Branching microtubule nucleation by its molecular mediators has never been directly observed in animal cells. By imaging augmin, γ-TuRC, and microtubules with high spatiotemporal resolution, Verma and Maresca quantitatively define the sequential steps of augmin-mediated branching microtubule nucleation in dividing Drosophila cells.
Abstract
Centrosome-mediated microtubule (MT) nucleation has been well characterized; however, numerous noncentrosomal MT nucleation mechanisms exist. The branching MT nucleation pathway envisages that the γ-tubulin ring complex (γ-TuRC) is recruited to MTs by the augmin complex to initiate nucleation of new MTs. While the pathway is well conserved at a molecular and functional level, branching MT nucleation by core constituents has never been directly observed in animal cells. Here, multicolor TIRF microscopy was applied to visualize and quantitatively define the entire process of branching MT nucleation in dividing Drosophila cells during anaphase. The steps of a stereotypical branching nucleation event entailed augmin binding to a mother MT and recruitment of γ-TuRC after 15 s, followed by nucleation 16 s later of a daughter MT at a 36° branch angle. Daughters typically remained attached throughout their ∼40-s lifetime unless the mother depolymerized past the branch point. Assembly of branched MT arrays, which did not require Drosophila TPX2, enhanced localized RhoA activation during cytokinesis.
Introduction
Microtubules (MTs) play a central role in many biological processes including cell division, cell movement, and intracellular transport. The efficacy of MT-dependent processes depends on spatiotemporal control of MT nucleation as well as the organization of polymerized MTs into specific 3D arrays. The centrosome is a major MT nucleation center and MT array organizer. However, the fact that centrosomes are absent from most mammalian oocytes/eggs and many plant species (Budde and Heald, 2003; Clift and Schuh, 2013; Severson et al., 2016) was an early indication of the existence of acentrosomal nucleation pathways. One such acentrosomal pathway is MT-dependent branching MT nucleation, which was initially described in plants, where the phenomenon was clearly visualized in cortical MT regrowth assays in green algae (Wasteneys and Williamson, 1989) and then quantitatively analyzed in Arabidopsis thaliana cortical interphase MT arrays and tobacco BY-2 cell-free lysates (Murata et al., 2005; Chan et al., 2009; Nakamura et al., 2010; Liu et al., 2014; Walia et al., 2014).
Centrosome-mediated MT nucleation requires γ-tubulin and its associated proteins known as the γ-tubulin ring complex (γ-TuRC; Zheng et al., 1995; Jeng and Stearns, 1999; Moritz et al., 2000; Kollman et al., 2010). γ-TuRC is not limited to the centrosome, as the complex localizes along the length of spindle MTs (Goshima et al., 2008; Zhu et al., 2008), and visualization of plant cortical arrays revealed that daughter MT branches were nucleated by mother-associated γ-TuRC (Murata et al., 2005). The mechanism and molecules responsible for recruiting γ-TuRC to mother MTs was unknown until a genome-wide RNAi screen in Drosophila melanogaster S2 cells identified five dim γ-tubulin proteins (DGT2–6) that were required for γ-tubulin localization to spindle MTs, but not centrosomes (Goshima et al., 2007). Dgt2–6 formed a stable complex called augmin that was required for proper spindle assembly and function (Goshima et al., 2007). Subsequently, augmin was determined to be a conserved octameric protein complex containing DGT2–9 subunits in Drosophila (Meireles et al., 2009; Uehara et al., 2009), and in humans, the following eight subunits of augmin were identified: Ccdc5 (HAUS1), Cep27 (HAUS2), hDgt3 (HAUS3), C14orf94 (HAUS4), hDgt5 (HAUS5), hDgt6 (HAUS6), UCHL5IP (HAUS7), and Hice1 (HAUS8; Lawo et al., 2009; Uehara et al., 2009). Hice1/HAUS8/Dgt4 has been shown to bind directly to MTs (Wu et al., 2008; Hsia et al., 2014), while the Dgt3, Dgt5, and hDgt6/HAUS6/Dgt6 subunits of augmin have been reported to bind to γ-TuRC via its subunit, NEDD1 (Haren et al., 2006; Lüders et al., 2006; Zhu et al., 2008; Uehara et al., 2009; Chen et al., 2017). Augmin is functionally well conserved, as depletion of augmin components in various cell types leads to reduction in spindle MT density, chromosome missegregation, midzone MT assembly defects, and increased incidence of cytokinesis failure (Zhu et al., 2008; Meireles et al., 2009; Uehara et al., 2009, 2016; Uehara and Goshima, 2010; Hayward et al., 2014).
Since the identification of augmin, the interphase cortical MT array in Arabidopsis has provided the best system to visualize the central players (MTs, augmin, and γ-TuRC) during branching nucleation (Liu et al., 2014; Wang et al., 2018). The formation of augmin-dependent branched MT arrays was also visualized in Xenopus laevis egg extracts (Petry et al., 2013), although the postulated steps of branching MT nucleation, (1) augmin binding, (2) recruitment of γ-TuRC to mother MTs, and (3) nucleation of daughter MTs, were not directly observed. In the egg extract model, autocatalytic MT nucleation mechanisms, including the augmin-mediated branching pathway, have been evoked to describe how very large spindles and asters can assemble in large cells such as eggs and oocytes (Brugués et al., 2012; Ishihara et al., 2014, 2016; Decker et al., 2018). RanGTP and its downstream target targeting protein for Xklp2 (TPX2), which mediate acentrosomal spindle assembly around DNA (Groen et al., 2009; Maresca et al., 2009; Oh et al., 2016), have been implicated in promoting branching MT nucleation (Petry et al., 2013; Alfaro-Aco et al., 2017; Thawani et al., 2019). Interestingly, these factors are unlikely to contribute to branching MT nucleation in interphase cortical MT arrays in plant cells, as both RanGTP and plant TPX2 are nuclear during interphase (Vos et al., 2008). There is also a discrepancy between measured branch angles across systems, with plants exhibiting larger augmin-mediated branch angles (∼40°) than those measured in Xenopus egg extracts (Murata et al., 2005; Chan et al., 2009; Nakamura et al., 2010; Petry et al., 2013; Liu et al., 2014; Walia et al., 2014). Present understanding of the branching MT nucleation pathway in animal cells is limited due to the absence of direct high-resolution imaging of daughter MT nucleation events by its molecular mediators. In this study, multicolor, live-cell total internal reflection fluorescence (TIRF) imaging in Drosophila S2 cells, the system in which augmin was first identified (Goshima et al., 2007, 2008), was applied to visualize the entire process of branching MT nucleation by augmin and γ-TuRC.
Results and discussion
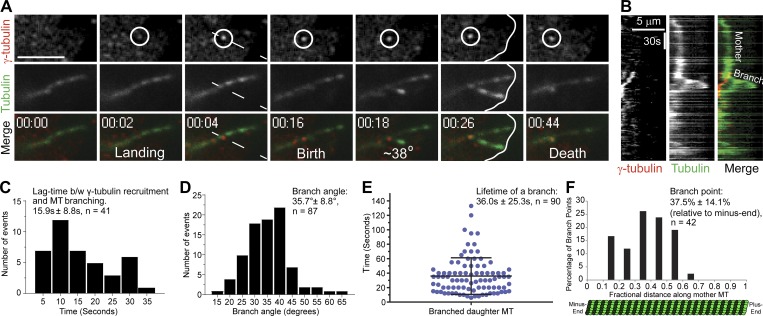
In a previous study, we observed the assembly of astral-like MT arrays that appeared to be generated by branching MT nucleation after anaphase onset in Drosophila S2 cells (Verma and Maresca, 2019). To test if these arrays were generated by bona fide branching MT nucleation events, a stable cell line coexpressing EGFP-α-tubulin and γ-tubulin-Tag-RFP-T was created and imaged with dual-color, high-resolution TIRF microscopy. Centrosomal and spindle MT populations of γ-tubulin were observed throughout mitosis, and the localization of γ-tubulin puncta to spindle MTs, while abundant, was transient and dynamic. Prior to anaphase onset, individual MTs could not be readily visualized by TIRF microscopy; however, after anaphase onset, more stable astral MTs entered the TIRF field, where they could be imaged for extended durations. Indeed, γ-tubulin puncta were observed to dynamically localize to astral MTs and to reside at the sites of daughter MT nucleation (Fig. 1, A and B; and Video 1). Quantification of bona fide branching nucleation events revealed that the lag time between the localization of a γ-tubulin puncta to a mother MT and nucleation of a daughter MT was 15.9 ± 8.8 s (mean ± SD, n = 41 events; Fig. 1 C). In contrast to branching MT nucleation events in Xenopus egg extracts, where reported branch angles were shallow (<10° for 52% of branching events; Petry et al., 2013), the branching nucleation events observed in anaphase Drosophila S2 cells exhibited a mean branching angle of 35.7° ± 8.8° (n = 87) and in the same orientation as the mother MT (Fig. 1 D).
