Retromer sorts diverse membrane proteins into recycling tubules/vesicles from the endosome. How it precisely selects its cargos is not well described. Suzuki et al. reveal that retromer interacts with a bipartite sorting signal, which facilitates precise cargo recognition.
Abstract
Retromer is an evolutionarily conserved protein complex, which sorts functionally diverse membrane proteins into recycling tubules/vesicles from the endosome. Many of the identified cargos possess a recycling signal sequence defined as ØX[L/M/V], where Ø is F/Y/W. However, this sequence is present in almost all proteins encoded in the genome. Also, several identified recycling sequences do not follow this rule. How then does retromer precisely select its cargos? Here, we reveal that an additional motif is also required for cargo retrieval. The two distinct motifs form a bipartite recycling signal recognized by the retromer subunits, Vps26 and Vps35. Strikingly, Vps26 utilizes different binding sites depending on the cargo, allowing retromer to recycle different membrane proteins. Thus, retromer interacts with cargos in a more complex manner than previously thought, which facilitates precise cargo recognition.
Introduction
Retromer is an evolutionarily conserved protein coat complex that mediates recycling of endosomal membrane proteins (Seaman et al., 1997, 1998; Cullen and Steinberg, 2018). Retromer is composed of five proteins, Vps5, Vps17, Vps26, Vps29, and Vps35, that together mediate the formation of cargo-containing recycling tubules/vesicles from the endosome (Seaman et al., 1998). The best-characterized retromer cargo is yeast Vps10 (Marcusson et al., 1994). Vps10, the first member of the Sortilin receptor family, is a transmembrane (TM) protein receptor for carboxypeptidase Y (CPY), which sorts CPY into vesicles at the Golgi (Fig. 1 A). After CPY-containing vesicles are transported to the endosome, the endosome matures and fuses with the vacuole, delivering soluble CPY to the vacuole lumen. Unlike CPY, which is released from the Vps10 receptor in the endosome, Vps10 is not delivered to the vacuole but is recycled from the endosome back to the Golgi by the retromer complex, making Vps10 available for additional rounds of CPY sorting (Fig. 1 A). In humans, loss of retromer function is associated with diseases such as Parkinson’s disease. Certain familial Parkinson’s disease patients have a mutation in VPS35, altering the localization of several retromer cargos (Vilariño-Güell et al., 2011; Zimprich et al., 2011; Follett et al., 2014; McGough et al., 2014; Zavodszky et al., 2014; McMillan et al., 2017). Additionally, several viral and bacterial pathogen effectors target the retromer to promote replication during infection (Personnic et al., 2016). For instance, the Legionella pneumophila effector RidL directly binds to Vps29, resulting in the inhibition of retrograde trafficking (Finsel et al., 2013; Romano-Moreno et al., 2017).
Figure 1.
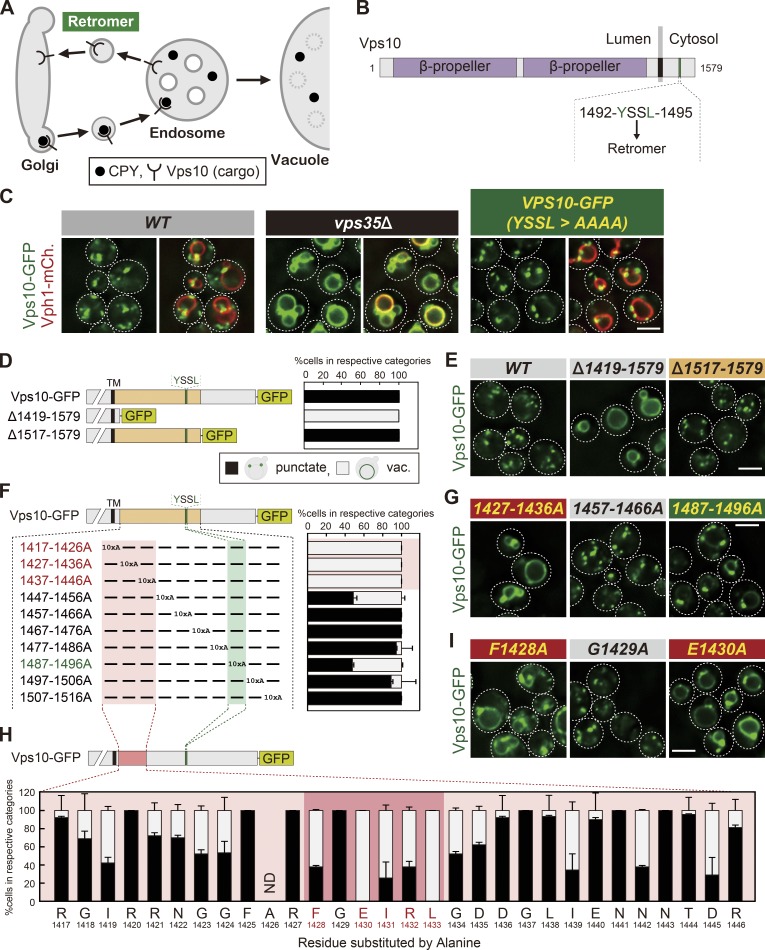
The FGEIRL motif of Vps10 is also important for its recycling. (A) A model of Vps10 recycling. (B) Schematic of Vps10. (C, E, G, and I) Vps10-GFP localization. (D, F, and H) Schematic of Vps10 mutational analysis and the percentage of each category of Vps10-GFP mutant localization from E (D), from G and Fig. S1 B (F), or from I and Fig. S1 C (H). Because the 1426th residue of Vps10 is Ala, A1426 was not substituted. ND, not determined; vac., vacuole. For all quantification shown in this figure, at least 30 cells were classified and the data were obtained from three independent experiments. Error bars represent SD. Scale bars: 2 µm.
Retromer is composed of two subcomplexes, the cargo-selective complex (CSC) and the sorting nexin (SNX)–bin, amphiphysin, and Rvs (BAR) dimer (Seaman et al., 1998). The CSC consists of Vps26, Vps29, and Vps35. The SNX-BAR dimer consists of Vps5 and Vps17. During cargo recycling, retromer is recruited to the endosomal membrane via the specific interaction of the Vps5-Vps17 Phox homology domains with phosphatidylinositol 3-phosphate (Burda et al., 2002). Cargo recognition is thought to be mediated primarily through Vps26 (Fjorback et al., 2012; Lucas et al., 2016; Cui et al., 2017), but Vps35 could also play a role (Nothwehr et al., 2000). Finally, the BAR domains of Vps5-Vps17 deform the endosomal membrane to form cargo-containing recycling tubules/vesicles (Seaman and Williams, 2002; Peter et al., 2004).
Although a large number of TM proteins are delivered to the endosomal membrane, retromer precisely selects its cargos and sorts them into recycling tubules/vesicles. The defined consensus sequence for retromer binding, ØX[L/M/V], where Ø is F/Y/W (Seaman, 2007; Cullen and Steinberg, 2018), is present on many proteins that are not retromer cargos. It is unlikely to mediate precise cargo recognition by itself. How retromer specifically recognizes each cargo is not known. Here, we reveal that an additional motif is required for cargo recycling, which explains the highly specific cargo recognition of the retromer complex.
