We describe two unrelated patients with inherited IFNAR1 deficiency who suffered from life-threatening infections following measles or yellow fever virus vaccination and were otherwise healthy.
Abstract
Vaccination against measles, mumps, and rubella (MMR) and yellow fever (YF) with live attenuated viruses can rarely cause life-threatening disease. Severe illness by MMR vaccines can be caused by inborn errors of type I and/or III interferon (IFN) immunity (mutations in IFNAR2, STAT1, or STAT2). Adverse reactions to the YF vaccine have remained unexplained. We report two otherwise healthy patients, a 9-yr-old boy in Iran with severe measles vaccine disease at 1 yr and a 14-yr-old girl in Brazil with viscerotropic disease caused by the YF vaccine at 12 yr. The Iranian patient is homozygous and the Brazilian patient compound heterozygous for loss-of-function IFNAR1 variations. Patient-derived fibroblasts are susceptible to viruses, including the YF and measles virus vaccine strains, in the absence or presence of exogenous type I IFN. The patients’ fibroblast phenotypes are rescued with WT IFNAR1. Autosomal recessive, complete IFNAR1 deficiency can result in life-threatening complications of vaccination with live attenuated measles and YF viruses in previously healthy individuals.
Introduction
There are currently 11 live attenuated viral (LAV) vaccines in use worldwide. All have been shown or suspected to cause life-threatening disease in rare individuals. So far, severe disease by only three LAV vaccines has been explained by inborn errors of immunity, in at least some patients. Live (oral) poliovirus vaccine can cause vaccine-associated paralytic poliomyelitis in patients with agammaglobulinemia due to a variety of inborn errors of T or B cells (Shaghaghi et al., 2014). Oral rotavirus vaccine can cause life-threatening dehydration in patients with severe combined immunodeficiencies (Pöyhönen et al., 2019). The measles vaccine strain in the measles, mumps, and rubella (MMR) vaccine can cause disseminated infections in patients with inborn errors of IFNAR2 (Duncan et al., 2015), STAT1 (Burns et al., 2016), or STAT2 (Hambleton et al., 2013; Moens et al., 2017), which control cellular responses to various IFNs (type I only in the case of IFNAR2, type I and type III IFNs for STAT2, and all three types for STAT1). Vaccine–strain mumps infections are extremely rare, with only one life-threatening case reported in a patient with RAG1 deficiency and a lack of T and B cell adaptive immunity (Morfopoulou et al., 2017). Patients with inborn errors of IRF7 (Ciancanelli et al., 2015), which controls the amplification of type I and III IFNs, or IRF9 (Hernandez et al., 2018), which controls the cellular responses to these IFNs, have been reported to suffer from an ill-defined adverse reaction to MMR vaccination. Most cases of severe MMR vaccine disease remain unexplained. None of the life-threatening adverse reactions to live vaccines for rubella virus, yellow fever (YF) virus (YFV), influenza virus, varicella zoster virus, vaccinia virus, or hepatitis A virus have been explained genetically. We studied two otherwise healthy patients who developed disseminated, life-threatening disease following LAV vaccination: a 9-yr-old Iranian boy following MMR vaccination at 1 yr, whose younger sister died at 1 yr following MMR vaccination, and a 14-yr-old Brazilian girl following YF vaccination at 12 yr.
Results
Two unrelated patients with adverse reactions to measles and YF vaccines
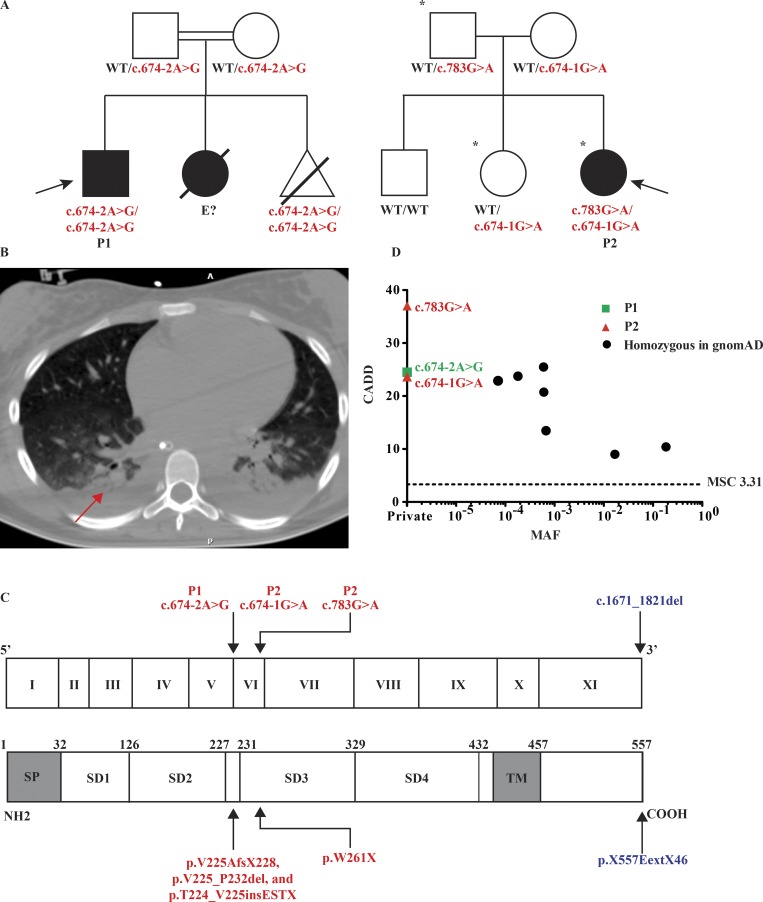
We studied two unrelated patients whose pedigrees are shown in Fig. 1 A. Full case reports are available in the Materials and methods. Briefly, patient 1 (P1) was born in Iran to consanguineous parents and presented at 1 yr of age with disseminated vaccine–strain measles (Fig. 1 A). Approximately 10 days after inoculation with the MMR vaccine, he presented with a generalized exanthem, fever, and neurological symptoms consistent with encephalitis. Computed tomography (CT) of the brain showed mild cerebral edema, and analysis of cerebrospinal fluid (CSF) demonstrated leukocytosis (350 lymphocytes/µl, 2 neutrophils/µl). PCR for measles was positive in both blood and CSF. He had no notable medical history. P1 is now 9 yr old, in good health, and has not manifested any other invasive infections. P1 had a younger sibling who died 4 wk after routine MMR vaccination at age 1 yr but for whom material was not available for genetic analysis. His mother electively terminated a third pregnancy following prenatal genetic identification of the same homozygous variant in IFNAR1. Patient 2 (P2) is a 14-yr-old Brazilian girl born to nonconsanguineous parents (Fig. 1 A). Otherwise healthy until age 12, she presented with fever, hypotension, vomiting, and lethargy 7 d following YF vaccination. Her condition deteriorated, and she developed renal and hepatic dysfunction and required intubation and mechanical ventilation. A CT scan of the chest demonstrated pleural effusions and atelectasis (Fig. 1 B). YFV RNA was detected in the patient’s blood by PCR. Sequencing confirmed that this was the vaccine–strain virus (Fig. S1; Nascimento Silva et al., 2011). A diagnosis of YF vaccine–associated viscerotropic disease (YEL-AVD) was made. She does not belong to any known risk groups for YEL-AVD (Seligman et al., 2014). 1 yr after recovery, she remains healthy. Notably, P2 had received two MMR vaccinations at 12 and 16 mo of age and other live vaccines (oral poliovirus and bacillus Calmette-Guérin [BCG]) without incident. She developed neutralizing antibodies to YFV in levels associated with seroprotection and seropositivity to measles, rubella, diphtheria, and tetanus.
Figure 1.
Three private IFNAR1 variants are present in two kindreds associated with life-threatening LAV vaccine disease. (A) Pedigrees of the IFNAR1-deficient families. Double lines connecting two parents indicate consanguinity. The probands are indicated by an arrow. Filled shapes indicate affected individuals while open shapes indicate unaffected individuals. Asterisks demarcate those individuals who received vaccination against YFV. The pregnancy of a third sibling in pedigree 1 (homozygous for the same mutation as P1) was terminated at 16 wk of gestational age. (B) CT scan of P2’s chest demonstrating pleural effusions and atelectasis following YF vaccination (red arrow). (C) Schematic illustration of the IFNAR1 gene with 11 coding exons and of the IFNAR1 protein with its four fibronectin type III subdomains (SD1–4). SP, signal peptide; TM, transmembrane domain. The exons are numbered in roman numerals (I–XI). The previously reported IFNAR1 variant is indicated in blue, while those of P1 and P2 are indicated in red. (D) Population genetics of homozygous coding missense and predicted loss-of-function IFNAR1 mutations taken from GnomAD and in-house cohorts. The patients’ variants are private and shown in red and green, while seven variants detected in GnomAD are shown in black. MAF, minor allele frequency; MSC, mutation significance cutoff.
Biallelic private nonsense or splicing IFNAR1 variations in the patients
We performed whole-exome sequencing (WES) analysis on both patients. The percentage of homozygosity was 2.9% and 0.25% in P1 and P2, respectively, consistent with the former being born to consanguineous parents. Principal component analysis confirmed the ethnicities of the two families (Belkadi et al., 2016). We then analyzed copy number variations and single nucleotide variations. We filtered out noncoding and synonymous single nucleotide variations, other than those at exonic and intronic essential splice sites. We additionally filtered out common variants (minor allele frequency > 0.01), variants predicted to be benign (combined annotation-dependent depletion [CADD] score below the mutation significance cutoff), and variants occurring in very frequently damaged genes (those with a gene damage index >13.84; Kircher et al., 2014; Itan et al., 2015, 2016). Finally, we tested a model of autosomal recessive (AR) inheritance and focused our attention on genes with homozygous or compound heterozygous lesions. In P1, we found a private homozygous mutation in IFNAR1, c.674-2A>G, among 48 variants in 39 genes (Table S1). In P2, 11 variants in 6 genes passed filtering criteria (Table S1), including two variants in IFNAR1, c.783G>A;p.W261X and c.674-1G>A (Fig. 1 C). None of these three IFNAR1 variants are found in online databases including 1000 Genomes, Bravo, and GnomAD (Genome Aggregation Database), which encompasses the Exome Aggregation Consortium database (Fig. 1 D). The IFNAR1 gene, together with IFNAR2, encodes the heterodimeric receptor for the type I IFNs (Uzé et al., 1990; Piehler et al., 2012). The three IFNAR1 mutations are predicted to be deleterious by either disrupting the same essential splice site (c.674-2A>G and c.674-1G>A, in P1 and P2, respectively) or creating a premature stop codon (p.W261X in P2). The three mutations were confirmed by Sanger sequencing of the patients and their relatives, which also verified that the mutations present in P2 were compound heterozygous (Figs. 1 A and S2A). We did not have genomic DNA from P1’s sister, who died in her first year of life following MMR vaccination. A total of only seven nonsynonymous or splicing IFNAR1 variants were found in homozygosity in GnomAD, all of which were missense (Fig. 1 D). Collectively, these findings suggested that the two patients had a novel and rare inborn error of immunity, AR IFNAR1 deficiency.
