Here, Marnef et al. found that DSBs induced on rDNA trigger transcriptional repression in a cohesin- and HUSH complex-dependent manner throughout the cell cycle. Their findings indicate that rDNA break localization at the nucleolar periphery is not a direct consequence of transcriptional repression but rather is an active process that shares features with the mobilization of persistent DSB in active genes and heterochromatin.
Keywords: DSB repair, HUSH, LINC, chromatin, cohesin, ribosomal DNA
Abstract
The ribosomal DNA (rDNA) represents a particularly unstable locus undergoing frequent breakage. DNA double-strand breaks (DSBs) within rDNA induce both rDNA transcriptional repression and nucleolar segregation, but the link between the two events remains unclear. Here we found that DSBs induced on rDNA trigger transcriptional repression in a cohesin- and HUSH (human silencing hub) complex-dependent manner throughout the cell cycle. In S/G2 cells, transcriptional repression is further followed by extended resection within the interior of the nucleolus, DSB mobilization at the nucleolar periphery within nucleolar caps, and repair by homologous recombination. We showed that nuclear envelope invaginations frequently connect the nucleolus and that rDNA DSB mobilization, but not transcriptional repression, involves the nuclear envelope-associated LINC complex and the actin pathway. Altogether, our data indicate that rDNA break localization at the nucleolar periphery is not a direct consequence of transcriptional repression but rather is an active process that shares features with the mobilization of persistent DSB in active genes and heterochromatin.
Double-strand breaks (DSBs) are highly harmful lesions that arise on DNA following exposition to damaging agents but also during normal cell metabolic activity. Importantly, genome-wide mapping of endogenous DSBs has indicated that they preferentially occur on the transcribing genome as an occasional consequence of topoisomerase activity or processing of DNA secondary structures (such as R loops and G quadruplexes) or due to replication fork stalling and/or collapse (for review, see Marnef et al. 2017; Tubbs and Nussenzweig 2017). Multiple repair pathways cope with such lesions, including nonhomologous end joining (NHEJ), which reseals DNA ends with no or minimal processing throughout the cell cycle phases, and homology-driven mechanisms, which take place in S and G2 and rely on the generation of ssDNA through a process called resection (for review, see Mladenov et al. 2016). Notably, these repair mechanisms trigger different mutational signatures when inaccurate, such as point mutations, translocations, or repeat amplification (Mladenov et al. 2016).
Recent work revealed that the initial genomic context in which a DSB occurs plays a critical role in the decision that will assign a specific repair pathway to the detected break (Clouaire and Legube 2015). We found previously that DSBs induced in transcribing loci are channeled to homologous recombination (HR) in G2 thanks to H3K36me3-dependent signaling (Aymard et al. 2014), while they tend to persist and relocate within DSB clusters during the G1 cell cycle phase (Aymard et al. 2017). Notably, movement into DSB clusters relies on ATM activity, the MRN complex, the nuclear envelope (NE)-embedded LINC complex, and the actin network (Aymard et al. 2017; Schrank et al. 2018; for review, see Guénolé and Legube 2017). Additionally, repair at active genes also necessitates prior transcriptional extinction of the damaged gene in a manner that depends on ATM activity, chromatin remodeling complexes (PBAF), and repressive histone modifications (Shanbhag et al. 2010; Kakarougkas et al. 2014; Ui et al. 2015). On the other hand, DSBs induced elsewhere in euchromatin (on inactive genes and intergenic regions) do not tend to move into clusters but rather are repaired by NHEJ in both G1 and G2 (Aymard et al. 2014).
DSBs located in heterochromatin that is tightly packaged, transcriptionally silent, and primarily composed of repetitive elements also display a specialized repair behavior (Noon et al. 2010; Goodarzi et al. 2011; Frohns et al. 2014). Notably, the use of HR involves the repositioning of the break at the periphery of the heterochromatin domain (Chiolo et al. 2011; Jakob et al. 2011; Tsouroula et al. 2016; Colmenares et al. 2017) and at the NE (Chiolo et al. 2011; Ryu et al. 2015; Colmenares et al. 2017; Caridi et al. 2018) in Drosophila in a manner that involves the actin network (Caridi et al. 2018).
The ribosomal DNA (rDNA) is another genomic region exhibiting a unique and specialized chromatin structure due to its repetitive nature, elevated level of secondary structures (R loops and G4), and considerable transcriptional activity (for review, see Lindström et al. 2018). In human cells, the ∼300 rDNA repeats are distributed between the short arms of the five acrocentric chromosomes, each of which contains a nucleolar organizer region (NOR), around which nucleoli form (McStay 2016). It is well described that inhibition of rRNA synthesis triggers a complete nucleolar reorganization, with the movement and segregation of the inactivated rDNA at the periphery of the nucleolus in the so-called “nucleolar caps,” with most caps corresponding to a single NOR (Floutsakou et al. 2013). This strong dependency of nucleolus morphology on active rDNA transcription led to the idea that the nucleolus is a self-organizing compartment ensured by the massive amount of RNAs produced and RNA-binding proteins, potentially through a liquid-demixing process, which is now supported by several pieces of evidence (Németh and Grummt 2018). From yeast to humans, the rDNA poses a challenge in terms of genome maintenance. Previous studies established that the rDNA is particularly susceptible to breakage, probably due to multiple secondary DNA structures, high RNA polymerase I (Pol I) occupancy, and tightly DNA-bound regulatory proteins, which generate a high incidence of replication fork stalling and/or collapse (Lindström et al. 2018). Additionally, its highly repetitive nature renders this locus particularly prone to rearrangements, generating translocations, extrachromosomal circles, and repeat contractions/expansions (Lindström et al. 2018). However, the DSB signaling and repair mechanisms that cope with this peculiar locus are only recently emerging in human cells.
Similarly to RNA Pol II transcribed genes, production of DSBs in the nucleolus rapidly elicits a local rDNA transcriptional shutdown in an ATM- and Nbs1-dependent manner (Kruhlak et al. 2007; Larsen et al. 2014; Harding et al. 2015; van Sluis and McStay 2015; Warmerdam et al. 2016). Such DSB-induced transcriptional repression is believed to trigger the segregation of the rDNA at the periphery of the nucleolus and the formation of the nucleolar caps (Kruhlak et al. 2007; Larsen et al. 2014; Harding et al. 2015; van Sluis and McStay 2015; Warmerdam et al. 2016). In terms of repair, the general picture is still blurry. Indeed, while the induction of DSBs on the rDNA array was reported to be largely repaired by HR within nucleolar caps even during G1 (van Sluis and McStay 2015), another report argues for a DNA-PK-dependent NHEJ repair pathway (Harding et al. 2015).
Here we reveal that the nucleolar cap formation observed upon rDNA damage is not the direct consequence of transcriptional arrest but rather reflects an active mechanism that allows the mobilization of resected DSBs to the nucleolar periphery to further undergo repair by HR. We identified the cohesin and human silencing hub (HUSH) complexes as involved in rDNA transcriptional repression upon damage, while DSB mobilization at the periphery of the nucleolus is ensured by an actin network and a LINC-dependent mechanism. Strikingly, the NE forms invaginations that contact the nucleolus, likely providing a safe environment for persistent DSB repair, as shown in yeast and Drosophila.
Results
Uncoupling between transcriptional arrest and nucleolar cap formation upon rDNA breakage
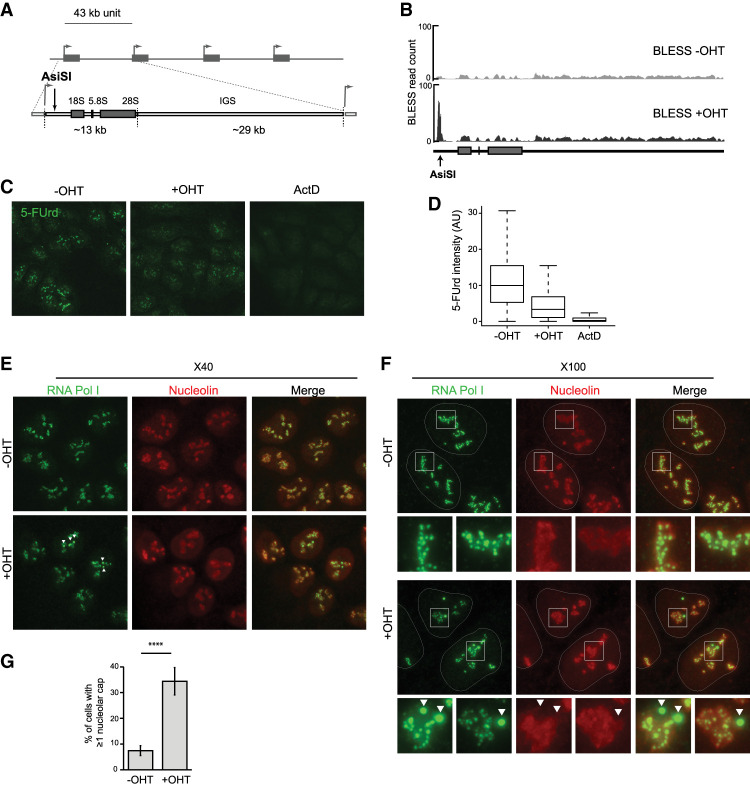
In order to investigate the rDNA breakage response and repair, we used the human DIvA stable cell line, in which relocation of the ectopic AsiSI restriction enzyme from the cytoplasm to the nucleus can be induced upon 4-hydroxytamoxifen (OHT) addition, allowing the formation of DSBs (Iacovoni et al. 2010). In addition to the DSBs induced elsewhere on the genome (174, as determined previously) (Clouaire et al. 2018), an AsiSI site (GCGATCGC) is predicted in the 5′ external transcribed spacer (5′ ETS) of the 47S rDNA units (Fig. 1A). We confirmed the appearance of a rDNA break at the AsiSI-annotated site after OHT addition by direct in situ break labeling, enrichment on streptavidin, and next-generation sequencing (BLESS) (Fig. 1B; Crosetto et al. 2013; Clouaire et al. 2018). Several studies showed previously that DSBs induced in the rDNA trigger inhibition of rRNA synthesis as well as nucleolar cap formation (Kruhlak et al. 2007; Larsen et al. 2014; Harding et al. 2015; van Sluis and McStay 2015; Warmerdam et al. 2016). AsiSI-induced rDNA breaks also triggered similar responses regarding rDNA transcriptional repression, as assessed by 5-fluorouridine (5-FUrd) incorporation followed by automated high-throughput microscopy (Fig. 1C,D), and nucleolar cap formation, as revealed by RNA Pol I (Fig. 1E–G; Supplemental Fig. S1A; Supplemental Movie S1) or UBF (Supplemental Fig. S1B,C) staining.
