Azoitei, Noh, et al. engineer fluorescent biosensors to measure activation at the subcellular level and with subsecond kinetics of GEF-H1, a Rho GTPase that regulates cytoskeletal dynamics. In combination with computational image time series analysis, the biosensors reveal the synergistic role of microtubule dynamics and Src phosphorylation in regulating GEF-H1 activity locally during cell migration.
Abstract
Rho family GTPases are activated with precise spatiotemporal control by guanine nucleotide exchange factors (GEFs). Guanine exchange factor H1 (GEF-H1), a RhoA activator, is thought to act as an integrator of microtubule (MT) and actin dynamics in diverse cell functions. Here we identify a GEF-H1 autoinhibitory sequence and exploit it to produce an activation biosensor to quantitatively probe the relationship between GEF-H1 conformational change, RhoA activity, and edge motion in migrating cells with micrometer- and second-scale resolution. Simultaneous imaging of MT dynamics and GEF-H1 activity revealed that autoinhibited GEF-H1 is localized to MTs, while MT depolymerization subadjacent to the cell cortex promotes GEF-H1 activation in an ~5-µm-wide peripheral band. GEF-H1 is further regulated by Src phosphorylation, activating GEF-H1 in a narrower band ~0–2 µm from the cell edge, in coordination with cell protrusions. This indicates a synergistic intersection between MT dynamics and Src signaling in RhoA activation through GEF-H1.
Introduction
Cell movement requires that the actin cytoskeleton be continuously reorganized by multiple signaling cascades, which must be precisely coordinated in space and time. Rho family GTPases fulfill essential functions in this process by activating downstream effectors that control actin and adhesion dynamics (Jaffe and Hall, 2005). Activation of GTPases is modulated in space and time by several classes of regulatory molecules, including guanine nucleotide exchange factors (GEFs). While the dynamics of Rho GTPases in polarized motility have been extensively characterized (Kraynov et al., 2000; Nalbant et al., 2004; Pertz et al., 2006; Aoki and Matsuda, 2009; Machacek et al., 2009), little is known about the subcellular distribution and timing of upstream activities that govern GTPase activation. The 69 members of the Dbl family GEFs comprise the largest group of Rho GTPase activators (Rossman et al., 2005). Studies in vitro and in living cells have revealed that each GTPase interacts with multiple upstream GEFs, and GEFs often interact with multiple downstream GTPases. This complexity undoubtedly contributes to the rich repertoire of spatially distributed and functionally distinct GTPase actions, but these interactions await characterization in time and space within living cells. Fluorescent biosensors are ideal tools to examine the activity of molecules at the subcellular level, in the context of real-time cellular behaviors. When combined with computational image analysis and statistical modeling, biosensors can provide quantitative insights into how molecules interact in space and time to generate specific biological behaviors (Machacek et al., 2009; Fusco et al., 2016).
The GTPase RhoA plays an essential role in cell migration; it can initiate either protrusion or retraction in different cell regions (Pertz et al., 2006; Machacek et al., 2009; Tkachenko et al., 2011). GEF-H1, a Dbl family GEF also known as ARHGEF2, was shown to be critical for controlling the activation of RhoA (Nalbant et al., 2009). Depletion of GEF-H1 leads to reduced RhoA activity in protrusions, decreased total migration, and increased focal adhesion lifetime (Nalbant et al., 2009). GEF-H1 is the only GEF reported to localize at microtubules (MTs), where it is inactive and becomes active upon MT dissociation (Krendel et al., 2002; Meiri et al., 2012). GEF-H1 is therefore thought to be an important integrator of MT and actin dynamics (Sandí et al., 2017).
To study the role and regulation of GEF-H1 in migrating cells, we designed a novel fluorescent biosensor that reports GEF-H1 activity. Quantitative analysis of live-cell imaging data revealed, with micrometer resolution and second-scale kinetics, the relationships between GEF-H1 and RhoA activities in modulating cell edge dynamics. Using the biosensor, we showed that GEF-H1 activity is controlled by both local changes in MT dynamics and by Src, a previously unknown regulator of this GEF, in distinct regions of the cell.
Results
Identification of an autoinhibitory domain (AID) in GEF-H1
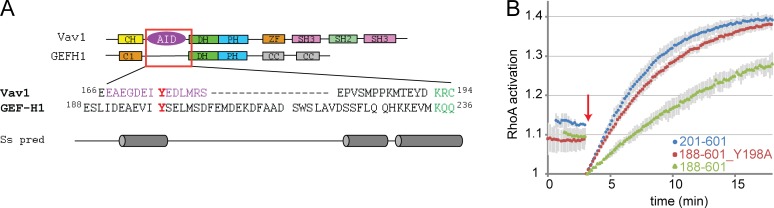
GEFs belonging to the Dbl family contain a Dbl homology (DH) domain, the primary catalytic site that interacts with the GTPase to stimulate nucleotide exchange (Rossman et al., 2005), and a Pleckstrin homology (PH) domain, with less conserved roles usually involved in localization. Dbl-family GEFs are typically autoinhibited through occlusion of the DH domain by an N- or C-terminal AID that is released upon GEF activation. To date, no AID has been reported for GEF-H1, although existing data show that truncation of regions N-terminal to the DH domain lead to GEF-H1 activation (Meiri et al., 2012). Our sequence and structural analysis of GEF-H1 revealed a putative helical fragment upstream of the DH domain with high sequence homology to the well-characterized AID of Vav1 (Yu et al., 2010; Fig. 1 A). The Vav1 AID contains a central tyrosine (Tyr174) that forms a hydrogen bond with an arginine on the DH domain, an interaction that is disrupted when Tyr phosphorylation by Src activates Vav1 (Figs. 1 A and S1 A). Similarly, the putative GEF-H1 AID contains a central tyrosine (Tyr198) surrounded by negatively charged and lipophilic residues.
Figure 1.
GEF-H1 contains an AID. (A) Domain organization and sequence alignment of GEF-H1 and Vav1, with AID of Vav1 shown in purple and the central Tyr residues in red. Secondary structure prediction (Ss pred) shown for GEF-H1 (gray). (B) RhoA exchange activity of recombinantly expressed GEF-H1 constructs with truncation or mutation of the AID. Red arrow indicates GEF addition. Curves show average of two independent experiments, and error bars display SD.
Since no structural information is available for the DH domain of GEF-H1, we performed homology modeling of the putative GEF-H1 AID–DH interaction using Rosetta (Das and Baker, 2008; Leaver-Fay et al., 2011). Our model showed that the GEF-H1 AID–DH complex closely resembles the known structure of the AID–DH of Vav1 (Yu et al., 2010; Fig. S1 A). Importantly, Tyr198 of GEF-H1 matched the position and recapitulated key interactions of the Tyr174 residue in the Vav1 AID that is targeted by Src phosphorylation. We attempted to measure directly the interaction of recombinant DH domains of Vav1 and GEF-H1 with the corresponding synthesized AID peptides. In both cases, however, binding was observed only at the highest concentration where the DH domains remained soluble (16 µM; data not shown), suggesting that the 1:1 AID–DH interactions are weak for both GEFs. Structural studies of the Vav1 AID–DH interaction (Yu et al., 2010) showed that additional interactions outside its AID contribute to autoinhibition. Although the affinity of the AID peptide was weak, in intact protein, AID binding to the DH can be augmented by constrained AID conformation and additional intra-protein contacts.
To further investigate whether the putative GEF-H1 AID has an inhibitory function, we expressed and purified recombinant GEF-H1 DH-PH domain variants that included or lacked residue Tyr198 (residues 188–601 and 201–601, respectively; Fig. S1 B). The 201–601 fragment showed higher GEF activity than the 188–601 fragment in an in vitro nucleotide exchange assay that measures the release of fluorescently labeled GDP from RhoA upon GEF addition (Fig. 1 B). Mutation of Tyr198 to alanine in the 188–601 construct led to increased GEF activity of the 188–601 construct (Fig. 1 B), supporting the existence of a GEF-H1 AID centered on tyrosine 198.
Design and validation of a GEF-H1 biosensor
GEF-H1 activation, defined here as a conformational change that exposes the RhoA binding site, would require dissociation of the AID from that binding site. To engineer a fluorescent biosensor that reports GEF-H1 activation, we inserted two fluorescent proteins at different positions in the hinge region between the AID and DH domains (Cerulean3 [Markwardt et al., 2011] and Ypet [Nguyen and Daugherty, 2005]; see Fig. S1 C for full biosensor sequence). These were connected by four copies of a previously described peptide linker (Pertz et al., 2006). In the inactive (autoinhibited) state of GEF-H1, interactions between the AID and DH domains could restrict the fluorescent proteins to a favorable orientation that produces efficient Förster resonance energy transfer (FRET), while in the active state of GEF-H1, dissociation of the AID from the DH domain could lead to increased flexibility of the fluorescent proteins or to a different conformation, in either case reducing the amount of FRET.
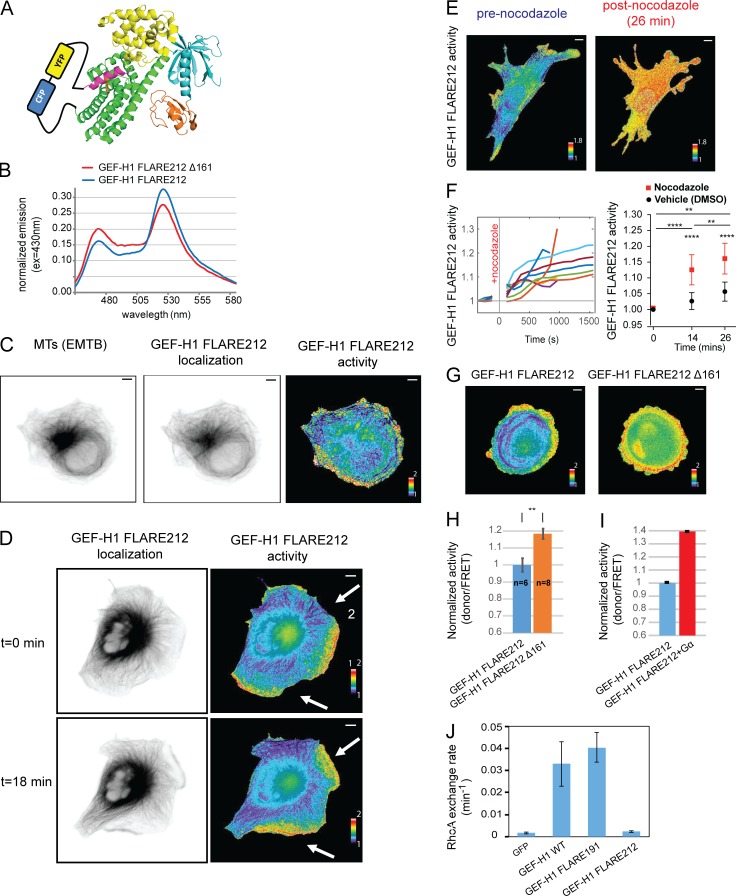
The fluorescent protein pair was inserted at five different locations in the hinge region between the AID and DH domains (after residues 203, 208, 212, 222, and 227; Fig. 2 A) in the context of both full-length GEF-H1 and a high-activity GEF-H1 variant generated by deleting the first 161 residues (Δ161; Meiri et al., 2012). Constructs were expressed in HEK293T cells, and the fluorescence emission spectra of cell suspensions were recorded upon excitation at 405 nm. For each insertion point, the dynamic range of each biosensor candidate was evaluated by measuring the difference in the donor/FRET ratios of full-length versus Δ161 variant. Insertion at position 212 yielded the highest dynamic range (45%; Fig. 2 B), and the resulting biosensor was named GEF-H1 FLARE212. There is no known mutation that traps GEF-H1 in its autoinhibited state, so the full-length version was used to evaluate the off state; this produced a lower dynamic range than that seen when examining full-length biosensor in living cells (see below).
Figure 2.
Design, characterization, and validation of the GEF-H1 FLARE biosensor. (A) Schematic of the biosensor design, using homology with the Vav1 structure (PDBID: 3ky9), and showing the FRET pair inserted in the hinge region between the AID and DH domains. Domains colored as in Fig. 1 A. (B) Emission spectra of unmodified and hyperactivated (Δ161) GEF-H1 FLARE212 expressed in suspended HEK293T cells (excitation at 405 nm). (C) Live MDA-MB-231 cells coexpressing the GEF-H1 FLARE212 and an MT marker (EMTB). GEF-H1 FLARE212 (middle) is localized primarily on MTs (left). The activity map (right) shows that inactive GEF-H1 is on MTs, while active GEF-H1 is at the cell edge. (D) Time-lapse microscopy of COS-7 cells expressing GEF-H1 FLARE212 shows that inactive GEF-H1 localized on MT and active GEF-H1 localized in protrusions (new protrusions occurring between t = 0 and t = 18 are indicated by white arrows). (E) Nocodazole treatment of MEFs expressing GEF-H1 FLARE212 led to a global increase of GEF-H1 activity. (F) Quantification of GEF-H1 FLARE212 activity from cells treated as in E and monitored for 14 and 26 min after treatment (left panel, n = 9 and n = 5, respectively). Statistical analysis of biosensor activity upon nocodazole treatment compared with vehicle control (DMSO, right panel; paired t test, P < 0.0001 [****] for before vs. after 14-min treatment, P < 0.01 [**] for before vs. after 26-min treatment and after 14-min vs. after 26-min treatment; P < 0.0001 [****] for vehicle vs. nocodazole at 14 and 26 min after treatment; right panel). Intensity values were normalized to the activity before treatment. (G) Truncation of the first 161 residues of GEF-H1 FLARE212 led to increased overall activity in MDA-MB-231 cells. (H) Statistical analysis of cells expressing the full-length (n = 6) or truncated GEF-H1 FLARE212 (n = 8; t test, P < 0.01 [**]). Intensity values were normalized to the activity of GEF-H1 FLARE212. Error bars indicate SD. (I) Coexpression of GEF-H1 FLARE and constitutively active Gα13 (Q226L) in suspended HEK293T cells led to higher activity. Intensity values were normalized to the activity of GEF-H1 FLARE212. Error bars indicate SD of three independent replicates. (J) RhoA exchange activity of GEF-H1 biosensor constructs as assayed by NMR. Activity of WT GEF-H1 and GFP shown for reference. Error bars indicate SD of three independent replicates. Scale bar = 10 µm.