Results and discussion
Two distinct motifs in Vps10 serve as a bipartite recycling signal
The retromer complex selectively recognizes cargos through a recycling sequence (Fig. 1 A). In mammalian cells, ØX[L/M/V], where Ø is F/Y/W, is defined as a consensus sequence for recognition by retromer. However, several identified yeast recycling sequences, such as YSSL of Vps10, FQFND of Ste13, YEF of Kex2, and WKY of Stv1, do not follow this rule (Fig. S1 A; Nothwehr et al., 1993; Cooper and Stevens, 1996; Redding et al., 1996; Finnigan et al., 2012). Also, while there are <30 known cargos in yeast (Bean et al., 2017), the ØX[L/M/V] sequence was found in almost all proteins (98.3% of yeast proteins; 5,841 of 5,916). We hypothesized that for retromer to specifically and accurately recognize the appropriate cargos, additional sequence information must be present. To test this idea, we performed mutational analysis of the CPY receptor Vps10 (Fig. 1 B). When Vps10-GFP was expressed from its native promoter in WT cells, it localized on punctate structures, which were previously reported to be Golgi or endosomes (Marcusson et al., 1994; Fig. 1 C). In the retromer-defective vps35Δ cells, retromer-mediated endosome-to-Golgi retrograde trafficking is impaired, and thus Vps10-GFP accumulated on the vacuole membrane. Although CPY sorting is severely blocked in vps35Δ cells (Seaman et al., 1997), the Vps10 recycling sequence mutant Y1492A exhibited only a partial defect in CPY sorting (Cooper and Stevens, 1996). Consistent with this observation, replacing the Vps10 recycling signal (1492-YSSL-1496) with alanine residues (YSSL>AAAA) showed only a partial recycling defect (Fig. 1 C), which raised the possibility that an additional sequence motif is required for its recycling. To define this motif, we first truncated the cytoplasmic tail of Vps10 and checked its localization (Fig. 1, D and E). The Δ1419-1579 mutant stably localized on the vacuole membrane, whereas the Δ1517-1579 mutant localized to punctate structures, mimicking full-length Vps10. This suggests that residues 1,419–1,516 on Vps10 are important for its retrieval. Next, we generated a series of Vps101417–1516 mutants in which 10 consecutive amino acids were replaced with alanine residues (Fig. 1, F and G; and Fig. S1 B). 1417-1426A, 1427-1436A, and 1437-1446A mutants exhibited a severe defect in Vps10 recycling. In contrast, 1487-1496A (which includes 1492-YSSL-1495) showed only a mild defect. The Δ1456-1459 mutant (1456-FYVF-1459) is known to cause the missorting of CPY (Cereghino et al., 1995). However, neither 1447-1456A nor 1457-1466A mutant exhibited striking defects. Next, we mutated single residues in the region 1,417–1,446 to alanine. Of the mutants tested, F1428A, E1430A, I1431A, R1432A, and L1433A mutants stabilized Vps10-GFP on the vacuole membrane (Fig. 1, H and I; and Fig. S1 C), suggesting that the 1428-FGEIRL-1433 region of Vps10 is also important for its recycling.
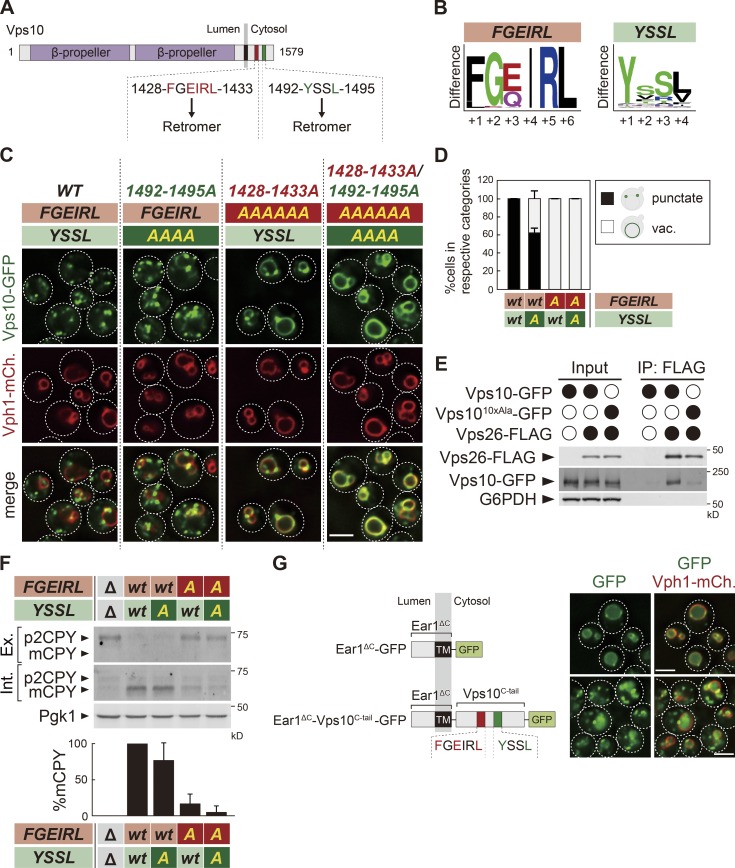
Based on the mutational analysis of Vps10, in addition to 1492-YSSL-1495, 1428-FGEIRL-1433 is required for its retrieval (Fig. 2 A). Both sequences are highly conserved among Vps10 homologues from related species (Fig. 2 B), suggesting that the cargo recognition mechanism via the two motifs is also conserved. To test the relationship of the two sequences, we replaced each sequence with alanine residues and examined Vps10-GFP localization (Fig. 2, C and D). While the 1492-1495A (YSSL>AAAA) mutant showed a partial defect in Vps10-GFP recycling, the 1428-1433A (FGEIRL>AAAAAA) mutant and the 1428-1433A/1492-1495A (FGEIRL>AAAAAA and YSSL>AAAA) double mutant exhibited a severe defect. To test whether these sequences are required for binding with the retromer, we performed coimmunoprecipitation experiments. While WT Vps10-GFP coprecipitated with Vps26-FLAG, the 1428-1433A/1492-1495A double mutant (Vps1010xAla-GFP) did not (Fig. 2 E). We also examined the effect on CPY sorting in these mutants. CPY sorting to the vacuole can be monitored by the appearance of the mature form of CPY. Consistent with Vps10-GFP localization, CPY maturation was moderately reduced in the 1492-1495A mutant, whereas it was strongly impaired in the 1428-1433A and 1428-1433A/1492-1495A mutants, with most of the precursor form of CPY secreted to the extracellular space (Fig. 2 F). Based on these observations, we propose that two distinct motifs in Vps10, 1428-FGEIRL-1433 and 1492-YSSL-1495, are both required for retromer recognition. However, the FGEIRL motif is essential for its recognition, whereas the YSSL enhances the recognition. Thus, these two distinct motifs form a bipartite recycling signal.
Figure 2.