Figure 1.
Transcription inhibition and cap formation following AsiSI-induced rDNA breaks. (A) Schematic representation of rDNA repeats and the position of the AsiSI site in the 5′ ETS. (B) BLESS signal at the AsiSI site in the presence or absence of OHT treatment. (C) High-content microscopy image (20× objective) of rRNA synthesis as visualized with 5-FUrd incorporation before and after OHT and actinomycin D (ActD) treatments. Actinomycin D was used as a control for the extinction of rRNA synthesis. (D) Box plot showing the quantification of 5-FUrd signal (>5000 cells) before OHT, after OHT, and after actinomycin D treatment. A representative experiment is shown. (E) DIvA cells either before or after OHT treatment were stained with antibodies against nucleolin (red) and RNA Pol I (green). Nucleolar caps are indicated with arrows. Images were acquired with a 40× objective. (F) Same as in E, but images were acquired with a 100× objective. Enlargements (white squares) of nucleoli that display nucleolar caps (arrow) or not are also shown. (G) The number of cells with at least one nucleolar cap, as measured by RNA Pol I staining before and after OHT treatment as indicated. The average and SD of four independent experiments are shown. (****) P < 0.0001.
After OHT treatment, nucleolar cap formation occurred in only a subset of DIvA cells (∼35%) (Fig.1G; Supplemental Fig. S1C), in contrast to what was reported previously following rDNA breaks generated by CRISPR/Cas9 (see also Supplemental Fig. S1D–F; van Sluis and McStay 2015). In addition, nucleolar segregation observed following AsiSI was only partial (Supplemental Fig. S1G,H), unlike following actinomycin D treatment (Supplemental Fig. S1E–H). Blocking AsiSI-induced rDNA repair with an inhibitor against DNA-PK (Caron et al. 2015) enhanced both the number of cells displaying at least one nucleolar cap (Supplemental Fig. S1E,F) and the number of cells displaying total nucleolar segregation (Supplemental Fig. S1G,H), hence mimicking the phenotypes observed with CRISPR/Cas9. Moreover, inducing rDNA breaks using simultaneously a CRISPR sgRNA and AsiSI also enhanced nucleolar cap formation compared with AsiSI alone (Supplemental Fig. S1I,J). Altogether, these data suggest that the discrepancy in nucleolar segregation observed upon AsiSI- and CRISPR/Cas9-induced breakage is likely due to the peculiar nature of Cas9-induced breaks (see the Discussion; Brinkman et al. 2018).
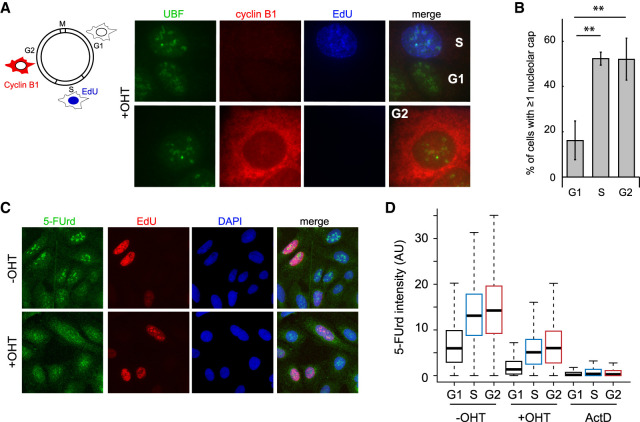
Given that only a subset of DIvA cells displayed nucleolar caps, we wondered whether this could be cell cycle-regulated. EdU labeling and Cyclin B1 costaining revealed that nucleolar caps are observed mainly in S/G2 cells (Fig. 2A,B) even though rDNA cleavage was detected efficiently by BLESS in G1 and G2 cells (Supplemental Fig. S2A). The fact that nucleolar caps were substantially reduced in G1 therefore gave us the opportunity to test whether nucleolar cap formation is a consequence of transcriptional arrest as proposed previously (van Sluis and McStay 2015). We observed that DSB-induced transcriptional arrest occurs similarly during G1, S, and G2, as measured by quantitative image-based cytometry (QIBC) coupled with 5-FUrd incorporation (Fig. 2C,D; Supplemental Fig. S2B), hence demonstrating that nucleolar cap formation is not solely the consequence of the rDNA transcriptional repression.
Figure 2.
Decoupling of transcription inhibition and cap formation during the cell cycle. (A, left panel) Schematic representation of the cell cycle and the methods used to visualize the G2 cells (cytoplasmic cyclin B1 staining in red), S-phase cells (nuclear EdU staining in blue), or G1 cells (absence of staining). (Right panel) Images of OHT-treated cells showing the presence of nucleolar caps (assessed by UBF staining) in EdU- or cyclin B1-positive cells. (B) The number of cells with at least one nucleolar cap in G1, S, or G2 phases as counted with UBF. The mean and SD from three independent experiments are shown. (**) P < 0.01. (C) Representative images with a 20× objective from the QIBC analysis of the 5-FUrd levels (transcription) using EdU labeling (DNA synthesis) and DAPI staining to distinguish cell cycle phases. (D) A representative box plot of the quantification of the 5-FUrd level (>1500 cells) across the cell cycle phase (G1 [black, S [blue], and G2 [red]) following OHT or actinomycin D (ActD) treatment (as indicated).
Extended resection occurs within the nucleolus at rDNA breaks
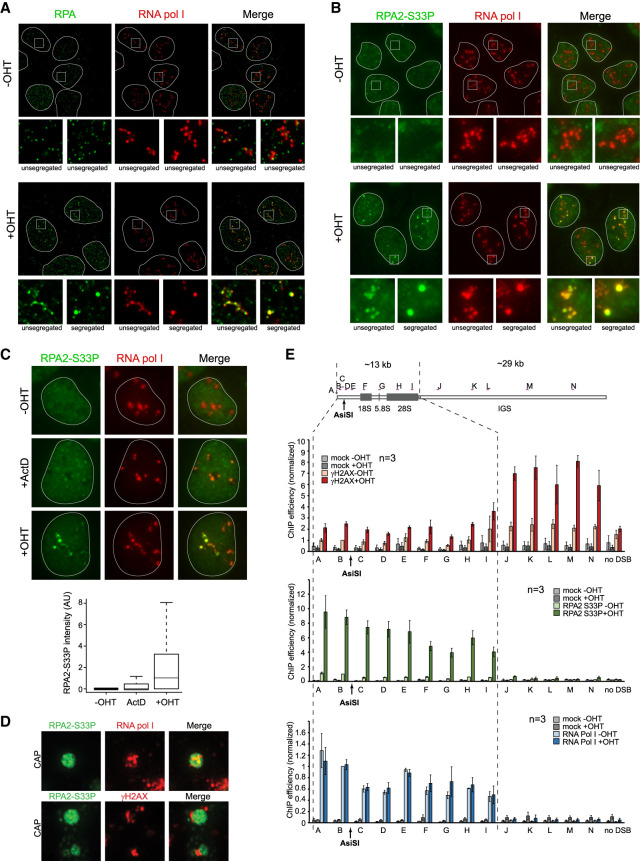
To investigate how rDNA DSB repair is orchestrated within the nucleolus, we further performed immunostaining against DSB signaling and repair proteins. As reported previously at CRISPR/Cas9- or I-PpoI-induced rDNA DSBs (Harding et al. 2015; Franek et al. 2016; Warmerdam et al. 2016), we found that AsiSI-rDNA breaks recruited γH2AX, ubiquitin, MDC1, 53BP1, and RIF1 only at the periphery of the nucleolus at nucleolar caps (Supplemental Fig. S3A–C,F). Similarly, proteins involved in HR (Rad51 and BRCA1) also accumulated at nucleolar caps (Supplemental Fig. S3D–F), indicating that DSB mobilization at the periphery of the nucleolus is a prerequisite for HR protein assembly. We therefore looked for the status of DNA end resection, which constitutes an early step of HR repair. Accumulation of RPA and phosphorylated RPA at Ser33 (RPA2-S33P), indicative of end resection, was clearly detected after DSB induction both inside the nucleolus and in nucleolar caps (Fig. 3A,B; Supplemental Fig. S3F). As expected, RPA2-S33P staining was detected mainly in S/G2 cells (Supplemental Fig. S3G). In contrast, actinomycin D treatment only mildly induced RPA staining, indicating that, in nucleoli, RPA accumulation is specific to DSB formation (Fig. 3C).
Figure 3.