For initial tests in living cells, we expressed GEF-H1 FLARE212 in COS-7, mouse embryonic fibroblasts (MEFs), and MDA-MB-231 cells. The biosensor organized as long fibers consistent with the reported localization of WT GEF-H1 on MT (Krendel et al., 2002; Meiri et al., 2012). Coexpression with a marker that labels the entire MT network (the MT binding domain of ensconsin, EMTB) confirmed that the biosensor colocalized with MT (Fig. 2 C). Analysis of GEF-H1 activity using previously described FRET ratio image analysis methods (Machacek et al., 2009; Hodgson et al., 2010) showed that GEF-H1 is inactive when on MTs and becomes active away from MTs (Fig. 2, C and D). Time-lapse imaging revealed that GEF-H1 is most active in protruding regions of the cells (Fig. 2 D and Video 1). Analysis of COS-7 images indicated that the biosensor was capable of producing a twofold difference in ratio (examining high- and low-ratio pixels, n = 5 cells; average change = 100 ± 6%; excluding highest and lowest 1% of values to eliminate spurious pixels).
Next, we verified that the biosensor responds to known biological stimuli similarly to WT GEF-H1. Treatments known to produce GEF-H1 activation in the cell, including nocodazole addition (Chang et al., 2008) or truncation of the GEF-H1 N-terminus (Meiri et al., 2012), resulted in release of the biosensor from MTs and elevated GEF-H1 FLARE212 activity (Fig. 2, E–H; and Video 2). Similarly, coexpression of constitutively active Gα13 led to increased biosensor activity (Meiri et al., 2014; Fig. 2 I).
We evaluated whether the GEF-H1 biosensor retained the GEF activity of the WT molecule. None of the four biosensor variants described above showed any significant GEF activity toward RhoA in a nuclear magnetic resonance (NMR) assay (Meiri et al., 2012; Gebregiworgis et al., 2018; Fig. 2 J and data not shown), probably due to the occlusion of the RhoA binding site on the DH domain by the inserted FRET pair. Catalytic activity is not a requirement for activity sensing, but a fully active probe could potentially replace endogenous GEF-H1 in cells or mice for future studies. To address this, we engineered another sensor variant where the fluorescent protein pair was inserted farther away from the DH domain, immediately before the AID (insertion at position 191). This sensor, GEF-H1 FLARE191, retained the ability to activate RhoA (Fig. 2 J) with activity similar to that of WT GEF-H1. Compared with GEF-H1 FLARE212, GEF-H1 FLARE191 had a lower dynamic range when measured in HEK293T cell suspensions (20% vs. 45%; Fig. S1 D). However, in single COS-7 cell imaging experiments, GEF-H1 FLARE191 showed a dynamic range similar to that of GEF-H1 FLARE 212 (n = 5 cells; average change = 127 ± 26%; highest and lowest 1% of pixels excluded; Fig. S1 E). When expressed in triple-negative breast cancer cells (MDA-MB-231) at concentrations required for quantitative mapping of activation dynamics by live-cell imaging, GEF-H1 FLARE191 had a significant effect on cell motility and caused clear changes in cell morphology. GEF-H1 FLARE212, which can report upstream stimuli but does not activate RhoA, did not affect protrusion/retraction (P/R) dynamics (Fig. S2, A and B). Therefore, all subsequent studies used GEF-H1 FLARE212.
The role of GEF-H1 and RhoA in controlling cell edge dynamics
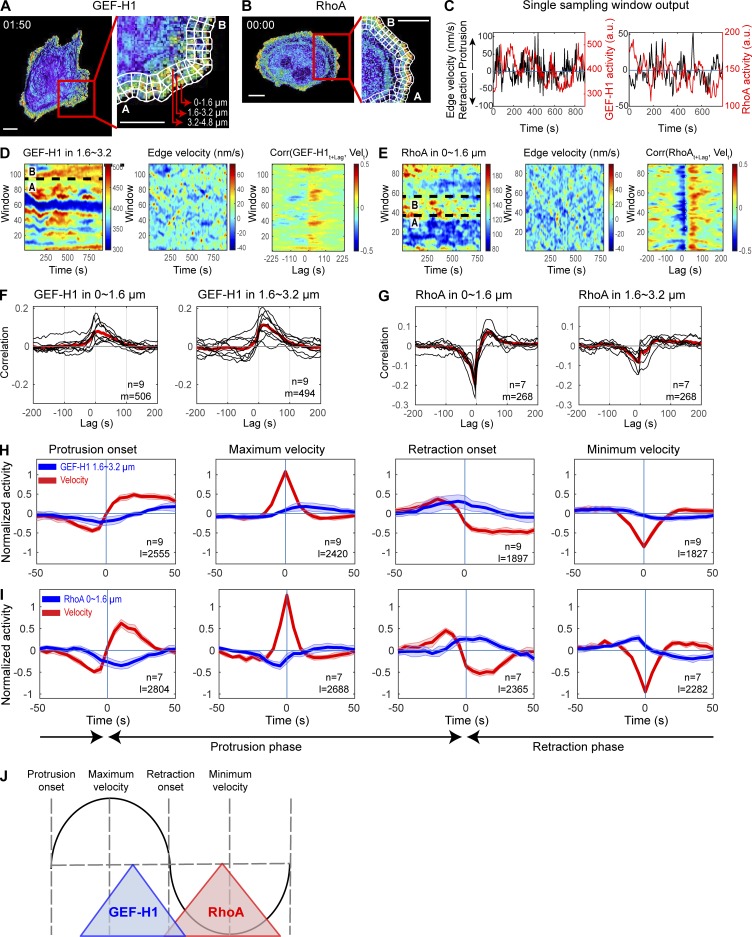
Equipped with the new biosensors, we investigated the spatial and temporal relationship between cell edge dynamics, activation of GEF-H1, and activation of RhoA. GEF-H1 has been shown to control the cytoskeletal dynamics, invasiveness, and migration of these cells when they are growing on stiff substrates (Heck et al., 2012; Liao et al., 2012). MDA-MB-231 cells expressing either GEF-H1 FLARE212 or an improved version of a previously described RhoA sensor (Pertz et al., 2006; see Materials and methods) were imaged using wide-field epifluorescence microscopy every 5 s over the course of 20–30 min (Videos 3 and 4). For quantification of biosensor activity in the context of cell motility, the edge of each cell was computationally divided into three layers of sampling windows (~1.6 × 1.6 µm2, at distances 1.6 µm, 3.2 µm, and 4.8 µm from the cell edge). The velocity of the edge adjacent to these windows, as well as the level of biosensor activity in each window, was tracked during time-lapse imaging (Machacek et al., 2009; Ma et al., 2018; Fig. 3, A–C; and Video 5). This partitioning allowed the quantification of biosensor activities relative to edge velocities at different distances from the cell edge over time. Biosensor activities were visualized using heatmaps displaying GEF activity as a function of time, distance from the edge, and position along the edge (Fig. 3, D and E). GEF-H1 and RhoA activity cofluctuated with spontaneous edge protrusion and retraction (Fig. 3, D and E).
Figure 3.
Spatiotemporal coordination of GEF-H1 and RhoA at the edge of migrating cells. (A and B) Individual time points from the migration studies used to generate panels C–E, showing MDA-MD-231 cells expressing the GEF-H1 (A) or RhoA (B) biosensors with superimposed sampling windows (right panels). Scale bar = 10 µm. (C) Time courses of biosensor activity (red) recorded in one sampling window and velocity (black) of the edge adjacent to the window. (D and E) Biosensor activity (left panel) and edge velocity (middle panel) maps of representative cells expressing the GEF-H1 (D) and RhoA (E) biosensors. Sections marked “A” and “B” in panels D and E and delimited with black dotted lines indicate ranges of windows corresponding to “A” and “B” in panels A and B. Right panels show in each sampling window the cross-correlation between the edge velocity and biosensor activity, as a function of the time lag between them. (F and G) Cross-correlation of edge velocity and GEF-H1 (F) or RhoA (G) as a function of their time lag. Black curves display the per-cell average cross-correlation (n = 9 for GEF-H1; n = 7 for RhoA). Red curves display average values. The total number of windows (m) from cells (n) imaged in multiple independent experiments (five for GEF-H1 and three for RhoA) is indicated. (H and I) Fluctuation of GEF-H1 (H) and RhoA (I) activity during major cell edge motion events. Fluctuation curves from different cells (n = 9 for GEF-H1; n = 7 for RhoA) are averaged (solid lines), and shaded confidence bands indicate ±2 × SEM. The total number of edge motion events sampled (l) is indicated. (J) Summary of GEF-H1 and RhoA activation during a protrusion and retraction cycle.
The spatiotemporal coupling of cell edge motions with the activation of GEF-H1 and RhoA was quantified using two orthogonal statistical analyses: a previously described cross-correlation analysis (Machacek et al., 2009) and a fluctuation analysis method that quantifies changes in activity around defined edge motion events such as the onset of protrusion or retraction (Lee et al., 2015). For each sampling window, Pearson’s correlations between edge velocity and biosensor activity were computed as a function of the lag between these two parameters (Fig. 3, D and E). For both GEF-H1 and RhoA, windows in quiescent edge regions displayed near-zero or weak cross-correlations (Fig. S2, C and D). Hence, quiescent windows were automatically excluded from subsequent analyses in an unbiased fashion (see Materials and methods). Within each cell, correlation curves were averaged at different distances from the edge, i.e., at different window layers. Each cell showed remarkably consistent patterns within the same layers across different cells, for both GEF-H1 and RhoA activity (Fig. 3, F and G; and Fig. S2, E and F). On average, GEF-H1 activity was best correlated with edge velocity with a delay of ~10–25 s; maximum edge velocity was followed by maximum GEF-H1 activity ~10–25 s later (~0.06–0.16 of one P/R cycle; see Materials and methods; Fig. 3 F). RhoA activity showed a strong negative correlation with velocity at a lag of −5 s, indicating that maximum RhoA activity is followed by the fastest retraction 5 s later (0.06 of one P/R cycle; Fig. 3 G). Unlike GEF-H1 activity, which spanned a band from the edge to 5 µm into the cell, the coupling of RhoA to edge velocity was strongest closer to the cell edge (~0–1.6 µm) and showed a steep decrease with distance from the edge (Fig. S2 F).
This cross-correlation analysis addressed only global coupling between edge motions and biosensor activity, but we wanted to determine the alignment of GEF-H1/RhoA fluctuation with edge velocity during discrete stages of protrusion and retraction. Motivated by the previously described registration of motion events (Lee et al., 2015), we developed a computational approach to quantify changes in biosensor activity around four important cell edge motion events: the initiation of protrusion, the time point of highest protruding speed, the initiation of retraction, and the time point of maximum retraction velocity. These four events were identified from smoothed velocity maps. Local GEF-H1/RhoA activities and edge velocities were sampled ±50 s around the respective motion events, and their fluctuation levels were standardized (see Materials and methods). This analysis revealed that increased GEF-H1 activity follows maximum protrusion velocity and occurs simultaneously with the initiation of retractions, while rising RhoA activity coincides with both retraction initiation and maximum retraction velocity (Fig. 3, H and I; and Fig. S2, G and H). Furthermore, this analysis showed that RhoA activity, unlike that of GEF-H1, is sustained throughout the retraction phase. Collectively, these analyses indicate that GEF-H1 is activated as protrusions slow down, and that GEF-H1 activity correlates with RhoA activation and subsequent initiation of cell edge retraction (Fig. 3 J).
Local changes in the MT cytoskeleton regulate GEF-H1 activity
Consistent with inferences from previous studies (Krendel et al., 2002; Meiri et al., 2012), our biosensor showed that GEF-H1 is inactive when bound to MTs. However, whether local changes in the MT network at the cell edge regulate GEF-H1 activity had not been demonstrated. Nocodazole-induced depolymerization of MTs resulted in elevated GEF-H1 activity (Fig. 2, E and F), suggesting that the polymerization state of MTs affects GEF-H1. However, nocodazole induces changes in the global MT network, altering cellular signaling on a scale unlikely to occur under physiological conditions. We therefore exploited the potential of our biosensor to probe signaling locally, by examining the relationship between MT dynamics and GEF-H1 activity at the edge of spontaneously migrating cells.
