed data, and prepared the manuscript. J. Roof carried out experiments. J.M. Dybas designed and performed computational proteomic analysis. L.A. Spruce and S.H. Seeholzer designed and carried out mass spectrometry experiments. M.P. Cancro and P.M. Oliver designed experiments, provided resources, provided intellectual input, and helped write the manuscript.
References
- Aki D., Zhang W., and Liu Y.C.. 2015. The E3 ligase Itch in immune regulation and beyond. Immunol. Rev. 266:6–26. 10.1111/imr.12301 [DOI] [PubMed] [Google Scholar]
- Berek C., Berger A., and Apel M.. 1991. Maturation of the immune response in germinal centers. Cell. 67:1121–1129. 10.1016/0092-8674(91)90289-B [DOI] [PubMed] [Google Scholar]
- Brochet X., Lefranc M.P., and Giudicelli V.. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36(Web Server, Web Server issue):W503-8 10.1093/nar/gkn316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancro M.P., and Smith S.H.. 2003. Peripheral B cell selection and homeostasis. Immunol. Res. 27:141–148. 10.1385/IR:27:2-3:141 [DOI] [PubMed] [Google Scholar]
- Clayton E., Bardi G., Bell S.E., Chantry D., Downes C.P., Gray A., Humphries L.A., Rawlings D., Reynolds H., Vigorito E., and Turner M.. 2002. A crucial role for the p110δ subunit of phosphatidylinositol 3-kinase in B cell development and activation. J. Exp. Med. 196:753–763. 10.1084/jem.20020805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran L.M., and Tarlinton D.M.. 2016. Regulation of germinal center responses, memory B cells and plasma cell formation-an update. Curr. Opin. Immunol. 39:59–67. 10.1016/j.coi.2015.12.008 [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., and Mann M.. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 13:2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D., Kung J., Holmes A.B., Wells V.A., Mo T., Basso K., and Dalla-Favera R.. 2015. The FOXO1 Transcription Factor Instructs the Germinal Center Dark Zone Program. Immunity. 43:1064–1074. 10.1016/j.immuni.2015.10.015 [DOI] [PubMed] [Google Scholar]
- Dubois E.L., Horowitz R.E., Demopoulos H.B., and Teplitz R.. 1966. NZB/NZW mice as a model of systemic lupus erythematosus. JAMA. 195:285–289. 10.1001/jama.1966.03100040091025 [DOI] [PubMed] [Google Scholar]
- Ersching J., Efeyan A., Mesin L., Jacobsen J.T., Pasqual G., Grabiner B.C., Dominguez-Sola D., Sabatini D.M., and Victora G.D.. 2017. Germinal Center Selection and Affinity Maturation Require Dynamic Regulation of mTORC1 Kinase. Immunity. 46:1045–1058.e6. 10.1016/j.immuni.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D., Elly C., Gao B., Fang N., Altman Y., Joazeiro C., Hunter T., Copeland N., Jenkins N., and Liu Y.C.. 2002. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3:281–287. 10.1038/ni763 [DOI] [PubMed] [Google Scholar]
- Goenka R., Matthews A.H., Zhang B., O’Neill P.J., Scholz J.L., Migone T.S., Leonard W.J., Stohl W., Hershberg U., and Cancro M.P.. 2014. Local BLyS production by T follicular cells mediates retention of high affinity B cells during affinity maturation. J. Exp. Med. 211:45–56. 10.1084/jem.20130505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z., Duncan G.S., Seagal J., Su Y.W., Hong C., Haight J., Chen N.J., Elia A., Wakeham A., Li W.Y., et al. . 2008. Fas receptor expression in germinal-center B cells is essential for T and B lymphocyte homeostasis. Immunity. 29:615–627. 10.1016/j.immuni.2008.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harless S.M., Lentz V.M., Sah A.P., Hsu B.L., Clise-Dwyer K., Hilbert D.M., Hayes C.E., and Cancro M.P.. 2001. Competition for BLyS-mediated signaling through Bcmd/BR3 regulates peripheral B lymphocyte numbers. Curr. Biol. 11:1986–1989. 10.1016/S0960-9822(01)00598-X [DOI] [PubMed] [Google Scholar]
- Huntington N.D., Xu Y., Puthalakath H., Light A., Willis S.N., Strasser A., and Tarlinton D.M.. 2006. CD45 links the B cell receptor with cell survival and is required for the persistence of germinal centers. Nat. Immunol. 7:190–198. 10.1038/ni1292 [DOI] [PubMed] [Google Scholar]
- Iwata T.N., Ramírez J.A., Tsang M., Park H., Margineantu D.H., Hockenbery D.M., and Iritani B.M.. 2016. Conditional Disruption of Raptor Reveals an Essential Role for mTORC1 in B Cell Development, Survival, and Metabolism. J. Immunol. 197:2250–2260. 10.4049/jimmunol.1600492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J., Kelsoe G., Rajewsky K., and Weiss U.. 1991. Intraclonal generation of antibody mutants in germinal centres. Nature. 354:389–392. 10.1038/354389a0 [DOI] [PubMed] [Google Scholar]
