In this study, Yeom et al. address how cells tune protease specificity to nutritional signals. They show that Salmonella enterica increases the specificity of the broadly conserved proteases Lon and ClpSAP by transforming the Lon activator and substrate HspQ into an inhibitor of the N-degron recognin ClpS, the adaptor of the ClpAP protease, thus providing insight into how cells modify the abundance of particular proteins while preserving the quality of their proteomes.
Keywords: acetyl-CoA, adaptor ClpS, ClpAP protease, HspQ, Lon protease, protein acetylation
Abstract
All cells use proteases to adjust protein amounts. Proteases maintain protein homeostasis by degrading nonfunctional toxic proteins and play regulatory roles by targeting particular substrates in response to specific signals. Here we address how cells tune protease specificity to nutritional signals. We report that Salmonella enterica increases the specificity of the broadly conserved proteases Lon and ClpSAP by transforming the Lon activator and substrate HspQ into an inhibitor of the N-degron recognin ClpS, the adaptor of the ClpAP protease. We establish that upon acetylation, HspQ stops being a Lon activator and substrate and that the accumulated HspQ binds to ClpS, hindering degradation of ClpSAP substrates. Growth on glucose promotes HspQ acetylation by increasing acetyl-CoA amounts, thereby linking metabolism to proteolysis. By altering protease specificities but continuing to degrade junk proteins, cells modify the abundance of particular proteins while preserving the quality of their proteomes. This rapid response mechanism linking protease specificity to nutritional signals is broadly conserved.
All organisms rely on proteins to carry out the vast majority of cellular work. The steady-state amounts of a given protein reflect the balance between synthesis and degradation. Which proteins encoded in a genome are produced at any given time and place is determined by signal-responsive transcription factors (Hengge-Aronis 1993; Stock et al. 2000; Groisman 2016) and by cis-acting sequences in leader mRNAs (Waters and Storz 2009; Mandin 2012) that alter mRNA abundance or translation upon binding metabolites, small RNAs, or regulatory proteins (Kaberdin and Bläsi 2006; Waters and Storz 2009; Mandin 2012). Here, we explore how cells choose which pre-existing proteins to degrade or spare degradation when experiencing a change in nutritional conditions.
ClpAP and Lon are adenosine triphosphate (ATP)-dependent proteases present in bacteria and eukaryotes (Charette et al. 1981; Gottesman et al. 1990; Wang et al. 1994; Shanklin et al. 1995; Weber-Ban et al. 1999; Fukui et al. 2002). The protease ClpAP consists of a chaperone—termed ClpA—that unfolds its substrates and shuffles them into ClpP, where proteolysis actually takes place (Fig. 1A; Thompson and Maurizi 1994). ClpA recognizes certain substrates directly but requires the adaptor ClpS to recognize others, most conspicuously those satisfying the N-degron pathway of proteolysis (Erbse et al. 2006; Kirstein et al. 2009; Nishimura et al. 2013). Lon is a single polypeptide displaying both chaperone and protease activities (Rotanova et al. 2004). The HspQ protein (referred to as YccV in certain organisms) is a Lon substrate that stimulates degradation of other Lon substrates (Fig. 1A; Puri and Karzai 2017). In contrast, ClpS is not degraded by ClpAP (Román-Hernández et al. 2011).
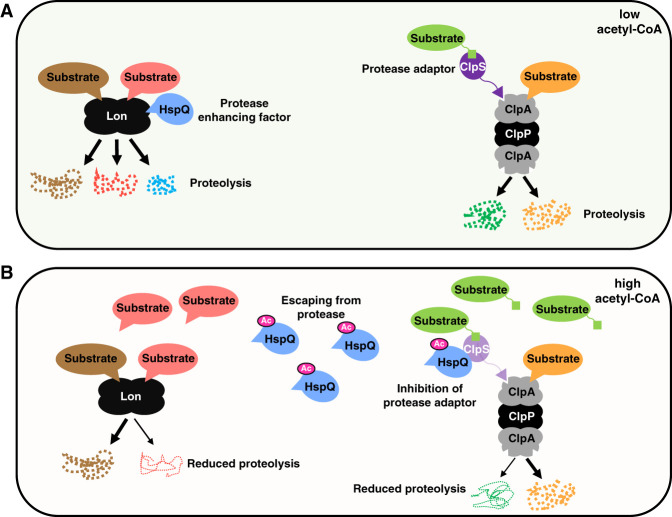
Figure 1.
Acetyl-CoA-promoted acetylation of the HspQ protein increases the specificity of the Lon and ClpAP proteases. (A) When bacteria experience low acetyl-CoA, the Lon substrate HspQ (blue) promotes degradation of some Lon substrates (red) but not others (brown). The ClpAP protease degrades certain substrates directly (orange) but requires the adaptor ClpS (purple) to degrade others (green). (B) When bacteria experience high acetyl-CoA, HspQ is acetylated (pink). Acetylated HspQ escapes degradation by Lon and does not stimulate degradation of a subset of Lon substrates (red). Thus, HspQ-dependent Lon substrates (red) accumulate, but those that are HspQ-independent continue to be degraded (brown). Accumulated HspQ binds to ClpS, inhibiting proteolysis by ClpSAP but not by ClpAP. Thus, the amounts of ClpS-dependent ClpAP substrates (green) increase, but those of ClpS-independent substrates (orange) do not.
Protein acetylation is one of the major posttranslational modifications that regulate protein function in all domains of life (Carabetta and Cristea 2017). In bacteria, protein acetylation by the protein acetyltransferase Pat regulates RNA metabolism, motility, DNA replication, gene expression, and virulence (Carabetta and Cristea 2017). Because Pat-dependent acetylation requires acetyl-CoA as the acetyl donor, it is sensitive to the abundance of acetyl-CoA (Carabetta and Cristea 2017) and, potentially, of acetyl-CoA-binding proteins. In contrast, when proteins acetylate spontaneously in a nonenzymatic fashion, their acetylation status is controlled by protein deacetylases (Weinert et al. 2013; Gibellini et al. 2014).
We now describe a mechanism that coordinately alters the specificity of the ClpAP and Lon proteases, enabling cells to stabilize protein subsets in response to nutritional signals (Fig. 1B). These signals promote acetylation of the HspQ protein, which prevents both HspQ's ability to enhance Lon's activity and HspQ degradation. The accumulated HspQ protein hinders proteolysis of ClpS-dependent ClpAP substrates (Fig. 1B) but not of junk proteins tagged for destruction by ClpAP in a ClpS-independent manner. While we conducted our experiments with the bacterium Salmonella enterica serovar Typhimurium and its proteins, the genes mediating control of the two proteases are broadly conserved, suggesting that the identified mechanism operates in other enteric bacteria.
Results
HspQ binds to the adaptor ClpS, thereby inhibiting proteolysis of ClpSAP substrates
We hypothesized that HspQ, an activator of the bacterial Lon protease (Puri and Karzai 2017), alters degradation by the ClpSAP protease because HspQ exhibits sequence similarity to a portion of ClpF, a chloroplast protein that, together with ClpS1, forms an adaptor that delivers substrates to the ClpCPR protease (Nishimura et al. 2015). Given that ClpS and ClpS1 display sequence similarity and that ClpS1 abundance decreases upon ClpF inactivation in chloroplasts (Nishimura et al. 2015), we reasoned that Salmonella ClpS abundance would be lower in the hspQ mutant than in the wild-type strain. Thus, we examined the stability of C-terminally HA-tagged ClpS expressed from its normal clpS promoter and chromosomal location in isogenic hspQ strains by Western blot using antibodies directed to the HA tag because antibodies directed to ClpS were not available. ClpS-HA was rapidly and similarly degraded in the two strains (Supplemental Fig. S1A). (Although it is presently unknown whether the hspQ-independent ClpS-HA instability is caused by the HA tag, such a tag did not destabilize the ClpsSAP substrate PhoP [Yeom et al. 2017].) In contrast, HspQ stability was lower in the clpS mutant than in wild-type Salmonella (Supplemental Fig. S1B), suggesting that ClpS stabilizes HspQ. This result is paradoxical given ClpS's role as adaptor of the ClpAP protease.
We determined that ClpS binds to HspQ because antibodies directed to the HA tag pulled down HspQ-Flag, and antibodies directed to the Flag tag pulled down ClpS-HA in a strain specifying the ClpS-HA and HspQ-Flag proteins from their normal promoters and chromosomal locations (Fig. 2A). These pull-downs were specific, as shown by the fact that they were not observed in wild-type Salmonella (with no epitope-tagged proteins) or strains specifying only one of the epitope-tagged proteins (Fig. 2A). In contrast, the anti-HA antibodies did not pull down HspQ-Flag, and anti-Flag antibodies did not pull down ClpA-HA in a strain expressing ClpA-HA and HspQ-Flag from their normal promoters and chromosomal locations (Fig. 2A). Thus, under the investigated conditions, HspQ associates with ClpS but not ClpA. In agreement with these results, bacterial two-hybrid experiments demonstrated HspQ binding to ClpS but not RssB, ClpA, or ClpP (Supplemental Fig. S1C). Moreover, pull-down assays with in vitro synthesized proteins demonstrated binding of HspQ-Flag to ClpS-HA but not RssB-HA (Supplemental Fig. S1D), the adaptor of the ClpXP protease (Zhou et al. 2001) used as negative control. Taken together, these results indicate that HspQ binds directly to ClpS.