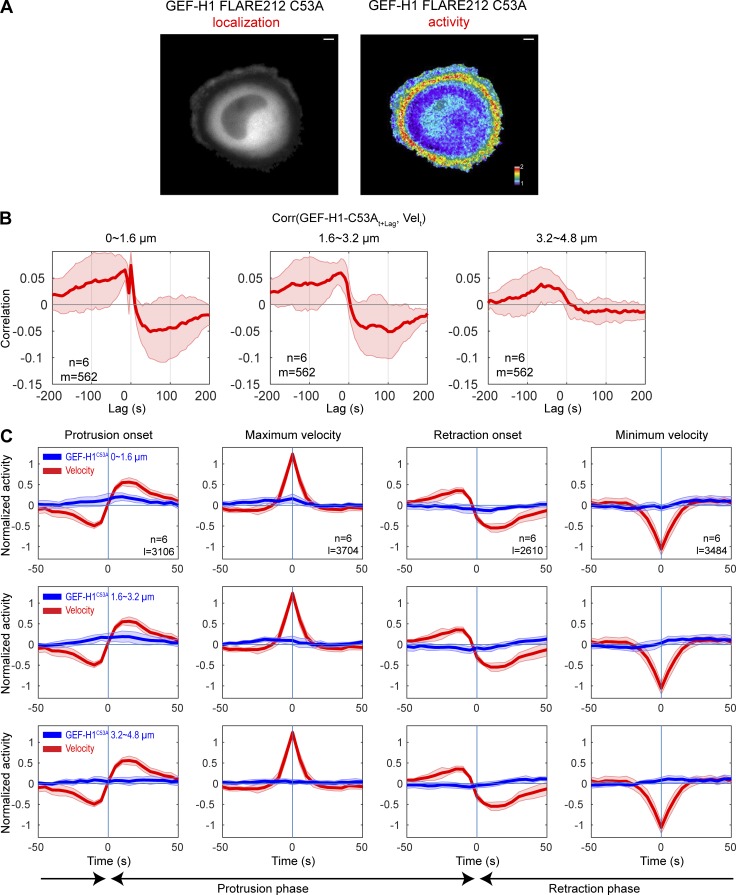
First, we examined the effect of introducing into the biosensor a mutation that is known to remove WT GEF-H1 from MTs (Krendel et al., 2002; C53A in the C1 domain). The GEF-H1 FLARE212 C53A sensor was indeed cytoplasmic, and its activity map showed marked differences from the unmodified biosensor. This biosensor was not localized on MTs (Fig. 4 A), and its activity distribution did not match that of GEF-H1 FLARE212 in either control (Fig. 4 A) or nocodazole-treated (Fig. 2 E) cells. The highest activity of the C53A biosensor was localized in a band well back from the edge. GEF-H1 FLARE212 C53A exhibited significant correlations at negative lags in the 0–3.2-µm region, indicating that while its activity still cofluctuated with edge motion, it now preceded edge protrusions, in contrast to the unmutated biosensor (Fig. 4, B and C). Overall, correlations were weaker than the ones measured for unmutated GEF-H1 FLARE212, and the correlation curve in the 3.2–4.8-µm region was close to zero. These results suggested that precise release of GEF-H1 from MTs is critical for proper activation in protrusions.
Figure 4.
GEF-H1 FLARE212 C35A shows shifted temporal coordination with edge motion. (A) Representative localization and activity maps of GEF-H1 FLARE212 C35A biosensor in migrating cells. Scale bar = 10 µm. (B) Averaged correlation curves between GEF-H1 FLARE212 C35A signal and edge velocity. The number of windows sampled (m) and total cells analyzed (n) are indicated. (C) Fluctuation of GEF-H1 FLARE212 C35A signal around edge motion events. Fluctuation curves from different cells (n = 6) are averaged (solid lines), and shaded confidence bands indicate ±2 × SEM. The total number of analyzed edge motion events (l) is indicated.
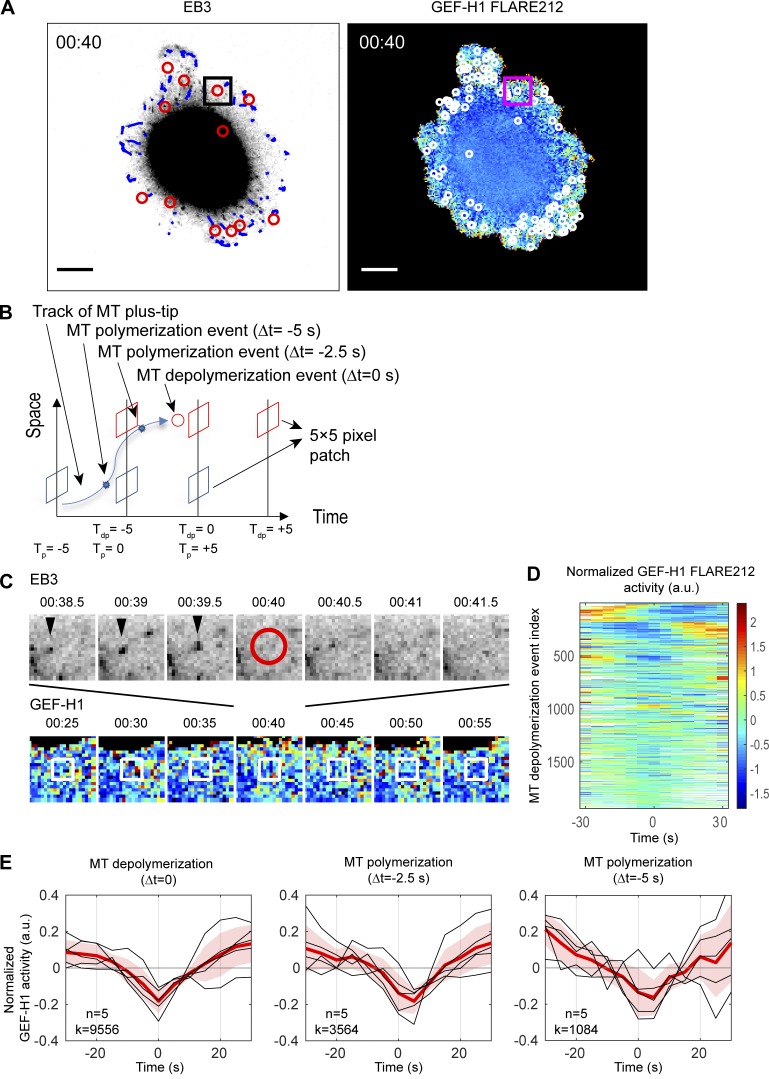
To determine how changes in MT dynamics affect GEF-H1 activity at the cell edge, MDA-MB-231 cells coexpressing mRuby-EB3 and the GEF-H1 FLARE212 were imaged during spontaneous migration. EB3 marks MT plus ends during polymerization, making it possible to track the trajectory of MT filaments using fast image acquisition and established computational analysis methods (Applegate et al., 2011; Roudot et al., 2017). Because imaging the biosensor required an exposure time of 1–1.5 s at the low concentrations required to not perturb edge behavior, it was impossible to capture EB3 and GEF-H1 images fast enough for MT tip tracking. Therefore, we developed an imaging protocol in which the EB3 channel was acquired every 500 ms, while FRET images of biosensor activity were acquired every 5 s. The EB3 channel was imaged for 180–250 time points before significant fluorophore bleaching occurred, enabling simultaneous measurements of the activity of GEF-H1 and the dynamics of MTs in migrating cells over the course of ~2 min (Fig. 5 A and Video 6).
Figure 5.
Local GEF-H1 activity is affected by MT dynamics. (A) Imaging of a MDA-MB-231 cell coexpressing mRuby-EB3 and GEF-H1 FLARE212. Right panel shows MT depolymerization events (red circles) and computationally tracked MT plus-tips (blue) over a 500-ms interval. Left panel shows MT depolymerization events (white circles) accumulated over a 5-s interval and mapped on the activity map of GEF-H1 FLARE212. Scale bar = 10 µm. (B) Schematic of GEF-H1 activity sampling before and after MT depolymerization (red) and polymerization (blue) events. Tdp = 0 (Tp = 0) denotes the time of MT depolymerization (polymerization) registered to the nearest GEF-H1 imaging frame. (C) Seven consecutive images of EB3 (top row) and GEF-H1 activity (bottom row) within the square area indicated in A to demonstrate sampling of local GEF-H1 activities around an MT depolymerization event shown in the center at 00:40 (red circle). White boxes indicate the fixed area around the MT depolymerization event over which GEF-H1 activity time courses are sampled. (D) Time courses of normalized local GEF-H1 activities aligned to the time of MT depolymerization (Tdp = 0) sampled from the cell shown in A. The time courses are ordered using hierarchical clustering for visualization purposes. White regions indicate values that cannot be calculated because an MT depolymerization event is near the beginning or the end of the imaging series. (E) Typical fluctuations of normalized local GEF-H1 activity 30 s before and after MT depolymerization (left) and polymerization (middle and right, at 2.5 or 5.0 s before MT shrinkage). For each plot, the per-cell average (red) of the local GEF-H1 activity time courses (n = 5, black) are taken, and the cell-to-cell variability is shown by ±2 × SEM (shaded bands). The total number (k) of MT (de)polymerization events at Δt = 0, −2.5, or −5 s detected from n = 5 cells is indicated.
We modified our previously described particle tracking algorithm (Roudot et al., 2017) to reconstruct MT growth patterns and discard artifacts induced by the pauses in EB3 channel acquisitions during biosensor imaging. On average, we were able to identify 1,700-2,700 MT tracks per cell, with a median track lifetime of 2.5 s (~1.5–63 s in range) and a median growth rate of 0.97 µm/s. We assumed that a growing track represented a MT polymerization event, while the disappearance of the EB3 signal from a track for more than two consecutive time points indicated the onset of MT depolymerization (Applegate et al., 2011).
GEF-H1 activity surrounding MT depolymerization events was sampled within a 5 × 5–pixel square (1.6 × 1.6 µm2), over a period spanning 30 s before and 30 s after each event (Fig. 5, B and C). Within these ranges, the GEF-H1 activity was normalized to highlight its fluctuation around the local mean (see Materials and methods). The temporal alignment of >1,500 normalized local GEF-H1 activities per cell revealed that GEF-H1 activities were consistently lowest at the time of MT depolymerization onset (Fig. 5, D and E). The same analysis was applied to the polymerizing region at the tips of MT filaments, 2.5 and 5.0 s before depolymerization onset (Fig. 5, B and E). This data showed that GEF-H1 activity was lower than its local mean activity at the time of polymerization and further decreased until the onset of MT depolymerization (Fig. 5 E). After the onset of MT depolymerization, GEF-H1 is locally activated for periods longer than 30 s.
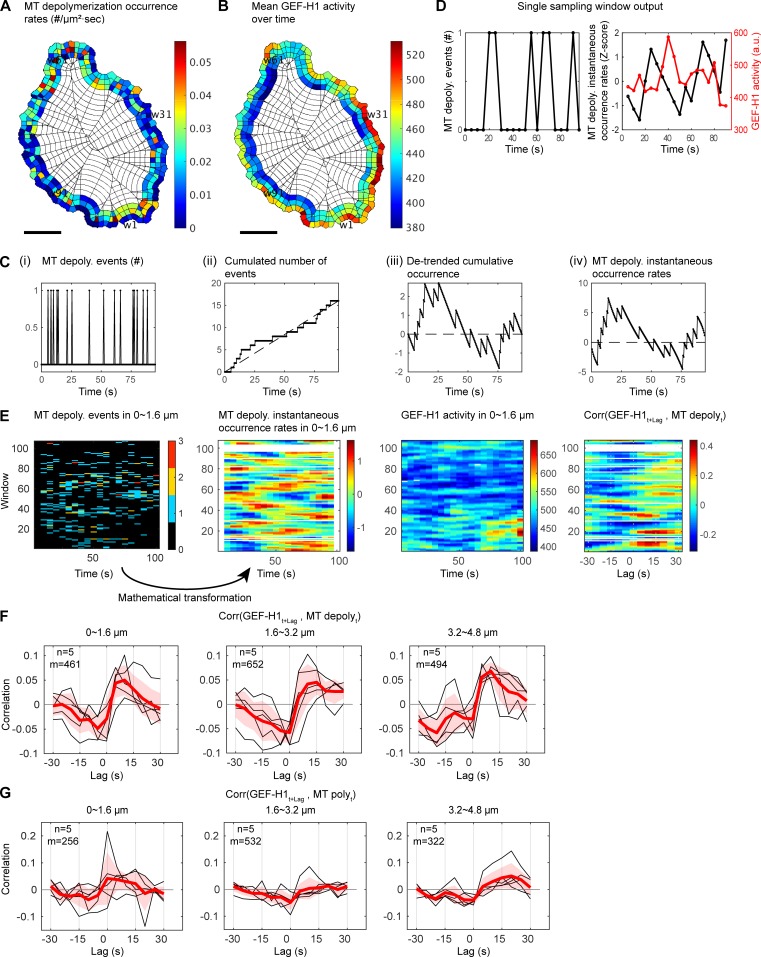
To establish the relationship between MT depolymerization and GEF-H1 activation more formally, we sought to develop a cross-correlation analysis between discrete events (MT depolymerization) and fluctuations in a continuous signal (GEF-H1 activity). Biosensor activity and the number of MT depolymerization events were sampled at the subcellular level using windows as before (Fig. 6, A and B). The series of MT depolymerization events in each window were then converted into a standardized local occurrence rate (Fig. 6, C–E; and Materials and methods), a measure that could be related to GEF-H1 activity using cross-correlation analysis. The averaged correlation curves within each cell revealed a consistent pattern over five cells. A significant positive correlation between the occurrence rate of MT depolymerization and GEF-H1 activity was measured with a time lag of 10–15 s, indicating that MT depolymerization is followed after ~10–15 s by maximum GEF-H1 activity (Fig. 6, E and F). Spatially, the effect of MT depolymerization events on GEF-H1 activity was homogeneous over the first 4.8 µm from the cell boundary (Fig. 6 F). We also observed consistent negative correlations with a lag of −5 to 0 s, suggesting that low GEF-H1 activity is related to MT polymerization. To confirm this, we performed the correlation analysis between GEF-H1 activity and MT polymerization events occurring 2.5 s before the depolymerization onset. Indeed, the cross-correlation curves showed negative correlations at 0 lag and 1.6–4.8-µm depth (Fig. 6 G). To evaluate the significance of our results, we randomized the MT depolymerization events as a negative control and found that the correlation with GEF-H1 activities was abrogated (Materials and methods and Fig. S3).
