Abstract
The application of next-generation sequencing (NGS) technology to the study of cancer genomes has been transformational. Not only has this technology revealed the genetic and epigenetic underpinnings of disease onset and progression, but also has redefined our clinical diagnosis and treatment paradigms. This rapid translation from discovery to clinical platform has occurred in the context of new pharmaceutical paradigms, enabling the use of NGS for the diagnosis and definition of therapeutic vulnerabilities of cancer. This review explores this transformation and identifies cutting-edge applications of NGS that will result in its additional utility in cancer care.
HISTORICAL UNDERPINNINGS OF CANCER GENOMICS
In 1902, even before the structure of DNA and its role in the cell was understood, Theodor Boveri postulated that a cancerous cell results from a scrambling of the chromosomes, leading to uncontrolled cell division (Boveri 2008). This hypothesis emerged from his studies of sea urchins, wherein he showed the need for all chromosomes to be present for proper embryonic development. Boveri further postulated that chromosomal abnormalities in cancer emerged as a result of radiation, physical or chemical insults, or microscopic pathogens. This early hypothesis about the role of the genome in cancer onset was supported by the work of T.H. Morgan, whose studies of Drosophila melanogaster showed that its chromosomes carried hereditary information, and further defined the concept of the gene as the fundamental unit of heredity (Morgan 1913).
As microscopy improved and techniques were developed for imaging-condensed chromosomes under the microscope, the ability to produce karyotypic evaluation of human chromosomes emerged. From these basic techniques, the work of Hungerford, Nowell, and Rowley (National Academy of Sciences 1960; Rudkin et al. 1964; Rowley 1973) showed that specific, aberrant chromosomes were always identified when examining chromosomes from patients with specific leukemia diagnoses. These included the Philadelphia chromosome (t9;22) of chronic myeloid leukemia (CML) and the characteristic translocation (t15;17) of acute promyelocytic leukemia (APL) (Rowley et al. 1977). The association of these cancer-specific chromosomes with the onset of leukemia was furthered by the advent of molecular cloning and DNA sequencing techniques in the late 1970s, which ultimately showed that chromosomal translocations resulted in gene fusions. In CML, the t9;22 led to the fusion of BCR and ABL genes (Shtivelman et al. 1986), and in APL, the t15;17 led to the fusion of PML with RARa (Goddard et al. 1991). Additional studies showed that both fusion proteins were necessary and sufficient drivers of oncogenesis, which ultimately helped to define therapeutic approaches to address the novel action of each fusion protein (Lugo et al. 1990; Fenaux et al. 1999). In essence, these early cancer genomics studies established a paradigm by which examining the chromosomes for the presence of known alterations that inferred specific cancer drivers both provided a means of diagnosis and a means of identifying specific therapeutics to alleviate cancer progression. Increasing knowledge of chromosomal-scale cancer drivers thereby ushered in the era of known cancer drug–gene interactions referred to as “targeted therapy.”
Although microscopic evaluation of cancer cells and chromosomes remains the standard of pathology today, there have been a variety of different technologies developed to aid in cancer diagnosis and the identification of alterations that drive cancer onset at increasing resolution. One microscopic technique that added resolution to the evaluation of chromosomes was fluorescent in situ hybridization (FISH) (Langer-Safer et al. 1982). This approach combines specific fluorescent probes that define juxtaposed loci, resulting from their involvement in either a chromosomal translocation or inversion with cultured cancer cells from a patient biopsy. Examination of the hybridized probes under the fluorescent microscope permits diagnostic confirmation of the specific alteration based on proximity of labeled probes at the specific chromosomal loci involved in the alteration. In some cancers, FISH confirmation informs therapeutic decision-making. As efforts in the 1990s helped to define the Human Genome Project by producing clone-based “maps” of each chromosome, techniques were developed to produce spotted arrays of isolated DNA from these clones onto treated glass slides. By labeling tumor and normal DNA from an individual patient with different fluors and hybridizing the mixed, labeled DNAs onto these cloned DNA “microarrays” followed by laser-based scanning, images of differential labeling intensity could be obtained that identified chromosomal losses, gains, and other structural changes, all at much higher resolution than by microscopic analysis (Kallioniemi et al. 1992; du Manoir et al. 1993).
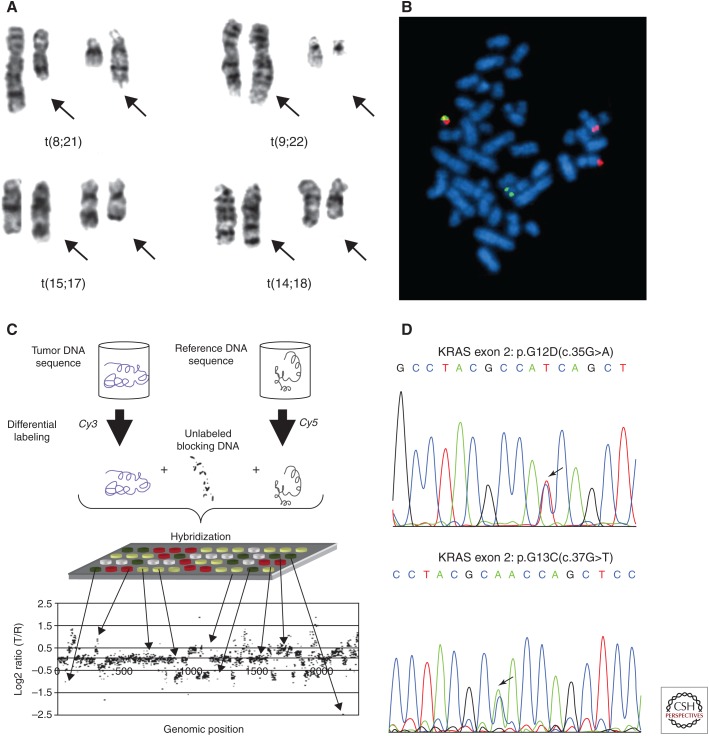
The completion of the Human Genome Project in 2004 (International Human Genome Sequencing Consortium 2004) introduced a template on which known cancer genes could be localized in silico and their sequences defined by computational matching. Using the technique of polymerase chain reaction (PCR) developed by Faloona, Mullis, and colleagues (Saiki et al. 1985), the exons of known cancer genes could be amplified from DNA extracts of the tumor material, and their sequences determined by fluorescent Sanger-sequencing instruments. Early efforts of this type were limited to the evaluation of small numbers of genes because of the limited amount of DNA available from a given cancer sample and the lack of scalability of the approach. Importantly, these early efforts helped to define new drug–gene interactions such as the mutations in EGFR that predicted response to new targeted therapies known as tyrosine kinase inhibitors being tested in the treatment of lung adenocarcinoma (Lynch et al. 2004; Paez 2004; Pao et al. 2004). Figure 1 shows the techniques that were developed and used to achieve increasing resolution on cancer-related alterations in DNA.