Figure 1.
Direct visualization of branching MT nucleation in Drosophila S2 cells. (A) Two-color TIRF imaging of an astral MT in a post-anaphase cell coexpressing EGFP-α-tubulin (green) and γ-tubulin-Tag-RFP-T (red). This representative branching event encompasses γ-tubulin landing on the mother MT through disassembly of the daughter MT, both of which contact the cortex (white line at 00:26). (B) Kymograph of the branching event from region (dashed line at 00:04) in A. (C) Histogram of the lag time between (b/w) γ-tubulin recruitment to the mother MT and birth of a daughter MT; n = 41. (D) Distribution of branch angles between mother and daughter MTs. The branch angles were measured 5–10 s after the appearance of daughter MTs; n = 87. (E) Dot plot shows distribution of lifetimes of daughter MTs; n = 90; lifetime is time from birth to death of the daughter MT. (F) Histogram shows distribution of branch origins along the length of mother MTs; n = 42. Time, min:s. Scale bars, 5 µm (A and B). Mean ± SD values are reported in all the histograms. Error bar on the dot plot indicates ± SD.
In most cases, if the mother MT did not depolymerize past the branch point, the daughter remained attached to the mother MT and exhibited dynamic instability. While variable, the mean lifetime of a daughter MT was 36.0 ± 25.3 s (n = 90; Fig. 1 E) and, over a typical lifetime, a dynamic daughter MT polymerized micrometers from the branch site (Fig. 1 B). The branch point distribution (n = 42) along the length of mother MTs spanned between the 11–61% fractional position (relative to the minus end), with a bias toward the minus end, as the median fractional position was 37.5 ± 14.1% (Fig. 1 F). Interestingly, the frequency of branching in the first third of the mother MT was reduced relative to the peak observed around the 37% fractional length. A caveat to this distribution is that it could be measured only along the length of the mother that was present in the TIRF field, which did not always extend to the minus end located near the spindle pole or centrosome. In some cases, branching MT nucleation of “granddaughter” MTs from daughters was observed; however, split branching of growing MT plus ends (Basnet et al., 2018) was not evident. We also occasionally observed de novo MT nucleation events from cytosolic γ-tubulin puncta, albeit at a lower frequency than branching MT nucleation, suggesting that the nucleating activity of γ-TuRC may be enhanced by binding to preexisting MTs (Kaye et al., 2018).
The 36° branch angle measured in S2 cells is consistent with branching MT nucleation in cortical interphase MT arrays in plants (Murata et al., 2005; Chan et al., 2009; Liu et al., 2014; Walia et al., 2014), although we did not observe obvious branching MT nucleation events from interphase MTs, which were stable and abundantly visible in the TIRF field. Thus, Drosophila augmin appears to be regulated in a cell cycle–dependent manner, most likely via phosphoregulation by mitotic kinases (Zhang et al., 2009; Johmura et al., 2011; Tsai et al., 2011). Interestingly, cell cycle regulation of augmin in plants is achieved through assembly of different complexes that incorporate distinct MT-binding Hice1/HAUS8-like subunits between interphase and mitosis (Lee et al., 2017). Multiple studies in plants have reported that during interphase, ∼80% of branches occur at a 40° angle, while ∼20% of daughters nucleate parallel to the mother MT. We do not rule out that parallel nucleation events also occur in anaphase S2 cells, but we could not confidently identify bona fide parallel branch events as defined by daughters originating from a mother-associated γ-TuRC. It is noteworthy that augmin depletion in Arabidopsis resulted in most nucleation events (63%) becoming parallel (Liu et al., 2014), indicative of the 40° branch angle being mediated by the interphase augmin complex. From an evolutionary perspective, it will be interesting to determine if the mitotic augmin complex in plants (Lee et al., 2017) shifts to shallow-angle nucleation events like those observed in metaphase Xenopus egg extracts (Petry et al., 2013; Thawani et al., 2019) or if the complex retains the ability to nucleate daughters at a 40° branch angle, comparable to the anaphase measurements reported here in Drosophila cells.
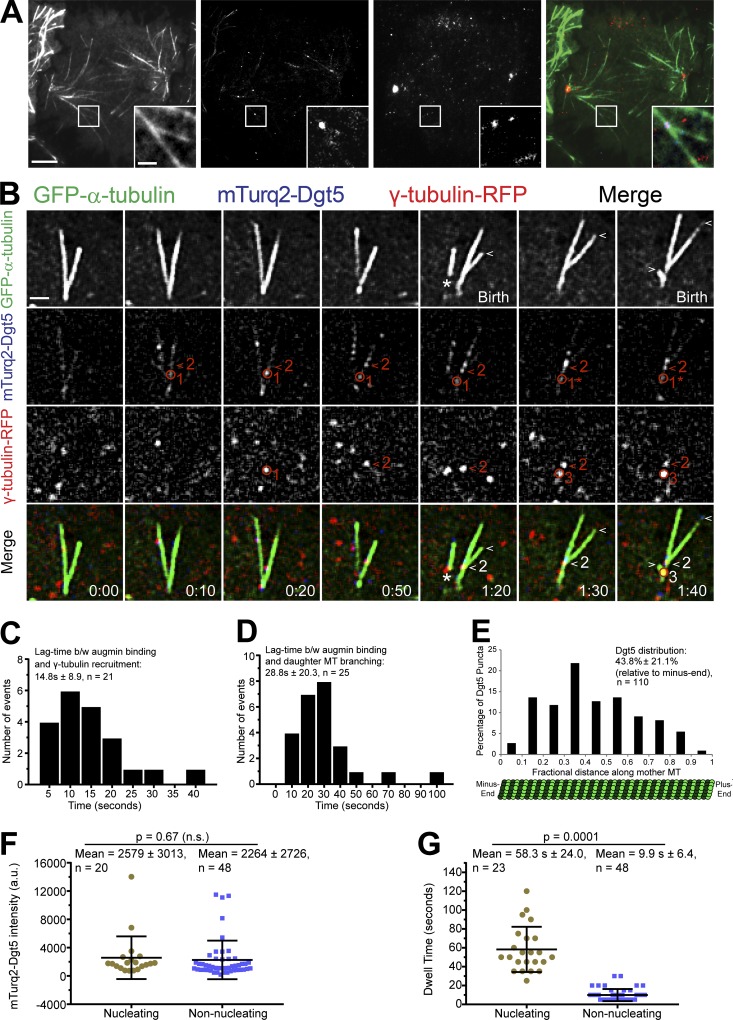
It has been proposed that the augmin complex targets γ-TuRC to preexisting MTs, which in turn initiates branching MT nucleation; however, this theoretical series of events has never been directly observed in animal cells. To visualize the properties of the augmin complex and γ-TuRC throughout the branching MT nucleation process in Drosophila S2 cells, we made a stable cell line coexpressing EGFP-α-tubulin, γ-tubulin-Tag-RFP-T, and mTurquoise2-Dgt5 (an augmin subunit) and performed three-color, live-cell TIRF microscopy. Dgt5 localized as discreet puncta to spindle MTs (Fig. 2 A) throughout mitosis. Dgt5 puncta were dynamic but typically remained localized to MTs slightly longer than γ-tubulin puncta and, while bound, Dgt5 often displayed minus end–directed movements (albeit many of these events were due to poleward movement of the MTs). Importantly, we were also able to observe branching nucleation events originating from colocalized puncta of Dgt5 and γ-tubulin (Figs. 2 B and S1; and Videos 2 and 3). Extensive imaging of the core components revealed a stepwise process of augmin-mediated branching MT nucleation whereby the augmin complex first associated with the mother MT and recruited γ-TuRC from the cytosol to nucleate daughter MTs (Fig. 2 B). The mean lag time between augmin complex binding and γ-TuRC recruitment was 14.8 ± 8.9 s (n = 21; Fig. 2 C), while the time between Dgt5 binding to the mother MT and daughter nucleation was 28.8 ± 20.3 s (n = 25; Fig. 2 D). Dgt5 and γ-tubulin typically remained at the branch point throughout the lifetime of the daughter unless the mother MT depolymerized past the branch point. Upon complete depolymerization of the daughter MT, the γ-tubulin puncta at the branch point dissociated from the mother MT (Fig. 1 B), while the augmin complex sometimes remained and was even capable of supporting another round of branching MT nucleation (Fig. 2 B and Video 3).
