Abstract
Objectives
Prosthetic joint infection (PJI) is the most common cause of arthroplasty failure. However, infection is often difficult to detect by conventional bacterial cultures, for which false-negative rates are 23% to 35%. In contrast, 16S rRNA metagenomics has been shown to quantitatively detect unculturable, unsuspected, and unviable pathogens. In this study, we investigated the use of 16S rRNA metagenomics for detection of bacterial pathogens in synovial fluid (SF) from patients with hip or knee PJI.
Methods
We analyzed the bacterial composition of 22 SF samples collected from 11 patients with PJIs (first- and second-stage surgery). The V3 and V4 region of bacteria was assessed by comparing the taxonomic distribution of the 16S rDNA amplicons with microbiome sequencing analysis. We also compared the results of bacterial detection from different methods including 16S metagenomics, traditional cultures, and targeted Sanger sequencing.
Results
Polymicrobial infections were not only detected, but also characterized at different timepoints corresponding to first- and second-stage exchange arthroplasty. Similar taxonomic distributions were obtained by matching sequence data against SILVA, Greengenes, and The National Center for Biotechnology Information (NCBI). All bacteria isolated from the traditional culture could be further identified by 16S metagenomics and targeted Sanger sequencing.
Conclusion
The data highlight 16S rRNA metagenomics as a suitable and promising method to detect and identify infecting bacteria, most of which may be uncultivable. Importantly, the method dramatically reduces turnaround time to two days rather than approximately one week for conventional cultures.
Cite this article: M-F. Chen, C-H. Chang, C. Chiang-Ni, P-H. Hsieh, H-N. Shih, S. W. N. Ueng, Y. Chang. Rapid analysis of bacterial composition in prosthetic joint infection by 16S rRNA metagenomic sequencing. Bone Joint Res 2019;8:367–377. DOI: 10.1302/2046-3758.88.BJR-2019-0003.R2.
Keywords: Prosthetic joint infection, 16S metagenomics, Synovial fluid, Bacterial composition, Polymicrobial infection
Article focus
This study investigated the use of 16S rRNA metagenomics for detecting bacterial pathogens in synovial fluid (SF) from patients with hip or knee prosthetic joint infection (PJI).
This study compared the performance of bacterial detection using different methods, including 16S metagenomics, traditional cultures, and targeted Sanger sequencing. Data highlighted 16S rRNA metagenomics as a suitable and promising method to detect and identify infecting bacteria, most of which may be uncultivable.
Key messages
This study demonstrates that 16S metagenomics is a method with high potential for PJI diagnosis in the future. This method could detect very low levels of bacterial infection in SF, even when the bacteria are dead, i.e. after the patient has received antibiotic treatment. This method reduces the time required for bacterial identification and also improves polymicrobial detection in PJI diagnosis.
Based on the performance against traditional cultures and targeted Sanger sequencing, the data highlight the potential of 16S metagenomics to diagnose PJIs, especially mixed infections. We provide a foundation for further development towards the 16S metagenomic diagnosis of PJI.
Strengths and limitations
16S rRNA metagenomics detects very low levels of bacterial infection but does not detect fungi or viruses in the body fluids of patients. Moreover, genus-level identification and quantification are generally more reliable than species-level identification.
16S rRNA metagenomics is strongly susceptible to contamination from reagents and sample processing, which may generate false positives (contaminated bacteria) or false negatives (underestimated infectious bacteria). Thus, all materials, reagents, and procedures should be strictly managed and standardized.
The future goals for application of 16S metagenomics to PJI diagnosis are established in a standardized protocol including specimen collection, DNA extraction, 16S polymerase chain reaction (PCR), next-generation sequencing (NGS) criteria setting, bioinformatic analysis, and final reports.
Introduction
Total joint arthroplasty is one of the most successful surgical procedures in modern medicine.1-4 The demand for primary total knee and total hip arthroplasty has been projected to grow by 673% and 174%, respectively, from 2005 to 2030 in the United States.1 Prosthetic joint infection (PJI) is the most common cause of knee arthroplasty failure,5 and accounts for 25.2% of failed total knee arthroplasties. It is also the third most common indication (14.7%) for revision hip arthroplasty.6-8 The diagnosis of PJI mainly depends on the combination of clinical tests, including serum C-reactive protein (CRP), peripheral blood leucocytes, synovial fluid (SF) white blood cells, bacterial cultures of preoperative SFs and intraoperative tissues, radiological, and other tests such as positron emission tomography.9,10 Identification of the bacteria is not only the benchmark for PJI diagnosis but also provides guidance for antibiotic choice in PJI treatment.11,12 However, the culture-negative rate of PJI is around 23% to 35%.13,14 Furthermore, prior antimicrobial use has been shown to decrease the sensitivity of culture in PJI, and 53% of patients received an antimicrobial agent before the diagnosis of culture-negative PJI.11,15-18 Accordingly, detection of the bacteria remains a challenge for the diagnosis of PJI.19
Polymerase chain reaction (PCR)-based methods may improve diagnosis of microorganism infection by reducing turnaround time and eliminating the requirement for culture. PCR assays of SF using pathogen-specific primers were reported to be 70% to 96% sensitive.20-22 However, this method detects only organisms that are tested for, and therefore will miss atypical or unexpected pathogens.23,24 High-throughput sequencing overcomes this issue by quantitative detection of unculturable, unsuspected, and non-viable pathogens without sacrificing speed.25 16S rRNA metagenomic analysis has been used successfully to analyze bacteria in clinical specimens.26-28 However, it is very rare for bacterial detection in PJI.29 In this study, we investigated the roles of 16S rRNA metagenomics in the detection of bacterial pathogens in SFs from patients with hip or knee PJI. We hypothesized that the bacterial V3 and V4 fragments would be amplified efficiently with all bacteria to generate amplicons for sequencing. We compare 16S rRNA metagenomics results before (first-stage surgery) and after debridement (second-stage surgery) in order to explore whether there is a difference in bacterial detection. We believe that this method can detect not only very low-level infections but also antibiotic-killed bacteria during preoperative antibiotic treatment. Accordingly, only live pathogenic bacteria can give positive results after bacterial culture-based methods. In the event of pathogenic bacteria being killed by preoperative antibiotics, bacterial culture-based methods may yield false-negative results. However, the 16S rRNA metagenomics method is capable of detecting bacterial nucleic acid regardless of whether the bacteria is alive or dead. We further attempted to optimize the protocol through comparison of the results from different database analyses including SILVA, Greengenes, and The National Center for Biotechnology Information (NCBI). This study also compared the performance of bacterial detection from different methods including 16S metagenomics, traditional cultures, and targeted Sanger sequencing.