Two distinct sequences of Vps10 serve as a bipartite recycling signal. (A) Schematic of Vps10. (B) Weblogo residue conservation of 1428-FGEIRL-1433 and 1492-YSSL-1495 of Vps10 among 85 Vps10 homologues from related species. (C) Vps10-GFP localization. (D) The percentage of each category of Vps10-GFP mutant localization from C. vac., vacuole. (E) The Vps26-Vps10 interaction in Vps10-GFP mutants. Vps26-FLAG was immunoprecipitated (IP) from cells expressing Vps10-GFP or Vps10-GFP with a mutation in a bipartite recycling signal (Vps1010xAla-GFP), and interacting Vps10-GFP was detected by immunoblotting using antibodies against FLAG, GFP, and G6PDH. (F) CPY sorting in vps10Δ cells expressing Vps10-GFP mutants. CPY sorting is examined by immunoblotting against CPY and Pgk1. Mature form of CPY in WT was set to 100%. Ex. and Int., extracellular and intracellular space, respectively; p2CPY, p2 precursor form of CPY; mCPY, mature CPY. (G) Ear1ΔC-GFP and Ear1ΔC-Vps10C-tail-GFP localization. For all quantification shown in this figure, at least 30 cells were classified and the data were obtained from three independent experiments. Error bars represent SD. Scale bars: 2 µm.
Finally, to address whether the bipartite recycling signal in Vps10 is sufficient for its recycling, we fused the C-tail of Vps10 (residues 1,416–1,523), which includes this recycling signal, to Ear1, an endosomal membrane protein (Léon et al., 2008). Truncation of the cytoplasmic tail of Ear1 (Ear1ΔC-GFP) resulted in its accumulation on the vacuole membrane, whereas fusion of the recycling sequences of Vps10 (Ear1ΔC-Vps10C-tail-GFP) restored its punctate localization (Fig. 2 G), suggesting that the 1428-FGEIRL-1433 and 1492-YSSL-1495 motifs of Vps10 are sufficient for recognition by the retromer.
Two separate motifs are also important for Ear1 retrieval
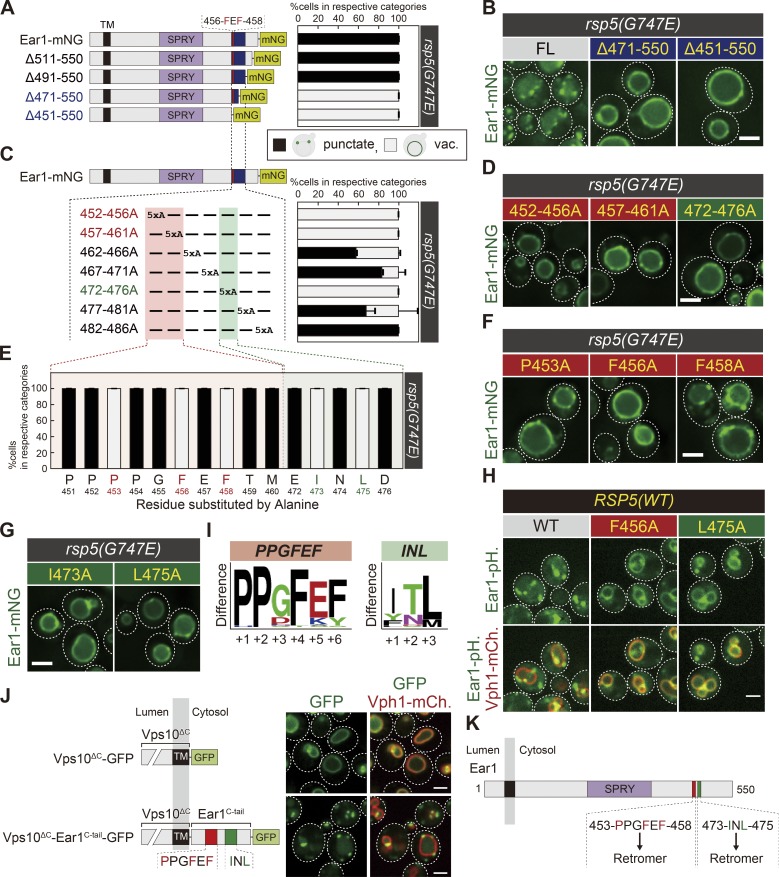
To assess if the two distinct motifs are also important for other retromer cargos, we performed mutational analysis of Ear1. Ear1 is a TM protein that localizes to the endosomal membrane (Léon et al., 2008; Fig. 3 A). The region 456-FEF-458 of Ear1 was previously identified as a recycling signal, which is required for its endosomal localization (Bean et al., 2017). Ear1 serves as an adaptor that recruits the E3 ubiquitin ligase Rsp5 to the endosome via its PY motifs (Fig. S1 D; Léon et al., 2008), allowing the ubiquitination of mislocalized membrane proteins on the endosomal membrane (Sardana et al., 2019). Since Ear1 itself is also ubiquitinated by Rsp5, leading to its degradation (Léon et al., 2008), we expressed Ear1-mNeonGreen in the rsp5(G747E) hypomorphic mutant (Fisk and Yaffe, 1999; Fig. S1 E). Ear1-mNeonGreen mainly localized to the endosomes in the rsp5(G747E) mutant but accumulated on the vacuole membrane in a rsp5(G747E) vps35Δ double mutant (Fig. S1 F), confirming that the endosomal localization of Ear1-mNeonGreen, is maintained due to its recycling by the retromer.
Figure 3.
Two separate sequences are also important for Ear1 retrieval. (A, C, and E) Schematic of Ear1-mNeonGreen mutational analysis and the percentage of each category of Ear1-mNeonGreen mutant localization in rsp5(G747E) mutants from B and Fig. S1 G (A), from D and Fig. S1 H (C), or from F, G, and Fig. S1, I and J (E). vac., vacuole. (B, D, F, and G) Ear1-mNeonGreen localization in rsp5 (G747E) mutants. FL, full-length. (H) Ear1-pHluorin localization in cells expressing WT Rsp5. (J) Vps10ΔC-GFP and Vps10ΔC-Ear1C-tail-GFP localization. (I) Weblogo residue conservation of 453-PPGFEF-458 and 473-INL-475 of Ear1 among 37 Ear1 homologues from related species. (K) Schematic of Ear1. For all quantification shown in this figure, at least 30 cells were classified and the data were obtained from three independent experiments. Error bars represent SD. Scale bars: 2 µm.
