Ben-Yishay et al. measure the interactions of the mRNA export factor NXF1 with NPC components in individual nuclear pores in intact human cells and show that NXF1 functions on the cytoplasmic side of the NPC for the release of mRNA into the cytoplasm.
Abstract
Translocation of mRNA through the nuclear pore complex (NPC) requires interactions with different NPC regions. To determine the interactions that are crucial for effective mRNA export in living cells, we examined mRNA export within individual pores by applying various types of mRNA export blocks that stalled mRNPs at different stages of transition. Focusing on the major mRNA export factor NXF1, we found that initial mRNP binding to the NPC did not require NXF1 in the NPC, whereas release into the cytoplasm did. NXF1 localization in the NPC did not require RNA or RNA binding. Superresolution microscopy showed that NXF1 consistently occupied positions on the cytoplasmic side of the NPC. Interactions with specific nucleoporins were pinpointed using FLIM-FRET for measuring protein–protein interactions inside single NPCs, showing that Dbp5 helicase activity of mRNA release is conserved in yeast and humans. Altogether, we find that specific interactions on the cytoplasmic side of the NPC are fundamental for the directional flow of mRNA export.
Introduction
Export of mRNAs from the nucleus is required for the normal function and regulation of the eukaryotic cell. Nuclear exit of mRNAs is a process using several classes of proteins and complexes that facilitate the transfer of an mRNA from the site of transcription through the nuclear pore complex (NPC) to translation in the cytoplasm. The NPC is a highly conserved eightfold symmetry channel (∼185–285 nm long and ∼120 nm wide in human cells) composed of proteins termed nucleoporins (Nups; Fahrenkrog and Aebi, 2003; Wälde and Kehlenbach, 2010; Fernandez-Martinez and Rout, 2012; Maimon et al., 2012; Adams and Wente, 2013), which demarcate three major domains: a nucleoplasmic ring–like region termed the “basket,” a central transmembranal channel, and a cytoplasmic filamentous domain. The central domain contains a core scaffold that serves as a barrier characterized by phenylalanine-glycine (FG) repeats; thus, the Nups carrying these repeats are termed FG-Nups (Fahrenkrog et al., 2002; Terry and Wente, 2009).
Cellular mRNAs are in dynamic association with multiple proteins and are exported to the cytoplasm as mRNA–protein complexes termed mRNPs. Various nuclear factors involved in mRNA export have been identified via studies on yeast mutants that accumulate nuclear polyA+ RNA, showing that mRNA exit is specifically dependent on the essential shuttling export receptor Mex67 in yeast or its orthologue NXF1/Tap in metazoans (Segref et al., 1997; Katahira et al., 1999). This protein acts as a heterodimer with Mtr2 (in yeast) or NXT1/p15 (in mammals; Suyama et al., 2000). Other key factors such as the DEAD-box protein 5 (Dbp5) helicase (DDX19B in vertebrates) and the transcription export (TREX) complex are needed to execute the export process (Snay-Hodge et al., 1998; Schmitt et al., 1999; Rodrigues et al., 2001; Lund and Guthrie, 2005). The protein composition of the mRNP changes during export (Dreyfuss et al., 2002; Müller-McNicoll and Neugebauer, 2013). NXF1 is recruited to the mRNP in the nucleus. At the nuclear envelope, it interacts with FG-Nups and mediates the transport of the mRNA through the NPC (Segref et al., 1997; Santos-Rosa et al., 1998; Kang and Cullen, 1999; Katahira et al., 1999; Bachi et al., 2000; Hurt et al., 2000; Strässer et al., 2000; Braun et al., 2001; Schmitt and Gerace, 2001; Strawn et al., 2001; Zenklusen et al., 2001; Viphakone et al., 2019). On the cytoplasmic side of the NPC, yeast Dbp5 disrupts the interaction between Mex67 and the mRNA, resulting in the release of the transcript to the cytoplasm (Lund and Guthrie, 2005; Tran et al., 2007; Hodge et al., 2011; Ledoux and Guthrie, 2011; Noble et al., 2011). This ensures the unidirectionality of the export process (Wente and Rout, 2010). Although the process of mRNA export and the structure of the NPC are similar in yeast and mammalian cells, they are not identical (Katahira et al., 1999; Köhler and Hurt, 2007; Yao et al., 2007; Bonnet and Palancade, 2014; Kosinski et al., 2016; Beck and Hurt, 2017; Kim et al., 2018; Rajoo et al., 2018; Shav-Tal and Tripathi, 2018), and it is still unclear whether this regulatory mechanism of Dbp5/DDX19B is conserved in mammalian cells (Okamura et al., 2015). For example, the ATPase activity of yeast Dbp5 is inhibited by Nup159, while in mammals, Nup214 (Nup159 homologue) enhances it (Lin et al., 2018). Human Dbp5 contains an inhibitory α helix that regulates its RNA and nucleotide binding that does not exist in fungi (Collins et al., 2009; Lin et al., 2018). There are no direct indications for the influence of Dbp5 on the interactions of NXF1 with the NPC or the mRNA itself. In fact, the factors removed by Dbp5 from the mRNP in higher eukaryotes have yet to be identified (Okamura et al., 2015; Lin et al., 2018).
Live-cell imaging has demonstrated that mRNA export dynamics through the NPC are highly rapid, occurring on the milliseconds scale (Grünwald and Singer, 2010; Mor and Shav-Tal, 2010; Mor et al., 2010; Siebrasse et al., 2012; Ma et al., 2013; Smith et al., 2015; Ben-Yishay et al., 2016). Rapid rates mean that mRNAs are usually not easily detectable in the NPC. Therefore, it has been challenging to measure and to characterize the interactions of mRNAs and export factors with NPC components in living cells and identify which regions of the NPC are important for the regulation of mRNA export. In this study, we used a variety of mRNA export blocks that stall mRNPs during transit (Kylberg et al., 2010; Mor et al., 2010; Hodge et al., 2011) to explore the interactions of endogenous NXF1 with various Nups within individual pores during mRNA export and assess how Dbp5 modulates these interactions in human cells. We applied high-resolution single mRNP tracking in living cells, stimulated emission depletion (STED) superresolution microscopy, and fluorescence lifetime imaging microscopy (FLIM)–Förster resonance energy transfer (FRET) measurements in individual NPCs to present an in-depth view of the interactions of NXF1 with the NPC in intact cells, particularly during the final stages of mRNA release to the cytoplasm.
Results
The position in the NPC at which mRNPs stall during export blockage can point to the site of activity of mRNA export factors
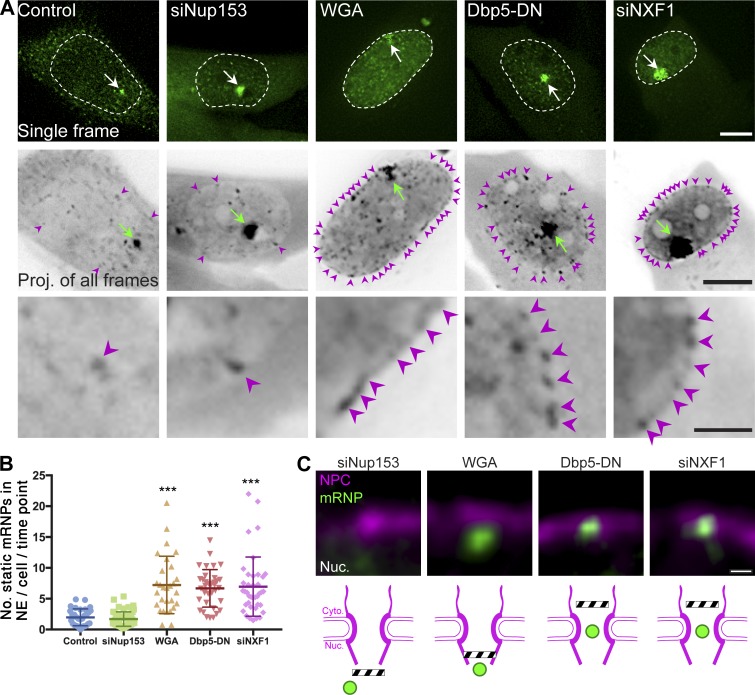
To reveal information about the interactions that are necessary for mRNA export, we examined at which NPC positions mRNPs stalled during different mRNA export blocks. Using live-cell imaging and tracking of single mRNPs, we quantified the mRNPs stuck at the nuclear periphery under the following conditions that block mRNA export: siRNA knockdown of Nup153 (Soop et al., 2005; Umlauf et al., 2013), use of WGA that binds to FG repeats (Davis and Blobel, 1986; Mohr et al., 2009; Kylberg et al., 2010; Mor et al., 2010), expression of a dominant-negative Dbp5 helicase (Dbp5-DN; Hodge et al., 2011), and siRNA knockdown of NXF1 (Johnson et al., 2009; Fig. S1). We followed the behavior of an mRNA tagged with the MS2 system (Mor et al., 2010), used for tracking of single mRNPs in living cells (Tutucci et al., 2018). The inducible gene that expresses this transcript transcribes a Cerulean-minidystrophin-MS2 mRNA that we previously used to examine mRNA nucleocytoplasmic transport and export dynamics in human cells (Carmo-Fonseca, 2010; Mor et al., 2010; Noble and Wente, 2010; Hodge et al., 2011). Time-lapse videos of U2OS cells stably expressing this gene along with the YFP-MS2 coat protein were acquired under the different mRNA export block conditions. The positions of the tagged mRNPs were then analyzed by performing an average projection of the movie into one image. Particles (i.e., mRNPs) that are not highly dynamic tend to stand out in an average time projection (Fig. 1 A). In addition, the average number of static mRNPs at the nuclear envelope per time point in each cell was measured for these movies (Fig. 1 B). Nontreated cells exhibited normal cellular mRNA distribution in which only few static transcripts were detected in the nucleoplasm, as previously shown (Mor et al., 2010). mRNA export inhibition treatments led to pronounced nuclear accumulation of the mRNPs, demonstrating various stalling patterns. When the levels of the nuclear basket component Nup153 were reduced, although the mRNPs accumulated in the nucleoplasm, only a few static mRNPs were detected at the nuclear periphery, as in control cells. WGA treatment, blocking mRNA export by expression of Dbp5-DN, or knocking down NXF1, significantly increased the stalled mRNP population at the nuclear envelope.
Figure 1.
mRNPs are blocked at different points of the NPC during various types of mRNA export blocks. (A) U2OS cells stably expressing YFP-MS2–tagged Cerulean-minidystrophin-MS2 mRNPs were induced to transcribe (4 h), either under normal conditions (control) or during mRNA export blocks, after siRNA knockdown of Nup153 or NXF1; transfection with Cerulean-Dbp5-DN; and treatment with WGA (administered together with digitonin for 5 min). Detection of cells that received the siRNA was performed by cotransfection with mCherry-POM121 and mainly by the export defect seen on the tagged mRNPs in the time-lapse videos. Top: Frames from representative movies showing single mRNPs (green dots). Dashed lines show nuclear borders, and arrows show sites of transcription. Middle: Average time projections showing the static mRNPs in each cell (black dots; marked by magenta arrowheads). Green arrows point to sites of transcription, seen in the projection as large black dots. Scale bars, 10 µm. Bottom: Enlarged areas of the average projection showing mRNPs stuck at the nuclear envelope. Scale bar, 1 µm. (B) Numbers of mRNPs anchored at the nuclear envelope (NE) were counted in control and mRNA export blocked cells. Each spot represents the average number of static mRNPs in the nuclear envelope per time point in one cell. The central line represents the average number of static mRNPs in the nuclear envelope per time point in the appropriate condition. Error bars show SD; ***, P < 0.001. See Materials and methods for the number of mRNPs and cells shown. Measurements were performed in at least three independent experiments. (C) Average time projections from Videos 1, 2, 3, and 4, showing the interactions of single mRNPs (green) with POM121-Cherry–tagged NPCs (magenta) under each export blockage condition. Scheme showing the suggested point of blockage is depicted under each treatment. See Materials and methods for the number of mRNPs and cells shown. Scale bar, 0.5 µm.
We next tracked single mRNPs in living cells, in spatial relation to the nuclear pores, under these mRNA export blocks (Fig. 1 C and Videos 1, 2, 3, and 4). When Nup153 levels were depleted, the mRNPs were mostly mobile and did not associate with the NPCs. When WGA was used, the mRNPs were stalled adjacent to the NPC marker. This was reminiscent of EM images of Balbiani ring mRNPs in WGA-treated cells stalled at a short distance from the nuclear envelope (Kylberg et al., 2010) and presumed to be attached to the nuclear basket but unable to penetrate into the NPC. In contrast, during Dbp5-DN expression or under NXF1 depletion conditions, the signal of the static mRNPs colocalized with the NPC signal. Moreover, we could detect mRNPs penetrating into the NPC zone, but not exiting into the cytoplasm. These data illustrate how different factors function at different locations of the export route at the NPC. The videos show that an mRNP can form productive interactions with the NPC and move into the channel, yet the mRNA export process cannot reach completion. This makes sense for the Dbp5-DN conditions, since Dbp5 is situated on the cytoplasmic side of the NPC at Nup214, and studies in yeast have shown that the interaction of Dbp5 with Nup159 (the yeast homologue of Nup214) and Gle1, and the activity of Nup42, all positioned at the cytoplasmic filaments of the NPC, are required for it to remodel the mRNP (Noble et al., 2011; Adams et al., 2017). Our results demonstrate the direct activity of human Dbp5 in releasing the mRNP from the cytoplasmic side of the NPC, which will be further tested below. Comparably, our data suggest that NXF1 depletion stalls mRNPs toward the cytoplasmic side of the NPC (seen also with other mRNPs tested; Fig. S2 and Videos 5, 6, and 7), as also observed in yeast (Smith et al., 2015), implying that a major site of action of NXF1 during mRNA export is during the final stages of mRNA transition through the pore. This is unexpected, since NXF1 is considered the central mRNA export factor that governs the interactions of mRNAs with the NPC.