The three mutant IFNAR1 alleles encode truncated proteins
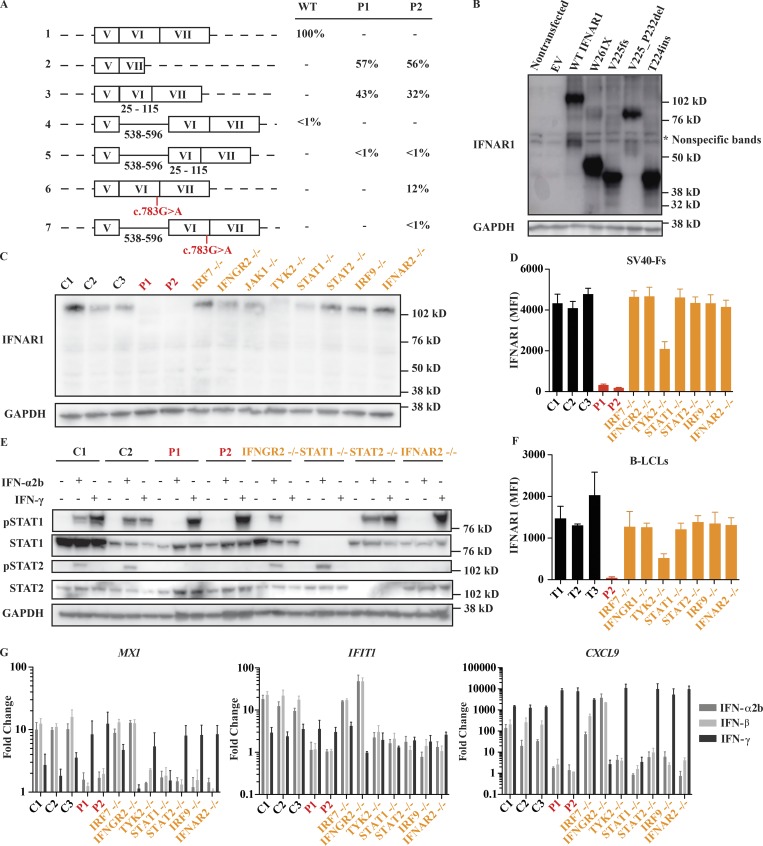
Because the c.674-1G>A and c.674-2A>G mutations occur at an essential splice acceptor site of exon 6, we hypothesized that they would lead to an aberrantly spliced transcript (Fig. 1 C). We therefore amplified the IFNAR1 cDNA from SV40 large T antigen-transformed fibroblasts (SV40-F cells) derived from both patients and a healthy control (Figs. 2 A and S2, B and C) and performed Sanger sequencing on individual cDNA clones. All IFNAR1 transcripts present in healthy control cells contained normal splicing at exon 5–6 and 6–7 junctions, whereas three alternative splice products were observed in P1 and P2. In both patients, ∼60% of transcripts lacked exon 6, which induces a frameshift and a premature stop at aa 228 (p.V225AfsX228); 30–40% of transcripts lacked the first 24 bp of exon 6 in P1 and P2, which results in the deletion of aa 225–232 (p.V225_P232del); a trace amount of transcripts contained a 56-bp segment retained from intron 5, which introduces a premature stop after three amino acids (p.T224_V225insESTX). For simplicity, we will refer to these mutations as V225fs, V225_P232del, and T224ins. A small percentage of transcripts from P2’s cells, ∼10%, were found to contain normal splicing at exon 5–6 and 6–7 junctions, yet the nonsense mutation c.783G>A, p.W261X (hereafter W261X) was present in all such transcripts. Western blot analysis of HEK293T cells overexpressing the four patient-derived transcripts and WT IFNAR1 transcripts showed that all four proteins are expressed but with a molecular weight lower than the WT protein (Fig. 2 B). These findings suggested that any proteins encoded by the three mutant IFNAR1 alleles are not functional.
Figure 2.
Impact of IFNAR1 variants on type I IFN signaling. (A) cDNA sequencing results demonstrate aberrant splicing of IFNAR1 mRNA from patients’ SV40-F cells. The percentage of positively identified transcripts is indicated. At least 150 transcripts were sequenced for each individual. The black numbers under the schematic representations indicate the positions of the included elements relative to their start sites. Results are representative of two experiments. (B) Western blot of IFNAR1 in HEK293T cells transiently transfected with IFNAR1 constructs; GAPDH was used as a loading control. A representative blot from three experiments is shown. (C) Western blot of endogenous IFNAR1 in SV40-F cells from three healthy controls (C1, C2, and C3), P1, P2, IRF7−/−, IFNGR2−/−, TYK2−/−, STAT1−/−, STAT2−/−, IRF9−/−, and IFNAR2−/− patients. A representative blot from three experiments is shown. (D and F) Mean fluorescence intensity (MFI) of IFNAR1 staining on SV40-F (D) and B-LCL (F) cells from three healthy controls (C1, C2, and C3 or T1, T2, and T3), P1, P2, IRF7−/−, IFNGR2−/−, IFNGR1−/−, TYK2−/−, STAT1−/−, STAT2−/−, and IRF9−/− patients as assessed by flow cytometry. Mean (n = 3) and SEM are shown. (E) Western blot analysis of phosphorylated STAT1 (pSTAT1) and pSTAT2 levels in SV40-F cells stimulated with 1,000 U/ml of IFN-α2b or IFN-γ for 20 min. Cells were from two healthy controls (C1, C2), P1, P2, IFNGR2−/−, STAT1−/−, STAT2−/−, and IFNAR2−/− patients. A representative blot from two experiments is shown. (G) Transcription levels of MX1, IFIT1, and CXCL9 assessed by qRT-PCR on SV40-F cells treated with 1,000 U/ml of either IFN-α2b, -β, or -γ for 2 h. Cells were from three healthy controls (C1, C2, and C3), P1, P2, IRF7−/−, IFNGR2−/−, TYK2−/−, STAT1−/−, STAT2−/−, IRF9−/−, and IFNAR2−/− patients. Mean (n = 3) and SEM are shown.
The patients’ cells do not express IFNAR1 and do not respond to type I IFNs
Quantitative RT-PCR (qRT-PCR) on patient cells showed a strong reduction of IFNAR1 mRNA compared with healthy controls, suggesting nonsense-mediated decay (Fig. S2 D). We tested if any IFNAR1 protein was expressed in patients’ cells by Western blotting and flow cytometry (Fig. 2, C, D, and F; and Fig. S3 A). We observed a complete lack of IFNAR1 expression in patients’ SV40-F and B-lymphoblastoid cell lines (B-LCLs) compared with healthy controls and patients with other defects in the type I IFN signaling pathway, such as IRF7−/−, STAT1−/−, STAT2−/−, and IRF9−/− patients (Fig. 2, C, D, and F; and Fig. S3 A). As previously reported, we detected severely decreased expression of IFNAR1 in TYK2-deficient cells (Ragimbeau et al., 2003). IFNAR2 surface expression was normal in both patients’ cells (Fig. S3, B and D). In summary, IFNAR1 expression was undetectable in both fibroblastic and lymphoid cell lines derived from both patients. We cannot exclude the remote but finite possibility that residual levels of IFNAR1 are expressed on the surface of other cells. We next tested type I IFN signaling in patients’ fibroblasts. Phosphorylation of STAT1 and STAT2 was abolished in patients’ SV40-F cells in response to IFN-α2b (Fig. 2 E). Moreover, patients’ SV40-F cells did not substantially up-regulate expression of IFN-stimulated genes (ISGs) MX1 and IFIT1 in response to IFN-α2b and IFN-β, whereas their CXCL9 induction in response to IFN-γ was intact and elevated to a level above that of healthy controls, in contrast to previous reports that had suggested the absence of type I IFN signaling could impair type II IFN signaling (Fig. 2 G; Takaoka et al., 2000). This result indicates that the patients have major defects in type I IFN signaling with impairment of MX1 and IFIT1 induction similar to other deficiencies in this pathway, including STAT1, STAT2, IRF9, and IFNAR2. Altogether, these data showed that the patients’ fibroblasts had IFNAR1 deficiency, with a complete lack of responses to IFN-α2b and -β. We did not exclude the possibility that these or other cells can respond to some of the 15 other type I IFNs, at least for some ISGs.