For mutational analysis of Ear1, we constructed a series of C-terminally truncated Ear1-mNeonGreen mutants and expressed them in the rsp5(G747E) strain. Ear1 Δ511-550 and Δ491-550 mutants localized to the endosome, whereas the Δ471-550 mutant accumulated on the vacuole membrane, even though this mutant still contains the previously identified Ear1 recycling signal (456-FEF-458; Fig. 3, A and B; and Fig. S1 G). The Δ451-550 mutant lacking 456-FEF-458 also localized to the vacuole membrane. Next, we generated a series of Ear1 mutants by replacing five consecutive amino acids with alanines in the region 452–486 and examined their localization. Among these, the 452-456A, 457-461A, and 472-476A mutants were blocked on the vacuole membrane (Fig. 3, C and D; and Fig. S1 H). Furthermore, mutating single residues to alanine in the region 452–461 revealed that, in addition to the F456A and F458A mutations, the P453A mutant localized to the vacuole membrane (Fig. 3, E and F; and Fig. S1 I), suggesting that the 453-PPGFEF-458 motif serves as the recycling signal in Ear1. Additionally, mutagenesis of the residues in the region 472–476 showed that the I473A and L475A mutants also exhibited vacuole membrane localization (Fig. 3, E and G; and Fig. S1 J), suggesting that 473-INL-475 is another motif required to maintain Ear1 localization to the endosome. To test whether both of these two discontinuous motifs were required for its endosomal localization in cells expressing WT Rsp5, we investigated the localization of Ear1 by using a pH-sensitive green fluorescence protein, pHluorin, whose fluorescence is quenched in the acidic pH of the vacuole lumen (Miesenböck et al., 1998). While WT Ear1-pHluorin localized to the endosome, the F456A and L475A mutants were blocked on the vacuole membrane (Fig. 3 H). Not surprisingly, both these motifs in Ear1 are strictly conserved across multiple fungal species (Fig. 3 I). Finally, to determine whether these two motifs are sufficient for Ear1 retrieval, we fused the C-tail of Ear1 (residues 451–550) to the truncated C-tail of Vps10 (Vps10ΔC). The Vps10ΔC-GFP truncation mutant localized to the vacuole membrane, whereas Vps10ΔC-Ear1C-tail-GFP restored the punctate localization of Vps10 (Fig. 3 J). We conclude that two discontinuous motifs in Ear1, 453-PPGFEF-458 and 473-INL-475, are sufficient for its recycling mediated by the retromer complex (Fig. 3 K). Interestingly, both of these motifs are essential for retromer recognition, unlike Vps10.
Retromer recognizes the endosomal cargo through Vps26 and Vps35
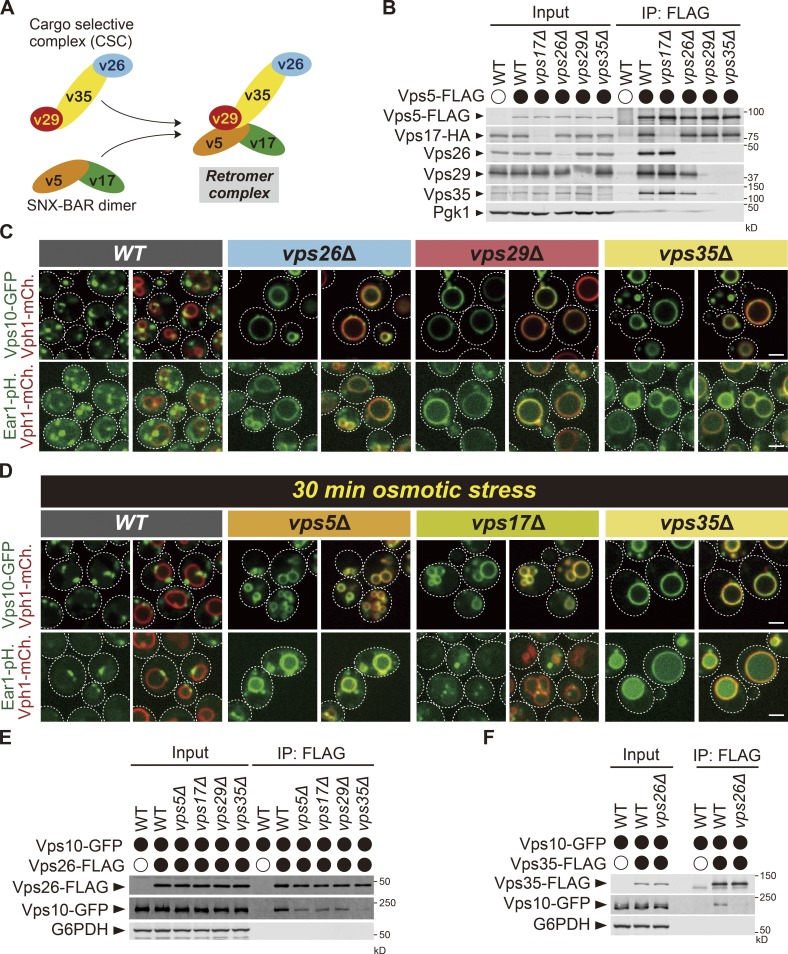
In yeast, retromer is composed of the CSC (Vps26-Vps29-Vps35 complex) and the SNX-BAR dimer (Vps5-Vps17 complex; Seaman et al., 1998; Fig. 4 A). Two models have been proposed for the assembly of the retromer complex (Fig. S2 A). Previous reports show that the CSC interacts with the SNX-BAR dimer through Vps29 and Vps35 (Reddy and Seaman, 2001; Seaman and Williams, 2002; Collins et al., 2005). Indeed, a Vps29 (L252E) mutant fails to interact with the SNX-BAR dimer, although it assembles normally with Vps26 and Vps35 (Collins et al., 2005). However, recent work based on the Cryo-EM structure of the Chaetomium thermophilum Vps5-Vps5-Vps26-Vps29-Vps35 complex by Kovtun et al. (2018) proposed another model where the CSC interacts with the SNX-BAR through Vps26. In the latter model, Vps26 forms the sole contact between the CSC and SNX-BAR dimer. Vps29 and Vps35 have no interaction with the SNX-BAR dimer. To explore this apparent discrepancy, we examined retromer assembly using yeast cells expressing genomically tagged functional Vps5-FLAG and Vps17-HA. In WT cells, Vps17-HA, Vps26, Vps29, and Vps35 coimmunoprecipitated with Vps5-FLAG (Fig. 4 B). Strikingly, we still observed coimmunoprecipitation of Vps29 and Vps35 with Vps5-FLAG in vps26Δ cells. On the other hand, the Vps26 association with Vps5-FLAG was abolished in vps29Δ or vps35Δ cells. These results support the previously published findings (Seaman and Williams, 2002; Collins et al., 2005) and demonstrate that the CSC interacts with the SNX-BAR dimer through Vps29 and Vps35 but not through Vps26. Based on this finding as well as the work by Collins et al. (2005) and others, we conclude that retromer assembly in Saccharomyces cerevisiae and in C. thermophilum may be rather different.
Figure 4.
The retromer complex recognizes the endosomal cargo through Vps26 and Vps35. (A) Model of the retromer complex in yeast. (B) The retromer assembly in retromer subunit mutants. Vps5-FLAG was immunoprecipitated (IP) from cells lacking Vps17, Vps26, Vps29, or Vps35, and interacting Vps17-HA, Vps26, Vps29, and Vps35 were detected by immunoblotting using antibody against FLAG, HA, Vps26, Vps29, Vps35, and Pgk1. (C) The localization of Vps10-GFP or Ear1-pHluorin (Ear1-pH) in WT, vps26Δ, vps29Δ, and vps35Δ cells. (D) The localization of Vps10-GFP or Ear1-pHluorin (Ear1-pH) in WT, vps5Δ, vps17Δ, and vps35Δ cells under osmotic stress. Cells expressing Vps10-GFP grown in YPD media were resuspended in water and incubated for 30 min. (E) The Vps26-Vps10 interaction in retromer subunit mutants. Vps26-FLAG was immunoprecipitated from Vps10-GFP–expressing cells lacking Vps5, Vps17, Vps29, or Vps35, and interacting Vps10-GFP was detected by immunoblotting using antibody against FLAG, GFP, and G6PDH. (F) The Vps35-Vps10 interaction in vps26Δ cells. Vps35-FLAG was immunoprecipitated from WT or vps26Δ cells, and interacting Vps10-GFP was detected by immunoblotting using antibody against FLAG, GFP, and G6PDH. Scale bars: 2 µm.