Extended resection along the transcribed region occurs within the nucleolus. (A) DIvA cells left untreated or treated for 4 h with OHT (as indicated) were stained with RNA Pol I (red) and RPA2 (green). Examples of RPA2 staining in normal and segregated (nucleolus with at least one cap) nucleoli are shown in magnification at the bottom. (B) Same as in A, except that total RPA2-S33P was stained (green). (C) Untreated (−OHT), actinomycin D-treated (ActD), or OHT-treated DIvA cells stained with RPA2-S33P (green) and RNA pol I (red). Box plot of RPA2-S33P nucleolar intensity in the indicated conditions. A representative experiment is shown. (D, top panel) A schematic representation of the primers used across the rDNA unit for quantitative PCR (qPCR). Chromatin immunoprecipitation (ChIP)-qPCR were performed using antibodies against gH2AX, RPA2-S33P, and RNA Pol I in DIvA cells left untreated or treated for 4 h with OHT (as indicated). The mean of normalized ChIP efficiency and SEM of three independent experiments are shown. (E) Superresolution images depicting the presence of RPA2-S33P (green) and RNA pol I (red) within nucleolar caps (top) or the mutual exclusion of RPA2-S33P (green) in caps and γH2AX (red) surrounding the caps (bottom).
Interestingly, superresolution microscopy showed a mutual exclusion of RPA2-S33P and γH2AX once relocated at nucleolar caps (Fig. 3D). In order to further characterize resection at rDNA breaks, we performed RPA2-S33P chromatin immunoprecipitation (ChIP). In agreement with superresolution microscopy, RPA2-S33P distribution was found to be antagonistic to that of γH2AX (Fig. 3E, top and middle panels). Of note, low γH2AX occupancy on the transcribed region is likely due to decreased nucleosome occupancy rather than decreased DDR kinase activity, as indicated by ChIP against H2AX (Supplemental Fig. S3H). Strikingly, RPA2-S33P was massively recruited across the 13 kb of the transcribed region, as indicated by RNA Pol I binding (Fig. 3E, bottom panel), indicating that resection takes place on the entire transcribed unit.
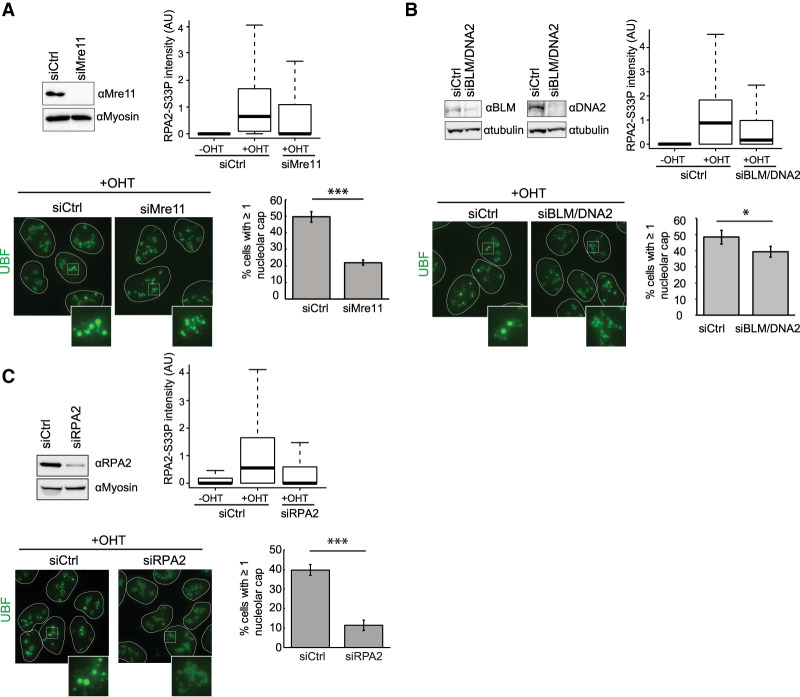
We further tested whether resection is a prerequisite for DSB mobilization in nucleolar caps. Blocking NHEJ in G1 cells using a DNA-PK inhibitor enhanced both resection (RPA2-S33P staining) and cap formation, suggesting a relationship between resection and DSB mobilization (Supplemental Fig. S4). Furthermore, depletion of the Mre11 component of the MRN complex (known to initiate resection), of BLM/DNA2 (mediating long-range resection) (Bordelet and Dubrana 2019), and of RPA2 all impaired nucleolar cap formation (Fig. 4A–C).
Figure 4.
Resection a is a prerequisite for nucleolar cap formation. (A, top panels) Western blots (left) and representative box plots of the level of RPA2-S33P nucleolar intensity (right) after Mre11 or control siRNA knockdown followed by 4 h of OHT treatment in DIvA cells (>100 nucleoli quantified). (Bottom panels) UBF staining was performed in OHT-treated Mre11 or control siRNA transfected DIvA cells. An example (left) and quantification of the percentage of cells displaying at least one nucleolar cap (right) are shown. The mean and SD from three independent experiments are shown. (***) P < 0.001. (B) Same as in A, except that BLM/DNA2 were knocked down by siRNAs. The mean and SD from three independent experiments are shown. (*) P < 0.05. (C) Same as in A, except that RPA2 was knocked down by siRNAs. The mean and SD from three independent experiments are shown. (***) P < 0.001.
Altogether, our data suggest that, in S/G2, rDNA DSBs undergo extensive resection spanning the entire transcribed unit within the interior of the nucleolus. This resection event is further required for the movement of rDNA DSBs at the periphery of the nucleolus, where both signaling (γH2AX, MDC1, and ubiquitin chain formation) and HR (Rad51 and BRCA1 binding) can occur.
The cohesin complex ensures transcriptional repression, DSB mobilization, and HR repair
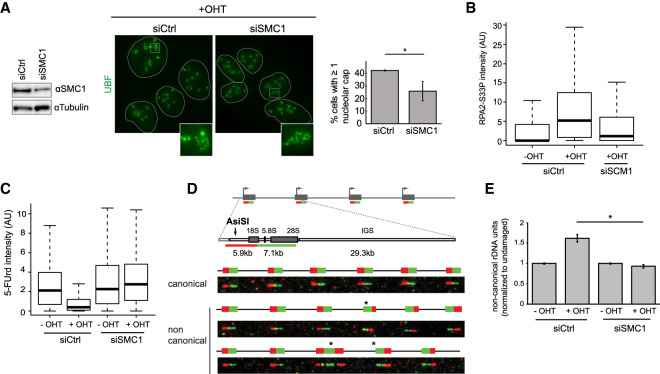
We further turned our attention toward the cohesin complex, given its pleiotropic involvements in rDNA stability and transcriptional regulation, DSB repair, and chromosome structure and mobility (Kobayashi et al. 2004; Caron et al. 2012; Gelot et al. 2016a; Herdman et al. 2017; Pal et al. 2018). Depletion of the SMC1, SCC1, and SMC3 cohesin subunits triggered a decrease in nucleolar cap formation following rDNA breakage (Fig. 5A; Supplemental Fig. S5A,B) without strongly affecting cell cycle distribution (Supplemental Fig. S5C). Cohesin depletion also prevented both resection (as measured by RPA2-S33P foci) (Fig. 5B) and transcriptional repression following rDNA damage (Fig. 5C; Supplemental Fig. S5D,F). At rDNA genes, HR frequently triggers rDNA arrays rearrangement, which can be visualized using DNA FISH on individual fibers (Fig. 5D). OHT-mediated rDNA DSB induction in control cells led to an increase of noncanonical units indicative of rDNA rearrangements, which was abolished upon SMC1 depletion using siRNA (Fig. 5E), in agreement with a function of cohesin in promoting HR at rDNA units.
Figure 5.
The cohesin complex regulates rDNA transcriptional repression, resection, DSB mobilization, and HR repair following rDNA breakage. (A, left) Western blot using SMC1 and tubulin antibodies in control or SMC1 siRNA transfected DIvA cells. (Middle) Control or SMC1-depleted DIvA cells treated for 4 h with OHT were stained using an UBF antibody to detect nucleoli and nucleolar caps. (Right) The number of cells with at least one nucleolar cap was quantified. The mean and SD from three independent experiments are shown. (*) P < 0.05. (B) Representative box plot of nucleolar RPA2-S33P intensity after SMC1 or control siRNA knockdown (>100 nucleoli quantified). (C) Representative box plot of the 5-FUrd signal before or after 4 h of OHT treatment in control or SMC1-depleted DIvA cells (>1000 cells). (D) Schematic representation of the rDNA repeats and the two fluorescent probes (red and green) used to measure rDNA rearrangements by DNA FISH combing (top), with examples of canonical and noncanonical rDNA units (indicated by an asterisk; bottom). (E) The level of noncanonical rDNA units measured by FISH combing before or after 24 h of OHT treatment in control or SMC1-depleted cells. (*) P < 0.05.
The above data therefore suggest that the cohesin complex acts at an early step in the rDNA DSB response and contributes to the DSB-induced transcriptional repression at rDNA, thereby influencing all downstream events; i.e., resection, DSB mobilization, and HR repair.
