Mastio et al. describe monocytic precursors of granulocytes. These precursors are barely detectable in steady state conditions and are not consequential for differentiation of granulocytes. However, they accumulate in cancer and substantially contribute to PMN-MDSC expansion.
Abstract
We have identified a precursor that differentiates into granulocytes in vitro and in vivo yet belongs to the monocytic lineage. We have termed these cells monocyte-like precursors of granulocytes (MLPGs). Under steady state conditions, MLPGs were absent in the spleen and barely detectable in the bone marrow (BM). In contrast, these cells significantly expanded in tumor-bearing mice and differentiated to polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs). Selective depletion of monocytic cells had no effect on the number of granulocytes in naive mice but decreased the population of PMN-MDSCs in tumor-bearing mice by 50%. The expansion of MLPGs was found to be controlled by the down-regulation of Rb1, but not IRF8, which is known to regulate the expansion of PMN-MDSCs from classic granulocyte precursors. In cancer patients, putative MLPGs were found within the population of CXCR1+CD15−CD14+HLA-DR−/lo monocytic cells. These findings describe a mechanism of abnormal myelopoiesis in cancer and suggest potential new approaches for selective targeting of MDSCs.
Graphical Abstract
Introduction
There is now ample evidence suggesting that the success of cancer immunotherapies and even more traditional methods of treatment, such as radiation and chemotherapy, heavily depend on myeloid cells in the tumor microenvironment (Coussens and Pollard, 2011; Galdiero et al., 2013). In cancer, one of the most prominent changes in the myeloid compartment is the expansion of pathologically activated immature myeloid cells, termed myeloid-derived suppressor cells (MDSCs), with the potent ability to suppress immune responses. Besides suppressing anti-tumor immunity, MDSCs also stimulate other aspects of tumor growth including tumor angiogenesis, tumor cell invasion, and formation of premetastatic niches (Condamine et al., 2015). MDSCs are directly implicated in negatively impacting patient responses to cancer therapies (Diaz-Montero et al., 2009; Arihara et al., 2013; Chen et al., 2014; Kawano et al., 2015; Romano et al., 2015; Lee et al., 2016; Tada et al., 2016; Wang et al., 2016; Wang and Yang, 2016), including immunotherapies (Kimura et al., 2013; Martens et al., 2016; Sade-Feldman et al., 2016; Weber et al., 2016; Butterfield et al., 2017; de Coaña et al., 2017).
In tumor-bearing (TB) mice, the population of MDSCs consists of two large groups of cells: the most abundant (>75%) population consists of pathologically activated neutrophils, CD11b+Ly6CintLy6G+ polymorphonuclear MDSCs (PMN-MDSCs), while the less abundant (<20%) population consists of CD11b+Ly6ChiLy6G− pathologically activated monocytes: monocytic MDSCs (M-MDSCs; Bronte et al., 2016). The current prevailing view is that M-MDSCs and PMN-MDSCs differentiate along the same pathways as monocytes and neutrophils, respectively, and their expansion in cancer is controlled by increased production of GM-CSF, CSF-1, and other growth factors (Gabrilovich et al., 2012). These pathways involve pluripotent hematopoietic stem cells, multipotent common myeloid progenitors (CMPs), and granulocyte-macrophage progenitors (GMPs). Recently, several populations of committed granulocytic precursors (GPs) were identified (Waight et al., 2013; Yáñez et al., 2015; Evrard et al., 2018). Down-regulation of the IRF8 transcription factor, which is important for the switch between monocytic and granulocytic cell differentiation (Kurotaki et al., 2014), was directly implicated in the expansion of PMN-MDSCs from GPs (Netherby et al., 2017).
We previously obtained evidence that M-MDSCs from TB mice were able to differentiate into PMN-MDSCs, suggesting that, in cancer, some monocytic cells can be redirected to differentiate into granulocytes (Youn et al., 2013). Down-regulation of the retinoblastoma 1 (Rb1) gene was implicated in this process. The concept of Rb1 involvement in PMN-MDSC differentiation was supported in more recent studies using a breast cancer mouse model (Casbon et al., 2015). However, only a portion of the M-MDSC population was found to be able to differentiate into granulocytes. This raised the question of whether the M-MDSC population contained some GPs rather than a demonstrated abnormal switch in lineage commitment. The contribution of these cells to the pool of PMN-MDSCs was also not known. In this study, we sought to address these questions. We have demonstrated that abnormal myelopoiesis in TB mice, and to some extent in cancer patients, manifests in the expansion of a defined population of monocyte-like precursors of granulocytes (MLPGs). We identified the phenotype and the nature of these cells and determined that, at least in two murine tumor models, these cells contributed up to half of the total pool of PMN-MDSCs.
Results
Identification of MLPGs in TB mice
To better characterize the population of M-MDSCs in TB mice, we used Rb1 reporter mice, which were generated by inserting the eGFP cassette at the start codon of the Rb1 gene. eGFP expression paralleled that of Rb1 in most of the organs surveyed, including the spleen and the lungs, along with Gr-1+CD11b+ cells and T and B lymphocytes (Burkhart et al., 2010). Inflammatory monocytes in the bone marrow (BM) of EL-4 TB mice were defined using a standard set of phenotypic markers: CD11b+Ly6ChiLy6G− (Bronte et al., 2016). These cells had a heterogeneous expression of Rb1, with most cells expressing high levels of Rb1 and ∼30% having relatively low expression levels (Fig. 1 A). BM and splenic CD11b+Ly6ChiLy6G− cells in TB mice had a substantially lower Rb1 expression than the cells with the same phenotype in naive mice (Fig. 1 B), reflecting a lower proportion of Rb1lo monocytes in naive mice (Fig. 1 C).
Figure 1.
Identification of monocytic precursors of granulocytes in TB mice. (A) Rb1 expression in CD11b+Ly6ChiLy6G− monocytic cells in BM of EL-4 TB Rb1-GFP reporter mice assessed by flow cytometry. Numbers in the plots indicate the percentage of gated cells. Typical example of three experiments is shown. (B) Rb1 expression in CD11b+Ly6ChiLy6G− monocytic cells in BM and spleens of TB and naive Rb1-GFP reporter mice. Mean and SD are shown of 13 TB and 13 tumor-free mice. P values were calculated by two-tailed Student’s t test. ****, P < 0.0001. (C) Rb1 expression in CD11b+Ly6ChiLy6G− monocytic cells in BM of naive Rb1-GFP reporter mice assessed by flow cytometry. Numbers in the plots indicate the percentage of gated cells. Typical example of three experiments is shown. (D) Phenotype of cells recovered after 3 d of culture with GM-CSF and TES of Rb1lo and Rb1hiCD11b+Ly6ChiLy6G− monocytic cells isolated from BM of EL-4 TB Rb1-GFP reporter mice. Numbers in the plots indicate the percentage of gated cells. Typical example of four experiments is shown. (E) Rb1 and CD117 expression in CD11b+Ly6ChiLy6G− monocytic cells from BM of EL-4 TB Rb1-GFP reporter mice assessed by flow cytometry. Numbers in the plots indicate the percentage of gated cells. Typical example of 13 experiments is shown. (F) Rb1 expression in CD117+ and CD117−CD11b+Ly6ChiLy6G− monocytic cells and CD11b+Ly6CintLy6G+ PMN-MDSCs in BM of EL-4 TB Rb1-GFP reporter mice. Mean and SD of three experiments are shown. *, P < 0.5; **, P < 0.01; ***, P < 0.001 by two-tailed Student’s t test. MFI, mean fluorescence intensity. (G) Antigen-specific proliferation of CD8+ T cells in the presence of Rb1lo and Rb1hiCD11b+Ly6ChiLy6G− monocytic cells sorted from BM of EL-4 TB Rb1-GFP reporter mice. Proliferation was measured in triplicate by 3H thymidine uptake. PMEL T cells were used as responder cells in this antigen-specific suppression assay. No st., no stimulation; Pept., stimulation of splenocytes with gp 100–derived peptide. Mean and SD of typical example of three experiments is shown. **, P < 0.01 by two-tailed Student’s t test. Three experiments with the same results were performed. (H) Phenotype of cells recovered after 3 d of culture with GM-CSF and TES or M-CSF of CD117+ and CD117−CD11b+Ly6ChiLy6G− monocytic cells sorted from BM of EL-4 TB mice. Numbers in the plots indicate the percentage of gated cells. Typical example of three experiments is shown. (I) Wright-Giemsa staining of cells differentiated with GM-CSF and TES from indicated precursors. Scale bar, 20 µm. Typical example of three experiments is shown. (J) In vivo differentiation of CD45.2+CD117+CD11b+Ly6ChiLy6G− monocytic cells transferred to sublethally irradiated EL-4 TB CD45.1+ recipient. Typical example of two experiments is shown. CPM, counts per minute; SSC, side scatter.
Rb1lo and Rb1hi CD11b+Ly6ChiLy6G− monocytes from the BM of TB mice were cultured for 3 d with GM-CSF and tumor explant supernatant (TES) to determine whether these two cell populations differentiate into similar or different cell types. More than 85% of the cells differentiated from Rb1lo monocytes had a CD11b+Ly6CintLy6G+ granulocytic phenotype. In contrast, only ∼5% of cells differentiated from Rb1hi monocytes had the same granulocytic phenotype (Fig. 1 D). Rb1 expression was closely associated with the expression of CD117 (c-kit). Most of the Rb1lo but not the Rb1hi monocytic cells expressed CD117 (Fig. 1 E). In TB mice, CD117+CD11b+Ly6ChiLy6G− monocytic cells had a low Rb1 expression, which was only marginally higher than the level found in CD11b+Ly6CintLy6G+ PMN-MDSCs, whereas the CD117−CD11b+Ly6ChiLy6G− monocytic cells had almost a threefold higher Rb1 expression than PMN-MDSCs (Fig. 1 F). Next, we investigated whether CD117+Rb1lo and CD117−Rb1hi CD11b+Ly6ChiLy6G− monocytic cells had the suppressive activity usually attributed to M-MDSCs. CD117+ monocytic cells had no T cell suppressive activity, whereas CD117− cells had a potent suppressive activity (Fig. 1 G). Thus, these two populations have different functional properties: CD117−CD11b+Ly6ChiLy6G− cells were bona fide M-MDSCs, whereas CD117+CD11b+Ly6ChiLy6G− cells were not suppressive but could differentiate to granulocytes.
We then performed an in vitro differentiation experiment with sorted BM monocytic cells based on CD117 expression. Within 3 d in culture with GM-CSF and TES, CD11b+Ly6ChiLy6G−CD117+ monocytic cells differentiated to cells with the phenotype (Fig. 1 H) and morphology (Fig. 1 I) of granulocytes, whereas the CD11b+Ly6ChiLy6G−CD117− monocytic cells remained Ly6G− and differentiated to CD115+ cells represented by F4/80+ macrophages and dendritic cells (Fig. 1, H and I). Similar results were obtained when these cells were cultured with CSF-1 (M-CSF; Fig. 1 H). To trace the fate of CD11b+Ly6ChiLy6G−CD117+ monocytic cells in vivo, these cells were sorted from the BM of EL-4 TB CD45.2+ mice and transferred to sublethally irradiated CD45.1+ EL-4 TB congenic recipients. 3 d after the transfer, almost all CD45.2+ cells had the phenotype of granulocytes (CD11b+Ly6CintLy6G+; Fig. 1 J). Taken together, these results indicated that CD11b+Ly6ChiLy6G−CD117+ monocytic cells with a low Rb1 expression represented the population of monocytic cells without suppressive activity but with potent ability to differentiate to granulocytes. We provisionally designated these cells as MLPGs. In contrast, CD11b+Ly6ChiLy6G−CD117− monocytic cells with a high Rb1 expression had a potent suppressive activity and differentiated into macrophages and dendritic cells, thus fitting the common definition of M-MDSCs.