In mammalian cells, the CSC and the SNX-BAR dimer do not form a stable complex (Harbour and Seaman, 2011; Cullen and Steinberg, 2018). Also, recycling of the mannose-6-phosphate receptor requires the SNX-BAR dimer but not the CSC (Kvainickas et al., 2017; Simonetti et al., 2017). Hence, we reevaluated the requirement of each retromer subunit for the retrieval of yeast retromer cargos. For this purpose, we examined Vps10-GFP or Ear1-pHluorin localization in cells lacking Vps5, Vps17, Vps26, Vps29, or Vps35. In vps26Δ, vps29Δ, and vps35Δ mutants, Vps10-GFP and Ear1-pHluorin accumulated on the vacuole membrane (Fig. 4 C), implying that the CSC is required for cargo recycling in yeast. Since the vacuole morphology is extremely fragmented in vps5Δ or vps17Δ cells (Köhrer and Emr, 1993; Horazdovsky et al., 1997; Nothwehr and Hindes, 1997), it was difficult to determine cargo localization in these mutants (Fig. S2 B). However, the vacuole fragmentation phenotype in vps5Δ or vps17Δ cells was partially rescued under osmotic stress (Fig. 4 D). By using these conditions, we found that Vps10-GFP required both Vps5 and Vps17 for its recycling; however, Ear1-pHluorin required only Vps5 but not Vps17.
Since Vps17 is not required for Ear1 retrieval, we tested if Vps10 and Ear1 are recycled by the same or distinct retromer complexes in WT cells (retromer complex with or without Vps17). For this purpose, we overexpressed Ear1 in Vps10-GFP–expressing cells and examined whether Ear1 competes with Vps10-GFP. In WT cells, overexpressing Ear1 led to missorting of Vps10-GFP to the vacuole membrane (Fig. S2, C and D). When we overexpressed the Ear1 PY motif mutant (Ear1PYmut.), Vps10-GFP still mislocalized to the vacuole membrane, suggesting that Ear1 binding with Rsp5 is dispensable to compete with Vps10-GFP. On the other hand, upon overexpression of the Ear1 recycling–defective mutant (F456A), Vps10-GFP exhibited punctate localization, suggesting that Vps10 and Ear1 are recycled by the same retromer complex in WT cells, although Ear1 is recycled even in the absence of Vps17. Based on these observations, we confirmed that yeast endosomal cargo retrieval requires both the CSC and the SNX-BAR dimer.
To determine which retromer subunits recognize cargos, we examined Vps10–Vps26 interaction in cells lacking Vps5, Vps17, Vps29, or Vps35. Vps10-GFP coprecipitated with Vps26-FLAG in vps5Δ, vps17Δ, and vps29Δ cells, but not in vps35Δ cells (Fig. 4 E). To test whether Vps26 is also required, we examined the Vps10–Vps35 interaction in vps26Δ cells. Vps10-GFP coprecipitated with Vps35-FLAG but not in vps26Δ cells (Fig. 4 F). We also asked whether the SNX-BAR dimer requires the association with the CSC for cargo binding. We analyzed the Vps5-Vps10 interaction in vps29Δ cells. The binding of Vps5-FLAG with Vps10-GFP was abolished in vps29Δ cells (Fig. S2, E and F). From these results, we propose that cargo recognition by the yeast retromer is mediated through Vps26 and Vps35.
Different sites in Vps26 are required for Vps10 or Ear1 recognition
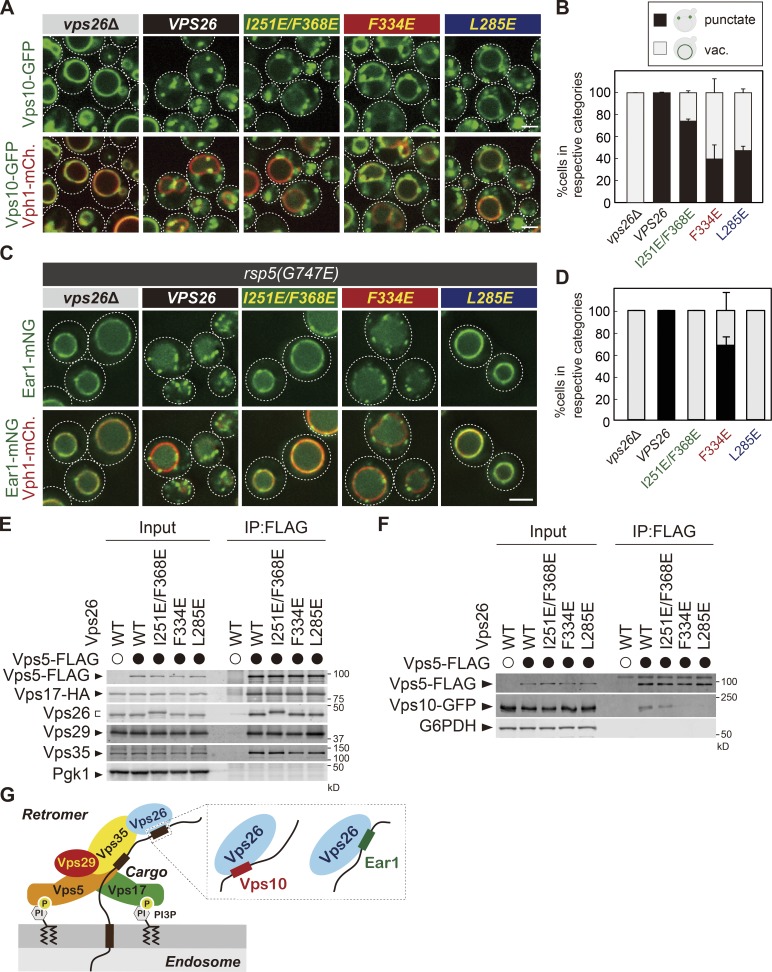
Since the recycling sequences identified in Vps10 and Ear1 are not similar to each other (Fig. S1 A), we wondered how retromer recognizes these different sequences. Our biochemical analysis revealed that both Vps26 and Vps35 are required for cargo recognition. Since the crystal structure of the mammalian Vps26-Vps35-Snx3 complex with its cargo, DMT1-II, is solved (Lucas et al., 2016), we further analyzed cargo recognition by Vps26. In the crystal structure, V168 and F287 residues of human Vps26 (I251 and F368 in yeast Vps26, respectively) recognize the human DMT1-II recycling signal sequence YLL. To test the function of the corresponding residues in yeast, we replaced the corresponding I251 and F368 to Glu in yeast Vps26 (Fig. S3 A) and examined the localization of Vps10-GFP (Fig. 5, A and B) and Ear1-mNeonGreen (Fig. 5, C and D). The I251E/F368E mutant exhibited a defect in Ear1-mNeonGreen recycling but not Vps10-GFP. Since Vps26 is required for Vps10 binding (Fig. 4 F), we mutated multiple conserved surface residues on Vps26 to identify residues required for cargo recognition (Collins et al., 2008) and found two mutants (F334E and L285E) that affect cargo localization. In the F334E mutant, Vps10-GFP localized to the vacuole membrane, whereas Ear1-mNeonGreen retained its punctate localization. The L285E mutant exhibited a defect in the recycling of both Vps10-GFP and Ear1-mNeonGreen. Coimmunoprecipitation experiments confirmed that these Vps26 mutants can still form a stable retromer complex (Fig. 5 E). To address whether these residues are required for cargo recognition, we examined cargo binding of retromer in vps26 mutants and found that while I251/F368E mutants only slightly decrease an affinity for Vps10-GFP, F334E and L285E mutants severely impaired binding (Fig. 5 F). These data indicate that Vps10 recycling requires an interaction with Vps26 residues L285 and F334, whereas Ear1 requires an interaction with I251, L285, and F368. Based on these findings, we conclude that different sites on Vps26 are required for Vps10 or Ear1 recognition and recycling. The residues of Vps26 required for cargo recognition, I251, L285, F334, and F368, are evolutionally conserved from yeast to humans (Fig. S3, B and C), raising the possibility that a similar mechanism is present in higher eukaryotes. Since both Vps10 and Ear1 require L285, they can compete for Vps26 binding (Fig. S2, C and D).