Transcriptional repression requires HUSH-mediated H3K9me3 deposition
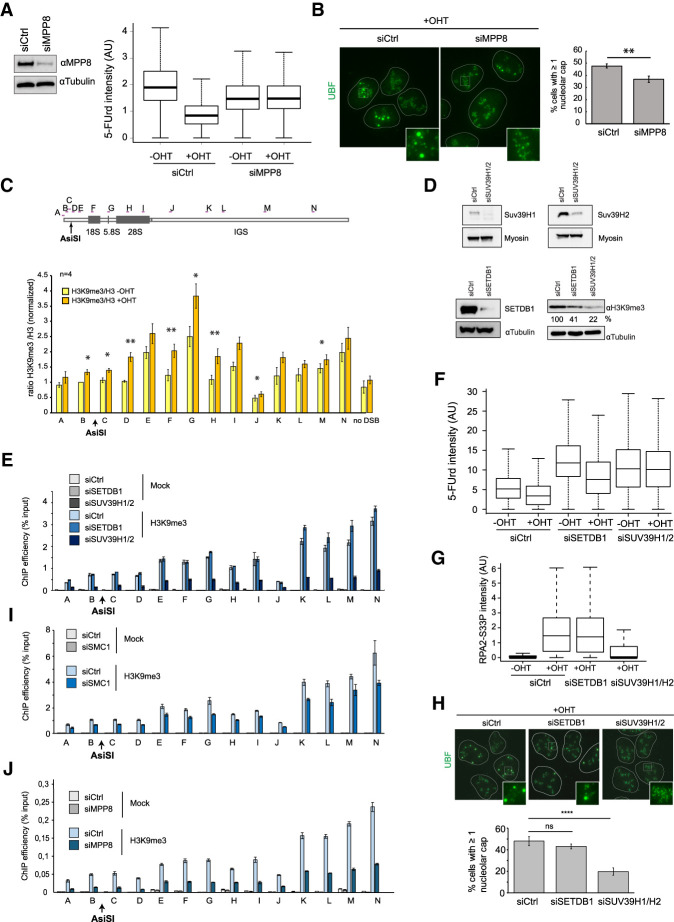
In order to identify potential effectors of the cohesin complex that could mediate transcriptional repression of damaged rDNA, we engineered a DIvA cell line overexpressing a Flag-GFP-tagged version of SMC1 (Supplemental Fig. S5G). Tandem purification of SMC1 interactors in the nucleus and nucleolus before and after DSB production followed by mass spectroscopy analysis (Supplemental Fig. S5H) allowed the identification of candidates, among which was periphilin 1, a subunit of the recently identified HUSH complex. This complex, composed of (at least) periphilin, MPP8, and TASOR, is involved in the epigenetic repression of retroviruses and LINE-1 retrotransposons via H3K9 trimethylation (H3K9me3) (Tchasovnikarova et al. 2015; Liu et al. 2018; Robbez-Masson et al. 2018). Indeed, depletion of MPP8 alleviated DSB-induced transcriptional repression of rDNA (Fig. 6A; Supplemental Fig. S6A), as observed after depletion of the cohesin complex. In agreement, MPP8 depletion also translated into decreased cap formation following rDNA damage (Fig. 6B), with no effect on the cell cycle (Supplemental Fig. S6B), suggesting that the HUSH complex contributes to rDNA transcription shutdown following rDNA breakage.
Figure 6.
The HUSH complex mediates transcriptional shutdown after DSB via H3K9me3 deposition. (A, left panel) Western blot using MPP8 and tubulin antibodies in control or MPP8 siRNA transfected DIvA cells. (Right panel) 5-FUrd level as measured by high-content microscopy (>5000 cells) in untreated or OHT-treated DIvA transfected with either control or MPP8 siRNA (as indicated). A representative experiment is shown. (B) UBF staining was performed following OHT treatment in control or MPP8-depleted cells as indicated. The left panel shows representative images, and the right panel shows the number of cells displaying at least one nucleolar cap. The mean and SD from four independent experiments are shown. (**) P < 0.01. (C) ChIP-qPCR was performed in untreated or OHT-treated DIvA cells using antibodies against H3K9me3 and H3. The mean and SEM of the ratio H3K9me3/H3 from four independent experiments are shown. (*) P < 0.05; (**) P < 0.01. (D) Western blots showing the level of SUV39H1 (top left), SUV39H2 (top right), or SETDB1 (bottom left) upon transfection of indicated siRNA as well as their influence on H3K9me3 levels (bottom right). (E) ChIP against H3K9me3 was performed in control, SETDB1, and SUV39H1/2 siRNA transfected untreated DIvA cells as indicated. ChIP efficiency (percentage of input) of a representative experiment is shown. (F) Representative box plot of 5-FUrd level as measured by high-content microscopy (>2000 cells) in untreated or OHT-treated DIvA transfected with either control, SETDB1, or SUV39H1/2 siRNA (as indicated). (G) Box plots of RPA2-S33P nucleolar intensity after depletion of SETDB1 or SUV39H1/2 followed by 4 h of OHT treatment in DIvA cells. A representative experiment is shown (>100 cells). (H) UBF staining was performed following OHT treatment in control, SETDB1-depleted, or SUV39H1/2-depleted cells as indicated. Representative images are shown (left panel), and the number of cells displaying at least one nucleolar cap was quantified (right panel). The mean and SD from four independent experiments are shown. (ns) Nonsignificant; (****) P < 0.0001. (I) ChIP against H3K9me3 was performed in control and SMC1 siRNA transfected untreated DIvA cells as indicated. ChIP efficiency (percentage of input) of a representative experiment is shown. (J) ChIP against H3K9me3 was performed in control and MPP8 siRNA transfected untreated DIvA cells as indicated. ChIP efficiency (percentage of input) of a representative experiment is shown.
Because the HUSH complex was suggested to mediate transcriptional silencing through H3K9me3, we assessed the levels of this chromatin mark on rDNA following DSB induction. An accumulation of H3K9me3 on rDNA was clearly detected, mostly in the transcribed region after DSB induction (Fig. 6C; Supplemental Fig. S6C). In mammalian cells, H3K9 trimethylation is catalyzed mostly by SUV39H1/2 and SETDB1 (Rea et al. 2000; Schultz et al. 2002); the latter is suggested to be involved in HUSH-dependent H3K9 trimethylation (Tchasovnikarova et al. 2015). Knockdown of both SUV39H1/2 and SETDB1 decreased global H3K9me3 levels in DIvA cells, as expected (Fig. 6D; Supplemental Fig. S6D). However, on rDNA, only depletion of SUV39H1/2 led to a detectable H3K9me3 decrease (Fig. 6E). The lack of SETDB1 effects at the rDNA appears to be a genuine response, as H3K9me3 level decreased upon SETDB1 depletion on zinc finger genes, as described previously (Supplemental Fig. S6E; Tchasovnikarova et al. 2015). In agreement with the above data, only SUV39H1/2 depletion, but not SETDB1, impaired DSB-induced transcription repression (Fig. 6F; Supplemental Fig. S6F), resection (Fig. 6G), and cap formation (Fig. 6H) without cell cycle changes (Supplemental Fig. S6G). Of importance, depletion of SMC1 or MPP8 also triggered decreased levels of H3K9me3 at rDNA (Fig. 6I,J), further strengthening the role of cohesin and the HUSH complex in mediating H3K9me3 deposition.
Collectively, these data suggest that following DSB production in rDNA, the cohesin complex mediates transcriptional shutdown of ribosomal genes in a manner that depends on the HUSH complex and SUV39H1/2-mediated H3K9me3.
The LINC complex subunit SUN1, the actin nucleator ARP3, and the myosin regulator UNC-45A promote DSB mobilization downstream from transcriptional repression
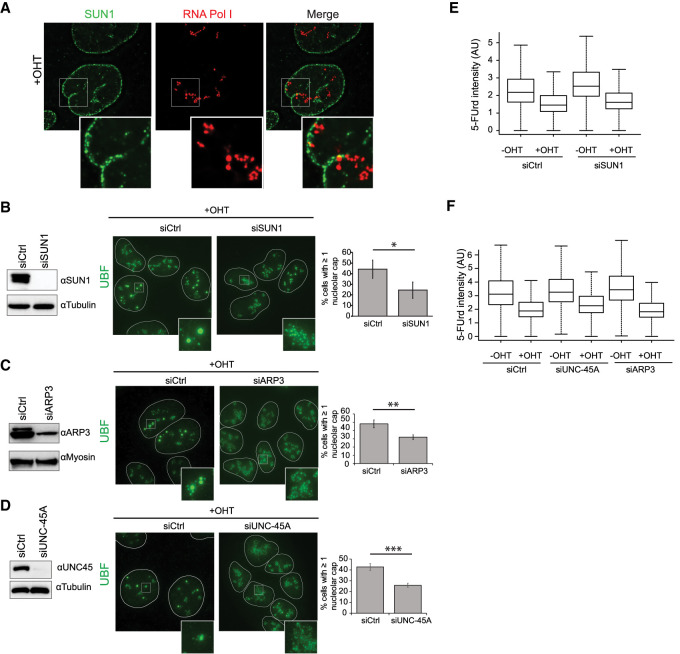
We next aimed at characterizing the mechanisms that ensure rDNA DSB mobilization at the periphery. Previous studies in yeast and mammals showed a role for the microtubule network in the movement of DSBs (Lottersberger et al. 2015; Lawrimore et al. 2017; Oshidari et al. 2018). However, a pretreatment with the microtubule-destabilizing drug nocodazole did not decrease the formation of nucleolar caps (Supplemental Fig. S7A,B). We next focused our attention on the LINC complex, which is embedded in the NE and mediates connections between the nucleoskeleton and the cytoskeleton. Indeed, the LINC complex plays roles in mobilization of damaged telomeres and IR-induced DSBs in mouse cells (Lottersberger et al. 2015) and also contributes to DSBs clustering in human nuclei (Aymard et al. 2017). Interestingly, SUN1, a subunit of the LINC complex localizing to the NE, was also reported recently to be present in nucleoli (Matsumoto et al. 2016; Moujaber et al. 2017). In agreement, we could indeed confirm that SUN1 forms patches close to RNA Pol I (Supplemental Fig. S7C; Supplemental Movie S2). Closer inspection revealed that SUN1-stained NE frequently forms invaginations that connect the nucleolus (Fig. 7A; Supplemental Fig. S7D). Neither DSB production nor actinomycin D treatment affected the number of NE invaginations, as observed using a Lamin B staining (Supplemental Fig. S7E,F; Supplemental Movie S3). However, SUN1 depletion decreased both invaginations and cap formation following rDNA DSB induction (Fig. 7B; Supplemental Fig. S7G), without affecting cell cycle distribution (Supplemental Fig. S7H), suggesting that the LINC complex contributes to rDNA mobilization in a manner that involves NE invaginations.
Figure 7.