The monocytic nature of MLPGs
CCR2+ differentiated monocytes with typical morphology represented most of the CD11b+Ly6ChiLy6G−CD117− M-MDSCs (Fig. 2, A and B). In contrast, CD11b+Ly6ChiLy6G−CD117+ MLPGs were mostly CCR2− (Fig. 2 A) and had morphology of immature cells with less condensed chromatin and a ring-shaped nucleus (Fig. 2 B). In the BM of TB mice, in addition to the populations of CD11b+Ly6G−Ly6Chi M-MDSCs and MLPGs, cells with CD11b+Ly6G−Ly6Cint phenotype were readily detectable (Fig. 2 C). The criterion to define Ly6C expression as high or intermediate was the level of Ly6C expression in Ly6G+ neutrophils. Expression of Ly6C above the level observed in neutrophils was considered as high, and at similar or lower levels than in neutrophils was considered as intermediate. To better characterize CD11b+Ly6G−Ly6Cint cells, we used the CCR2 marker of mature monocytes and the SiglecF marker of eosinophils/neutrophils (Stockwell et al., 2017). While CD11b+Ly6G−Ly6Chi cells were largely CCR2+SiglecF−, the CD11b+Ly6G−Ly6Cint cells were largely CCR2−, with >30% SiglecF+ cells (Fig. 2 C). A previous study identified a population of Ly6C+CD117+LKS−CD34+FcγRhi cells as committed granulocytic progenitors (Yáñez et al., 2015). Ly6C expression in these cells could be characterized as intermediate. In contrast to that granulocyte progenitor population, in our experiments, MLPGs expressed high Ly6C and CD11b and had a morphology of more advanced GPs, with a ring-shaped nucleus (Fig. 2 B). Also, the morphology of MLPGs was different from that of more upstream granulocyte progenitors such as GMPs (Fig. 2 B). The fact that GMPs do not express CD11b and had low or no expression of Ly6C indicates that MLPGs and GMPs are quite distinct cells. CD11b+Ly6CintCCR2−SiglecF−Ly6G−CD117+ cells had similar characteristics as the late GPs, specifically the preneutrophil (preNeu) population, described recently in another study (Evrard et al., 2018). To confirm that CD11b+Ly6CintCCR2−SiglecF−Ly6G−CD117+ indeed represented GPs, these cells were sorted from the BM of TB mice and were cultured for 3 d with GM-CSF. Practically all cells generated from those precursors had the phenotype and morphology of granulocytes (Fig. S1 A). To verify these findings in vivo, CD45.2+CD11b+Ly6CintCCR2−SiglecF−Ly6G−CD117+ cells from the BM of EL-4 TB mice were transferred to sublethally irradiated congenic CD45.1+ EL-4 TB recipients. Within 3 d after the transfer, most donor cells became CD11b+Ly6CintLy6G+ granulocytes (Fig. S1 B).
Figure 2.
The monocytic nature of MLPGs. (A) CCR2 expression in CD117− and CD117+CD11b+Ly6ChiLy6G− M-MDSCs and MLPGs in BM of EL-4 TB mice assessed by flow cytometry. Numbers in the plots indicate the percentage of gated cells. Typical example of three experiments is shown. (B) May-Grünwald-Giemsa staining of CD117−CD11b+Ly6ChiLy6G− M-MDSCs, CD117+CD11b+Ly6ChiLy6G− MLPGs, CD117+CD11b+Ly6CintLy6G− GPs, and Lin−Sca-1−CD117+CD34+CD16/32hi GMPs sorted from BM of EL-4 TB mice. Scale bar, 20 µm. (C) CCR2 and SiglecF expression in Ly6Chi and Ly6CintCD11b+Ly6G− populations in BM of EL-4 TB mice assessed by flow cytometry. Numbers in the plots indicate the percentage of gated cells. Typical example of three experiments is shown. (D) Comparison of the transcriptomes of CD11b+Ly6ChiLy6G−CD117+ MLGP and CD11b+Ly6CintLy6G−CD117+ GPs from BM of EL-4 TB mice with transcriptomic profiles of different populations of hematopoietic cells and progenitors from the Immgen website. Cell types from the Immgen Consortium whose expression profiles correlated the most with MLPGs and GPs are shown. (E) tSNE clustering of granulocytes and monocytes with MLPGs and GPs. *, intermediate; 6C, Ly6C; Arth, arthritic; bl, blood; GN, granulocytes; II, MHCII; Mo, monocytes; PC, peritoneal cavity; SSC, side scatter; SF/SynF, synovial fluid; Thio, thioglycolate; UrAc, uric acid.
The next question was whether MLPGs could differentiate into GPs. To address this question, we sorted MLPGs (with >95% purity) from the BM of CD45.2+ EL-4 TB mice and transferred them to sublethally irradiated CD45.1+ EL-4 TB recipient mice. 3 h after the transfer, only a few CD45.2+Ly6CloLy6G+ donor PMNs were detected in recipient spleens, and practically all donor cells were still Ly6Chic-kit (CD117)+. 24 h later, all Ly6Chi donor cells had disappeared. Half of the donor cells were Ly6G+ PMNs. However, no more Ly6Cloc-kit+ cells were detected (Fig. S1 C). After 48 h, all donor cells became PMNs. In contrast, a GP population was clearly detectable in CD45.1+ host cells (Fig. S1 C). These data suggest that MLPG differentiation to PMNs in vivo does not include the GP stage.
The major question about MLPGs was whether these cells indeed belonged to the monocytic lineage or were cells of granulocytic lineage that had high expression of the Ly6C marker. To address this question, we compared the whole transcriptome of MLPGs and GPs from the BM of EL-4 TB mice with those of different cell populations defined by the Immgen Consortium. When all expressed genes in MLPGs were analyzed, their expression profile correlated with monocytes, macrophage dendritic cell progenitors (MDPs), and common dendritic cell progenitors. In contrast, GPs correlated only with the genes expressed in granulocytes (Fig. 2 D). Based on clustering analysis of granulocytes and monocytes from Immgen along with GPs and MLPGs, MLPGs showed clear association with monocytes, while GPs clustered with granulocytes (Fig. 2 E). The top genes differentially expressed between MLPGs and GPs that overlapped with genes specific to granulocytes and monocytes are shown in Fig. S2 A. We compared the expression profile of MLPGs and GPs with populations of BM and spleen CD11b+Ly6CintLy6G+ neutrophils/PMN-MDSCs and CD11b+Ly6ChiLy6G− monocytes/M-MDSCs isolated from naive and EL4-TB mice that have been characterized previously (Youn et al., 2012). The gene expression of GPs clustered closely with the profile of neutrophils/PMN-MDSCs, whereas MLPGs correlated closely with monocytes/M-MDSCs (Fig. S2 B). We also compared the genomic signatures of different populations of GPs, including GMPs, as well as the preNeu population (Evrard et al., 2018), with GPs and MLPGs from our study. The RNA sequencing (RNA-seq) data for GMPs, preNeu, and immature neutrophils were downloaded from Gene Expression Omnibus (accession no. GSE109467; Evrard et al., 2018). Genes were overlapped with microarray data using Entrez IDs, and differentially expressed genes between GPs and MLPGs (n = 248) with false discovery rate (FDR) <5% were normalized to average within each dataset. Principal component analysis was used to cluster the data using principal components obtained from GP/MLPG samples. We observed that GPs were close to preNeu and GMPs, whereas MLPGs were quite distinct (Fig. S2 C).
The chemokine receptor CX3CR1 is a marker specific for the monocytic lineage. Using reporter mice with a Cx3cr1-driven expression of egfp, we assessed the expression of CX3CR1 in the MLPGs from the BM of TB mice. As a positive control for CX3CR1 expression, we used CD11b+Ly6CloLy6G−CD117− spleen patrolling monocytes, and as negative control, CD11b+Ly6CintLy6G+ neutrophils. MLPGs expressed CX3CR1 at levels comparable to that of M-MDSCs, whereas GPs expressed CX3CR1 at levels similar to those found in PMN-MDSCs (Fig. 3 A). Dot-plot analysis of CX3CR1 expression in c-kit− M-MDSCs and c-kit+ MLPGs demonstrated a homogeneous expression of CX3CR1 among MLPGs, without cells having especially high or low CX3CR1 expression. The overall pattern of CX3CR1 expression was similar between M-MDSCs and MLPGs (Fig. 3 B). This supports the conclusion that MLPGs are a homogeneous population of cells. In addition, BM-derived MLPGs failed to form colonies in myeloid colony-forming assays. Taken together, these results indicated that MLPGs belong to the monocytic lineage of cells and can differentiate almost exclusively to granulocytes.
Figure 3.
The nature of MLPGs. (A) CX3CR1 expression in CD117−CD11b+Ly6ChiLy6G− M-MDSCs, CD117+CD11b+Ly6ChiLy6G− MLPGs, CD117+CD11b+Ly6CintLy6G− GPs, and CD11b+Ly6CintLy6G+ PMN-MDSCs in BM of CX3CR1-GFP+ EL-4 TB mice. As a positive control, CD117−CD11b+Ly6CintLy6G− monocytes were used. Top: Typical example of staining. Bottom: Mean and SD from five mice per group are shown. ****, P < 0.0001 by two-tailed Student’s t test. MFI, mean fluorescence intensity. (B) Pattern of CX3CR1 expression in MLPGs in EL-4 TB mice. CX3CR1 expression in CD11b+Ly6ChiLy6G− monocytic cells in BM of EL-4 TB CX3CR1 reporter mice assessed by flow cytometry. Numbers in the plots indicate the percentage of gated cells. Expression of CX3CR1 is shown for individual mice (n = 5). (C and D) Differentiation of CD45.2+ GMPs isolated from the BM of EL-4 TB mice after adoptive transfer to sublethally irradiated CD45.1+ EL-4 TB recipient mice. CD45.2+ donor cells were evaluated by flow cytometry in spleens of CD45.1+ recipient mice after 24 h. (C) Phenotype of cells before transfer. (D) Phenotype of cells differentiated from the donor’s GMPs 24 h after transfer. Two experiments with the same results were performed. SSC, side scatter.
To clarify the origin of MLPGs, we tested whether GMPs can produce neutrophils through the MLPG stage. GMPs were sorted from the BM of CD45.2+ EL4 TB mice using established phenotypic criteria (Fig. 3 C). Sorted GMPs were then injected i.v. into sublethally irradiated CD45.1+ EL-4 TB recipient mice. 24 h later, among the donor cells, ∼30% of CD11b+Ly6ChiLy6G− cells had the phenotype of MLPGs, suggesting that the MLPGs were downstream of the GMPs (Fig. 3 D).
CD115 is a marker of monocytic cells. We studied the expression of CD115 in MLPGs from EL-4 TB mice. CD115− cells represented 90% and CD115+ 10% of all MLPGs (Fig. 4 A). We hypothesized that CD115+ and CD115− MLPGs may represent different stages of differentiation to granulocytes. To test this hypothesis, CD115+ and CD115− MLPGs were sorted from the BM of EL-4 TB mice and cultured for 3 d in the presence or absence of TES. More than 80% of cells that differentiated from CD115− MLPGs in the presence of TES were CD115−Ly6G+ granulocytes, with CD115+Ly6G− monocytic cells representing only a small fraction of cells (Fig. 4 B). Absence of TES in culture did not affect those proportions. In contrast, cells differentiated from CD115+ MLPGs in the presence of TES were equally split between CD115+Ly6G− monocytic and CD115−Ly6G+ granulocytic cells (Fig. 4 B). All monocytic cells also expressed the macrophage marker F4/80. In the absence of TES, the proportion of monocytic/macrophage cells differentiated from CD115+ MLPGs increased to 70% and granulocytic cells reduced to 10% (Fig. 4 B). These data indicate that CD115− MLPGs represent committed precursors, whereas differentiation of CD115+ MLPGs could be reverted to monocyte/macrophage lineage in the absence of tumor-derived factors. Expression of CX3CR1 in CD115+ MLPGs was similar to that in monocytes, whereas in CD115− it was significantly lower but still threefold higher than in neutrophils (Fig. 4 C).