A prominent NXF1 population is present at the NPC
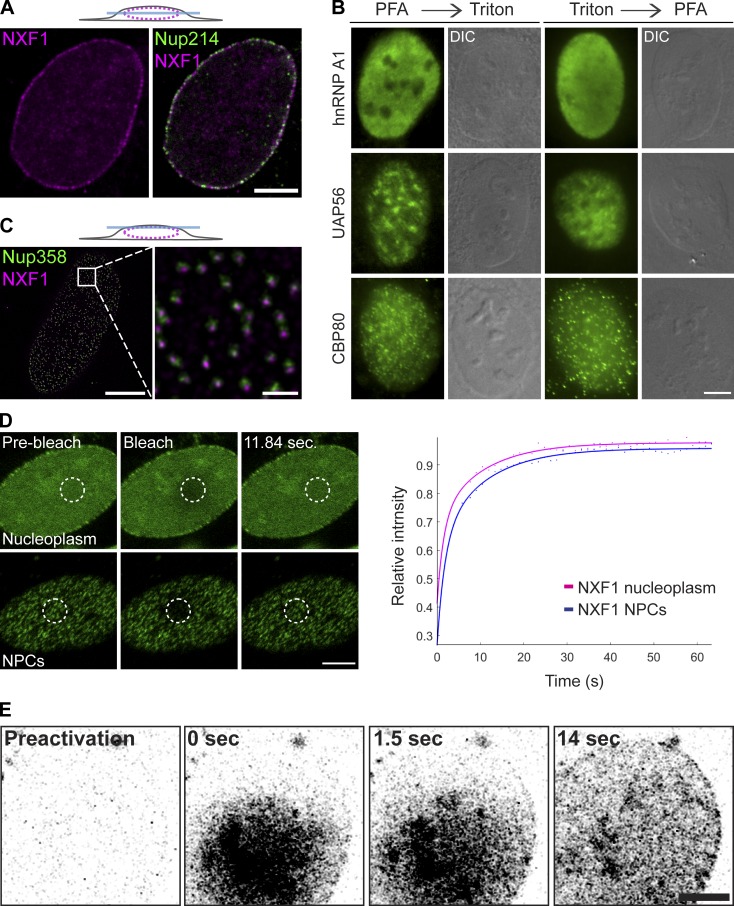
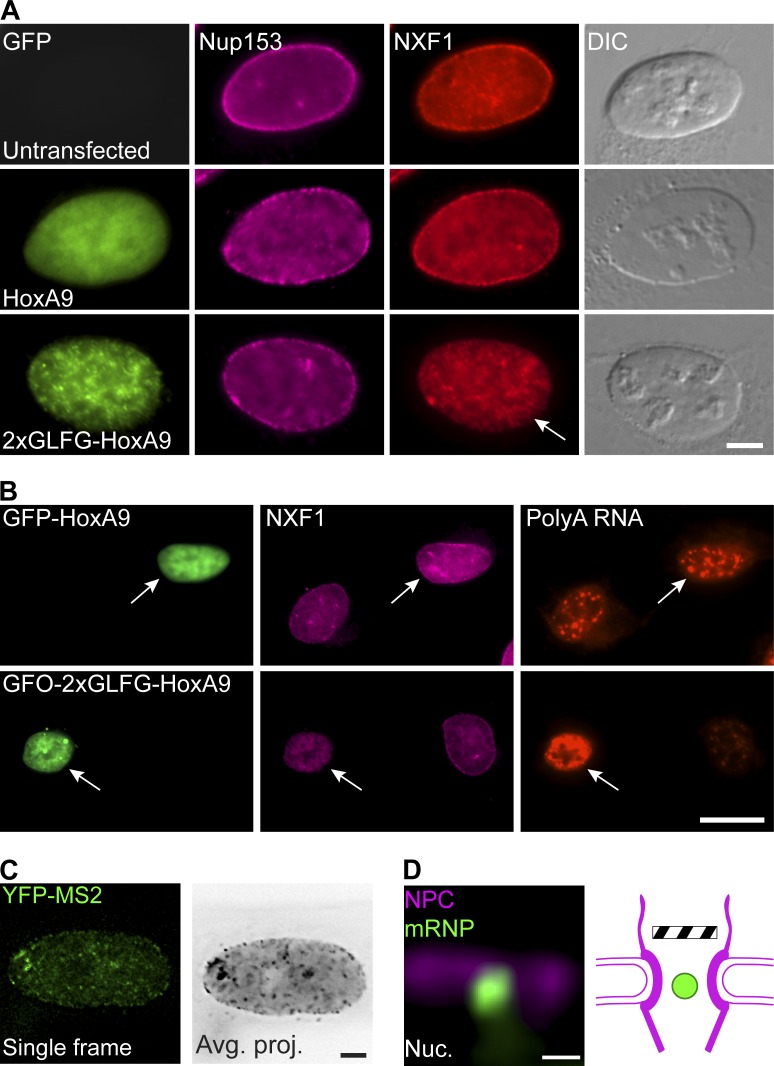
Since the depletion of NXF1 levels causes an mRNA export block as expected but still allows the mRNP to penetrate into the NPC, we decided to examine the point of action of NXF1 at the NPC. As previously demonstrated (Bachi et al., 2000), antibody staining of NXF1 showed nucleoplasmic distribution of the factor, but a prominent population was obvious in the NPCs (Fig. 2 A). Enrichment of an RNA-binding factor at NPCs is not common (Fig. 2 B) but has been observed for import and export factors (Umlauf et al., 2013; Shao et al., 2015). This suggests that NXF1 does not solely act as a typical RNA-binding protein that shuttles between the nucleus and the cytoplasm as part of the mRNP complex. Using STED superresolution microscopy, we could focus on the top plane of the nucleus to detect individual NPCs and found that most NPCs (>98%) contained an NXF1 signal (Fig. 2 C). This could suggest that all NPCs are engaged in mRNA export. However, measuring the dynamics of GFP-NXF1 by FRAP while focusing on the NPCs at the surface of the nucleus showed a recovery time of ∼10 s (Fig. 2 D), which is faster than the recovery times measured for Nups (Rabut et al., 2004) and significantly slower than the rapid time frame of mRNA export (Ben-Yishay et al., 2016). The two time frames of NXF1 and the mRNP at the NPC do not correlate, implying that NXF1 can be present in the NPC even if the mRNP has passed through, which suggests that NXF1 may be present in the NPC even when mRNA is not being exported and may serve several mRNA export cycles as concluded for Dbp5 (Hodge et al., 2011). The unique dynamics of NXF1 at the NPC were further corroborated by use of a photoactivatable NXF1. Photoactivation of PAGFP-NXF1 in the nucleoplasm showed that the factor reached the NPCs within several seconds under normal and export-blockage conditions (Fig. 2 E, Fig. 8 D, and Video 8). Since the time required for an mRNP to travel from the site of transcription to the NPCs is in the range of minutes (Ben-Ari et al., 2010; Mor et al., 2010), this suggests that a fraction of nucleoplasmic NXF1 can reach the NPCs independently of mRNA. This was tested below.
Figure 2.
NXF1 is constantly present at the NPC. (A) Coimmunostaining of endogenous NXF1 and Nup214, imaged by confocal microscopy focusing on the middle plane of the nucleus. (B) Immunofluorescence of hnRNP A1, UAP56, and CBP80 in cells permeabilized after fixation (left) or before fixation (right; see Materials and methods). DIC, differential interference contrast. (C) Colocalization of NXF1 (magenta) with the NPCs (Nup358, green) in U2OS cells imaged by STED, focusing on the upper surface of the nucleus. Scale bars, 5 µm (left) and 0.5 µm (right). (D) FRAP experiment on U2OS cells stably expressing GFP-NXF1. Cells were imaged and photobleached in the nucleoplasm or at the NPCs. Fluorescence recovery was tracked over time and plotted. See Materials and methods for the number of cells shown. Fit of average of three repeated experiments is presented (script FRAP_fit in the online supplemental materials). (E) Photoactivatable PAGFP-NXF1 was expressed in cells. A nucleoplasmic region was activated, and the NXF1 signal (black) accumulating in the NPCs was tracked over time. Frames from Video 8. Scale bars, 5 µm.
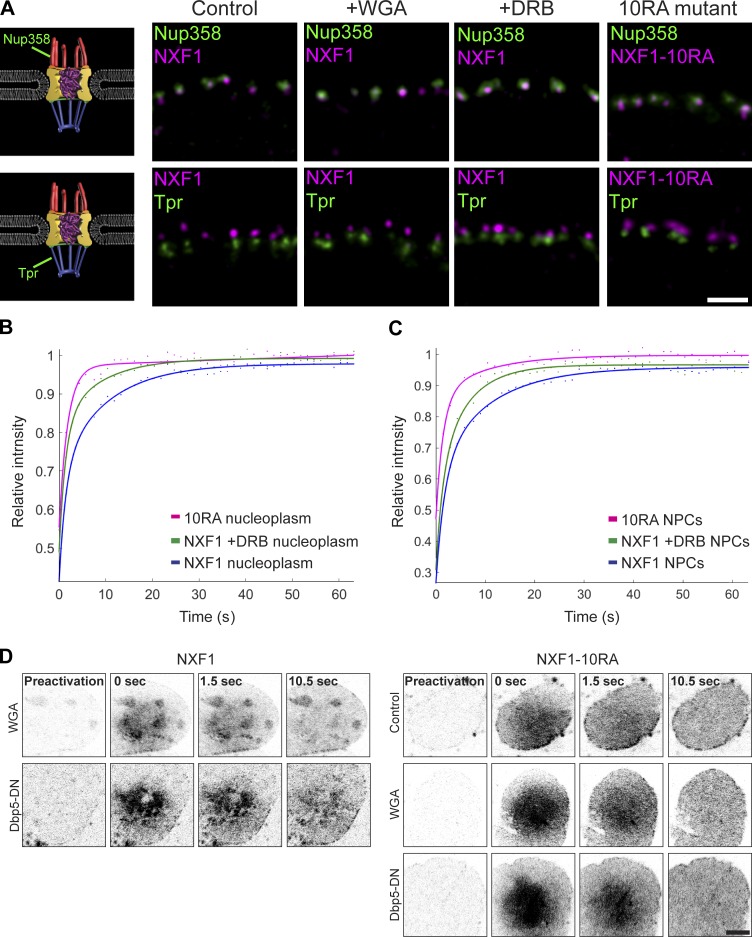
Figure 8.
The position and dynamics of mRNA-free NXF1 in NPCs. (A) STED images showing the position of endogenous NXF1 relative to endogenous Nup358 (top) or Tpr (bottom) in control, WGA-treated, and DRB-treated cells. NXF1-10RA mutant (anti-Myc) was also tested. (B and C) FRAP experiment on U2OS cells expressing GFP-NXF1-10RA or GFP-NXF1 under untreated and DRB-treated conditions. Cells were imaged and photobleached in the nucleoplasm (B) or at the NPCs (C). Fluorescence recovery was tracked over time and plotted. See Materials and methods for the number of cells shown. Fit of average of three repeated experiments is presented. (D) Photoactivatable PAGFP-NXF1 (left) and PAGFP-NXF1-10RA (right) expressed in cells under WGA or Dbp5-DN mRNA export block conditions. A nucleoplasmic region was photoactivated, and the signal accumulating in the NPCs was tracked over time. Scale bar, 5 µm.
Detection of specific NXF1 interactions with Nups within individual NPCs
To obtain higher resolution measurements of NXF1 interactions with specific Nups within the NPC, we implemented a FLIM-FRET approach. FLIM measures the change in the decay rate of the fluorescence of the donor fluorophore during FRET and is therefore intensity independent. As a result of the energy transfer to the acceptor, the FRET process can be monitored as a decrease of the fluorescence lifetime (LT) of the donor in comparison to the LT of the stand-alone donor (where there is no energy transfer or FRET). Therefore, if the molecules are interacting and FRET occurs, then a shortening of donor lifetime is observed due to energy transfer to the acceptor.