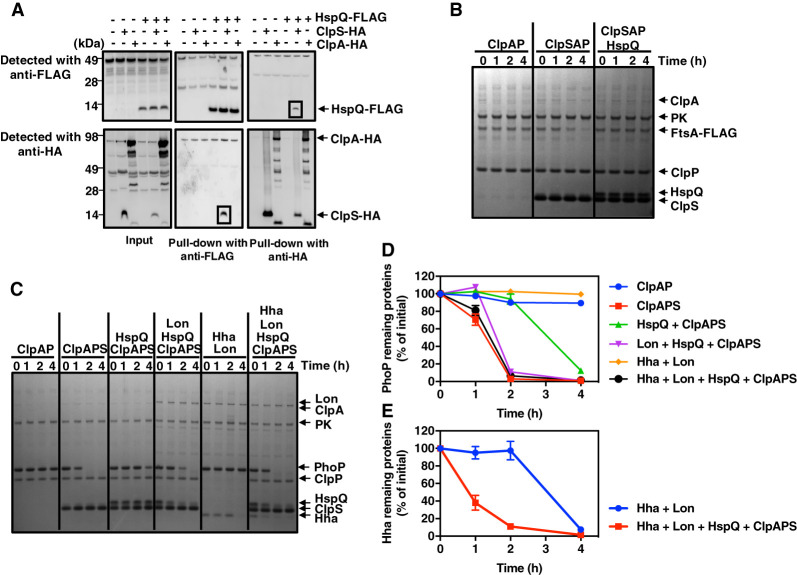
Figure 2.
The Lon-enhancing factor HspQ protein binds to ClpS, inhibiting proteolysis by ClpSAP. (A) Pull-down of lysed extracts prepared from wild-type (14028s), hspQ-Flag (JY674), clpS-HA (JY691), hspQ-Flag clpS-HA (JY692), clpA-HA (JY694), and hspQ-Flag clpA-HA (JY695) Salmonella. Squares indicate specific HspQ–ClpS interactions. Data are representative of two independent experiments, which gave similar results. (B) SDS-PAGE analysis for in vitro degradation of ClpSAP substrate FtsA-Flag. FtsA-Flag (0.2 µM) was mixed with 0.08 µM ClpA and 0.2 µM ClpP in the absence or presence of 0.08 µM ClpS or 0.5 µM HspQ. All reactions were carried out for the indicated times at 30°C in the presence of an ATP regeneration system and started by the addition of substrates. After incubation, protein amounts were determined by Coomassie-staining following separation on a 4%–12% SDS-PAGE gel. Data are representative of two independent experiments, which gave similar results. (C) SDS-PAGE analysis for time-course in vitro degradation of ClpSAP substrate PhoP. PhoP (0.5 µM), 0.5 µM Hha, and 0.5 µM HspQ were mixed with 0.08 µM ClpA, 0.2 µM ClpP, 1.0 µM ClpS, and/or 0.2 µM Lon. Reactions were carried out for the indicated times at 30°C in the presence of an ATP regeneration system and started by the addition of substrates. After incubation, protein amounts were determined by Coomassie staining following separation on a 4%–12% SDS-PAGE gel. Data are representative of two independent experiments, which gave similar results. (D) Degradation of the PhoP protein in C was determined by quantification of bands. Relative PhoP levels were calculated from two independent experiments. (E) Degradation of the His-Hha protein in C was determined by quantification of bands. Relative His-Hha amounts were calculated from two independent experiments. See also Supplemental Figures S1, S2, and S6.
How does HspQ binding to ClpS impact proteolysis of ClpS-dependent substrates of ClpAP? The purified HspQ protein hindered degradation of FtsA-Flag (Fig. 2B) and PhoP (Supplemental Fig. S1E) when incubated with the purified ClpS, ClpA, and ClpP proteins. Unlike the FtsA-Flag and PhoP proteins, HspQ was not degraded (Fig. 2B; Supplemental Fig. S1E), demonstrating that it is not a ClpSAP substrate. Cumulatively, the results in this section identify HspQ as an antiadaptor of the protease adaptor ClpS.
Lon promotes proteolysis of ClpSAP substrates by degrading HspQ
Because Lon degrades HspQ in Yersinia (Puri and Karzai 2017) and HspQ inhibits ClpS-dependent proteolysis in Salmonella (Fig. 2B), we reasoned that Lon decreases the abundance of ClpSAP substrates. To explore this possibility, we first determined that the purified Salmonella HspQ protein stimulates degradation of the Lon substrate Hha (Supplemental Fig. S2A,B) and is itself degraded by the Lon protease (Supplemental Fig. S2A,B), like its Yersinia counterpart (Puri and Karzai 2017). We then established that HspQ abundance is higher in the lon mutant than in wild-type, clpA, and clpX Salmonella (Supplemental Fig. S2C), supporting the notion that in vivo, HspQ is a substrate of Lon but not of ClpAP or ClpXP.
We determined that HspQ protects the purified PhoP protein from proteolysis by ClpSAP (Fig. 2C,D) and, in agreement with our previous report (Yeom et al. 2017), that PhoP is degraded by ClpSAP but not by ClpAP (Fig. 2C,D). By degrading HspQ, Lon overcame the HspQ protection of PhoP (Fig. 2C,E). When Lon, ClpS, ClpA, ClpP, HspQ, Hha, and PhoP were all present in the reaction, we observed the disappearance of HspQ, Hha, and PhoP but not ClpS (Fig. 2C–E). In the absence of ClpSAP, Lon degraded Hha but not PhoP (Fig. 2C). Lon did not degrade PhoP even when incubated in the absence of Hha and the presence of HspQ (Supplemental Fig. S2D). As shown above (Supplemental Fig. S2A,B), Lon degraded Hha even in the absence of HspQ (Fig. 2C,E). These in vitro results raised the following question: How does HspQ avoid being degraded by Lon so it can inhibit proteolysis of ClpS-dependent ClpAP substrates in vivo?
Acetylation prevents HspQ from being degraded by Lon and enhancing degradation of other Lon substrates
High-resolution tandem mass spectrometry (MS/MS) analysis revealed that HspQ is acetylated at Lys96 (Supplemental Fig. S3A,B). In bacteria, proteins can be acetylated from acetyl-CoA by the protein acetyltransferase Pat or from acetylphosphate without the participation of an enzyme (Fig. 3A; Weinert et al. 2013). Immediately adjacent to hspQ is the STM14_1223 gene (Supplemental Fig. S3C), specifying a protein predicted to bind a CoA-containing metabolite (Supplemental Fig. S3D,E). Thus, we reasoned that STM14_1223 promotes HspQ acetylation and designated its gene product Qad (HspQ acetyl donor) because of the results described below.
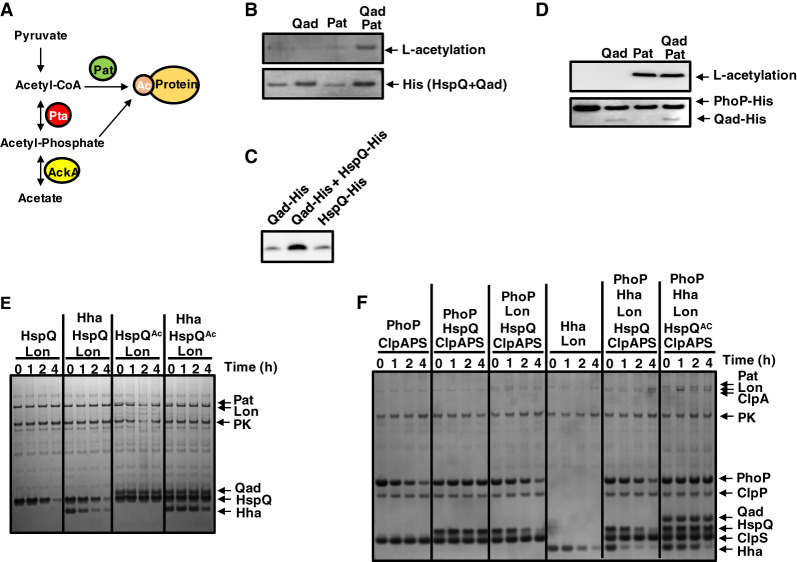
Figure 3.
Acetylated by Pat, Qad, and acetyl-CoA, HspQ is neither a Lon substrate nor a Lon activator but inhibits proteolysis of ClpS-dependent ClpAP substrate PhoP. (A) Diagram of the pathways for protein acetylation in bacteria. (B,D) Western blot analysis of in vitro acetylation of HspQ (B) or PhoP (D) using Qad, Pat, and acetyl-CoA. (C) The purified HspQ-His and Qad-His proteins exhibit similar mobility. Data are representative of two independent experiments, which gave similar results. (E) SDS-PAGE analysis for time-course in vitro degradation of Hha with acetylated HspQ. Hha (0.5 µM) was mixed with 0.2 µM Lon in the absence or presence of 0.5 µM acetylated HspQ. HspQ acetylation was achieved by preincubating HspQ with 0.5 µM Qad, 2 mM acetyl-CoA, and 0.2 µM Pat for 3 h at 37°C. Reactions were carried out for the indicated times at 30°C in the presence of an ATP regeneration system and started by the addition of substrates. After incubation, protein amounts were determined by Coomassie staining following separation on a 4%–12% SDS-PAGE gel. Data are representative of two independent experiments, which gave similar results. (F) SDS-PAGE analysis for time-course in vitro degradation of 0.5 µM PhoP and 0.5 µM Hha in the absence or presence of 0.5 µM acetylated HspQ. Substrates were mixed with 0.08 µM ClpA, 0.2 µM ClpP, 1.0 µM ClpS, and/or 0.2 µM Lon. HspQ acetylation was achieved by preincubating HspQ with 0.5 µM Qad, 2 mM acetyl-CoA, and 0.2 µM Pat for 3 h at 37°C. Reactions were carried out at 30°C for the indicated times in the presence of an ATP regeneration system and started by the addition of substrates. After incubation, protein amounts were determined by Coomassie staining following separation on a 4%–12% SDS-PAGE gel. Data are representative of two independent experiments, which gave similar results. See also Supplemental Figures S3–S5 and S8.