Materials and Methods
Patients and sampling
Between November 2016 and March 2017, 11 hip/knee PJI patients (three female patients, eight male patients) were enrolled in this study. PJI was defined by fulfilling one of the following three criteria: 1) sinus tract communicating with the prosthesis; 2) pathogen isolated from two or more samples obtained from the infected prosthetic joint; 3) four of the following six criteria exist: a) elevated serum erythrocyte sedimentation rate (ESR) and serum CRP concentration; b) elevated synovial leucocyte count; c) elevated synovial polymorphonuclear neutrophil percentage (PMN%); d) presence of purulence in the affected joint; e) isolation of a microorganism in one culture of prosthetic tissue or fluid; and f) greater than five neutrophils per high-power field in five high-power fields observed from histological analysis of prosthetic tissue at ×400 magnification.30,31 All of the PJI patients were treated with two-stage exchange arthroplasty. In brief, resection arthroplasty for PJI included radical debridement, removal of prosthesis, implantation of antibiotic-loaded bone cement, and administration of systemic antimicrobial agents for the control of joint infection (first-stage surgery). Delayed reimplantation of the prosthesis was performed after successful antimicrobial therapy, which was defined by the absence of signs of infection and the return of ESR and serum CRP levels to normal (second-stage surgery).32 During the same enrolment period, three patients (two hip and one knee) with aseptic loosening who were scheduled for revision arthroplasty were enrolled as a control group. Specimens of joint fluid measuring at least 2 ml were collected by needle aspiration prior to entering the joint to minimize contamination by blood. Patients with malignant tumours, those who had received immunosuppressive agents, and those who had a history of allergy to vancomycin or ceftazidime were excluded. The study was approved by the local institutional review board, and was compliant with accepted ethical standards at our hospital. Informed consent was obtained from all patients before initiating this study.
Specimen preparation, sequencing, and bacterial culture
Synovial fluid specimens were delivered to the laboratory immediately after aspiration and centrifuged at 10 000 ×g. The resulting pellet was extracted with EasyPrep HY Genomic DNA Extraction Kit (TE-GD01; BIOTOOLS Co., Ltd., New Taipei City, Taiwan). Bacterial V3 and V4 fragments were amplified with primer 341F (CCTAYGGGRBGCASCAG) and primer 806R (GGACTACNNGGGTATCTAAT) to generate amplicons of 466 bp.33 Paired-end reads from the amplicons were assembled into tags in FLASH v.1.2.7, clustered into operational taxonomic units (OTUs) at 97% similarity using Uparse v7.0.1001 (http://drive5.com/uparse/), and identified with regard to genus and species using Ribosomal Database Project (RDP) classifier v2.2 against SILVA (v128), Greengenes (13_8), and NCBI databases. These analyses were conducted on 15 August 2017.33
Specimens were analyzed by 16S rRNA metagenomic analysis on an Illumina HiSeq 2000 Sequencing system (Illumina, Inc., San Diego, California) and the results were compared with 16S rDNA amplification and Sanger sequencing results, as well as with bacterial culture results, which represent the current benchmark for the diagnosis of PJI. Patient characteristics are summarized in Table I. Of note, we initially tested SFs from three uninfected patients to obtain control results. However, 16S PCR products were not obtained from these three specimens, and these specimens were therefore excluded because they were unable to be used for 16S rRNA metagenomic analysis.
Table I.
Patient characteristics
| Parameter | Aseptic | First-stage surgery | Second-stage surgery |
|---|---|---|---|
| Number of patients | 3 | 11 | N/A |
| Sex, male:female, n (%) | 0:3 (0:100) | 8:3 (73:27) | N/A |
| Mean age at surgery, yrs (sd; range) | 68 (6; 63 to 77) | 63 (10.2; 40 to 76) | N/A |
| Type of joint prosthesis, knee:hip, n (%) | 1:2 (33:67) | 8:3 (73:27) | N/A |
| Mean serum CRP, mg/dl (sd) | 3.3 (1.3) | 65.2 (72.2) | 24.9 (40.9) |
| Mean synovial fluid white blood cells, cells/dl (sd) | 333 (451) | 22 393 (18 133) | 5967 (8110) |
| Mean synovial fluid neutrophils, % (sd) | 42 (26) | 79 (16) | 60 (35) |
CRP, C-reactive protein; N/A, not applicable
For Sanger sequencing, the 16S rDNA amplicons were cloned using T&A Cloning Kit and DH5a competent cells (both Yeastern Biotech Co. Ltd, Taipei, Taiwan). For each sample, at least ten clones were picked, sequenced, and compared against the genetic sequence database, GenBank, using the Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information (NCBI), U.S. National Library of Medicine, Bethesda, Maryland). Specimens of periarticular tissue and joint fluid were sent for bacterial culture in both the PJI and control groups. In brief, SFs and deeper layers of the synovial membrane were cultured in BD BACTEC Peds Plus or BD BACTEC Plus Aerobic (Becton, Dickinson and Company (BD), Sparks, Maryland) and incubated at 37°C under aerobic and anaerobic conditions. The patient's deep tissue was placed directly into the bacterial culture container, and then sterile normal saline was added to avoid tissue drying. Cultures were examined daily for two weeks, and isolated bacteria were identified by matrix-assisted laser desorption/ionization (MALDI)-time of flight (TOF) mass spectrometry on an Ultraflex III TOF/TOF system (Bruker Corporation, Billerica, Massachusetts).
Data analysis
Data are reported as the mean (sd), and were analyzed in GraphPad Prism (GraphPad Software Inc., San Diego, California).
Results
Distribution of bacterial taxa over two-stage exchange arthroplasty
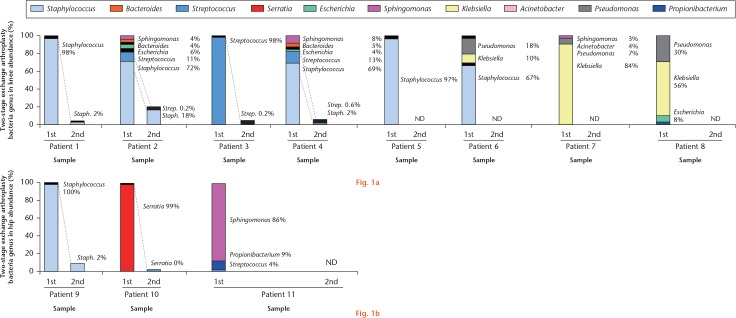
We identified ten major pathogen species in 11 infected patients, including Staphylococcus, Streptococcus, Klebsiella, Serratia, Escherichia, Pseudomonas, Bacteroides, Acinetobacter, Propionibacterium, and Sphingomonas (Fig. 1). The polymicrobial composition of SF from PJI revealed a significant change between the first- and second-stage surgeries. Changes in relative abundance between the first- and second-stage surgeries were assessed prior to, and three months after, the first-stage surgery. These data demonstrate that first-stage resection arthroplasty and sequential antibiotic treatment provide a very effective way to eliminate the microorganism that caused the PJIs. This observation may help to guide patient management and treatment selection between second-stage surgery and additional debridement.