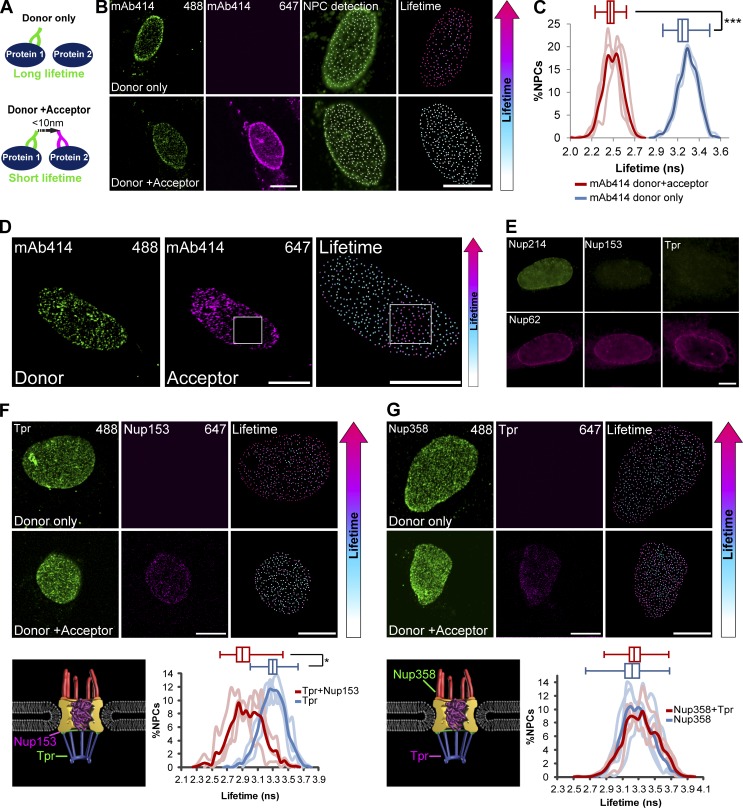
To prove that interactions within the NPC can be detected at single-pore resolution by FLIM-FRET, we performed a control immunofluorescence experiment with the well-known antibody mAb414 that binds to the FXFG motif present in several Nups (Nup358, Nup214, Nup62, and Nup153; Davis and Blobel, 1986). FLIM-FRET was measured between two secondary antibodies, each with a different fluorophore that together form a FRET pair, which were bound to the same primary mAb414 antibody (Fig. 3 A). The high concentration of FG repeats in the NPC provides many binding sites for mAb414, and we expected high FRET levels. Cells were first imaged by standard fluorescence confocal microscopy to identify single pores and then immediately imaged to measure the photon LT for each pixel. Lifetime values were assigned for each individual NPC, and measurements were obtained from thousands of pores. LT measurements were also performed when no acceptor fluorophore was present (donor only) so that the donor molecule could not transfer energy (Fig. 3 B). Indeed, high FRET levels were detected, which are presented as a shift to the left of the LT plots (Fig. 3 C). As a control, we performed photobleaching of the acceptor in a region of NPCs. This led to loss of FRET, as demonstrated by longer donor LT values in the photobleached area (Fig. 3 D). To exclude the possibility of antibody accessibility bias, cells were treated with digitonin, which does not permeabilize the nuclear envelope. Antibodies to Nups found within the internal part of the NPC channel stained the NPCs (Fig. 3 E), showing that antibodies can penetrate the NPC from the cytoplasmic side.
Figure 3.
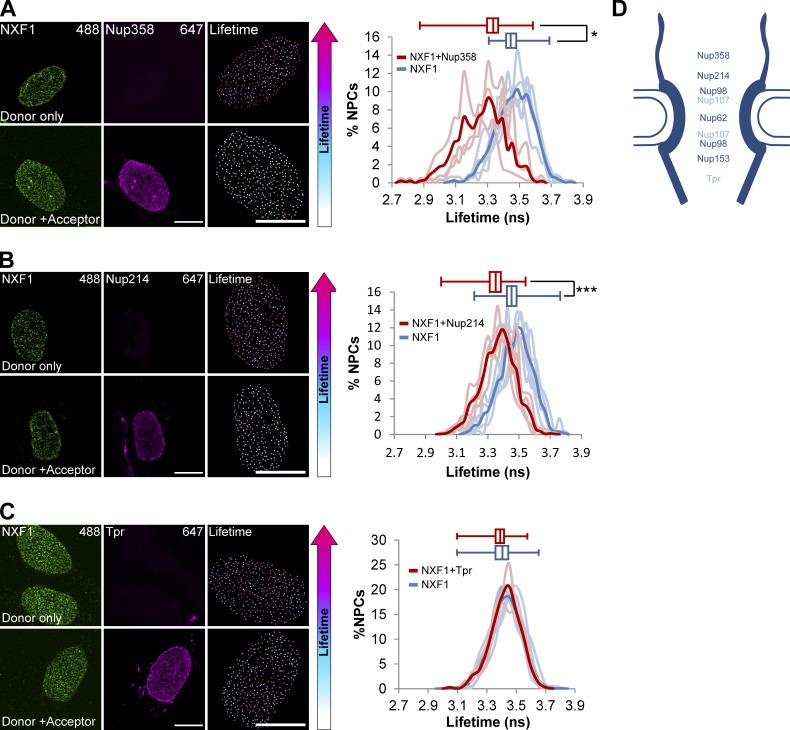
Detecting interactions within single NPCs using FLIM-FRET. (A) Scheme describing a FLIM-FRET experiment. A pair of proteins is immunostained with either one (donor only) or two primary antibodies (donor + acceptor) and two secondary antibodies (Alexa Fluor 488 as the donor fluorophore and Alexa Fluor 647 as the acceptor). If the distance between the proteins is <10 nm, then FRET occurs and the LT of the donor shortens. (B) NPCs were immunostained using mAb414. Cells were either stained with one secondary antibody (donor only, Alexa Fluor 488) or two secondary antibodies (donor + acceptor, Alexa Fluor 488 and Alexa Fluor 647). Single NPCs were detected using Imaris, and the lifetime values for the pixels of each individual NPC were measured. All detected NPCs were pseudo-colored according to the donor’s LT values. Right-hand boxes show an enlarged image of the nucleus. (C) Histograms showing the LT values of the mAb414-donor in single NPCs in donor-only cells (blue; n = 907 NPCs, 3 cells) compared with donor + acceptor cells (red; n = 1,179 NPCs, 4 cells) in one representative experiment. Light lines represent data from single cells, and the bold plot contains data from all cells. Box plots of the LT values show the reduction in the lifetimes when the acceptor is introduced (center line, median; box limits, upper and lower quartiles; whiskers, minimum to maximum range). ***, P < 0.001. FLIM-FRET measurements were performed in three independent experiments. ns, nanoseconds. (D) Control acceptor-bleach experiment. Mab414 labeled NPCs were stained with two secondary antibodies. The acceptor was photobleached in the marked area (box), and the LT was measured in all NPCs. There was no FRET in the bleached region. (E) Antibodies can penetrate into the NPC from the cytoplasmic side. Cells were permeabilized with digitonin. Nup214 and Nup62 inside the NPC were stained, but nuclear basket Nups Nup153 and Tpr were not. (F and G) Immunostaining and LT analysis of (F) Tpr (donor) along with Nup153 (acceptor) or (G) Nup358 (donor) along with Tpr (acceptor). Detected NPCs were pseudocolored according to the donor’s LT values, and analysis is as in C above. Right-hand boxes show an enlarged image of the nucleus. The histogram shows the LT values of donor cells (blue) compared with donor plus acceptor cells (red). Light lines represent data from single cells, and the bold plot contains data from all cells. See Materials and methods for the number of NPCs and cells shown. *, P < 0.05. Scale bars, 10 µm.
We then measured interactions between two different NPC components within individual NPCs in intact cells. FLIM-FRET was detected between Tpr and Nup153 at the nuclear basket (Fig. 3 F). In contrast, there was no energy transfer between two relatively distant molecules, Tpr and Nup358, located on either side of the NPC (Fig. 3 G). Next, we acquired FLIM-FRET measurements to measure interactions between NXF1 and mAb414, and significant FRET was detected as expected (Fig. S3 A). Then a series of experiments was performed in which FLIM-FRET between NXF1 (as donor) and specific Nups (as acceptors) was measured. While there was no evident LT decrease with Tpr and Nup107 as acceptors, interactions were measured between NXF1 and Nup153, Nup62, Nup98, Nup214, and Nup358 (Fig. 4 and Fig. S3, B–E). Using this FLIM-FRET approach, we were able to detect, for the first time in intact cells, interactions between NXF1 and various Nups at the endogenous level within single NPCs.
Figure 4.
Interactions of NXF1 with Nups in single NPCs detected using FLIM-FRET. (A–C) Immunostaining and LT analysis of NXF1 (donor) along with (A) Nup358, (B) Nup214, and (C) Tpr (acceptor). Cells were immunostained with either donor-only or donor + acceptor primary antibodies and with two secondary antibodies (Alexa Fluor 488 donor and Alexa Fluor 647 acceptor). Histograms show the LT values of the NXF1-donor in single NPCs in donor-only cells (blue) compared with donor + acceptor cells (red), in a representative experiment. Box plots of the LT values show the reduction in lifetimes when the Nup358 or Nup214 acceptor are introduced. Light lines represent data from single cells, and the bold plot contains data from all cells. ns, nanoseconds. See Materials and methods for the number of NPCs and cells shown. ***, P < 0.001; *, P < 0.05. Scale bars, 10 µm. (D) Summary of the FLIM-FRET measurements between NXF1 and indicated Nups (see Fig. S3). Dark blue indicates Nups that were found to interact with NXF1.
The main fraction of NXF1 is situated on the cytoplasmic side of the NPC
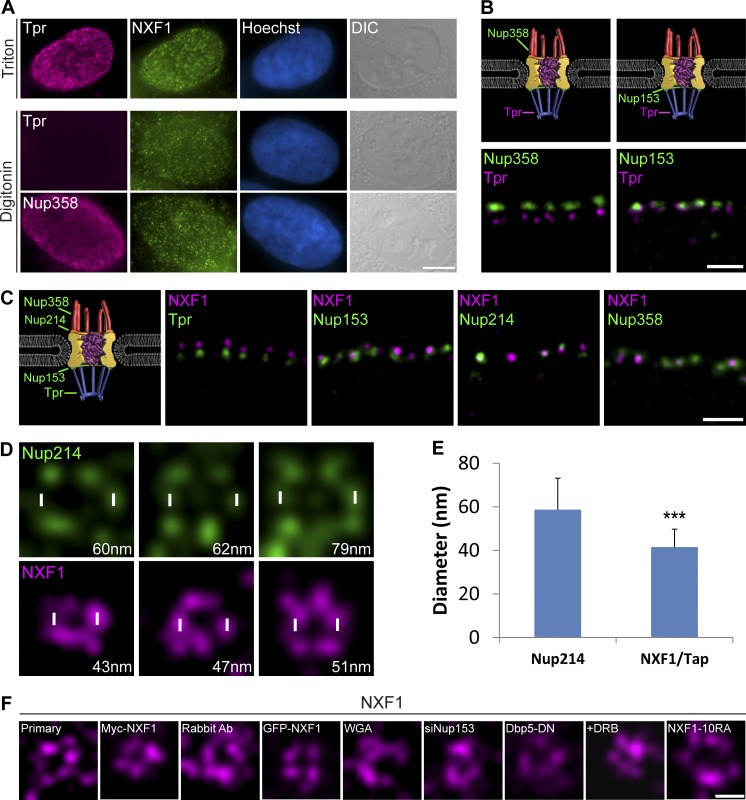
Since the FLIM-FRET analysis showed that NXF1 can interact with several positions in the NPC, we wished to determine where NXF1 is situated within the pore. First, we performed immunofluorescence with two types of detergents. Using Triton X-100, which permeabilizes the cell membrane and nuclear envelope, we could detect NXF1 in individual NPCs as well as the inner nuclear staining of Tpr, a nuclear basket protein. In contrast, digitonin conditions, which do not permeabilize the nuclear envelope, did not show Tpr staining on the nuclear side of the NPC but did show staining of NXF1 on the cytoplasmic side of the NPC, as also observed with Nup358 (Fig. 5 A), meaning that a prominent NPC population of NXF1 is situated on the cytoplasmic side of the pore.
Figure 5.
NXF1 is situated mainly on the cytoplasmic side of the NPC. (A) U2OS cells were permeabilized with either Triton X-100 (top) or digitonin (bottom) and coimmunostained with anti-NXF1 together with anti-Tpr (nuclear basket marker) or anti-Nup358 (cytoplasmic side marker). Hoechst DNA stain, blue; differential interference contrast (DIC), gray. Scale bar, 5 µm. (B) STED images of regions of the nuclear envelope of U2OS cells immunostained with antibodies to Tpr and Nup358 (left) or Nup153 (right). (C) Representative STED images of regions of the nuclear envelope immunostained with antibodies to NXF1 together with antibodies against Tpr, Nup153, Nup214, or Nup358 (n = 780, 1,428, 556, and 900 NPCs; 13, 26, 16, and 16 cells, respectively). Measurements were performed in three independent experiments. Scale bars, 0.5 µm. (D) Representative STED images showing a top view of either Nup214 (top) or NXF1 (bottom; in different experiments) within single NPCs imaged at the nuclear surface. Diameter of the circular pattern created by the proteins is shown in the bottom-right corner of each image (script NPC_diameter in the online supplemental material). (E) Plot showing the average diameter of the circular patterns of Nup214 and NXF1. Error bars represent SD. See Materials and methods for the number of NPCs and cells shown. ***, P < 0.001. Measurements were performed in three independent experiments. (F) Representative STED images showing a top view of NXF1 staining in single NPCs imaged at the nuclear surface under the following conditions: using a fluorescently labeled primary anti-NXF1 antibody; Myc-tagged NXF1; rabbit antibody to NXF1; GFP-NXF1; export blocks WGA, siNup153, and Dbp5-DN; DRB treatment; and NXF1-10RA.
Next, we used STED superresolution microscopy to examine whether NXF1 is situated along the NPC or if it is localized to a certain part of each pore. STED provides the resolution to distinguish among the cytoplasmic, central, and nuclear parts of the NPC (Fig. 5 B; Schermelleh et al., 2008; Adams and Wente, 2013). When we examined the position of NXF1 in respect to Nup markers of these three NPC regions, we found that the NXF1 signal was localized to one area of each NPC rather than being spread along the entire pore (Fig. 5 C). In the vast majority of NPCs, NXF1 was in close proximity to the cytoplasmic side markers Nup358 and Nup214. A significantly smaller portion of the pores exhibited a colocalization of NXF1 with the nucleoplasmic side (marked by Nup153), while in none of the pores was NXF1 adjacent to the nuclear basket component Tpr. This suggests that the population of NXF1 molecules in the NPC does not play a significant role during the initial points of mRNA export.
