Bertero et al. observe that lamin A/C haploinsufficiency in human cardiomyocytes markedly alters electrophysiology, contractility, gene expression, and chromosomal topology. Contrary to expectations, however, changes in chromatin compartments involve just few regions, and most dysregulated genes lie outside these hotspots.
Abstract
Mutations in A-type nuclear lamins cause dilated cardiomyopathy, which is postulated to result from dysregulated gene expression due to changes in chromatin organization into active and inactive compartments. To test this, we performed genome-wide chromosome conformation analyses in human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs) with a haploinsufficient mutation for lamin A/C. Compared with gene-corrected cells, mutant hiPSC-CMs have marked electrophysiological and contractile alterations, with modest gene expression changes. While large-scale changes in chromosomal topology are evident, differences in chromatin compartmentalization are limited to a few hotspots that escape segregation to the nuclear lamina and inactivation during cardiogenesis. These regions exhibit up-regulation of multiple noncardiac genes including CACNA1A, encoding for neuronal P/Q-type calcium channels. Pharmacological inhibition of the resulting current partially mitigates the electrical alterations. However, chromatin compartment changes do not explain most gene expression alterations in mutant hiPSC-CMs. Thus, global errors in chromosomal compartmentation are not the primary pathogenic mechanism in heart failure due to lamin A/C haploinsufficiency.
Introduction
Chromatin organization in the 3D space has emerged as a key layer of mammalian gene expression control. The development of powerful technologies to map chromatin architecture, in particular genome-wide chromosome conformation capture (Hi-C; Lieberman-Aiden et al., 2009), has revealed that such 3D structure is complex, nonrandom, and hierarchical (Yu and Ren, 2017). Short- and long-range intra-chromosomal (cis) DNA interactions are generally constrained within topologically associating domains (TADs; Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012). TADs tend to interact based on their epigenetic status and transcriptional activity, thus dividing chromosomes into two types of large-scale compartments generally called active and inactive (A and B, respectively; Simonis et al., 2006; Lieberman-Aiden et al., 2009; Rao et al., 2014).
Moving beyond the third dimension, there has been growing interest in understanding the functional changes in chromatin organization during development and in disease (Dekker et al., 2017). An emerging body of work has shown that TADs are largely invariant across cell types (Dixon et al., 2012, 2015; Fraser et al., 2015; Schmitt et al., 2016; Won et al., 2016; Bertero et al., 2019). However, these same studies have established that pluripotent stem cell differentiation leads to a substantial degree of A/B compartment reorganization (e.g., ∼20% of the genome), and that this is associated with important developmental changes in gene expression. In the context of disease, disruption of TADs due to copy number variations or point mutations leads to congenital developmental disorders and cancers (Katainen et al., 2015; Lupiáñez et al., 2015; Redin et al., 2017; Sun et al., 2018). In contrast, whether dysregulation of A/B compartments plays a role in functional changes of gene expression leading to human disease is still unclear (Krumm and Duan, 2018).
The nuclear lamina has been proposed as a regulator of chromatin compartmentalization in development and disease (Buchwalter et al., 2019). The lamina lies along the inner nuclear membrane and is a complex mesh of nuclear intermediate filaments (A- and B-type lamins) and lamin-associated proteins. This structure provides key mechanical support to the nucleus, and is an important hub for the control of intracellular signaling (Dobrzynska et al., 2016). Moreover, the nuclear lamina interacts with large chromatin regions, aptly named lamin-associated domains (LADs; van Steensel and Belmont, 2017), which show heterochromatic features such as low gene density, enrichment for repressive histone marks, and poor transcriptional activity (Guelen et al., 2008). Several studies have established that such peripherally located LADs strongly correlate with the B compartment, while the A compartment is predominantly in the nuclear interior (Kind et al., 2015; Luperchio et al., 2017; Stevens et al., 2017; Zheng et al., 2018). Both lamins and lamin-associated proteins, such as lamin B receptor, can directly interact with chromatin (Yuan et al., 1991; Ye and Worman, 1994, 1996; Taniura et al., 1995). Based on this and on the results of loss-of-function studies, it has been proposed that the nuclear lamina tethers LADs at the nuclear periphery (Solovei et al., 2013; Harr et al., 2015; Luperchio et al., 2017). However, the precise role of A- and B-type lamins in chromatin compartmentalization is still being debated (Amendola and van Steensel, 2015; Zheng et al., 2015, 2018; Adriaens et al., 2018).
Elucidating the function of A-type lamins (lamin A and lamin C, which result from alternative splicing of the LMNA gene) is particularly important because of their involvement in human disease. LMNA mutations lead to a wide spectrum of conditions collectively referred to as laminopathies, which most often affect striated muscles (Capell and Collins, 2006; Bertrand et al., 2011). The majority of patients with striated muscle laminopathies develop dilated cardiomyopathy (DCM; Captur et al., 2018), and mutations in LMNA are among the most common causes of familial DCM, depending on the ethnicity of the population (Akinrinade et al., 2015; Haas et al., 2015; Tobita et al., 2018). Compared with other types of DCM, LMNA-DCM (which throughout the rest of the article we refer to as “cardiac laminopathy”) is quite atypical and more severe, as it is characterized by early onset of life-threatening cardiac electrical abnormalities such as severe conduction system disease and/or atrial and ventricular arrhythmias (van Rijsingen et al., 2012; Kumar et al., 2016; Hasselberg et al., 2018).
Over the nearly 20 yr since the first report linking mutations in LMNA to human disease (Bonne et al., 1999), three central nonmutually exclusive mechanisms have been hypothesized to underpin the pathogenesis of cardiac laminopathy: (1) impaired nuclear mechanoresistance via the nucleo–cytoplasmic network, or “mechanical hypothesis”; (2) alteration of lamin A/C–controlled intracellular signaling pathways, or “signaling hypothesis”; and (3) dysregulation of heterochromatin organization leading to gene expression alterations, or “chromatin hypothesis” (Worman and Courvalin, 2004; Cattin et al., 2013). While evidence supporting the first two hypotheses has accumulated over the years, and therapies targeting intracellular signaling alterations are being preclinically developed (Cattin et al., 2013; Captur et al., 2018), the possible involvement of chromatin dysregulation in cardiac laminopathy is still far from established (Adriaens et al., 2018). Indeed, while there have been reports of changes in the nuclear positioning of selected loci in patients with cardiac laminopathy (Meaburn et al., 2007; Mewborn et al., 2010), the functional consequences of such alterations on the disease pathogenesis are unclear. Moreover, these studies have relied on fibroblasts instead of cardiomyocytes, the primary cell type involved in cardiac laminopathy. Most importantly, to the best of our knowledge, the 3D chromatin organization changes associated with cardiac laminopathy have not yet been tested at a genome-wide level.
To address these limitations, we performed Hi-C and gene expression (RNA sequencing [RNA-seq]) analyses to examine the changes in 3D chromatin architecture induced by a haploinsufficient LMNA mutation in cardiomyocytes derived from human induced pluripotent stem cells (hiPSC-CMs). We hypothesized that decreased expression of A-type lamins would lead to broad functional alterations in A/B compartmentalization, leading to aberrant gene expression. However, our findings indicate that while lamin A/C haploinsufficiency functionally affects selected aspects of 3D chromatin organization in human cardiomyocytes, altered A/B compartmentalization does not represent the primary mechanism directly leading to gene expression changes and disease pathogenesis.
Results
Generation of an in vitro model of cardiac lamin A/C haploinsufficiency
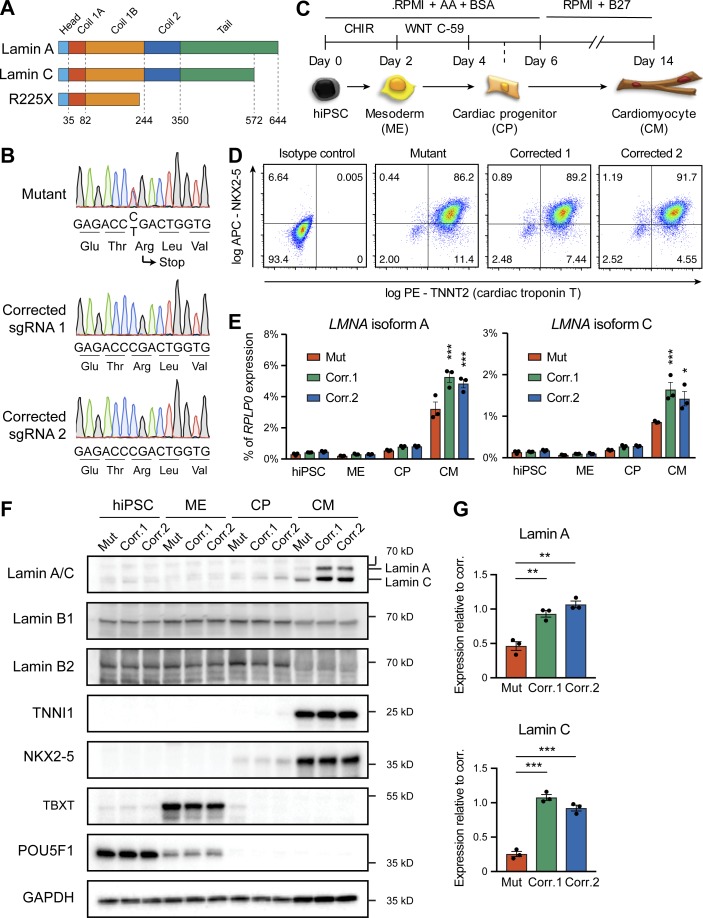
To investigate the role of chromatin dynamics in cardiac laminopathy, we took advantage of hiPSCs bearing a heterozygous nonsense mutation in LMNA predicted to cause premature truncation of both lamin A and lamin C splicing isoforms (c.672C>T, resulting in p.Arg225*, which we will refer to as R225X; Fig. 1 A). This hiPSC line was previously derived from a 56-yr-old male patient who developed severe cardiac conduction disease evolving into heart failure, a condition that segregated within the family with autosomal-dominant inheritance of the R225X mutation (Siu et al., 2012). This same mutation has been reported in multiple other cohorts with similar symptoms (Jakobs et al., 2001; van Tintelen et al., 2007a; Saga et al., 2009), establishing it as a bona fide genetic cause of cardiac laminopathy.
Figure 1.
Generation of lamin A/C haploinsufficient hiPSC-CMs. (A) Predicted effect of the LMNA R225X mutation on the two splicing products lamin A and C. (B) Sanger sequencing of LMNA exon 4 in hiPSCs with heterozygous R225X mutation (top), or in hiPSCs obtained after CRISPR/Cas9-based scarless correction of the mutation (bottom). (C) Schematic of the protocol for step-wise directed differentiation of hiPSC-CMs. CHIR, CHIR-99021; AA, ascorbic acid. (D) Quantification of cardiac differentiation efficiency by flow cytometry for TNNT2 and NKX2-5 on hiPSC-CMs at day 14 of differentiation. (E) RT-qPCR analyses at the indicated stages of hiPSC-CM differentiation (see panel C). Differences versus mutant were calculated by two-way ANOVA with post hoc Holm–Sidak binary comparisons (*, P < 0.05; ***, P < 0.001; n = 3 differentiations; average ± SEM). (F) Representative Western blot for A- and B-type lamins and differentiation markers during iPSC-CM differentiation. (G) Quantification of lamin A/C expression in hiPSC-CMs from Western blot densitometries. Differences versus mutant were calculated by one-way ANOVA with post hoc Holm–Sidak binary comparisons (**, P < 0.01; ***, P < 0.001; n = 3 differentiations; average ± SEM). Throughout the figure (and in all other figures), Mut or Mutant indicates LMNA R225X hiPSCs, and Corr.1/2 or Corrected 1/2 indicates the two isogenic corrected control LMNA R225R hiPSC lines.
Since variability among hiPSC lines can profoundly influence both molecular and cellular phenotypes, we generated isogenic control hiPSCs by correcting the R225X mutation back to the wild-type allele, a strategy currently considered the gold standard to determine the association between genotype and phenotype in hiPSC-CMs (Bellin et al., 2013; Kodo et al., 2016; Mosqueira et al., 2018). By leveraging existing methods (Yusa, 2013), we designed a two-step scarless gene editing strategy relying on CRISPR/Cas9-facilitated homologous recombination of a targeting vector containing the wild-type allele in the 3′ homology arm, and an excisable piggyBac drug resistance cassette (Fig. S1, A and B). To control for the potential variability between hiPSC sublines and for potential off-target mutations, we obtained two isogenic control hiPSCs using distinct single guide RNA (sgRNA) sequences (Fig. 1 B; and Fig. S1, C and D). The resulting hiPSCs, which we subjected to extensive genomic quality control (Fig. S1, E–G; and Table S1), will be referred to as Corrected 1 and Corrected 2 (Corr.1 and Corr.2, respectively).
We differentiated hiPSCs into hiPSC-CMs using an established protocol based on the temporal modulation of WNT signaling using small molecules (Burridge et al., 2014; Bertero et al., 2019; Fig. 1 C). Both mutant and corrected hiPSCs could be differentiated with high efficiency, as measured by flow cytometry for TNNT2 (Fig. 1 D, Video 1, and Video 2; 91.6% ± 3.2%, 94.4% ± 2.5%, and 91.3% ± 3.1% for Mutant, Corr.1, and Corr.2, respectively; average ± SEM of n = 4 independent differentiations). Western blot, quantitative RT-PCR (RT-qPCR), flow cytometry, and immunofluorescence analyses confirmed that all hiPSC lines underwent the expected developmental progression through mesoderm and cardiac progenitors before reaching a cardiomyocyte phenotype (Fig. 1 F and Fig. S2, A–F). hiPSC-CMs coexpressed NKX2-5 and TNNT2 (Fig. 1 D), excluding sinoatrial node fate (Birket et al., 2015; Protze et al., 2017). Furthermore, hiPSC-CMs from all lines showed comparable expression of the mature ventricular marker cardiac troponin I (Fig. S2, G and H). Interestingly, mutant hiPSC-CMs presented well-organized sarcomeres and did not show any obvious sign of nuclear blebbing or invagination, indicating that such abnormal phenotypes often observed in patients may develop only after long-term stress in vivo (Fig. S2, F and H; also see Fig. S3 D described below). Collectively, the R225X lamin A/C mutation does not substantially delay or otherwise impair cardiac differentiation from hiPSCs.
Lamin A/C was up-regulated specifically in hiPSC-CMs, and mutant lines showed significantly reduced levels of both mRNA and protein compared with both corrected controls (Fig. 1, E–G). Of note, no detectable protein truncation was detected by Western blot (Fig. S3 A), and lamin A/C expression in mutant hiPSC-CMs also proved to be reduced when compared with cardiomyocytes generated from an unrelated hiPSC line derived from a healthy individual (Fig. S3 B). Finally, other minor LMNA isoforms were nearly undetectable by RT-qPCR in both control and mutant hiPSC-CMs, excluding possible compensatory mechanisms (Fig. S3 C). These gene expression data agree with earlier findings from analysis of skin fibroblasts bearing the R225X heterozygous mutation (Siu et al., 2012), and indicate that such premature nonsense mutation leads to lamin A/C haploinsufficiency, presumably due to nonsense-mediated decay of both the lamin A and lamin C transcripts. On the other hand, B-type lamins were expressed at comparable levels and localized normally in mutant and corrected hiPSC-CMs (Fig. 1 F and Fig. S3, D and E), excluding compensatory mechanisms due to the reduction of A-type lamins. Collectively, we established a robust in vitro model to study cardiac laminopathy due to lamin A/C haploinsufficiency in developing cardiomyocytes.
Lamin A/C haploinsufficiency alters hiPSC-CM automaticity and prolongs membrane depolarization
Before exploring the effect of lamin A/C haploinsufficiency on chromatin dynamics, we wished to confirm a phenotypic effect on cardiac physiology in developing hiPSC-CMs. Since electrical abnormalities are the primary and most characteristic manifestations of cardiac laminopathy (van Rijsingen et al., 2012; Kumar et al., 2016; Hasselberg et al., 2018), we began by assessing the electrophysiological properties of mutant hiPSC-CMs. For this, we first used multi-electrode arrays (MEAs) to measure the extracellular electric field potential elicited by spontaneously contracting monolayers of hiPSC-CMs. Mutant cells showed a number of alterations compared with both corrected controls. Most notably, the beat rate proved highly erratic and prone to prolonged pauses (Fig. 2 A). Moreover, even when the analysis was focused on the periods showing the highest consistency, the beat rate was still more irregular and reduced (Fig. 2 B). The amplitude of field potential changes was elevated more than twofold compared with controls, which is indicative of stronger depolarizing ion currents (Fig. 2 B). Along the same lines, the field potential duration (FPD), which indicates the interval between depolarization and repolarization, was prolonged by ∼60% (Fig. 2 B). This finding held true even when the FPD was corrected for the beat period (FPDc; Fig. 2 B), an established way to robustly compare the FPD across multiple conditions (Asakura et al., 2015; Rast et al., 2016). Of note, despite all of these alterations, the conduction velocity across the monolayer was not affected (Fig. 2 B), indicating that intercellullar electrical coupling was preserved in mutant cells.