The purified HspQ protein was acetylated in the presence of acetyl-CoA and the purified Pat and Qad proteins (Fig. 3B). Whereas Pat was essential for the reaction (Fig. 3B), HspQ was still acetylated, albeit minimally, when Qad was absent (Fig. 3B). (Please note that the HspQ-His and Qad-His proteins exhibit the same mobility on the gels used in these experiments [Fig. 3C].) The PhoP protein was similarly acetylated by Pat and acetyl-CoA (Ren et al. 2016) in the presence and absence of Qad (Fig. 3D), arguing that Qad serves as acetyl donor primarily, if not solely, to HspQ.
Acetylation protects HspQ from degradation by Lon because HspQ was not proteolyzed when Pat, Qad, and acetyl-CoA were included in the reaction (Fig. 3E; Supplemental Fig. S4A). The HspQ acetylated mimetic with Lys96 substituted by glutamine (HspQK96Q) was fully resistant to Lon (Supplemental Fig. S5A,B), whereas the nonacetylated mimetic with Lys96 substituted by arginine (HspQK96R) was a better Lon substrate than wild-type HspQ (Supplemental Fig. S5A,B).
Acetylated HspQ failed to enhance proteolysis of the Lon substrate Hha (Fig. 3E; Supplemental Fig. S4B), and this was also the case for the acetylated mimetic (HspQK96Q) (Supplemental Fig. S5A). In contrast, the nonacetylated mimetic (HspQK96R) stimulated Hha proteolysis more than the wild-type HspQ protein (Supplemental Fig. S5A). Thus, acetylation prevents HspQ degradation by Lon's and HspQ's ability to enhance proteolysis of other Lon substrates.
Accumulated HspQ inhibits ClpS-dependent proteolysis
Next, we investigated whether acetylation impacts HspQ's ability to bind to the ClpS protein. We found that anti-Flag antibodies pulled down similar amounts of ClpS-His when this protein was incubated with Flag-tagged wild-type HspQ or the K96R and K96Q variants (Supplemental Fig. S5C). Likewise, anti-His antibodies pulled down similar amounts of Flag-tagged wild-type, K96R, and K96Q HspQ proteins when incubated in the presence of ClpS-His (Supplemental Fig. S5C). These results indicate that ClpS binds similarly to the acetylated and nonacetylated forms of HspQ.
The wild-type, K96R, and K96Q HspQ proteins were equally proficient at inhibiting PhoP proteolysis by ClpSAP in vitro (Supplemental Fig. S5D). As expected, PhoP was degraded in the presence of the ClpS, ClpA, and ClpP proteins (Supplemental Figs. S1E, S5D; Yeom et al. 2017) but not when ClpS was omitted from the reaction (Supplemental Figs. S1E, S5D). Both acetylated and nonacetylated forms of HspQ inhibited PhoP proteolysis by ClpSAP (Supplemental Fig. S5D).
To examine how acetylation impacts HspQ's ability to reduce proteolysis by Lon when both ClpSAP and Lon are together in the reaction, we investigated degradation of PhoP and Hha in the presence and absence of Qad, Pat, and acetyl-CoA. By degrading nonacetylated HspQ, Lon decreases HspQ's abundance, resulting in more PhoP proteolysis by ClpSAP than in the presence of acetylated HspQ (Fig. 3F; Supplemental Fig. S4C). (As shown above, PhoP is not a Lon substrate [Supplemental Fig. S2D].) The nonacetylated form of HspQ enhanced degradation of the Lon substrate Hha, whereas the acetylated form did not (Fig. 3F), in agreement with the results obtained in the absence of ClpSAP (Supplemental Fig. S2A, S5A). Because acetylated HspQ is not degraded by Lon (Fig. 3E), it accumulated to higher abundance than nonacetylated HspQ (Fig. 3F), allowing for enhanced PhoP protection from ClpSAP (Fig. 3F; Supplemental Fig. S4C). In the absence of acetyl-CoA, HspQ did not enhance Hha degradation or protect PhoP from ClpSAP (Supplemental Fig. S4D). In other words, HspQ acetylation increases the abundance of ClpSAP substrates because Lon's inability to degrade acetylated HspQ furthers the amount of HspQ available to inhibit ClpS-dependent proteolysis (Fig. 1).
Qad- and Pat-dependent acetylation of HspQ prevents HspQ degradation by Lon in vivo
To test the model described in the previous section, we investigated the consequences of acetylation on HspQ abundance by using sets of isogenic Salmonella strains. We established that HspQ acetylation and abundance are directly correlated and that qad inactivation has no effect on HspQ abundance if HspQ cannot be acetylated at K96; that is, the K96R and K96Q HspQ variants were present in lower and higher amounts, respectively, than the wild-type HspQ protein (Fig. 4A) when the hspQ gene was expressed from its normal promoter and chromosomal location. Moreover, the abundance of the K96Q and K96R HspQ variants was unaltered upon qad inactivation (Fig. 4A). Furthermore, the qad and pat single mutants and the qad pat double mutant had lower HspQ abundance than wild-type Salmonella (Fig. 4B). HspQ abundance was restored to wild-type levels in the qad mutant by the qad-expressing plasmid pqad but not by vector control (Fig. 4C), demonstrating that the qad mutation does not impair expression of the convergently transcribed hspQ gene (Supplemental Fig. S3C). The qad-expressing plasmid did not restore normal HspQ abundance to the qad pat double mutant (Fig. 4C), indicating that Qad operates in a Pat-dependent manner. Critically, inactivation of the lon gene in the qad mutant increased HspQ amounts to levels higher than those of the wild-type strain (Fig. 4D), providing genetic evidence that Qad-dependent acetylation prevents HspQ degradation by Lon.
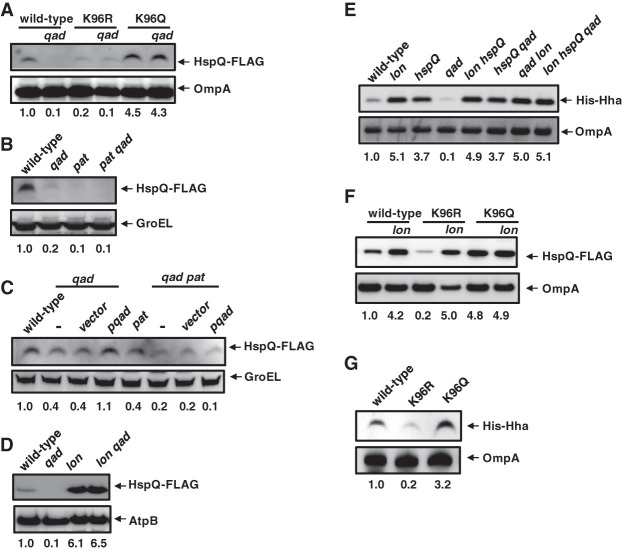
Figure 4.
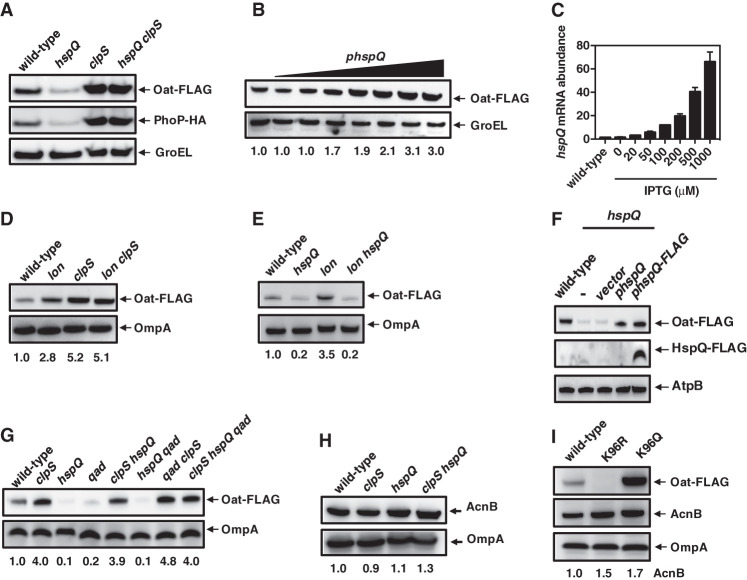
Acetylated HspQ is neither a Lon substrate nor a Lon activator in vivo. (A) Western blot analysis of crude extracts from hspQ-Flag (JY674), hspQ-Flag qad (JY740), hspQK96R-Flag (JY892), hspQK96R-Flag qad (JY893), hspQK96Q-Flag (JY894), and hspQK96Q-Flag qad (JY895) Salmonella. (B) Western blot analysis of crude extracts from hspQ-Flag (JY674), hspQ-Flag qad (JY740), hspQ-Flag pat (JY898), and hspQ-Flag pat qad (JY899) Salmonella. (C) Western blot analysis of crude extracts from hspQ-Flag (JY674), hspQ-Flag qad (JY740), and hspQ-Flag qad pat (JY899) Salmonella with the plasmid vector (vector) or the qad-expressing plasmid (pqad) using 100 µM IPTG. (D) Western blot analysis of crude extracts from hspQ-Flag (JY674), hspQ-Flag qad (JY740), hspQ-Flag lon (JY703), and hspQ-Flag lon qad (JY745) Salmonella. (E) Western blot analysis of crude extracts from oat-Flag (JY655), oat-Flag lon (JY1009), oat-Flag hspQ (JY686), oat-Flag qad (JY774), oat-Flag lon hspQ (JY1011), hspQ qad (JY773), oat-Flag qad lon (JY1012), and lon hspQ qad (JY1013) Salmonella with a His-hha-expressing plasmid (pHis-hha). his-hha transcription from pHis-hha was induced with 100 µM IPTG. (F) Western blot analysis of crude extracts from hspQ-Flag (JY674), hspQ-Flag lon (JY703), hspQK96R-Flag (JY892), hspQK96R-Flag lon (JY2000), hspQK96Q-Flag (JY894), and hspQK96Q-Flag lon (JY2001) Salmonella. (G) Western blot analysis of crude extracts from hspQ-Flag (JY674), hspQK96R-Flag (JY892), and hspQK96Q-Flag (JY894) Salmonella with a His-hha-expressing plasmid (pHis-hha). his-hha transcription from pHis-hha was induced with 100 µM IPTG. Numbers below blots show quantification of the top band in the various strains. Words with italicized letters indicate gene names. For Western blot analysis, samples were analyzed with antibodies directed to acetylated lysine; the Flag, HA, or His epitopes; or the AtpB, OmpA, or GroEL proteins. Data are representative of three independent experiments, which gave similar results. See also Supplemental Figures S3 and S5.