Figure 1.
Techniques developed to detect cancer-related alterations of genomic DNA at increasing resolution. (A) Chromosomal metaphase analysis identifies driver translocations that result in fusion proteins. (From Rowley 2008; reprinted, with permission, from the American Society of Hematology © 2008.) (B) Fluorescence in situ hybridization (FISH) identifies a t(9;22) translocation. (Reprinted from Wikimedia Commons under the terms of the GNU Free Documentation License.) (C) Microarray-based comparative genome hybridization identifies chromosomal copy number alterations in tumor DNA. (Figure reprinted from Alqallaf and Hajjiah 2011 under the terms of the Creative Commons Attribution-Non-Commercial-ShareAlike-3.p License.) (D) Polymerase chain reaction (PCR) amplification and Sanger sequencing identifies heterozygous KRAS driver mutations. (Reprinted from Gao et al. 2016a in accordance with the Creative Commons Attribution Non-Commercial (CC BY-NC 4.0) License.)
Over time, Vogelstein's group scaled this PCR amplification and Sanger-sequencing approach to address nearly every known human gene and identify point mutations, as described in a landmark manuscript that first described the colorectal and breast cancer genome “landscapes” (Sjöblom et al. 2006). Although admittedly limited in sample numbers studied, this seminal work defined the notion of characterizing all the mutations in a cancer genome to identify cancer “drivers” and thereby identify points of therapeutic vulnerability (Papadopoulos et al. 2006). Subsequently, the three large-scale U.S.-based genome centers funded by the National Human Genome Research Institute collaborated with a group of lung cancer oncologists and pathologists to combine microarray-based chromosomal copy number analysis with mutations identified by combined PCR and Sanger sequencing of genes in known cancer pathways to characterize the somatic landscapes of a cohort of 188 human lung adenocarcinomas (Weir et al. 2007; Ding et al. 2008). Whereas these studies were impactful, it was clear that this approach overall was not scalable because of the need for large amounts of tumor tissue that would yield sufficient DNA to assay, and the insensitivity of Sanger sequencing to detect low-level mutations (those present in only a fraction of the cancer cells) or to survey tumors with a large proportion of normal cells intermingled. Finally, because pseudogenes and other challenges related to homology in gene families did not permit the design of uniquely amplifying primer pairs, the ability to PCR amplify specific loci with high fidelity was simply not possible, thereby eliminating the inclusion of these genes into variant discovery.
NEXT-GENERATION SEQUENCING OF CANCER GENOMES
These challenges were largely addressed by the development of new sequencing technologies that eliminated the conventional PCR amplification steps required to produce templates for Sanger sequencing, thereby reducing the number of preparatory steps and the amount of DNA needed to characterize each tumor genome. Collectively referred to as next-generation sequencing (NGS), these technologies permitted the construction of a sequencing library using only a few discrete steps (Mardis 2011). Each library construction produced DNA fragments from a genome of interest with common adapter sequences ligated at each fragment end, which were suitable for universal priming and surface-borne amplification, followed by sequencing in situ. In other words, next-generation instruments were capable of sequencing millions of whole-genome fragments at once using a stepwise approach of nucleotide addition and detection.
This “massively parallel” approach to sequence data generation dramatically decreased the time and cost of data generation at a steady pace from its introduction in 2005 until the present day (Mardis 2017). In spite of these significant technology advances, cancer samples present a unique set of challenges that must be taken into consideration to produce the optimal data set from NGS methods (Mardis 2015). One challenge arises from the fact that any solid tumor sample consists of a mixture of tumor and normal cells in which the normal cells include blood vessels, stromal cells, and infiltrating immune cells. Ideally for sequencing studies, the majority of cells in the cancer sample should be tumor cells, so they contribute the majority of the genomic DNA or RNA into an extraction. If not, there are physical methods that can enhance the tumor cell content into extraction, including laser capture microdissection or flow sorting. Another challenge is contributed by preservation methods like formalin fixation and paraffin embedding, which have been used for more than 100 years by pathology to preserve the cellular architecture for microscopic examination of the tissue. However, formalin cross-links the phosphodiester backbone of DNA and RNA resulting in fragmentation over time. Paraffin at temperatures above 60°C also can degrade RNA during tissue embedding. Cumulatively, this preservation method requires extensive quality checking of extracted DNA and RNA before NGS-based assay (Arreaza et al. 2016). Tissue availability for NGS studies is another limitation that impacts discovery and clinical testing, and will be discussed later. In principle, most cancer samples have a set of assays in standard-of-care anatomical and molecular pathology that must be completed for diagnosis. If a resection sample is limited in size, or if one or more core biopsy samples constitute the patient material available for assay, there may be minimal tissue available for nucleic acid extraction and NGS assay. As such, methods to construct NGS libraries from very low input DNA have been developed to compensate for this reality (Picelli et al. 2014a).