Figure 2.
Electrophysiological properties of lamin A/C haploinsufficient hiPSC-CMs. (A) Representative traces from MEA recordings of spontaneous electrical activity in hiPSC-CM monolayers. (B) Representative quantifications of electrophysiological properties from MEA analyses. Differences versus mutant were calculated by one-way ANOVA with post hoc Holm–Sidak binary comparisons (**, P < 0.01; ***, P < 0.001; n = 5–16 wells; average ± SEM). (C) Representative voltage recordings by whole-cell patch clamp during evoked action potentials in individual hiPSC-CMs. (D) Quantifications of electrophysiological properties from whole-cell patch clamp analyses (APD90, action potential duration at 90% repolarization). Differences versus mutant were calculated by one-way ANOVA with post hoc Holm–Sidak binary comparisons (*, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 26–30 cells from two differentiations; average ± SEM). (E) Representative optical recordings of calcium fluxes with Fluo-4 in hiPSC-CM monolayers electrically paced at 1 Hz (Fmax, peak fluorescence; F0, baseline fluorescence). (F) Representative quantifications of calcium fluxes properties. Differences versus mutant were calculated by unpaired t test (***, P < 0.001; n = 69–70 cells; average ± SEM).
We also performed whole-cell patch clamp recordings of voltage changes occurring during action potentials firing in individual cardiomyocytes. While a number of parameters were unaffected in mutant cells (such as maximum diastolic potential, action potential amplitude, mean diastolic potential, and repolarization amplitude; Fig. 2 D and data not shown), the action potential duration and exponential time constant for the action potential decay were increased (Fig. 2, C and D). This observation provided a cell-autonomous explanation for the increased FPDc in cell monolayers.
Finally, we measured the effects of excitation abnormalities in mutant cells on their intracellular calcium dynamics using the fluorescent calcium reporter Fluo-4. Electrically paced mutant cell monolayers showed stronger and longer calcium peaks (Fig. 2, E and F; Video 3; and Video 4), in agreement with MEA and patch clamp data. Overall, these findings indicate that lamin A/C haploinsufficiency in developing cardiomyocytes leads to altered automaticity and prolonged membrane depolarization, leading to more robust calcium transients.
Systolic hyperfunction and diastolic dysfunction in lamin A/C haploinsufficient hiPSC-CMs
Since changes in intracellular calcium concentrations are the primary determinant of hiPSC-CM contractility, we examined the effect of lamin A/C haploinsufficiency on this process. First, we measured the cellular displacement associated with cardiac contraction in electrically paced hiPSC-CM monolayers. Mutant cells showed ∼50% stronger contractions and a delayed relaxation time (Fig. 3, A and B; Video 5; and Video 6), consistent with their stronger and prolonged calcium fluxes.
Figure 3.
Contractile properties of lamin A/C haploinsufficient hiPSC-CMs. (A) Representative measurements of cellular displacement during contraction of hiPSC-CM monolayers electrically paced at 1 Hz. (B) Representative quantifications of cell contractility from analyses of optical recordings. Differences versus mutant were calculated by one-way ANOVA with post hoc Holm–Sidak binary comparisons (*, P < 0.05; n = 5–6 FOVs; average ± SEM). (C) Representative measurements of twitch force during contraction of 3D-EHTs electrically paced at 1 Hz. (D) Representative quantifications of tissue contractility from analyses of optical recordings. Differences versus mutant were calculated by one-way ANOVA with post hoc Holm–Sidak binary comparisons (*, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 4–5 3D-EHTs; average ± SEM).
To validate these findings in a more physiologically relevant model, we performed biomechanical assessments of contractility in electrically paced 3D engineered heart tissues (3D-EHTs), an established model to promote cardiac maturation by providing topological, mechanical, and multicellular cues (Ruan et al., 2016; Leonard et al., 2018). In agreement with observations in 2D monolayers, mutant 3D-EHTs showed approximately twofold stronger and prolonged contractions with a markedly impaired relaxation kinetic (Fig. 3, C and D; Video 7; and Video 8). Collectively, we concluded that lamin A/C haploinsufficiency leads to systolic hyperfunction and diastolic dysfunction in both early and maturing cardiomyocytes.
Lamin A/C haploinsufficiency causes dysregulation of specific ion channel genes and moderate gene expression changes
To explore the molecular mechanisms that might explain the electrophysiological and contractile phenotypes observed in lamin A/C haploinsufficient hiPSC-CMs, we first monitored the expression of the genes encoding key ion-handling proteins involved in the generation of action potentials and in excitation–contraction coupling (Amin et al., 2010; Eisner et al., 2017). RT-qPCR analyses excluded a dysregulation of most such genes (Fig. S3 F). However, mutant cells showed a significant up-regulation of CACNA1C and down-regulation of KCNQ1 (Fig. 4 A). CACNA1C encodes for the pore-forming subunit of L-type calcium channels, which mediate ICa,L, the main source of inward depolarizing current during phase 2 (plateau) of the action potential (Bodi et al., 2005). On the other hand, IKS, the outward potassium current resulting from the potassium channel encoded by KCNQ1, antagonizes ICa,L in phase 2 by initiating membrane repolarization (Peroz et al., 2008). Thus, the combined effects of CACNA1C up-regulation and KCNQ1 down-regulation could explain the prolonged membrane depolarization observed in mutant cells.
Figure 4.
Gene expression changes in lamin A/C haploinsufficient hiPSC-CMs. (A) RT-qPCR analyses in hiPSC-CMs at day 14 of differentiation. Differences versus mutant were calculated by one-way ANOVA with post hoc Holm–Sidak binary comparisons (*, P < 0.05; n = 3 differentiations; average ± SEM). (B) Hierarchical clustering of hiPSC-CMs analyzed by RNA-seq based on all expressed genes. r, replicate. (C) Overlap in genes up- or down-regulated in mutant hiPSC-CMs versus hiPSC-CMs from the two corrected control lines (change greater than twofold and q-value <0.05; Table S2). (D) Selected results from gene ontology (GO) and pathway enrichment analyses of genes consistently up- or down-regulation in mutant hiPSC-CMs. KEGG, Kyoto Encyclopedia of Genes and Genomes. (E) Linear dimensionality reduction by principal component (PC) analysis of RNA-seq data of mutant and corrected hiPSC-CMs, and hESC-CMs sampled at different time points of differentiation.
We then expanded these gene expression analyses genome-wide by performing RNA-seq (Table S2). While hiPSC-CMs from the two corrected control lines clustered closely and showed remarkably similar gene expression profiles (Fig. 4 B and Table S2), mutant cells clustered separately due to substantial gene up- and down-regulation (Fig. 4, B and C; and Table S2). To increase the robustness of our subsequent analyses, we only considered genes as dysregulated if they had significant expression changes in mutant hiPSC-CMs versus both corrected controls (185 up-regulated and 140 down-regulated with fold-change >2 and q value <0.05; Fig. 4 C and Table S2). Ontology and pathway enrichment analyses on these gene lists revealed that up-regulation in mutant cells was significantly associated with (1) focal adhesion, MAPK, and TGFβ pathways; (2) transcriptional activation, positive regulation of cardiac differentiation, and inhibition of apoptosis; and (3) noncardiac lineage expression (such as genes preferentially expressed in fibroblast and smooth muscle; Fig. 4 D and Table S3). In contrast, down-regulated genes were enriched in cardiac transcripts and factors involved in cardiomyopathy and phosphoinositide 3-kinase pathways (Fig. 4 D and Table S3). We also compared these RNA-seq data with those we recently obtained from human embryonic stem cells (hESCs) profiled at different time points of cardiac differentiation (Bertero et al., 2019). Linear dimensionality reduction by principal component analysis showed that both mutant and corrected hiPSC-CMs clustered closely to hESC-derived cardiomyocytes (hESC-CMs; Fig. 4 E). This indicated that despite dysregulation of >300 genes and of multiple signaling pathways, mutant hiPSC-CMs were not globally developmentally delayed from a transcriptional standpoint.
Lamin A/C haploinsufficiency strengthens the separation between chromosome territories and between chromatin compartments
Having established that lamin A/C haploinsufficiency results in clear changes in both cellular physiology and gene expression in developing hiPSC-CMs, we tested whether some of these phenotypes could be explained by changes in chromatin topology. To explore this aspect at a genome-wide level, we took advantage of in situ DNase Hi-C to capture all pairwise interactions between any two genomic regions (Ramani et al., 2016). Hi-C data from mutant and corrected hiPSC-CMs were of high quality and reproducible across biological replicates, with samples clustering separately based on their genotype (Fig. 5 A and Table S4). We then began exploring the global properties of chromatin topology and noticed that mutant cells showed an increased ratio of genomic interactions within the same chromosome (in cis) over those involving different chromosomes (in trans; Fig. 5 B). Moreover, trans interactions were distributed differently: while all hiPSC-CMs showed the expected pattern of preferential self-association between small, gene-rich chromosomes (especially chromosomes 16, 17, 19, 20, and 22) and between large, gene-poor chromosomes (such as chromosomes 1–8; Fig. S4 A; Lieberman-Aiden et al., 2009), this property was even more striking in mutant cells (Fig. 5 C). On the contrary, interactions between small and large chromosomes were less frequent (Fig. 5 C). Thus, lamin A/C haploinsufficiency reinforces the separation between chromosome territories and their segregation based on size and gene density.
Figure 5.
Global properties of chromatin topology in lamin A/C haploinsufficient hiPSC-CMs. (A) Hierarchical clustering of hiPSC-CMs analyzed by in situ DNase Hi-C based on similarity scores between the genomic contact matrices calculated with HiCRep. (B) Proportion of trans or cis interactions involving distances <20 kb (cis short) or >20 kb (cis long; Table S4). Differences versus mutant were calculated by two-way ANOVA with post hoc Holm–Sidak binary comparisons (*, P < 0.05; n = 2 differentiations; average ± SEM). (C) Representative heatmaps of differential contact matrices between autosomes (ordered by size). (D) Representative heatmaps of differential cis interactions between A and B compartments. (E) Probability of cis genomic contacts over increasing genomic distance for regions in homotypic (A-A or B-B) or heterotypic (A-B) chromatin compartments (gray background: 95% confidence bands). obs/exp, observed/expected.
We then analyzed the separation of chromatin domains into the A or B compartment. For this, we computed the first principal component (PC1) from the contact matrix using bins of 500 kb (Table S5), a well-established method to determine chromatin compartments based on their preferential association (Lieberman-Aiden et al., 2009; Imakaev et al., 2012). In agreement with our previous observations in hESC-CMs (Bertero et al., 2019), trans interactions between chromatin domains favored A-A compartments, and this was not affected by lamin A/C haploinsufficiency (Fig. S4 B). In cis, mutant cells showed stronger interactions between A-A compartments (Fig. 5 D and Fig. S4 B), particularly due to increased short-range contacts within 0.5–1 Mb (Fig. 5 E). In contrast, interactions between heterotypic regions (A-B) were reduced in mutant cells (Fig. 5 D and Fig. S4 B), particularly for long-range contacts >10 Mb (Fig. 5 E). Collectively, lamin A/C haploinsufficiency reinforces the separation between the active and inactive chromatin compartments.
Incomplete transitions from the active to inactive chromatin compartment in lamin A/C haploinsufficient hiPSC-CMs
To further assess the effect of lamin A/C haploinsufficiency on chromatin compartmentalization, we identified genomic bins with significantly different A/B compartment scores and switching from active to inactive or vice versa between at least two conditions (Table S5). We noticed that the vast majority of compartment transitions were observed for mutant hiPSC-CMs versus each corrected control (Fig. 6 A), and that B to A inversions were more common than A to B ones (42 and 27, respectively; Table S5). We observed that 63% of A to B transitions in mutant cells involved the X chromosome, while B to A changes showed a notable concentration on chromosome 19 but were otherwise evenly spread across 13 additional chromosomes (Fig. 6 B, Fig. S5 A, and Table S5). Overall, compartment changes involved ∼1.2% of the genome, indicating that chromatin compartment dysregulation in mutant cells is not widespread but is actually highly restricted.
Figure 6.
Chromatin compartment transitions in lamin A/C haploinsufficient hiPSC-CMs. (A) Heatmap of all significantly different A/B compartment scores (Hi-C matrix PC1; P < 0.05 by one-way ANOVA; n = 2 differentiations; Table S5) in 500-kb bins that changed PC1 sign between two or more conditions. Positive and negative PC1 indicate A and B compartmentalization, respectively. (B) Representative log-transformed contact probability maps for chromosome 19. TADs are visible as squares along the diagonal. TADs within the same compartment interact off the diagonal as indicated by the symmetrical checkerboard patterns. Two genomic regions that show different compartmentalization in mutant hiPSC-CMs are indicated by dashed boxes to highlight the differences in contact probabilities with other genomic regions off the diagonal. (C) Linear dimensionality reduction by principal component (PC) analysis of A/B compartment scores from Hi-C data of mutant and corrected hiPSC-CMs, and hESC-CMs sampled at different time points of differentiation. (D) Significance of the overlap between changes in A/B compartments in mutant hiPSC-CMs and those occurring during hESC-CM differentiation. The number of genomic bins within each of the categories is indicated, and P values were calculated by χ2 tests. (E) Representative genomic tracks of chromatin compartmentalization for chromosome 19 and a section of chromosome 5. Positive and negative Hi-C matrix PC1 scores are shown in red and blue, and indicate 500-kb genomic bins in the A and B compartments, respectively. Genomic regions that transition from A to B during hESC-CM differentiation but remain in A in mutant hiPSC-CMs (noted as B to A) are indicated by dashed boxes. Selected genes found within such regions are listed (refer to Fig. 8 and Fig. S5).
To gain insight into the relationship between dysregulated regions and normal chromatin compartment dynamics during cardiomyocyte differentiation, we integrated Hi-C data from mutant and corrected hiPSC-CMs with those we previously generated at different time points throughout hESC-CM differentiation (Bertero et al., 2019). Linear dimensionality reduction of A/B compartment scores for all samples confirmed that mutant hiPSC-CMs cluster separately from both corrected controls (Fig. 6 C). Moreover, this analysis revealed that based on the first principal component (which explained 39% of the variance and ordered hESC samples based on their developmental progression), mutant cells were mildly developmentally delayed from a chromatin compartmentalization standpoint (Fig. 6 C, PC1). Accordingly, we observed a strong and significant enrichment for domains that normally transition from A to B during cardiac differentiation but remain in A in mutant cells (Fig. 6 D). Notable examples of such behavior involved two 0.5- and 2.5-Mb-long portions of chromosome 19 (corresponding to 19p13.13 and 19q13.33, respectively), and a 1-Mb region on chromosome 5 (5q31.3; Fig. 6, B and E). We also observed a weaker enrichment for the opposite dynamic (impaired B to A transitions; Fig. 6 D), but we note that this analysis was limited to autosomes since compartment transitions of the X chromosome could not be assessed in female hESC-CMs due to the confounding factor of X inactivation.
We previously showed that transition from the B to A compartment during cardiogenesis often reflects relocalization of loci from the nuclear periphery to the nuclear interior (Bertero et al., 2019), in agreement with the notion that the B compartment contains the vast majority of LADs (Luperchio et al., 2017). Thus, we speculated that the opposite could also be true, and that lack of transition from the A to B compartment in mutant hiPSC-CMs might reflect impairment of translocation to peripheral LADs. We tested this by combined immunofluorescence for the nuclear lamina and 3D-DNA FISH for three loci contained within genomic locations showing aberrant compartmentalization in mutant hiPSC-CMs: CACNA1A, LRRC4B, and PCDHGB4 (Fig. 6 E). Analysis of hiPSCs versus hiPSC-CMs showed that both CACNA1A and LRRC4B moved closer to the nuclear lamina during differentiation, but that this transition was less marked in mutant hiPSC-CMs (Fig. 7, A and B). Interestingly, PCDHGB4 did not change localization either during differentiation or in mutant hiPSC-CMs (Fig. 7 B), indicating that aberrant compartmentalization of this chromatin region does not result from changes in association to the nuclear lamina, and thus it must reflect some other mechanism. As controls, we selected VAV1 and LGALS14, which transition from the A to B compartment both in mutant and corrected hiPSC-CMs (Fig. 7 C) and are found on the same chromosome arms as CACNA1A and LRRC4B, respectively. These regions demonstrated the expected localization change during differentiation both in mutant and corrected hiPSC-CMs (Fig. 7 B), establishing that the impairment observed for CACNA1A and LRRC4B does not merely result from differentiation-associated changes in the territory of chromosome 19. In sum, lamin A/C haploinsufficiency in developing hiPSC-CMs results in highly selective dysregulation of chromatin compartmentalization, particularly for a handful of genomic hotspots that fail to transition from the active compartment in the nuclear interior to the inactive compartment associated to the nuclear lamina. We will refer to these as lamin A/C–sensitive B domains.