Figure 2.
Direct visualization of the key mediators of branching MT nucleation. (A) Representative TIRF micrographs showing expression and localization of γ-tubulin-Tag-RFP-T (red) and mTurquoise2-Dgt5 (blue) in a mid-anaphase Drosophila S2 cell. Inset shows a representative MT branching event where mTurquoise2-Dgt5 and γ-tubulin-Tag-RFP-T colocalized. (B) Still frames from a TIRF time-lapse of MT branching events in a cell coexpressing GFP-α-tubulin (green), mTurquoise2-Dgt5 (blue), and γ-tubulin-TagRFP (red). The events include the following: (a) Localization of two Dgt5 puncta at 10 s on mother MT (one indicated by the red circle and marked 1, and another indicated by the red arrowhead and marked 2 in the mTurquoise2 channel). (b) Dgt5 puncta (1) recruits γ-tubulin (1) (RFP channel, 0:20). The γ-tubulin puncta (1) dissociates from Dgt5 (1) within 30 s without a branch event; although the Dgt5 puncta (1) remains associated with the mother. (c) Dgt5 (2) recruits γ-tubulin (2) (RFP channel, 0:50), and this complex nucleates a daughter within 30 s (1:20). (d) Dgt5 puncta (now denoted 1*) recruits a second γ-tubulin (3) (RFP channel, 1:30), which nucleates a branch within 20 s (1:40). The first daughter MT grows for 20 s (plus end indicated by white arrowheads in GFP channel) while the second daughter MT was born shortly before the mother depolymerizes. The asterisk (GFP channel, 1:20) indicates a rare event in which a daughter with minus end–associated γ-tubulin and Dgt5 dissociates from an intact mother. The unattached daughter depolymerized completely within 20 s. (C) Average lag time between (b/w) mTurquoise2-Dgt5 (augmin complex subunit) binding to the mother MT and recruitment of γ-tubulin-TagRFP-T; n = 21. (D) Histogram of lag time between Dgt5 recruitment to the mother MT and birth of a daughter MT; n = 25. (E) Histogram shows distribution of Dgt5 puncta along the length of mother MTs; n = 110. (F) Dot plot of fluorescence intensities of Dgt5 puncta that supported nucleation of branched MTs (n = 20) versus those that do not support nucleation of branched MTs (n = 48). (G) Dot plot of dwell times of Dgt5 puncta that supported nucleation of branched MTs (n = 23) versus those that do not support nucleation of branched MTs (n = 48). Time: min:s. Scale bars, 5 µm (A), 1 µm (A inset and B). Mean ± SD values are reported. Two-tailed P values of Student’s t tests are reported; n.s., not significant or P > 0.05.
Dgt5 puncta (n = 110) were distributed along a broader length of the mother MT (5–93% fractional distance) than the branch point distribution (Fig. 1 F), but the median fractional distance of 43% for Dgt5 was comparable to the branch point median (37%), and the first third of the mother MT also exhibited reduced Dgt5 compared with the peak at ∼40% (Fig. 2 E). The branch point and augmin distributions suggest that there may be a spatial regulatory mechanism that reduces augmin binding, and hence branching, in the first third of mother MTs that emanate from centrosomes and/or spindle poles. Interestingly, aurora A kinase, which can generate a polar activity gradient that has been shown to spatially regulate other important mitotic phenomena (Chmátal et al., 2015; Ye et al., 2015; Mangal et al., 2018), reduces the binding affinity of augmin for MTs by phosphorylating Hice1 (Tsai et al., 2011). There was not a significant difference between the intensity of Dgt5 puncta that supported branching versus those that did not (Fig. 2 F). These data do not preclude the possibility that augmin oligomers recruit γ-TuRC. Further investigation of the number of Dgt5 molecules in the puncta will be informative, especially since augmin binds to NEDD1 (Zhu et al., 2008; Uehara et al., 2009; Chen et al., 2017), of which there is thought to be multiple copies in a single γ-TuRC (Tovey and Conduit, 2018). The mean dwell time of Dgt5 puncta at nucleation sites was 58.3 ± 24.0 s (n = 23) versus 9.9 ± 6.4 s (n = 48) for Dgt5 puncta that did not support daughter nucleation (Fig. 2 G). These dwell times are comparable to measurements made for nucleating versus nonnucleating augmin and γ-TuRC components in plant cells (Nakamura et al., 2010; Liu et al., 2014; Walia et al., 2014), as well as to a subset of purified augmin complexes on MTs in vitro (Hsia et al., 2014). The observations that nucleation-supporting Dgt5 puncta are not significantly brighter than nonnucleating puncta yet reside on the MT for 5.9 times longer (which is in accordance with the fact that augmin and γ-TuRC typically remain at the branch point for the daughter’s lifetime) suggest that the affinity of augmin for MTs may be increased by γ-TuRC binding, either directly or through the introduction of another regulatory factor such as an enzyme or MT-binding factors.
TPX2 is a highly conserved protein, and its activity has been associated with multiple roles in mitosis, such as spindle assembly (Wittmann et al., 2000; Gruss et al., 2001, 2002; Ma et al., 2010; Wadsworth, 2015), MT nucleation, and MT stabilization (Groen et al., 2009; Petry et al., 2013; Reid et al., 2016; Alfaro-Aco et al., 2017; Roostalu and Surrey, 2017). In Xenopus egg extracts, branching MT nucleation events were observed following the addition of 5–10-fold molar excess TPX2 and RanGTP, and branching MT nucleation was compromised in extracts depleted of endogenous TPX2 (Petry et al., 2013; Thawani et al., 2019). One model of augmin-mediated branching MT nucleation proposes that γ-TuRC is recruited to augmin complex via TPX2, because the three components coimmunoprecipitated from egg extracts (Petry et al., 2013). TPX2 contains a composite binding sequence that shows resemblance to the yeast Spc110/Pcp1 motif and centrosomin motif (CM1)/γ-TuRC nucleation activator motif (γ-TuNA), deletions of which resulted in reduced or no branching MT nucleation in the Xenopus egg extract branching assay (Alfaro-Aco et al., 2017).
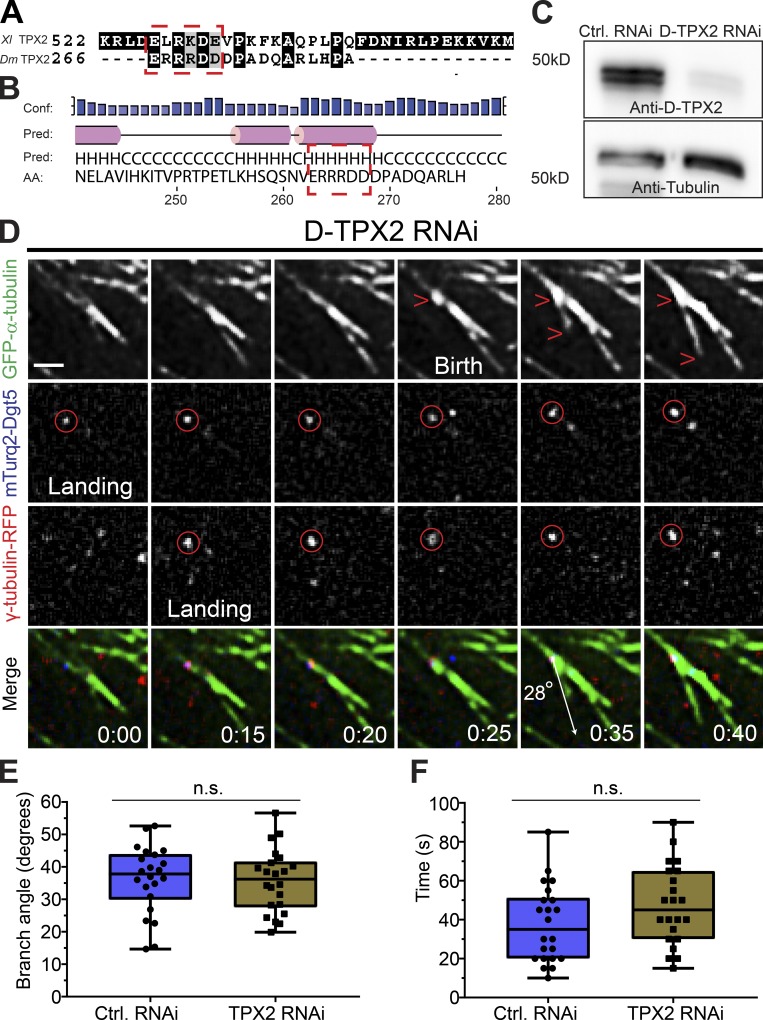
Sequence alignment between Xenopus TPX2 and Drosophila D-TPX2 indicated that D-TPX possesses a partially conserved CM1/γ-TuNA motif “ERRRDD,’’ while the other conserved motif, Spc110/Pcp1 motif, is absent (Fig. 3 A). Further investigation into the predicted secondary structure of D-TPX2 revealed that its C-terminus (190–325 amino acids) contains five short stretches of α-helices, and the partially conserved γ-TuNA motif lies within the predicted α-helices (Fig. 3 B), as is the case for Xenopus TPX2 (Alfaro-Aco et al., 2017). To further investigate the role of D-TPX2 in branching MT nucleation events in Drosophila S2 cells, TIRF microscopy was performed in stable cell lines expressing mTurquoise2-Dgt5, γ-tubulin-Tag-RFP-T, and EGFP-α-tubulin depleted of D-TPX2 by RNAi. Consistent with previous reports from live S2 cells, spindle assembly was largely unperturbed following D-TPX2 depletion (>95%; Fig. 3 C; Goshima, 2011). Therefore, we were able to readily analyze the effects of D-TPX2 depletion on branching MT nucleation events after anaphase onset. D-TPX2 depletion did not alter key parameters of branching MT nucleation (Fig. 3, D–F). The same branch angle (Fig. 3 E, control RNAi, 36.2° ± 10.4°, n = 22; TPX2 RNAi, 35.7° ± 9.7°, n = 22) and lifetime of daughter MTs (Fig. 3 F, control RNAi, 37.7 ± 19.8 s, n = 22; TPX2 RNAi, 47.2 ± 20.5 s, n = 22) were measured in control and D-TPX2–depleted cells. The branch frequency, as defined by the number of branching events observed in the TIRF field over a 3-min imaging window after anaphase onset, was also indistinguishable between control and TPX2-depleted cells (control RNAi, branch/1.3 min; TPX2 RNAi, branch/1.5 min, n = 44 branching events in control and TPX2 RNAi cells). Taken together, these results indicate that D-TPX2 is dispensable for branching MT nucleation in Drosophila despite D-TPX2 possessing a putative CM1/γ-TuNA–like motif.