The first demonstration of the use of NGS for sequencing and comparing the whole-genome sequencing (WGS) of a matched tumor (an acute myeloid leukemia [AML]) and normal (skin punch) was published in 2008 (Ley et al. 2008). This pioneering work showed that accurate interpretation of somatic mutations was ideally performed in comparison with a matched normal tissue from the same patient as a result of the large numbers of variants detected. Indeed, all large-scale cancer genomics discovery efforts have used this paradigm, wherein solid tumors are typically compared with DNA from peripheral blood monocytes (PBMCs) readily obtained from whole blood, and DNA from hematologic malignancies (e.g., leukemias and lymphomas) are compared with DNA isolates from skin punch, buccal swab, or sputum samples. Importantly, these early efforts predominately used the normal DNA in a subtractive approach, whereas later analyses focused on defining the germline or constitutional DNA contribution to cancer susceptibility by studying the normal DNA as a stand-alone analysis (Zhang et al. 2015; Huang et al. 2018; Wang et al. 2018). The importance of normal comparator DNA will be discussed later in a section focused on NGS-based clinical assays of cancer samples.
Although the initial NGS devices only permitted sequencing of whole genomes (which was initially cost-prohibitive for human genomes), the development of clever preparatory methods enabled the selection of exons of known genes from a whole-genome NGS library (Gnirke et al. 2009; Hodges et al. 2009; Bainbridge et al. 2010). Collectively known as “hybrid capture” methods, these approaches used synthetic DNA or RNA probes that were complementary to known coding exon sequences and were derivatized with biotin. When combined at an excess concentration with a whole-genome NGS library under appropriate conditions, the biotinylated “bait” sequences drive the formation of hybrids with complementary fragments from the WGS library. On mixing the hybrid capture reaction with streptavidin-linked magnetic beads, the probe-selected fragments are removed from solution by virtue of the biotin–streptavidin binding of the probes. Subsequent steps to denature the bead-isolated hybrids release the selected “exome” library fragments in a form suitable for amplification and sequencing. In effect, hybrid capture methods permit either a focus on selected genes, or an unbiased evaluation of all known genes. In either case, interpreting the impact of the altered nucleotide sequence on the final protein sequence in the tumor (“somatic mutations”) is reasonably straightforward with annotation software like VEP (McLaren et al. 2016) or Annovar (Wang et al. 2010). More challenging with the breadth of the NGS survey of many genes, however, is the interpretation of how somatic variants change the function of the protein. Absent high-throughput methods to evaluate this question of functional impact, most cancer genomics studies instead accumulate information about the frequency of somatic alterations in genes as a means of inferring their importance in oncogenesis.
Beyond the use of NGS to decode cancer exomes and genomes, preparatory library construction methods were devised to sequence RNA using NGS platforms as well. So-called “RNA-seq” (RNA-sequencing) methods provided a means of evaluating not only the downstream expression of mutations identified from DNA, but also provided a readout of altered methylation, chromatin packaging, and other epigenomic mechanisms that were known to be altered in cancer cells. Later methodological developments have yielded NGS-based methods for evaluating methylation (methyl-seq) (Kernaleguen et al. 2018) and chromatin packaging (Giresi et al. 2007; Thurman et al. 2012; Buenrostro et al. 2013) of genomic DNA, although accurate interpretation of these data is optimal when compared with that from adjacent, nonmalignant tissue. Taken together, the increasing use and utility of NGS to characterize cancer genomes and transcriptomes, coupled with innovative bioinformatic methods to interpret the data, ushered in a decade of cancer genomics discovery that has radically changed our understanding of cancer as a disease of the genome.
OVERVIEW OF LARGE-SCALE DISCOVERY EFFORTS
Initial NGS-based cancer genomics studies, similar to the first NGS whole-genome study mentioned above, focused on individual patient samples in the context of WGS. This singular focus was largely a result of the high cost of NGS coverage for the whole-genome approach, which was necessary as hybrid capture had yet to reach widespread usage. In spite of the case study nature of these reports, however, they were impactful by emphasizing the power of unbiased inquiry made possible by NGS. For example, the second whole-genome NGS cancer study of a second AML patient (Mardis et al. 2009) identified a mutation in the TCA cycle gene IDH1, which encodes isocitrate dehydrogenase. PCR-based examination of this gene in a cohort of AML patients (n = 188) showed the prevalence of this mutation and identified its putative prognostic value. Another important cancer WGS study published the same year examined the genomic comparison of a primary lobular breast tumor and its metastatic lesion, which emerged 9 years later in the same patient (Shah et al. 2009), demonstrating the shared genomic landscape of the two cancers, including detection of structural variants in the cancer genomes. This pioneering study also evaluated tumor RNAs from both primary and metastasis, and showed their potential to reveal RNA editing as a mutational mechanism. Taken cumulatively, the aforementioned PCR/Sanger-sequencing studies and initial NGS-based reports generated enthusiasm in the biomedical research community for the potential of an unbiased NGS-based discovery to characterize the genomic underpinnings of cancers at nucleotide resolution, using multiple analytes and studying cancers from different tissue sites.