HspQ's acetylation status controls the abundance of Lon and ClpSAP substrates in vivo
To test how HspQ acetylation impacts Lon- and ClpSAP-mediated proteolysis in vivo, we investigated the abundance of Lon and ClpSAP substrates using sets of isogenic strains. First, we determined that inactivation of the hspQ gene increased Hha abundance nearly to the levels of the lon mutant (Fig. 4E), reflecting that HspQ is necessary for efficient Hha proteolysis by Lon. Hha abundance was lower in the qad mutant than in the wild-type strain (Fig. 4E), in agreement with the notion that nonacetylated HspQ stimulates Hha proteolysis, whereas acetylated HspQ does not. The low Hha abundance present in the qad mutant was corrected upon inactivation of the hspQ or lon genes (Fig. 4E), consistent with Qad decreasing Lon degradation of Hha by acetylating HspQ. The qad lon double mutant and the lon hspQ qad triple mutant harbored the same high Hha amounts as the lon single mutant (Fig. 4E). This result argues that qad and hspQ alter Hha abundance only when Lon is present. Likewise, the fact that the lon hspQ double mutant had the same high Hha amounts as the lon single mutant (Fig. 4E) indicates that HspQ controls Hha amounts via Lon.
In addition, we established that HspQ acetylation is directly correlated with the abundance of both HspQ and Hha; that is, the abundance of the acetylated mimetic K96Q HspQ variant was unaltered upon lon inactivation (Fig. 4F). In contrast, the abundance of the wild-type HspQ and nonacetylatable K96R HspQ variant increased upon lon inactivation (Fig. 4F). The abundance of Lon substrate Hha was higher in the strain expressing the K96Q HspQ variant than in that expressing the wild-type HspQ protein, which in turn was higher than in the strain expressing the K96R HspQ variant (Fig. 4G).
Second, we found the in vivo abundance of the ClpSAP substrates PhoP and Oat to be lower in the hspQ mutant than in wild-type Salmonella (Fig. 5A). When the hspQ gene was expressed from a heterologous inducible promoter, Oat amounts increased with the concentration of inducer (Fig. 5B), reflecting the increase in hspQ mRNA abundance (Fig. 5C). A clpS-null mutant and an hspQ clpS double mutant had similar amounts of PhoP and Oat proteins, which were higher than those present in the wild-type and hspQ strains (Fig. 5A), supporting previous findings that PhoP and Oat are ClpS-dependent substrates of ClpAP (Yeom et al. 2017) and the notion that hspQ operates in a clpS-dependent manner. In addition, the abundance of the ClpSAP substrate Oat was higher in the lon mutant than in wild-type Salmonella (Fig. 5D), approaching the abundance observed in the clpS single mutant and lon clpS double mutant (Fig. 5D). The increased Oat abundance exhibited by the lon mutant was eliminated upon inactivation of the hspQ gene (Fig. 5E), indicating the Lon alters Oat abundance via HspQ. (The Flag tag in HspQ does not impact Oat abundance because Oat abundance was restored to the hspQ mutant by a plasmid expressing the wild-type hspQ gene or one encoding HspQ-Flag [Fig. 5F].)
Figure 5.
Acetylated HspQ inhibits ClpS-dependent proteolysis of ClpAP substrates in vivo. (A) Western blot analysis of crude extracts from oat-Flag phoP-HA (JY872), oat-FLAG phoP-HA hspQ (JY873), oat-Flag phoP-HA clpS (JY874), and oat-Flag phoP-HA hspQ clpS (JY875) Salmonella. (B,C) Western blot analysis of crude extracts (B) and mRNA abundance of the hspQ gene (C) from oat-Flag (JY655) Salmonella with no plasmid or with a hspQ-expressing plasmid (phspQ; pUHE-hspQ) using 0, 20, 50, 100, 200, 500, or 1000 µM IPTG. (D) Western blot analysis of crude extracts from oat-Flag (JY655), oat-Flag lon (JY1009), oat-Flag clpS (JY657), and oat-Flag lon clpS (JY1010) Salmonella. (E) Western blot analysis of crude extracts from oat-Flag (JY655), oat-Flag hspQ (JY686), oat-Flag lon (JY1014), and oat-Flag lon hspQ (JY1016) Salmonella. (F) Western blot analysis of crude extracts from oat-Flag (JY655) and oat-Flag hspQ (JY686) Salmonella with the plasmid vector (vector), the hspQ-expressing plasmid (phspQ), or the hspQ-Flag-expressing plasmid (phspQ-Flag) using 100 µM IPTG. (G) Western blot analysis of crude extracts from oat-Flag (JY655), oat-Flag clpS (JY657), oat-Flag hspQ (JY686), oat-Flag qad (JY774), oat-Flag clpS hspQ (JY687), oat-Flag hspQ qad (JY773), oat-Flag qad clpS (JY775), and oat-Flag clpS hspQ qad (JY772) Salmonella. (H) Western blot analysis of crude extracts from oat-Flag (JY655), oat-Flag clpS (JY657), oat-Flag hspQ (JY686), and oat-Flag clpS hspQ (JY687) Salmonella. (I) Western blot analysis of crude extracts from hspQ-Flag oat-Flag (JY865), hspQK96R-Flag oat-Flag (JY2002), and hspQK96Q-Flag oat-Flag (JY2003) Salmonella. Words with italicized letters indicate gene names. For Western blot analysis, samples were analyzed using antibodies directed to the Flag or HA epitopes or the AcnB, AtpB, OmpA, or GroEL proteins. Data are representative of at least two independent experiments, which gave similar results. See also Supplemental Figure S6.
Oat amounts were lower in the qad and hspQ single mutants than in wild-type Salmonella (Fig. 5G), reflecting that qad is necessary for HspQ acetylation (Fig. 4B) and that acetylated HspQ is not a Lon substrate (Fig. 4E). The qad clpS double mutant and the clpS hspQ qad triple mutant harbored the same high Oat amounts as the clpS single mutant (Fig. 5G), indicating that qad and hspQ alter Oat abundance via clpS. The clpS hspQ double mutant exhibited high Oat amounts similar to the clpS single mutant (Fig. 5G), reinforcing the notion that HspQ controls Oat amounts via ClpS.
The in vivo abundance of ClpS-independent ClpAP substrates is independent of HspQ
The HspQ protein appears to protect ClpS-dependent substrates of ClpAP exclusively because the ClpS-independent ClpAP substrate AcnB exhibited wild-type abundance in the hspQ and clpS single mutants and the clpS hspQ double mutant (Fig. 5H). In agreement with these results, the clpS mutant exhibited wild-type AcnB stability, whereas no AcnB degradation was observed in clpA or clpX mutants (Supplemental Fig. S6A). In contrast, the abundance of the control ClpS-dependent ClpAP substrate Oat (Yeom et al. 2018) was higher in the clpS mutant than in wild-type Salmonella (Supplemental Fig. S6B,C) and lower in the hspQ and qad single mutants than in the wild-type strain (Supplemental Fig. S6B).
The abundance of GFP-LAA, a GFP variant with the complete ssrA-encoded degradation motif at the C terminus that renders GFP-LAA a ClpS-independent ClpAP substrate (Farrell et al. 2005), was the same in wild-type, qad, hspQ, and clpS strains (Supplemental Fig. S6B). In agreement with these results, the stability of GFP-LAA was the same in wild-type and hspQ strains (Supplemental Fig. S6D). In contrast, GFP-LAA was stable in the clpP mutant (Supplemental Fig. S6D). Furthermore, HspQ acetylation did not impact ClpAP-mediated proteolysis because AcnB abundance was similar in strains expressing wild-type, K96Q, or K96R HspQ proteins (Fig. 5I). This is in contrast to Oat abundance, which was higher in bacteria expressing the K96Q HspQ variant than in those expressing the wild-type HspQ protein, which in turn was higher than in bacteria expressing the K96R HspQ variant (Fig. 5I). As the SsrA tag marks nonfunctional proteins for degradation by the ClpAP and ClpXP proteases (Farrell et al. 2005), quality control of the proteome continues unimpeded when HspQ is not acetylated or absent.