Figure 3.
Branching MT nucleation is unaffected by D-TPX2 depletion. (A) Protein sequences of Xenopus (Xl) TPX2 and Drosophila (Dm) TPX2 (D-TPX2) were aligned using T-Coffee multiple alignment software (Notredame et al., 2000). Red dashed rectangle indicates a partially conserved CM1/γ-TuNA motif in D-TPX2. (B) Secondary structure prediction of D-TPX2 was generated using PSIPRED bioinformatics software (Buchan and Jones, 2019). Partially conserved CM1/γ-TuNA motif is expected to lie within α-helices (indicated with red dashed square). Conf, confidence; pred, predicted. (C) Western blot showing depletion of endogenous TPX2 with tubulin as a loading control (Ctrl). (D) A representative MT branching event in a D-TPX2–depleted cell coexpressing GFP-α-tubulin (green), mTurquoise2-Dgt5 (blue), and γ-tubulin-TagRFP (red). Time point 00:00 indicates landing of a Dgt5 molecule, which recruits γ-tubulin at 0:15 s. The Dgt5-γ-tubulin complex nucleates a MT branch at 0:35 s. Scale bar, 1 µm. (E) Distribution of branch angles in control and D-TPX2–depleted cells; n = 44. (F) Lifetime of branched daughter MTs in control and D-TPX2–depleted cells; n = 44. Time: min:s. Box plots indicate full range of variation (from minimum to maximum), the interquartile range, and the median. Two-tailed P values of Student’s t tests are reported; n.s., not significant or P > 0.05.
While TPX2 is required for branching in Xenopus egg extracts, the fact that it is not required in Drosophila S2 cells is not surprising when one considers that (1) there is a strong effect of augmin depletion on spindle MT density and morphology in S2 cells (Goshima et al., 2007, 2008), but (2) spindle assembly is largely unperturbed in D-TPX2–depleted cells (Goshima, 2011). While Drosophila is distinct from Xenopus in this regard, our direct visualization shows that branching nucleation occurs via a multistep sequential process, as was recently proposed in a sequential model of branching nucleation in Xenopus egg extracts (Thawani et al., 2019). However, unlike the Xenopus system in which the first step was proposed to be TPX2 binding, D-TPX2 appears to be dispensable for the sequential branching process in Drosophila cells, although we cannot exclude the possibility that other factors fulfill the role of TPX2. The RanGTP gradient around mitotic chromatin (Kalab and Heald, 2008) has also been implicated in promoting branching MT nucleation (Petry et al., 2013). We propose that RanGTP is not required for branching MT nucleation in Drosophila S2 cells because (1) branching nucleation occurs at a significant distance (>5 µm) from segregating chromosomes in anaphase and (2) robust branching nucleation continues throughout telophase after the nuclear envelope has reformed.
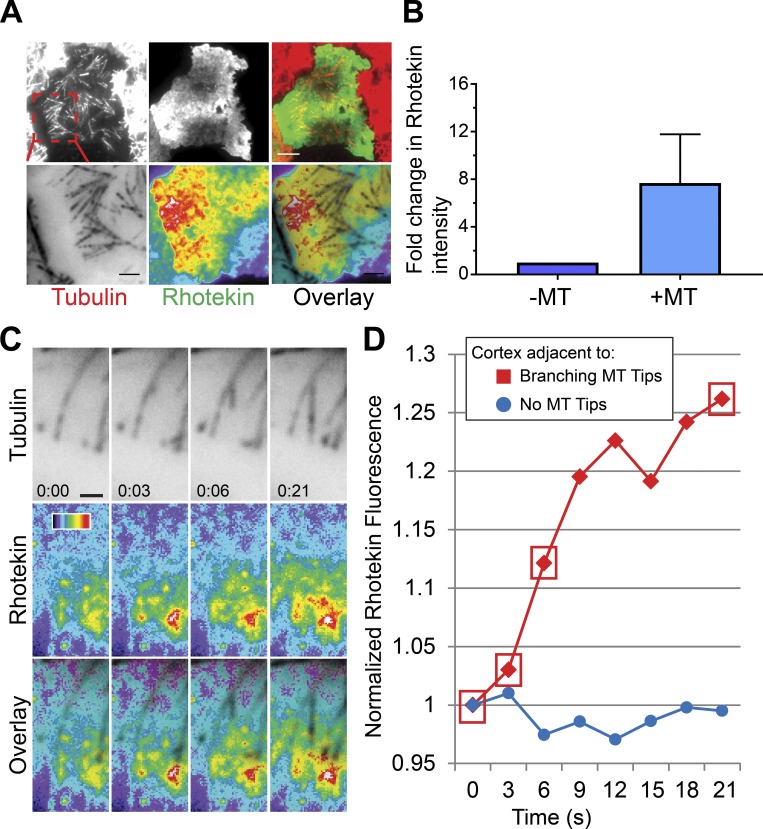
Augmin-dependent branching MT nucleation contributes to furrow ingression and abscission during cytokinesis (Uehara et al., 2016). To better understand how branching may contribute to cytokinesis, we performed live-cell microscopy on dividing cells coexpressing Tag-RFP-T-α-tubulin and EGFP-Rhotekin, a reporter for RhoA-GTP (Bement et al., 2005; Benink and Bement, 2005). During anaphase, we often observed amplification of Rhotekin in the vicinity of large branched MT arrays (Fig. 4, A and B; and Video 4). Quantification of Rhotekin fluorescence intensity revealed that localized RhoA-GTP increased by ∼30% proximal to the mother and daughter MTs during the branching MT event, while a nearby cortical area where astral MTs were absent exhibited no change in Rhotekin fluorescence during the same time period (Fig. 4, C and D). We recently showed that the plus ends of astral MTs recruit the RhoGEF ECT2 and activate RhoA upon physically contacting the plasma membrane (Verma and Maresca, 2019). Thus, we propose that branching MT nucleation acts as a mechanism to amplify cortical RhoA activation by generating new MT plus ends oriented toward the plasma membrane. We propose that reduced RhoA activation by branching astral MTs could contribute to cytokinesis defects observed in augmin-depleted cells, which heretofore has been attributed to compromised midzone assembly (Uehara et al., 2009, 2016).
Figure 4.