Figure 7.
Alterations in peripheral localization of lamin A/C–sensitive loci. (A) Representative immunoFISH for the nuclear lamina (lamin B1), the cardiac marker α-actinin, and the CACNA1C locus in mutant and corrected hiPSCs and hiPSC-CM (nuclei counterstained with DAPI). Scale bars, 5 µm. (B) Quantification of the distance between the indicated loci and the nuclear lamina in diploid cells based on immunoFISH data. Violin plots report the whole range, and horizontal lines indicate the first quartile, median, and third quartile. Statistical analysis by Brown–Forsythe and Welch ANOVA test followed by the Holm–Sidak multiple comparisons versus hiPSC for the same line or versus mutant, as indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001; n = individual loci, as indicated). (C) As in Fig. 6 E, but reporting chromatin compartmentalization changes for two genomic regions not affected by lamin A/C haploinsufficiency and used as negative control for immunoFISH experiments (B).
Dysregulation of lamin A/C–sensitive B domains leads to ectopic expression of noncardiac genes
We then assessed the functional consequences of compartment dysregulation due to lamin A/C haploinsufficiency. Strikingly, we observed almost no overlap between genes within lamin A/C–sensitive domains and genes significantly and strongly up- or down-regulated in mutant cells (Fig. S5 B). Accordingly, there were no significant changes in the average expression of genes found in lamin A/C–sensitive domains in mutant versus corrected controls (Fig. 8 A). Nevertheless, we noticed a small number of genes located in lamin A/C–sensitive B domains that were expressed at very low levels in corrected hiPSC-CMs and up-regulated in mutant cells (29 genes with an average change greater than twofold; Fig. 8 A and Fig. S5 C). These genes were significantly enriched for three chromosome locations (Fig. 8 B), two of which corresponded to the lamin A/C–sensitive hotspots 5q31.3 and 19q13.33 (Fig. 6 E), and were associated with neuronal development (Fig. 8 B and Table S6).
Figure 8.
Correlation between altered chromatin compartmentalization and gene expression changes in lamin A/C haploinsufficient hiPSC-CMs. (A) Violin plots showing the expression of genes found with lamin A/C–sensitive domains. Boxplots indicate the first quartile, median, and third quartile, while whiskers are from the 5th to 95th percentiles. In the left panel, note that the tail of genes expressed at very low levels in corrected hiPSC-CMs is less pronounced in mutant cells. (B) Selected results from ontology enrichment analyses of up-regulated genes in domains aberrantly found in the A compartment in mutant hiPSC-CMs (average fold-change greater than two; Fig. S5 C). (C and D) RT-qPCR validation of gene expression changes in hiPSC-CMs matured by culture in vitro for 30 d (C) or by generation of 3D-EHTs (D). Differences versus mutant were calculated by one-way ANOVA with post hoc Holm–Sidak binary comparisons (*, P < 0.05; **, P < 0.01; n = 4 differentiations for panel C, and n = 3 3D-EHT batches for panel D; average ± SEM). RPKM, read kilobase per million mapped reads.
Of note, most genes in the group just described had not been determined as differentially expressed based on RNA-seq analysis using Cufflinks, since lowly expressed genes are subject to strong negative penalization when calculating the q score due to the challenges in robustly assessing their expression (Trapnell et al., 2010, 2012). This explains their absence in the lists used for the overlap shown in Fig. S5 B. To increase our confidence with these results, we validated the expression of several genes within this class by RT-qPCR. Moreover, to exclude that such gene expression differences were simply explained by a differentiation delay in mutant hiPSC-CMs, we analyzed their expression during cardiac differentiation as well as in hiPSC-CMs matured either by longer 2D culture or by the generation of 3D-EHTs. Remarkably, nearly all genes tested were not expressed during normal cardiogenesis, and showed strongest up-regulation in mutant hiPSC-CMs and 3D-EHTs (Fig. 8, C and D; and Fig. S5, D–F). These findings confirmed that impaired transition to the B compartment of selected lamin A/C–sensitive domains leads to up-regulation of multiple noncardiac genes that would otherwise be transcriptionally repressed during cardiomyocyte differentiation.
Ectopic P/Q-type and potentiated L-type calcium currents contribute to electrophysiological abnormalities of lamin A/C haploinsufficient hiPSC-CMs
Genes found in lamin A/C–sensitive B domains and up-regulated in mutant hiPSC-CMs included multiple factors with either unknown function, such as the putative uncharacterized protein C19orf81, or with established roles in the nervous system but not normally expressed in the heart (Fig. 8, C and D; and Fig. S5, D–F). This group included genes from all of the three protocadherin clusters on chromosome 5 (α, β, and γ, exemplified by PCDHA10, PCDHB15, and PCHDGB4; Chen and Maniatis, 2013), LRRC4B (encoding for the postsynaptic cell adhesion molecule NGL-3; Maruo et al., 2017), SYT3 (involved in postsynaptic endocytosis; Awasthi et al., 2018), and CACNA1A (which encodes for the pore-forming subunit of neuronal P/Q-type calcium channels; Rajakulendran et al., 2012).
CACNA1A appeared particularly interesting given the prolonged action potential duration observed in mutant hiPSC-CM populations. Indeed, the depolarizing ICa,P and ICa,Q currents resulting from the protein product of CACNA1A are known to be strong and long-lasting, even more so than ICa,L currents typical of hiPSC-CMs (Catterall et al., 2005; Nimmrich and Gross, 2012). Thus, we tested whether ICa,P and ICa,Q currents contributed to the electrophysiological abnormalities of mutant cells by inhibiting such currents using two structurally unrelated, highly specific inhibitors derived from spider venoms: ω-conotoxin MVIIC and ω-agatoxin TK (Adams et al., 1993; McDonough et al., 1996; Nimmrich and Gross, 2012). MEA experiments demonstrated that both toxins led to a modest but significant decrease in the FPDc in monolayers of mutant hiPSC-CMs, while they did not affect their depolarization amplitude (Fig. 9, A and B). In contrast, neither toxin had a significant effect on the FPDc in corrected controls (Fig. 9, A and B), confirming that ICa,P and ICa,Q currents do not play a role in cardiac depolarization in physiological conditions, and establishing that the toxins had no overt nonspecific effects on hiPSC-CM electrophysiology at the doses tested. These experiments indicated that ectopic expression of CACNA1A in mutant cells and the resulting P/Q-type calcium currents contribute to the prolonged depolarization observed in lamin A/C haploinsufficient hiPSC-CMs.
Figure 9.
Role of P/Q- and L-type calcium currents in electrophysiological abnormalities of lamin A/C haploinsufficient hiPSC-CMs. (A and B) Representative quantifications of electrophysiological properties from MEA analyses in standard culture conditions or after treatment with the indicated inhibitors for P/Q-type calcium channels (ω-conotoxin MVIIC: 2 µM; ω-agatoxin TK: 0.5 µM). Differences versus mutant were calculated by one-way ANOVA with post hoc Holm–Sidak binary comparisons (*, P < 0.05; **, P < 0.01; n = 3–8 wells; average ± SEM). (C and D) RT-qPCR validation of gene expression changes in hiPSC-CMs matured by culture in vitro for 30 d (C) or by generation of 3D-EHTs (D). Differences versus mutant were calculated by one-way ANOVA with post hoc Holm–Sidak binary comparisons (*, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 4 differentiations for panel C, and n = 3 3D-EHT batches for panel D; average ± SEM). (E) As in panels A and B, but hiPSC-CMs were treated with increasing doses of the L-type calcium channel blocker verapamil (*, P < 0.05; ***, P < 0.001; n = 1–4 wells; average ± SEM).
As mentioned above, we also observed CACNA1C up-regulation and KCNQ1 down-regulation in mutant hiPSC-CMs at day 14 of differentiation (Fig. 4 A). RT-qPCR analyses in hiPSC-CMs matured for a longer period in 2D monolayers or in 3D-EHTs indicated that KCNQ1 down-regulation was specific to early hiPSC-CMs (Fig. 9, C and D). On the other hand, up-regulation of CACNA1C was maintained in more mature hiPSC-CMs (Fig. 9, C and D). Of note, CACNA1C is a gene always found in the A chromatin compartment throughout hiPSC-CM differentiation (Bertero et al., 2019), and such localization was unaltered in mutant hiPSC-CMs (Table S5). Given the established role of ICa,L in the development and maintenance of the cardiac action potential (Catterall et al., 2005), we tested if its inhibition with the L-type calcium blocker verapamil could revert the electrophysiological abnormalities of mutant hiPSC-CMs. Remarkably, low concentrations of verapamil markedly reduced both the FPDc and the spike amplitude of mutant hiPSC-CM monolayers, while they had little or no effect on corrected controls (Fig. 9 E). Overall, we conclude that the combination of ectopic P/Q-type calcium currents due to aberrant chromatin compartment dynamics, and of enhanced L-type calcium currents via other epigenetic mechanisms, lead to the electrophysiological abnormalities of lamin A/C haploinsufficient hiPSC-CMs (Fig. 10).
Figure 10.
Proposed model for the chromatin compartmentalization–dependent and –independent effects of lamin A/C haploinsufficieny in developing hiPSC-CMs. Compartmentalization and pheripheral localization of few genomic hotspots is dysregulated (exemplified by the magenta and green loci), while a large number of genes are dysregulated independently of chromatin compartment changes (exemplified by the red locus).
Discussion
Disease modeling of cardiac laminopathy in developing cardiomyocytes
Because the earliest manifestation of cardiac laminopathy is severe electrical abnormalities in the myocardium (van Rijsingen et al., 2012; Kumar et al., 2016; Hasselberg et al., 2018), our observations of aberrant electrophysiological properties in mutant hiPSC-CMs may be noteworthy. While alterations in cardiac rhythm could have been anticipated given the clinical manifestations of this disease, we were surprised to note increased FPDc and action potential duration. The FPD is the in vitro analogue of the QT interval measured by electrocardiogram (which indicates the interval between ventricular depolarization and repolarization). Genetic or acquired prolongation of QT interval is a strong risk factor for development of severe arrhythmias (Vandael et al., 2017), and QT interval prolongation has been reported in patients with LMNA mutations causing cardiac laminopathy (Pan et al., 2009), Emery–Dreifuss muscular dystrophy (Russo et al., 2012), and Hutchinson–Gilford progeria syndrome (Merideth et al., 2008). Although prolonged QT has not been widely described as a hallmark of cardiac laminopathy, we speculate that this could be an underappreciated early clinical phenotype. Prolonged QT and subsequent increase in calcium influx might predispose patients to cardiac arrhythmias and/or could be an early marker of a broader conduction system disease.
A striking phenotype of the laminopathic cardiomyocytes was their enhanced contractility in monolayers and in EHTs. Increased contractility can be explained by the prolonged action potentials and stronger calcium fluxes that these myocytes exhibit. At a first glance, greater contractility might appear to conflict with the clinical phenotype of some laminopathy patients who develop systolic heart failure. However, in contrast to the early onset and highly penetrant malignant conduction disease, left ventricular dilatation and systolic failure are experienced by just a fraction of cardiac laminopathy patients and only after years after the initial diagnosis (van Berlo et al., 2005; Kumar et al., 2016; Tobita et al., 2018). Moreover, heart failure is a complex disease that involves multiple cell types as well as the extracellular matrix (Metra and Teerlink, 2017). Thus, we propose that the systolic hyperfunction and diastolic dysfunction of lamin A/C haploinsufficient hiPSC-CMs might reflect an early and cell-autonomous phenotype of cardiac laminopathy, which over the years can evolve into an organ-level disease characterized by decreased systolic function. A possible mechanism for this could be the chronic calcium overload in laminopathic cardiomyocytes, as this is a well-established stimulus that can lead to DCM due to activation of signaling pathways such as calcineurin/NFAT (Molkentin et al., 1998; Nakamura et al., 2008; Zhang et al., 2016). Additionally, it was previously shown that laminopathic cardiomyocytes are highly sensitive to apoptosis (Ho et al., 2011; Lee et al., 2017), which could over the years contribute to the development of severe cardiac fibrosis that is typical in cardiac laminopathy patients (van Tintelen et al., 2007b; Fontana et al., 2013; Tobita et al., 2018). A corollary of these intriguing hypotheses is that medical interventions during the early phases of the disease, such as the use of calcium antagonists (such as our use of verapamil here) and/or anti-apoptotic agents (Lee et al., 2014; Nie et al., 2018), may be able to prevent the development of heart failure in cardiac laminopathy patients.
Chromatin architecture changes in lamin A/C haploinsufficient cardiomyocytes
The major hypothesis going into this study was that lamin A/C haploinsufficiency would induce widespread gene dysregulation due to inappropriate A/B compartmentalization, the so-called “chromatin hypothesis” for the pathogenesis of cardiac laminopathy (Worman and Courvalin, 2004; Cattin et al., 2013). This hypothesis was not robustly supported. RNA-seq analysis showed that only ∼325 genes were strongly up- or down-regulated in laminopathic cardiomyocytes. Analysis of A/B compartment changes revealed that only ∼1.2% of the genome changed compartments in LMNA mutants, and these aberrations were concentrated in hotspots on chromosomes 5 and 19. Surprisingly, the overlap between strong dysregulation in gene expression and compartment aberrations was minimal (<2%; Fig. S5 B). As discussed below, the CACNA1A gene is ectopically expressed and resides in an A compartment that fails to silence during differentiation. Thus, while examples can be found that may contribute to disease, most of the transcriptional dysregulation appears to result from factors other than errors in compartmentalization. Overall, these findings do not support the “chromatin hypothesis” for the pathogenesis of cardiac laminopathy. On the other hand, our results agree with previous findings from mouse embryonic stem cells, in which depletion of B-type nuclear lamins results in minimal changes in A/B compartmentalization (Amendola and van Steensel, 2015; Zheng et al., 2015, 2018). Our results establish that while A-type lamins seem to participate in chromatin organization in developing cardiomyocytes, lamin A/C haploinsufficiency leads to only modest alterations in A/B compartmentalization even in mechanically active cells.
Interestingly, we identify CACNA1A as a disease-associated gene linked to alterations of A/B compartmentalization. CACNA1A is normally expressed throughout the nervous system with the highest expression on Purkinje neurons in the cerebellum and in cerebellar granular cells (Nimmrich and Gross, 2012). Its ectopic expression and the modest contribution of the resulting P/Q-type calcium currents on FPDc prolongation in lamin A/C haploinsufficient hiPSC-CMs suggest that this may represent a therapeutic target to ameliorate the electrical abnormalities in the myocardium of cardiac laminopathy patients. Further assessments of CACNA1A expression in patient-derived primary samples will be of high interest.
If chromatin compartment alterations do not directly explain the majority of gene expression changes observed in lamin A/C haploinsufficient hiPSC-CMs, what is driving these changes? An important example of this behavior is CACNA1C, which is up-regulated in mutant cells while not changing compartmentalization. At least two nonexclusive possibilities come to mind. First, it remains possible that alterations in chromatin organization play a primary role, albeit not in the form of outright changes in A/B compartmentalization. For instance, it was reported that B-type lamins can indirectly affect the expression of genes within the nuclear interior by affecting the interaction between TADs as a consequence of distal alterations in LAD compaction (Zheng et al., 2018). It is thus possible that A-type lamins might have a similar role, which will be interesting to test in detail in future studies. A second possibility for compartment-independent gene expression changes might be the result of changes in intracellular signaling pathways such as MAPK and mTOR. Indeed, such pathways have well-established links with the nuclear lamina (Dobrzynska et al., 2016) and are up-regulated in animal models of cardiac laminopathy (Muchir et al., 2007, 2012; Choi et al., 2012).
Emerging evidence suggests that nonsense/haploinsufficency mutations in LMNA may have a different pathogenesis than missense mutations. We have recently collaborated to study a heterozygous K219T missense mutation in LMNA (Salvarani et al., 2019). Interestingly, this mutation leads to distinct electrophysiological abnormalities, namely reduced peak sodium current and diminished conduction velocity, which are caused by down-regulation of SCN5A as result of closer proximity to the nuclear lamina and increased H3K27me3. In the current study, we have not observed a similar reduction of SCN5A expression in lamin A/C haploinsufficient hiPSC-CMs (Fig. S3 F). Furthermore, our Hi-C data indicate that the SCN5A gene is found in a chromatin domain, which is always part of the A compartment both throughout normal cardiac differentiation (Bertero et al., 2019) and in lamin A/C haploinsufficient hiPSC-CMs (Table S5). These observations prompt the intriguing hypothesis that haploinsufficient and missense mutations in LMNA might lead to cardiac laminopathy via distinct molecular mechanisms.