Fig. 1.
Relative abundance of bacterial taxa in individual samples. a) Samples from first- or second-stage knee surgery, and b) first- or second-stage hip surgery are plotted along the horizontal axis. Relative abundances inferred from 16S metagenomic sequencing are plotted on the vertical axis. ND, not detected.
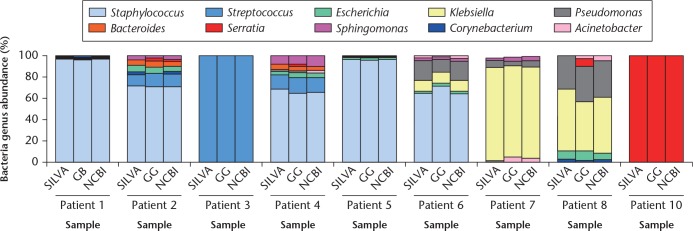
Although SILVA is a comprehensive, up-to-date, and quality-controlled database of rRNA genes for 16S metagenomics, Greengenes and NCBI are also often used.34 Thus, we matched individual patient data against all three databases (Fig. 2), and obtained similar results, with variability observed only among less abundant genera. For example, Serratia were detected in patients 2, 4, and 8 only against Greengenes. Genera with relative abundances higher than 0.5% are listed in Table II, in which those with abundances of 0.6% to 5.0%, and 6% to 100%, are marked by a dagger symbol and an asterisk, respectively. The results were essentially the same across databases for genera with an abundance higher than 5%, implying that analysis based on major genera is robust.
Fig. 2.
Similarities of bacterial composition in each specimen as annotated against SILVA, Greengenes (GG), and the National Center for Biotechnology Information (NCBI) databases. Samples are plotted along the horizontal axis, and relative abundances are plotted on the vertical axis.
Table II.
Taxonomic abundances as annotated against SILVA, Greengenes, and NCBI databases. Only genera with a relative abundance ⩾ 0.5% are listed
| Age (yrs)/sex/ type/number | SILVA |
Greengenes |
NCBI |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genera | % of reads | Normalized reads | Raw reads | Genera | % of reads | Normalized reads | Raw reads | Genera | % of reads | Normalized reads | Raw reads | |
| 76/M/RK/Patient 1 | Staphylococcus* | 97.5 | 10 193 | 65 200 | Staphylococcus* | 96.5 | 580 | 1381 | Staphylococcus* | 97.5 | 10 127 | 65 200 |
| Sphingomonas† | 0.7 | 80 | 463 | Sphingomonas† | 1.5 | 9 | 28 | Sphingomonas† | 0.7 | 76 | 464 | |
| Escherichia† | 0.6 | 42 | 314 | Escherichia† | 0.7 | 4 | 7 | N/A | N/A | N/A | N/A | |
| N/A | N/A | N/A | N/A | Bacteroides† | 0.8 | 5 | 10 | Bacteroides† | 0.7 | 71 | 372 | |
| 71/F/RK/Patient 2 | Staphylococcus* | 71.7 | 914 | 1777 | Staphylococcus* | 70.9 | 180 | 1766 | Staphylococcus* | 71.2 | 840 | 1777 |
| Streptococcus* | 10.9 | 146 | 275 | Streptococcus* | 12.2 | 31 | 274 | Streptococcus* | 11.9 | 140 | 275 | |
| Escherichia* | 6.1 | 74 | 141 | Escherichia* | 5.1 | 13 | 139 | Escherichia† | 4.9 | 58 | 141 | |
| Bacteroides† | 3.9 | 42 | 101 | Bacteroides* | 5.1 | 13 | 4 | Bacteroides† | 4.2 | 50 | 104 | |
| Sphingomonas† | 3.9 | 40 | 85 | Sphingomonas† | 2.0 | 5 | 85 | Sphingomonas† | 3.6 | 43 | 85 | |
| Corynebacterium† | 1.2 | 15 | 30 | Corynebacterium† | 0.8 | 2 | 30 | Corynebacterium† | 1.7 | 20 | 31 | |
| Pseudomonas† | 1.2 | 11 | 25 | N/A | N/A | N/A | N/A | Pseudomonas† | 0.8 | 10 | 25 | |
| N/A | N/A | N/A | N/A | Serratia | 3.1 | 8 | 53 | Serratia† | 1.3 | 15 | 39 | |
| 60/F/LK/Patient 3 | Streptococcus* | 98.1 | 8161 | 22 396 | Streptococcus* | 98.1 | 1773 | 22 292 | Streptococcus* | 97.9 | 8216 | 22 396 |
| Staphylococcus† | 1.2 | 100 | 277 | Staphylococcus† | 1.1 | 19 | 277 | Staphylococcus† | 1.2 | 102 | 277 | |
| 56/M/RK/Patient 4 | Staphylococcus* | 68.7 | 348 | 671 | Staphylococcus* | 64.5 | 71 | 662 | Staphylococcus* | 66.0 | 324 | 671 |
| Streptococcus* | 13.0 | 76 | 138 | Streptococcus* | 14.5 | 16 | 137 | Streptococcus* | 13.4 | 66 | 138 | |
| Sphingomonas* | 8.1 | 43 | 85 | Sphingomonas* | 8.2 | 9 | 84 | Sphingomonas* | 9.6 | 47 | 85 | |