The LINC complex, ARP3, and UNC-45A mediate rDNA DSB mobilization at the nucleolar periphery. (A) Example of NE invagination (SUN1 in green) connecting the nucleolus (RNA Pol I in red); a magnification is shown at the bottom. (B, left panel) Western blot using SUN1 and tubulin antibodies in control or SUN1 siRNA transfected DIvA cells. (Middle panel) Representative image of UBF staining in OHT-treated DIvA cells in control or SUN1-depleted cells. (Right panel) The number of cells displaying at least one nucleolar cap. The mean and SD from three independent experiments are shown. (*) P < 0.05. (C) Same as in B, except that a siRNA against ARP3 was used. The mean and SD from three independent experiments are shown. (**) P < 0.01. (D) Same as in B, except that a siRNA against UNC-45A was used. The mean and SD from five independent experiments are shown. (***) P < 0.001. (E) 5-FUrd level measured by high-content microscopy (>5000 cells) in untreated or OHT-treated DIvA transfected with either control or SUN1 siRNA (as indicated). A representative experiment is shown. (F) Same as in E, except that siRNA against ARP3 and UNC-45A were used. A representative experiment is shown.
Interestingly, persistent and heterochromatic DSBs were shown to move toward the NE in yeast and Drosophila cells, respectively (Horigome et al. 2014; Ryu et al. 2015), via nuclear actin polymerization in Drosophila (Caridi et al. 2018). We hence further asked whether rDNA DSB mobilization also necessitates the actin network. Indeed, depletion of UNC-45A (also known as general cell UNC-45 [GC-UNC-45], a myosin regulator) (Lehtimäki et al. 2017) and ARP3 (an actin nucleator-mediating branched actin network), both previously involved in heterochromatic DSB mobilization in flies (Caridi et al. 2018), also decreased DSB-induced cap formation (Fig. 7C,D) without major changes to cell cycle distribution (Supplemental Fig. S7H). Strikingly, depletion of all three factors (SUN1, UNC-45A, and ARP3) did not affect transcriptional repression following DSB induction (Fig. 7E,F; Supplemental Fig. S7I). Collectively, these data suggest that the NE/LINC complex and actin network (ARP3 and UNC-45A) ensure rDNA DSB mobilization outside of the nucleolus downstream from transcriptional inhibition.
Discussion
The rDNA is one of the most fragile loci on the genome and is also particularly prone to experience major rearrangements during replication and repair (Lindström et al. 2018). Highly specialized repair mechanisms are likely implemented in order to ensure its stability across cell divisions. Here we uncovered that repair of DSBs produced on the rDNA requires transcriptional extinction via a cohesin/HUSH-dependent pathway, followed in S/G2 cells by end resection in the interior of the nucleolus. Resected DNA ends are further mobilized at the nucleolar periphery, where they undergo HR repair and accumulation of γH2AX, MDC1, and ubiquitin chains. Interestingly, a recent study reported that the Nbs1 component of the MRN complex as well as low levels of γH2AX signal can nevertheless be observed within the interior of the nucleolus following rDNA break induction (Korsholm et al. 2019). Moreover, ATM activation following DSB in rDNA precedes DSB mobilization (this study; Harding et al. 2015; Korsholm et al. 2019). Hence, altogether, this suggests that while DSB signaling by PI3 kinases can take place at the interior of the nucleolus, signaling is further amplified at the periphery of the nucleolus once resection has occurred, in agreement with our recent findings that γH2AX accumulates more at DSBs repaired by HR (Clouaire et al. 2018).
Of importance, we further found that the NE forms invaginations that connect the nucleolus and that both the LINC complex and the actin network contribute to rDNA DSB mobilization (Supplemental Fig. S8).
Uncoupling DSB mobilization from transcriptional repression in the context of rDNA damage
In agreement with previous studies using either the CRISPR/Cas9 or the I-PpoI homing endonuclease to induce DSBs on the rDNA array, we showed that AsiSI-induced rDNA breaks trigger both rDNA transcriptional arrest and nucleolar segregation as visualized by cap formation (i.e., DSB movement) (Larsen et al. 2014; Harding et al. 2015; van Sluis and McStay 2015; Pefani et al. 2018). However, as opposed to CRISPR/Cas9- or I-PpoI-mediated breaks that promote nucleolar segregation across all cell cycle stages, caps are rather restricted to S/G2 cells when induced by AsiSI despite comparable levels of DSB formation and transcriptional repression that were observed throughout the cell cycle. We could further verify that nucleolar segregation occurred in nearly all cells following (1) actinomycin D treatment, (2) CRISPR/Cas9-mediated cleavage induced using nanoblades (Mangeot et al. 2019), and (3) AsiSI activation in presence of an inhibitor of DNA-PK that strongly blocks AsiSI DSB repair (Supplemental Fig. S1E–H; Caron et al. 2015). This indicated that, in principle, DIvA cells are able to undergo robust nucleolar segregation and that blocking AsiSI DSB repair somehow mimics CRISPR/Cas9- or I-PpoI-induced rDNA cleavage. In addition, we showed that DSBs produced by AsiSI elsewhere on the genome do not compete with the rDNA response, since CRISPR/Cas9-induced breaks on the rDNA also triggered robust nucleolar segregation in the presence of activated AsiSI (Supplemental Fig. S1I,J). Altogether, this suggests that the milder phenotypes observed upon AsiSI-mediated breakage compared with Cas9-mediated break induction—both transcriptional repression (data not shown) and nucleolar cap formation (Supplemental Fig. S1)—could be due to either a lower amount of rDNA copies cleaved by AsiSI (sensitive to DNA methylation) compared with Cas9 and/or discrepancies in repair efficiency. Of note, recent work on CRISPR/Cas9-induced DSBs suggested that such breaks are particularly refractory to repair (Brinkman et al. 2018) and may be handled by the Fanconi anemia pathway (Richardson et al. 2018). In addition, in contrast to type II restriction enzyme (such as AsiSI), the I-PpoI homing endonuclease recognizes particularly long DNA sequences in a manner that severely bends and alters the DNA structure (Chevalier and Stoddard 2001), which may also affect repair efficiency. Altogether, we propose that the inherent persistent nature of CRISPR/Cas9- or I-PpoI-induced breaks may hence account for the extreme nucleolar segregation and HR usage reported previously at all cell cycle stages (van Sluis and McStay 2015) and that the restriction of DSB movement to S and G2 cells is a bona fide rDNA DSB response.
DSB mobilization at the nucleolar periphery has been proposed previously to be a direct consequence of DSB-induced rDNA transcriptional extinction (van Sluis and McStay 2015). Indeed, the nucleolus is a self-organizing membrane-free organelle in which rRNAs act as seeds initiating a phase separation process leading to nucleolus formation (Falahati et al. 2016). Extinction of rDNA transcription rapidly triggers a drastic reorganization in which rDNA is expelled at the periphery by a phase separation event (as observed with actinomycin D). However, our findings indicate that rDNA DSB mobilization at the nucleolar periphery is not only a consequence of a phase separation driven by rDNA transcriptional arrest. Indeed, DSBs are mobilized only in S/G2 stage, while transcriptional repression occurs throughout the cell cycle. Moreover, depletion of the LINC/actin pathway impairs mobilization without affecting the upstream transcriptional inhibition. Of note, this rDNA mobilization pathway is specific to DSB formation, since depletion of SMC1, Suv39H1/2, Sun1, UNC45, and Arp3 did not affect rDNA segregation observed after actinomycin D (data not shown). Importantly, so far, we could not identify any conditions under which transcriptional arrest is impaired and DSBs are still mobilized, which might suggest that transcriptional repression is a prerequisite for DSB movement.
Mechanisms ensuring DSB mobilization: NE invagination, LINC, and the actin network
Interestingly, the mobilization of rDNA DSBs shares common features with the mobilization of those induced in heterochromatin and active genes and of persistent DSBs. First, in mammals, DSBs induced in pericentromeric heterochromatin display relocalization to the periphery of the heterochromatin focus during S/G2 in a manner that depends on resection (Tsouroula et al. 2016), as shown here for a DSB induced in rDNA. Second, in Drosophila, heterochromatic DSBs move toward the periphery of the heterochromatic focus and further to the NE in a manner that depends on Suv39H1-mediated H3K9me3, on the LINC complex, and on the actin network (Chiolo et al. 2011; Ryu et al. 2015; Colmenares et al. 2017; Caridi et al. 2018). Third, studies in yeast have shown that persistent DSBs are relocalized to the NE via the SUN domain-containing proteins Mps3 or Sad1 (Oza et al. 2009; Horigome et al. 2014; Swartz et al. 2014). Fourth, we reported previously that both the actin network and the LINC complex contribute to clustering of DSBs induced in active genes in G1 (Aymard et al. 2017). Of importance all of the above-mentioned DSBs were described previously as refractory to fast repair (for review, see Marnef et al. 2017; Fortuny and Polo 2018). We thus propose that breaks in rDNA may also display impaired or delayed repair kinetics—in line with its repetitive nature, high RNA Pol I occupancy, and prevalence of secondary structures (R loops and G4)—and hence use similar mobilization pathways to undergo safe repair.
Notably, while the interaction of persistent DSBs with the NE has been reported in Drosophila and yeast, there was no evidence of such phenomena in mammals despite a reported function of the LINC complex in DSB repair (Lottersberger et al. 2015; Aymard et al. 2017). Here we report that the NE forms invaginations that contact the nucleolus (in line with previous reports showing strong relationship with the NE and nucleolus) (Hernandez-Verdun et al. 2010) and that depletion of the LINC complex, which reduces NE invaginations (Supplemental Fig. S7), also affects rDNA DSB relocalization to the nucleolus periphery. Hence, our data suggest that NE/persistent DSB anchorage also occur in mammals and that, for rDNA breaks, this interaction contributes to the localization of the DSB at the nucleolar periphery. Alternatively, or in addition, the LINC complex and NE invaginations could also directly regulate resection, which could further foster DSB mobilization to the periphery of the nucleolus.