In conclusion, our work establishes that while lamin A/C haploinsufficient hiPSC-CMs show marked alterations in electrophysiology, contractility, and chromosomal topology, phenotypic changes cannot, for the most part, be directly explained by alterations in chromatin compartmentalization. With this in mind, it is important to mention that this study does not come without limitations. We acknowledge that modeling diseases of the adult heart in immature hiPSC-CMs suffers from inherent drawbacks. It is possible that functional chromatin dysregulation could be more important in adult myocytes subjected to high levels of mechanical stress in vivo. Addressing this aspect will require substantial advances in our ability to mature hiPSC-CMs and/or improvements of genome-wide chromatin conformation capture technologies in order to reliably measure chromatin architecture from small numbers of myocytes isolated from precious primary samples. Furthermore, replication of our findings in additional hiPSC lines with nonsense/haploinsufficient mutations will be important, as our study focused on only one such mutant line. Finally, future studies will be required to test whether other types of mutations (such as missense changes like the aforementioned K219T mutation) result in more substantial genome-wide alterations in chromatin topology than what was observed following lamin A/C haploinsufficiency. All considered, our work provides a stepping stone toward understanding the relevance of the “chromatin hypothesis” in the pathogenesis of cardiac laminopathy.
Materials and methods
hiPSC culture and differentiation
hiPSCs were cultured and differentiated into cardiomyocytes (hiPSC-CMs) with minor modifications of previously described methods (Burridge et al., 2014; Bertero et al., 2019). hiPSCs were cultured on plates precoated with 0.5 µg/cm2 recombinant human Laminin-521 matrix (diluted in PBS with Ca2+ and Mg2+; rhLaminin521; Biolamina) with daily changes of antibiotic-free Essential 8 (E8) media (Thermo Fisher Scientific). Cells were passaged as small clumps with Versene (Thermo Fisher Scientific), and 10 µM Y-27632 (Tocris) was added for the first 16 h.
For hiPSC-CM generation, cells were passaged as single cells with Versene, and seeded at a density of 2.5 × 105 per well of a 12-well plate precoated with 2 µg/cm2 rhLaminin521 (denoted day −2). After 24 h, medium was changed to E8 with 1 µM CHIR-99021 (Cayman), denoted day −1. On day 0, medium was changed to RBA media (RPMI-1640 with glutamine [Thermo Fisher Scientific] supplemented with 500 µg/ml BSA and 213 µg/ml ascorbic acid [both from Sigma-Aldrich]) supplemented with 4 µM CHIR-99021. At day 2, medium was changed to RBA with 2 µM WNT-C59 (Selleckchem). On day 4, medium was changed to RBA. On day 6, medium was changed to RPMI-B27 media (RPMI with 1× B-27 supplement; both from Thermo Fisher Scientific), with further media changes every other day. Beating was first observed between day 7 and day 9, and cells were cultured until day 14 before collection (unless otherwise indicated). hiPSC-CMs to be used for assays at later stages of differentiation were preconditioned with a 30-min heat shock at 42°C on day 13 and cryopreserved at day 14 following single-cell dissociation using 0.25% wt/vol trypsin (Thermo Fisher Scientific) in Versene.
Frozen hiPSC-CM stocks were thawed and seeded at a density of 2 × 105 cells/cm2 onto rhLamin521 precoated dishes (2 µg/ml) in RPMI-B27, which was supplemented with 10 µM Y-27632 and 5% FBS (Thermo Fisher Scientific) for the first 16 h. hiPSC-CMs were then cultured in RPMI-B27 with media changes every other day. After 1 wk (day 21 of differentiation), hiPSC-CMs were dissociated to single cells using 0.05% trypsin-EDTA (Thermo Fisher Scientific) and seeded at the desired density for the downstream assays.
Gene editing
hiPSCs with a heterozygous c.672C>T mutation in the LMNA gene (resulting in p.Arg225*, or R225X) were generated through lentiviral reprogramming of dermal fibroblasts from a 56-yr-old male patient with severe cardiac laminopathy (Siu et al., 2012). Cells were obtained at passage 29, adapted to culture in E8/rhLaminin521, and banked at passage 34. These hiPSC stocks were confirmed to be Mycoplasma-negative (MycoAlert Detection Kit; Lonza) and proved euploid by conventional G-banding karyotyping (Diagnostic Cytogenetics). Cell identity was tested by Sanger sequencing of a genomic PCR product for exon 4 of LMNA (PCR primers: 5′-GGCTGGGTGATGACAGACTT-3′ and 5′-TACTGCTCCACCTGGTCCTC-3′; sequencing primer: 5′-GCCCTAGTGGACAGGGAGTT-3′), which confirmed the expected c.672C>T heterozygous mutation.
The mutation was corrected into the wild-type allele by adapting a previously described two-step method for scarless genome editing relying on CRISPR/Cas9-facilitated homologous recombination of a targeting vector containing the wild-type allele in one homology arm, and an excisable piggyBac drug resistance cassette (Fig. S1 A; Yusa et al., 2011; Yusa, 2013). Since the c.672C>T mutation lies close to the 3′ splice acceptor site of exon 4 (Fig. S1 B), we reasoned that any intergenic mutation could have poorly predictable effects on LMNA splicing. To avoid any kind of genomic scar, we identified a suitable endogenous “TTAA” site in the third intron of LMNA and located 151 bp upstream to the c.672C>T mutation in exon 4 (Fig. S1 B). Further, we designed sgRNAs spanning such TTAA sites, so that only the endogenous allele could be cut by CRISPR/Cas9. This strategy allowed us to avoid inserting any additional mutation onto one of the homology arms in the targeting vector (such as those classically used to disrupt the protospacer adjacent motif [PAM] site). Two sgRNAs were designed using the Zhang lab CRISPR design tool (Ran et al., 2013) and had a score higher than 75%, indicating a very high in silico predicted specificity (sgRNA1: 5′-CTACCAGCCCCACTTTAACC-3′ and sgRNA2: 5′-TCAGCTCCCAGGTTAAAGTG-3′, sequences without PAM site). To further decrease the risk of CRISPR/Cas9 off-target activity, we adopted the enhanced specificity Streptococcus pyogenes Cas9 (eSpCas9) developed by Slaymaker et al. (2016). The sgRNA was cloned into the eSpCas9(1.1) plasmid (71784; Addgene) using a standard method based on restriction digestion with BbsI followed by ligation of a double-stranded oligo (Ran et al., 2013). The resulting plasmids were named eSpCas9(1.1)_LMNA_sgRNA1 and eSpCas9(1.1)_LMNA_sgRNA2. The sequences were confirmed by Sanger sequencing, and the sgRNAs were validated to have a high on-target activity as measured by T7E1 assay in HEK293 cells (which was comparable to that observed using wild-type SpCas9).
The LMNA targeting vector was constructed starting from the MV-PGK-Puro-TK_SGK-005 plasmid (Transposagen), which contains a piggyBac transposon encoding for a PGK-EM7 promoter-driven dual positive/negative selection cassette (puromycin N-acetlytransferase, ensuring resistance to puromycin, and truncated Herpes simplex virus thymidine kinase, conferring sensitivity to ganciclovir or its analogue fialuridine). First, the piggyBac cassette was excised using NsiI and BsiWI and isolated. Then, a backbone with ends suitable for the subsequent overlap-based assembly was obtained from this same plasmid after removal of the piggyBac cassette using NotI and AscI. Finally, these two fragments were reassembled together with two PCR products representing the 5′ and 3′ homology arms to the LMNA gene. The two homology arms were ∼1 kb long and were amplified from genomic DNA of RUES2 hESCs using the following primers: 5′-GGTCCCGGCATCCGATACCCAATGGCGCGCCCGTACTTCAGGCTTCAGCAGT-3′ and 5′-AAAGAGAGAGCAATATTTCAAGAATGCATGCGTCAATTTTACGCAGACTATCTTTCTAGGGTTAACCTGGGAGCTGAGTGC-3′ (for the 5′ homology arm); 5′-AATTTTACGCATGATTATCTTTAACGTACGTCACAATATGATTATCTTTCTAGGGTTAAAGTGGGGCTGGTAGTG-3′ and 5′-CGAATGCGTCGAGATATTGGGTCGCGGCCGCCCTGTCACAAATAGCACAGCC-3′ (for the 3′ homology arm). For this, Q5 High-Fidelity DNA Polymerase (New England Biolabs) was used according to the manufacturer’s instructions. The four-way assembly reaction was performed using the NEBuilder HiFi DNA Assembly Kit (New England Biolabs) according to the manufacturer’s instructions, and the resulting targeting plasmid was named pbLMNA_R225R. Sanger sequencing confirmed that the 3′ homology arm contained the wild-type R225R allele, while the remaining genomic sequence of both homology arms was identical to that of the R225X hiPSC line as no single-nucleotide polymorphisms were identified. The cloning strategy was designed so that during PCR, the “TTAA” site was inserted both at the end of the 5′ homology arm and at the start of the 3′ homology arm, ensuring that the piggyBac cassette contained within could be excised using transposase while leaving behind a single “TTAA” matching the original genomic sequence (Fig. S1 A).
For the first gene targeting step, 7.5 × 104 hiPSCs were seeded in each well of a 6-well plate and immediately transfected using GeneJuice (Millipore) according to the manufacturer’s instructions. Briefly, for each well, 3 µl of GeneJuice was mixed with 100 µl of Opti-MEM (Thermo Fisher Scientific) and incubated for 5 min at RT. 1 µg of DNA was added to the transfection solution (equally divided between pbLMNA_R225R and either eSpCas9(1.1)_LMNA_sgRNA1 or eSpCas9(1.1)_LMNA_sgRNA2), which was further incubated for 15 min at RT and finally added to the cell suspension. After 16 h from transfection, cells were washed with Dulbecco’s PBS and cultured for another 3 d. Gene-targeted cells were selected by adding 1 µg/ml puromycin to the media for 4 d, after which the dose was reduced to 0.5 µg/ml. 10 µM Y-27362 was added for the first 48 h of selection. Puromycin was then maintained at all times until the second gene targeting step to prevent silencing of the piggyBac transgene. After 7 d from the transfection, 10–15 individual and well-separated colonies could be identified in each well of a 6-well plate, indicating that they likely arose from clonal expansion of a single gene-edited hiPSC. Colonies were manually picked following gentle treatment with Versene to facilitate their detachment from the matrix, and individually expanded as individual lines. Clones were screened by genomic PCR using LongAmp Taq Polymerase (New England Biolabs) according to the manufacturer’s instructions, except that all reactions were performed using an annealing temperature of 63°C and an extension time of 2 min. The primer sequences are reported in Table S7, and the genotyping strategies are illustrated in Fig. S1. Briefly, junctional PCRs for both the 5′ and 3′ integration site (5′- and 3′-INT) were used to confirm site-specific integration, while locus PCRs were used to monitor the presence of residual wild-type alleles. This allowed discrimination of homozygous clones from heterozygous ones or mixed cell populations. Finally, PCRs of the targeting vector backbone (5′- and 3′-BB) were performed to exclude random integration of the plasmid elsewhere in the genome. Homozygous clones with only on-target integration events were selected (3 out of 18 and 2 out of 24 for sgRNA1 and sgRNA2, respectively). These positive clones were further characterized by Sanger sequencing of the 5′- and 3′-INT PCR products to confirm the presence of the wild-type R225R allele in homozygosity (found in all of the five lines) and exclude other unwanted mutations elsewhere in the locus (absent in all five lines). Two clones (one for each sgRNA) were karyotyped by standard G-banding, which confirmed their euploidy, and were therefore selected for the second gene-targeting step. These clones were named pb R225R g1 and pb R225R g2 (Fig. S1 C).
To remove the piggyBac and restore the LMNA locus to its original form, pb R225R g1 and pb R225R g2 hiPSCs were transfected as described above but using 1 µg of excision-only piggyBac transposase expression vector (PBx; Transposagen). Puromycin was removed from the media the day before transfection, and subsequently omitted. After 3 d from the transfection, the populations were passaged as single cells, and 104 cells were seeded per 10-cm plate in the presence of 10 µM Y-27362. The next day, negative selection of cells still possessing the piggyBac cassette was initiated by adding 200 nM fialuridine. 10 µM Y-27362 was added for the subsequent 48 h. Selection was complete after 5 d, at which point 10–50 individual and well-separated colonies could be identified in each 10-cm dish. Individual colonies were isolated, clonally expanded, and screened by genomic PCR as described above to identify those with homozygous reconstitution of the wild-type allele (5 out of 30 and 6 out of 39 for sgRNA1 and sgRNA2, respectively). These were further characterized by sequencing to ensure that the sequence surrounding the “TTAA” site was faithfully reconstituted upon piggyBac excision (confirmed in a subset of four lines, two for each sgRNA; Fig. S1, C and D).
To exclude the prevalence of off-target mutations as result of CRISPR/Cas9 gene editing, we predicted potential off-targets for each sgRNA using Cas-OFFinder (Bae et al., 2014) and experimentally examined a subset of putative high-risk sites. For this, we determined all possible off-targets allowing for up to three mismatches between the sgRNA and the genomic target, or up to one mismatch while allowing a single base pair bulge in either molecule. In both cases, we considered both canonical SpCas9 PAM sites (NGG, where N is any nucleotide), and noncanonical SpCas9 PAM sites (NAG) that may allow cleaving albeit at lower frequency (Hsu et al., 2013). Table S1 reports the resulting list of potential off-targets and their annotation to nearby RefSeq genes. Notably, no overlap was observed between the nearest genes to possible off-targets of sgRNA1 and sgRNA2, indicating a very low risk of inducing mutations affecting the same gene. We experimentally validated the sequence of potential off-targets that (1) had two or fewer mismatches (no bulge) or (2) had three mismatches (no bulge) or one mismatch and one bulge, and fell within an exon, the proximal promoter (<1 kb from a transcription start site), the poly-adenylation site (<1 kb from a transcription end site), or a splice donor/acceptor site (<100 bp from an exon) of genes expressed during hPSC-CM differentiation (fragments per kilobase of transcript per million mapped reads is >1; Bertero et al., 2019). These criteria identified three possible high-risk off-targets for each sgRNA, which were assessed by Sanger sequencing of genomic PCR products (primers reported in Table S8). No such off-target mutation was observed, confirming the high specificity of the gene editing design. We also confirmed silencing of the reprogramming transgenes that had been delivered by lentiviral transduction to generate the R225X hiPSC line by RT-qPCR, as previously described (Siu et al., 2012). No amplification was observed in any of the hiPSC lines analyzed after 40 PCR cycles, confirming transgene silencing and lack of reactivation following clonal selection. Finally, two clones (one for each sgRNA), were karyotyped by standard G-banding, which confirmed their euploidy, and therefore selected for subsequent functional experiments. These clones were named R225R g1-15 and R225R g1-38, and are referred to in the text and figures as Corr.1 and Corr.2.
Parental R225X hiPSCs (referred to in the text and figures as Mutant or Mut) were cultured in parallel throughout the whole gene editing procedure to provide a passage-matched control, and were rebanked at passage 49 together with Corr.1 and Corr.2. These cells were subjected to the same genomic quality controls as the corrected lines. Mutant and Corrected hiPSCs to be used for derivation of hiPSC-CMs were cultured between passage 50 to 60 before resorting to a new frozen stock.
MEA
MEA analyses were performed on hiPSC-CM monolayers at day 30 of differentiation. hiPSC-CMs at day 21 of differentiation were seeded at a density of 5 × 104 cells per well of 48-well MEA plates (CytoView MEA 48; Axion Byosystems) precoated with rhLaminin521 (2 µg/ml). Cells were cultured in RPMI-B27 with media changes every other day. After 9 d (day 30 of differentiation), cells were prepared for MEA analysis by changing the culture media to Tyrode’s buffer (140 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 0.33 mM NaH2PO4, 5 mM D-glucose, and 10 mM Hepes; pH adjusted to 7.36) prewarmed at 37°C. After 10 min of equilibration in Tyrode’s buffer at 37°C, MEA data were acquired for 5 min using the Maestro MEA system (Axion Biosystems) using standard recording settings for spontaneous cardiac field potentials. Data acquisition and automated data analysis were performed using Axis software version 2.1. Standard acquisition settings have 130 × gain, and record from 1 to 25,000 Hz, with a low-pass digital filter of 2 kHz for noise reduction. Automated data analysis was focused on the 30 most stable beats within the recording period. The beat detection threshold was 100 µV, and the FPD detection used an inflection search algorithm with the threshold set at 1.5 × noise to detect the T wave. The FPD was corrected for the beat period according to the Fridericia’s formula: FPDc = FPD/(beat period)1/3 (Asakura et al., 2015; Rast et al., 2016). Reported results for individual wells were calculated by averaging all of the electrodes. In certain instances, poor signal quality and/or irregularity of field potential behavior prevented the calculation of certain parameters (such as FPD). The presented data constitute all recorded values that could be reliably measured by the software based on automatic quality-control thresholds.
For pharmacological studies of P/Q- and L-type calcium current inhibition, Tyrode’s buffer was supplemented with 2 µM ω-conotoxin MVIIC, 0.5 µM ω-agatoxin TK, or 0.001–0.5 µM verapamil (all from Tocris). hiPSC-CMs were incubated at 37°C for 20 min or 10 min (for ω-conotoxin and ω-agatoxin, or verapamil, respectively) before MEA data acquisition.