| Bacteroides† | 4.9 | 21 | 50 | Bacteroides† | 3.6 | 4 | 50 | Bacteroides† | 3.5 | 17 | 51 | |
| Escherichia† | 3.6 | 23 | 37 | Escherichia† | 4.5 | 5 | 36 | Escherichia† | 3.9 | 19 | 37 | |
| Pseudomonas† | 1.3 | 9 | 24 | Pseudomonas† | 0.9 | 1 | 24 | Pseudomonas† | 2.4 | 12 | 24 | |
| N/A | N/A | N/A | N/A | Serratia† | 1.8 | 2 | 9 | N/A | N/A | N/A | N/A | |
| 74/M/LK/Patient 5 | Staphylococcus* | 96.9 | 6821 | 18 760 | Staphylococcus* | 95.3 | 1549 | 18 582 | Staphylococcus* | 96.0 | 6669 | 18 759 |
| Escherichia† | 2.0 | 139 | 395 | Escherichia† | 2.4 | 39 | 391 | Escherichia† | 2.1 | 145 | 395 | |
| N/A | N/A | N/A | N/A | Bacteroides† | 0.9 | 14 | 61 | Bacteroides† | 0.6 | 41 | 113 | |
| 73/M/LK/Patient 6 | Staphylococcus* | 65.1 | 560 | 717 | Staphylococcus* | 71.5 | 138 | 706 | Staphylococcus* | 64.6 | 560 | 717 |
| Pseudomonas* | 17.9 | 158 | 199 | Pseudomonas* | 11.9 | 23 | 159 | Pseudomonas* | 17.4 | 151 | 205 | |
| Klebsiella* | 10.3 | 79 | 99 | Klebsiella* | 10.4 | 20 | 102 | Klebsiella* | 10.6 | 92 | 123 | |
| Sphingomonas† | 2.3 | 24 | 29 | Sphingomonas† | 1.6 | 3 | 28 | Sphingomonas† | 2.4 | 21 | 29 | |
| Acinetobacter† | 2.3 | 21 | 25 | Acinetobacter† | 1.6 | 3 | 24 | Acinetobacter† | 2.7 | 23 | 25 | |
| Escherichia† | 1.6 | 18 | 23 | Escherichia† | 2.6 | 5 | 19 | Escherichia† | 2.0 | 17 | 23 | |
| 62/M/RK/Patient 7 | Klebsiella* | 84.2 | 1788 | 4520 | Klebsiella* | 84.8 | 436 | 4468 | Klebsiella* | 84.3 | 1794 | 4482 |
| Pseudomonas* | 6.6 | 118 | 314 | Pseudomonas† | 3.9 | 20 | 259 | Pseudomonas* | 5.6 | 120 | 313 | |
| Acinetobacter† | 4.2 | 107 | 269 | Acinetobacter† | 1.9 | 30 | 267 | Acinetobacter† | 4.7 | 100 | 269 | |
| Sphingomonas† | 2.7 | 59 | 132 | Sphingomonas† | 2.5 | 13 | 131 | Sphingomonas† | 2.5 | 53 | 132 | |
| Staphylococcus† | 1.7 | 39 | 111 | Staphylococcus* | 5.8 | 10 | 108 | Staphylococcus† | 2.2 | 47 | 111 | |
| N/A | N/A | N/A | N/A | Serratia† | 0.6 | 3 | 82 | N/A | N/A | N/A | N/A | |
| 62/M/LK/Patient 8 | Klebsiella* | 56.1 | 227 | 356 | Klebsiella* | 46.4 | 51 | 351 | Klebsiella* | 52.3 | 229 | 356 |
| Pseudomonas* | 29.9 | 130 | 216 | Pseudomonas* | 32.7 | 36 | 206 | Pseudomonas* | 33.8 | 148 | 216 | |
| Escherichia* | 7.7 | 28 | 46 | Escherichia* | 9.1 | 10 | 45 | Escherichia* | 6.2 | 27 | 46 | |
| Acinetobacter† | 3.0 | 20 | 36 | Acinetobacter† | 2.7 | 3 | 34 | Acinetobacter† | 4.3 | 19 | 36 | |
| Corynebacterium† | 3.0 | 11 | 20 | Corynebacterium† | 1.8 | 2 | 22 | Corynebacterium† | 2.7 | 12 | 20 | |
| N/A | N/A | N/A | N/A | Serratia* | 7.3 | 8 | 39 | N/A | N/A | N/A | N/A | |
| 63/M/RH/Patient 10 | Serratia* | 99.1 | 12 894 | 45 891 | Serratia* | 98.7 | 926 | 926 | Serratia* | 98.9 | 12 828 | 46 760 |
Abundance of 6% to 100%
Abundance of 0.6% to 5.0%
RK, right knee; LK, left knee; RH, right hip; N/A, not applicable; NCBI, National Center for Biotechnology Information
Comparison of bacterial cultures, targeted Sanger sequencing, and 16S metagenomics
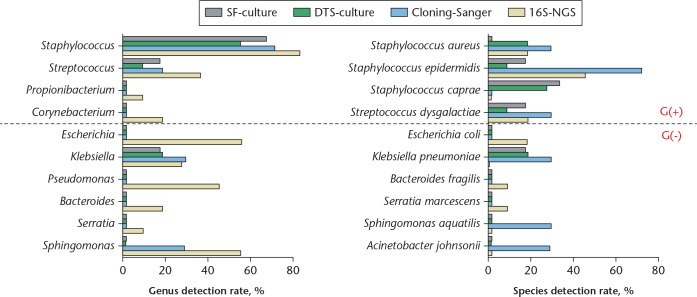
As bacterial cultures are the benchmark test to detect infections, deeper layers of the synovial membrane or SF were also collected during surgery and inoculated into blood culture bottles. Bacteria from positive cultures were identified by MALDI-TOF mass spectrometry and are listed in Table III. Bacteria were not detected by mass spectrometry in two cultures, resulting in a positive rate of about 82% to 85%. One species was detected in each of the remaining cultures, including Staphylococcus aureus in two patients, Staphylococcus caprae in three patients, Klebsiella pneumoniae in three patients, and Staphylococcus epidermidis and Streptococcus dysgalactiae in one patient each (Table III).
Table III.