Figure 5.
Different sites in Vps26 are required for Vps10 or Ear1 recognition. (A) The localization of Vps10-GFP in vps26 mutants. (B) The percentage of each category of Vps10-GFP mutant localization from A. vac, vacuole. (C) The localization of Ear1-mNeonGreen (Ear1-mNG) in rsp5(G747E) vps26 mutants. (D) The percentage of each category of Ear1-mNeonGreen mutant localization from C. (E) The retromer assembly in vps26 mutants. Vps5-FLAG was immunoprecipitated (IP) from cells expressing vps26 mutants, and interacting Vps17-HA, Vps26, Vps29, and Vps35 were detected by immunoblotting using antibody against FLAG, HA, Vps26, Vps29, Vps35, and Pgk1. (F) The Vps5-Vps10 interaction in vps26 mutants. Vps5-FLAG was immunoprecipitated from cells expressing vps26 mutants, and interacting Vps10-GFP was detected by immunoblotting using antibody against FLAG, GFP, and G6PDH. (G) The model of retromer cargo recognition. For all quantification shown in this figure, at least 30 cells were classified and the data were obtained from three independent experiments. Error bars represent SD. Scale bars: 2 µm.
A bipartite recycling signal sequence ensures precise cargo recognition by the retromer complex
It had been believed that retromer recognizes its cargo through the consensus sequence defined as ØX[L/M/V] (Cullen and Steinberg, 2018). However, since almost all proteins coded in the yeast genome have this sequence, how the retromer complex selectively recognizes its cargo remained elusive. In the present study, we reveal that in addition to the previously defined recycling sequence, a second recognition motif is also important for cargo retrieval. These two sequences form a bipartite recycling signal recognized by the retromer subunits Vps26 and Vps35 (Fig. 5 G). Thus, retromer interacts with its cargo more specifically than previously thought, which explains the selectivity of cargo recognition by the retromer.
In this study, we demonstrate that a bipartite recycling signal is recognized by Vps26 and Vps35. Strikingly, I251 and F368 on Vps26, which recognizes DMT1-II (Lucas et al., 2016), are required for Ear1 recycling but dispensable for Vps10 recycling. Alternatively, Vps10 requires F334 on Vps26. Thus, the retromer complex has multiple cargo binding sites for interacting with different cargos. Interestingly, F334 on Vps26 locates close to Vps35 (Fig. S3 D). Vps10 might interact with Vps26 and Vps35, coincidently. There are several questions to be answered. (1) How many cargo binding sites does the retromer have? (2) How does Vps35 interact with the cargo? (3) Which sequences are recognized by each binding pocket? Further studies will be necessary to address these questions.
Materials and methods
Yeast strains and plasmids
S. cerevisiae strains used in this study are listed in Table S1. Plasmids used in this study are listed in Table S2. Standard protocols were used for yeast manipulation (Kaiser et al., 1994). Cells were cultured at 30°C to midlog phase in YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] bacto peptone, and 2% [wt/vol] glucose) or YNB medium (0.17% [wt/vol] yeast nitrogen base without amino acids and ammonium sulfate, 0.5% [wt/vol] ammonium sulfate, and 2% [wt/vol] glucose) supplemented with the appropriate nutrients.
CPY sorting assay
Cells were grown in YPD medium for 3 h at 30°C and separated to intracellular and extracellular fractions by centrifugation. The fractions were mixed with trichloroacetic acid at a final concentration of 15%, and the mixtures were incubated for 30 min at 4°C. After centrifugation at 17,400 g for 10 min at 4°C, the fractions were washed once with 100% acetone and then lysed in SDS-PAGE sample buffer (75 mM Tris-HCl, pH 7.5, 2% [wt/vol] SDS, 10% glycerol, 100 mM DTT, and bromophenol blue) by beading with 0.5-mm zirconia beads (Yasui Kikai) for 15 min. The lysates then were heated at 98°C for 5 min. After centrifugation at 10,000 g for 1 min at room temperature, supernatants were analyzed by SDS-PAGE and immunoblotted using anti-CPY and anti-Pgk1.
Antibodies
For immunoblotting, mouse monoclonal anti-FLAG (clone 1E6; Wako), mouse monoclonal anti-HA (12CA5; Roche), rabbit polyclonal anti-Vps26 (Reddy and Seaman, 2001), rabbit polyclonal anti-Vps29 (Seaman et al., 1998), rabbit polyclonal anti-Vps35 (Seaman et al., 1998), rabbit polyclonal anti-G6PDH (Sigma-Aldrich), rabbit polyclonal anti-CPY (Klionsky et al., 1988), and mouse monoclonal anti-Pgk1 (459250; Invitrogen) were used at dilution factors of 1:500, 1:1,000, 1:2,000, 1:5,000, 1:5,000, 1:20,000, 1:5,000, and 1:10,000 respectively.
Immunoprecipitation
Anti-FLAG–conjugated magnetic beads were prepared according to the manufacturer’s protocol. In brief, N-hydroxysuccinimide ester ferrite-glycidyl methacrylate beads (Tamagawa Seiki) were treated with methanol and then incubated with anti-DYKDDDK antibody (Wako) at 4°C for 30 min. The magnetic beads were mixed with 1.0 M 2-aminoethanol, pH 8.0, at 4°C for 16–20 h to quench the conjugation reaction, washed three times with the bead wash buffer (10 mM Hepes-NaOH, pH 7.2, 50 mM KCl, 1 mM EDTA, and 10% glycerol), and stored in wash buffer containing 1 mg/ml BSA (A7030; Sigma-Aldrich).