Whole-cell patch clamp
Whole-cell patch clamp recordings were obtained from individual hiPSC-CMs at day 30 of differentiation. To promote a mature phenotype and enhance cell viability during the invasive patch procedure, we cultured hiPSC-CMs onto biomimetic anisotropic nanopatterns (Macadangdang et al., 2015; Carson et al., 2016). hiPSC-CMs at day 21 of differentiation were seeded at a density of 4.5 × 105 cells per well of 35-mm glass-bottom FluoroDish with nanopatterned surfaces precoated with rhLaminin521 (2 µg/ml). Anisotropically nanofabricated substrata with 800-nm topographic features were fabricated via UV-assisted capillary force lithography as previously described (Macadangdang et al., 2015). First, liquid polyurethane acrylate (PUA) prepolymer was drop-dispensed onto a silicon master mold. A transparent polyester film was then placed on top of the dispensed PUA. After exposure to UV radiation (λ = 250–400 nm), the film was peeled away from the silicon master, creating a PUA mold. A polyurethane-based prepolymer (NOA76; Norland Products) was then drop-dispensed onto standard glass coverslips, and the PUA mold was placed on top. The mold was then exposed to UV radiation for curing. After curing, the PUA mold was peeled off, leaving behind anisotropically nanofabricated substrata for cell culture. Dishes were sterilized and activated by gas plasma treatment before coating with rhLaminin521. After 9 d, cells were assayed by whole-cell patch clamp on the 37°C heated stage of an inverted DIC microscope (Nikon) connected to an EPC10 patch clamp amplifier and computer running Patchmaster software version 2 × 73.2 (HEKA). Cells on patterned coverslips were loaded onto the stage and bathed in Tyrode’s buffer. An intracellular recording solution (120 mM L-aspartic acid, 20 mM KCl, 5 mM NaCl, 1 mM MgCl2, 3 mM Mg2+-ATP, 5 mM EGTA, and 10 mM Hepes) was used in conjunction with borosilicate glass patch pipettes (World Precision Instruments) with a resistance in the range of 2–6 MΩ. Offset potentials were nulled before formation of a gigaΩ seal, and fast and slow capacitance was compensated for in all recordings. Membrane potentials were corrected by subtraction of a 15-mV liquid junction potential calculated by the HEKA software. Current injection was controlled by the software and used to hold patched cells at an artificial resting membrane potential of −70 mV. Cells that required >100 pA of current to achieve a −70-mV resting membrane potential were excluded from analysis as excessive application of current was taken as indication of poor patch quality and/or membrane integrity. To generate a single action potential, a 5-ms depolarizing current pulse of 50 nA was then applied and the resulting voltage change recorded in current clamp mode. Action potential rise times were calculated as the time taken to reach 90% maximum action potential amplitude from 10% of the maximum amplitude. The exponential time constant (τ) was calculated from 90% to 10% repolarization of the action potential. Action potential duration was calculated as the time delay between 10% of the maximum depolarization and 90% repolarization from the maximum action potential amplitude. Gap-free recordings of spontaneous cardiomyocyte activity were then collected for 30 s with 0 pA current injection to provide a measure of the maximum diastolic membrane potential held by the cell without current input.
Assessment of intracellular calcium fluxes
Calcium fluxes were assessed in hiPSC-CM monolayers at day 30 of differentiation. hiPSC-CMs at day 21 of differentiation were seeded at a density of 5 × 105 cells per well of a six-well plate precoated with rhLaminin521 (2 µg/ml). After 9 d, cells were prepared for imaging by incubation for 30 min at 37°C with 1 µM Fluro-4, AM (Thermo Fisher Scientific) diluted in culture media. Cells were rinsed in fresh media for 30 min at 37°C, and equilibrated in Tyrode’s buffer prewarmed at 37°C for 10 min. hiPSC-CMs were paced at 1 Hz using a C-Dish for a six-well plate connected to a C-Pace EM cell stimulator (both from IonOptix) providing biphasic field stimulation (pulses of 10 V/cm for 20 ms). Videos of Fluo-4 fluorescence (excitation/emission of 494 and 516) were recorded at 20 frames per second (fps) for at least five contractions using a Nikon Ti-E epi-fluorescent microscope with a 20× objective and 1× coupler between the microscope and Hamamatsu flash V3 camera. Videos were obtained for at least 20 random fields of view (FOVs). A custom Matlab program was used to define the region of interest (ROI; containing an individual hiPSC-CM), threshold the Fluo-4 intensity based on the surrounding nonfluorescent background, and track the average ROIFluo-4 fluorescence (F) over time. The relaxation time constant τ was determined by fitting the formula F(t) = Ae−t/τ + B to the decay phase of the Fluo-4 transient profile, where t is time in seconds and A and B are fitted constants.
Contraction correlation quantification (CCQ) analysis of cardiac contractility in hiPSC-CM monolayers
CCQ analysis was performed on hiPSC-CM monolayers at day 30 of differentiation. hiPSC-CMs at day 21 of differentiation were seeded at a density of 106 cells per well of a six-well plate precoated with rhLaminin521 (2 µg/ml). After 9 d, cells were paced at 1 Hz as just described for the measurement of calcium fluxes. Bright-field videos of at least 10 contractions in multiple random FOVs were recorded at 30 fps using a Nikon TS100 microscope with a 20× objective and 1× coupler between the microscope and a Canon VIXIA HF S20 camera. Videos were analyzed by CCQ using a custom Matlab script, as previously described (Macadangdang et al., 2015). Briefly, this method utilizes particle image velocimetry and digital image correlation algorithms to provide relevant contractile endpoints from bright field video recordings. A reference video frame is divided into a grid of windows of a set size. Each window is run through a correlation scheme with a second frame, providing the new location for that window in the second frame. This displacement is converted into a vector map, which provides contraction angles and, when spatially averaged, contraction magnitudes. The correlation equation used provides a Gaussian correlation peak with a probabilistic nature that provides sub-pixel accuracy.
Generation and biomechanical characterization of 3D-EHTs
3D-EHTs were generated and characterized with minor changes to a previously described method (Leonard et al., 2018). Racks of polydimethylsiloxane (PDMS) posts were fabricated by pouring uncured PDMS (Sylgard 184 mixed at a 1:10 curing agent to base ratio) into a custom acrylic mold. Glass capillary tubes (1.1 mm in diameter; Drummond) were cut to length and inserted into the holes on one side of the mold before curing to render one post in each pair rigid. Post racks were baked overnight at 65°C before being peeled from the molds. Racks consisted of six pairs of posts that were evenly spaced to fit along one row of a standard 24-well plate. Fabricated posts were 12.5 mm long and 1.5 mm in diameter with a cap structure (2.0 mm in diameter for the topmost 0.5 mm) to aid in the attachment of 3D-EHTs. The center-to-center post spacing (corresponding to precompacted 3D-EHT length) was 8 mm. Prior to casting 3D-EHTs, all 3D-printed parts and PDMS posts were sterilized in a UVO Cleaner (342; Jetlight) for 7 min, submerged in 70% ethanol, and rinsed with sterile deionized water. Rectangular 2% wt/vol agarose/PBS casting troughs (12 mm in length, 4 mm in width, and ∼4 mm in depth) were generated in the bottom of 24-well plates by using custom 3D-printed spacers (12 mm × 4 mm in cross-section and 13 mm long) as negative molds. PDMS post racks were positioned upside down with one rigid-flexible post pair centered in each trough (leaving a 0.5-mm gap between the tip of the post and the bottom of the casting trough). Each tissue consisted of a 97-µl fibrinogen-media solution (89 µl of RPMI-B-27; 5.5 µl of 2× DMEM with 20% FBS; and 2.5 µl of 200 mg/ml bovine fibrinogen; Sigma-Aldrich) containing 5 × 105 hiPSC-CMs and 5 × 104 supporting HS27a human bone marrow stromal cells (ATCC), which was chilled and mixed with 3 µl of cold thrombin (at 100 U/ml; Sigma-Aldrich) just before pipetting into the agarose casting troughs. The 3D-EHT mixtures were incubated for 90 min at 37°C, at which point the fibrin gels were sufficiently polymerized around the posts to be lubricated in media and transferred from the casting troughs into a 24-well plate with fresh 3D-EHT media (RPMI-B-27 with penicillin/streptomycin, and 5 mg/ml aminocaproic acid; Sigma-Aldrich). 3D-EHTs were supplied with 2.5 ml/well of fresh 3D-EHT media three times per week.
In situ force measurements were performed after 4 wk from 3D-EHT casting. To pace 3D-EHTs, post racks were transferred to a custom-built 24-well plate with carbon electrodes connected through an electrical stimulator (S88X; Astro Med Grass Stimulator) to provide biphasic field stimulation (5 V/cm for 20-ms durations) during imaging (Leonard et al., 2018). 3D-EHTs were equilibrated in Tyrode’s buffer (containing 1.8 mM Ca2+) preheated to 37°C and paced at 1 Hz, which was greater than the average spontaneous twitch frequency of the tissues. Videos of at least 10 contractions were recorded inside a 37°C heated chamber using a monochrome CMOS camera (SMN-B050-U; Mightex) with a board lens (TBL 8.4–2 5MP; The Imaging Source). The camera lens configuration allowed for a capture rate of 65 fps with 8.3 µm/pixel resolution and a FOV of 1,536 × 400 pixels, which was sufficient to capture images of the whole 3D-EHT from rigid to flexible post. A custom Matlab program was used to threshold the images and track the centroid of the flexible post relative to the centroid of the rigid post. The twitch force profile, Ftwitch(t) = kpost × Δpost(t), was calculated from the bending stiffness kpost and deflection of the flexible post Δpost at all time points (t), where kpost = 0.95 μN/µm was determined from beam bending theory using the dimensions of the posts and taking the Young’s modulus of PDMS to be 2.5 MPa (Sniadecki and Chen, 2007). The twitch force and twitch kinetics were calculated from the twitch force profiles using a custom Matlab program.
RT-qPCR
RT-qPCR was performed as previously described for 2D cell monolayers (Bertero et al., 2019) or 3D-EHTs (Leonard et al., 2018). 2D monolayers were lysed in RLT buffer supplemented with 1% 2-mercaptoethanol before RNA purification using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions and including the on-column DNase digestion step. cDNA was synthetized by reverse transcription of 500 ng of total RNA using M-MLV RT (Invitrogen) and random hexamer priming according to the manufacturer’s protocol and including RNase OUT (Invitrogen). RT-qPCR was performed in technical duplicate with SYBR Select Master Mix (Applied Biosystems) using 10 ng of cDNA and 400 nM forward and reverse primers. Reactions were run for 40 cycles on a 7900HT Fast Real-Time PCR System (4329001; Applied Biosystems), all according to the manufacturers’ instructions. Gene expression relative to the housekeeping gene RPLP0 was calculated using the ΔCt method as 2(Ct gene − Ct housekeeping). Where indicated, gene expression was further normalized to a control condition (which was set at the arbitrary value of 1). Primers were designed using PrimerBlast and confirmed to amplify a single product. A complete list can be found in Table S9.
For 3D-EHTs, RNA was extracted using RNeasy Plus Micro Kit (QIAGEN) according to the manufacturer’s instructions except for the following modifications. Individual 3D-EHTs were predigested using 2 mg/ml Proteinase K in 100 µl Dulbecco’s PBS for 10 min at 56°C in agitation. Cells were then lysed by adding 350 µl of RLT Plus Buffer supplemented with 1% 2-mercaptoethanol, and cleared through the gDNA Eliminator Mini Spin Column. The RNA lysate was finally prepared for binding to the RNeasy MinElute Spin Column by adding 250 µl of 200-proof ethanol. All subsequent steps were performed according to the supplier’s recommendations. 10 µl of eluted RNA (corresponding to 50–100 ng) was subjected to reverse transcription, and 2 ng of cDNA was used as a template for RT-qPCR, all as described for 2D monolayers.
Flow cytometry
Flow cytometry was performed at different differentiation time points from day 0 through day 30 on either live cells for surface markers (day 6 and day 10 of differentiation) or PFA-fixed cells for intracellular markers (day 0, day 2, day 14, and day 30 of differentiation). Single-cell suspensions were obtained by incubation in TrypLE (Thermo Fisher Scientific) for 5–10 min at 37°C. Following one wash in RPMI supplemented with 10% FBS and 2.5 U/ml DNase (this and all subsequent centrifugation steps performed at 300 g for 4 min at RT), cells were incubated for 5 min at RT with LIVE/DEAD Fixable Violet Dead Cell Stain Kit (L34955; Thermo Fisher Scientific) diluted 1:2,500 in PBS. Cells were washed once with PBS 1% BSA (FACS buffer) and either used directly for staining or fixed with 4% PFA in PBS for 10 min followed by two washes in FACS buffer. Primary antibody incubation was performed for 30 min at RT using ∼1 M cells per condition in 100 µl. Antibodies were diluted either in FACS buffer (for live cells) or in PBS supplemented with 5% FBS, 0.75% saponin, and 0.01% Triton X-100 (permeabilization and blocking buffer, for fixed cells). Surface marker antibodies were preconjugated with fluorophores: mouse monoclonal anti-PDGFRA-APC (clone PRa292; FAB1264A; R&D Systems), 1:10 dilution; mouse monoclonal anti-CD82-PE (clone ASL-24; 342104; Biolegend), 1:20 dilution; mouse monoclonal anti-CD56-PE (clone B159; 555516; BD Biosciences), 1:5 dilution; mouse monoclonal IgG1-APC (clone 11711; IC0002A; R&D Systems), 1:10 dilution; and mouse monoclonal IgG1-PE (clone MOPC-21; 55749; BD Biosciences), 1:5 dilution. After staining with these antibodies, cells were washed twice in FACS buffer and directly analyzed. Intracellular marker antibodies were not conjugated: rabbit polyclonal anti-OCT4 (ab19857; Abcam), 1:200 dilution; goat polyclonal anti-TBXT (AF2085; R&D Systems), 1:40 dilution; mouse monoclonal anti-TNNT2 (clone 13–11; MA5-12960; Invitrogen), 1:200 dilution; rabbit monoclonal anti–NKX2-5 (clone E1Y8H; 8792S; Cell Signaling), 1:200 dilution; rabbit polyclonal anti-TTN (TTN-1; Myomedix), 1:300 dilution; mouse monoclonal anti-ACTC1 (clone EA-53; ab9465; Abcam), 1:300 dilution; rabbit polyclonal anti-TNNI3 (clone EP1106Y; ab52862; Abcam), 1:100 dilution; rabbit normal IgG (27295; Cell Signaling), 1:200 dilution; goat normal IgG (AB-108-C; R&D Systems), 1:200 dilution; and mouse normal IgG (14–4714-85; eBioscience), 1:200 dilution. After staining with these antibodies, cells were washed twice with PBS supplemented with 5% FBS and 0.75% saponin (wash buffer) and incubated for 30 min at RT (protected from light) with donkey secondary antibodies against the appropriate species and conjugated to Alexa Fluor 488 or 647 (Invitrogen), all diluted 1:250 in wash buffer. Cells were further washed once with wash buffer and once with FACS buffer before analysis. Samples were run on a BD FACSCanto II equipped with 405-, 488-, and 647-nm lasers, and data from at least 20,000 valid events were acquired with the BD FACSDIVA software. Analysis was performed with FloJo version 10 software. After identifying single cells based on forward and side scatter properties, samples were gated to be negative for the LIVE/DEAD staining (live cells). Positivity for lineage markers was defined by additional gates based on the signal from isotype antibody negative control stainings, as depicted in the figures.
Immunofluorescence
Immunofluorescence was performed on hiPSC-CMs after replating onto glass-bottom 24-well culture dishes (P24-1.5H-N; CellVis). Dishes were prepared by (1) incubation with 0.5 M acetic acid for 30 min at RT; (2) three washes with deionized water; (3) incubation with 0.1% polyethylenimine overnight at 4°C; (4) two washes with PBS and one wash with deionized water; and finally (5) coating with 2 µg/ml rhLaminin521 (diluted in PBS with Ca2+ and Mg2+) overnight at 4°C (all solutions filter-sterilized before use). hiPSC-CMs were seeded at a density of 5 × 104 cells per well, and cultured for 1 wk before fixation with 4% PFA in PBS for 10 min at RT followed by three washes with PBS. Cells were permeabilized and blocked in PBS with 0.1% Triton X-100 and 5% normal donkey serum for 30 min, and subsequently incubated overnight at 4°C with primary antibodies diluted in antibody buffer (PBS with 0.1% Triton X-100 and 1% normal donkey serum). The following antibodies were used: goat polyclonal anti-lamin A/C (sc-6215; Santa Cruz), 1:50 dilution; rabbit polyclonal anti–lamin B1 (PA5-19468; Invitrogen), dilution 1:300; rabbit polyclonal anti-TTN (TTN-1; Myomedix), 1:300 dilution; mouse monoclonal anti-ACTC1 (clone EA-53; ab9465; Abcam), 1:300 dilution; and rabbit polyclonal anti-TNNI3 (clone EP1106Y; ab52862; Abcam), 1:200 dilution. After three washes with PBS for 5 min, secondary antibodies (donkey anti-rabbit Alexa Fluor 488, donkey anti-goat Alexa Fluor 568, and donkey anti-mouse Alexa Fluor 647, all used at 1 µg/ml) were diluted in antibody buffer and incubated for 1 h at RT in the dark. Nuclei were counterstained with DAPI (5 µg/ml in PBS) for 3 min at RT, and finally washed three times in PBS. Stainings were imaged in PBS at 37°C using a Nikon Eclipse Ti epi-fluorescent microscope using a Nikon CFI PlanApo VC 60× Water Immersion lens (NA 1.2). Sample emission was captured sequentially through NIKON filter sets (UV-2E/C DAPI/Hosechst; FITC HYQ; TRITC HYQ; and CY5 HYQ) by a Hamamatsu ORCA-Flash4.0 V3 Digital CMOS camera and the NIS-Elements AR software (version 5.11), which was also used to create composite images.