Figure 4.
Differentiation of MLPGs. (A) Example of gating strategy and proportion of CD115+ cells among CD11b+Ly6ChiLy6G−CD117+ MLPGs from BM of EL4-TB mice. Three mice were evaluated. The results of individual experiments, mean and SD, are shown. (B) CD115− and CD115+ MLPGs were sorted from the BM of EL-4 TB mice and cultured for 3 d with GM-CSF with or without TES (20% vol/vol). The frequencies of CD115+Ly6G− monocytic cells and of CD115−Ly6G+ granulocytic cells obtained upon differentiation of both subsets of MLPGs are shown. The results of individual experiments (n = 5), mean and SD, are shown. **, P < 0.01; ****, P < 0.0001 by two-tailed Student’s t test. (C) CX3CR1 expression was measured using naive reporter mice in the indicated cell populations. The results of individual experiments (n = 3), mean and SD, are shown. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 by two-tailed Student’s t test. MFI, mean fluorescence intensity. (D) Example of phenotype of cells recovered after 3 d of culture with GM-CSF of CD11b+Ly6ChiLy6G−CD117− monocytes and CD11b+Ly6ChiLy6G−CD117+ MLPGs sorted from BM of naive mice. Numbers in the plots indicate the percentage of gated cells. (E) Numbers of cells generated from naive or EL-4 BM MLPGs after 3-d culture in complete RPMI with GM-CSF. Results of five experiments are shown. *, P < 0.05. (F) Antigen-specific proliferation of CD8+ T cells in the presence of Ly6G+ cells isolated after cultures of naive or EL-4 TB BM MLPGs. Cultures were done with GM-CSF and additional TES for EL-4 BM MLPGs. Proliferation was measured in triplicate by 3H thymidine uptake. PMEL T cells were used as responder cells in this antigen-specific suppression assay. Mean and SD are shown. Two experiments with similar results were performed. *, P < 0.05; ***, P < 0.001 by two-tailed Student’s t test. No stim, no stimulation; cpm, counts per minute; SSC, side scatter.
Although naive mice contained a very small amount of MLPGs in the BM, these cells could differentiate into granulocytes in vitro (Fig. 4 D). However, on a per-cell basis, the number of granulocytes generated from MLPGs in TB mice was significantly (P = 0.04) higher than from MLPGs in naive mice (Fig. 4 E). We tested the ability of MLPGs from control or TB mice to generate immunosuppressive PMN-MDSCs. MLPGs were isolated from the BM of naive or EL-4 TB mice and cultured for 3 d in the presence of GM-CSF without or with TES, respectively. At the end of the culture, granulocytes generated from the precursors were purified and tested in an antigen-specific suppression assay. Granulocytes generated from MLPGs of EL-4 TB mice had a stronger suppressive activity than granulocytes generated from MLPGs of control mice (Fig. 4 F).
Expansion of MLPGs in TB mice and their contribution to PMN-MDSC accumulation
Next, we compared the presence of MLPGs in BM and spleens of naive and TB mice using different transplantable and genetically engineered tumor models (GEMs). The presence of MLPGs in the BM of naive C57BL/6 and BALB/c mice was very low (Fig. 5, A and B), and in the spleens, these cells were practically undetectable (Fig. 5 C). In all tested transplantable tumor models in C57BL/6 mice (EL-4 lymphoma, LL2 lung adenocarcinoma, and B16F10 melanoma; Fig. 5 A) and in Balb/c mice (CT26 colon carcinoma; Fig. 5 B), the number of MLPGs in BM was increased two- to sixfold, and these cells were readily detectable in spleens (Fig. 5 C). This was associated with a substantial accumulation of PMN-MDSCs in spleens (Fig. 5, A and B). A modest increase in BM MLPGs in the B16F10 model of melanoma was associated with a relatively modest up-regulation of spleen PMN-MDSCs (Fig. 5 A). In addition, the Ret melanoma (Kato et al., 1998), transgenic adenocarcinoma mouse prostate (TRAMP) prostate (Greenberg et al., 1995), and KrasG12DTrp53R172HPdx-1 (KPC) pancreatic (Bayne et al., 2012) GEMs were used. In KPC mice, a significant (P < 0.01) up-regulation of BM and spleen MLPGs was associated with a substantial increase of spleen PMN-MDSCs (Fig. 5 D). In TRAMP mice, the little changes in BM and spleen MLPGs were associated with a minimal accumulation of PMN-MDSCs (Fig. 5 E), whereas a modest BM and spleen MLPG increase in Ret melanoma mice was associated with a small accumulation of spleen PMN-MDSCs (Fig. 5 F). We performed a regression analysis of the absolute numbers of BM MLPGs and spleen PMN-MDSCs across the different C57BL/6 tumor models described above. The absolute number of BM MLPGs closely and positively correlated with the expansion of spleen PMN-MDSCs (P = 0.007; Fig. 5 G). The increased presence of MLPGs was observed not only in cancer but also in an experimental model of colitis, which was associated with neutrophil expansion (Fig. S3 A).
Figure 5.
Population of MLPGs in TB mice. (A) Total cell number of MLPGs in BM and PMNs/PMN-MDSCs in spleens of naive or TB mice 3–4 wk after subcutaneous inoculation of the indicated tumor cells to C57BL/6 mice. Results of individual mice are shown (5–12 mice per group). (B) Total cell number of MLPGs in BM and PMNs/PMN-MDSCs in spleens of naive or CT26 TB mice BALB/c mice 3–4 wk after subcutaneous inoculation of the tumor cells. Results of individual mice are shown (3–5 mice per group). (C) Total number of MLPGs in spleens of B16F10 and CT26 TB mice 3–4 wk after subcutaneous inoculation of the tumor cells. Results of individual mice are shown (6–9 mice per group). (D) Total cell number of MLPGs in BM and spleens, and PMNs/PMN-MDSCs in spleens of control (PC) or KPC mice. Results of individual mice are shown (three mice per group). (E) Total cell number of MLPGs in BM and spleens, and PMNs/PMN-MDSCs in spleens of control or TRAMP mice. Results of individual mice are shown (three mice per group). (F) Total cell number of MLPGs in BM and spleens, and PMNs/PMN-MDSCs in spleens of control or RET melanoma mice. 3–11 mice were used in each experiment, and the number of mice evaluated is shown in each panel. Values of mean and SD are shown. *, P ≤ 0.05; **, P ≤ 0.01; ***, P < 0.001; ****, P ≤ 0.0001 by two-tailed Student’s t test. (G) Correlation between total cell numbers of MLPGs in BM and total cell numbers of PMN-MDSCs in spleens in all C57BL/6 TB mice described above. Spearman coefficient was used to calculate correlation.
Since MLPGs were detected in naive mice (albeit in much smaller numbers), we assessed the characteristics of these cells. MLPGs from tumor-free mice also had a ring-shaped nucleus, a morphology similar to the one seen for MLPGs from TB mice (Fig. S3 B). Similarly to MLPGs from TB mice, naive MLPGs expressed very little CCR2 (Fig. S3 C), were positive for CX3CR1 (Fig. S3 D), and did not suppress T cell response (Fig. S3 E).
Substantial accumulation of MLPGs in spleens of TB mice raised the question of the role of the spleens in the expansion of PMN-MDSCs in TB mice. To address this question, mice underwent either splenectomies or sham surgery followed by a long recovery period (10 wk) to eliminate the effect of surgical stress on granulopoiesis. After that time, mice were injected s.c. with LL2, B16F10, or KPC (G43.7) tumor cells. In all three models, splenectomy did not affect the tumor growth (Fig. S4 A). On day 21 after inoculation of LL2 cells, the presence of different populations of myeloid cells was evaluated. There were no differences in the presence of MLPGs in the BM (Fig. S4 B), as well as no differences observed in MDSCs and other myeloid cells in the BM, blood, tumor, or liver (Fig. S4 C). A similar effect was seen in the B16F10 melanoma model (Fig. S4 D). We isolated tumor-associated macrophages (TAMs), PMN-MDSCs, and M-MDSCs from tumors and evaluated their suppressive activity. All three populations from splenectomized and control mice (sham surgery) suppressed T cell function equally (Fig. S4 E). Thus, PMN-MDSC expansion in TB mice was not restricted to the spleen, with instead the BM playing the major role in the process.
What could be the contribution of MLPGs to the total pool of neutrophils and PMN-MDSCs? We used CX3CR1CreER/GFP × ROSA26-DTRfl/fl mice to selectively deplete monocytic cells. Treatment of these mice with tamoxifen (Tm) causes an up-regulation of the diphtheria toxin (DT) receptor by cells expressing CX3CR1, rendering these cells susceptible to death after diphtheria toxin injection. Since classic GPs, neutrophils, and PMN-MDSCs do not express CX3CR1, we expected that only MLPG-derived granulocytes would be affected in these mice. Control and EL-4 TB mice were treated with Tm for 5 d, followed by two administrations of DT (every other day) in addition to Tm gavage (Fig. 6 A). In EL-4 or LL2 TB mice, Tm was administered after the mice exhibited palpable tumors. DT treatment resulted in the depletion of monocytic cells in TB and tumor-free mice (Fig. 6 B). Of note, to confirm monocyte depletion, we compared DT-treated and untreated Cre+ mice, since these mice have Cx3cr1-driven expression of Cre and GFP. In all other experiments described below, DT-treated Cre− mice served as control. This treatment substantially decreased the number of BM MLPGs but did not affect GPs (Fig. S5, A and B). In naive mice, targeting MLPGs did not change the presence of granulocytes in the spleens (Fig. 6, C and D) or BM (Fig. 6, E and F). Moreover, the number of these cells had a trend to increase. In contrast, in EL-4 and LL2 TB mice, MLPG depletion significantly reduced the proportion and total number of PMN-MDSCs in the spleen (Fig. 6, C and D). Overall, the total number of PMN-MDSCs was decreased by >50%. In the BM of TB mice, targeting MLPGs also caused a decrease in granulocytic cells; however, it did not reach statistical significance (Fig. 6, E and F). It was likely due to the fact that the BM normally contains a large population of granulocytes that makes changes in TB mice difficult to detect. As expected, the population of M-MDSCs was reduced in the spleen, but not in the BM of TB mice treated with DT (Fig. 6 G). Lack of the effect in BM may be the result of a faster turnover of these cells in the BM compared with the spleen. After the treatment with DT, Cre+ TB mice had significantly smaller tumors than Cre− mice (Fig. 6 H).
Figure 6.