Using STED to examine NXF1 distribution within individual NPCs on the surface of the nucleus showed a punctate circular pattern spanning an ∼41-nm diameter on average, which was significantly smaller than the average ∼59-nm-diameter ring formed by Nup214 (Fig. 5, D and E; Löschberger et al., 2012; Maimon et al., 2012; Göttfert et al., 2013). This ring-shaped pattern of NXF1 demonstrates that NXF1 coats the periphery of the NPC channel rather than concentrating in the center. The peripheral position of NXF1 was maintained under the various mRNA export block conditions and during transcription inhibition (Fig. 5 F). Together, this analysis yields a three-dimensional view of NXF1 localization in individual NPCs, suggesting that this export factor tends to occupy positions within the periphery of the pore ring and mainly on its cytoplasmic side. This concurs with the live-cell imaging of mRNPs that showed that even when NXF1 was absent, mRNPs could still enter the NPC but were not released into the cytoplasm. The position of NXF1 on the cytoplasmic side of the NPC suggests that its main point of activity during mRNA export is at the final step of mRNA export. Namely, the cytoplasmic side of the NPC is a crucial site of function for NXF1 during the release of the mRNA into the cytoplasm.
NXF1 localizes in the NPC independently of mRNA
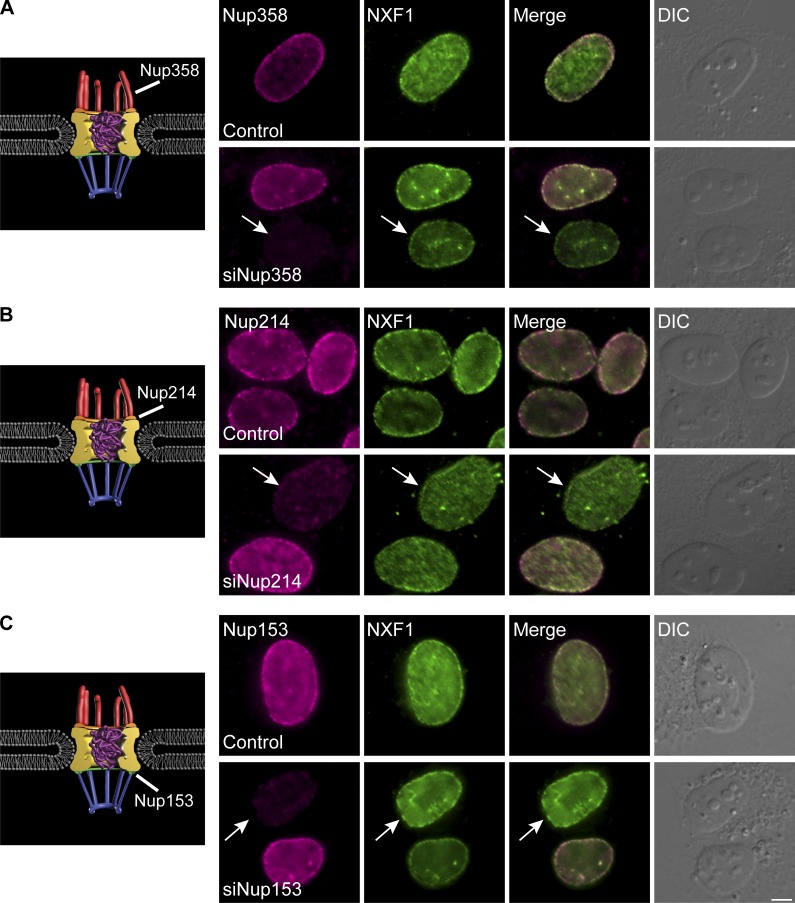
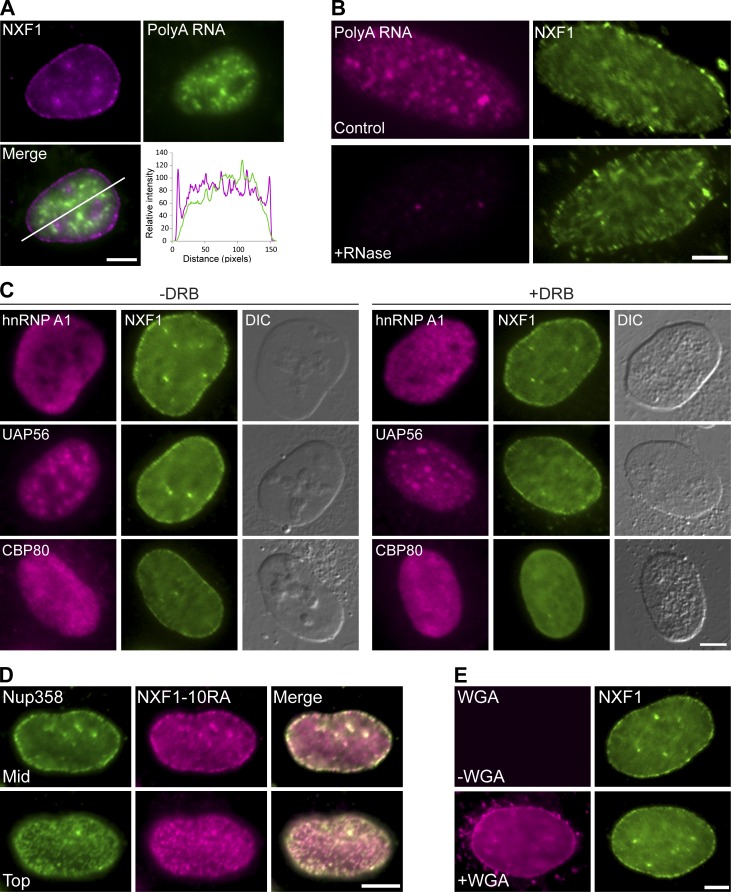
We then examined whether NXF1 was removed or repositioned from the NPC when some of the Nups interacting with it were depleted (Fig. 6 and Fig. S4, A and B). NXF1 remained in the NPC even when Nup358, Nup214, or Nup153 were knocked down. The fact that depleting these Nups can block mRNA export to some extent, particularly the Nup153 knockdown that prevented the entry of mRNPs into the nuclear pore (Fig. 1 C), led us to examine whether NXF1 actually requires mRNA for its localization at the NPC. Examining the colocalization profile of NXF1 with polyA+ RNA in the nucleus showed that RNA transcripts do not accumulate at the NPC, while NXF1 does (Fig. 7 A). The removal of RNA from the cells by treatment of cells with an RNase before fixation (Fig. 7 B), or transcription inhibition by 5,6-dichloro-1-β-D-ribofuranosylbenzimidazol (DRB), which inhibits RNA polymerase II activity (Fig. 7 C), showed that NXF1 was still present in the NPCs, while other RNA-binding proteins were not. A mutant NXF1 that cannot bind RNA (NXF1-10RA; Hautbergue et al., 2008) localized to the NPC (Fig. 7 D). Blocking mRNA export with WGA (Fig. 7 E) or by siRNA knockdown of AlyREF and UAP56 together (adaptors for NXF1 binding to the mRNP; Hautbergue et al., 2009) did not remove NXF1 from the NPC (Fig. S4, C–E). Finally, STED imaging showed that mRNA-free NXF1, either the NXF1-10RA mutant or under WGA export block, or under DRB transcription inhibition conditions, continued to localize at the periphery of the cytoplasmic side of the NPC channel (Figs. 5 F and 8 A).
Figure 6.
NXF1 is present at the NPC when Nups are depleted. (A–C) Immunofluorescence of (A) Nup358, (B) Nup214, or (C) Nup153 (magenta) and NXF1 (green) in control (top) and siRNA-treated cells (bottom). Arrows point at Nup-depleted cells. Scale bar, 5 µm.
Figure 7.
NXF1 is present at the NPC independently of mRNA. (A) PolyA+ RNA detection by RNA FISH in U2OS cells. NXF1 (magenta) is detected in the NPCs while the mRNA (green) is not. The plot shows the intensity of both channels along the line in the merge. (B) PolyA+ RNA detection by RNA FISH in control cells and cells treated with DRB followed by RNase A digestion (RNase was added after permeabilization with 0.5% Triton, 2 min) and stained for NXF1. (C) Immunofluorescence of hnRNP A1, UAP56, CBP80, and NXF1 in control or DRB-treated U2OS cells. Decomposition of nucleoli is seen in the differential interference contrast (DIC) images indicating DRB transcription inhibition. (D) U2OS cell expressing the mutant Myc-NXF1-10RA protein that cannot bind mRNA (anti-Myc) and anti-Nup358 immunofluorescence for NPC detection. Cells were imaged at the middle (mid) or the top planes of the nucleus (top), and merged images show the presence of NXF1-10RA mutant in the NPCs. (E) NXF1 in control cells (top) or WGA-treated cells (bottom). Scale bars, 5 µm.
FRAP analysis demonstrated significantly faster dynamics of the NXF1-10RA mutant compared with wild-type NXF1 in the nucleoplasm and NPCs. This means that NXF1 bound to mRNA can be distinguished from NXF1 that cannot bind mRNA based on its dynamics and that NXF1 participating in mRNA export stalls at the NPC. To further test this, we reduced nuclear mRNA levels by DRB transcription inhibition, expecting this to change NXF1 dynamics at the NPC since more NXF1 free of mRNA should be present. Indeed, this treatment resulted in faster dynamics of NXF1, both in the nucleoplasm and the NPCs (Fig. 8, B and C). Similarly to PAGFP-NXF1 (Fig. 2 E), nucleoplasmic PAGFP-NXF1-10RA was detected in the NPCs within seconds of photoactivation, both under normal conditions and under Dbp5-DN or WGA export blocks (Fig. 8 D). This means that free NXF1 can shuttle between the nucleoplasm and the NPC regardless of mRNA binding or export. Altogether, these findings indicate that the NXF1 population detected in the pores is not necessarily engaged in mRNA transport at all times and that the localization of NXF1 to the cytoplasmic side of the NPC can be independent of mRNA binding.
We then searched for conditions under which NXF1 is removed from the NPCs but is still present in the nucleus to examine if mRNPs can continue to interact with the NPC. This only occurred when FG repeats were overexpressed in the nucleoplasm. These FG domains, Nup98-2xGLFG/HoxA9, taken from Nup98 (Xu and Powers, 2013), served as nucleoplasmic binding sites for NXF1, thereby titrating it out from the pores (Fig. 9 A) and causing an mRNA export block due to the unavailability of NXF1 for export (Fig. 9 B). Tracking of the mRNPs as performed in Fig. 1 B showed significant accumulation of stalled mRNPs at the NPC. Live-cell imaging of single mRNPs under these conditions indicated that denying NXF1 access to the NPC did not prevent mRNP docking at the pore or their transit inwards. Nevertheless, just like NXF1 knockdown conditions, only the final release to the cytoplasm was inhibited (Fig. 9, C and D; and Video 9). This supports the conclusion that NXF1 presence at the pore is required for the final stages of mRNA export.
Figure 9.
Removal of NXF1 from NPCs. (A) U2OS cells were transfected with either GFP-HoxA9 (control) or GFP-2xGLFG-HoxA9 and immunostained with antibodies to NXF1 and Nup153. Overexpressed FG domains dissociated NXF1 (but not Nup153) from the NPC (arrow). Scale bar, 5 µm. (B) U2OS cells transfected with either GFP-HoxA1 (as control) or GFP-2xGLFG-HoxA9 were stained for PolyA+ RNA FISH and immunostained with anti-NXF1. Arrows point at transfected cells, indicating that overexpression of FG domains removes NXF1 from the NPC and inhibits mRNA export. Scale bar, 20 µm. (C) U2OS cells stably expressing YFP-MS2–tagged Cerulean-minidystrophin-MS2 mRNPs were transfected with mCerulean-2xGLFG-HoxA9 and induced to transcribe (4 h). Left: Frame from a representative live-cell video showing single mRNPs (green dots; large dots are sites of transcription). Right: Movie average time projection showing the static mRNPs (black dots). Number of static mRNPs in the nuclear envelope was calculated as in Fig. 1 (mRNPs tracked: 1,558 mRNPs, 22 cells). Measurements were performed in three independent experiments. Scale bar, 10 µm. (D) Average time projection (Video 9) showing an mRNP (green) stably inserted within the POM121-Cherry–tagged signal (NPCs, red) in a cell expressing mCerulean-2xGLFG-HoxA9. Scale bar, 0.5 µm.
The interaction of NXF1 with Nup358 requires Dbp5 activity
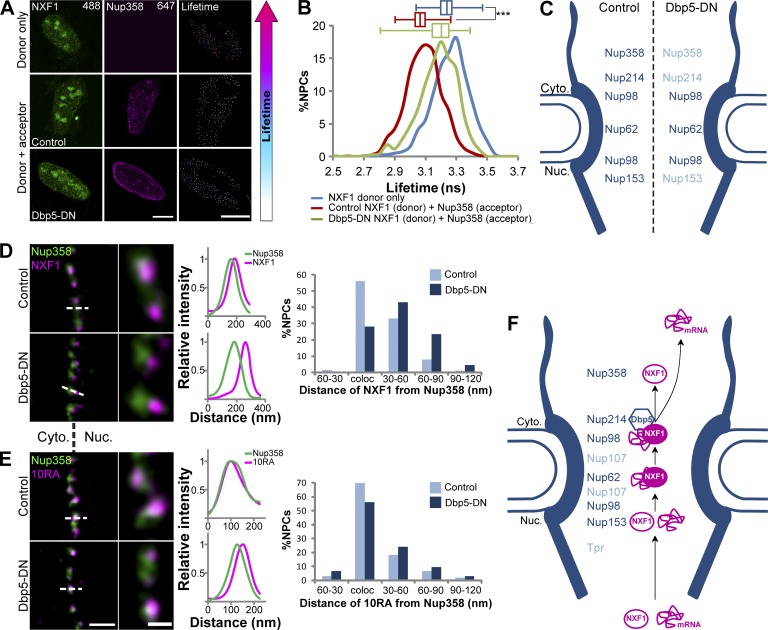
To examine the function of NXF1 at the pore, which is vital for mRNA export, we focused on the export block caused by Dbp5-DN. In yeast, this mutant helicase cannot bind mRNA and therefore does not allow the release of NXF1 from the transcript (Hodge et al., 2011). We found that in human cells, blocking of mRNA export by the expression of Dbp5-DN did not remove NXF1 or NXF1-10RA from the NPC (Fig. S5, A–C). However, FLIM-FRET measurements of NXF1 interaction with Nups under Dbp5-DN conditions showed that NXF1 interactions were retained for Nup98 and Nup62 (data not shown), whereas the interactions of NXF1 with Nup358 and Nup214 were significantly decreased (Fig. 10, A–C; and Fig. S5 D). Since most NXF1 localizes to the cytoplasmic side, we focused on the NXF1–Nup358 interaction.