The patients’ fibroblast-intrinsic type I IFN immunity to viruses is impaired
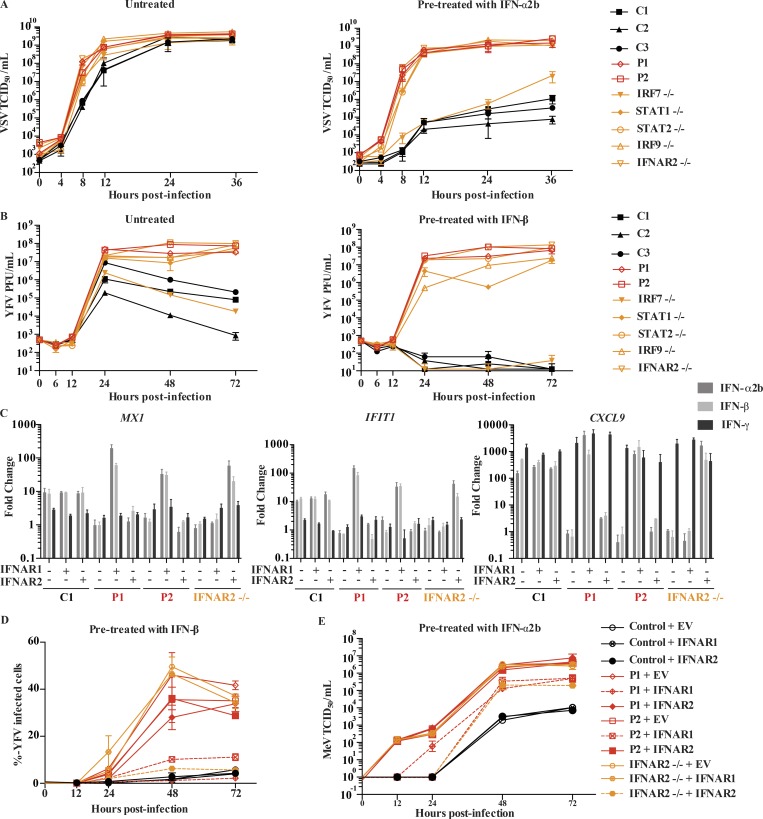
To test if patient cells were able to control viral infections in the presence of type I IFNs, we pretreated SV40-F cells from both patients with IFN-α2b and infected them with vesicular stomatitis virus (VSV), a virus commonly used in testing type I IFN responses (Fig. 3 A). When pretreated with IFN-α2b, healthy control cells were able to control VSV replication 24 h after infection. In contrast, viral replication could not be suppressed in patient cells treated with IFN-α2b, a finding also seen in IFNAR2-, STAT1-, STAT2-, and IRF9-deficient patient cells. We observed similar results with YFV-17D, a vaccine strain highly similar to that responsible for P2’s infection (Fig. 3 B). Interestingly, IRF7-deficient cells were able to control YFV-17D replication to the same degree as healthy controls, in the presence or absence of exogenous IFN-β (Fig. 3 B). To reassess if the patients’ cellular phenotypes were caused by the lack of normal IFNAR1, we transduced patients’ primary fibroblasts with WT IFNAR1, IFNAR2, or an empty vector (EV). Only transduction with IFNAR1 was able to rescue the defect in IFNAR1 expression levels (Fig. S3, C and E) and patients’ responses to type I IFN as assessed by induction of MX1 and IFIT1 following IFN-α2b or -β stimulation (Fig. 3 C). Importantly, transduction with WT IFNAR1 rescued viral titers, thus rescuing, at least partially, the phenotype of increased VSV, measles virus, and YFV susceptibility in P1 and P2’s fibroblasts (Fig. 3, D and E; and Fig. S3, F and H). Then, to assess the extent to which the cellular phenotype replicated the patients’ infectious phenotype, we also infected patients’ cells with influenza A virus (IAV, A/CA/07/2009/H1N1 strain) and herpes simplex virus-1 (HSV-1), two viruses the patients had been exposed to without developing severe disease (Fig. S4, A–F). Again, the patients’ SV40-F cells supported higher levels of viral replication than healthy controls’ for both HSV-1 (Fig. S4, A–D) and IAV (Fig. S4, E and F). We also assessed the ability of patients’ SV40-F cells to control viral replication of YFV-Asibi and Zika viruses (Fig. S4, G–J). Consistent with other viruses, both patients displayed an inability to control viral replication, even following pretreatment with exogenous IFN-β, compared with cells from a healthy control (Fig. S4, G–J). Collectively, these data indicate that IFNAR1 deficiency results in a broad defect in fibroblast-intrinsic immunity.
Figure 3.
IFNAR1 is required for type I IFN–mediated cell intrinsic immunity to viral infections. (A) VSV titers in SV40-F cells unstimulated (left) or pretreated (right) with 1,000 U/ml IFN-α2b for 16 h, followed by VSV infection at an MOI of 3. Cells from three healthy controls were included (C1, C2, and C3), as well as those from P1, P2, IRF7−/−, STAT1−/−, STAT2−/−, IRF9−/−, and IFNAR2−/− patients. Mean (n = 3) and SEM are shown. (B) YFV titers in SV40-F cells unstimulated (left) or pretreated (right) with 1,000 U/ml IFN-β for 16 h, followed by infection with YFV-17D at MOI = 0.05. Mean and range (n = 2) are shown. Cells from three healthy controls were included (C1, C2, and C3), as well as those from P1, P2, IRF7−/−, STAT1−/−, STAT2−/−, IRF9−/−, and IFNAR2−/− patients. (C) Transcription levels of MX1, IFIT1, and CXCL9 assessed by qRT-PCR on SV40-F cells treated with 1,000 U/ml of IFN-α2b, -β, or -γ for 2 h. Cells were from a healthy control, P1, P2, and an IFNAR2−/− patient that were transduced with WT IFNAR1, WT IFNAR2, or EV. Mean (n = 3) and SEM are shown. (D) SV40-F cells were infected with YFV-17D at an MOI of 0.05, and the percentage of cells positive for YFV viral proteins was assessed by flow cytometry. Cells were from a healthy control, P1, P2, and an IFNAR2−/− patient and were transduced with WT IFNAR1, WT IFNAR2, or EV and stimulated with 1,000 U/ml IFN-β for 16 h before infection. Mean and range (n = 2) are shown. (E) SV40-F cells were infected with MeV at an MOI of 0.1, and viral titer was assessed at the given time points. Cells were from a healthy control, P1, P2, and an IFNAR2−/− patient and were transduced with WT IFNAR1, WT IFNAR2, or EV and stimulated with 1,000 U/ml IFN-α2b for 16 h before infection. Mean and SEM (n = 3) are shown. TCID, tissue culture infectious dose.
Finally, in the course of viral infection, we quantified the production of IFN-β, known as the fibroblastic IFN because it is the predominant type I IFN produced by these cells. We observed that elevated amounts of IFN-β were produced in IFNAR1-deficient cells upon infection by YFV-17D, compared with healthy control cells, indicating that the observed antiviral defects arose from abolished type I IFN signaling and not from impaired IFN-β production (Fig. S3 G). These findings also confirmed that IFN-β production, unlike that of other type I IFNs, does not require IFNAR1- and IRF7-dependent amplification (Honda et al., 2006). Altogether, these data showed that cell-intrinsic type I IFN immunity to VSV, YF vaccine–strain virus, IAV, HSV-1, and measles vaccine–strain virus was abolished in the patients’ cells. These findings strongly suggest that AR IFNAR1 deficiency underlies measles vaccine disease in P1 (and by inference in his deceased sister) and YEL-AVD in P2 by disruption of cell-intrinsic type I IFN immunity to viruses.
Discussion
We herein describe AR IFNAR1 deficiency in two unrelated families. We demonstrate that this monogenic error of immunity can underlie life-threatening, isolated measles vaccine disease and YEL-AVD in previously healthy children and adolescents. Patients with complete forms of AR IFNAR2, STAT1, and STAT2 deficiencies have also presented with life-threatening complications following MMR vaccination (Table 1; Hambleton et al., 2013; Duncan et al., 2015; Shahni et al., 2015; Burns et al., 2016; Moens et al., 2017), whereas IRF7- or IRF9-deficient patients are apparently less susceptible to measles vaccine infection (Ciancanelli et al., 2015; Hernandez et al., 2018). Interestingly, patients deficient in JAK1 or TYK2, whose cells show impaired but not abolished response to type I IFNs, have not been reported to experience disease following MMR vaccination (Minegishi et al., 2006; Boisson-Dupuis et al., 2012; Kreins et al., 2015; Eletto et al., 2016). Collectively, these data differ from experimental work done in mice, where STAT1 appears dispensable for defense against both WT and attenuated measles viruses (Hahm et al., 2005; O’Donnell et al., 2012). They suggest that a set of genes jointly regulated by human IFNAR1, IFNAR2, STAT1, and STAT2, and perhaps by IRF7 and IRF9, are critical for control of vaccine–strain measles virus (Hahm et al., 2005; O’Donnell et al., 2012). Furthermore, the fact that IFNAR1 and IFNAR2 deficiencies both result in measles virus vaccine–related disease, while deficiency in IL-10RB, a subunit of the type III IFN receptor, has not been reported to produce this infectious phenotype, suggests that type I IFN signaling is more critical for defense against measles vaccine–strain virus than type III IFN signaling (Glocker et al., 2009; Neven et al., 2013; Karaca et al., 2016; Gao et al., 2018).
Table 1. Patients suffering from MMR vaccine-related disease.
| Gene mutated | Age (mo) at first MMR vaccination | Age (yr) at last known follow-up | Clinical manifestations |
|---|---|---|---|
| STAT2 P1 (Hambleton et al., 2013) | 18 | 5 | P1: 6 d following MMR vaccination, the patient developed fever, rash, conjunctivitis, and lymphadenopathy, followed by hepatitis and pneumonitis requiring supplemental oxygen. Patient recovered from MMR infection. |
| STAT2 P2, P3 (Shahni et al., 2015) | 12, 13 | 3, 2.5 | P2: 1 wk after MMR vaccination, the patient developed a febrile illness. Readmitted 1 mo later with opsoclonus-myoclonus and lymphocytes in the CSF, readmitted again at 2.5 yr with impaired renal and liver function, opsoclonus-myoclonus, and seizures. Patient recovered from MMR infection. |
| P3: 1 wk after MMR vaccination, the patient developed a febrile illness, anemia, and lymphopenia. CSF was positive for mumps virus by PCR. Readmitted at 15 mo due to persistent fever and malaise, eventually developed septic shock and metabolic acidosis. Patient recovered from MMR infection. | |||
| STAT2 P4, P5 (Moens et al., 2017) | 24, 18 | 7a, 11 | P4: 1 wk after MMR vaccination, the patient developed a morbilliform rash, tonsillitis, conjunctivitis, lymphadenopathy, hepatosplenomegaly, and arthritis. Patient recovered from MMR infection. Patient experienced frequent severe viral infections during the first 3 yr and died at 7 yr of fulminant viral infection. |
| P5: 1 wk after MMR vaccination, the patient developed disseminated measles, complicated by hepatitis and pneumonitis. Patient recovered from MMR infection. | |||
| IFNAR2 (Duncan et al., 2015) | 13 | 1.3a | 6 d after MMR vaccination, the patient developed a marked injection-site reaction and generalized morbilliform skin rash, recurrent maculopapular rash, fever, and intractable seizures. Patient died on day 81 after vaccination. |
| IRF7 (Ciancanelli et al., 2015) | 12 | 9 | 1 wk after MMR vaccination, the patient developed a rash suggestive of measles. Patient recovered from MMR infection. Patient also suffered from a life-threatening influenza infection at 3 yr of age. |
| IRF9 (Hernandez et al., 2018) | 16 | 5 | 2 wk after MMR vaccination, the patient developed an idiopathic biliary perforation. Patient recovered from MMR infection. Patient also suffered a life-threatening influenza infection at 2 yr of age, parainfluenza virus at age 4, and recurrent fevers without a causative pathogen identified throughout her childhood. |
| STAT1 (Burns et al., 2016) | 13 | 2 | 1 wk after MMR vaccination, patient developed fever, irritability, rash, diarrhea, encephalitis, a hemophagocytic lymphohistiocytosis-like syndrome, and was positive for human herpesvirus 6. Patient was readmitted 7 d later with refusal to walk, unsteady gait, and inflammatory cells in the CSF. Patient recovered from MMR infection. |
| IFNAR1 P1 (this report) | 12 | 9 | 10 d after MMR vaccination, patient developed a generalized exanthema, fever, and encephalitis. Patient recovered from MMR infection. |
Deaths.