Contribution of MLPGs to PMN-MDSC accumulation in TB mice. (A) Schema of depletion of monocytic cells in CX3CR1CreER+ ROSA-DTRfl/fl EL-4 TB mice. (B) Example of flow cytometry analysis performed in blood samples from naive or EL-4 TB CX3CR1CreER+ ROSA-DTRfl/fl mice treated with Tm with or without subsequent DT injection. Note the efficient depletion of CD11b+CX3CR1+Ly6Clo/− patrolling monocytes. (C and D) Results of individual mice are shown in the figures (6–10 mice per group). Percentage (C) and total cell numbers (D) of PMNs/PMN-MDSCs in spleens of naive, EL-4, or LL-2 TB CX3CR1CreER− and CX3CR1CreER+ ROSA-DTRfl/fl mice. (E and F) Results of individual mice are shown in the figures (five to six mice per group). Percentage (E) and total cell numbers (F) of PMNs/PMN-MDSCs in BM of naive, EL-4, or LL-2 TB CX3CR1CreER− and CX3CR1CreER+ ROSA-DTRfl/fl mice. Result from each tested mouse as well as mean and SD are shown. Individual P values (*, P < 0.05; **, P < 0.01; ***, P < 0.001) by two-tailed Student’s t test are shown. (G) M-MDSC population in CX3CR1CreER− and CX3CR1Cre-ER+ Rosa-DTRfl/fl LL2 TB mice. Total cell numbers of M-MDSCs in spleens and BM are shown. Result from each tested mouse (n = 7) as well as mean and SD are shown. Individual P values (*, P < 0.05) by two-tailed Student’s t test are shown. (H) Tumor size in CX3CR1CreER− and CX3CR1Cre-ER+ Rosa-DTRfl/fl EL-4 TB mice at day 16 at the time of sacrifice. Result from each tested mouse (n = 8–10) as well as mean and SD are shown. Individual P values (*, P < 0.05) by two-tailed Student’s t test are shown. (I) EG7 tumors were established s.c. in CX3CR1CreER− and CX3CR1Cre-ER+ Rosa-DTRfl/fl mice. 10 d after tumor inoculation, when tumor became palpable, mice were treated with Tm (4 mg per day) for 8 d and DT injections (1 µg in 200 µl of PBS) on days 6 and 8. 1 d after that, spleen CD8+ T cells were isolated and stimulated with the OVA-derived SIINFEKL peptide, and their IFN-γ production was measured in an ELISpot assay (n = 3). P values were calculated by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P ≤ 0.0001. SSC, side scatter.
To assess the impact of the DT treatment on tumor specific immune response, we established OVA-expressing EL4 (EG7) tumors s.c. in CX3CR1CreER− and CX3CR1Cre-ER+ Rosa-DTRfl/fl mice. 10 d later, when tumor became palpable, mice were treated with Tm for 8 d and DT on days 6 and 8. 1 d after that, spleen CD8+ T cells were isolated and stimulated with the OVA-derived SIINFEKL peptide, and their IFN-γ production was measured in an ELISpot assay. We observed a strong response to the peptide in Cre+ mice, whereas the response in Cre− mice was barely detectable (Fig. 6 I). However, this effect may not necessarily argue for a specific role of PMN-MDSCs, since the population of M-MDSCs in Cre+ mice was also decreased. These results suggest that MLPGs do not contribute to the generation of neutrophils in naive mice but are responsible for the accumulation of ≤50% of PMN-MDSCs in EL-4 and LL2 TB mice.
Mechanisms of MLPG expansion in TB mice
Next, we investigated the possible specific role of Rb1 in MLPGs. We assessed the number of MLPGs in the BM and spleens of tumor-free Rb1−/− (Rb1 KO) mice, and mice with a deletion of all three members of the pocket protein family (Rb1, p107, and p130; triple KO [TKO] mice). The total numbers of MLPGs in both Rb1 KO and TKO mice increased more than twofold in the BM and >6 times in the spleens (Fig. 7, A and B); these levels were very similar to those observed in various tumor models, suggesting that Rb1 could be a dominant factor in regulating MLPG accumulation. It is known that the IRF8 transcription factor plays an important role in the switch between monocytic and granulocytic differentiation (Holtschke et al., 1996; Wang et al., 2014). Its down-regulation was previously implicated in the accumulation of PMN-MDSCs (Stewart et al., 2009; Waight et al., 2013; Paschall et al., 2015). MLPGs, in contrast to monocytes, did not express irf8 (Fig. 7 C). No difference in the expression of Rb1 was found between MLPGs and GPs (Fig. 7 D). We compared the effect of Rb1 and IRF8 deletion on the accumulation of MLPGs and GPs. Rb1 and IRF8 KO mice demonstrated a similar substantial accumulation of neutrophils in the BM (Fig. 7 E). However, the effect of their deletion on the populations of MLPGs and GPs was opposite. Rb1 deletion caused a significant (P = 0.03) increase in the population of MLPGs while not affecting GPs, whereas IRF8 deletion resulted in a massive expansion of GPs but and a decrease of MLPGs (Fig. 7 F). We asked whether Rb1-deficient GMPs produced more MLPGs. To address this question, we injected CD45.1+ WT GMPs together with CD45.2+ Rb1−/− GMPs to sublethally irradiated CD45.1+CD45.2+ recipient mice. On day 4, we assessed the frequencies of MLPGs generated from CD45.1+ WT GMPs, CD45.2+ Rb1−/− GMPs, and host CD45.1+CD45.2+ MLPGs. The frequency of MLPGs generated from Rb1−/− GMPs was the same as from WT GMPs (Fig. S5, C and D). These results not only underscore a specific role of Rb1 in the regulation of MLPG expansion but also demonstrated distinct molecular mechanisms regulating the expansion of MLPGs and GPs.
Figure 7.
Rb1 regulates MLPG expansion while IRF8 controls GP accumulation. Results of individual mice are shown (5–11 mice per group). (A) Total cell numbers of MLPGs in BM (left) and spleens (right) in WT or Rb1 KO naive mice. (B) Total cell numbers of MLPGs in BM (left) and spleens (right) in control mice (p107+/−) or Rb TKO naive mice. (C) Expression of irf8 in MLPGs. Expression of irf8 (normalized to β-actin) in BM MLPGs from naive mice. Monocytes and neutrophils are presented as controls. Gene expression was evaluated by quantitative RT-PCR in three independent experiments, each performed in triplicate. Mean and SD of each measurement in triplicate is shown. (D) Rb1 expression in CD117+CD11b+Ly6ChiLy6G− MLPGs and CD117+CD11b+Ly6CintLy6G− GPs in BM of EL-4 and LL2 TB Rb1-GFP reporter mice. Individual results and mean and SD are shown. P values were calculated by two-sided Student’s t test. MFI, mean fluorescence intensity. (E) Total cell numbers of neutrophils in BM of WT mice and Rb1 KO or IRF8 KO mice. (F) Total cell numbers of MLPGs and GPs in BM of WT mice and Rb1 KO or IRF8 KO mice. Mean and SEM are shown. In experiments with Rb1 KO mice, n = 6, and with IRF8 KO mice, n = 4. P values were calculated by two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (G) Growth of subcutaneous EL-4 tumor injected into WT and Rb1 KO mice. Results in individual mice (n = 5) are shown. (H) Total cell numbers of BM and spleen MLPGs and GPs in WT and Rb1 KO EL-4 TB mice. Values in individual mice (n = 5) and mean and SD are shown. P values were calculated by two-tailed Student’s t test. **, P < 0.01; ****, P < 0.0001. (I) Growth of subcutaneous MC38 tumors injected into WT and Rb1 KO mice. Results in individual mice (n = 4) are shown. P value was calculated by two-way ANOVA.
We next asked whether tumors could affect populations of MLPGs and GPs in Rb1 KO mice. We found a modest increase in tumor burden in EL-4 TB Rb1 KO mice compared with EL-4 TB Rb1 WT mice (Fig. 7 G). The presence of MLPGs in BM in WT TB mice, which was already elevated relative to the control level in WT naive mice (as seen in Fig. 3), was not further increased in Rb1 KO EL-4 TB mice (Fig. 7 H). However, in the spleens, the total cell number of MLPGs and GPs was markedly expanded. This was associated with an increased population of PMN-MDSCs (Fig. 7 H). We asked whether the lack of substantial increase in the tumor burden in EL-4 TB Rb1 KO mice was the result of the relative low immunogenicity of the EL-4 tumor model. To test this hypothesis, we used a more immunogenic model, the MC38 tumor model, and observed a substantially higher tumor growth in Rb1 KO mice than WT mice (Fig. 7 I).
To elucidate the mechanism of MLPG accumulation in TB mice, we compared the proliferation and survival of MLPGs from TB mice with those from naive mice. MLPGs from EL-4 TB mice had a higher proliferation than MLPGs isolated from naive mice (as assessed by BrdU uptake or 3H thymidine incorporation; Fig. 8 A). MLPGs from EL-4 TB mice had significantly (P = 0.0008) less apoptosis (Annexin V+ cells) after overnight culture than MLPGs from naive mice (Fig. 8 B). The addition of TES decreased apoptosis in both the naive and EL-4 TB MLPGs. However, even in the presence of TES, MLPGs from TB mice demonstrated significantly (P = 0.04) less apoptosis (Fig. 8 C). MLPGs from tumor-free Rb1 KO mice demonstrated higher proliferation in vitro (Fig. 8 D) and in vivo (Fig. 8 E) than MLPGs from WT mice. In contrast, apoptosis of MLPGs from Rb1 KO mice was only slightly reduced compared with MLPGs from naive mice (Fig. 8 F). Thus, the decreased apoptosis together with the increased proliferation contributed to the expansion of MLPGs in cancer. Since Rb1 did not affect the survival of MLPGs, there are likely other factors contributing to MLPG expansion in cancer.
Figure 8.
Mechanism of MLPG expansion in TB mice. (A) Proliferation of BM MLPGs from naive and EL-4 TB mice in vitro assessed by BrdU uptake (left) and 3H thymidine incorporation (right) after 20 h of culture in complete RPMI without cytokines (n = 4). (B) Frequency of Annexin V+ BM MLPGs from naive and EL-4 TB mice after 20 h of culture in complete RPMI without cytokines (n = 7). (C) Frequency of Annexin V+ BM MLPGs from naive and EL-4 TB mice cultured in the presence of 5% vol/vol EL-4 TES for 20 h in complete RPMI without cytokines (n = 4). (D) Proliferation of BM WT and Rb1 KO MLPGs in vitro, assessed by BrdU uptake after 20 h of culture in complete RPMI without cytokines (n = 4). (E) Proliferation of BM WT and Rb1 KO MLPGs in vivo, assessed by BrdU uptake. Frequency of BrdU+ cells was evaluated 4 h after i.p. injection of BrdU (n = 3). (F) Frequency of Annexin V+ BM WT and Rb1 KO MLPGs 20 h after culture in complete RPMI without cytokines (n = 6). (G) Numbers of cells generated from EL-4 BM CD11b+Ly6ChiLy6G−CD117− M-MDSCs and CD11b+Ly6ChiLy6G−CD117+ MLPGs after 3-d culture in RPMI (supplemented or not with FBS) with or without addition of the indicated cytokines. Dashed line, starting number of cells (n = 6); Lip, lipids. (H) Frequency of live cells from the experiments described in G (n = 6). (I) Frequency of Ly6G+ cells generated from the MLPG cultures described in G (n = 6). (J) Numbers of cells generated from EL-4 BM MLPGs cultured for 3 d in serum-free media (SFM) with or without the indicated cytokines. Complete RPMI was used as a positive control (n = 4). P values were calculated by two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Mean and SD are shown.
We investigated the cytokine requirements for survival and differentiation of MLPGs in vitro. In the absence of cytokines and serum, practically no live cells were found after 3 d of culture of MLPGs (Fig. 8, G and H). However, the presence of FBS was sufficient to support the survival and expansion of MLPGs and their differentiation to granulocytes (Fig. 8, G–I). Moreover, the addition of G-CSF, M-CSF, or SCF did not change the total number and the proportion of granulocytic cells generated in culture; only GM-CSF provided modest improvement (Fig. 8 G). In contrast, FBS was not sufficient to support survival of M-MDSCs, and these cells required the presence of GM-CSF and M-CSF (Fig. 8, G and H). We investigated the role of individual growth factors in MLPG survival and differentiation using serum-free medium. In the absence of FBS, MLPG survival and expansion was not supported by any of the tested cytokines alone (Fig. 8 J). The presence of lipids in serum was critical for MLPG survival and differentiation, since lipid-depleted FBS did not support MLPG survival and differentiation. In contrast to serum-free medium, GM-CSF alone supported MLPG survival and expansion in lipid-depleted medium (Fig. 8 G). Thus, yet unidentified lipid factors in combination with GM-CSF were required to support differentiation of MLPGs.