Immunofluorescence combined with 3D-DNA FISH (ImmunoFISH)
ImmunoFISH was performed as previously described (Bertero et al., 2019), with minor modifications (procedure performed at RT unless mentioned otherwise). DNA FISH probes were generated using bacterial artificial chromosomes (BACs) from the BACPAC Resources Center: CH17-4L20 (CACNA1A), CH17-291L21 (LRRC4B), CH17-197I17 (PCDHGB4), CH17-122P20 (LGASLS14), and CH17-54F13 (VAV1). Following purification using the QIAGEN Plasmid Midi Kit, each BAC was validated by PCR using two primer sets specific for the expected genomic region. BACs were labeled with SpectrumGreen (Vysis) for 4 h at 15°C using the Vysis Spectrum Nick Translation Kit according to the manufacturer’s recommendations. Probes were purified with the QIAGEN PCR purification kit and then precipitated together with 40 µl Salmon Sperm DNA (Thermo Fisher Scientific) and 60 µl Human Cot-1 DNA (Thermo Fisher Scientific) by adding 15 µl sodium acetate (3 M; pH 5.5) and 390 µl 100% ethanol. After incubation for 2 h at −20°C, probes were spun at 16,000 g for 30 min at 4°C and resuspended in 50 µl hybridization buffer (50% formamide, 2× SSC, and 10% dextran sulfate).
Poly-L-lysine slides (Sigma-Aldrich) were precoated with 0.5 or 2 µg/ml rhLaminin521 (for hiPSCs and hiPSC-CMs, respectively), and 4 × 104 cells were seeded on an ∼2-cm2 spot outlined by a hydrophobic layer for 4 h at 37°C. After fixation with 4% PFA in PBS and quenching in 0.1 M Tris-HCl, pH 7.4 (10 min each), cells were washed twice in PBS, equilibrated in 20% glycerol/PBS for 20 min, and stored overnight in 50% glycerol/PBS at −20°C. Slides were washed twice in PBS for 5 min and processed according to the immunofluorescence protocol just described, except that nuclei were not counterstained with DAPI. The following primary antibodies were used: rabbit polyclonal anti–lamin B1 (PA5-19468; Invitrogen), dilution 1:300; and mouse monoclonal anti-ACTC1 (clone EA-53; ab9465; Abcam), dilution 1:300. After immunofluorescence, slides were refixed with 3% PFA in PBS for 10 min, washed two times in 2× SSC, denatured in 50% formamide 2× SSC, pH 7.2, for 30 min at 80°C, and washed three times with ice-cold 2× SSC. DNA FISH probes were also denatured for 10 min at 80°C, followed by snap cooling on ice. Coverslips containing 10 µl of DNA FISH probes were placed onto the slides (one probe per slide), after which slides were sealed with rubber cement, air-dried in the dark for 30 min, and hybridized overnight at 42°C. The rubber cement was gently peeled off to allow removal of the coverslip. Slides were then washed thrice in 50% formamide and 50% 2× SSC, pH 7.2, at 42°C. Slides were incubated with DAPI (5 µg/ml in 2× SSC) for 2 min to counterstain nuclei, and finally washed twice with PBS. Coverslips were mounted using 10 µl of Slow Fade Gold (Thermo Fisher Scientific) and sealed with nail polish.
Slides were imaged at 37°C using a Yokogawa CSU-W1 spinning disk confocal mounted on an inverted Nikon Eclipse Ti base and a Nikon CFI PlanApo VC 60× Water Immersion lens (NA 1.2). Samples were excited via full field exposure from 100 mW 405-, 490-, and 561-nm lasers. Sample emission was captured sequentially through Chroma Technology filter sets (ET455/50, ET525/35, and ET605/52) by an Andor iXon EM+EMCCD camera and the NIS-Elements AR software (version 5.11). Z-stacks were acquired at a step size of 0.3 µm to cover a z-distance of 15 µm (51 stacks). 10–15 randomly selected FOVs were imaged at a separation of three FOVs from each other. Imaris software was used for automated image analysis (version 9.2.0; Bitplane). After removal of background signal using a Gaussian filter (9 µm for lamin B1 and 1 µm for FISH signals, based on the average size of each feature), nuclear lamina boundaries were defined with the surface feature (surface detail of 1 µm; all other parameters as default). For hiPSC-CM analysis, nuclear lamina features for the few contaminating noncardiomyocytes (indicated by lack of costaining for cardiac α-actinin) were removed at this stage. FISH signal was masked based on the nuclear lamina boundaries to retain only the nuclear signal, and then loci were identified using the spot calling feature (1 µm wide, based on the average size of FISH signal; all other parameters as default). The distance transformation function was then used to calculate inside distances from the nuclear lamina and distances from the FISH spots. Statistics were exported from Imaris and processed using a custom Python script to extract and analyze information only for diploid nuclei (containing exactly two green FISH spots). At least 150 cells per condition were analyzed (corresponding to at least 10 FOVs), and the exact number is indicated in each figure. Distances between loci and the nuclear lamina were calculated relative to the center of 1-µm-wide FISH spots. The resulting data proved to follow a Gaussian distribution (based on Kolmogorov–Smirnov normality tests) with unequal SD among groups (based on the Bartlett’s test). Therefore, statistical analysis was performed using the Brown–Forsythe and Welch ANOVA test, followed by the Holm–Sidak correction of multiple paired comparisons across preselected groups of interest (mutant versus corrected, and hiPSC-CMs versus hiPSCs).
Western blot
Protein lysates were obtained using ice-cold 1× RIPA buffer containing protease and phosphatase inhibitors (9806; Cell Signaling) and freshly supplemented with 1 mM PMSF. After incubation for 30 min on ice, the lysate was clarified from insoluble material by centrifugation at 16,000 g for 10 min at 4°C. The protein concentration was assessed using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. After addition of Laemmli Sample Buffer (Bio-Rad) to a final concentration of 1×, and 2-mercaptoethanol to a final concentration of 2.5%, the samples were denatured by heating at 95°C for 5 min. For electrophoretic separation, 20 µg of protein for each sample was loaded onto 7.5% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad) and run at 100 V for 60 min using 1× Tris/glycine/SDS running buffer (Bio-Rad). Proteins were transferred onto Immobilion-P polyvinylidene fluoride membranes (Millipore Sigma-Aldrich) by means of tank blotting in 1× Tris/glycine (Bio-Rad) supplemented with 20% methanol; transfer was performed at 100 V for 60 min at 4°C. Membranes were blocked in PBS supplemented with 0.1% Tween-20 (PBST) and 4% Blotting-Grade Blocker (Bio-Rad) for 1 h at RT. Primary antibody incubation of whole membranes was performed overnight at 4°C under agitation, and antibodies were diluted in the same blocking buffer. The following antibodies were used: goat polyclonal anti-lamin A/C (sc-6215; Santa Cruz), 1:500 dilution; rabbit polyclonal anti–lamin B1 (PA5-19468; Invitrogen), dilution 1:500; mouse monoclonal anti–lamin B2 (clone E-3; 33–2100; Invitrogen), dilution 1:200; mouse monoclonal anti-TNNI (clone OTI8H8; NBP2-46170; Novus Biologicals), dilution 1:500; goat polyclonal anti-NKX2-5 (AF2444; R&D Systems), dilution 1:500; goat polyclonal anti-TBXT (AF2085; R&D Systems), dilution 1:500; rabbit polyclonal anti-POU5F1 (19857; Abcam), dilution 1:1,000; and mouse monoclonal anti-GAPDH (clone 6C5; ab8245; Abcam), dilution 1:2,000. Membranes were washed three times in PBST for 10 min at RT, incubated for 1 h at RT with species-appropriate HRP-conjugated secondary antibodies (Thermo Fisher Scientific) diluted 1:10,000 in blocking buffer, and washed three times in PBST for 10 min at RT. Chemiluminescent reaction was initiated by incubation with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific), and images were acquired using a ChemiDoc Imaging System (Bio-Rad) in “high resolution” mode. Before reprobing with a new antibody, membranes were treated with Restore Plus Western blot stripping buffer (Thermo Fisher Scientific), washed three times, and reblocked. Densitometric quantification of Western blots was performed using ImageJ, and protein abundance estimation was normalized on the levels of GAPDH within each lysate.
RNA-seq
RNA-seq was performed on 2–3 × 106 hiPSC-CMs at day 14 of differentiation on three biological replicates (independent differentiations) per cell line. RNA-seq libraries were prepared from 100 ng of total RNA (obtained as described above for RT-qPCR) using the TruSeq Stranded Total RNA LT Kit with Ribo-Zero H/M/R (all Illumina), according to the manufacturer’s instructions. RNA-seq libraries were paired-end sequenced on an Illumina NextSeq 500 in a high output run with 150 cycles (75 for each end), achieving ∼40 million paired-end reads per sample. Reads were mapped to hg38 using STAR (Dobin et al., 2013) and then quantified and processed through the Cufflinks suite (Trapnell et al., 2012), all using default parameters. Differential expressed genes exhibited a q-value <0.05 for a pairwise comparison and were expressed at least 1 read kilobase per million mapped reads in one time point. Hierarchical clustering was performed with the CummeRbund suite in R. Ontology enrichment analysis was done using EnrichR (Chen et al., 2013). For comparision with previously described RNA-seq data of hESC-CM differentiation (Bertero et al., 2019), all datasets were collectively renormalized and quantified using Cufflinks. Principal component analysis was performed using the R function “prcomp.”
In situ DNase Hi-C
Hi-C based on in situ proximity ligation of DNase I–digested nuclei (DNase Hi-C) was performed with minor modifications of a previously described method (Ramani et al., 2016; Bertero et al., 2019). The assay was performed on ∼2 × 106 hiPSC-CMs at day 14 of differentiation on two biological replicates (independent differentiations) per cell line. Unless otherwise indicated, all molecular biology reagents were obtained from Thermo Fisher Scientific and reactions were performed at RT. Cells were fixed in the dish with fresh RPMI-1640 supplemented with 2% formaldehyde while in gentle orbital rotation for 10 min, and subsequently quenched with 25 mM glycine for 5 min at RT followed by 15 min at 4°C. Cells were then treated with 0.05% trypsin for 10 min at 37°C, rinsed in RPMI-1640 with 10% FBS, and scraped off the plate. Cells were washed once with PBS, flash-frozen in liquid nitrogen, and stored at −80°C. After rapid thawing, samples were resuspended in 500 µl of ice-cold cell lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, and 0.5% Igepal CA-630, supplemented with the protease inhibitor cocktail from Sigma-Aldrich), incubated for 20 min on ice, and dounce-homogenized 60–80 times with a tight pestle. Extracted nuclei were centrifuged at 2,500 g for 1 min at RT (standard spinning protocol), resuspended in 300 µl 0.5× DNase I digestion buffer with 0.2% SDS and 20 mM MnCl2, and incubated at 37°C for 60 min with periodic gentle vortexing. Then, 300 µl 0.5× DNase I digestion buffer with 2% Triton X-100, 20 mM MnCl2, and 0.4 µg/µl RNase A was added and incubated for another 10 min. Chromatin was finally digested with 7 U of DNase I during a 7-min incubation at RT. The reaction was stopped with 30 µl of 0.5 M EDTA and 15 µl 10% SDS. The efficiency of DNase I digestion was confirmed by comparing the DNA shearing patterns of small samples of undigested and digested nuclei using a 6% PAGE gel (following proteinase K digestion). Nuclei were spun down and resuspended in 150 µl RNase and DNase-free water. 300 µl of AMPure XP beads (Beckman) was added to irreversibly bind nuclei; going forward, nuclei were therefore cleaned up by magnetic purification. In situ reactions were performed first for end repair (15 U of T4 DNA Polymerase and 30 U of Klenow Fragment for 1 h at RT) and dA-tailing (75 U of Klenow exo-minus for 1 h at 37°C), each in 200 µl, and followed by inactivation with 5 µl of 10% SDS and cleanup. Nuclei were then subjected to overnight ligation of custom T- and blunt-bridge biotin-tagged adapters (each at 8 µM; sequences detailed in Ramani et al., 2016) using 25 U of T4 DNA ligase and 5% polyethylene glycol in a 100-µl reaction incubated at 16°C. The reaction was stopped with 5 µl of 10% SDS, and nuclei were washed twice with AMPure buffer (20% polyethylene glycol in 2.5 M NaCl) followed by two washes with 80% ethanol to remove un-ligated adapter. Nuclei were then treated for 1 h at 37°C with 100 U of T4 PNK in a 100-µl reaction to phosphorylate the adapters. Proximity ligation of DNA ends was performed for 4 h at RT using 30 U of T4 DNA ligase in a 1-ml reaction maintained in gentle agitation. Nuclei were resuspended in 1× NEBuffer 2 (New England Biolabs) with 1% SDS, and digested with 800 µg of Proteinase K overnight at 62°C. DNA was precipitated by adding 60 µg glycogen, 50 µl 3 M Na-acetate (pH 5.2), and 500 µl isopropanol followed by incubation for 2 h at −80°C and centrifugation at 16,000 g for 30 min at 4°C. DNA was resuspended in 100 µl water and purified with 100 µl AMPure beads according to the manufacturer’s instructions, and resuspended in 100 µl elution buffer (EB; 10 mM Tris-HCl, pH 8.5). Biotin pull-down was performed on the purified DNA to isolate ligation products containing the biotin adapters. 100 µl Myone C1 beads were mixed with the DNA for 30 min at RT under gentle rotation. Samples were washed four times with bind and wash buffer (5 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 1 M NaCl, and 0.05% Tween-20) and twice with EB. DNA was then treated on the beads to perform end-repair (200-µl reaction with the Fast DNA End Repair Kit for 10 min at 18°C) and dA-Tailing (50-µl reaction with 25 U of Klenow exo-minus in NEBuffer 2 and 1.2 mM dATP for 30 min at 37°C). Custom Y-adapters for Illumina sequencing (each at 2 µM; sequences detailed in Ramani et al., 2016) were then ligated using 20 U of T4 DNA ligase for 1 h at RT in a 50-µl reaction using the Rapid Ligation Buffer. Beads were washed four times with bind and wash buffer and twice with EB after each of these reactions. Finally, libraries were amplified by 12 PCR cycles with Kapa HiFi ReadyStart Master Mix with custom barcode-containing primers (sequences detailed in Ramani et al., 2016). Libraries were purified with 0.8× Ampure XP beads according to the manufacturer’s instructions, quantified with a Qubit and the DNA high-sensitivity reagent, and pooled at equimolar ratios in preparation for next-generation sequencing. Samples were paired-end sequenced on three runs with an Illumina NextSeq 500 at high output with 150 cycles (75 for each end), resulting in ∼150 M paired-end reads per sample.
Hi-C data analysis
Fastq files were mapped to the hg38 genome using Burrows-Wheeler Aligner (BWA-MEM) with default parameters, mapping each end of the read pairs individually. The mapped files were processed through HiC-Pro (Servant et al., 2015), filtering for mapping quality (MAPQ) score >30 and excluding pairs <1 kb apart, to generate valid pairs and iterative correction of Hi-C data (ICE)–balanced matrices at 500-kb resolution. Hi-C quality control metrics from HiC-Pro are reported in Table S4. Samples were clustered based on HiC-Rep scores calculated using a resolution of 500 kb with a maximum distance of 5 Mb and a smoothing parameter h of 1 (Yang et al., 2017). Hi-C contact matrix heatmaps for cis or trans interactions were generated with Cooler (https://github.com/mirnylab/cooler) using default parameters and logarithmic interaction probabilities, without diagonals. A/B compartmentalization was computed by eigenvalue decomposition of the contact maps using HOMER (Heinz et al., 2010) with 500-kb resolution and no additional windowing (super-resolution was also set at 500 kb). The sign of the first eigenvector (PC1) was selected based on the expression of ∼5,000 genes constitutively expressed across hESC-CM differentiation (Bertero et al., 2019), so that positive and negative values indicate A and B compartmentalization, respectively. Saddle plots of intercompartment interaction enrichment were generated by assigning each genomic bin to its corresponding percentile value based on PC1 and dividing the genome into 10 deciles. Each interaction was normalized to the average score at the corresponding distance (for cis interactions) or to the average of all contacts (for trans interactions) and assigned to a pair of deciles based on the PC1 scores of the two bins. The data were plotted in heatmaps representing the log2 average value for pairs of deciles, while the change between mutant and corrected hiPSC-CMs is the log2 value of the difference in such values. To calculate the interaction probability at varying genomic distances based on compartmentalization, each interaction was assigned to A-A, B-B, or A-B based on the pairs of bins involved, and then the average interaction score for a given distance was normalized to the average interaction score for all pairs of contacts at that distance. The data were plotted on a logarithmic scale and LOESS smoothed using the R function “geom_smooth.” Visualization of sample similarity by PC1 scores was performed with the R function “prcomp.” Gene tracks were generated using Integrative Genomics Viewer (Thorvaldsdóttir et al., 2013). Changes in A/B compartmentalization were determined by a one-way ANOVA of PC1 scores across the two replicates for the three cell lines, using a significance cutoff of P < 0.05 combined by the need for the average PC1 to change sign across at least one pair of condition. Consistent changes in A/B compartmentalization were further selected if the average PC1 score for mutant hiPSC-CMs changed sign compared with the average PC1 score of each corrected hiPSC-CMs.