Figure 10.
Effects of Dbp5 on NXF1 interactions with the NPC. (A) FLIM-FRET analysis of NXF1 interactions with Nups in U2OS cells during an mRNA export block caused by Dbp5-DN. Since the cytoplasmic GFP-Dbp5 signal is lost during the fixation procedure, we used a nucleolar protein (GFP-NOL7) as a transfection marker (NOL7 signal was not included in the analysis; see Materials and methods and Fig. S5). Cells transfected with GFP-NOL7 only (control) or GFP-Dbp5-DN along with GFP-NOL7 (10:1 ratio) were immunostained with either donor-only (anti-NXF1) or donor + acceptor (anti-NXF1 + anti-Nup358) primary antibodies and with two secondary antibodies (Alexa Fluor 488 as donor and Alexa Fluor 647 as acceptor). Analysis was made only on single NPCs detected by Imaris. Detected NPCs were pseudocolored according to the donors’ LT values. Right-hand boxes show the enlarged nucleus area. Scale bar, 10 µm. (B) Histogram showing the LT values of the NXF1-donor in single NPCs in donor-only cells (blue, n = 1,694 NPCs, 6 cells) compared with donor + acceptor control cells (red, n = 1,328 NPCs, 6 cells) and donor + acceptor cells expressing Dbp5-DN (green, n = 786 NPCs, 3 cells). ***, P < 0.001. Box plots show the LT values (center line, median; box limits, upper and lower quartiles; whiskers, minimum to maximum range). (C) Summary of the FLIM-FRET measurements between NXF1 and indicated Nups in control and Dbp5-DN–expressing cells. Dark blue indicates Nups found to interact with NXF1, and light blue indicates loss of interaction due to Dbp5-DN. (D and E) Example for analysis of the distance frequencies between NXF1 (D) or NXF1-10RA (E) and Nup358 signals (script Nup_dis in the online supplemental material). Left: STED images showing the localization of NXF1 relative to Nup358 in control (top) and Dbp5-DN–expressing cells (bottom). An enlarged area is shown on the right. Measured distances (dashed line) were plotted in a histogram (right). See Materials and methods for the number of NPCs and cells shown. Measurements were performed in three independent experiments. Scale bars, 0.3 µm (left) and 0.1 µm (right). (F) Suggested model illustrating the interactions among the mRNA, NXF1, and NPC during mRNA export, as described in the Discussion.
The loss of the interaction between NXF1 and Nup358 under the Dbp5-DN export block, while NXF1 is present in the whole cell, led us to postulate that the NXF1–Nup358 interaction must require the prior shuttling of NXF1 into the nucleus, followed by the exit of the mRNA and NXF1. To test this, we examined the position of NXF1 in the NPC by STED under the Dbp5-DN export block. To pinpoint the exact location of the main NXF1 population at the pore, we measured the distances between NXF1 and Nup358 in individual NPCs (Fig. 10 D). We found that under conditions of the Dbp5-DN mRNA export block, the NXF1 protein signal was shifted inwards into the NPC channel, leaving the Nup358 position devoid of NXF1. In contrast, the positioning at Nup358 of the mutant NXF1-10RA that cannot bind mRNA was not affected by the mRNA export block (Fig. 10 E). These results imply that (a) human Dbp5 acts on mRNA-bound NXF1 and (b) that endogenous NXF1 (and not NXF1-10RA) must undergo mRNA export in order to interact with Nup358. Dbp5 situated at Nup214 releases the mRNA from NXF1, which then proceeds to move to the Nup358 position. Together with the live-cell data (Fig. 1 C), we conclude that this activity taking place on the cytoplasmic side of the NPC is a crucial point at which NXF1 is required to govern the final release of mRNA into the cytoplasm.
Discussion
The passage of mRNA through the NPC is very rapid (Ben-Yishay et al., 2016). This poses technical challenges for detecting mRNPs during export under live-cell imaging conditions and hinders the examination of specific molecular interactions occurring within individual NPCs. In this study, we employed imaging techniques, both in fixed and living cells, to define the NPC domain that is functionally crucial for mRNA release into the cytoplasm. Using various mRNA export blocks, we characterized the role of proteins involved in different stages of mRNA transition through the NPC. We identified the limiting step of mRNA export at the cytoplasmic side of the NPC, requiring the activity of Dbp5 and the presence of NXF1. Importantly, we showed that NXF1 molecules are predominantly located and required on the cytoplasmic side of the NPC.
The MS2 system is the major mRNA tagging system that has allowed the tracking of single mRNP export in living cells and is the source of most of the dynamic data available on the subject. The extensive use of this tagging system on a large variety of genes in many organisms, even endogenously tagged in living mice, demonstrates the robustness of the system for examining mRNA dynamics, keeping in mind that the added tag is exogenous. We find that in unperturbed cells the mRNAs reach the cytoplasm and are translated as expected. Only when mRNA export is blocked are the unique stalling phenotypes observed.
Using STED, we found that almost all NPCs contained NXF1 at any given time, meaning that each NPC has the potential to engage in mRNA export and that NPCs probably do not specialize in the export of specific cargos (Blobel, 1985; Dworetzky and Feldherr, 1988; Huang et al., 1994; Ben-Yishay et al., 2016). In vitro studies and recombinant protein studies have demonstrated that NXF1 can interact with a number of Nups: Nup153, Nup98, Nup62, Nup88, Nup214, and Nup358 (Katahira et al., 1999; Bachi et al., 2000; Forler et al., 2004). Recombinant NXF1 has been detected on both sides of the NPC in yeast and in isolated Xenopus laevis oocyte nuclei (Santos-Rosa et al., 1998; Bachi et al., 2000). However, the position and interactions of endogenous NXF1 within the NPC in intact cells have not been examined at high resolution. Using a FLIM-FRET approach, we mapped NXF1–Nup interactions in individual NPCs in intact cells and found that when a mutant Dbp5 protein (Hodge et al., 2011) was expressed in the cells, some of these interactions were sustained, while interactions with Nup358 and Nup214 were lost. This information, together with the STED data showing that NXF1 localizes on the cytoplasmic side of most NPCs, suggest that this is the major site of action of NXF1 during mRNA export. Previous studies have identified a cytoplasmic docking step for mRNPs as part of the mRNA export process, suggesting remodeling events occurring on the mRNP, including the removal of certain protein factors (Lund and Guthrie, 2005; Smith et al., 2015). This is supported by our live-cell imaging movies showing that the mRNPs could penetrate into the NPC yet were not released when NXF1 levels were depleted. Furthermore, using STED, we peered into NPCs from the nucleus “roof” and detected a ring-like NXF1 signal coating the periphery of the NPC channel. This is in agreement with previous studies concluding that mRNPs pass along the NPC periphery and do not enter the central part of the channel (Huang et al., 1994; Iborra et al., 1998, 2000; Ma et al., 2013).
NXF1 was present at the NPC under all conditions tested, namely, when mRNA export or transcription were blocked, if some Nups and RNA binding adaptors were depleted, or even when no mRNA was present in the cells. The latter was also observed in yeast cells (Katahira et al., 1999). Even when WGA was used or when Nup153 was depleted and mRNPs could not interact with or enter the pore, NXF1 was still found in the NPC. Finally, a mutated version of NXF1 that cannot bind mRNA was also observed in the NPCs. RNA FISH to polyA+ RNA does not show accumulation in NPCs, corroborating the rapid dynamics of mRNA passing through the NPC (∼200 ms; Ben-Yishay et al., 2016), yet the association times of NXF1 with the NPC were in the range of 10 s. These two time frames do not correlate. Moreover, the photoactivation data showed that NXF1 reaches the NPC within seconds. Altogether, this implies that NXF1 can be present in the NPC even if the mRNP has passed through and that there is a fraction of NXF1 in the NPC that is not necessarily transferring mRNA at a given time.
However, NXF1 binds the mRNA in the nucleus and is part of the mRNP. The function of the NPC-bound NXF1 might be similar to what has been suggested for Dbp5 in yeast (not known yet for human cells). Two models have been put forth describing the localization of Dbp5 at the NPC (Heinrich et al., 2017). Either Dbp5 arrives together with the transcript or Dbp5 is already situated at the NPC where it functions. It was proposed that use of mRNA export blocks could help decide between the models. Our study using mRNA export blocks shows that mRNPs could interact with and insert into the NPC even when Dbp5 was defective, suggesting that Dbp5 waits for mRNA in the NPC (Hodge et al., 2011). According to our data showing that NXF1 can be positioned at the NPC independently of mRNA binding, in addition to the live-cell videos showing that mRNPs can enter into the NPC independently of NXF1, one can speculate that, like Dbp5, a fraction of the NXF1 population also waits for the mRNA at the pore. Comparably, mutant Mex67 yeast cells showed mRNPs reaching the cytoplasmic side of the NPC but unable to complete export (Smith et al., 2015). Indeed, even when titrating NXF1 out of the NPCs by overexpressing FG domains in the nucleoplasm, as previously performed in yeast and in human cells (Strawn et al., 2001; Wickramasinghe et al., 2010), mRNPs could enter the NPCs but remained stuck inside, once again showing that NXF1 is required for the later stages of mRNA export.
The mode of action of Dbp5 has been studied mainly in yeast. Although the general mechanism is considered as evolutionally conserved, some differences between mammalian and fungal Dbp5 structure and regulation have been described (Lin et al., 2018). Here, we showed that Dbp5 in human cells also functions on the cytoplasmic side of the NPC and is required for the final movement of NXF1 to the Nup358 position.
It was previously suggested that in Drosophila melanogaster Nup358 is the site from which NXF1 returns to the nucleus after completion of an mRNA export cycle (Forler et al., 2004). We demonstrated that the Nup358 position in mammalian cells is important for NXF1 that has undergone mRNA export. When mRNA export was blocked using the mutant Dbp5-DN, NXF1-10RA still continued to localize to Nup358. In contrast, under the Dbp5-DN mRNA export block, the NXF1 population was not present at Nup358 anymore and held a more inner position and showed reduced interactions with Nup214. This demonstrates that Dbp5 action in yeast and humans is conserved and that Dbp5 affects only mRNA-bound NXF1 to release the mRNA into the cytoplasm and NXF1 to Nup358.
In summary, we propose the following flow of events during mRNA export (Fig. 10 F). The mRNP enters the NPC in a Nup153-dependent manner. NXF1, present in all the NPCs, interacts with the mRNA and with several Nups within the NPC channel, probably along the peripheral ring of the NPC. The mRNP bound with NXF1 encounters Dbp5 situated at Nup214. The helicase separates the mRNA from NXF1, releasing the mRNA into the cytoplasm. NXF1 then moves to Nup358 and from there can presumably recycle back into the nucleus for another round of mRNA export. This study highlights the molecular specialization of the different regions of the NPC in the process of mRNA export.
Materials and methods
Plasmids
The GFP-NXF1 plasmid was obtained from Maria Carmo-Fonseca (Faculdade de Medicina da Universidade de Lisboa, Lisbon, Portugal; Custódio et al., 2004). To generate PA-GFP-NXF1, the NXF1 coding region was inserted (EcoRI and BamHI) into a PA-GFP-C1 plasmid (Shav-Tal et al., 2004). NXF1-10RA construct was obtained from Stuart Wilson (The University of Sheffield, Sheffield, England; Hautbergue et al., 2008). The GFP/PA-GFP coding region was PCR amplified with 5′-ATAGCGGCCGCTATGGTGAGCAAGGGCGAGGA-3′ (sense) and 5′-ATAAGCGGCCGCTCTAGATCCGGTGGATCCCG-3′ (antisense) primers and inserted (NotI) into the NXF1-10RA coding region. GFP-Nup98-2xGLFG/HoxA9 and GFP-HoxA9 plasmids were obtained from Maureen Powers (Emory University, Atlanta, GA; Xu and Powers, 2013). The GFP coding region was replaced with the Cerulean coding region (AgeI and EcoRI). To generate Cerulean-Dbp5-DN, the Dbp5-DN coding region (Hodge et al., 2011) was inserted (BglII and SalI) into a mCerulean-C1 plasmid (Clontech). GFP/RFP-Nol7 (Kinor and Shav-Tal, 2011) and POM121-mCherry (Mor et al., 2010) were previously described.