Carbon metabolism controls proteolysis by Lon and ClpSAP
Given that HspQ acetylation increases the stability of some Lon substrates and of ClpS-dependent substrates of ClpAP, we wondered which physiological conditions promote HspQ acetylation. Because Pat, Qad, and acetyl-CoA are responsible for HspQ acetylation (Fig. 3B), conditions that increase the amounts of Pat, Qad, and/or acetyl-CoA may favor HspQ acetylation, thus hindering proteolysis of both HspQ-dependent Lon substrates and ClpS-dependent ClpAP substrates.
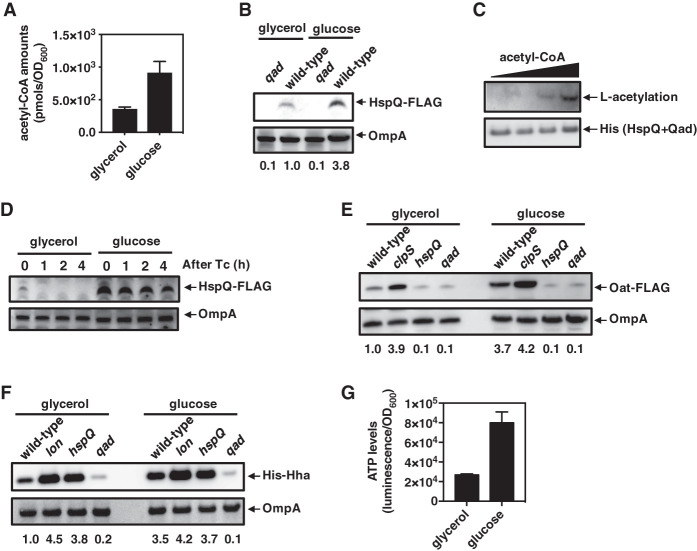
We determined that wild-type Salmonella produces more acetyl-CoA when grown on glucose than on glycerol (Fig. 6A), as reported in Escherichia coli (Takamura and Nomura 1988). Growth on glucose resulted in larger HspQ amounts than growth on glycerol (Fig. 6B). This result is in agreement with acetylated HspQ being resistant to degradation by Lon (Fig. 3E), HspQ acetylation increasing with acetyl-CoA amounts (Fig. 6C), and growth on glucose resulting in more acetyl-CoA than growth on glycerol (Fig. 6A). Both the steady-state amount and the stability of HspQ-Flag were higher in bacteria grown on glucose than on glycerol (Fig. 6D). The increased abundance of acetylated HspQ in turn resulted in larger amounts of both the ClpSAP substrate Oat (Fig. 6E) and the Lon substrate Hha (Fig. 6F).
Figure 6.
Growth on glucose promotes HspQ accumulation by increasing acetyl-CoA amounts. (A,G) Acetyl-CoA (A) and ATP (G) amounts present in wild-type Salmonella (14028s) grown on glucose or glycerol. Acetyl-CoA and ATP amounts were normalized to growth yield (OD600). The mean and SD from three independent experiments is shown. (B) Western blot analysis of crude extracts prepared from hspQ-Flag (JY674) and hspQ-Flag qad (JY740) Salmonella grown on glucose or glycerol. (C) Western blot analysis of in vitro acetylation of HspQ with different acetyl-CoA amounts (0.2, 0.5, 1.0, or 2 mM). Data are representative of two independent experiments, which gave similar results. (D) Stability of the HspQ-Flag protein in hspQ-Flag (JY674) Salmonella. Protein synthesis was inhibited with 50 µg/mL tetracycline. Samples were removed at the indicated times and analyzed by Western blotting with antibodies directed to the Flag epitope or OmpA protein. (E) Western blot analysis of crude extracts prepared from oat-Flag (JY655), oat-Flag clpS (JY657), oat-Flag hspQ (JY686), and oat-Flag qad (JY740) Salmonella grown on glucose or glycerol. (F) Western blot analysis of crude extracts prepared from oat-Flag (JY655), oat-Flag lon (JY1009), oat-Flag hspQ (JY686), and oat-Flag qad (JY774) Salmonella with a His-hha-expressing plasmid (pHis-hha; pUHE-His-hha) grown on glucose or glycerol. his-hha transcription from pHis-Hha was induced with 100 µM IPTG. For Western blot analysis, samples were analyzed using antibodies directed to acetylated lysine, theFlag or His epitopes, or the OmpA protein. Data are representative of two independent experiments, which gave similar results. See also Supplemental Figure S7.
Glucose appears to exert its effects by promoting HspQ acetylation from acetyl-CoA bound to Qad because, first, the qad mutant had equally low HspQ amounts when grown on glucose or glycerol (Fig. 6B). Second, the hspQ and qad single mutants displayed similar Oat amounts when grown on either carbon source (Fig. 6E). Third, the hspQ and qad single mutants exhibited similar Hha abundance regardless of the carbon source (Fig. 6F). Control experiments demonstrated that a clpS mutant had similarly large Oat amounts under both growth conditions (Fig. 6E), and the same was true for Hha abundance when lon was mutated (Fig. 6F). Fourth, the carbon source used to grow Salmonella had no effect on the mRNA abundances of the oat (Supplemental Fig. S7A) or hha (Supplemental Fig. S7B) genes, arguing against the observed differences in protein abundance resulting from effects on mRNA amounts. Fifth, the greater abundance of Oat and Hha during growth on glucose versus glycerol was observed despite growth on glucose resulting in higher ATP concentrations (Fig. 6G) and ATP stimulating proteolysis by the ATP-dependent proteases ClpSAP and Lon (Charette et al. 1981; Weber-Ban et al. 1999; Dougan et al. 2002). These results demonstrate how nutritional conditions that increase acetyl-CoA abundance decrease proteolysis of specific substrates of two different proteases.
Discussion
We uncovered a mechanism by which a protein coordinately narrows the specificities of two different proteases (Fig. 1). This mechanism operates when cells experience nutritional conditions that favor the chemical modification of this protein (Fig. 1). The uncovered mechanism is in contrast to the “Russian doll” effect of protease adaptors and substrates acting on a single protease to establish a degradation hierarchy (Joshi et al. 2015). The rapid response mechanism described here adjusts the stability of specific protease substrates without compromising proteome quality because degradation of nonfunctional proteins continues unimpeded (Supplemental Fig. S6B).
The Qad-dependent acetylation of the HspQ protein is predicted to alter the abundance of a significant portion of the Salmonella proteome by narrowing the substrate spectrum of the ClpSAP and Lon proteases. This is because the substrates of Lon (Takaya et al. 2005; Madrid et al. 2007) and ClpSAP (Yeom et al. 2018) include several regulatory DNA-binding proteins as well as enzymes that alter the abundance of key metabolites such as polyamines.
The acetylation-dependent control of ClpSAP and Lon is likely to operate in multiple species apart from Salmonella given the high shared identity in the deduced amino acid sequences of the hspQ and qad genes among members of the family Enterobacteriaceae (Supplemental Fig. S8; Supplemental Table S1). For example, the shared amino acid identity between the Qad proteins of E. coli and Salmonella is 91%, and the amino acid identity between the HspQ proteins is 92%. Most species retain the lysine that is acetylated in the Salmonella HspQ protein (Supplemental Fig. S8; Supplemental Table S1). However, the Yersinia pestis HspQ protein harbors a histidine at position 96 (Supplemental Fig. S8; Supplemental Table S1). This raises the possibility of allelic differences in acetylation sites of protease regulators, such as HspQ, being responsible for changes in protein stability. These changes in turn may give rise to phenotypic differences among closely related organisms, which are typically ascribed to differences in gene content (Ochman et al. 2000).
The use of protein acetylation to implement nutritional governance of proteolytic pathways appears to be a broadly conserved mechanism. For example, insulin promotes acetylation of TUG, a protein that sequesters the glucose transporter GLUT4 in human fat and muscle (Belman et al. 2015). Acetylation of the TUG protein has two consequences: It inhibits TUG binding to the acyl-CoA-binding protein ACBD3 and favors TUG endoproteolytic cleavage, thereby freeing GLUT4 to migrate to the plasma membrane and import glucose. Thus, acetylation has different effects on different substrates because it decreases proteolysis of HspQ in Salmonella (Fig. 3) but stimulates proteolysis of TUG in mammalian cells (Belman et al. 2015).
It may be possible for cells to increase the amount of acetylated HspQ even in the absence of Qad based on the abundances of the HspQ protein, acetyl-CoA, and/or other metabolites. For instance, HspQ amounts increase with temperature between 25°C and 45°C in Yersinia (Puri and Karzai 2017). Likewise, HspQ may use acetylphosphate as acetyl donor under conditions in which cells accumulate acetylphosphate instead of acetyl-CoA (Weinert et al. 2013). In mitochondria, inactivation of a deacetylase increases both Lon acetylation and abundance (Gibellini et al. 2014), raising the possibility of HspQ acetylation status being controlled by the deacetylase CobB, which is regulated by NAD+ in Salmonella (Schmidt et al. 2004; Cantó et al. 2009).