Statistical analyses
Unless specifically described elsewhere in the methods, all statistical analyses were performed using Prism 7 (GraphPad). The type and number of replicates, the statistics plotted, the statistical test used, and the test results are described in the figure legends. All statistical tests used were two-tailed. No experimental samples were excluded from the statistical analyses. Sample size was not predetermined through power calculations, and no randomization or investigator blinding approaches were implemented during the experiments and data analyses. When a representative experiment is reported, this exemplifies the results obtained in at least two independent biological replications.
Data and code availability
Hi-C and RNA-seq data are available in GEO under accession no. GSE126460. The custom code for Hi-C analyses was previously described (Bertero et al., 2019) and is available on github (https://github.com/pfields8/Fields_et_al_2018/).
Online supplemental material
Fig. S1 shows scarless correction of the LMNA R225X heterozygous mutation in hiPSCs. Fig. S2 shows generation of lamin A/C haploinsufficient hiPSC-CM. Fig. S3 shows characterization of lamin A/C haploinsufficient hiPSC-CMs. Fig. S4 shows global properties of chromatin topology in lamin A/C haploinsufficient hiPSC-CMs. Fig. S5 shows chromatin compartment transitions and associated gene expression changes in lamin A/C haploinsufficient hiPSC-CMs. Table S1 shows CRISPR/Cas9 off-target analysis. Table S2 shows differential gene expression analyses of RNA-seq data. Table S3 shows gene ontology analyses of RNA-seq data. Table S4 shows QC metrics of DNase Hi-C experiments. Table S5 shows A/B compartment analyses of Hi-C data. Table S6 shows ontology enrichment analyses of genes in lamin A/C–sensitive B compartments. Table S7 shows genotyping strategies for the characterization of CRISPR/Cas9-edited hiPSCs. Table S8 shows genomic PCR primers for CRISPR/Cas9 off-target analysis. Table S9 shows RT-qPCR primers. Video 1 shows spontaneous contractions of mutant hiPSC-CMs. Video 2 shows spontaneous contractions of corrected hiPSC-CMs. Video 3 shows calcium fluxes indicated by Fluo-4 fluorescence in monolayers of mutant hiPSC-CMs electrically paced at 1 Hz. Video 4 shows calcium fluxes indicated by Fluo-4 fluorescence in monolayers of corrected hiPSC-CMs electrically paced at 1 Hz. Video 5 shows CCQ analyses in monolayers of mutant hiPSC-CMs electrically paced at 1 Hz. Video 6 shows CCQ analyses in monolayers of corrected hiPSC-CMs electrically paced at 1 Hz. Video 7 shows contractions of 3D-EHTs from mutant hiPSC-CMs. Video 8 shows contractions of 3D-EHTs from corrected hiPSC-CMs.
Supplementary Material
Acknowledgments
We thank the members of the Murry laboratory and of the University of Washington Center for Nuclear Organization and Function, especially Choli Lee for assistance with next-generation sequencing; Galip Gurkan Yardimci for advice on Hi-C analysis; and Katie Mitzelfelt, Kai-Chun Yang, Elaheh Karbassi, Xiulan Yang, and Hans Reinecke for helpful discussions. We are also grateful to Jesse Macadangdang for assistance with CCQ analysis; the Mike and Lynn Garvey Cell Imaging Lab at the University of Washington and its director, Dale Hailey, for assistance with sample imaging and analysis; and the University of Washington Cell Analysis Facility for support with flow cytometry analyses.
A. Bertero is funded by an European Molecular Biology Organization Long-Term Fellowship (ALTF 448-2017). P.A. Fields is funded by the National Institutes of Health through an Experimental Pathology of Cardiovascular Disease 1073 training grant (T32 HL007312). This work is part of the National Institutes of Health 4D Nucleome consortium (NIH U54 DK107979 to C.E. Murry, W.S. Noble, and J. Shendure), with additional support from National Science Foundation grants CBET-1509106 and CMMI-1661730 (to N.J. Sniadecki); National Institutes of Health grants F32 HL126332 (A. Leonard), R01 HL135143 (D-H. Kim and C.E. Murry), R01 HL14643601 (D-H. Kim and C.E. Murry), P01 GM081619 (C.E. Murry), and R01 HL128362 (C.E. Murry); and the Foundation Leducq Transatlantic Network of Excellence (C.E. Murry).
The authors declare no competing financial interests.
Author contributions: A. Bertero: lead methodology, investigation, formal analysis, data curation, visualization, writing – original draft, and writing – review and editing; supporting conceptualization. P.A. Fields: supporting software and formal analysis (Hi-C and RNA-seq); supporting writing – review and editing. A.S.T. Smith: supporting investigation and formal analysis (patch clamp); supporting writing – review and editing. A. Leonard: supporting methodology, software, and formal analysis (3D-EHT contractility). K. Beussman: supporting software and formal analysis (Fluo-4). H-F. Tse: supporting resources (hiPSCs). N.J. Sniadecki, D-H. Kim, J. Shendure, and W.S. Noble: supporting supervision, funding acquisition, and resources. L. Pabon: supporting supervision, project administration, and writing – review and editing. C.E. Murry: lead conceptualization, funding acquisition, supervision, project administration, resources, and writing – review and editing. All authors have read, commented on, and approved the final manuscript.
References
- Adams M.E., Mintz I.M., Reily M.D., Thanabal V., and Bean B.P.. 1993. Structure and properties of omega-agatoxin IVB, a new antagonist of P-type calcium channels. Mol. Pharmacol. 44:681–688. [PubMed] [Google Scholar]
- Adriaens C., Serebryannyy L.A., Feric M., Schibler A., Meaburn K.J., Kubben N., Trzaskoma P., Shachar S., Vidak S., Finn E.H., et al. . 2018. Blank spots on the map: some current questions on nuclear organization and genome architecture. Histochem. Cell Biol. 150:579–592. 10.1007/s00418-018-1726-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinrinade O., Ollila L., Vattulainen S., Tallila J., Gentile M., Salmenpera P., Koillinen H., Kaartinen M., Nieminen M.S., Myllykangas S., et al. . 2015. Genetics and genotype-phenotype correlations in Finnish patients with dilated cardiomyopathy. Eur. Heart J. 36:2327–2337. 10.1093/eurheartj/ehv253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola M., and van Steensel B.. 2015. Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep. 16:610–617. 10.15252/embr.201439789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin A.S., Tan H.L., and Wilde A.A.M.. 2010. Cardiac ion channels in health and disease. Heart Rhythm. 7:117–126. 10.1016/j.hrthm.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Asakura K., Hayashi S., Ojima A., Taniguchi T., Miyamoto N., Nakamori C., Nagasawa C., Kitamura T., Osada T., Honda Y., et al. . 2015. Improvement of acquisition and analysis methods in multi-electrode array experiments with iPS cell-derived cardiomyocytes. J. Pharmacol. Toxicol. Methods. 75:17–26. 10.1016/j.vascn.2015.04.002 [DOI] [PubMed] [Google Scholar]
- Awasthi A., Ramachandran B., Ahmed S., Benito E., Shinoda Y., Nitzan N., Heukamp A., Rannio S., Martens H., Barth J., et al. . 2018. Synaptotagmin-3 drives AMPA receptor endocytosis, depression of synapse strength, and forgetting. Science. 363:eaav1483. [DOI] [PubMed] [Google Scholar]
- Bae S., Park J., and Kim J.S.. 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 30:1473–1475. 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin M., Casini S., Davis R.P., D’Aniello C., Haas J., Ward-van Oostwaard D., Tertoolen L.G.J., Jung C.B., Elliott D.A., Welling A., et al. . 2013. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 32:3161–3175. 10.1038/emboj.2013.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero A., Fields P.A., Ramani V., Bonora G., Yardimci G.G., Reinecke H., Pabon L., Noble W.S., Shendure J., and Murry C.E.. 2019. Dynamics of genome reorganization during human cardiogenesis reveal an RBM20-dependent splicing factory. Nat. Commun. 10:1538 10.1038/s41467-019-09483-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand A.T., Chikhaoui K., Yaou R.B., and Bonne G.. 2011. Clinical and genetic heterogeneity in laminopathies. Biochem. Soc. Trans. 39:1687–1692. 10.1042/BST20110670 [DOI] [PubMed] [Google Scholar]
- Birket M.J., Ribeiro M.C., Verkerk A.O., Ward D., Leitoguinho A.R., den Hartogh S.C., Orlova V.V., Devalla H.D., Schwach V., Bellin M., et al. . 2015. Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nat. Biotechnol. 33:970–979. 10.1038/nbt.3271 [DOI] [PubMed] [Google Scholar]
- Bodi I., Mikala G., Koch S.E., Akhter S.A., and Schwartz A.. 2005. The L-type calcium channel in the heart: the beat goes on. J. Clin. Invest. 115:3306–3317. 10.1172/JCI27167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne G., Di Barletta M.R., Varnous S., Bécane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A., et al. . 1999. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 21:285–288. 10.1038/6799 [DOI] [PubMed] [Google Scholar]
- Buchwalter A., Kaneshiro J.M., and Hetzer M.W.. 2019. Coaching from the sidelines: the nuclear periphery in genome regulation. Nat. Rev. Genet. 20:39–50. 10.1038/s41576-018-0063-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M., et al. . 2014. Chemically defined generation of human cardiomyocytes. Nat. Methods. 11:855–860. 10.1038/nmeth.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell B.C., and Collins F.S.. 2006. Human laminopathies: nuclei gone genetically awry. Nat. Rev. Genet. 7:940–952. 10.1038/nrg1906 [DOI] [PubMed] [Google Scholar]
- Captur G., Arbustini E., Bonne G., Syrris P., Mills K., Wahbi K., Mohiddin S.A., McKenna W.J., Pettit S., Ho C.Y., et al. . 2018. Lamin and the heart. Heart. 104:468–479. 10.1136/heartjnl-2017-312338 [DOI] [PubMed] [Google Scholar]
- Carson D., Hnilova M., Yang X., Nemeth C.L., Tsui J.H., Smith A.S.T., Jiao A., Regnier M., Murry C.E., Tamerler C., and Kim D.-H.. 2016. Nanotopography-Induced Structural Anisotropy and Sarcomere Development in Human Cardiomyocytes Derived from Induced Pluripotent Stem Cells. ACS Appl. Mater. Interfaces. 8:21923–21932. 10.1021/acsami.5b11671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall W.A., Perez-Reyes E., Snutch T.P., and Striessnig J.. 2005. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol. Rev. 57:411–425. 10.1124/pr.57.4.5 [DOI] [PubMed] [Google Scholar]
- Cattin M.-E., Muchir A., and Bonne G.. 2013. ‘State-of-the-heart’ of cardiac laminopathies. Curr. Opin. Cardiol. 28:297–304. 10.1097/HCO.0b013e32835f0c79 [DOI] [PubMed] [Google Scholar]
- Chen W.V., and Maniatis T.. 2013. Clustered protocadherins. Development. 140:3297–3302. 10.1242/dev.090621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A.. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.C., Muchir A., Wu W., Iwata S., Homma S., Morrow J.P., and Worman H.J.. 2012. Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci. Transl. Med. 4:144ra102 10.1126/scitranslmed.3003875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Belmont A.S., Guttman M., Leshyk V.O., Lis J.T., Lomvardas S., Mirny L.A., O’Shea C.C., Park P.J., Ren B., et al. 4D Nucleome Network . 2017. The 4D nucleome project. Nature. 549:219–226. 10.1038/nature23884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., and Ren B.. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 485:376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W., et al. . 2015. Chromatin architecture reorganization during stem cell differentiation. Nature. 518:331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzynska A., Gonzalo S., Shanahan C., and Askjaer P.. 2016. The nuclear lamina in health and disease. Nucleus. 7:233–248. 10.1080/19491034.2016.1183848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner D.A., Caldwell J.L., Kistamás K., and Trafford A.W.. 2017. Calcium and Excitation-Contraction Coupling in the Heart. Circ. Res. 121:181–195. 10.1161/CIRCRESAHA.117.310230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana M., Barison A., Botto N., Panchetti L., Ricci G., Milanesi M., Poletti R., Positano V., Siciliano G., Passino C., et al. . 2013. CMR-verified interstitial myocardial fibrosis as a marker of subclinical cardiac involvement in LMNA mutation carriers. JACC Cardiovasc. Imaging. 6:124–126. 10.1016/j.jcmg.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Fraser J., Ferrai C., Chiariello A.M., Schueler M., Rito T., Laudanno G., Barbieri M., Moore B.L., Kraemer D.C., Aitken S., et al. FANTOM Consortium . 2015. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol. Syst. Biol. 11:852 10.15252/msb.20156492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., and van Steensel B.. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 453:948–951. 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- Haas J., Frese K.S., Peil B., Kloos W., Keller A., Nietsch R., Feng Z., Müller S., Kayvanpour E., Vogel B., et al. . 2015. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur. Heart J. 36:1123–35a. 10.1093/eurheartj/ehu301 [DOI] [PubMed] [Google Scholar]
- Harr J.C., Luperchio T.R., Wong X., Cohen E., Wheelan S.J., and Reddy K.L.. 2015. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol. 208:33–52. 10.1083/jcb.201405110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselberg N.E., Haland T.F., Saberniak J., Brekke P.H., Berge K.E., Leren T.P., Edvardsen T., and Haugaa K.H.. 2018. Lamin A/C cardiomyopathy: young onset, high penetrance, and frequent need for heart transplantation. Eur. Heart J. 39:853–860. 10.1093/eurheartj/ehx596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.C.Y., Zhou T., Lai W.H., Huang Y., Chan Y.C., Li X., Wong N.L.Y., Li Y., Au K.W., Guo D., et al. . 2011. Generation of induced pluripotent stem cell lines from 3 distinct laminopathies bearing heterogeneous mutations in lamin A/C. Aging (Albany N.Y.). 3:380–390. 10.18632/aging.100277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. . 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31:827–832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., Dekker J., and Mirny L.A.. 2012. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods. 9:999–1003. 10.1038/nmeth.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs P.M., Hanson E.L., Crispell K.A., Toy W., Keegan H., Schilling K., Icenogle T.B., Litt M., and Hershberger R.E.. 2001. Novel lamin A/C mutations in two families with dilated cardiomyopathy and conduction system disease. J. Card. Fail. 7:249–256. 10.1054/jcaf.2001.26339 [DOI] [PubMed] [Google Scholar]
- Katainen R., et al. . 2015. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet. 47:818–821. 10.1038/ng.3335 [DOI] [PubMed] [Google Scholar]
- Kind J., Pagie L., de Vries S.S., Nahidiazar L., Dey S.S., Bienko M., Zhan Y., Lajoie B., de Graaf C.A., Amendola M., et al. . 2015. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 163:134–147. 10.1016/j.cell.2015.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodo K., Ong S.-G., Jahanbani F., Termglinchan V., Hirono K., InanlooRahatloo K., Ebert A.D., Shukla P., Abilez O.J., Churko J.M., et al. . 2016. iPSC-derived cardiomyocytes reveal abnormal TGF-β signalling in left ventricular non-compaction cardiomyopathy. Ber. Nat. Cell Biol. 18:1031–1042. 10.1038/ncb3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A., and Duan Z.. 2018. Understanding the 3D genome: Emerging impacts on human disease. Semin. Cell Dev. Biol. 90:62–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Baldinger S.H., Gandjbakhch E., Maury P., Sellal J.M., Androulakis A.F.A., Waintraub X., Charron P., Rollin A., Richard P., et al. . 2016. Long-Term Arrhythmic and Nonarrhythmic Outcomes of Lamin A/C Mutation Carriers. J. Am. Coll. Cardiol. 68:2299–2307. 10.1016/j.jacc.2016.08.058 [DOI] [PubMed] [Google Scholar]
- Lee P.J.H., Rudenko D., Kuliszewski M.A., Liao C., Kabir M.G., Connelly K.A., and Leong-Poi H.. 2014. Survivin gene therapy attenuates left ventricular systolic dysfunction in doxorubicin cardiomyopathy by reducing apoptosis and fibrosis. Cardiovasc. Res. 101:423–433. 10.1093/cvr/cvu001 [DOI] [PubMed] [Google Scholar]