Figure 6.
GEF-H1 activity is temporally correlated with spontaneous MT polymerization/depolymerization events. (A and B) Spatial distributions of MT depolymerization event occurrence rates (n/µm2 · s; A) and GEF-H1 FLARE212 activity (B), shown as the temporal average in each sampling window. Scale bar = 10 µm. (C) Mathematical transformation of sparse event time series into instantaneous occurrence rate time series. (Ci) A serially observed number of MT depolymerization events within a small sampling window over 190 frames (0.5 s/frame). (Cii) Time series of cumulative events displays a linear trend, which is characteristic for a Poisson point process. (Ciii) To measure cofluctuation of discrete events with another continuous signal, the cumulative events time series is detrended. (Civ) The detrended signal has a time-dependent variance (larger in the middle and smaller at the beginning and ending of the observation period). The time-varying factor in the variance is normalized to obtain a time series that is suitable for cross-correlation analysis. The transformed signal is interpreted as an excess instantaneous occurrence rate time series (see Materials and methods for details). (D) Time courses of MT depolymerization occurrence within every 5-s time interval in one sampling window (left) and its instantaneous occurrence rate (black) and GEF-H1 activity (red) recorded in the same window (right). (E) Activity maps for the number of MT depolymerization events, their instantaneous occurrence rates, and GEF-H1 activities (from left to right) recorded over time in sampling windows ~0–1.6 µm from the cell edge. The right panel shows in each sampling window the cross-correlation between the GEF-H1 activity and the MT depolymerization occurrence rate. (F) Cross-correlations between the instantaneous occurrence rate of MT depolymerization and the lagged GEF-H1 activity at ~0–1.6 µm (left), ~1.6–3.2 µm (middle), and ~3.2–4.6 µm (right) from the edge. For each case, per-cell average cross-correlation curves (n = 5, black) are taken (red), and the cell-to-cell variability is shown by ±2 × SEM (shaded bands). The total number of windows with at least one MT depolymerization event sampled (m) is indicated. (G) Same as F with MT polymerization events occurring at 2.5 s before shrinkage.
Src controls GEF-H1 activity at the cell edge
Overall, MT dynamics correlated only weakly with cell edge movements, and only in discrete locations at the cell boundary (Fig. S4). However, our results showed that active GEF-H1 was strongly coupled to edge dynamics. This implied that regulatory steps in addition to MT depolymerization are critical for the proper distribution of GEF-H1 activity after its release from MTs. The newly identified AID of GEF-H1 is centered on residue Tyr198, which corresponds to a site of Src phosphorylation in the GEF Vav1. Given that Src is active at the cell edge and that it plays a major role in controlling edge morphodynamics (Playford and Schaller, 2004; Chu et al., 2014), we decided to investigate whether Src regulates GEF-H1 to control protrusions.
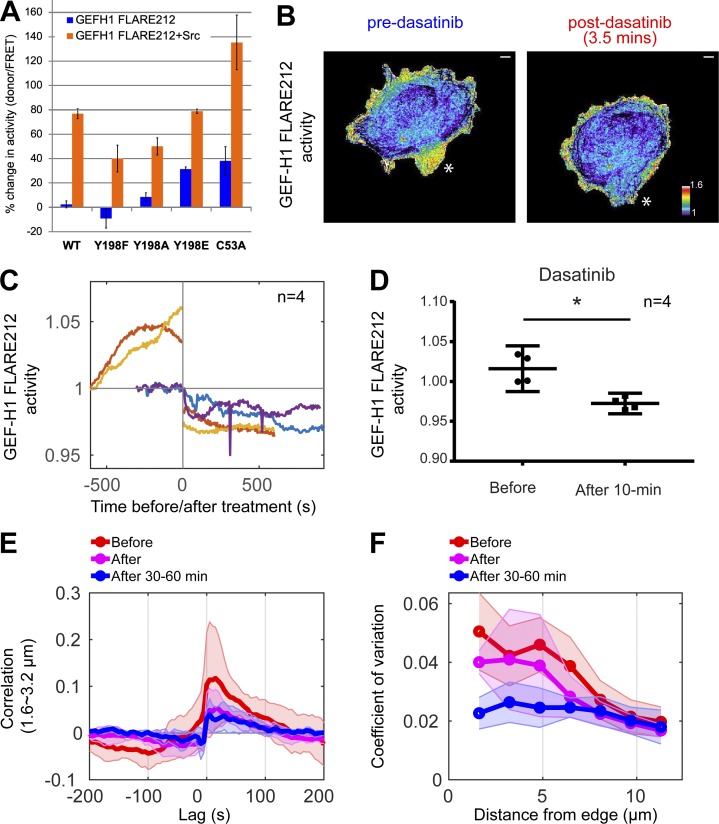
Coexpression of constitutively active Src (CA-Src, Y538F mutant) with the biosensor led to a 75% increase in GEF-H1 activity as measured by a high-content multicellular assay (Slattery and Hahn, 2014; Fig. 7 A). Mutating Tyr198 to Phe or Ala so that it could no longer be phosphorylated (Y198F and Y198A) reduced the effect of CA-Src on GEF-H1 activity by 35–48% (Fig. 7 A), and a phosphomimetic Y198E biosensor mutant had a 30% higher basal activity than the WT biosensor in the absence of Src. Phosphoproteomic analysis confirmed that Y198 was phosphorylated in cells coexpressing WT GEF-H1 and CA-Src (Fig. S5, A and B), but not in cells expressing WT GEF-H1 alone, and further revealed two other putative Src phosphorylation sites (Tyr125 and Tyr 434; data not shown). Interestingly, the C53A biosensor variant with reduced MT binding had 38% higher basal activity than the WT sensor, consistent with reduced activity for GEF-H1 bound to MT. Upon CA-Src coexpression, this biosensor achieved maximal activity levels that were 60% higher than for the unmodified biosensor (Fig. 7 A), suggesting that Src phosphorylation and MT dissociation play additive roles in GEF-H1 activation.
Figure 7.
Src activates GEF-H1. (A) Effect of CA-Src overexpression on the activity of GEF-H1 FLARE212 mutants, measured in populations of suspended HEK293T cells. Results are averages of at least three independent experiments (WT, n = 12; Y198F, n = 7; Y198A, n = 5; Y198E, n = 3; C53A, n = 6). Error bars indicate SD. (B) Example showing the effect of dasatinib (Src inhibitor) on GEF-H1 activity at a cell protrusion, indicated by the white star. Scale bar = 10 µm. (C) Src inhibition with 10 nM dasatinib decreases average GEF-H1 activity in motile cells. Curves show individual cell data where GEF-H1 activity was measured at 10-s intervals before and after treatment. Mean biosensor activity levels are normalized to the first acquisition time point. (D) Statistical analysis of the dasatinib-induced decrease in GEF-H1 activity (paired t test, P = 0.043). (E) Cross-correlation curves of GEF-H1 activity and cell edge motion in the ~1.6–3.2-µm windows before, immediately after, and 30–60 min after dasatinib. (F) Modulation of GEF-H1 activity for the three stages of the treatment at different distances from the edge, quantified by temporal coefficients of variation. Curves of individual cells are averaged (solid lines, n = 4 for each condition), and shaded confidence bands indicate ±2 × SEM in E and F.
To understand the timing and the subcellular regions where GEF-H1 was regulated by Src during migration, we inhibited endogenous Src and monitored GEF-H1 activity during live-cell imaging experiments. MDA-MB-231 cells expressing GEF-H1 FLARE were imaged before and after treatment with the Src inhibitor dasatinib (Fig. 7 B and Video 7). A low concentration of dasatinib (10 nM) was used to ensure that cells remained motile in order to allow quantification of the relationship between GEF-H1 and cell edge dynamics using cross-correlation analysis, although GEF-H1 activity could be further decreased with higher dasatinib concentrations (1 µM; Fig. S5 C). To ensure that Src was indeed inhibited at this low dose, we measured the effect of dasatinib on cell edge movement. We previously showed that Src activation increases cell edge dynamics (Karginov et al., 2014; Dagliyan et al., 2016); therefore, we expected that dasatinib addition would reduce cell edge motions. Indeed, in four of the five cells analyzed, 10 nM dasatinib led to decreased fluctuation of cell edge velocity (Fig. S5 C) and significant decreases in GEF-H1 activity (Fig. 7, C and D). Further analysis of these four cells showed that Src inhibition dramatically diminished the normal degree of modulation of GEF-H1 near the cell edge and significantly reduced cross-correlation between GEF-H1 activity and edge dynamics (0.12 to 0.05 at 1.6–3.2-µm depth), an effect further augmented by exposure of cells to dasatinib for 30–60 min (Fig. 7, E and F).
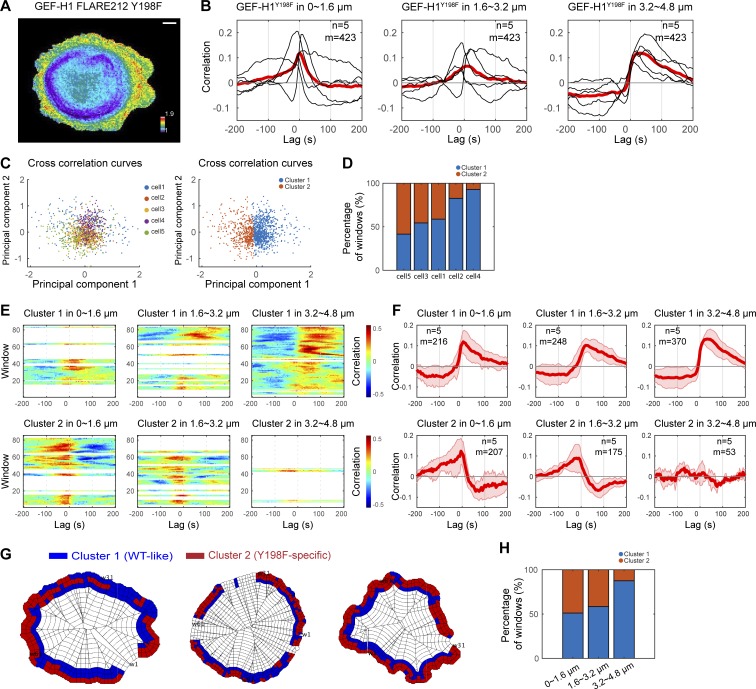
To corroborate the role of Src phosphorylation in regulating GEF-H1, we examined the activity distribution of the phosphorylation-deficient Y198F biosensor mutant in living cells (Fig. 8 A and Video 8). This biosensor showed marked differences from the WT biosensor in the relationship between activity and cell edge dynamics, particularly in the 0–3.2-µm region. Although correlation curves of individual cells were heterogeneous, the mutant biosensor showed a shorter lag in the 0–3.2-µm region but resembled WT biosensor in the 3.2–4.8-µm band (Fig. 8 B). We examined the source of the cell-to-cell heterogeneity. The individual sampling windows in the region 0–4.8 µm from the edge could be clustered into two types of couplings between GEF-H1-Y198F and edge dynamics: (a) a subset located primarily within 0–3.2-µm depth, for which activity best correlated with edge motion with a time lag of −20 to −5 s (i.e., GEF activity rises before protrusion velocity), and (b) a subset located primarily at 3.2–4.8 µm from the edge, where the correlation was similar to WT GEF-H1 (~10–25-s time lag; Fig. 8, C–G). The timing and spatial distribution of the two correlation patterns differed significantly among the five cells included in the analysis, explaining the cell-to-cell heterogeneity of correlation curves averaged over all the sampling windows. However, once separated at the subcellular level, the correlations displayed a consistent coupling pattern associated with GEF-H1 Y198F activity within windows located <3.2 µm from the cell edge (Fig. 8, F–H). Overall, our results identify Src as a new regulator of GEF-H1 and show that Src phosphorylation of Y198 is critical for both localization and kinetics of GEF-H1 activity at the periphery of the cell.
Figure 8.
Regulation by Src is critical for GEF-H1 activity at the cell edge. (A) Activity map of GEF-H1 FLARE212 Y198F in migrating MDA-MB-231 cells. Scale bar = 10 µm. (B) Cross-correlation analysis of GEF-H1 FLARE212 Y198F activity and edge dynamics at three different distances from the cell edge. Black curves represent data from individual cells, with the red curve showing the average value. The total number of windows sampled (m) in n = 5 cells is indicated. (C) A 2D projection of the cross-correlation curves of sampling windows from multiple cells using principal component analysis. Each data point represents the correlation curve of each window at −100- to 100-s lag. The correlation curves are divided into two clusters using the k-means method. (D) A bar graph showing cell-to-cell heterogeneity in the composition of the two different correlation patterns, which are the main source of heterogeneity in the correlation curves shown in B. (E) Correlation maps of edge velocity and lagged GEF-H1 FLARE212 Y198F activity split in two clusters of windows by clustering correlation curves. (F) Cross-correlation analysis of GEF-H1 FLARE212 Y198F activity and edge dynamics in the two clusters within the three different cell edge regions. The correlation pattern of cluster 1 is similar to GEF-H1 FLARE212, and the pattern for cluster 2 is specific to the Y198F mutant. Solid red curves represent averages (n = 5), and shaded confidence bands indicate ±2 × SEM. For each condition and cluster, the total number of windows sampled (m) in n = 5 cells is indicated. (G) Subcellular distribution of the windows in the two clusters at the edge of migrating MDA-MB-231 cells. (H) Composition of the two correlation patterns at different distances from the cell edge.