Our data also establish IFNAR1 deficiency as the first genetic etiology for YEL-AVD, and as its first explanation (Seligman et al., 2014). Despite the development of the YF vaccine in the 1930s, YFV remains a global health threat partially due to deficient rates of vaccination. As the eponymous flavivirus, the study of YFV and its vaccine is of interest to studies of other flaviviruses. Chimeric YF viruses in which prM-E sequences are introduced into the YF vaccine virus backbone are the basis of ongoing vaccine trials against dengue and Japanese encephalitis viruses (Monath et al., 2015), with controversial results in clinical trials (Vaccine, 2018; Halstead, 2018). Although still considered a safe and effective vaccine, the uncertainty regarding the causes of YEL-AVD have encouraged attempts to develop an inactivated alternative vaccine (Monath et al., 2011; Monath and Vasconcelos, 2015; Pereira et al., 2015). The difficulties encountered while attempting to develop safer vaccines related to YFV-17D for the past decade highlight that a better understanding of host defenses against the virus is necessary. Recent studies have demonstrated that the YFV-17D virus promotes the induction of critical type I IFN mediators such as STAT1 and IRF7 in humans (Gaucher et al., 2008; Querec et al., 2009; Fernandez-Garcia et al., 2016). Importantly, YFV-17D viremia was typically detectable in healthy vaccine recipients 5–7 days following vaccination, and no deficiency of adaptive immunity has been found in individuals with YEL-AVD, suggesting that the disease is most likely the result of a failure of innate or intrinsic immunity (Reinhardt et al., 1998; Pulendran et al., 2008; Silva et al., 2010). Consistent with this suggestion, recent work in mice has shown that type I IFN signaling is critical for control of YFV-17D (Erickson and Pfeiffer, 2013). Moreover, deficiency in this pathway can be exacerbated by defects in type III IFN signaling (Douam et al., 2017). Our data provide proof-of-principle that single gene inborn errors of innate immunity, in a broad sense, and specifically of type I IFN cell-intrinsic immunity, can underlie life-threatening adverse reaction to the YF vaccine.
Strikingly, these two patients harboring biallelic IFNAR1 variants, now 9 and 14 yr of age, have not had clinically significant viral infections other than the LAV-associated complications at an early age. In contrast, a recently reported patient with combined partial IFNAR1 and complete IFNGR2 deficiencies (Hoyos-Bachiloglu et al., 2017) suffered from CMV infection, in addition to BCG disease. In contrast, P1 and P2 were IgG+ and IgM− for CMV, and CMV DNA was not detected in their plasma by PCR, confirming that they had been exposed to CMV. The other patient’s combined deficiency in IFNAR1 (partial) and IFNGR2 (complete) may have contributed to CMV disease, consistent with its occurrence in a few patients with isolated, complete IFNGR1 or IFNGR2 deficiency (Holland et al., 1998; Dorman et al., 1999; Cunningham et al., 2000; Rosenzweig et al., 2004).
Three hypotheses could explain the robust anti-viral defense in the two patients with AR complete IFNAR1 deficiency. A first hypothesis is that the IFNAR1 splice allele is hypomorphic, i.e., leaky, in some cell types that were not tested. An alternative hypothesis is that IFNAR1 is redundant for some type I IFNs, in some cells that have a complete deficiency and were not tested, at least for some ISGs, as previously shown for IFNAR2 (de Weerd et al., 2013). A third hypothesis is that IFNAR1 and all type I IFNs are redundant for immunity against most viruses, at least in certain conditions of infection. It is probably relevant that patients with IFNAR2, STAT2, or IRF9 deficiency, which impair responses to type I IFNs and type III IFNs for STAT2 and IRF9, are apparently also normally resistant to a broad range of viruses, including CMV, and have survived until 5–11 yr of age (Hambleton et al., 2013; Duncan et al., 2015; Shahni et al., 2015; Moens et al., 2017; Hernandez et al., 2018), whereas all patients with complete STAT1 deficiency, which impairs responses to type I, II, and III IFNs, died by 2 yr of various viral infections (Dupuis et al., 2003; Chapgier et al., 2006; Vairo et al., 2011; Burns et al., 2016), unless they had received both hematopoietic stem cell transplantation and IgG substitution (Burns et al., 2016). Future studies are required to understand the mechanisms of protective immunity to viruses and to develop safer vaccines against viral diseases in patients with inborn errors of type I and/or type III IFNs.
Materials and methods
Study oversight
The study was approved by the institutional review boards of Rockefeller University, Institut National de la Santé et de la Recherche Médicale, University Hospitals Leuven, and the National Institute of Infectious Diseases, Fundação Oswaldo Cruz, Rio de Janeiro, Brazil. Written informed consent was obtained from all patients or their guardians.
Case reports
P1 was born in Iran to consanguineous parents. He was evaluated at the age of 1 yr for disseminated vaccine–strain measles. He had previously received hepatitis B virus, BCG, and influenza vaccinations without any adverse effect. 10 d after inoculation with the MMR vaccine, he presented with a generalized exanthem, fever, and neurological symptoms consistent with encephalitis. Laboratory findings showed moderate lymphocytosis with 7,800 total leukocytes/µl, 29% neutrophils, 45% lymphocytes, 10% monocytes, 4% eosinophils, and mild hyponatremia. CT of the brain showed mild cerebral edema, and analysis of CSF demonstrated leukocytosis (350 lymphocytes/µl, 2 neutrophils/µl). PCR for measles was positive in both blood and CSF, with negative bacterial cultures. Antibodies against measles and mumps were at the lower limit of the normal range, isohemagglutinin titers were normal, and antibody responses to pneumococcal and hepatitis B virus antigens were also normal. Additionally, P1 was IgG+ and IgM− for CMV, and CMV DNA was not detectable in his plasma. Lymphocyte subpopulations (T, B, and natural killer cells) were in the normal range. His medical history was characterized by frequent viral upper respiratory tract infections with a hospital admission for bronchiolitis but was otherwise not significant. His growth and development were normal. His younger sister also developed meningoencephalitis 1 wk after receiving MMR vaccination and died of complications 4 wk later. P1 is now 9 yr old, in good health, and has not manifested any other invasive infection (Fig. 1 A).
P2 is a 14-yr-old Brazilian girl born to nonconsanguineous parents. Otherwise healthy until age 12, she presented with fever, hypotension, vomiting, and lethargy 7 d following YF vaccination. Laboratory findings after admission demonstrated leukocytosis with total leukocytes at 28,200/µl, and thrombocytopenia with a platelet count of 21,000/µl. Consistent with renal and hepatic dysfunction, the patient was icteric on exam with an international normalized ratio of 2.0 and a partial thromboplastin time of 66.4. She demonstrated alanine aminotransferase and aspartate aminotransferase levels of 187 IU/liter and 623 IU/liter, respectively, and had a creatinine of 2.71 mg/dl. Her condition rapidly deteriorated, with bradycardia and diminished consciousness, with a Glasgow coma score of 8, necessitating admission to the intensive care unit, intubation, and mechanical ventilation. A CT of the chest demonstrated pleural effusions and atelectasis (Fig. 1 B). Neutralizing antibodies against YFV were detectable by plaque reduction neutralization test at a dilution titer of 1:640, as were antibodies against herpesviruses 1 and 2, while antibodies against hepatitis A, B, and C virus, as well as dengue virus and leptospirosis, were negative. P2’s serum was similarly IgG+ and IgM− against CMV antigens, with CMV DNA not detectable in her plasma by PCR. YFV was detected in the patient’s blood by PCR analysis, and sequencing confirmed that this was the vaccine–strain virus, not WT YFV (Fig. S1). A diagnosis of YEL-AVD was made. She was treated with an aggressive course of i.v. fluids, plasma and platelet transfusions, vasopressors, and hydrocortisone and was prophylactically given i.v. broad-spectrum antibiotics. Her hypotension and thrombocytopenia resolved over the next week and, 2 wk after her admission, she was discharged from the intensive care unit and made a full recovery. 1 yr after this episode, she remains healthy. Her circulating immunoglobulin levels and leukocyte subsets are normal. Notably, the patient received two MMR vaccines at 12 and 16 mo of age, and other live vaccines (oral poliovirus and BCG), without incident (Fig. 1 A).
WES
Exome capture was performed with the SureSelect Human All Exon 50 Mb kit (Agilent Technologies). Paired-end sequencing was performed on a HiSeq 2000 (Illumina) generating 100-base reads. We aligned the sequences with the GRCh38 reference build of the human genome using the Burrows-Wheeler Aligner (Li and Durbin, 2009). Downstream processing and variant calling were performed with the Genome Analysis Toolkit, SAMtools, and Picard (Li et al., 2009; McKenna et al., 2010). Substitution and InDel calls were made with GATK Unified Genotyper. All variants were annotated using an annotation software system that was developed in-house (Ng and Henikoff, 2001; Adzhubei et al., 2010; Kircher et al., 2014).