- Lee Y.K., Lau Y.M., Cai Z.J., Lai W.H., Wong L.Y., Tse H.F., Ng K.M., and Siu C.W.. 2017. Modeling Treatment Response for Lamin A/C Related Dilated Cardiomyopathy in Human Induced Pluripotent Stem Cells. J. Am. Heart Assoc. 6:e005677 10.1161/JAHA.117.005677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A., Bertero A., Powers J.D., Beussman K.M., Bhandari S., Regnier M., Murry C.E., and Sniadecki N.J.. 2018. Afterload promotes maturation of human induced pluripotent stem cell derived cardiomyocytes in engineered heart tissues. J. Mol. Cell. Cardiol. 118:147–158. 10.1016/j.yjmcc.2018.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. . 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326:289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luperchio T.R., Sauria M.E.G., Wong X., Gaillard M., Tsang P., and Pekrun K.. 2017. Chromosome Conformation Paints Reveal the Role of Lamina Association in Genome Organization and Regulation. bioRxiv. doi:. (Preprint posted March 30, 2017) 10.1101/122226 [DOI] [Google Scholar]
- Lupiáñez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R., et al. . 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 161:1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macadangdang J., Guan X., Smith A.S.T., Lucero R., Czerniecki S., Childers M.K., Mack D.L., and Kim D.-H.. 2015. Nanopatterned Human iPSC-based Model of a Dystrophin-Null Cardiomyopathic Phenotype. Cell. Mol. Bioeng. 8:320–332. 10.1007/s12195-015-0413-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo T., Mandai K., Miyata M., Sakakibara S., Wang S., Sai K., Itoh Y., Kaito A., Fujiwara T., Mizoguchi A., and Takai Y.. 2017. NGL-3-induced presynaptic differentiation of hippocampal neurons in an afadin-dependent, nectin-1-independent manner. Genes Cells. 22:742–755. 10.1111/gtc.12510 [DOI] [PubMed] [Google Scholar]
- McDonough S.I., Swartz K.J., Mintz I.M., Boland L.M., and Bean B.P.. 1996. Inhibition of calcium channels in rat central and peripheral neurons by omega-conotoxin MVIIC. J. Neurosci. 16:2612–2623. 10.1523/JNEUROSCI.16-08-02612.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn K.J., Cabuy E., Bonne G., Levy N., Morris G.E., Novelli G., Kill I.R., and Bridger J.M.. 2007. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell. 6:139–153. 10.1111/j.1474-9726.2007.00270.x [DOI] [PubMed] [Google Scholar]
- Merideth M.A., Gordon L.B., Clauss S., Sachdev V., Smith A.C.M., Perry M.B., Brewer C.C., Zalewski C., Kim H.J., Solomon B., et al. . 2008. Phenotype and course of Hutchinson-Gilford progeria syndrome. N. Engl. J. Med. 358:592–604. 10.1056/NEJMoa0706898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metra M., and Teerlink J.R.. 2017. Heart failure. Lancet. 390:1981–1995. 10.1016/S0140-6736(17)31071-1 [DOI] [PubMed] [Google Scholar]
- Mewborn S.K., Puckelwartz M.J., Abuisneineh F., Fahrenbach J.P., Zhang Y., MacLeod H., Dellefave L., Pytel P., Selig S., Labno C.M., et al. . 2010. Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLoS One. 5:e14342 10.1371/journal.pone.0014342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J.D., Lu J.R., Antos C.L., Markham B., Richardson J., Robbins J., Grant S.R., and Olson E.N.. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 93:215–228. 10.1016/S0092-8674(00)81573-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosqueira D., Mannhardt I., Bhagwan J.R., Lis-Slimak K., Katili P., Scott E., Hassan M., Prondzynski M., Harmer S.C., Tinker A., et al. . 2018. CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. Eur. Heart J. 39:3879–3892. 10.1093/eurheartj/ehy249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A., Pavlidis P., Decostre V., Herron A.J., Arimura T., Bonne G., and Worman H.J.. 2007. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J. Clin. Invest. 117:1282–1293. 10.1172/JCI29042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A., Wu W., Choi J.C., Iwata S., Morrow J., Homma S., and Worman H.J.. 2012. Abnormal p38α mitogen-activated protein kinase signaling in dilated cardiomyopathy caused by lamin A/C gene mutation. Hum. Mol. Genet. 21:4325–4333. 10.1093/hmg/dds265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T.Y., Iwata Y., Arai Y., Komamura K., and Wakabayashi S.. 2008. Activation of Na+/H+ exchanger 1 is sufficient to generate Ca2+ signals that induce cardiac hypertrophy and heart failure. Circ. Res. 103:891–899. 10.1161/CIRCRESAHA.108.175141 [DOI] [PubMed] [Google Scholar]
- Nie J., Duan Q., He M., Li X., Wang B., Zhou C., Wu L., Wen Z., Chen C., Wang D.W., et al. . 2018. Ranolazine prevents pressure overload-induced cardiac hypertrophy and heart failure by restoring aberrant Na+ and Ca2+ handling. J. Cell. Physiol. 234:11587–11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmrich V., and Gross G.. 2012. P/Q-type calcium channel modulators. Br. J. Pharmacol. 167:741–759. 10.1111/j.1476-5381.2012.02069.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J., et al. . 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 485:381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Richards A.A., Zhu X., Joglar J.A., Yin H.L., and Garg V.. 2009. A novel mutation in LAMIN A/C is associated with isolated early-onset atrial fibrillation and progressive atrioventricular block followed by cardiomyopathy and sudden cardiac death. Heart Rhythm. 6:707–710. 10.1016/j.hrthm.2009.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peroz D., Rodriguez N., Choveau F., Baró I., Mérot J., and Loussouarn G.. 2008. Kv7.1 (KCNQ1) properties and channelopathies. J. Physiol. 586:1785–1789. 10.1113/jphysiol.2007.148254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze S.I., Liu J., Nussinovitch U., Ohana L., Backx P.H., Gepstein L., and Keller G.M.. 2017. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 35:56–68. 10.1038/nbt.3745 [DOI] [PubMed] [Google Scholar]
- Rajakulendran S., Kaski D., and Hanna M.G.. 2012. Neuronal P/Q-type calcium channel dysfunction in inherited disorders of the CNS. Nat. Rev. Neurol. 8:86–96. 10.1038/nrneurol.2011.228 [DOI] [PubMed] [Google Scholar]
- Ramani V., Cusanovich D.A., Hause R.J., Ma W., Qiu R., Deng X., Blau C.A., Disteche C.M., Noble W.S., Shendure J., and Duan Z.. 2016. Mapping 3D genome architecture through in situ DNase Hi-C. Nat. Protoc. 11:2104–2121. 10.1038/nprot.2016.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., and Aiden E.L.. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 159:1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast G., Kraushaar U., Buckenmaier S., Ittrich C., and Guth B.D.. 2016. Influence of field potential duration on spontaneous beating rate of human induced pluripotent stem cell-derived cardiomyocytes: Implications for data analysis and test system selection. J. Pharmacol. Toxicol. Methods. 82:74–82. 10.1016/j.vascn.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Redin C., Brand H., Collins R.L., Kammin T., Mitchell E., Hodge J.C., Hanscom C., Pillalamarri V., Seabra C.M., Abbott M.-A., et al. . 2017. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet. 49:36–45. 10.1038/ng.3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J.L., Tulloch N.L., Razumova M.V., Saiget M., Muskheli V., Pabon L., Reinecke H., Regnier M., and Murry C.E.. 2016. Mechanical Stress Conditioning and Electrical Stimulation Promote Contractility and Force Maturation of Induced Pluripotent Stem Cell-Derived Human Cardiac Tissue. Circulation. 134:1557–1567. 10.1161/CIRCULATIONAHA.114.014998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo V., Rago A., Politano L., Papa A.A., Di Meo F., Russo M.G., Golino P., Calabrò R., and Nigro G.. 2012. Increased dispersion of ventricular repolarization in Emery Dreifuss muscular dystrophy patients. Med. Sci. Monit. 18:CR643–CR647. 10.12659/MSM.883541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga A., Karibe A., Otomo J., Iwabuchi K., Takahashi T., Kanno H., Kikuchi J., Keitoku M., Shinozaki T., and Shimokawa H.. 2009. Lamin A/C gene mutations in familial cardiomyopathy with advanced atrioventricular block and arrhythmia. Tohoku J. Exp. Med. 218:309–316. 10.1620/tjem.218.309 [DOI] [PubMed] [Google Scholar]
- Salvarani N., Crasto S., Miragoli M., Bertero A., Paulis M., Kunderfranco P., Serio S., Forni A., Lucarelli C., Dal Ferro M., et al. . 2019. The K219T-Lamin A/C mutation induces myocardial conduction defects through epigenetic inhibition of SCN5A in a human model of cardiac laminopathy. Nat. Commun. 10:2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A.D., Hu M., Jung I., Xu Z., Qiu Y., Tan C.L., Li Y., Lin S., Lin Y., Barr C.L., and Ren B.. 2016. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Reports. 17:2042–2059. 10.1016/j.celrep.2016.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.-J., Vert J.-P., Heard E., Dekker J., and Barillot E.. 2015. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16:259 10.1186/s13059-015-0831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., and Cavalli G.. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 148:458–472. 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., and de Laat W.. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38:1348–1354. 10.1038/ng1896 [DOI] [PubMed] [Google Scholar]
- Siu C.W., Lee Y.K., Ho J.C.Y., Lai W.H., Chan Y.C., Ng K.M., Wong L.Y., Au K.W., Lau Y.M., Zhang J., et al. . 2012. Modeling of lamin A/C mutation premature cardiac aging using patient-specific induced pluripotent stem cells. Aging (Albany N.Y.). 4:803–822. 10.18632/aging.100503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., and Zhang F.. 2016. Rationally engineered Cas9 nucleases with improved specificity. Science. 351:84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniadecki N.J., and Chen C.S.. 2007. Microfabricated Silicone Elastomeric Post Arrays for Measuring Traction Forces of Adherent Cells. Methods Cell Biol. 83:313–328. [DOI] [PubMed] [Google Scholar]
- Solovei I., Wang A.S., Thanisch K., Schmidt C.S., Krebs S., Zwerger M., Cohen T.V., Devys D., Foisner R., Peichl L., et al. . 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 152:584–598. 10.1016/j.cell.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Stevens T.J., Lando D., Basu S., Atkinson L.P., Cao Y., Lee S.F., Leeb M., Wohlfahrt K.J., Boucher W., O’Shaughnessy-Kirwan A., et al. . 2017. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 544:59–64. 10.1038/nature21429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.H., Zhou L., Emerson D.J., Phyo S.A., Titus K.R., Gong W., Gilgenast T.G., Beagan J.A., Davidson B.L., Tassone F., and Phillips-Cremins J.E.. 2018. Disease-Associated Short Tandem Repeats Co-localize with Chromatin Domain Boundaries. Cell. 175:224–238.e15. 10.1016/j.cell.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniura H., Glass C., and Gerace L.. 1995. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J. Cell Biol. 131:33–44. 10.1083/jcb.131.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., and Mesirov J.P.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14:178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobita T., Nomura S., Fujita T., Morita H., Asano Y., Onoue K., Ito M., Imai Y., Suzuki A., Ko T., et al. . 2018. Genetic basis of cardiomyopathy and the genotypes involved in prognosis and left ventricular reverse remodeling. Sci. Rep. 8:1998 10.1038/s41598-018-20114-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., and Pachter L.. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., and Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berlo J.H., de Voogt W.G., van der Kooi A.J., van Tintelen J.P., Bonne G., Yaou R.B., Duboc D., Rossenbacker T., Heidbüchel H., de Visser M., et al. . 2005. Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: do lamin A/C mutations portend a high risk of sudden death? J. Mol. Med. (Berl.). 83:79–83. 10.1007/s00109-004-0589-1 [DOI] [PubMed] [Google Scholar]
- Vandael E., Vandenberk B., Vandenberghe J., Willems R., and Foulon V.. 2017. Risk factors for QTc-prolongation: systematic review of the evidence. Int. J. Clin. Pharm. 39:16–25. 10.1007/s11096-016-0414-2 [DOI] [PubMed] [Google Scholar]
- van Rijsingen I.A.W., Arbustini E., Elliott P.M., Mogensen J., Hermans-van Ast J.F., van der Kooi A.J., van Tintelen J.P., van den Berg M.P., Pilotto A., Pasotti M., et al. . 2012. Risk factors for malignant ventricular arrhythmias in lamin a/c mutation carriers. J. Am. Coll. Cardiol. 59:493–500. 10.1016/j.jacc.2011.08.078 [DOI] [PubMed] [Google Scholar]
- van Steensel B., and Belmont A.S.. 2017. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell. 169:780–791. 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tintelen J.P., Hofstra R.M.W., Katerberg H., Rossenbacker T., Wiesfeld A.C.P., du Marchie Sarvaas G.J., Wilde A.A.M., van Langen I.M., Nannenberg E.A., van der Kooi A.J., et al. Working Group on Inherited Cardiac Disorders, line 27/50, Interuniversity Cardiology Institute of The Netherlands . 2007a High yield of LMNA mutations in patients with dilated cardiomyopathy and/or conduction disease referred to cardiogenetics outpatient clinics. Am. Heart J. 154:1130–1139. 10.1016/j.ahj.2007.07.038 [DOI] [PubMed] [Google Scholar]
- van Tintelen J.P., Tio R.A., Kerstjens-Frederikse W.S., van Berlo J.H., Boven L.G., Suurmeijer A.J.H., White S.J., den Dunnen J.T., te Meerman G.J., Vos Y.J., et al. . 2007b Severe myocardial fibrosis caused by a deletion of the 5′ end of the lamin A/C gene. J. Am. Coll. Cardiol. 49:2430–2439. 10.1016/j.jacc.2007.02.063 [DOI] [PubMed] [Google Scholar]
- Won H., de la Torre-Ubieta L., Stein J.L., Parikshak N.N., Huang J., Opland C.K., Gandal M.J., Sutton G.J., Hormozdiari F., Lu D., et al. . 2016. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 538:523–527. 10.1038/nature19847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman H.J., and Courvalin J.-C.. 2004. How do mutations in lamins A and C cause disease? J. Clin. Invest. 113:349–351. 10.1172/JCI20832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Zhang F., Yardımcı G.G., Song F., Hardison R.C., Noble W.S., Yue F., and Li Q.. 2017. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res. 27:1939–1949. 10.1101/gr.220640.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., and Worman H.J.. 1994. Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J. Biol. Chem. 269:11306–11311. [PubMed] [Google Scholar]
- Ye Q., and Worman H.J.. 1996. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem. 271:14653–14656. 10.1074/jbc.271.25.14653 [DOI] [PubMed] [Google Scholar]
- Yu M., and Ren B.. 2017. The Three-Dimensional Organization of Mammalian Genomes. Annu. Rev. Cell Dev. Biol. 33:265–289. 10.1146/annurev-cellbio-100616-060531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Simos G., Blobel G., and Georgatos S.D.. 1991. Binding of lamin A to polynucleosomes. J. Biol. Chem. 266:9211–9215. [PubMed] [Google Scholar]
- Yusa K. 2013. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nat. Protoc. 8:2061–2078. 10.1038/nprot.2013.126 [DOI] [PubMed] [Google Scholar]
- Yusa K., Rashid S.T., Strick-Marchand H., Varela I., Liu P.-Q., Paschon D.E., Miranda E., Ordóñez A., Hannan N.R.F., Rouhani F.J., et al. . 2011. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 478:391–394. 10.1038/nature10424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ai X., Nakayama H., Chen B., Harris D.M., Tang M., Xie Y., Szeto C., Li Y., Li Y., et al. . 2016. Persistent increases in Ca(2+) influx through Cav1.2 shortens action potential and causes Ca(2+) overload-induced afterdepolarizations and arrhythmias. Basic Res. Cardiol. 111:4 10.1007/s00395-015-0523-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Kim Y., and Zheng Y.. 2015. Identification of lamin B-regulated chromatin regions based on chromatin landscapes. Mol. Biol. Cell. 26:2685–2697. 10.1091/mbc.E15-04-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Hu J., Yue S., Kristiani L., Kim M., Sauria M., Taylor J., Kim Y., and Zheng Y.. 2018. Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery. Mol. Cell. 71:802–815.e7. 10.1016/j.molcel.2018.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Hi-C and RNA-seq data are available in GEO under accession no. GSE126460. The custom code for Hi-C analyses was previously described (Bertero et al., 2019) and is available on github (https://github.com/pfields8/Fields_et_al_2018/).