Cells and transfections
Human U2OS cells were maintained in low-glucose DMEM (Biological Industries) containing 10% FBS (HyClon). Mouse embryonic fibroblasts (MEFs) were maintained in high-glucose DMEM (GIBCO BRL). For transient transfections, U2OS cells were transfected using the PolyJet transfection reagent (SignaGem Laboratories) and fixed with 4% PFA 24 h after the transfection. For live cell imaging, U2OS cells stably expressing the Cerulean-minidystrophin-MS2 gene (Mor et al., 2010) and the YFP-MS2 coat protein were cotransfected with Cerulean-Dbp5-DN or Cerulean-Nup98-2xGLFG/HoxA9 together with the POM121-mCherry plasmid. 24 h after transfection, Cerulean-minidystrophin-MS2 transcription was induced with 1 µg/ml Ponasterone A (Enzo Life Sciences) for 4 h before live-cell imaging. Stable transfections were performed by calcium phosphate. Antibiotics selections were performed with either G418 (for GFP-NXF1) or Zeocin (for YFP-MS2). Other cell lines used were the Cerulean-1/2-mini-dystrophin-MS2 gene (Mor et al., 2010), doxycycline-induced E6 gene in U2OS cells (0.01 mg/ml, 6 h; Brody et al., 2011), and β-actin MS2-knockin MEFs (Lionnet et al., 2011).
For WGA export inhibition, U2OS cells were incubated for 5 min with 30 µg/ml digitonin (Sigma) together with 5 µg/ml Cy5-WGA (Invitrogen). Digitonin was washed out, and cells were incubated for 4 h before fixation. For live-cell imaging, U2OS cells stably expressing the Cerulean-minidystrophin-MS2 gene and the YFP-MS2 coat protein were transfected with the POM121-mCherry plasmid 24 h before WGA treatment, and transcription was induced together with WGA treatment.
For transcription inhibition, cells were treated with 100 µM DRB (Sigma) for 3 h. For RNase digestion, cells were treated with 5 µg/ml actinomycin D (Sigma) for 3 h and then permeabilized with 0.5% Triton X-100 in PBS for 2 min and digested with RNase A (100 µg/ml in PBS with 3 mM MgCl2; Sigma) for 45 min at room temperature.
siRNA knockdowns
U2OS cells were transfected with siRNAs to Nup153 5′-GGCUACAAAGAUACUUCAACAAGAA-3′ (duplex 2, 25 nM + 500 ng POM121-Cherry DNA), Nup358 5′-CUGAAGAACCUGAUUCUAUUACCAA-3′ (duplex 1, 25 nM), Nup214 5′-CGGAGAGACAGCAUCAUUACUACC-3′ (duplex 1, 16.6 nM), UAP56 5′-GGAAGGUAAAUACUACAGUGAGGAG-3′ (duplex 2, 16.6 nM), negative control 5′-CGUUAAUCGCGUAUAAUACGCGUA-3′ (TriFECTa siRNA kit; IDT), NXF1 5′-GAUGGGACCUCAAAGAACUGGUUC-3′ (catalog number 1299001, oligo ID HSS173670; Invitrogen; 60 nM with 500 ng POM121-Cherry DNA), and AlyREF 5′-GACAUGUCUCUGGACGACAUCAUUA-3′ (catalog number 1299001, oligo ID HSS173451; Invitrogen) using Lipofectamine 2000 (Invitrogen). 72 h after siRNA transfection, cells were fixed with 4% PFA and taken to RNA FISH followed by immunofluorescence. For live-cell imaging, U2OS cells stably expressing the Cerulean-minidystrophin-MS2 gene, Cerulean-1/2-mini-dystrophin-MS2, or CFP-SKL-E6-MS2 gene and the YFP-MS2 coat protein were cotransfected with the siRNAs together with POM121-mCherry 72 h after siRNA+DNA transfection. Cerulean-minidystrophin-MS2 and Cerulean-1/2-minidystrophin-MS2 transcription was induced with 1 µg/ml Ponesterone A for 4 h before live-cell imaging. CFP-SKL-E6-MS2 was induced with 0.01 mg/ml doxycycline (Sigma). MEFs expressing the endogenous β-actin-MS2 gene were transfected with siRNA to mouse NXF1 5′-GCUUACUUUGUAGAGCUGACACUAA-3′ (duplex 2, 25 nM TriFECTa siRNA kit). 24 h later, cells were transfected with POM121-mCherry and YFP-MS2 using Polyjet reagent. The day before imaging, cells were starved for serum for 15 h, and serum was added 2 h before live-cell imaging to induce β-actin transcription.
Immunofluorescence
Cells were grown on coverslips, washed with PBS, and fixed for 20 min in 4% PFA. Cells were then permeabilized in 0.5% Triton X-100 for 2.5 min. Cells were washed twice with PBS, blocked with 5% BSA for 20 min, and immunostained for 1 h with a primary antibody. After three washes with PBS, the cells were incubated for 1 h with secondary fluorescent antibodies. For detection of NXF1 at the NPC, cells were treated with 0.1% Triton X-100 for 5 min on ice before fixation.
Primary antibodies
The primary antibodies used were rabbit anti-Nup153 (ab84872), mouse anti-NXF1 (ab50609), rabbit anti-NXF1 (ab129160), rabbit anti-Nup214 (ab70497), rabbit anti-Nup358 (ab64276), rabbit anti-UAP56 (ab47955), mouse anti-Nup62 (ab610497), rabbit anti-Tpr (ab84516), rabbit anti-Nup107 (ab73290), rat anti-Nup98 (ab50610; Abcam), mouse anti-Tpr (sc271317), mouse anti-hnRNP A1 (sc32301), mouse anti-Myc (sc789; Santa Cruz), rabbit anti-UAP56 (SAB2700772), mouse anti-Aly (A9979; Sigma), and mAb414 (MMS-120P; Covance). Primary antibody labeling of mouse anti-NXF1 was performed using the Mix-n-Stain CF 488A Antibody labeling kit (MX488AS100; Sigma).
Secondary antibodies
For wide-field microscopy, the secondary antibodies used were Alexa Fluor 488 goat anti-rabbit (ab150077), Alexa Fluor 488 goat anti-mouse (ab150113), anti-mouse Cy3 (ab97035), anti-rabbit Cy3 (ab6939; Abcam), Chromeo 505 goat anti-rabbit (15040; Active Motif), Alexa Fluor 647 goat anti-mouse (A21235), and Alexa Fluor 594 goat anti-rabbit (A11072; Molecular Probes).
For FLIM-FRET experiments, the secondary antibodies used were Alexa Fluor 488 goat anti-mouse IgG (H+L), Alexa Fluor 647 goat anti-mouse IgG (H+L), Alexa Fluor 488 goat anti-rabbit IgG (H+L), Alexa Fluor 647 donkey anti-rabbit IgG (H+L), and Alexa Fluor 647 chicken anti rat IgG (H+L; Molecular Probes).
For STED microscopy, the secondary antibodiess used were Fab′ donkey anti-rabbit 488 and Fab′ donkey anti-mouse Rhodamine Red-x antibodies (Jackson ImmunoResearch).
FISH
Cells were grown on coverslips and fixed for 20 min in 4% PFA and overnight with 70% ethanol at 4°C. The next day, cells were washed with 1× PBS and treated for 2.5 min with 0.5% Triton X-100. Cells were washed with 1× PBS and incubated for 10 min in 15% formamide (4% SSC; Sigma). Cells were hybridized overnight at 37°C in 15% formamide with a specific fluorescently labeled Cy3, Atto488, or Cy5 DNA oligo(dT) probe (∼10 ng probe, 50 mer). The next day, cells were washed twice with 15% formamide for 15 min and then washed for 2 h with 1× PBS. Nuclei were counterstained with Hoechst 33342, and coverslips were mounted in mounting medium. In some cases, immunofluorescence was performed after RNA FISH using the standard protocol.
Fluorescence microscopy and live-cell imaging
Wide-field fluorescence images were obtained using the Cell^R system based on an Olympus IX81 fully motorized inverted microscope (60× PlanApo objective, 1.42 NA) fitted with an Orca-AG CCD camera (Hamamatsu) driven by the Cell^R software. Live-cell imaging was performed using the Cell^R system. For time-lapse imaging, cells were plated on glass-bottom tissue culture plates (Greiner Bio-One) in medium containing 10% FBS at 37°C. The microscope is equipped with an incubator that includes temperature and CO2 control (Life Imaging Services).
Image processing and analysis
Movies were deconvolved using Huygens Essential II with a time-series option (Scientific Volume Imaging). Counting of mRNPs and tracking of mRNP movement was performed by identifying the nuclear periphery in ImageJ and detecting and tracking static mRNPs with the Imaris (Bitplane) “spot function.” Static mRNPs were defined as mRNPs moving <250 nm in 800 ms for at least four adjacent frames along the video. The average number of static mRNPs per time point along the video was calculated and plotted.
Calculation of colocalization percentage between NXF1 and NPCs was performed using the Imaris spot function to identify each individual NPC and using the MATLAB colocalization tool in Imaris software.
For analysis of the distance frequencies between NXF1 or NXF1-10RA and Nup358, a line was manually drawn across each NPC. The intensity of both channels along the lines was measured by LAS X software and outputted as an Excel file representing a single pore. A MATLAB script (Nup_dis) interpolated the data obtained from each Excel file to find the maximum intensity position of each channel and measured the distance between them. That distance was defined as the distance between the proteins within a specific NPC. Finally, all measured distances from all NPCs analyzed for each condition were plotted in a histogram.
For analysis of NPC diameter, cells were imaged by STED microscopy from the upper plane of the nucleus to detect the ring-shaped pattern of NPCs. Two lines were manually drawn across each NPC. The fluorescence intensity along the lines was measured by LAS X software and outputted as two Excel files (each file represents one line). A MATLAB script (NPC_diameter) interpolated the data obtained from each Excel file to find two maximum intensity positions along the drawn line. Each maximum position represents one end of the pore ring, and the distance between them was measured. The average between two measured distances along one NPC was defined as its diameter. Finally, the average diameter of all NPCs was measured.
FRAP and photoactivation
Cells were maintained in Leibovitz’s L-15 phenol red–free (Invitrogen) containing 10% FCS at 37°C. Image sequences were obtained on an Olympus FV1000 inverted scanning confocal microscope, running the FV1000 acquisition software, equipped with a heated plate (37°C) and a 60×, 1.35-NA oil immersion objective. Cells were scanned using an argon 488-nm laser for detection of GFP/PA-GFP–labeled NXF1/10RA. For FRAP, the nucleoplasm or the nuclear pores were bleached using the 488-nm laser. Five prebleach images were acquired. Postbleach images were acquired at a frequency of 45 images every 1.435 s. For analysis of the fluorescence recovery, FRAP data were normalized and calculated. For each time point, the background taken from a region of interest (ROI) outside of the cell was subtracted from all other measurements. T(t) and I(t) were measured for each time point as the average intensity of the nucleus and the average intensity in the bleached ROI, respectively. Five images were collected before bleaching, and these initial conditions are referred to as Ti = nuclear intensity and Ii = intensity in ROI before bleaching. Ic(t) is the corrected intensity of the bleached ROI at time t (14): Ic(t) =(I(t) Ti)/(Ii Ti); also see Statistical analysis. Recovery curves were fit in MATLAB (FRAP_fit script). For photoactivation, the nucleoplasm of U2OS cells stably expressing PAGFP-NXF1 or PAGFP-10RA was pulsed with a 405-nm laser for 400 ms. Five pre-photoactivation images were acquired. Post-photoactivation images were acquired at a frequency of 90 images every 1.49 s.
STED
Superresolution imaging was performed on a Leica SP8 inverted microscope equipped with a STED module, a pulsed white-light laser, and gating. Fab′ fragment secondary antibodies were used to create as physically a small fluorescent source as possible. The objective used was a STED dedicated 100× 1.4 NA, with Leica immersion oil, at room temperature. Mounting medium was homemade 80% glycerol with p-Phenylenediamine antifade (Sigma), and the coverglasses were high-precision #1.5 (Thermo Scientific). Dual-color experiments were performed by between-line sequential imaging using the 660-nm depletion laser set at 90% (slider) of 90% laser power for Alexa Fluor 488 and 50% slider for Rhodamine Red-x. This allowed for more accurate spatial imaging (as opposed to using the 592-nm depletion laser for Alexa Fluor 488, which would necessitate between frame imaging). Gate settings were 0.3 ns for the Rhodamine Red-x and 2.5 ns for Alexa Fluor 488. When imaging GFP-NXF1 or the labeled NXF1 primary antibody, the 592-nm depletion laser was used at 10% slider of 90% laser power and gating set at 2.5 ns. For the top-view imaging of NXF1 and Nup214, resulting images were submitted to the Leica smoothing algorithm. Colocalization and NXF1 position mapping images were deconvolved with Huygens Professional (Scientific Volume Imaging) using the CMLE algorithm, with signal to noise ratio of 12 and 40 iterations, using the Huygens STED module.
FLIM-FRET measurements and analysis
FRET results from direct interactions between donor and acceptor molecules that lead to a decrease in the LT of the donor molecules. FRET is based on the ability of a donor fluorophore to transfer energy from its excited state to an adjacent acceptor fluorophore, resulting in acceptor fluorescence. The energy transfer is distance dependent and requires that the two fluorophores reside in close proximity of less than ∼10 nm of each other, which signifies an interaction between molecules. FRET itself is an intensity-based method and can introduce some bias, so we therefore used FLIM. FLIM is used for measuring fluorophore lifetime τ, or the exponential rate at which a fluorescent molecule decays and emits a photon after excitation. FRET efficiency follows the equation:
where Td is the measured lifetime of the donor-only cells and Td + a is the measured lifetime of the donor and acceptor cells, all in the range of nanoseconds.
Fluorescence images were obtained on an inverted Olympus IX-81 confocal fluorescence microscope coupled to a FluoView-1000 confocal setup (Olympus) using an oil UPLSAPO 60× objective with NA 1.35. Lifetime data were obtained by coupling the system to a time-resolved MicroTime200 (PicoQuant) system that uses a 20-MHz, 470-nm pulsed picosecond diode laser (LDH-P-C-470B; PicoQuant). After collecting the emitted light by the objective, the excitation light is filtered by an appropriate dichroic mirror (405/488 nm), transmitted through a confocal pinhole (D = 120 µm), and sent to a single photon avalanche detector (SPCM-AQRH 13; Perkin Elmer) through a 520/35-nm band-pass filter (FF01-520/35-25; Semrock). The system works in a time-correlated single-photon counting method, namely, each time that an excitation laser pulse is generated, a stop-watch with a time-resolution of 64 picoseconds starts counting. It stops when the single photon avalanche detector detects an emitted photon signal. To generate a decay curve of the number of events across time, the process is repeated many times and for all the pixels in the image, to obtain lifetime measurements for each pixel and to create a map of intensity distributions.