Genetics
WES analysis of each patient revealed a total of 16,797 variants in P1 and 50,634 variants in P2. We filtered out all variations found in 1000 Genomes database, GnomAD, and our own database of 4,892 exomes for infectious diseases at a frequency of >1%, leaving 48 and 11 nonsynonymous coding variants in P1 and P2’s exomes, respectively: 30 and 6, respectively, were homozygous, and 18 and 5 were heterozygous. The heterozygous variants inherited from one parent were not considered in a model of complete penetrance. Excluding those in IFNAR1, none of the variants affected genes known to be related to immunity. The nonsense c.783G>A mutation had a CADD score of 37, and the two splicing mutations, c.674-1G>A and c.674-2A>G, had CADD scores of 23.6 and 24, respectively. A genomic measure of individual homozygosity was plotted for P1 and P2, two European individuals from consanguineous families, and 37 individuals from nonconsanguineous families from our in-house WES database. Homozygosity was computed as the proportion of the autosomal genome belonging to runs of homozygosity, which were defined as ranging ≥1 Mb of length and containing at least 100 SNPs, and were estimated using the homozyg option of PLINK software (Purcell et al., 2007). The centromeres were excluded because they are long genomic stretches devoid of SNPs, and their inclusion might inflate estimates of homozygosity if both flanking SNPs are homozygous. The length of the autosomal genome was fixed at 2,673,768 kb as previously described (McQuillan et al., 2008). We estimated the selective pressure acting on IFNAR1 to be 2.762 (indicative of positive selection), by estimating the neutrality index (Stoletzki and Eyre-Walker, 2011) at the population level: (PN/PS)/(DN/DS), where PN and PS are the number of nonsynonymous and synonymous alleles, respectively, at the population level (1000 Genomes Project) and DN and DS are the number of nonsynonymous and synonymous fixed sites, respectively, for the coding sequence of IFNAR1.
Cells
Peripheral blood mononuclear cells were isolated by Ficoll-Paque density gradient (Lymphoprep, Proteogenix) from the blood of patients and healthy donors. SV40-immortalized dermal fibroblasts and Vero cells were maintained in DMEM supplemented with 10% FBS. B-LCLs were grown in RPMI 1640 medium supplemented with 10% FBS. Primary fibroblasts were grown in DMEM/F-12 (1:1) containing l-glutamine and Hepes supplemented with 10% fetal calf serum, amphotericin B (0.5 µg/ml), penicillin (100 U/ml), and streptomycin (100 µg/ml).
YFV plaque reduction neutralization test
Serum samples were used to quantify the levels of neutralizing antibodies specific to the 17DD-YF virus using the microplaque reduction neutralization test (micro-PRNT50). The assays were performed at Laboratório de Tecnologia Virológica, Bio-Manguinhos (LATEV, Laboratory of Virological Technology of Instituto Oswaldo Cruz [Fiocruz-RJ], Brazil). First, a viral suspension was prepared with YF 213/77 #002/16, diluted previously with the objective to obtain ∼30 lysis plaques per well. Before neutralization, the serum sample was inactivated at 56°C for 30 min and submitted to serial twofold dilutions (1:5 to 1:640), and then ∼30 PFU of YFV suspension was added to the samples in 96-well tissue culture plates. After addition of virus, the plates were incubated for 1 h at 37°C in an incubator with 5% CO2. A 50-µl cell suspension with 1,600,000 cells/ml were inoculated in all 96 wells of the plate and incubated at 37°C for 3 h under 5% CO2 for adsorption/sedimentation. The medium was then discarded and carboxymethylcellulose (2.5%) added, and the cells were incubated for 6 d at 37°C with 5% CO2. After incubation, the plates were fixed in formaldehyde (5%) for ≥1 h at room temperature and stained with 2% crystal violet for 30 min at room temperature. Both steps were followed by cleansing with running water.
The arithmetic mean of all viral plaques obtained without serum was estimated. From the mean, the 50% endpoint of the number of plaques was calculated. Afterwards, using the dilution with plaque numbers immediately above and below the endpoint, the serum dilution that would result in the 50% endpoint was estimated by linear regression. The result were expressed in reciprocals of dilution.
Measurement of antibodies against diphtheria, tetanus, rubella, and measles
An ELISA standardized at the Immunological Technology Laboratory of Bio-Manguinhos (LATIM, Fiocruz-RJ) was used for antibody measurement. Standard diphtheria and tetanus curves were prepared using in-house sera titrated against National Institute for Biological Standard Controls International References. The diphtheria and tetanus antigen for coating ELISA plates were obtained from National Institute for Biological Standard Controls. Anti-diphtheria and anti-tetanus ELISA titers were considered to be protective if they were equal to or higher than the cutoff of 0.1 IU/ml. Serum titers were calculated by four-parameter logistic curve using SoftMax Pro, version 5.2 (Molecular Devices; Martins et al., 2008).
The ELISAs for rubella and measles antibodies were performed at Fiocruz-RJ, utilizing the kits Anti-Measles Virus ELISA (IgG) and Anti-Rubella Virus ELISA (IgG; Euroimmun) according to manufacturer instructions. Anti-measles and anti-rubella ELISA titers were considered to be positive if they were equal to or higher than the cutoff of 275 and 11 IU/ml, respectively.
Plasmids
The cDNA of IFNAR1 was cloned into pGEMT cloning vector (Promega). Site-directed mutagenesis was performed to obtain the indicated mutant constructs. All IFNAR1 constructs were then subcloned into pCAGGS for overexpression studies. For lentiviral vector production, lentiviral vector transfer plasmids pCH_EF1a_IFNaR1_IRES_Bsd and pCH_EF1a_IFNaR2_IRES_Bsd were generated by cloning the codon-optimized coding sequence for human IFNAR1 isoform 1 (NM_000629 or NP_000620) and human IFNAR2 into the multiple cloning site of the HIV-based lentiviral vector transfer plasmid pCH_EF1a_MCS_IRES_Bsd. Both cDNAs were ordered as a gBlock (IDT Haasrode). Sequences were cloned in-frame with the IRES-Bsd sequence. All constructs were resequenced to ensure no adventitious mutations were generated during the cloning.
Production of IFNAR1 and IFNAR2 lentiviral vectors
HIV-based viral vectors were produced by the Leuven Viral Vector Core as previously described (Ibrahimi et al., 2009), by triple transient transfection of 293T cells with a VSV glycoprotein G envelope encoding plasmid and a packaging plasmid together with the respective transfer plasmids using polyethylenimine (Polysciences), resulting in LV_IFNAR1, LV_IFNAR2, and LV_control (EV), respectively. After collecting the supernatant, the medium was filtered using a 0.45-µm filter (Corning) and concentrated using a Vivaspin 50,000 mol wt column (Vivascience). The vector containing concentrate was aliquoted and stored at −80°C.
Generation of stably reconstituted cell lines
HIV-based vectors were used to transduce primary fibroblasts in a serial dilution series. Cells were cultured and subjected to blasticidin selection (5 µg/ml). IFNAR1 expression was corroborated by qRT-PCR, Western blot, and cell surface staining.
Western blotting
Fibroblasts, with or without pretreatment with IFN-α2b (Schering) or IFN-γ (Imukin, Boehringer Ingelheim) for the specified times, were lysed in NP-40 lysis buffer (280 mM NaCl, 50 mM Tris, pH 8, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, and 0.5% NP-40) supplemented with 1 mM DTT, PhosSTOP (Roche), and complete protease inhibitor cocktail (Roche). 40 ug protein lysate per lane was resolved by SDS-PAGE and transferred to polyvinylidene fluoride membrane, which was probed with unconjugated primary antibodies and HRP-conjugated secondary antibodies. An anti-GAPDH antibody (Santa Cruz) was used as a loading control. Endogenous IFNAR1 was probed with an antibody recognizing the amino-terminus at a dilution of 1:1,000 (64G12, generously provided by Sandra Pellegrini; Pasteur Institute, Paris, France), while a polyclonal anti-IFNAR1 was used to detect overexpressed protein (AP8550c; Abgent). SuperSignal West Femto Chemiluminescent substrate (Thermo Fisher Scientific) was used to visualize HRP activity, and this signal was detected by an Amersham Imager 600 (GE Life Sciences).
Flow cytometry
For measuring surface expression on patients’ cells, B-LCL and SV40-F cells (5 × 105 cells per well) were plated in 96-well plates and surface-stained with either purified mouse anti-IFNAR1 AA3 (provided by Sandra Pellegrini) or PE mouse anti-IFNAR2 (PBL Assay Science) antibodies. Cells stained with AA3 were then washed once with PBS and incubated with a biotinylated rat anti-mouse secondary antibody (Thermo Fisher Scientific) for 30 min before being washed once with PBS and incubated for 30 min with PE-conjugated streptavidin (Thermo Fisher Scientific). The cells were then washed twice with PBS and analyzed by flow cytometry. Data were acquired on an LSRII flow cytometer (BD), and the results were analyzed with FlowJo (TreeStar).
For YFV infections, transduced fibroblasts were submitted to IFN-β pretreatment and YFV-17D infection as described below. Cells were detached after incubation for 10 min at room temperature in Accumax (07921; Stemcell). Cells were then fixed in 4% paraformaldehyde for 20 min at room temperature before permeabilization with BD perm buffer (554723; BD Biosciences). BD perm buffer was used for antibody incubations and washes. Cells were incubated with primary antibodies for 1 h on ice, washed, incubated with secondary antibodies for 30 min at room temperature, washed, and resuspended in PBS with 1% FBS before analysis using a BD LSRII flow cytometer. The primary antibody was a mouse anti-YFV (sc-58083, RRID:AB_630447; Santa Cruz Biotechnology) diluted 1:250, while the secondary antibody was a goat anti-mouse Alexa Fluor 647 (A-21203; Invitrogen) diluted 1:1,000.
qRT-PCR
SV40-F cells stimulated with 1,000 IU/ml IFN-α2b, IFN-β, or IFN-γ for 2 or 8 h, or primary fibroblasts treated for 6 h, were lysed with RNA lysis buffer and treated with DNase, and RNA was purified according to the manufacturer’s protocol (Zymo Research). RT-PCR was performed using random hexamers and the Superscript III reverse strand synthesis kit according to the manufacturer’s recommendations (Thermo Fisher Scientific). qRT-PCR was performed with Applied Biosystems Taqman assays using the β-glucuronidase housekeeping gene (GUS) for normalization for SV40-F cells or β-actin for primary fibroblasts. Results are expressed using the ΔΔCt method, as described by the manufacturer.