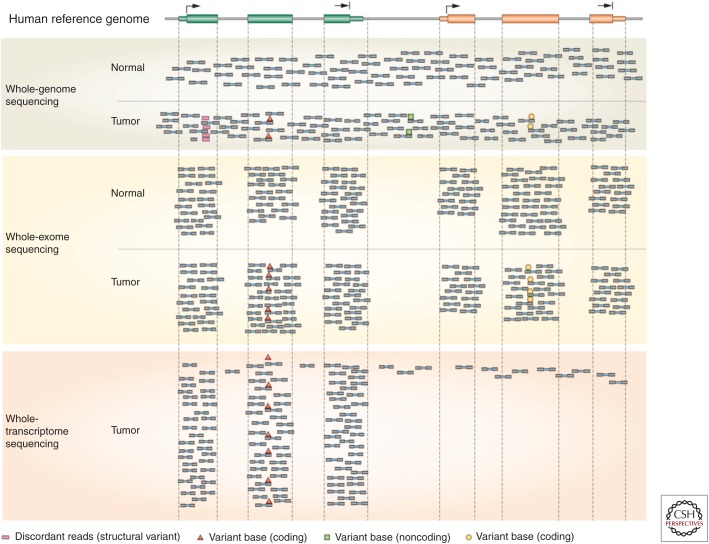
As a result of this momentum, international large-scale cancer genomics discovery efforts using NGS-based methods and computational analyses have been underway during the past decade. The largest studies have been government-funded consortia like The Cancer Genome Atlas (TCGA) (cancergenome.nih.gov), funded jointly by the National Cancer Institute and National Human Genome Research Institute of the National Institutes of Health and the International Cancer Genome Consortium (ICGC) (icgc.or), a suite of large-scale studies in different countries worldwide that were funded by the governments of each country. Most of these studies were conducted by large-scale genome centers with the unique capability to achieve economies of scale and the infrastructure necessary to produce and analyze data from hundreds of tumor/normal paired samples in a rapid and cost-effective manner. Our enhanced understanding of cancer genomics from these studies has been transformational, but of equal importance, the innovative approaches to NGS data analysis needed to make sense of these large data sets have been critical to achieve this new knowledge. Although the cumulative breadth of these computational analyses is beyond the scope of this article, the success of a large-scale cancer genomics-based discovery absolutely required innovative approaches for algorithmic examination of data, permitting multiple types of variation to be gleaned from the same set of NGS data (Fig. 2). For example, the ability to identify single-point mutations, insertion/deletion mutations, and structural alterations (copy number changes, translocations, inversions), as well as loss of heterozygosity and aneuploidy analyses all can be obtained from a whole-genome data set comparing tumor and normal DNA, by the application of different algorithmic treatments. As mentioned earlier, the analysis of the normal DNA also can reveal inherited or de novo cancer susceptibility alterations. Similarly, RNA-seq data analysis can produce information about gene expression levels, alternative splicing, allelic silencing or differential allelic expression, fusion gene evidence, and RNA editing. More recently, data analysis approaches that integrate DNA and RNA NGS data are emerging (Vandin et al. 2012; Hou and Ma 2014), providing a unique view of cancer biology at the pathway or network level, as well as immunogenomic classifications that characterize the neoantigen landscape of the tumor (Hundal et al. 2016; O'Donnell et al. 2018) as well as the immune microenvironment (Liu et al. 2017). These capabilities further emphasize the importance of multianalyte studies as opposed to solely DNA-based characterizations for obtaining a more complete picture of the biological programs in play for individual tumors, which may, in turn, better inform clinical therapeutic decision-making.
Figure 2.
Aligned next-generation sequencing (NGS) read coverage from whole-genome, whole-exome, and whole-transcriptome (RNA-seq) data. This figure shows the alignment of paired-end NGS sequencing reads along a stretch of the human genome containing two genes. Several scenarios are shown, including tumor-unique (somatic) single-nucleotide variant identification (red triangles) in a gene, concordant between whole-genome, exome, and RNA-seq data; discordant paired-end read mapping (red squares) in tumor whole-genome data, indicative of a somatic structural alteration (translocation or inversion); a single-base alteration (green squares) found in noncoding DNA by whole-genome sequencing; and a somatic single-nucleotide variant (yellow circles) found in a gene that is not expressed in the tumor. (From Mardis 2012; reprinted, with permission, from Nature Reviews Gastroenterology and Hepatology © 2012.)
As important as large-scale cancer genomics has been to advancing our knowledge of how genomic changes lead to cancer onset, that knowledge is still incomplete. For example, most large-scale discovery projects used exome sequencing instead of WGS because of cost considerations and relative ease of analysis. As a result, noncoding variation is poorly understood, as is information about recurrent complex structural variations such as inter- and intrachromosomal inversions and translocations. A recent set of papers has been published that provides cumulative analysis of WGS data from cancers sequenced by TCGA and ICGC projects, shedding some new light on these types of analyses and results for around 2200 cancers across many different tissue sites (www.cell.com/pb-assets/consortium/pancanceratlas/pancani3/index.html). Also, because of the frequency with which primary solid tumors are resected by surgery and banked, these studies focused almost entirely on primary disease samples. In contrast, most patients die of metastatic disease, which is infrequently biopsied and even less frequently removed by surgery, except in the few tumor types in which surgery to resect metastases is potentially curative (ovarian, liver metastases from recurrent colorectal cancer, and central nervous system [CNS] cancers). Hence, our understanding of the genomic landscape of metastatic disease remains limited. One recent large-scale genomic study of metastatic cancers, which characterized multiple analytes and included integrated data analysis, showed that the active biological pathways in metastatic disease may be quite concordant regardless of the primary tissue site (Robinson et al. 2017). However, more work remains to better characterize the transition from primary to metastatic disease in the context of different therapeutic modalities, including chemotherapy and radiation therapy as well as targeted therapies. This information can prove quite valuable in the context of understanding the types of acquired resistance mechanisms that can arise during the use of targeted therapies (Juric et al. 2015), or how chemotherapy can influence the prevalence of different clonal populations (Ding et al. 2012). In the former case, knowing the resistance mechanism(s) to a given therapy can inform monitoring of the patient by techniques known collectively as “liquid biopsy,” as discussed later in this article and elsewhere in this collection.