Finally, our findings suggest that cells achieve specificity in protein degradation by modifying the abundance of proteins that interact with proteases directly or bind proteins that interact with proteases. These interactions are typically governed by chemical modifications of the protein partners, often in response to environmental or cellular signals. Because they act on premade proteins, this mechanism allows for faster changes in the proteome than can be accomplished by traditional transcriptional responses.
Materials and methods
Bacterial strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are listed in Supplemental Table S2. All S. enterica serovar Typhimurium strains were derived from strain 14028s (Fields et al. 1986) and constructed by phage P22-mediated transductions as described (Davis et al. 1980). DNA oligonucleotides used in this study are listed in Supplemental Table S3. Bacteria were grown at 37°C in Luria-Bertani broth (LB) and N-minimal medium (pH 7.7) (Snavely et al. 1991) supplemented with 0.1% casamino acids, 38 mM glycerol or glucose, and the indicated concentrations of MgCl2. E. coli DH5α was used as the host for the preparation of plasmid DNA. Ampicillin was used at 50 µg/mL, kanamycin was used at 50 µg/mL, chloramphenicol was used at 25 µg/mL, and tetracycline was used at 12.5 µg/mL except for the protein stability assays, when it was used at 50 µg/mL.
Construction of chromosomal mutants and plasmids
Chromosomal mutants were constructed using the one-step disruption method (Datsenko and Wanner 2000) with minor modifications. To construct the hspQ mutant (JY683), a cat cassette was introduced into the hspQ gene as follows: A cat gene fragment was amplified from plasmid pKD3 using primer pairs 16046/16047 and then introduced into wild-type Salmonella 14028s harboring plasmid pKD46.
To construct the qad mutant (JY889), a kan cassette was introduced into the qad gene as follows: A kan gene fragment was amplified from plasmid pKD4 using primer pairs 16924/16925 and then introduced into wild-type Salmonella 14028s harboring plasmid pKD46.
To construct the lon mutant (EG16039), a cat cassette was introduced into the lon gene as follows: A cat gene fragment was amplified from plasmid pKD3 using primer pairs 3815/3816 and then introduced into wild-type Salmonella 14028s harboring plasmid pKD46.
To construct the clpP mutant (JY186), a cat cassette was introduced into the clpP gene as follows: A cat gene fragment was amplified from plasmid pKD3 using primer pairs 20000/20001 and then introduced into wild-type Salmonella 14028s harboring plasmid pKD46.
To construct the pat mutant (JY898), a cat cassette was introduced into the pat gene as follows: A cat gene fragment was amplified from plasmid pKD3 using primer pairs 16926/16927 and then introduced into hspQ-Flag (JY674) Salmonella 14028s harboring plasmid pKD46. Strain JY899 was made by transducing the qad::kan insertion into strain JY898 using a P22 lysate generated in strain JY889.
To construct strains specifying a C-terminally Flag-tagged HspQ protein (JY674, JY692, JY695, and JY740), a cat cassette was introduced at the 3′ end of hspQ: A cat gene fragment was amplified from pKD3 using primers 16044/16045 for hspQ-Flag and then introduced into wild-type Salmonella 14028s, clpS-HA (JY691) (Yeom et al. 2018), clpA-HA (JY694) (Yeom et al. 2018), and qad::kan (JY889) strains, each harboring plasmid pKD46. The resulting strains was kept at 30°C and transformed with pCP20 to remove the cat cassette.
To construct strains specifying C-terminally Flag-tagged HspQ derivatives in which Lys96 was substituted by an arginine (JY892) or a glutamine (JY894), a cat cassette was introduced at the 3′ end of hspQ: A cat gene fragment was amplified from pKD3 using primers 16928/16045 and 16929/16045 for hspQ-Flag, respectively, and then introduced into wild-type Salmonella 14028s harboring plasmid pKD46. Strains JY893 and JY895 were made by transducing the qad::kan insertion into strains JY892 and JY894, respectively, using a P22 lysate generated in strain JY889.
Strain JY872 was made by transducing the phoP-HA::cat (Shin and Groisman 2005) insertion into strain JY655 using a P22 lysate generated in strain EG13917. The resulting strains were kept at 30°C and transformed with pCP20 to remove the cat cassette. Strains JY696, JY775, and JY874 were made by transducing the clpS::cat insertion (Yeom et al. 2017) into strains JY674, JY774, and JY872, respectively, using a P22 lysate generated in strain JY570. The resulting strains were kept at 30°C and transformed with pCP20 to remove the cat cassette. Strains JY701 and JY902 were made by transducing the clpA::cat insertion (Yeom et al. 2017) into strains JY674 and JY655, respectively, using a P22 lysate generated in strain JY199. Strain JY741 was made by transducing the clpX::cat insertion (Tu et al. 2006) into strain JY674 using a P22 lysate generated in strain EG18499. Strains JY703, JY754, JY1009, JY1010, JY1011, JY1012, JY1013, and JY1014 were made by transducing the lon::cat insertion into strains JY674, JY740, JY655, JY657, JY686, JY774, JY773, and JY696, respectively, using a P22 lysate generated in strain EG16039.
Strains JY686, JY687, JY699, JY700, JY873, and JY875 were made by transducing the hspQ::cat insertion into strains JY655 (Yeom et al. 2018), JY657 (Yeom et al. 2018), JY694 (Yeom et al. 2018), JY691 (Yeom et al. 2018), JY872, and JY874, respectively, using a P22 lysate generated in strain JY683.
Strains JY705, JY772, JY773, and JY774 were made by transducing the qad::kan insertion into strains JY872, JY687, JY686, and JY655, respectively, using a P22 lysate generated in strain JY889.
Strains JY2000 and JY2001 were made by transducing the lon::cat insertion into strains JY892 and JY894, respectively, using a P22 lysate generated in strain EG16039. The resulting strains were kept at 30°C and transformed with pCP20 to remove the cat cassette. Strains JY2002 and JY2003 were made by transducing the hspQK96R-Flag::cat and hspQK96Q-FlagG::cat insertions into strains JY655 using a P22 lysate generated in strains JY892 and JY894, respectively.
A plasmid expressing HspQ was constructed as follows: The hspQ gene was amplified from wild-type Salmonella (14028s) using primer pairs 16052/16053. The PCR product was digested with BamHI and HindIII and then introduced between the BamHI and HindIII sites of pUHE21-2lacIq (Soncini et al. 1996).
A plasmid expressing Qad was constructed as follows: The qad gene was amplified from wild-type Salmonella (14028s) using primer pairs 16286/16287. The PCR product was digested with BamHI and HindIII and then introduced between the BamHI and HindIII sites of pUHE21-2lacIq (Soncini et al. 1996).
A plasmid expressing His-Hha was constructed as follows: The hha gene was amplified from wild-type Salmonella (14028s) using primer pairs 16629/16630. The PCR product was digested with BamHI and HindIII and then introduced between the BamHI and HindIII sites of pUHE21-2lacIq (Soncini et al. 1996).
A plasmid expressing the SsrA degradation motif-added GFP (GFP-LAA) was constructed as follows: The gfp-laa gene was amplified from pFPV25 plasmid DNA using primer pairs 1550/3071. The PCR product was digested with EcoRI and HindIII and then introduced between the EcoRI and HindIII sites of pFPV25 (Valdivia and Falkow 1996). GFP-LAA had a full SsrA tag sequence (RPAANDENYALAA) at the C terminus.
A plasmid expressing HspQ-His6 was constructed as follows: The hspQ gene was amplified from wild-type Salmonella (14028s) using primer pairs 16934/16935. The PCR product was digested with NdeI and HindIII and then introduced between the NdeI and HindIII sites of pET-28a(+).
Plasmids harboring genes specifying Qad-His6 and Pat-His6 were constructed as follows: The qad and pat genes were amplified from wild-type Salmonella (14028s) using primer pairs 16284/16285 and 16313/16314, respectively. The PCR products were digested with BamHI and XhoI and then introduced between the BamHI and XhoI sites of pET-28a(+).
Purification of the ClpA, ClpP, ClpS, FtsA, HspQ, Lon, Qad, Pat, and PhoP proteins
For purification of the Lon protein, 5 mL of saturated cultures of E. coli BL21 (DE3) harboring plasmid pUHE-His-Lon was used to inoculate 1 L of LB medium. Cells were grown to logarithmic phase (OD600 ≈ 0.3) at 37°C, and expression was induced by addition of 0.2 mM IPTG followed by growth for an additional 6 h at 30°C. Cells were collected and washed twice with 1× TBS (50 mM Tris-HCl at pH 8.0, 138 mM NaCl, 2.7 mM KCl). Next, cells were resuspended with 1× TBS and subjected to a French press at 18,000 psi. Clarified lysates were loaded onto an Ni-NTA gravity column equilibrated in equilibration buffer containing 10 mM imidazole, washed with 20 column volumes of the same, and eluted in 250 mM imidazole in the same buffer. Eluted proteins were exchanged with 1× TBS buffer followed by TBS buffer containing 10% glycerol and concentrated using an Amicon Ultra-3 (MW 3000; Millipore) filter.