- Jin H.S., Park Y., Elly C., and Liu Y.C.. 2013. Itch expression by Treg cells controls Th2 inflammatory responses. J. Clin. Invest. 123:4923–4934. 10.1172/JCI69355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathania M., Khare P., Zeng M., Cantarel B., Zhang H., Ueno H., and Venuprasad K.. 2016. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-γt ubiquitination. Nat. Immunol. 17:997–1004. 10.1038/ni.3488 [DOI] [PubMed] [Google Scholar]
- Layman A.A.K., Deng G., O’Leary C.E., Tadros S., Thomas R.M., Dybas J.M., Moser E.K., Wells A.D., Doliba N.M., and Oliver P.M.. 2017. Ndfip1 restricts mTORC1 signalling and glycolysis in regulatory T cells to prevent autoinflammatory disease. Nat. Commun. 8:15677 10.1038/ncomms15677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., and Tamayo P.. 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1:417–425. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhang Y., Wei Y., Wang Z., Zhu G., Fang Y., Zhai B., Xu R., Han G., Chen G., et al. . 2019. The E3 ubiquitin ligase Itch is required for B-cell development. Sci. Rep. 9:421 10.1038/s41598-018-36844-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr N.J., Molleston J.P., Strauss K.A., Torres-Martinez W., Sherman E.A., Squires R.H., Rider N.L., Chikwava K.R., Cummings O.W., Morton D.H., and Puffenberger E.G.. 2010. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am. J. Hum. Genet. 86:447–453. 10.1016/j.ajhg.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.-F., Singh M., and Cerny J.. 2001. Canonical germinal center B cells may not dominate the memory response to antigenic challenge. Int. Immunol. 13:643–655. 10.1093/intimm/13.5.643 [DOI] [PubMed] [Google Scholar]
- Luo W., Weisel F., and Shlomchik M.J.. 2018. B Cell Receptor and CD40 Signaling Are Rewired for Synergistic Induction of the c-Myc Transcription Factor in Germinal Center B Cells. Immunity. 48:313–326.e5. 10.1016/j.immuni.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver N.J., Michalek R.D., and Rathmell J.C.. 2013. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 31:259–283. 10.1146/annurev-immunol-032712-095956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matesic L.E., Haines D.C., Copeland N.G., and Jenkins N.A.. 2006. Itch genetically interacts with Notch1 in a mouse autoimmune disease model. Hum. Mol. Genet. 15:3485–3497. 10.1093/hmg/ddl425 [DOI] [PubMed] [Google Scholar]
- McLetchie S., Raybuck A., Cho S.H., Lin J., and Boothby M.R.. 2017. mTORC1 in B cells regulates antibody responses and promotes mitochondrial and metabolic fitness. J. Immunol. 198:195.13. https://www.jimmunol.org/content/198/1_Supplement/195.13
- Merrell K.T., Benschop R.J., Gauld S.B., Aviszus K., Decote-Ricardo D., Wysocki L.J., and Cambier J.C.. 2006. Identification of anergic B cells within a wild-type repertoire. Immunity. 25:953–962. 10.1016/j.immuni.2006.10.017 [DOI] [PubMed] [Google Scholar]
- Morel L. 2010. Genetics of SLE: evidence from mouse models. Nat. Rev. Rheumatol. 6:348–357. 10.1038/nrrheum.2010.63 [DOI] [PubMed] [Google Scholar]
- Moser E.K., Field N.S., and Oliver P.M.. 2018. Aberrant Th2 inflammation drives dysfunction of alveolar macrophages and susceptibility to bacterial pneumonia. Cell. Mol. Immunol. 15:480–492. 10.1038/cmi.2016.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P.M., Cao X., Worthen G.S., Shi P., Briones N., MacLeod M., White J., Kirby P., Kappler J., Marrack P., and Yang B.. 2006. Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity. 25:929–940. 10.1016/j.immuni.2006.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panner A., Nakamura J.L., Parsa A.T., Rodriguez-Viciana P., Berger M.S., Stokoe D., and Pieper R.O.. 2006. mTOR-independent translational control of the extrinsic cell death pathway by RalA. Mol. Cell. Biol. 26:7345–7357. 10.1128/MCB.00126-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish C.R., Glidden M.H., Quah B.J., and Warren H.S.. 2009. Use of the intracellular fluorescent dye CFSE to monitor lymphocyte migration and proliferation. Curr. Protoc. Immunol. Chapter 4:9 10.1002/0471142735.im0409s84 [DOI] [PubMed] [Google Scholar]
- Parravicini V., Field A.C., Tomlinson P.D., Basson M.A., and Zamoyska R.. 2008. Itch-/- alphabeta and gammadelta T cells independently contribute to autoimmunity in Itchy mice. Blood. 111:4273–7282. 10.1182/blood-2007-10-115667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry W.L., Hustad C.M., Swing D.A., O’Sullivan T.N., Jenkins N.A., and Copeland N.G.. 1998. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 18:143–146. 10.1038/ng0298-143 [DOI] [PubMed] [Google Scholar]