CLINICAL APPLICATION OF CANCER GENOMICS
Coincident with the emerging knowledge of somatic alterations, drug development efforts have focused on the screening of compounds, as well as novel synthetic efforts, to identify inhibitory molecules that can interact with the resulting protein drivers. As these compounds have been tested in clinical trials and received FDA approval, genomics-based assays have provided a diagnostic to identify additional patients who might benefit from these targeted therapies. In particular, because cancer genomics discovery has taught us that mutations in cancer genes are not tissue-type-specific, and multiple different driver genes/mutations may be active in any cancer, developing NGS tests for multiple genes that can be applied to cancers from different tissue sites is logical, and permits a judicious use of tumor tissue that is often in short supply, as discussed above. Several large studies have been published that apply this paradigm of using broad NGS assays to identify therapeutic vulnerabilities to adult (Roychowdhury et al. 2011; Zehir et al. 2017) and pediatric (Mody et al. 2015; Oberg et al. 2016) patients. Although clinically effective, the challenge of NGS-based clinical testing is that often variants are identified that have not been previously characterized with respect to their likelihood to (1) drive the cancer onset, or (2) respond to indicated therapies, leaving the question of response to therapy unknown. Another challenge that relates to the shared nature of cancer driver genes/mutations is that FDA approval of drugs, historically, has been tissue-site-specific, and whereas an assay indicates a positive indication of therapeutic vulnerability, if the therapy is not FDA approved for that tissue site, it is considered “off label” and typically is not covered by insurance. This is an increasingly common problem that is now being somewhat addressed by new clinical trial designs such as “basket” or “umbrella” trials, which enroll patients with the same vulnerability across multiple tumor sites for a defined set of genomic drivers (basket) and a corresponding set of targeted therapies (umbrella) (Berger and Mardis 2018).
Another clinical impact from large-scale cancer genomics discovery projects has been an improved estimate of the genetic susceptibility to cancer, which now can be identified in about 10%–12% of individuals who go on to develop cancer. This level of germline contribution to risk was obtained from an NGS assay of tens of thousands of cancer patients, both adult and pediatric, many without a family history of cancer that might suggest the role of genetic susceptibility. In particular, the clinical impact of this finding has changed practice in two ways. First, clinical NGS assays that sequence known cancer genes to identify genetic susceptibility now are ordered for patients who present with cancer at a young age (typically before age 40) as a standard of care, regardless of family history. Second, the presence of pathogenic germline mutations in BRCA1/2 genes has been shown to predict improved outcomes for patients treated with the combination of chemotherapy and a PARP inhibitor therapy in the setting of ovarian cancer (Matulonis 2018). Furthermore, microsatellite instability, which can be identified either in the germline or cancer DNA, leads to increased mutational burden and has been shown to predict patients’ likely response to checkpoint blockade inhibitors, one class of immunotherapies (Le et al. 2015; Bever and Le 2017), as is now FDA approved. These findings collectively make an argument for the NGS assay of germline, as well as cancer DNA, to inform therapeutic decision-making.
One more recent clinical application of cancer genomics that stems from cancer genomics and the outcomes of patients receiving checkpoint blockade inhibitor therapy is the predictive value of estimating the tumor mutational burden (TMB). Several clinical trials have now shown a correlation between response to checkpoint blockade immunotherapy and high TMB, evaluated retrospectively (Devarakonda et al. 2018; Fabrizio et al. 2018). The concept of using NGS-based TMB to predict response is controversial, however. This controversy arises from multiple areas that lack standardization across NGS assays in terms of (1) the numbers and types of genes surveyed, (2) the depth of NGS data read depth or “coverage” obtained, (3) the algorithmic mutation caller(s) used to identify variants, and (4) whether the TMB cut point has been evaluated with sufficiently robust data from samples with therapy-related outcomes, and/or has been confirmed retrospectively with multiple data sets. As a result, NGS-based estimates of TMB are not yet being used as a defined clinical parameter to determine whether patients should receive checkpoint blockade inhibitors.
CUTTING-EDGE APPLICATIONS IN CANCER GENOMICS
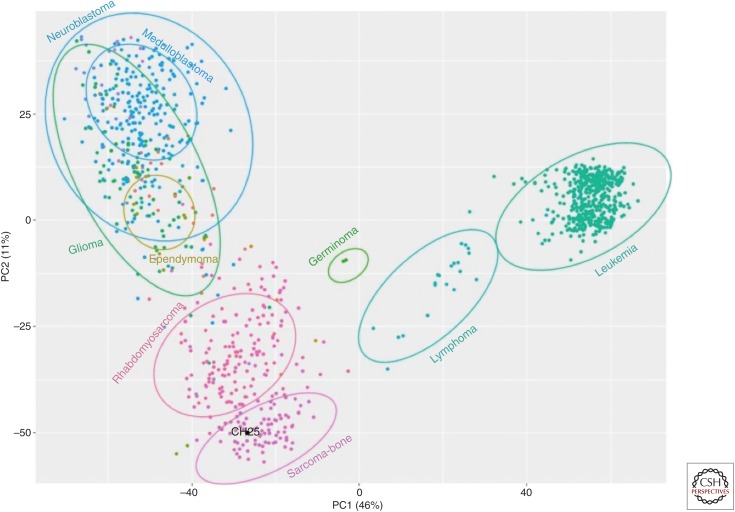
The application of NGS methods to the study of cancers has evolved rapidly, as have myriad computational approaches to the data. Several current applications are worth mentioning as they are bringing increasingly important information about cancer cell heterogeneity and biology to our knowledge of the disease. One such application involves the use of emerging single-cell preparatory methods to study cancer cell genomics from DNA sequencing data and cancer biology from RNA-seq data. Methods have been developed that focus either on amplified genomic DNA (Gao et al. 2016b; Kim et al. 2018) or 3′-based sampling of RNA isolates (Puram et al. 2017; Filbin et al. 2018) from tumors treated to produce single cells or single nuclei (Picelli et al. 2014b). Analytical methods that address these data sets are aimed at structural variant detection and mutational analysis from DNA of tumor single cells as well as the evaluation of intratumoral heterogeneity (De Mattos-Arruda and Caldas 2015; Sumanasuriya et al. 2017). From RNA-seq data, comparative gene expression evaluation typically involves some form of principal components analysis (PCA) to identify cells with similar expression profiles, comparative pathway analysis among the different groups of cells obtained by PCA, immune cell content and typing-based evaluation of the tumor microenvironment, and others (Fig. 3; Brummelman et al. 2018; De Simone et al. 2018; Zhu et al. 2018). Generally speaking, these types of evaluations, whether they are from DNA or RNA data (or both), have provided new information from studies of recurrent tumors (primary and metastasis from the same patient), in some cases with a known therapy interval between the two tumors.