Virus assays
VSV infections
SV40-F cells were infected with VSV at a multiplicity of infection (MOI) of 3. The inoculum was absorbed onto the cells for 30 min at 25°C, washed twice with PBS, and cultured in DMEM with 10% FBS at 37°C. Virus samples were collected at the indicated time points. Viral titers were determined by endpoint dilution on Vero cells using the Reed and Muench calculation (Reed and Muench, 1938). Where indicated, cells were pretreated with IFN-α2b for 16 h before virus infection. Primary fibroblasts were infected with Indiana stock VSV (MOI = 9.3), with or without pretreatment with 10,000 U/ml IFN-α2b for 24 h before infection. The inoculum was left on the cells, and cell survival was monitored with Resazurin assay kit (ab112119; Abcam) at 1, 2, and 3 d after infection.
IAV and HSV-1 infections
SV40-fibroblasts were infected with influenza virus (A/California/4/2009) as previously described (Hernandez et al., 2018). Briefly, a viral inoculum was absorbed onto the cells (MOI = 0.5) for 30 min at 25°C. Cells were then washed twice with HBSS and cultured at 37°C in the presence of 0.1 µg/ml TPCK-trypsin (Sigma-Aldrich). Virus samples were collected at the indicated times after infection, and influenza titers were determined by plaque assay on MDCK cells. For HSV-1-GFP infection, SV40-F cells were plated in a 96-well dish and infected with HSV-1-GFP (MOI = 0.01) in DMEM supplemented with 2% FCS. GFP fluorescence was then assessed at 0, 8, 24, 48, and 72 h after infection. HSV-1-GFP was a gift from Dr. P. Desai (Johns Hopkins University School of Medicine, Baltimore, MD; Desai and Person, 1998). For HSV-1 (KOS strain; ATCC), SV40-Fs were seeded at a density of 105 cells per well in a 24-well plate and infected (MOI = 0.01) in DMEM plus 2% FCS. Supernatants were collected at the given time points after infection, and median tissue culture infectious dose values were calculated following the method of Reed and Muench, after inoculation of Vero cells in 96-well plates (Reed and Muench, 1938).
YFV infections
Preparation of virus stocks
YFV-17D viral stocks were derived from pACNR-2015FLYF-17Da plasmid, a derivative of the previously described pANCR-FLYF-Dx containing the full-length infectious YFV-17D genome under an SP6 promoter (Bredenbeek et al., 2003). In pANCR-2015FLYF-17Da, the linearization site for preparation of RNA was changed to AfIII, an XhoI site within the YF coding region was restored, and adventitious mutations that had occurred during bacterial propagation of pANCR-FLYF-17Dx in the laboratory were corrected. Asibi viral stocks were derived from an analogous full-length Asibi cDNA infectious clone designated pACNR-2015FLYF-Asibi. In vitro–generated RNA transcripts were electroporated in Huh7.5 cells (Blight et al., 2002). Virus was harvested 24 h after transfection, and titers of 6 × 105 PFU/ml (17D) and 6 × 103 FFU/ml (Asibi) were determined by plaque or focus forming assay on C3 fibroblasts. Single-use aliquots were stored frozen at −80°C until use.
Infection of cells
Before infection, patient-derived fibroblasts were pretreated with or without 1,000 U/ml IFN-β (PBL Assay Science) for 16 h. 17D (or Asibi) was then inoculated to ∼50,000 (or 20,000) fibroblasts in 24-well (or 48-well) plates. For 17D infections, medium was discarded, and cells were inoculated with 250 µl of 17D diluted in Optimem (MOI = 0.05) for 2 h at 37°C. Asibi infections were similar to 17D, except cells were inoculated with 100 µl of Asibi diluted in Optimem (MOI = 0.03). Cells were then washed twice with culture medium and incubated at 37°C with 0.5 ml of fresh culture medium. At the indicated time points after infection, media containing the virus produced by the fibroblasts were harvested and titered on Huh7.5 cells by plaque-forming assay.
Plaque assay
Harvested medium was serially diluted 1:10 in Optimem. Approximately 0.5 million Huh7.5 cells were inoculated with 400 µl of diluted virus in 6-well plates, for 2 h at 37°C. After removal of the inoculum, a 3-ml overlay of 1.2% Avicel in DMEM supplemented with 1× penicillin/streptomycim and 2% FCS was added, and cells were incubated for 3 d (17D) or 4 d (Asibi) at 37°C. After formaldehyde fixation, the cells were stained with crystal violet, and the plaques were enumerated.
IFN-β measurement
The concentration of IFN-β in the supernatant of infected cells was measured by ELISA using the VeriKine-HS Human IFN-β Serum ELISA Kit (PBL Assay Science), according to the manufacturer’s instructions, using 50 µl of medium.
ZIKV infections
Infection of cells
Before the infection, the ZIKV stock titer of 106 FFU/ml was determined by focus forming assay on C1 fibroblasts. IFN-β pretreatment, virus infection (ZIKV MOI = 0.05), and quantification of virus production were conducted on patient-derived fibroblasts as described above for Asibi, except Vero cells were inoculated with 300 µl of virus for the plaque-forming assays.
MeV infections
Approximately 50,000 patient-derived SV40-F cells were seeded in 48-well plates and pretreated or not with 1,000 U/ml IFN-α2b (PBL Assay Science) for 16 h. Cells were incubated for 1 h at 37°C with measles virus diluted in DMEM with 10% FCS (MOI = 0.1). After virus inoculation, cells were washed twice with PBS and incubated at 37°C with 0.2 ml of fresh culture medium. At the indicated time points after infection, medium containing the virus produced by the fibroblasts was harvested, and median tissue culture infectious dose values were calculated following the method of Reed and Muench after inoculation of Vero cells in 96-well plates (Reed and Muench, 1938).
Online supplemental material
Fig. S1 shows sequence alignments from the YFV isolated from P2’s serum and WT YFV or the YFV-17D strains used in their vaccine. Fig. S2 provides additional information on the genetic analysis of P1 and P2, including Sanger sequencing, additional information on mRNA sequencing, and qRT-PCR analysis of IFNAR1 mRNA levels in patient SV40-Fs. Fig. S3 presents additional evidence of abrogated IFN signaling in patient cells, including IFNAR1 expression in B-LCLs, IFNAR2 expression in patient cells, IFN-β production following infection, IFNAR1 expression in transduced SV40-Fs, and VSV replication in the transduced SV40s. Fig. S4 shows the replication of a number of viruses in patient SV40-Fs, including HSV-1, IAV, YFV-Asibi, and Zika virus. Table S1 lists the rare homozygous or possible compound heterozygous variants revealed by WES analysis of P1 and P2.
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of Ion Gresser, who passed away in April 2019 (Casanova, J.-L. 2019. J. Interferon Cytokine Res. https://doi.org/10.1089/jir.2018.29015.mem). Ion was a giant in the field of IFN. Among many achievements, he discovered the IFNAR1 studied in this paper. This paper is also dedicated to the memory of Dr. Reinaldo de Menezes Martins. Dr. Martins, who passed away in January 2019, made immense contributions to the development and study of vaccines, and thereby contributed to thousands of lives being saved. We warmly thank our patients and their families. We thank T. Kochetkov, L. Shang, Y. Liang, D. Papandrea, B. Razooky, A. O’Connell, and Y. Nemirovskaya for their contributions and all members of the laboratories for fruitful discussions. We thank Sandra Pellegrini for providing IFNAR1 antibodies. We thank Irina Thiry for the production of lentiviral particles and transduction of primary fibroblasts. We also thank Jose German Casas Martin for assistance with confocal microscopy.
This work was supported by National Center for Research Resources and National Center for Advancing Translational Sciences grant UL1TR001866; the National Institute of Allergy and Infectious Diseases (NIAID) for Cooperative Center on Human Immunology grants U19AI111825 and U19AI057229; National Institute of Allergy and Infectious Diseases grants R21AI137371 and R01AI124690; National Vaccine Program Office of the US Department of Health and Human Services grant VSRNV000006; the St. Giles Foundation; the Agence Nationale de la Recherche under the “Investments for the Future” program grant ANR-10-IAHU-01 and the Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (ANR-10-LABX-62-IBEID); Institut National de la Santé et de la Recherche Médicale; National Immunization Program; Institute of Technology in Immunobiology (Bio-Manguinhos), Ministry of Health, Brazil; KU Leuven research grant (GOA/13/013); Caps-It research infrastructure (project ZW13-02) financially supported by the Hercules Foundation and Rega Foundation, KU Leuven; Fonds Wetenschappelijk Onderzoek Vlaanderen grant G0C8517N; and Université Paris Descartes. N. Hernandez was supported by the Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. J. Le Pen was supported in part by funds from a Francois Wallace Monahan Postdoctoral Fellowship at the Rockefeller University and by a European Molecular Biology Organization Long-Term Fellowship (ALTF 380-2018).
The authors declare no competing financial interests.