Human monocytic cells contain MLPGs
We asked whether the human M-MDSCs could contain MLPGs. To address this question, we sorted M-MDSCs and monocytes from patients with advanced non–small cell lung cancer based on established phenotypic criteria (CD15−CD14+HLA-DR−/low for M-MDSCs and CD15−CD14+HLA-DRhigh for monocytes; Bronte et al., 2016). Gene expression profiles were evaluated using RNA-seq. Analysis of individual genes significantly differentially expressed in CD15−CD14+HLA-DR−/low cells compared with monocytes (619 genes with FDR <5%) revealed a number of up-regulated genes associated with neutrophil function: IL-8, MNDA (myeloid cell nuclear differentiation antigen), CSF3R (G-CSF receptor), LYZ (lysozyme), and NCF4 and NCF2 (neutrophil cytosolic factors 4 and 2). At the same time, CD15−CD14+HLA-DR−/low cells demonstrated significant down-regulation of several genes associated with monocytes/macrophages: CSF1R (M-CSF receptor), NR4A1 (Nur77), CX3CR1, ITGA4 (CD49d), and CD52 (Fig. 9 A). Function enrichment analysis revealed significant overrepresentation in CD15−CD14+HLA-DR−/low cells of genes associated with various neutrophil functions (Fig. 9 B). These results may suggest that CD15−CD14+HLA-DR−/low cells in patients with cancer may contain precursors of neutrophils.
Figure 9.
MLPGs in humans. (A) Analysis of 619 genes significantly differentially expressed (FDR <5%) in M-MDSCs compared with monocytes from lung cancer patients. Expression heatmap for a gene subset specific to neutrophils and monocytes/macrophages and top significant genes. Mono, monocyte. (B) Functions significantly enriched among significant genes as identified by Ingenuity Pathway Analysis. (C) Proportion of CXCR1+ cells among healthy donor (HD) and prostate cancer patient CD14+CD15−HLA-DRhi monocytes (Mon) and among CD14+CD15−HLA-DRlo/− M-MDSCs from the same patients. Results from individual patients are shown (n = 8). Mean and SD are shown. ****, P < 0.0001 by two-sided Student’s t test. Mon, monocyte. (D) CXCR1+CD14+CD15−HLA-DRlo/− and CXCR1− CD14+CD15−HLA-DRlo/− M-MDSCs were sorted from the peripheral blood of cancer patients and used in a three-way allogeneic mixed leukocyte reaction using dendritic cells (DC) and T cells from unrelated donors. T cell proliferation was measured in triplicate, and mean and SD are shown. P values were calculated by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.0001. (E) Differentiation of neutrophils from the indicated monocytic populations after 4-d culture on OP-9 feeder cells with G-CSF and GM-CSF. A typical example is shown. May-Grünwald-Giemsa staining on the left; staining for naphthol-AS-chloroacetate esterase (napththol-AS-chlor.acet. Est) activity on the right. Four experiments with the same results were performed. Scale bar, 20 µm. (F) Frequency of neutrophils differentiated from monocytes and M-MDSCs isolated from cancer patients after 4 d culture on OP-9 feeder cells with G-CSF and GM-CSF. Values for each patient are shown (n = 4). In all experiments, mean and SD values are shown. P values were calculated by Student’s t test. *, P < 0.05; **, P < 0.01. cpm, counts per minute.
One notable gene that was up-regulated on M-MDSCs was Cxcr1. This is a chemokine receptor expressed on neutrophils and usually not associated with monocytes. We evaluated CXCR1 expression by flow cytometry and determined that monocytes from healthy individuals and patients with prostate cancer had very low expression of this protein, whereas almost 20% of M-MDSCs from the same patients were positive for CXCR1 (Fig. 9 C). Next, we sorted CD15−CD14+HLA-DRlo/−CXCR1− and CXCR1+ M-MDSCs from three patients with lung and pancreatic cancers and assessed the ability of these cells to suppress T cell proliferation in an allogeneic mixed leukocyte reaction. CXCR1− cells had a potent suppressive activity, whereas CXCR1+ cells had less or were not suppressive at all (Fig. 9 D). Thus, CXCR1− M-MDSCs were bona fide M-MDSCs, whereas CXCR1+ cells did not fit the basic criteria of M-MDSCs. Since mouse MLPGs had the same feature (lack of suppressive activity despite phenotypic criteria of M-MDSCs), we investigated the ability of CXCR1+ M-MDSCs to differentiate to neutrophils. CD15−CD14+HLA-DRhi monocytes and CXCR1+ and CXCR1− M-MDSCs were sorted from the peripheral blood of patients with non–small cell lung cancer and cultured for 4 d on OP9 feeder cells in the presence of GM-CSF and G-CSF (Anani and Shurin, 2017). Practically all cells differentiated from monocytes and CXCR1−CD14+CD15−HLA-DR−/lo M-MDSCs had morphological features of macrophages. In contrast, a substantial proportion of cells with the morphology of neutrophils were detected after the culture of CD15−CD14+HLA-DR−/lowCXCR1+ cells (Fig. 9 E). To verify that these cells were indeed neutrophils, we evaluated the activity of naphthol AS-D chloroacetate esterase, a specific marker of human neutrophils. Practically no esterase-positive cells were found among cells differentiated from monocytes and CXCR1− M-MDSCs, whereas esterase-positive cells were readily detectable among cells differentiated from CXCR1+ monocytic cells (Fig. 9 E). Overall, ∼20% of cells differentiated from CXCR1+CD14+CD15−HLA-DR−/lo cells were represented by neutrophils, whereas neutrophils were practically undetectable among cells differentiated from monocytes and bona fide CXCR1− M-MDSCs (Fig. 9 F). These results suggest that human CXCR1+CD14+CD15−HLA-DR−/lo monocytic cells are enriched for MLPGs.
Discussion
In this report, we describe the existence of a population of committed GPs that belongs to the monocytic lineage, named MLPGs. These cells were either present at very low numbers or completely absent in tumor-free mice in the steady state, but were markedly expanded in the BM and spleens of TB mice. They expressed the CD11b and Ly6Chi markers and lacked expression of Ly6G, which would define them as classic inflammatory monocytes in control mice or M-MDSCs in TB mice (Bronte et al., 2016). However, MLPGs lacked suppressive activity and were characterized by the expression of c-kit (CD117). C-kit expression is associated with progenitors of granulocytes or monocytes/macrophages such as CMPs (Lin−IL7Rα−c-Kit+Sca-1−CD34+CD16/32lo) and GMPs (Lin−IL7Rα−c-Kit+Sca-1−CD34+CD16/32hi; Akashi et al., 2000). CD117 was also present on committed neutrophil precursors (GPs) that can be either Ly6Clo or Ly6Cint. Using fate mapping experiments and gene expression profile analysis, we demonstrated that CD11b+Ly6CintLy6G−CD117+SiglecF−CCR2− cells were indeed committed GPs, which were very similar to the preNeu granulocyte progenitors described previously (Evrard et al., 2018). The main challenge was to determine whether MLPGs were indeed monocytic cells rather than a subset of GPs with a high Ly6C expression.
Several lines of evidence support the conclusion of the monocytic origin of MLPGs distinct from that of GPs. (i) MLPGs had a very distinct genomic profile from GPs. It correlated with monocytes, MDPs and common dendritic cell progenitors, whereas the genomic profile of GPs correlated only with the profiles of granulocytes. Moreover, after filtering for only genes differentially expressed between MLPGs and GPs, the MLPG profile correlated only with those of monocytes/M-MDSCs, whereas the GP gene expression profile correlated only with those of granulocytes/PMN-MDSCs. (ii) The genomic profile of MLPGs was also distinct from that of GMPs. (iii) MLPGs expressed the monocytic cell–specific marker CX3CR1 at a level similar to that found in monocytes/M-MDSCs, whereas GPs had very low expression of this marker, as in neutrophils/PMN-MDSCs. (iv) MLPGs differentiated to neutrophils similarly in the presence of GM-CSF and M-CSF. (v) Deletion of the Irf8 transcription factor caused a massive accumulation of GPs but resulted in a decreased amount of MLPGs. In contrast, Rb1 deletion resulted in the accumulation of MLPGs but did not affect GPs.
Under steady state conditions, granulocyte differentiation in mice proceeds through a well-defined CMP–GMP–GP pathway, which is negatively regulated by IRF8. Monocytes differentiate from GMPs, MDPs, or common myelolymphoid progenitors, which are positively regulated by IRF8 (Kurotaki et al., 2014). It appears that under steady state conditions, there is a very small (and likely inconsequential for the generation of granulocytes) population of monocytic cells that can differentiate into granulocytes. In our study, MLPGs were downstream of GMPs. In a recent study, differentiation of monocytes from GMPs independently of MDPs was described (Yáñez et al., 2017). Interestingly monocytes derived from GMPs had expression of genes specific for neutrophils (Yáñez et al., 2017). It is tempting to suggest that the “neutrophil-like” subset of monocytes from that study may contain MLPGs. In contrast to control mice, in cancer, the MLPG population expanded extensively and substantially contributed to PMN-MDSC accumulation (Fig. 10). It appears that down-regulation of Rb1 could be a major mechanism of this expansion. Although the molecular mechanisms of this effect warrant further elucidation, more active proliferation of MLPGs could contribute to this phenomenon. This would be consistent with the overall role of Rb1 as a checkpoint in the cell cycle (Calo et al., 2010). MLPGs from Rb1 KO and TB mice showed similar enhanced proliferation compared with WT and naive tumor-free mice. However, it may be only part of the mechanism, since Rb1 did not affect MLPG survival but only proliferation, whereas in TB mice, MLPGs demonstrated both better survival and proliferation than cells from naive mice.
Figure 10.
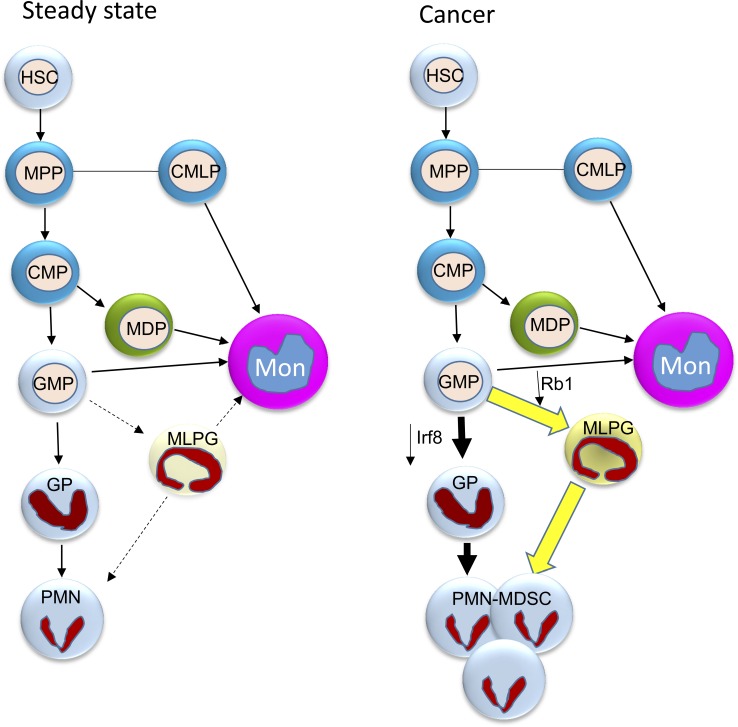
Schema of neutrophil differentiation in cancer. In the steady state, neutrophil differentiation is the end point of the MPP–CMP–GMP–GP pathway. This pathway is negatively regulated by the IRF8 transcription factor. In steady state, the MLPG contribution to the neutrophil and monocyte pool is minimal. In cancer, down-regulation of IRF8 and Rb1 leads to expansion of both PMN-MDSC progenitors: GPs and their monocytic counterparts MLPGs. MLPGs, in some models, may contribute to 50% of the PMN-MDSC pool. CMLP, common myelolymphoid progenitor; HSC, hematopoietic stem cell; Mon, monocyte; MPP, multipotent progenitor.