Figure 3.
Illustration of a principal components analysis (PCA) plot showing patient-specific RNA-seq-based clustering. In this example, RNA-seq data from a cancer patient (black dot) is grouped according to principal components–based clustering relative to RNA-seq data from other cancer patients. Dots are colored and clustered according to pathology-based diagnosis of cancer type.
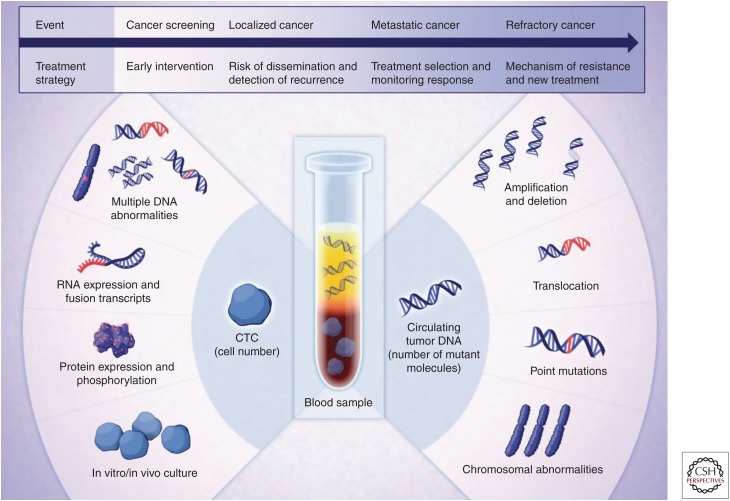
The interval between primary and recurrent cancer has traditionally been monitored by imaging-based evaluation of patients at periodic intervals. In the genomics era, and especially with increasing numbers of cancer driver mutations now known and/or characterized from an individual patient's cancer sample, new methods for blood or other bodily fluids (cerebral spinal fluid, urine) to be assayed are emerging. As a result of the release of cell contents during apoptosis, all cells in the body can release DNA into the circulation or adjacent bodily fluid as so-called “circulating free DNA” or cfDNA. Also, cancers infrequently shed cells into the circulation, known as circulating tumor cells (CTCs) (Fig. 4). The technique of sampling bodily fluids for the presence of CTCs, or specific DNA sequences released from cancer cells is broadly referred to as “liquid biopsy,” and some evidence exists to show that these methods can provide information about recurrent cancer in advance of imaging-based modalities. Although these initial data are encouraging regarding the clinical application of cfDNA monitoring (Sakaeva et al. 2016; Taylor et al. 2016; Garcia-Murillas and Turner 2018; Khan et al. 2018; Mansukhani et al. 2018), it also has been shown that more advanced-stage cancers release more DNA into the circulation than do early-stage cancers. Nevertheless, as the approaches to monitor cfDNA increase in sensitivity, early detection may be possible (Taylor et al. 2016) and is the source of significant commercial development efforts.
Figure 4.
Liquid biopsy can be used to evaluate circulating tumor cells (CTCs) or circulating free DNA (cfDNA). This figure shows the types of analyses that can be used to characterize CTCs (left) or cfDNA (right) as a means of monitoring patients with known genomic alterations in their cancer cell DNA. (From Haber and Velculescu 2014; reprinted, with permission, from the American Association for Cancer Research © 2014.)
CONCLUSIONS
Cancer genomics has changed our understanding of how modifications to DNA at the genomic and epigenomic levels can lead to uncontrolled cell division and drive neoplasia in most tissue sites in which cancers have been diagnosed. Extending our understanding of these changes as they impact cancer biology has been enabled by sequencing of various classes of RNA in the cancer cell, integration of these data with identified DNA alterations, and abstraction of this information to the cellular pathways that drive cancer when they are dysregulated. No doubt, our knowledge of the cancer genome will continue to grow, and the associated data will be mined with new tools and approaches of increasing sophistication. Translating the methods and analyses to the clinic already is benefitting cancer patients by identifying the therapeutic vulnerabilities in their cancer and permitting a more genome-guided approach to treatment. Although this is being predominantly applied in the metastatic setting, there are targeted therapies now approved for first line of treatment and increasing reports of the clinical benefit obtained by tumor DNA analysis by NGS that will drive insurance reimbursement over time. This review sets the stage for numerous other works in the literature that outline specific approaches to the clinical translation of cancer genomics in greater detail.