Branching MT nucleation amplifies RhoA activation during cytokinesis. (A) The active RhoA reporter Rhotekin is enriched near the MT plus ends of branched MT arrays (top); enlarged view of the branched MT arrays (red dashed box) that activate cortical RhoA (bottom). (B) Fold change in Rhotekin fluorescence intensity near the MT plus ends with respect to a nearby cortical region devoid of astral MTs. Data were pooled from three independent experiments (n = 3). (C) Still frames from a multicolor TIRF time-lapse showing a branching event that locally increases cortical RhoA activation, visualized with Rhotekin. (D) Rhotekin fluorescence increases ∼30% (red) proximal to the mother and daughter MTs during the branching nucleation event shown in C. A nearby cortical region devoid of astral MTs (blue) exhibited no change in Rhotekin fluorescence during the same time period. Red boxes correspond to the time points in C. Error bars indicate SD. Time: min:s. Color wedge, pixel values 150–1,200. Scale bars, 5 µm (A), 1 µm (C).
The lag time from γ-tubulin binding to nucleation (15.9 ± 8.8 s) and the branch angle (35.7° ± 8.8°) in anaphase Drosophila S2 cells are nearly identical to branching parameters quantified in plants (Murata et al., 2005; Chan et al., 2009; Nakamura et al., 2010; Liu et al., 2014; Walia et al., 2014). Interestingly, smaller (<30°) branch angles have been measured in Xenopus egg extracts (Petry et al., 2013), observed by electron tomography of metaphase spindles in human U2OS cells and inferred from tracking EB3 comets in human HeLa and RPE1 cells (David et al., 2019). This discrepancy may stem from fundamental mechanistic and/or structural differences between the fly/plant and vertebrate branching pathways. However, we propose an alternative explanation for the observed differences in branch angles stemming from two considerations: (1) the structural organization and physical properties of the augmin complex, and (2) the cellular environment in which a daughter is born.
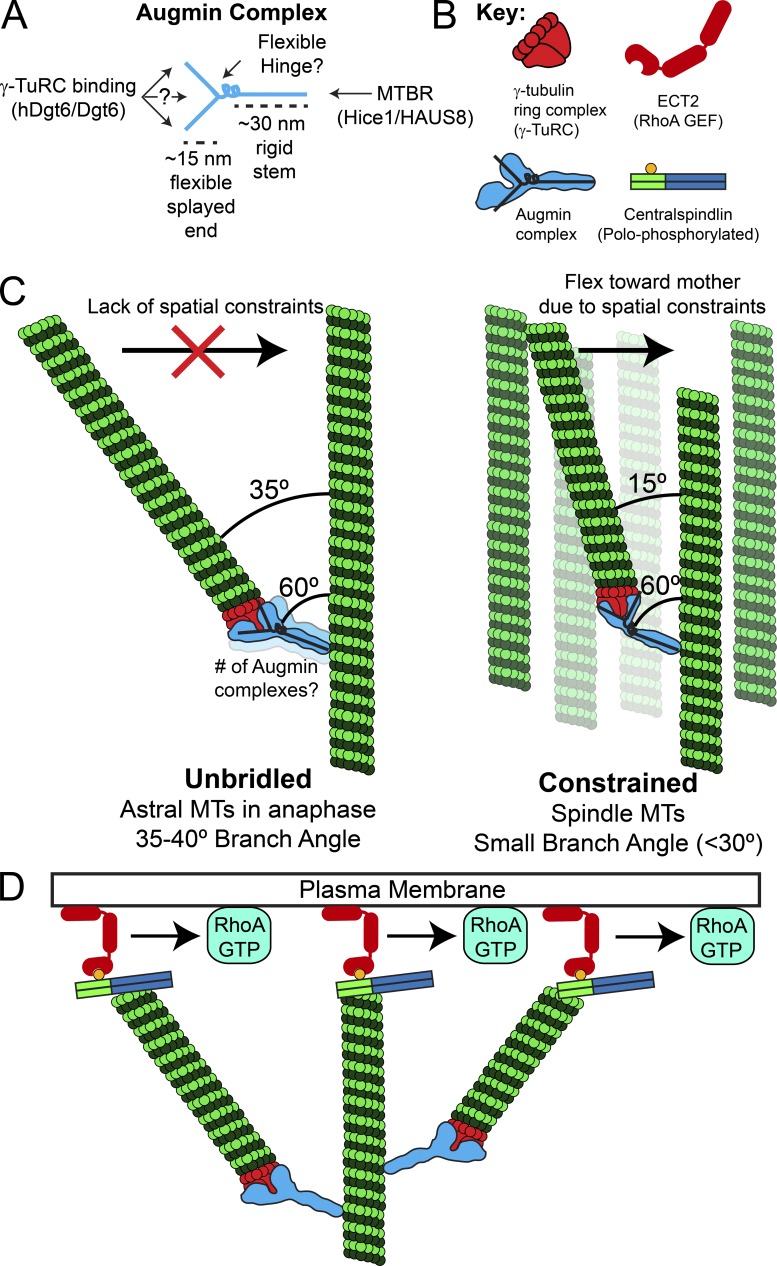
Negative stain EM of the reconstituted octameric complex revealed that human augmin is an ∼40–45-nm-long, Y-shaped structure with an ∼30-nm-long stem and ∼15-nm flexible splayed end that can adopt multiple conformations (Hsia et al., 2014; Fig. 5 A). Structural analyses of a human augmin subcomplex indicated that one end of a stem-like structure contains the MT binding region of Hice1/HAUS8 (Hsia et al., 2014). It is compelling to postulate that the splayed Y-shaped end of the complex contains γ-TuRC binding activities. In sum, the augmin complex may contain a rigid 30-nm-long stem that binds MTs at one end and a flexible splayed end that recruits γ-TuRC. Further angular flexibility could be achieved via a flexible hinge, analogous to a wrist, between the MT-binding “arm” and γ-TuRC-binding “hand” (Fig. 5, A and B). Interestingly, negative stain EM of the Xenopus augmin complex revealed a stem-like structure bound at a nearly perpendicular angle to the MT, with a fair number of MT-bound particles exhibiting hinge-like bends (Song et al., 2018).
Figure 5.
Model of augmin-mediated branching MT nucleation and its role in cytokinesis. (A) Based on negative stain EM of the augmin complex, we propose that the augmin complex contains a rigid ∼30-nm-long stem that binds MTs at one end and an ∼15-nm flexible splayed end that recruits γ-TuRC through Dgt6 and possibly other interfaces. MTBR, MT binding region in Hice 1/HAUS8. (B) Key to molecular schematics in the figure. Note the overlaid “Y” on the augmin complex schematic, which is drawn based on negative stain EM of the complex. (C) A flexible hinge region in the augmin complex and/or flexibility in the splayed Y-end of augmin may allow a daughter MT to sample a broad range of possible branch angles relative to its mother. The branch angle is impacted by the local cellular environment such that “unbridled” daughter MTs with fewer spatial constraints (e.g., astral MTs in anaphase) would have larger branch angles than daughters nucleated in the spatially constrained environment of the spindle and midzone MT array. (D) Branched MT arrays may amplify RhoA activation by generating more MT plus ends that are capable of recruiting cortical ECT2 via direct interaction with plus end–bound (polo-phosphorylated) centralspindlin.
Augmin complex flexibility would allow a daughter MT to sample a broad range of possible branch angles relative to its mother. The branch angle will almost certainly be impacted by MT-based motors and cross-linkers, but the cellular environment in which a daughter is nucleated is also likely to have an impact on the branch angle. Specifically, we propose that daughters born into “open” cytosol with fewer spatial constraints, as is the case for the equatorial astral MTs in early anaphase observed here, will be more likely to exhibit larger branch angles than daughters nucleated in the spatially constrained environments of axonal bundles (Sánchez-Huertas et al., 2016), the spindle, or midzone MT arrays (Fig. 5 C). If this is true, then the branch angle of “unbridled” daughters would be closer to the angle at which an augmin complex binds to the mother MT. On the other hand, a daughter that encounters spatial constraints, for example, in a spindle or axon with many MTs oriented nearly parallel to the mother, would flex inward due to flexible or hinge-like parts of the augmin complex. Interestingly, mother and daughter MTs of small-angle branches visualized by electron tomography in human metaphase spindles were linked by 29-nm “rods” that bound the mother at a 60° angle (Kamasaki et al., 2013). While the molecular identity of these rods was not determined, it was reasonably speculated that the rods may be augmin (Kamasaki et al., 2013). In fact, the rods could be the 30-nm stem of the augmin complex.