Our data demonstrated that MLPGs do not contribute to granulocyte differentiation in naive mice but are important precursors of PMN-MDSCs in TB mice, based on the following findings. (i) All transplantable tumor models and most GEMs of cancer demonstrated accumulation of MLPGs in BM as well as in the spleen, the site of extramedullary hematopoiesis in mice. (ii) The strength of this accumulation corresponded to the levels of PMN-MDSC expansion in these models. (iii) Depletion of monocytic cells by targeting CX3CR1 reduced the presence of PMN-MDSCs by half in EL-4 and LL2 TB mice, without affecting granulocyte numbers in control mice.
MLPGs from TB may be endowed with some intrinsic ability to generate immunosuppressive PMN-MDSCs. Further experiments would be required to figure out the suppressive mechanism of MLPG-derived PMN-MDSCs. Dramatic expansion of MLPGs in spleens of TB mice supported the important role of extramedullary hematopoiesis in MDSC accumulation. We tested the relative contribution of the spleen to MDSC accumulation in tumors. Previous studies demonstrated that splenectomy reduced the presence of MDSCs in tumors (Lecut et al., 2009; Kawano et al., 2015; Levy et al., 2015; Schug et al., 2015). This would directly support the concept of our study, where MLPGs were found to be dramatically expanded in spleen. However, in one study, a splenectomy was performed 21 d after tumor inoculation and significantly reduced tumor growth (Levy et al., 2015). In another study, a splenectomy was performed 10 d before tumor inoculation (Kawano et al., 2015), whereas in several other studies splenectomy was performed at the time of tumor inoculation (Lecut et al., 2009; Schug et al., 2015). We were concerned with the possibility that such invasive surgery, as well as removal of the spleen at the time of myeloid cell expansion, may not accurately recapitulate the possible contribution of the spleen to the expansion of MLPGs and PMN-MDSCs. Therefore, we performed splenectomy 10 wk before tumor inoculation to allow for full recovery. Our data indicated that in the absence of the spleen, BM provided full support for the expansion of functionally potent M-MDSCs and PMN-MDSCs in tumors. Thus, although the role of the spleen in MDSC production is unquestionably important, the BM can provide sufficient support for MDSC accumulation.
Thus, this study demonstrated the existence of monocytic precursors of granulocytes that are expanded in cancer, not only in mouse models, but also in cancer patients. Mechanistic studies in mouse models demonstrated that this myeloid differentiation pathway is controlled by the down-regulation of Rb1, which substantially contributed to the accumulation of PMN-MDSCs. This opens a new opportunity for selective targeting of abnormal myelopoiesis in cancer.
Materials and methods
Human subjects and samples
Samples of peripheral blood from healthy volunteers were collected at the Wistar Institute, while samples from patients were collected at the Helen F. Graham Cancer Center, part of the Christiana Care Health System. The study was approved by the Institutional Review Boards of the Christiana Care Health System at the Helen F. Graham Cancer Center and the Wistar Institute. All patients signed approved consent forms. Six healthy volunteers and 11 patients with previously untreated stage II–III melanoma or non–small cell lung cancer were enrolled. This cohort included four women and six men, ages 34–72 yr.
Mice and tumor models
All experiments with mice were approved by the Wistar Institute Animal Care and Use Committee. The mice were kept under pathogen-free conditions. Female C57BL/6 CD45.1+, C57BL/6 CD45.2+, and Balb/c mice (age 6–8 wk) were purchased from Charles River Laboratories. OT-I TCR-transgenic mice (C57Bl/6-Tg(TCRaTCRb)1100mjb; 4–6 wk old), female Pmel TCR-transgenic mice (B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest; 4–6 wk old), CX3CR1CreER mice (B6.129P2(Cg)-Cx3cr1tm2.1(cre/ERT2)Litt/Wgan), and ROSA-DTRfl/fl (CBy.B6-Gt(ROSA)26Sortm1(HBEGF)Awai) were purchased from the Jackson Laboratory. TRAMP mice were bred in Dr. L.R. Languino’s laboratory (Thomas Jefferson University, Philadelphia, PA; Chapman et al., 2011). CX3CR1CreERROSA-DTRfl/fl mice were generated by crossing a male CX3CR1CreER with a female ROSA-DTRfl/fl. Ret mice were obtained from Dr. Umansky (German Cancer Center, Heidelberg, Germany). Transgenic KPC mice (Bayne et al., 2012), Rb1 KO (Youn et al., 2013), and Rb TKO (Viatour et al., 2008) mice were described previously. IRF8−/− mice on a C57BL/6 background were generated as previously described (Holtschke et al., 1996) and originally were a kind gift of Dr. K. Ozato (National Institutes of Health, Bethesda, MD). Rb1-GFP reporter mice were kindly provided by Dr. J. Sage (Stanford University, Stanford, CA). They were generated by inserting an eGFP cassette at the start codon of Rb (Burkhart et al., 2010). The eGFP reporter construct also included two polyadenylation signals that prevent downstream expression of the coding Rb transcript from the bacterial artificial chromosome. These mice were backcrossed to C57BL/6 background for 10 generations. For the experiments described in this study, age- and sex-matched littermates were used as controls. To induce CX3CR1+ monocytic cell depletion in CX3CR1CreERROSA-DTRfl/fl mice, mice received Tm (Sigma-Aldrich) as a solution in corn oil (Sigma-Aldrich) and ethanol at 20 mg/ml. Animals received five 4-mg doses of Tm (200 µl) by gavage for five consecutive days. At day 6, the coinjection of Tm and DT (Sigma-Aldrich) started. Mice received two i.p. doses of DT (1 µg in 200 µl of PBS) every other day.
For splenectomy, the surgical site was sterilely prepared and draped after anesthesia. A 1-cm paramedian incision was made over the left upper quadrant. The spleen was identified, small clips were applied to the vascular pedicles, and the spleen was removed. For sham operations, a laparotomy incision was made, but no splenic tissue was resected. For adoptive transfers experiments, mice were sublethally irradiated with a single dose of 450 cGy.
Cell lines
Cell lines including EL-4 lymphoma, Lewis lung carcinoma, B16F10 melanoma, CT26 colon carcinoma, and OP9 stromal cells were obtained from ATCC. All except OP9 cells were maintained in DMEM supplemented with 10% FBS (Sigma-Aldrich) and 1% penicillin-streptomycin (Thermo Fisher Scientific) at 37°C with 5% CO2. OP9 cells were maintained in MEMα without nucleosides (Thermo Fisher Scientific) supplemented with 20% FBS and 1% penicillin-streptomycin at 37°C with 5% CO2. Tumor cells were injected subcutaneously at 5 × 105 cells (diluted in 100 µl of PBS) per mouse to form tumors 1.5 cm in diameter within 3 wk of injection.
Reagents
A list of reagents is provided in Table S1.
Cell isolation and culture
For the BM, one leg (one tibia and one femur) was used (absolute numbers given are for one leg). When a higher number of cells was required (especially for progenitor populations such MLPGs, GPs, and GMPs), all bones were used (legs, pelvises, sternum, and spine). Bones cleaned of muscular tissues are crushed with a sterile mortar and pestle in a cold solution of cell suspension buffer (CSB): 1× PBS (Thermo Fisher Scientific), 1% FBS, and 2 mM EDTA (Thermo Fisher Scientific). The cell suspension was then filtered through a 70-µm strainer (Thermo Fisher Scientific) placed on a conical 50-ml Falcon tube. For spleens, the organ was put in a 70-µm strainer placed on a conical 50-ml Falcon tube and cut into small pieces. These pieces were then ground against the cell strainer using the plunger of a 5-ml syringe and washed several times with cold CSB. BM and spleen cell suspensions were then centrifuged at 1,500 rpm at 4°C, the supernatant was removed, and red blood cells were lysed in ammonium chloride lysis buffer for 5 min at room temperature. Cells were washed and resuspended in cold CSB. Single-cell suspensions from tumor tissues were prepared using the mouse tumor dissociation kit (Miltenyi Biotec) according to the manufacturer’s recommendations, with an additional red blood cell lysis step as described above.
Sorted cells (≤105 cells per well for MLPGs) were cultured in 24-well plates in 1 ml total volume in the presence of 20 ng/ml recombinant GM-CSF (Thermo Fisher Scientific). In some experiments, 20 ng/ml G-CSF (Peprotech) or M-CSF (Peprotech) were also used. Cells were cultured in complete RPMI: RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% FBS, 1% penicillin-streptomycin, and 1× 2-mercaptoethanol (Thermo Fisher Scientific). 5, 10, or 20% vol/vol TES from EL-4 tumors was used in some experiments. For some experiments, cells were kept in complete RPMI only, without addition of cytokines. For 3H thymidine incorporation for MLPG proliferation, cells were kept in complete RPMI without extra cytokines in 230 µl total volume including 1 µCi of 3H thymidine (PerkinElmer) in a U-bottom 96-well plate.
In vivo transfer of MLPGs and GMPs
EL-4 TB female C57Bl/6 CD45.1+ recipient mice were irradiated with a single dose of 450 cGy and set aside for a few hours to recover. During the recovery time, EL-4 TB female C57Bl/6 CD45.2+ donor mice were used to isolate the different progenitors from the BM. After sorting, collected CD45.2+ progenitor cells were resuspended in 100 µl of PBS and injected i.v. into the irradiated EL-4 TB CD45.1+ recipient mice. In most experiments with MLPG and GP differentiation, we used ∼2 × 106 cells. For GMP differentiation after 24 h, we used 5 × 105 cells.
Preparation of TESs
TESs were prepared from excised nonulcerated EL-4 tumors ∼1.5 cm in diameter. A small tumor piece (5–10 mm2) was harvested, minced into pieces <3 mm in diameter, and resuspended in complete RPMI without extra cytokines. After 16–18 h of incubation at 37°C with 5% CO2, the cell-free supernatant was collected using 0.22-µm filters (EMD Millipore) and kept at −80°C.
Microarrays of mouse cells
1–2 million cells were lysed in 1 ml of Tri-Reagent (Sigma-Aldrich), and RNA was extracted using phase separation and isopropanol precipitation as recommended by the vendor. RNA quality was validated using the Agilent Bioanalyzer and the RNA Nano chip (Agilent), and quantity was determined using the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). A constant amount (100 ng) of total RNA was amplified using the Illumina approved RNA amplification kit (Epicentre). Samples were processed in batches of six on Illumina Mouse WG6v2 microarrays for mRNA expression. Data were exported for analysis using Illumina GenomeStudio software. GenomeStudio was used to export expression levels and detection P values for each probe of each sample. Signal intensity data were quantile normalized, log2-transformed, and mean centered, and genes that showed insignificant detection P values (P > 0.05) in all samples were removed from further analysis. Expression level comparisons between two groups were done using two-sample significance analysis of microarrays (SAM; Zhang, 2007), and correction for multiple testing to estimate FDR was done as described (Storey and Tibshirani, 2003), with FDR <5% used as a significance threshold. Expression data for granulocytes, monocytes, and stem cells were downloaded from the Immgen Consortium, and overlap was done based on Entrez ID mean centered for further analysis. Two-dimensional clustering was done using t-distributed stochastic neighbor embedding (tSNE; Kim and Cohen, 2016). Matlab package with initial solution parameter set at 20 and perplexity parameter set at 30 was used.