Footnotes
Editors: W. Richard McCombie, Elaine R. Mardis, James A. Knowles, and John D. McPherson
Additional Perspectives on Next-Generation Sequencing in Medicine available at www.perspectivesinmedicine.org
REFERENCES
- Alqallaf A, Hajjiah A. 2011. Identifying variations within unstable regions of the genome reveal autism associated patterns. In Autism—A neurodevelopmental journey from genes to behaviour: IntechOpen, London, UK. [Google Scholar]
- Arreaza G, Qiu P, Pang L, Albright A, Hong LZ, Marton MJ, Levitan D. 2016. Pre-analytical considerations for successful next-generation sequencing (NGS): Challenges and opportunities for formalin-fixed and paraffin-embedded tumor tissue (FFPE) samples. Int J Mol Sci 17: 1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge MN, Wang M, Burgess DL, Kovar C, Rodesch MJ, D'Ascenzo M, Kitzman J, Wu YQ, Newsham I, Richmond TA, et al. 2010. Whole exome capture in solution with 3 Gbp of data. Genome Biol 11: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Mardis ER. 2018. The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol 15: 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever KM, Le DT. 2017. An expanding role for immunotherapy in colorectal cancer. J Natl Compr Canc Netw 15: 401–410. [DOI] [PubMed] [Google Scholar]
- Boveri T. 2008. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci 121: 1–84. [DOI] [PubMed] [Google Scholar]
- Brummelman J, Mazza EMC, Alvisi G, Colombo FS, Grilli A, Mikulak J, Mavilio D, Alloisio M, Ferrari F, Lopci E, et al. 2018. High-dimensional single cell analysis identifies stem-like cytotoxic CD8T cells infiltrating human tumors. J Exp Med 215: 2520–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mattos-Arruda L, Caldas C. 2015. Cell-free circulating tumour DNA as a liquid biopsy in breast cancer. Mol Oncol 10: 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone M, Rossetti G, Pagani M. 2018. Single cell T cell receptor sequencing: Techniques and future challenges. Front Immunol 9: 1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarakonda S, Rotolo F, Tsao MS, Lanc I, Brambilla E, Massod A, Olaussen KA, Fulton R, Sakashita S, McLeer-Florin A, et al. 2018. Tumor mutation burden as a biomarker in resected non-small-cell lung cancer. J Clin Oncol 36: 2995–3006. 10.1200/JCO.2018.78.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. 2008. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Manoir S, Speicher MR, Joos S, Schröck E, Popp S, Döhner H, Kovacs G, Robert-Nicoud M, Lichter P, Cremer T. 1993. Detection of complete and partial chromosome gains and losses by comparative genomic in situ hybridization. Hum Genet 90: 590–610. [DOI] [PubMed] [Google Scholar]
- Fabrizio DA, George TJ Jr, Dunne RF, Frampton G, Sun J, Gowen K, Kennedy M, Greenbowe J, Schrock AB, Hezel AF, et al. 2018. Beyond microsatellite testing: Assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition. J Gastrointest Oncol 9: 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenaux P, Chastang C, Chevret S, Sanz M, Dombret H, Archimbaud E, Fey M, Rayon C, Huguet F, Sotto JJ, et al. 1999. A randomized comparison of all transretinoic acid (ATRA) followed by chemotherapy and ATRA plus chemotherapy and the role of maintenance therapy in newly diagnosed acute promyelocytic leukemia. The European APL Group. Blood 94: 1192–1200. [PubMed] [Google Scholar]
- Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, Neftel C, Frank N, Pelton K, Herbert CM, et al. 2018. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360: 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wu H, Wang L, Zhang H, Duan H, Lu J, Liang Z. 2016a. Validation of targeted next-generation sequencing for RAS mutation detection in FFPE colorectal cancer tissues: Comparison with Sanger sequencing and ARMS-Scorpion real-time PCR. BMJ Open 6: e009532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, Tsai PC, Casasent A, Waters J, Zhang H, et al. 2016b. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet 48: 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Murillas I, Turner NC. 2018. Assessing HER2 amplification in plasma cfDNA. In Methods in molecular biology, pp. 161–172. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. 2007. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard A, Borrow J, Freemont P, Solomon E. 1991. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science 254: 1371–1374. [DOI] [PubMed] [Google Scholar]
- Haber DA, Velculescu VE. 2014. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov 4: 650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Rooks M, Xuan Z, Bhattacharjee A, Benjamin Gordon D, Brizuela L, Richard McCombie W, Hannon GJ. 2009. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat Protoc 4: 960–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou JP, Ma J. 2014. DawnRank: Discovering personalized driver genes in cancer. Genome Med 6: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, et al. 2018. Pathogenic germline variants in 10,389 adult cancers. Cell 173: 355–370.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, Griffith M. 2016. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med 8: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945. [DOI] [PubMed] [Google Scholar]
- Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, et al. 2015. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature 518: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. 1992. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258: 818–821. [DOI] [PubMed] [Google Scholar]
- Kernaleguen M, Daviaud C, Shen Y, Bonnet E, Renault V, Deleuze JF, Mauger F, Tost J. 2018. Whole-genome bisulfite sequencing for the analysis of genome-wide DNA methylation and hydroxymethylation patterns at single-nucleotide resolution. Methods Mol Biol 1767: 311–349. [DOI] [PubMed] [Google Scholar]
- Khan KH, Cunningham D, Werner B, Vlachogiannis G, Spiteri I, Heide T, Mateos JF, Vatsiou A, Lampis A, Damavandi MD, et al. 2018. Longitudinal liquid biopsy and mathematical modeling of clonal evolution forecast time to treatment failure in the PROSPECT-C phase II colorectal cancer clinical crial. Cancer Discov 8: 1270–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, Crosetto N, Foukakis T, Navin NE. 2018. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 173: 879–893.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer-Safer PR, Levine M, Ward DC. 1982. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci 79: 4381–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. 2015. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372: 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. 2008. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456: 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Shirley Liu X, Mardis ER. 2017. Applications of immunogenomics to cancer. Cell 168: 600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo TG, Pendergast AM, Muller AJ, Witte ON. 1990. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science 247: 1079–1082. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- Mansukhani S, Barber LJ, Kleftogiannis D, Moorcraft SY, Davidson M, Woolston A, Proszek PZ, Griffiths B, Fenwick K, Herman B, et al. 2018. Ultra-sensitive mutation detection and genome-wide DNA copy number reconstruction by error-corrected circulating tumor DNA sequencing. Clin Chem 10.1373/clinchem.2018.289629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. 2011. A decade's perspective on DNA sequencing technology. Nature 470: 198–203. [DOI] [PubMed] [Google Scholar]