We propose that branch angle flexibility may allow the branching MT nucleation pathway to make diverse functional contributions throughout mitosis. During spindle assembly and metaphase, shallow branch angles would contribute to the establishment and maintenance of kinetochore–MT attachments (David et al., 2019). However, the functional contribution of branching is likely different in the context of pioneering astral MTs during anaphase/telophase. We recently characterized an MT-based RhoA activation pathway that functions during anaphase, telophase, and cytokinesis via recruitment of the RhoGEF ECT2 to cortical sites contacted by MT plus ends enriched with the centralspindlin complex (Verma and Maresca, 2019; Fig. 5, B and D). Interestingly, MT arrays with larger branch angles (>35°) produced more plus ends that contacted the cortex, thereby amplifying localized RhoA activation (Figs. 4 A and 5 D). We posit that larger branch angle nucleation from equatorial astral MTs would allow for the activation of cortical RhoA over micrometers, the size/scale in which a cleavage furrow is initiated and an area significantly larger than a submicrometer-sized kinetochore. Furthermore, autocatalytic amplification of astral MT arrays (Ishihara et al., 2016; Decker et al., 2018) via larger branch angle nucleation would more efficiently (relative to shallow branching) fill the MT array expansion volume observed in large cells (100–1,000 µm) during early embryonic cell divisions (Wühr et al., 2008; Mitchison et al., 2012; Ishihara et al., 2014). Unlike actin network organization that uses distinct nucleation mechanisms and molecules for branched versus parallel arrays (Svitkina, 2013), angular flexibility of branching MT nucleation may allow a single complex to effectively assemble both branched and near-parallel MT arrays.
More than 30 yr ago, Salmon and colleagues demonstrated the power of directly observing a cellular phenomenon and quantifying its parameters by visualizing MT dynamic instability in vitro and in cells (Cassimeris et al., 1988; Walker et al., 1988). The work transformed thinking around MT dynamics and laid the foundation for decades of research that continues today (Heald and Khodjakov, 2015). To date, the branching MT nucleation pathway has been visualized and quantified in plants but not in animals, a significant knowledge gap given the central importance of this pathway to animal cell division (Lawo et al., 2009; Uehara et al., 2009, 2016). Much like the quantification of MT dynamic instability parameters accomplished many years ago, we hope that the direct observation of branching MT nucleation and quantification of its parameters in living animal cells will inform future mechanistic models of cellular processes that depend on dynamic MTs.
Materials and methods
DNA constructs
The γ-tubulin gene (CG3157) with its endogenous promoter was amplified from the genomic DNA of Drosophila. The resulting PCR product was cloned between 5′ KpnI and 3′ EcoRI sites of the pMT/V5-His vector (Invitrogen). In-frame Tag-RFP-T gene was then introduced at the 3′ end of γ-tubulin gene between 5′ EcoRI and 3′ NotI sites. The Drosophila Dgt5gene (augmin complex subunit, CG 8828) was PCR amplified from cDNA clone LD 47477 with a 5′ SpeI site and a 3′ EcoRI site. The resulting PCR product was then inserted into the 5′ SpeI and 3′ EcoRI sites of the pMT/V5 His-B vector (Invitrogen) containing in-frame mTurquoise2 gene at the 5′ end, cloned between 5′ NcoI and 3′ SpeI sites, and the Mis12 promoter at the 5′ end, cloned between a single Kpn1 site.
Cell culture and generation of stable cell line
Drosophila S2 cells were grown in Schneider’s medium (Life Technologies) supplemented with 10% heat-inactivated FBS and 0.5× antibiotic/antimycotic cocktail (Sigma-Aldrich) and maintained at 25°C. Cell lines were generated by transfecting the DNA constructs containing the gene of interest with Effectene transfection reagent (Qiagen), following the manufacturer’s protocol. 4 d after transfection, expression of EGFP/Tag-RFP-T/mTurquoise2-tagged proteins was checked by fluorescence microscopy. To make a stable cell line, cells were selected in the presence of Blasticidin S HCl (Thermo Fisher Scientific) and/or Hygromycin B (Sigma-Aldrich) until there was no observable cell death. Thereafter, cell lines were either frozen down or maintained in the S2 medium at 25°C without Blasticidin or Hygromycin B.
RNAi experiments
Approximately 500 base pairs of the D-Tpx2 gene (also known as Ssp1/mei-38 in Drosophila, CG14781) was amplified from cDNA clone RE11134 by PCR to contain flanking T7 promoter sequences. Double-stranded RNA was synthesized from the D-TPX2 PCR product at 37°C using the T7 RiboMax Express Large-Scale RNA Production System (Promega), following the manufacturer’s protocol. For RNAi experiments, cells at ∼25% confluence were incubated in a 35 × 10-mm tissue culture dish for 1 h. Thereafter, medium was carefully aspirated off the dish, and 1 ml of serum-free Schneider’s medium containing 20 µg of double-stranded RNA was added to the dish. After 1 h, 1 ml of fresh medium containing FBS was added to the dish and incubated for 4 d at 25°C. Depletion of endogenous D-TPX2 was confirmed by Western blot.
Western blotting
Equal amounts of proteins were loaded into an 8% SDS-PAGE gel. After the gel was run, proteins were transferred to a nitrocellulose membrane using the Trans-Blot Turbo transfer system (Bio-Rad) for 10 min. Subsequently, the membrane was incubated in 5% milk (wt/vol, made in Tris-buffered saline with 0.1% Tween [TBST]) for 1 h. Following block, the membrane was incubated with either α-D-TPX2 antibody at 1:2,000 dilution or α-tubulin antibody (DM1-A; Sigma-Aldrich) at 1:10,000 dilution for 1 h, followed by three 10-min washes in TBST and secondary antibody incubation at 1:5,000 dilution for 1 h. Following secondary antibody incubation, membrane was washed three times for 10 min in TBST and developed with ECL reagent (Millipore). The blot was imaged with a G:BOX system controlled by GeneSnap software (Syngene). Images were further quantified to estimate the knockdown efficiency using Fiji/ImageJ software (Schindelin et al., 2012). To obtain the intensity values, identical regions were drawn over each band, and their integrated intensity was recorded. Intensity values were normalized to their respective loading controls (tubulin) to estimate the knockdown efficiency.
Live-cell TIRF microscopy
Cells at ∼50% confluence, expressing the gene of interest, were seeded on a 35-mm glass-bottom dish (Cellvis) coated with concanavalin A for 30–60 min. Before imaging, the total volume was brought up to 2 ml with fresh Schneider’s medium containing FBS. Multicolor, live-cell TIRF videos of all the constructs (EGFP-α-tubulin and γ-tubulin-Tag-RFP-T; EGFP-α-tubulin, γ-tubulin-Tag-RFP-T, and mTurquoise2-Dgt5; Tag-RFP-T-α-tubulin and Rhotekin-EGFP) were acquired on a Nikon Ti-E microscope equipped with a 100× 1.49-NA differential interference contrast Apochromat oil-immersion objective, a Hamamatsu ORCA-Flash 4.0 LT digital complementary metal-oxide semiconductor camera (C11440), four laser lines (447, 488, 561, and 641 nm), and MetaMorph software (Molecular Devices). Metamorph was used to control the imaging systems.
Measurement of augmin intensity, distribution along the length of MTs, and dwell time
Identical regions were drawn over the mTurquoise2-Dgt5 puncta to measure the fluorescence intensity of Dgt5 that nucleated a branched MT and those that did not. The local background was estimated by placing the same region to a nearby location on the mother MT lacking Dgt5. Background intensity values were subtracted from mTurquoise2-Dgt5 intensity values to estimate the actual mTurquoise2-Dgt5 fluorescence intensity. The distribution of mTurquoise2-Dgt5 puncta along the length of the mother MT was measured by drawing a line from the minus end of the mother MT to the Dgt5 puncta (note that this measurement was done for the MTs that were visible in the TIRF field). The distance of Dgt5 puncta from the minus end of the MT was divided by the total length of the mother MT to calculate the fractional distance of Dgt5 puncta on mother MTs. Frame rates of the live-cell imaging videos were used to calculate the dwell time. Metamorph software was used to measure the intensity values, distribution of mTurquoise2-Dgt5 puncta along the length of MTs, and dwell time. Histograms and dot plots were generated using Excel (Microsoft) or Prism software (GraphPad), and figures were assembled using Illustrator (Adobe Systems). Statistical analysis was performed using Prism software.
Online supplemental material
Fig. S1 shows an MT branching event from a colocalized augmin and γ-tubulin puncta during anaphase. Video 1 shows two-color TIRF imaging of γ-tubulin and MTs during an MT branching event. Videos 2 and 3 show augmin-mediated MT branching. Video 4 shows activation of RhoA near the branched MT arrays.