Our data also indicate that rDNA movement relies on the actin network, as reported for Drosophila heterochromatic breaks (Caridi et al. 2018) and in line with the contribution of actin regulation in DSB repair (Belin et al. 2015; Spichal et al. 2016; Aymard et al. 2017; Schrank et al. 2018). One possibility is that rDNA DSB mobilization involves the formation of nuclear/nucleolar actin filaments as found in other contexts (Belin et al. 2015; Caridi et al. 2018; Schrank et al. 2018). Alternatively, since cytoplasmic filamentous actin can be found in NE invaginations (Jorgens et al. 2017), one could envisage that cytoplasmic actin cables/filaments regulate NE deformation and the dynamics of invaginations, which could further contribute to the relocalization of the rDNA DSBs outside of the nucleolus thanks to a physical link between the DSBs and NE-embedded LINC complex. Altogether our data suggest that a conserved mechanism involving the LINC complex and the actin network probably exists to mobilize all “persistent” or “difficult to repair” DSBs to the NE, providing a safe recombination environment.
Cohesin/HUSH/H3K9me3-driven DSB-induced rDNA transcriptional repression
DSB-induced rDNA transcriptional repression has been shown to require ATM activity, Nbs1 (our data not shown; Kruhlak et al. 2007; Larsen et al. 2014; Harding et al. 2015; van Sluis and McStay 2015; Warmerdam et al. 2016; Pefani et al. 2018), and, more recently, the MST2-dependent phosphorylation of H2B on Ser14 (Pefani et al. 2018). Here we found that the cohesin complex also regulates RNA Pol I transcription in response to DSBs. Interestingly, conserved mechanisms may account for RNA Pol I and Pol II regulation in cis to DSBs, since both ATM and the cohesin complex were identified as key players in RNA Pol II transcriptional arrest in cis to DSBs (Shanbhag et al. 2010; Meisenberg et al. 2019). This raises the interesting possibility that ATM may regulate transcription at least in part by regulating the cohesin biology in cis to DSBs, as the cohesin complex is an ATM target (Kim et al. 2002). Our data also indicate that the HUSH complex, shown previously to display H3K9me3 methylation activity (Tchasovnikarova et al. 2015; Liu et al. 2018; Robbez-Masson et al. 2018), participates in DSB-induced RNA Pol I transcriptional repression. Notably, SUV39H1, SETDB1, and also the histone demethylase KDM4A, all known to regulate H3K9me3 levels, have been involved in DSB repair by HR (Chiolo et al. 2011; Ayrapetov et al. 2014; Alagoz et al. 2015; Colmenares et al. 2017), and loss of H3K9me3 triggers aberrant recombination between repetitive sequence (Peng and Karpen 2007). On the other hand, the cohesin complex is a key regulator of HR; it prevents the end joining of distant DSB ends (for review, see Dorsett and Ström 2012; Gelot et al. 2016b) and suppresses aberrant recombination at rDNA (Kobayashi et al. 2004; Kobayashi and Ganley 2005). Our study provides a mechanistic link between the cohesin complex and the regulation of H3K9me3 levels and suggests that cohesin contributes to the regulation of H3K9me3 levels by recruiting the HUSH complex, further allowing transcriptional repression, resection, and DSB mobilization for recombination.
In summary, here we report that as for heterochromatic DSBs, rDNA DSB repair relies on a spatiotemporal separation, where resection within the nucleolus is followed by the mobilization of the break outside its environment. Since mobilization involves players also required for the relocalization of other “persistent” DSBs such as the LINC complex, our data suggest that conserved DSB mobilization-dependent mechanisms exist from yeast to humans in order to relocate harmful breaks to a NE compartment, allowing safe repair.
Materials and methods
Cell culture and treatments
DIvA cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (Invitrogen) in the presence of 1 µg/mL puromycin (Invivogen). Unless stated otherwise, DSBs were induced by the addition of 300 nM OHT (Sigma, H7904) for 4 h. For DNA-PK inhibition, 2 µM NU7441 (Selleckchem) was added 1 h prior to OHT addition and maintained during the OHT-mediated break induction. When indicated, 100 ng/mL actinomycin D (Sigma) was added to cells for a total of 1 h. Nocodazole (3 µM) was added 1 h prior to OHT addition. siRNA-mediated depletion of proteins (Supplemental Table S1) was performed using the cell line Nucleofector kit V (Program X-001, Amaxa) according to the manufacturer's instructions. Details for CRISPR–Cas9-induced breakage and cell synchronization are in the Supplemental Material.
BLESS analysis on rDNA
BLESS sequences from asynchronized cells or G1 and G2 cell populations were retrieved from our previous work (Aymard et al. 2017; Clouaire et al. 2018; Mourad et al. 2018). Barcodes were removed using Cutadapt, and BLESS data were aligned on a custom-built genome composed of hg19 plus rRNA unit U13369.1 from GenBank using bwa in paired-end mode (bwa aln and bwa sampe). Fragments were reconstituted from paired reads using Rsamtools and GenomicAlignments (R packages). Fragments with lengths >500 bp were dropped, and the remaining fragments were deduplicated (fragments with the exact same start and end positions were considered as duplicates). For the G1 and G2 BLESS data analysis, proximal positions were extracted from fragments based on their relative positions to the DSB.
Immunofluorescence
Cells were plated on glass coverslips and fixed in 4% paraformaldehyde/PBS for 20 min, permeabilized in 0.5% Triton X-100/PBS for 5 min unless stated otherwise (Supplemental Table S2), and blocked with 3% BSA/PBS for 30 min before incubation with primary antibodies (see Supplemental Table S2) in 3% BSA/PBS overnight at 4°C. For CSK pre-extraction, cells were permeabilized using 0.5% Triton X-100 for 5 min followed by 10 min in CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES at pH 6.8), fixation in 4% PFA/PBS, and blocking with 3% BSA/PBS. For buffer II pre-extraction (20 mM NaCl, 0.5% NP-40, 20 mM HEPES at pH 7.5, 5 mM MgCl2, 1 mM DTT), buffer II was added to cells for 20 min on ice followed by fixation in 4% PFA/PBS and blocking with 3% BSA/PBS. The appropriate Alexa-conjugated secondary antibodies diluted in 3% BSA/PBS (mouse or rabbit Alexa 594 or Alexa 488 used at a 1:1000 dilution [Invitrogen]) were then added to cells for 1 h at room temperature followed by DAPI or Hoechst 33342 staining. Coverslips were mounted in Citifluor (Citifluor, AF-1), and images were acquired using a wide-field microscope (Leica, DM6000) equipped with a cooled charge-coupled device camera (CoolSNAP HQ2) using 40× or 100× objectives and MetaMorph.
Images of total RPA and SUN1 immunostainings were processed by nonlinear deconvolution with Huygens software (Scientific Volume Imaging). Classical maximum likelihood estimation was used to estimate positive fluorescent signal after deconvolution. An adaptive estimation of background was done for each image, the signal to noise ratio was set to 40 (RPA) or 20 (SUN1), and 50 iterations were used during the minimization.
For superresolution microscopy, we imaged cells using a fast structure illumination microscopy (SIM) setup with a binary phase optical modulator (QXGA SLM Fourth Dimension) under a 100× objective lens (CFI SR APO 100XH ON 1,49 DT 0,12 Nikon) mounted on a Nikon Eclipse Ti-E and a Hamamatsu SCMOS camera (ORCA FLASH4.0 LT). A series of 800 3D Speckle illuminations was made on the object plane thanks to the random phase modulation from the SLM conjugate with the pupil plane of the microscope objective. A 150-nm Z interval was acquired on each image. Superresolution images were processed by a joint reconstruction strategy using random speckle illuminations (Negash et al. 2016; Labouesse et al. 2017). A positivity constraint was applied based on the realistic hypothesis that the fluorescent signal was only positive.
Quantifications of immunofluorescence
For DSB mobilization, the number of cells displaying at least one nucleolar cap was counted manually using the 40× objective in >150 cells per experiments and for a minimum of three independent experiments using either RNA pol I or UBF as markers. The number of caps (RNA Pol I staining) per nucleus was counted manually in >80 cells. Resection was quantified using RPAS33P antibody in >100 cells acquired with a 40× objective and analyzed using Columbus software (Perkin Elmer). Nucleoli were detected thanks to the RNA Pol I (using method A), and the sum of RPA2S33 spot (method D) intensity inside the nucleolar region was determined. At least three independent experiments were performed; only one representative experiment is shown as a box plot. In Supplemental Figure S3F, the presence of repair protein in either unsegregated or segregated nucleoli was quantified in cells with at least one nucleolar cap. The number of invaginations per cell was quantified manually using Lamin B1 staining in >70 cells and in three independent experiments.
High-throughput microscopy
The Operetta automated high-content screening microscope (PerkinElmer) was used for the rDNA transcription and/or cell cycle analyses. DIvA cells were plated in 96-well plates and treated with OHT for 4 h. For the visualization of nascent rRNA synthesis, 2 mM 5-FUrd (Sigma) was added 20–40 min before the end of the OHT treatment. For the detection of S-phase cells, EdU was added 10–20 min prior to the end of the OHT treatment. Cells were fixed and permeabilized as mentioned above, and saturation was done in 1%BSA/PBS for 30 min. For EdU-treated cells, a Click-iT reaction (Alexa 488; Invitrogen) was performed according to the manufacturer's instruction before the addition of an anti-BrdU primary antibody (BU-33; Sigma) at 1:500 in 1% BSA/PBS in the presence of 160 U/m RNasinL (Promega) overnight at 4°C. Secondary antimouse Alexa 647 (1:500 in 1% BSA/PBS; Molecular Probes, A21235) was incubated for 30 min at 37°C in the presence of 80 U/m RNasinL followed by DAPI staining. Image acquisition with a 20× objective lens was automated to obtain at least 36 fields per well, allowing the visualization of a total of >1000 cells (three wells were acquired for each condition). Each picture was analyzed with the integrated Columbus software. Briefly, the DAPI-stained nuclei were selected (method B), and the size and roundness of nuclei were used as parameters to eliminate false positive compounds. The 5-FUrd staining attributed to the nucleoli was delineated using the find spot method D, and the sum of the 5-FUrd intensity was measured and is represented as box plots. For cell cycle analysis, the sum of the DAPI intensity and the mean of the EdU intensity were plotted in order to separate G1, S, and G2 cells. The sum of the 5-FUrd intensity was subsequently determined in each of these cell population.