For purification of the HspQ, Qad, and Pat proteins, 5 mL of saturated cultures of E. coli BL21 (DE3) (Studier and Moffatt 1986) harboring plasmid pET-28a(+)-HspQ, pET-28a(+)-Qad, or pET-28a(+)-Pat was used to inoculate, separately, 500 mL of LB medium. Cells were grown to logarithmic phase (OD600 ≈ 0.3) at 37°C, and expression was induced by addition of 1.0 mM IPTG followed by growth for an additional 16 h at 30°C. Cells were collected and washed twice with 1× TBS (50 mM Tris-HCl atpH 8.0, 138 mM NaCl, 2.7 mM KCl). Next, cells were resuspended with 1× TBS and subjected to a French press at 18,000 psi. Clarified lysates were loaded onto an Ni-NTA gravity column equilibrated in equilibration buffer containing 10 mM imidazole, washed with 20 column volumes of the same, and eluted in 250 mM imidazole in the same buffer. Eluted proteins were exchanged with 1× TBS buffer followed by TBS buffer containing 10% glycerol and concentrated using an Amicon Ultra-3 (MW 3000; Millipore) filter. The His tag of HspQ was cleaved by ∼2 U/mL thrombin for 16 h at 4°C and then concentrated using an Amicon Ultra-3 (MW 3000; Millipore) filter.
Purification of the PhoP, FtsA, ClpS, ClpA, and ClpP was performed as described (Yeom et al. 2018).
Western blot assay
Cells were grown in N-minimal medium containing 10 µM or 10 mM MgCl2. Crude extracts were prepared in B-PER reagent (Pierce) with 100 µg/mL lysozyme and EDTA-free protease inhibitor (Roche). Samples were loaded onto 4%–12% NuPAGE gels (Life Technologies) and transferred to nitrocellulose membrane using the iBot machine (Life Technologies). Membranes were blocked with 3% skim milk solution at room temperature for 2 h. Next, samples were analyzed using antibodies directed to the HA, His, or Flag tags or the OmpA, GFP, GroEL, or AtpB proteins. Rabbit anti-HA, anti-Flag, and anti-His antibodies were used at 1:2000 dilution. Mouse anti-GFP antibody was used at 1:5000 dilution. Rabbit anti-OmpA was used as control at 1:5000 dilution. Mouse anti-AtpB and anti-GroEL were used as control at 1:5000 dilution. Secondary horseradish peroxidase-conjugated antirabbit or antimouse antiserum (GE healthcare) was used at 1:5000 dilution. The blots were developed with the Amersham ECL Western blotting detection reagents (GE Healthcare) or SuperSignal West Femto chemiluminescent system (Pierce).
In vitro substrate degradation assay
In vitro substrate degradation assays were performed as described earlier with some modifications (Puri and Karzai 2017). The assay was performed in a solution containing 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 20 mM MgCl2, 10% glycerol, and 1 mM DTT. Purified proteins were used as 0.2 or 1.0 µM ClpS, 0.2 or 0.1 µM ClpA, 0.2 µM ClpP, 0.2 µM Lon, 0.5 µM HspQ, 0.5 µM PhoP, 0.2 µM FtsA-Flag, and 0.5 µM Hha. All reaction components except the ATP regeneration system were mixed and incubated for 5 min at 30°C. Reactions were carried out for the indicated times at 30°C in the presence of an ATP regeneration system (2 mM ATP, 20 µg/mL pyruvate kinase, 4 mM pyruvate phosphate) started by addition of substrates. Samples were removed from the reactions at the indicated times, and reactions were stopped by the addition of sample buffer. After separation by SDS-PAGE, proteins were detected by Coomassie blue staining (Invitrogen).
In vivo protein degradation assay
To measure protein stability, cells were grown in 10 mL of N-minimal medium containing 10 µM MgCl2 for 5 h. Cells were treated with 50 µg/mL tetracycline, and 1.5 mL samples were removed at the indicated times and harvested at 4°C. Pelleted cells were kept on dry ice for 30 min. Samples were then resuspended in B-PER reagent (Pierce) with 100 µg/mL lysozyme and EDTA-free protease inhibitor (Roche). After addition of the same volume of SDS sample buffer, samples were separated on 4%–12% SDS–polyacrylamide gel and analyzed by Western blotting.
In vivo pull-down assay
Interactions among the HspQ-Flag, ClpS-HA, and ClpA-HA proteins were investigated using a strain expressing a C-terminally Flag-tagged HspQ protein, C-terminally HA-tagged ClpS protein, or C-terminally HA-tagged ClpA protein from their normal chromosomal locations. Cells were grown overnight in N-minimal medium containing 10 mM Mg2+. One milliliter of the overnight culture was washed in N-minimal medium without Mg2+ and resuspended in 1 mL of the same medium. Bacteria (1/50 dilution) were inoculated in 20 mL of N-minimal medium containing 10 µM Mg2+ and grown for 6 h. Crude extracts were prepared as described above and incubated with anti-HA magnetic beads (Pierce) or anti-Flag magnetic beads (Sigma) overnight at 4°C. After washing the beads with 1× TBS buffer (Sigma), binding proteins were eluted in 100 µL of SDS sample buffer without reducing agents, separated on 4%–12% SDS–polyacrylamide gel, and analyzed by Western blotting using antibodies directed to the Flag or HA tags described above.
Pull-down assay with proteins synthesized in vitro
Pull-down assay was performed with proteins produced using an in vitro transcription/translation system as described (Lee et al. 2013) with some modifications. Proteins were produced using the cell-free PURExpress in vitro protein synthesis system (New England Biolabs) for 3 h at 37°C. DNA templates were prepared according to the manufacturer's instructions. To synthesize the hspQ-Flag, clpS-HA, and rssB-HA genes, we used primers 16936/16937 for hspQ-Flag, 15949/16938 for clpS-HA, and 16939/16940 for rssB-HA. At the end of the reaction, samples were diluted with 20 vol in 1× TBS buffer. Diluted reactions were mixed in 500 µL of 1× TBS and incubated for 2 h at room temperature. Next, samples were pulled down with either anti-HA, anti-His, or anti-Flag antibodies for 2 h at room temperature. Pulled-down samples were analyzed by Western blotting using anti-HA, anti-His or anti-Flag antibodies.
In vitro protein acetylation assay
In vitro protein acetylation assays were performed as described (Qin et al. 2016) with some modifications. The assay was performed in a solution containing 50 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, 10% glycerol, 1 mM DTT, and 10 mM sodium butyrate. Purified proteins were used as 0.2 µM HspQ, 0.5 µM Qad, and 0.2 µM Pat. Reactions were carried out for 3 h at 37°C in the presence of 2 mM acetyl-CoA. Reactions were stopped by incubation on ice for 10 min. Samples were separated on 4%–12% SDS–polyacrylamide gel and analyzed by Western blotting.
Detection of acetylated HspQ protein by high-resolution MS/MS analysis
Protein posttranslational modifications (acetylation, oxidation, methylation, and decarboxylation) were determined by liquid chromatography (LC)-MS/MS analysis. The purified HspQ protein was separated by SDS-PAGE, and proteins were detected by Coomassie blue staining (Invitrogen). The band corresponding to HspQ was excised from the gel and digested with trypsin. Protein samples were desalted and applied in a Thermo Scientific LTQ Orbitrap XL connected to a Waters nanoACQUITY ultrahigh-performance LC (UPLC) system and equipped with a Waters Symmetry C18 180 µm × 20 mm trap column and a 1.7-µm 75 µm × 250 mm nanoAcquity UPLC column at 35°C. Detection of posttranslational modifications and peptide identification were carried out using the Mascot search algorithm (Matrix Science, version 2.4.0).
Measurement of acetyl-coA amounts
Acetyl-CoA amounts (PicoProbe acetyl CoA assay kit, Abcam) was determined as per the manufacturer's protocol. Cells were grown in N-minimal medium containing 10 µM MgCl2 in 38 mM glucose or glycerol for 6 h at 37°C.
Quantitative RT-PCR
To measure mRNA abundance, cells were grown in N-minimal medium containing 10 µM MgCl2 in 38 mM glucose or glycerol for 6 h at 37°C. Total RNA was purified by using RNeasy kit (Qiagen) with on-column DNase treatment, and cDNA was synthesized by using VILO supermix (Life Technologies). Quantification of transcripts was carried out by quantitative RT-PCR using SYBR Green PCR master mix (Applied Biosystems) in a QuantStudio 6 Flex real-time PCR system (Applied Biosystems). mRNA abundance was determined by using a standard curve obtained from PCR products generated with serially diluted genomic DNA, and results were normalized to the abundance of the ompA gene mRNA. Data shown are an average from at least three independent experiments. The primers used in quantitative RT-PCR assay are listed in Supplemental Table S3.
Bacterial two-hybrid analysis to examine protein–protein interactions
We used the BACTH system (Battesti and Bouveret 2012) with the following constructs: The hspQ, rssB, clpS, clpA, and clpP genes were PCR-amplified, and the PCR fragments were cloned between the XbaI and KpnI sites of the pUT18C vectors and pKT25 to generate genes specifying the corresponding fusion proteins. Recombinant plasmids carrying the pKT25-hspQ and pUT18c-rssB, clpS, clpA, or clpP genes were cotransformed into strain BTH101. Transformants were plated on LB agar plates containing 100 µg/mL ampicillin and 50 µg/mL kanamycin and incubated for 24 h at 30°C. To quantify the interaction between hybrid proteins, bacteria were grown overnight at 30°C as recommended in the BACHT protocol (Battesti and Bouveret 2012) in LB Amp Kan liquid medium supplemented with 0.5 mM IPTG. All samples were spotted onto LB agar plates supplemented with 100 µg/mL ampicillin, 50 µg/mL kanamycin, 40 µg/mL X-Gal, and 0.5 mM IPTG.