To examine the retromer complex formation, cells expressing Vps5-FLAG and Vps17-HA grown to mid-log phase were washed twice with the wash buffer (20 mM Hepes-KOH, pH 7.2, 0.2 M sorbitol, 50 mM AcOK, and 2 mM EDTA) and harvested. The cells were lysed in IP buffer (20 mM Hepes-KOH, pH 7.2, 0.2 M sorbitol, 50 mM AcOK, 2 mM EDTA, and 1× protease inhibitor cocktail [Roche]) by beating with 0.5-mm zirconia beads (Yasui Kikai) for 1 min. IP buffer containing 1.0% Triton X-100 was added to the lysate (final concentration of 0.5%), and the samples were rotated at 4°C for 10 min. The solubilized lysates were cleared at 500 g for 5 min at 4°C, and the resultant supernatants were subjected to high-speed centrifugation at 17,400 g for 10 min. The cleared supernatants were incubated with preequilibrated anti-FLAG–conjugated magnetic beads and rotated at 4°C for 2 h. After the beads were washed with wash buffer containing 0.5% Triton X-100, the bound proteins were eluted by incubating the beads in SDS-PAGE sample buffer at 98°C for 5 min.
To examine the cargo–retromer interaction, cells expressing Vps5-FLAG, Vps26-FLAG, or Vps35-FLAG grown to mid-log phase were washed twice with the wash buffer and harvested. The cells were lysed in IP buffer by beating with 0.5-mm zirconia beads for 2 min. IP buffer containing 2.0% saponin was added to the lysate (final concentration of 1.0%), and the samples were rotated at 4°C for 60 min. The solubilized lysates were cleared at 500 g for 5 min at 4°C, and the resultant supernatants were subjected to high-speed centrifugation at 17,400 g for 10 min. The cleared supernatants were incubated with preequilibrated anti-FLAG–conjugated magnetic beads and rotated at 4°C for 4 h. After the beads were washed with wash buffer containing 1.0% saponin, the bound proteins were eluted by incubating the beads in elution buffer (0.1 M glycine-HCl, pH 3.0, and 1% Triton X-100) at 4°C for 30 min. Eluted samples was mixed with 2× SDS-PAGE sample buffer, then incubated at 42°C for 5 min.
Fluorescence microscopy
Fluorescence microscopy was performed using a CSU-X spinning-disk confocal microscopy system (Intelligent Imaging Innovations) or a DeltaVision Elite system (GE Healthcare Life system). The CSU-X spinning-disk confocal microscopy system was equipped with a DMI 6000B microscope (Leica), 100×/1.45-NA objective, and a QuantEM electron-multiplying charge-coupled device camera (Photometrics). Imaging was done at room temperature in YNB medium using GFP and mCherry channels with different exposure times according to the fluorescence intensity of each protein. Images were analyzed and processed with SlideBook 6.0 software (Intelligent Imaging Innovations).
The DeltaVision Elite system is equipped with an Olympus IX-71 inverted microscope, DV Elite complementary metal-oxide semiconductor camera, a 100×/1.4-NA oil objective, and a DV Light SSI 7 Color illumination system with Live Cell Speed Option with DV Elite filter sets. Imaging was done at room temperature in YNB medium using GFP and mCherry channels with different exposure times according to the fluorescence intensity of each protein. Image acquisition and deconvolution (conservative setting; five cycles) were performed using DeltaVision software softWoRx 6.5.2 (Applied Precision).
Quantitative analysis of Vps10-GFP and Ear1-mNeonGreen localizations
The Vps10-GFP or Ear1-mNeonGreen localizations were classified in two categories: punctate structures and vacuole membrane localization. Cells having both punctate structures and vacuole membrane localization were classified in the vacuole membrane localization category. For each experiment, at least 30 cells were classified and the data were obtained from three independent experiments. Error bars were obtained from three individual experiments. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows the localization of Vps10-GFP and Ear1-mNeonGreen mutants. Fig. S2 shows analysis of cargo recycling in retromer mutants. Fig. S3 shows analysis of cargo recognition by Vps26. Table S1 lists S. cerevisiae strains used in this study. Table S2 lists plasmids used in this study.
Supplementary Material
Acknowledgments
We appreciate Richa Sardana and Matthew G. Baile for critical reading of the manuscript. We also thank all Emr laboratory members for helpful discussions.
S.W. Suzuki is supported by Japan Society for the Promotion of Science Research Fellowship for Research Abroad. This work was supported by a Cornell University research grant (CU563704) to S.D. Emr.
The authors declare no competing financial interests.
Author contributions: S.W. Suzuki and Y.-S. Chuang contributed equally and either has the right to put himself first in bibliographic documents. S.W. Suzuki designed and performed experiments and wrote the manuscript. Y.-S. Chuang, M. Li, and M.N.J. Seaman performed experiments. S.D. Emr designed experiments and wrote the manuscript.
References
- Bean B.D., Davey M., and Conibear E.. 2017. Cargo selectivity of yeast sorting nexins. Traffic. 18:110–122. 10.1111/tra.12459 [DOI] [PubMed] [Google Scholar]
- Burda P., Padilla S.M., Sarkar S., and Emr S.D.. 2002. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 115:3889–3900. 10.1242/jcs.00090 [DOI] [PubMed] [Google Scholar]
- Cereghino J.L., Marcusson E.G., and Emr S.D.. 1995. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol. Biol. Cell. 6:1089–1102. 10.1091/mbc.6.9.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B.M., Skinner C.F., Watson P.J., Seaman M.N., and Owen D.J.. 2005. Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nat. Struct. Mol. Biol. 12:594–602. 10.1038/nsmb954 [DOI] [PubMed] [Google Scholar]
- Collins B.M., Norwood S.J., Kerr M.C., Mahony D., Seaman M.N., Teasdale R.D., and Owen D.J.. 2008. Structure of Vps26B and mapping of its interaction with the retromer protein complex. Traffic. 9:366–379. 10.1111/j.1600-0854.2007.00688.x [DOI] [PubMed] [Google Scholar]
- Cooper A.A., and Stevens T.H.. 1996. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J. Cell Biol. 133:529–541. 10.1083/jcb.133.3.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T.Z., Peterson T.A., and Burd C.G.. 2017. A CDC25 family protein phosphatase gates cargo recognition by the Vps26 retromer subunit. eLife. 6:e24126 10.7554/eLife.24126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., and Steinberg F.. 2018. To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19:679–696. 10.1038/s41580-018-0053-7 [DOI] [PubMed] [Google Scholar]
- Finnigan G.C., Cronan G.E., Park H.J., Srinivasan S., Quiocho F.A., and Stevens T.H.. 2012. Sorting of the yeast vacuolar-type, proton-translocating ATPase enzyme complex (V-ATPase): identification of a necessary and sufficient Golgi/endosomal retention signal in Stv1p. J. Biol. Chem. 287:19487–19500. 10.1074/jbc.M112.343814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsel I., Ragaz C., Hoffmann C., Harrison C.F., Weber S., van Rahden V.A., Johannes L., and Hilbi H.. 2013. The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe. 14:38–50. 10.1016/j.chom.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Fisk H.A., and Yaffe M.P.. 1999. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145:1199–1208. 10.1083/jcb.145.6.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjorback A.W., Seaman M., Gustafsen C., Mehmedbasic A., Gokool S., Wu C., Militz D., Schmidt V., Madsen P., Nyengaard J.R., et al. . 2012. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J. Neurosci. 32:1467–1480. 10.1523/JNEUROSCI.2272-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follett J., Norwood S.J., Hamilton N.A., Mohan M., Kovtun O., Tay S., Zhe Y., Wood S.A., Mellick G.D., Silburn P.A., et al. . 2014. The Vps35 D620N mutation linked to Parkinson’s disease disrupts the cargo sorting function of retromer. Traffic. 15:230–244. 10.1111/tra.12136 [DOI] [PubMed] [Google Scholar]