Infecting species as detected by different methods. Only genera with relative abundance ⩾5% are listed
| Age (yrs)/sex/type/number | 16S metagenomic analysis | Targeted Sanger sequencing | Bacterial culture | |
|---|---|---|---|---|
| Genus | Species | |||
| 76/M/RK/Patient 1 | Staphylococcus | Staphylococcus aureus; Escherichia coli | Staphylococcus aureus | DTS: Staphylococcus aureus (3/3) |
| 71/F/RK/Patient 2 | Staphylococcus; Streptococcus; Escherichia | Staphylococcus aureus; Escherichia coli | Staphylococcus aureus; Sphingomonas aquatilis | DTS: Staphylococcus aureus (3/3) |
| 60/F/LK/Patient 3 | Streptococcus | Streptococcus dysgalactiae | Streptococcus dysgalactiae | DTS: Streptococcus dysgalactiae (1/4); BB-SY: Streptococcus dysgalactiae |
| 56/M/RK/Patient 4 | Staphylococcus; Streptococcus; Sphingomonas | Staphylococcus epidermidis; Bacteroides fragilis | Staphylococcus epidermidis/S. capitis; Streptococcus dysgalactiae | DTS: Staphylococcus caprae (3/3); BB-SY: Staphylococcus caprae |
| 74/M/LK/Patient 5 | Staphylococcus | Staphylococcus epidermidis | Staphylococcus epidermidis/S. caprae | DTS: Staphylococcus caprae (3/3); BB-SY: Staphylococcus caprae |
| 73/M/LK/Patient 6 | Staphylococcus; Pseudomonas; Klebsiella | Staphylococcus epidermidis | Staphylococcus epidermidis; Acinetobacter johnsonii | DTS: Staphylococcus epidermidis; BB-SY: Staphylococcus epidermidis |
| 62/M/RK/Patient 7 | Klebsiella; Pseudomonas | ND | Klebsiella pneumoniae | DTS: Klebsiella pneumoniae; BB-SY: Klebsiella pneumoniae |
| 62/M/LK/Patient 8 | Klebsiella; Pseudomonas; Escherichia | ND | ND | DTS: Klebsiella pneumoniae; BB-SY: NBG |
| 61/F/RH/Patient 9 | Staphylococcus | Staphylococcus epidermidis | ND | DTS: Staphylococcus caprae (3/3); SY: Staphylococcus caprae |
| 63/M/RH/Patient 10 | Serratia | Serratia marcescens | ND | DTS: NBG |
| 40/M/LH/Patient 11 | Sphingomonas; Propionibacterium | Streptococcus dysgalactiae; Staphylococcus epidermidis | ND | DTS: NBG (3/3) |
| Detection rate, n (%) | 11/11 (100) | 9/11 (82) | 7/11 (64) | DTS: 9/11 (82); BB-SY: 6/7 (85) |
RK, right knee; LK, left knee; DTS, deep tissue culture; BB-SY, synovial fluid culture on BD BACTEC Peds Plus (Becton, Dickinson and Company (BD), Sparks, Maryland); ND, not detected; NBG, no bacterial growth; RH, right hip; LH, left hip; SY, synovial fluid culture on BD BACTEC Plus Aerobic
The data highlight 16S rRNA metagenomics as sufficient to identify mixed pathogens in a single specimen. Indeed, this method identifies not only the same genera detected in bacterial cultures, but also others that were not detectable in the bacterial cultures. For example, patients 1, 3, 5, 9, and 10 were infected with only one strain based on an abundance cutoff of 5% (Table III), whereas mixed infections with one to seven genera were detected in the other six patients. At a cutoff of 0.5%, only patients 9 and 10 can be considered infected with a single strain (Table II).
The most common Gram-positive infecting genus was Staphylococcus (82%; Fig. 3), whereas Escherichia spp., Klebsiella spp., Pseudomonas spp., and Sphingomonas spp. were the most common Gram-negative pathogens. Finally, we assessed the impact of antibiotic treatment, and found that infection had recurred after three months of debridement in patient 1. Patients 2 and 5 tested positive in intraoperative cultures from second-stage surgery; patients 3, 4, 6, 9, and 10 were infection-free; and patient 11 was infected with fungus, while patients 7 and 8 had died due to sepsis from PJIs (Table IV). Collectively, the data demonstrate that 16S metagenomic analysis detects low-abundance 16S rRNA genes in specimens such as SF.
Fig. 3.
Detection rates by 16S metagenomic analysis, targeted Sanger sequencing, and blood cultures. Data are for ten species and ten genera detected by different methods. G(+), Gram-positive; G(-), Gram-negative; SF-culture, synovial fluid culture on BD BACTEC Plus Aerobic (Becton, Dickinson and Company (BD), Sparks, Maryland); DTS-culture, deep tissue culture; Cloning-Sanger, targeted Sanger sequencing; 16S-NGS, 16S metagenomic analysis-next-generation sequencing.
Table IV.
Association between detected microorganisms, antibiotic treatment, and recurrent infection
| Age (yrs)/sex/type/number | First-stage surgery |
Infection status at 3 mths |
|||||
|---|---|---|---|---|---|---|---|
| 16S metagenomic analysis |
Bacterial culture |
Antibiotics pre-surgery |
Antibiotics in ALBCs |
Infection status |
Bacterial culture |
||
| Genera (against SILVA) | % of reads | ||||||
| 76/M/RK/Patient 1 | Staphylococcus | 97.5 | DTS: Staphylococcus aureus (3/3) | None | Vancomycin; ceftazidime | Recurrent | DTS: Citrobacter koseri; SY: Citrobacter koseri, Serratia marcescens |
| 71/F/RK/Patient 2 | Staphylococcus; Streptococcus; Escherichia | 71.7; 10.9; 6.1 | DTS: Staphylococcus aureus (3/3) | Amoxycillin | Vancomycin; ceftazidime | PIOC | DTS: Staphylococcus haemolyticus, Moraxella osloensis |
| 60/F/LK/Patient 3 | Streptococcus | 98.1 | DTS: Streptococcus dysgalactiae (1/4); BB-SY: Streptococcus dysgalactiae | None | Vancomycin; ceftazidime | Infection-free | NBG |
| 56/M/RK/Patient 4 | Staphylococcus; Streptococcus; Sphingomonas | 68.7; 13.0; 8.1 | DTS: Staphylococcus caprae (3/3); BB-SY: Staphylococcus caprae | Dicloxacillin | Vancomycin; ceftazidime | Infection-free | NBG |
| 74/M/LK/Patient 5 | Staphylococcus | 96.9 | DTS: Staphylococcus caprae (3/3); BB-SY: Staphylococcus caprae | None | Vancomycin; ceftazidime | PIOC | DTS: Staphylococcus aureus |
| 73/M/LK/Patient 6 | Staphylococcus; Pseudomonas; Klebsiella | 65.1; 17.9; 10.3 | DTS: Staphylococcus epidermidis; BB-SY: Staphylococcus epidermidis | None | Teicoplanin; ceftazidime; gentamicin | Infection-free | NBG |
| 62/M/RK/Patient 7 | Klebsiella; Pseudomonas | 84.2; 6.6 | DTS: Klebsiella pneumoniae; BB-SY: Klebsiella pneumoniae | None | Vancomycin; ceftazidime | Died* | Not performed because the patient had died |
| 62/M/LK/Patient 8 | Klebsiella; Pseudomonas; Escherichia | 56.1; 29.9; 7.7 | DTS: Klebsiella pneumoniae; BB-SY: NBG | None | Vancomycin; ceftazidime | Died* | Not performed because the patient had died |
| 61/F/RH/Patient 9 | Staphylococcus | 100 | DTS: Staphylococcus caprae (3/3); SY: Staphylococcus caprae | None | Vancomycin; ceftazidime; gentamicin | Infection-free | NBG |
| 63/M/RH/Patient 10 | Serratia | 99.1 | DTS: NBG | None | Vancomycin; ceftazidime | Infection-free | NBG |
| 40/M/LH/Patient 11 | Sphingomonas; Propionibacterium | 85.7; 8.9 | DTS: NBG (3/3) | Ciprofloxacin | Vancomycin; ceftazidime; gentamicin | Infection | DTS: Candida albicans; SY: Candida albicans; WD: Staphylococcus haemolyticus, Staphylococcus epidermidis |
From sepsis due to prosthetic joint infection
ALBC, antibiotic-loaded poly(methyl methacrylate) bone cement; RK, right knee; LK, left knee; DTS, deep tissue culture; SY, synovial fluid culture on BD BACTEC Plus Aerobic (Becton, Dickinson and Company (BD), Sparks, Maryland); PIOC, positive intraoperative cultures; BB-SY, synovial fluid culture on BD BACTEC Peds Plus; NBG, no bacterial growth; RH, right hip; LH, left hip; WD, wound tissue
Discussion
Current methods in PJI diagnosis
Bacterial cultures of SF and prosthetic tissue are the benchmark test for PJI diagnosis, however some bacteria are difficult to grow or are even uncultivable.19 In addition, cultures have high false-negative rates and are time consuming, as they might need one to two weeks for bacteria growth, especially in patients with low-grade infections.35,36 Targeted cloning and subsequent Sanger sequencing of 16S rDNA has also been used in the past decade, for example to identify disease-associated bacteria in clinical specimens, such as dorsal tongue and bronchoalveolar lavage fluid,37 enamel and dentin lesions,38 and pus from brain abscesses.39,40 Remarkably, this approach identified many bacteria that were not previously detected in these specimens, some of which were unculturable.37,39,40 Although this method generally overcomes the limitations of bacterial cultures, it is also time consuming and expensive because many bacteria, typically 46 to 125 per subject, have to be analyzed. In contrast, next-generation sequencing (NGS) and 16S metagenomics can now be used to characterize mixed infections and to identify infecting pathogens. The first commercial NGS platform was released in 2005, and metagenomics analysis of human and environmental microbiota has since flourished.41-43 However, its application in clinical diagnosis is not as well developed, although it has already been used to detect antimicrobial resistance genes in septic patients.44 Metagenomic analysis also detected varicella zoster virus in cerebrospinal fluids from patients with multiple sclerosis, even though the virus was never previously associated with the disease.45 We have now used 16S rRNA metagenomics to investigate bacteria in the SF of infected prosthetic joints. To this end, we have also developed a new protocol to assess pathogen composition and eliminate contaminating signals.