Discussion
We have described fluorescent biosensors that report the interaction of a newly identified GEF-H1 AID with the DH domain. For GEF-H1 to become active and bind RhoA, the dissociation of the AID and DH domains must occur, so this conformational change can serve as an indicator of GEF-H1 conversion to an activated state. Our biosensors responded with the same fluorescence change to each of the known GEF-H1 activation mechanisms we tested in living cells (nocodazole-induced MT depolymerization, Gα, and truncation of 161 N-terminal residues). Consistent with previous studies, the biosensor also showed binding of the inactive conformation to MTs, whereas the active conformation was found in protrusions of motile cells. We made biosensors that did and did not retain GEF activity (GEF-H1 FLARE191 and GEF-H1 FLARE212, respectively). GEF-H1 FLARE212 can be expressed at levels that result in a higher signal-to-noise ratio without affecting GEF-controlled cellular behaviors. GEF-H1 FLARE191, which retains GEF function, could potentially be used to replace endogenous GEF, even by expressing it from the normal GEF-H1 gene locus, producing biosensor levels that closely match the physiological changes in expression of the native molecule.
The biosensor revealed that in cell protrusions, GEF-H1 activity peaks after maximal forward velocity and correlates with the initiation of retractions. RhoA, the target of GEF-H1, is active during both the initiation and propagation of retraction. This suggests that while GEF-H1 may switch on RhoA initially, additional GEFs are likely important to maintain active RhoA at the cell edge, and inactivation by GAPs or GDIs may play additional roles (Hodge and Ridley, 2016). Our analysis showed a considerable lag between peak GEF-H1 and RhoA activities, implying that additional regulatory steps may be important to translate GEF-H1 activity to the site of RhoA interaction. This conjecture is supported by the much broader spatial distribution of active GEF-H1 (0–4.8 µm) relative to active RhoA, which is restricted to the very edge of the cell (0–1.6 µm).
The biosensor provided direct evidence that GEF-H1 is bound in an inactive state to MTs, as was previously inferred from global disruption of MT networks with nocodazole (Krendel et al., 2002). Our studies demonstrated that localized changes of MT filaments produce spatially constrained GEF-H1 activity. To demonstrate the direct coupling between local MT polymerization dynamics and GEF activation, we developed a novel statistical approach to correlate discrete, local changes in MT filaments with the continuous signal of GEF-H1 activity at the leading edge. In motile cells, local MT polymerization events correlated with low GEF-H1 activity, while MT disassembly led to higher activity.
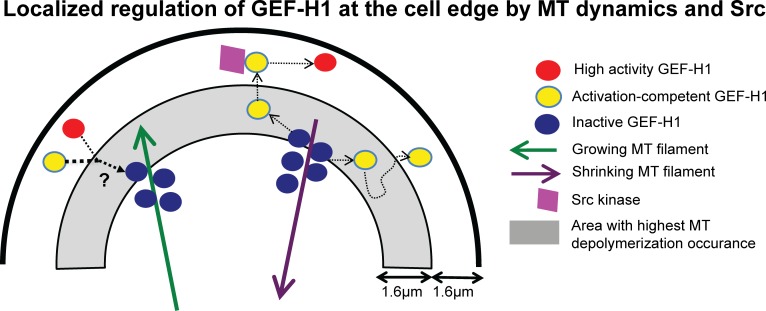
There are at least two models, not mutually exclusive, that can explain the positive coupling between MT disassembly and GEF-H1 activation: the release from MTs could be accompanied by a switch from an inactive to an active conformation, and/or it could permit interactions with other regulators that are necessary for GEF-H1 to attain its highest catalytic state and its proper localization in protrusions. Although the former model cannot be refuted at the resolution achieved by live-cell imaging, our analyses provide strong evidence for additional interactions that control the activation state of GEF-H1 after release from MTs (Fig. 9). We identified Src as one such regulator. The majority of the MT depolymerization events occurred in a band ~1.6–3.2 µm from the cell edge. The Y198F-mutated GEF-H1 biosensor, which lacks the AID Src phosphorylation site, indicated that Src controls GEF-H1 activity predominantly in a band ~0–3.2 µm from the cell edge (Fig. 9), where temporal coordination of GEF-H1 activation was altered (Fig. 8, G and H). Hence, after release from MTs, additional transport, likely by diffusion, is necessary for GEF-H1 to interact with Src and become fully activated. This was further supported by the observed differences between the coupling of MT disassembly and edge motion versus the coupling of GEF-H1 activity and edge motion. Unlike GEF-H1 activity, MT dynamics did not strongly correlate with edge dynamics.
Figure 9.
Localized regulation of GEF-H1 at the cell edge by MT dynamics and Src. GEF-H1 is released at the cell edge by depolymerizing MTs. Upon its release, phosphorylation by Src further activates GEF-H1 and localizes it at the cell edge. While MT depolymerization occurs throughout the cell, the highest number of MT disassembly events was observed in the area ~1.6–3.2 µm from the cell boundary. Control of GEF-H1 by Src occurs in discrete locations of the region ~0–1.6 µm from the edge. Polymerizing MTs recapture and deactivate GEF-H1.
Interestingly, the Y198F-mutated GEF-H1 biosensor also revealed that within the ~0–3.2-µm Src-controlled band, there were discrete locations where Src coordinated GEF-H1 with cell edge movement. Src is recruited to integrin-mediated focal adhesions by FAK (Mitra and Schlaepfer, 2006), and the FAK–Src complex promotes protrusion by activating GEFs that act on Rac1 and Cdc42 (Huveneers and Danen, 2009). As cell spreading slows down, focal adhesions mature, stress fibers form, and RhoA activity increases. Src plays a key role in this process by activating different RhoA GEFs in response to tension (Dubash et al., 2007; Lim et al., 2008). Since GEF-H1 was previously shown to become activated when force is applied to cells (Guilluy et al., 2011), the subcellular regions where Src activity is critical to coordinate GEF-H1 with edge dynamics may correspond to parts of the cell that undergo tension during adhesion reinforcement and contractility. Regardless of the exact molecular mechanisms, this observation lends further support to the notion that GEF-H1 activity is modulated not by MT disassembly alone, but also by signaling events that respond to more precise spatiotemporal cues.
While our focus was on Src, it is likely that other molecules also play important roles in controlling GEF-H1. Previously, GEF-H1 activity was shown to be regulated by interaction with 14-3-3 proteins through phosphorylation by protein kinase A (Zenke et al., 2004; Meiri et al., 2012) and by Erk (Fujishiro et al., 2008; Guilluy et al., 2011). Future studies, enabled by quantitative image analysis of biosensors in live cells and multiplexed biosensor imaging, will assess how these molecules synergize with MTs and Src to regulate the spatiotemporal dynamics of GEF-H1 activity in cell migration.
Materials and methods
Plasmids and DNA
GEF-H1 biosensors were cloned into pTriEx4 vector (Novagen). Mutations were introduced by Q5 Mutagenesis (NEB) using synthesized oligonucleotides from IDT. Cherry-EMTB was generated by replacing cherry with GFP in the previously described 3xGFP-EMTB construct (Faire et al., 1999). Gα13 Q226L was cloned in the pCDNA3.1 vector. CA-Src (Src Y538F mutation) was cloned in the pUSE vector.
Expression and purification of recombinant GEF-H1 and RhoA
Genes encoding residues 188–601 and 201–601 of GEF-H1 were cloned into the pet28a vector (Novagen) at NcoI/XhoI sites. Tyr198 in the 188–601 construct was mutated to alanine using Q5 Mutagenesis (New England Biolabs [NEB]). Constructs were transformed into BL21 cells (NEB). 5-ml Luria-Bertani broth starter cultures inoculated with individual colonies were grown with shaking at 37°C overnight, and expanded the next day into 1 liter Luria broth (Genesee Scientific) supplemented with antibiotics. Cultures were then grown at 37°C until they reached an absorbance of 0.6–0.8 at 600 nm, when protein expression was induced with 500 nM IPTG. Protein expression was induced at room temperature for 24 h. Cultures were subsequently spun down at 4°C for 30 min (4,000 rpm) and frozen at −80°C until processing. Cell pellets were resuspended in 10 ml lysis buffer (50 mM Tris, 150 mM NaCl, 1 mM DTT, 5 mM MgCl2, 0.1% NP-40, 2 mM MgSO4, and 10% glycerol, pH 8.6) supplemented with protease inhibitors (cOmplete Protease Inhibitor Cocktail; Sigma-Aldrich) and lysozyme (Sigma-Aldrich). Cells were then sonicated on ice, and the lysate was clarified by spinning down at 4°C for 30 min at 14,000 rpm. The supernatant was applied to equilibrated His GraviTrap columns (GE Healthcare). Columns were washed with 20 ml of wash buffer (50 mM Tris, 150 mM NaCl, 1 mM DTT, 5 mM MgCl2, 10% glycerol, and 30 mM imidazole, pH 8.6) and eluted in 1-ml fractions with elution buffer (50 mM Tris, 150 mM NaCl, 1 mM DTT, 5 mM MgCl2, 10% glycerol, and 500 mM imidazole, pH 8.6). Collected fractions were analyzed by SDS-PAGE, and buffer (20 mM Hepes, 100 mM NaCl, 5 mM MgCl2, and 2 mM DTT, pH 7.0) was exchanged using Zeba desalting spin columns (Thermo Fisher Scientific), aliquoted, supplemented with 5% glycerol, frozen in liquid nitrogen, and stored at −80°C.
RhoA was cloned in the pProEx HTA plasmid and transformed into BL-21 cells. Upon inoculation and expansion at 37°C with shaking, 1-liter cultures were induced with 1 mM IPTG once they reached an OD of 0.6–0.8 at 600 nm. Cultures were then grown at 18°C for 24 h, pelleted by centrifugation at 4°C (30 min, 4,000 rpm), and frozen at −80°C until further processing. Cells were resuspended in 20 ml lysis buffer (25 mM Tris, 300 mM NaCl, 5% glycerol, 10 mM imidazole, 1% NP-40, 50 µM GDP, 5 mM MgCl2, and 1 mM DTT, pH 7.5) supplemented with lysozyme and protease inhibitors. Cells were lysed by sonication at 4°C, and the lysate was clarified by centrifugation (30 min, 14,000 rpm). The supernatant was applied to equilibrated His GraviTrap columns. Columns were washed with 20 ml wash buffer (25 mM Tris, 300 mM NaCl, 5% glycerol, and 10 mM imidazole, pH 7.5), and RhoA was eluted in elution buffer (25 mM Tris, 300 mM NaCl, 5% glycerol, 1 M imidazole, and 0.02% NP-40, pH 7.5). Fractions were analyzed by SDS-PAGE, and buffer was exchanged (25 mM Tris, 300 mM NaCl, and 5% glycerol, pH 7.5) and frozen at −80°C.
RhoA nucleotide exchange assay
Reaction mixtures contained 400 nM purified RhoA and 10 µM BODIPY FL GDP (Thermo Fisher Scientific) in 1 ml assay buffer (20 mM Tris, 150 mM NaCl, 2 mM DTT, and 5% glycerol, pH 7.5). Samples were read on a spectrofluorometer (Fluorolog; Jobin Yvon), by recording emission at 511 nm upon 500 nm excitation. Upon baseline stabilization, GEF-H1 constructs were added to the reaction mixture at a final concentration of 200 nM.
FRET measurements of GEF-H1 biosensor candidates using a fluorometer
Emission spectra of GEF-H1 biosensor candidates were obtained using a Fluorolog fluorometer (Horiba). HEK293T cells grown in 6-well plates (Nunc) were transfected with biosensor DNA using Lipofectamine Plus Reagent (Invitrogen) using conditions suggested by the manufacturer. After 24 h, cells were detached with brief trypsin (Cellgro) treatment and resuspended in cold PBS (Sigma-Aldrich) + 1% FBS (Hyclone), washed, and then resuspended in cold PBS. Samples were excited at 430 nm, and spectra were obtained from 460 to 600 nm.
FRET measurements using a multicellular high-content assay
Measurements to quantify the FRET change of GEF-H1, GEF-H1 C53A, GEF-H1 Y198A, GEF-H1 Y198F, and GEF-H1 Y198E biosensors upon coexpression with either Gα13 Q226L (for GEF-H1 only) or CA-Src (Y535F) were done using a high-content multicellular plate–based assay previously described (Slattery and Hahn, 2014). Briefly, ∼25,000 HEK293T cells were seeded in each well of a 96-well plate and transfected using Lipofectamine Plus reagent. For activation by CA-Src experiments, cells were transfected with 15 ng of GEF-H1 FLARE212 variants and different amounts of CA-Src (twofold dilutions, 0.2–100 ng). For each well, CFP emission and YFP (FRET) emission data were collected upon CFP excitation on an Olympus IX-81 microscope with automated stage and autofocus. Data were analyzed with MatLab as previously described (Slattery and Hahn, 2014). Reported FRET changes were calculated from cells transformed with 100 ng CA-Src (Fig. 7 A). To measure activation of GEF-H1 FLARE212 by Gα13 Q226L, FRET values were calculated from cells transfected with 150 ng GEF-H1 biosensor and 350 ng Gα13 Q226L. Biosensors samples with and without Gα13 Q226L were analyzed, and the average activity (donor/FRET) was calculated (GEF-H1 biosensor alone vs. GEF-H1 biosensor + Gα13 Q226L: 0.31 vs. 0.44), normalized by the activity value of GEF-H1 biosensor alone and reported in Fig. 2 I.