Supplementary Material
Acknowledgments
We acknowledge Patricia Wadsworth (University of Massachusetts, Amherst, MA) for sharing many insightful conversations. We thank Gohta Goshima (Nagoya University, Nagoya, Japan) for sharing the α-D-TPX2 (Ssp1/Mei-38) serum. We acknowledge Ted Salmon for inspiring this work and for consistently demonstrating the power of observation and quantification of cellular phenomena using high-resolution light microscopy.
This work was supported by a National Institutes of Health grant (GM107026) to T.J. Maresca.
The authors declare no competing financial interests.
Author contributions: V. Verma contributed to formal analysis, investigation, methodology, resource development, data validation, visualization, and original drafting, as well as review and editing of the manuscript. T.J. Maresca conceptualized the project, acquired financial support, administered the project, supervised research, and contributed to visualization, formal analysis, and original drafting and editing of the manuscript.
References
- Alfaro-Aco R., Thawani A., and Petry S.. 2017. Structural analysis of the role of TPX2 in branching microtubule nucleation. J. Cell Biol. 216:983–997. 10.1083/jcb.201607060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basnet N., Nedozralova H., Crevenna A.H., Bodakuntla S., Schlichthaerle T., Taschner M., Cardone G., Janke C., Jungmann R., Magiera M.M., et al. . 2018. Direct induction of microtubule branching by microtubule nucleation factor SSNA1. Nat. Cell Biol. 20:1172–1180. 10.1038/s41556-018-0199-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement W.M., Benink H.A., and von Dassow G.. 2005. A microtubule-dependent zone of active RhoA during cleavage plane specification. J. Cell Biol. 170:91–101. 10.1083/jcb.200501131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benink H.A., and Bement W.M.. 2005. Concentric zones of active RhoA and Cdc42 around single cell wounds. J. Cell Biol. 168:429–439. 10.1083/jcb.200411109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugués J., Nuzzo V., Mazur E., and Needleman D.J.. 2012. Nucleation and transport organize microtubules in metaphase spindles. Cell. 149:554–564. 10.1016/j.cell.2012.03.027 [DOI] [PubMed] [Google Scholar]
- Buchan D.W.A., and Jones D.T.. 2019. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 47:W402–W407. 10.1093/nar/gkz297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde P.P., and Heald R.. 2003. Centrosomes and kinetochores, who needs 'Em? The role of noncentromeric chromatin in spindle assembly. Curr. Top. Dev. Biol. 56:85–113. 10.1016/S0070-2153(03)01008-1 [DOI] [PubMed] [Google Scholar]
- Cassimeris L., Pryer N.K., and Salmon E.D.. 1988. Real-time observations of microtubule dynamic instability in living cells. J. Cell Biol. 107:2223–2231. 10.1083/jcb.107.6.2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Sambade A., Calder G., and Lloyd C.. 2009. Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell. 21:2298–2306. 10.1105/tpc.109.069716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.W.C., Chen Z.A., Rogala K.B., Metz J., Deane C.M., Rappsilber J., and Wakefield J.G.. 2017. Cross-linking mass spectrometry identifies new interfaces of Augmin required to localise the γ-tubulin ring complex to the mitotic spindle. Biol. Open. 6:654–663. 10.1242/bio.022905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L., Yang K., Schultz R.M., and Lampson M.A.. 2015. Spatial Regulation of Kinetochore Microtubule Attachments by Destabilization at Spindle Poles in Meiosis I. Curr. Biol. 25:1835–1841. 10.1016/j.cub.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D., and Schuh M.. 2013. Restarting life: fertilization and the transition from meiosis to mitosis. Nat. Rev. Mol. Cell Biol. 14:549–562. 10.1038/nrm3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A.F., Roudot P., Legant W.R., Betzig E., Danuser G., and Gerlich D.W.. 2019. Augmin accumulation on long-lived microtubules drives amplification and kinetochore-directed growth. J. Cell Biol. 218:2150–2168. 10.1083/jcb.201805044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker F., Oriola D., Dalton B., and Brugués J.. 2018. Autocatalytic microtubule nucleation determines the size and mass of Xenopus laevis egg extract spindles. eLife. 7:e31149 10.7554/eLife.31149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G. 2011. Identification of a TPX2-like microtubule-associated protein in Drosophila. PLoS One. 6:e28120 10.1371/journal.pone.0028120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., and Stuurman N.. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421. 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., and Vale R.D.. 2008. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181:421–429. 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen A.C., Maresca T.J., Gatlin J.C., Salmon E.D., and Mitchison T.J.. 2009. Functional overlap of microtubule assembly factors in chromatin-promoted spindle assembly. Mol. Biol. Cell. 20:2766–2773. 10.1091/mbc.e09-01-0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O.J., Carazo-Salas R.E., Schatz C.A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., and Mattaj I.W.. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 104:83–93. 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- Gruss O.J., Wittmann M., Yokoyama H., Pepperkok R., Kufer T., Silljé H., Karsenti E., Mattaj I.W., and Vernos I.. 2002. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4:871–879. 10.1038/ncb870 [DOI] [PubMed] [Google Scholar]
- Haren L., Remy M.H., Bazin I., Callebaut I., Wright M., and Merdes A.. 2006. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172:505–515. 10.1083/jcb.200510028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward D., Metz J., Pellacani C., and Wakefield J.G.. 2014. Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev. Cell. 28:81–93. 10.1016/j.devcel.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., and Khodjakov A.. 2015. Thirty years of search and capture: The complex simplicity of mitotic spindle assembly. J. Cell Biol. 211:1103–1111. 10.1083/jcb.201510015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia K.C., Wilson-Kubalek E.M., Dottore A., Hao Q., Tsai K.L., Forth S., Shimamoto Y., Milligan R.A., and Kapoor T.M.. 2014. Reconstitution of the augmin complex provides insights into its architecture and function. Nat. Cell Biol. 16:852–863. 10.1038/ncb3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K., Nguyen P.A., Groen A.C., Field C.M., and Mitchison T.J.. 2014. Microtubule nucleation remote from centrosomes may explain how asters span large cells. Proc. Natl. Acad. Sci. USA. 111:17715–17722. 10.1073/pnas.1418796111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K., Korolev K.S., and Mitchison T.J.. 2016. Physical basis of large microtubule aster growth. eLife. 5:e19145 10.7554/eLife.19145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng R., and Stearns T.. 1999. Gamma-tubulin complexes: size does matter. Trends Cell Biol. 9:339–342. 10.1016/S0962-8924(99)01621-9 [DOI] [PubMed] [Google Scholar]
- Johmura Y., Soung N.K., Park J.E., Yu L.R., Zhou M., Bang J.K., Kim B.Y., Veenstra T.D., Erikson R.L., and Lee K.S.. 2011. Regulation of microtubule-based microtubule nucleation by mammalian polo-like kinase 1. Proc. Natl. Acad. Sci. USA. 108:11446–11451. 10.1073/pnas.1106223108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., and Heald R.. 2008. The RanGTP gradient - a GPS for the mitotic spindle. J. Cell Sci. 121:1577–1586. 10.1242/jcs.005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T., O’Toole E., Kita S., Osumi M., Usukura J., McIntosh J.R., and Goshima G.. 2013. Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J. Cell Biol. 202:25–33. 10.1083/jcb.201304031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye B., Stiehl O., Foster P.J., Shelley M.J., Needleman D.J., and Fürthauer S.. 2018. Measuring and modeling polymer concentration profiles near spindle boundaries argues that spindle microtubules regulate their own nucleation. New J. Phys. 20:055012 10.1088/1367-2630/aac2a5 [DOI] [Google Scholar]
- Kollman J.M., Polka J.K., Zelter A., Davis T.N., and Agard D.A.. 2010. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 466:879–882. 10.1038/nature09207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S., Bashkurov M., Mullin M., Ferreria M.G., Kittler R., Habermann B., Tagliaferro A., Poser I., Hutchins J.R., Hegemann B., et al. . 2009. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19:816–826. 10.1016/j.cub.2009.04.033 [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Hiwatashi Y., Hotta T., Xie T., Doonan J.H., and Liu B.. 2017. The Mitotic Function of Augmin Is Dependent on Its Microtubule-Associated Protein Subunit EDE1 in Arabidopsis thaliana. Curr. Biol. 27:3891–3897. 10.1016/j.cub.2017.11.030 [DOI] [PubMed] [Google Scholar]
- Liu T., Tian J., Wang G., Yu Y., Wang C., Ma Y., Zhang X., Xia G., Liu B., and Kong Z.. 2014. Augmin triggers microtubule-dependent microtubule nucleation in interphase plant cells. Curr. Biol. 24:2708–2713. 10.1016/j.cub.2014.09.053 [DOI] [PubMed] [Google Scholar]