Alexa Fluor 488 (donor 1:2,000) and Alexa Fluor 647 (acceptor 1:200) were used as a FRET pair (R0 = 56). Staining was performed such that the acceptor was fully saturated. The experiments with GFP-Dbp5 for export blockage required the cotransfection of another nuclear GFP-fusion protein to detect the transfected cells, since most of the GFP-Dbp5 signal was washed out of the cytoplasm during the fixation procedure. We used the nucleolar protein GFP-NOL7 (Kinor and Shav-Tal, 2011) as an indicator of the export blocked cells. GFP-Dbp5 and GFP-NOL7 were transfected at a 10:1 ratio. The GFP signals did not interfere with the FLIM-FRET measurements.
The PicoQuant system exports the data into bin files. An ImageJ script divides each bin file into two images: an intensity map and an average lifetime (LT) map. We superimposed these two images to detect the NPCs and then used Imaris software to quantify the lifetime values at each individual NPC for thousands of NPCs. The distribution of lifetimes from the hundreds of NPCs measured is presented in histograms. For acceptor photobleaching experiments, a square ROI at the top plane of the nucleus was bleached with a 647-nm laser before lifetime measurements (n = 9 cells from three different experiments).
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from cells using the AurumTM Total RNA mini kit (Bio-Rad). After reverse transcription using the qScript cDNA Synthesis Kit (Quanta Biosciences), cDNA was amplified using the following primer pairs: NXF1 sense, 5′-CTTGAGGAAGATGATGGAGAT-3′; NXF1 antisense, 5′-GTATCACCCCGACGGTTAGG-3′; Nup153 sense, 5′-CCAGAAGCTGAACGTTCTCA-3′; Nup153 antisense, 5′-GTGAAGGGGAAAGTGTTCCA-3′; Aly sense, 5′-ACAGCAGGCCAAAACAACTT-3′; Aly antisense, 5′-CAGCAGTTTCCCACCTGTCT-3′; UAP56 sense, 5′-ATCCGTCAGAAGTCCAGCAT-3′; UAP56 antisense, 5′-TGTGGCCAAGACAAACACTG-3′; Nup358 sense, 5′-TGGCTGCATTGTGCTATCTC-3′; Nup358 antisense, 5′-GCCATCATTTCCAGCAGATT-3′; Nup214 sense, 5′-AGTCCTCAGTCTTGCCCTCA-3′; Nup214 antisense, 5′-GAGGGGTTATCCTGGGTGAT-3′; DBP5 sense, 5′-CGGCATTGATGTTGAACAAG-3′; DBP5 antisense, 5′-CGGTGCAGGTAGGTCTCATT-3′; tubulin sense, 5′-GCCTGGACCACAAGTTTGAC-3′; tubulin antisense, 5′-TGAAATTCTGGGAGCATGAC-3′; 18S sense, 5′-TGTGCCGCTAGAGGTGAAATT-3′; and 18S antisense, 5′-TGGCAAATGCTTTCGCTTT-3′.
Real-time qRT-PCR was performed using PerfeCTa SYBR Green FastMix (Quanta Biosciences) on a CFX-96 system (Bio-Rad). Analysis was performed with the Bio-Rad CFX manager. Relative levels of mRNA expression were measured as the ratio of the comparative threshold cycle to internal controls (tubulin and 18S RNA).
Western blotting
Cells were washed with cold PBS and placed on ice for 15 min after resuspending in immunoprecipitation lysis buffer (Pierce) containing 10 mM Na-fluoride, 1 mM Na-orthovanadate, protease inhibitor cocktail (Sigma), and 1 mM PMSF. The resulting lysate was centrifuged at 10,000 rpm for 10 min at 4°C. 30 µg protein was run on 7.5% SDS-polyacrylamide gels and transferred to a nitrocellulose membrane (0.45 µm) for 2.5 h at 250 mA. The membrane was blocked with 5% BSA and then probed with a primary antibody for 2 h at RT, followed by incubation with HRP-conjugated goat anti-rabbit IgG (Sigma) for 1 h at RT. Immunoreactive bands were detected by the Enhanced Chemiluminescence kit (ECL; Pierce). Primary antibodies used were mouse anti-Nup153 (ab24700), rabbit anti-NXF1 (ab129160), and rabbit anti-tubulin (ab4074; Abcam). Experiments were performed three times and quantified in ImageJ. After the selection of the bands, the values of the intensities were obtained. The intensities of the Nup153 and NXF1 bands were divided by the corresponding tubulin band intensities.
Statistical analysis
A two-tailed t test was performed in the following experiments: NPC diameter, qRT-PCR, and FLIM-FRET measurements. For live-cell video analysis, all treatments were compared using a one-way Welch’s ANOVA followed by a Games–Howell post hoc analysis. Box plot analysis was performed using Prism software (GraphPad).
For FRAP experiments, linear mixed-effects modeling was used to test the effect of different treatments on the log2 relative intensity of fluorescence recovery as a function of time. Specifically, a second-degree polynomial linear mixed-model regression was fitted, with time as the continuous predictor and treatment as a fixed effect. Experiments were defined as random effect. Post hoc analysis was performed in terms of linear contrasts between treatments, and P values were corrected for multiple testing using the false discovery rate procedure.
Following are the numbers of mRNPs, NPCs, and cells examined in the different experiments:
Fig. 1 B: total mRNPs tracked: control, 640 mRNPs, 31 cells; siNup153, 1116 mRNPs, 51 cells; WGA, 2,301 mRNPs, 29 cells; Dbp5-DN, 2,064 mRNPs, 35 cells; and siNXF1, 3,239 mRNPs, 39 cells.
Fig. 1 C: n = 6 cells, 13 mRNPs (siNup153); 11 cells, 44 mRNPs (WGA); 7 cells, 20 mRNPs (Dbp5-DN); and 9 cells, 31 mRNPs (siNXF1).
Fig. 2 D: nucleoplasm, n = 28 cells, middle plane of the nucleus; NPCs, n = 38 cells, top plane of the nucleus.
Fig. 3 F: n = 1,423 NPCs, 3 cells, (Tpr, donor only); 838 NPCs, 3 cells (Tpr + Nup153, donor + acceptor).
Fig. 3 G: n = 3,405 NPCs, 4 cells (Nup358, donor only); 2,705 NPCs, 4 cells (Nup358 + Tpr, donor + acceptor).
Fig. 4 A: n = 1,828 NPCs, 5 cells (NXF1, donor only); 1,720 NPCs, 4 cells (NXF1 + Nup214, donor + acceptor).
Fig. 4 B: n = 1,840 NPCs, 4 cells (NXF1, donor only); 1,008 NPCs, 3 cells (NXF1 + Tpr, donor + acceptor).
Fig. 4 C: n = 1,604 NPCs, 5 cells (NXF1, donor only); 899 NPCs, 3 cells (NXF1 + Nup358, donor + acceptor).
Fig. 5 E: n = 71 NPCs, 7 cells (Nup214), 71 NPCs, 8 cells (NXF1).
Fig. 8 B: NXF1: nucleoplasm, n = 29 cells; NPCs, 38 cells; DRB nucleoplasm, 21 cells, DRB NPC, 21 cells; NXF1-10RA: nucleoplasm, n = 22 cells; NPCs, 41 cells.
Fig. 10 D: n = 900 NPCs, 16 cells (NXF1-Nup358 control); 1,500 NPCs, 22 cells (NXF1-Nup358 Dbp5-DN).
Fig. 10 E: n = 1,415 NPCs, 18 cells (NXF1-10RA-Nup358 control); 844 NPCs, 14 cells (NXF1-10RA-Nup358 Dbp5-DN).
Online supplemental material
Fig. S1 shows quantifications of knockdown levels and RNA-FISH staining for mRNA export blocks. Fig. S2 shows NXF1 knockdown experiments on different cell lines. Fig. S3 shows FLIM-FRET measurements. Fig. S4 shows quantifications of knockdown levels. Fig. S5 shows FLIM-FRET experiments under mRNA export block conditions. Videos 1, 2, 3, and 4 show single mRNP live-cell imaging of the images shown in Fig. 1 C. Videos 5, 6, and 7 show single mRNP live-cell imaging of the images shown in Fig. S2. Video 8 shows the photoactivation images shown in Fig. 2 E. Video 9 shows single mRNP live-cell imaging of the images shown in Fig. 9 D. The Nup_dis MATLAB script interpolates the data obtained from Excel files representing the intensity of two fluorescent channels, and finds the distance between their maximum intensity positions. The NPC_diameter MATLAB script interpolates the data obtained from two Excel files representing fluorescent intensity along a line to find two maximum intensity positions in each file and calculates the distance between them and the average between the two calculated distances. The FRAP_fit MATLAB script obtains times and average intensities of FRAP recovery experiments and provides a fit (nonlinear least squares) of the data.
Supplementary Material
Acknowledgments
This study is dedicated to the memory of Elisa Izaurralde.
We thank Maria Carmo-Fonseca for GFP-NXF1, Maureen Powers for the FG-Nups, Stuart Wilson for the Tap-10RA construct, Techia Atias for generating Cerulean-Dbp5-DN, Boaz Kowal for generating Cerulean-2xGLFG-HoxA9, Shira Avivi for generating GFP-NXF1-10RA, and Gabe Philip Faber for image processing.
This study was supported by the European Research Council (GENEXP; Y. Shav-Tal) and the Israel Science Foundation (2252/15 and 1278/18; Y. Shav-Tal).
The authors declare no competing financial interests.
Author contributions: Conceptualization, R. Ben-Yishay, A. Mor, and Y. Shav-Tal; Investigation, R. Ben-Yishay, A. Askenzai-Titelman, A. Mor, A. Shraga, N. Kinor, A. Schwed-Gross, N. Kozer, and A. Jacob; Methodology, R. Ben-Yishay and A. Mor; Formal Analysis, R. Ben-Yishay, A. Askenzai-Titelman, A. Shraga, A. Mor, and N. Kozer; Software, P. Kumar and Y. Garini; Resources, A. Jacob Y. Garini and Y. Shav-Tal; Validation, R. Ben-Yishay and A. Askenzai-Titelman; Project Administration, R. Ben-Yishay and Y. Shav-Tal; Writing – Original Draft, R. Ben-Yishay and Y. Shav-Tal; Writing – Review & Editing, R. Ben-Yishay and Y. Shav-Tal; Supervision, Y. Garini and Y. Shav-Tal; Funding Acquisition, Y. Shav-Tal.