16S rRNA metagenomics as a potential method for PJI diagnosis
Based on our data, 16S rRNA metagenomics appears to be more sensitive than bacterial cultures in detecting pathogens at the genus and species level. It also reduces turnaround time from approximately one week (at least five days) for bacterial cultures to two days, where the procedure consists of one hour of DNA extraction, two hours of PCR, 40 hours of NGS, and four hours of bioinformatics analysis. Universal bacterial primers also detect very low-abundance pathogens, especially in patients who received antibiotics before surgery. In addition, 16S metagenomics detects polymicrobial infections and quantifies infecting pathogens based on true abundance, and not culturability, growth in culture, antibiotic resistance, and dominance. According to our experimental results (Fig. 1), the 16S rRNA-based method also tested some atypical pathogens in PJI such as Acinetobacter and Sphingomonas. Although we detected Acinetobacter in specimens from our patient, it is one of many bacterial infections and accounts for less than 4% of total bacteria. Therefore, we reasonably speculate that, in the case of multiple bacterial infections, presence of Acinetobacter is not common because earlier technology has not detected them. On the other hand, the sample from patient 11 did not show growth of bacteria, but instead growth of Candida albicans. Moreover, the 16S rRNA-based method identified Sphingomonas from the samples of this patient. Based on these results, we reasonably speculated that patient 11 may have both Sphingomonas and C. albicans infections. Taxonomic analysis of 16S reads following metagenomic sequencing is typically based on SILVA, Greengenes, and RDP.46 SILVA is a comprehensive database of bacterial rRNA genes and is the largest and most widely used of the three.46 Remarkably, we obtained similar results by matching patient data with SILVA, Greengenes, and NCBI databases,47 especially for the major genera detected.
To date, there is little literature on the diagnosis of PJI using NGS. We tried to compare the differences between a few research methods. Our experimental method uses 16S rRNA primer to amplify the bacterial gene, and then analyze bacterial infections by the Illumina HiSeq Sequencing platform. We also compared the sequencing results of the infections by the comparison of three databases (SILVA, Greengenes, and NCBI). Tarabichi et al29 combined the primer for the 16S rRNA gene and internal transcribed spacer gene to amplify bacterial and fungal genes simultaneously, and then analyze microorganism infections using the Ion Torrent PGM sequencing platform (Thermo Fisher Scientific, Waltham, Massachusetts). For Street et al’s48 strategy, after sonication fluid is collected, the Illumina MiSeq Sequencing platform analysis is performed directly without amplification of the PCR. Although the experimental design and analysis methods of each study are different, establishing a suitable NGS standard procedure for joint fluids in patients with PJI is a future method for PJI diagnosis.
Points to consider in 16S rRNA metagenomics
Nevertheless, some issues need to be resolved. First, 16S rRNA metagenomics is specific for bacteria, and will not detect fungi or viruses. Second, genus-level identification and quantification are generally more reliable than species-level identification. Indeed, although we detected four Gram-positive genera (Staphylococcus, Streptococcus, Propionibacterium, and Corynebacterium) and six Gram-negative genera (Escherichia, Klebsiella, Pseudomonas, Bacteroides, Serratia, and Sphingomonas), we identified only ten species, including Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus caprae, Streptococcus dysgalactiae, Escherichia coli, Klebsiella pneumoniae, Bacteroides fragilis, Serratia marcescens, Sphingomonas aquatilis, and Acinetobacter johnsonii. Third, 16S rRNA metagenomics is strongly susceptible to contamination from reagents and sample processing,49 which may generate false positives or false negatives.24,50 For instance, analysis of sonicated samples may be more sensitive than analysis of whole tissues,36,51 but additional procedures and reagents for sonication and inefficient DNA extraction also increase the risk of contamination.21,23,52 Indeed, removal of interference from contaminating DNA is a major challenge. Thus, all materials, reagents, and procedures should be strictly managed and standardized.49 Finally, we amplified 16S rDNA directly from SFs to minimize contamination from human DNA, which may account for > 90% of reads even if microbiome DNA is enriched prior to sequencing.48 Selection of a suitable cutoff value is also a serious issue. For example, low cutoff values such as 0.5% or 0.1% identify too many genera as being present, probably including spurious or irrelevant taxons. However, a strict cutoff value, such as 5%, may eliminate too many genera and prevent detection of mixed infections. In this study, our data indicate that if we were to select 5% as the cutoff value, the results would coincide with clinical observations of PJI. If we were to select 0.5% as the cutoff value, we would be able to identify a very low level of bacterial infection with species unculturable in bacterial culture. According to the comparison of 16S metagenomics and bacterial culture, 16S metagenomics could distinguish the genus when the specimens exhibit a very low level of bacterial infection. The future goals for application of 16S metagenomics to PJI diagnosis are not only to set a suitable cutoff value but also to establish a standardized protocol including specimen collection, DNA extraction, 16S PCR, NGS criteria setting, bioinformatic analysis, and final reports. NGS holds great promise in detecting pathogenic bacteria in clinical samples. We believe that this study highlights the potential of 16S metagenomics to diagnose PJIs, especially mixed infections. Understanding the bacterial composition of polymicrobial infection is of great benefit for the future development of novel antimicrobial metal orthopaedic implants.53-55 A comprehensive understanding of the composition of infectious bacteria can also provide appropriate protective measures for the surgeon before the surgical procedure is performed.56
Acknowledgments
We acknowledge the bioinformatics service provided by Dr. Yu-Lun Kuo (Bioinformatics Scientist, BIOTOOLS Co., Ltd., New Taipei City, Taiwan).