Author contributions: N. Hernandez, Q. Zhang, E. Jouanguy, L. Abel, X. Bossuyt, I. Meyts, M. Shahrooei, A. Shirkani, M. Changi-Ashtiani, A. Cobat, and H. Rokni-Zadeh analyzed WES data. Y. Seeleuthner assisted with bioinformatics analysis of homozygosity in patient gDNA. N. Hernandez, G. Bucciol, L. Moens, and J. Le Pen assessed IFNAR1 expression, and N. Hernandez performed Western blot analyses. N. Hernandez, G. Bucciol, L. Moens, and J. Le Pen performed qRT-PCR experiments. N. Hernandez performed IAV and HSV infections. N. Hernandez, G. Bucciol, and L. Moens performed VSV infections, and, along with D. Jochmans, R. Boudewijns, and J. Neyts, measles virus infections. J. Le Pen and A. Jurado performed YFV-Asibi and ZIKV infections and, along with D. Zijlmans, YFV-17D infections. I. Tietjen and H.H. Hoffman prepared viral stocks. R. Pholien assisted with microscopy work. R. Gijsbers assisted with the production of lentiviral vectors. M. Shahrooei, E. Goudouris, E.H. Sayar, I. Reisli, A. Lefevre-Utile, M. Momeniland, L. Lorenzo-Diaz, C. Enemchukwu, M. Shahrooei, M.M. Siqueira, S.M.B. de Lima, D.C. de Souza Matos, A. Homma, M.L.S. Maia, T.A. da Costa Barros, P.M.N. de Oliveira, E.C. Mesquita, R.M. Martins, S.J. Seligman, Q. Zhang, E. Jouanguy., M.R. MacDonald, C.M. Rice, and J.-L. Casanova recruited patients and coordinated clinical study protocol and sample collection. T.A. de Costa Barros performed analysis of anti-tetanus and diphtheria antibodies. Q. Zhang, E. Jouanguy., M.R. MacDonald, C.M. Rice, I. Meyts, X. Bossuyt, and J.-L. Casanova planned the experimental work and supervised the data analysis. S. Drutman, S. Belkaya, L. Pöyhönen, L. Abel, P. Hertzog, N. Marr, and S.-Y. Zhang contributed to study design and analysis. N. Hernandez, Q. Zhang, E. Jouanguy, and J.-L. Casanova prepared the manuscript. All authors discussed and revised the manuscript.
References
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., and Sunyaev S.R.. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7:248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkadi A., Pedergnana V., Cobat A., Itan Y., Vincent Q.B., Abhyankar A., Shang L., El Baghdadi J., Bousfiha A., Alcais A., et al. Exome/Array Consortium . 2016. Whole-exome sequencing to analyze population structure, parental inbreeding, and familial linkage. Proc. Natl. Acad. Sci. USA. 113:6713–6718. 10.1073/pnas.1606460113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight K.J., McKeating J.A., and Rice C.M.. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014. 10.1128/JVI.76.24.13001-13014.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dupuis S., Kong X.-F., Okada S., Cypowyj S., Puel A., Abel L., and Casanova J.-L.. 2012. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr. Opin. Immunol. 24:364–378. 10.1016/j.coi.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P.J., Kooi E.A., Lindenbach B., Huijkman N., Rice C.M., and Spaan W.J.M.. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84:1261–1268. 10.1099/vir.0.18860-0 [DOI] [PubMed] [Google Scholar]
- Burns C., Cheung A., Stark Z., Choo S., Downie L., White S., Conyers R., and Cole T.. 2016. A novel presentation of homozygous loss-of-function STAT-1 mutation in an infant with hyperinflammation-A case report and review of the literature. J. Allergy Clin. Immunol. Pract. 4:777–779. 10.1016/j.jaip.2016.02.015 [DOI] [PubMed] [Google Scholar]
- Chapgier A., Wynn R.F., Jouanguy E., Filipe-Santos O., Zhang S., Feinberg J., Hawkins K., Casanova J.-L., and Arkwright P.D.. 2006. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J. Immunol. 176:5078–5083. 10.4049/jimmunol.176.8.5078 [DOI] [PubMed] [Google Scholar]
- Ciancanelli M.J., Huang S.X.L., Luthra P., Garner H., Itan Y., Volpi S., Lafaille F.G., Trouillet C., Schmolke M., Albrecht R.A., et al. . 2015. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 348:448–453. 10.1126/science.aaa1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham J.A., Kellner J.D., Bridge P.J., Trevenen C.L., Mcleod D.R., and Davies H.D.. 2000. Disseminated bacille Calmette-Guérin infection in an infant with a novel deletion in the interferon-gamma receptor gene. Int. J. Tuberc. Lung Dis. 4:791–794. [PubMed] [Google Scholar]
- Desai P., and Person S.. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 72:7563–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weerd N.A., Vivian J.P., Nguyen T.K., Mangan N.E., Gould J.A., Braniff S.-J., Zaker-Tabrizi L., Fung K.Y., Forster S.C., Beddoe T., et al. . 2013. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 14:901–907. 10.1038/ni.2667 [DOI] [PubMed] [Google Scholar]
- Dorman S.E., Uzel G., Roesler J., Bradley J.S., Bastian J., Billman G., King S., Filie A., Schermerhorn J., and Holland S.M.. 1999. Viral infections in interferon-gamma receptor deficiency. J. Pediatr. 135:640–643. 10.1016/S0022-3476(99)70064-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douam F., Soto Albrecht Y.E., Hrebikova G., Sadimin E., Davidson C., Kotenko S.V., and Ploss A.. 2017. Type III Interferon-Mediated Signaling Is Critical for Controlling Live Attenuated Yellow Fever Virus Infection In Vivo. MBio. 8:. 10.1128/mBio.00819-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C.J.A., Mohamad S.M.B., Young D.F., Skelton A.J., Leahy T.R., Munday D.C., Butler K.M., Morfopoulou S., Brown J.R., Hubank M., et al. . 2015. Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci. Transl. Med. 7:307ra154 10.1126/scitranslmed.aac4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I.Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P., et al. . 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388–391. 10.1038/ng1097 [DOI] [PubMed] [Google Scholar]
- Eletto D., Burns S.O., Angulo I., Plagnol V., Gilmour K.C., Henriquez F., Curtis J., Gaspar M., Nowak K., Daza-Cajigal V., et al. . 2016. Biallelic JAK1 mutations in immunodeficient patient with mycobacterial infection. Nat. Commun. 7:13992 10.1038/ncomms13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson A.K., and Pfeiffer J.K.. 2013. Dynamic viral dissemination in mice infected with yellow fever virus strain 17D. J. Virol. 87:12392–12397. 10.1128/JVI.02149-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Garcia M.D., Meertens L., Chazal M., Hafirassou M.L., Dejarnac O., Zamborlini A., Despres P., Sauvonnet N., Arenzana-Seisdedos F., Jouvenet N., and Amara A.. 2016. Vaccine and Wild-Type Strains of Yellow Fever Virus Engage Distinct Entry Mechanisms and Differentially Stimulate Antiviral Immune Responses. MBio. 7:e01956-15 10.1128/mBio.01956-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Yuan Y.-Y., Lin Q.-F., Xu J.-C., Wang W.-Q., Qiao Y.-H., Kang D.-Y., Bai D., Xin F., Huang S.-S., et al. . 2018. Mutation of IFNLR1, an interferon lambda receptor 1, is associated with autosomal-dominant non-syndromic hearing loss. J. Med. Genet. 55:298–306. 10.1136/jmedgenet-2017-104954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher D., Therrien R., Kettaf N., Angermann B.R., Boucher G., Filali-Mouhim A., Moser J.M., Mehta R.S., Drake D.R. III, Castro E., et al. . 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 205:3119–3131. 10.1084/jem.20082292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker E.-O., Kotlarz D., Boztug K., Gertz E.M., Schäffer A.A., Noyan F., Perro M., Diestelhorst J., Allroth A., Murugan D., et al. . 2009. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361:2033–2045. 10.1056/NEJMoa0907206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm B., Trifilo M.J., Zuniga E.I., and Oldstone M.B.A.. 2005. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 22:247–257. 10.1016/j.immuni.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Halstead S.B. 2018. Safety issues from a Phase 3 clinical trial of a live-attenuated chimeric yellow fever tetravalent dengue vaccine. Hum. Vaccin. Immunother. 14:2158–2162. 10.1080/21645515.2018.1445448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambleton S., Goodbourn S., Young D.F., Dickinson P., Mohamad S.M.B., Valappil M., McGovern N., Cant A.J., Hackett S.J., Ghazal P., et al. . 2013. STAT2 deficiency and susceptibility to viral illness in humans. Proc. Natl. Acad. Sci. USA. 110:3053–3058. 10.1073/pnas.1220098110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Melki I., Jing H., Habib T., Huang S.S.Y., Danielson J., Kula T., Drutman S., Belkaya S., Rattina V., et al. . 2018. Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J. Exp. Med. 215:2567–2585. 10.1084/jem.20180628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland S.M., Dorman S.E., Kwon A., Pitha-Rowe I.F., Frucht D.M., Gerstberger S.M., Noel G.J., Vesterhus P., Brown M.R., and Fleisher T.A.. 1998. Abnormal regulation of interferon-gamma, interleukin-12, and tumor necrosis factor-alpha in human interferon-gamma receptor 1 deficiency. J. Infect. Dis. 178:1095–1104. 10.1086/515670 [DOI] [PubMed] [Google Scholar]
- Honda K., Takaoka A., and Taniguchi T.. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 25:349–360. 10.1016/j.immuni.2006.08.009 [DOI] [PubMed] [Google Scholar]
- Hoyos-Bachiloglu R., Chou J., Sodroski C.N., Beano A., Bainter W., Angelova M., Al Idrissi E., Habazi M.K., Alghamdi H.A., Almanjomi F., et al. . 2017. A digenic human immunodeficiency characterized by IFNAR1 and IFNGR2 mutations. J. Clin. Invest. 127:4415–4420. 10.1172/JCI93486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi A., Vande Velde G., Reumers V., Toelen J., Thiry I., Vandeputte C., Vets S., Deroose C., Bormans G., Baekelandt V., et al. . 2009. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum. Gene Ther. 20:845–860. 10.1089/hum.2008.188 [DOI] [PubMed] [Google Scholar]