ChIP
ChIP experiments were performed as in Iacovoni et al. (2010). The amount of chromatin and antibodies used are detailed in Supplemental Table S2. ChIP efficiencies were calculated as a percentage of input of DNA immunoprecipitated. For quantitative PCR (qPCR) analyses, the input as well as immunoprecipitation samples were analyzed with the primers listed in Supplemental Table S3. For Supplemental Figs. 3E and S6C, ChIP efficiencies (percentage of input) obtained with each primer pair were further normalized to the value obtained for primer B in the undamaged condition. The ratio of H3K9m3 over H3 was calculated before normalization with primer B for Figure 6C.
DNA FISH combing and analysis
The combing and FISH probing of the rDNA fibers were carried out before or after 24 h of OHT treatment (Genomic Vision). The detection and rDNA fibers and the attribution of rDNA-specific probes (Caburet et al. 2005) was carried out using Genomic Vision's provided Web interface. Analysis of rearrangements was performed using a script that was designed to count the total number of units (as defined when comprised between two “IGS” regions) and assess the occurrence of changes when comparing one unit with the next. The source code was deposited on GitHub (https://github.com/LegubeDNAREPAIR/rDNA/blob/master/get_break_event.py).
Western blotting
Cells were incubated in RIPA buffer (0.5% deoxycholate, 1% NP-40, 50 mM Tris at pH 8, 150 mM NaCl, 0.1% SDS) for 20 min on ice and centrifuged at 13,000 rpm for 10 min before SDS loading buffer addition to the supernatant. In the case of H3 and H3K9me3 immunoblots, cells were sonicated three times for 10 sec in lysis buffer (1% SDS, 1% Triton, 10 mM Tris at pH 7.5, 450 mM NaCl) before the addition of SDS loading buffer. Protein extracts were resolved on 3%–8% NuPAGE Tris-acetate gels or 4%–12% Bis-Tris (Invitrogen) according to the manufacturer's instructions and transferred onto PVDF membranes (Invitrogen). The latter were blocked in TBS containing 0.1% Tween 20 (Sigma) and 3% nonfat dry milk for 1 h followed by an overnight incubation at 4°C with primary antibodies (Supplemental Table S2). The appropriate horseradish peroxidase-coupled secondary antibodies were used to reveal the proteins (antimouse at 1:10,000 [Sigma, A2554] and antirabbit at 1:10,000 [Sigma, A0545]) using a luminol-based enhanced chemiluminescence HRP substrate (Super Signal West Dura Extended Duration Substrate, Thermo Scientific).
Acknowledgments
We thank T. Clouaire and N. Puget for their advice throughout the project and comments on the manuscript. We thank E. Watrin (Rennes) for the Flag-GFP-SMC1 construct. We thank the high-throughput microscopy platform of the Centre de Biologie Intégrative (Toulouse), and the mass spectrometry platform from the Institut de Pharmacologie et de Biologie Structurale (Toulouse). Funding in E.P.R.’s laboratory was provided by Labex Ecofect (ANR-11-LABX-0048), Fondation Innovations en Infectiologie (Finovi), and the European Research Council (ERC-StG-LS6-805500) under the European Union's Horizon 2020 research and innovation programs. Funding in G.L.’s laboratory was provided by grants from the European Research Council (ERC-2014-CoG 647344), the Agence Nationale pour la Recherche (ANR-14-CE10-0002-01 and ANR-13-BSV8-0013), the Institut National Contre le Cancer (INCA), and the Ligue Nationale Contre le Cancer (LNCC).
Author contributions: A.M., A.-L.F., C.A., V.D., and E.G. performed experiments. V.R. performed bioinformatic analyses of BLESS data sets. T.M. performed superresolution microscopy. P.E.M. and E.P.R. provided nanoblades for CRISPR/Cas9-induced breakage. A.M. and G.L. conceived, supervised, and analyzed experiments. A.M. and G.L. wrote the manuscript. All authors commented and edited the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.324012.119.
Freely available online through the Genes & Development Open Access option.
References
- Alagoz M, Katsuki Y, Ogiwara H, Ogi T, Shibata A, Kakarougkas A, Jeggo P. 2015. SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Res 43: 7931–7944. 10.1093/nar/gkv722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, et al. 2014. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 21: 366–374. 10.1038/nsmb.2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Aguirrebengoa M, Guillou E, Javierre BM, Bugler B, Arnould C, Rocher V, Iacovoni JS, Biernacka A, Skrzypczak M, et al. 2017. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat Struct Mol Biol 24: 353–361. 10.1038/nsmb.3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. 2014. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci 111: 9169–9174. 10.1073/pnas.1403565111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Lee T, Mullins RD. 2015. DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-1/2 that promotes efficient DNA repair. Elife 4: e07735. 10.7554/eLife.07735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordelet H, Dubrana K. 2019. Keep moving and stay in a good shape to find your homologous recombination partner. Curr Genet 65: 29–39. 10.1007/s00294-018-0873-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, de Haas M, Holland HA, Akhtar W, van Steensel B. 2018. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol Cell 70: 801–813.e6. 10.1016/j.molcel.2018.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caburet S, Conti C, Schurra C, Lebofsky R, Edelstein SJ, Bensimon A. 2005. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res 15: 1079–1085. 10.1101/gr.3970105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi CP, D'Agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, et al. 2018. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559: 54–60. 10.1038/s41586-018-0242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron P, Aymard F, Iacovoni JS, Briois S, Canitrot Y, Bugler B, Massip L, Losada A, Legube G. 2012. Cohesin protects genes against γH2AX Induced by DNA double-strand breaks. PLoS Genet 8: e1002460. 10.1371/journal.pgen.1002460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron P, Choudjaye J, Clouaire T, Bugler B, Daburon V, Aguirrebengoa M, Mangeat T, Iacovoni JS, Álvarez-Quilón A, Cortés-Ledesma F, et al. 2015. Non-redundant functions of ATM and DNA-PKcs in response to DNA double-strand breaks. Cell Rep 13: 1598–1609. 10.1016/j.celrep.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier BS, Stoddard BL. 2001. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res 29: 3757–3774. 10.1093/nar/29.18.3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. 2011. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744. 10.1016/j.cell.2011.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Legube G. 2015. DNA double strand break repair pathway choice: a chromatin based decision? Nucleus 6: 107–113. 10.1080/19491034.2015.1010946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Rocher V, Lashgari A, Arnould C, Aguirrebengoa M, Biernacka A, Skrzypczak M, Aymard F, Fongang B, Dojer N, et al. 2018. Comprehensive mapping of histone modifications at DNA double-strand breaks deciphers repair pathway chromatin signatures. Mol Cell 72: 250–262.e6. 10.1016/j.molcel.2018.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Swenson JM, Langley SA, Kennedy C, Costes SV, Karpen GH. 2017. Drosophila histone demethylase KDM4A has enzymatic and non-enzymatic roles in controlling heterochromatin integrity. Dev Cell 42: 156–169.e5. 10.1016/j.devcel.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, et al. 2013. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods 10: 361–365. 10.1038/nmeth.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Ström L. 2012. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol 22: R240–R250. 10.1016/j.cub.2012.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. 2016. Nucleation by rRNA dictates the precision of nucleolus assembly. Curr Biol 26: 277–285. 10.1016/j.cub.2015.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floutsakou I, Agrawal S, Nguyen TT, Seoighe C, Ganley ARD, McStay B. 2013. The shared genomic architecture of human nucleolar organizer regions. Genome Res 23: 2003–2012. 10.1101/gr.157941.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortuny A, Polo SE. 2018. The response to DNA damage in heterochromatin domains. Chromosoma 127: 291–300. 10.1007/s00412-018-0669-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franek M, Kovaříková A, Bártová E, Kozubek S. 2016. Nucleolar reorganization upon site-specific double-strand break induction. J Histochem Cytochem 64: 669–686. 10.1369/0022155416668505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohns A, Frohns F, Naumann SC, Layer PG, Löbrich M. 2014. Inefficient double-strand break repair in murine rod photoreceptors with inverted heterochromatin organization. Curr Biol 24: 1080–1090. 10.1016/j.cub.2014.03.061 [DOI] [PubMed] [Google Scholar]
- Gelot C, Guirouilh-Barbat J, Le Guen T, Dardillac E, Chailleux C, Canitrot Y, Lopez BS. 2016a. The cohesin complex prevents the end joining of distant DNA double-strand ends. Mol Cell 61: 15–26. 10.1016/j.molcel.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Gelot C, Guirouilh-Barbat J, Lopez BS. 2016b. The cohesin complex prevents the end-joining of distant DNA double-strand ends in S phase: consequences on genome stability maintenance. Nucleus 7: 339–345. 10.1080/19491034.2016.1194159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Kurka T, Jeggo PA. 2011. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 18: 831–839. 10.1038/nsmb.2077 [DOI] [PubMed] [Google Scholar]
- Guénolé A, Legube G. 2017. A meeting at risk: unrepaired DSBs go for broke. Nucleus 8: 589–599. 10.1080/19491034.2017.1380138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SM, Boiarsky JA, Greenberg RA. 2015. ATM dependent silencing links nucleolar chromatin reorganization to DNA damage recognition. Cell Rep 13: 251–259. 10.1016/j.celrep.2015.08.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdman C, Mars J-C, Stefanovsky VY, Tremblay MG, Sabourin-Felix M, Lindsay H, Robinson MD, Moss T. 2017. A unique enhancer boundary complex on the mouse ribosomal RNA genes persists after loss of Rrn3 or UBF and the inactivation of RNA polymerase I transcription. PLoS Genet 13: e1006899. 10.1371/journal.pgen.1006899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DLJ. 2010. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA 1: 415–431. 10.1002/wrna.39 [DOI] [PubMed] [Google Scholar]