Footnotes
Author contributions: M-F. Chen: Designed the research, Acquired, analyzed and interpreted the data, Wrote the first draft of the manuscript, Revised the manuscript critically.
C-H. Chang: Designed the research, Acquired, analyzed and interpreted the data.
C. Chiang-Ni: Analyzed and interpreted the data.
P-H. Hsieh: Designed the research, Acquired the data.
H-N. Shih: Designed the research, Acquired the data.
S. W. N. Ueng: Designed the research, Acquired the data.
Y. Chang: Designed the research, Acquired, analyzed and interpreted the data, Wrote the first draft of the manuscript, Revised the manuscript critically.
Conflict of interest statement: Each author certifies that he or she has no commercial associations (e.g., consultancies, stock ownership, equity interest, patent/licensing arrangements, etc.) that might pose a conflict of interest in connection with the submitted article.
Ethical review statement: The study protocol was approved by our institutional review board (IRB number, 105-1046C), and was compliant with accepted ethical standards at Chang Gung Memorial Hospital. The written informed consent was obtained from all patients prior to their participation in the study. This study was carried out in accordance with the ethical standards in the 1964 Declaration of Helsinki.
Follow us @BoneJointRes
Funding statement
The authors have received, during the study period, grant funding support from the Ministry of Science and Technology, Taiwan (MOST 108-2320-B-182A-020 -MY3 and MOST 107-2314-B-182A-036 -MY3), and from Chang Gung Memorial Hospital (CRRPG3H0051, CRRPG3H0052, CRRPG3H0053, CMRPG3H1311, CMRPG3H1312, and CMRPG3H1313). The funders did not participate in study design, data collection, data interpretation and analysis, decision to publish, or preparation of the manuscript.
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
References
- 1. Kurtz S, Ong K, Lau E, Mowat F, Halpern M. Projections of primary and revision hip and knee arthroplasty in the United States from 2005 to 2030. J Bone Joint Surg [Am] 2007;89-A:780-785. [DOI] [PubMed] [Google Scholar]
- 2. Lenguerrand E, Whitehouse MR, Beswick AD, et al. Revision for prosthetic joint infection following hip arthroplasty: Evidence from the National Joint Registry. Bone Joint Res 2017;6:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Illingworth KD, Mihalko WM, Parvizi J, et al. How to minimize infection and thereby maximize patient outcomes in total joint arthroplasty: a multicenter approach: AAOS exhibit selection. J Bone Joint Surg [Am] 2013;95-A:e50. [DOI] [PubMed] [Google Scholar]
- 4. Hotham WE, Malviya A. A systematic review of surgical methods to restore articular cartilage in the hip. Bone Joint Res 2018;7:336-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hooper G. The challenge of the increasing demand for joint replacement. N Z Med J 2016;129:8-9. [PubMed] [Google Scholar]
- 6. Bozic KJ, Kurtz SM, Lau E, et al. The epidemiology of revision total knee arthroplasty in the United States. Clin Orthop Relat Res 2010;468:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bozic KJ, Kurtz SM, Lau E, et al. The epidemiology of revision total hip arthroplasty in the United States. J Bone Joint Surg [Am] 2009;91-A:128-133. [DOI] [PubMed] [Google Scholar]
- 8. Gwam CU, Mistry JB, Mohamed NS, et al. Current epidemiology of revision total hip arthroplasty in the United States: National Inpatient Sample 2009 to 2013. J Arthroplasty 2017;32:2088-2092. [DOI] [PubMed] [Google Scholar]
- 9. Peel TN, Buising KL, Choong PF. Diagnosis and management of prosthetic joint infection. Curr Opin Infect Dis 2012;25:670-676. [DOI] [PubMed] [Google Scholar]
- 10. Saleh A, George J, Faour M, Klika AK, Higuera CA. Serum biomarkers in periprosthetic joint infections. Bone Joint Res 2018;7:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morgenstern M, Vallejo A, McNally MA, et al. The effect of local antibiotic prophylaxis when treating open limb fractures: a systematic review and meta-analysis. Bone Joint Res 2018;7:447-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsang STJ, McHugh MP, Guerendiain D, et al. Underestimation of Staphylococcus aureus (MRSA and MSSA) carriage associated with standard culturing techniques: one third of carriers missed. Bone Joint Res 2018;7:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bejon P, Berendt A, Atkins BL, et al. Two-stage revision for prosthetic joint infection: predictors of outcome and the role of reimplantation microbiology. J Antimicrob Chemother 2010;65:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qu X, Zhai Z, Wu C, et al. Preoperative aspiration culture for preoperative diagnosis of infection in total hip or knee arthroplasty. J Clin Microbiol 2013;51:3830-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berbari EF, Marculescu C, Sia I, et al. Culture-negative prosthetic joint infection. Clin Infect Dis 2007;45:1113-1119. [DOI] [PubMed] [Google Scholar]
- 16. Shahi A, Deirmengian C, Higuera C, et al. Premature therapeutic antimicrobial treatments can compromise the diagnosis of late periprosthetic joint infection. Clin Orthop Relat Res 2015;473:2244-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yuenyongviwat V, Ingviya N, Pathaburee P, Tangtrakulwanich B. Inhibitory effects of vancomycin and fosfomycin on methicillin-resistant Staphylococcus aureus from antibiotic-impregnated articulating cement spacers. Bone Joint Res 2017;6:132-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Samara E, Moriarty TF, Decosterd LA, et al. Antibiotic stability over six weeks in aqueous solution at body temperature with and without heat treatment that mimics the curing of bone cement. Bone Joint Res 2017;6:296-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Osmon DR, Berbari EF, Berendt AR, et al. Diagnosis and management of prosthetic joint infection: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis 2013;56:e1-e25. [DOI] [PubMed] [Google Scholar]