- Harbour M.E., and Seaman M.N.. 2011. Evolutionary variations of VPS29, and their implications for the heteropentameric model of retromer. Commun. Integr. Biol. 4:619–622. 10.4161/cib.16855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky B.F., Davies B.A., Seaman M.N., McLaughlin S.A., Yoon S., and Emr S.D.. 1997. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol. Biol. Cell. 8:1529–1541. 10.1091/mbc.8.8.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., and Mitchell A.. 1994. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Lab Press, Cold Spring Harbor, NY. [Google Scholar]
- Klionsky D.J., Banta L.M., and Emr S.D.. 1988. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol. Cell. Biol. 8:2105–2116. 10.1128/MCB.8.5.2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer K., and Emr S.D.. 1993. The yeast VPS17 gene encodes a membrane-associated protein required for the sorting of soluble vacuolar hydrolases. J. Biol. Chem. 268:559–569. [PubMed] [Google Scholar]
- Kovtun O., Leneva N., Bykov Y.S., Ariotti N., Teasdale R.D., Schaffer M., Engel B.D., Owen D.J., Briggs J.A.G., and Collins B.M.. 2018. Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature. 561:561–564. 10.1038/s41586-018-0526-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvainickas A., Jimenez-Orgaz A., Nägele H., Hu Z., Dengjel J., and Steinberg F.. 2017. Cargo-selective SNX-BAR proteins mediate retromer trimer independent retrograde transport. J. Cell Biol. 216:3677–3693. 10.1083/jcb.201702137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S., Erpapazoglou Z., and Haguenauer-Tsapis R.. 2008. Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol. Biol. Cell. 19:2379–2388. 10.1091/mbc.e08-01-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M., Gershlick D.C., Vidaurrazaga A., Rojas A.L., Bonifacino J.S., and Hierro A.. 2016. Structural Mechanism for Cargo Recognition by the Retromer Complex. Cell. 167:1623–1635.e14. 10.1016/j.cell.2016.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcusson E.G., Horazdovsky B.F., Cereghino J.L., Gharakhanian E., and Emr S.D.. 1994. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 77:579–586. 10.1016/0092-8674(94)90219-4 [DOI] [PubMed] [Google Scholar]
- McGough I.J., Steinberg F., Jia D., Barbuti P.A., McMillan K.J., Heesom K.J., Whone A.L., Caldwell M.A., Billadeau D.D., Rosen M.K., and Cullen P.J.. 2014. Retromer binding to FAM21 and the WASH complex is perturbed by the Parkinson disease-linked VPS35(D620N) mutation. Curr. Biol. 24:1670–1676. 10.1016/j.cub.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan K.J., Korswagen H.C., and Cullen P.J.. 2017. The emerging role of retromer in neuroprotection. Curr. Opin. Cell Biol. 47:72–82. 10.1016/j.ceb.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenböck G., De Angelis D.A., and Rothman J.E.. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 394:192–195. 10.1038/28190 [DOI] [PubMed] [Google Scholar]
- Nothwehr S.F., and Hindes A.E.. 1997. The yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J. Cell Sci. 110:1063–1072. [DOI] [PubMed] [Google Scholar]
- Nothwehr S.F., Roberts C.J., and Stevens T.H.. 1993. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J. Cell Biol. 121:1197–1209. 10.1083/jcb.121.6.1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr S.F., Ha S.A., and Bruinsma P.. 2000. Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J. Cell Biol. 151:297–310. 10.1083/jcb.151.2.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personnic N., Bärlocher K., Finsel I., and Hilbi H.. 2016. Subversion of Retrograde Trafficking by Translocated Pathogen Effectors. Trends Microbiol. 24:450–462. 10.1016/j.tim.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Peter B.J., Kent H.M., Mills I.G., Vallis Y., Butler P.J., Evans P.R., and McMahon H.T.. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 303:495–499. 10.1126/science.1092586 [DOI] [PubMed] [Google Scholar]
- Redding K., Brickner J.H., Marschall L.G., Nichols J.W., and Fuller R.S.. 1996. Allele-specific suppression of a defective trans-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins. Mol. Cell. Biol. 16:6208–6217. 10.1128/MCB.16.11.6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy J.V., and Seaman M.N.. 2001. Vps26p, a component of retromer, directs the interactions of Vps35p in endosome-to-Golgi retrieval. Mol. Biol. Cell. 12:3242–3256. 10.1091/mbc.12.10.3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano-Moreno M., Rojas A.L., Williamson C.D., Gershlick D.C., Lucas M., Isupov M.N., Bonifacino J.S., Machner M.P., and Hierro A.. 2017. Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL. Proc. Natl. Acad. Sci. USA. 114:E11151–E11160. 10.1073/pnas.1715361115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana R., Zhu L., and Emr S.D.. 2019. Rsp5 Ubiquitin ligase-mediated quality control system clears membrane proteins mistargeted to the vacuole membrane. J. Cell Biol. 218:234–250. 10.1083/jcb.201806094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N. 2007. Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J. Cell Sci. 120:2378–2389. 10.1242/jcs.009654 [DOI] [PubMed] [Google Scholar]
- Seaman M.N., and Williams H.P.. 2002. Identification of the functional domains of yeast sorting nexins Vps5p and Vps17p. Mol. Biol. Cell. 13:2826–2840. 10.1091/mbc.02-05-0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N., Marcusson E.G., Cereghino J.L., and Emr S.D.. 1997. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J. Cell Biol. 137:79–92. 10.1083/jcb.137.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N., McCaffery J.M., and Emr S.D.. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665–681. 10.1083/jcb.142.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti B., Danson C.M., Heesom K.J., and Cullen P.J.. 2017. Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR. J. Cell Biol. 216:3695–3712. 10.1083/jcb.201703015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño-Güell C., Wider C., Ross O.A., Dachsel J.C., Kachergus J.M., Lincoln S.J., Soto-Ortolaza A.I., Cobb S.A., Wilhoite G.J., Bacon J.A., et al. . 2011. VPS35 mutations in Parkinson disease. Am. J. Hum. Genet. 89:162–167. 10.1016/j.ajhg.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E., Seaman M.N., Moreau K., Jimenez-Sanchez M., Breusegem S.Y., Harbour M.E., and Rubinsztein D.C.. 2014. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat. Commun. 5:3828 10.1038/ncomms4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A., Benet-Pagès A., Struhal W., Graf E., Eck S.H., Offman M.N., Haubenberger D., Spielberger S., Schulte E.C., Lichtner P., et al. . 2011. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 89:168–175. 10.1016/j.ajhg.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.