RNA-seq of human cells
Cell pellets containing 500,000 to 1 million cells were lysed in 300 µl of Tri-Reagent, and RNA was extracted using the Direct-Zol RNA Mini-Prep Kit (Zymo Research), including an on-column DNaseI treatment. RNA quality was validated using the Agilent Tapestation and the High Sensitivity RNA Screentape (Agilent), and quantity was determined using the Qubit 2.0 Fluorometer (Thermo Fisher Scientific). 3′ mRNA-seq libraries were generated from DNaseI-treated total RNA using the QuantSeq FWD Library Preparation kit (Lexogen), according to the manufacturer’s directions. Overall library size was determined using the Agilent Tapestation and the DNA 5000 Screentape, and libraries were quantitated using real-time PCR (Kapa Biosystems). Libraries were pooled and denatured, and high-output, single-read, 75–base pair next-generation sequencing was done on a NextSeq 500 (Illumina). Data were aligned using bowtie2 (Wan et al., 2004) and RSEM v1.2.12 software (Li and Dewey, 2011) against mm10 genome and gene-level read counts, and reads per kilobase of transcript per million mapped read values on gene level were estimated for ensemble transcriptome. DESeq2 (Love et al., 2014) was used to estimate significance between any two experimental groups. Overall changes were considered significant if they passed FDR <5% thresholds. Gene set enrichment analysis was done using Ingenuity Pathway Analysis software (Qiagen) based on “Functions” and “Upstream Regulators.” Only categories with ≥20 genes enriched at P < 10−10 with significantly predicted activation scores (|Z-score| > 2) were reported. Data were submitted to the Gene Expression Omnibus under accession nos. GSE131516 for mouse microarray and GSE131552 for human RNA-seq.
May-Grünwald-Giemsa and Wright-Giemsa staining
105 to 2 × 105 cells were resuspended in 100 µl of PBS and spun down at 800 rpm for 5 min using a Shandon Cytospin 2 machine and slides. May-Grünwald (Sigma-Aldrich) and Giemsa stain (Sigma-Aldrich) were used according to the manufacturer’s recommendations. For Wright-Giemsa pictures, the Harleco Wright-Giemsa stain pack (EMD Millipore) was used according to the manufacturer’s recommendations. The cells were imaged with a Nikon Eclipse E600 microscope.
Flow cytometry
All incubations were performed for 15 min at 4°C in the dark, and centrifugations were done at 1,500 rpm at 4°C for 5 min, unless recommended otherwise by the manufacturer. Usually, ≤106 cells were incubated with Fc-block (BD Biosciences; clone 2.4G2; 553142) in 50 µl of CSB, then washed in CSB and spun down before cell surface staining with additional antibodies. After the last incubation, cells were washed in CSB, spun down, and resuspended in 400 µl of CSB before acquisition. Cells were run on an LSRII flow cytometer (BD Biosciences), and data were analyzed by FlowJo (Tristar). For sorting, both FACS Aria II (BD Biosciences) and MoFlo Astrios EQ (Beckman Coulter) were used. The antibodies, dyes, and kits used are described in Table S1.
Human M-MDSC/monocyte isolation, culture, and staining
1.07 × 105 OP9 cells were plated in a 12-well plate the day before sorting. The low-density PBMC fraction was isolated by centrifugation over a Ficoll-Paque Plus density gradient (GE Healthcare Life Sciences). The PBMC layer was collected, and cells were washed and stained with CD14-APC-Cy7, HLA-DR-APC, CD15-PE, and Aqua Live/Dead Fixable 405. After sorting, MEMα without nucleosides was removed from the OP9 cells wells and replaced with IMDM (Thermo Fisher Scientific) supplemented with 20% FBS and 1% penicillin–streptomycin. Up to 3.5 × 105 sorted cells were added per well in a total volume of 3 ml of IMDM in the presence of 100 ng/ml of G-CSF and 25 ng/ml of GM-CSF (Peprotech) or 100 ng/ml of M-CSF. At day 3, 2 ml of fresh media supplemented with fresh cytokines were added on the top. At day 4, cells were harvested by pipetting up and down. Human CD45+ cells were separated from murine OP9 cells by magnetic separation using biotinylated anti-human CD45 antibody (Miltenyi Biotec), streptavidin-coated microbeads (Miltenyi Biotec), and magnetic-activated cell sorting columns (Miltenyi Biotec) according to the manufacturer’s instructions. Cells were then stained with the May-Grünwald-Giemsa kit or the naphthol AS-D chloroacetate esterase (Sigma-Aldrich) kit or stained for flow cytometry.
Suppression assay
TAMs, M-MDSCs, and PMN-MDSCs were isolated from tumor tissues by either sorting (TAMs and M-MDSCs) or magnetic separation (PMN-MDSCs). For PMN-MDSC isolation, cells were labeled with biotinylated anti-Ly6G antibody (Miltenyi Biotec), incubated with streptavidin-coated microbeads (Miltenyi Biotec), and separated on magnetic-activated cell sorting columns. Ly6G+ cells obtained after MLPG cultures were isolated in the same way. After isolation, cells were plated in U-bottom 96-well plates in triplicate in complete RMPI without extra cytokines. They were cocultured at different ratios with total splenocytes from Pmel or OT-1 transgenic mice in the presence of cognate peptides: OT-1, SIINFEKL; Pmel, EGSRNQDWL. Cells were incubated for 48 h, and then 3H thymidine (PerkinElmer) was added (1 µl/well) and incubated overnight. Samples were counted with a TopCount NXT instrument (PerkinElmer).
Quantitative real-time PCR (qRT-PCR)
RNA was extracted using Total RNA Kit I (Omega) according to the manufacturer’s instructions, including an on-column DNase digestion. cDNA was generated with high-capacity cDNA reverse transcription kit (Applied Biosystems). qRT-PCR was performed using Power SYBR green PCR master mix (Applied Biosystems) in 96-well plates in triplicate. Plates were read on a QuantStudio 6 Flex real-time PCR system (Applied Biosystems). The murine primers used were Actb: forward, 5′-CCTTCTTGGGTATGGAATCCTGT-3′, and reverse, 5′-GGCATAGAGGTCTTTACGGATGT-3′; Irf8: forward, 5′-AGAACAAAAATGCAAGCTGGGC-3′, and reverse, 5′-GCTCCTCTTGGTCATACCCAT-3′.
Statistical analysis
Statistical analyses were performed using two-tailed Student’s t test or Mann–Whitney U test and Prism 5 software (GraphPad). Paired t test was used since data were normally distributed. All the data are presented as mean ± SD unless noted otherwise, and P values <0.05 were considered significant.
Online supplemental material
Fig. S1 describes differentiation of MLPGs and GPs in vitro and in vivo. Fig. S2 depicts the gene signatures of MLPGs and GPs. Fig. S3 shows characteristics of MLPGs in naive and tumor-free mice. Fig. S4 describes the contribution of spleens in PMN-MDSC accumulation in TB mice. Fig. S5 depicts data on MLPGs and GPs in mice with targeted depletion of monocytic cells. Table S1 presents a list of reagents used in this study.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants CA084488 and CA140043 and the Wistar Institute Animal and Bioinformatics core facilities under Cancer Center Support Grant P30 CA010815.
The authors declare no competing financial interests.
Author contributions: D.I. Gabrilovich conceived the idea; J. Mastio performed most of the experiments; T. Condamine, G. Dominguez, L. Donthireddy, F. Veglia, C. Lin, F. Wang, S. Fu, S. Lavilla-Alonso, A.T. Polo, and E.N. Tcyganov performed experiments; A.V. Kossenkov performed bioinformatics analysis; J. Zhou, P. Viatour, C. Mulligan Jr., B. Nam, J. Bennett, G. Masters, M. Guarino, A. Kumar, Y. Nefedova, R.H. Vonderheide, L.R. Languino, and S.I. Abrams provided transgenic and KO mice, clinical samples, and other resources; J. Mastio and D.I. Gabrilovich wrote the original draft; G. Dominguez, S.I. Abrams, Y. Nefedova, and R.H. Vonderheide provided review and editing; D.I. Gabrilovich supervised the work and was responsible for acquisition of funding.