- 20. Achermann Y, Vogt M, Leunig M, Wüst J, Trampuz A. Improved diagnosis of periprosthetic joint infection by multiplex PCR of sonication fluid from removed implants. J Clin Microbiol 2010;48:1208-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gomez E, Cazanave C, Cunningham SA, et al. Prosthetic joint infection diagnosis using broad-range PCR of biofilms dislodged from knee and hip arthroplasty surfaces using sonication. J Clin Microbiol 2012;50:3501-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Janz V, Schoon J, Morgenstern C, et al. Rapid detection of periprosthetic joint infection using a combination of 16s rDNA polymerase chain reaction and lateral flow immunoassay: a pilot study. Bone Joint Res 2018;7:12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cazanave C, Greenwood-Quaintance KE, Hanssen AD, et al. Rapid molecular microbiologic diagnosis of prosthetic joint infection. J Clin Microbiol 2013;51:2280-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fenollar F, Roux V, Stein A, Drancourt M, Raoult D. Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J Clin Microbiol 2006;44:1018-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goldberg B, Sichtig H, Geyer C, Ledeboer N, Weinstock GM. Making the leap from research laboratory to clinic: challenges and opportunities for next-generation sequencing in infectious disease diagnostics. MBio 2015;6:e01888-e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abayasekara LM, Perera J, Chandrasekharan V, et al. Detection of bacterial pathogens from clinical specimens using conventional microbial culture and 16S metagenomics: a comparative study. BMC Infect Dis 2017;17:631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Willner D, Low S, Steen JA, et al. Single clinical isolates from acute uncomplicated urinary tract infections are representative of dominant in situ populations. MBio 2014;5:e01064-e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xia LP, Bian LY, Xu M, et al. 16S rRNA gene sequencing is a non-culture method of defining the specific bacterial etiology of ventilator-associated pneumonia. Int J Clin Exp Med 2015;8:18560-18570. [PMC free article] [PubMed] [Google Scholar]
- 29. Tarabichi M, Shohat N, Goswami K, Parvizi J. Can next generation sequencing play a role in detecting pathogens in synovial fluid? Bone Joint J 2018;100-B:127-133. [DOI] [PubMed] [Google Scholar]
- 30. Parvizi J. New definition for periprosthetic joint infection. Am J Orthop (Belle Mead NJ) 2011;40:614-615. [PubMed] [Google Scholar]
- 31. Parvizi J, Zmistowski B, Berbari EF, et al. New definition for periprosthetic joint infection: from the Workgroup of the Musculoskeletal Infection Society. Clin Orthop Relat Res 2011;469:2992-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wheat LJ, Freifeld AG, Kleiman MB, et al. Clinical practice guidelines for the management of patients with histoplasmosis: 2007 update by the Infectious Diseases Society of America. Clin Infect Dis 2007;45:807-825. [DOI] [PubMed] [Google Scholar]
- 33. Klindworth A, Pruesse E, Schweer T, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Balvočiūtė M, Huson DH. SILVA, RDP, Greengenes, NCBI and OTT - how do these taxonomies compare? BMC Genomics 2017;18(Suppl 2):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peel TN, Dylla BL, Hughes JG, et al. Improved diagnosis of prosthetic joint infection by culturing periprosthetic tissue specimens in blood culture bottles. MBio 2016;7:e01776-e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Puchner SE, Döring K, Staats K, et al. Sonication culture improves microbiological diagnosis of modular megaprostheses. J Orthop Res 2017;35:1383-1387. [DOI] [PubMed] [Google Scholar]
- 37. Bahrani-Mougeot FK, Paster BJ, Coleman S, et al. Molecular analysis of oral and respiratory bacterial species associated with ventilator-associated pneumonia. J Clin Microbiol 2007;45:1588-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aas JA, Griffen AL, Dardis SR, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 2008;46:1407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Al Masalma M, Armougom F, Scheld WM, et al. The expansion of the microbiological spectrum of brain abscesses with use of multiple 16S ribosomal DNA sequencing. Clin Infect Dis 2009;48:1169-1178. [DOI] [PubMed] [Google Scholar]
- 40. Al Masalma M, Lonjon M, Richet H, et al. Metagenomic analysis of brain abscesses identifies specific bacterial associations. Clin Infect Dis 2012;54:202-210. [DOI] [PubMed] [Google Scholar]
- 41. Zhernakova A, Kurilshikov A, Bonder MJ, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016;352:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang H, Huang X, Fang S, et al. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci Rep 2016;6:27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hong X, Chen J, Liu L, et al. Metagenomic sequencing reveals the relationship between microbiota composition and quality of Chinese Rice Wine. Sci Rep 2016;6:26621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gyarmati P, Kjellander C, Aust C, et al. Metagenomic analysis of bloodstream infections in patients with acute leukemia and therapy-induced neutropenia. Sci Rep 2016;6:23532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perlejewski K, Bukowska-Ośko I, Nakamura S, et al. Metagenomic analysis of cerebrospinal fluid from patients with multiple sclerosis. Adv Exp Med Biol 2016;935:89-98. [DOI] [PubMed] [Google Scholar]
- 46. Yilmaz P, Parfrey LW, Yarza P, et al. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Nucleic Acids Res 2014;42(Database issue):D643-D648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Street TL, Sanderson ND, Atkins BL, et al. Molecular diagnosis of orthopedic-device-related infection directly from sonication fluid by metagenomic sequencing. J Clin Microbiol 2017;55:2334-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marín M, Garcia-Lechuz JM, Alonso P, et al. Role of universal 16S rRNA gene PCR and sequencing in diagnosis of prosthetic joint infection. J Clin Microbiol 2012;50:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trampuz A, Piper KE, Jacobson MJ, et al. Sonication of removed hip and knee prostheses for diagnosis of infection. N Engl J Med 2007;357:654-663. [DOI] [PubMed] [Google Scholar]
- 52. Hartley JC, Harris KA. Molecular techniques for diagnosing prosthetic joint infections. J Antimicrob Chemother 2014;69(Suppl 1):i21-i24. [DOI] [PubMed] [Google Scholar]
- 53. Itabashi T, Narita K, Ono A, et al. Bactericidal and antimicrobial effects of pure titanium and titanium alloy treated with short-term, low-energy UV irradiation. Bone Joint Res 2017;6:108-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou J, Zhou XG, Wang JW, Zhou H, Dong J. Treatment of osteomyelitis defects by a vancomycin-loaded gelatin/β-tricalcium phosphate composite scaffold. Bone Joint Res 2018;7:46-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martinez-Perez M, Perez-Jorge C, Lozano D, et al. Evaluation of bacterial adherence of clinical isolates of Staphylococcus sp. using a competitive model: an in vitro approach to the "race for the surface" theory. Bone Joint Res 2017;6:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheng T, Zhang XL, Hu JJ, Li B, Wang Q. The role of routine screening in blood-borne pathogens in Chinese patients undergoing joint arthroplasty. Bone Joint Res 2017;6:566-571. [DOI] [PMC free article] [PubMed] [Google Scholar]