References
- Adams R.L., and Wente S.R.. 2013. Uncovering nuclear pore complexity with innovation. Cell. 152:1218–1221. 10.1016/j.cell.2013.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R.L., Mason A.C., Glass L., Aditi, and Wente S.R.. 2017. Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells. Traffic. 18:776–790. 10.1111/tra.12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi A., Braun I.C., Rodrigues J.P., Panté N., Ribbeck K., von Kobbe C., Kutay U., Wilm M., Görlich D., Carmo-Fonseca M., and Izaurralde E.. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 6:136–158. 10.1017/S1355838200991994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., and Hurt E.. 2017. The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 18:73–89. 10.1038/nrm.2016.147 [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y., Brody Y., Kinor N., Mor A., Tsukamoto T., Spector D.L., Singer R.H., and Shav-Tal Y.. 2010. The life of an mRNA in space and time. J. Cell Sci. 123:1761–1774. 10.1242/jcs.062638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yishay R., Ashkenazy A.J., and Shav-Tal Y.. 2016. Dynamic Encounters of Genes and Transcripts with the Nuclear Pore. Trends Genet. 32:419–431. 10.1016/j.tig.2016.04.003 [DOI] [PubMed] [Google Scholar]
- Blobel G. 1985. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. USA. 82:8527–8529. 10.1073/pnas.82.24.8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet A., and Palancade B.. 2014. Regulation of mRNA trafficking by nuclear pore complexes. Genes (Basel). 5:767–791. 10.3390/genes5030767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun I.C., Herold A., Rode M., Conti E., and Izaurralde E.. 2001. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 276:20536–20543. 10.1074/jbc.M100400200 [DOI] [PubMed] [Google Scholar]
- Brody Y., Neufeld N., Bieberstein N., Causse S.Z., Böhnlein E.M., Neugebauer K.M., Darzacq X., and Shav-Tal Y.. 2011. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 9:e1000573 10.1371/journal.pbio.1000573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M. 2010. Follow that messenger: live-imaging a journey out of the nucleus. Dev. Cell. 18:880–882. 10.1016/j.devcel.2010.05.021 [DOI] [PubMed] [Google Scholar]
- Collins R., Karlberg T., Lehtiö L., Schütz P., van den Berg S., Dahlgren L.G., Hammarström M., Weigelt J., and Schüler H.. 2009. The DEXD/H-box RNA helicase DDX19 is regulated by an alpha-helical switch. J. Biol. Chem. 284:10296–10300. 10.1074/jbc.C900018200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custódio N., Carvalho C., Condado I., Antoniou M., Blencowe B.J., and Carmo-Fonseca M.. 2004. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA. 10:622–633. 10.1261/rna.5258504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L.I., and Blobel G.. 1986. Identification and characterization of a nuclear pore complex protein. Cell. 45:699–709. 10.1016/0092-8674(86)90784-1 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim V.N., and Kataoka N.. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195–205. 10.1038/nrm760 [DOI] [PubMed] [Google Scholar]
- Dworetzky S.I., and Feldherr C.M.. 1988. Translocation of RNA-coated gold particles through the nuclear pores of oocytes. J. Cell Biol. 106:575–584. 10.1083/jcb.106.3.575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B., and Aebi U.. 2003. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 4:757–766. 10.1038/nrm1230 [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B., Maco B., Fager A.M., Köser J., Sauder U., Ullman K.S., and Aebi U.. 2002. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 140:254–267. 10.1016/S1047-8477(02)00524-5 [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J., and Rout M.P.. 2012. A jumbo problem: mapping the structure and functions of the nuclear pore complex. Curr. Opin. Cell Biol. 24:92–99. 10.1016/j.ceb.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forler D., Rabut G., Ciccarelli F.D., Herold A., Köcher T., Niggeweg R., Bork P., Ellenberg J., and Izaurralde E.. 2004. RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol. Cell. Biol. 24:1155–1167. 10.1128/MCB.24.3.1155-1167.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttfert F., Wurm C.A., Mueller V., Berning S., Cordes V.C., Honigmann A., and Hell S.W.. 2013. Coaligned dual-channel STED nanoscopy and molecular diffusion analysis at 20 nm resolution. Biophys. J. 105:L01–L03. 10.1016/j.bpj.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald D., and Singer R.H.. 2010. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature. 467:604–607. 10.1038/nature09438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue G.M., Hung M.L., Golovanov A.P., Lian L.Y., and Wilson S.A.. 2008. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. USA. 105:5154–5159. 10.1073/pnas.0709167105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue G.M., Hung M.L., Walsh M.J., Snijders A.P., Chang C.T., Jones R., Ponting C.P., Dickman M.J., and Wilson S.A.. 2009. UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr. Biol. 19:1918–1924. 10.1016/j.cub.2009.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S., Derrer C.P., Lari A., Weis K., and Montpetit B.. 2017. Temporal and spatial regulation of mRNA export: Single particle RNA-imaging provides new tools and insights. BioEssays. 39:1600124 10.1002/bies.201600124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge C.A., Tran E.J., Noble K.N., Alcazar-Roman A.R., Ben-Yishay R., Scarcelli J.J., Folkmann A.W., Shav-Tal Y., Wente S.R., and Cole C.N.. 2011. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 25:1052–1064. 10.1101/gad.2041611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Deerinck T.J., Ellisman M.H., and Spector D.L.. 1994. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J. Cell Biol. 126:877–899. 10.1083/jcb.126.4.877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E., Strässer K., Segref A., Bailer S., Schlaich N., Presutti C., Tollervey D., and Jansen R.. 2000. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J. Biol. Chem. 275:8361–8368. 10.1074/jbc.275.12.8361 [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Jackson D.A., and Cook P.R.. 1998. The path of transcripts from extra-nucleolar synthetic sites to nuclear pores: transcripts in transit are concentrated in discrete structures containing SR proteins. J. Cell Sci. 111:2269–2282. [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Jackson D.A., and Cook P.R.. 2000. The path of RNA through nuclear pores: apparent entry from the sides into specialized pores. J. Cell Sci. 113:291–302. [DOI] [PubMed] [Google Scholar]
- Johnson L.A., Li L., and Sandri-Goldin R.M.. 2009. The cellular RNA export receptor TAP/NXF1 is required for ICP27-mediated export of herpes simplex virus 1 RNA, but the TREX complex adaptor protein Aly/REF appears to be dispensable. J. Virol. 83:6335–6346. 10.1128/JVI.00375-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., and Cullen B.R.. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 13:1126–1139. 10.1101/gad.13.9.1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Strässer K., Podtelejnikov A., Mann M., Jung J.U., and Hurt E.. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18:2593–2609. 10.1093/emboj/18.9.2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.J., Fernandez-Martinez J., Nudelman I., Shi Y., Zhang W., Raveh B., Herricks T., Slaughter B.D., Hogan J.A., Upla P., et al. . 2018. Integrative structure and functional anatomy of a nuclear pore complex. Nature. 555:475–482. 10.1038/nature26003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinor N., and Shav-Tal Y.. 2011. The dynamics of the alternatively spliced NOL7 gene products and role in nucleolar architecture. Nucleus. 2:229–245. 10.4161/nucl.2.3.15893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A., and Hurt E.. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761–773. 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- Kosinski J., Mosalaganti S., von Appen A., Teimer R., DiGuilio A.L., Wan W., Bui K.H., Hagen W.J., Briggs J.A., Glavy J.S., et al. . 2016. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science. 352:363–365. 10.1126/science.aaf0643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kylberg K., Björk P., Fomproix N., Ivarsson B., Wieslander L., and Daneholt B.. 2010. Exclusion of mRNPs and ribosomal particles from a thin zone beneath the nuclear envelope revealed upon inhibition of transport. Exp. Cell Res. 316:1028–1038. 10.1016/j.yexcr.2009.10.016 [DOI] [PubMed] [Google Scholar]
- Ledoux S., and Guthrie C.. 2011. Regulation of the Dbp5 ATPase cycle in mRNP remodeling at the nuclear pore: a lively new paradigm for DEAD-box proteins. Genes Dev. 25:1109–1114. 10.1101/gad.2062611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.H., Correia A.R., Cai S.W., Huber F.M., Jette C.A., and Hoelz A.. 2018. Structural and functional analysis of mRNA export regulation by the nuclear pore complex. Nat. Commun. 9:2319 10.1038/s41467-018-04459-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T., Czaplinski K., Darzacq X., Shav-Tal Y., Wells A.L., Chao J.A., Park H.Y., de Turris V., Lopez-Jones M., and Singer R.H.. 2011. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods. 8:165–170. 10.1038/nmeth.1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löschberger A., van de Linde S., Dabauvalle M.C., Rieger B., Heilemann M., Krohne G., and Sauer M.. 2012. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. Cell Sci. 125:570–575. 10.1242/jcs.098822 [DOI] [PubMed] [Google Scholar]
- Lund M.K., and Guthrie C.. 2005. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell. 20:645–651. 10.1016/j.molcel.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Ma J., Liu Z., Michelotti N., Pitchiaya S., Veerapaneni R., Androsavich J.R., Walter N.G., and Yang W.. 2013. High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat. Commun. 4:2414 10.1038/ncomms3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon T., Elad N., Dahan I., and Medalia O.. 2012. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 20:998–1006. 10.1016/j.str.2012.03.025 [DOI] [PubMed] [Google Scholar]
- Mohr D., Frey S., Fischer T., Güttler T., and Görlich D.. 2009. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 28:2541–2553. 10.1038/emboj.2009.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A., and Shav-Tal Y.. 2010. Dynamics and kinetics of nucleo-cytoplasmic mRNA export. Wiley Interdiscip. Rev. RNA. 1:388–401. 10.1002/wrna.41 [DOI] [PubMed] [Google Scholar]
- Mor A., Suliman S., Ben-Yishay R., Yunger S., Brody Y., and Shav-Tal Y.. 2010. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 12:543–552. 10.1038/ncb2056 [DOI] [PubMed] [Google Scholar]
- Müller-McNicoll M., and Neugebauer K.M.. 2013. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 14:275–287. 10.1038/nrg3434 [DOI] [PubMed] [Google Scholar]
- Noble K.N., and Wente S.R.. 2010. Nuclear mRNA on the move. Nat. Cell Biol. 12:525–527. 10.1038/ncb0610-525 [DOI] [PubMed] [Google Scholar]
- Noble K.N., Tran E.J., Alcázar-Román A.R., Hodge C.A., Cole C.N., and Wente S.R.. 2011. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 25:1065–1077. 10.1101/gad.2040611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura M., Inose H., and Masuda S.. 2015. RNA Export through the NPC in Eukaryotes. Genes (Basel). 6:124–149. 10.3390/genes6010124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Doye V., and Ellenberg J.. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121. 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- Rajoo S., Vallotton P., Onischenko E., and Weis K.. 2018. Stoichiometry and compositional plasticity of the yeast nuclear pore complex revealed by quantitative fluorescence microscopy. Proc. Natl. Acad. Sci. USA. 115:E3969–E3977. 10.1073/pnas.1719398115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J.P., Rode M., Gatfield D., Blencowe B.J., Carmo-Fonseca M., and Izaurralde E.. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA. 98:1030–1035. 10.1073/pnas.98.3.1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno H., Simos G., Segref A., Fahrenkrog B., Panté N., and Hurt E.. 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 18:6826–6838. 10.1128/MCB.18.11.6826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P.M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M.C., Agard D.A., Gustafsson M.G., et al. . 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 320:1332–1336. 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt I., and Gerace L.. 2001. In vitro analysis of nuclear transport mediated by the C-terminal shuttle domain of Tap. J. Biol. Chem. 276:42355–42363. 10.1074/jbc.M103916200 [DOI] [PubMed] [Google Scholar]
- Schmitt C., von Kobbe C., Bachi A., Panté N., Rodrigues J.P., Boscheron C., Rigaut G., Wilm M., Séraphin B., Carmo-Fonseca M., and Izaurralde E.. 1999. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 18:4332–4347. 10.1093/emboj/18.15.4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma K., Doye V., Hellwig A., Huber J., Lührmann R., and Hurt E.. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256–3271. 10.1093/emboj/16.11.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X., Kawauchi K., Shivashankar G.V., and Bershadsky A.D.. 2015. Novel localization of formin mDia2: importin β-mediated delivery to and retention at the cytoplasmic side of the nuclear envelope. Biol. Open. 4:1569–1575. 10.1242/bio.013649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., and Tripathi T.. 2018. Yeast and Human Nuclear Pore Complexes: Not So Similar After All. Trends Cell Biol. 28:589–591. 10.1016/j.tcb.2018.06.004 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y., Darzacq X., Shenoy S.M., Fusco D., Janicki S.M., Spector D.L., and Singer R.H.. 2004. Dynamics of single mRNPs in nuclei of living cells. Science. 304:1797–1800. 10.1126/science.1099754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebrasse J.P., Kaminski T., and Kubitscheck U.. 2012. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc. Natl. Acad. Sci. USA. 109:9426–9431. 10.1073/pnas.1201781109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C., Lari A., Derrer C.P., Ouwehand A., Rossouw A., Huisman M., Dange T., Hopman M., Joseph A., Zenklusen D., et al. . 2015. In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p. J. Cell Biol. 211:1121–1130. 10.1083/jcb.201503135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge C.A., Colot H.V., Goldstein A.L., and Cole C.N.. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17:2663–2676. 10.1093/emboj/17.9.2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soop T., Ivarsson B., Björkroth B., Fomproix N., Masich S., Cordes V.C., and Daneholt B.. 2005. Nup153 affects entry of messenger and ribosomal ribonucleoproteins into the nuclear basket during export. Mol. Biol. Cell. 16:5610–5620. 10.1091/mbc.e05-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K., Bassler J., and Hurt E.. 2000. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 150:695–706. 10.1083/jcb.150.4.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L.A., Shen T., and Wente S.R.. 2001. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J. Biol. Chem. 276:6445–6452. 10.1074/jbc.M008311200 [DOI] [PubMed] [Google Scholar]
- Suyama M., Doerks T., Braun I.C., Sattler M., Izaurralde E., and Bork P.. 2000. Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO Rep. 1:53–58. 10.1093/embo-reports/kvd009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L.J., and Wente S.R.. 2009. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell. 8:1814–1827. 10.1128/EC.00225-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.J., Zhou Y., Corbett A.H., and Wente S.R.. 2007. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell. 28:850–859. 10.1016/j.molcel.2007.09.019 [DOI] [PubMed] [Google Scholar]
- Tutucci E., Livingston N.M., Singer R.H., and Wu B.. 2018. Imaging mRNA In Vivo, from Birth to Death. Annu. Rev. Biophys. 47:85–106. 10.1146/annurev-biophys-070317-033037 [DOI] [PubMed] [Google Scholar]
- Umlauf D., Bonnet J., Waharte F., Fournier M., Stierle M., Fischer B., Brino L., Devys D., and Tora L.. 2013. The human TREX-2 complex is stably associated with the nuclear pore basket. J. Cell Sci. 126:2656–2667. 10.1242/jcs.118000 [DOI] [PubMed] [Google Scholar]
- Viphakone N., Sudbery I., Griffith L., Heath C.G., Sims D., and Wilson S.A.. 2019. Co-transcriptional Loading of RNA Export Factors Shapes the Human Transcriptome. Mol. Cell. 10.1016/j.molcel.2019.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wälde S., and Kehlenbach R.H.. 2010. The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 20:461–469. 10.1016/j.tcb.2010.05.001 [DOI] [PubMed] [Google Scholar]
- Wente S.R., and Rout M.P.. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2:a000562 10.1101/cshperspect.a000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe V.O., McMurtrie P.I., Mills A.D., Takei Y., Penrhyn-Lowe S., Amagase Y., Main S., Marr J., Stewart M., and Laskey R.A.. 2010. mRNA export from mammalian cell nuclei is dependent on GANP. Curr. Biol. 20:25–31. 10.1016/j.cub.2009.10.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., and Powers M.A.. 2013. In vivo analysis of human nucleoporin repeat domain interactions. Mol. Biol. Cell. 24:1222–1231. 10.1091/mbc.e12-08-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W., Roser D., Köhler A., Bradatsch B., Bassler J., and Hurt E.. 2007. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell. 26:51–62. 10.1016/j.molcel.2007.02.018 [DOI] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra P., Strahm Y., and Stutz F.. 2001. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell. Biol. 21:4219–4232. 10.1128/MCB.21.13.4219-4232.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.