Cell lines and transfection
HEK293T, COS-7, and MDA-MB-231 cell lines were obtained from the American Type Culture Collection. COS-7 cells were transiently transfected using FuGENE 6 (Promega) following the manufacturer’s instructions. MDA-MB-231 cells were transiently transfected using TransIT-BrCa (Mirus) following the manufacturer’s instructions. For nocodazole experiments, GEF-H1 biosensor was stably expressed in MEFs (Clontech) by retroviral infection under a tet-off system. Briefly, HEK293T cells were transfected with p-Babe-puro (Hofmann et al., 1996) vector containing the GEF-H1 biosensor and with retroviral packaging plasmid pCL-Eco (Naviaux et al., 1996). Medium was replaced the next day, and after 24–48 h, filtered supernatant containing viral particles was added to MEFs treated with polybrene (Sigma-Aldrich). After 24–48 h, cells expressing the biosensor were selected by adding puromycin to the growth medium. The RhoA biosensor was stably expressed in MDA-MB-231 cells using a tet-off system and a similar retroviral infection protocol. The RhoA biosensor used in this study is a modified version of our previously described biosensor (Machacek et al., 2009). In the new version, the donor CyPEt was replaced with Cerulean3, and the acceptor Ypet was attached to the C-terminus of the Rhotekin effector domain for improved brightness. For MT-tracking experiments, GEF-H1 biosensor was transiently transfected in MDA-MB-231 cells stably expressing EB3-Ruby under a tet-off system. For experiments investigating the colocalization of GEF-H1 FLARE212 and MTs, cherry-EMTB and the biosensor were transiently transfected into MDA-MB-231 cells. All cells were cultured in DMEM without l-glutamine (Thermo Fisher Scientific) and supplemented with 1% GlutaMAX (Thermo Fisher Scientific) and 10% FBS (Hyclone).
Single-cell imaging and image processing
Cells were transiently transfected 24 h before imaging experiments. For stable tet-off cell lines, doxycycline was removed from the medium 72 h before imaging experiments, and the culture medium was subsequently replaced twice a day to induce protein expression. COS-7 and MEF cells were plated on glass coverslips coated with fibronectin by overnight incubation at 37°C with a 50 µg/ml fibronectin solution (Sigma-Aldrich) in PBS. MDA-MB-231 cells were plated on glass coverslips coated overnight at 37°C with 1 µg/ml collagen I (Sigma-Aldrich) in PBS. Cells were imaged in Ham’s F12 (Kaighn’s modification; Caisson Laboratories) supplemented with 5% FBS (HyClone) and 15 mM Hepes (Thermo Fisher Scientific), with a 40× 1.25-NA Silicone oil objective on an IX-81 inverted microscope (Olympus) using Metamorph software. For drug treatment experiments, the imaging series was interrupted, and either nocodazole (5 µM final; 0.25% DMSO final in a 2-ml volume imaging chamber) or dasatinib (10 nM, 0.0001% DMSO final; or 1 µM final, 0.1% DMSO final) were added to the imaging chamber before imaging was resumed. Cells were illuminated with a 100-W Hg arc lamp through a neutral density (1.5) filter. For emission, the following filter sets were used (Semrock): CFP: (ex)FF-434/17, (em)FF-482/35; FRET: (ex)FF-434/17, em(FF-550/49); YFP: (ex) FF-510/10, em(FF-550/49), using a dual band dichroic FF462/523. CFP and FRET images were simultaneously captured using a TuCam system (Andor) fitted with an imaging flat FF509-FDi01 dichroic and two Flash4 sCMOS cameras (Hamamatsu) or two HQ2 CCD cameras (Photometrics). For mCherry imaging, the dichroic was switched to a custom 440/505/595/740 dichroic with FF586/15 mCherry excitation and the dual-band emission filter ET eYFP/mCherry (Chroma). Images were analyzed to determine biosensor activity using MatLab as previously described (Machacek et al., 2009; Hodgson et al., 2010). Given the design of the GEF-H1 biosensor, where active GEF-H1 is in the lower FRET state and inactive GEF-H1 is in the higher FRET state, biosensor activity is calculated and reported as the donor/FRET ratio. Biosensor dynamic ranges for image scales were determined after excluding the lowest and highest 1% of ratio values, to eliminate the effects of spurious pixels. To compare the activity levels of the full-length and Δ161 GEF-H1 FLARE212 biosensor, average intensity values were determined from the activity maps of each processed cell, and group averages and SDs were subsequently calculated (full-length sensor, n = 6, average intensity ± SD: 345 ± 36; Δ161 biosensor, n = 8, average intensity ± SD: 408 ± 41). The average intensity of the Δ161 biosensor was normalized to the full-length value and reported in Fig. 2 H.
Immunoprecipitation and phosphoproteomic analysis
FLAG-tagged WT GEF-H1 FLARE alone, or coexpressed with CA-Src, was transfected in a 10-cm dish of HEK293T cells. 24 h after transfection, the cells were washed three times with 1 ml cold PBS and lysed with 1 ml lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol, with protease inhibitors) in the presence of 1× sodium pervanadate solution (for 200× mixed and incubated for 10 min at room temperature: 100 µl of 200 mM sodium orthovanadate, 800 µl of PBS, and 20 µl of 30% H2O2). After lysis for 30 min at 4°C, the lysate was clarified by centrifugation at 6,000 g for 10 min and incubated for 4 h at 4°C with 2 µl of anti-FLAG M2 antibody (Sigma-Aldrich). The mixture was then applied to Protein G magnetic beads (Thermo Fisher Scientific) to bind the M2 antibody and subsequently pull down FLAG-GEF-H1. Beads were washed, mixed with SDS loading dye, and boiled for 5 min at 90°C. The supernatant was analyzed by SDS-PAGE gel, and the band of interest was excised and submitted for phosphoproteomics analysis to the University of North Carolina Michael Hooker Proteomics Center. The gel band was digested using AspN and trypsin, phosphopeptide enrichment was performed, and both the enriched and nonenriched samples were analyzed by liquid chromatography/tandem mass spectrometry on a Thermo Easy nLC 1200-QExacative HF instrument. In samples where GEF-H1 was coexpressed with CA-Src, Y198 was identified automatically as a high-probability phospho-site and subsequently validated manually by confirming the presence of a peak in the spectrum that corresponded to the DEAEVIY-phospho fragment. The DEAEVIY-phospho ion was observed at different masses, with and without methionine oxidation and without water, confirming its presence in the spectrum. Importantly, no fragments corresponding to putative phosphorylated serines in the GEF-H1 AID (underlined in DEAEVIYSELMS) were observed. When GEF-H1 was expressed alone, no fragments corresponding to Y198 phosphorylation were identified.
NMR-based GEF of GEF-H1 biosensors
HEK293T cells were grown in DMEM (Invitrogen) in a 5% CO2 environment at 37°C. A 6-cm plate was seeded to 80% confluence and grown overnight before transfection of plasmid DNAs (2.5 µg total) using LipoD293 (SignaGen) according to the manufacturer’s instructions. Transfected cells were maintained for ∼48 h in fully supplemented medium before they were washed with PBS, and then the cells were harvested in a minimal volume of lysis buffer (150 µl of 2% Triton X-100, 10% glycerol, 50 mM Hepes, pH 7.5, 100 mM NaCl, Complete Protease Inhibitor cocktail [Pierce], and phosphatase inhibitor cocktail [Pierce]). After a 10-min incubation on ice, the lysates were gently centrifuged, yielding supernatants with total protein concentrations of ∼7 µg/µl, as measured by Bradford reagent.
To assess the catalytic activity of the biosensors, we transfected GEF-H1 FLARE212, GEF-H1 FLARE191, WT GEF-H1 tagged at the C-terminus with the FRET pair (Cer-Ypet), and EGFP alone (in the vector pEGFP-C1). The fluorescence intensity of YFP (measured by a Shimadzu RF-5301PC spectrofluorophotometer using excitation and emission wavelengths of 516 nm and 530 nm, respectively) in the lysate was used to normalize the loading of the biosensors and the WT constructs. On the basis of fluorescence, the expression of these constructs was similar, with a standard deviation of ±15%. Because the fluorescence properties of EGFP are distinct and EGFP alone was expressed more highly than the chimeras, this lysate was normalized to that of WT on the basis of total protein amount (Bradford).
GEF activities of the FLARE biosensors were measured directly in the cell lysates using a real-time NMR-based assay (Marshall et al., 2012; Meiri et al., 2012) that monitors the heights of peaks that are specific to either the GDP- or GTP-bound form of [15N]RhoA in 15N-1H heteronuclear single quantum coherence (HSQC) spectra. These nucleotide exchange assays were performed on a Bruker 600-MHz NMR spectrometer equipped with a 1.7-mm microcryoprobe. [15N]RhoA-GDP (residues 1–181) was prepared at a concentration of 0.3 mM in NMR buffer (20 mM Hepes, 100 mM NaCl, 5 mM MgCl2, 2 mM Tris(2-carboxyethyl)phosphine, and 10% D2O, pH 7). To a 35-µl sample of [15N]RhoA was added ∼1 µl of lysate (normalized by fluorescence) together with GTPγS to a final concentration of 3 mM (10-fold excess over RhoA). Successive 15N-1H HSQC spectra (10 min/spectrum) were collected at 25°C, and the half-life of nucleotide exchange was determined as the time point at which the GDP-bound and GTP-bound peaks exhibited equal intensities, which was used to calculate exchange rates based on single-phase exponential decay (rate = ln 2/half-life).
Homology modeling of the GEF-H1 AID and DH domain interactions
The sequence of the DH domain of GEF-H1 (UniProtKB entry Q92974-1) was analyzed with HMMER (Potter et al., 2018) to identify closely related homologues of known structure. The DH domain of p115 RhoGEF (Protein Data Bank ID 3ODO) was the highest scoring match. Therefore the sequence of the GEF-H1 DH domain was threaded onto the DH structure of p115 RhoGEF per the sequence alignment using Rosetta (Das and Baker, 2008; Leaver-Fay et al., 2011). The protein backbone of the resulting model was allowed to sample different conformations with the fastrelax subroutine of Rosetta (Tyka et al., 2011), to generate a final GEF-H1 DH model that was free of clashes and that matched the Rosetta metrics (energy, ramachandran angle distribution, core packing) of the DH domains of p115 RhoA and Vav1. To model the interactions of the newly identified AID with the GEF-H1 DH domain, first the DH and AID domains of Vav1 (Protein Data Bank ID 3KY9) were aligned onto the GEF-H1 DH domain model. The sequence of the GEF-H1 AID was threaded onto the structure of the Vav1 AID as per the alignment (Fig. 1 A) to generate a starting GEF-H1-AID complex. The complex was subsequently refined by iteratively allowing the AID peptide to dock onto the DH domain, while sampling alternative AID backbone and amino acid side-chain conformations (Davis et al., 2006; Chaudhury et al., 2011; Fleishman et al., 2011). The model with the lowest Rosetta total energy and binding energy is shown in Fig. S1 A.
Cell edge tracking and definition of a cell frame of reference for activity fluctuation analysis
To study dynamic subcellular activities of the biosensors in relation to edge motion, we computationally tracked the cell boundary movement over time and subsequently defined a cell shape–invariant coordinate system allowing registration of movement and signaling. The cell boundaries were segmented using intensity thresholding of the donor channel. To calculate locally the displacement of the cell edge, we morphed the segmented cell outlines between consecutive time points using the morphodynamic profiling algorithm previously described (Ma et al., 2018).
Upon definition of the cell edge motion, the segmented cell masks were partitioned into hundreds of sampling windows of size 5 × 5 pixels (∼1.6 × 1.6 µm2) using contour lines and ridges in the Euclidean distance transform map to the cell edge. One of the windows within the outermost layer at the first time point was set to be the origin. The location was propagated through time frames using the information of edge displacements calculated as above. This resulted in a cell shape–invariant coordinate system for the sampling of biosensor activity and MT depolymerization event density over time (Figs. 3 A and 6, A and B). Within each sampling window, biosensor activities were averaged. The subcellular activities in window rows at a fixed distance from the moving edge were represented as a space-versus-time heatmap, with the spatial axis ranging from the first to the last window of the row (Fig. 3, D and E, left panels). Similarly, for the window row at the cell edge, instantaneous P/R velocities were sampled and represented as a space-versus-time edge motion heatmap (Fig. 3, D and E, center panels). These shape-invariant maps then allowed us to cross-correlate fluctuations in edge motion with fluctuations in biosensor activity. Cross-correlation analyses produced a peak magnitude as a surrogate of the strength of coupling between motion and signaling activity, as well as the peak time lag as a measure of the delay of the signal relative to motion or vice versa. Cross-correlation curves for individual windows were averaged within each cell to generate a representative coupling profile. The profiles for different cells enabled us to compute the standard confidence interval (± 2 × SEM) of averaged profile reflecting cell-to-cell variation (Figs. 4 B and S2). Additional details of the correlation procedures were described previously (Machacek et al., 2009).