- Lüders J., Patel U.K., and Stearns T.. 2006. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8:137–147. 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- Ma N., Tulu U.S., Ferenz N.P., Fagerstrom C., Wilde A., and Wadsworth P.. 2010. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol. Biol. Cell. 21:979–988. 10.1091/mbc.e09-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangal S., Sacher J., Kim T., Osório D.S., Motegi F., Carvalho A.X., Oegema K., and Zanin E.. 2018. TPXL-1 activates Aurora A to clear contractile ring components from the polar cortex during cytokinesis. J. Cell Biol. 217:837–848. 10.1083/jcb.201706021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., Groen A.C., Gatlin J.C., Ohi R., Mitchison T.J., and Salmon E.D.. 2009. Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr. Biol. 19:1210–1215. 10.1016/j.cub.2009.05.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles A.M., Fisher K.H., Colombié N., Wakefield J.G., and Ohkura H.. 2009. Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J. Cell Biol. 184:777–784. 10.1083/jcb.200811102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., Wühr M., Nguyen P., Ishihara K., Groen A., and Field C.M.. 2012. Growth, interaction, and positioning of microtubule asters in extremely large vertebrate embryo cells. Cytoskeleton (Hoboken). 69:738–750. 10.1002/cm.21050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Braunfeld M.B., Guénebaut V., Heuser J., and Agard D.A.. 2000. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2:365–370. 10.1038/35014058 [DOI] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., and Hasebe M.. 2005. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7:961–968. 10.1038/ncb1306 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Ehrhardt D.W., and Hashimoto T.. 2010. Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12:1064–1070. 10.1038/ncb2110 [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., and Heringa J.. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–217. 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- Oh D., Yu C.H., and Needleman D.J.. 2016. Spatial organization of the Ran pathway by microtubules in mitosis. Proc. Natl. Acad. Sci. USA. 113:8729–8734. 10.1073/pnas.1607498113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S., Groen A.C., Ishihara K., Mitchison T.J., and Vale R.D.. 2013. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 152:768–777. 10.1016/j.cell.2012.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid T.A., Schuster B.M., Mann B.J., Balchand S.K., Plooster M., McClellan M., Coombes C.E., Wadsworth P., and Gardner M.K.. 2016. Suppression of microtubule assembly kinetics by the mitotic protein TPX2. J. Cell Sci. 129:1319–1328. 10.1242/jcs.178806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J., and Surrey T.. 2017. Microtubule nucleation: beyond the template. Nat. Rev. Mol. Cell Biol. 18:702–710. 10.1038/nrm.2017.75 [DOI] [PubMed] [Google Scholar]
- Sánchez-Huertas C., Freixo F., Viais R., Lacasa C., Soriano E., and Lüders J.. 2016. Non-centrosomal nucleation mediated by augmin organizes microtubules in post-mitotic neurons and controls axonal microtubule polarity. Nat. Commun. 7:12187 10.1038/ncomms12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A.F., von Dassow G., and Bowerman B.. 2016. Oocyte Meiotic Spindle Assembly and Function. Curr. Top. Dev. Biol. 116:65–98. 10.1016/bs.ctdb.2015.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.G., King M.R., Zhang R., Kadzik R.S., Thawani A., and Petry S.. 2018. Mechanism of how augmin directly targets the γ-tubulin ring complex to microtubules. J. Cell Biol. 217:2417–2428. 10.1083/jcb.201711090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T.M. 2013. Ultrastructure of protrusive actin filament arrays. Curr. Opin. Cell Biol. 25:574–581. 10.1016/j.ceb.2013.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thawani A., Stone H.A., Shaevitz J.W., and Petry S.. 2019. Spatiotemporal organization of branched microtubule networks. eLife. 8:e43890 10.7554/eLife.43890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovey C.A., and Conduit P.T.. 2018. Microtubule nucleation by γ-tubulin complexes and beyond. Essays Biochem. 62:765–780. 10.1042/EBC20180028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.Y., Ngo B., Tapadia A., Hsu P.H., Wu G., and Lee W.H.. 2011. Aurora-A phosphorylates Augmin complex component Hice1 protein at an N-terminal serine/threonine cluster to modulate its microtubule binding activity during spindle assembly. J. Biol. Chem. 286:30097–30106. 10.1074/jbc.M111.266767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.Y., Ngo B., Tapadia A., Hsu P.H., Wu G., and Lee W.H.. 2011. Aurora-A phosphorylates Augmin complex component Hice1 protein at an N-terminal serine/threonine cluster to modulate its microtubule binding activity during spindle assembly. J. Biol. Chem. 286:30097–30106. 10.1074/jbc.M111.266767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., and Goshima G.. 2010. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J. Cell Biol. 191:259–267. 10.1083/jcb.201004150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Nozawa R.S., Tomioka A., Petry S., Vale R.D., Obuse C., and Goshima G.. 2009. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 106:6998–7003. 10.1073/pnas.0901587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Kamasaki T., Hiruma S., Poser I., Yoda K., Yajima J., Gerlich D.W., and Goshima G.. 2016. Augmin shapes the anaphase spindle for efficient cytokinetic furrow ingression and abscission. Mol. Biol. Cell. 27:812–827. 10.1091/mbc.E15-02-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V., and Maresca T.J.. 2019. Microtubule plus-ends act as physical signaling hubs to activate RhoA during cytokinesis. eLife. 8:e38968 10.7554/eLife.38968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos J.W., Pieuchot L., Evrard J.L., Janski N., Bergdoll M., de Ronde D., Perez L.H., Sardon T., Vernos I., and Schmit A.C.. 2008. The plant TPX2 protein regulates prospindle assembly before nuclear envelope breakdown. Plant Cell. 20:2783–2797. 10.1105/tpc.107.056796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P. 2015. TPX2. Curr. Biol. 25:R1156–R1158. 10.1016/j.cub.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia A., Nakamura M., Moss D., Kirik V., Hashimoto T., and Ehrhardt D.W.. 2014. GCP-WD mediates γ-TuRC recruitment and the geometry of microtubule nucleation in interphase arrays of Arabidopsis. Curr. Biol. 24:2548–2555. 10.1016/j.cub.2014.09.013 [DOI] [PubMed] [Google Scholar]
- Walker R.A., O’Brien E.T., Pryer N.K., Soboeiro M.F., Voter W.A., Erickson H.P., and Salmon E.D.. 1988. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 107:1437–1448. 10.1083/jcb.107.4.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Wang C., Liu W., Ma Y., Dong L., Tian J., Yu Y., and Kong Z.. 2018. Augmin Antagonizes Katanin at Microtubule Crossovers to Control the Dynamic Organization of Plant Cortical Arrays. Curr. Biol. 28:1311–1317.e1313. [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Williamson R.E.. 1989. Reassembly of microtubules in Nitella tasmanica: assembly of cortical microtubules in branching clusters and its relevance to steady-state microtubule assembly. J. Cell Sci. 93:705–714. [Google Scholar]
- Wittmann T., Wilm M., Karsenti E., and Vernos I.. 2000. TPX2, A novel xenopus MAP involved in spindle pole organization. J. Cell Biol. 149:1405–1418. 10.1083/jcb.149.7.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lin Y.T., Wei R., Chen Y., Shan Z., and Lee W.H.. 2008. Hice1, a novel microtubule-associated protein required for maintenance of spindle integrity and chromosomal stability in human cells. Mol. Cell. Biol. 28:3652–3662. 10.1128/MCB.01923-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M., Chen Y., Dumont S., Groen A.C., Needleman D.J., Salic A., and Mitchison T.J.. 2008. Evidence for an upper limit to mitotic spindle length. Curr. Biol. 18:1256–1261. 10.1016/j.cub.2008.07.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye A.A., Deretic J., Hoel C.M., Hinman A.W., Cimini D., Welburn J.P., Maresca T.J., and Aurora A.. 2015. Aurora A Kinase Contributes to a Pole-Based Error Correction Pathway. Curr. Biol. 25:1842–1851. 10.1016/j.cub.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen Q., Feng J., Hou J., Yang F., Liu J., Jiang Q., and Zhang C.. 2009. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J. Cell Sci. 122:2240–2251. 10.1242/jcs.042747 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wong M.L., Alberts B., and Mitchison T.. 1995. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 378:578–583. 10.1038/378578a0 [DOI] [PubMed] [Google Scholar]
- Zhu H., Coppinger J.A., Jang C.Y., Yates J.R. III, and Fang G.. 2008. FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J. Cell Biol. 183:835–848. 10.1083/jcb.200807046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.