Data deposition
The images of the original gels corresponding to the figures in this study were deposited in the Mendeley database and are available at http://dx.doi.org/10.17632/ftykgzf2tn.1.
Acknowledgments
We thank Xinyu Hong for genomic analysis of hspQ-qad region, and Jennifer Aronson for comments on the manuscript. This research was supported by National Institutes of Health grant AI49561 to E.A.G.
Author contributions: J.Y. and E.A.G. conceived the study, designed and performed the research, analyzed the data, and wrote the paper.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.325241.119.
References
- Battesti AL, Bouveret E. 2012. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58: 325–334. 10.1016/j.ymeth.2012.07.018 [DOI] [PubMed] [Google Scholar]
- Belman JP, Bian RR, Habtemichael EN, Li DT, Jurczak MJ, Alcázar-Román A, McNally LJ, Shulman GI, Bogan JS. 2015. Acetylation of TUG protein promotes the accumulation of GLUT4 glucose transporters in an insulin-responsive intracellular compartment. J Biol Chem 290: 4447–4463. 10.1074/jbc.M114.603977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056–1060. 10.1038/nature07813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta VJ, Cristea IM. 2017. Regulation, function, and detection of protein acetylation in bacteria. J Bacteriol 199: e00107–e00117. 10.1128/JB.00107-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette MF, Henderson GW, Markovitz A. 1981. ATP hydrolysis-dependent protease activity of the lon (capR) protein of Escherichia coli K-12. Proc Natl Acad Sci 78: 4728–4732. 10.1073/pnas.78.8.4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97: 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RW, Botstein D, Roth JR. 1980. Advanced bacterial genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Dougan DA, Reid BG, Horwich AL, Bukau B. 2002. ClpS, a substrate modulator of the ClpAP machine. Mol Cell 9: 673–683. 10.1016/S1097-2765(02)00485-9 [DOI] [PubMed] [Google Scholar]
- Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B. 2006. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439: 753–756. 10.1038/nature04412 [DOI] [PubMed] [Google Scholar]
- Farrell CM, Grossman AD, Sauer RT. 2005. Cytoplasmic degradation of ssrA-tagged proteins. Mol Microbiol 57: 1750–1761. 10.1111/j.1365-2958.2005.04798.x [DOI] [PubMed] [Google Scholar]
- Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci 83: 5189–5193. 10.1073/pnas.83.14.5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T, Eguchi T, Atomi H, Imanaka T. 2002. A membrane-bound archaeal Lon protease displays ATP-independent proteolytic activity towards unfolded proteins and ATP-dependent activity for folded proteins. J Bacteriol 184: 3689–3698. 10.1128/JB.184.13.3689-3698.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibellini L, Pinti M, Beretti F, Pierri CL, Onofrio A, Riccio M, Carnevale G, De Biasi S, Nasi M, Torelli F, et al. 2014. Sirtuin 3 interacts with Lon protease and regulates its acetylation status. Mitochondrion 18: 76–81. 10.1016/j.mito.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Gottesman S, Squires C, Pichersky E, Carrington M, Hobbs M, Mattick JS, Dalrymple B, Kuramitsu H, Shiroza T, Foster T, et al. 1990. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc Natl Acad Sci 87: 3513–3517. 10.1073/pnas.87.9.3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA. 2016. Feedback control of two-component regulatory systems. Ann Rev Micro 70: 103–124. 10.1146/annurev-micro-102215-095331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72: 165–168. 10.1016/0092-8674(93)90655-A [DOI] [PubMed] [Google Scholar]
- Joshi KK, Bergé M, Radhakrishnan SK, Viollier PH, Chien P. 2015. An adaptor hierarchy regulates proteolysis during a bacterial cell cycle. Cell 163: 419–431. 10.1016/j.cell.2015.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberdin VR, Bläsi U. 2006. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol Rev 30: 967–979. 10.1111/j.1574-6976.2006.00043.x [DOI] [PubMed] [Google Scholar]
- Kirstein J, Molière N, Dougan DA, T ürgay K. 2009. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol 7: 589–599. 10.1038/nrmicro2185 [DOI] [PubMed] [Google Scholar]
- Lee EJ, Pontes MH, Groisman EA. 2013. A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium's own F1Fo ATP synthase. Cell 154: 146–156. 10.1016/j.cell.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid C, Balsalobre C, García J, Juárez A. 2007. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol Microbiol 63: 7–14. 10.1111/j.1365-2958.2006.05497.x [DOI] [PubMed] [Google Scholar]
- Mandin P. 2012. Genetic screens to identify bacterial sRNA regulators. Methods Mol Biol 905: 41–60. 10.1007/978-1-61779-949-5_4 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Asakura Y, Friso G, Kim J, Oh SH, Rutschow H, Ponnala L, van Wijk KJ. 2013. ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 25: 2276–2301. 10.1105/tpc.113.112557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Apitz J, Friso G, Kim J, Ponnala L, Grimm B, van Wijk KJ. 2015. Discovery of a unique Clp component, ClpF, in chloroplasts: a proposed binary ClpF–ClpS1 adaptor complex functions in substrate recognition and delivery. Plant Cell 27: 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–304. 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- Puri N, Karzai AW. 2017. HspQ functions as a unique specificity-enhancing factor for the AAA+ Lon protease. Mol Cell 66: 672–683.e4. 10.1016/j.molcel.2017.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin R, Sang Y, Ren J, Zhang Q, Li S, Cui Z, Yao YF. 2016. The bacterial two-hybrid system uncovers the involvement of acetylation in regulating of Lrp activity in Salmonella typhimurium. Front Microbiol 7: 1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Sang Y, Tan Y, Tao J, Ni J, Liu S, Fan X, Zhao W, Lu J, Wu W, et al. 2016. Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS Pathog 12: e1005458. 10.1371/journal.ppat.1005458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román-Hernández G, Hou JY, Grant RA, Sauer RT, Baker TA. 2011. The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease. Mol Cell 43: 217–228. 10.1016/j.molcel.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotanova TV, Melnikov EE, Khalatova AG, Makhovskaya OV, Botos I, Wlodawer A, Gustchina A. 2004. Classification of ATP-dependent proteases Lon and comparison of the active sites of their proteolytic domains. Eur J Biochem 271: 4865–4871. 10.1111/j.1432-1033.2004.04452.x [DOI] [PubMed] [Google Scholar]
- Schmidt MT, Smith BC, Jackson MD, Denu JM. 2004. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem 279: 40122–40129. 10.1074/jbc.M407484200 [DOI] [PubMed] [Google Scholar]
- Shanklin J, DeWitt ND, Flanagan JM. 1995. The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell 7: 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Groisman EA. 2005. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J Biol Chem 280: 4089–4094. 10.1074/jbc.M412741200 [DOI] [PubMed] [Google Scholar]
- Snavely MD, Miller CG, Maguire ME. 1991. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem 266: 815–823. [PubMed] [Google Scholar]
- Soncini FC, García Véscovi E, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol 178: 5092–5099. 10.1128/jb.178.17.5092-5099.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69: 183–215. 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189: 113–130. 10.1016/0022-2836(86)90385-2 [DOI] [PubMed] [Google Scholar]
- Takamura Y, Nomura G. 1988. Changes in the intracellular concentration of acetyl-Coa and malonyl-Coa in relation to the carbon and energy-metabolism of Escherichia coli K12. J Gen Microbiol 134: 2249–2253. [DOI] [PubMed] [Google Scholar]
- Takaya A, Kubota Y, Isogai E, Yamamoto T. 2005. Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol Microbiol 55: 839–852. 10.1111/j.1365-2958.2004.04425.x [DOI] [PubMed] [Google Scholar]
- Thompson MW, Maurizi MR. 1994. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem 269: 18201–18208. [PubMed] [Google Scholar]
- Tu X, Latifi T, Bougdour A, Gottesman S, Groisman EA. 2006. The PhoP/PhoQ two-component system stabilizes the alternative σ factor RpoS in Salmonella enterica. Proc Natl Acad Sci 103: 13503–13508. 10.1073/pnas.0606026103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Falkow S. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol 22: 367–378. 10.1046/j.1365-2958.1996.00120.x [DOI] [PubMed] [Google Scholar]
- Wang N, Maurizi MR, Emmert-Buck L, Gottesman MM. 1994. Synthesis, processing, and localization of human Lon protease. J Biol Chem 269: 29308–29313. [PubMed] [Google Scholar]
- Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136: 615–628. 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. 1999. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401: 90–93. 10.1038/43481 [DOI] [PubMed] [Google Scholar]
- Weinert BT, Iesmantavicius V, Wagner SA, Scholz C, Gummesson B, Beli P, Nystrom T, Choudhary C. 2013. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell 51: 265–272. 10.1016/j.molcel.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Yeom J, Wayne KJ, Groisman EA. 2017. Sequestration from protease adaptor confers differential stability to protease substrate. Mol Cell 66: 234–246.e5. 10.1016/j.molcel.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom J, Gao X, Groisman EA. 2018. Reduction in adaptor amounts establishes degradation hierarchy among protease substrates. Proc Natl Acad Sci 115: E4483–E4492. 10.1073/pnas.1722246115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. 2001. The RssB response regulator directly targets σS for degradation by ClpXP. Genes Dev 15: 627–637. 10.1101/gad.864401 [DOI] [PMC free article] [PubMed] [Google Scholar]