To relate GEF-H1 and RhoA cross-correlation lag values to the timing of edge protrusion or retraction, we computed the length of the P/R cycles for GEF-H1– and RhoA-expressing cells. In each cell, an autocorrelation function of the edge velocity was used to determine the time lag with the largest negative autocorrelation in absolute value, which corresponded to half of one P/R cycle (Machacek et al., 2009). For cells expressing GEF-H1 FLARE212, the median P/R cycle was 160 s (n = 9, range of 90–320 s), while for cells expressing the RhoA biosensor the median cycle was 90 s (n = 7, range of 80–230 s). The two median P/R cycles showed no significant difference (P = 0.07, Wilcoxon rank sum test). Temporal coordination of GEF-H1 and RhoA was then compared by calculating time lags relative to median P/R cycles (Fig. 3 J).
To ensure that biosensor expression did not affect cell behavior, we examined whether the biosensor expression levels in different cells showed systematic relations with (a) the parameters of the cross-correlation analysis between biosensor activity and edge motion and (b) the average P/R velocities. The GEF-H1 biosensor expression levels were computed from images in the donor channel (CFPex→CFP→em) corrected for uneven illumination of the field of view (shade or flat-field correction). For each frame in a time series, we computed a ratio between the mean fluorescence intensity of the cellular region and the fluorescence intensity of the background region generated by imaging media alone. The biosensor expression level of one cell was then determined by averaging all such ratios in a time series. This procedure allows us to determine “average signal over background” as a way to compare expression across cells that were imaged with different exposures times (100–1,500 ms).
Statistical identification of quiescent subcellular regions and biosensor activity signals with trends
For the cross-correlation analyses, both quiescent subcellular regions and biosensor activities with strong low-frequency signals were excluded. By nature of the analysis, cross-correlations were weaker in quiescent edge regions than in dynamic edge regions (Fig. S2, C and D). To automatically identify the quiescent regions, we statistically tested whether the velocity time series in a window corresponded to white noise using the Ljung–Box test (Ljung and Box, 1978). We used the MatLab (MathWorks) function lbqtest() and examined the significance of autocorrelations of ≤20 lags of the velocity time series. A subset of edge boundaries was identified as white noise signals at the standard significance level of 0.05, and visual inspection of these indicated that they matched with the quiescent regions in GEF-H1/RhoA time-lapse images. This procedure eliminated 2–46% of the windows as quiescent (Fig. S2, I and J). When GEF-H1 was imaged upon dasatinib treatment, most cell boundaries became quiescent, so we could not filter out quiescent edges. In this case, we compared the dasatinib-treated and untreated conditions without filtering out quiescent edges (Fig. 7 E).
We also excluded biosensor activities with strong low-frequency signals from the input for the cross-correlation analysis. In a few cells, GEF-H1and RhoA activities reported by the respective biosensors displayed very slow dynamics compared with other cells. This was often accompanied by slow edge protrusion and retraction over wide cell edge sectors and a systematic increase or decrease of biosensor activity over the full duration of the video. Time series with systematic trends are known to produce spurious correlations (Yule, 1926; Dean and Dunsmuir, 2016). We employed the augmented Dickey–Fuller test (Dickey and Fuller, 1979) to check the existence of trends for biosensor activity time series. We used MatLab function adftest() with the autoregressive model and a drift coefficient.
Fluctuation profiling around edge motion events
Cross-correlation analysis assumes that the temporal relationship between edge motion and biosensor activity remains stable over the full duration of the video. To check the validity of the assumption, we quantified the average fluctuation patterns before and after four characteristic edge motion events: P/R initiations and maximum P/Rs. The method entails two steps: (1) identifying motion events in the time frames and subcellular locations when and where the four edge motion events occurred; and (2) local sampling of the signals before and after the motion events and summarizing across cells.
To identify the edge motion events, we first smoothed the edge velocity map by representing the motion time series of an individual window by a smoothing spline, computed with a MatLab function csaps() and manually chosen smoothing parameters. We identified the time points and locations where the smoothed velocities are positive (negative) as a protrusion (retraction) phase. P/R periods <25 s were filtered. Within each sampling window, the beginning time points of P/R phases were then detected as the P/R onsets. Within each P/R period, the time points with the maximum/minimum smoothed velocities were also detected, respectively. Then, we examined typical biosensor activities around the four detected motion events.
To capture the local signaling dynamics reported by the GEF-H1/RhoA biosensor activities, we computed the standardized Z-scores of the activity time series for individual windows. We then locally sampled the standardized activities within 50 s before and after the events. As a control for the stability of the motion event identification, we also sampled the edge velocity time series and visualized in the same way. The averaged curves showed narrow confidence bands (Fig. 3, H and I), indicating that the detection of motion events and their registration was reasonable.
Profiling GEF-H1 activity patterns around MT (de)polymerization events
The EB3 channel was imaged every 0.5 s to track MT plus-tips, while GEF-H1 biosensor images were acquired every 5 s. Before quantifying the coupling between MT dynamics and GEF signals, MT depolymerization events detected at the time scale of the EB3 channel were registered to the nearest future GEF-H1 imaging frame, while the MT polymerization events were registered to the nearest GEF-H1 frame. To examine local GEF-H1 activity patterns around the MT (de)polymerization events, we first examined the absolute locations of the events without considering cell edge movement. Only events within 9.7 µm (30 pixels) from the cell edge were considered, which excludes perinuclear areas where plus-tip tracks are unstable and the GEF-H1 activities unclear. We considered 5 × 5–pixel-sized (1.6 × 1.6 µm2) patches centered at each MT (de)polymerization event (Fig. 5, B and C). At each patch, local GEF-H1 activities within ±30 s were sampled directly from the biosensor images and standardized into Z-scores to focus only on the fluctuations around local mean activity. For each cell, we sampled ∼2,000 such event-centered GEF-H1 activity time courses (Fig. 5 D). The individual time courses then were averaged within each cell to generate a representative profile of the GEF-H1 activity pattern about MT (de)polymerization. The profiles for different cells indicated a remarkably consistent pattern, allowing us to compute average profiles with confidence intervals reflecting cell-to-cell variation (Fig. 5 E).
Transformation of sparse event time series into continuous time series of instantaneous event occurrence rates
To determine the coupling between MT depolymerization events and GEF-H1 activities, we accumulated the depolymerization events over the 5 s between GEF-H1 frames in the sampling windows of the cell frame of reference. The resulting time courses of MT depolymerization events were sparse. On average, we observed one event every 25 s per sampling window (Fig. 6, A and E). To test whether there is any systematic relation between these sparse, discrete signals and the continuous GEF-H1 activity, we developed a mathematical transformation of the discrete event time series into a continuous time series of instantaneous occurrence rates. Given the series of sparse incidents, we first compiled a time series of the cumulative events (Fig. 6 C). Most of the cumulative event time series showed a nearly linear trend, supporting the notion that the event occurrence follows a Poisson point process, i.e., events occur at a constant rate and the waiting times between events are independent, following an exponential distribution (Streit, 2010). We then subtracted the expected linear trend from the actual cumulative event time series to extract fluctuations about the baseline process (Fig. 6 C). Under the assumption of the Poisson point process, the detrended time series has time-dependent variance that increases until the midpoint of the time series and then decreases again to zero in the last time point. Specifically, the variance is calculated as follows: let Nt denote the total number of events that occur up to and including time t ≥ 0. After observing the number of occurrences NT until the end time point T, the cumulated number of events can be detrended as
under the assumption that the underlying stochastic process {Nt : 0 ≤ t ≤ T} follows a Poisson point process with an occurrence rate λ > 0. Accounting for the variance of the detrended series follows as
To calculate the covariance, we used the property that the number of events during disjoint time intervals is independent. To account for this time-dependent variance, we normalized the detrended cumulative time series,
to result in a continuous time series with an expectation value of 0 and a constant variance, which can be interpreted as a time series of excess event occurrence rates. This mathematical transformation is generally applicable for event time series where the occurrence rate is constant.
Randomization of MT dynamics
We designed a computational negative control to check the significance of the coupling patterns between MT dynamics and GEF-H1 activity. To accomplish this, for each cell used in the analysis, we generated time-lapse images of spatially shifted MT depolymerization events by randomly shifting the position of the observed events by a fixed distance of d = 1, 3, 6, 9, 12, or 15 pixels, which corresponds to shifts ranging from ∼0.32 to 4.84 µm. For each distance, we generated 50 sets of five videos of shifted MT depolymerization events corresponding to the five imaged cells and computed 50 average GEF-H1 activity profiles around MT depolymerization events as well as 50 correlation curves between MT depolymerization occurrence rate and GEF-H1 activity. As the shifting distance increased, GEF-H1 activity before and after the MT depolymerization became more uniform, particularly beyond 0.97 µm (3 pixels; Fig. S3 A). Furthermore, the cross-correlation curves between MT depolymerization occurrence rates and GEF-H1 activity showed monotonic decreases toward zero. Of note, shifts of depolymerization events by 0.32–0.97 µm exhibited weaker but significant correlations with the delayed GEF-H1 activation (Fig. S3, B and C). Overall, these computations demonstrated that the GEF-H1/MT relationship for spatially unperturbed depolymerization events could not arise from random data patterns but is rather generated by bona fide coupling of MT disassembly and GEF-H1 activation. Furthermore, the results show that this coupling occurs within an ∼1-µm radius from the disassembly event and is thus likely controlled by the diffusion of the released signaling molecules.
Online supplemental material
Fig. S1 shows the computational model of the GEF-H1 AID–DH domain interaction, the SDS-PAGE gels of recombinantly expressed and purified GEF-H1 constructs, the full sequence of the GEF-H1 FLARE212 biosensor, and the excitation/emission spectra of GEF-H1 FLARE191 biosensor together with its activity in live cells. Fig. S2 shows that biosensor expression levels do not affect edge motions and reports the relationship between GEF-H1/RhoA activity and cell edge motions in both dynamic and quiescent regions of the cell edge. Fig. S3 shows the correlation of randomized MT depolymerization events and GEF-H1 activity. Fig. S4 shows the correlation of MT dynamics with cell edge motion. Fig. S5 shows the phosphoproteomics data for GEF-H1 phosphorylation by Src and the effect of dasatinib on GEF-H1 FLARE212 activity. Video 1 shows the localization and activity of GEF-H1 FLARE212 in a COS-7 cell. Video 2 shows the effect of nocodazole treatment on GEF-H1 FLARE212 activity in a MEF. Video 3 shows the localization and activity of GEF-H1 FLARE212 in a motile MDA-MB-231 cell. Video 4 shows the localization and activity of RhoA biosensors in a motile MDA-MB-231 cell. Video 5 shows cell edge tracking in a MDA-MB-231 cell expressing GEF-H1 FLARE212. Video 6 shows the reconstruction and tracking of MT filaments in a MDA-MB-231 cell coexpressing GEF-H1 FLARE212 and EB3. Video 7 shows the effect of dasatinib treatment on GEF-H1 FLARE212 activity in motile MDA-MB-231 cells. Video 8 shows the localization and activity of GEF-H1 FLARE212 Y198F mutant in a motile MDA-MB-231 cell.
Supplementary Material
Acknowledgments
We thank Emily Wilkerson and Laura Herring from the University of North Carolina Michael Hooker Proteomic Center for the phosphoproteomic analysis. The Cherry-EMTB plasmid was a gift from Takashi Watanabe (University of North Carolina at Chapel Hill, Chapel Hill, NC). The CA-Src plasmid and the EMTB-mRuby cell line were gifts from Onur Dagliyan (University of North Carolina at Chapel Hill). Jinqi Ren (University of North Carolina at Chapel Hill) assisted with the RhoA BODIPY-GDP exchange assay. Lindsey Holloway assisted with the recombinant protein expression and purification.
This work was funded by the following grants from the National Institutes of Health: R01GM071868 (to G. Danuser), R35GM122596 (to K.M. Hahn), and R01HL133668 (to K.M. Hahn).
The authors declare no competing financial interests.
Author contributions. M.L. Azoitei, J. Noh, G. Danuser, and K.M. Hahn designed the study; M.L. Azoitei developed and characterized the biosensors, performed the GEF-H1 DH-AID modeling and the GEF assays, carried out the immunoprecipitation experiments, did all the live cell imaging experiments, and subsequently quantified biosensor activity; J. Noh designed the mathematical transformation of event time series, developed statistical methods for activity map analysis, and wrote software for image analysis; D.J. Marston contributed to the biosensor design and assisted with live cell imaging and biosensor analysis methods; J. Sondek contributed to the biosensor design; P. Roudot developed a method for the detection of MT depolymerization; T.A. Daugird and S.L. Lisanza assisted with plasmid construction and protein purification; C.B. Marshall, M.J. Sandí, R. Rottapel, and M. Ikura performed the GEF assay by NMR; M.L. Azoitei, J. Noh, G. Danuser, and K.M. Hahn wrote the manuscript; and all coauthors contributed to the final version.