- Itan Y., Shang L., Boisson B., Patin E., Bolze A., Moncada-Vélez M., Scott E., Ciancanelli M.J., Lafaille F.G., Markle J.G., et al. . 2015. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. USA. 112:13615–13620. 10.1073/pnas.1518646112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan Y., Shang L., Boisson B., Ciancanelli M.J., Markle J.G., Martinez-Barricarte R., Scott E., Shah I., Stenson P.D., Gleeson J., et al. . 2016. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat. Methods. 13:109–110. 10.1038/nmeth.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca N.E., Aksu G., Ulusoy E., Aksoylar S., Gozmen S., Genel F., Akarcan S., Gulez N., Hirschmugl T., Kansoy S., et al. . 2016. Early Diagnosis and Hematopoietic Stem Cell Transplantation for IL10R Deficiency Leading to Very Early-Onset Inflammatory Bowel Disease Are Essential in Familial Cases. Case Reports Immunol. 2016:5459029 10.1155/2016/5459029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreins A.Y., Ciancanelli M.J., Okada S., Kong X.-F., Ramírez-Alejo N., Kilic S.S., El Baghdadi J., Nonoyama S., Mahdaviani S.A., Ailal F., et al. . 2015. Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 212:1641–1662. 10.1084/jem.20140280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins R. de M., Camacho L.A.B., Marcovistz R., Noronha T.G., Maia M.L., dos Santos E.M., Barbosa G.G., Silva A.M., Souza P.C., Lemos M.C., and Homma A.. 2008. Immunogenicity, reactogenicity and consistency of production of a Brazilian combined vaccine against diphtheria, tetanus, pertussis and Haemophilus influenzae type b. Mem. Inst. Oswaldo Cruz. 103:711–718. 10.1590/S0074-02762008000700014 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M.A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan R., Leutenegger A.-L., Abdel-Rahman R., Franklin C.S., Pericic M., Barac-Lauc L., Smolej-Narancic N., Janicijevic B., Polasek O., Tenesa A., et al. . 2008. Runs of homozygosity in European populations. Am. J. Hum. Genet. 83:359–372. 10.1016/j.ajhg.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y., Saito M., Morio T., Watanabe K., Agematsu K., Tsuchiya S., Takada H., Hara T., Kawamura N., Ariga T., et al. . 2006. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 25:745–755. 10.1016/j.immuni.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Moens L., Van Eyck L., Jochmans D., Mitera T., Frans G., Bossuyt X., Matthys P., Neyts J., Ciancanelli M., Zhang S.-Y., et al. . 2017. A novel kindred with inherited STAT2 deficiency and severe viral illness. J. Allergy Clin. Immunol. 139:1995–1997.e9. 10.1016/j.jaci.2016.10.033 [DOI] [PubMed] [Google Scholar]
- Monath T.P., and Vasconcelos P.F.C.. 2015. Yellow fever. J. Clin. Virol. 64:160–173. 10.1016/j.jcv.2014.08.030 [DOI] [PubMed] [Google Scholar]
- Monath T.P., Fowler E., Johnson C.T., Balser J., Morin M.J., Sisti M., and Trent D.W.. 2011. An inactivated cell-culture vaccine against yellow fever. N. Engl. J. Med. 364:1326–1333. 10.1056/NEJMoa1009303 [DOI] [PubMed] [Google Scholar]
- Monath T.P., Seligman S.J., Robertson J.S., Guy B., Hayes E.B., Condit R.C., Excler J.L., Mac L.M., Carbery B., and Chen R.T.. Brighton Collaboration Viral Vector Vaccines Safety Working Group (V3SWG) . 2015. Live virus vaccines based on a yellow fever vaccine backbone: standardized template with key considerations for a risk/benefit assessment. Vaccine. 33:62–72. 10.1016/j.vaccine.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfopoulou S., Mee E.T., Connaughton S.M., Brown J.R., Gilmour K., Chong W.K., Duprex W.P., Ferguson D., Hubank M., Hutchinson C., et al. . 2017. Deep sequencing reveals persistence of cell-associated mumps vaccine virus in chronic encephalitis. Acta Neuropathol. 133:139–147. 10.1007/s00401-016-1629-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento Silva J.R., Camacho L.A.B., Siqueira M.M., Freire M.S., Castro Y.P., Maia M.L., Yamamura A.M., Martins R.M., and Leal M.L.. Collaborative Group for the Study of Yellow Fever Vaccines . 2011. Mutual interference on the immune response to yellow fever vaccine and a combined vaccine against measles, mumps and rubella. Vaccine. 29:6327–6334. 10.1016/j.vaccine.2011.05.019 [DOI] [PubMed] [Google Scholar]
- Neven B., Mamessier E., Bruneau J., Kaltenbach S., Kotlarz D., Suarez F., Masliah-Planchon J., Billot K., Canioni D., Frange P., et al. . 2013. A Mendelian predisposition to B-cell lymphoma caused by IL-10R deficiency. Blood. 122:3713–3722. 10.1182/blood-2013-06-508267 [DOI] [PubMed] [Google Scholar]
- Ng P.C., and Henikoff S.. 2001. Predicting deleterious amino acid substitutions. Genome Res. 11:863–874. 10.1101/gr.176601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell L.A., Conway S., Rose R.W., Nicolas E., Slifker M., Balachandran S., and Rall G.F.. 2012. STAT1-independent control of a neurotropic measles virus challenge in primary neurons and infected mice. J. Immunol. 188:1915–1923. 10.4049/jimmunol.1101356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R.C., Silva A.N.M.R., Souza M.C.O., Silva M.V., Neves P.P.C.C., Silva A.A.M.V., Matos D.D.C.S., Herrera M.A.O., Yamamura A.M.Y., Freire M.S., et al. . 2015. An inactivated yellow fever 17DD vaccine cultivated in Vero cell cultures. Vaccine. 33:4261–4268. 10.1016/j.vaccine.2015.03.077 [DOI] [PubMed] [Google Scholar]
- Piehler J., Thomas C., Garcia K.C., and Schreiber G.. 2012. Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol. Rev. 250:317–334. 10.1111/imr.12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyhönen L., Bustamante J., Casanova J.-L., Jouanguy E., and Zhang Q.. 2019. Life-threatening infections due to live attenuated vaccines: early manifestations of inborn errors of immunity. J. Clin. Immunol. 39:376–390. 10.1007/s10875-019-00642-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B., Miller J., Querec T.D., Akondy R., Moseley N., Laur O., Glidewell J., Monson N., Zhu T., Zhu H., et al. . 2008. Case of yellow fever vaccine--associated viscerotropic disease with prolonged viremia, robust adaptive immune responses, and polymorphisms in CCR5 and RANTES genes. J. Infect. Dis. 198:500–507. 10.1086/590187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., and Sham P.C.. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B., et al. . 2009. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10:116–125. 10.1038/ni.1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragimbeau J., Dondi E., Alcover A., Eid P., Uzé G., and Pellegrini S.. 2003. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. EMBO J. 22:537–547. 10.1093/emboj/cdg038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., and Muench H.. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27:493–497. 10.1093/oxfordjournals.aje.a118408 [DOI] [Google Scholar]
- Reinhardt B., Jaspert R., Niedrig M., Kostner C., and L’age-Stehr J.. 1998. Development of viremia and humoral and cellular parameters of immune activation after vaccination with yellow fever virus strain 17D: a model of human flavivirus infection. J. Med. Virol. 56:159–167. [DOI] [PubMed] [Google Scholar]
- Rosenzweig S.D., Dorman S.E., Uzel G., Shaw S., Scurlock A., Brown M.R., Buckley R.H., and Holland S.M.. 2004. A novel mutation in IFN-γ receptor 2 with dominant negative activity: biological consequences of homozygous and heterozygous states. J. Immunol. 173:4000–4008. 10.4049/jimmunol.173.6.4000 [DOI] [PubMed] [Google Scholar]
- Seligman S.J., Cohen J.E., Itan Y., Casanova J.-L., and Pezzullo J.C.. 2014. Defining risk groups to yellow fever vaccine-associated viscerotropic disease in the absence of denominator data. Am. J. Trop. Med. Hyg. 90:267–271. 10.4269/ajtmh.13-0542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaghaghi M., Parvaneh N., Ostad-Rahimi P., Fathi S.M., Shahmahmoodi S., Abolhassani H., and Aghamohammadi A.. 2014. Combined immunodeficiency presenting with vaccine-associated paralytic poliomyelitis: a case report and narrative review of literature. Immunol. Invest. 43:292–298. 10.3109/08820139.2013.859156 [DOI] [PubMed] [Google Scholar]
- Shahni R., Cale C.M., Anderson G., Osellame L.D., Hambleton S., Jacques T.S., Wedatilake Y., Taanman J.-W., Chan E., Qasim W., et al. . 2015. Signal transducer and activator of transcription 2 deficiency is a novel disorder of mitochondrial fission. Brain. 138:2834–2846. 10.1093/brain/awv182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.L., Espírito-Santo L.R., Martins M.A., Silveira-Lemos D., Peruhype-Magalhães V., Caminha R.C., de Andrade Maranhão-Filho P., Auxiliadora-Martins M., de Menezes Martins R., Galler R., et al. . 2010. Clinical and immunological insights on severe, adverse neurotropic and viscerotropic disease following 17D yellow fever vaccination. Clin. Vaccine Immunol. 17:118–126. 10.1128/CVI.00369-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N., and Eyre-Walker A.. 2011. Estimation of the neutrality index. Mol. Biol. Evol. 28:63–70. 10.1093/molbev/msq249 [DOI] [PubMed] [Google Scholar]
- Takaoka A., Mitani Y., Suemori H., Sato M., Yokochi T., Noguchi S., Tanaka N., and Taniguchi T.. 2000. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science. 288:2357–2360. 10.1126/science.288.5475.2357 [DOI] [PubMed] [Google Scholar]
- Uzé G., Lutfalla G., and Gresser I.. 1990. Genetic transfer of a functional human interferon alpha receptor into mouse cells: cloning and expression of its cDNA. Cell. 60:225–234. 10.1016/0092-8674(90)90738-Z [DOI] [PubMed] [Google Scholar]
- Vaccine 2018. Dengue vaccine: WHO position paper, September 2018 - Recommendations. Vaccine. 10.1016/j.vaccine.2018.09.063 [DOI] [PubMed] [Google Scholar]
- Vairo D., Tassone L., Tabellini G., Tamassia N., Gasperini S., Bazzoni F., Plebani A., Porta F., Notarangelo L.D., Parolini S., et al. . 2011. Severe impairment of IFN-γ and IFN-α responses in cells of a patient with a novel STAT1 splicing mutation. Blood. 118:1806–1817. 10.1182/blood-2011-01-330571 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.