References
- Akashi K., Traver D., Miyamoto T., and Weissman I.L.. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- Anani W., and Shurin M.R.. 2017. Targeting Myeloid-Derived Suppressor Cells in Cancer. Adv. Exp. Med. Biol. 1036:105–128. 10.1007/978-3-319-67577-0_8 [DOI] [PubMed] [Google Scholar]
- Arihara F., Mizukoshi E., Kitahara M., Takata Y., Arai K., Yamashita T., Nakamoto Y., and Kaneko S.. 2013. Increase in CD14+HLA-DR -/low myeloid-derived suppressor cells in hepatocellular carcinoma patients and its impact on prognosis. Cancer Immunol. Immunother. 62:1421–1430. 10.1007/s00262-013-1447-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne L.J., Beatty G.L., Jhala N., Clark C.E., Rhim A.D., Stanger B.Z., and Vonderheide R.H.. 2012. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 21:822–835. 10.1016/j.ccr.2012.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte V., Brandau S., Chen S.-H., Colombo M.P., Frey A.B., Greten T.F., Mandruzzato S., Murray P.J., Ochoa A., Ostrand-Rosenberg S., et al. 2016. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 7:12150 10.1038/ncomms12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart D.L., Ngai L.K., Roake C.M., Viatour P., Thangavel C., Ho V.M., Knudsen E.S., and Sage J.. 2010. Regulation of RB transcription in vivo by RB family members. Mol. Cell. Biol. 30:1729–1745. 10.1128/MCB.00952-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield L.H., Zhao F., Lee S., Tarhini A.A., Margolin K.A., White R.L., Atkins M.B., Cohen G.I., Whiteside T.L., Kirkwood J.M., and Lawson D.H.. 2017. Immune Correlates of GM-CSF and Melanoma Peptide Vaccination in a Randomized Trial for the Adjuvant Therapy of Resected High-Risk Melanoma (E4697). Clin. Cancer Res. 23:5034–5043. 10.1158/1078-0432.CCR-16-3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Quintero-Estades J.A., Danielian P.S., Nedelcu S., Berman S.D., and Lees J.A.. 2010. Rb regulates fate choice and lineage commitment in vivo. Nature. 466:1110–1114. 10.1038/nature09264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casbon A.J., Reynaud D., Park C., Khuc E., Gan D.D., Schepers K., Passegué E., and Werb Z.. 2015. Invasive breast cancer reprograms early myeloid differentiation in the bone marrow to generate immunosuppressive neutrophils. Proc. Natl. Acad. Sci. USA. 112:E566–E575. 10.1073/pnas.1424927112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., et al. BRIM-3 Study Group . 2011. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364:2507–2516. 10.1056/NEJMoa1103782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.F., Kuan F.C., Yen T.C., Lu M.S., Lin P.Y., Chung Y.H., Chen W.C., and Lee K.D.. 2014. IL-6-stimulated CD11b+ CD14+ HLA-DR- myeloid-derived suppressor cells, are associated with progression and poor prognosis in squamous cell carcinoma of the esophagus. Oncotarget. 5:8716–8728. 10.18632/oncotarget.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condamine T., Ramachandran I., Youn J.I., and Gabrilovich D.I.. 2015. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu. Rev. Med. 66:97–110. 10.1146/annurev-med-051013-052304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens L.M., and Pollard J.W.. 2011. Leukocytes in mammary development and cancer. Cold Spring Harb. Perspect. Biol. 3:a003285 10.1101/cshperspect.a003285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Coaña Y.P., Wolodarski M., Poschke I., Yoshimoto Y., Yang Y., Nyström M., Edbäck U., Brage S.E., Lundqvist A., Masucci G.V., et al. 2017. Ipilimumab treatment decreases monocytic MDSCs and increases CD8 effector memory T cells in long-term survivors with advanced melanoma. Oncotarget. 8:21539–21553. 10.18632/oncotarget.15368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Montero C.M., Salem M.L., Nishimura M.I., Garrett-Mayer E., Cole D.J., and Montero A.J.. 2009. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol. Immunother. 58:49–59. 10.1007/s00262-008-0523-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard M., Kwok I.W.H., Chong S.Z., Teng K.W.W., Becht E., Chen J., Sieow J.L., Penny H.L., Ching G.C., Devi S., et al. 2018. Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity. 48:364–379.e8. 10.1016/j.immuni.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Gabrilovich D.I., Ostrand-Rosenberg S., and Bronte V.. 2012. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 12:253–268. 10.1038/nri3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero M.R., Garlanda C., Jaillon S., Marone G., and Mantovani A.. 2013. Tumor associated macrophages and neutrophils in tumor progression. J. Cell. Physiol. 228:1404–1412. 10.1002/jcp.24260 [DOI] [PubMed] [Google Scholar]
- Greenberg N.M., DeMayo F., Finegold M.J., Medina D., Tilley W.D., Aspinall J.O., Cunha G.R., Donjacour A.A., Matusik R.J., and Rosen J.M.. 1995. Prostate cancer in a transgenic mouse. Proc. Natl. Acad. Sci. USA. 92:3439–3443. 10.1073/pnas.92.8.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtschke T., Löhler J., Kanno Y., Fehr T., Giese N., Rosenbauer F., Lou J., Knobeloch K.P., Gabriele L., Waring J.F., et al. 1996. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 87:307–317. 10.1016/S0092-8674(00)81348-3 [DOI] [PubMed] [Google Scholar]
- Kato M., Takahashi M., Akhand A.A., Liu W., Dai Y., Shimizu S., Iwamoto T., Suzuki H., and Nakashima I.. 1998. Transgenic mouse model for skin malignant melanoma. Oncogene. 17:1885–1888. 10.1038/sj.onc.1202077 [DOI] [PubMed] [Google Scholar]
- Kawano M., Mabuchi S., Matsumoto Y., Sasano T., Takahashi R., Kuroda H., Kozasa K., Hashimoto K., Isobe A., Sawada K., et al. 2015. The significance of G-CSF expression and myeloid-derived suppressor cells in the chemoresistance of uterine cervical cancer. Sci. Rep. 5:18217 10.1038/srep18217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A., and Cohen M.S.. 2016. The discovery of vemurafenib for the treatment of BRAF-mutated metastatic melanoma. Expert Opin. Drug Discov. 11:907–916. 10.1080/17460441.2016.1201057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., McKolanis J.R., Dzubinski L.A., Islam K., Potter D.M., Salazar A.M., Schoen R.E., and Finn O.J.. 2013. MUC1 vaccine for individuals with advanced adenoma of the colon: a cancer immunoprevention feasibility study. Cancer Prev. Res. (Phila.). 6:18–26. 10.1158/1940-6207.CAPR-12-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki D., Yamamoto M., Nishiyama A., Uno K., Ban T., Ichino M., Sasaki H., Matsunaga S., Yoshinari M., Ryo A., et al. 2014. IRF8 inhibits C/EBPα activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat. Commun. 5:4978 10.1038/ncomms5978 [DOI] [PubMed] [Google Scholar]
- Lecut C., Frederix K., Johnson D.M., Deroanne C., Thiry M., Faccinetto C., Marée R., Evans R.J., Volders P.G., Bours V., and Oury C.. 2009. P2X1 ion channels promote neutrophil chemotaxis through Rho kinase activation. J. Immunol. 183:2801–2809. 10.4049/jimmunol.0804007 [DOI] [PubMed] [Google Scholar]
- Lee S.E., Lim J.Y., Ryu D.B., Kim T.W., Yoon J.H., Cho B.S., Eom K.S., Kim Y.J., Kim H.J., Lee S., et al. 2016. Circulating immune cell phenotype can predict the outcome of lenalidomide plus low-dose dexamethasone treatment in patients with refractory/relapsed multiple myeloma. Cancer Immunol. Immunother. 65:983–994. 10.1007/s00262-016-1861-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy L., Mishalian I., Bayuch R., Zolotarov L., Michaeli J., and Fridlender Z.G.. 2015. Splenectomy inhibits non-small cell lung cancer growth by modulating anti-tumor adaptive and innate immune response. OncoImmunology. 4:e998469 10.1080/2162402X.2014.998469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., and Dewey C.N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens A., Wistuba-Hamprecht K., Geukes Foppen M., Yuan J., Postow M.A., Wong P., Romano E., Khammari A., Dreno B., Capone M., et al. 2016. Baseline Peripheral Blood Biomarkers Associated with Clinical Outcome of Advanced Melanoma Patients Treated with Ipilimumab. Clin. Cancer Res. 22:2908–2918. 10.1158/1078-0432.CCR-15-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netherby C.S., Messmer M.N., Burkard-Mandel L., Colligan S., Miller A., Cortes Gomez E., Wang J., Nemeth M.J., and Abrams S.I.. 2017. The Granulocyte Progenitor Stage Is a Key Target of IRF8-Mediated Regulation of Myeloid-Derived Suppressor Cell Production. J. Immunol. 198:4129–4139. 10.4049/jimmunol.1601722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschall A.V., Zhang R., Qi C.F., Bardhan K., Peng L., Lu G., Yang J., Merad M., McGaha T., Zhou G., et al. 2015. IFN regulatory factor 8 represses GM-CSF expression in T cells to affect myeloid cell lineage differentiation. J. Immunol. 194:2369–2379. 10.4049/jimmunol.1402412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A., Parrinello N.L., Vetro C., Forte S., Chiarenza A., Figuera A., Motta G., Palumbo G.A., Ippolito M., Consoli U., and Di Raimondo F.. 2015. Circulating myeloid-derived suppressor cells correlate with clinical outcome in Hodgkin Lymphoma patients treated up-front with a risk-adapted strategy. Br. J. Haematol. 168:689–700. 10.1111/bjh.13198 [DOI] [PubMed] [Google Scholar]
- Sade-Feldman M., Kanterman J., Klieger Y., Ish-Shalom E., Olga M., Saragovi A., Shtainberg H., Lotem M., and Baniyash M.. 2016. Clinical Significance of Circulating CD33+CD11b+HLA-DR- Myeloid Cells in Patients with Stage IV Melanoma Treated with Ipilimumab. Clin. Cancer Res. 22:5661–5672. 10.1158/1078-0432.CCR-15-3104 [DOI] [PubMed] [Google Scholar]
- Schug Z.T., Peck B., Jones D.T., Zhang Q., Grosskurth S., Alam I.S., Goodwin L.M., Smethurst E., Mason S., Blyth K., et al. 2015. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 27:57–71. 10.1016/j.ccell.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart T.J., Greeneltch K.M., Reid J.E., Liewehr D.J., Steinberg S.M., Liu K., and Abrams S.I.. 2009. Interferon regulatory factor-8 modulates the development of tumour-induced CD11b+Gr-1+ myeloid cells. J. Cell. Mol. Med. 13(9b, 9B):3939–3950. 10.1111/j.1582-4934.2009.00685.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascón S., Hatzios S.K., Kagan V.E., et al. 2017. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 171:273–285. 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., and Tibshirani R.. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 100:9440–9445. 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K., Kitano S., Shoji H., Nishimura T., Shimada Y., Nagashima K., Aoki K., Hiraoka N., Honma Y., Iwasa S., et al. 2016. Pretreatment Immune Status Correlates with Progression-Free Survival in Chemotherapy-Treated Metastatic Colorectal Cancer Patients. Cancer Immunol. Res. 4:592–599. 10.1158/2326-6066.CIR-15-0298 [DOI] [PubMed] [Google Scholar]
- Viatour P., Somervaille T.C., Venkatasubrahmanyam S., Kogan S., McLaughlin M.E., Weissman I.L., Butte A.J., Passegué E., and Sage J.. 2008. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 3:416–428. 10.1016/j.stem.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waight J.D., Netherby C., Hensen M.L., Miller A., Hu Q., Liu S., Bogner P.N., Farren M.R., Lee K.P., Liu K., and Abrams S.I.. 2013. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J. Clin. Invest. 123:4464–4478. 10.1172/JCI68189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan P.T., Garnett M.J., Roe S.M., Lee S., Niculescu-Duvaz D., Good V.M., Jones C.M., Marshall C.J., Springer C.J., Barford D., and Marais R.. Cancer Genome Project . 2004. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 116:855–867. 10.1016/S0092-8674(04)00215-6 [DOI] [PubMed] [Google Scholar]
- Wang J., and Yang J.. 2016. Identification of CD4+CD25+CD127- regulatory T cells and CD14+HLA-DR-/low myeloid-derived suppressor cells and their roles in the prognosis of breast cancer. Biomed. Rep. 5:208–212. 10.3892/br.2016.694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., An G., Xie S., Yao Y., and Feng G.. 2016. The clinical and prognostic significance of CD14(+)HLA-DR(-/low) myeloid-derived suppressor cells in hepatocellular carcinoma patients receiving radiotherapy. Tumour Biol. 37:10427–10433. 10.1007/s13277-016-4916-2 [DOI] [PubMed] [Google Scholar]
- Wang H., Yan M., Sun J., Jain S., Yoshimi R., Abolfath S.M., Ozato K., Coleman W.G. Jr., Ng A.P., Metcalf D., et al. 2014. A reporter mouse reveals lineage-specific and heterogeneous expression of IRF8 during lymphoid and myeloid cell differentiation. J. Immunol. 193:1766–1777. 10.4049/jimmunol.1301939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J., Gibney G., Kudchadkar R., Yu B., Cheng P., Martinez A.J., Kroeger J., Richards A., McCormick L., Moberg V., et al. 2016. Phase I/II Study of Metastatic Melanoma Patients Treated with Nivolumab Who Had Progressed after Ipilimumab. Cancer Immunol. Res. 4:345–353. 10.1158/2326-6066.CIR-15-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez A., Ng M.Y., Hassanzadeh-Kiabi N., and Goodridge H.S.. 2015. IRF8 acts in lineage-committed rather than oligopotent progenitors to control neutrophil vs monocyte production. Blood. 125:1452–1459. 10.1182/blood-2014-09-600833 [DOI] [PubMed] [Google Scholar]
- Yáñez A., Coetzee S.G., Olsson A., Muench D.E., Berman B.P., Hazelett D.J., Salomonis N., Grimes H.L., and Goodridge H.S.. 2017. Granulocyte-Monocyte Progenitors and Monocyte-Dendritic Cell Progenitors Independently Produce Functionally Distinct Monocytes. Immunity. 47:890–902.e4. 10.1016/j.immuni.2017.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J.I., Collazo M., Shalova I.N., Biswas S.K., and Gabrilovich D.I.. 2012. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J. Leukoc. Biol. 91:167–181. 10.1189/jlb.0311177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J.I., Kumar V., Collazo M., Nefedova Y., Condamine T., Cheng P., Villagra A., Antonia S., McCaffrey J.C., Fishman M., et al. 2013. Epigenetic silencing of retinoblastoma gene regulates pathologic differentiation of myeloid cells in cancer. Nat. Immunol. 14:211–220. 10.1038/ni.2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. 2007. A comprehensive evaluation of SAM, the SAM R-package and a simple modification to improve its performance. BMC Bioinformatics. 8:230 10.1186/1471-2105-8-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.