References
- Aoki K., and Matsuda M.. 2009. Visualization of small GTPase activity with fluorescence resonance energy transfer-based biosensors. Nat. Protoc. 4:1623–1631. 10.1038/nprot.2009.175 [DOI] [PubMed] [Google Scholar]
- Applegate K.T., Besson S., Matov A., Bagonis M.H., Jaqaman K., and Danuser G.. 2011. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J. Struct. Biol. 176:168–184. 10.1016/j.jsb.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.C., Nalbant P., Birkenfeld J., Chang Z.F., and Bokoch G.M.. 2008. GEF-H1 couples nocodazole-induced microtubule disassembly to cell contractility via RhoA. Mol. Biol. Cell. 19:2147–2153. 10.1091/mbc.e07-12-1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S., Berrondo M., Weitzner B.D., Muthu P., Bergman H., and Gray J.J.. 2011. Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS One. 6:e22477 10.1371/journal.pone.0022477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu P.H., Tsygankov D., Berginski M.E., Dagliyan O., Gomez S.M., Elston T.C., Karginov A.V., and Hahn K.M.. 2014. Engineered kinase activation reveals unique morphodynamic phenotypes and associated trafficking for Src family isoforms. Proc. Natl. Acad. Sci. USA. 111:12420–12425. 10.1073/pnas.1404487111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagliyan O., Tarnawski M., Chu P.H., Shirvanyants D., Schlichting I., Dokholyan N.V., and Hahn K.M.. 2016. Engineering extrinsic disorder to control protein activity in living cells. Science. 354:1441–1444. 10.1126/science.aah3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R., and Baker D.. 2008. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 77:363–382. 10.1146/annurev.biochem.77.062906.171838 [DOI] [PubMed] [Google Scholar]
- Davis I.W., Arendall W.B. III, Richardson D.C., and Richardson J.S.. 2006. The backrub motion: how protein backbone shrugs when a sidechain dances. Structure. 14:265–274. 10.1016/j.str.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Dean R.T., and Dunsmuir W.T.M.. 2016. Dangers and uses of cross-correlation in analyzing time series in perception, performance, movement, and neuroscience: The importance of constructing transfer function autoregressive models. Behav. Res. Methods. 48:783–802. 10.3758/s13428-015-0611-2 [DOI] [PubMed] [Google Scholar]
- Dickey D.A., and Fuller W.A.. 1979. Distribution of the Estimators for Autoregressive Time-Series with a Unit Root. J. Am. Stat. Assoc. 74:427–431. [Google Scholar]
- Dubash A.D., Wennerberg K., García-Mata R., Menold M.M., Arthur W.T., and Burridge K.. 2007. A novel role for Lsc/p115 RhoGEF and LARG in regulating RhoA activity downstream of adhesion to fibronectin. J. Cell Sci. 120:3989–3998. 10.1242/jcs.003806 [DOI] [PubMed] [Google Scholar]
- Faire K., Waterman-Storer C.M., Gruber D., Masson D., Salmon E.D., and Bulinski J.C.. 1999. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J. Cell Sci. 112:4243–4255. [DOI] [PubMed] [Google Scholar]
- Fleishman S.J., Leaver-Fay A., Corn J.E., Strauch E.M., Khare S.D., Koga N., Ashworth J., Murphy P., Richter F., Lemmon G., et al. 2011. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One. 6:e20161 10.1371/journal.pone.0020161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishiro S.H., Tanimura S., Mure S., Kashimoto Y., Watanabe K., and Kohno M.. 2008. ERK1/2 phosphorylate GEF-H1 to enhance its guanine nucleotide exchange activity toward RhoA. Biochem. Biophys. Res. Commun. 368:162–167. 10.1016/j.bbrc.2008.01.066 [DOI] [PubMed] [Google Scholar]
- Fusco L., Lefort R., Smith K., Benmansour F., Gonzalez G., Barillari C., Rinn B., Fleuret F., Fua P., and Pertz O.. 2016. Computer vision profiling of neurite outgrowth dynamics reveals spatiotemporal modularity of Rho GTPase signaling. J. Cell Biol. 212:91–111. 10.1083/jcb.201506018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebregiworgis T., Marshall C.B., Nishikawa T., Radulovich N., Sandí M.J., Fang Z., Rottapel R., Tsao M.S., and Ikura M.. 2018. Multiplexed Real-Time NMR GTPase Assay for Simultaneous Monitoring of Multiple Guanine Nucleotide Exchange Factor Activities from Human Cancer Cells and Organoids. J. Am. Chem. Soc. 140:4473–4476. 10.1021/jacs.7b13703 [DOI] [PubMed] [Google Scholar]
- Guilluy C., Swaminathan V., Garcia-Mata R., O’Brien E.T., Superfine R., and Burridge K.. 2011. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat. Cell Biol. 13:722–727. 10.1038/ncb2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck J.N., Ponik S.M., Garcia-Mendoza M.G., Pehlke C.A., Inman D.R., Eliceiri K.W., and Keely P.J.. 2012. Microtubules regulate GEF-H1 in response to extracellular matrix stiffness. Mol. Biol. Cell. 23:2583–2592. 10.1091/mbc.e11-10-0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge R.G., and Ridley A.J.. 2016. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 17:496–510. 10.1038/nrm.2016.67 [DOI] [PubMed] [Google Scholar]
- Hodgson L., Shen F., and Hahn K.. 2010. Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr. Protoc. Cell Biol. Chapter 14:Unit 14.11.1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A., Nolan G.P., and Blau H.M.. 1996. Rapid retroviral delivery of tetracycline-inducible genes in a single autoregulatory cassette. Proc. Natl. Acad. Sci. USA. 93:5185–5190. 10.1073/pnas.93.11.5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S., and Danen E.H.. 2009. Adhesion signaling - crosstalk between integrins, Src and Rho. J. Cell Sci. 122:1059–1069. 10.1242/jcs.039446 [DOI] [PubMed] [Google Scholar]
- Jaffe A.B., and Hall A.. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269. 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Karginov A.V., Tsygankov D., Berginski M., Chu P.H., Trudeau E.D., Yi J.J., Gomez S., Elston T.C., and Hahn K.M.. 2014. Dissecting motility signaling through activation of specific Src-effector complexes. Nat. Chem. Biol. 10:286–290. 10.1038/nchembio.1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov V.S., Chamberlain C., Bokoch G.M., Schwartz M.A., Slabaugh S., and Hahn K.M.. 2000. Localized Rac activation dynamics visualized in living cells. Science. 290:333–337. 10.1126/science.290.5490.333 [DOI] [PubMed] [Google Scholar]
- Krendel M., Zenke F.T., and Bokoch G.M.. 2002. Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat. Cell Biol. 4:294–301. 10.1038/ncb773 [DOI] [PubMed] [Google Scholar]
- Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W., et al. 2011. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487:545–574. 10.1016/B978-0-12-381270-4.00019-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Elliott H.L., Oak Y., Zee C.T., Groisman A., Tytell J.D., and Danuser G.. 2015. Functional hierarchy of redundant actin assembly factors revealed by fine-grained registration of intrinsic image fluctuations. Cell Syst. 1:37–50. 10.1016/j.cels.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.C., Ruan J.W., Lua I., Li M.H., Chen W.L., Wang J.R., Kao R.H., and Chen J.H.. 2012. Overexpressed hPTTG1 promotes breast cancer cell invasion and metastasis by regulating GEF-H1/RhoA signalling. Oncogene. 31:3086–3097. 10.1038/onc.2011.476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y., Lim S.T., Tomar A., Gardel M., Bernard-Trifilo J.A., Chen X.L., Uryu S.A., Canete-Soler R., Zhai J., Lin H., et al. 2008. PyK2 and FAK connections to p190Rho guanine nucleotide exchange factor regulate RhoA activity, focal adhesion formation, and cell motility. J. Cell Biol. 180:187–203. 10.1083/jcb.200708194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung G.M., and Box G.E.P.. 1978. Measure of Lack of Fit in Time-Series Models. Biometrika. 65:297–303. 10.1093/biomet/65.2.297 [DOI] [Google Scholar]
- Ma X., Dagliyan O., Hahn K.M., and Danuser G.. 2018. Profiling cellular morphodynamics by spatiotemporal spectrum decomposition. PLOS Comput. Biol. 14:e1006321 10.1371/journal.pcbi.1006321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G.L., Hahn K.M., and Danuser G.. 2009. Coordination of Rho GTPase activities during cell protrusion. Nature. 461:99–103. 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwardt M.L., Kremers G.J., Kraft C.A., Ray K., Cranfill P.J., Wilson K.A., Day R.N., Wachter R.M., Davidson M.W., and Rizzo M.A.. 2011. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 6:e17896 10.1371/journal.pone.0017896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C.B., Meiri D., Smith M.J., Mazhab-Jafari M.T., Gasmi-Seabrook G.M., Rottapel R., Stambolic V., and Ikura M.. 2012. Probing the GTPase cycle with real-time NMR: GAP and GEF activities in cell extracts. Methods. 57:473–485. 10.1016/j.ymeth.2012.06.014 [DOI] [PubMed] [Google Scholar]
- Meiri D., Marshall C.B., Greeve M.A., Kim B., Balan M., Suarez F., Bakal C., Wu C., Larose J., Fine N., et al. 2012. Mechanistic insight into the microtubule and actin cytoskeleton coupling through dynein-dependent RhoGEF inhibition. Mol. Cell. 45:642–655. 10.1016/j.molcel.2012.01.027 [DOI] [PubMed] [Google Scholar]
- Meiri D., Marshall C.B., Mokady D., LaRose J., Mullin M., Gingras A.C., Ikura M., and Rottapel R.. 2014. Mechanistic insight into GPCR-mediated activation of the microtubule-associated RhoA exchange factor GEF-H1. Nat. Commun. 5:4857 10.1038/ncomms5857 [DOI] [PubMed] [Google Scholar]
- Mitra S.K., and Schlaepfer D.D.. 2006. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 18:516–523. 10.1016/j.ceb.2006.08.011 [DOI] [PubMed] [Google Scholar]
- Nalbant P., Hodgson L., Kraynov V., Toutchkine A., and Hahn K.M.. 2004. Activation of endogenous Cdc42 visualized in living cells. Science. 305:1615–1619. 10.1126/science.1100367 [DOI] [PubMed] [Google Scholar]
- Nalbant P., Chang Y.C., Birkenfeld J., Chang Z.F., and Bokoch G.M.. 2009. Guanine nucleotide exchange factor-H1 regulates cell migration via localized activation of RhoA at the leading edge. Mol. Biol. Cell. 20:4070–4082. 10.1091/mbc.e09-01-0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux R.K., Costanzi E., Haas M., and Verma I.M.. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A.W., and Daugherty P.S.. 2005. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 23:355–360. 10.1038/nbt1066 [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R.L., and Hahn K.M.. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 440:1069–1072. 10.1038/nature04665 [DOI] [PubMed] [Google Scholar]
- Playford M.P., and Schaller M.D.. 2004. The interplay between Src and integrins in normal and tumor biology. Oncogene. 23:7928–7946. 10.1038/sj.onc.1208080 [DOI] [PubMed] [Google Scholar]
- Potter S.C., Luciani A., Eddy S.R., Park Y., Lopez R., and Finn R.D.. 2018. HMMER web server: 2018 update. Nucleic Acids Res. 46(W1):W200–W204. 10.1093/nar/gky448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman K.L., Der C.J., and Sondek J.. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6:167–180. 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- Roudot P., Liya Ding K., Jaqaman K., Kervrann C., and Danuser G.. 2017. Piecewise-Stationary Motion Modeling and Iterative Smoothing to Track Heterogeneous Particle Motions in Dense Environments. IEEE Trans. Image Process. 26:5395–5410. 10.1109/TIP.2017.2707803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandí M.J., Marshall C.B., Balan M., Coyaud É., Zhou M., Monson D.M., Ishiyama N., Chandrakumar A.A., La Rose J., Couzens A.L., et al. 2017. MARK3-mediated phosphorylation of ARHGEF2 couples microtubules to the actin cytoskeleton to establish cell polarity. Sci. Signal. 10:eaan3286 10.1126/scisignal.aan3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery S.D., and Hahn K.M.. 2014. A High-Content Assay for Biosensor Validation and for Examining Stimuli that Affect Biosensor Activity. Curr. Protoc. Cell Biol. 65:14.15.1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit R.L. 2010. Poisson Point Processes: Imaging, Tracking and Sensing. Springer, New York; 273 pp. [Google Scholar]
- Tkachenko E., Sabouri-Ghomi M., Pertz O., Kim C., Gutierrez E., Machacek M., Groisman A., Danuser G., and Ginsberg M.H.. 2011. Protein kinase A governs a RhoA-RhoGDI protrusion-retraction pacemaker in migrating cells. Nat. Cell Biol. 13:660–667. 10.1038/ncb2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyka M.D., Keedy D.A., André I., Dimaio F., Song Y., Richardson D.C., Richardson J.S., and Baker D.. 2011. Alternate states of proteins revealed by detailed energy landscape mapping. J. Mol. Biol. 405:607–618. 10.1016/j.jmb.2010.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Martins I.R., Li P., Amarasinghe G.K., Umetani J., Fernandez-Zapico M.E., Billadeau D.D., Machius M., Tomchick D.R., and Rosen M.K.. 2010. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 140:246–256. 10.1016/j.cell.2009.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yule G.U. 1926. Why do we sometimes get nonsense-correlations between time-series? - A study in sampling and the nature of time-series. J. R. Stat. Soc. 89:1–69. 10.2307/2341482 [DOI] [Google Scholar]
- Zenke F.T., Krendel M., DerMardirossian C., King C.C., Bohl B.P., and Bokoch G.M.. 2004. p21-activated kinase 1 phosphorylates and regulates 14-3-3 binding to GEF-H1, a microtubule-localized Rho exchange factor. J. Biol. Chem. 279:18392–18400. 10.1074/jbc.M400084200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.