- Mardis ER. 2012. Applying next-generation sequencing to pancreatic cancer treatment. Nat Rev Gastroenterol Hepatol 9: 477–486. [DOI] [PubMed] [Google Scholar]
- Mardis E. 2015. Cancer genomics. F1000Res 10.12688/f1000research.6645.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. 2017. DNA sequencing technologies: 2006–2016. Nat Protoc 12: 213–218. [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulonis UA. 2018. Management of newly diagnosed or recurrent ovarian cancer. Clin Adv Hematol Oncol 16: 426–437. [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. 2016. The Ensembl Variant Effect Predictor. Genome Biol 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mody RJ, Wu YM, Lonigro RJ, Cao X, Roychowdhury S, Vats P, Frank KM, Prensner JR, Asangani I, Palanisamy N, et al. 2015. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA 314: 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. 1913. Heredity of body color in Drosophila. Z Indukt Abstamm Vererbungsl 10: 292–293. [Google Scholar]
- National Academy of Sciences. 1960. Abstracts of papers presented at the Autumn Meeting, 14–16 November 1960, Philadelphia, Pennsylvania. Science 132: 1488–1501.17739576 [Google Scholar]
- Oberg JA, Glade Bender JL, Sulis ML, Pendrick D, Sireci AN, Hsiao SJ, Turk AT, Dela Cruz FS, Hibshoosh H, Remotti H, et al. 2016. Implementation of next generation sequencing into pediatric hematology-oncology practice: Moving beyond actionable alterations. Genome Med 8: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell TJ, Rubinsteyn A, Bonsack M, Riemer AB, Laserson U, Hammerbacher J. 2018. MHCflurry: Open-source class I MHC binding affinity prediction. Cell Syst 7: 129–132.e4. [DOI] [PubMed] [Google Scholar]
- Paez JG. 2004. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. 2004. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci 101: 13306–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos N, Kinzler KW, Vogelstein B. 2006. The role of companion diagnostics in the development and use of mutation-targeted cancer therapies. Nat Biotechnol 24: 985–995. [DOI] [PubMed] [Google Scholar]
- Picelli S, Björklund AK, Reinius B, Sagasser S, Winberg G, Sandberg R. 2014a. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 24: 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. 2014b. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9: 171–181. [DOI] [PubMed] [Google Scholar]
- Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, et al. 2017. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171: 1611–1624.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Lonigro RJ, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V, et al. 2017. Integrative clinical genomics of metastatic cancer. Nature 548: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. 1973. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243: 290–293. [DOI] [PubMed] [Google Scholar]
- Rowley JD. 2008. Chromosomal translocations: Revisited yet again. Blood 112: 2183–2189. [DOI] [PubMed] [Google Scholar]
- Rowley JD, Golomb HM, Vardiman J, Fukuhara S, Dougherty C, Potter D. 1977. Further evidence for a non-random chromosomal abnormality in acute promyelocytic leukemia. Int J Cancer 20: 869–872. [DOI] [PubMed] [Google Scholar]
- Roychowdhury S, Iyer MK, Robinson DR, Robinson DR, Lonigro RJ, Wu YM, Cao X, Kalyana-Sundaram S, Sam L, Balbin OA, et al. 2011. Personalized oncology through integrative high-throughput sequencing: A pilot study. Sci Transl Med 3: 111ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudkin CT, Hungerford DA, Nowell PC. 1964. DNA contents of chromosome PH1 and chromosome 21 in human chronic granulocytic leukemia. Science 144: 1229–1231. [DOI] [PubMed] [Google Scholar]
- Saiki R, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. 1985. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230: 1350–1354. [DOI] [PubMed] [Google Scholar]
- Sakaeva DD, Gordiev M, Blokhina M. 2016. Monitoring of EGFRm level in cfDNA in patients with advanced NSCLC on the gefitinib therapy. Ann Oncol 27: 416–454. [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461: 809–813. [DOI] [PubMed] [Google Scholar]
- Shtivelman E, Lifshitz B, Gale RP, Roe BA, Canaani E. 1986. Alternative splicing of RNAs transcribed from the human abl gene and from the bcr-abl fused gene. Cell 47: 277–284. [DOI] [PubMed] [Google Scholar]
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274. [DOI] [PubMed] [Google Scholar]
- Sumanasuriya S, Lambros MB, de Bono JS. 2017. Application of liquid biopsies in cancer targeted therapy. Clin Pharmacol Ther 102: 745–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor F, Ottolini B, Bradford J, Woll P, Teare D, Shaw J, Cox A. 2016. A preliminary study to compare cfDNA levels in lung cancer cases and high risk controls to evaluate the role of cfDNA levels in early lung cancer detection. Eur J Surg Oncol 42: S243–S244. [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandin F, Clay P, Upfal E, Raphael BJ. 2012. Discovery of mutated subnetworks associated with clinical data in cancer. Pac Symp Biocomput 2012: 55–66. [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wilson CL, Easton J, Thrasher A, Mulder H, Liu Q, Hedges DJ, Wang S, Rusch MC, Edmonson MN, et al. 2018. Genetic risk for subsequent neoplasms among long-term survivors of childhood cancer. J Clin Oncol 36: 2078–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. 2007. Characterizing the cancer genome in lung adenocarcinoma. Nature 450: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. 2017. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, Hedges D, Ma X, Zhou X, Yergeau DA, et al. 2015. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 373: 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Lei J, Devlin B, Roeder K. 2018. A unified statistical framework for single cell and bulk RNA sequencing data. Ann Appl Stat 12: 609–632. [DOI] [PMC free article] [PubMed] [Google Scholar]