- Ramos-Hernández N., Ramon H.E., Beal A.M., Laroche A., Dekleva E.A., and Oliver P.M.. 2013. Ndfip1 enforces a requirement for CD28 costimulation by limiting IL-2 production. J. Immunol. 191:1536–1546. 10.4049/jimmunol.1203571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi S., Ganju P., and Sehgal D.. 2008. Systematic design and testing of nested (RT-)PCR primers for specific amplification of mouse rearranged/expressed immunoglobulin variable region genes from small number of B cells. J. Immunol. Methods. 339:205–219. 10.1016/j.jim.2008.09.017 [DOI] [PubMed] [Google Scholar]
- Sabouri Z., Perotti S., Spierings E., Humburg P., Yabas M., Bergmann H., Horikawa K., Roots C., Lambe S., Young C., et al. . 2016. IgD attenuates the IgM-induced anergy response in transitional and mature B cells. Nat. Commun. 7:13381 10.1038/ncomms13381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S., Chu V.T., Yasuda T., Franklin A., Graf R., Calado D.P., Li S., Imami K., Selbach M., Di Virgilio M., et al. . 2015. PI3 Kinase and FOXO1 Transcription Factor Activity Differentially Control B Cells in the Germinal Center Light and Dark Zones. Immunity. 43:1075–1086. 10.1016/j.immuni.2015.10.021 [DOI] [PubMed] [Google Scholar]
- Schultz C.L., and Coffman R.L.. 1991. Control of isotype switching by T cells and cytokines. Curr. Opin. Immunol. 3:350–354. 10.1016/0952-7915(91)90037-2 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M., and Calame K.. 2005. Regulation of plasma-cell development. Nat. Rev. Immunol. 5:230–242. 10.1038/nri1572 [DOI] [PubMed] [Google Scholar]
- Sonoda E., Pewzner-Jung Y., Schwers S., Taki S., Jung S., Eilat D., and Rajewsky K.. 1997. B cell development under the condition of allelic inclusion. Immunity. 6:225–233. 10.1016/S1074-7613(00)80325-8 [DOI] [PubMed] [Google Scholar]
- Su G.H., Chen H.M., Muthusamy N., Garrett-Sinha L.A., Baunoch D., Tenen D.G., and Simon M.C.. 1997. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 16:7118–7129. 10.1093/emboj/16.23.7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traba J., Miozzo P., Akkaya B., Pierce S.K., and Akkaya M.. 2016. An Optimized Protocol to Analyze Glycolysis and Mitochondrial Respiration in Lymphocytes. J. Vis. Exp. (117). 10.3791/54918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuprasad K., Elly C., Gao M., Salek-Ardakani S., Harada Y., Luo J.L., Yang C., Croft M., Inoue K., Karin M., and Liu Y.C.. 2006. Convergence of Itch-induced ubiquitination with MEKK1-JNK signaling in Th2 tolerance and airway inflammation. J. Clin. Invest. 116:1117–1126. 10.1172/JCI26858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Vinuesa C.G., Linterman M.A., Goodnow C.C., and Randall K.L.. 2010. T cells and follicular dendritic cells in germinal center B-cell formation and selection. Immunol. Rev. 237:72–89. 10.1111/j.1600-065X.2010.00937.x [DOI] [PubMed] [Google Scholar]
- Wurster A.L., Rodgers V.L., White M.F., Rothstein T.L., and Grusby M.J.. 2002. Interleukin-4-mediated protection of primary B cells from apoptosis through Stat6-dependent up-regulation of Bcl-xL. J. Biol. Chem. 277:27169–27175. 10.1074/jbc.M201207200 [DOI] [PubMed] [Google Scholar]
- Xiao N., Eto D., Elly C., Peng G., Crotty S., and Liu Y.C.. 2014. The E3 ubiquitin ligase Itch is required for the differentiation of follicular helper T cells. Nat. Immunol. 15:657–666. 10.1038/ni.2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Batten M., Mackay C.R., and King C.. 2009. Lineage specification and heterogeneity of T follicular helper cells. Curr. Opin. Immunol. 21:619–625. 10.1016/j.coi.2009.09.013 [DOI] [PubMed] [Google Scholar]
- Zhang M., Veselits M., O’Neill S., Hou P., Reddi A.L., Berlin I., Ikeda M., Nash P.D., Longnecker R., Band H., and Clark M.R.. 2007. Ubiquitinylation of Ig beta dictates the endocytic fate of the B cell antigen receptor. J. Immunol. 179:4435–4443. 10.4049/jimmunol.179.7.4435 [DOI] [PubMed] [Google Scholar]
- Zhang S., Readinger J.A., DuBois W., Janka-Junttila M., Robinson R., Pruitt M., Bliskovsky V., Wu J.Z., Sakakibara K., Patel J., et al. . 2011. Constitutive reductions in mTOR alter cell size, immune cell development, and antibody production. Blood. 117:1228–1238. 10.1182/blood-2010-05-287821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Collins S.L., Lutz M.A., Allen A.N., Kole T.P., Zarek P.E., and Powell J.D.. 2007. A role for mammalian target of rapamycin in regulating T cell activation versus anergy. J. Immunol. 178:2163–2170. 10.4049/jimmunol.178.4.2163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.