- Horigome C, Oma Y, Konishi T, Schmid R, Marcomini I, Hauer MH, Dion V, Harata M, Gasser SM. 2014. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol Cell 55: 626–639. 10.1016/j.molcel.2014.06.027 [DOI] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. 2010. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J 29: 1446–1457. 10.1038/emboj.2010.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Conrad S, Voss K-O, Zink D, Durante M, Löbrich M, Taucher-Scholz G. 2011. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res 39: 6489–6499. 10.1093/nar/gkr230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgens DM, Inman JL, Wojcik M, Robertson C, Palsdottir H, Tsai W-T, Huang H, Bruni-Cardoso A, López CS, Bissell MJ, et al. 2017. Deep nuclear invaginations are linked to cytoskeletal filaments - integrated bioimaging of epithelial cells in 3D culture. J Cell Sci 130: 177–189. 10.1242/jcs.190967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Künzel J, Löbrich M, Jeggo PA, Downs JA. 2014. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol Cell 55: 723–732. 10.1016/j.molcel.2014.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-T, Xu B, Kastan MB. 2002. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev 16: 560–570. 10.1101/gad.970602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ganley ARD. 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584. 10.1126/science.1116102 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441–453. 10.1016/S0092-8674(04)00414-3 [DOI] [PubMed] [Google Scholar]
- Korsholm LM, Gál Z, Lin L, Quevedo O, Ahmad DA, Dulina E, Luo Y, Bartek J, Larsen DH. 2019. Double-strand breaks in ribosomal RNA genes activate a distinct signaling and chromatin response to facilitate nucleolar restructuring and repair. Nucleic Acids Res 10.1093/nar/gkz518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montaño C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. 2007. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447: 730–734. 10.1038/nature05842 [DOI] [PubMed] [Google Scholar]
- Labouesse S, Negash A, Idier J, Bourguignon S, Mangeat T, Liu P, Sentenac A, Allain M. 2017. Joint reconstruction strategy for structured illumination microscopy with unknown illuminations. IEEE Trans Image Process 26: 2480–2493. 10.1109/TIP.2017.2675200 [DOI] [PubMed] [Google Scholar]
- Larsen DH, Hari F, Clapperton JA, Gwerder M, Gutsche K, Altmeyer M, Jungmichel S, Toledo LI, Fink D, Rask M-B, et al. 2014. The NBS1–Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat Cell Biol 16: 792–803. 10.1038/ncb3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J, Barry TM, Barry RM, York AC, Friedman B, Cook DM, Akialis K, Tyler J, Vasquez P, Yeh E, et al. 2017. Microtubule dynamics drive enhanced chromatin motion and mobilize telomeres in response to DNA damage. Mol Biol Cell 28: 1701–1711. 10.1091/mbc.e16-12-0846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtimäki JI, Fenix AM, Kotila TM, Balistreri G, Paavolainen L, Varjosalo M, Burnette DT, Lappalainen P. 2017. UNC-45a promotes myosin folding and stress fiber assembly. J Cell Biol 216: 4053–4072. 10.1083/jcb.201703107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström MS, Jurada D, Bursac S, Orsolic I, Bartek J, Volarevic S. 2018. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 37: 2351–2366. 10.1038/s41388-017-0121-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Lee CH, Swigut T, Grow E, Gu B, Bassik MC, Wysocka J. 2018. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 553: 228–232. 10.1038/nature25179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottersberger F, Karssemeijer RA, Dimitrova N, de Lange T. 2015. 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell 163: 880–893. 10.1016/j.cell.2015.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeot PE, Risson V, Fusil F, Marnef A, Laurent E, Blin J, Mournetas V, Massourides E, Sohier TJM, Corbin A, et al. 2019. Genome editing in primary cells and in vivo using viral-derived ‘nanoblades’ loaded with Cas9–sgRNA ribonucleoproteins. Nat Commun 10: 45. 10.1038/s41467-018-07845-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Cohen S, Legube G. 2017. Transcription-coupled DNA double-strand break repair: active genes need special care. J Mol Biol 429: 1277–1288. 10.1016/j.jmb.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Sakamoto C, Matsumori H, Katahira J, Yasuda Y, Yoshidome K, Tsujimoto M, Goldberg IG, Matsuura N, Nakao M, et al. 2016. Loss of the integral nuclear envelope protein SUN1 induces alteration of nucleoli. Nucleus 7: 68–83. 10.1080/19491034.2016.1149664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B. 2016. Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev 30: 1598–1610. 10.1101/gad.283838.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenberg C, Pinder SI, Hopkins SR, Wooller SK, Benstead-Hume G, Pearl FMG, Jeggo PA, Downs JA. 2019. Repression of transcription at DNA breaks requires cohesin throughout interphase and prevents genome instability. Mol Cell 73: 212–223.e7. 10.1016/j.molcel.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E, Magin S, Soni A, Iliakis G. 2016. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: cell cycle and proliferation-dependent regulation. Semin Cancer Biol 37–38: 51–64. 10.1016/j.semcancer.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Moujaber O, Omran N, Kodiha M, Pié B, Cooper E, Presley JF, Stochaj U. 2017. Data on the association of the nuclear envelope protein Sun1 with nucleoli. Data Brief 13: 115–123. 10.1016/j.dib.2017.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourad R, Ginalski K, Legube G, Cuvier O. 2018. Predicting double-strand DNA breaks using epigenome marks or DNA at kilobase resolution. Genome Biol 19: 34. 10.1186/s13059-018-1411-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negash A, Labouesse S, Sandeau N, Allain M, Giovannini H, Idier J, Heintzmann R, Chaumet PC, Belkebir K, Sentenac A. 2016. Improving the axial and lateral resolution of three-dimensional fluorescence microscopy using random speckle illuminations. J Opt Soc Am A Opt Image Sci Vis 33: 1089–1094. 10.1364/JOSAA.33.001089 [DOI] [PubMed] [Google Scholar]
- Németh A, Grummt I. 2018. Dynamic regulation of nucleolar architecture. Curr Opin Cell Biol 52: 105–111. 10.1016/j.ceb.2018.02.013 [DOI] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Löbrich M, Stewart GS, Jeggo PA, Goodarzi AA. 2010. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 12: 177–184. 10.1038/ncb2017 [DOI] [PubMed] [Google Scholar]
- Oshidari R, Strecker J, Chung DKC, Abraham KJ, Chan JNY, Damaren CJ, Mekhail K. 2018. Nuclear microtubule filaments mediate non-linear directional motion of chromatin and promote DNA repair. Nat Commun 9: 2567. 10.1038/s41467-018-05009-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23: 912–927. 10.1101/gad.1782209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Postnikoff SD, Chavez M, Tyler JK. 2018. Impaired cohesion and homologous recombination during replicative aging in budding yeast. Sci Adv 4: eaaq0236. 10.1126/sciadv.aaq0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefani DE, Tognoli ML, Pirincci Ercan D, Gorgoulis V, O'Neill E. 2018. MST2 kinase suppresses rDNA transcription in response to DNA damage by phosphorylating nucleolar histone H2B. EMBO J 37: e98760. 10.15252/embj.201798760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9: 25–35. 10.1038/ncb1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599. 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- Richardson CD, Kazane KR, Feng SJ, Zelin E, Bray NL, Schäfer AJ, Floor SN, Corn JE. 2018. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat Genet 50: 1132–1139. 10.1038/s41588-018-0174-0 [DOI] [PubMed] [Google Scholar]
- Robbez-Masson L, Tie CHC, Conde L, Tunbak H, Husovsky C, Tchasovnikarova IA, Timms RT, Herrero J, Lehner PJ, Rowe HM. 2018. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res 28: 836–845. 10.1101/gr.228171.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, Karpen GH, Chiolo I. 2015. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol 17: 1401–1411. 10.1038/ncb3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, Gottesman ME, Gautier J. 2018. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559: 61–66. 10.1038/s41586-018-0237-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932. 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141: 970–981. 10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spichal M, Brion A, Herbert S, Cournac A, Marbouty M, Zimmer C, Koszul R, Fabre E. 2016. Evidence for a dual role of actin in regulating chromosome organization and dynamics in yeast. J Cell Sci 129: 681–692. 10.1242/jcs.175745 [DOI] [PubMed] [Google Scholar]
- Swartz RK, Rodriguez EC, King MC. 2014. A role for nuclear envelope-bridging complexes in homology-directed repair. Mol Biol Cell 25: 2461–2471. 10.1091/mbc.e13-10-0569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchasovnikarova IA, Timms RT, Matheson NJ, Wals K, Antrobus R, Göttgens B, Dougan G, Dawson MA, Lehner PJ. 2015. Gene silencing. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348: 1481–1485. 10.1126/science.aaa7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsouroula K, Furst A, Rogier M, Heyer V, Maglott-Roth A, Ferrand A, Reina-San-Martin B, Soutoglou E. 2016. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol Cell 63: 293–305. 10.1016/j.molcel.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Tubbs A, Nussenzweig A. 2017. Endogenous DNA damage as a source of genomic instability in cancer. Cell 168: 644–656. 10.1016/j.cell.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui A, Nagaura Y, Yasui A. 2015. Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Mol Cell 58: 468–482. 10.1016/j.molcel.2015.03.023 [DOI] [PubMed] [Google Scholar]
- van Sluis M, McStay B. 2015. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev 29: 1151–1163. 10.1101/gad.260703.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmerdam DO, van den Berg J, Medema RH. 2016. Breaks in the 45S rDNA lead to recombination-mediated loss of repeats. Cell Rep 14: 2519–2527. 10.1016/j.celrep.2016.02.048 [DOI] [PubMed] [Google Scholar]